Abstract
A genetic screen for mutations synthetically lethal with temperature sensitive alleles of nop2 led to the identification of the nucleolar proteins Nop12p and Nop13p in Saccharomyces cerevisiae. NOP12 was identified by complementation of a synthetic lethal growth phenotype in strain YKW35, which contains a single nonsense mutation at codon 359 in an allele termed nop12-1. Database mining revealed that Nop12p was similar to a related protein, Nop13p. Nop12p and Nop13p are not essential for growth and each contains a single canonical RNA recognition motif (RRM). Both share sequence similarity with Nsr1p, a previously identified, non-essential, RRM-containing nucleolar protein. Likely orthologs of Nop12p were identified in Drosophila and Schizosaccharomyces pombe. Deletion of NOP12 resulted in a cold sensitive (cs) growth phenotype at 15°C and slow growth at 20 and 25°C. Growth of a nop12Δ strain at 15 and 20°C resulted in impaired synthesis of 25S rRNA, but not 18S rRNA. A nop13 null strain did not produce an observable growth phenotype under the laboratory conditions examined. Epitope-tagged Nop12p, which complements the cs growth phenotype and restores normal 25S rRNA levels, was localized to the nucleolus by immunofluorescence microscopy. Epitope-tagged Nop13p was distributed primarily in the nucleolus, with a lesser portion localizing to the nucleoplasm. Thus, Nop12p is a novel nucleolar protein required for pre-25S rRNA processing and normal rates of cell growth at low temperatures.
INTRODUCTION
Ribosome synthesis is a remarkable and complex process requiring hundreds of different gene products and consuming much of the cell’s metabolic resources. Central to this process is synthesis of rRNA in the nucleolus. In rapidly growing yeast, synthesis of 35S pre-rRNA by RNA polymerase I accounts for ∼60% of total cellular transcription (1). In yeast, 18S, 5.8S and 25S rRNAs are synthesized from a 35S pre-rRNA by means of a series of co- and post-transcriptional processing steps. 5S rRNA is transcribed from a separate transcription unit by RNA polymerase III. 18S rRNA is incorporated into the small 40S subunit, whereas 5S, 5.8S and 25S rRNAs are incorporated into the large 60S subunit. Seventy-eight ribosomal proteins are incorporated into the two subunits in yeast. The process by which subunits are assembled in the nucleolus involves formation of RNA–protein interactions during pre-rRNA transcription and processing and results in a final architecture that places mature rRNAs at the heart of the ribosomal subunits (2).
Processing of pre-rRNA has been scrutinized for over two decades and our knowledge of RNA precursor–product relationships in this pathway is highly detailed (reviewed in 3,4). Covalent changes that occur during processing fall into two categories: (i) removal of spacer sequences not found in mature rRNAs and (ii) modification of ribose sugars and nucleotide bases that are found in mature rRNAs. Removal of external and internal transcribed spacer (ETS and ITS) sequences involves a complex series of endonucleolytic and exonucleolytic reactions that are absolutely required for ribosome synthesis. As a result, a multitude of processing endonucleases and exonucleases are in generous supply within the same nuclear subregion where pre-rRNAs are rapidly transcribed. While ETS and ITS sequences are degraded, mature rRNA sequences are protected from degradative mechanisms. Thus, the nucleolus must harbor mechanisms to protect pre-rRNAs from unwanted degradation, which is crucial to production of mature rRNAs. Protection undoubtedly entails binding of ribosomal proteins to pre-rRNAs in nascent ribosomal particles in the nucleolus, much in the same way ribosomal proteins stabilize mature rRNAs in the cytoplasm. Association of rRNA sequences with ribosomal proteins may also play a direct role in precursor processing by delimiting the extent to which spacer sequences are removed at 5′ and 3′ endpoints of mature rRNAs. Moreover, pre-rRNAs adopt specific and complex secondary and tertiary conformations during transcription and processing, despite their considerable length and size (mature 25S is 3396 nt in length and 1.11 MDa in size) and the nucleolus harbors mechanisms that facilitate RNA folding events, such as nucleolar ATP-dependent DExH-box helicases (5). In addition, interactions between ribosomal proteins and rRNAs are likely to assist in the formation and stabilization of particular secondary and tertiary rRNA structures.
In addition to the roles that ribosomal proteins play in protecting pre-rRNAs and stabilizing folded rRNA structures, it is possible that nucleolar RNA-binding proteins perform similar functions, perhaps on a more transient basis. Pre-RNAs may be stabilized immediately after transcription and before association with ribosomal proteins through interactions with nucleolar RNA-binding proteins that may function in either a specific or global, non-specific manner. Nucleolar RNA-binding proteins may bind and trap particular pre-rRNA folding intermediates and in so doing participate in establishment of rRNA conformation. Interactions of nucleolar RNA-binding proteins with pre-rRNAs may also assist in 5′ and 3′ end formation of mature rRNAs by delimiting extent of spacer sequence removal. In general, nucleolar RNA-binding proteins could be involved in various aspects of pre-rRNA processing, by broad analogy to hnRNA-binding proteins that participate in different aspects of pre-mRNA metabolism (6).
One of the best known and most widespread RNA-binding motifs is the canonical RNA recognition motif [RRM or RNA-binding domain (RBD); 7,8]. Three previously identified yeast nucleolar proteins have been found to contain RRMs: Nop4p/Nop77p (9,10), Nsr1p (11) and Sbp1p (12). Mutational analysis of the RRMs in Nop4p indicates that each of the four RRMs, one of which is non-canonical, is essential for function (13). Detailed functional information for RRMs in Nsr1p and Sbp1p is not yet available. Other RRM-containing proteins, such as Npl3p/Nop3p/Nab1p/Mtr13p (14–17) and Yra1p (18,19), have been localized to the nucleolus, but are present in the nucleoplasm as well. The majority of known RRM-containing nuclear proteins, including Npl3p and Yra1p, are involved in pre-mRNA splicing and mRNA export and not directly involved in pre-rRNA processing. It is clear from studies of RRM function in splicing factors and export factors that RRMs play a key role in RNA recognition during mRNA splicing, packaging and export (20). Structural studies have begun to reveal the molecular basis for RRM function (20). It is clear that RRMs typically act as RNA recognition modules that exercise their conserved function in a variety of different protein structural and functional contexts (20). Generally speaking, RRMs can function in the formation long-lived RNA–protein interactions within stable ribonucleoprotein complexes, or in more short-lived RNA–protein interactions during RNA processing and transport. Although significant progress has been made in understanding RRM function during pre-mRNA processing, less is known about RRM function during pre-rRNA processing.
In this report, we describe the identification of two novel RRM-containing proteins: Nop12p and Nop13p. NOP12 was identified in a genetic screen for mutations synthetically lethal with the nop2-4 ts allele, which is described in Hong et al. (21). Nop12p is nucleolar, whereas Nop13p is primarily nucleolar but is also present in the nucleoplasm to a lesser extent. Deletions of NOP12 and NOP13 are not lethal, but a nop12 strain is cold sensitive (cs) and defective in pre-25S rRNA processing at low temperatures.
MATERIALS AND METHODS
Media and reagents
Yeast were grown in YPD or SD medium plus supplements (22). Bacteria were grown in LB medium as described (22). Red/white colony sectoring (23) was scored on SD media containing 25% of the normal concentrations of histidine and adenine (22). 5-Fluoroorotic acid (5-FOA) was added to medium at a final concentration of 1 mg/ml (24). In general, for the PCR methods described below, Pfu polymerase (Stratagene) was used for PCR-based gene clonings, whereas Taq polymerase (Promega) was used in PCR reactions done for analytical purposes.
Strains and plasmids
YJPA22 was constructed by crossing YBH21 (nop2::LEU2, ade2, his3, leu2, trp1, ura3, pRS313/nop2-9; 21) and YCH128 (ade2, ade3, leu2, lys2, ura3; a gift from C. F. Hardy, Washingon University). A temperature sensitive meiotic segregant that was white, ade–, his–, leu+, lys+, trp–, ura– was transformed to ade+, his+, ura+ with pPW67 (NOP2, ADE3, URA3) (Table 1). Following growth in the presence of histidine to permit loss of pRS313/nop2-9, a sect–, temperature-resistant isolate sensitive to 5-FOA was selected and termed YJPA22. YJPA22 was transformed with pJPA83, pJPA84, pJPA85, pJPA86, pJPA89 or pJPA90 (Table 1) to yield YJPA23, YJPA24, YJPA25, YJPA26, YJPA29 and YJPA30, respectively. YKW157 was created by replacing pPW67 in YJPA24 with pKW12 by plasmid shuffling on medium containing 5-FOA. During efforts to construct nop2 strains appropriate for synthetic lethal (sl) screens, we were able to integrate nop2-4 and nop2-5 into the YCH128 genome at nop2::TRP1 using linearized integrating vector pRS304 and confirm integration by PCR (data not shown). However, none of the hundreds of ura3–, 5-FOAR colonies screened exhibited temperature sensitivity. This may be indicative of a strong selection against nop2-4 and nop2-5 at unit copy in the genome. Certain conditional lethal alleles of SNM1 are not maintained in single copy in the genome (25). Thus, we used a plasmid-based approach for sl screens.
Table 1. Yeast strains and plasmids.
| Strain |
Description |
Source |
|
| BY4141 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ | Research Genetics | |
| BY4742 | MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ | Research Genetics | |
| BY4773 | BY4741/BY4742 | EUROSCARFa | |
| YJPA22 | nop2::LEU2, ade2, ade3, his3, leu2, trp1, ura3, pPW67 | This study | |
| YJPA24 | YJPA22, pJPA84 | This study | |
| YKW35 | YJPA22, nop12-1 (Lys359→Ter) | This study | |
| YKW157 | YKW35, pKW12 | This study | |
| YKW180 | BY4743, pKW42 (expresses Nop12p–HA) | This study | |
| YKW182 | Y11732, pKW39 (expresses Nop12p) | This study | |
| YKW183 | Y11732, pKW42 (expresses Nop12p–HA) | This study | |
| YKW184 | BY4743, pKW49 (expresses Nop13p–HA) | This study | |
| YKW189 | Y11732, pKW44 | This study | |
| YKW190 | Y11732, pKW46 | This study | |
| YKW191 | Y11732, pRS313 | This study | |
| Y01732 | BY4741, nop12::kanMX4 | EUROSCARF | |
| Y11732 | BY4742, nop12::kanMX4 | EUROSCARF | |
| Y02038 | BY4741, nop13::kanMX4 | EUROSCARF | |
| Plasmid | Relevant functional DNA | Vector | Source |
| pJPA30 | NOP2 | pRS314 | (11) |
| pJPA83 | nop2-3 | pRS314 | This study |
| pJPA84 | nop2-4 | pRS314 | This study |
| pJPA85 | nop2-5 | pRS314 | This study |
| pJPA86 | nop2-6 | pRS314 | This study |
| pJPA89 | nop2-9 | pRS314 | This study |
| pJPA90 | nop2-10 | pRS314 | This study |
| pKW12 | NOP2, ADE3 | pRS313 | This study |
| pKW28 | Sau3AI fragment from Ch XV(coordinates 249460–259373) | YCp50 | This study |
| pKW30 | Sau3AI fragment from Ch XV(coordinates 248599–257589) | YCp50 | This study |
| pKW36 | RPS15 | pRS313 | This study |
| pKW38 | RPP2A | pRS313 | This study |
| pKW39 | NOP12 | pRS313 | This study |
| pKW42 | HA-tagged NOP12 | pRS313 | This study |
| pKW44 | nop12-1 (Lys359→Ter by PCR) | pRS313 | This study |
| pKW46 | HA-tagged nop12-1 (Lys359→Ter + HA by PCR) | pRS313 | This study |
| pKW49 | HA-tagged NOP13 | pRS313 | This study |
| pPW67 | NOP2, ADE3, 2 µ origin | pTSV31A | This study |
aEUROSCARF, European Saccharomyces cerevisiae archive for functional analysis
Plasmids pJPA83, pJPA84, pJPA85, pJPA86, pJPA89 and pJPA90 were constructed by inserting KpnI fragments from pBH55, pBH60, pBH67, pBH72, pBH74 and pBH77 (21), respectively, into pRS314 (26). pPW67 was generated by cloning NOP2 with 715 nt of 5′ and 456 nt of 3′ flanking sequence on a SacI fragment from pJPA40 (27) into the SacI site of pTSV31. To create pKW12, ADE3 in pTSV31 was amplified by PCR using primers KW1, 5′-ATGCGGCCGCTACTTACGTGAGCTAAAGCACAGATT-3′ and KW2, 5′-ATGAGCTCGTCCAATACCGTTTTTGACTTATC-3′ and cloned into the SacI and NotI sites of pBH49 (21). pKW28 was isolated by complementation of the sl phenotype and consists of a Sau3A fragment (249460–259373 of chromosome XV) cloned into the BamHI site of YCp50. RPS15 and RPP2A were amplified by PCR using primer pairs p15U2, 5′-ATGGATCCGCTATTGTTGCTTTCCGTGTA-3′ and p15L2, 5′-CGGGATCCTTGGTTCTGTTGTTGTGTATG-3′; and p2AU, 5′-ATGGATCCTGTACTTCTAGTTCGTCTTAT-3′ and p2AL, 5′-TCGGATCCTTCTAACCAGTAAAACAATCG-3′, respectively. Following amplification, the products were subcloned into the BamHI site of pRS313, to yield pKW36 and pKW38, respectively. pKW39 contains a NOP12 PCR product from primers p41U2, 5′-AAGGATCCAAAATACTGTGAGGCATTACTATCTTG-3′ and p41L2 5′-ATGGATCCTGCTACTACTTCCCGTTTCATCCCA-3′ cloned into the BamHI site of pRS313 (26). pKW42 consists of an HA-tagged NOP12 PCR product from primers p41HA5′, 5′-CGCCAGGATCCGCACTTTATTAAGAAAACAGTCG-3′ and p41HA3′, 5′-GCGCAGGATCCCTAAGCATAATCAGGAACATCATATGGATATTTCTTGCTCTTTTCAGCTTG-3′ cloned into the BamHI site of pRS313. The resulting C-terminal sequence is ...QAEKSKKYPYDVPDYA468 (introduced HA epitope tag amino acids are underlined). pKW44 carries a nop12-1-like allele that consists of the NOP12 promoter and the coding region for amino acids 1–358. pKW44 was created by cloning the PCR product obtained with primers p41HA5′ and pMu3′, 5′-GCGCCGGGATCCTTAAGTTGGTTTCTTTGTATTTTCG-3′, into the BamHI site of pRS313. pKW46 was constructed identically to pKW44 with the exception that primer pMuHA3′, 5′-GCGCAGGATCCCTAAGCATAATCAGGAACATCATATGGATAAGTTGGTTTCTTTGTATTTTCGT-CT-3′ was used in place of primer pMu3′, resulting in the introduction of the HA epitope tag after T358, to yield the C-terminal sequence TKKPTYPYDVPDYA367. pKW49 contains the HA-tagged NOP13 PCR product from primers p75-5GE2, 5′-CGCCAGGATCCATGACGTACTCGCCATACCTAACT-3′ and p75–3HA2, 5′-GCGCAGGATCCCTAAGCATAATCAGGAACATCATATGGATAGTCAAACTTCACTTTCTTA-CCTTGTG-3′ cloned into the BamHI site of pRS313.
For all experiments requiring the use of strains bearing plasmid-based gene constructs, freshly prepared transformants were used. In all cases this entailed colony purification of transformants and performing experiments with duplicate isolates. We found that plasmid-bearing, colony-purified strain isolates stored as patches for 2–3 weeks on SD minimal selective media at 4°C did not yield consistent results, even when grown again under selective conditions. Problems with strains stored in this manner for 2–3 weeks were clearly apparent in immunofluorescence localization experiments, in which the number of cells expressing HA-epitope tagged nucleolar protein declined roughly in proportion to length of storage (data not shown). Such problems were invariably overcome by preparation of fresh transformants, which yielded consistent results.
Synthetic lethal screen and characterization
UV irradiation was used to mutagenize ∼400 000 c.f.u. of YJPA24 (Fig. 1A) on Petri dishes to ∼10% survival. Of 203 sect– colonies obtained, 76 were sensitive to 5-FOA, of which 73 were capable of spontaneously losing pJPA84. Following re-transformation with pJPA84, 52 colonies remained sect– and 5-FOAS. One of these, YKW35, exhibited a ‘tight’ sect– phenotype. YJPA24 was selected for the sl screen because a prior screen of mutagenized YJPA25, which exhibits more robust sectoring, yielded isolates with ‘leaky’ sect– phenotypes. In general, YJPA22-based strains offered the advantage that synthetic lethality could be scored on the basis of colony sectoring as well as growth on 5-FOA-containing medium, which was used to discriminate between tight and leaky phenotypes.
Figure 1.
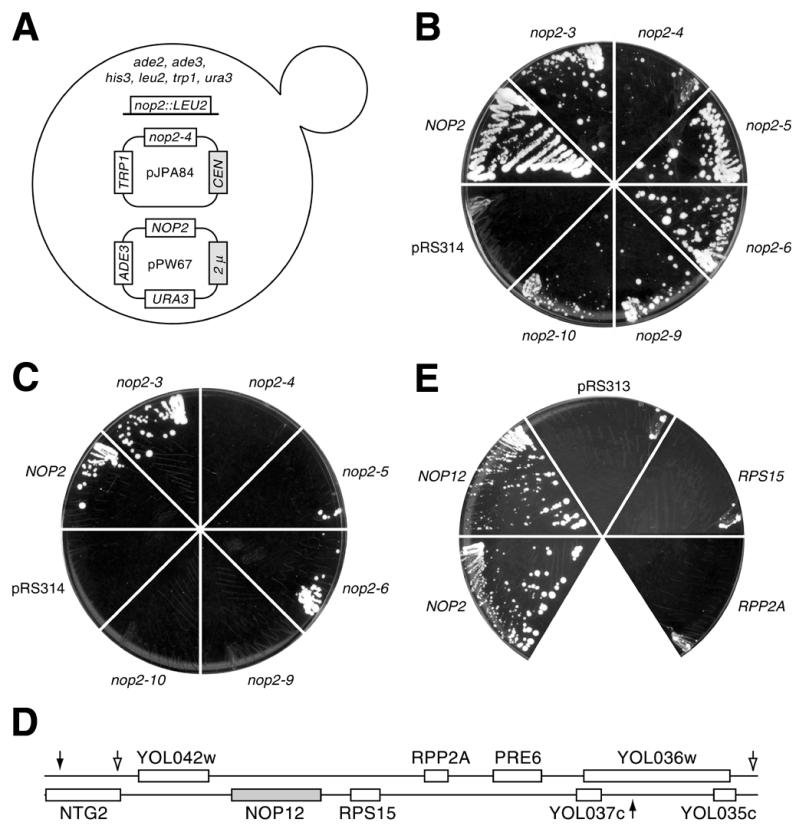
nop12-1 is synthetically lethal with nop2-4. (A) Diagram of strain YJPA24 used for the sl screen. Retention of pJPA84 is selected on minimal medium minus tryptophan. Spontaneous loss of pPW67 at permissive temperature (25°C) results in white sectors in red colonies and confers resistance to 5-FOA. (B) Growth at 25°C on medium containing 5-FOA. YJPA22 was transformed with pRS314, or plasmids bearing NOP2 or different nop2 ts alleles. NOP2 supports loss of pPW67, whereas pRS314 does not. Temperature sensitive nop2 alleles support spontaneous loss of pPW67 and growth to different extents. (C) Allele specificity. The nop12-1 allele in YKW35 is synthetically lethal with nop2-4, nop2-9 and nop2-10, but not with nop2-3, nop2-5 or nop2-6. YKW35 transformants that carry nop2-4, nop2-9 or nop2-10 cannot spontaneously lose pPW67 and remain viable. (D) Complementation of synthetic lethality. Region of chromosome XV that complements the sl phenotype in YKW157. Open arrows denote endpoints of insert in pKW28. Filled arrows denote insert in pKW30. RPS15 is also known as RPS21. (E) NOP12 supports growth on medium containing 5-FOA. YKW35 bearing nop2-4 (on pJPA30) can afford to lose pPW67 and remain viable on medium containing 5-FOA if transformed with NOP2 (pBH49) or NOP12 (pKW39), but not with control plasmid pRS313, or plasmids bearing RPS15 (pKW36) or RPP2A (pKW38).
To complement the sl phenotype in YKW35, plasmid shuffling was used to replace pPW67 with pKW12, to yield YKW157. A yeast genomic library in YCp50 (Rose, Bank A, from ATCC) was used to complement the sect– phenotype and 51 sect+ isolates were obtained. Plasmid DNAs were extracted as described (28), used to transform Escherichia coli and AmpR E.coli colonies were analyzed by PCR to exclude those containing NOP2 (15 out of 51). Plasmid DNAs from E.coli colonies were retransformed into YKW157. Eight plasmid DNAs conferred strong sectoring, all of which fell into two classes, based on restriction mapping. Insert ends were sequenced using primers 5′-CGTGGCCGGGGGACTGT-3′ and 5′-GCCGGAAGCGAGAAGAATCATAAT-3′ flanking the BamHI site in the tet gene of YCp50 and two overlapping inserts of ∼9.0 and ∼9.9 kb from chromosome XV were identified.
To identify the mutation(s) in the nop12-1 allele, primers p41U2 and p41L2 were used to amplify the coding region plus 1.7 and 0.5 kb of 5′- and 3′-flanking regions, respectively. Three PCR products from three separate reactions were sequenced in their entirety. In each case, the ORF plus ∼100 nt of flanking sequence was sequenced on both strands using Big-Dye terminators (Perkin Elmer).
Immunofluorescence localization
YKW180 and YKW184 were grown in SD medium without histidine to OD600 ≤ 0.25, centrifuged at 250 g for 10 min and fixed in 3% freshly hydrolyzed paraformaldehyde, 50 mM NaPO4, pH 7 for 30 min at ∼25°C. Addition of 3% formaldehyde directly to SD media yielded poor fixation and cell morphology (data not shown). Slide preparation, primary incubation with anti-HA mAb 16B12 (IgG1; 1/200 dilution; BAbCo) and anti-Nop1p mAb A66 (IgG3; 1/20 000 dilution; 29), secondary incubation with isotype-specific fluorochrome-conjugated anti-mouse IgG antibodies (1/10 dilution; Southern Biotechnology Associates) and counterstaining with 4′,6-diamidino-2-phenylindole (DAPI) were performed as described (30). Data were collected on a Zeiss Axiophot microscope equipped with a 100× Neofuar objective lens and a CCD-based digital camera using the single color channel mode.
Northern and western blotting
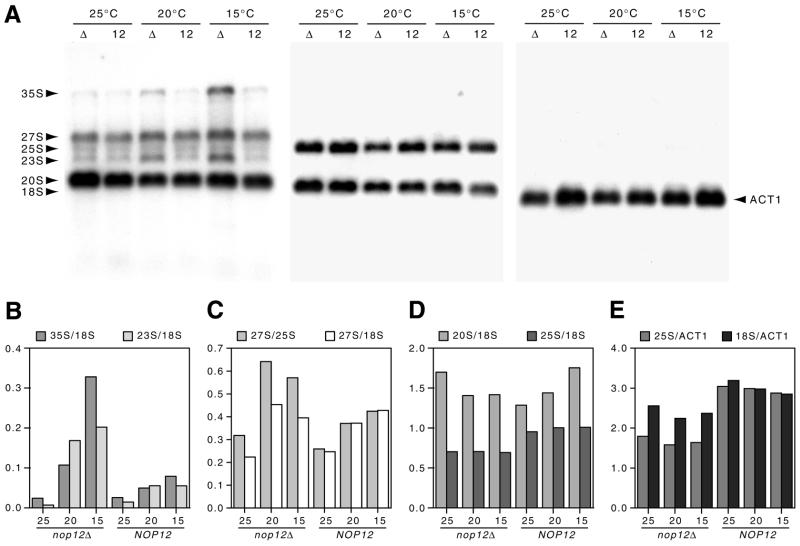
Western blots of whole cell extracts prepared as previously described (30) were probed with anti-HA tag mAb 16B12 at 1/10 000 and processed for chemiluminescent detection as described (30). Total RNA was prepared as described (30) and separated on formaldehyde gels (22). Following three 10 min washes in ddH2O, RNAs were transferred to positively charged nylon membrane (Bio-Rad) in 7.5 mM NaOH. Detection of pre-rRNAs by northern hybridization was done using the following 5′-32P-end-labeled oligonucleotide probes: 18S, 5′-AGCCATTCGCAGTTTCACTG-3′; 20S, 5′-GCACAGAAATCTCTCACCGT-3′; 25S, 5′-TACTAAGGCAATCCGGTTGG-3′; 27S, 5′-CATCCAATGAAAAGGCCAGC-3′. Simultaneous hybridizations were done by combining 18S and 25S probes, or 20S and 27S probes, in one hybridization mixture. To detect ACT1 mRNA, a region of the ACT1 gene from position 21–723 was used as template to prepare random-primed labeled probe with a kit (New England Biolabs). Northern blots were stripped in between hybridizations by 1 min exposure to 10 mM Tris–HCl, 1 mM EDTA pH 8 at 100°C. For quantitation of hybridization results, data were collected with a Storm PhosphorImager (Molecular Dynamics) and analyzed using ImageQuant image analysis software. Local background values (from a grid rectangle immediately above or below an identically sized band rectangle) were subtracted from specific signal values due to hybridization of probe. Ratios presented in Figure 5 are quotients of data obtained in this manner and do not reflect any further mathematical manipulation. Thus, the absolute values of these ratios are determined solely by the specific data values corrected only for local background on the northern blot.
Figure 5.
Northern analysis of steady-state levels of pre-rRNAs and rRNAs. (A) Northern blots of total RNA were prepared from the nop12Δ strain (Δ) or the isogenic NOP12 strain (12). Early log cultures at 25°C were continued at 25°C or shifted to either 20 or 15°C for 5 h prior to RNA extraction. The blot was hybridized sequentially with probes to ITS1 and ITS2 (which detect 35S, 27S and 20S pre-rRNAs); 25S and 18S rRNAs and ACT1 mRNA. (B–E) Quantification of hybridization results. Data for individual bands were acquired with a PhosphorImager, corrected for local background and used to calculate ratios (without any further computational manipulation). Thus, the absolute values on the ordinate axis are arbitrary and presented without units.
Pulse–chase labeling analysis
Pulse–chase analysis of rRNA precursor–product relationships was peformed as described (30). Pulse-labeling and chase incubations were carried out at 15°C. Because of this low temperature, pulse-labeling was done for 5 min and chase times were extended for up to 60 min. Isolated RNAs were separated on formaldehyde gels (22), transferred to positively charged nylon membrane under alkaline conditions (22) and visualized by fluorography using En3Hance spray as directed by the manufacturer (DuPont NEN).
RESULTS
nop12-1 is synthetically lethal with nop2-4
To pursue a better understanding of Nop2p function and mechanisms of rRNA processing in general, we sought to identify mutations in novel genes that were synthetically lethal with temperature sensitive mutations in nop2 (21). Strain YJPA22 was constructed in order to generate a colony-sectoring phenotype under permissive conditions (25°C) that allow spontaneous loss of plasmid pPW67, which carries NOP2 and ADE3 (Table 1). Loss of pPW67 causes formation of white sectors in red colonies. In addition to colony sectoring, sensitivity to the drug 5-FOA was used to score spontaneous loss of pPW67 as loss of pPW67 confers resistance to 5-FOA. Thus, YJPA22 was designed so that plasmid-borne nop2 ts alleles could be easily transformed in and tested using a combination of red/white colony sectoring and sensitivity to 5-FOA. In these studies, resistance to 5-FOA was a more reliable indicator of synthetic lethality, although not as convenient as the sectoring assay.
Transformation of YJPA22 with plasmids bearing different nop2 ts alleles allowed us to compare the extent to which different alleles supported growth in the absence of wild-type NOP2 on plasmid pPW67. nop2-4 and nop2-10 supported spontaneous loss of pPW67 only poorly (Fig. 1B). In contrast, nop2-3, nop2-5, nop2-6 and nop2-9 supported higher rates of loss of pPW67 and more robust growth in the presence of 5-FOA (Fig. 1B). Formation of white sectors, due to loss of pPW67, gave qualitatively similar results, with nop2-4 and nop2-10 colonies sectoring least (data not shown). YJPA24, which carries nop2-4 (Fig. 1A), was used for a sl screen and was selected because we viewed the weak sectoring phenotype as an indication that this allele would afford higher sensitivity to synthetically lethal mutations. A previous screen using YJPA25, which carries nop2-5 and sectors well, was not successful in identifying mutations synthetically lethal with nop2-5 (data not shown).
YJPA24 was mutagenized by UV irradiation and screened for loss of red/white colony sectoring on minimal medium that selected for retention of pJPA84. Sect– colonies were tested for sensitivity to 5-FOA (to exclude mutations in ADE3 on pPW67). Trp– isolates were retransformed with pJPA84 and retested (to exclude intragenic mutations within nop2-4 on pJPA84). One mutant strain, YKW35, exhibited a tight sl phenotype and was transformed with pKW12 (NOP2, ADE3), which allowed loss of pPW67, resulting in the creation of strain YKW157 (Table 1). YKW157 was transformed with a yeast genomic library in YCp50 (31) and sect+ transformants were isolated. Two insert-containing YCp50 plasmids, pKW28 and pKW30, yielded the most pronounced sectoring, were isolated more than once each and contained overlapping inserts consisting of a region of chromosome XV (Fig. 1D). Eight ORFs were common to the region of overlap: YOL042w, YOL041c (NOP12), RPS15, RPP2A, PRE6 and YOL037c (Fig. 1D). YOL042w is predicted to encode a protein of unknown function that is similar to Ccr4p, an RNA Pol II transcription factor (32). If YOL037c encodes a protein, it would not be similar to protein entries in current databases. PRE6 encodes an essential subunit of the 20S proteasome (33). Initially, we found it interesting and somewhat confounding that the three remaining genes could be considered potential suppressors of the sl phenotype. RPP2A (≠ RPP2/POP7, which encodes a subunit of RNases P and MRP) and RPS15 (= RPS21) encode, respectively, the non-essential 60S acidic ribosomal protein P2A (also known as L44) (34) and the essential 40S ribosomal protein S15 (also known as S21) (35). YOL041c was predicted to encode a protein similar to the nucleolar protein Nsr1p, which functions in pre-18S rRNA processing (36). To determine which of these three ORFs complemented the synthetic lethality, each was subcloned and tested for ability to support loss of pPW67 in YKW35 (Fig. 1E). Only one, YOL041c, permitted loss of pPW67 similar to the positive control, NOP2 (Fig. 1E). RPS15 and RPP2A did not permit loss of pPW67 beyond that observed with pRS313, the negative control (Fig. 1E). YOL041c encodes a 51.9 kDa protein with a pI of 9.4. For reasons given below this ORF was designated NOP12.
To ascertain the nature of the nop12-1 mutation that is responsible for the sl interaction, the nop12-1 allele was sequenced. One change at the nucleotide level was found: A1075→T. This results in the introduction of a nonsense codon (TAA) at position 359, which normally encodes lysine (AAA) (Fig. 2). We conclude that the nop12-1 allele is recessive to NOP12, based on the finding that NOP12 complements the sl phenotype observed in strain YKW35 (Fig. 1E).
Figure 2.
CLUSTAL alignment of Nop12p (encoded by YOL041c), Nop13p (encoded by YNL175c) and Nsr1p (11). Identical amino acids are boxed. RRM elements RNP-1 (underlined octapeptide) and RNP-2 (underlined hexapeptide) are indicated. RNP-1 and RNP-2 elements in Nsr1p that are not present in Nop12p are indicated (dashed underline). Amino acid residues within the conserved RRM that are present in either Nop12p or Nop13p and both Nsr1p and mammalian nucleolin are denoted with a filled circle. Lysine359, which is mutated to a stop codon in nop12-1, is denoted with a black box.
Nop12p shares sequence similarity with Nop13p (encoded by YNL175c) and Nsr1p
BLASTP searches of GenBank yielded highest scores for relatively short stretches of similarity between Nop12p and other proteins containing RRM motifs. In all cases, amino acids within RRM motifs RNP1 and RNP2 were the most highly conserved regions (data not shown). Most common were RRM motifs present in poly(A) binding proteins from multiple species (data not shown). To analyze and extend the results from database mining using BLAST (37), pairwise and multiple protein sequence alignments were done using Lipman–Pearson and CLUSTAL methods (38) (using default parameters).
Two proteins not from Saccharomyces cerevisiae exhibited similarity over most or all of their lengths. A Drosophila protein (Entrez Protein Record 7298360; Drosophila gene product CG12288) was found to be 24% identical to Nop12p (96 identities in a Lipman–Pearson alignment extending over 88% of Nop12p from residues 39–443). The Drosophila protein is predicted to be 48.0 kDa in size, with a pI of 10.1. A putative RNA-binding protein from Schizosaccharomyces pombe (Entrez Protein Record 7492926) was found to be 38% identical to Nop12p (175 identities in a Lipman–Pearson alignment extending over the entire length of Nop12p from residues 1–459). The S.pombe protein is predicted to be 49.4 kDa, with a pI of 9.4. Considering the similarities to Nop12p, which is 51.9 kDa with a pI of 9.4, the S.pombe protein is likely to be a Nop12p ortholog.
Saccharomyces cerevisiae gene products exhibiting significant similarity over the majority of the length of Nop12p include the predicted protein encoded by YNL175c, Nsr1p and Mrd1p. For reasons described below we refer to the YNL175c gene product as Nop13p. The multiple RNA-binding domain (Mrd), Mrd1p, is a 101.1 kDa protein that is 17.4% identical to Nop12p over 50% of the length of Mrd1p (78 identities in a Lipman–Pearson alignment extending over 97% of length of Nop12p from residues 12–458). Nop13p and Nsr1p exhibit significant similarity to Nop12p and are similar to each other. Nop13p has a predicted size of 45.7 kDa and pI of 9.3. Nop13p is 20% identical to Nop12p (73 identities in a Lipman–Pearson alignment extending over the entire length of Nop13p from residues 1–363). Nsr1p is 18.8% identical to Nop12p (55 identities in a Lipman–Pearson alignment extending over 64% of the length of Nop12p from residues 46–337). The sequence similarities between Nop12p, Nop13p and Nsr1p are depicted in Figure 2. Nop12p contains a single RRM, whereas Nsr1p has two RRMs (Fig. 2). Nop13p contains one canonical RRM and one RRM that departs from the consensus at position 188, which is M rather than L, I, V or A (Fig. 2). The RRM motifs are highly conserved in all three proteins and there is considerable similarity in regions surrounding the RRM to regions near an RRM in nucleolin, which has been previously reported to be similar to Nsr1p (39). Although Nop12p and Nop13p are similar, the corresponding genes do not appear to have arisen from an ancient gene duplication event (40).
Deletion of NOP12 causes a cs growth phenotype
To determine if deletion of NOP12 resulted in a growth phenotype, strains in which NOP12, NOP13 or NSR1 were replaced by KanR were obtained from sources associated with the Saccharomyces Genome Deletion Project (41) (Table 1). Deletion of NOP12 resulted in impaired growth at 25 and 15°C when compared to an isogenic wild-type strain (Fig. 3A). No significant reduction in growth in the nop12Δ strain was observed at 30 or 37°C. The impairment of growth due to nop12Δ was similar phenotypically to the impairment of growth in an isogenic nsr1Δ strain, with the nop12Δ strain growing somewhat better than the nsr1Δ strain at 25°C (Fig. 3A). Deletion of NOP13 did not result in an observable temperature-dependent growth phenotype.
Figure 3.
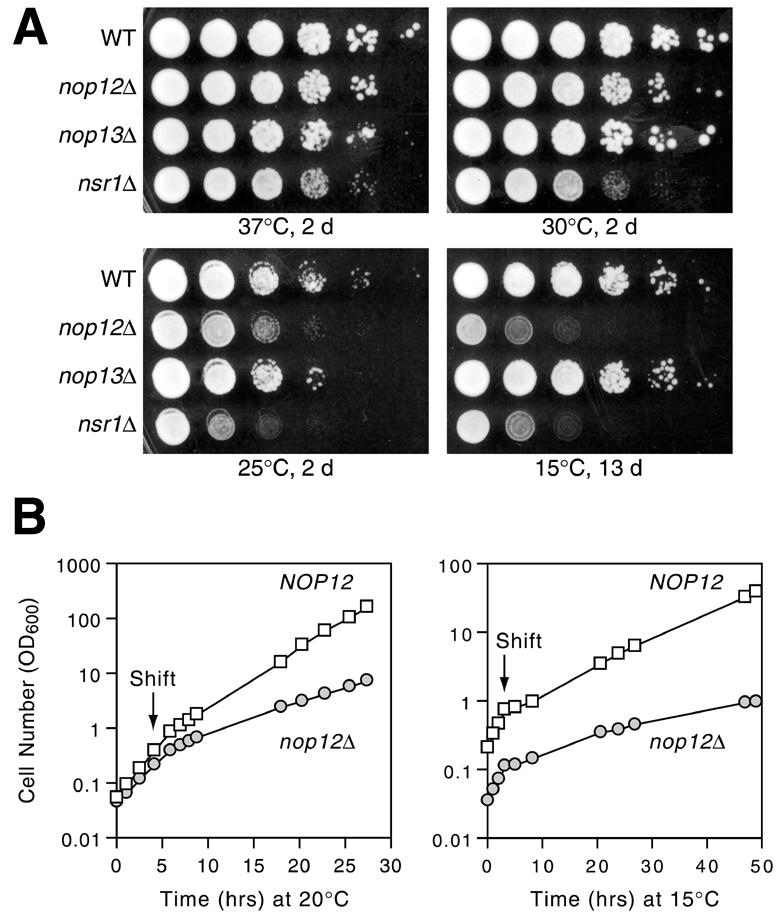
Cold sensitivity in a nop12Δ strain. (A) Replica platings of serial dilutions of wild-type (WT), nop12Δ, nop13Δ and nsr1Δ strains grown for 2 or 13 days at the indicated temperatures on YPD medium. (B) Growth in liquid culture of wild-type (NOP12) and nop12Δ strains at 30°C and after shift to 20 and 15°C. Cultures were diluted during the time course to maintain OD600 < 0.5.
We have extended the analysis of the cold sensitivity of the nop12Δ strain by examining two double knock-out mutants. A nop12Δ, nop13Δ double knock-out exhibited a cs growth phenotype on plates that is indistinguishable from the phenotype of the nop12Δ single knock-out alone (data not shown). This is not unexpected given that nop13Δ does not confer a cs phenotype. A nop12Δ, nsr1Δ double knock-out exhibits a cs growth phenotype on plates that is indistinguishable from the nsr1Δ single knock-out (data not shown). Our interpretation of this result is that the defect in 18S rRNA synthesis in the nsr1Δ mutant is more severe than the defect in 25S rRNA synthesis in the nop12Δ mutant.
In preparation to investigate the metabolic basis for cs growth in the nop12Δ strain, we measured the rate of onset of growth rate reduction at 15 and 20°C (Fig. 3B). Shift of a logarithmically-growing culture from 30 to 20°C resulted in a 2.9-fold reduction in growth rate in the nop12Δ strain, with doubling time increasing from 1.8 to 5.3 h. The wild-type control strain showed a 1.9-fold reduction in growth rate under the same conditions, with doubling time increasing from 1.5 to 2.8 h. The reduction in growth rate in the nop12Δ strain occurred within ∼4 h of shift and there was no perceptible growth lag after shift to 20°C (Fig. 3B). Shift of a logarithmically growing culture from 30 to 15°C resulted in a 6.9-fold reduction in growth rate in the nop12Δ strain, from a doubling time of 1.9 to 13.2 h (Fig. 3B). In comparison, the wild-type strain showed a 4.5-fold reduction in growth rate, going from a doubling time of 1.7 to 7.6 h (Fig. 3B). Immediately after shift to 15°C, growth slowed considerably for ∼2 h, as a response to cold shock (Fig. 3B). By 4 h after the shift to 15°C, cells had recovered from cold stress and were growing exponentially, albeit slowly. We sought to avoid lower temperatures, such as 10°C, even though cold sensitivity studies have been done at these temperatures (42), to avoid exacerbating cold stress-related effects.
Synthesis of 25S rRNA is impaired in a nop12Δ strain at 15°C
Reduction in growth rates of the nop12Δ strain at 20 and 15°C is consistent with a requirement for Nop12p in an aspect of ribosome biogenesis. This hypothesis was tested by pulse–chase analysis of rRNA synthesis in the nop12Δ strain and the isogenic control strain. Each strain was cultured at 30°C, shifted to 15°C for 5 h and pulse-labeled and chased at 15°C. In order to visualize mature rRNAs, chase incubations were done for longer times than are typically done for incubations at 25 or 30°C (30). Pulse–chase analysis revealed a specific decrease in 25S rRNA synthesis in the nop12Δ strain at 15°C (Fig. 4). The amount of mature 25S rRNA in the nop12Δ strain was ∼40% of the amount of the 25S rRNA present in the NOP12 strain, based on quantification of band intensities using NIHImage software. In addition, 35S and 27S pre-rRNAs were more persistent in the nop12Δ strain and were visible after 5, 10, 20 and 60 min of chase (Fig. 4). At 60 min of chase, there was little difference between the nop12Δ and wild-type strains with respect to levels of 18S rRNA or 20S pre-rRNA (Fig. 4). However, at 5 and 10 min of chase, the amount of mature 18S rRNA was diminished in the nop12Δ strain compared to the control. This is likely to be due to delayed processing of 35S pre-rRNA (Fig. 4). Consistent with this, early chase timepoints show a small amount of 23S intermediate, resulting from a failure to cleave at A0, A1 and A2 during synthesis of 18S. Although synthesis of 18S rRNA appears delayed, NOP12 does not appear to be required for normal levels of 18S rRNA and the delay in its synthesis may be a secondary effect. Essentially the same results have been obtained in similar pulse–chase experiments done using different pulse and chase times and temperatures. Thus, we conclude that NOP12 is required for synthesis of normal levels of 25S rRNA at low temperature.
Figure 4.
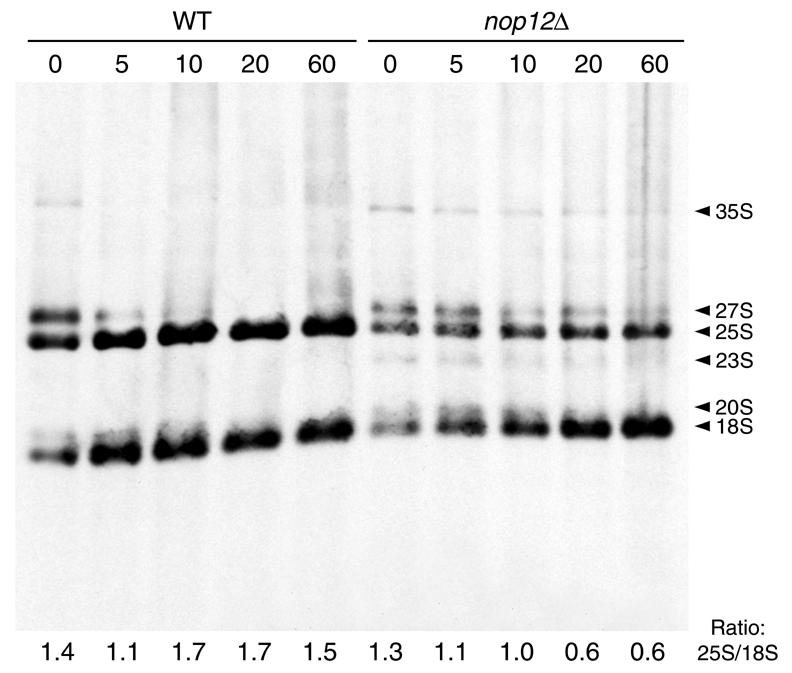
Pulse–chase analysis of rRNA synthesis. Wild-type and nop12Δ strains were cultured at 30°C, shifted to 15°C for 5 h, pulse-labeled with [3H-methyl]methionine for 5 min at 15°C and chased for the indicated times (in min) at 15°C. RNAs were separated by gel electrophoresis, transferred to nylon membrane and visualized by fluorography. Positions of pre-rRNAs and mature rRNAs are indicated. Ratios of band intensities (25S/18S) as determined by NIHImage software are shown below each lane.
To examine steady-state levels of rRNAs and their precursors, northern blot analysis was done. Cultures of wild-type and nop12Δ strains were grown to early log phase and either maintained at 25°C, or shifted to 20 or 15°C for 5 h, after which total RNAs were isolated. End-labeled oligonucleotides complementary to ITS1 and ITS2 were used to simultaneously detect 35S, 27S and 20S pre-rRNAs (Fig. 5A). End-labeled oligonucleotides were used to detect mature 25S and 18S rRNAs and an anti-ACT1 probe was used to detect yeast actin mRNA. Hybridization results were quantitated using a PhosphorImager and ratios between hybridization signals are presented graphically in Figure 5B.
The 35S pre-rRNA accumulated in the nop12Δ strain at 20°C and to a greater extent at 15°C (Fig. 5B). The 23S intermediate accumulated at both 20 and 15°C as well, consistent with a delay in processing of 35S. In the NOP12 control strain, levels of 35S and 23S increased by only a small amount (Fig. 5B). Levels of 27S pre-rRNA, the immediate precursor to 25S rRNA, also increased at 20 and 15°C in the nop12Δ strain compared to the control (Fig. 5C). Although there was a trend toward increase in levels of 27S pre-rRNA at reduced temperatures in the control strain, the increase in 27S levels was much greater in the absence of NOP12 (Fig. 5C). The increase in levels of 27S pre-rRNAs appeared at the expense of 25S rRNA levels, as shown by differences between 25S/18S ratios (Fig. 5D) and 25S/ACT1 ratios (Fig. 5E) in the nop12Δ and NOP12 strains. In contrast, levels of 20S pre-rRNA did not increase in the nop12Δ strain at reduced temperature but, rather, decreased (Fig. 5D). The net result is a significant decrease in the 25S/18S ratio in the nop12Δ strain compared to the control (Fig. 5D). A similar reduction in 25S levels was observed with ethidium bromide staining. Finally, comparison of 18S rRNA and ACT1 mRNA levels showed a high degree of uniformity over different temperatures (Fig. 5E), which validates the use of 18S rRNA levels as a basis of comparison in the computation of ratios.
Complementation of cold sensitive phenotype by epitope-tagged Nop12p
In order to develop a molecular handle for Nop12p, the coding sequence for the HA-1 epitope tag was introduced immediately upstream of the termination codon in NOP12. When expressed under the control of its cognate promoter from a centromeric plasmid, Nop12p–HA complements the cs growth defect observed at 20 and 15°C in the nop12Δ strain (Fig. 6A). Nop12p–HA restores levels of 25S rRNA to normal. This is apparent in ethidium bromide stained profiles of RNA prepared from strains shifted to 15°C and grown for 5 h (Fig. 6B). Expression of Nop12p–HA, as well as HA-tagged Nop13p, was confirmed by western blotting (Fig. 6C). HA-tagged Nop12p and Nop13p migrate at apparent molecular masses of 59 and 51 kDa, respectively, which is in reasonable agreement with the predicted sizes of 53 and 47 kDa (including epitope), respectively.
Figure 6.
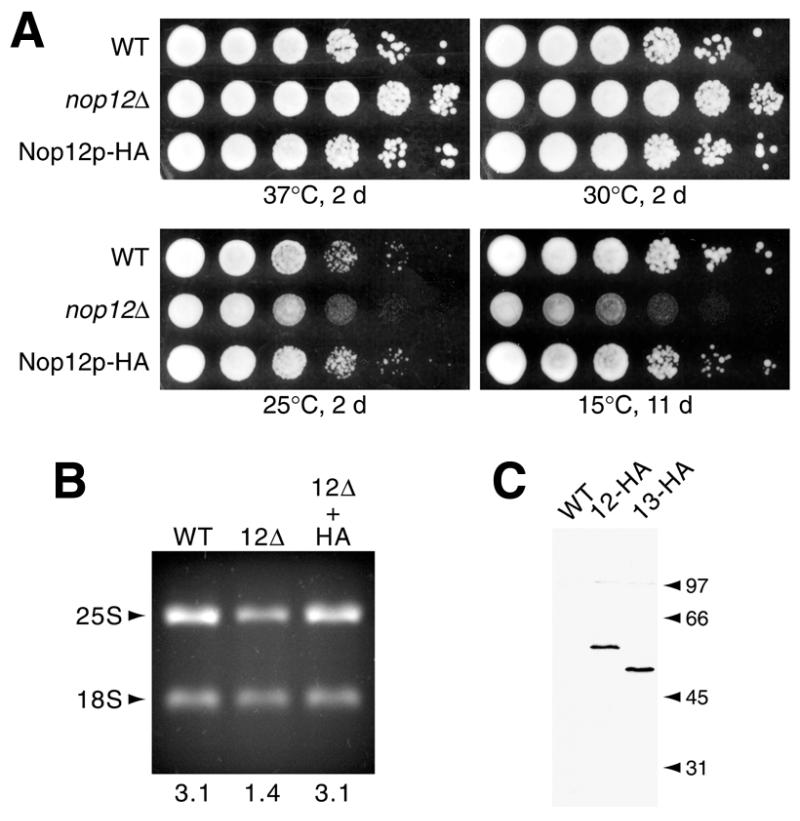
Nop12p–HA complements the nop12Δ cs phenotype. (A) Replica platings of serial dilutions were grown for 2 or 11 days at the indicated temperatures on YPD. The strains compared are wild-type (WT), nop12Δ and the nop12Δ strain transformed with a centromeric plasmid expressing HA-tagged Nop12p from its endogenous promoter (pKW42). (B) Ethidium bromide-stained rRNAs. Total RNAs were isolated from the strains described above grown at 15°C for 5 h and were separated on a formaldehyde agarose gel. Ratios of band intensities (25S/18S) as determined by NIHImage software are shown below each lane. (C) Western blot. Total cell protein extracts prepared from wild-type cells, or cells expressing HA epitope tagged Nop12p or Nop13p, were analyzed by western blotting with mAb 16B12, followed by chemiluminescent detection. Positions of molecular standards are indicated (in kDa).
Immunofluorescence localization of epitope-tagged Nop12p and Nop13p
The subcellular distribution of Nop12p–HA was determined using indirect immunofluorescence localization and was compared to the intracellular distribution of the nucleolar protein Nop1p (yeast fibrillarin). Isotype-specfic secondary antibodies were used in conjunction with primary monoclonal antibodies of different subclasses. Nop12p–HA under control of its endogenous promoter on a centromeric plasmid was expressed in almost all cells and in virtually all cells co-localized precisely with Nop1p (Fig. 7). Not all cells expressed the same level of Nop12p–HA, but the observed signal intensities extended over a relatively narrow range of only a few-fold (Fig. 7a). Subtle variation in staining intensities within a given panel in Figure 7 is due in large part to the fact that nuclei in different cells lie in slightly different focal planes in a given field. The Nop12p–HA staining pattern was distinctly crescent shaped, which is typical of the nucleolus in yeast. In addition, the nucleolus is typically offset from the chromatin (stained with DAPI), which is apparent in cells in which the orientation of the nucleus allows this side-by-side arrangement to be discerned.
Figure 7.
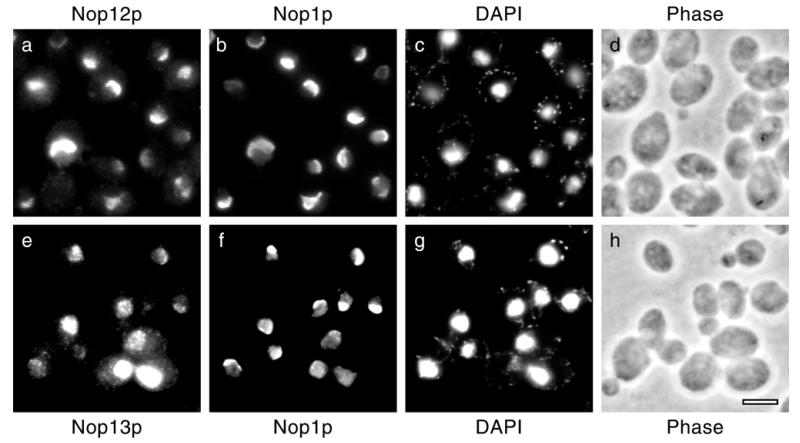
Immunofluorescence localization. Yeast cells expressing epitope tagged Nop12p or Nop13p were prepared for microscopy and incubated with antibodies as described in Materials and Methods. Nop12p–HA (a) and Nop13p–HA (e) were localized simultaneously with Nop1p (b and f). Counterstaining with DAPI revealed the distribution of chromatin (c and g). Phase contrast images (d and h) of the same fields of cells are shown. Bar, 5 µm.
The intracellular distribution of Nop13p–HA was also determined using this approach. Although Nop13p–HA was expressed from its endogenous promoter on a centromeric plasmid, like Nop12p–HA, there was a broader range of staining intensities, with some cells overexpressing Nop13p–HA (Fig. 7e). Nevertheless, Nop13p–HA was localized primarily to the nucleolus based on concentration of staining intensity in a region offset from chromatin (Fig. 7g) and significant colocalization with Nop1p (Fig. 7f). Clearly, however, Nop13p–HA is not exclusively colocalized with Nop1p, in contrast to the case with Nop12p. Moreover, the Nop13p–HA staining pattern was punctate, unlike the Nop12p pattern, which shows little punctate character (Fig. 7a). Overexpression of Nop13p–HA resulted in a bright nuclear staining pattern, reflecting the confinement of Nop13p–HA to the nucleus but not the nucleolus (Fig. 7e). In most cells overexpressing Nop13p–HA, the distribution of Nop1p was more diffuse and less restricted to a typical crescent-shaped nucleolar region (Fig. 7e and data not shown). Thus, Nop12p and Nop13p are novel nucleolar proteins in S.cerevisiae.
The nop12-1 gene product fails to complement cold sensitive phenotype
Two key observations regarding NOP12 function have been made: (i) the nop12-1 allele is synthetically with nop2-4 and (ii) the nop12 null allele confers a cs phenotype. We were interested in determining if these two phenotypes shared a common basis and wished to test if nop12-1 could complement the cs phenotype. PCR was used to create two ‘nop12-1-like’ alleles under control of the NOP12 cognate promoter in plasmids. Plasmid pKW44 carries a ‘nop12-1’ ORF that encodes amino acids 1–358. pKW46 carries a ‘nop12-1–HA’ allele that encodes amino acids 1–358 immediately followed by an HA epitope tag followed by a stop codon. These two plasmids were transformed into the nop12Δ strain and examined.
Interestingly, the cs phenotype observed in the nop12Δ strain was not complemented by either the ‘nop12-1’ or the ‘nop12-1–HA’ alleles (Fig. 8A). At 25 and 15°C, ‘nop12-1’ and ‘nop12-1–HA’ showed the same amount of growth as observed with the control strain transformed with vector pRS313 alone (Fig. 8A). As was shown above, plasmid bearing NOP12 plus a C-terminal epitope tag coding region, NOP12–HA, restored normal growth at 25 and 15°C (compare Figs 8A and 6A). To extend our analysis, we wished to investigate the basis for the inability of the ‘nop12-1–HA’ allele to complement the cs phenotype. Potentially, the ‘nop12-1–HA’ allele could be non-functional because it encodes a truncated gene product that is unstable or mislocalized. The stretch of amino acids 359–469 removed by the C-terminal truncation is highly basic (pI >11) and contains potential nuclear localization sequence motifs, which may play a role in nucleolar localization. Immunofluorescence localization experiments revealed that the Nop12-1p–HA protein was not detected at levels comparable to Nop12p–HA (Fig. 8B). Long CCD camera exposure times were capable of detecting low levels of HA-specific signal in the nucleolus (data not shown). Based on a comparison of exposure times, we estimate that the nucleolar signal attributable to Nop12-1p–HA was <10% of the signal intensity level generated by Nop12p–HA (data not shown). Although signal levels were low, no relative increase in cytoplasmic or nucleoplasmic signal was observed in the ‘nop12-1–HA’ strain (data not shown). This suggests that ‘nop12-1–HA’ encodes a protein with a significantly shortened half-life and suggests that shortened protein half-life may underlie the sl phenotype conferred by the nop12-1 allele in strain YKW35.
Figure 8.
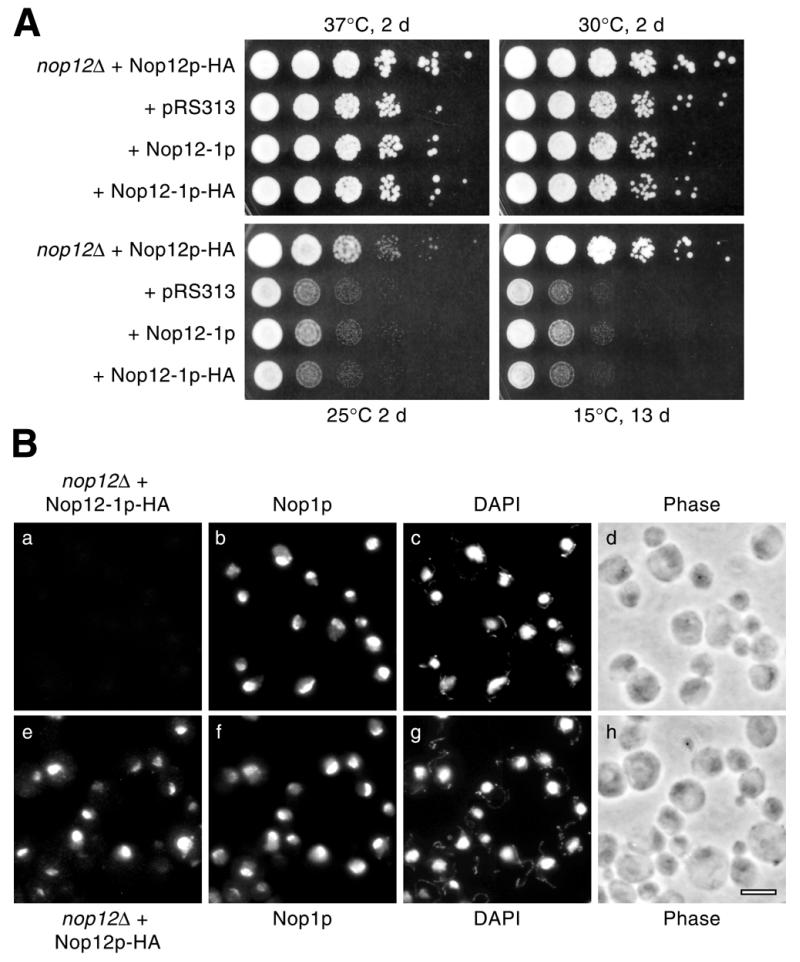
Characterization of the nop12-1 allele. (A) A nop12 null strain was transformed with the following plasmids: pKW42 (expresses Nop12p + C-terminal HA epitope tag); pRS313 (vector); pKW44 (carrying ‘nop12-1’, which encodes Nop12-1p); and pKW46 (carrying ‘nop12-1–HA’, which encodes Nop12-1p–HA). Replica platings of serial dilutions were grown for 2 or 13 days at the indicated temperatures on YPD. (B) Immunofluorescence localization in nop12Δ strains transformed with pKW46 or pKW42 was done as described in Figure 7. Nop12-1p–HA (a) and Nop12p–HA (e) were localized simultaneously with Nop1p (b and f). Counter-staining with DAPI (c and g) and phase contrast images (d and h) of the same fields of cells are shown. Bar, 5 µm.
DISCUSSION
Mutagenesis by UV irradiation generated a point mutation in NOP12 that resulted in a sl interaction with the ts allele nop2-4, which is described in the accompanying paper by Hong et al. (21). Synthetic lethality was also observed between nop12-1 and the ts alleles nop2-9 and nop2-10, but not with the ts alleles nop2-3, nop2-5 and nop2-6 (21). At present, we do not fully understand the allele-specific nature of the sl interaction. This may be due to the fact that nop2-4, -9 and -10 confer more pronounced defects in pre-rRNA processing and ribosome subunit synthesis than nop2-3, -5 and -6 (at both permissive and non-permissive temperatures, see 21). Another possibility is that nop2-3, -5 and -6 support better growth of the reporter strain than nop2-4, -9 and -10 (in the absence of NOP2 plasmid pPW67; Fig. 1B). The nop12-1 allele may impair function below a critical threshold that lies between these two groups of nop2 conditional alleles. Mutations in these two groups of nop2 alleles do not map in such a way as to suggest that mutation of a specific region of NOP2 is responsible for the synthetic lethality with nop12-1.
The allele specific nature of the sl interaction implies that Nop12p and Nop2p function within the same pathway, rather than in parallel pathways. Consistent with this, we find that Nop12p is a nucleolar protein that functions in synthesis of 25S rRNA, like Nop2p. NOP12 is not an essential gene, but it is required for normal levels of 25S rRNA synthesis at 15°C and to a lesser extent at 20°C. NOP2 is an essential gene that is also required for pre-25S rRNA processing (27). Nop2p functions at the step of conversion of 27S pre-rRNA to mature 25S rRNA (27). At 15°C in the absence of NOP12, 35S pre-rRNA accumulates to levels above those present in an isogenic wild-type strain and processing of 27S pre-rRNA is likewise retarded. Similar defects in pre-rRNA processing are observed upon depletion or inactivation of Nop2p (21,27).
Although Nop12p function appears to be essential in S.cerevisiae only during growth at low temperature, this function appears to have been conserved during evolution, by inference from the identification of likely Nop12p orthologs in S.pombe and Drosophila. Nop12p is also similar to Nop13p, which is 20% identical to Nop12p over its entire length. Nop13p localizes primarily to the nucleolus, with some Nop13p distributed in the nucleoplasm as well. NOP13 is not essential and a nop13 null allele does not bestow an observable phenotype. The highest degree of similarity between Nop12p and proteins deposited in GenBank, including Nop13p, lies within RRM sequences. Many of the proteins containing RRMs similar to the RRM in Nop12p do not function in rRNA synthesis, which is not surprising given the different functional and structural contexts in which RRMs are found in different proteins. One RRM-containing protein that ranked particularly high in sequence alignments was the RBR29/SGN1 (YIR001c) gene product, which yields a punctate cytoplasmic staining pattern by immunofluorescence localization when expressed as a C-terminally-HA epitope-tagged construct (K. Wu and J. P. Aris, unpublished results).
The nop12-1 allele contained a single nucleotide substitution resulting in a nonsense mutation at codon 359, which is followed by 100 codons in the wild-type ORF. Codon 359 lies downstream of the RRM, a key element (RNP-1) of which is found between K320 and K328 in Nop12p. HA epitope-tagged Nop12p rescued the cs phenotype of a nop12Δ strain. However, Nop12-1p–HA, which consists of an HA epitope tag plus stop codon inserted after T358, was unable to rescue the cs nop12Δ phenotype. Moreover, although Nop12p–HA was localized to the nucleolus by immunofluorescence, Nop12-1p–HA was detected only at very low levels, indicating that truncation of 100 C-terminal amino acids destabilizes the resulting protein and renders it unable to complement the cs phenotype in the nop12Δ strain. This effect on Nop12p function also provides a very plausible explanation for the synthetic lethality between nop12-1 and nop2-4, nop2-9 and nop2-10.
Like a nop12 null mutant, an nsr1 disruption yields a cs phenotype (36,39). NSR1 expression is upregulated in response to cold shock in yeast (42). Cold sensitivity is a phenotype often associated with defects in ribosome biogenesis. A large number of mutant alleles of genes involved in ribosome biogenesis have been shown to be cs, including, but not limited to: DRS1, DRS2, MPP10, NSR1, SNR17a (encodes U3) and SPB4, (39,43–47). A compelling interpretation of the connection between cold sensitivity and ribosome biogenesis defects is that low temperature ‘traps’ pre-rRNA folding intermediates. This could be due to ‘troughs’ of low free energy that occur in certain conformations of pre-rRNA intermediates. Low temperature may impose an activation energy barrier, which requires the assistance of factors that act in trans. This requirement should not exist at higher temperatures, which promotes ‘sampling’ of alternative conformations. At low temperature, factors such as RNA helicases may enhance conformational sampling in an ATP-dependent fashion. Nucleolar RNA-binding proteins may also influence pre-rRNA conformation. Perhaps trapped intermediates are stabilized by binding to a nucleolar RNA-binding protein such as Nop12p until folding resumes. Or, single-stranded pre-rRNA segments bound to Nop12p may be precluded from participation in formation of an undesirable secondary structure. Based on this line of reasoning, we hypothesize that without Nop12p function to facilitate steps in pre-rRNA folding and assembly, pre-rRNA processing is slowed at low temperature. Future studies of Nop12p should increase our knowledge of the function of RRM-containing nucleolar proteins in pre-rRNA processing. In general, elucidation of molecular interactions of RRM-containing nucleolar proteins should shed light on the significance of interactions between rRNAs and non-ribosomal proteins during pre-rRNA processing, folding and subunit assembly. However, we can not exclude potential roles in other nucleolar processes. Recently, it has come to light that the nucleolus is involved in maturation of non-rRNA precursors, e.g. tRNA precursors (48), regulation of the cell cycle (49,50) and control of aging in yeast (51).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Christopher Hardy and Alan Bender for advice on construction of strain YPA22 in the initial phase of this project. Maurice Swanson generously provided an aliquot of the yeast genomic library in YCp50. This work was supported in part by NIH grant GM48586 to J.P.A.
References
- 1.Warner J.R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci., 24, 437–440. [DOI] [PubMed] [Google Scholar]
- 2.Cate J.H., Yusupov,M.M., Yusupova,G.Z., Earnest,T.N. and Noller,H.F. (1999) X-ray crystal structures of 70S ribosome functional complexes. Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- 3.Venema J. and Tollervey,D. (1995) Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast, 11, 1629–1650. [DOI] [PubMed] [Google Scholar]
- 4.Kressler D., Linder,P. and de La Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae.Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Cruz J., Kressler,D. and Linder,P. (1999) Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci., 24, 192–198. [DOI] [PubMed] [Google Scholar]
- 6.Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- 7.Draper D.E. (1995) Protein-RNA recognition. Annu. Rev. Biochem., 64, 593–620. [DOI] [PubMed] [Google Scholar]
- 8.Draper D.E. (1999) Themes in RNA-protein recognition. J. Mol. Biol., 293, 255–270. [DOI] [PubMed] [Google Scholar]
- 9.Sun C. and Woolford,J.L. (1994) The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J., 13, 3127–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berges T., Petfalski,E., Tollervey,D. and Hurt,E.C. (1994) Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J., 13, 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee W.C., Xue,Z.X. and Melese,T. (1991) The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J. Cell Biol., 113, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark M.W., Yip,M.L., Campbell,J. and Abelson,J. (1990) SSB1 of the yeast Saccharomyces cerevisiae is a nucleolar-specific, silver-binding protein that is associated with the snR10 and snR11 small nuclear RNAs. J. Cell Biol., 111, 1741–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun C. and Woolford,J.L. (1997) The yeast nucleolar protein Nop4p contains four RNA recognition motifs necessary for ribosome biogenesis. J. Biol. Chem., 272, 25345–25352. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y., Liang,S. and Tartakoff,A.M. (1996) Heat shock disassembles the nucleolus and inhibits nuclear protein import and poly(A)(+) RNA export. EMBO J., 15, 6750–6757. [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson S.M., Datar,K.V., Paddy,M.R., Swedlow,J.R. and Swanson,M.S. (1994) Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J. Cell Biol., 127, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell I.D. and Tollervey,D. (1992) NOP3 is an essential yeast protein which is required for pre-rRNA processing. J. Cell Biol., 119, 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry M., Borland,C.Z., Bossie,M. and Silver,P.A. (1996) Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics, 142, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strasser K. and Hurt,E. (2000) Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J., 19, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Portman D.S., O’Connor,J.P. and Dreyfuss,G. (1997) YRA1, an essential Saccharomyces cerevisiae gene, encodes a novel nuclear protein with RNA annealing activity. RNA, 3, 527–537. [PMC free article] [PubMed] [Google Scholar]
- 20.Varani G. and Nagai,K. (1998) RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct., 27, 407–445. [DOI] [PubMed] [Google Scholar]
- 21.Hong B., Wu,K., Brockenbrough,J.S., Wu,P. and Aris,J.P. (2001) Temperature sensitive nop2 alleles defective in synthesis of 25S rRNA and large ribosomal subunits in Saccharomyces cerevisiae. Nucleic Acids Res., 29, 2927–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ausubel F.A., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (2000) Current Protocols in Molecular Biology. Greene Publishing and Wiley-Interscience, New York.
- 23.Bender A. and Pringle,J.R. (1991) Use of a screen for synthetic lethal and multicopy suppresser mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boeke J.D., Truehart,J., Natsoulis,G. and Fink,G.R. (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol., 154, 164–175. [DOI] [PubMed] [Google Scholar]
- 25.Cai T., Reilly,T.R., Cerio,M. and Schmitt,M.E. (1999) Mutagenesis of SNM1, which encodes a protein component of the yeast RNase MRP, reveals a role for this ribonucleoprotein endoribonuclease in plasmid segregation. Mol. Cell. Biol., 19, 7857–7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong B., Brockenbrough,J.S., Wu,P. and Aris,J.P. (1997) Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol., 17, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strathern J.N. and Higgins,D.R. (1991) Recovery of plasmids from yeast into Escherichia coli: shuttle vectors. Methods Enzymol., 194, 319–329. [DOI] [PubMed] [Google Scholar]
- 29.Aris J.P. and Blobel,G. (1988) Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol., 107, 17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu P., Brockenbrough,J.S., Metcalfe,A.C., Chen,S. and Aris,J.P. (1998) Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J. Biol. Chem., 273, 16453–16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose M.D., Novick,P., Thomas,J.H., Botstein,D. and Fink,G.R. (1987) A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene, 60, 237–243. [DOI] [PubMed] [Google Scholar]
- 32.Draper M.P., Liu,H.Y., Nelsbach,A.H., Mosley,S.P. and Denis,C.L. (1994) CCR4 is a glucose-regulated transcription factor whose leucine-rich repeat binds several proteins important for placing CCR4 in its proper promoter context. Mol. Cell. Biol., 14, 4522–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinemeyer W., Trondle,N., Albrecht,G. and Wolf,D.H. (1994) PRE5 and PRE6, the last missing genes encoding 20S proteasome subunits from yeast? Indication for a set of 14 different subunits in the eukaryotic proteasome core. Biochemistry, 33, 12229–12237. [DOI] [PubMed] [Google Scholar]
- 34.Vilella M.D., Remacha,M., Ortiz,B.L., Mendez,E. and Ballesta,J.P. (1991) Characterization of the yeast acidic ribosomal phosphoproteins using monoclonal antibodies. Proteins L44/L45 and L44′ have different functional roles. Eur. J. Biochem., 196, 407–414. [DOI] [PubMed] [Google Scholar]
- 35.Takasawa S., Tohgo,A., Unno,M., Yonekura,H. and Okamoto,H. (1992) Structural determination of Saccharomyces cerevisiae rig gene and identification of its product as ribosomal protein S21. FEBS Lett., 307, 318–323. [DOI] [PubMed] [Google Scholar]
- 36.Lee W.C., Zabetakis,D. and Melese,T. (1992) NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol. Cell. Biol., 12, 3865–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 38.Higgins D.G. and Sharp,P.M. (1988) CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene, 73, 237–244. [DOI] [PubMed] [Google Scholar]
- 39.Kondo K. and Inouye,M. (1992) Yeast NSR1 protein that has structural similarity to mammalian nucleolin is involved in pre-rRNA processing. J. Biol. Chem., 267, 16252–16258. [PubMed] [Google Scholar]
- 40.Wolfe K.H. and Shields,D.C. (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature, 387, 708–713. [DOI] [PubMed] [Google Scholar]
- 41.Winzeler E.A., Shoemaker,D.D., Astromoff,A., Liang,H., Anderson,K., Andre,B., Bangham,R., Benito,R., Boeke,J.D., Bussey,H. et al. (1999) Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- 42.Kondo K., Kowalski,L.R. and Inouye,M. (1992) Cold shock induction of yeast NSR1 protein and its role in pre-rRNA processing. J. Biol. Chem., 267, 16259–16265. [PubMed] [Google Scholar]
- 43.Hughes J.M.X. (1996) Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal-subunit RNA. J. Mol. Biol., 259, 645–654. [DOI] [PubMed] [Google Scholar]
- 44.de la Cruz J., Kressler,D., Rojo,M., Tollervey,D. and Linder,P. (1998) Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA, 4, 1268–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S.J. and Baserga,S.J. (1997) Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, mpp10. Proc. Natl Acad. Sci. USA, 94, 13536–13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ripmaster T.L., Vaughn,G.P. and Woolford,J.L. (1993) DRS1 to DRS7, novel genes required for ribosome assembly and function in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 7901–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ripmaster T.L., Vaughn,G.P. and Woolford,J.L.,Jr (1992) A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc. Natl Acad. Sci. USA, 89, 11131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertrand E., Houser-Scott,F., Kendall,A., Singer,R.H. and Engelke,D.R. (1998) Nucleolar localization of early tRNA processing. Genes Dev., 12, 2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shou W., Seol,J.H., Shevchenko,A., Baskerville,C., Moazed,D., Chen,Z.W., Jang,J., Charbonneau,H. and Deshaies,R.J. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell, 97, 233–244. [DOI] [PubMed] [Google Scholar]
- 50.Straight A.F., Shou,W., Dowd,G.J., Turck,C.W., Deshaies,R.J., Johnson,A.D. and Moazed,D. (1999) Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell, 97, 245–256. [DOI] [PubMed] [Google Scholar]
- 51.Johnson F.B., Sinclair,D.A. and Guarente,L. (1999) Molecular biology of aging. Cell, 96, 291–302. [DOI] [PubMed] [Google Scholar]
- 52.de Beus E., Brockenbrough,J.S., Hong,B. and Aris,J.P. (1994) Yeast NOP2 encodes an essential nucleolar protein with homology to a human proliferation marker. J. Cell Biol., 127, 1799–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]