Abstract
Biochemical and cryo-electron microscopy studies have just been published revealing interactions among proteins of the yeast replisome that are important for highly coordinated synthesis of the two DNA strands of the nuclear genome. These studies reveal key interactions important for arranging DNA polymerases α, δ, and ε for leading and lagging strand replication. The CMG (Mcm2-7, Cdc45, GINS) helicase is central to this interaction network. These are but the latest examples of elegant studies performed in the recent past that lead to a much better understanding of how the eukaryotic replication fork achieves efficient DNA replication that is accurate enough to prevent diseases yet allows evolution.
Keywords: cryo-electron microscopy, DNA polymerases, DNA replication, initiation of replication, replisome
Introduction
Nuclear DNA replication in eukaryotic cells proceeds at an optimal rate approaching 2,000 bases per minute while generating less than one single base error per million nucleotides polymerized [1, 2]. This fast and accurate replication of the two undamaged DNA strands of the nuclear genome is primarily performed by three members of the B family of DNA polymerases. DNA polymerase α performs limited synthesis to initiate new DNA chains, and DNA polymerases δ and ε then take over to perform the vast majority of replication [3, 4]. DNA synthesis by these polymerases always occurs in the 5′ to 3′ direction on the two antiparallel DNA strands, through formation of a replication fork that replicates the “leading strand” of DNA and then slightly thereafter replicates the lagging strand. Replication requires that the three polymerases interact with numerous, evolutionarily conserved accessory proteins [1]. These factors assist in initiating replication from hundreds to thousands of origins, in separating the two DNA strands, in DNA synthesis, and in terminating DNA replication [5, 6]. This allows efficient and fast replication of nuclear genomes that vary in size by more than 100-fold among eukaryotes, e.g. from 12 million base pairs in budding yeast to three billion base pair in humans, and that also vary widely in DNA sequence composition and in biological functions. Moreover, replication must be closely coordinated with the repair of mismatches and of ribonucleotides incorporated during replication, with re-packaging of DNA in chromatin, with transcription, and with other cellular processes.
What mechanisms operate to achieve efficient and accurate nuclear DNA replication? Answers partly depend on understanding the roles of the three polymerases in replicating the leading strand and lagging strand DNA templates. As B family members, the catalytic subunits of Pol α, Pol δ, and Pol ε share high homology [7]. Pol α contains four subunits, two of which cooperate to synthesize RNA primers to initiate replication. The DNA polymerase catalytic subunit of Pol α lacks a proofreading exonuclease and is therefore less accurate than the other two DNA polymerases. Pol δ and Pol ε are also multi-subunit enzymes, and their catalytic subunits also harbor 3′ to 5′ proofreading exonuclease activity. This activity renders them among the most accurate DNA polymerases known, an ideal property for major nuclear replicases [3, 8]. However, their accessory subunits differ, as does their processivity. Pol ε synthesizes DNA in a highly processive manner, while Pol δ is much less processive alone but is highly processive in the presence of the replication clamp PCNA. Moreover, Pol δ can perform strand-displacement synthesis needed for maturation of the lagging strand, whereas Pol ε cannot [9].
The properties of Pols δ and ε are consistent with genetic studies on their roles in leading strand and lagging strand DNA replication. For example, an error prone variant of yeast Pol ε produces specific mismatches and incorporates increased ribonucleoside triphosphates during DNA synthesis in vitro [10, 11], and it does so in patterns that match Pol ε’s specificity for the same events during spontaneous leading-strand DNA replication in vivo [12, 13]. The analogous situation is true for variants of budding and fission yeast Pol δ, whose specificities for strand-specific incorporation of mismatches and rNTPs is largely consistent with lagging strand replication [12–14]. This model for the division of labor between Pol ε and Pol δ at the nuclear DNA replication fork is strongly supported by numerous biochemical studies [1–3, 9]. Collectively, the data suggest that under normal conditions in the absence of exogenous stress to cells, Pol ε is the primary replicase for leading strand DNA synthesis and Pol δ is the primary replicase for lagging strand DNA synthesis [9]. Here we consider the organization of the nuclear DNA replication fork in relationship to this model. This being said, a recently published study of replication in budding yeast [15] has led to an alternative model wherein Pol δ is suggested to be the major replicase for both DNA strands. In that model, Pol ε’s role is limited to proofreading of errors made by Pol δ during leading strand replication. That model is consistent with early work on SV40 origin-dependent viral replication that is considered in a thoughtful recent review by Stillman [4].
Reconstituting the eukaryotic nuclear DNA replication fork in vitro
Insights into how the three replicases are delivered to the correct strand of the replication fork to perform fast and accurate replication has been greatly facilitated by reconstituting the replication fork using purified proteins. This approach requires purifying many proteins, which is a nontrivial feat that has taken time to accomplish and is now bearing fruit. These studies show that the minimal eukaryotic replication fork contains several dozen conserved proteins (see [16], Table 1 in [1] and Fig. 1 in [17]). In addition to the three multi-subunit DNA polymerases, these proteins include the CMG complex that circles the leading strand template and unwinds the double helix [18, 19]. CMG is an 11-subunit complex consisting of the 6-subunit MCM helicase, the four-subunit GINS complex and Cdc45. GINS and Cdc45 are required for functional helicase activity of the CMG complex [20, 21], and they mediate interactions with Pol ε and several other replication factors. At least two additional proteins are also required. Mcm10, together with the single-stranded DNA binding protein RPA, catalyzes the initial rearrangement of CMG from a complex that encircles double stranded DNA to that encircling single-stranded DNA form (Fig. 1). Mcm10 also stabilizes the CMG complex and stimulates replication elongation [22, 23]. Yet another protein, Ctf4, is proposed to coordinate leading and lagging strand DNA replication by binding to CMG on the leading strand and to Pol α on the lagging strand [24]. The minimal replication system also requires the 5-subunit RFC complex, which loads the PCNA clamp. PCNA encircles the DNA and interacts with Pols δ and ε to enhance the processivity of DNA synthesis, i.e. the number of nucleotides polymerized each time Pol δ or Pol ε binds to a primer-template. Other proteins present at the replication fork include those needed for maturation of lagging strand Okazaki fragments such as FEN1 and DNA ligase [9]. Several proteins increase the stability of the fork and ensure proper coordination, including Ctf4, the Mrc1 protein, and a complex of Csm1 and Tof1 proteins. Still other proteins are required immediately after completing replication, including those that repair mismatches [25] and ribonucleotides [13] incorporated by the DNA polymerases, and histones and the chaperons that load them onto newly replicated DNA to reorganize the genome.
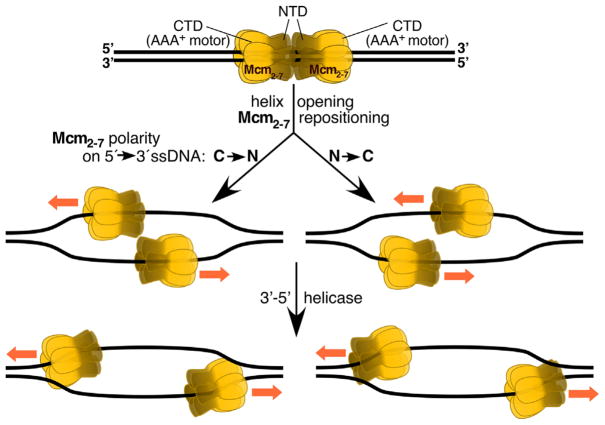
Figure 1.
Rearrangement of the MCM double hexamer during helicase activation. The double hexamer that initially surrounds double-strand DNA is reconfigured during activation to yield two active helicases surrounding single stranded DNA. In one mode (left), the Mcm2-7 hexamers become positioned in a C–>N direction on each of the 5′ to 3′ strands and move away from each other. In the other mode (right), the Mcm2-7 hexamers become positioned in a N to C direction on the 5′ to 3′ strands and move past each other toward the fork junctions.
Understanding how all these proteins work together to replicate the two nuclear DNA strands is now rapidly advancing. Here we highlight several studies just published on interactions of the replicases with the CMG complex.
Delivering the replicases to the right place
Replisome assembly is a complex, multi-step process [1] that is initiated in the G1 phase of the cell cycle when two Mcm2-7 helicase complexes are loaded onto double stranded DNA at replication origins. Each hexameric Mcm2-7 complex contains two rings (Fig. 1), an N-tier ring composed of N-terminal domains and a C-tier AAA+ ring of C-terminal domains, and these assemble at origins as double hexamers. Upon entry into S phase, the regulated loading of GINS, Cdc45, and Pol ε converts these static pre-replication complexes into mobile CMG complexes that surround single-stranded DNA, in a rearrangement that also requires RPA and Mcm10. Although the exact mechanism of this rearrangement is still poorly understood, two cyro-electron microscopy (cryoEM) studies have just been published [26, 27] that now offer new insights into the structure and function of the eukaryotic nuclear replisome.
One question addressed in these studies is the polarity of the helicase on the DNA. The CMG complex is a 3′-5′-helicase. In principle, CMG could accomplish this activity either with the C-terminal AAA+ motor pulling the CMG along ssDNA (Fig. 1, left), or with the motor propelling the CMG along ssDNA (right). In the former case, the motor is closest to the fork junction. Studies with the archaeal MCM complex and the Drosophila ssDNA-CMG complex support this orientation [28, 29]. However, the opposite orientation is now suggested by a new cryoEM study by O’Donnell and coworkers of the yeast CMG complex loaded onto a model replication fork [26]. Each AAA+ motor is proposed to hydrolyze ATP in order to push each N-tier, thereby allowing the two CMGs to pass one another while opening the DNA helix. This then allows the two CMGs to bind to the complementary 5′ to 3′ single-strand DNAs, which can then be used as templates for bidirectional DNA replication (Fig. 1, right). How can these apparently contrary results be reconciled? It is possible but unlikely that different organisms (archaea, Drosophila, yeast) use different mechanisms. Or are both directionalities possible? Interestingly, ambiguity about the directionality of enzymes on ssDNA is not an uncommon problem. It can occur when enzymes are loaded onto naked ssDNA. However, coating of the ssDNA with RPA may impose a unique directionality upon these enzymes. Examples are the checkpoint clamp 9-1-1 and Dna2 nuclease, which show promiscuous directionality on naked ssDNA but unique directionality on RPA-coated DNA [30, 31]. The CMG studies were carried out on naked ssDNA.
How the CMG moves has obvious consequences for the localization of the DNA polymerases with which it interacts. If the N-collar of the CMG is proximal to the fork junction, then this is also the case for Pol α that binds this collar. Pol ε, which interacts with GINS and Cdc45 and with the C-terminal AAA+ domain of Mcm5, would be distal from the fork (see Fig. 8A in [26]). This interaction involves the non-catalytic C-terminal domain of the large subunit of Pol ε, which is among the largest DNA polymerases known. The interaction would thereby allow the N-terminal catalytic domain of this same subunit to perform leading strand synthesis, a conclusion consistent with genetic studies.
Complementing and extending the above study is a study in press by Costa, Diffley and coworkers [27] that begins by describing the negative-stain structure of Pol ε. Earlier studies of the 4-subunit Pol ε complex suggested it is a two-domain assembly connected by a flexible linker [32]. The new study combines the EM model of the 4-subunit enzyme and of its sub-complexes with crosslinking data [18] to arrive at an improved model. The authors propose that Pol ε contains two large flexibly tethered domains shaped like a dumb bell, which are defined by the non-catalytic C-terminal domain and the N-terminal polymerization domain of the large subunit. This structure can be modeled onto the EM structure of the CMG-Pol ε complex that is also presented in this study. Although the exact CMG-Pol ε structures from the Diffley and Costa groups differ somewhat from the structure by the Li and O’Donnell groups, both models suggest that the catalytic subunit can undergo large rotary motions with respect to the C-terminal domain, which stays fixed to the CMG [18, 27]. Both studies also suggest that this arrangement may allow the non-catalytic C-terminal domain to remain bound at a replication fork while Pol ε’s N-terminal catalytic domain would be free to dissociate from the leading strand template-primer to permit loading of other proteins important for genomic integrity. In Fig. 2, we use the CMG directionality proposed by O’Donnell and coworkers [26]. However, it should be pointed out that, because of the large sizes of Pol α and Pol ε and the large flexible domain rotations possible by both enzymes [33, 34], and because of the natural flexibility of ssDNA, an opposite orientation of the CMG complex may in principle also sustain priming and elongation of replication.
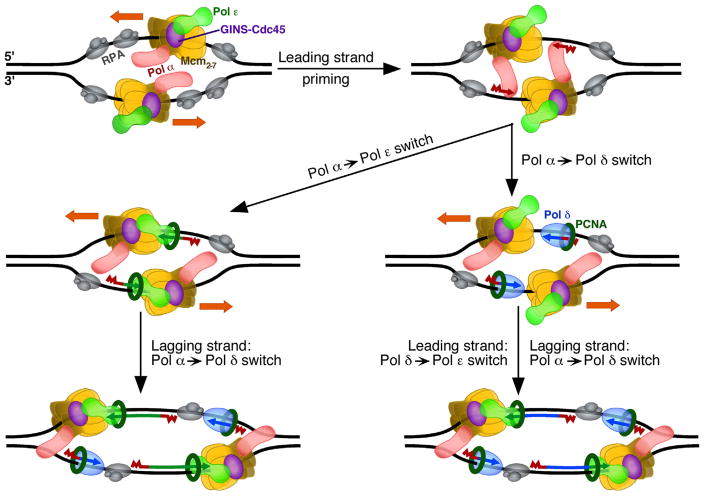
Figure 2.
Possible roles for Pol δ and Pol ε in initiating leading-strand DNA replication. In the model shown here, priming of the leading strands is carried out by the CMG-Pol ε-Pol α complex bound to opposite strands (CMG =Mcm2-7-Cdc45-GINS). In the minimal pathway (left), the primer is captured and elongated by PCNA-Pol ε (left). Alternatively (right), the primer is first captured and elongated by PCNA-Pol δ, and after collision of the elongating complex with CMG, Pol δ is ejected and Pol ε then loaded to carry out further elongation. Note that the orientation of the Mcm2-7 helicase is shown as N → C on the 5′ to 3′ strands. An opposite orientation (see Fig. 1) may also support either mechanism for starting leading strand elongation. Not all known accessory factors are shown in this diagram. RPA, replication protein A.
Reducing the delivery of the incorrect polymerase
Substantial recent evidence indicates that Pol ε is recruited to the leading strand through interactions with CMG, and that Pol δ is recruited to the lagging strand through its interaction with PCNA. A new study just published by Schauer and O’Donnell [35] now adds to this understanding by describing “eviction processes” that prevent these polymerases from inadvertently being used incorrectly. Early SV40 DNA replication studies by Tsurimoto and Stillman [36] indicated that the clamp loader RFC inhibited Pol α in the elongation of primers made by its primase activity, beyond a size of ~30 nucleotides, and permitted the association of Pol δ with loaded PCNA. This function of RFC extends to inhibiting Pol ε from binding to the lagging strand primer-template, and to facilitate Pol δ association with PCNA for the synthesis of Okazaki fragments. On the leading strand, this inhibition is prevented by Pol ε interaction with CMG, while Pol δ does not bind well (if at all) to CMG. This leads the authors to suggest that, similar to bacterial replicases, any Pol δ bound on the leading strand undergoes “collision release” upon encountering the more-slowly moving CMG complex, thereby ejecting Pol δ from the leading strand.
Results supporting Pol ε as the primary replicase for the leading strand do not exclude that Pol δ can also have one or more transient but important roles in leading strand replication. A great example is the biochemical study published in January of this year by the Diffley group [37], suggesting that Pol δ is involved in initiating leading strand replication at origins. They find that without Pol δ, leading strand synthesis is compromised and the distribution of leading strand lengths is broader, consistent with more unidirectional forks or asymmetric initiation of the two forks. This leads the authors to propose a model (Fig. 2, right) wherein, following synthesis of a primer by Pol α at origins, the PCNA-Pol δ complex rapidly synthesizes DNA until it catches up with the more slowly moving Pol ε-CMG complex. At this point, “collision release” of Pol δ occurs, allowing PCNA to stimulate Pol ε to rapidly and continuously synthesize the leading strand while promoting more rapid DNA unwinding by CMG. Interestingly, this idea that Pol δ contributes to synthesis of the leading strand at replication origins was suggested two years ago by Carr and coworkers [38] based on their analysis of ribonucleotide incorporation by variants of Schizosaccharomyces pombe Pols δ and ε. This three-polymerase model for initiating replication differs from a simpler, yet non-exclusive model, in which Pol α-synthesized primers are directly handed off to Pol ε to conduct leading strand replication (Fig. 2, left).
Diffley and coworkers [37] also propose that the participation of Pol δ in initiating replication at origins may reflect a wider role for Pol δ in re-establishing coupled leading and lagging strand replication in any situation where the 3′-end of the leading strand becomes uncoupled from the advancing fork. This may be highly relevant to circumstances that cause replication stress, e.g. following DNA damage or structural blocks in an undamaged genome that are problematic for normal fork progression. A great example of the latter is a recent study by Carr and coworkers [39], indicating that Pol δ replicates both the leading and lagging strands following replication re-start at the Rst1 locus in fission yeast.
The rate of the minimal replication fork is relatively normal
Recent studies have shown that a minimal reconstituted replication fork synthesizes undamaged DNA several-fold more slowly than do replication forks in cells [40, 41]. The new study by Diffley and coworkers [37] reports that the fork rate is further stimulated by three other proteins. One of these is Mrc1, a protein suggested by the authors to directly stimulate the replisome by increasing the rate of unwinding of DNA by CMG. Further stimulating the rate is the two-protein complex of Cms3/Tof1, which is suggested to increase the rate of association of Mrc1 to the fork. PCNA also stimulated the replication rate, including the rate of leading strand synthesis by Pol ε. The authors suggest that, by forming a complex with Pol ε and CMG to create a bridge between the 3′-end of the growing leading strand and the unwinding replication fork, PCNA helps prevent the uncoupling of DNA unwinding from leading strand DNA synthesis. Interestingly, in a MRC1 deletion strain of yeast, Pol ε and CMG continue to progress even when DNA synthesis is inhibited by hydroxurea [42], implying that Mrc1 is important for normal coupling of DNA unwinding to DNA replication. Uncoupling of DNA unwinding from leading strand DNA replication is likely to be highly relevant to Pol ε’s role in defining cellular responses to DNA damage.
Coordinating nucleosome disassembly/re-assembly with DNA replication
Eukaryotic nuclear DNA replication requires that nucleosomes be disassembled ahead of the fork and then reassembled onto newly replicated DNA behind the fork. Our understanding of the complex coordination between replication and nucleosome positioning now benefits from two articles published in January of this year, one by the Remus group [43] and another by Diffley et al. [44]. Like the O’Donnell and Diffley groups, Remus and coworkers reconstituted efficient replication using a large number of proteins purified from budding yeast. In addition to studying replication of free DNA, they provide evidence that DNA containing nucleosomes can be replicated in vitro. They show that the presence of nucleosomes on DNA before replication helps to facilitate preferential loading of the MCM helicase to replication origins, an interpretation also made by Diffley et al. [44]. Efficient, bidirectional leading and lagging strand replication by the three polymerases then proceeds, and new DNA is made that contains nucleosomes, including Okazaki fragments that are matured into a continuous lagging strand. This Okazaki fragment maturation process in vitro strongly supports an excellent study by Smith and Whitehouse [45], whose recent work indicate that nucleosomes deposited on the lagging strand behind the fork limit strand displacement synthesis by Pol δ during Okazaki fragment maturation. Kurat et al. [44] present additional evidence that the rate of replication of DNA containing chromatin factors is enhanced by several chromatin-associated proteins. These include the histone chaperone protein FACT, its associated Nhp6 protein, the nucleosome re-modeling proteins INO80 or ISW1A, and the lysine acetyltransferases Gnc5 and Esa1. They also present evidence that chromatin promotes regular priming of lagging strand replication by Pol α. This may be relevant to the studies by O’Donnell and coworkers mentioned above, indicating that Pol α is bound to the N-tier of CMG (Fig. 2). This arrangement is suggested to minimize the amount of single-stranded DNA required for replication, and to place a histone binding motif that is localized to the N-terminus of Mcm2 right at the fork, where it may interact with parental H3-H4 tetramers.
Final comments
The studies highlighted here are but a part of the outstanding work that has been done in the past few years on minimal replication systems. Additional insights are contained in these and other recent articles that are not discussed here. A continuation of such structure-function studies in combination with genetics has great potential for increasing our understanding of the mechanisms of eukaryotic nuclear replication and their connections to a wide variety of other cellular processes that help to assure genome stability while still allowing rare mistakes to drive evolution. These studies are also highly relevant to understanding the cellular processes that drive human diseases, including cancers.
Acknowledgments
We thank Scott Lujan and Jessica Williams for helpful comments.
The authors have declared no conflict of interest.
Footnotes
This article has been contributed to by US Government employees and their work is in the public domain in the USA.
References
- 1.Bell S, Labib K. Chromosome duplication in Saccharomyces cerevisiae. Genetics. 2016;203:1027–67. doi: 10.1534/genetics.115.186452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgers PM, Kunkel TA. Eukaryotic DNA replication fork. Annu Rev Biochem. 2017;86 doi: 10.1146/annurev-biochem-061516-044709. in press, https://doi.org/10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lujan SA, Williams JS, Kunkel TA. DNA polymerases divide the labor of genome replication. Trends Cell Biol. 2016;26:640–54. doi: 10.1016/j.tcb.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stillman B. Reconsidering DNA polymerases at the replication fork in eukaryotes. Mol Cell. 2015;59:139–41. doi: 10.1016/j.molcel.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewar JM, Budzowska M, Walter JC. The mechanism of DNA replication termination in vertebrates. Nature. 2015;525:345–50. doi: 10.1038/nature14887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey R, Priego Moreno S, Gambus A. Termination of DNA replication forks: “breaking up is hard to do”. Nucleus. 2015;6:187–96. doi: 10.1080/19491034.2015.1035843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson E, Macneill SA. The eukaryotic replicative DNA polymerases take shape. Trends Biochem Sci. 2010;35:339–47. doi: 10.1016/j.tibs.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Kunkel TA. Evolving views of DNA replication (in)fidelity. Cold Spring Harb Symp Quant Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stodola JL, Burgers PM. Mechanism of lagging strand DNA replication in eukaryotes. In: Masai H, Foiani M, editors. DNA Replication: From Old Principles to New Discovery. Heidelberg: Springer; 2017. in press. [DOI] [PubMed] [Google Scholar]
- 10.Pursell ZF, Isoz I, Lundstrom E-B, Johansson E, et al. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–30. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nick McElhinny SA, Watts BE, Kumar D, Watt DL, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci USA. 2010;107:4949–54. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lujan SA, Clausen AR, Clark AB, MacAlpine HK, et al. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014;24:1751–64. doi: 10.1101/gr.178335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams JS, Lujan SA, Kunkel TA. Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat Rev Mol Cell Biol. 2016;17:350–63. doi: 10.1038/nrm.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PMJ, et al. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–44. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson RE, Klassen R, Prakash L, Prakash S. A major role of DNA polymerase δ in replication of both the leading and lagging DNA strands. Mol Cell. 2015;59:163–75. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D, O’Donnell M. The eukaryotic replication machine. Enzymes. 2016;39:191–229. doi: 10.1016/bs.enz.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Yurieva O, O’Donnell M. Reconstitution of a eukaryotic replisome reveals the mechanism of asymmetric distribution of DNA polymerases. Nucleus. 2016;7:360–8. doi: 10.1080/19491034.2016.1205774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, Shi Y, Georgescu RE, Yuan Z, et al. The architecture of a eukaryotic replisome. Nat Struct Mol Biol. 2015;22:976–82. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abid Ali F, Costa A. The MCM helicase motor of the eukaryotic replisome. J Mol Biol. 2016;428:1822–32. doi: 10.1016/j.jmb.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 20.Simon AC, Sannino V, Costanzo V, Pellegrini L. Structure of human Cdc45 and implications for CMG helicase function. Nat Commun. 2016;7:11638. doi: 10.1038/ncomms11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker MW, Botchan MR, Berger JM. Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit Rev Biochem Mol Biol. 2017;52:107–44. doi: 10.1080/10409238.2016.1274717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Looke M, Maloney MF, Bell SP. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev. 2017;31:291–305. doi: 10.1101/gad.291336.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baxley RM, Bielinsky AK. Mcm10: a dynamic scaffold at eukaryotic replication forks. Genes. 2017;8:E73. doi: 10.3390/genes8020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villa F, Simon AC, Ortiz Bazan MA, Kilkenny ML, et al. Ctf4 Is a Hub in the eukaryotic replisome that links multiple CIP-box proteins to the CMG helicase. Mol Cell. 2016;63:385–96. doi: 10.1016/j.molcel.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunkel TA, Erie DA. Eukaryotic mismatch repair in relation to DNA replication. Annu Rev Genet. 2015;49:291–313. doi: 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georgescu R, Yuan Z, Bai L, de Luna Almeida Santos R, et al. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc Natl Acad Sci USA. 2017;114:E697–706. doi: 10.1073/pnas.1620500114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou JC, Janska A, Goswami P, Renault L, et al. CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proc Nat Acad Sci USA. 2017;114:4141–6. doi: 10.1073/pnas.1700530114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothenberg E, Trakselis MA, Bell SD, Ha T. MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J Biol Chem. 2007;282:34229–34. doi: 10.1074/jbc.M706300200. [DOI] [PubMed] [Google Scholar]
- 29.Abid Ali F, Renault L, Gannon J, Gahlon HL, et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun. 2016;7:10708. doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majka J, Binz SK, Wold MS, Burgers PM. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem. 2006;281:27855–61. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- 31.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–6. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asturias FJ, Cheung IK, Sabouri N, Chilkova O, et al. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol. 2006;13:35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- 33.Klinge S, Nunez-Ramirez R, Llorca O, Pellegrini L. 3D architecture of DNA Pol alpha reveals the functional core of multi-subunit replicative polymerases. EMBO J. 2009;28:1978–87. doi: 10.1038/emboj.2009.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baranovskiy AG, Babayeva ND, Zhang Y, Gu J, et al. Mechanism of concerted RNA-DNA primer synthesis by the human primosome. J Biol Chem. 2016;291:10006–20. doi: 10.1074/jbc.M116.717405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schauer GD, O’Donnell ME. Quality control mechanisms exclude incorrect polymerases from the eukaryotic replication fork. Proc Natl Acad Sci USA. 2017;114:675–80. doi: 10.1073/pnas.1619748114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis. J Biol Chem. 1991;266:1961–8. [PubMed] [Google Scholar]
- 37.Yeeles YT, Janska A, Early A, Diffley JF. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol Cell. 2017;65:105–16. doi: 10.1016/j.molcel.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daigaku Y, Keszthelyi A, Muller CA, Miyabe I, et al. A global profile of replicative polymerase usage. Nat Struct Mol Biol. 2015;22:192–8. doi: 10.1038/nsmb.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyabe I, Mizuno K, Keszthelyi K, Daiigaku Y, et al. Polymerase delta replicates both strands after homologous recombination-dependent fork restart. Nat Struct Mol Biol. 2015;22:932–8. doi: 10.1038/nsmb.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Georgescu RE, Langston L, Yao NY, Yurieva O, et al. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21:664–70. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeeles JT, Deegan TD, Janska A, Early A, et al. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–5. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katou Y, Kanoh Y, Bando M, Noguch H, et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–83. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 43.Devbhandari S, Jiang J, Kumar C, Whitehouse I, et al. Chromatin constrains the initiation and elongation of DNA replication. Mol Cell. 2017;65:131–41. doi: 10.1016/j.molcel.2016.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurat CF, Yeeles JT, Patel H, Early A, et al. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol Cell. 2017;65:117–30. doi: 10.1016/j.molcel.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–8. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]