Abstract
Nucleotide excision repair (NER) removes a variety of DNA lesions. Using a yeast cell-free repair system, we have analyzed the repair synthesis step of NER. NER was proficient in yeast mutant cell-free extracts lacking DNA polymerases (Pol) β, ζ or η. Base excision repair was also proficient without Polβ. Repair synthesis of NER was not affected by thermal inactivation of the temperature-sensitive mutant Polα (pol1-17), but was reduced after thermal inactivation of the temperature-sensitive mutant Polδ (pol3-1) or Polɛ (pol2-18). Residual repair synthesis was observed in pol3-1 and pol2-18 mutant extracts, suggesting a repair deficiency rather than a complete repair defect. Deficient NER in pol3-1 and pol2-18 mutant extracts was specifically complemented by purified yeast Polδ and Polɛ, respectively. Deleting the polymerase catalytic domain of Polɛ (pol2-16) also led to a deficient repair synthesis during NER, which was complemented by purified yeast Polɛ, but not by purified yeast Polη. These results suggest that efficient repair synthesis of yeast NER requires both Polδ and Polɛ in vitro, and that the low fidelity Polη is not accessible to repair synthesis during NER.
INTRODUCTION
Nucleotide excision repair (NER) is an important mechanism for removing a wide spectrum of different base lesions in DNA. In humans, defects in NER can lead to the hereditary disease xeroderma pigmentosum (XP) (1). XP patients exhibit photosensitivity and a highly increased incidence of skin cancers (1). Therefore, NER plays a crucial defensive role against cytotoxicity, mutagenesis and carcinogenesis induced by a variety of DNA damaging agents.
Conceptually, NER can be divided into five biochemical steps: damage recognition, incision, excision, repair synthesis and DNA ligation. Depending on whether the repaired DNA strand is transcribed by RNA polymerase II or not, NER is further differentiated by two subpathways: global genome repair and transcription-coupled repair (2,3). In the yeast Saccharomyces cerevisiae, the first three steps of global genome repair require at least the following proteins and protein complexes: Rad7, Rad16, Rad14, replication protein A (RPA), Rad4, Rad23, TFIIH, Rad2, Rad1 and Rad10 (4–7). In the transcription-coupled repair in yeast, Rad7 and Rad16 are replaced by the presumptive coupling factor Rad26 (8,9). The last step of yeast NER requires DNA ligase I encoded by the CDC9 gene (10). The presence of DNA ligase IV cannot substitute for the NER function of ligase I (10).
Based on an in vitro system reconstituted from purified mammalian NER proteins, the repair synthesis step of NER involves RPA, replication factor C and proliferating cell nuclear antigen (PCNA) (11,12). Either purified DNA polymerase (Pol) δ or Polɛ can fill the DNA gap in this reconstituted system (11,12). Using permeabilized human fibroblasts exposed to UV radiation, Nishida et al. (13) found that Polɛ is required for repair synthesis. In a separate study, Zeng et al. (14) reported that repair of UV-irradiated DNA in HeLa nuclear extract was inhibited by antibodies against human Polδ. In contrast, repair synthesis of NER in oocyte extracts of Xenopus laevis was reported to require both Polα and Polβ (15).
In yeast, the requirement for RPA and PCNA in NER has been demonstrated in a cell-free system (16). Yeast cells contain DNA Polα, β, γ, δ, ɛ, ζ and η. Most recently, the eighth S.cerevisiae DNA polymerase was identified as the TRF4 gene product and was named Polκ (17), which is required for sister chromatid cohesion. It should be noted that this polymerase bears no relation to the recently identified human Polκ encoded by the DINB1 gene (18,19). By analyzing molecular weight changes in cellular DNA after UV radiation, Budd and Campbell (20) concluded that Polδ and Polɛ are involved in repair of UV-induced damage in yeast. However, this study did not differentiate between NER and base excision repair (BER) (20). BER has been detected with UV-irradiated DNA in yeast (21), and a requirement for Polɛ in yeast BER has been reported (22). Therefore, further biochemical analysis would shed more light on the DNA polymerase requirement for yeast NER.
Most recently, it was found that DNA Polη is an extraordinarily low fidelity polymerase (23–25). Furthermore, the expression of yeast Polη is induced by UV radiation (26,27). These observations raised the question of whether Polη is accessible to repair synthesis during NER. Participation or interference of Polη in repair synthesis of NER would significantly affect the repair fidelity. Using a cell-free system, we have performed biochemical analyses of yeast NER with respect to the DNA polymerase requirement for and Polη accessibility to the repair synthesis step. In this report, we show that efficient repair synthesis in yeast cell-free extracts requires both Polδ and Polɛ, and present evidence suggesting that the low fidelity DNA Polη is not accessible to repair synthesis during yeast NER in vitro.
MATERIALS AND METHODS
Materials
Osmium tetroxide and cis-diamminedichloroplatinum (II) (cisplatin) were purchased from Sigma (St Louis, MO). N-acetoxy-N-2-acetylaminofluorene (AAAF) was obtained from the Midwest Research Institute (Kansas City, MO). The S.cerevisiae wild-type strains used were CL1265-7C (28), SX46A (29), TC102 (30), BY4741 (MATa his3 leu2 met15 ura3) and CWY231 (MATa ade1 his2 leu2-3,112 trp1-1 bar1Δ ura3Δns) (31). The S.cerevisiae mutant strains used were AMY32 (rev3Δ) (28), BY4741rad30Δ (rad30Δ) SK-2-1β (pol4Δ) (32), TAY237 (pol2-16) (31), and the temperature-sensitive mutants 488 (pol1-17) (22), YHA302 (pol2-18) (33) and ts370 (cdc2-1/pol3-1) (22).
Purified yeast DNA Polα containing associated primase activity was provided by David C. Hinkle (University of Rochester, Rochester, NY). Purified yeast Polδ and Polɛ were obtained from Akio Sugino (Osaka University, Osaka, Japan). One unit of DNA Polα incorporates 1 nmol of total nucleotide per hour at 30°C, using activated salmon sperm DNA as the substrate. One unit of DNA Polδ or Polɛ incorporates 1 nmol of total nucleotide per 30 min at 30°C, using poly(dA).oligo(dT) as the substrate (34). Yeast DNA Polη (Rad30 protein) was purified to near homogeneity as previously described (35).
Damaged DNA substrates
Single-stranded oligonucleotide U-mse1 (30mer) containing a uracil residue at position 13 and its complementary strand (30mer) were synthesized by Operon (Alameda, CA). The nucleotide sequence of the uracil-containing strand is 5′-GGATGGCATGCAUTAACCGGAGGCCGCGCG-3′. Equal molar amounts of the two oligonucleotides were mixed and annealed by incubating for 5 min at 85°C in TES (10 mM Tris–HCl pH 7.5, 1 mM EDTA and 100 mM NaCl) buffer followed by cooling slowly to room temperature.
To prepare cisplatin-damaged DNA, plasmid pUC18 (100 µg/ml) in TE (10 mM Tris–HCl pH 7.5 and 1 mM EDTA) buffer was incubated with cis-dichlorodiamineplatinum (II) at a drug/nucleotide ratio of 0.005 at 37°C for 20 h in the dark. After adding NaCl to 0.5 M, the DNA was purified by centrifugation in a linear 5–20% sucrose gradient as described by Wang et al. (21). Purified DNA was precipitated in ethanol, dissolved in TE buffer, and stored at –20°C. To prepare osmium tetroxide-damaged DNA, plasmid pUC18 (100 µg) was treated with the agent at 70°C for 90 min in TES buffer (300 µl). The DNA was then purified by centrifugation in a linear 5–20% sucrose gradient to remove nicked plasmids (21). To prepare DNA containing N-acetyl-2-aminofluorene (AAF) adducts, pUC18 (100 µg) was incubated at 37°C for 3 h in 1 ml of TE buffer containing 3 µM AAAF (the activated form of AAF) and 20% ethanol. The DNA was then purified by centrifugation in a linear 5–20% sucrose gradient (21).
In vitro DNA repair
Yeast cell-free extracts were prepared according to our previously reported methods (36,37). The same extracts were used for both in vitro NER and BER. However, NER and BER were carried out under different reaction conditions with different DNA lesions. In vitro NER and BER were performed as described by Wang et al. (21,36,37) and are described briefly below.
A standard NER reaction mixture (50 µl) contained 200 ng each of damaged pUC18 DNA and undamaged pGEM3Zf DNA, 45 mM HEPES–KOH pH 7.8, 7.4 mM MgCl2, 0.9 mM dithiothreitol, 0.4 mM EDTA, 2 mM ATP, 20 µM each dATP, dGTP and dTTP, 4 µM dCTP, 1 µCi of [α-32P]dCTP (3000 Ci/mmol), 40 mM phosphocreatine (disodium salt), 2.5 µg of creatine phosphokinase, 4% glycerol, 100 µg/ml bovine serum albumin, 5% polyethylene glycol 8000 and 250–300 µg of yeast cell-free extracts. After incubation at 26°C for 2 h, EDTA and RNase A were added to 20 mM and 20 µg/ml, respectively, and incubated at 37°C for 10 min. SDS and proteinase K were added to 0.5% and 200 µg/ml, respectively, and incubated at 37°C for 30 min. Plasmid DNA was purified by phenol/chloroform extraction, and linearized with HindIII restriction endonuclease. DNA was separated by electrophoresis on a 1% agarose gel and repair synthesis was visualized by autoradiography.
A standard BER reaction mixture (50 µl) contained 200 ng each of osmium tetroxide-damaged pUC18 and undamaged pGEM3Zf DNA, 45 mM HEPES–KOH pH 7.8, 7.4 mM MgCl2, 0.9 mM dithiothreitol, 0.4 mM EDTA, 2 mM ATP, 20 µM each dATP, dGTP and dCTP, 4 µM dTTP, 1 µCi of [α-32P]dTTP (3000 Ci/mmol), 40 mM phosphocreatine, 2.5 µg of creatine phosphokinase, 4% glycerol, 100 µg/ml bovine serum albumin and 50 µg of yeast cell-free extracts. After incubation at 30°C for 2 h, the DNA was purified by phenol/chloroform extraction, separated by electrophoresis, and visualized by autoradiography of the gel as described above for NER assays. For BER of the uracil-containing substrate U-mse1, 2 pmol of the 30mer duplex DNA was used in place of the damaged pUC18 DNA in the standard BER assay described above, and incubated at 23°C for 2 h. Reactions were stopped by phenol/chloroform extraction and the DNA was recovered by precipitation in ethanol. Repair products were separated by electrophoresis on a 20% denaturing polyacrylamide gel. Repair synthesis was visualized by autoradiography of the wet gel.
RESULTS
NER in yeast cell-free extracts lacking Polζ or Polη
Yeast Polζ and Polη are two lesion bypass DNA polymerases encoded by the non-essential genes REV3 and RAD30, respectively (26–28,35,38,39). To examine whether these two DNA polymerases affect repair synthesis of yeast NER, we performed in vitro NER in rev3 and rad30 deletion mutant extracts, using plasmid DNA containing cisplatin or AAF adducts. We have shown previously that under the conditions used cisplatin and AAF DNA adducts are repaired specifically by the NER pathway in yeast cell-free extracts (6,36,37,40,41). NER was monitored by radiolabeling the repair patch during DNA repair synthesis (36,37). As shown in Figure 1A, repair synthesis of NER in cisplatin-damaged DNA was not affected by deleting the REV3 gene. Repair synthesis of NER in AAF-adducted DNA was also not affected by deleting the RAD30 gene (Fig. 1B). These results indicate that yeast Polζ and Polη are not required for NER in vitro.
Figure 1.
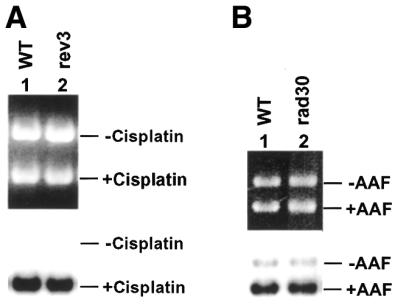
NER in Polζ and Polη mutant extracts. (A) In vitro NER of cisplatin-damaged DNA was performed in yeast cell-free extracts of the wild-type (WT) strain CL1265-7C (lane 1) and its isogenic Polζ (rev3) deletion mutant strain AMY32 (lane 2). (B) In vitro NER of AAF-adducted DNA was performed in yeast cell-free extracts of the wild-type (WT) strain BY4741 (lane 1) and its isogenic Polη (rad30) deletion mutant strain BY4741rad30Δ (lane 2). Top, ethidium bromide-stained gel; bottom, autoradiograph of the gel. +Cisplatin and +AAF, damaged pUC18 DNA; –Cisplatin and –AAF, undamaged pGEM3Zf DNA as the internal control.
Yeast Polβ is not required for NER or BER in vitro
DNA Polβ is an important repair polymerase for BER in mammalian cells (42). Yeast Polβ encoded by the POL4 gene is not essential for growth (32,43,44). Thus, pol4 deletion mutants have been isolated (32,43,44). To examine whether Polβ plays an important role in yeast NER or BER, we performed repair in pol4 deletion mutant extracts. In vitro NER assays were carried out using AAF-damaged plasmid DNA as the repair substrate. As shown in Figure 2, deleting the POL4 gene did not affect repair synthesis of NER in yeast extracts. These results suggest that Polβ is not required for NER in yeast.
Figure 2.
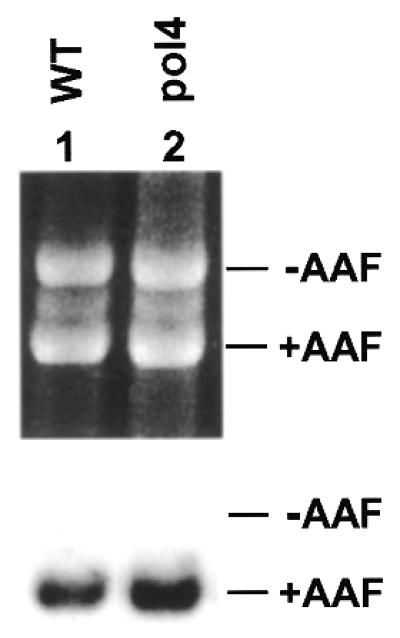
NER in Polβ mutant extracts. In vitro NER of AAF-adducted DNA was performed in yeast cell-free extracts of the wild-type (WT) strain TC102 (lane 1) and the Polβ (pol4) deletion mutant strain SK-2-1β (lane 2). Top, ethidium bromide-stained gel; bottom, autoradiograph of the gel. +AAF, damaged pUC18 DNA; –AAF, undamaged pGEM3Zf DNA as the internal control.
Depending on whether the initiating DNA glycosylase is with or without an associated AP lyase, two modes of BER are known. For example, repair of oxidative base damage in DNA is initiated by a glycosylase with associated AP lyase, while repair of uracil residues in DNA is initiated by a glycosylase without associated AP lyase. To examine both modes of BER in yeast pol4 deletion mutant extracts we used osmium tetroxide-damaged plasmid DNA that contained thymine glycol as the major damage and uracil-containing short duplex DNA as the repair substrates. Under the conditions used, BER was specifically measured without interference by NER (45). As shown in Figure 3A, BER of osmium tetroxide-damaged DNA was not affected by deleting the POL4 gene. BER of uracil residues in DNA in yeast pol4 mutant extracts was then compared with that in two different wild-type yeast extracts. As shown in Figure 3B, uracil repair was not significantly affected without yeast Polβ. These results suggest that yeast Polβ is not required for BER in vitro.
Figure 3.
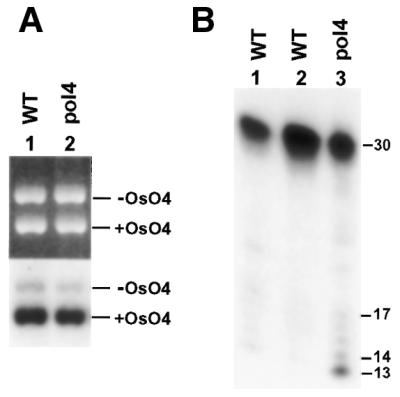
BER in Polβ mutant extracts. (A) In vitro BER of OsO4-damaged DNA was performed in yeast cell-free extracts of the wild-type (WT) strain TC102 (lane 1) and the Polβ deletion mutant (pol4) strain SK-2-1β (lane 2). +OsO4, damaged pUC18 DNA; –OsO4, undamaged pGEM3Zf DNA as the internal control. Top, ethidium bromide-stained gel; bottom, autoradiograph of the gel. (B) In vitro BER of the uracil-containing 30mer duplex DNA was performed in yeast cell-free extracts of the wild-type (WT) strains SX46A (lane 1) and TC102 (lane 2), or in yeast cell-free extracts of the Polβ deletion mutant (pol4) strain SK-2-1β (lane 3). Repair products were separated by electrophoresis on a 20% denaturing polyacrylamide gel and visualized by autoradiography. DNA size markers in nucleotides are indicated on the right.
Deficient NER in pol2-18 and pol3-1 mutant extracts
To identify the DNA polymerase(s) required for yeast NER, we examined the repair synthesis step in pol1-17, pol2-18 and pol3-1 mutant extracts. The pol1-17, pol2-18 and pol3-1 mutant cells are temperature-sensitive for growth. Previously, Boulet et al. (46) showed that the Polα activity in pol1-17 cells and the Polδ activity in pol3-1 cells were reduced to undetectable levels after shifting the growth temperature from 24 to 36°C for 2 h. Araki et al. (33) showed that partially purified mutant Polɛ (pol2-18) was very sensitive to temperature shift with a half-life of <1 min at 45°C. Therefore, the pol1-17, pol2-18 and pol3-1 mutant cells were grown at 23°C to late logarithmic phase and then at 37°C for 2 h before preparation of cell-free extracts. We expected that this growth condition should lead to inactivation of the mutant DNA polymerases in the corresponding mutant cells. To validate this expectation, we determined the effect of the temperature shift on the growth of these mutant cells. One hour after shifting to 37°C, all three mutants stopped growing. From 1 to 2 h at 37°C, viable cells of pol1-17, pol2-18 and pol3-1 were slightly reduced from 8.4 × 107 to 8.2 × 107, 2.2 × 107 to 1.7 × 107, and 2.3 × 107 to 1.9 × 107 per ml, respectively. In contrast, from 1 to 2 h at 37°C, wild-type cells continued to grow from 3.2 × 107 to 4.2 × 107 per ml. These results suggest that the temperature-sensitive Polα, δ and ɛ were inactivated in the mutant cells after incubation for 2 h at 37°C.
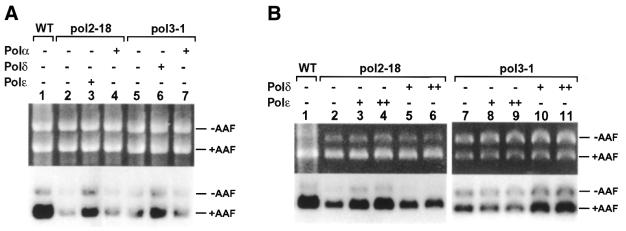
NER of AAF adducts in plasmid DNA was performed in the mutant cell extracts at 23°C. As shown in Figure 4 (lanes 1 and 2), repair synthesis was readily detected in wild-type or pol1-17 mutant extracts. In contrast, repair synthesis activities in pol2-18 (Fig. 4, lane 3) and pol3-1 (Fig. 4, lane 4) mutant extracts were significantly reduced. However, residual repair synthesis was observed in both pol2-18 and pol3-1 mutant extracts when compared with the undamaged internal control (Figs 4, lanes 3 and 4, 5A, lanes 2 and 5, and 5B, lanes 2 and 7). To show direct participation of DNA Polδ and ɛ in yeast NER, we complemented pol2-18 and pol3-1 mutant extracts with the purified yeast polymerases. Deficient repair synthesis in pol2-18 mutant extracts was partially complemented by 0.05 U of purified yeast Polɛ (Fig. 5A, lane 3). In contrast, purified yeast Polα (80 U) had no effect (Fig. 5A, lane 4). Similarly, deficient repair synthesis in pol3-1 mutant extracts was partially complemented by 0.25 U of purified yeast Polδ (Fig. 5A, lane 6), but not by purified yeast Polα (80 U) (Fig. 5A, lane 7). Furthermore, the catalytic subunit of Polɛ alone was also able to partially complement pol2-18 mutant extracts (Fig. 5B, lanes 3 and 4), but not pol3-1 mutant extracts (Fig. 5B, lanes 5 and 6). The catalytic subunit of Polδ alone was able to partially complement pol3-1 mutant extracts (Fig. 5B, lanes 10 and 11), but not pol2-18 mutant extracts (Fig. 5B, lanes 8 and 9). These results suggest that both Polδ and Polɛ are required for yeast NER in vitro.
Figure 4.
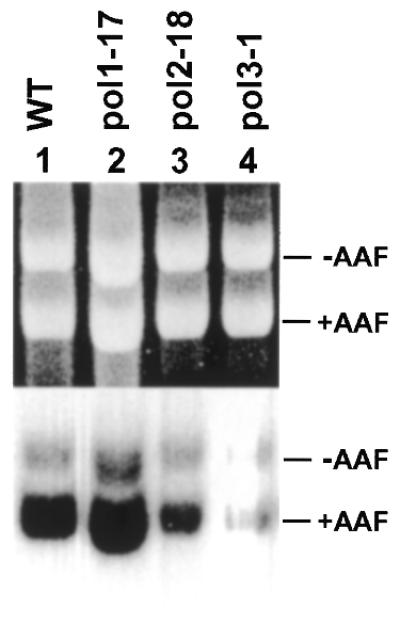
NER in temperature-sensitive mutant extracts of Polα, Polδ and Polɛ. The mutant yeast cells were grown at the permissive temperature (23°C) to late logarithmic phase of growth. The cultures were then shifted to the restrictive temperature (37°C) for 2 h. Cell-free extracts were prepared for in vitro NER. The wild-type extracts were prepared from cells grown at 37°C. In vitro NER assays were performed at 23°C for 2 h. WT, the wild-type SX46A; pol1-17, the Polα temperature-sensitive mutant 488; pol2-18, the Polɛ temperature-sensitive mutant YHA302; and pol3-1, the Polδ temperature-sensitive mutant ts370. +AAF, damaged pUC18 DNA; –AAF, undamaged pGEM3Zf DNA as the internal control. Top, ethidium bromide-stained gel; bottom, autoradiograph of the gel.
Figure 5.
Complementation of deficient NER in pol2-18 and pol3-1 mutant cell extracts. (A) Deficient repair synthesis of NER in the pol2-18 and pol3-1 mutant extracts was complemented by adding purified yeast DNA Polα, δ or ɛ to the repair reactions as indicated. The amounts of DNA polymerases added were: Polα, 80 U; Polδ, 0.25 U; and Polɛ, 0.05 U. (B) Deficient repair synthesis of NER in the pol2-18 and pol3-1 mutant extracts was complemented by adding the purified catalytic subunit of yeast Polδ or Polɛ as indicated. The His6-tagged catalytic subunits of Polδ (125 kDa) and Polɛ (256 kDa) were overexpressed in yeast cells and purified to near homogeneity. Both proteins were active in the polymerase activity. The amounts of proteins added were: Polδ catalytic subunit, 12 ng (+) and 20 ng (++); Polɛ catalytic subunit, 30 ng (+) and 50 ng (++). WT, the wild-type SX46A. +AAF, damaged pUC18 DNA; –AAF, undamaged pGEM3Zf DNA as the internal control. Top, ethidium bromide-stained gel; bottom, autoradiograph of the gel.
Participation of both Polδ and Polɛ in yeast NER raised the question of whether complete filling of each DNA gap requires both polymerases. To address this question, we performed NER reaction in pol2-18 and pol3-1 mutant extracts. After repair, the plasmid DNA was purified and loaded directly onto a 1% agarose gel without prior digestion with the HindIII restriction endonuclease. This modification of the standard in vitro NER assay allowed examination of the ligation step of the repair (10). If the DNA gap was not completely filled, subsequent DNA ligation would not occur. As expected for NER in the wild-type extracts, the vast majority of the repair patches were in the ligated form of the plasmid DNA (Fig. 6, lane 1). Similarly, the repair patches synthesized by the residual repair activities in pol2-18 and pol3-1 mutant extracts were mostly contained in the ligated form of the plasmid (Fig. 6, lanes 2 and 3). These results indicate that some DNA gaps were completely filled during residual NER in the mutant extracts. Therefore, we conclude that complete synthesis of a DNA gap during yeast NER can be performed by either Polδ or Polɛ, although repair synthesis is more efficient when both polymerases are present.
Figure 6.
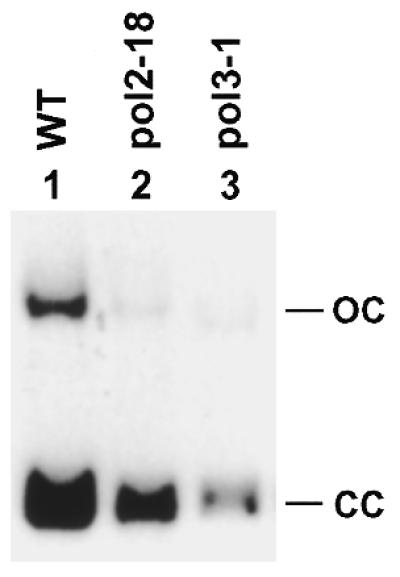
DNA ligation during residual NER in Polδ and Polɛ mutant extracts. In vitro NER reactions were performed at 26°C for 2 h using AAF-damaged pUC18 plasmid DNA. The DNA was then purified and loaded directly onto a 1% agarose gel without prior digestion with HindIII restriction endonuclease. Electrophoresis was performed in the presence of 0.5 µg/ml ethidium bromide to separate the ligated form (CC, closed circle) of plasmid pUC18 from the unligated form (OC, opened circle). Autoradiograph of the gel is shown. WT, wild-type SX46A.
The catalytic domain of yeast Polɛ is required for NER in vitro
Recently, Kesti et al. (31) showed that the catalytic domain of Polɛ is dispensable for viability of yeast cells. To determine whether the catalytic domain of Polɛ is also dispensable for NER, we performed repair assays in pol2-16 mutant extracts. The sequence encoding amino acids 176–1134 of yeast Polɛ that contains the polymerase and the 3′→5′ exonuclease functions has been deleted in pol2-16 mutant cells (31). As shown in Figure 7 (compare lanes 1 and 2), repair synthesis in pol2-16 mutant extracts was deficient. Again, residual repair synthesis activity in the mutant extracts was observed (Fig. 7, lane 2). Furthermore, the repair patches synthesized by the residual repair activity in pol2-16 mutant extracts were mostly contained in the ligated form of the plasmid DNA (Fig. 7, lane 2). The deficient repair synthesis in pol2-16 mutant extracts was complemented by 0.08 U of purified yeast Polɛ (Fig. 7, lane 4). In contrast, 0.4 U of purified yeast Polδ was unable to complement the deficient repair synthesis in pol2-16 mutant extracts (Fig. 7, lane 3). These results show that the catalytic domain of yeast Polɛ is required for NER in vitro.
Figure 7.
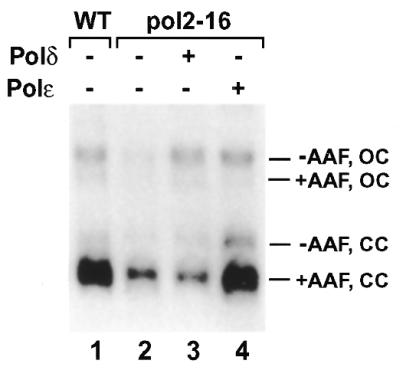
Deficient repair synthesis of NER in pol2-16 mutant extracts. In vitro NER was performed in yeast cell-free extracts of the wild-type strain CWY231 (lane 1) and its isogenic pol2-16 mutant strain TAY 237 (lane 2). Separately, purified yeast Polδ (0.4 U) or Polɛ (0.08 U) was added to the in vitro NER reaction for complementation, as indicated in lanes 3 and 4, respectively. After reaction, the DNA was purified and loaded directly onto a 1% agarose gel without prior digestion with HindIII restriction endonuclease. Electrophoresis was performed in the presence of 0.5 µg/ml ethidium bromide to separate the ligated forms (CC, closed circle) of the damaged pUC18 (+AAF) and the undamaged pGEM3Zf (–AAF) from the unligated forms (OC, opened circle) of the plasmids. Autoradiograph of the gel is shown.
The low fidelity Polη is not accessible to repair synthesis of NER in yeast extracts
Polη synthesizes DNA with an extraordinarily low fidelity (23,25,47). In yeast, Polη expression is induced by UV radiation (26,27). Thus, it is of particular interest to determine whether Polη is accessible to the DNA gap for repair synthesis during NER, especially when the amount of Polη is increased upon DNA damage. To do this, we performed repair synthesis assays in yeast pol2-16 mutant extracts. As presented above, deficient repair synthesis in this mutant extract could be complemented by purified yeast Polɛ (Fig. 7), suggesting that some DNA gaps were accumulated during NER in pol2-16 extracts. When we added excess amounts of purified yeast Polη (14-fold molar excess over DNA) during NER in pol2-16 mutant extracts, deficient repair synthesis could not be complemented (Fig. 8). These results suggest that Polη is not accessible to repair synthesis of NER in yeast cell-free extracts even when the polymerase is present in excess amount.
Figure 8.
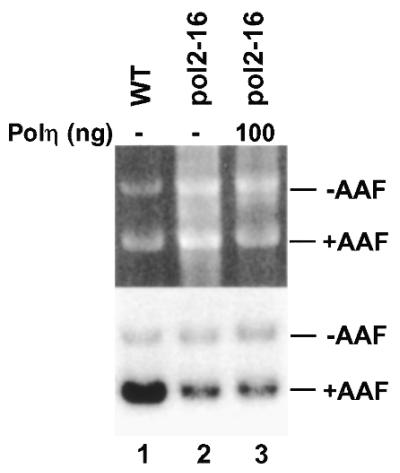
Effect of purified yeast Polη on NER in pol2-16 mutant extracts. Standard in vitro NER assays were performed in the pol2-16 mutant extracts without (lane 2) or with (lane 3) 100 ng of purified yeast Polη. Lane 1, in vitro NER in yeast cell-free extract of the isogenic wild-type strain CWY231. +AAF, damaged pUC18 DNA; –AAF, undamaged pGEM3Zf DNA as the internal control. Top, ethidium bromide-stained gel; bottom, autoradiograph of the gel.
DISCUSSION
Using biochemical analyses of in vitro repair in cell-free extracts, we show that the repair synthesis step of the yeast NER pathway requires both Polδ and Polɛ. Loss of either Polδ or Polɛ activity reduced repair synthesis during NER in vitro. Nevertheless, residual repair synthesis was present, probably due to functional Polɛ in the Polδ mutant extract or functional Polδ in the Polɛ mutant extract. It is unlikely that the residual repair synthesis activity was derived from partial inactivation of Polδ or Polɛ in the mutant extracts, because such residual activity was also observed even when the catalytic domain of Polɛ was deleted. Therefore, efficient NER in yeast cell-free extracts requires the participation of both Polδ and Polɛ at the repair synthesis step. It is not clear mechanistically how the presence of both polymerases would mediate efficient repair synthesis of NER. One possibility is that both polymerases may be parts of a large protein complex for repair synthesis during NER. Loss of one polymerase may destabilize and/or reduce the activity of the repair synthesis complex. Alternatively, each repair synthesis complex may contain only one of the two polymerases. The presence of mutant Polδ or Polɛ may form an inactive complex that competes for the functional complex at the repair synthesis step.
Our results indicate that during NER repair synthesis can occur without Polδ or Polɛ, although less efficiently. These results are consistent with earlier reports that either Polδ or Polɛ alone can carry out repair synthesis in an in vitro system reconstituted from purified human NER proteins (11,12). Overlapping or partially overlapping functions of Polδ and Polɛ in repair synthesis of NER provide an explanation that the pol3-1 (cdc2-1), pol2-18 and pol2-16 mutant cells are not significantly sensitive to UV radiation (20,31) (X.Wu and Z.Wang, unpublished results). With significant repair synthesis activity still present in the mutant cells, it is expected that UV damage can be repaired by NER, even if it may take somewhat longer to do so. It is noted that some pol3 mutant alleles lead to cellular sensitivity to a DNA damaging agent, such as methyl methanesulfonate (MMS) (48). Since post-replication repair requires Polδ (49), it is likely that the sensitivity is a consequence of defective post-replication repair rather than defective NER or BER.
Mammalian Polβ plays a major role in BER (42). However, our in vitro results indicate that yeast Polβ is not required for BER, regardless of whether the repair is initiated by a glycosylase with or without associated AP lyase. Polβ is also not required for NER. Consistent with this conclusion, yeast cells without Polβ are not sensitive to UV radiation or MMS (32,44). In fact, yeast Polβ is expressed at a very low level during vegetative growth (32). Its expression, however, is stimulated during meiosis (32). Recently, Wilson and Lieber (50) showed that yeast Polβ is involved in non-homologous DNA end joining, a special mode of DNA recombination. Therefore, the BER function of Polβ appears to be acquired later during the evolution of higher eukaryotes. The repair synthesis step of BER in yeast is catalyzed by Polɛ (22). During yeast BER of uracil-containing DNA, the 5′ deoxyribose phosphate moiety resulted from Apn1 incision is removed by the nuclease activities of Rad27 (51). The resulting gapped DNA can then be filled in by Polɛ and the nick is ligated by DNA ligase I (Cdc9 protein) (10). In higher eukaryotes, the Rad27 homolog is known as FEN1 and DNase IV (52,53), and Polδ–Fen1 combination for BER is also reported (54,55). The Polɛ–Fen1 combination for BER may also occur in higher eukaryotes (42,56). Unlike yeast, however, this repair mechanism is thought to play only a minor role in human BER (42,54,56).
Recently, it was demonstrated that the yeast RAD30 gene codes for DNA Polη (26,27,39,57). Originally, Polη was identified as an error-free lesion bypass polymerase opposite TT dimers (39,58). Later, in vitro bypass of other DNA lesions by Polη was also demonstrated (35,59,60). Surprisingly, Polη copies DNA from undamaged templates with an extraordinarily low fidelity (23,25). For cells to maintain genomic stability, mechanisms must exist to exclude Polη from DNA replication. One mechanism is to regulate its expression. Indeed, transcription of yeast Polη is UV-inducible (26,27). In this study, we asked whether Polη is also excluded from NER in yeast. We found that excess amounts of purified yeast Polη were unable to complement deficient repair synthesis in pol2-16 mutant extracts. This result suggests that Polη cannot participate in repair synthesis during NER. Even purified Polδ could not complement deficient repair synthesis in pol2-18 and pol2-16 mutant extracts, and purified Polɛ could not complement deficient repair synthesis in pol3-1 mutant extracts. Thus, it appears that following damage excision the resulting DNA gap is not exposed. Participation of Polδ and Polɛ in repair synthesis of NER is likely to involve a recruitment mechanism, which may be coordinated with an earlier reaction in the NER pathway. This process is probably carried out through protein–protein interactions. Possible interactions include Rad2–PCNA, PCNA–Polδ and PCNA–DNA ligase I, which have been reported (29,30,61). Thus, NER is most likely operated by protein complexes from damage recognition to DNA ligation in a coordinated manner.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Akio Sugino for providing yeast strains YHA302 and 488 and purified yeast DNA Polδ and Polɛ, David Hinkle for purified yeast DNA Polα, Christopher Lawrence for yeast strains CL1265-7C and AMY32, Judith Campbell for yeast strains TC102 and SK-2-1β, and Curt Wittenberg for yeast strains CWY231 and TAY237. These studies were supported by a research grant CA67978 from NIH. D.G. is a recipient of the Kentucky Opportunity Fellowship.
References
- 1.Cleaver J.E. and Kraemer,K.H. (1989) Xeroderma pigmentosum. In Scriver,C.R., Beaudet,A.L., Sly,W.S. and Valle,D. (eds), The Metabolic Basis of Inherited Disease, 6th Edn. McGraw–Hill Book Co., New York, NY, pp. 2949–2971.
- 2.Hanawalt P.C. (1994) Transcription-coupled repair and human disease. Science, 266, 1957–1958. [DOI] [PubMed] [Google Scholar]
- 3.de Laat W.L., Jaspers,N.G. and Hoeijmakers,J.H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- 4.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society of Microbiology Press, Washington, DC.
- 5.Bhatia P.K., Wang,Z. and Friedberg,E.C. (1996) DNA repair and transcription. Curr. Opin. Genet. Dev., 6, 146–150. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z., Wei,S., Reed,S.H., Wu,X., Svejstrup,J.Q., Feaver,W.J., Kornberg,R.D. and Friedberg,E.C. (1997) The RAD7, RAD16 and RAD23 genes of S. cerevisiae: requirement for transcription-independent nucleotide excision repair in vitro and interactions between the gene products. Mol. Cell. Biol., 17, 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prakash S. and Prakash,L. (2000) Nucleotide excision repair in yeast. Mutat. Res., 451, 13–24. [DOI] [PubMed] [Google Scholar]
- 8.Verhage R.A., van Gool,A.J., de Groot,N., Hoeijmakers,J.H., van de Putte,P. and Brouwer,J. (1996) Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol. Cell. Biol., 16, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Gool A.J., Verhage,R., Swagemakers,S.M.A., van de Putte,P., Brouwer,J., Troelstra,C., Bootsma,D. and Hoeijmakers,J.H.J. (1994) RAD26, the functional S.cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J., 13, 5361–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X., Braithwaite,E. and Wang,Z. (1999) DNA ligation during excision repair in yeast cell-free extracts is specifically catalyzed by the CDC9 gene product. Biochemistry, 38, 2628–2635. [DOI] [PubMed] [Google Scholar]
- 11.Aboussekhra A., Biggerstaff,M., Shivji,M.K.K., Vilpo,J.A., Moncollin,V., Podust,V.N., Protic,M., Hubscher,U., Egly,J.-M. and Wood,R.D. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell, 80, 859–868. [DOI] [PubMed] [Google Scholar]
- 12.Shivji M.K.K., Podust,V.N., Hubscher,U. and Wood,R.D. (1995) Nucleotide excision repair DNA synthesis by DNA polymerase ɛ in the presence of PCNA, RFC, and RPA. Biochemistry, 34, 5011–5017. [DOI] [PubMed] [Google Scholar]
- 13.Nishida C., Reinhard,P. and Linn,S. (1988) DNA repair synthesis in human fibroblasts requires DNA polymerase δ. J. Biol. Chem., 263, 501–510. [PubMed] [Google Scholar]
- 14.Zeng X.R., Jiang,Y., Zhang,S.J., Hao,H. and Lee,M.Y. (1994) DNA polymerase δ is involved in the cellular response to UV damage in human cells. J. Biol. Chem., 269, 13748–13751. [PubMed] [Google Scholar]
- 15.Oda N., Saxena,J.K., Jenkins,T.M., Prasad,R., Wilson,S.H. and Ackerman,E.J. (1996) DNA polymerases α and β are required for DNA repair in an efficient nuclear extract from Xenopus oocytes. J. Biol. Chem., 271, 13816–13820. [DOI] [PubMed] [Google Scholar]
- 16.Huang W., Feaver,W.J., Tomkinson,A.E. and Friedberg,E.C. (1998) The N-degron protein degradation strategy for investigating the function of essential genes: requirement for replication protein A and proliferating cell nuclear antigen proteins for nucleotide excision repair in yeast extracts. Mutat. Res., 408, 183–194. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z., Castano,I.B., De Las Penas,A., Adams,C. and Christman,M.F. (2000) Pol κ: A DNA polymerase required for sister chromatid cohesion. Science, 289, 774–779. [DOI] [PubMed] [Google Scholar]
- 18.Gerlach V.L., Aravind,L., Gotway,G., Schultz,R.A., Koonin,E.V. and Friedberg,E.C. (1999) Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc. Natl Acad. Sci. USA, 96, 11922–11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogi T., Kato,T.,Jr, Kato,T. and Ohmori,H. (1999) Mutation enhancement by DINB1, a mammalian homologue of the Escherichia coli mutagenesis protein dinB. Genes Cells, 4, 607–618. [DOI] [PubMed] [Google Scholar]
- 20.Budd M.E. and Campbell,J.L. (1995) DNA polymerases required for repair of UV-induced damage in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 2173–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z., Wu,X. and Friedberg,E.C. (1992) Excision repair of DNA in nuclear extracts from the yeast Saccharomyces cerevisiae. Biochemistry, 31, 3694–3702. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z., Wu,X. and Friedberg,E.C. (1993) DNA repair synthesis during base excision repair in vitro is catalyzed by DNA polymerase ɛ and is influenced by DNA polymerases α and δ in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda T., Bebenek,K., Masutani,C., Hanaoka,F. and Kunkel,T.A. (2000) Low fidelity DNA synthesis by human DNA polymerase η. Nature, 404, 1011–1013. [DOI] [PubMed] [Google Scholar]
- 24.Washington M.T., Johnson,R.E., Prakash,S. and Prakash,L. (1999) Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J. Biol. Chem., 274, 36835–36838. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Yuan,F., Xin,H., Wu,X., Rajpal,D., Yang,D. and Wang,Z. (2000) Human DNA polymerase κ synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res., 28, 4147–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roush A.A., Suarez,M., Friedberg,E.C., Radman,M. and Siede,W. (1998) Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol. Gen. Genet., 257, 686–692. [DOI] [PubMed] [Google Scholar]
- 27.McDonald J.P., Levine,A.S. and Woodgate,R. (1997) The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics, 147, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison A., Christensen,R.B., Alley,J., Beck,A.K., Bernstine,E.G., Lemontt,J.F. and Lawrence,C.W. (1989) REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol ., 171, 5659–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang P., Mo,J.Y., Perez,A., Leon,A., Liu,L., Mazloum,N., Xu,H. and Lee,M.Y. (1999) Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase δ. J. Biol. Chem., 274, 26647–26653. [DOI] [PubMed] [Google Scholar]
- 30.Levin D.S., Bai,W., Yao,N., O’Donnell,M. and Tomkinson,A.E. (1997) An interaction between DNA ligase I and proliferating cell nuclear antigen: Implications for Okazaki fragment synthesis and joining. Proc. Natl Acad. Sci. USA, 94, 12863–12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kesti T., Flick,K., Keranen,S., Syvaoja,J.E. and Wittenberg,C. (1999) DNA polymerase ɛ catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell, 3, 679–685. [DOI] [PubMed] [Google Scholar]
- 32.Budd M.E. and Campbell,J.L. (1995) Purification and enzymatic and functional characterization of DNA polymerase beta-like enzyme, POL4, expressed during yeast meiosis. Methods Enzymol., 262, 108–130. [DOI] [PubMed] [Google Scholar]
- 33.Araki H., Ropp,P.A., Johnson,A.L., Johnston,L.H., Morrison,A. and Sugino,A. (1992) DNA polymerase II, the probable homolog of mammalian DNA polymerase ɛ, replicates chromasomal DNA in the yeast Saccharomyces cerevisiae. EMBO J., 11, 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamatake R.K., Hasegawa,H., Clark,A.B., Bebenek,K., Kunkel,T.A. and Sugino,A. (1990) Purification and characterization of DNA polymerase II from the yeast Saccharomyces cerevisiae. Identification of the catalytic core and a possible holoenzyme form of the enzyme. J. Biol. Chem., 265, 4072–4083. [PubMed] [Google Scholar]
- 35.Yuan F., Zhang,Y., Rajpal,D.K., Wu,X., Guo,D., Wang,M., Taylor,J.-S. and Wang,Z. (2000) Specificity of DNA lesion bypass by the yeast DNA polymerase η. J. Biol. Chem., 275, 8233–8239. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z., Wu,X. and Friedberg,E.C. (1993) Nucleotide-excision repair of DNA in cell-free extracts of the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 90, 4907–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z., Wu,X. and Friedberg,E.C. (1996) A yeast whole cell extract supports nucleotide excision repair and RNA polymerase II transcription in vitro. Mutat. Res., 364, 33–41. [DOI] [PubMed] [Google Scholar]
- 38.Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science, 272, 1646–1649. [DOI] [PubMed] [Google Scholar]
- 39.Johnson R.E., Prakash,S. and Prakash,L. (1999) Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science, 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z., Svejstrup,J.Q., Feaver,W.J., Wu,X., Kornberg,R.D. and Friedberg,E.C. (1994) Transcription factor b (TFIIH) is required during nucleotide-excision repair in yeast. Nature, 368, 74–76. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z., Buratowski,S., Svejstrup,J.Q., Feaver,W.J., Wu,X., Kornberg,R.D., Donahue,T.F. and Friedberg,E.C. (1995) Yeast TFB1 and SSL1 genes, which encode subunits of transcription factor IIH, are required for nucleotide excision repair and RNA polymerase II transcription. Mol. Cell. Biol., 15, 2288–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindahl T., Karran,P. and Wood,R.D. (1997) DNA excision repair pathways. Curr. Opin. Genet. Dev., 7, 158–169. [DOI] [PubMed] [Google Scholar]
- 43.Shimizu K., Santocanale,C., Ropp,P.A., Longhese,M.P., Plevani,P., Lucchini,G. and Sugino,A. (1993) Purification and characterization of a new DNA polymerase from budding yeast Saccharomyces cerevisiae, a probable homolog of mammalian DNA polymerase β. J. Biol. Chem., 268, 27148–27153. [PubMed] [Google Scholar]
- 44.Prasad R., Widen,S.G., Singhal,R.K., Watkins,J., Prakash,L. and Wilson,S.H. (1993) Yeast open reading frame YCR14C encodes a DNA β-polymerase-like enzyme. Nucleic Acids Res., 21, 5301–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z., Wu,X. and Friedberg,E.C. (1995) The detection and measurement of base and nucleotide excision repair in cell-free extracts of the yeast Saccharomyces cerevisiae. Methods, 7, 177–186. [Google Scholar]
- 46.Boulet A., Simon,M., Faye,G., Bauer,G.A. and Burgers,P.M. (1989) Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J., 8, 1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson R.E., Washington,M.T., Prakash,S. and Prakash,L. (2000) Fidelity of human DNA polymerase η. J. Biol. Chem., 275, 7447–7450. [DOI] [PubMed] [Google Scholar]
- 48.Blank A., Kim,B. and Loeb,L.A. (1994) DNA polymerase δ is required for base excision repair of DNA methylation damage in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 91, 9047–9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torres-Ramos C.A., Prakash,S. and Prakash,L. (1997) Requirement of yeast DNA polymerase δ in post-replicational repair of UV-damaged DNA. J. Biol. Chem., 272, 25445–25448. [DOI] [PubMed] [Google Scholar]
- 50.Wilson T.E. and Lieber,M.R. (1999) Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase β (Pol4)-dependent pathway. J. Biol. Chem., 274, 23599–23609. [DOI] [PubMed] [Google Scholar]
- 51.Wu X. and Wang,Z. (1999) Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res., 27, 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrington J.J. and Lieber,M.R. (1994) Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev., 8, 1344–1355. [DOI] [PubMed] [Google Scholar]
- 53.Robins P., Pappin,D.J., Wood,R.D. and Lindahl,T. (1994) Structural and functional homology between mammalian DNase IV and the 5′-nuclease domain of Escherichia coli DNA polymerase I. J. Biol. Chem., 269, 28535–28538. [PubMed] [Google Scholar]
- 54.Klungland A. and Lindahl,T. (1997) Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J., 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim K., Biade,S. and Matsumoto,Y. (1998) Involvement of flap endonuclease 1 in base excision DNA repair. J. Biol. Chem., 273, 8842–8848. [DOI] [PubMed] [Google Scholar]
- 56.Fortini P., Pascucci,B., Parlanti,E., Sobol,R.W., Wilson,S.H. and Dogliotti,E. (1998) Different DNA polymerases are involved in the short- and long-patch base excision repair in mammalian cells. Biochemistry, 37, 3575–3580. [DOI] [PubMed] [Google Scholar]
- 57.Masutani C., Kusumoto,R., Yamada,A., Dohmae,N., Yokoi,M., Yuasa,M., Araki,M., Iwai,S., Takio,K. and Hanaoka,F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- 58.Masutani C., Araki,M., Yamada,A., Kusumoto,R., Nogimori,T., Maekawa,T., Iwai,S. and Hanaoka,F. (1999) Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J., 18, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masutani C., Kusumoto,R., Iwai,S. and Hanaoka,F. (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J., 19, 3100–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y., Yuan,F., Wu,X., Rechkoblit,O., Taylor,J.-S., Geacintov,N.E. and Wang,Z. (2000) Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res., 28, 4717–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gary R., Ludwig,D.L., Cornelius,H.L., MacInnes,M.A. and Park,M.S. (1997) The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J. Biol. Chem., 272, 24522–24529. [DOI] [PubMed] [Google Scholar]