Abstract
Aim:
Develop and apply a comprehensive and accurate next-generation sequencing based assay to help clinicians to match oncology patients to therapies.
Materials & methods:
The performance of the CANCERPLEX® assay was assessed using DNA from well-characterized routine clinical formalin-fixed paraffin-embedded (FFPE) specimens and cell lines.
Results:
The maximum sensitivity of the assay is 99.5% and its accuracy is virtually 100% for detecting somatic alterations with an allele fraction of as low as 10%. Clinically actionable variants were identified in 93% of patients (930 of 1000) who underwent testing.
Conclusion:
The test's capacity to determine all of the critical genetic changes, tumor mutation burden, microsatellite instability status and viral associations has important ramifications on clinical decision support strategies, including identification of patients who are likely to benefit from immune checkpoint blockage therapies.
Keywords: : analytical validation, diagnostics, gene-panels, next-generation sequencing (NGS), precision oncology
Cancer can be caused by a diverse set of genetic abnormalities, such as point mutations, gene and chromosomal copy number changes and structural variants. Some are well known to occur within oncogenes or tumor suppressor genes that facilitate tumor initiation and progression while others are less common or less well understood. Along with the early discovery of individual cancer driver genes, several large-scale genomic studies have annotated the broad genomic landscape within common human tumors and cataloged many cancer-associated genomic changes. In parallel, a rapidly growing list of cancer drugs has been approved for use in patients with specific genetic alterations. In addition, an avalanche of targeted therapies is currently being tested in multiple genomically matched clinical trials. Using the genomic profile of a patient's tumor to determine the most optimal treatment protocol is the goal of precision/personalized medicine, and the salutary benefits of such therapies upon survival have been well documented [1–4]. The success of herceptin (a drug targeting HER2/neu, the product of ERBB2 gene) in breast cancers with ERBB2 gene amplification [5] has been reproduced in many cancers with other genetic alterations that are effectively treated by targeted therapies [6–8]. Thus, comprehensive genomic profiling is likely to become the standard of clinical practice in determining the optimal treatment for individual cancer patients [9–12].
To address the needs of modern precision oncology and to realize the benefits of routine tumor genome profiling for patients, we report utilization of CANCERPLEX®, a comprehensive next-generation sequencing (NGS) based analytical system that can identify and prioritize potential treatment strategies for solid tumors. CANCERPLEX is based on the rapid and accurate genetic analysis of clinical FFPE tissue, including core needle biopsies and cell blocks prepared from fine-needle aspirations, malignant pleural effusions and ascites. The assay includes efficient extraction of FFPE DNA followed by sequencing of 435 important cancer genes that are altered in a wide range of solid cancers. The assay identifies oncogenic driver events that predict response or resistance to treatments and, thus, can impact therapeutic strategies. Mutation burden, microsatellite instability (MSI) and presence of oncogenic viruses are additional biomarkers that CANCERPLEX can detect, which enables oncologists to reach more informed therapeutic decisions. The test was developed by KEW, Inc. (Cambridge, MA, USA) to support clinical decisions by oncologists. KEW laboratory is accredited by the College of American Pathologists (CAP) and has Clinical Laboratory Improvement Amendments (CLIA).
Materials & methods
Reference material & tumor tissue
For analytical validation of the assay, we used a panel of characterized DNA from the HapMap consortium (Coriell Institute for Medical Research, NJ, USA), cancer cell lines purchased directly from the American Type Culture Collection, and patient tumor and normal FFPE samples. Patient FFPE samples consisted of discarded and deidentified tumor specimens purchased from BioServe (MD, USA) or obtained from clinical operations. Normal FFPE samples of tonsil and endometrial tissue were acquired from UMass Cancer Center Tissue and Tumor Bank (MA, USA) (Supplementary Table 13).
Pathologist review of tissue sections & genomic extraction of tumor DNA
For each hematoxylin and eosin stained tissue section, regions of high tumor purity were selected for macrodissection and the marked hematoxylin and eosin slides were then digitally scanned and documented. For FFPE blocks, tissue macrodissection was done using 1-mm biopsy punches. Genomic DNA was extracted from tumor tissue using methods previously described [13]. For quality control (QC) purposes, extracted genomic DNA (gDNA) was evaluated by measuring the A260/A230 ratio (NanoDrop, Thermo Fisher Scientific, DE, USA) and by measuring DIN with TapeStation (Agilent Technologies, CA, USA). There was no cut-off on DNA Integrity Number (DIN) though less gDNA can be used when DIN >3.5. The Quant-iT PicoGreen dsDNA Assay was used to determine DNA concentration (Thermo Fisher Scientific, MA, USA).
Selection of targets
Genes were selected by comprehensive mining of the US FDA databases, NCCN, ASCO and ESMO Clinical Practice Guidelines in Oncology, COSMIC, TCGA and R&D pipelines of large pharmaceutical companies. In addition to gene-coding sequences, probes were added to address the accurate solving of selected chromosomal translocations, broad copy-number profiling, splice sites and untranslated regions (promoter of TERT, 3′UTR of CD274) as well as detection of oncoviruses (human papillomavirus [HPV] 16, HPV18 and Epstein–Barr virus [EBV]), MSI and mutation burden.
Library preparation & Illumina-based NGS
Illumina-NGS ready libraries were produced using a proprietary protocol, which utilizes the KAPA/Roche HyperPlus kit (KK8514, Kapa Biosystems, MA, USA). The protocol was optimized for real-world clinical specimens to produce DNA fragments in the 100–200 bp range. Input of gDNA ranged from 50 to 150 ng. Modifications included the ligation of a unique molecular counter. Unique molecular counters are used to computationally remove sequencing artifacts, including PCR duplicates. Prepared libraries were tagged with molecular barcodes and individually enriched for CANCERPLEX genes using custom probes. Patient samples were never pooled. Following target enrichment, 160-bp sequencing was performed using Illumina instruments to 500× mean depth. A denatured PhiX library was added to each run as a sequencing QC.
Sequence data extraction
For data analysis, we developed a proprietary bioinformatics pipeline that utilizes GATK Best Practices [14]. To deal with FFPE-associated biases, a diverse panel of normal samples was generated. DNA extracted from 40 normal, nontumor FFPE tissue specimens was sequenced the same way as the clinical samples. The resulting panel of normal samples data were used as a baseline reference for calling mutations.
Sample QC
Read and base quality score and GC content were analyzed with FastQC tool prior to alignment. After alignment, QC metrics were calculated for each sample including: read counts (total bases, total, unique and duplicate reads, mapped and uniquely mapped reads, percentages of reads mapped/mapped to the region of interest [ROI]); coverage (mean, uniformity, percentage of bases with zero coverage); and library complexity. Sequencing data identity was assured by molecular barcodes, process controls, and also based on a composite of tools including determining the percentage of concordant single-nucleotide variants (SNVs) with respect to previously sequenced samples, inferring sex and iAdmix predictions.
Variant calling
SNVs, small insertions and deletions (indels) calling was restricted to genomic regions intended to be captured by the assay (ROI). Copy number variants (CNVs) were called for exons as well as globally. The threshold for gains was at >2.5-fold and for loss at <0.75-fold. Fused genes (structural variants) were detected if at least one end mapped to any of the following genes: AKT3, ALK, BRAF, EGFR, ETV1, ETV4, ETV5, ETV6, FGFR2, FGFR3, MET, NOTCH1, NOTCH2, NRG1, PDGFRA, RAF1, RET, ROS1 and TMPRSS2. Variants were filtered or flagged according to technical quality (e.g., coverage, allelic fraction, number of supporting reads and strand bias). SNVs and indels were annotated using SnpEff and the output was adapted per Human Genome Variation Society recommendations.
Other types of analyses
To validate SNVs and indels, pools of highly characterized Coriell DNA samples were prepared with allele frequencies ranging from 0.1 to 1.0. The performance of the assay was based upon the successful detection of the variants known to be present in the Coriell samples. To determine whether the depth of coverage affects the assay's sensitivity, a series of diluted samples were sequenced in consecutive windows of sequencing depth until 200× coverage was reached. To validate intra-assay reproducibility and to demonstrate consistency of test results between different aliquots of the same sample and between separate assay batches, libraries were prepared from three different samples on different days and/or by two different operators. Sequencing results were analyzed for variant concordance.
CNVs and structural rearrangements were validated using FFPE tumor samples and cancer cell lines that had been previously demonstrated (using FISH, immunohistochemistry (IHC) and/or qPCR) to harbor the ERBB2 gene amplification or the EML4-ALK rearrangement. The minimum tumor content requirement was determined by assessing the impact of sequencing depth on the sensitivity of the test to detect the EML4-ALK gene rearrangement. The H2228 cell line, which carries the EML4-ALK gene fusion, was diluted by FFPE normal to generate series of samples with a tumor content ranging from 0.1 to 0.5. Samples underwent multiple sequencing runs and the number of chimeric reads as well as overall coverage was determined.
The tumor mutation burden (TMB), defined as the rate of peptide-changing SNVs per Mb, was determined for all tumors. To estimate TMB, SNVs with a mutation allelic fraction (MAF) of at least 0.1 after standard filtering and with high or moderate putative impact were retained.
Determination of MSI was based on an extended loci panel. In addition to the Bethesda panel, we analyze a collection of up to 950 regions consisting of tandem repeats of one, two or three nucleotides of minimum length of ten bases. The number of indels within the ROI was calculated and tumors were classified as high MSI (MSI-H) or microsatellite stable. The MSI status of 15 clinical FFPE samples was determined using PCR to amplify the Bethesda markers and IHC to confirm the presence of mismatch repair (MMR) proteins. For validation, CANCERPLEX testing was performed on the 15 characterized FFPE samples and the number of indels within the ROI was calculated. Based upon these results, the MSI cut-off was set (Supplementary Table 10). Primers for PCR validation of MSI status were designed as previously described [15]. The PCR was set up using Type-it Microsatellite PCR Kit by QIAGEN (MD, USA). PCR products were resolved by on-chip electrophoresis on an Agilent Bioanalyzer 2100.
Tumors were also analyzed for the presence of HPV-16, HPV-18 and EBV (HHV-4) viral sequences. The reference genomes used were: GI:310698439, GI:9626069 and NC_007605, respectively. The percentage of total number of reads mapped to the viral genomes was calculated and samples were designated as positive based on empirical cut offs of 0.02, 0.01 and 0.0005% of reads that mapped to HPV-16, HPV-18 and EBV genomes, respectively. To validate EBV status of DNA samples, PCR primers were designed as previously described [16]. The forward and reverse primer sequences for EBV were 5′-CCCGCCTACACACCAACTAT-3′ and 5′-AGTCTGGGAAGACAACCACA-3′. PCR conditions were 95°C for 3 min followed by 28 cycles of 98°C for 20 s, 60°C for 15 s and 72°C for 30 s, followed by the final extension at 72°C for 5 min.
Variant assessment in GENEKEEPER
GENEKEEPER is a curation tool and proprietary database developed by KEW, Inc. that prioritizes variants based upon their clinical relevance. Only nonsynonymous variants relative to canonical transcripts retrieved from the UniProt database are submitted for review. Also, variants were deprioritized if they were present under certain conditions (e.g., at allele frequency >1%) in dbSNP, 1000 Genomes, ExAC databases, and then reprioritized with COSMIC. Next, allele frequencies for each mutation were used to fit a model to determine whether the variant is likely germline heterozygous or somatic. Finally, results underwent manual molecular pathologist review validating somatic versus possible germline status of a variant as described before [17]. The catalog of FDA-approved drugs, NCCN treatment guidelines, multiple mutation databases and current scientific literature are additional resources used to determine if the variant protein is a target of an FDA-approved drug, a target of a drug in clinical development or confers resistance to known treatments. Clinical trials are identified using the ClinicalTrials.gov and other tools (e.g., Thomson Reuter Cortellis and Pharma Intelligence Trialtrove).
Results
Validation of the assay
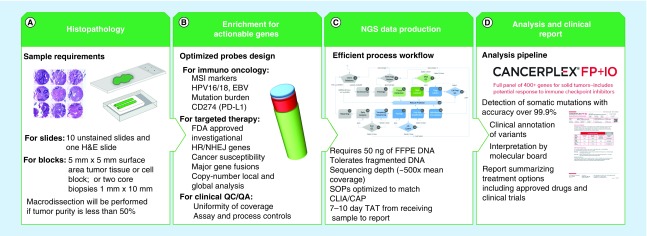
We have developed a highly accurate and actionable NGS-based cancer diagnostic assay to provide physicians with the genetic information they need to guide personalized, patient treatment strategies. The process workflow includes the review of tissue specimens by a pathologist and the macrodissection of tumor cells in specimens with <50% tumor purity (Figure 1A). Probes have been custom designed to enrich for genetic markers that predict response to immuno-oncology drugs as well as for genetic alterations that are associated with FDA-approved and investigational targeted treatments (Figure 1B). Sequencing reagents and protocols have been optimized to routinely produce 500× mean coverage from as little as 50 ng of FFPE DNA (Figure 1C). We also developed a proprietary bioinformatics pipeline with an accuracy of over 99.9% for detecting somatic mutations and a custom analysis database that is used to match clinically relevant genetic alterations with appropriate gene-targeted therapies or clinical trials (Figure 1D).
Figure 1. . Schematic presentation of CANCERPLEX® workflow.
The workflow is optimized to produce highly accurate and clinically relevant results from FFPE tumor tissue. (A) Slides are cut from tumor samples embedded in paraffin blocks, after which H&E staining is performed. Specimens that are composed of <50% tumor cells are dissected to extract regions with the greatest purity and viability of tumors cells. (B) Target enrichment probes have been custom designed to produce uniform coverage across all exons of the 435 targeted genes as well as regions of the genome that are likely to predict response to immuno-oncology drugs. (C) An overview of the process workflow is shown from sample acquisition to production of the patient's final report. (D) The raw sequencing data are uploaded to KEW proprietary informatics pipeline for identification of multiple classes of genetic variation, including SNVs, indels, CNVs, translocations, MSI and viral integration. The clinical significance of each genetic variant is determined by reviewing the scientific literature and cancer-related gene variant databases and FDA-approved and/or investigational therapies are reported.
CNV: Copy-number variant; FFPE: Formalin-fixed paraffin-embedded; H&E: Hematoxylin and eosin; indel: Insertion and deletion; MSI: Microsatellite instability; NGS: Next-generation sequencing; SNV: Single-nucleotide variant; TAT: Turnaround time.
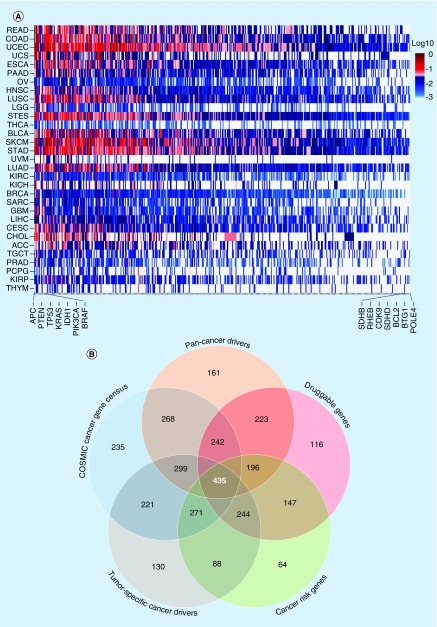
Most tumor tissue specimens are heterogeneous having mixtures of normal and tumor cells, and tumor cells might also be mixtures of different subclones. Academic studies are often based on highly pure tumors and use up to 100× mean coverage of whole exomes to detect mutations (TCGA). To identify genetic variants that are critical for clinical decisions in ‘real-world’ tumor samples, it is necessary to sequence at significantly higher depths. To facilitate cost–effectiveness, we examined whether a subset of all the coding genes would provide the comprehensive information needed for clinical decision support at current state of care. After design and testing of several subsets, we decided upon a panel including 435 genes to profile for genetic changes in solid tumor samples (Supplementary Table 1 for the list of genes). This test is called CANCERPLEX. These genes are known to have somatic alterations in one or more tumor types (Figure 2A & Supplementary Table 14). The 435 genes include all clinically relevant targets of FDA-approved therapies, targets of investigational therapies in current clinical trials, DNA repair genes, cancer biomarkers and targets of prospective clinical trials (Figure 2B). CANCERPLEX protocols and reagents were optimized to maximize sequencing uniformity and depth of coverage across ROI (Supplementary Table 15). The average coverage for the targeted sequences is 500× (Supplementary Figure 1A) and the majority of sequenced bases (∼80%) were aligned to ROI (Supplementary Figure 1B). Sequencing libraries produced uniform coverage, regardless of the regional GC content, (Supplementary Figure 1C), and high coverage was attained across all 435 genes that are part of the panel (Supplementary Figure 2).
Figure 2. . Selection of CANCERPLEX® targets.
(A) Heat map depicting the mutation frequency of the 435 genes on the CANCERPLEX gene panel across 31 different cancer types (y-axis). Red indicates genes with the highest mutation frequencies (1÷0.05) and blue indicates genes with lower mutation frequencies. Genes with the highest and lowest overall mutations frequencies are shown on the x-axis. (B) Venn diagram showing the different categories of clinically relevant cancer genes that make up the gene panel. The number of genes that are represented in each category are shown and the overlaps represent the union of sets (total number less double counting).
We developed the gene panel assay with the goal of identifying all the genomic features that inform treatment decisions and with the highest possible accuracy. We have thoroughly evaluated the performance of the assay using reference samples that represent all classes of genetic variation, including SNPs, indels, CNVs, rearrangements, HPV/EBV and MSI. FFPE clinical samples were also included to assess the ability of the assay to detect genetic variation in complex and heterogeneous tumors. We created a series of samples with mutant allelic fractions (MAF) ranging from 0.1 to 1.0 and found that the assay has excellent performance with a mean sensitivity of 99.5% and accuracy of 99.9% to detect SNPs with MAF = 10% (Supplementary Table 2). Indels with an allelic fraction of 10% were detected with a mean sensitivity of 100% and accuracy of 100% (Supplementary Table 3). For cancer cell lines that were independently validated by Sanger sequencing, the accuracy was similarly high at 98.7% for SNPs and 100% for indels (Supplementary Tables 4 & 5). The variant allele fraction as determined by CANCERPLEX correlated with expected results (Supplementary Table 6). The results of inter- and intra-assay reproducibility studies revealed a high level of precision; the concordance between replicate samples was 92.3–97.7% (Supplementary Table 7). The minimum tumor content requirement was determined by assessing the impact of sequencing depth on the sensitivity of the assay to detect SNVs. For tumor purity 20%, acceptable high sensitivity detection of SNVs (>98%) was achieved when coverage was increased to between 150 and 200× (Supplementary Table 8), setting the minimum requirement of tumor content at as low as 20%. We routinely use 500× coverage for the clinical samples.
Characteristics of tumor specimens
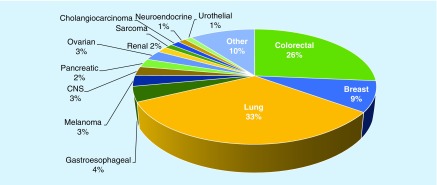
We have examined 1000 clinical samples sent from diverse groups of oncologists. We were able to extract adequate amounts of DNA from different types of samples, including FFPE blocks, core biopsies, fine-needle aspirations and effusions. The majority of the specimens were derived from advanced stage tumors (T4, 78%), while the remaining tumors were either unknown stage (TX, 14%) or from less advanced stage tumors (8%). We examined samples from lung cancer (31%), colorectal cancer (35%), gastric and esophageal cancer (15%) and breast cancer (9%). All other tumor types constituted the remaining 20% of tumors (Figure 3).
Figure 3. . Tumor types across 1000 clinical samples tested.
The percentage of tumors of each type is provided.
Genetic alterations detected by CANCERPLEX
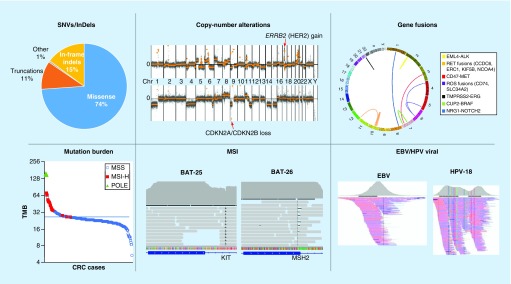
CANCERPLEX is an assay that uses NGS data to determine many different types of somatic genetic changes in tumors. The types of changes that could be detected are shown in Figure 4. All tumor samples had somatic mutations. Among point mutations, missense mutations were the most frequent (81%), while mutations that result in indels or prematurely truncated proteins made up the vast majority (18%) of remaining mutations. (Figure 4 top, left panel).
Figure 4. . The types of somatic genetic changes that have been identified using CANCERPLEX®.
Top left: Pie graph depicting the types of functional somatic mutations detected in 1000 clinical tumors. Top middle: Global copy number profiles from representational tumors showing high-level amplification of 17q12, which involves the ERBB2 gene (red arrow, top panel) and a homozygous deletion of chromosome 9p21, which involves the CDKN2A and CDKN2B genes (red arrow, bottom panel). Top right: Summary Circos plot of CANCERPLEX data presenting chromosome rearrangements detected in the 1000 cases. The outer ring depicts the human chromosomes end-to-end forming a circle. Each color-coded line represents a unique gene fusion, which may or may not have been described previously. The fusion partner genes are described in the box to the right of the plot. Bottom left: A plot was generated displaying the TMB (y-axis) for 262 colorectal tumors (x-axis). Tumors that exhibited MSI are shown in red and tumors that exhibited POLE deficiency are shown in green. Bottom middle: Representational images from IGV displaying indels in regions of the genome that include the BAT-25 and BAT-26 loci. Reference sequence is shown below. Bottom right: NGS output displaying representational images from an EBV positive tumor and an HPV-positive tumor. The regions with the highest overall coverage are shown (gray peak) as well as the overlap between sequencing reads (pink and blue lines).
EBV: Epstein–Barr virus; HPV: Human papillomavirus; IGV: Integrative genomics viewer; MSI: Microsatellite instability; TMB: Tumor mutation burden.
We have developed analytical methods to detect copy number alterations in oncogenes, such as ERBB2, and tumor suppressors, such as CDKN2A. (Figure 4 top, middle panel). Results of validation testing revealed that the detection of key pathogenic CNVs was highly concordant with results obtained using standard techniques, including qPCR and FISH (Table 1 & Supplementary Table 9). Additionally, we show that the global CNV profile generated by CANCERPLEX is consistent with the profile generated by the Affymetrix SNP 6.0 array (Supplementary Figure 3A & B). We used IHC testing to confirm that a focal deletion detected in glioblastoma tumor cells at the 9p21.3 loci correlated with lack of CDKN2A staining and that a focal amplification in colorectal tumor cells at the 17q13 loci correlated with positive staining for the HER2 protein (Supplementary Figure 3C–F). Taken together these results affirm the power of the assay to detect cancer-associated CNVs.
Table 1. . Orthogonal validation of CANCERPLEX results.
| Type of independent testing | Tested by | Detected by CANCERPLEX | Concordant/tested samples | Concordance | Comments on discordant cases |
|---|---|---|---|---|---|
| KRAS hotspot variants |
PCR |
SNVs |
18/19 |
95% |
Different variant detected at the same codon |
| NRAS hotspot variants |
PCR |
SNVs |
2/2 |
100% |
|
| BRAF hotspot variants |
PCR |
SNVs |
7/7 |
100% |
|
| EGFR hotspot variants |
PCR |
SNVs/indels |
29/29 |
100% |
|
| Expression of DNA MRR enzymes |
IHC |
SNVs and indels in MRR genes |
43/43 |
100% |
|
| Her2 amplification |
FISH/IHC |
ERRB2 gain (CNV) |
78/83 |
94% |
Different specimens tested (different timepoints, after treatment) or ERBB2 gain detected below cut-off (tumor heterogeneity) |
| MET amplification |
FISH |
MET gain (CNV) |
1/1 |
100% |
– |
| ALK rearrangement |
FISH, PCR |
ALK rearrangement |
24/24 |
100% |
– |
| ROS1 rearrangement | FISH | ROS1 rearrangement | 9/9 | 100% | – |
The detected 217 key driver events are highly concordant with results obtained using techniques other than Illumina NGS. Samples underwent CANCERPLEX testing and the concordance between methods was then determined.
CNV: Copy number variant; IHC: Immunohistochemistry; indel: Insertion and deletion; MRR: Mismatch repair; NGS: Next-generation sequencing; SNV: Single-nucleotide variant.
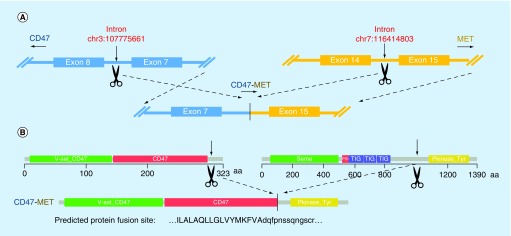
Chromosomal rearrangements occasionally result in the formation of functional hybrid proteins that drive tumorigenesis. The assay was able to detect a number of rearrangements that have been previously shown to produce oncogenic fusion proteins, such as ALK and ROS [18,19] as well as several rearrangements that have not been reported previously but which are likely to produce functional gene fusions (Figure 4 top, right panel). A schematic representation of a CD47-MET gene rearrangement and the predicted fusion protein consisting of the N-terminal region of the CD47 protein and the kinase domain of the MET protein is shown in Figure 5.
Figure 5. . CD47-MET gene rearrangement and formation of a potentially functional gene fusion protein.
The genomic structure surrounding the breakpoint within the intron 14 of the MET gene and intron 7 of the CD47 gene is shown. A chromosomal inversion leads to fusion of intron 7 of the CD47 gene to intron 14 of the MET gene. The domain organization of each protein is shown as well as the domain structure of the predicted fusion protein. The sequence surrounding the protein fusion site is predicted to be in-frame and is shown below.
MSI is due to the inactivation of MMR genes, such as MLH1 and MSH2, resulting in the loss or gain of short tandem DNA repeats (microsatellite repeats) [20,21]. The CANCERPLEX assay utilizes 950 microsatellite repeats including markers that are currently used in a clinical setting, and thus has great statistical power to identify tumors that exhibit MSI (Figure 4 bottom, middle panel). Independent testing methods, including IHC and/or PCR were used to determine the MSI status of a subset of colorectal patients. The CANCERPLEX test correctly identified the MSI status of every colorectal patient, with the same sensitivity as IHC but with greater sensitivity than PCR (Supplementary Table 10). The limit of detection was determined to be 10% tumor content, based upon results of tests from samples spiked with serial dilutions of a MSI-H tumor DNA (data not shown).
The TMB, defined as the rate of SNVs per Mb, can be determined from data generated from either whole-exome sequencing (WES) or targeted-gene panels [22,23]. MMR deficiency is found in roughly 15–20% of sporadic colorectal cancers (MSI-H colorectal cancers) and those resulting from germline mutations leading to hereditary nonpolyposis colorectal cancer or Lynch syndrome [24]. Tumors that exhibit MMR deficiency harbor hundreds to thousands of mutations, whereas MMR-proficient tumors typically have dozens [21]. The TMB and MSI status was determined for 262 colorectal tumors sequenced using CANCERPLEX and the results are shown in Figure 4 bottom, left panel.
CANCERPLEX includes probes that correspond to HPV and EBV biomarkers. Infection with these agents confers an increased risk of developing several different types of cancer, including head and neck and cervical cancer [25,26] and have implications on tumor classification, prognosis and treatment strategies [27,28]. The proficiency of the assay to detect these viruses was evaluated by testing a set of previously characterized virus-positive and -negative human DNAs, including clinical FFPE tumors, HeLa cells and two DNA samples derived from EBV-transformed cell lines. Results of validation testing demonstrated that the assay correctly identified EBV and HPV16/18 status in 100% of tested samples (Figure 4 bottom, right & Supplementary Table 11) and based on in silico dilutions, we established the limit of detection at 5% tumor content.
Clinical relevance
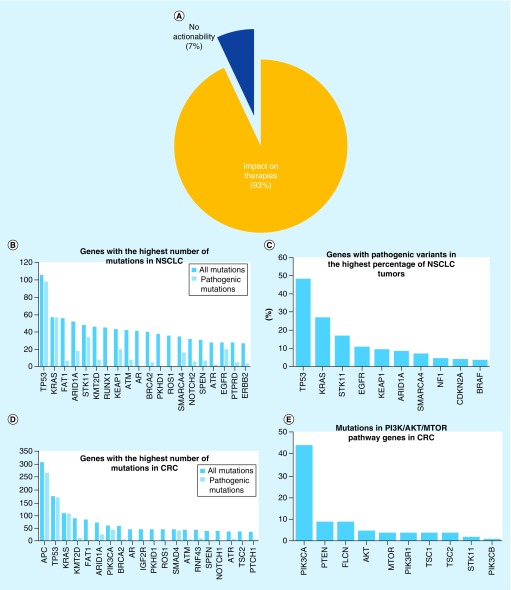
We assessed the usefulness of the 435-gene panel in two ways. First we used the TCGA data to compare the mutations in genes obtained by WES and from the 435-gene panel. We found that all cancer gene mutations that could be detected by WES were also detected by the 435-gene panel and at a comparable mutation frequency across a wide range of tumor types (Figure 2A). Second, the genetic/genomic profile generated by CANCERPLEX was used to assess potential therapeutic opportunities for the patients. We find that the results from 93% of the samples are actionable in determining an optimal clinical treatment decision for the patient (Figure 6A). Actionability is defined as having a genetic change for which there is either an FDA-approved therapy for the patient's disease or a different disease or therapies in clinical development. Aberrations that influenced therapeutic decisions, for example, by conferring resistance to therapies, are also reported.
Figure 6. . Clinical validity of CANCERPLEX®.
(A) Percentage of 1000 tumors in which clinically relevant mutations were detected. (B) The 20 most frequently mutated genes in 211 NSCLC adenocarcinoma tumors are shown. Dark bars denote the total number of mutations detected in each gene, light blue bars depict only pathogenic or likely pathogenic mutations. (C) The ten genes in NSCLC adenocarcinoma that had the highest frequency of pathogenic or likely pathogenic mutations are presented on the x-axis. The percentage of NSCLC adenocarcinoma tumors that harbored pathogenic or likely pathogenic mutations in each gene is presented on the y-axis. (D) The 20 most frequently mutated genes in 262 CRC tumors are shown. Dark bars denote the total number of mutations detected in each gene, light blue bars depict only pathogenic or likely pathogenic mutations. (E) The ten most frequently mutated genes in CRC that are either known to or predicted to activate the PI3K-AKT-MTOR pathway.
CRC: Colorectal cancer; NSCLC: Non-small-cell lung carcinoma.
In the clinical settings, if the detected pathogenic variant is coming from the ‘cancer risk’ gene group (e.g., BRCA1, BRCA2 and Lynch Syndrome genes), CANCERPLEX reporting system recommends a retest using dedicated genetic counseling services. We have examined the genetic changes in two of the tumor types with the largest number of specimens. In non-small-cell lung carcinoma (NSCLC) adenocarcinoma, mutations were most frequently detected in TP53, KRAS, ARID1A and FAT1 (Figure 6B). These results are consistent with TCGA data [29]. Pathogenic variants associated with targeted therapies that were detected in the highest percentage of NSCLC patients include TP53 (48%), KRAS (27%), STK11 (17%) and EGFR (11%) (Figure 6C). In colorectal tumors, mutations were most frequently detected in APC, TP53, KRAS and KMT2D. (Figure 6D). These results are also consistent with TCGA data [30]. In addition, we identified multiple genes in the PI3K-AKT-MTOR signaling pathway that were predicted to promote constitutive pathway activation (Figure 6E). MTOR-targeted inhibitors, including the FDA-approved drug everolimus, block the tumor-promoting signals generated by mutations in each of these genes. Many additional drugs that target each node of the PI3K-AKT-MTOR pathway are currently being evaluated in clinical trials to treat patients with aberrant PI3K-AKT-MTOR signaling [31].
Using TMB & MSI to predict candidates for immunotherapy
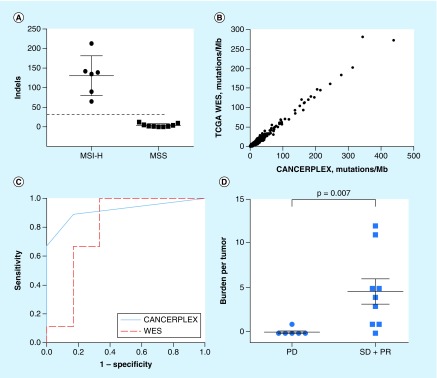
CANCERPLEX detects all mutations in the 435 genes as well as in confirmed microsatellite loci. This information can be used to deduce the mutation burden and the MSI status in each tumor (Figure 7A). It has been shown that TMB can be used to identify patients who are more likely to respond to immune checkpoint inhibitors that target programmed death 1 (PD-1) or PD-L1 [32,33]. Le et al. has shown previously that TMB, as determined by WES, correlated with how a cohort of 16 patients with advanced cancer responded to pembrolizumab [35]. Additional studies have shown that panel-based tumor sequencing can predict the hypermutated phenotype as accurately as WES [23,34]. To determine if mutation burden, as determined by the gene panel, also predicts response to checkpoint inhibitors, we performed downsampling on the WES data provided by Le et al., including only the 435 genes represented on CANCERPLEX, and found that assessment of TMB using the 435 gene panel correlated with the results obtained using WES and accurately predicted response to pembrolizumab (Figure 7B–D). Among the tested cohort of 262 colorectal cancer patients, 25.7% qualified as candidates for immunotherapy based upon TMB/MSI results.
Figure 7. . Microsatellite instability and tumor mutation burden readouts.
(A) 15 colorectal tumors, whose MSI status had been previously determined, underwent CANCERPLEX testing (Supplementary Table 9). The number of indels was calculated for each tumor and based upon these results, the MSI threshold was set (dashed line). (B) Mutation burden as calculated by a 435-gene panel (CANCERPLEX; x-axis) correlates with mutation burden as calculated by WES (y-axis). (C) ROC analysis of CANCERPLEX for CRC responders versus nonresponders to pembrolizumab (downsampling published data [35]). Red dashed line ROC curve using WES data, blue line ROC curve using data produced from CANCERPLEX testing. (D) Predictive accuracy of CANCERPLEX-mutational load (y-axis) for durable clinical benefit is comparable to that estimated by WES (downsampling published data [35]). Progressive disease (PD), stable disease (SD), partial response (PR).
CRC: Colorectal cancer; indel: Insertion and deletion; MSI: Microsatellite instability; ROC: Receiver operating characteristic; WES: Whole-exome sequencing.
Gene fusions provide an opportunity for precision medicine
Gene rearrangements can result in the expression of fusion proteins that drive tumors with unique clinical features [36–38]. Patients harboring tumors that express oncogenic fusion proteins tend to be particularly sensitive to targeted inhibition of the fusion protein. For instance, the tyrosine kinase inhibitor (TKI) crizotinib is approved to treat ALK-positive NSCLC based upon results of clinical trials demonstrating superior progression-free survival and response rates compared with standard chemotherapy [39,40]. Importantly, targeted inhibitors of the ALK and ROS1 kinases are approved for NSCLC patients with tumors that express these gene fusions, while a number of other targeted drugs are approved for other indications. We designed probes that target intronic regions flanking previously reported breakpoints for 19 oncogenes involved in genomic rearrangements that are likely to have clinical implications. Validation testing using DNA from cell lines expressing the EML-ALK gene fusion demonstrated that results from the CANCERPLEX assay were highly concordant with results obtained using FISH and that the assay's sensitivity and accuracy to detect the EML4-ALK gene rearrangement was 100% (Supplementary Table 9). We performed a limit of detection and linearity analysis and found that fusion junction points were detected with 100% accuracy when tumor cellularity was ≥20% and mean coverage was at least 300× (Supplementary Table 12). Out of the 238 NSCLC cases that underwent CANCERPLEX testing, ALK, ROS1 or RET gene fusions were detected in 5.0% (12 of 238), which is similar to the reported frequency of these three gene fusions in lung adenocarcinoma [41–43].
Case studies
NGS-based comprehensive tumor profiling enables the identification of therapeutically actionable mutations in tumor types they are not commonly associated with. For instance, we detected a BRAF-V600E mutation in a patient with rhabdoid meningioma. Importantly, dabrafenib, a BRAF-V600 specific inhibitor, has been used previously with impressive results to treat a child with BRAF-mutant positive rhabdoid meningioma [44]. In a patient with melanoma, we identified a less common mutation in the BRAF gene (BRAF-L597R). Although the BRAF-V600 specific drugs dabrafenib and vemurafenib and the MEK inhibitor trametinib are not approved to treat patients with BRAF non-V600 mutations, there are two case reports that describe significant clinical response to vemurafenib and trametinib in patients with BRAF-L597R mutant melanoma [45,46].
NGS-based comprehensive tumor profiling tests enable clinicians to optimize patient treatments by identifying therapeutically relevant mutations that would be missed if pathology-based classifications, individual gene testing, hot-spot testing or small panels were used. Tumor mutation profiling can also be important for patients that are likely to acquire mutations that confer resistance to standard therapies. For instance, we identified androgen receptor mutations in prostate cancers that act as agonists in the presence of enzalutamide (H875Y and T878A) or bicalutamide (W742C) and that confer resistance to bicalutamide (S889G). In a single patient with castrate-resistant prostate cancer, three mutations were detected that have previously been shown to cause acquired resistance to bicalutamide and enzalutamide treatments [47]. Patients with acquired drug resistance mutations may retain sensitivity to alternative treatments. Patients expressing the W742C mutation, for example, remain sensitive to the antiandrogen, hydroxyflutamide [48]. Hormone therapies are also commonly prescribed to patients with ER-positive breast cancer, however, 30% of these patients exhibit de novo resistance and significantly more patients will acquire resistance [49,50]. Several mutations found in the ESR1 gene confer resistance to aromatase inhibitors while retaining sensitivity to antiestrogen treatments [51–53]. We identified three mutations, Y537N, D538G and E380Q, that confer this type of selective endocrine resistance in 9.5% of ER-positive breast cancers.
Trastuzumab, a monoclonal antibody against the ERBB2 (HER2) receptor that was first approved by the FDA to treat a subset of patients with ERBB2-overexpressing breast cancer is now approved, in combination with chemotherapeutic agents, to treat metastatic gastric or gastroesophageal junction adenocarcinoma. ERBB2 can also be activated by somatic mutations that cause constitutive kinase activity [54]. However, unlike ERBB2 amplifications, ERBB2 gain-of-function mutations would be missed by methods used in pathology labs to classify the ERBB2 status of breast tumors. We have detected previously described ERBB2-activating mutations in several cases of breast, colorectal, gastroesophageal and lung cancers and in single cases of cervical, bladder and small intestinal cancer. Importantly, FDA-approved, small molecule TKIs that target the ERBB2 protein have shown promise in clinical trials to treat lung adenocarcinoma patients with ERBB2-activating mutations [55,56].
Discussion
Our understanding of the biology of many types of cancer is significantly enhanced by our understanding of the genetic/genomic changes in tumors. Use of genetic testing is often required prior to prescribing drugs (e.g., EGFR TKIs for certain NSCLC, crizotinib for NSCLC patients with ALK or ROS rearrangements), and testing can help the development of optimal therapies for a large number of tumors. NGS-based comprehensive tumor profiling provides the essential basis for obtaining the information to make such therapeutic decisions. A major obstacle to the success of these assays is reaching the sensitivity and specificity levels required to make reliable variant calls from clinical specimens that are often of dubious quality, quantity and/or tumor content. The CANCERPLEX assay has overcome these challenges by adopting a robust workflow that includes dissecting regions of the FFPE specimen enriched for tumor cells, customizing reagents and protocols to augment the sensitivity and specificity of the assay, sequencing data analysis pipeline to efficiently filter out technical artifacts and to extract clinically relevant features. CANCERPLEX demonstrated sensitivity of 99.5% and specificity of 99.9% for MAF ≥0.1. Detection of key oncogenic CNVs and chromosomal rearrangements was 100% concordant with the results obtained using independent well-established techniques.
The 435-gene panel provides significant advantages over individual gene testing, hot-spot testing or small panels. For instance, in 4 colorectal and 8 CNS tumors that had been previously characterized as wild-type using a small targeted gene panel, 30 additional clinically actionable mutations were identified in 9 of the 12 tumors using the 435-gene panel [57]. Importantly, expanded gene panels also enable the identification of additional genetic/genomic events, including TMB and MSI status, that may provide patients with treatment options that would have otherwise been missed.
The ability to mobilize an individual patient's immune system to treat cancer has long been a goal and that goal is now being recognized with the approval of several drugs that can stimulate the immune system for treating several different cancers [58]. Identifying biomarker(s) that predict which cancer patients will respond to checkpoint inhibitors is essential. PD-1 and its ligand (PD-L1) are the targets of most immune checkpoint drugs and PD-L1 expression is commonly used as a biomarker for selecting patients who are most likely to respond to immunotherapy [59]. More recent clinical data have demonstrated that PD-L1 expression alone is insufficient to accurately predict patient response to immune-oncology drugs [60]. TMB and DNA repair deficiencies (MSI) have evolved as more reliable biomarkers for selecting candidates for immunotherapy [35,61–62]. Virus-associated tumors express neo-antigens making these tumors additional, excellent targets for immunotherapy [63].
The CANCERPLEX assay was designed to facilitate the identification of cancer patients most likely to respond to immunotherapies by incorporating probes that detect amplification of the PD-L1, PD-L2 and JAK2 gene locus (9p24.1), regions of DNA implicated in MSI and integration of the HPV and EBV viral genomes. Additionally, we have adopted a formula that utilizes only highly informative, nonsynonymous mutations to calculate TMB. We have found that calculating TMB using the comprehensive gene panel predicts response to pembrolizumab just as effectively as using WES. The inclusion of these predictive biomarkers in comprehensive tumor profiling will enable the identification of patients who are likely to achieve significant benefit from immunotherapy who would have otherwise been missed.
Conclusion
Herein we report our experience with CANCERPLEX, an NGS-based assay that can accurately identify actionable genomic alterations within a clinically reasonable turnaround time and, thus, could guide personalized treatment strategies in upto 93% of clinical cases. Our methodology successfully addresses the challenges of sequencing samples with limited tumor content or low purity. CANCERPLEX was designed as a high-performance assay that leverages all clinically relevant information derived from genomic profiling to generate personalized patient reports. The benefit of adopting comprehensive molecular profiling into routine cancer care is a reality for common cancers and continues to expand to additional tumor types as knowledge is gained from a broad spectrum of targeted clinical trials.
Summary points.
CANCERPLEX® is a next-generation sequencing based analytical platform that characterizes somatic changes in FFPE tissue from solid tumors.
Tumor sequencing is performed with high uniform coverage (500× depth) across all exons of 435 cancer-related genes.
The assay was designed to detect various types of genomic alterations, in other words, single-nucleotide variants, small insertions/deletions, copy number variants and structural variants.
The test evaluates additional biomarkers, such as potential microsatellite instability, infection with oncoviruses (HPV16, HPV18 and EBV) and the tumor's mutation burden, that may have an impact on tumor classification, prognostic assessment and therapeutic decisions.
We applied an integrated analytical validation approach and confirmed high sensitivity and specificity metrics for all tested genomic aberrations and biomarkers in a range of variant allele fractions.
Our experience in a sample cohort of 1000 patients shows that 93% of the cases demonstrate at least one genomic alteration that can affect treatment strategies.
CANCERPLEX reports known somatic aberrations in frequencies consistent with the findings of large-scale cancer studies, for example, TCGA, and reveals potential novel oncogenic driver changes, such as the CD47-MET gene rearrangement.
The assay estimates tumor mutation burden and microsatellite instability to predict patients that are likely to respond to immunotherapy.
Final annotation, curation and reporting of results highlight the relevance of genomic changes to the clinical management of the disease in the form of an individualized patient's report.
We present case studies that demonstrate how patients with low-frequency or drug-resistant somatic alterations can benefit from the comprehensive gene-panel profiling.
Supplementary Material
Acknowledgements
The authors would like to thank Raju Kucherlapati for his careful reading of the manuscript and many insightful comments.
Footnotes
Financial & competing interests disclosure
The authors work for KEW, Inc., a biotechnology company. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Mendes D, Alves C, Afonso N, et al. The benefit of HER2-targeted therapies on overall survival of patients with metastatic HER2-positive breast cancer--a systematic review. Breast Cancer Res. 2015;17:140. doi: 10.1186/s13058-015-0648-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J. Clin. Oncol. 2009;27(26):4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 6.Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 2012;367(18):1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang CH, Yu CJ, Shih JY, et al. Specific EGFR mutations predict treatment outcome of stage IIIB/IV patients with chemotherapy-naive non-small-cell lung cancer receiving first-line gefitinib monotherapy. J. Clin. Oncol. 2008;26(16):2745–2753. doi: 10.1200/JCO.2007.15.6695. [DOI] [PubMed] [Google Scholar]
- 8.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 9.Normanno N, Tejpar S, Morgillo F, De Luca A, Van Cutsem E, Ciardiello F. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat. Rev. Clin. Oncol. 2009;6(9):519–527. doi: 10.1038/nrclinonc.2009.111. [DOI] [PubMed] [Google Scholar]
- 10.Macconaill LE, Van Hummelen P, Meyerson M, Hahn WC. Clinical implementation of comprehensive strategies to characterize cancer genomes: opportunities and challenges. Cancer Discov. 2011;1(4):297–311. doi: 10.1158/2159-8290.CD-11-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin EI, Tseng LH, Gocke CD, et al. Mutational profiling of colorectal cancers with microsatellite instability. Oncotarget. 2015;6(39):42334–42344. doi: 10.18632/oncotarget.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yost SE, Smith EN, Schwab RB, et al. Identification of high-confidence somatic mutations in whole genome sequence of formalin-fixed breast cancer specimens. Nucleic Acids Res. 2012;40(14):e107. doi: 10.1093/nar/gks299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43(11.10):11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odenthal M, Barta N, Lohfink D, et al. Analysis of microsatellite instability in colorectal carcinoma by microfluidic-based chip electrophoresis. J. Clin. Pathol. 2009;62(9):850–852. doi: 10.1136/jcp.2008.056994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jebbink J, Bai X, Rogers BB, Dawson DB, Scheuermann RH, Domiati-Saad R. Development of real-time PCR assays for the quantitative detection of Epstein-Barr virus and cytomegalovirus, comparison of TaqMan probes, and molecular beacons. J. Mol. Diagn. 2003;5(1):15–20. doi: 10.1016/S1525-1578(10)60446-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagahashi M, Wakai T, Shimada Y, et al. Genomic landscape of colorectal cancer in Japan: clinical implications of comprehensive genomic sequencing for precision medicine. Genome Med. 2016;8(1):136–149. doi: 10.1186/s13073-016-0387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263(5151):1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 19.Mano H. Non-solid oncogenes in solid tumors: EML4-ALK fusion genes in lung cancer. Cancer Sci. 2008;99(12):2349–2355. doi: 10.1111/j.1349-7006.2008.00972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonay TB, Koletou M, Wagner A. A survey of tandem repeat instabilities and associated gene expression changes in 35 colorectal cancers. BMC Genomics. 2015;16:702. doi: 10.1186/s12864-015-1902-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woerner SM, Benner A, Sutter C, et al. Pathogenesis of DNA repair-deficient cancers: a statistical meta-analysis of putative real common target genes. Oncogene. 2003;22(15):2226–2235. doi: 10.1038/sj.onc.1206421. [DOI] [PubMed] [Google Scholar]
- 22.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes whole-exome sequencing derived tumor mutation burden as a biomarker that can predict response to immunotherapy in melanoma patients.
- 23.Campesato LF, Barroso-Sousa R, Jimenez L, et al. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to PD-1 blockade in clinical practice. Oncotarget. 2015;6(33):34221–34227. doi: 10.18632/oncotarget.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Extends the possibility of estimating tumor mutation burden from gene-panel next-generation sequencing data to predict clinical benefit of immunotherapies.
- 24.Poulogiannis G, Frayling IM, Arends MJ. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology. 2010;56(2):167–179. doi: 10.1111/j.1365-2559.2009.03392.x. [DOI] [PubMed] [Google Scholar]
- 25.Ko YH. EBV and human cancer. Exp. Mol. Med. 2015;47:e130. doi: 10.1038/emm.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bansal A, Singh MP, Rai B. Human papillomavirus-associated cancers: a growing global problem. Int. J. Appl. Basic Med. Res. 2016;6(2):84–89. doi: 10.4103/2229-516X.179027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho J, Kang MS, Kim KM. Epstein-Barr virus-associated gastric carcinoma and specific features of the accompanying immune response. J. Gastric Cancer. 2016;16(1):1–7. doi: 10.5230/jgc.2016.16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asati V, Mahapatra DK, Bharti SK. PI3K/Akt/mTOR and Ras/Raf/MEK/ERK signaling pathways inhibitors as anticancer agents: structural and pharmacological perspectives. Eur. J. Med. Chem. 2016;109:314–341. doi: 10.1016/j.ejmech.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 32.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Showcases that mutation burden, as determined by whole-exome sequencing, correlates with response to pembrolizumab in non-small-cell lung cancer, which, according to our results, extends to CANCERPLEX's gene panel approach.
- 33.Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J. Clin. Oncol. 2016;34(19):2206–2211. doi: 10.1200/JCO.2016.66.6552. [DOI] [PubMed] [Google Scholar]
- 34.Johnson DB, Frampton GM, Rioth MJ, et al. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol. Res. 2016;4(11):959–967. doi: 10.1158/2326-6066.CIR-16-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Confirms that panel-based tumor sequencing can assess mutation burden and help stratify responders to programmed death 1 pharmacologic blockade.
- 35.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Q, Zhan P, Zhang X, Lv T, Song Y. Clinicopathologic characteristics of patients with ROS1 fusion gene in non-small cell lung cancer: a meta-analysis. Transl. Lung Cancer Res. 2015;4(3):300–309. doi: 10.3978/j.issn.2218-6751.2015.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuta K, Kohno T, Yoshida A, et al. RET-rearranged non-small-cell lung carcinoma: a clinicopathological and molecular analysis. Br. J. Cancer. 2014;110(6):1571–1578. doi: 10.1038/bjc.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N. Engl. J. Med. 2013;368(25):2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 40.Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl. J. Med. 2014;371(23):2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 41.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 42.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 2012;30(8):863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 2012;18(3):378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 44.Mordechai O, Postovsky S, Vlodavsky E, et al. Metastatic rhabdoid meningioma with BRAF V600E mutation and good response to personalized therapy: case report and review of the literature. Pediatr. Hematol. Oncol. 2015;32(3):207–211. doi: 10.3109/08880018.2014.936058. [DOI] [PubMed] [Google Scholar]; • This is a case study showing the benefit of treating established oncogenic driver alterations, such as BRAF V600E mutation, in nontypical tumor type in a precision-medicine manner.
- 45.Bahadoran P, Allegra M, Le Duff F, et al. Major clinical response to a BRAF inhibitor in a patient with a BRAF L597R-mutated melanoma. J. Clin. Oncol. 2013;31(19):e324–e326. doi: 10.1200/JCO.2012.46.1061. [DOI] [PubMed] [Google Scholar]
- 46.Bowyer SE, Rao AD, Lyle M, et al. Activity of trametinib in K601E and L597Q BRAF mutation-positive metastatic melanoma. Melanoma Res. 2014;24(5):504–508. doi: 10.1097/CMR.0000000000000099. [DOI] [PubMed] [Google Scholar]; • The above two studies refer to case reports on patients with low-frequency genetic changes that responded to treatment, which is approved for use in patients with a similar mutation (BRAF V600E).
- 47.Lallous N, Volik SV, Awrey S, et al. Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients. Genome Biol. 2016;17:10. doi: 10.1186/s13059-015-0864-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hara T, Miyazaki J, Araki H, et al. Novel mutations of androgen receptor: a possible mechanism of bicalutamide withdrawal syndrome. Cancer Res. 2003;63(1):149–153. [PubMed] [Google Scholar]
- 49.Riggins RB, Schrecengost RS, Guerrero MS, Bouton AH. Pathways to tamoxifen resistance. Cancer Lett. 2007;256(1):1–24. doi: 10.1016/j.canlet.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis JS, Jordan VC. Selective estrogen receptor modulators (SERMs): mechanisms of anticarcinogenesis and drug resistance. Mutat. Res. 2005;591(1–2):247–263. doi: 10.1016/j.mrfmmm.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 51.Robinson DR, Wu YM, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 2013;45(12):1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ingle JN, Suman VJ, Rowland KM, et al. Fulvestrant in women with advanced breast cancer after progression on prior aromatase inhibitor therapy: North Central Cancer Treatment Group Trial N0032. J. Clin. Oncol. 2006;24(7):1052–1056. doi: 10.1200/JCO.2005.04.1053. [DOI] [PubMed] [Google Scholar]
- 53.Chigira T, Nagatoishi S, Tsumoto K. Differential binding of prohibitin-2 to estrogen receptor alpha and to drug-resistant ERalpha mutants. Biochem. Biophys. Res. Commun. 2015;463(4):726–731. doi: 10.1016/j.bbrc.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Herter-Sprie GS, Greulich H, Wong KK. Activating mutations in ERBB2 and their impact on diagnostics and treatment. Front. Oncol. 2013;3:86. doi: 10.3389/fonc.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Greve J, Teugels E, Geers C, et al. Clinical activity of afatinib (BIBW 2992) in patients with lung adenocarcinoma with mutations in the kinase domain of HER2/neu. Lung Cancer. 2012;76(1):123–127. doi: 10.1016/j.lungcan.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 56.De Greve J, Moran T, Graas MP, et al. Phase II study of afatinib, an irreversible ErbB family blocker, in demographically and genotypically defined lung adenocarcinoma. Lung Cancer. 2015;88(1):63–69. doi: 10.1016/j.lungcan.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 57.Hedayat A, Peterson J, Tse J, Eifert C, Gallagher T, Deharvengt S. USCAP Annual Meeting. San Antonio; TX, USA: 4–10 March 2017. Assessing the Clinical Utility of Expanded vs. Small Targeted Solid Tumor NGS Panels. Presented at. [Google Scholar]
- 58.Menon S, Shin S, Dy G. Advances in cancer immunotherapy in solid tumors. Cancers (Basel) 2016;8(12) doi: 10.3390/cancers8120106. pii:E106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masucci GV, Cesano A, Hawtin R, et al. Validation of biomarkers to predict response to immunotherapy in cancer: volume I - pre-analytical and analytical validation. J. Immunother. Cancer. 2016;4:76. doi: 10.1186/s40425-016-0178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grigg C, Rizvi NA. PD-L1 biomarker testing for non-small cell lung cancer: truth or fiction? J. Immunother. Cancer. 2016;4:48. doi: 10.1186/s40425-016-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Madore J, Strbenac D, Vilain R, et al. PD-L1 negative status is associated with lower mutation burden, differential expression of immune-related genes, and worse survival in stage III melanoma. Clin. Cancer Res. 2016;22(15):3915–3923. doi: 10.1158/1078-0432.CCR-15-1714. [DOI] [PubMed] [Google Scholar]
- 62.Falchook GS, Leidner R, Stankevich E, et al. Responses of metastatic basal cell and cutaneous squamous cell carcinomas to anti-PD1 monoclonal antibody REGN2810. J. Immunother. Cancer. 2016;4:70. doi: 10.1186/s40425-016-0176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ott PA, Hodi FS. The B7-H1/PD-1 pathway in cancers associated with infections and inflammation: opportunities for therapeutic intervention. Chin. Clin. Oncol. 2013;2(1):7. doi: 10.3978/j.issn.2304-3865.2012.11.05. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.