Abstract
Background
Duchenne muscular dystrophy (DMD) is the most common disease in children caused by mutations in the DMD gene, and DMD and Becker muscular dystrophy (BMD) are collectively called dystrophinopathies. Dystrophinopathies show a complex mutation spectrum. The importance of mutation databases, with clinical phenotypes and protein studies of patients, is increasingly recognized as a reference for genetic diagnosis and for the development of gene therapy.
Methods
We used the data from the Japanese Registry of Muscular Dystrophy (Remudy) compiled during from July 2009 to March 2017, and reviewed 1497 patients with dystrophinopathies.
Results
The spectrum of identified mutations contained exon deletions (61%), exon duplications (13%), nonsense mutations (13%), small deletions (5%), small insertions (3%), splice-site mutations (4%), and missense mutations (1%). Exon deletions were found most frequently in the central hot spot region between exons 45–52 (42%), and most duplications were detected in the proximal hot spot region between exons 3–25 (47%). In the 371 patients harboring a small mutation, 194 mutations were reported and 187 mutations were unreported.
Conclusions
We report the largest dystrophinopathies mutation dataset in Japan from a national patient registry, “Remudy”. This dataset provides a useful reference to support the genetic diagnosis and treatment of dystrophinopathy.
Electronic supplementary material
The online version of this article (10.1186/s13023-017-0703-4) contains supplementary material, which is available to authorized users.
Keywords: Duchenne and Becker muscular dystrophies, Genetic diagnosis, MLPA, Nucleotide sequencing
Background
Dystrophinopathies are an X-linked disease caused by mutations in the DMD gene (OMIM 300377) located at Xp21.2. DMD is a large gene, spanning more than 2.2 Mb of genomic DNA, and containing 79 exons and lengthy introns, which produces a 14-kb mRNA transcript [1]. Dystrophinopathies are the most common muscle disease in children which affects one in 3600–6000 live male births [2, 3], and has been classically classified into two forms: Duchenne muscular dystrophy (DMD; OMIM 310200) and Becker muscular dystrophy (BMD; OMIM 300376). DMD shows a severe phenotype clinically characterized by rapid progression in early childhood, and loss of ambulation during the second decade of life, while BMD shows a milder form with patients being ambulant after 16 years of age. Patients with an intermediate phenotype are sometimes referred to as having intermediate muscular dystrophy (IMD). Nevertheless, the classification into the three forms is not always easy.
To date, approximately 70% of mutations found in DMD patients are deletions/duplications of one or more exons, while the remaining 30% are caused by small mutations at the nucleotide level. Multiplex ligation-dependent probe amplification (MLPA), which can examine the duplications and/or deletions of all 79 exons, has been developed and widely used. [4–6] However, the remaining 30% of patients harboring small mutations are undiagnosed and need to perform sanger sequencing for diagnosis.
Recently, promising mutation-specific molecular therapies have been developed. For instance, exon skipping is expected to be applicable to patients that have one or more exons deletions in DMD. Furthermore, read-through therapy of a nonsense codon to produce full-length dystrophin is applicable to patients with DMD harboring nonsense mutations. Nonsense mutations are present in approximately 15% of all DMD patients [7–9]. However, the precise analysis of the DMD mutation is required for the application of these molecular therapies to patients.
In Japan, a hospital based dataset of 127 patients harboring small mutations by Takeshima et al. in 2010 [10], and a registry based dataset in Japanese Registry of Muscular Dystrophy (Remudy) of exon deletions and duplications by Nakamura et al. [11] have been compiled. Here, we extend these previous reports not only by increasing the number of the patients (1497 patients), but also by comprehensive genetic analysis of exonic and small nucleotide mutations and immunostaining of dystrophin on muscle biopsies to deduce genotype-phenotype correlations in patients with dystrophinopathies.
Methods
Registry-based datasets
Remudy was developed in 2009 in collaboration with the Translational Research in Europe-Assessment and Treatment of Neuromuscular Diseases (TREAT-NMD) Network of Excellence [11–13]. The Remudy database for male patients with dystrophinopathies includes clinical and molecular genetic data, as well as all mandatory and highly encouraged items for the TREAT-NMD global patient registry. To classify into DMD, BMD or IMD, the attending physicians reviewed their clinical information. Then, our clinical and genetic curators independently evaluated physicians’ classification by reviewing the clinical information, and pathological data (including dystrophin immune-staining, if applicable), and report of genetic analysis for DMD. In the present study, we used the registry data compiled during from July 2009 to March 2017.
Analysis of small mutations
When the mutations were not detected by MLPA, but dystrophin immunostaining supported the diagnosis of dystrophinopathies, the nucleotide sequence of all exons and their flanking intronic regions in DMD was determined by IonPGM (Thermo Fisher Scientific, MA, USA) [14] or Sanger method. Missense variants were filtered with an allele frequency under 0.01 in the Human Genetics Variation Browser (HGVD; http://www.hgvd.genome.med.kyoto-u.ac.jp/), the Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org/), and the NHLBI Exome Sequencing Project (ESP6500; http://evs.gs.washington.edu/EVS/). In silico analysis of the effect of the amino acid change was conducted using Polymorphism Phenotyping version 2 (PolyPhen 2; http://genetics.bwh.harvard.edu/pph2/) [15, 16].
Dystrophin immunostaining
Immunohistochemical analyses for dystrophin on patient’s muscle sections were performed using mouse monoclonal antibodies against dystrophin C-terminus (NCL-DYS2), rod (NCL-DYS1), and N-terminus (NCL-DYS3) (all from Novocastra Lab) by standard procedures as reported previously [17].
Results
In July 2009 through March 2017, a total of 1497 dystrophinopathies patients were registered in Remudy. Among them, 1167, 295, and 35 patients were respectively diagnosed having DMD, BMD, and IMD. The genetic diagnosis was made by MLPA and/or multiplex-PCR in 1092 patients with exon deletion in 901 patients (61% of 1497 dystrophinopathies patients), exon duplications in 188 (13%), and both exon deletions and duplications in 3 (0.2%). All patients harboring single exon deletion were performed sanger sequencing to confirm the deletion.
Among the remaining 405 patients, DNA sequencing revealed small mutations in 371.
One patient was found to have pericentric inversion of the X chromosome by chromosome testing. One patients were diagnosed as having facioscalpulohumeral muscular dystrophy after registration. However, the remaining 32 patients remained genetically undiagnosed.
Large deletion/duplication mutation patterns and hot spot analysis
The deletion or duplication patterns and their frequencies were plotted, in Fig. 1a and b. The cumulative number of subjects with deletions or duplications of each exon was plotted as shown in Fig. 2a and b. The most common exon deletion pattern was the deletion of exons 45–47 (68/901, 7.5%) (Fig. 1a). Most exon deletions in patients with dystrophinopathies were observed in the hotspot region between exons 45 and 52 (42%) (Fig. 2a). A second frequent-deletion hotspot region towards the 5′-end was distinguished in the region between exons 3 and 21 (28%) (Fig. 2a). On the other hand, the most common exon duplication pattern was the duplication of exon 2 (14/188, 7.4%) (Fig. 1b). The largest duplications in the patients with dystrophinopathies were observed in the hotspot region between exons 3 and 25 (47%) (Fig. 2b).
Fig. 1.
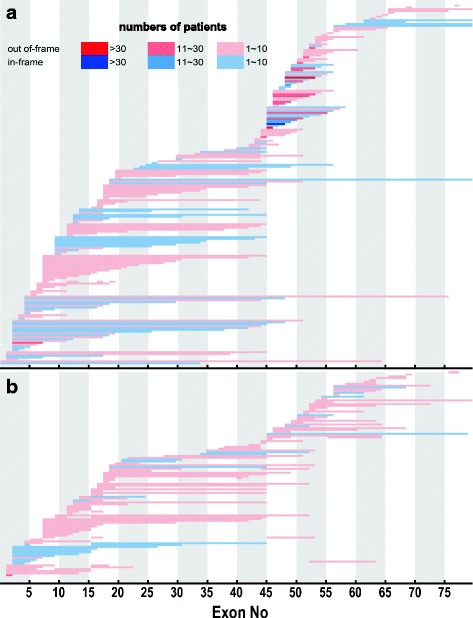
Patterns of exon deletion and exon duplication in the dystrophin gene in patients with dystrophinopathy. a Exon deletion. Each bar represents a deleted exon observed in a patient. b Exon duplication. Each bar represents a duplicated exon observed in a patient
Fig. 2.
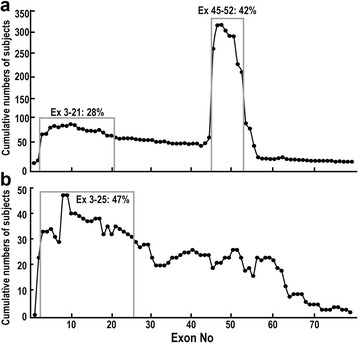
Cumulative numbers of subjects with a deletion/duplication per exon. a Deletions. A hot spot region is visible between exon 45 and exon 52. b Duplications. A hot spot region is visible between exon 3 and exon 25
Small mutations
The spectrum of identified mutations included nonsense mutations (186/371, 50%); small deletions (77/371, 21%); small insertions (39/371, 11%); splice-site mutations (61/371, 16%); and missense mutations (8/371, 2%) (Fig. 3a). The location of small mutations in patients with DMD and BMD patients is shown in Fig. 3b. Detailed results of the small mutations in the 371 patients, with 312 kinds of mutations, are shown in Additional file 1: Table S1. Among the 312 mutations, 149 were reported and 163 were unreported by HGMD (Human Gene Mutation Database) [18].
Fig. 3.

a Mutation spectrum of small mutations in the dystrophin gene in patients with dystrophinopathy. b Distribution of small mutations in DMD
Nonsense mutations
Nonsense mutations were the most common small mutations, accounting for 50% of the patients (186/371) (Fig. 3a). Although the mutations were almost uniformly distributed throughout the DMD, 6 patients shared the common mutation p.Arg3381* (c.10141C > T). Interestingly, 17 of 186 patients showed faint and patchy patterns of dystrophin immunostaining on skeletal muscles (Fig. 4, Additional file 1: Table S2). Clinical diagnosis of the 17 patients was DMD in 3 patients, IMD in 2, and BMD in 12, respectively.
Fig. 4.

Distribution of the number of patients with nonsense mutations and faint and patchy dystrophin immunostaining
Missense variations
Only 8 patients (6 unrelated and 2 siblings) had 7 distinct missense variants (2% of the 371 patients with small mutations) (Table 1). Among them, 3 presented DMD phenotype while 3 BMD. Among the 7 patients who received muscle biopsy, the 2 patients showed total dystrophin deficiency, while 5 showed faint and patchy expression pattern on immunohistochemistry. None of the 7 missense variants was registered in any of HGVD, ExAC, and ESP6500. All variants were predicted to be probably or possibly damaging by PolyPhen2 (Table 1).
Table 1.
Patients with DMD/BMD harboring missense mutations
| Patient | Phetnotype | Dystrophin immunostaining | Exon | DNA change | Protein (predicted) | Frequency | PolyPhen2 prediction | |||
|---|---|---|---|---|---|---|---|---|---|---|
| HGVD | ESP6500 | ExAC | ||||||||
| 5 | BMD | faint&patchy | 3 | c.152 T > G | p.Leu51Arg | no | no | no | 0.999 | probably |
| 19 | BMD | faint&patchy | 6 | c.434G > C | p.Arg145Pro | no | no | no | 0.998 | probably |
| 22*2 | BMD | not done | 6 | c.481A > C | p.Thr161Pro | no | no | no | 0.628 | possibly |
| 23*2 | BMD | faint&patchy | 6 | c.481A > C | p.Thr161Pro | no | no | no | 0.628 | possibly |
| 112 | DMD | negative | 22 | c.2949G > T | p.Gln9832His | no | no | no | 0.991 | probably |
| 317 | BMD | faint&patchy | 68 | c.9896A > G | p.His3299Arg | no | no | no | 0.999 | probably |
| 320 | DMD | faint&patchy | 68 | c.9937 T > C | p.Cys3313Arg | no | no | no | 0.999 | probably |
| 325 | DMD | negative | 69 | c.10011C > G | p.Cys3337Trp | no | no | no | 1 | probably |
*2 represents brothers
Discussion
This study retrospectively evaluated the results of gene analysis for 1497 patients with dystrophinopathies with clinical features and dystrophin immunostaining. Among the 1497 patients, 1092 (73%) were diagnosed by the MLPA method; including 901 patients with exon deletions (901/1497, 61%) and 188 patients with exon duplications (188/1497, 13%).
This is the report of the largest number of patients all previous dystrophinopathies reports from Asian countries [19–24]. Most of the previously reports from Asian countries [19–21] demonstrated lower deletion and higher duplication rates. This may be explained by the fact that much smaller numbers of patients were studied including female carriers. Nevertheless, Suh et al. [24], who reported the rates of deletions and duplications respectively, as 71.8%/ and 16.4% among 130 patients in Korea, which is similar not only to our results but also to the results reported from Western countries [25, 26]. In addition, exon deletion and duplication hotspots in our study (Fig. 2) are similar to those in the previous reports [20–22, 25, 26], suggesting that ethnicity is not a factor to cause any difference in the proportion of exon deletions and duplications in DMD.
Based on our results of exon deletions/duplications, we estimated the applicability of exon skipping therapies (Tables 2 and 3). Skipping of exon 51, which has been approved by FDA, and of exon 53 skipping, which is in clinical trials, cans theoretically be applied to the largest group of the patients with exon deletions, as suggested in previous reports [27].
Table 2.
Overview of therapeutic exon 51 or 53 skipping for a series of DMD patients with exon deletion
| skipped exon | Deleted exon | Number of patients |
|---|---|---|
| 51 | 13–50 | 0 |
| 29–50 | 0 | |
| 43–50 | 0 | |
| 45–50 | 23 | |
| 48–50 | 25 | |
| 49–50 | 29 | |
| 50 | 12 | |
| 52 | 18 | |
| 47–50 | 0 | |
| 52–63 | 0 | |
| total 107 | ||
| 53 | 10–53 | 0 |
| 43–52 | 0 | |
| 45–52 | 35 | |
| 47–52 | 0 | |
| 48–52 | 33 | |
| 49–52 | 19 | |
| 50–52 | 6 | |
| 52 | 18 | |
| total 111 |
Table 3.
Overview of the applicability of exon 51 or 53 skipping for DMD exon deletions
| ekipping exon | Rate of applicable patients in all mutated patients | Rate of applicable patients in patients with exon deletion |
|---|---|---|
| 51 | 7.1% | 11.9% |
| 53 | 7.4% | 12.3% |
Among 185 patients (13%) who harbor a nonsense mutation, interestingly, 17 patients showed faint and patchy dystrophin expression pattern on immunohistochemistry (Fig .4 and Additional file 1: Table S2). Nonsense mutations found in patients with BMD patients have reported to be present in exons 27, 29, 31, 37, 49, and 72 [28–30], all of which are in-frame exons, resulting in dystrophin positivity and milder phenotypes. Similarly 15 of the 17 patients with nonsense mutations and faint and patchy dystrophin staining in this study had mutations in in-frame exons. BMD phenotype in these patients may well be explained by the skipping of the correspondent exon, but cDNA analysis is necessary to be conclusive. From these results, there are 168 patients (12%) who are treated for nonsense codon read-through treatment.
Seven distinct missense variants were identified in 8 patients (Table 1). They may well be pathogenic since they are not reported in the HGVD, ESP6500, or ExAC and are predicted to be possibly or probably damaging in proteins by PolyPhen2. Especially, the 4 variants located in the N-terminus of dystrophin may be more likely to have pathogenic significance as previous reports showed the missense mutations in the actin binding domain of dystrophin, which is in the N-terminus region, induce thermodynamic instability of dystrophin molecules and protein aggregation [31, 32]. Needless to say, however, there still remains a possibility that patients may have unidentified causative mutations in other regions such as deep intronic regions or some missense mutations have an effect on splicing.
Conclusion
Our report provides the largest DMD mutation dataset in Japan, which could be used as a reference for genetic diagnosis and will also help in further elucidating the nature of the disease.
Additional files
Clinical and dystrophin immunostaining in the patients with a small mutation. Table S2. Patients with nonsense mutation and faint&patchy dystrophin staining. (PDF 286 kb)
Acknowledgements
We express our heartfelt gratitude for patients, families, physicians, and the Japanese muscular dystrophy association to support the national dystrophinopathies registry.
We also express our appreciation and condolences to deceased Dr. Mitsuru Kawai, former director of National hospital organization Higashisaitama hospital, who was a founder and godfather of Remudy.
Ethic approval and consent to participate
All clinical information and materials used in the present study were obtained for diagnostic purposes with written informed consent. The study was approved by the Ethics Committee of National Center of Neurology and Psychiatry (NCNP).
Funding
This study was supported partly by the Intramural Research Grant (23–4, 26–7, 28–6 and 29–4) for Neurological and Psychiatric Disorders of NCNP, Research and Development Grants for Practical Research Project for Rare/Intractable Diseases (16ek0109066h0003), Research Grants for Nervous and Mental Disorders (20–11) from the Ministry of Health, Labour and Welfare of Japan.
Availability of data and materials
The datasets generated and analyzed during the current study are available in the Japanese Registry of Muscular Dystrophy (Remudy: http://www.remudy.jp/). The details of mutation data of the study are available upon request.
Author’s contributions
MO and SN participated in planning this study, analyzed and interpreted the data, and wrote manuscript. KG, YH and SM participated in the genetic analysis, and reviewed the genetic and molecular data as curators. HK and MMY collected and reviewed the clinical data as curators. HN, EK, and IN supervised the planning of this study. All authors read and approved of the final manuscript.
Abbreviations
- BMD
Becker muscular dystrophy
- DMD
Duchenne muscular dystrophy
- ESP6500
Exome Sequencing Project
- ExAC
Exome Aggregation Consortium
- HGMD
Human gene mutation database
- HGVD
Human Genetics Variation Browser
- MLPA
Multiplex Ligation-dependent Probe Amplification
- Remudy
Registry of Muscular Dystrophy
- TREAT-NMD
Translational Research in Europe-Assessment and Treatment of Neuromuscular Diseases
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13023-017-0703-4) contains supplementary material, which is available to authorized users.
Contributor Information
Mariko Okubo, Email: ohkubom@ncnp.go.jp.
Kanako Goto, Email: kgoto@ncnp.go.jp.
Hirofumi Komaki, Email: komakih@ncnp.go.jp.
Harumasa Nakamura, Email: hnakamura@ncnp.go.jp.
Madoka Mori-Yoshimura, Email: yosimur@ncnp.go.jp.
Yukiko K. Hayashi, Email: y-hayashi@tokyo-med.ac.jp
Satomi Mitsuhashi, Email: satomi11@me.com.
Satoru Noguchi, Email: noguchi@ncnp.go.jp.
Ichizo Nishino, Phone: +81-42-341-2711, Email: nishino@ncnp.go.jp.
References
- 1.Ahn AH, Kunkel LM. The structural and functional diversity of dystrophin. Nat Genet. 1993;3:283–291. doi: 10.1038/ng0493-283. [DOI] [PubMed] [Google Scholar]
- 2.Emery AE. Population frequencies of inherited neuromuscular disease-a world survey. Neuromuscl Disord. 1991;1:19–29. doi: 10.1016/0960-8966(91)90039-U. [DOI] [PubMed] [Google Scholar]
- 3.Bushby K, Finkel R, Brinkrant DJ, Case LE, Clemens PR, Cripe L, et al. Diagnosis and management of Duchenne muscular dystrophy, part1:diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2010;9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- 4.Gatta V, Scarciolla O, Gaspari AR, Palka C, De Angelis MV, Di Mizuno A, et al. Identification of deletions and duplications of the DMD gene in affected males and carrier females by multiples ligation probe amplification (MLPA) Hum Genet. 2005;117:92–98. doi: 10.1007/s00439-005-1270-7. [DOI] [PubMed] [Google Scholar]
- 5.Lalic T, Vossen R, Coffa J, Schouten J, Guc-Scekic M, Radivojevic D, et al. Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet. 2005;13:1231–1234. doi: 10.1038/sj.ejhg.5201465. [DOI] [PubMed] [Google Scholar]
- 6.Todorova A, Todorov T, Georgieva B, Lukova M, Guergueltcheva V, Kremensky I, et al. MLPA analysis/complete sequencing of DMD gene in a group of Bulgarian Duchenne/Becker muscular dystrophy patients. Neuromuscul Disord. 2008;18:667–670. doi: 10.1016/j.nmd.2008.06.369. [DOI] [PubMed] [Google Scholar]
- 7.MaliK V, Rodino-Klapac LR, Viollet L, Wall C, King W, Al-Dahhak R, et al. Gentamicin-induced Readthrough of stop codons in Duchenne muscular dystrophy. Ann Neurol. 2010;67:771–780. doi: 10.1002/ana.22024. [DOI] [PubMed] [Google Scholar]
- 8.Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–93. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 9.Finkel RS, Flanigan KM, Wong B, Bonnemann C, Sampson K, Sweeney HL, et al. Phase 2a study of Ataluren-mediated Dystrophin production in patients with nonsense mutation Duchenne muscular dystrophy. PLoS One. 2013;8 doi: 10.1371/journal.pone.0081302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeshima Y, Yagi M, Okizuka Y, Awano H, Zhang Z, Yamauchi Y, et al. Mutation spectrum of dystrophin gene in 442 Duchenne/Becker muscular dystrophy cases from one Japanese referral center. J Hum Genet. 2010;55:379–388. doi: 10.1038/jhg.2010.49. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura H, Kiumura E, Mori-Yoshimura M, Komaki H, Matsuda Y, Goto K, et al. Characteristics of Japanese Duchenne and Becker muscular dystrophy patients in a novel Japanese national registry of muscular dystrophy patients (Remudy) Orphanet J Rare. 2013;8:60. doi: 10.1186/1750-1172-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bladen CL, Rafferty K, Straub V, Monges S, Moresco A, Dawkins H, et al. The TREAT-NMD Duchenne muscular dystrophy registries: conception, design and utilization by industry and academia. Hum Mutat. 2013;34:1449–1457. doi: 10.1002/humu.22390. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi F, Yonemoto N, Nakamura H, Shimizu R, Komaki H, Mori-Yoshimura M, et al. Prednisolone improves walking in Japanese Duchenne muscular dystrophy patients. J Neurol. 2013;260:3023–3029. doi: 10.1007/s00415-013-7104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okubo M, Minami N, Goto K, Goto Y, Noguchi S, Mitsuhashi S, Nishino I. Genetic diagnosis of Duchenne/Becker muscular dystrophy using next-generation sequencing: validation analysis of DMD mutations. J of Hum Genet. 2016;61:483–489. doi: 10.1038/jhg.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Human Genetic Variation Browser, Japanese genetic variation consortium, a reference database of genetic variations in Japanese population. http://www.genome.med.kyoto-u.ac.jp/SnpDB
- 16.Polyphen-2 prediction of functional effects of human nsSVPs. http://genetics.bwh.harvard.edu/pph2/.
- 17.Sahashi K, Ibi T, Suoh H, Nakao N, Tashiro M, Marui K, et al. Immunostaining of Dystrophin and Utrophin in skeletal muscle Dystrophinopathies. Intern Med. 1994;33:277–283. doi: 10.2169/internalmedicine.33.277. [DOI] [PubMed] [Google Scholar]
- 18.Human Gene Mutation Database https://portal.biobase-international.com/cgi-bin/portal/login.cgi
- 19.Wang X, Zheng W, Yan M, Huang S, Chen T-J, Zhong N. Similarity of DMD gene deletion and duplication in the Chinese patients compared to global populations. Behav Brain Funct. 2008;4:20. doi:10.1186/1744-9081-4-20. [DOI] [PMC free article] [PubMed]
- 20.Hwa HL, Chang YY, Chen CH, Kao YS, Jong YJ, Chao MC, et al. Multiplex ligation-dependent probe amplification identification of deletions and duplications of the Duchenne muscular dystrophy gene in Taiwanese subjects. J Formos Med Assoc. 2007;106:339–346. doi: 10.1016/S0929-6646(09)60318-1. [DOI] [PubMed] [Google Scholar]
- 21.Lee BL, Nam SH, Lee JH, Ki CS, Lee M, Lee J. Genetic analysis of dystrophin gene for affected male and female carries with Duchenne/Becker muscular dystrophy in Korea. J Korean Med Sci. 2012;27:274–280. doi: 10.3346/jkms.2012.27.3.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C, Ma H, Zhang F, Chen L, Xing X, Wang S, et al. Screening of Duchenne muscular dystrophy (DMD) mutations and investigating its mutational mechanism in Chinese patients. PLoS One. 2014;9:e108038. doi: 10.1371/journal.pone.0108038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manjunath M, Kiran P, Preethish-Kumar V, Nalini A, Singh RJ, Gayathri N. A comparative study of mPCR, MLPA, and muscle biopsy results in a cohort of children with Duchenne muscular dystrophy: a first study. Neurol India. 2015;63:58–62. doi: 10.4103/0028-3886.152635. [DOI] [PubMed] [Google Scholar]
- 24.Suh MR, Lee KA, Kim EY, Jung J, Choi WA, Kang SW. Multiplex ligation-dependent probe amplification in X-linked recessive muscular dystrophy in Korean subjects. Yonsei Med J. 2017;58:613–618. doi: 10.3349/ymj.2017.58.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daniel GA, Mioni F, Muller CR, Vitiello L, Mostacciculo ML, Grimm T. Patterns of deletions of the dystrophin gene in different European populations. Hum Genet. 1993;91:342–346. doi: 10.1007/BF00217354. [DOI] [PubMed] [Google Scholar]
- 26.Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations : one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/S1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 27.Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar L, Deutekom JV, Ommen GJ, Dunnen JT. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mut. 2009;30:293–299. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- 28.A. Nishida, N Kataoka, Y Takeshima, M Yagi, H Awano, M Ota, K Itoh, M Hagiwara, M Matsuo. Chemical treatment enhances skipping of a mutated exon in the dystrophin gene. Nature Communications. 2011; doi: 10.1038/mcp,,s1306. [DOI] [PMC free article] [PubMed]
- 29.Shiga N, Takeshima Y, Sakamoto H. K Inoue, Y Yokota, M Yokoyama, M Matsuo. Disruption of the splicing enhancer sequence within exon 27 of the Dystrophin gene by a nonsense mutation induces partial skipping of the exon and is responsible for Becker muscular dystrophy. J Clin Invest. 1997;100:2204–2210. doi: 10.1172/JCI119757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deburgrave N, Daoud F, Llense S, Barbot JC, Recan D, Peccate C, Burghes AHM, Beroud C, Garcia L, Kaplan JC, Chelly J, Leturcq F. Protein and mRNA based phenotype-genotype correlations in DMD/BMD with point mutations and molecular basis for BMD with nonsense and frameshift mutations in the DMD gene. Human Mutat. 2007;28:183–195. doi: 10.1002/humu.20422. [DOI] [PubMed] [Google Scholar]
- 31.Henderson DM, Lee A, Ervasti JM. Disease-causing missense mutations in actin binding domain 1 of dystrophin induce thermodynamic instability and protein aggregation. PNAS. 2010;107:9632–9637. doi: 10.1073/pnas.1001517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh SM, Bandi S, DD., Shah GA, Mallela KMG. Missense mutation Lys18Asn in Dystrophin that triggers X-Linked dilated cardiomyopathy decreases protein stability, increased protein unfolding, and perturbs protein structure, but dose not affect protein function. PLOS ONE. 2014;9:e110439. doi: 10.1371/journal.pone.0110439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical and dystrophin immunostaining in the patients with a small mutation. Table S2. Patients with nonsense mutation and faint&patchy dystrophin staining. (PDF 286 kb)
Data Availability Statement
The datasets generated and analyzed during the current study are available in the Japanese Registry of Muscular Dystrophy (Remudy: http://www.remudy.jp/). The details of mutation data of the study are available upon request.


