Abstract
Several enzymes in cellular bioenergetics metabolism require NAD+ as an essential cofactor for their activity. NAD+ depletion following ischemic insult can result in cell death and has been associated with over-activation of poly-ADP-ribose polymerase PARP1 as well as an increase in NAD+ consuming enzyme CD38. CD38 is an NAD+ glycohydrolase that plays an important role in inflammatory responses. To determine the contribution of CD38 activity to the mechanisms of post-ischemic brain damage we subjected CD38 knockout (CD38KO) mice and wild-type (WT) mice to transient forebrain ischemia. The CD38KO mice showed a significant amelioration in both histological and neurologic outcome following ischemic insult. Decrease of hippocampal NAD+ levels detected during reperfusion in WT mice was only transient in CD38KO animals, suggesting that CD38 contributes to post-ischemic NAD+ catabolism. Surprisingly, pre-ischemic poly-ADP-ribose (PAR) levels were dramatically higher in CD38KO animals compared to WT animals and exhibited reduction post-ischemia in contrast to the increased levels in WT animals. The high PAR levels in CD38 mice were due to reduced expression levels of poly-ADP-ribose glycohydrolase (PARG). Thus, the absence of CD38 activity can not only directly affect inflammatory response, but also result in unpredicted alterations in the expression levels of enzymes participating in NAD+ metabolism. Although the CD38KO mice showed significant protection against ischemic brain injury, the changes in enzyme activity related to NAD+ metabolism makes the determination of the role of CD38 in mechanisms of ischemic brain damage more complex.
Keywords: Nicotinamide dinucleotide, Poly-ADP-ribose, Ischemia, Damage, Brain, Mouse
Introduction
Over-activation of PARP1, an NAD+ consuming enzyme, due to post-ischemic DNA damage is associated with depletion of tissue NAD+ pools that can lead to bioenergetics failure and cell death [1]. Maintenance of physiological NAD+ levels is critical for cell energy metabolism since NAD+ and NADH are essential cofactors for glycolytic and mitochondrial bioenergetic metabolism. There are several other enzymes that use NAD+ as a substrate [2, 3]. The bifunctional ectoenzyme and receptor CD38 is an NAD+ glycohydrolase that uses NAD+ to generate second messengers, cyclic ADP-ribose and ADP-ribose [4, 5]. These metabolites function as signaling molecules by mobilizing calcium from intracellular stores [6] or activating calcium permeable plasma membrane channels [7]. CD38 is expressed in various tissues and cell types [5], including hematopoietic-derived cells such as monocytes, dendritic cells, lymphocytes, and microglia. Furthermore, CD38 is considered a major enzyme that controls cytosolic NAD+ levels [8].
It was shown that after focal cerebral ischemia CD38 contributes to post-ischemic chemokine production and aggravates cerebral injury by promoting immune cell infiltration [9]. Since CD38 regulates microglial activation and response to chemokines it has an important role in neuroinflamation after traumatic brain injury (TBI) [10].
We have shown that this enzyme can very rapidly deplete brain tissue NAD+ stores and is activated following global cerebral ischemia [11]. To determine the functional significance and contribution of the CD38 activity to mechanisms of post-ischemic brain damage we utilized both wild-type (WT) and CD38 knock out (CD38KO) mice that were subjected to transient global cerebral ischemia.
Material and Methods
Animals
Both wild-type (WT; C57Bl6) and CD38-deficient mice (CD38KO) were obtained from Jackson laboratory. Male, 3 month old mice were used for experiments. All animal protocols were approved by the Animal care and Use Committee of the University of Maryland Baltimore, in accordance with the National Institute of Health Guidelines for the Care and Use of Laboratory Animals. The animals were maintained in a 12-h light/dark cycle, and were housed in a group of 2–5 mice per cage.
The animals were randomly divided into several experimental groups. Forty-eight mice (24 WT and 24 CD38KO) were used to determining the histological and neurologic outcome following the ischemic insult. Another group of 24 mice were subjected to a sham procedure or transient forebrain ischemia where hippocampal tissue was collected at 2, 4, and 24 h of recovery. The tissue samples were processed for high performance liquid chromatography (HPLC) to determine the NAD+ and ADP-ribose levels. In the next experimental set (24 mice), samples were used for western blots, or the mice were perfusion-fixed and their brains processed for histology after 2, 4, and 24 h of reperfusion.
Transient Forebrain Ischemia
Transient global cerebral ischemia was induced by common carotid arteries occlusion and reduction of mean arterial blood pressure (MABP) as described in Onken et al. [12]. To reduce the variability in blood glucose levels before the ischemic insult the animals were fasted for several hours before surgery with free access to tap water. During surgery the animals were ventilated with 1.5–2.0 % isoflurane in N2O:O2 (70:30). The head and core temperature were monitored by placing a needle probe subcutaneously on the skull and using a rectal probe. Both the head and body temperature was maintained at 37.0 ± 0.5 °C during ischemia and at the beginning of reperfusion until the animals were disconnected from the ventilator. The forebrain ischemia was induced by clamping the common carotid arteries (CCA) that was preceded by an increase in isoflurane levels to 5 %, which caused a decrease in MABP to about 30 mmHg [12]. Following the 10 min ischemic period, reperfusion was induced by reducing the isoflurane to 0 % and removing the CCA clamps. Then the animals were moved into a temperature-control incubator for about 3 h before they were moved into pre-heated cages for another 24 h [12, 13]. Animals in the sham group underwent the same surgical procedure without the occlusion of common carotid arteries. Before the onset of ischemia, tail arterial blood samples were collected to determine the animals blood gases (pH, PaO2 and PaCO2) and blood glucose levels.
Regional Cerebral Blood Flow (rCBF) Measurement
To monitor relative changes in regional cerebral blood flow during ischemia and reperfusion we used a laser-Doppler flowmeter (ADInstruments) equipped with a pencil probe affixed to the animal’s skull [12]. Pre-ischemic, intra-ischemic and during first 10 min of recovery rCBF was recorded in both WT and CD38KO animals.
Histology
For histology purpose, mice were perfusion-fixed via transcardial perfusion. Mice were deeply anesthetized with a mixture of ketamine (60 %), xylazine (30 %), acepromazine (10 %) via intraperitoneal injection. The whole body was flushed with PBS plus 0.1 unit/ml heparin and followed by fixation with warm 4 % paraformaldehyde. After perfusion-fixation, the brains were retrieved and post-fixed in cold 4 % paraformaldehyde overnight. The next day, brains were transferred into 30 % sucrose solution and stored at 4 °C for at least 3 days before sectioning. The brains were cut coronally at 40 μm thickness on a freezing microtome (Thermo scientific, MICROM HM450). The cut tissue sections were stored in cryoprotectant at −20 °C until use.
Cresyl Violet Staining
Tissue sections collected at 240 μm intervals were mounted on a slide coated with vectabond and dried for an hour [12]. For the defatting process, slides were immersed in acetone for 10 min and dipped in distilled water three times and air-dried for 1 h. The slides were immersed in filtered cresyl violet (FD Neurotechnologies, PS102-02) for 3 min and dipped in distilled water to remove excess cresyl violet. Next, the slides were dehydrated by 95 % ethanol with 0.1 % glacial acetic acid for 1 min then moved into 100 % ethanol for 2 min twice. The slides were put in xylene for 5 min then coverslipped with DPX mounting media avoiding any bubbles inside the coverslip.
Stereologic Quantifications
Mouse brain sections stained with cresyl violet were used for counting of hippocampal CA1 uninjured neurons. For each animal, four sections per hippocampus starting at 1.2 mm posterior to bregma encompassing the lesion volume of CA1 sub-region (−1.2, −1.44, −1.68, −1.92 mm from bregma) were counted using the optical fractionator method of stereology. The Stereo Investigator program assigned 50 × 50 μm optical dissector every 150 μm in x, y plane in the region of interest and an investigator who was blinded to the genotype of the animal or experimental group counted the surviving CA1 neurons. Following counting of the neurons, the volume of the hippocampal CA1 region was measured with the Cavalieri estimator. The cellular density was calculated by dividing the number of neurons by the volume of CA1 with unit of counts/mm3.
Immunofluorescence Labeling
After washing off the cryoprotectant from the tissue sections, antigen retrieval with 0.01 M citrate buffer was performed, followed by an hour in blocking solution (1:30 Normal goat serum and 1 % BSA fraction V in TBS (SIGMA cat #T6664) with 2 % Triton-X (TBS-2T)). Primary antibody anti-poly-ADP-ribose (PAR) (10H from Enzo Life sciences, 1:500) diluted in TBS-2T was applied to the tissues and shaken at room temperature for 1 h then put on a rocker at 4 °C for 40–48 h. Following washes in TBS, tissues were incubated in goat anti-mouse (Alexa Fluor 594; 1:600) secondary antibody for 1 h in the dark, washed again, and put in Hoechst 33342 for 15 min for counter staining. The sections were examined with Nikon Eclipse E800 microscope equipped with an Optronics camera and Spot digital microscope camera (Diagnostic Instruments, Inc.).
Double Immunolabeling
Following immunofluorescence labeling of PAR, sections were incubated with an additional primary antibody (S100β from Dako, 1:20,000 or Iba1 from Wako, 1:200) in KPBST (150 mM NaCl, 400 μM K2HPO4, 100 μM KH2PO4, 0.4 % Triton-X). Sections with S100β were incubated in the dark for 1 h at room temperature while Iba1 sections were prepared using the PELCO Biowave microwave tissue processor (TED PELLA, Inc.) (3 × 5 min at 10 % power). Following washes in KPBS, tissues were incubated in goat anti-rabbit secondary antibody (Alexa Fluor 488, 1:600) in the dark for 1 h at room temperature and washed again.
Laser Scanning Confocal Microscopy
To determine co-localization of poly-ADP-ribosylated proteins (anti-PAR immunolabeling) with cell-type specific markers following immunostaining the brain sections were imaged on a Zeiss LSM 510 laser scanning confocal microscope using a Plan-Apochromat 20× lens. Single planes of 1024 × 1024 pixels were recorded. Lasers 488 and 555 were used to visualize Alexa Fluor 488, and Alexa Fluor 594, respectively.
Western Blots
After dissection, hippocampi were combined and homogenized in lysis buffer (NaCl 150 mM, Tris 10 mM, 1 % Triton X-100, 0.5 % nonidet p-40) with protease inhibitor cocktail (EMD Milipore) and protein concentrations were measured using the Lowry assay. Per sample, 50 μg of protein were heated at 75 °C, loaded into Bio Rad 10 well mini-protean TGX precast gels and separated through sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Gels were transferred to immobilon PVDF-FL membranes using the Trans-Blot Turbo system (Bio-Rad). Membranes were incubated in Odyssey blocking buffer (Li-cor Biosciences) for 1 h before being incubated in primary antibody overnight at 4 °C, anti-PAR (10H), 1:1000 Enzo; anti-Nampt (PBEF), 1:1000 abcam; anti-PARG, 1:666 abcam; anti-β-actin, 1:10,000 Cell Signaling was used as a loading control. Membranes were washed with PBS with 0.1 % tween-20, and then incubated for 30 min at room temperature with the appropriate infrared fluorophore conjugated secondary antibody (Li-cor). The Odyssey infrared image system (Li-cor) was used to scan the membranes and quantify bands.
To confirm the specificity of PAR (10H) antibody we treated animals with PARP1 inhibitor nicotinamide [14, 15] (Nam, 500 mg/kg i.p.) or vehicle (PBS). 1 h after the Nam administration the animals were decapitated, their hippocampi were removed and processed for western blots.
HPLC Analysis of Nucleotides NAD+ Measurement
NAD+ and ADP-ribose were separated and measured using an Agilent Chemstation 1100 high-performance liquid chromatography (HPLC) machine and a reverse phase Biobasic C18 Column (5 μm; 250 × 4.6 mm; Thermo Scientific). A concentration gradient similar to Broetto-Biazon et al. (2009) was created with 50 mM sodium phosphate pH 6 and 50 % of 50 mM sodium phosphate in HPLC grade methanol pH 7. The gradient consisted of following steps (in % methanol): 0 min, 0 %; 2.5 min, 0.5 %; 5 min, 3 %; 7 min, 5 %; 8 min, 12 %; 10 min, 15 %; 12 min, 20 %; 20 min, 30 %. Standards to make the calibration curve for NAD+ and ADP-ribose were diluted with 50 mM sodium phosphate pH 7. The authenticity of NAD+ and ADP-ribose in the samples was confirmed by co-elution with NAD+ and ADP-ribose standards.
Mouse hippocampi were dissected on ice and homogenized in 500 μl of 7 % perchloric acid (PCA), centrifuged for 10 min at 7100 g. The supernatant was removed and adjusted to pH 7, while the pellet was used to determine protein concentration by the Lowry assay. All samples were filtered by 0.22 μm PVDF filter and were diluted 1:1 in 50 mM sodium phosphate pH 7 before being loaded and run on HPLC. The temperature was kept at 35 °C, and the injection volume was constant at 80 μl. Between runs the column was washed with water and methanol.
Y-Maze Spontaneous Alternation Test
The Y-maze tests the spatial working memory of mice and utilizes the propensity of mice to explore new environments. We used the Y-maze test as described in Wu et al. [16]. In brief, each mouse was put at the end of one arm of black plastic Y-maze and allowed to freely explore for 5 min. An investigator who was blinded to the genotype and experimental group of mice recorded the sequence and number of all arm entries for each animal wiping the Y-maze with 70 % ethanol in between runs. When the mouse entered all three arms consecutively, the number of alternation (A) was marked and this was used to calculate the alternation rate along with the total number of arm entries (E).
This test was performed before and 6 days following forebrain ischemia.
Statistical Analysis
Data among individual experimental groups were compared by one-way ANOVA followed by appropriate post hoc test. Values are presented as mean ± SEM. The p values of <0.05 were considered to be statistically significant.
Results
Effect of CD38 on Post-ischemic Histological and Neurologic Outcome
CD38 is an enzyme that uses NAD+ as a substrate to generate cyclic ADP-ribose and ADP-ribose [5]. We have shown that CD38 activity increases in brain tissue following ischemic insult [11] and effectively depletes brain tissue of NAD+ [17]. This prompted us to use CD38 knockout animals to examine the role of this enzyme in cell death mechanisms following transient forebrain ischemia. Interestingly, control, CD38KO sham mice show higher number of CA1 neurons when compared to the WT sham group (p = 0.0034; Student t test) (Fig. 1b). As Fig. 1 shows in WT mice about 50 % of CA1 neurons survived following the ischemic insult (Fig. 1b). However, ischemia in the CD38KO mice resulted in only a 26 % decrease in CA1 surviving neurons when compared to CD38KO sham animals, suggesting that CD38 contributes to cell death mechanisms. Similarly, we detected an improvement in neurologic outcome of post-ischemic CD38KO animals when compared to WT mice. Hippocampal damage was assessed in the Y-maze test. Wild-type animals manifested reduced spontaneous alternations (Fig. 2a) following ischemic insult suggesting hippocampal injury [18]. The CD38KO mice showed the same trend, however the changes were not significant (Fig. 2a). CD38 knockout animals also displayed moderate hyperactivity after the ischemic insult (increased total number of arm entry, Fig. 2b).
Fig. 1.
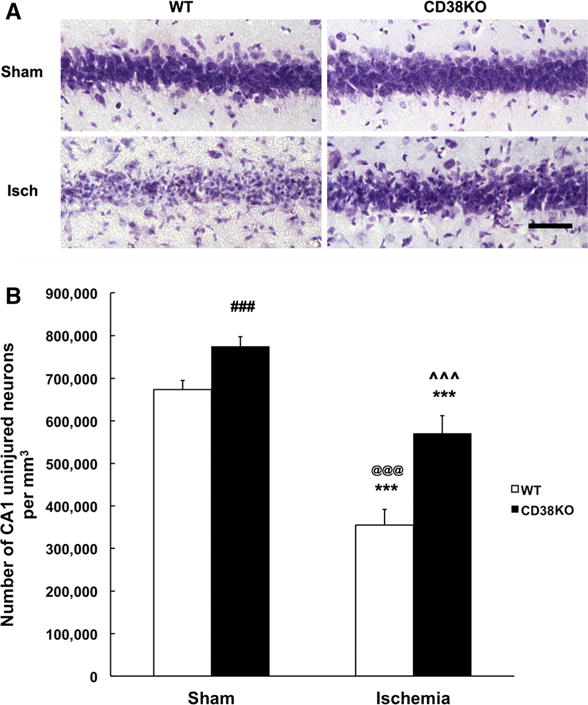
CD38KO mice show marked reduction in CA1 neuronal death following transient global cerebral ischemia. a Cresyl violet staining of CA1 hippocampal neurons in WT and CD38KO sham operated animals and in animals subjected to ischemic insult and 6 days of recovery. Scale bar represents 50 μm. b Stereological quantification of neuronal death. There is about 50 % loss of CA1 neurons in the WT mice while in CD38KO mice the ischemia-induced neuronal death was only 26 % (***p < 0.001 when compared to corresponding sham animals, @@@p < 0.001 when compared to CD38KO sham group, ˆˆˆp < 0.001 when compared to WT ischemia group, ANOVA followed by Bonferroni’s test, n = 12). Interestingly, the sham control (sham) CD38KO animals have significantly higher number of CA1 neurons when compared to WT sham animals (###p < 0.005, Student t test, n = 12)
Fig. 2.
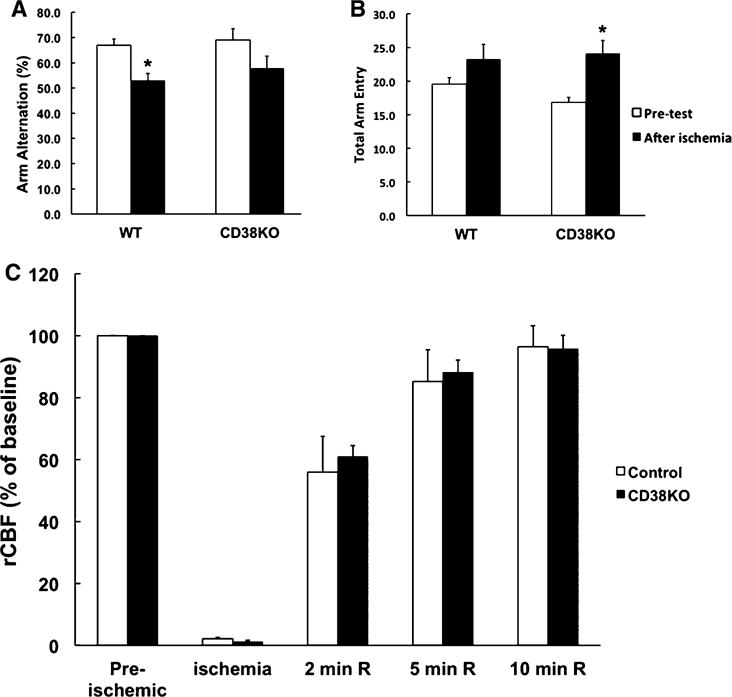
CD38KO animals do not show significant neurological deficit following transient forebrain ischemia. a Y-maze arm entry alternation for WT and CD38KO animals before and 6 days after forebrain ischemia. Ischemic insult significantly reduced the arm entries alternations only in WT animals. *p < 0.05, ANOVA followed by Student–Newman–Keuls test, n = 12. b CD38KO mice also showed significant hyperactivity that was reflected in higher total number of arm entries. In WT animals there was a tendency for increase, however it was not significant. *p < 0.05, ANOVA followed by Student–Newman–Keuls test, n = 12. c Ischemia-induced changes in regional cerebral blood flow (rCBF). In both WT and CD38KO animals during ischemia the rCBF was reduced to about 5 % of the pre-ischemic values and gradually recovers during the first 10 min of reperfusion (n = 6)
Ischemia-Induced Changes in rCBF are not Affected in CD38 Knockout Animals
To ensure that the improved histological outcome in the CD38KO mice is not due to alterations in rCBF during ischemia or reperfusion we monitored the rCBF with a laser-doppler probe. As Fig. 2c shows there were no significant changes in rCBF levels between WT and CD38KO animals. Similarly, no significant differences were in other physiological parameters (animals temperature, blood glucose levels, and blood pH, PaO2 and PaCO2), between the WT and CD38KO animal groups (data not shown).
CD38-Dependent Changes in Brain NAD+ and Poly-ADP-Ribosylation Levels
Next, we decided to examine whether CD38 contributes to post-ischemic NAD+ degradation and its affect on poly-ADP-ribose (PAR) formation. Following ischemic insult, there was a significant reduction of NAD+ pools in both WT and the CD38KO mice (Fig. 3a). This trend continued in WT animals reaching about 40 % reduction of NAD+ levels at 24 h of recovery. Interestingly, in the CD38KO mice, NAD+ levels were recovered at 4 h after ischemia. This was followed by a secondary moderate decrease at 24 h when compared to control (Fig. 3a). Since the NAD+ levels are determined by the activity of enzymes of both the catabolic and NAD+ re-synthesis pathways, we also assessed the changes in the expression levels of the rate-limiting enzyme of the NAD+ salvage pathway, nicotinamide phosphoribosyltransferase (Nampt). Hippocampal Nampt levels (Fig. 3b) were increased at 2 h of recovery in both WT and CD38KO mice. At later reperfusion times Nampt expression normalized and was not significantly different from the pre-ischemic levels in both genetic strains.
Fig. 3.
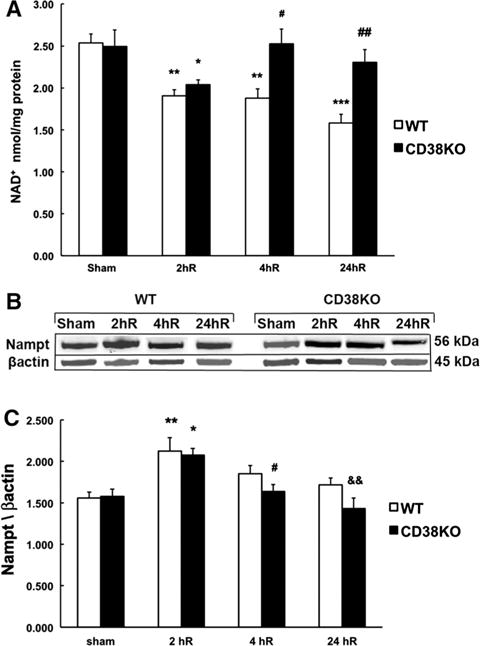
CD38 contributes to delayed post-ischemic NAD+ catabolism. a At 2 h of recovery (2hR) both WT and CD38KO animals show a reduction in hippocampal NAD+ levels, however, following 4 h of reperfusion in CD38KO mice the NAD+ recovered to pre-ischemic levels and was further reduced in WT animals. *p < 0.05, **p < 0.01, ***p < 0.001 when the post-ischemic groups are compared to corresponding sham group, #p < 0.05, ##p < 0.01 when compared to WT ischemia group, ANOVA followed by Student-Newman-Keul test, n = 6. b, c There was no significant difference in expression levels of the rate-limiting NAD+ synthesis enzyme Nampt between sham or post-ischemic CD38KO animals and WT animals. However, the Nampt expression was significantly increased in both WT and CD38KO mice at 2 h of recovery. *p < 0.05, **p < 0.01, when compared to corresponding sham groups, #p < 0.05 when compared to 2hR CD38KO, &&p < 0.01 when compared to 2hR CD38KO, ANOVA followed by Student-Newman-Keul test, n = 6
Surprisingly, although CD38 is not involved directly in PAR metabolism, the control CD38KO animals showed 140 % higher PAR levels when compared to WT animals (Fig. 4a, b). The changes in PAR immunoreactivity following ischemic insult were even more surprising. Although, as expected, in WT animals the PAR levels increased at 2 h of recovery, CD38KO mice interestingly exhibited a reduction in PAR levels. In WT animals the poly-ADP-ribosylation returned at later 4 and 24 h of recovery to pre-ischemic levels. However, hippocampal tissue of CD38KO mice showed significantly higher PAR levels when comparing to WT animals at these recovery times. As Fig. 4c, d shows the PAR bands intensity was significantly reduced in hippocampal samples from animals treated with PARP1 inhibitor nicotinamide (Nam), confirming the specificity of PAR (10H) antibody to poly-ADP-ribosylated proteins (see also [19]).
Fig. 4.
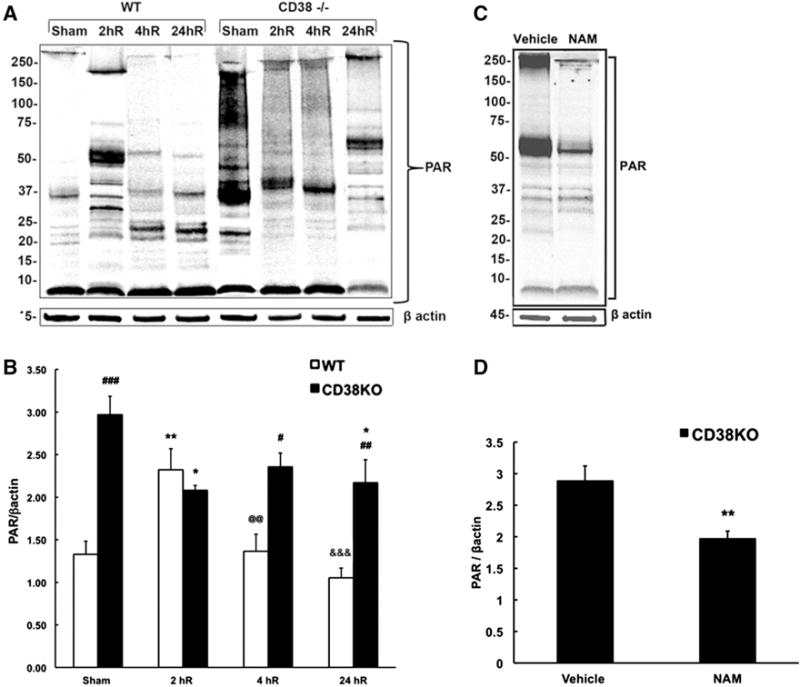
CD38 animals show dramatically higher poly-ADP-ribosylated (PAR) proteins levels when compared to WT animals. a, b There is a significant increase in PAR levels at 2 h (2hR) recovery time in WT mice that then decreased back to sham levels at 4 and 24 h of recovery (4hR, 24hR). The sham CD38KO animals show dramatic increase in PAR levels when compared to sham WT mice. Surprisingly, in CD38KO animals the poly-ADP-ribosylation was reduced following ischemia. b Quantification of western blot images. β actin was used as the loading control. *p < 0.05, **p < 0.01; when compared to corresponding sham, #p < 0.05, ##p < 0.01, ###p < 0.001 when compared to corresponding WT, @@p < 0.01 when compared to 2hR WT, &&&p < 0.001 when compared to 2hR WT, ANOVA followed by Student–Newman–Keuls test, n = 6. c, d Effect of PARP1 inhibitor nicotinamide (Nam) on hippocampal PAR levels. c Immunoblots show marked reduction of band intensities in samples from Nam treated animals. d Nam reduced the hippocampal PAR levels by about 30 %. **p < 0.01 when compared to vehicle, student t test, n = 8
Interestingly, the PAR immuostaining was exclusively localized in the cytosolic perinuclear regions of neurons (Fig. 5) in both WT and CD38KO animals. In post-ischemic hippocampal sections we detected very few cells with nuclear PAR staining at 2 and 24 h of recovery. Interestingly, after ischemic insult in CD38KO animals the perinuclear staining in CA1 neurons was markedly reduced and non-neuronal cells showed an increase in cytosolic staining (Fig. 5). Double staining of hippocampal sections with PAR antiserum and microglial (Iba1) or astrocytic (S100β) antiserum marker revealed that the post-ischemic non-neuronal increase in cytosolic PAR levels occurs preferentially in astrocytes (Fig. 6b, c). The post-ischemic poly-ADP-ribosylation of astrocytic proteins was observed in all hippocampal sub-regions (Fig. 6c).
Fig. 5.
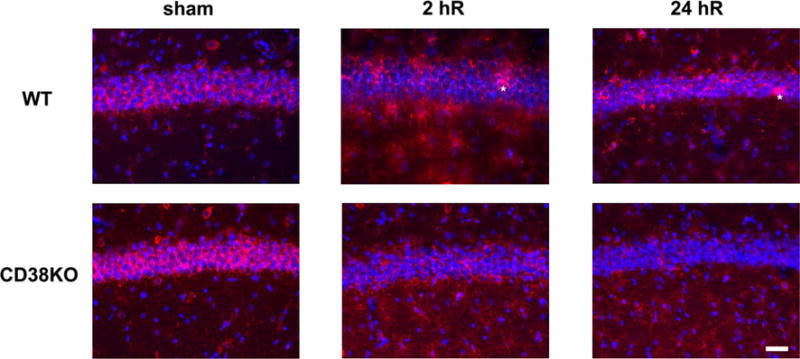
Immunostaining of CA1 hippocampal cells with PAR antibody in WT and CD38KO animals. The PAR immunostaining was localized preferentially to the perinuclear region of CA1 neurons. CD38KO animals show stronger PAR immunoreactivity in CA1 neurons in sham animals when compared to WT mice. CD38KO post-ischemic tissue displayed lower PAR immunoreactivity when compared to sham or WT hippocampal CA1 neurons. After the ischemic insult very few neurons showed a nuclear PAR immunoreactivity (asterisk) in the WT mice. However, during reperfusion the PAR immunostaining was also visible in non-neuronal cells, particularly in CD38KO animals. The cellular nuclei are stained with Hoechst 33342. Scale bar represents 50 μm
Fig. 6.
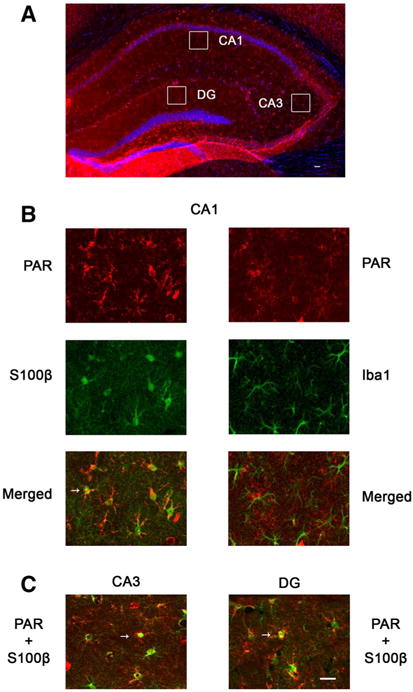
Post-ischemic poly-ADP-ribosylation in hippocampal non-neuronal cells. a Overview of CD38KO hippocampal section immunostaining with PAR antibody at 24 h of recovery. The cellular nuclei are visualized with Hoechst 33342 staining (blue). Squares represent areas of collected images from double immunostaining shown in panels (b, c). b Double immunostaining of CD38KO hippocampal tissue with PAR and S100β or Iba1 antibody (CA1 sub-region). The increased PAR immunoreactivity of non-neuronal cells in all hippocampal sub-regions of post-ischemic hippocampus is localized in astrocytes (b, c, arrows). Similar non-neuronal PAR staining was also observed in WT animals at 2 and 24 h of recovery (Fig. 5). Scale bars represent 50 μm (Color figure online)
Since the levels of poly-ADP-ribosylation are determined by the activity of both PARP1 and the PAR catabolizing enzyme poly-ADP-ribose glycohydrolase (PARG), we next examined PARG levels. As Fig. 7a, b shows, in the control sham operated animals the PARG levels were lower in CD38KO mice when compared to WT mice suggesting that the PAR hydrolysis was less effective, thus leading to higher cellular poly-ADP-ribosylation levels. The ischemic insult decreased the hippocampal PARG expression at 2 h of recovery in both WT and CD38KO animals. This reduction of PARG levels was then reversed at later 24 h of reperfusion.
Fig. 7.
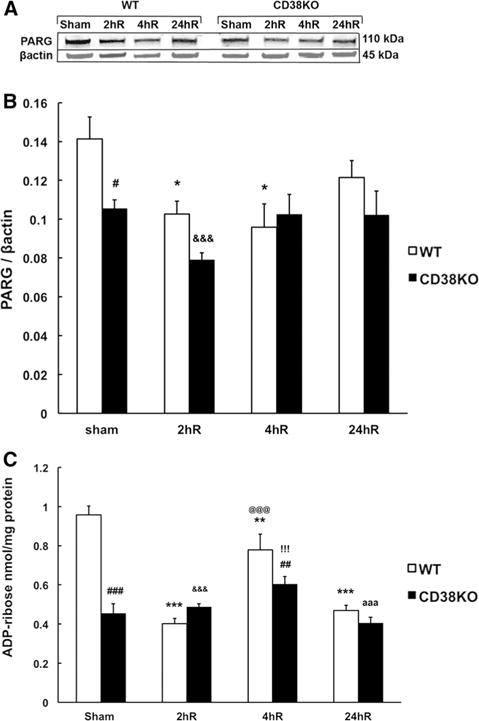
Effect of ischemia on PARG expression and ADP-ribose (ADPR) levels in WT and CD38KO animals. a, b In sham animals the PARG expression levels are significantly lower in CD38KO animals when compared to WT mice. After the ischemic insult at 2 and 4 h of recovery (2hR and 4hR) the PARG levels are reduced when compared to sham group. *p < 0.05 when compared to WT sham, #p < 0.05 when compared to WT, &&&p < 0.001 when compared to WT sham, ANOVA followed by Student–Newman–Keuls test n = 6. c Correspondingly, the product of PARG activity, ADPR, is lower in CD38KO sham animals when compared to WT animals. Following ischemia in WT animals the ADPR levels were lower when compared to sham animals while ischemia in CD38KO animals did not cause significant changes in hippocampal ADPR levels. **p < 0.01, ***p < 0.001 when compared to sham animals, ##p < 0.01, ###p < 0.001 when compared to corresponding WT group, &&&p < 0.001 when compared to WT sham, @@@p < 0.001 when compared to WT 2hR, !!!p < 0.001 when compared to WT sham, aaap < 0.001 when compared to WT sham, ANOVA followed by Student–Newman–Keuls test, n = 6
Degradation of poly-ADP-ribose by PARG generates mono-ADP-ribose (ADPR). Therefore, ADPR levels in hippocampal tissue were assessed in both WT and CD38KO animals. As Fig. 7c shows the ADPR levels were 50 % lower in sham CD38KO animals when compared to sham WT mice. Interestingly, in WT ischemic animals the ADPR was reduced at 2 h of recovery by about 50 %. 4 h after ischemia there was a partial recovery that was followed by a return to 50 % levels at 24 h when compared to pre-ischemic values. There were no dramatic changes in hippocampal ADPR levels in CD38KO animals during post-ischemic period.
Discussion
We report here that CD38 deficient mice show improved ability to recover from transient global cerebral ischemia. Stereologic assessment of uninjured CA1 neurons revealed reduced cell death in CD38KO animals when compared to WT mice. The ischemic insult in WT mice caused a moderate but significant impairment in the animal’s cognitive performance at 6 days of recovery, which was not seen in the CD38 deficient mice. These results suggest that CD38 function may play an important role in repair/recovery processes following global cerebral ischemia.
Similar protective effects in CD38KO animals were observed following focal cerebral ischemia [9]. The reduced migration and accumulation of T-cells and especially macrophages was suggested as the mechanism responsible for neuroprotection [9]. However, when the CD38-deficient animals were subjected to TBI the neurological severity score was significantly worse and also novel object recognition performance after injury was significantly impaired [10]. In agreement with the focal ischemia model, the amount of activated microglia/macrophages at the injury site was lower in CD38-deficient mice when compared to WT mice. The conflicting results in CD38 deficient animals following different acute brain injury models suggest a more complex change in brain metabolism than just the absence of CD38 activity.
Several mechanisms may account for the effect of CD38 on recovery processes. Since CD38 is one of the major enzymes that use NAD+ as a substrate, in the present study, we focused on changes in cellular NAD+ metabolism following ischemic insult. CD38KO animals showed a tendency for slightly elevated NAD+ levels, although these were not significantly different from values determined in the WT mice. Previous studies suggested that the CD38 deficiency results in significantly higher NAD+ levels in the brain when compared to WT animals [8, 20]. Our results did not confirm these reports, however we used only the hippocampi, not the whole brain tissue for analysis. Thus, it is possible that the NAD+ levels are not homogenous throughout the whole brain, and the total NAD+ content can vary based on the brain region.
As expected ischemic insult caused a reduction of hippocampal NAD+ pools in both CD38 deficient and WT animals at 2 h recovery. Similar increased consumption of NAD+ during the first hours of reperfusion was observed in mice following middle cerebral artery occlusion (see [21, 22]). At later 4 and 24 h of reperfusion the hippocampal NAD+ further decreased in the WT mice, however in CD38KO mice the NAD+ recovered to pre-ischemic levels. This suggests that at later recovery times CD38 significantly contributes to NAD+ catabolism.
The reduced post-ischemic NAD+ levels coincided with increased PAR formation in the WT mice. Surprisingly, the PAR metabolism in CD38 deficient mice was strikingly different. The baseline level of PAR in sham CD38KO animals was about 140 % higher when compared to WT mice, suggesting that the CD38 deficiency leads to changes in the ratio between PAR generation and degradation activity under physiologic conditions. One possible explanation is a lower activity of poly-ADP-ribose glycohydrolase (PARG), which degrades the PAR into free ADP-ribose residues [23, 24]. PARG is encoded by one gene, however there are several splicing variants of this enzyme expressed in the brain [25]. There are two major forms of PARG in mouse [24, 26]. The 110 kDa PARG (PARG110) has been found in the cytoplasm and nucleus, and the 60-kDa form (PARG60) is localized only in the cytoplasm [19, 27]. The PARG110 is the major poly-ADP-ribose degrading enzyme [28]. In agreement with the high levels of PAR in CD38 deficient animals we detected about 25 % reduction of hippocampal PARG110 expression. The PARG60 bands were hardly detectable preventing reliablequantification. Immunohistochemical staining for PAR in control animals showed only cytosolic perinuclear localization of poly-ADP-ribosylated proteins (see also [19]). Since PARG110 translocation to the nucleus is initiated by elevated levels of PAR [29] it is feasible that in the CD38 deficient animals at least some of the PARG110 is located in the nucleus.
Interestingly, at 4 and 24 h of recovery in WT animals we detected only very few CA1 neurons with positive nuclear localization of PAR. We did not detect nuclear PAR staining in the CD38KO animals that would support the notion of nuclear PARG110 localization. Furthermore, the very short half-life of PAR in vivo (as short as 1 min, [30]) reduces the possibility to detect the positive nuclear staining for PAR. However, nuclear PAR localization was observed following focal ischemia insult [19, 21].
The changes in post-ischemic hippocampal PAR levels of CD38KO animals are even more puzzling. One would expect an increase in poly-ADP-ribosylation during the reperfusion period, similar to the WT animals. However, we detected a decrease in PAR levels at 2 h of recovery, followed to a slight increase trending toward pre-ischemic values. The immunohistochemical staining for PAR suggests that this could be due to reduced PAR levels in the neuronal perinuclear cytosolic compartment. Since PARG60 is localized in the cytosol and at least a fraction of the PARG110 is translocated into nucleus after the injury [29], the lower cytosolic PAR levels suggest that there is higher activity of PARG60 at early recovery time points in the post-ischemic hippocampus of CD38KO animals. A similar observation was reported by Burns et al. [31].
As mentioned earlier ADP-ribose is the product of PARG activity. In sham animals there was a marked difference in the baseline levels of ADP-ribose between WT and CD38 deficient animals. The reduction of ADP-ribose by 50 % in hippocampal tissue of WT mice can be a result of significantly lower expression of PARG110 in these animals after ischemia. Interestingly, the levels of ADP-ribose did not change in CD38KO animals when compared to pre-ischemic levels. This could be due to ADP-ribose having a fast enzymatic degradation [32], making a simple measurement of its steady-state levels is not sufficient to evaluate the rate of its formation.
The present study indicates that one needs to exercise caution when using CD38KO animals as a genetic model to study the effects and role of CD38 enzyme in pathophysiologic mechanisms. Our data surprisingly revealed that by disrupting the CD38 gene the animals not only show absence of CD38 activity and suppression of the directly related metabolic pathways, but also as side effect the expression levels of enzymes participating in NAD+ metabolism are affected. CD38KO animals despite showing markedly high levels of poly-ADP-ribosylation of cytosolic proteins are more resistance to ischemic brain damage than WT animals. This is in contrast to data indicating that poly-ADP-ribose accumulation is toxic to cells and that PAR polymer is a death signal [33]. Furthermore, our data suggest that the higher PAR levels are due to lower expression of the 110 kDa PARG (see also [19]). The selective reduction of this PARG isomer was shown to attenuate PARP1-induced cell death, however the mechanisms of this effect remain to be determined [31].
In conclusion, the alterations in NAD+ metabolism in CD38KO mice raise more questions about CD38-dependent mechanisms that contribute to ischemic cell death. Further experiments are needed, preferably using conditional CD38 knockout animal models or CD38 specific inhibitor, to shed more light on the role of CD38 in pathophysiology of neurodegenerative disease.
Acknowledgments
The project described was supported by Award Number I01BX000917 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development to TK and by NIH/NINDS Grant R01NS082308 to D. J. L.
References
- 1.Baxter P, Chen Y, Xu Y, Swanson RA. Mitochondrial dysfunction induced by nuclear poly(ADP-ribose) polymerase-1: a treatable cause of cell death in stroke. Transl Stroke Res. 2014;5:136–144. doi: 10.1007/s12975-013-0283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owens K, Park JH, Schuh R, Kristian T. Mitochondrial dysfunction and NAD+ metabolism alterations in the pathophysiology of acute brain injury. Transl Stroke Res. 2013;4:618–634. doi: 10.1007/s12975-013-0278-x. [DOI] [PubMed] [Google Scholar]
- 3.Conforti L, Gilley J, Coleman MP. Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nature Rev Neurosci. 2014;15:394–409. doi: 10.1038/nrn3680. [DOI] [PubMed] [Google Scholar]
- 4.Schuber F, Lund FE. Structure and enzymology of ADP-ribosyl cyclases: conserved enzymes that produce multiple calcium mobilizing metabolites. Curr Mol Med. 2004;4:249–261. doi: 10.2174/1566524043360708. [DOI] [PubMed] [Google Scholar]
- 5.Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, Vaisitti T, Aydin S. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev. 2008;88:841–886. doi: 10.1152/physrev.00035.2007. [DOI] [PubMed] [Google Scholar]
- 6.Lee HC. Multiplicity of Ca2+ messengers and Ca2+ stores: a perspective from cyclic ADP-ribose and NAADP. Curr Mol Med. 2004;4:227–237. doi: 10.2174/1566524043360753. [DOI] [PubMed] [Google Scholar]
- 7.Perraud AL, Fleig A, Dunn CA, Bagley LA, Launay P, Schmitz C, Stokes AJ, Zhu Q, Bessman MJ, Penner R, Kinet JP, Scharenberg AM. ADP-ribose gating of the calcium-permeable LTRPC2 channel revealed by Nudix motif homology. Nature. 2001;411:595–599. doi: 10.1038/35079100. [DOI] [PubMed] [Google Scholar]
- 8.Aksoy P, White TA, Thompson M, Chini EN. Regulation of intracellular levels of NAD: a novel role for CD38. Biochem Biophys Res Commun. 2006;345:1386–1392. doi: 10.1016/j.bbrc.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 9.Choe CU, Lardong K, Gelderblom M, Ludewig P, Leypoldt F, Koch-Nolte F, Gerloff C, Magnus T. CD38 exacerbates focal cytokine production, postischemic inflammation and brain injury after focal cerebral ischemia. PloS One. 2011;6:e19046. doi: 10.1371/journal.pone.0019046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy A, Bercovich-Kinori A, Alexandrovich AG, Tsenter J, Trembovler V, Lund FE, Shohami E, Stein R, Mayo L. CD38 facilitates recovery from traumatic brain injury. J Neurotrauma. 2009;26:1521–1533. doi: 10.1089/neu.2008.0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kristian T, Balan I, Schuh R, Onken M. Mitochondrial dysfunction and nicotinamide dinucleotide catabolism as mechanisms of cell death and promising targets for neuroprotection. J Neurosci Res. 2011;89:1946–1955. doi: 10.1002/jnr.22626. [DOI] [PubMed] [Google Scholar]
- 12.Onken M, Berger S, Kristian T. Simple model of forebrain ischemia in mouse. J Neurosci Methods. 2012;204:254–261. doi: 10.1016/j.jneumeth.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Owens K, Park JH, Gourley S, Jones H, Kristian T. Mitochondrial dynamics: cell-type and hippocampal region specific changes following global cerebral ischemia. J Bioenerg Biomembr. 2015;47:13–31. doi: 10.1007/s10863-014-9575-7. [DOI] [PubMed] [Google Scholar]
- 14.Klaidman LK, Mukherjee SK, Hutchin TP, Adams JD. Nicotinamide as a precursor for NAD+ prevents apoptosis in the mouse brain induced by tertiary-butylhydroperoxide. Neurosci Lett. 1996;206:5–8. doi: 10.1016/0304-3940(96)12446-0. [DOI] [PubMed] [Google Scholar]
- 15.Liu D, Gharavi R, Pitta M, Gleichmann M, Mattson MP. Nicotinamide prevents NAD+ depletion and protects neurons against excitotoxicity and cerebral ischemia: NAD+ consumption by SIRT1 may endanger energetically compromised neurons. Neuromolecular Med. 2009;11:28–42. doi: 10.1007/s12017-009-8058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, Stoica BA, Luo T, Sabirzhanov B, Zhao Z, Guanciale K, Nayar SK, Foss CA, Pomper MG, Faden AI. Isolated spinal cord contusion in rats induces chronic brain neuroinflammation, neurodegeneration, and cognitive impairment. Involvement of cell cycle activation. Cell Cycle. 2014;13:2446–2458. doi: 10.4161/cc.29420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balan IS, Fiskum G, Kristian T. Visualization and quantification of NAD(H) in brain sections by a novel histoenzymatic nitrotetrazolium blue staining technique. Brain Res. 2010;1316:112–119. doi: 10.1016/j.brainres.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurinami H, Shimamura M, Ma T, Qian L, Koizumi K, Park L, Klann E, Manfredi G, Iadecola C, Zhou P. Prohibitin viral gene transfer protects hippocampal CA1 neurons from ischemia and ameliorates postischemic hippocampal dysfunction. Stroke. 2014;45:1131–1138. doi: 10.1161/STROKEAHA.113.003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cozzi A, Cipriani G, Fossati S, Faraco G, Formentini L, Min W, Cortes U, Wang ZQ, Moroni F, Chiarugi A. Poly(ADP-ribose) accumulation and enhancement of postischemic brain damage in 110-kDa poly(ADP-ribose) glycohydrolase null mice. J Cereb Blood Flow Metab. 2006;26:684–695. doi: 10.1038/sj.jcbfm.9600222. [DOI] [PubMed] [Google Scholar]
- 20.Young GS, Choleris E, Lund FE, Kirkland JB. Decreased cADPR and increased NAD+ in the CD38−/− mouse. Biochem Biophys Res Commun. 2006;346:188–192. doi: 10.1016/j.bbrc.2006.05.100. [DOI] [PubMed] [Google Scholar]
- 21.Endres M, Wang ZQ, Namura S, Waeber C, Moskowitz MA. Ischemic brain injury is mediated by the activation of poly(ADP-ribose)polymerase. J Cereb Blood Flow Metab. 1997;17:1143–1151. doi: 10.1097/00004647-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Eliasson MJ, Sampei K, Mandir AS, Hurn PD, Traystman RJ, Bao J, Pieper A, Wang ZQ, Dawson, Snyder SH, Dawson VL. Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 23.Brochu G, Duchaine C, Thibeault L, Lagueux J, Shah GM, Poirier GG. Mode of action of poly(ADP-ribose) glycohydrolase. Biochim Biophys Acta. 1994;1219:342–350. doi: 10.1016/0167-4781(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 24.Davidovic L, Vodenicharov M, Affar EB, Poirier GG. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp Cell Res. 2001;268:7–13. doi: 10.1006/excr.2001.5263. [DOI] [PubMed] [Google Scholar]
- 25.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 26.Brochu G, Shah GM, Poirier GG. Purification of poly(ADP-ribose) glycohydrolase and detection of its isoforms by a zymogram following one- or two-dimensional electrophoresis. Anal Biochem. 1994;218:265–272. doi: 10.1006/abio.1994.1177. [DOI] [PubMed] [Google Scholar]
- 27.Di Meglio S, Denegri M, Vallefuoco S, Tramontano F, Scovassi AI, Quesada P. Poly(ADPR) polymerase-1 and poly(ADPR) glycohydrolase level and distribution in differentiating rat germinal cells. Mol Cell Biochem. 2003;248:85–91. doi: 10.1023/a:1024136927637. [DOI] [PubMed] [Google Scholar]
- 28.Winstall E, Affar EB, Shah R, Bourassa S, Scovassi AI, Poirier GG. Poly(ADP-ribose) glycohydrolase is present and active in mammalian cells as a 110-kDa protein. Exp Cell Res. 1999;246:395–398. doi: 10.1006/excr.1998.4321. [DOI] [PubMed] [Google Scholar]
- 29.Poitras MF, Koh DW, Yu SW, Andrabi SA, Mandir AS, Poirier GG, Dawson VL, Dawson Spatial and functional relationship between poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in the brain. Neuroscience. 2007;148:198–211. doi: 10.1016/j.neuroscience.2007.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Gonzalez R, Althaus FR. Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mutat Res. 1989;218:67–74. doi: 10.1016/0921-8777(89)90012-8. [DOI] [PubMed] [Google Scholar]
- 31.Burns DM, Ying W, Kauppinen, Zhu K, Swanson RA. Selective down-regulation of nuclear poly(ADP-ribose) glycohydrolase. PloS One. 2009;4:e4896. doi: 10.1371/journal.pone.0004896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oei SL, Ziegler M. ATP for the DNA ligation step in base excision repair is generated from poly(ADP-ribose) J Biol Chem. 2000;275:23234–23239. doi: 10.1074/jbc.m002429200. [DOI] [PubMed] [Google Scholar]
- 33.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]


