Abstract
In kinetoplastid protozoa, mitochondrial (mt) mRNAs are post-transcriptionally edited by insertion and deletion of uridylate residues, the information being provided by guide (g)RNAs. Currently popular mechanisms for the editing process envisage a series of consecutive ‘cut-and-paste’ reactions, carried out by a complex RNP machinery. Here we report on the purification, cloning and functional analysis of two gRNA-binding proteins of 28.8 (gBP29) and 26.8 kDa (gBP27) from mitochondria of the insect trypanosome Crithidia fasciculata. gBP29 and gBP27 proved to be similar, Arg + Ala-rich proteins, with pI values of ∼10.0. gBP27 has no homology to known proteins, but gBP29 is the C.fasciculata orthologue of gBP21 from Trypanosoma brucei, a gRNA-binding protein that associates with active RNA editing complexes. As measured in UV cross-linking assays, His-tagged recombinant gBP29 and gBP27 bind to radiolabelled poly(U) and synthetic gRNAs, while competition experiments suggest a role for the gRNA 3′-(U)-tail in binding to these proteins. Immunoprecipitates of mt extracts generated with antibodies against gBP29 also contained gBP27 and vice versa. The immunoprecipitates further harbored a large proportion of the cellular content of four different gRNAs and of edited and pre-edited NADH dehydrogenase subunit 7 mRNAs, but only small amounts of mt rRNAs. In addition, the bulk of gBP29 and gBP27 co-eluted with gRNAs from gel filtration columns in the high molecular weight range. Together, these results suggest that the proteins are part of a large macromolecular complex(es). We infer that gBP29 and gBP27 are components of the C.fasciculata editing machinery that may interact with gRNAs.
INTRODUCTION
In kinetoplastid mitochondria, the nucleotide sequence of mRNAs is post-transcriptionally altered by U insertion/deletion editing under the direction of guide (g)RNAs (reviewed in refs 1–4). The development of in vitro RNA editing systems derived from mitochondrial (mt) extracts of Trypanosoma brucei (5–7) and Leishmania tarentolae (8) has allowed the elucidation of important mechanistic features of the editing process. RNA editing appears to be the result of successive enzymatic cleavage–ligation reactions catalysed by a gRNA-dependent endonuclease(s), a terminal uridylyl transferase (TUTase), a U exonuclease and an RNA ligase, respectively. This shows that the original enzyme cascade model for RNA editing (9) was essentially correct. Somewhat contrary to expectation, it has been demonstrated that insertion and deletion activities are distinct with respect to optimal reaction conditions and are, for example, inversely affected by the presence of adenosine nucleotides (10,11).
All available evidence suggests that editing reactions are mediated by large multicomponent complexes. In mt extracts from T.brucei, two classes of gRNA-containing complexes seem to exist, sedimenting on glycerol gradients at 35–40S and ∼20S, respectively (12). These complexes both contain the four key enzymatic activities mentioned above, albeit that the relative amounts of TUTase found in each complex differ between different laboratories (12–15). Fully edited RNAs are predominantly associated with the 35–40S fractions, whereas pre-edited RNAs appear to be more abundant in the smaller complexes. It has been suggested that these complexes represent native editing complexes, the broad distribution presumably reflecting the size of the associated mRNA and the extent to which the mRNA is edited (4,12,15). The bulk of the deletion editing activity sediments in a broad peak around 20S on the gradients (15) and, indeed, an eight polypeptide complex of ∼20S was purified from T.brucei mt extracts that was capable of one full round of insertional and deletional editing (11,16). This complex was devoid of g+mRNA, but it possessed all the required enzymatic activities and may represent a core editosomal particle. Since only a small percentage of the added substrate RNAs is edited at only one site in the current in vitro systems, it can be expected that other proteins are required to increase the efficiency and/or accuracy of the editing process. In mt extracts of L.tarentolae two classes of gRNA-containing RNPs have been detected of 10 and ∼20S (17,18). In view of their smaller size, greater heterogeneity and significant differences in enzyme and RNA contents, the functional relationship to the mt RNPs from T.brucei is not immediately obvious.
At the individual protein level, a number of candidate editing proteins have been identified and characterised by molecular cloning. So far, the list includes two potential editing ligases of ∼48 and 52 kDa (19–22), of which the 52 kDa species is essential for editing, as demonstrated by targeted gene replacement experiments (19). No published records are currently available on the other key enzymes of the editing process and only a gRNA-independent endoribonuclease was cloned in L.tarentolae (23). Missel et al. (24) showed that disruption of the two alleles of T.brucei mHel61p, a potential RNA helicase, resulted in a specific reduction of edited (but not of non-edited) mRNA levels. However, an RNA helicase does not seem to be an integral part of the 20S editing activity (15,16) and mt extracts of mHel61p– cells are fully active in in vitro editing assays. A 45 kDa protein associated with T.brucei 35–40S complexes has been characterised, RNA editing-associated protein 1 (REAP-1; 25). REAP-1 contains a repetitive amino acid domain of unknown function and anti-REAP-1 antibodies inhibit in vitro editing. The specific role in RNA editing of this protein also remains to be elucidated.
The potential to bind to synthetic gRNA variants and/or to associate with gRNA-containing RNPs has been an incentive to clone a large number of proteins. In L.tarentolae, this has led to the finding that gRNA-binding proteins of 18, 51 and 110 kDa (26,27) are, in fact, ATPase subunit b (28), aldehyde dehydrogenase (26) and glutamate dehydrogenase (27), respectively. The biological significance of the gRNA-binding properties of these abundant metabolic enzymes remains to be established (for a more extensive discussion see refs 4,26,27). In T.brucei, an 83/90 kDa gRNA/poly(U)-binding protein (29,30) has serendipitously been cloned in a search for the trypanosome homologue of nucleolin (31). This protein contains five RGG motifs, which are also found in other RNA-binding proteins (32), and was therefore termed TBRGG1. TBRGG1 co-localizes with in vitro editing activity on glycerol gradients (31). By far the best characterised gRNA-binding protein is gBP21, an arginine-rich mt protein from T.brucei that binds to gRNAs with nanomolar affinity (33), although the specific gRNA structural elements that are essential for this binding have not yet been identified. gBP21 has been found to be associated with editing complexes, since polyclonal α-gBP21 antibodies inhibit in vitro editing reactions (34) and immunoprecipitate an in vitro RNA editing activity (35). In addition, the immunoprecipitates contain gRNA-dependent endonuclease, TUTase and RNA ligase, together with gRNAs and both edited and unedited mRNA (35). Recently, it was shown that gBP21 possesses RNA annealing activity (36). Together, these results strongly suggest a role for gBP21 in the editing process, in which it may facilitate the interaction between gRNAs and pre-mRNAs. Nevertheless, gBP21 knockout bloodstream T.brucei cells are viable and contain edited mRNA, indicating that it is a non-essential role (34). Recently, a third gRNA-binding protein was cloned from T.brucei, which contains a cold shock domain at the N-terminus and also seems to use the gRNA (U)-tail as a major determinant for binding (RBP16; see 37,38).
We have purified and characterised proteins present in the mt extract of the insect trypanosomatid Crithidia fasciculata that can be UV cross-linked to synthetic gRNAs equipped with a (U)-tail and to poly(U) (29). In this report we describe the characterisation of a UV cross-linking activity that migrated on PAGE gels at ∼28 kDa and we show that this activity in fact consisted of two separate poly(U)- and gRNA-binding proteins of similar mass and biochemical properties, i.e. gBP29, the C.fasciculata orthologue of gBP21, and gBP27, a novel polypeptide. Although the lack of an in vitro system for RNA editing from C.fasciculata hampered direct verification, our results suggest that both gBP29 and gBP27 play a role in RNA editing.
MATERIALS AND METHODS
Cell growth, isolation of RNA and mitochondrial vesicles
Crithidia fasciculata (Steinert strain; see ref. 39) was grown as described (40) in batches of 10 l with shaking and aeration to a density of 1 × 108 cells/ml. Mitochondrial vesicles were isolated according to Birkenmeyer and Ray (41). Total RNA was isolated using the hot phenol extraction method (42), DNA being removed by digestion with DNase I (5 µg/ml) according to Tullis and Rubin (43).
Electrophoresis, blotting, hybridisation and PCR
The electrophoresis of DNA and RNA on agarose gels, Southern and northern blotting and hybridisation were performed following standard protocols (44–46).
To amplify DNA fragments we used the following PCR protocol: 1 ng recombinant DNA was mixed with 25 pmol primers, 200 µM dNTPs, 1 mM MgCl2, 1 U Taq polymerase in 1× PCR reaction buffer (both Promega). This reaction mixture was incubated at 95°C for 5 min, followed by 30 cycles of 1 min at 95°C, 1.5 min at 50°C and 2 min at 72°C, followed by 10 min at 72°C. The annealing temperature varied in each experiment, depending on the length and GC content of each of the primers used: 4°C per G or C plus 2°C per A or T, minus 5°C. To amplify mRNA fragments (RT–PCR) the PCR reaction was preceded by a first-strand cDNA synthesis reaction: 1 µg total RNA from C.fasciculata was denatured for 2 min at 70°C, quickly cooled on ice and added to an RT mix containing 40 pmol downstream primer, 10 U AMV reverse transcriptase (Promega) and 10 U RNasin (Promega) in Promega RT buffer in a total volume of 30 µl. This reaction mixture was incubated for 1 h at 42°C, followed by 5 min at 95°C; 5 µl of this reaction was used for PCR.
Purification of gBP29 and gBP27
Mitochondrial vesicles, isolated from 200 l of culture (totalling 2 × 1013 cells), were lysed for 5 min on ice in 20 ml of buffer A (25 mM HEPES–KOH pH 7.6, 20 mM sucrose, 10 mM MgCl2, 2 mM EDTA, 5 mM 2-mercaptoethanol, 10% v/v glycerol and 0.1% v/v Triton X-100) containing 60 mM KCl, in the presence of protease inhibitors [0.2 mM phenylmethylsulfonyl fluoride (PMSF) and 5 µg/ml each chymostatin, leupeptin, antipain and pepstatin]. The extract was clarified by centrifugation at 16 000 g for 20 min. The pellet was extracted with 100 ml of buffer A for 5 min on ice and clarified again. The supernatants (containing 2000 mg protein) were pooled, adjusted to 250 mM KCl and loaded batchwise onto a 45 ml heparin–Sepharose column (Pharmacia), which was equilibrated in buffer A containing 250 mM KCl. The column was washed with 3 vol buffer A containing 250 mM KCl until no more protein eluted. Bound protein was eluted with a 160 ml linear gradient of buffer A containing 250–1000 mM KCl (with 0.02 mM PMSF and 5 µg/ml each chymostatin, leupeptin, antipain and pepstatin). Fractions of 2 ml were collected and tested in a UV cross-linking assay (see below). Fractions 22–36, containing the protein(s) of ∼28 kDa active in a UV cross-linking assay (5 mg total protein), were pooled, dialysed against buffer A containing 200 mM KCl and loaded onto a 10 ml poly(U)–Sepharose column (Pharmacia), equilibrated in the same buffer. Bound protein was eluted with a 50 ml linear gradient of buffer A containing 250–1000 mM KCl, followed by an additional 10 ml of buffer A with 1 M KCl. Fractions of 2 ml were collected and tested in the UV cross-linking assay. Fractions containing the protein(s) of 28 kDa active in this assay (0.26 mg total protein) were pooled and concentrated by dialysis against saturated solutions of polyethyleneglycol 20 000 in buffer A. In a last purification step, the proteins were applied to a preparative 8% (w/v) PAGE gel with 0.05% (w/v) SDS and 25 mg/l Serva Blue G in cathode buffer. Protein(s) migrating at a position corresponding to 28 kDa was eluted (without staining) in 25 mM Tris–HCl pH 8.0, 1 mM EDTA and 5% (w/v) acetonitrile.
Peptide generation and microsequencing
Aliquots of 190 µg excised protein(s) were digested with Lys-C (Boehringer Mannheim) in a protease:target weight ratio of 1:100, in 25 mM Tris–HCl pH 8.0, 1 mM EDTA and 5% acetonitrile for 12 h at 37°C. The resulting peptide mixture was loaded onto a µRC P C2/C18 column. Peptides were eluted with a gradient ranging from 0.05% (w/v) trifluoroacetic acid (TFA) in water to 0.04% TFA in 70% (w/v) acetonitrile and collected by peak-based collection, all with the aid of the Pharmacia SMART system. Peptide-containing fractions were spotted onto Prosorb membranes (ABI). Sequencing was performed with a Perkin Elmer/Applied Biosystems Procise 494 protein sequencer or a Beckman-Porton LF 3200 protein sequencer. The peptide sequences obtained are indicated in Figures 2 and 3A.
Figure 2.
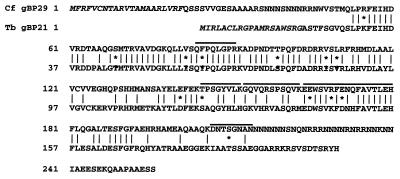
Inferred amino acid sequence of gBP29. Amino acid sequence alignment of C.fasciculata gBP29 (top lines) and T.brucei gBP21 (bottom lines). Identical amino acids are indicated by vertical lines. Asterisks indicate conservative amino acid substitutions: K, R; S, T; D, E; F, Y; V, I, L. Dashes are introduced for optimal alignment. The putative mitochondrial import signals are given in italic and Lys-C generated peptide sequences of gBP29 are indicated above the lines.
Denaturing protein gels and western blotting
Proteins were separated on 10 or 12% (w/v) SDS–PAGE gels as described by Laemmli (47). After electrophoresis, gels were fixed in 30% (v/v) methanol, 10% (v/v) acetic acid, dried and exposed to X-OMAT-AR film (Kodak) or silver stained. For immunodetection, proteins were blotted onto Hybond-P membranes (Amersham Life Science), according to the manufacturer’s instructions as previously described (44). This was followed by standard immunodetection procedures, utilising rabbit sera and secondary antibodies coupled to horseradish peroxidase. Detection of horseradish peroxidase activity was performed as described by the manufacturer (Amersham Life Science).
Oligodeoxyribonucleotides
The following oligodeoxyribonucleotides were used for amplification of (parts of) C.fasciculata gBP29 and gBP27 cD-A and T.brucei gBP25. (N = G, A, T or C; Y = C or T; R = G or A; B = not A; S = C or G; W = A or T): C146, TGYCAYACNYTNCAYTG (target gBP27 ORF); C165, GNGCNGAYGTNGAYGA (target gBP27 ORF); C178, CCGCCGTCCACTCCAC (target gBP27 ORF); C181, CCAAACGACAACAACGCACAAC (target gBP27 3′-UTR); C188, GGATCCGGCCGTTTCGCCTGCGC (target gBP27 ORF); C191, GGATCCGCGCGCCTCGTCCGCTTC (target gBP29 ORF); C201, AAGCAGCTBCTBGTBWSBCAGTT (target gBP29 ORF); C204, TCSARSGTSACSGCGAAYTGGTT (target gBP29 ORF); C217, GGAATTCGCTATATAAGTATCAGTTTCTGTAC [target spliced leader (C.fasciculata)]; TB6, TGCGTTAGTATAACGTGAGTG (gBP25 ORF); TB7, TCTCCCACCTTTTCTGAACG (target gBP25 ORF); TB8, GTCCTTGATTACCATCGCCA (target gBP25 ORF); TB9, CATCTATTGCGAGTACTTCAT (target gBP25 ORF); H33, GGAATTCTTTTTTTTTTTTTTTTTTTT [target poly(A) tail].
The following oligodeoxyribonucleotides were used for primer extension analyses: C34, CATAAGGATAGCAAATGTTC (target ND7 mRNA); C231, TTAATTGACTAAAATATATTAAGTA (target 9S rRNA); C232, GTAATATAATATTCAAATAACTAT (target 12S rRNA).
Cloning of gBP29, gBP27 and gBP25 from T.brucei
The obtained amino acid sequence NQFAVTLE (Fig. 2) was used to develop the degenerate primer C204. In combination with primer C217, which contains part of the spliced leader sequence of C.fasciculata, the 5′-part of the gBP29 cDNA was amplified in a PCR reaction, using oligo(dT) (=H33)-primed cDNA of C.fasciculata. The obtained 690 bp fragment was cloned into the pGEM-T vector (Promega), sequenced by the procedure of Sanger et al. (48) and used as a probe to screen a C.fasciculata cDNA library (49). Positive clones were picked and the nucleotide sequence of both strands was determined by the procedure of Sanger with –21M13 and universal M13 fluorescent primers on an Applied Biosystems 377A automated DNA sequencer following ABI protocols. The gBP29 cDNA sequence has been submitted to GenBank under accession no. AF156855.
The gBP27 cDNA was cloned in two parts. The 3′-part of the cDNA was amplified in a nested RT–PCR reaction, using primers C146 and C165, which were based on the obtained amino acid sequences CHTLHC and ADVDE, respectively (Fig. 3A). In the first round of this procedure, a PCR reaction was performed with primers C146 and H33 on cDNA that had been primed with H33. The products of this reaction were then used as starting material for a nested PCR, using primers C165 and H33. The resulting band of 504 bp was cloned into pGEM-T and sequenced. Part of this cDNA sequence was then used to design primer C178. The 5′-part of the gBP27 cDNA was amplified in a PCR reaction with primers C178 and C217, which contains part of the spliced leader sequence of C.fasciculata, using H33-primed cDNA as starting material. The resulting 1369 bp fragment was cloned into the pGEM-T vector and sequenced. The gBP27 cDNA sequence has been submitted to GenBank under accession no. AF157559.
Figure 3.
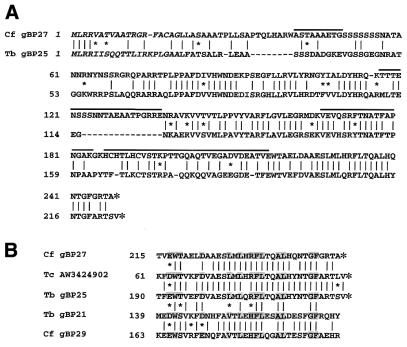
Inferred amino acid sequence of gBP27 and T.brucei gBP25. (A) Amino acid sequence alignment of C.fasciculata gBP27 (top lines) and its putative T.brucei orthologue gBP25 (bottom lines). Identical amino acids are indicated by vertical lines. Small asterisks indicate conservative amino acid substitutions: K, R; S, T; D, E; F, Y; V, I, L. Large asterisks represent stop codons. Dashes are introduced for optimal alignment. The putative mitochondrial import signals are given in italic and Lys-C generated peptide sequences of gBP27 are indicated above the lines. (B) Sequence alignment of a C-terminal section of C.fasciculata gBP29 and gBP27, T.brucei gBP21 and gBP25 and a translated T.cruzi EST AW3424902. Identical and conserved amino acids found in all sequences have been highlighted.
The sequence of the T.brucei orthologue of gBP27 cDNA (gBP25) was obtained via amplification of overlapping cDNA sections using the spliced leader-derived primer, H33 and primers TB6–9, the sequence of which was derived from expressed sequence tags from T.brucei present in the Parasite Genome Database at EBI (AI707247, AI707324, AI881040, AI707274 and AI707444; ftp://ftp.ebi.ac.uk/pub/databases/parasites/blastdbs/). The amplified fragments were cloned into the pGEM-T vector and sequenced. The gBP25 cDNA sequence has been submitted to GenBank under accession no. AF373808.
Expression cloning of gBP29 and gBP27 and generation of antibodies
gBP29 cDNA, starting with Ala16 and lacking most of the putative mitochondrial import signal, was amplified from C.fasciculata total RNA using primers C191 and H33. The amplified fragment was digested with BamHI and PstI, generating a fragment of 705 bp, which was cloned into the expression vector pQE9 (Qiagen) digested with BamHI and PstI, leading to plasmid pQE9-gBP29. The gBP27 ORF, starting with Gly13, was amplified from C.fasciculata total RNA using primers C181 and C188. The resulting fragment was cloned into the pGEM-T vector (Promega) and clones with the correct orientation were digested with BamHI and PstI. This 934 bp BamHI–PstI fragment was then cloned into pQE9 digested with BamHI and PstI, leading to plasmid pQE9-gBP27. The plasmids pQE9-gBP29 and pQE9-gBP27 were introduced into Escherichia coli SG13009 [pREP4] cells (Qiagen) and transfectants were grown at 37°C in L broth containing 100 µg/ml ampicillin and 25 µg/ml kanamycin. For large-scale expression, cells were grown in L broth containing 100 µg/ml ampicillin and 25 µg/ml kanamycin at 27°C, until the A600 reached 0.5. The expression of recombinant protein was then induced by addition of isopropyl-1-thio-β-d-galactopyranoside (IPTG) to a final concentration of 1 mM, for 1.0–1.5 h at 27°C. Cells were harvested by centrifugation at 5000 g for 10 min at 4°C. For purification of denatured proteins we used the guanidine hydrochloride procedure as described by the manufacturer (Qiagen), which uses a Ni–NTA column (HisTrap; Pharmacia). An aliquot of 100 µg purified recombinant protein was used to immunise rabbits. For purification of native proteins the cells were resuspended in sonication buffer (50 mM Na phosphate, pH 7.8, 300 mM NaCl) and sonicated four times for 30 s in a Soniprep 150 (Sanyo) on ice. The sonicate was centrifuged at 10 000 g for 20 min at 4°C and the supernatant loaded onto a HisTrap column (Pharmacia). The column was washed with a Na phosphate buffer, pH 7.4, containing 10 mM imidazole, until no more protein eluted. The recombinant, His-tagged proteins were eluted in a step-wise fashion with Na phosphate buffer containing increasing concentrations (60–500 mM) of imidazole, the elution profile being monitored by SDS–PAGE and western blot analysis. Routinely, the bulk of the proteins eluted between 100 and 400 mM imidazole. rec gBP29- or gBP27-containing fractions were pooled and dialysed against 6 mM HEPES–KOH pH 7.5, 50 mM KCl, 2.1 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, 6% (v/v) glycerol and 1 mM ATP. Analysis by SDS–PAGE as described above indicated the preparations to be >25% pure.
Affinity purification of antibodies
Antibodies specific for gBP27 and gBP29 were purified from rabbit sera as described by Pringle et al. (50), using western blots (Hybond-P membranes; Amersham Life Science) obtained following electrophoresis of 250 µl rec gBP27- or gBP29-containing E.coli extracts.
UV cross-linking assays
The purification of gBP29 and gBP27 from C.fasciculata mitochondria was monitored with a UV cross-linking assay, utilising radiolabelled poly(U), essentially as described by Leegwater et al. (29). The UV cross-linking assay was also used to assess the binding specificity of recombinant gBP29 and gBP27, utilising poly(U) and the following radiolabelled (g)RNA molecules: gND7[FS] (directing editing of the frameshift region of ND7 mRNA; 46), GGAACAGCAUUAGUCUAAUCUAUCAGAAUGCUACUCAAAAAUUUAU-AUUAUUUUUUUUUUUUUUUU; gND7[5′] (directing editing of the 5′-region of ND7 mRNA; 46), GGACGGCUGAUAUAAGUGCAAAAAGGCAAUAAAGACAAAAUAAAUAUA-UUUUUUUUUUUUUUU; RNA63 (the sequence corresponding to nt 72–9 of pGEM-3; Promega), GGAGACCGGAAUUCGAGCUCGGUACCCGGGGAUCCUCUAGAGUCGACCUGCAGGCAUGCAAGC.
The (g)RNAs used were analysed by the mfold algorithm v.3.1 (M.Zuker, http://mfold.wustl.edu/~folder/mfold). They appeared to contain little secondary structure, with ΔG° values (27°C) ranging between –3.0 and –6.0 kcal/mol. RNA63 displayed a slightly higher potential for secondary structure formation (ΔG° = –17.0 kcal/mol)
RNA was synthesised in vitro with a T7-Megashortscript kit (Ambion) according to the manufacturer’s instructions. In each transcription reaction, 35 pmol T7 promotor-containing oligonucleotide (AATTTAATACGACTCACTATAG) was used in combination with 17.5 pmol antisense oligodeoxyribonucleotides consisting of a sequence complementary to the T7 promoter oligonucleotide in juxtaposition to a sequence complementary to one of the RNA sequences shown above. Each reaction included 40 µCi [α-32P]UTP, 0.1 mM unlabelled UTP and 7.5 mM each GTP, CTP and ATP. The radiolabelled RNAs were purified on a 6% (w/v) polyacrylamide/7 M urea gel and dissolved in 10 mM Tris–HCl pH 7.5. The conditions used resulted in RNAs with the following specific radioactivities: gND7[FS], 2 × 106 d.p.m./pmol; gND7[5′], 1.5 × 106 d.p.m./pmol; RNA63, 0.66 × 106 d.p.m./pmol. Just prior to the binding assay, the RNA molecules were heated to 75°C for 2 min and slowly cooled to room temperature. In a representative RNA binding assay, 5 pmol purified rec gBP29 or gBP27 and 0.5 pmol labelled (g)RNA were mixed in 100 µl of binding buffer (10 mM Tris–HCl pH 8.0, 150 mM sucrose, 1 mM EDTA, 5 mM MgCl2, 50 mM KCl, 2 mM DTT and 0.5% Triton X-100) and incubated for 30 min at 27°C.
Other His6-tagged recombinant proteins, such as human His6-ubiquitin B or a His6-tagged fragment of the human LDL receptor (exon 5–exon 8), did not display (g)RNA-binding activity in this assay.
Renografin gradients
Crithidia fasciculata cells were lysed hypotonically and extracts were separated on a 20–35% (w/v) renografin gradient, essentially as described (51). After centrifugation, the gradients were divided into 36 fractions of 1 ml, laurylmaltoside was added to 0.02% (w/v) and the fractions were dialysed against 2 × 2 l of 25 mM Tris–HCl pH 7.6, 5 mM EDTA. Of each fraction, 4–10 µl was used for detection of gBP29, gBP27, ATPase and glyceraldehyde 3-phosphatedehydrogenase (GAPDH) on western blots. For detection of 9S rRNA and 12S rRNA on northern blots, 100 µl of each fraction was used.
RNA ligase and TUTase assays
RNA ligase and TUTase activity were determined as described (15).
Gel filtration chromatography
Mitochondrial vesicles, isolated from 1.5 l of culture (∼1.5 × 1011 cells), were lysed as described above. The extract was clarified by centrifugation at 13 000 g in an Eppendorf centrifuge for 15 min and loaded onto a 150 ml Sephacryl S-500 column (Pharmacia). The column was run at 0.5 ml/min and fractions of 1.2 ml were collected; 880 µl of every fifth fraction was used for detection of gRNA and mRNA by northern blotting. An aliquot of 250 µl, containing 0–50 µg protein, was used for detection of gBP29 and gBP27 protein by western blotting and 50 µl, containing 0–10 µg protein, for detection of RNA ligase and TUTase activities. Protein concentrations were determined according to Bradford (52).
Immunoprecipitation
Immunoprecipitation of gBP29-containing complexes and of gBP27-containing complexes was performed essentially according to Rutjes et al. (53), with the difference that fixed Staphylococcus aureus (Zymed) was used as carrier material instead of protein A–agarose beads. After precipitation, the pellet was washed five times with IPP50 buffer (10 mM Tris–HCl pH 8.0, 50 mM KCl and 0.1% w/v NP-40). In a typical experiment, 60 µl of a Staph A suspension was used with 20 µl of the antibodies and 100 µl of the mt extract. When the immunoprecipitates were analysed for proteins on a western blot, the proteins were directly dissolved in Laemmli sample buffer. When TUTase and RNA ligase activities were determined, proteins were extracted with IPP500 (10 mM Tris–HCl pH 8.0 and 500 mM KCl) and dialysed against 2 × 1 l of 20 mM HEPES–KOH pH 7.9, 50 mM KCl, 10 mM MgAc2, 0.5 mM DTT and 10% (v/v) glycerol. RNA for northern blot and primer extension (54) analyses was isolated from the immunoprecipitates by phenol extraction.
RESULTS
Purification and cloning of gBP29 and gBP27
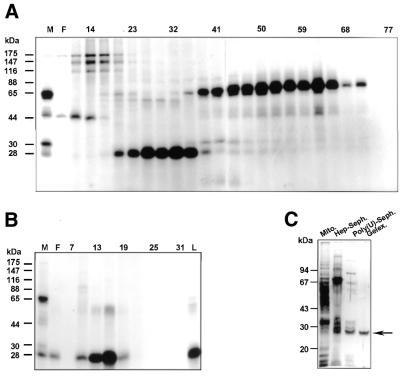
Previously, we described the identification of mt proteins from C.fasciculata that can be UV cross-linked to radiolabelled gRNAs and to poly(U) (29). In SDS–PAGE these proteins appeared to have molecular weights of 88, 65 and 30 kDa, although upon repeated freezing and thawing of the mt extracts and during the purification procedure the 30 kDa protein was gradually processed to a protein migrating at 28 kDa [Fig. 1A, compare lane M and lanes 6–14 (fractions 20–38); for more details see Discussion]. We used the ability to UV cross-link to radiolabelled poly(U) as an assay to purify the 30/28 kDa proteins from 2000 mg isolated mitochondria by chromatography on columns of heparin–Sepharose (Fig. 1A) and poly(U)–Sepharose (Fig. 1B), respectively. The purification procedure (described in Materials and Methods) resulted in a 3300-fold enrichment of the 30/28 kDa cross-linking activity (with 30% recovery) and in a clearly visible 28 kDa protein band on a silver stained PAGE gel of the column fractions containing the UV cross-linking activity (Fig. 1C). As a final purification step, we extracted the 28 kDa protein band from a preparative PAGE gel (Fig. 1C, lane 4) and used it for microsequencing. Comparison of the nine peptide sequences obtained with the GenBank/NCBI database showed that four of them possessed a high degree of similarity to sequences of gBP21 of T.brucei (33). The other five peptide sequences showed no clear similarity to any known protein in the databases.
Figure 1.
Purification of 30/28 kDa proteins that bind to poly(U) and gRNAs. (A) Mitochondrial extracts were loaded onto a heparin–Sepharose column and bound proteins were eluted with a KCl gradient (see Materials and Methods). An aliquot of 7.5 µl of every third heparin–Sepharose fraction, containing 0.1–1.0 µg protein, was tested in a UV cross-linking assay, followed by separation on a 10% (w/v) SDS–PAGE gel. Labelled proteins were then visualised by autoradiography. Lane M, 13 µg mitochondrial extract; lane F, 6.8 µg flow-through. (B) Heparin–Sepharose fractions containing the 28 kDa UV cross-linking activity were pooled, loaded onto a poly(U)–Sepharose column and bound proteins were eluted with a KCl gradient. An aliquot of 7.5 µl of every third poly(U)–Sepharose fraction, containing 10–100 ng protein, was tested in a UV cross-linking assay, followed by separation on a 10% SDS–PAGE gel. Labelled proteins were visualised by autoradiography. Lane M, 13 µg mitochondrial extract; lane F, 1.0 µg flow-through; lane L, 1.0 µg load. (C) Protein samples of different steps in purification of the 28 kDa UV cross-linking activity were separated on a 10% SDS–PAGE gel and silver stained. Mito., 500 ng mitochondrial extract; Hep-Seph., 200 ng pooled heparin–Sepharose fractions containing the 28 kDa UV cross-linking activity; Poly(U)-Seph., 80 ng pooled poly(U)–Sepharose fractions containing the 28 kDa UV cross-linking activity; Gelex., 25 ng PAGE gel-purified 28 kDa UV cross-linking activity. The arrow indicates the position of the 28 kDa protein active in a UV cross-linking assay. Molecular masses of marker proteins are indicated on the left-hand side of each protein gel.
These results suggested that the 28 kDa UV cross-linking activity consists of a mixture of (at least) two co-purifying proteins of the same size and physical properties. In order to further assess this, we set out to clone the cDNAs encoding these proteins. Firstly, we used the peptide sequences with high similarity to T.brucei gBP21 to design degenerate primers that were used in RT–PCR-based cDNA cloning procedures with C.fasciculata total RNA. This resulted in the cloning and sequencing of a 2064 nt cDNA, which contained an ORF encoding a protein of 256 amino acids with a predicted molecular weight of 28.8 kDa. The central 140 amino acids encoded by the ORF displayed 66% identity to the corresponding portion of T.brucei gBP21 (Fig. 2), the sequence containing five peptide sequences that were obtained by microsequencing, one of which was derived from the non-conserved C-terminus of the protein (Fig. 2). This allowed us to conclude that we had indeed cloned the C.fasciculata orthologue of gBP21, which was named gBP29 (gRNA-binding protein of 29 kDa), by virtue of its capacity to bind to synthetic gRNAs (see below). Like its T.brucei counterpart, gBP29 is a hydrophillic basic protein (pI 10.0), with a relatively high Arg + Ala content. Compared to the T.brucei protein, gBP29 has N- and C-terminal extensions rich in asparagine residues that lower the overall identity to 46% (Fig. 2). Except for the high similarity with the gRNA-binding protein gBP21 from T.brucei, no other similarities to proteins in the GenBank/NCBI database could be found.
Northern blot analysis using the ORF-containing region of the cDNA as probe resulted in a prominent hybridizing band of ∼1500 nt and a much fainter band of ∼2900 nt (data not shown). By performing RT–PCR analyses with part of the 39 nt C.fasciculata spliced leader (SL) sequence as the 5′-primer, we could identify an RNA species in which the SL was spliced onto position 763 of the gBP29 cDNA sequence. This species most likely represented the mature mRNA and its size would indeed be ∼1500 nt, taking the position of the splice acceptor site and the possible length of the poly(A) tail into account. The larger RNA of ∼2900 nt, from which the 2 kb sequenced cDNA must have been derived, most likely represented a primary transcript. Probes derived from the area upstream of the gBP29 ORF hybridized to a major product of ∼1300 nt (and to the precursor), indicating that the 2900 nt RNA may represent another example of a dicistronic transcript in trypanosomatids (compared with RGG1 from T.brucei; 31). Since the 2 kb cDNA lacks the 5′-most 900 nt of the precursor and since it does not contain long, recognisable ORFs upstream of the gBP29 ORF, determination of the nature of the protein encoded by the 5′-cistron must await further analysis.
Next, the four as yet unassigned peptide sequences were used for the design of degenerate primers which were used in a nested RT–PCR strategy. This resulted in cloning of a second cDNA containing an ORF which encodes a protein of 248 amino acids with a calculated molecular weight of 26.8 kDa (Fig. 3A). Like gBP29, the protein was a basic (pI 10.1) Arg + Ala-rich protein. Probing of a northern blot with the cDNA resulted in a single hybridizing band of 1400 nt, the expected size for the mRNA (data not shown). Because all four remaining orphan peptide sequences were present in the encoded protein (indicated in Fig. 3), we can conclude that we purified a mixture of (no more than) two proteins of nearly identical size and physical properties: gBP29 and the protein of 26.8 kDa, which was named gBP27 once it had been determined that it was also a gRNA-binding protein (see below). Comparison with the GenBank/NCBI database did not show any similarity to known proteins or protein motifs. The Parasite Genome Database appeared to contain overlapping expressed sequence tags (EST) from T.brucei (AI707247, AI707324, AI881040, AI707274 and AI707444) and T.cuzi (AI562349 and AW324902), which upon translation revealed a high identity to sections of the gBP27 sequence. With the aid of oligonucleotide primers derived from the T.brucei ESTs, we have determined the complete coding sequence of the corresponding cDNA. The inferred protein comprises 224 amino acids with a predicted molecular weight of 25.1 kDa. The C-terminal 160 amino acids of this sequence display a 60% identity to the corresponding region of gBP27, with an overall identity for the full-length protein of 50%, strongly suggesting that it is the T.brucei orthologue of gBP27 (T.brucei gBP25; Fig. 3A).
RNA-binding properties of recombinant gBP29 and gBP27
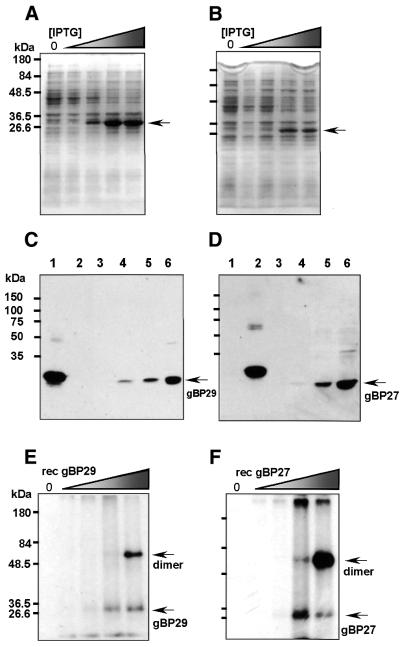
The ORFs of gBP29 and gBP27 (downstream of the putative mt targeting signal, indicated in italic in Figs 2 and 3A; see also Fig. 6) were cloned into the expression vector pQE9, followed by transformation into E.coli and production of His6-tagged recombinant (rec) proteins (Fig. 4A and B). We used the purified recombinant proteins to immunise rabbits and, as evidenced by western blotting, we obtained antibodies that specifically recognised the corresponding recombinant protein in E.coli cell extracts (compare lanes 1 and 2 in Fig. 4C and D, which show no cross-reactivity between α-gBP29 and rec gBP27, and vice versa). The antisera also recognised proteins of 29 and 27 kDa in freshly isolated mt extracts of C.fasciculata (Fig. 4C and D). In addition, we purified the recombinant proteins under native conditions and tested them in UV cross-linking assays. As shown in Figure 4E and F, both rec gBP29 and rec gBP27 could be cross-linked to radiolabelled poly(U). Increasing the amount of recombinant protein in the cross-linking reaction led to formation of dimers, which is in agreement with the experiments shown in Figure 1A and B, in which bands migrating at the position expected for dimers of the 28/30 kDa cross-linking proteins are also visible. The formation of dimers is probably caused by the binding of two protein molecules to the same poly(U) molecule, since dimer formation did not occur when shorther gRNA molecules were used as substrate in this assay (see below). Taken together, these data demonstrate that the 30/28 kDa UV cross-linking activity that was purified from the C.fasciculata mt lysate is in fact composed of two different poly(U)-binding proteins, i.e. gBP29 and gBP27.
Figure 6.
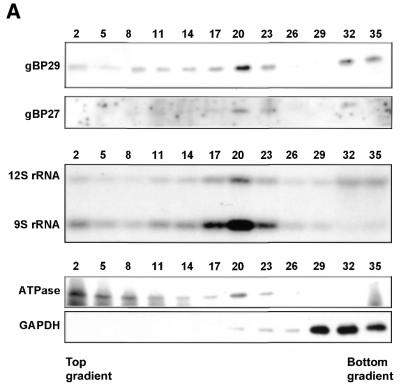
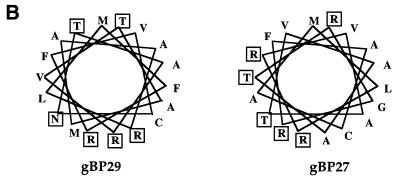
Mitochondrial localization of gBP29 and gBP27. (A) Total cell extracts of C.fasciculata were submitted to renografin gradient centrifugation (20–35% w/v) and 36 fractions were collected (fraction 1 was taken from the top of the gradient, etc.). Protein extracts from indicated fractions were submitted to SDS–PAGE (10% polyacrylamide), followed by western blotting with antisera against gBP29 and gBP27 (upper two panels) and mt ATPase and GAPDH (lower two panels). The same fractions were tested for the presence of mitochondrial 9S and 12S rRNA by northern blot hybridization (middle panel). (B) Helical wheel projection of the putative mitochondrial import signal of gBP29 (left) and gBP27 (right). Charged and hydrophilic amino acids are boxed.
Figure 4.
Expression of recombinant gBP29 and gBP27, antibody generation and UV cross-linking activity. (A and B) Expression of rec gBP29 (A) and gBP27 (B) in E.coli was induced by addition of increasing concentrations of IPTG (shaded wedge) to the culture medium. After induction, cell extracts were prepared and separated on a 10% SDS–PAGE gel, which was stained with Coomassie Brilliant Blue. Concentrations of IPTG used: lanes 1, 0 mM; lanes 2, 0.01 mM; lanes 3, 0.05 mM; lanes 4, 0.25 mM; lanes 5, 1.25 mM. The arrows indicate the positions of rec gBP29 (A) and rec gBP27 (B). (C and D) rec gBP29, rec gBP27 and C.fasciculata mt extract were run on a 10% SDS–PAGE gel and transferred to PVDF membranes. Proteins were visualized by ECL with polyclonal α-gBP29 (C) and α-gBP27 (D) antibodies. Amounts of protein loaded: lanes 1, 8 ng E.coli extract expressing rec gBP29; lanes 2, 8 ng E.coli extract expressing rec gBP27; lanes 3–6, 0.24, 1.19, 2.38 and 11.9 µg C.fasciculata mt extract, respectively. The arrows indicate the positions of gBP29 (C) and gBP27 (D). (E and F) Increasing amounts of purified native rec gBP29 (E) and gBP27 (F) were cross-linked to radiolabelled poly(U) (see Materials and Methods), separated on a 10% SDS–PAGE gel and visualised by autoradiography. Amounts of protein loaded in each lane: 0, 0.5, 5, 50 and 500 ng purified rec gBP29 (E) or purified rec gBP27 (F). Molecular masses of marker proteins are indicated in kDa on the left-hand side of each protein gel.
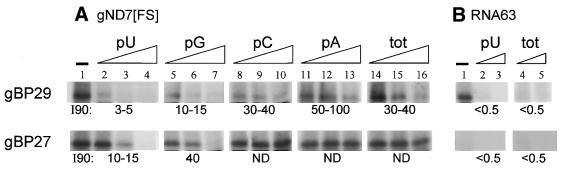
In order to further assess the (g)RNA-binding characteristics of rec gBP29 and gBP27, we measured the capacity of the proteins to bind to synthetic radiolabelled (g)RNAs. We used two different gRNA species (gND7[FS] and gND7[5′]; 46) and a control RNA with a vector-derived sequence of similar length (RNA63), in the absence and presence of various unlabelled competitor RNAs. The results of representative experiments are shown in Figure 5. In the absence of competitor RNAs, both rec gBP29 and gBP27 could be cross-linked to radiolabelled gRNAs (gND7[FS], Fig. 5A, lane 1; gND7[5′], results not shown). The binding of gBP29 to gND7[FS] was inhibited by 90% by addition of a 3- to 5-fold mass excess of poly(U) [this is defined as the I90 value for poly(U); Fig. 5A, lanes 2–4] or unlabelled gND7[FS] (data not shown). The addition of poly(G), poly(C), poly(A) or total human RNA was also inhibitory, but substantially more competitor was required to reach similar levels of inhibition, with 3- to 20-fold higher I90 values (lanes 5–16 and results not shown). In the absence of competitor RNAs, gBP29 could also be cross-linked to RNA63 (Fig. 5B, lane 1), but this binding was almost completely inhibited by small amounts of poly(U) RNA or total human RNA (Fig. 5B, lanes 2–5). Poly(U) (Fig. 5A, lanes 2–4) and unlabelled gRNAs (not shown) are also efficient competitors for binding of gRNA to gBP27. Of the other competitors, only poly(G) turned out to be inhibitory under the conditions of our experiments (with an I90 of ∼40), binding being unaffected by the addition of a 30-fold mass excess of the other competitors (Fig. 5A, lanes 5–16, and results not shown). Even in the absence of competitors, the level of binding of gBP27 to RNA63 turned out to be low (Fig. 5B, lanes 1–5). Although similar amounts of gBP29 and gBP27 bound similar levels of gRNA, 90% inhibition of gRNA binding to gBP27 required 2- to 3-fold more poly(U) and poly(G) (Fig. 5A), but also unlabelled gRNA (not shown), than that to gBP29. The most likely explanation for these observations is that our preparations of gBP27 were 2- to 3-fold more active than those of gBP29.
Figure 5.
RNA binding characteristics of recombinant gBP29 and gBP27. (A) Aliquots of 5 pmol purified gBP29 (upper) or gBP27 (lower) were cross-linked to 0.5 pmol radiolabelled gND7[FS] in the presence of varying amounts of competitor RNAs. The samples were analysed on a 12.5% SDS–PAGE gel, followed by autoradiography. Lane 1, no competitor. Lanes 2–16, before the addition of protein to the mixture, 12, 60 or 300 ng of different competitor RNAs were added: lanes 2–4, poly(U) (pU); lanes 5–7, poly(G) (pG); lanes 8–10, poly(C) (pC); lanes 11–13, poly(A) (pA); lanes 14–16, total RNA from CHP100 cells, a human neuroblastoma cell line (tot). The radioactivity present in cross-linked gBP29 and gBP27 was quantified in a Storm phosphorimager apparatus and the level of binding relative to the experiments without competitor was calculated. The figure shows an autoradiograph of a representative experiment; the numbers given for each competitor RNA represent the mass excess required for ∼90% inhibition of binding (I90), as deduced from a number of different experiments (n = 4 for gBP29; n = 3 for gBP27). The I90 values of gBP27 competition experiments with poly(C), poly(A) and total RNA were not determined (ND); with the highest amounts of these competitors tested (150-fold mass excess) binding levels were still between 20 and 30%. (B) rec gBP29 and gBP27 were cross-linked to 0.5 pmol radiolabelled RNA63, without competitor (lane 1) or with 5 and 20 ng poly(U) (lanes 2 and 3) or 5 and 20 ng total human RNA (lanes 4 and 5). For other details see (A).
We infer from these experiments that gBP29 and gBP27 are indeed gRNA-binding proteins and that the gRNA U-tail may be one of the binding determinants. Nevertheless, gBP29 seems to have a rather broad specificity, given that other RNAs also competed for binding, albeit less efficiently than poly(U). In addition, without competitor, gBP29 also bound to control RNA63, although the affinity of gBP29 for gRNA appeared to be much higher than that for RNA63, since full inhibition of binding to gRNA required >300 times more competitor RNA. gBP27, on the other hand, hardly bound to RNA63 and behaved more like a specific gRNA and poly(U)-binding protein, in view of the fact that only unlabelled gRNA and poly(U) [and to some extent poly(G)] were efficient competitors.
Mitochondrial localization of gBP29 and gBP27
Leegwater et al. (29) showed that the 28/30 kDa cross-linking proteins are mitochondrially localised. To verify the mitochondrial localisation of gBP29 and gBP27, we submitted a crude organellar preparation to renografin floating density gradient analysis. We tested fractions from the gradients for the presence of gBP29 and gBP27 by western blotting and found these proteins to peak around fraction 20 (Fig. 6A, upper two panels). The fractions were also tested for the presence of mt markers, such as the 9S and 12S mt rRNAs (as judged by northern blot analysis; third panel from the top) and the mt ATPase, as visualised with an α-ATPase antiserum on western blots, of which the most prominent reacting band (the F1 β-subunit; 28) is shown. Both the mt rRNAs and the ATPase subunits were also found to peak around fraction 20. Assuming that the rRNAs are located in the mt matrix whereas the mt ATPase is situated in the mt inner membrane, we concluded that the mt vesicles peak around fraction 20 and that these mt vesicles contain both gBP29 and gBP27. A substantial amount of the ATPase was found in the top fractions of the gradient. This is most probably due to the fact that the F1 part of the ATPase complex dissociates easily from the membrane-associated F0 part (55). The glycosomal protein GAPDH (bottom panel) was found at the bottom of the gradients, while cytosolic pyruvate kinase (not shown) was present mostly in the top fractions. The small amount of gBP29, gBP27, rRNA and ATPase that was also found at the bottom of the gradients in some experiments (Fig. 6A) is most likely derived from denatured material that was pelleted during centrifugation.
Comparison of the N-terminal amino acid sequences of gBP29 and gBP27 with the mt targeting sequences of a number of C.fasciculata mt proteins (56,57) revealed a significant homology (data not shown). gBP29 and gBP27 start with Met-Phe-Arg and Met-Leu-Arg, respectively, sequences frequently found at the N-terminus of mt pre-proteins in trypanosomatids. Moreover, the N-terminus of both gBP29 and gBP27, containing several hydrophobic residues, three or four arginine residues, but lacking acidic residues, could be folded into an amphipathic helix, the hallmark of a mt import signal (Fig. 6B; 31,33).
Together, our results strongly suggest that, like T.brucei gBP21 (33,35), C.fasciculata gBP29 and gBP27 are mitochondrially localized.
gBP29 and gBP27 are part of high molecular weight complexes
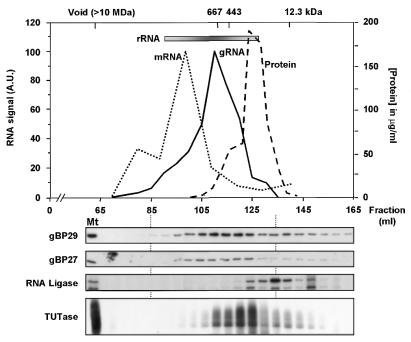
To determine whether gBP29 and gBP27 are part of a larger complex(es), we fractionated mitochondrial extracts of C.fasciculata on a Sephacryl S-500 gel filtration column. We found that both proteins co-eluted over a broad range, as judged by western blot analysis, clearly peaking in fractions corresponding to 400–750 kDa, before the bulk of proteins (Fig. 7, panels 2 and 3). In addition, in the high molecular weight range the elution profile of the proteins was very similar to that of gRNAs, as judged from northern blot hybridizations with a mixture of oligonucleotide probes specific for four gRNAs, i.e. gND7[FS], gND7[5′], gCyb-II and gMURF2-II (Fig. 7, top panel). The leading edge of the gBP29/gBP27/gRNA peak coincided with the (ND7) mRNA peak (Fig. 7, top panel), while the bulk of the 9S and 12S rRNAs eluted slightly ahead of the gRNA peak. We also determined the elution profiles of RNA ligase and TUTase, two key enzymes of the editing process that can be easily assayed (15). As shown in Figure 7 (panel 4), the majority of the RNA ligase activity, as assayed by self-adenylylation of two proteins of 43 and 49 kDa, was found to elute in the low molecular weight range, with only small amounts present in gBP29- and gBP27-containing fractions. In contrast, the TUTase activity peaked at a higher molecular weight position in fractions eluting between gBP29/gBP27 and the RNA ligase (Fig 7, bottom panel).
Figure 7.
Fractionation of mt extract on a Sephacryl S-500 gel filtration column. Mitochondrial extracts were fractionated on a Sephacryl S-500 column as described in Materials and Methods. Fractions were tested by northern blot hybridisation (upper) for the presence of ND7 mRNA (dotted line; A.U. = arbitrary units), the combined presence of ND7[FS], ND7[5′], Cyb-II and MURF2-II gRNAs (solid line) and for 9S + 12S rRNA (inserted bar; the elution profile of 9S + 12S rRNA is indicated by shading). The protein concentration of every fifth fraction was determined by Bradford assay (dashed line). Marker proteins used to calibrate the Sephacryl column were: thyroglobulin (647 kDa), apoferritin (443 kDa) and cytochrome c (12.3 kDa). Every fifth fraction was tested for the presence of gBP29 and gBP27 by western blotting (panels 2 and 3 from the top), for RNA ligase activity by self-adenylylation of two proteins of 43 and 49 kDa, respectively (panel 4), and for TUTase activity by uridylate addition to tRNAs (bottom). Mt, mitochondrial extract.
gBP29 and gBP27 co-immunoprecipitate with gRNA and with edited and pre-edited mRNA
To further investigate the existence of complexes containing gBP29 and gBP27 and to determine the identity of (some of) the other components present in these complexes, we performed immunoprecipitation studies with α-gBP29 and α-gBP27 polyclonal antibodies and mt extracts and/or fractions from the Sephacryl column. The immunoprecipitates were screened on western blots and, as shown in Figure 8A, immunoprecipitates generated with α-gBP29 antibodies not only proved to contain gBP29, but also gBP27, while control immunoprecipitates generated with pre-immune serum did not contain either protein. As also shown in Figure 4, α-gBP27 antibodies did not cross-react with rec gBP29, and vice versa. A similar experiment performed with α-gBP27 antibodies produced a similar result (Fig. 8B): immunoprecipitates generated with α-gBP27 not only contained gBP27 but also gBP29 and also in this case the control experiment with pre-immune serum was negative. In combination with the results of the gel filtration experiment of Figure 7, these experiments led to the conclusion that in the mt extract gBP29 and gBP27 were both present in a large complex.
Figure 8.
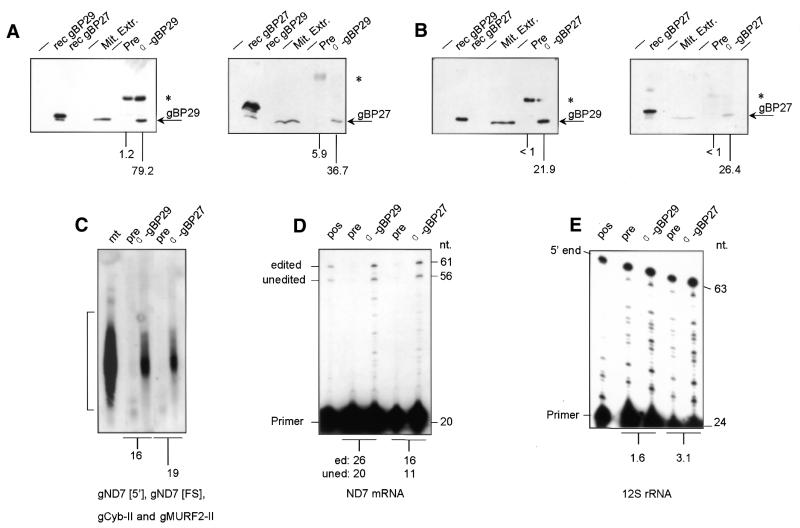
Co-immunoprecipitation analysis. (A and B) Mt extracts of C.fasciculata were incubated with Staph A coated with α-gBP29 serum (A), α-gBP27 serum (B) or pre-immune serum (Pre) (A and B), followed by washing and centrifugation as described in Materials and Methods. Immunoprecipitates were analysed for the presence of either gBP29 (left-hand panels) or gBP27 (right-hand panels) by PAGE and western blotting. In addition, 8 ng E.coli extracts expressing either gBP29 (rec gBP29) or gBP27 (rec gBP27) and the mt extract used as starting material for the immunoprecipitation reaction (Mit. Extr.) were applied to the PAGE gel. Arrows indicate the positions of gBP29 and gBP27; asterisks mark the positions of IgG heavy chains that are occasionally visible. The numbers under the lanes indicate the amounts of gBP29 or gBP27 found in the immunoprecipitate, as a percentage of the total amount present in the mt extract used in the experiment. (C–E) Immunoprecipitates obtained with pre-immune serum (pre), α-gBP29 antibodies and α-gBP27 antibodies were tested for the presence of gND7[FS], gND7[5′], gCyb-II and gMURF2-II by northern blot analysis (C), the presence of ND7 mRNA (edited or unedited at the frameshift position) (D) and 12S rRNA (E) by primer extension analysis. The position of the radiolabelled oligonucleotide (primer) used in the extension reaction is indicated, as is that of the extension products. Primer extension was carried out in the presence of ddGTP instead of dGTP, resulting in a product of 61 nt derived from ND7 RNA edited by insertion of five U residues at the frameshift position and a 56 nt fragment derived from unedited ND7 RNA. Pos., positive control of the primer extension reaction with C.fasciculata total RNA. The numbers under (C)–(E) indicate the ratios of the amounts of RNA present in the immunoprecipitates generated with α-gBP29 and α-gBP27, respectively, and those generated with pre-immune serum.
We also isolated RNAs from the immunoprecipitates and tested them for the presence of gRNAs, mRNA and rRNAs by northern blotting and primer extension analyses. We found gRNAs to be present in both the α-gBP29-generated and the α-gBP27-generated immunoprecipitates, in amounts 16- to 30-fold higher than in control immunoprecipitates (depending on the experiment). Of the total amount of the gRNAs present in the lysate, 25–50% was found in the immunoprecipitates. When the immunoprecipitates were tested for the presence of mRNA, we obtained similar results (Fig. 8D): α-gBP29 as well as α-gBP27 antibodies specifically immunoprecipitated ∼25% of the total amount of both unedited and edited ND7 mRNA present in the lysate (as analysed for editing at the frameshift position by poisoned primer extension experiments; 49). However, when tested for the presence of 9S and 12S mt rRNA, we detected only a 2-fold difference in rRNA content between immunoprecipitates generated with α-gBP29 and α-gBP27 and those obtained with control immunoprecipitates (Fig. 8E). With both α-gBP29 and α-gBP27 only ∼2% of the total amount of 12S rRNA present in the lysates was immunoprecipitated. Last but not least, we screened the immunoprecipitates for the presence of TUTase and RNA ligase. The immunoprecipitates generated with both α-gBP29 and α-gBP27 contained RNA ligase and TUTase activity in amounts very similar to those present in control immunoprecipitates (results not shown). As a whole, our results suggest that (some of) the complexes that contain gBP29 and gBP27 also contain gRNAs and edited and pre-edited mRNA. However, they appear to be (mostly) devoid of rRNAs, RNA ligase and TUTase (see Discussion).
DISCUSSION
In this paper we describe the molecular cloning and characterisation of two mt gRNA-binding proteins from C.fasciculata, gBP29 and gBP27, which co-purified in a procedure that was aimed at the characterisation of proteins of 30/28 kDa from mt extracts that can be UV cross-linked to radiolabelled gRNAs and poly(U) (Fig. 1A, lane M). The results show that our original interpretation that the 28 kDa protein is a proteolytic product of the 30 kDa protein is only partly true and that there is a second 28 kDa UV cross-linking protein (gBP27), whose size and chromatographic behaviour on the columns used are identical to those of the processed 30 kDa protein (gBP29). The use of antibodies has indeed confirmed this scenario, showing, for example, a clear size difference between native gBP29 and gBP27 in freshly isolated mt extracts (Fig. 4C and D; compare with Fig. 1A, lane M), whereas the processed form of gBP29, as present in heparin–Sepharose and poly(U)–Sepharose column fractions, and gBP27 migrated to virtually the same position on SDS–PAGE (results not shown; Fig. 1A and B). The reason for the precisely overlapping elution profiles of gBP29 and gBP27 on heparin–Sepharose and poly(U)–Sepharose columns remains unclear, but the results of the experiments shown in Figures 4 and 5 suggest that gBP29 and gBP27 have very similar RNA-binding characteristics. An alternative, albeit not mutually exclusive, explanation would be that the proteins co-purified in a complex, possibly together with other proteins and/or RNA. Preliminary experiments failed to show any direct interaction between rec gBP29 and gBP27 (results not shown) and further work is obviously required to settle this issue.
The high degree of identity of the central region (Fig. 2) identifies gBP29 as the C.fasciculata orthologue of gBP21, a 21 kDa gRNA-binding protein with RNA annealing activity from T.brucei (33,36). Indeed, studies with His-tagged recombinant protein showed rec gBP29 to also be a gRNA-binding protein (Fig. 5). The gRNA structural elements that determine binding to gBP21 are not yet known, but the 3′-U-tail does not seem to be important for gRNA binding to rec gBP21 (33,35,36). Although the results of the competition experiments shown in Figure 5 suggest a role for the U-tail in binding of gRNA to the C.fasciculata rec protein, gBP29 appears to have a rather broad binding specificity and our dataset is too limited to exclude a role for other gRNA structural elements. It is interesting to note, though, that the binding of gRNA by the native trypanosomatid version of gBP21 is completely inhibited by addition of relatively small amounts of poly(U), whereas 3-fold higher concentrations of poly(A), tRNAPhe or total yeast RNA have no or little effect (58). These experiments also point at the gRNA U-tail as a binding determinant for gBP21, implying that the apparent difference in the binding characteristics between rec gBP29 and rec gBP21 does not reflect a real difference in the binding specificities of native gBP29 and gBP21, but instead could be related to differences in the way the recombinant versions of these proteins were constructed and isolated. As an alternative explanation, species-specific differences in gRNA-binding characteristics between gBP29 and gBP21 could exist, since even in the conserved region, gBP29 and gBP21 are only 66% identical (Fig. 2). In addition, gBP29 contains stretches of multiple asparagine residues in its N- and C-terminal regions that are lacking in gBP21. Asparagine-rich regions have been suggested to be involved in protein–protein contacts (59,60) and, since they are absent in gBP21, they are most likely not essential for gRNA binding per se. It could be envisaged though that their presence has a modulatory effect on the RNA-binding properties of the protein. Obviously, more work is required to settle this issue. Our studies further identified rec gBP27 as a potential gRNA-binding protein, the competition experiments of Figure 5 suggesting that the U-tail is also a binding determinant for this protein. It is reassuring to note that the combined binding characteristics of C.fasciculata gBP29 and gBP27 are similar to those of the recombinant proteins, given that the UV cross-linking of gRNAs to the 30/28 kDa proteins in the mt lysate (which are a mixture of gBP29 and gBP27) completely depended on the presence of a U-tail, with poly(U) as an efficient competitor also in these assays (29).
The sequences of the cloned cDNAs do not provide further clues to the function of gBP29 and gBP27. Both have a high alanine content (11.3 and 14.6%, respectively) and are hydrophillic proteins without identifiable membrane-spanning regions. They are also basic and arginine-rich (9.8 and 8.5%, respectively), a characteristic of other RNA-binding proteins (61), but the sequences do not contain any of the known RNA-binding motifs (cf. gBP21; 33). Crithidia fasciculata gBP27 displays a high degree of similarity to the putative T.brucei orthologue (gBP25, Fig. 3A), particularly in the C-terminal two-thirds of the protein, demonstrating that we have identified a conserved trypanosomatid protein. Fractionation of mt extracts under native conditions by gel filtration on Sephacryl S-500 demonstrated that gBP29 and gBP27 co-eluted with each other and with gRNAs in the high molecular weight range, implying that both proteins are present in a large complex(es) (Fig. 7). This was further analysed in the experiments shown in Figure 8, which showed that gBP29 and gBP27 co-immunoprecipitated and that gRNAs and edited and pre-edited ND7 mRNAs, but not mt rRNAs, were also present in the immunoprecipitates, irrespective of whether α-gBP29 antibodies or α-gBP27 antibodies were used. A comparison of the Sephacryl S-500 elution profiles of all these components makes it very likely that the gBP29 + gBP27-containing RNP complexes are heterogeneous in composition, given the broad molecular mass range in which gBP29 and gBP27 eluted from the Sephacryl column (30–1000 kDa).
What is the evidence that gBP29 and gBP27 play a role in RNA editing? Despite numerous attempts, we have been unable to develop an in vitro editing system for C.fasciculata. We have no clear explanation for this, but it may be related to the fact that a rather harsh and lengthy procedure has to be employed for the preparation of mt extracts from C.fasciculata. As a consequence, one or more components required for the editing process might be inactivated. This could be reflected in, for example, the apparent difference between C.fasciculata and T.brucei gRNP particles. Although gRNA- and mRNA-containing complexes, the size of which would correspond to 19S and 35–40S, may exist in C.fasciculata mt extacts (Fig. 7), they apparently do not contain (active) TUTase and RNA ligase (Fig. 8). Therefore, the evidence for a role in editing of gBP29 and gBP27 is indirect, but the arguments in favour of such a role are not easily dismissed. Firstly, as discussed above, the UV cross-linking results show that rec gBP29 and rec gBP27 prefentially bind to gRNAs. Most importantly, however, native gBP29 and gBP27 are present in complexes that also contain 25–50% of the total amount of four gRNAs and ∼25% of edited and pre-edited mRNA. Our experiments further demonstrated that these RNP complexes contain very little (∼2%) of the 9S and 12S rRNAs, in spite of the abundance of these RNAs in the mt lysate and the fact that they also possess a 3′-terminal U extension (62). This emphasises that the complexes observed are not the result of some form of artifactual aggregation occurring during preparation of the mt extract and suggests that the RNP complexes containing gBP29 and gBP27 are not rRNPs. As far as we know, this is the first time that this issue has been addressed in a quantitative manner in studies of gRNA-binding proteins and gRNP particles (see for example refs 12,15,31,35,37). These results imply that in vivo a possible interaction between gBP29 and gBP27 and the U-tail of rRNAs is prevented, for example by binding of ribosomal proteins to the tail, prior to or during assembly of the ribosome. Finally, gBP29 is the C.fasciculata orthologue of T.brucei gBP21, a protein whose association with editing complexes and capacity to stimulate gRNA:pre-mRNA duplex formation has been clearly demonstrated (34–36). Since gBP27 is also associated with g+mRNP complexes, we infer that gBP27 is also an editosomal component.
The biochemical properties of gBP29 and gBP27 appear to be similar. This is further underlined by a striking resemblance of the hydrophillicity profiles (data not shown) and a 43% similarity of a 28 amino acid region in the C-terminal regions of the proteins (Fig. 3B). Although we cannot as yet assign a function to this region, all amino acid residues that are conserved between gBP29 and gBP27 are also present in the corresponding sections of T.brucei gBP21 and gBP25 and the T.cruzi EST, underlining their possible functional importance. The specific role(s) in the editing process of gBP29 and gBP27 and the details of their interaction with gRNAs remain to be elucidated, but it could be speculated that two proteins that are so similar may have at least partially overlapping functions. Molecular redundancy has been mentioned as a possible explanation for the apparently normal levels of edited RNAs in a T.brucei gBP21 knockout strain (34,63) and it could be speculated that gBP25, the T.brucei version of gBP27, is capable of taking over the chores of the missing gBP21 in the editing process. The construction of strains deficient in both gBP25 and gBP21 may shed further light on this issue.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Thijs Hendriks for expert help in generation of the α-gBP29 and α-gBP27 antibodies, Paul Michels (ICP, Bruxelles) for the gift of L.mexicana GAPDH and PK antisera, Annett de Haan for skillful experimental assistance, Paul Sloof for stimulating discussions and Wendy van Noppen for editorial comments on the manuscript. The research was supported by the Netherlands Foundation for Chemical Research (SON), which is subsidised by the Netherlands Foundation for Scientific Research (NWO). The Procise 494 protein sequencer was in large part funded by the NWO Medical Sciences Branch (NWO-MW).
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +1 31 20 5665131; Fax: +1 31 20 6915519; Email: Present address:Cornelis K. D. Breek, Institute of Plant Molecular Sciences, Clusius Laboratory, Leiden University, Wassenaarseweg 64, 2333 AL Leiden, The Netherlands AF156855, AF157559, AF373808
References
- 1.Arts G.J. and Benne,R. (1996) Mechanism and evolution of RNA editing in kinetoplastida. Biochim. Biophys. Acta, 1307, 39–54. [DOI] [PubMed] [Google Scholar]
- 2.Alfonzo J.D., Thiemann,O. and Simpson,L. (1997) The mechanism of U insertion/deletion RNA editing in kinetoplastid mitochondria. Nucleic Acids Res., 25, 3751–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stuart K., Allen,T.E., Heidmann,S. and Seiwert,S.D. (1997) RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev., 61, 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajduk S.L. and Sabatini,R.S. (1998) Mitochondrial mRNA editing in kinetoplastid mitochondria. In Grosjean,H. and Benne,R. (eds) Modification and Editing of RNA. ASM press, Washington, DC, pp. 377–393.
- 5.Seiwert S.D., Heidmann,S. and Stuart,K. (1996) Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell, 84, 831–841. [DOI] [PubMed] [Google Scholar]
- 6.Kable M.L., Seiwert,S.D., Heidmann,S. and Stuart,K. (1996) RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science, 273, 1189–1195. [DOI] [PubMed] [Google Scholar]
- 7.Cruz-Reyes J. and Sollner-Webb,B. (1996) Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease and RNA ligase activities. Proc. Natl Acad. Sci. USA, 93, 8901–8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrne E.M., Connell,G.J. and Simpson,L. (1996) Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J., 15, 6758–6765. [PMC free article] [PubMed] [Google Scholar]
- 9.Blum B., Bakalara,N. and Simpson,L. (1990) A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell, 60, 189–198. [DOI] [PubMed] [Google Scholar]
- 10.Cruz-Reyes J., Rusché,L.N., Piller,K.J. and Sollner-Webb,B. (1998) T. brucei RNA editing: adenosine nucleotides inversely affect U-deletion and U-insertion reactions at mRNA cleavage. Mol. Cell, 1, 401–409. [DOI] [PubMed] [Google Scholar]
- 11.Cruz-Reyes J., Rusché,L.N. and Sollner-Webb,B. (1998) Trypanosoma brucei U insertion and U deletion activities co-purify with an enzymatic editing complex but are differentially optimized. Nucleic Acids Res., 26, 3634–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollard V.W., Harris,M.E. and Hajduk,S.L. (1992) Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J., 11, 4429–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adler B.K. and Hajduk,S.L. (1997) Guide RNA requirement for editing-site-specific endonucleolytic cleavage of preedited mRNA by mitochondrial ribonucleoprotein particles in Trypanosoma brucei. Mol. Cell. Biol., 17, 5377–5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piller K.J., Rusché,L.N., Cruz-Reyes,J. and Sollner-Webb,B. (1997) Resolution of the RNA editing gRNA-directed endonuclease from two other endonucleases of Trypanosoma brucei mitochondria. RNA, 3, 279–290. [PMC free article] [PubMed] [Google Scholar]
- 15.Corell R.A., Read,L.K., Riley,G.R., Nellissery,J.K., Allen,T.E., Kable,M.L., Wachal,M.D., Seiwert,S.D., Myler,P.J. and Stuart,K.D. (1996) Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol. Cell. Biol., 16, 1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rusché L.N., Cruz-Reyes,J., Piller,K.J. and Sollner-Webb,B. (1997) Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J., 16, 4069–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peris M., Frech,G.C., Simpson,A.M., Bringaud,F., Byrne,E., Bakker,A. and Simpson,L. (1994) Characterization of two classes of ribonucleoprotein complexes possibly involved in RNA editing from Leishmania tarentolae mitochondria. EMBO J., 13, 1664–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peris M., Simpson,A.M., Grunstein,J., Liliental,J.E., Frech,G.C. and Simpson,L. (1997) Native gel analysis of ribonucleoprotein complexes from a Leishmania tarentolae mitochondrial extract. Mol. Biochem. Parasitol., 85, 9–24. [DOI] [PubMed] [Google Scholar]
- 19.Schnaufer A., Panigrahi,A.K., Panicucci,B., Igo,R.P.Jr, Salavati,R. and Stuart,K. (2001) An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science, 291, 2159–2162. [DOI] [PubMed] [Google Scholar]
- 20.Panigrahi A.K., Gygi,S.P., Ernst,N.L., Igo,R.P.Jr, Palazzo,S.S., Schnaufer,A., Weston,D.S., Carmean,N., Salavati,R., Aebershold,R. and Stuart,K. (2001) Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol., 21, 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rusché L.N., Huang,C.E., Piller,K.J., Hemann,M., Wirtz,E. and Sollner-Webb,B. (2001) The two RNA ligases of the Trypanosoma brucei RNA editing complex: cloning the essential band IV gene and identifying the band V gene. Mol. Cell. Biol., 21, 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McManus M.T., Shimamura,M., Grams,J. and Hajduk,S.L. (2001) Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA, 7, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alfonzo J.D., Thiemann,O.H. and Simpson,L. (1998) Purification and characterization of MAR1. A mitochondrial associated ribonuclease from Leishmania tarentolae. J. Biol. Chem., 273, 30003–30011. [DOI] [PubMed] [Google Scholar]
- 24.Missel A., Souza,A.E., Nörskau,G. and Göringer,H.U. (1997) Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol., 17, 4895–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madison-Antenucci S., Sabatini,R.S., Pollard,V.W. and Hajduk,S.L. (1998) Kinetoplastid RNA-editing-associated protein 1 (REAP-1): a novel editing complex protein with repetitive domains. EMBO J., 17, 6368–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bringaud F., Peris,M., Zen,K.H. and Simpson,L. (1995) Characterization of two nuclear-encoded protein components of mitochondrial ribonucleoprotein complexes from Leishmania tarentolae. Mol. Biochem. Parasitol., 71, 65–79. [DOI] [PubMed] [Google Scholar]
- 27.Bringaud F., Stripecke,R., Frech,G.C., Freedland,S., Turck,C., Byrne,E.M. and Simpson,L. (1997) Mitochondrial glutamate dehydrogenase from Leishmania tarentolae is a guide RNA-binding protein. Mol. Cell. Biol., 17, 3915–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speijer D., Breek,C.K.D., Muijsers,A.O., Hartog,A.F., Berden,J.A., Albracht,S.P.J., Samyn,B., Van Beeumen,J. and Benne,R. (1997) Characterization of the respiratory chain from cultured Crithidia fasciculata. Mol. Biochem. Parasitol., 85, 171–186. [DOI] [PubMed] [Google Scholar]
- 29.Leegwater P., Speijer,D. and Benne,R. (1995) Identification by UV cross-linking of oligo(U)-binding proteins in mitochondria of the insect trypanosomatid Crithidia fasciculata. Eur. J. Biochem., 227, 780–786. [DOI] [PubMed] [Google Scholar]
- 30.Read L.K., Göringer,H.U. and Stuart,K. (1994) Assembly of mitochondrial ribonucleoprotein complexes involves specific guide RNA (gRNA)-binding proteins and gRNA domains but does not require preedited mRNA. Mol. Cell. Biol., 14, 2629–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanhamme L., Perez-Morga,D., Marchal,C., Speijer,D., Lambert,L., Geuskens,M., Alexandre,S., Ismaïli,N., Göringer,U., Benne,R. and Pays,E. (1998) Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem., 273, 21825–21833. [DOI] [PubMed] [Google Scholar]
- 32.Kiledjian M. and Dreyfuss,G. (1992) Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J., 11, 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Köller J., Müller,U.F., Schmid,B., Missel,A., Kruft,V., Stuart,K. and Göringer,H.U. (1997) Trypanosoma brucei gBP21—an arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem., 272, 3749–3757. [DOI] [PubMed] [Google Scholar]
- 34.Lambert L., Müller,U.F., Souza,A.E. and Göringer,H.U. (1999) The involvement of gRNA-binding protein gBP21 in RNA editing—an in vitro and in vivo analysis. Nucleic Acids Res., 27, 1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen T.E., Heidmann,S., Reed,R., Myler,P.J., Göringer,H.U. and Stuart,K.D. (1998) Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol. Cell. Biol., 18, 6014–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller U.F., Lambert,L. and Göringer,H.U. (2001) Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J., 20, 1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayman M.L. and Read,L.K. (1999) Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem., 274, 12067–12074. [DOI] [PubMed] [Google Scholar]
- 38.Pelletier M., Miller,M.M. and Read,L.K. (2000) RNA-binding properties of the mitochondrial Y-box protein RBP16. Nucleic Acids Res., 28, 1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yasuhira S. and Simpson,L. (1995) Minicircle-encoded guide RNAs from Crithidia fasciculata. RNA, 1, 634–643. [PMC free article] [PubMed] [Google Scholar]
- 40.Kleisen C.M., Borst,P. and Weijers,P.J. (1975) Properties of the intact multicircular complex from Crithidia fasciculata. Biochim. Biophys. Acta, 390, 155–167. [PubMed] [Google Scholar]
- 41.Birkenmeyer L. and Ray,D.S. (1986) Replication of kinetoplast DNA in isolated kinetoplasts from Crithidia fasciculata. Identification of minicircle DNA replication intermediates. J. Biol. Chem., 261, 2362–2368. [PubMed] [Google Scholar]
- 42.Borst P. and Fase-Fowler,F. (1979) The maxicircle of Trypanosoma brucei kinetoplast DNA. Biochim. Biophys. Acta, 565, 1–12. [DOI] [PubMed] [Google Scholar]
- 43.Tullis R.H. and Rubin,H. (1980) Calcium protects DNase I from proteinase K: a new method for the removal of contaminating RNase from DNase I. Anal. Biochem., 107, 260–264. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Van der Spek H., Speijer,D., Arts,G.J., Van den Burg,J., Van Steeg,H., Sloof,P. and Benne,R. (1990) RNA editing in transcripts of the mitochondrial genes of the insect trypanosome Crithidia fasciculata. EMBO J., 9, 257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van der Spek H., Arts,G.J., Zwaal,R.R., Van den Burg,J., Sloof,P. and Benne,R. (1991) Conserved genes encode guide RNAs in mitochondria of Crithidia fasciculata. EMBO J., 10, 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 48.Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van der Spek H., Van den Burg,J., Croiset,A., Van den Broek,M., Sloof,P. and Benne,R. (1988) Transcripts from the frameshifted MURF3 gene from Crithidia fasciculata are edited by U insertion at multiple sites. EMBO J., 7, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pringle J.R., Adams,A.E.M., Drubin,D.G. and Haarer,B.K. (1991) Immunofluorescence methods for yeast. Methods Enzymol., 194, 565–602. [DOI] [PubMed] [Google Scholar]
- 51.Feagin J.E., Jasmer,D.P. and Stuart,K. (1987) Developmentally regulated addition of nucleotides within apocytochrome b transcripts in Trypanosoma brucei. Cell, 49, 337–345. [DOI] [PubMed] [Google Scholar]
- 52.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248. [DOI] [PubMed] [Google Scholar]
- 53.Rutjes S.A., Vree Egberts,W.T.M., Jongen,P., Van den Hoogen,F., Pruijn,G.J.M. and Van Venrooij,W.J. (1997) Anti-Ro52 antibodies frequently co-occur with anti-Jo-1 antibodies in sera from patients with idiopathic inflammatory myopathy. Clin. Exp. Immunol., 109, 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arts G.J., Van der Spek,H., Speijer,D., Van den Burg,J., Van Steeg,H., Sloof,P. and Benne,R. (1993) Implications of novel guide RNA features for the mechanism of RNA editing in Crithidia fasciculata. EMBO J., 12, 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker J.E., Collinson,I.R., Van Raaij,M.J. and Runswick,M.J. (1995) Structural analysis of ATP synthase from bovine heart mitochondria. Methods Enzymol., 260, 163–190. [DOI] [PubMed] [Google Scholar]
- 56.Xu W.C., Hines,J.C., Engel,M.L., Russell,D.G. and Ray,D.S. (1996) Nucleus-encoded histone H1-like proteins are associated with kinetoplast DNA in the trypanosomatid Crithidia fasciculata. Mol. Cell. Biol., 1, 564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hines J.C. and Ray,D.S. (1998) The Crithidia fasciculata KAP1 gene encodes a highly basic protein associated with kinetoplast DNA. Mol. Biochem. Parasitol., 94, 41–52. [DOI] [PubMed] [Google Scholar]
- 58.Köller J.K., Nörskau,G., Paul,A.S., Stuart,K. and Göringer,H.U. (1994) Different Trypanosoma brucei guide RNA molecules associate with an identical complement of mitochondrial proteins in vitro. Nucleic Acids Res., 22, 1988–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siomi H. and Dreyfuss,G. (1997) RNA-binding proteins as regulators of gene expression. Curr. Opin. Genet. Dev., 7, 345–353. [DOI] [PubMed] [Google Scholar]
- 60.Bröhl S., Lisowsky,T., Riemen,G. and Michaelis,G. (1994) A new nuclear suppressor system for a mitochondrial RNA polymerase mutant identifies an unusual zinc-finger protein and a polyglutamine domain protein in Saccharomyces cerevisiae. Yeast, 10, 719–731. [DOI] [PubMed] [Google Scholar]
- 61.Brendel V. and Karlin,S. (1989) Association of charge clusters with functional domains of cellular transcription factors. Proc. Natl Acad. Sci. USA, 86, 5698–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Priest J.W. and Hajduk,S.L. (1995) The trypanosomatid Rieske iron-sulfur proteins have a cleaved presequence that may direct mitochondrial import. Biochim. Biophys. Acta, 1269, 201–204. [DOI] [PubMed] [Google Scholar]
- 63.Thomas J.H. (1993) Thinking about genetic redundancy. Trends Genet., 9, 395–399. [DOI] [PubMed] [Google Scholar]