Abstract
An acute bout of exercise is sufficient to induce changes in skeletal muscle gene expression that are ultimately responsible for the adaptive responses to exercise. Although much research has described the intracellular signaling responses to exercise that are linked to transcriptional regulation, the epigenetic mechanisms involved are only just emerging. This review will provide an overview of epigenetic mechanisms and what is known in the context of exercise. Additionally, we will explore potential interactions between metabolism during exercise and epigenetic regulation, which serves as a framework for potential areas for future research. Finally, we will consider emerging opportunities to pharmacologically manipulate epigenetic regulators and mechanisms to induce aspects of the skeletal muscle exercise adaptive response for therapeutic intervention in various disease states.
Early studies have identified acute reductions in DNA methylation and increases in histone acetylation in skeletal muscle in response to exercise. Metabolic changes in muscle may heavily influence epigenetic processes.
Regular exercise evokes a number of adaptive responses in skeletal muscle that contribute to alterations in muscle function and physical fitness. Indeed, skeletal muscle is a highly plastic tissue that can rapidly adapt to the energetic and mechanical demands placed on it through repeated bouts of exercise. In response to endurance exercise, skeletal muscle increases its capacity to produce energy through aerobic metabolism by enhancing delivery of substrates and oxygen secondary to increasing capillary density (Clausen and Trap-Jensen 1970). The capacity for skeletal muscle substrate uptake is also enhanced through a greater abundance of sarcolemmal glucose and fatty acid transporters (Glatz et al. 2002). In parallel, the capacity to oxidize these substrates to produce ATP is enhanced through increased mitochondrial size and number (Holloszy 1967). In contrast, strength exercise increases the power that a muscle can exert by increasing its contractile units, cross-sectional area, and the connective tissue structures that assist in force transmission and muscle stabilization (Nader et al. 2014). Furthermore, strength exercise also increases the ability of skeletal muscle to produce energy through anaerobic pathways (Egan and Zierath 2013).
Many of these phenotypic adaptations are driven by transient alterations in the expression of genes involved in various aspects of skeletal muscle function (Egan and Zierath 2013). The expression of some genes increases during exercise while others increase in the postexercise period. These transient changes in gene expression result in persistent alterations in the levels of the proteins that they encode, which ultimately produce a change in skeletal muscle phenotype (Egan and Zierath 2013). A wealth of research has described many of the intracellular signaling mechanisms that drive exercise-mediated changes in gene expression, which include activation of kinase cascades, transcription factors, and transcriptional coregulators (Egan and Zierath 2013). It is well established that another layer of biological regulation, termed epigenetics, also controls gene expression. Although there is conjecture on the exact definition of the term (Henikoff and Greally 2016), epigenetics is generally considered to describe heritable changes in gene regulation that occur independently of changes in DNA sequence, but instead are caused by chemical modification of the various components of chromatin, which regulates the access of the transcriptional machinery to DNA (Kouzarides 2007).
This review will provide a brief overview of the most commonly studied epigenetic mechanisms before describing the nascent field of exercise epigenetics. We will also speculate on potential interactions between exercise metabolism and epigenetic regulation before providing an assessment of the potential usage of drugs that act by modulating epigenetic modifiers to induce aspects of the exercise adaptive response in skeletal muscle.
OVERVIEW OF EPIGENETIC MECHANISMS
Epigenetic modification of the fundamental components of chromatin, which includes the histone protein core and DNA, influences how surrounding genes are regulated. These modifications can be broadly categorized into those affecting DNA, and those affecting histone proteins (Kouzarides 2007). The following section will provide an overview of the best-characterized epigenetic modifications and how they impact on transcriptional regulation.
DNA Methylation
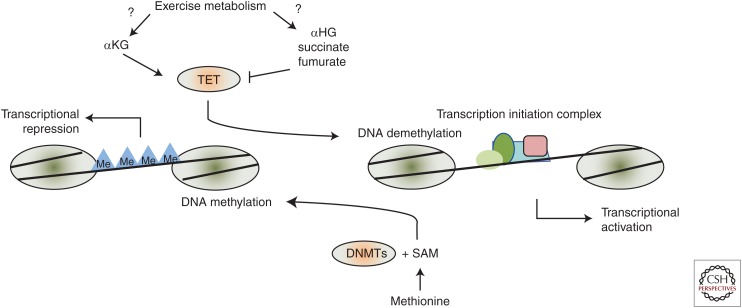
DNA methylation involves the enzymatic addition of a methyl group to the fifth carbon of a cytosine residue, yielding 5-methylcytosine (Nakao 2001). This process is catalyzed by a family of enzymes known as DNA methyltransferases (DNMTs) (Suzuki and Bird 2008), and is regarded as one of the fundamental epigenetic mechanisms to regulate gene transcription. DNMTs are encoded in many genomes, from bacteria to plants and mammals. The evolutionary conservation of these enzymes suggests that DNA methylation provides a selective advantage to the organism. In mammals, DNA methylation typically occurs at CpG-rich regions of the genome, known as CpG islands, which are enriched in the promoters and first exons of genes (Klose and Bird 2006). When the CpG island becomes methylated, the transcription of the cognate genes will be blocked (Lister et al. 2009; Ziller et al. 2013). However, DNA methylation also occurs outside CpG islands in mammals, and this also contributes the regulation of gene transcription, including in skeletal muscle (Barres et al. 2009). DNA methylation is generally associated with a reduction in gene expression, thought to be mediated by affecting the binding of transcriptional activators to promoter regions (Fig. 1) (Nakao 2001), although the precise mechanism involved is still under investigation.
Figure 1.
DNA methylation and the regulation of transcription. DNA methylation (Me), controlled by DNA methyltransferases (DNMTs) using S-adenosylmethionine (SAM) as the methyl donor, prevents transcriptional activators such as the transcriptional initiation complex from gaining access to gene regulatory regions. Demethylation of DNA, initiated by the ten-eleven translocation (TET) enzymes, exposes DNA regulatory regions to transcriptional activators. The TET enzymes are activated by the α-ketoglutarate (αKG) metabolites and inhibited by metabolites with similar structures, including 2-hydroxyglutarate (2HG), succinate, and fumurate. It is unclear whether exercise alters the profile of these metabolites in cellular compartments that might affect TET activity.
DNA methylation patterns and the resultant effects on gene transcription are a critical part of normal development programs, and these processes are controlled by the precise regulation of the DNMTs. Aberrant DNA methylation, often associated with altered levels or dysfunction of the DNMTs, has been observed in a range of developmental diseases, such as neural tube defects (Chang et al. 2011), as well as adult diseases, including a range of cancers (Robertson 2005; Hamidi et al. 2015), but also in complex chronic diseases such as schizophrenia (Gavin and Sharma 2010) and autism spectrum disorder (Jiang et al. 2004; Nagarajan et al. 2006).
DNMT1 is regarded as a maintenance enzyme whose primary role is to preserve methylation patterns during DNA replication and mitosis, which would otherwise alter methylation patterns (Denis et al. 2011). DNMT3A and DNMT3B are the principal methyltransferases involved in establishing de novo DNA methylation patterns in somatic and differentiated cell types (Shen and Laird 2013; Bedi et al. 2014). Emerging evidence suggests that DNA methylation patterns can be dynamically changed in response to various physiological challenges, including diet (Amaral et al. 2014; Silva-Martinez et al. 2016), exposure to toxins (Tryndyak et al. 2016), and exercise (Voisin et al. 2015). These changes are induced by altered activity of the DNMTs in concert with changes in active demethylation of DNA, which involves sequential modification of the methylated cytosine by hydroxylation, deamination, and/or oxidation (Bhutani et al. 2011). The base is then replaced by DNA repair mechanisms. These processes are mediated by the ten-eleven translocation (TET), AID (activation-induced cytosine deaminase)/APOBEC (apolipoprotein B mRNA editing enzyme component 1) and the base excision repair (BER) enzymes, respectively (Bhutani et al. 2011). Dynamic changes to DNA methylation patterns in response to a range of physiological stimuli are thought to be integral to both physiological adaptation and disease pathology.
Histone Modifications
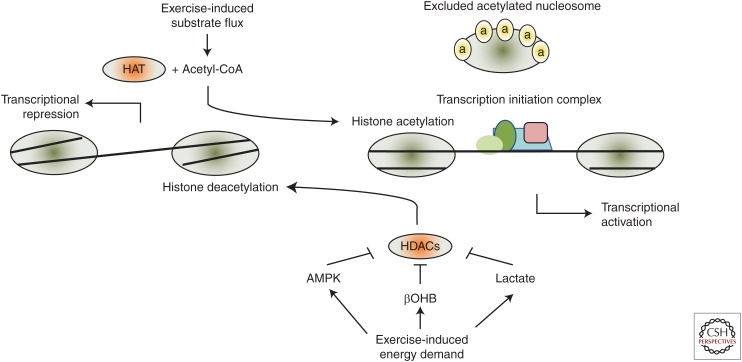
Histone proteins form the nucleosome core, which comprises an octamer of two copies each of histone 2A, 2B, 3, and 4 (Li et al. 2007). The double-stranded DNA helix wraps around this stable core, which provides structural stability to DNA and has also been hypothesized to protect DNA from damaging agents and stimuli (Ljungman and Hanawalt 1992). The physical relationship between DNA and surrounding histone proteins determines how surrounding genes are expressed, with a closed chromatin formation favoring transcriptional repression while an open chromatin structure favors transcriptional activation (Fig. 2) (Li et al. 2007). Functionally, this is linked to the ability of the transcriptional machinery, including the basal transcription factors and RNA polymerase, to gain access to gene regulatory regions to initiate transcription (Kouzarides 2007). A closed or condensed chromatin structure is the result of strong electrostatic interactions between the negatively charged DNA backbone and positively charged histone proteins. Alterations in histone protein charge through posttranslational modifications are often sufficient to disrupt the interaction with DNA, resulting in an open chromatin confirmation (Kouzarides 2007).
Figure 2.
Histone acetylation, transcription, and potential interactions with exercise metabolism. In their deacetylated state, catalyzed by histone deacetylase (HDAC) enzymes, histones retain a close interaction with DNA, resulting a compacted chromatin structure that is associated with transcriptional repression. Increased histone acetyltransferase (HAT) activity and acetyl-CoA availability increase histone acetylation, resulting in a loose chromatin structure that can result in transient nucleosome exclusion and exposure of regulatory DNA regions to transcriptional activators such as the transcriptional initiation complex. Inhibition of HDAC activity has similar effects. Exercise metabolism could regulate this system through multiple mechanisms, including by influencing acetyl-CoA availability and through regulation of factors that inhibit HDAC activity. These include increased AMP-activated protein kinase (AMPK) activity and increased concentrations of the metabolites lactate and β-hydroxybutyrate (βOHB).
Although not exhaustive, characterized posttranslational histone modifications on a number of amino acid sites include phosphorylation, acetylation, methylation, ubiquitination, sumoylation, and ADP-ribosylation (Bannister and Kouzarides 2011). A wide range of regulatory enzymes, the activity of which serves as a control nexus that links cellular signaling pathways with epigenetic control of gene expression, governs these posttranslational histone modifications (Feinberg et al. 2016). One of the best-characterized histone modifications is lysine acetylation, particularly to histones 3 (H3) and 4 (H4). Lysine amino acid (K) side-chains carry a positive charge when unmodified; however, acetylation neutralizes this positive charge, thereby disrupting the electrostatic interaction with DNA (Ettig et al. 2011). This favors an open chromatin confirmation and transcriptional activation. Indeed, K9 and K14 acetylation on H3 is associated with transcriptional activation (Fig. 2). Histone acetylation state is determined by the balance between histone acetyltransferase (HAT) and histone deacetylase (HDAC) activities (Saha and Pahan 2006). There are numerous HAT and HDAC isoforms, each with different substrate specificities, signaling pathway responsiveness and gene regulatory functions. These epigenetic modifiers are often recruited to specific nucleosome sites through interactions with DNA-bound transcription factors, with various HDAC and HAT isoforms having different affinities for different transcription factors (Kouzarides 2007). This diversity allows control of histone acetylation at specific loci, ensuring specificity in the gene expression response. Histone acetylation can work in concert with other histone modifications to provide precise control over the transcriptional response (Bannister and Kouzarides 2011). For example, acetylation of K14 on H3 appears dependent on prior phosphorylation of serine 10 (Lo et al. 2000). Furthermore, H3 acetylation is often coupled with H3 methylation at K36, which appears to control transcriptional elongation (Wagner and Carpenter 2012). Such interactions have coined the term “the histone code” to describe broader regulatory modifications controlling specific gene expression patterns. However, adding further complexity to this notion is the fact that subtle variation in modifications to the same histone site can confer opposing effects on gene transcription. For example, methylation of H3 at K9 has the opposite effect to acetylation, inducing transcriptional repression through inhibition of the p300 HAT (Stewart et al. 2005). These studies highlight the complexity in the “histone code” concept and how it might control context-specific gene expression responses. For a full review on details of other histone posttranslational modifications, their modifying enzymes, the “histone code” concept, and how these modifications interact, readers are encouraged to consult the following excellent reviews: Bannister and Kouzarides (2011), Carone and Rando (2012), and Rando (2012).
The following sections will detail the emerging data describing the epigenetics of exercise, which at present is principally described through alterations in DNA methylation and histone acetylation.
SKELETAL MUSCLE DNA METHYLATION AND REGULATING ENZYMES IN RESPONSE TO EXERCISE
There are a limited number of studies that have examined the effects of exercise on DNA methylation in skeletal muscle. The available data suggest that acute exercise causes reduced methylation in skeletal muscle in a gene-specific manner. For example, Barres et al. (2012) showed that high-intensity exercise (80% VO2 max) caused an immediate reduction in the methylation levels in the promoter regions of PGC1α, PDK4, and PPARδ in human skeletal muscle, which was associated with increased transcription of these genes. Interestingly, the same exercise modality at a lower intensity (40% VO2 max) had no effect on DNA methylation, suggesting that exercise intensity may be a critical factor in influencing DNA methylation. A recent study in mice has replicated some of these findings, showing that exercise reduced PGC1α promoter methylation, which was associated with increased basal transcription of this gene (Lochmann et al. 2015). Another study has found that 120 min of cycling in healthy subjects (60% VO2 max) increased methylation at the promoters of FABP3 and COX4L1, which was associated with decreased expression of these genes (Lane et al. 2015).
These studies show that alterations in DNA methylation can occur more rapidly than previously thought and clearly indicate that DNA methylation may be a crucial component of the immediate physiological response to an acute bout of exercise. The mechanism of alterations in DNA methylation patterns in skeletal muscle following a bout of exercise are unknown, but are likely to involve precise regulation of the DNMTs and the enzymes involved in demethylation of DNA. Although little is known in this area, in vitro knockdown of DNMT3B was shown to prevent saturated fatty acid-induced methylation of the promoter of PGC1α in a muscle cell line (Barres et al. 2009), and TET1 was induced by hypoxia and associated with the regulation of glycolytic gene expression in various cell lines (Tsai et al. 2014). More research is required to understand the role of DNA methylation and demethylation in the adaptive response to acute exercise, but it appears clear that this system is likely to play a role. Of particular interest will be an understanding of whether such changes are retained over time, and how rapidly they are lost before a further bout of exercise is required to have the same beneficial effect.
There is some evidence in the literature describing the effects of exercise training on DNA methylation. Nitert et al. (2012) studied first-degree relatives of patients with T2D and found altered DNA methylation in skeletal muscle in pathways, including MAPK, insulin, and calcium signaling, and in several genes known to have metabolic functions, including PRKAB1. After a 6-month exercise intervention in these healthy but sedentary subjects, which consisted primarily of endurance exercise, decreased DNA methylation in skeletal muscle was observed for retinol metabolism and calcium signaling pathways and a number of genes with known metabolic function such as MEF2A and NDUFC2 (Nitert et al. 2012). The reduced DNA methylation was associated with increased gene expression in many pathways, but particularly those associated with mitochondrial function. Furthermore, Alibegovic et al. (2010) showed that 9 days of enforced bed rest tended to increase DNA methylation in the promoter region of PPARGC1A, a key regulator of mitochondrial biogenesis and function, and that this was associated with down-regulation of many genes involved in mitochondrial function and particularly oxidative phosphorylation. When the same subjects underwent 4 weeks of exercise retraining (supervised cycle ergometry at 70% of VO2 max), DNA methylation in the PPARGC1A promoter tended to decrease (Alibegovic et al. 2010).
More research is required to understand the role of DNA methylation and demethylation in the adaptive response to exercise, but it appears clear that this system is likely to play a prominent role. Of particular interest will be an understanding of whether such changes are retained over time, and how rapidly they are lost before a further bout of exercise is required to have the same beneficial effect.
SKELETAL MUSCLE HISTONE MODIFICATIONS AND REGULATING ENZYMES IN RESPONSE TO EXERCISE
Just as histone acetylation is the most widely studied histone modification in epigenetics, it is one of the few histone modifications that have been examined in the context of responses to exercise. We have previously examined skeletal muscle global H3 K9/14 and K36 acetylation, which are associated with transcriptional initiation and elongation, respectively, in response to 60 min of cycling in humans (McGee et al. 2009). This study showed that H3 K36 acetylation was increased immediately after exercise, while there was no change in H3 K9/14 acetylation. The physiological relevance of alterations in global acetylation is not entirely clear given that acetylation at specific gene loci are most closely associated with gene-specific transcription. However, other studies have examined histone acetylation at specific gene loci in response to exercise. For example, repeated bouts of swimming in rats increased H3 K9/14 acetylation at the GLUT4 promoter in triceps muscle (Smith et al. 2008). Further understanding of the role of chromatin remodeling in the regulation of exercise adaptations will require future studies to examine multiple histone modifications with approaches such as ChIP-seq. As these technologies are becoming more accessible, so too are the opportunities to decipher the molecular regulation of muscle adaptation.
Histone modifications are controlled by a diverse number of enzymes that are also regulated by various signaling mechanisms. These include HATs and HDACs, histone methyltransferases (HMTs) and demethylases (HDMs), and histone kinases and histone ligases (Bannister and Kouzarides 2011), to name a few. Many of these chromatin remodeling enzymes exist as larger multipeptide transcriptional regulatory complexes and control of their activity is regulated through complex cofactor exchanges, localization, and posttranslational modifications (Bannister and Kouzarides 2011). Just as histone acetylation has served as a starting point for understanding how epigenetic modifications might influence skeletal muscle adaptation to exercise, a number of studies have also examined the regulation of chromatin remodeling enzymes that control histone acetylation in response to exercise. For example, we have previously found that 60 min of cycling exercise in humans reduces the nuclear abundance of the class IIa HDACs in skeletal muscle (McGee and Hargreaves 2004; McGee et al. 2009), which has been implicated in the regulation of muscle-specific genes via repression of the MEF2 transcription factor (McKinsey et al. 2000). This is consistent with phosphorylation-dependent nuclear export of the class IIa HDACs being a major mechanism controlling class IIa HDAC repressive activity. Once exported from the nucleus, MEF2 is derepressed, which activates MEF2-dependent transcription (McKinsey et al. 2000). An interesting aspect of class IIa HDAC biology is that they do not possess intrinsic deacetylase activity themselves, but instead require a corepressor complex that includes N-Cor/SMRT and HDAC3 (Fischle et al. 2002). A reduction in HDAC activity and an increase in HAT activity has also been observed in the hippocampus of rats following a single session of treadmill exercise (Elsner et al. 2011).
Perhaps the most obvious link between exercise-regulated signal transduction and epigenetic regulation comes from the kinases that are able to phosphorylate the class IIa HDACs, resulting in class IIa HDAC nuclear export. We found that the exercise-responsive AMP-activated protein kinase (AMPK) is a class IIa HDAC kinase. Phosphorylation of HDAC5 by AMPK was also shown to increase H3 K9/14 acetylation and transcription of the GLUT4 gene (McGee et al. 2008). The importance of this signaling nexus to exercise adaptations was highlighted by our identification of redundancy and compensation in class IIa regulation in the absence of AMPK signaling (McGee et al. 2014). Indeed, a number of other exercise-responsive kinases are also class IIa HDAC kinases, such as the calcium/calmodulin dependent protein kinase II (CaMKII) (Rose and Hargreaves 2003; Backs et al. 2006). Through the use of the KN93 inhibitor, CaMKII has been implicated in the acetylation of H3 at the GLUT4 promoter during swimming exercise in rats (Smith et al. 2008). Another interesting link between these exercise-responsive kinases and regulation of histone modifications is that many of these same kinases are also H3 kinases (McGee and Hargreaves 2008; Bungard et al. 2010). This is particularly intriguing given that phosphorylation of H3 at S10 is required before acetylation of K9 and 14 can occur in a number of different cellular contexts (Cheung et al. 2000). This suggests a coordinated signaling mechanism involving multiple histone modifications to regulate transcriptional responses.
Another link between exercise and epigenetic regulation comes from the class III HDACs, termed sirtuins. Unlike the class I and II HDACs that are Zn2+-dependent, the Sirtuin family are dependent on NAD+ for full activity, making this group of HDACs exquisitely sensitive to alterations in cellular metabolism and redox state (Chang and Guarente 2014). There are seven identified sirtuins, each with different subcellular localizations and substrate specificities (Houtkooper et al. 2012). Once activated, the Sirtuins enhance a number of aspects of metabolism, including enhanced oxidative flux and capacity, by deacetylating both histone and nonhistone proteins (Houtkooper et al. 2012). The effects of exercise on various sirtuin isoforms are difficult to assess in activity assays, as allosteric activation by NAD+ is rapidly lost. However, a number of sirtuin targets are deacetylated during exercise and there is evidence that sirtuin activity during exercise is controlled by AMPK (Canto et al. 2009). Knockout of various sirtuin isoforms in mice has had limited effects on metabolic adaptations to exercise (Menzies et al. 2013), possibly indicating that further redundancy in this adaptive response exists. Whether sirtuin activation during exercise contributes to adaptive responses through direct histone deacetylation or through deacetylation of nonhistone proteins such as PGC-1α or Foxo1 (Canto et al. 2009) also remains to be established. Although no studies to our knowledge have examined whether exercise modulates other chromatin remodeling enzymes that control methylation, ubiquitination, etc., it would seem likely that there is a coordinated response to control exercise adaptions by regulating a number of chromatin remodeling enzymes. Indeed, just as the field of epigenetics is starting to unravel aspects of this “histone code” and how it regulates specific transcriptional responses, the field of exercise physiology will also have to design appropriate studies to unravel this code and its potential contribution to skeletal muscle adaptations to exercise.
EXERCISE-MEDIATED EPIGENETIC ALTERATIONS IN OTHER TISSUES
Transcriptional adaptive responses to exercise occur in a number of tissues in addition to skeletal muscle. For example, global gene expression alterations immediately following treadmill running in mice are more profound in the liver than in skeletal muscle (Hoene et al. 2010). Therefore, epigenetic mechanisms must also be engaged in nonmuscle tissues during exercise. Intriguingly, a number of studies have examined histone modifications in various brain regions in response to exercise. Forced swimming induces H3 K14 acetylation and serine 10 phosphorylation in spatially distinct regions of the dentate gyrus in a time dependent manner (Chandramohan et al. 2008). In addition, voluntary exercise in rats increases H3 acetylation at the brain-derived neurotrophic factor (BDNF) promoter in the hippocampus (Gomez-Pinilla et al. 2011). These studies were among the first to associate changes in histone modifications with alterations in exercise-induced gene expression. However, whether these modifications are a result of exercise per se or a psychological stress response to exercise remains unclear. There is a dearth of information on how epigenetic mechanisms might regulate gene expression programs in noncontracting tissues during exercise, although local alterations in energy balance, blood flow, and hypoxia are all candidate stimuli. Evidence is emerging that circulating factors could be involved in systemic regulation of epigenetic processes during exercise. For example, exercise-conditioned plasma reduced the nuclear abundance of DNMT3B in isolated peripheral blood mononuclear cells (Horsburgh et al. 2015). Although the exact circulating factors involved remain to be determined, a number of bona fide myokines that are released from contracting muscle during exercise have been identified (Whitham and Febbraio 2016). How such circulating factors might regulate epigenetic processes in response to exercise are only just emerging. However, other studies have noted altered DNA methylation in response to both endurance and resistance exercise in various blood cells (Denham et al. 2016; King-Himmelreich et al. 2016), further supporting the idea that circulating factors might be important. Interestingly, the genomic loci and genes regulated in blood cells by these different exercise modalities are divergent. This suggests that specific systemic signaling mechanisms are used to ensure an appropriate physiological response to the particular exercise modality.
SKELETAL MUSCLE METABOLISM AND EPIGENETIC INTERACTIONS AND THEIR POTENTIAL ROLE IN EXERCISE ADAPTIVE RESPONSES
Although epigenetic mechanisms regulate the expression of metabolic gene networks in response to exercise, there is emerging evidence that metabolism can influence epigenetic processes by controlling the availability of substrates used for epigenetic modifications and/or the activity of epigenetic modifiers. The major substrate for DNA methylation is S-adenosylmethionine (SAM), which is produced in the methionine-recycling pathway (Sauter et al. 2013). Changes in the cellular levels of SAM result in altered levels of DNA methylation, with concomitant effects on gene transcription (Ables et al. 2016). Ultimately, the methyl groups required for DNA methylation are derived from a range of dietary sources, and altering the availability of dietary methyl donors such as folate and selenium has been shown to affect DNA methylation and gene expression, including improving diseases such as neural tube defects associated with DNA hypomethylation (Chango and Pogribny 2015). Although there are numerous studies that have investigated the impact of increasing or decreasing dietary methionine, which has impacts including extending life span in a number of species following dietary methionine restriction (Ables et al. 2016), there is no information regarding whether any of the phenotypic differences seen may be mediated by altered levels of DNA methylation. Similarly, the effects of exercise on the methionine-recycling pathway are currently unknown. Although ATP is required for the conversion of SAM to methionine (Sauter et al. 2013), it is difficult to conceive that the ATP demand during exercise could acutely compromise methionine availability and methylation reactions. In contrast, the activity of the demethylating TET enzymes is increased by the tricarboxylic acid (TCA) cycle intermediate metabolite α-ketoglutarate, whereas accumulation of metabolites with similar structural features, such as succinate, fumurate, and 2-hydroxyglutarate inhibit TET activity (Figueroa et al. 2010). Although the effect of exercise on the abundance of these metabolites is unclear, particularly in cellular compartments relevant to TET regulation, this could suggest that metabolite fluxes are critical for coordinating skeletal muscle DNA-methylation responses to exercise.
The availability of the two-carbon metabolite acetyl-CoA can also be rate limiting for acetylation reactions (Galdieri and Vancura 2012). Acetyl-CoA is principally derived from pyruvate oxidation and through β-oxidation of fatty acids, both of which occur in the mitochondria. However, acetyl-CoA is unable to diffuse across phospholipid membranes and mitochondrial citrate is exported from the TCA cycle via the citrate transporter to the cytosol and the nucleus. Nucleocytosolic citrate can then be converted to back to acetyl-CoA by ATP-citrate lyase (ACL) and used in acetylation reactions of histone and nonhistone proteins (Choudhary et al. 2014). This pathway appears to be particularly important for linking metabolic flux with metabolic gene expression. Evidence for this comes from experiments in which increases in the expression of genes such as GLUT4, HK2, PFK-1, and LDH in response to increasing glucose concentrations are abrogated with ACL knockdown (Wellen et al. 2009). At present, it is unknown how the energetic demands of exercise might influence histone acetylation secondary to increased TCA cycle flux. Given that exercise increases skeletal muscle acetyl-CoA levels via increased substrate catabolism (Constantin-Teodosiu et al. 1991), it is tempting to speculate that an increased acetyl-CoA pool would contribute to increased skeletal muscle histone acetylation in concert with altered activity/function of HDACs and HATs to enhance the transcription of specific genes.
In addition to substrate availability directly controlling epigenetic modifications, a number of metabolites are known to regulate the activity of histone modifiers. Indeed, lactate has been observed to inhibit HDACs in the context of cancer (Latham et al. 2012). It is unknown whether lactate accumulation during intense exercise contributes to the increased local histone acetylation that has been observed during endurance-based exercise. Given that lactate concentrations of 10–50 mM are required to reduce total HDAC activity ∼50%–80% (Latham et al. 2012), it appears unlikely that lactate is a key regulator of HDAC activity during predominantly aerobic endurance exercise. However, during intense supramaximal exercise, muscle lactate concentrations can reach ∼60 mM (Jacobs et al. 1983). This suggests that lactate production during intense exercise could acutely reduce HDAC activity and contribute to epigenetic regulation in addition to canonical signaling pathways that control epigenetic modifiers, which are activated during supramaximal exercise (Gibala et al. 2009). Ketone bodies such as βOHB can also inhibit HDAC activity (Shimazu et al. 2013). Ketone bodies are principally generated in the liver in response to elevated rates of β-oxidation and plasma βOHB increases during prolonged endurance exercise (Bordin et al. 1992). Circulating βOHB is taken up by a number of tissues, particularly the brain, where it can be converted back to acetyl-CoA for oxidation. Whether increased systemic βOHB contributes to exercise-mediated transcriptional regulation in nonmuscle tissues, through direct regulation of HDACs or through generation of acetyl-CoA for acetylation reactions, remains to be determined. However, altered gene expression is a well-characterized response to exercise in noncontracting tissues (Hoene and Weigert 2010; Catoire et al. 2012) and is thought to contribute to the many whole body positive health effects of exercise.
Collectively, these studies indicate that altered metabolism during exercise could have a profound impact on the epigenome (Figs. 1 and 2). Future studies will be required to mechanistically determine whether this is the case.
EPIGENETIC THERAPIES TO INDUCE EXERCISE ADAPTATIONS
As the epigenetics of exercise becomes better understood, opportunities are presented to experimentally manipulate aspects of the exercise adaptive response. Clearly, this has many potential therapeutic implications for managing chronic diseases underpinned by inactivity, which include obesity, type 2 diabetes, and cardiovascular and liver diseases.
Following from our observations that the repressive actions of the class IIa HDACs are reduced by exercise (McGee and Hargreaves 2004; McGee et al. 2009), as signified by disruption of the class II HDAC corepressor complex and their nuclear export, we have recently examined whether manipulating class IIa HDAC activity could induce exercise adaptations in skeletal muscle. Genetic manipulation of HDAC4 and 5, so that they could not form a corepressor complex, increased MEF2 transcriptional activity, the expression of a program of exercise-responsive metabolic genes and fatty acid oxidation (Gaur et al. 2016). We next identified an existing HDAC inhibitor, termed scriptaid, which was also able to disrupt the class IIa HDAC corepressor complex. Acute administration of scriptaid to mice increased the expression of exercise-responsive metabolic genes in skeletal muscle, whereas chronic administration increased exercise performance, whole-body energy expenditure, and lipid oxidation and reduced plasma glucose and lipids (Gaur et al. 2016). A number of other studies using various HDAC inhibitors have reported similar enhancements in various aspects of muscle and whole-body metabolism, including insulin sensitivity (Tan et al. 2015) and enhanced oxidative capacity (Galmozzi et al. 2013). The exact mechanism of action of some of these broad-spectrum HDAC inhibitors remains to be determined, but could include inhibition of HDAC3, the class I HDAC that provides deacetylase activity to the class IIa HDAC corepressor complex (Fischle et al. 2002; Hudson et al. 2015). Systemic administration of these inhibitors could have compound-specific effects in tissues other than skeletal muscle. Although this could raise concerns about off-target effects, the increase in histone acetylation in other tissues during exercise, such as in the brain (Chandramohan et al. 2008; Gomez-Pinilla et al. 2011), indicates that some of these effects might not be considered off-target. Furthermore, many positive benefits might be derived from systemic HDAC inhibition. For example, HDAC inhibitors have been used to prevent pathological cardiac hypertrophy (Kong et al. 2006), psychiatric diseases (Meylan et al. 2016), and cancer (Falkenberg and Johnstone 2014) in preclinical and clinical studies. Indeed, the wide-ranging positive benefits of HDAC inhibition are consistent with these enzymes as important regulators of the exercise adaptive response in a number of tissues.
These studies highlight the value in understanding the epigenetic processes regulating skeletal muscle adaptations to exercise. As the field continues to progress and defines the roles of other epigenetic modifiers such as HATs, HMTs, and HMDs and ubiquitin ligases in the exercise adaptive response, there will be more opportunities for therapeutic intervention in chronic diseases driven by inactivity. Given the huge burden imparted by such diseases on modern societies, such research should be a priority for the field.
CONCLUDING REMARKS
Epigenetic regulation of gene expression is a major control point for gene expression responses across all organisms in response to environmental stimuli. Methylation of DNA and posttranslational modification of histones are the major epigenetic control mechanisms that have been characterized to date under experimental conditions. The interaction between DNA methylation and the various histone modifications at specific gene loci is only just beginning to be understood; however, it is expected that these complex spatially and temporally dependent interactions will become clearer as the technology to probe these interactions becomes more accessible.
At present, very little is known about the epigenetics that contribute to transcriptional responses and phenotypic adaptations to exercise. Early studies have established roles for acute reductions in DNA methylation and increases in histone acetylation as contributors to this response. Additional studies also implicate regulation of the class IIa HDACs as being important in exercise adaptations. Another emerging area of research will be how alterations in muscle metabolism impact on epigenetics and exercise adaptations, given that a number of metabolites are known to be rate limiting for epigenetic modification reactions or directly regulate the activity of epigenetic modifiers. Finally, an exciting area for future research will be whether our understanding of the epigenetics of exercise can be leveraged to provide therapeutic targets and ultimately therapies for diseases underpinned by inactivity, such as obesity, type 2 diabetes, and cardiovascular diseases. Future studies should systematically examine the effects of exercise modality, intensity, and duration when examining interactions between epigenetic mechanisms to provide an exercise-epigenetic roadmap. Incorporating new omics technologies, such as unbiased proteomics encompassing posttranslational modifications, to existing epigenetic pipelines will make this a realistic prospect that could have wide-ranging implications for understanding the health benefits of exercise that could be harnessed for medicine.
ACKNOWLEDGMENTS
The authors thank Prof. Mark Hargreaves (The University of Melbourne) and members of their laboratories for ongoing discussions on the topic of this review. The authors are supported by grants from the National Health and Medical Research Council (NHMRC), the Diabetes Australia Research Program (DARP), the National Breast Cancer Foundation (NBCF), and the Centre for Molecular and Medical Research (C-MMR) at Deakin University.
Footnotes
Editors: Juleen R. Zierath, Michael J. Joyner, and John A. Hawley
Additional Perspectives on The Biology of Exercise available at www.perspectivesinmedicine.org
REFERENCES
- Ables GP, Hens JR, Nichenametla SN. 2016. Methionine restriction beyond life-span extension. Ann NY Acad Sci 1363: 68–79. [DOI] [PubMed] [Google Scholar]
- Alibegovic AC, Sonne MP, Hojbjerre L, Bork-Jensen J, Jacobsen S, Nilsson E, Faerch K, Hiscock N, Mortensen B, Friedrichsen M, et al. 2010. Insulin resistance induced by physical inactivity is associated with multiple transcriptional changes in skeletal muscle in young men. Am J Physiol Endocrinol Metab 299: E752–E763. [DOI] [PubMed] [Google Scholar]
- Amaral CL, Crisma AR, Masi LN, Martins AR, Hirabara SM, Curi R. 2014. DNA methylation changes induced by a high-fat diet and fish oil supplementation in the skeletal muscle of mice. J Nutrigenet Nutrigenomics 7: 314–326. [DOI] [PubMed] [Google Scholar]
- Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. 2006. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest 116: 1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR. 2009. Non-CpG methylation of the PGC-1α promoter through DNMT3B controls mitochondrial density. Cell Metab 10: 189–198. [DOI] [PubMed] [Google Scholar]
- Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, Caidahl K, Krook A, O’Gorman DJ, Zierath JR. 2012. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab 15: 405–411. [DOI] [PubMed] [Google Scholar]
- Bedi U, Mishra VK, Wasilewski D, Scheel C, Johnsen SA. 2014. Epigenetic plasticity: A central regulator of epithelial-to-mesenchymal transition in cancer. Oncotarget 5: 2016–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. 2011. DNA demethylation dynamics. Cell 146: 866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordin D, Bottecchia D, Bettini V, Aragno R, Sartorelli L. 1992. Effect of middle-intensity exercise on carnitine and β-hydroxybutyrate plasmatic concentration in men and women. J Sports Med Phys Fitness 32: 394–399. [PubMed] [Google Scholar]
- Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL. 2010. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science 329: 1201–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. 2009. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458: 1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone BR, Rando OJ. 2012. Rewriting the epigenome. Cell 149: 1422–1423. [DOI] [PubMed] [Google Scholar]
- Catoire M, Mensink M, Boekschoten MV, Hangelbroek R, Muller M, Schrauwen P, Kersten S. 2012. Pronounced effects of acute endurance exercise on gene expression in resting and exercising human skeletal muscle. PLoS ONE 7: e51066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramohan Y, Droste SK, Arthur JS, Reul JM. 2008. The forced swimming-induced behavioural immobility response involves histone H3 phospho-acetylation and c-Fos induction in dentate gyrus granule neurons via activation of the N-methyl-d-aspartate/extracellular signal-regulated kinase/mitogen- and stress-activated kinase signalling pathway. Eur J Neurosci 27: 2701–2713. [DOI] [PubMed] [Google Scholar]
- Chang HC, Guarente L. 2014. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab 25: 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Zhang T, Zhang Z, Bao R, Fu C, Wang Z, Bao Y, Li Y, Wu L, Zheng X, et al. 2011. Tissue-specific distribution of aberrant DNA methylation associated with maternal low-folate status in human neural tube defects. J Nutr Biochem 22: 1172–1177. [DOI] [PubMed] [Google Scholar]
- Chango A, Pogribny IP. 2015. Considering maternal dietary modulators for epigenetic regulation and programming of the fetal epigenome. Nutrients 7: 2748–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell 5: 905–915. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. 2014. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol 15: 536–550. [DOI] [PubMed] [Google Scholar]
- Clausen JP, Trap-Jensen J. 1970. Effects of training on the distribution of cardiac output in patients with coronary artery disease. Circulation 42: 611–624. [DOI] [PubMed] [Google Scholar]
- Constantin-Teodosiu D, Carlin JI, Cederblad G, Harris RC, Hultman E. 1991. Acetyl group accumulation and pyruvate dehydrogenase activity in human muscle during incremental exercise. Acta Physiol Scand 143: 367–372. [DOI] [PubMed] [Google Scholar]
- Denham J, Marques FZ, Bruns EL, O’Brien BJ, Charchar FJ. 2016. Epigenetic changes in leukocytes after 8 weeks of resistance exercise training. Eur J Appl Physiol 116: 1245–1253. [DOI] [PubMed] [Google Scholar]
- Denis H, Ndlovu MN, Fuks F. 2011. Regulation of mammalian DNA methyltransferases: A route to new mechanisms. EMBO Rep 12: 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B, Zierath JR. 2013. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab 17: 162–184. [DOI] [PubMed] [Google Scholar]
- Elsner VR, Lovatel GA, Bertoldi K, Vanzella C, Santos FM, Spindler C, de Almeida EF, Nardin P, Siqueira IR. 2011. Effect of different exercise protocols on histone acetyltransferases and histone deacetylases activities in rat hippocampus. Neuroscience 192: 580–587. [DOI] [PubMed] [Google Scholar]
- Ettig R, Kepper N, Stehr R, Wedemann G, Rippe K. 2011. Dissecting DNA-histone interactions in the nucleosome by molecular dynamics simulations of DNA unwrapping. Biophys J 101: 1999–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg KJ, Johnstone RW. 2014. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 13: 673–691. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Koldobskiy MA, Gondor A. 2016. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 17: 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. 2002. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell 9: 45–57. [DOI] [PubMed] [Google Scholar]
- Galdieri L, Vancura A. 2012. Acetyl-CoA carboxylase regulates global histone acetylation. J Biol Chem 287: 23865–23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmozzi A, Mitro N, Ferrari A, Gers E, Gilardi F, Godio C, Cermenati G, Gualerzi A, Donetti E, Rotili D, et al. 2013. Inhibition of class I histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue. Diabetes 62: 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur V, Connor T, Sanigorski A, Martin SD, Bruce CR, Henstridge DC, Bond ST, McEwen KA, Kerr-Bayles L, Ashton TD, et al. 2016. Disruption of the class IIa HDAC corepressor complex increases energy expenditure and lipid oxidation. Cell Rep 16: 2802–2810. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP. 2010. Histone modifications, DNA methylation, and schizophrenia. Neurosci Biobehav Rev 34: 882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibala MJ, McGee SL, Garnham AP, Howlett KF, Snow RJ, Hargreaves M. 2009. Brief intense interval exercise activates AMPK and p38 MAPK signaling and increases the expression of PGC-1α in human skeletal muscle. J Appl Physiol 106: 929–934. [DOI] [PubMed] [Google Scholar]
- Glatz JF, Bonen A, Luiken JJ. 2002. Exercise and insulin increase muscle fatty acid uptake by recruiting putative fatty acid transporters to the sarcolemma. Curr Opin Clin Nutr Metab Care 5: 365–370. [DOI] [PubMed] [Google Scholar]
- Gomez-Pinilla F, Zhuang Y, Feng J, Ying Z, Fan G. 2011. Exercise impacts brain-derived neurotrophic factor plasticity by engaging mechanisms of epigenetic regulation. Eur J Neurosci 33: 383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidi T, Singh AK, Chen T. 2015. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics 7: 247–265. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Greally JM. 2016. Epigenetics, cellular memory and gene regulation. Curr Biol 26: R644–R648. [DOI] [PubMed] [Google Scholar]
- Hoene M, Weigert C. 2010. The stress response of the liver to physical exercise. Exerc Immunol Rev 16: 163–183. [PubMed] [Google Scholar]
- Hoene M, Franken H, Fritsche L, Lehmann R, Pohl AK, Haring HU, Zell A, Schleicher ED, Weigert C. 2010. Activation of the mitogen-activated protein kinase (MAPK) signalling pathway in the liver of mice is related to plasma glucose levels after acute exercise. Diabetologia 53: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Holloszy JO. 1967. Biochemical adaptations in muscle. Effects of exercise on mitochondrial oxygen uptake and respiratory enzyme activity in skeletal muscle. J Biol Chem 242: 2278–2282. [PubMed] [Google Scholar]
- Horsburgh S, Todryk S, Toms C, Moran CN, Ansley L. 2015. Exercise-conditioned plasma attenuates nuclear concentrations of DNA methyltransferase 3B in human peripheral blood mononuclear cells. Physiol Rep 3: e12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. 2012. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13: 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson GM, Watson PJ, Fairall L, Jamieson AG, Schwabe JW. 2015. Insights into the recruitment of class IIa histone deacetylases (HDACs) to the SMRT/NCoR transcriptional repression complex. J Biol Chem 290: 18237–18244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs I, Tesch PA, Bar-Or O, Karlsson J, Dotan R. 1983. Lactate in human skeletal muscle after 10 and 30 s of supramaximal exercise. J Appl Physiol Respir Environ Exerc Physiol 55: 365–367. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Sahoo T, Michaelis RC, Bercovich D, Bressler J, Kashork CD, Liu Q, Shaffer LG, Schroer RJ, Stockton DW, et al. 2004. A mixed epigenetic/genetic model for oligogenic inheritance of autism with a limited role for UBE3A. Am J Med Genet A 131: 1–10. [DOI] [PubMed] [Google Scholar]
- King-Himmelreich TS, Schramm S, Wolters MC, Schmetzer J, Moser CV, Knothe C, Resch E, Peil J, Geisslinger G, Niederberger E. 2016. The impact of endurance exercise on global and AMPK gene-specific DNA methylation. Biochem Biophys Res Commun 474: 284–290. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. 2006. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci 31: 89–97. [DOI] [PubMed] [Google Scholar]
- Kong Y, Tannous P, Lu G, Berenji K, Rothermel BA, Olson EN, Hill JA. 2006. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation 113: 2579–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Lane SC, Camera DM, Lassiter DG, Areta JL, Bird SR, Yeo WK, Jeacocke NA, Krook A, Zierath JR, Burke LM, et al. 2015. Effects of sleeping with reduced carbohydrate availability on acute training responses. J Appl Physiol 119: 643–655. [DOI] [PubMed] [Google Scholar]
- Latham T, Mackay L, Sproul D, Karim M, Culley J, Harrison DJ, Hayward L, Langridge-Smith P, Gilbert N, Ramsahoye BH. 2012. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res 40: 4794–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. 2007. The role of chromatin during transcription. Cell 128: 707–719. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman M, Hanawalt PC. 1992. Efficient protection against oxidative DNA damage in chromatin. Mol Carcinog 5: 264–269. [DOI] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell 5: 917–926. [DOI] [PubMed] [Google Scholar]
- Lochmann TL, Thomas RR, Bennett JP Jr., Taylor SM. 2015. Epigenetic modifications of the PGC-1α promoter during exercise induced expression in mice. PLoS ONE 10: e0129647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee SL, Hargreaves M. 2004. Exercise and myocyte enhancer factor 2 regulation in human skeletal muscle. Diabetes 53: 1208–1214. [DOI] [PubMed] [Google Scholar]
- McGee SL, Hargreaves M. 2008. AMPK and transcriptional regulation. Front Biosci 13: 3022–3033. [DOI] [PubMed] [Google Scholar]
- McGee SL, van Denderen BJ, Howlett KF, Mollica J, Schertzer JD, Kemp BE, Hargreaves M. 2008. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes 57: 860–867. [DOI] [PubMed] [Google Scholar]
- McGee SL, Fairlie E, Garnham AP, Hargreaves M. 2009. Exercise-induced histone modifications in human skeletal muscle. J Physiol 587: 5951–5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee SL, Swinton C, Morrison S, Gaur V, Campbell DE, Jorgensen SB, Kemp BE, Baar K, Steinberg GR, Hargreaves M. 2014. Compensatory regulation of HDAC5 in muscle maintains metabolic adaptive responses and metabolism in response to energetic stress. FASEB J 8: 3384–3395. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Lu J, Olson EN. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408: 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies KJ, Singh K, Saleem A, Hood DA. 2013. Sirtuin 1-mediated effects of exercise and resveratrol on mitochondrial biogenesis. J Biol Chem 288: 6968–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan EM, Halfon O, Magistretti PJ, Cardinaux JR. 2016. The HDAC inhibitor SAHA improves depressive-like behavior of CRTC1-deficient mice: Possible relevance for treatment-resistant depression. Neuropharmacology 107: 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader GA, von Walden F, Liu C, Lindvall J, Gutmann L, Pistilli EE, Gordon PM. 2014. Resistance exercise training modulates acute gene expression during human skeletal muscle hypertrophy. J Appl Physiol 116: 693–702. [DOI] [PubMed] [Google Scholar]
- Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. 2006. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics 1: e1–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao M. 2001. Epigenetics: Interaction of DNA methylation and chromatin. Gene 278: 25–31. [DOI] [PubMed] [Google Scholar]
- Nitert MD, Dayeh T, Volkov P, Elgzyri T, Hall E, Nilsson E, Yang BT, Lang S, Parikh H, Wessman Y, et al. 2012. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes 61: 3322–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ. 2012. Combinatorial complexity in chromatin structure and function: Revisiting the histone code. Curr Opin Genet Dev 22: 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD. 2005. DNA methylation and human disease. Nat Rev Genet 6: 597–610. [DOI] [PubMed] [Google Scholar]
- Rose AJ, Hargreaves M. 2003. Exercise increases Ca2+-calmodulin-dependent protein kinase II activity in human skeletal muscle. J Physiol 553: 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha RN, Pahan K. 2006. HATs and HDACs in neurodegeneration: A tale of disconcerted acetylation homeostasis. Cell Death Differ 13: 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M, Moffatt B, Saechao MC, Hell R, Wirtz M. 2013. Methionine salvage and S-adenosylmethionine: Essential links between sulfur, ethylene and polyamine biosynthesis. Biochem J 451: 145–154. [DOI] [PubMed] [Google Scholar]
- Shen H, Laird PW. 2013. Interplay between the cancer genome and epigenome. Cell 153: 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD, et al. 2013. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 339: 211–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Martinez GA, Rodriguez-Rios D, Alvarado-Caudillo Y, Vaquero A, Esteller M, Carmona FJ, Moran S, Nielsen FC, Wickstrom-Lindholm M, Wrobel K, et al. 2016. Arachidonic and oleic acid exert distinct effects on the DNA methylome. Epigenetics 11: 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Kohn TA, Chetty AK, Ojuka EO. 2008. CaMK activation during exercise is required for histone hyperacetylation and MEF2A binding at the MEF2 site on the Glut4 gene. Am J Physiol Endocrinol Metab 295: E698–E704. [DOI] [PubMed] [Google Scholar]
- Stewart MD, Li J, Wong J. 2005. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol 25: 2525–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. 2008. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet 9: 465–476. [DOI] [PubMed] [Google Scholar]
- Tan HW, Sim AY, Huang SL, Leng Y, Long YC. 2015. HC toxin (a HDAC inhibitor) enhances IRS1-Akt signalling and metabolism in mouse myotubes. J Mol Endocrinol 55: 197–207. [DOI] [PubMed] [Google Scholar]
- Tryndyak V, de Conti A, Doerge DR, Olson GR, Beland FA, Pogribny IP. 2016. Furan-induced transcriptomic and gene-specific DNA methylation changes in the livers of Fischer 344 rats in a 2-year carcinogenicity study. Arch Toxicol 10.1007/s00204-016-1786-8. [DOI] [PubMed] [Google Scholar]
- Tsai YP, Chen HF, Chen SY, Cheng WC, Wang HW, Shen ZJ, Song C, Teng SC, He C, Wu KJ. 2014. TET1 regulates hypoxia-induced epithelial-mesenchymal transition by acting as a co-activator. Genome Biol 15: 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisin S, Eynon N, Yan X, Bishop DJ. 2015. Exercise training and DNA methylation in humans. Acta Physiol (Oxf) 213: 39–59. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. 2012. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham M, Febbraio MA. 2016. The ever-expanding myokinome: Discovery challenges and therapeutic implications. Nat Rev Drug Discov 15: 719–729. [DOI] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. 2013. Charting a dynamic DNA methylation landscape of the human genome. Nature 500: 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]