In this study, Yang et al. find that the timing of Ccnb1 mRNA translation in mouse oocytes is dependent on the presence of transcripts with different 3′ UTRs. Their results reveal an additional layer of translation control through alternative polyadenylation usage required to fine-tune the timing of meiosis progression.
Keywords: APA, cyclins, meiosis, oocyte, translation
Abstract
The final stages of female gamete maturation occur in the virtual absence of transcription, with gene expression driven by a program of selective unmasking, translation, and degradation of maternal mRNAs. Here we demonstrate that the timing of Ccnb1 mRNA translation in mouse oocytes is dependent on the presence of transcripts with different 3′ untranslated regions (UTRs). This 3′ UTR heterogeneity directs distinct temporal patterns of translational activation or repression. Inclusion or exclusion of cis-acting elements is responsible for these divergent regulations. Our findings reveal an additional layer of translation control through alternative polyadenylation usage required to fine-tune the timing of meiosis progression.
A common property of female germ cells from most species is the initiation of meiosis during fetal life followed by a prolonged arrest in a specialized prophase termed dictyate (Eppig 1996; Gosden and Lee 2010). During puberty and follicle recruitment, oocytes enter a growth phase when they accumulate massive amounts of mRNAs; however, many of these maternal transcripts are stored without engaging in translation (Clarke 2012). Toward the end of this growth phase, oocytes become competent to re-enter the meiotic cell cycle. At this transition, oocytes of several species, including rodents, have assembled all of the machinery required for meiotic re-entry, including the metaphase-promoting factor (MPF) (see below), and are in a poised state ready to quickly re-enter meiosis (Eppig 1996). They will re-enter the cell cycle upon withdrawal of the inhibitory signal originating from somatic cells (Norris et al. 2009; Vaccari et al. 2009; Zhang et al. 2010b). In other species, a variable time interval is necessary from the hormonal trigger to meiotic re-entry and involves the synthesis of critical components necessary for meiotic progression, including Cyclins, the regulatory subunits of CDK1 kinase (Ferrell 1999). The complex of Cyclin B and CDK1 constitutes the MPF, the master regulator of both meiosis and mitosis (Morgan 2007). An inactive pre-MPF is assembled prior to meiotic re-entry through translational regulations.
It has long been known that synthesis of Cyclins is critical at different stages of meiotic progression, with synthesis required before germinal vesicle (GV) breakdown (GVBD) in those species where insufficient pre-MPFs have been assembled. In rodents, a stockpile of cyclins sufficient for meiotic re-entry is established during growth. Thus, Cyclin mRNA translation participates in the maintenance of the steady state of pre-MPFs prior to meiotic re-entry and during spindle assembly in prometaphase.
Extensive studies in Xenopus oocytes have demonstrated that cyclin synthesis is dependent on the activity of the RNA-binding protein cytoplasmic polyadenylation element (CPE) binding 1 (CPEB1) (Richter 2007). CPEB1 recognizes a CPE present in the 3′ untranslated region (UTR) of cyclin mRNAs and is found in complex with a scaffold protein (Symplekin) and a protein (CPSF) that binds to the polyadenylation signal (PAS) sequence (Ivshina et al. 2014). This complex recruits additional components that prevent poly(A) tail elongation. Under these conditions, CPEB1 association with an mRNA results in repression of translation. Upon re-entry into the cell cycle, CPEB1 becomes phosphorylated, leading to dissociation of the repressive components and the recruitment of factors that lead to mRNAs polyadenylation, ultimately increasing translation. This cascade of events has been investigated extensively in frog oocytes (Ivshina et al. 2014), and studies in rodents also indicate that CPEB1 is involved in translational activation of Cyclin B1 (Tay et al. 2000).
To explain the dual CPEB1 function and account for the presence of multiple CPEs in the 3′ UTR of cyclin mRNA, it has been proposed that a combinatorial code is functioning during Xenopus meiotic prophase as well as during maturation defining the timing of translational activation (Pique et al. 2008). In this posit, more than one CPE is required for CPEB function as a repressor, and the distance between two CPEs determines the level of repression. A CPE in close proximity to a PAS domain is critical for polyadenylation and translational activation (Pique et al. 2008), with the relative position of the CPEs dictating early or late translation during meiotic progression. Here we provide evidence that the temporal pattern of Ccnb1 mRNA translation in mouse oocytes is also dependent on the presence of multiple Ccnb1 mRNAs with 3′ UTRs of variable length. The differential effect of the 3′ UTRs on translational repression and activation defines the pattern of ribosome loading onto the Ccnb1 at different times during maturation and the consequent protein accumulation. Thus, an additional layer of complexity controls the translation of Ccnb1 in mammals, a property critical for proper progression through the cell cycle.
Results and Discussion
Although de novo protein synthesis is not required for the G2/M transition in rodent oocytes (Ferrell 1999), the steady-state levels of Ccnb1 and other cell cycle components are critical to maintain meiotic arrest (Holt et al. 2010). After re-entry into meiosis, de novo CCNB1 synthesis is thought to be indispensable for maintaining MPF activity, for chromosome condensation and spindle assembly, and for the additional steps leading to development of the oocyte into a fertilizable egg. We used a RiboTag strategy to investigate the pattern of ribosome loading onto maternal mRNA to gain insight into the translational regulations operating during mouse oocyte meiosis (Sousa Martins et al. 2016). While performing the quality controls of RNA sequencing (RNA-seq) data of transcripts expressed in mouse oocytes during maturation, we noted that the read coverage of the Ccnb1 transcript was unevenly distributed, with a consistent pattern along the ∼930 nucleotides (nt) of the 3′ UTR (Fig. 1A). Reads clustered around the boundary between the end of the coding region and the beginning of the 3′ UTR, with read density decreasing in the middle and end of the 3′ UTR. More importantly, reads from libraries monitoring ribosome association of the maternal mRNA (RiboTag immunoprecipitation) suggested a time dependence in their distribution, with reads close to the stop codon being more abundant in GV oocytes and coverage at the end of the 3′ UTR increasing with the progression through meiosis (Fig. 1A). Analysis of the GC content of this region could not explain the difference in sequencing efficiency (Love et al. 2016) or any other obvious property of the sequence, including the presence of possible cryptic splicing junctions that we monitored by PCR amplification with different primers. Thus, we explored the additional possibility that heterogeneity of the 3′ UTRs and the presence of multiple PASs are the cause of uneven read coverage.
Figure 1.
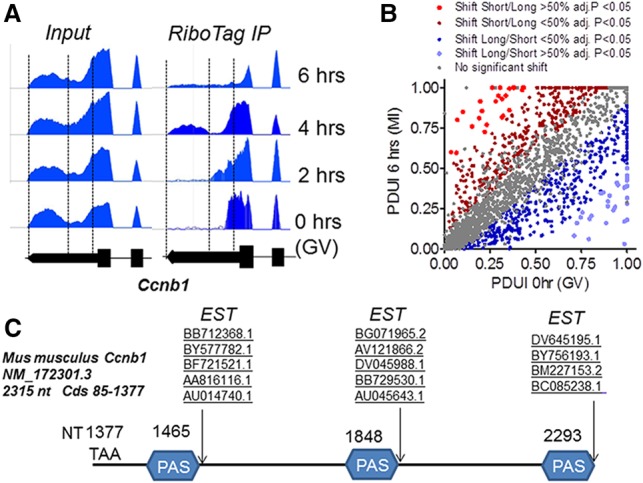
Analysis of the mouse Ccnb1 3′ UTR. (A) Snapshot of Integrated Genome Browser (IGB) mapping RNA-seq reads of input and RiboTag immunoprecipitation libraries to the Ccnb1 3′ UTR. Pools of ∼140 oocytes from RiboTagfl/fl-ZP3-CRE mice were isolated and cultured under maturing conditions for 0–6 h. After harvesting, an aliquot of the oocyte extract was used as input, and the rest was used for immunoprecipitation with HA antibodies. Input RNA and RNA recovered in the immunoprecipitation pellet were extracted and used for library preparation. Reads were mapped to the Ccnb1 locus using TopHat and visualized using IGB. Vertical lines mark the three potential PASs reported in C. (B) Analysis of the 3′ UTR of mRNA present in the oocyte using the dynamic analyses of alternative polyadenylation from the RNA-seq (DaPars) algorithm. The PDUI parameters were calculated as described in the Materials and Methods and plotted for 0–6 h, with different colors representing different statistical significance and direction of the shift in PAS usage. (C) The 3′ UTR of Ccnb1 (NM172301.3) was used to search for deposited expressed sequence tags (ESTs), and the boundary of hits is reported in the scheme. Together with a boundary located 19 nt from the last PAS and corresponding to the deposited sequence, additional boundaries at 19–23 nt from the first PAS and 10 nt from the second PAS were identified.
Given the presence of similar read patterns in many other 3′ UTRs, a second RiboTag immunoprecipitation/RNA-seq data set with duplicate biological observations at 0 h (GV) and 6 h after meiotic re-entry (prometaphase I) was analyzed. To broaden the analysis to all transcripts in the oocytes, we applied the dynamic analyses of alternative polyadenylation (APA) from RNA-seq (DaPars) algorithm (Xia et al. 2014) to search for the presence of proximal PASs in oocyte mRNAs in an unbiased fashion. In the context of RiboTag immunoprecipitation, we reasoned that this algorithm may provide information on the differential translation of mRNAs with 3′ UTRs of different lengths during oocyte maturation. This algorithm predicts that 2276 transcripts present in the oocyte contain a proximal PAS (Supplemental Table 1), a finding consistent with the tenet that a large number of mammalian transcripts undergo APA (Tian and Manley 2017). When the algorithm is applied to transcripts recovered in the RiboTag immunoprecipitation at 0 and 6 h, a significant shift (>10%) in 3′ UTR association with the ribosome was predicted in 798 transcripts (adjusted P < 0.05). No significant bias toward distal or proximal PAS preferential association with the ribosomes was noted with progression through meiosis (Supplemental Fig. 1B; Supplemental Table 1). Manual validation of the DaPars data revealed that the algorithm predicted the presence and use of a proximal PAS in the Ccnb1 mRNA as well a shift toward distal PAS usage during maturation (Supplemental Fig. S1), as observed with the first data set. A proximal PAS was also identified in Ccnb2 even though no significant shift in usage was detected (Supplemental Table 1). Other cell cycle components (including Pcna, Bub1b, and Wee2) or genes with other functions (including Eif2s2 or Ybx2/Msy2) behaved in a manner similar to Ccnb1, with the long 3′ UTR increasing in ribosome association at 6 h. Ccno and Ppt2 are instead representative of a group of transcripts in which association with ribosomes shifts from the distal to the proximal PAS during maturation (Supplemental Fig. S1). Other cell cycle-encoding transcripts, including c-Mos and Cdc20, were not identified as bearing a proximal PAS in the 3′ UTR. Of note, a group of transcripts, including that coding for Cdk, is expressed with a short 3′ UTR in the oocytes different from those deposited. Thus, this global analysis predicts the presence and differential ribosome association of mRNAs with multiple PASs in the oocyte.
Next, we further characterized the properties of the Ccnb1 mRNA as a prototypic transcript with 3′ UTRs used differentially during maturation. The mouse Ccnb1 3′ UTR is considerably longer (938 nt) than that of Xenopus (86 nt) or zebrafish (210 nt). The beginning of the 3′ UTR is highly conserved among mammals, with some notable differences (see below). It then diverges toward the 3′ end (Supplemental Fig. S3). Mapping-deposited mouse expressed sequence tags (ESTs) to the Ccnb1 3′ UTR showed that many ESTs covered the 3′ UTR up to the end, consistent with the deposited RefSeq NM_172301.3 (Fig. 1C) in the proximity of a PAS consensus. However, several additional ESTs suggested the presence of two additional boundaries (Fig. 1C; Supplemental Fig. S2). These boundaries were confirmed by mapping the PAS motifs with at least two additional PASs closely corresponding to the boundaries identified with the EST mapping. Comparison of the mouse sequence with that of rats showed a complete conservation of the three PAS sequences (Supplemental Fig. S3). The location of the first PASs in mice was the only one conserved among humans, cows, monkeys, and pigs (Tremblay et al. 2005; Zhang et al. 2010a). Two additional PAS consensuses were also identified in the human Ccnb1 3′ UTR, although not in the same location as in mice. The presence of multiple PASs in the mouse Ccnb1 had not been noted during functional studies of 3′ UTR (Tay et al. 2000).
To define whether the three PASs are functional in mouse oocytes, an anchor PCR strategy was used (Fig. 2). RNA was extracted from oocytes at 0 h (GV), 2 h (GVBD), and 6 h (prometaphase) after re-entry into meiosis; reverse transcribed; and amplified with a primer in the coding sequence together with an oligo(dT) anchor primer. Three amplification products were clearly detected in GV oocytes, corresponding to ∼150, ∼550, and ∼985 nt, and their presence was confirmed by amplification with additional primers internal to the 3′ UTR (Fig. 2B). The mobility of these products was similar to that of amplicons generated with a 3′ UTR plasmid cDNA as template as well as primers located 5′ of the stop codon and 3′ to the PAS elements at the three EST boundaries (Fig. 2B; Supplemental Figs. S2, S3). The identity of the sequences amplified with anchored primers was further confirmed by subcloning and sequencing of the quantitative PCR (qPCR) fragments. Although no change or decrease in size was detected for the short form during maturation, the length of the two other amplified fragments increased during oocyte maturation, implying an increase in the poly(A) tail length (Fig. 2C). Thus, RNA-seq data, the genome-wide proximal PAS search, mining of deposited ESTs, and anchored PCR assays all consistently point to the presence of transcripts that include the Ccnb1-coding region and three 3′ UTRs of different lengths in the oocyte.
Figure 2.
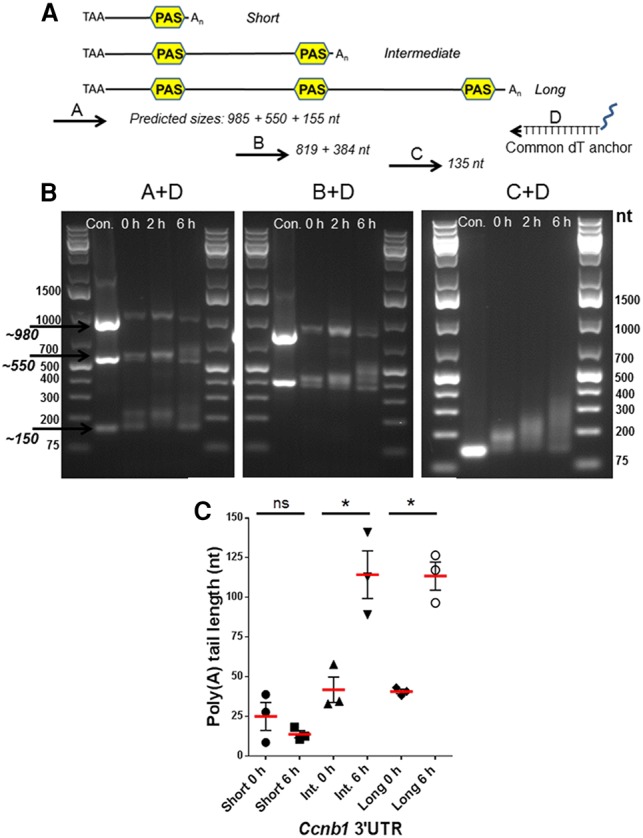
Anchored PCR of the 3′ UTR of Ccnb1 expressed during mouse oocyte maturation. (A) Scheme reporting the location of the primers used for anchored PCR on the Ccnb1 3′ UTR and sizes of predicted amplicons with different primer pairs. (B) RNA was extracted from oocytes incubated for 0, 2, and 6 h of maturation. Anchored PCR conditions were as detailed in the Materials and Methods. A representative experiment of the three performed is reported. In C, the increase in length of the amplified fragment was calculated using ImageJ. The mean ± SEM from the experiments is included in the graph.
Evidence of differential polyadenylation during oocyte maturation opens the possibility that the translation of the three transcripts may follow different time courses during meiotic progression. The CCNB1 protein is present at low levels in GV oocytes and during GVBD; it begins to increase at 2–3 h during prometaphase and by 8 h has reached levels sevenfold to eightfold higher than in GVs (Fig. 3B). To test how the different 3′ UTRs affect mRNA translation and protein accumulation, the short, intermediate, and long 3′ UTRs were fused to a Renilla luciferase reporter and injected into GV oocytes (Fig. 3C,D) together with a control firefly reporter. After a recovery period of 12–16 h, oocytes were allowed to re-enter meiosis, and reporter accumulation was measured at different time points through the meiotic cell cycle progression. The reporter driven by the short 3′ UTR was translated at a significantly higher rate in GV oocytes compared with the other two forms (Fig. 3C; Supplemental Fig. S5). However, no significant changes in the rate of translation directed by the short 3′ UTR were observed upon maturation, suggesting a constitutive translation independent of the cell cycle (Fig. 3D; Supplemental Figs. S4, S5). Conversely, the translation of the reporters driven by the intermediate and long forms increased significantly during maturation using two different experimental protocols (Fig. 3D; Supplemental Fig. S5). The time course of reporter accumulation driven by the intermediate 3′ UTR was slightly faster than the long 3′ UTR. These findings open the possibility that the overall pattern of CCNB1 synthesis during mouse oocyte meiotic re-entry is the outcome of combined constitutive as well as cell cycle-activated translations.
Figure 3.
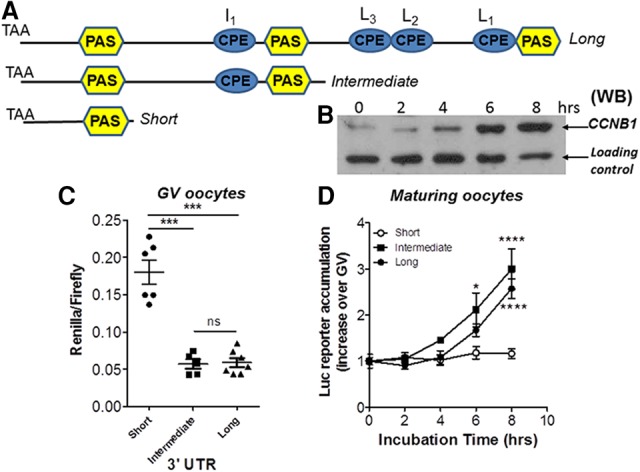
Differential translation of the three different 3′ UTRs present in mouse oocytes: exogenous reporter accumulation. (A) Schematic representation of the 3′ UTR present in the mouse Ccnb1 mRNA. PAS sequences are in yellow. Consensus CPEs are in cyan. (B) Western blot analysis of CCNB1 protein accumulation during oocyte maturation. (C) Reporter constructs with the Renilla luciferase coding region fused to the three 3′ UTRs was injected into GV oocytes together with firefly luciferase mRNA to control for the injection volume. After 12–14 h of recovery, the oocytes were harvested, and luciferase activity was measured as detailed in the Materials and Methods. The data are reported as the ratio Renilla/firefly luciferase, and each point represents a different biological replicate. The reporter accumulation of the short form was significantly different from that of the intermediate or long form. P = 0.0078 unpaired t-test with Welsh correction. (D) Oocytes were injected as in B. After recovery, the oocytes were allowed to mature to metaphase, and groups of oocytes were collected at different times and used for luciferase assay. The data are reported as ratio of luciferase activity at each time point over the activity in GVs. Each point is the mean ± SEM of four independent experiments for the short, three independent experiments for the intermediate, and five independent experiments for the long.
If the reporter accumulation mirrors the behavior of the endogenous transcripts, the ribosome loading onto the three endogenous species would be predicted to be different at the prophase-to-metaphase transition. To test this possibility, oocytes expressing the RiboTag were used to monitor ribosome loading onto the three transcripts. After immunoprecipitation, mRNA was extracted, and transcript amounts were calculated by the number of copies recovered in the immunoprecipitation pellet after subtraction of the nonspecific immunoprecipitation (Fig. 4). The concentration for each variant was also calculated by subtraction (see the Materials and Methods for details). Using this approach, we show that the short and long forms are the most abundant and that the former is the one recovered the most in the ribosome immunoprecipitation of GV oocytes (Fig. 4B). Its ribosome association increased up to threefold during maturation. Conversely, the recovery of the two other forms increased between 10-fold and 15-fold when comparing RiboTag immunoprecipitation at 6 h of maturation versus GV oocytes (Fig. 4C; Supplemental Fig. 6).
Figure 4.
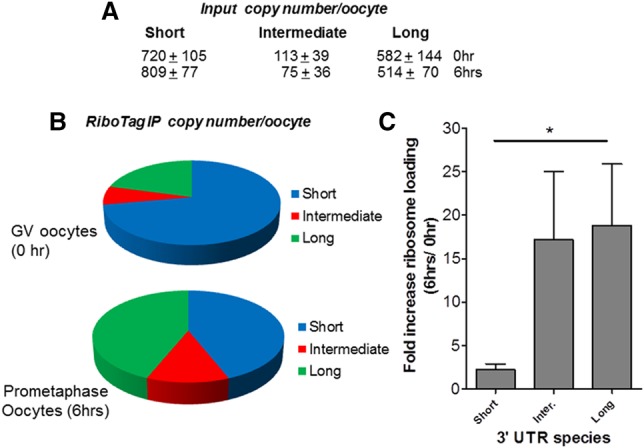
Differential translation of the three different Ccnb1 3′ UTRs present in mouse oocytes: ribosome loading of the endogenous mRNAs with the three 3′ UTRs. Pools of ∼150–200 oocytes from RiboTagfl/fl-ZP3-CRE mice were isolated and cultured under maturing conditions for 0 and 6 h. (A) At the end of the incubation, an aliquot of the oocyte extract was used as input, and the rest was used for immunoprecipitation with HA antibody or IgG. Input RNA and RNA recovered in the immunoprecipitation pellet were extracted and used for qPCR analysis using three sets of primers that amplify all 3′ UTRs, the intermediate and long 3′ UTRs, or the long 3′ UTR exclusively. Copy number per oocyte was calculated after correction of the data for the differences in primer efficiency as detailed in the Materials and Methods. The relative abundance of the three forms in GVs or after 6 h of maturation is reported in B. In C, bars indicate the fold increase at 6 h versus GVs for the three forms recovered in the ribosome immunoprecipitation pellet. P < 0.05 short versus long. n = 5.
Collectively, the above experiments strongly indicate a differential translational regulation of the three transcripts during maturation. Since the Ccnb1 translation is dependent on the RNA-binding protein CPEB1 expressed in the oocyte (Tay et al. 2000), we analyzed the contribution of the several CPEs present to the differential regulation of the three transcripts. Ccnb1 is recovered in the immunoprecipitation pellet after RNA immunoprecipitation (RIP)-qPCR with CPEB1 antibody (Chen et al. 2011), and the increased polyadenylation is consistent with a CPEB1-dependent translational regulation. Consensus CPEs were identified in the intermediate (I1) and long (L1, L2, L3) 3′ UTR forms but not in the short form (see Fig. 3A). Mutagenesis of each individual putative CPE as well as collective removal of all CPEs had divergent effects on reporter translation. Specifically, mutagenesis of the single CPE in the intermediate form caused an up-regulation of the translation driven by the intermediate 3′ UTR in GVs but completely prevented the translational activation of this form (Fig. 5C). Individual mutation of the CPE L1, L2, or L3 present in the unique long 3′ UTR did not significantly affect reporter accumulation in GV oocytes, and maximal accumulation was similar to that induced by the wild-type 3′ UTR. However, cumulative mutations of all three consensus CPEs caused a significant increase in translation of the reporter in GV oocytes. With these mutants, translation still increased with re-entry in the cell cycle even though it did not reach the levels of the long wild-type control (Fig. 5E). A complete absence of activated translation of the long form was achieved only when the additional CPE located upstream of the second PAS was also mutated (Fig. 5E). When I1 was the only CPE remaining in the long form, the timing of translational activation was accelerated, with a significant increase observed as early as 2 h (Fig. 5D). Thus, the CPE located upstream of the second PAS is critical for activation of the intermediate 3′ UTR but also contributes to the activation of the long 3′ UTR.
Figure 5.
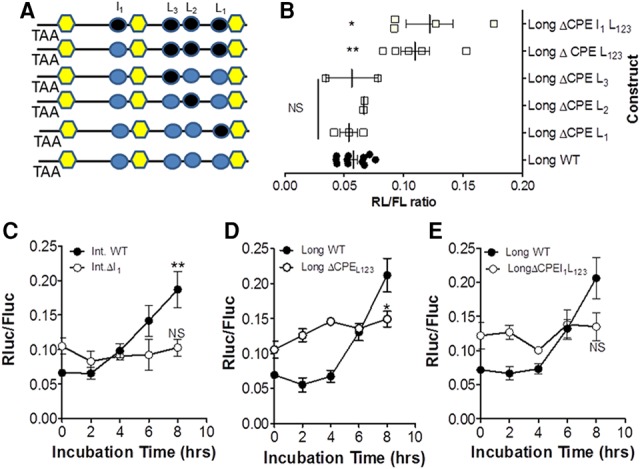
Translation of the Ccnb1 mRNAs with intermediate and long 3′ UTRs is dependent on the presence of CPEs: dual repressor/activation function. (A) Mutagenesis of CPEs was performed as detailed in the Materials and Methods. (Black) Mutated CPEs; (cyan) wild type. Constructs with the mutated CPEs or with wild-type 3′ UTRs were injected into oocytes, and incubation and luciferase assays were as described in Figure 3. (B) The effect of individual or cumulative CPE mutations on translation in GV oocytes. The basal levels of the CPEB1ΔL1,2,3 and CPEB1ΔI1,L1,2,3 are significantly different from wild type. P < 0.05. (C) Time course of reporter accumulation driven by wild-type intermediate 3′ UTRs or UTRs with mutation in the I1 CPE. (D) The effect of mutation of the three CPEs present in the long form. (E) The effect of mutation of the three CPEs present in the long 3′ UTRs as well as the CPE in the intermediate 3′ UTRs.
These CPE mutagenesis studies confirm the concept developed in Xenopus of a dual role of CPEB1 during oocyte maturation, functioning as a repressor of translation in quiescent oocytes and a translational activator during late prometaphase. Mutagenesis of the single CPE present in the intermediate form and the four CPEs present in the long form cause an increased accumulation of the reporter in GV oocytes, confirming a repressive role of these elements. Consistent with the finding in Xenopus, we show that repression of translation requires multiple CPE 3′ UTRs. The relief of repression when a single CPE upstream of the second PAS is mutated is inconsistent with the view that two or more CPEs are required for repression (Pique et al. 2008). However, we cannot exclude the possibility that additional nonconsensus or cryptic CPEs are functioning in the intermediate or short form. Also, we could not assess whether the CPE distance from the PAS element had a significant effect on translational activation. In Xenopus oocytes, an interaction between CPEB and CPSF is indispensable for the translational regulation of the reporter (Bava et al. 2013).
In summary, our findings indicate that alternative PAS usage in approximately one-third of the mRNAs present in GV oocytes generates transcripts with 3′ UTRs of different lengths. Bioinformatics analysis of the data generated after RiboTag immunoprecipitation is consistent with the view that ribosome association with a subgroup of transcripts with short and long 3′ UTRs switches during oocyte re-entry into the cell cycle. This observation is likely relevant to mRNA translational regulation during meiosis, as documented with prototypic cell cycle Ccnb1 mRNA. We show that additional layers of complexity in the translational regulation of Ccnb1 mRNA direct the accumulation of this protein essential for the meiotic cell cycle in mammals. Multiple PAS usage generates Ccnb1 transcripts with 3′ UTRs of different lengths. Given the presence of different regulatory elements in the three 3′ UTRs, these transcripts are repressed or translated at different rates during the prophase-to-metaphase transition, as indicated by the ribosome loading, reporter translation, and CPE mutagenesis. In quiescent GV oocytes, the accumulation of Ccnb1 protein is likely driven by the translation of the short form. The absence of an increase in polyadenylation, the constitutive translation of the reporter driven by this 3′ UTR, and the small changes in ribosome loading compared with the other forms during maturation are consistent with a cell cycle-independent translational rate. Currently, it is unknown what drives the constitutive translation of this form. In this context, it should be noted that a perfect Pumilio-binding element (PBE) is present in this short 3′ UTR and is conserved from rodents to humans. A modulatory role of PBE elements has been reported in Xenopus oocytes (Pique et al. 2008). Conversely, translation of the Ccnb1 mRNAs with intermediate and long 3′ UTRs is repressed in a CPEB1-dependent manner in GV oocytes. Upon re-entry into the cell cycle, the intermediate and long forms are polyadenylated, and ribosome loading increases, indicating activated translation and becoming significant contributors to Ccnb1 synthesis during the prometaphase progression. Thus, the combinatorial code described in frog oocytes is enriched by an additional layer of regulation driven by APA. We did not investigate the mechanisms underlying the differential PAS usage, which is likely determined during oocyte growth, when transcription is still active. We note that CPEB1 itself has been implicated in APA in a Hodgkin's lymphoma-derived cell line (Bava et al. 2013). Thus, it is possible that CPEB1 is also involved in the differential cleavage of the Ccnb1 mRNA during oocyte development.
The presence of multiple 3′ UTRs driving Ccnb1 mRNA translation is also likely important during mitosis, as our preliminary anchor PCR shows the presence of multiple 3′ UTRs also in mouse embryonic stem cells (Supplemental Fig. 7). In addition, the presence of multiple 3′ UTRs has already been noted in bovine (Tremblay et al. 2005) and porcine ((Zhang et al. 2010a) oocytes, and we show here that multiple PAS elements are also found in the human Ccnb1 3′ UTR, suggesting that regulatory mechanisms that we described are functioning in all mammalian species. Last, it has been proposed that Ccnb1 mRNA is located in a P-granule compartment in the quiescent state in zebrafish and mouse oocytes and that its translation is activated after release from this compartment (Kotani et al. 2013). Thus, it is possible that the different 3′ UTRs contribute to the subcellular targeting and the differential regulation of the Ccnb1 mRNA species, warranting further investigation of the localization of these variants in mice.
Materials and methods
Mice and oocyte collection
All experimental procedures involving the mouse genetic models used were approved by the Institutional Animal Care and Use Committee of the University of California at San Francisco (protocol AN101432). C57BL/6 female mice (22–24 d old) were used in the experiments. Oocyte collection and in vitro maturation were performed following the protocol described previously (Chen et al. 2011). Generation of Zp3cre-RiboTag mice was performed as described (Sousa Martins et al. 2016) using a CRE recombinase driven by the oocyte-specific ZP3 promoter (de Vries et al. 2000).
Oocyte culture and microinjection
Oocyte isolation and microinjection were performed in HEPES modified minimum essential medium Eagle (Sigma-Aldrich, M2645) supplemented with 2 µM milrinone. Oocytes were cultured in supplemented MEM-α (Gibco, 12561-056) containing 0.2 mM pyruvate, 75 µg/mL penicillin, 10 µg/mL streptomycin, and 3 mg/mL BSA at 37°C in 5% CO2. Denuded oocytes were injected with 5–10 pL of 12.5 ng/µL RNA reporter using a FemtoJet Express programmable microinjector. After injection, oocytes were incubated overnight (10–16 h) in supplemented MEM-α with 2 µM milrinone.
Oligo(dT)-anchored PCR
The poly(A) test was conducted using the method described by Salles and Strickland (1995) with minor modifications. Total RNA was isolated from groups of 50 oocytes using RNeasy Plus microkit (Qiagen, 74034). Reagents from the SuperScript III first strand synthesis system (Invitrogen, 18080-051) were used for reverse transcription. PCR was performed with each forward primer and reverse oligo(dT) anchor primer in Table 2. PCR products were subjected to electrophoresis in 1.5% agarose gel and stained with GelRed (Biotium, 41003).
Reporter mRNA preparation and luciferase assay
The Renilla luciferase reporter plasmids were constructed as described previously (Chen et al. 2011). Point mutations in the pRL-TK-Ccnb1 3′ UTR to ablate the canonical CPE were introduced referencing the previously published procedure (Sousa Martins et al. 2016) using a Choo-Choo cloning kit (MCLAB, CCK-20). CPE “TTTTAT” and “TTTTAAT” sequences were replaced with “TCCGAC” and “CGACTCC,” respectively. Luciferase mRNA reporters were transcribed in vitro with mMachine T7 kit (Ambion, AM1344). Firefly luciferase mRNA, the control for microinjection, was polyadenylated using a poly(A) tailing kit (Ambion, AM1350). Luciferase activity was assessed with the dual-luciferase reporter assay kit (Promega, E1910) and detected with the SpectraMaxL Luminometer (Molecular Devices). Data are reported as ratios of the luminescence of Renilla luciferase to that of firefly luciferase.
Supplementary Material
Acknowledgments
We are indebted to Royce Harner, Federica Franciosi, and Estella Orpilla for technical assistance, and Xuan Luong for critical reading of the manuscript. We acknowledge helpful discussions with Dr. Raul Mendez (University of Barcelona) and Dr. Richard Schultz (University of Pennsylvania). Y.Y. is supported by China Scholarship Council, Ministry of Education, People's Republic of China. This work was supported by National Institutes of Health R01 GM097165 and GM116926 to M.C.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.296871.117.
References
- Bava FA, Eliscovich C, Ferreira PG, Minana B, Ben-Dov C, Guigo R, Valcarcel J, Mendez R. 2013. CPEB1 coordinates alternative 3′-UTR formation with translational regulation. Nature 495: 121–125. [DOI] [PubMed] [Google Scholar]
- Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M. 2011. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev 25: 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke HJ. 2012. Post-transcriptional control of gene expression during mouse oogenesis. Results Probl Cell Differ 55: 1–21. [DOI] [PubMed] [Google Scholar]
- de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. 2000. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis 26: 110–112. [PubMed] [Google Scholar]
- Eppig JJ. 1996. Coordination of nuclear and cytoplasmic oocyte maturation in eutherian mammals. Reprod Fertil Dev 8: 485–489. [DOI] [PubMed] [Google Scholar]
- Ferrell JE Jr. 1999. Xenopus oocyte maturation: new lessons from a good egg. Bioessays 21: 833–842. [DOI] [PubMed] [Google Scholar]
- Gosden R, Lee B. 2010. Portrait of an oocyte: our obscure origin. J Clin Invest 120: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt JE, Weaver J, Jones KT. 2010. Spatial regulation of APCCdh1-induced cyclin B1 degradation maintains G2 arrest in mouse oocytes. Development 137: 1297–1304. [DOI] [PubMed] [Google Scholar]
- Ivshina M, Lasko P, Richter JD. 2014. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu Rev Cell Dev Biol 30: 393–415. [DOI] [PubMed] [Google Scholar]
- Kotani T, Yasuda K, Ota R, Yamashita M. 2013. Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. J Cell Biol 202: 1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Hogenesch JB, Irizarry RA. 2016. Modeling of RNA-seq fragment sequence bias reduces systematic errors in transcript abundance estimation. Nat Biotechnol 34: 1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D. 2007. The cell cycle: principles of control. New Science Press Ltd, London, UK. [Google Scholar]
- Norris RP, Ratzan WJ, Freudzon M, Mehlmann LM, Krall J, Movsesian MA, Wang H, Ke H, Nikolaev VO, Jaffe LA. 2009. Cyclic GMP from the surrounding somatic cells regulates cyclic AMP and meiosis in the mouse oocyte. Development 136: 1869–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. 2008. A combinatorial code for CPE-mediated translational control. Cell 132: 434–448. [DOI] [PubMed] [Google Scholar]
- Richter JD. 2007. CPEB: a life in translation. Trends Biochem Sci 32: 279–285. [DOI] [PubMed] [Google Scholar]
- Salles FJ, Strickland S. 1995. Rapid and sensitive analysis of mRNA polyadenylation states by PCR. PCR Methods Appl 4: 317–321. [DOI] [PubMed] [Google Scholar]
- Sousa Martins JP, Liu X, Oke A, Arora R, Franciosi F, Viville S, Laird DJ, Fung JC, Conti M. 2016. DAZL and CPEB1 regulate mRNA translation synergistically during oocyte maturation. J Cell Sci 129: 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay J, Hodgman R, Richter JD. 2000. The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev Biol 221: 1–9. [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay K, Vigneault C, McGraw S, Sirard MA. 2005. Expression of cyclin B1 messenger RNA isoforms and initiation of cytoplasmic polyadenylation in the bovine oocyte. Biol Reprod 72: 1037–1044. [DOI] [PubMed] [Google Scholar]
- Vaccari S, Weeks JL II, Hsieh M, Menniti FS, Conti M. 2009. Cyclic GMP signaling is involved in the luteinizing hormone-dependent meiotic maturation of mouse oocytes. Biol Reprod 81: 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. 2014. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun 5: 5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DX, Cui XS, Kim NH. 2010a. Molecular characterization and polyadenylation-regulated expression of cyclin B1 and Cdc2 in porcine oocytes and early parthenotes. Mol Reprod Dev 77: 38–50. [DOI] [PubMed] [Google Scholar]
- Zhang M, Su YQ, Sugiura K, Xia G, Eppig JJ. 2010b. Granulosa cell ligand NPPC and its receptor NPR2 maintain meiotic arrest in mouse oocytes. Science 330: 366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


