Abstract
Carnitine octanoyltransferase (COT) produces three different transcripts in rat through cis- and trans-splicing reactions, which may lead to the synthesis of two proteins. Generation of the three COT transcripts in rat does not depend on sex, development, fat feeding, the inclusion of the peroxisome proliferator diethylhexyl phthalate in the diet or hyperinsulinemia. In addition, trans-splicing was not detected in COT of other mammals, such as human, pig, cow and mouse, or in Cos7 cells from monkey. Rat COT exon 2 contains two purine-rich sequences. Mutation of the rat COT exon 2 upstream box does not affect the trans-splicing in vitro between two truncated constructs containing exon 2 and its adjacent intron boundaries. In contrast, mutation of the downstream box from the rat sequence (GAAGAAG) to a random sequence or the sequence observed in the other mammals (AAAAAAA) decreased trans-splicing in vitro. In contrast, mutation of the AAAAAAA box of human COT exon 2 to GAAGAAG increases trans-splicing. Heterologous reactions between COT exon 2 from rat and human do not produce trans-splicing. HeLa cells transfected with minigenes of rat COT sequences produced cis- and trans-spliced bands. Mutation of the GAAGAAG box to AAAAAAA abolished trans-splicing and decreased cis-splicing in vivo. We conclude that GAAGAAG is an exonic splicing enhancer that could induce natural trans-splicing in rat COT.
INTRODUCTION
The precise removal of introns from pre-mRNA by splicing is an important step in the expression of genes. It requires the interaction of several proteins and small ribonucleoprotein particles, which all function in concert. These components recognize exon–intron boundaries at both 5′ and 3′ splice sites and then produce the necessary reactions to ligate the exons involved. Two kinds of splicing are observed in nature. Cis-splicing, by which introns from the same pre-mRNA are removed, and trans-splicing, in which two pre-mRNAs are processed to produce a mature transcript that contains exons from both precursors. The trans-splicing process has been described mostly in trypanosome, nematodes, plant/algal chloroplasts and plant mitochondria (1).
A family of serine–arginine-rich RNA binding proteins, known as SR proteins, plays a critical role in the formation of active spliceosomes. The SR proteins recognize exonic splicing enhancers (ESEs) promoting both constitutive and regulated splicing by bringing the flanking splice sites. Trans-splicing efficiency may correlate with the presence of these ESE sequences (2,3). Trans-splicing in vitro only occurs when the 3′ exon contains ESE sequences (4).
In the course of our research, natural trans-splicing was observed in rat carnitine octanoyltransferase (COT) pre-mRNAs (5). This mechanism produces three different transcripts: one produced by cis-splicing and two produced by trans-splicing, in which exons 2 and 3 are repeated. Western blot analysis of peroxisomal proteins revealed two translated proteins with molecular masses which may correspond to the cis-spliced as well as the trans-spliced RNAs in which exons 2 and 3 are repeated. Recently, trans-splicing corresponding to other genes has been observed in mammals. In some cases, mature mRNAs containing a mixture of exons from different genes have been observed; this transcript heterogeneity may contribute to phenotypic variation in mammals (6–12).
In this report we show that, in contrast to rat, other mammals such as human, mouse, pig, cow and Cos7 cells of monkey do not produce detectable trans-splicing of the COT gene. We also show that trans-splicing in rat COT is not due to any metabolic, nutritional, hormonal or developmental condition. Trans-splicing in rat is attributable in part to the occurrence of an ESE sequence (GAAGAAG). Trans-splicing is not found in other mammals, which have a conserved sequence, AAAAAAA, instead. Splicing experiments in vitro, in which the rat putative ESE sequence (GAAGAAG) was mutated either to a random sequence or to AAAAAAA, revealed a marked decrease in trans-splicing of the COT pre-mRNA. Moreover, when the human COT exon 2 box AAAAAAA was mutated to GAAGAAG in vitro a marked increase in trans-splicing was observed. In vivo experiments also showed that mutation of the GAAGAAG strongly decreased trans-splicing. This shows that the evolution from AAAAAAA to GAAGAAG, which does not affect the protein sequence, enabled the rat to produce trans-splicing of the COT pre-mRNA.
MATERIALS AND METHODS
Rat treatment
Groups of adult rats, weighing 200–250 g (three for each treatment) were fed for 7 days with a fat diet or a diet supplemented with diethylhexylphthalate (DEHP) as described (13), or injected i.p. with insulin (40 U/kg body weight) and killed at 30 min.
Northern blot analysis and RNase H digestion of total RNA
Total RNA from different samples used was extracted with guanidine isothiocyanate and purified by centrifugation through a CsCl cushion (14). Twenty micrograms of total liver RNA was incubated with the primer E4.r-2 (5′-CTTCTTAGCAGCTGCCAGTAG-3′) and digested with 0.8 U RNase H as described (5). The 32P-labeled cDNA probe used corresponds to positions 31 to 1700 bp in the rat COT cDNA sequence (15). mRNA analysis by northern blot was performed as described (5).
RT and PCR
RT was performed in 20 µl of a mixture containing 1 µg of total liver RNA from human, cow, mouse, pig or Cos7 cultured monkey cells or 1 µg of liver and intestine RNA from rats. Random hexamers were used as primers to synthesize the cDNAs. The reaction was performed as described (5). The conditions for the PCR were 94°C for 45 s; 55°C for 45 s and 72°C for 45 s for 35 cycles. The bands were purified with Quiaex kit (Stratagene) and sequenced with an automatic sequencer ABI prism.
Constructs, pre-mRNA synthesis and trans-splicing reaction in vitro
Constructs A and B were prepared as described (5). Construct A1 was also prepared as described (5) but with less polylinker region from Bluescript vector. To this end, a fragment was amplified with E2.f (5′-TTTTCTTACTGTGACTATACCATGG-3′) and I2.r (5′-CTACACTAGAAGATTATGAACATAC-3′) and subcloned in a plasmid Bluescript SK+ previously cut with Ecl136II, an isoschizomer of SacI that produces blunt ends. All amplifications were performed with Pfu polymerase. Construct C was prepared by PCR using megaprimer for asymmetric PCR as described (16) taking construct B as template. The first PCR amplification was carried out with primers T7 and MutESE1.f (5′-CAATTGGCTAAGTCAATTGTCTACCGAACATTCCAGTACCAGG-3′). The second PCR was performed with primer T3 and with the purified product of the first amplification (megaprimer). Construct D was also prepared by PCR taking construct B as template and primers I1.f (5′-CATGGGGTGCTAAATCCACA-3′) and MutESE2.r (5′-CTGACTCAAGGTACAGATGCAGTGATTCTTC-3′) and subcloned into Bluescript SK+ digested with EcoRV. The sequences underlined above indicate the mutated nucleotides. To reduce their polylinker regions, constructs C and D were cut with XhoI and EcoRI, blunt ended with the Klenow fragment and cloned in Bluescript SK+ previously digested with SacI and blunt ended with T4 DNA polymerase. Construct E was prepared by PCR taking construct B as template. The primers used were I1.f and MutESE3.r (5′-CTGACTCAAGGTATTTTTTTAGTGATTCTTC-3′). Construct F was prepared by the megaprimer technique taking construct A as template. Primers for the first amplification were E2.f and MutESE3.r. In the second amplification primer I2.r was used. The constructs G, H, I and J were obtained by PCR amplifications using genomic DNA obtained from human liver. The primers were designed using the human COT cDNA sequence AF168793 (17) and the genomic sequence ACC005045 from the DDBJ/EMBL/GenBank database that contains the human COT genomic sequence. PCR was performed in a total volume of 50 µl with 300 ng of genomic DNA, 1 mM of each primer and Pfu polymerase. The conditions for PCR were 94°C for 5 min followed by 30 cycles of 94°C for 45 s, 53°C for 45 s and 72°C for 1 min. Constructs G and H were obtained with primers hE2.f (5′-GTTTCCTATTGTGATTTTATC-3′) and hI2.r (5′-TATATACTAAAACATTATTAAC-3′), and hE2.r (5′-CTGATTCAAGGTATTTTTTTAATG-3′) and hI1.f (5′-GCTGGTACCAAGTCCTAGATATGC-3′), respectively. Construct I was obtained with primers hI1.f and MutESE4.r (5′-CTGATTCAAGGTTGTTGTTGAATGATTCTTCAAG-3′). Construct J was obtained using a megaprimer asymmetric PCR; in the first PCR primers hE2.f and mutESE4.r were employed to produce the megaprimer, the second PCR was performed using the megaprimer and primer hI2.r. All the blunt-ended PCR fragments (E–J) were subcloned in pBluescript SK+, previously digested with Ecl136II.
For the trans-splicing experiments, construct B was linearized with EcoRI, constructs C and D with HindIII and constructs A and E–J with NotI and all were transcribed with T3 RNA polymerase. In vitro trans-splicing assays were carried out as described (5). The products of trans-splicing were loaded on a denaturing 6% polyacrylamide/7 M urea gel. Autoradiographs of the labeled bands in the gels were scanned with a Molecular Dynamics laser densitometer using the internal integration to determine peak areas with the ImageQuant program and also with the PhosphorImager technique. The percentage inhibition was calculated from the values of the integration areas of the bands.
Preparation of COT minigene constructs, cell culture and transfections
A minigene containing several regions of the rat COT gene (construct E2E3COT, seen in Fig. 8) was created by Pfu polymerase (Clontech) PCR amplification of 1 ng from genomic DNA λGA5 clone (5). To amplify different fragments of this minigene, several primers were used. Primers I1.f (5′-CATGGGGGTGCTAAATCCACA-3′) and I2.r (5′-CTACACTAGAAGATTATGAAGATAC-3′) amplify a fragment containing 125 nt of the 3′ region of intron 1 (which contains the branch point), the whole of exon 2 and 25 nt of the 5′ splice site of intron 2. Primers I2.f (5′-GCATTGCAGTTTTCACATTGCC-3′) and COT291.r (5′-CCAGTTTCTTTTTCCTTTAGCCC-3′) amplify a fragment containing 115 nt of the 3′ splice site of intron 2 (containing the branch point) and the whole of exon 3. Products were cloned into the EcoRV site in plasmid pcDNA3 (Invitrogen) and sequenced. A mutated minigene of E2E3COT (E2E3COT-ESEmut) was obtained by using the site-directed mutagenesis kit from Stratagene. The primers used were COTE2G/A.f (5′-CCGTTCCTTCGCTTGAAGAATCACTAAAAAAATACCTTGAGTCAG-3′) and COTE2G/A.r (5′-CTGACTCAAGGTATTTTTTTAGTGATTCTTCAAGCGAAGGAACGG-3′). The introduction of mutations of the E2E3COT-ESEmut minigene was confirmed by sequencing.
Figure 8.
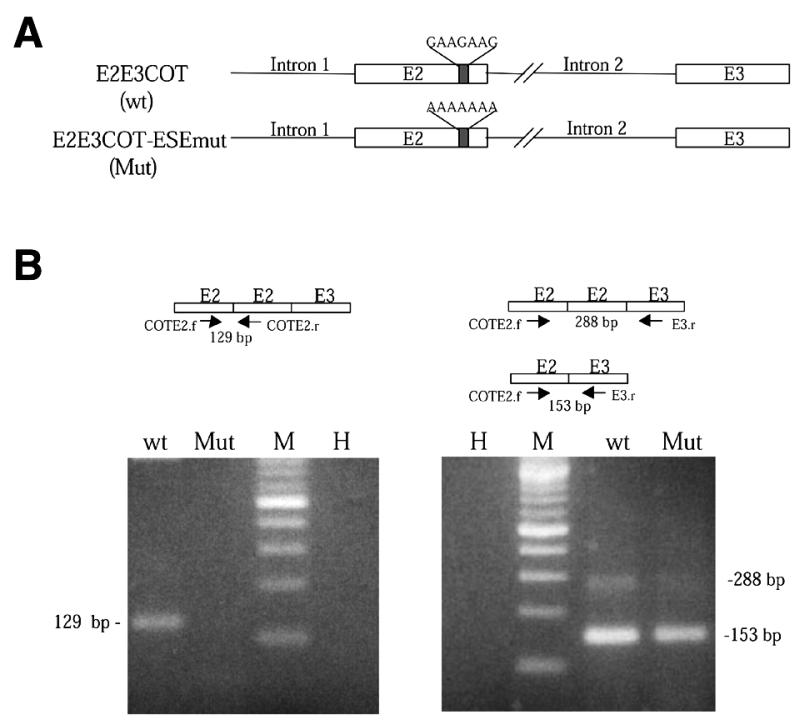
Mutation on the ESE box of rat exon 2 from GAAGAAG to AAAAAAA inhibits trans-splicing in vivo. (A) Schematic representation of the COT minigenes E2E3COT (wt) and mutated E2E3COT-ESEmut. (B) Representative agarose gels of the RT–PCR amplification of DNA-free total RNA from HeLa cells transfected with E2E3COT and E2E3COT-ESEmut minigenes. cDNAs were amplified with rat COT primers COTE2.f (5′-GGACTCTCTTCCGCCCTTGC-3′) and COTE2.r (5′-GGAATGTTCGTTCTTCAATTGAC-3′) (left), and with COTE2.f and E3.r (5′-TGATGCAATGTCTTGCCAAC-3′) (right). The set of primers COTE2.f and COTE2.r can only detect trans-spliced rat COT mRNAs. wt or Mut indicate the amplification cDNAs products of transfected HeLa cells with wild-type COT minigene E2E3COT or mutated COT minigene E2E3COT-ESE mut. M indicates the molecular DNA markers and H the products of amplified cDNAs from untransfected HeLa cells. The size of PCR products corresponds to the exonic representation.
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) fetal bovine serum, 100 i.u./ml penicillin and 100 µg/ml streptomycin. The cells were cultured in 65 mm plates. Cells (1.6 × 106) were transiently transfected with 5 µg of either E2E3COT plasmid or E2E3COT-ESEmut plasmid. Plasmid dosage was kept constant by the addition of an appropriate amount of DNA salmon sperm. Plasmid pCMVbGAL (cytomegalovirus promoter β-galactosidase; 3 µg) was included as an internal control in co-transfections. Transfection experiments were performed by the calcium phosphate method as previously described (14,18). After incubation for a further 24 h the cells were collected and RNA was extracted by trizol reagent (Bethesda Research Laboratories). Each series of experiments was performed at least four times and each transfection was performed in triplicate. RNA samples were treated with DNase I to eliminate the plasmidic DNA from transfection as described (5). The cis- and trans-spliced mRNA of rat COT was analyzed by RT–PCR.
RESULTS
Of the various mammals studied only the rat produced trans-splicing
The occurrence of trans-splicing in exons 2 and 3 of the rat COT pre-mRNAs (5) produced two different transcripts, with the following compositions: exon1–exon2–exon2–exon3 etc. and exon1–exon2–exon3–exon2–exon3 etc., in addition to the cis-spliced transcript (exon1–exon2–exon3 etc.). This phenomenon was observed both in vivo and in vitro. We thus attempted to determine: (i) whether trans-splicing of the COT gene occurs in other mammals, and (ii) the molecular basis for this phenomenon in the rat.
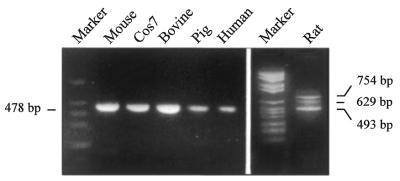
We first amplified mRNAs by RT–PCR from several mammals, among them human, pig, cow, mouse and Cos cells from monkey (Fig. 1). A single band was observed in all cases. This band was obtained whether the primers used were from rat (not shown) or cow. The band of each species was excised from the gel and, after sequencing, it was found to correspond to the cis-spliced transcript. The sequences of exon 2 of the COT gene from mouse, Cos7 cells and pig are shown in Table 1 together with those of human, cow and rat. The amplification of the rat mRNAs, using primers from either cow (not shown) or rat, shows the three-band pattern that corresponds to cis- and trans-spliced transcripts, as expected.
Figure 1.
RT–PCR amplification of COT mRNAs in various mammals. RT of total liver RNA from mouse, cow (bovine), pig, human, rat and Cos7 (monkey) cultured cells was performed. The primers used were from the bovine COT cDNA: bE2.f (5′-TGCATGGTACCATAAAATGGAAAATCAACTGGC-3′) and bE5.r (5′-CGGAATTGATTCATATCTAGAGGAGTATTTCC-3′) except for the rat cDNA which was amplified with homologous rat primers E2.f (5′-TTTTCTTACTGTGACTATACCATGG-3′) and E5.r (5′-GGTTCATGTCTAGAGGAG-3′). The bands were sequenced and correspond to cis-splicing at 493 (in rat) and 478 bp (in other mammals), and trans-splicing (629 and 754 bp) only in rat. The small difference in size of cis-spliced bands among rat and other mammals is due to the differences in the primers used.
Table 1. Alignment of nucleotide sequences in exon 2 from the COT cDNA from various mammals.
Sequences were obtained from the GenBank database of rat (U26033), human (H00195) and bovine (U65745). PCR amplified bands (shown in Fig. 1) from mouse (AF144397), pig (AF144398) and Cos7 monkey cultured cells (AF144399) were excised from the gel and sequenced. The numbers correspond to those of the GenBank database. All sequences end at the last nucleotide of rat COT exon 2. Purine-rich sequences in COT exon 2 are enclosed in squares. The alignments were performed using the Wisconsin Package Version 9.1 (Genetics Computer Group, Madison, WI).
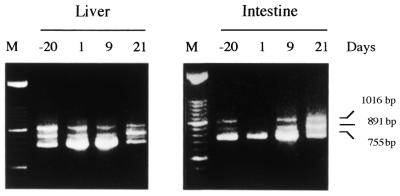
Trans-splicing in rat COT depends on the gene sequence and not on sex, development or metabolic condition
Once it had been established that only the rat produced trans-splicing (Fig. 1), we examined whether the trans-splicing observed for rat COT could be either tissue-dependent or produced as a result of metabolic alteration, or dependent on sex or development. RT–PCR amplification of transcripts from rat liver and intestine gave the three-transcript pattern (Fig. 2), which was also observed in other tissues such as testis, kidney, heart and brown adipose tissue (data not shown). We then performed RT amplifications of total RNAs from rats at different developmental stages: fetuses of 20 days, and suckling rats of 1, 9 and 21 days of age. The COT mRNA levels during the suckling period are transcriptionally regulated (19). These developmental studies in liver and intestine also showed the three-band pattern (Fig. 2). The variability in the relative proportion of the three transcripts had been observed before (5), but no explanation has been found. Both male and female adult rats (six animals in each case) also gave three mature transcripts of the COT gene (data not shown). Groups of three rats were treated with insulin or fed with a fat diet, or with a diet supplemented with DEHP, a peroxisomal proliferator, as described in Materials and Methods. These treatments produced small changes in the COT mRNA levels with respect to controls (Fig. 3A) except DEHP treatment, which produced a 12-fold increase. Total RNAs were incubated with a specific primer and digested with RNase H (Fig. 3B). Once more, the three mature transcripts corresponding to cis- and trans-splicing were also observed. All these results taken together suggest that trans-splicing in rat COT depends on the gene sequence rather than on any metabolic, nutritional, hormonal or developmental condition.
Figure 2.
Developmental trans-splicing analysis in rat liver and intestine by RT–PCR. Total RNA was extracted from liver and intestine of 20-day rat fetus (lane –20), 1-, 9- and 21-day-old rats. RT–PCR was performed. The primers used in the amplification were E1.f (5′-GAGTGCAGAGAGCCAAGCCGGG-3′) and E7.r (5′-GGCCCAACAGGTTCATTCCAG-3′). The bands observed correspond to cis-splicing (755 bp) and trans-splicing (891 and 1016 bp).
Figure 3.
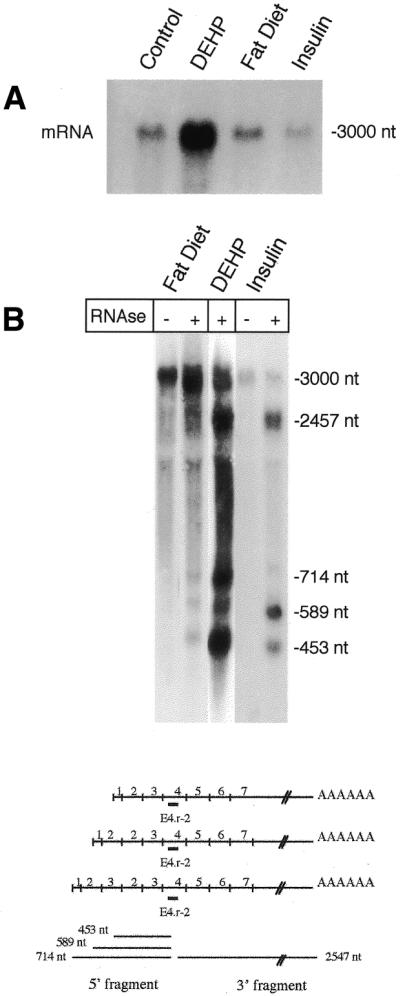
Northern blot analysis and RNase H digestion of total RNA from liver of treated rats. (A) Northern blot of liver COT mRNA levels of rats treated as described in Materials and Methods. (B) Representative results of three experiments in which 20 µg of total RNA from livers of treated rats was incubated with the primer E4.r-2 and digested with RNase H (+). The lanes marked with – correspond to samples not digested with RNase H. The diagram shows the expected products formed after binding the primer E4.r-2 to RNA and digesting with RNase H as described (5).
Identification of possible ESEs in exon 2 which may promote trans-splicing in experiments in vitro
The occurrence of an ESE sequence in the acceptor exon is necessary for artificial trans-splicing to occur (2,3). Splicing enhancers facilitate the assembly of protein complexes on pre-mRNAs containing a 3′ splice site, and these complexes are sufficiently stable to interact with 5′ splice sites located in other RNA molecules. By examination of rat COT exon 2, at least two purine-rich sequences were identified at positions 79 (GAAGAACGAA, named upstream box) and 147 (GAAGAAG, named downstream box) (Table 1), which are similar to ESE sequences previously described (20–22).
To assess the individual roles of the downstream and upstream boxes in trans-splicing, different truncated pre-mRNAs were prepared: a wild-type donor pre-mRNA containing only exon 2 and the 5′ splice site of intron 2 (construct A), and a wild-type acceptor pre-mRNA, containing the branch-point region, the 3′ splice site of intron 1 and exon 2 (construct B) (Fig. 4). We then separately mutated the upstream and the downstream boxes of the rat exon 2 in construct B. The sequence GAAGAA of the upstream box (construct B) was changed to GTCTAC (construct C), and the sequence GAAGAAG of the downstream box was changed to GCATCTG (construct D).
Figure 4.
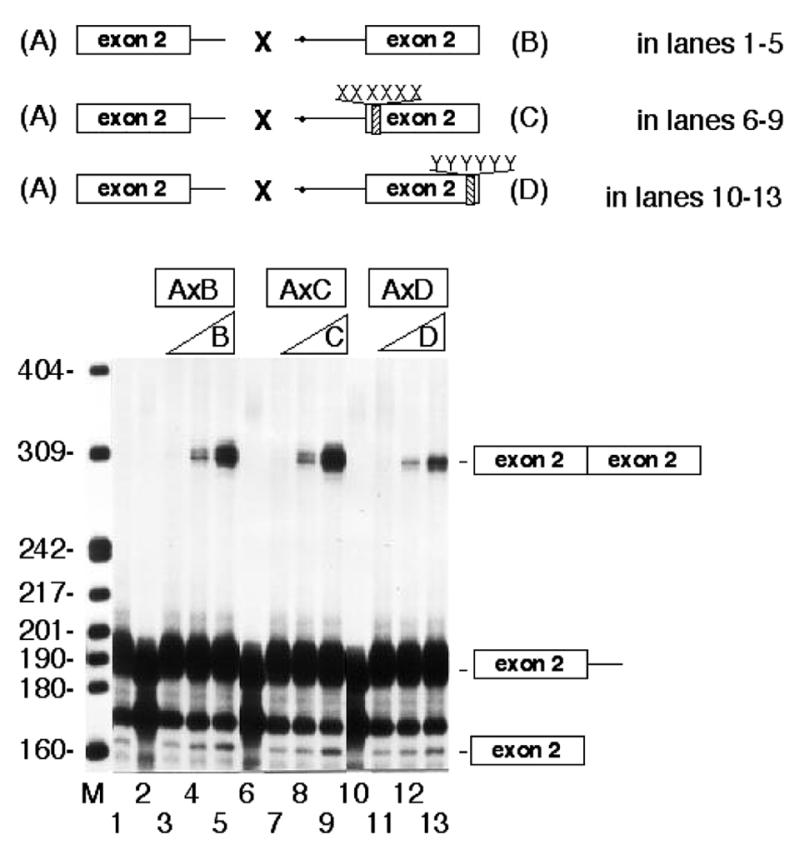
Random mutation on the downstream box of rat exon 2 inhibits trans-splicing in vitro. Trans-splicing assays in vitro contained 0.9 ng (16 fmol) of radiolabeled wild-type donor (A) pre-mRNA (lanes 1–13) and increasing amounts, 1, 10 and 25 ng (11, 111 and 277 fmol), of either wild-type acceptor B (lanes 3–5), or mutated purine-rich boxes, acceptor C (lanes 7–9) and acceptor D (lanes 11–13). The letters XXXXXX in acceptor C indicate that the wild-type sequence GAAGAA of the upstream box was mutated to GTCTAC, and YYYYYY denote that the wild-type downstream box sequence GAAGAAG was mutated to GCATCTG. Product bands were mapped by RT–PCR. Splicing products and intermediates are depicted on the right. Lane M contains the molecular weight markers. Lane 1 corresponds to an incubation in the absence of any acceptor but in the presence of ATP. Lanes 2, 6 and 10 represent controls without ATP but with 50 ng of acceptor. Bands in lanes 7–9 and in 11–13 represent an average of 105 and 60% with respect to bands seen in lanes 3–5, respectively. The branch-points are shown by circles in the acceptor constructs.
Figure 4 shows trans-splicing assays with the combination of donor construct A with acceptor constructs B, C and D. The donor pre-mRNA (construct A) was 32P-labeled during transcription in vitro and the unlabeled acceptors were added to the trans-splicing reaction. The combination of donor A with acceptor C (random mutated upstream box) produced slight changes in the trans-splicing product in vitro when compared with the A×B product (lanes 7–9 and lanes 3–5). However, the trans-splicing between donor A and acceptor D (random mutated downstream box) was reduced by 60% (compare lanes 11–13 with lanes 3–5). Mutation of both upstream and downstream boxes did not modify trans-splicing when compared with results of the downstream ESE box (data not shown). These results show that the downstream box behaves as an ESE sequence, while the upstream box does not seem to contribute to the process.
Mutation of downstream rat ESE sequence to the sequence observed in other mammals strongly decreases trans-splicing in vitro
We first compared the composition of the rat COT exon 2 sequence with the corresponding sequences from those mammals in which trans-splicing did not occur (Table 1). The upstream box in these mammals differed only slightly (0, 1 or 2 nucleotides changed) from that in the rat. In contrast, the downstream box was invariably AAAAAAA, whereas in rat it was GAAGAAG. Taken together, these results emphasize the relevance of the downstream ESE box in trans-splicing. We attempted to assess whether the change of GAAGAAG (present in rat exon 2) to AAAAAAA (seen in all the other mammals studied) is responsible for the absence of trans-splicing in these species. To this end, three types of experiments were performed. In the first, in vitro trans-splicing was carried out with constructs of rat exon 2 in which the downstream box GAAGAAG was mutated to AAAAAAA, to see whether this mutation decreased trans-splicing. In the second sets of experiments, the AAAAAAA box of human exon 2 was mutated to GAAGAAG, to see whether there was a gain of function. The third set of experiments showed that rat COT minigenes transfected in HeLa cells produced trans-splicing in vivo, and that mutation of the GAAGAAG box abolished the phenomenon.
To perform the first experiment two truncated pre-mRNAs were also prepared: construct E, in which the downstream box in rat construct B was mutated to AAAAAAA, and construct F, in which the rat donor construct A was also mutated in the downstream box to AAAAAAA. Figure 5 shows that the in vitro combination of donor F and acceptor E, which reflects what is produced in vivo in mammals other than the rat, inhibits trans-splicing (50%) as compared with the wild-type trans-splicing (donor A and acceptor B) (see lanes 8–10 and 3–5).
Figure 5.
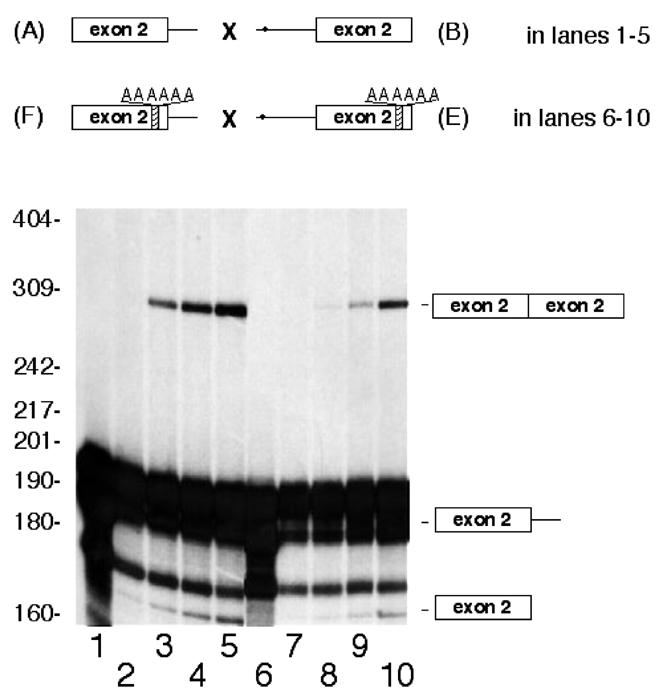
Mutation on the downstream ESE box of rat exon 2 from GAAGAAG to AAAAAAA inhibits trans-splicing in vitro. Trans-splicing assays in vitro contained 0.9 ng (16 fmol) of 32P-radiolabeled A pre-mRNA (lanes 1–5) or 32P-radiolabeled F pre-mRNA (lanes 6–10) and increasing amounts, 25, 50 and 100 ng (280, 555 and 1108 fmol), of either acceptor B (lanes 3–5) or acceptor E (lanes 8–10). The letters AAAAAAA in the F donor and E acceptor indicate that the wild-type downstream ESE box GAAGAAG was mutated to AAAAAAA. The molecular weight markers are shown to the left. Lanes 1 and 6 correspond to incubations in the absence of any acceptor but in the presence of ATP. Lanes 2 and 7 represent controls without ATP but with 200 ng of constructs B and E, respectively. Bands in lanes 8–10 represent an average of 50% of bands seen in lanes 3–5.
Mutation of human downstream box sequence to the sequence observed in rat strongly increased trans-splicing in vitro
To analyze the role of the rat COT downstream ESE sequence, four new truncated pre-mRNAs were prepared: a donor human pre-mRNA containing only human exon 2 and the 5′ splice site of intron 2 (construct G), and an acceptor human pre-mRNA, containing the branch point region, the 3′ splice site of intron 1 and exon 2 (construct H) (Fig. 6). We then mutated the downstream AAAAAAA box of the human exon 2 in constructs G and H so that this human sequence was changed to GAAGAAG (seen in rat) (constructs J and I, respectively). The combination of constructs G and H corresponding to the natural human exon 2 only produced trans-splicing when a high amount of acceptor was present (lanes 8–11). The combination of mutated human exon 2 as a donor with non-mutated human acceptor exon 2 (J×H) only slightly increased the trans-splicing reaction (16%) (see lanes 19–22). The combination of donor construct G with acceptor I, in which the rat ESE box was present in human exon 2 (lanes 3–6), increased trans-splicing (60%) compared with combination of constructs G×H as in the rat, suggesting that the rat ESE box induces trans-splicing when present in the acceptor exon 2. Similar trans-splicing was produced when constructs J×I (both containing the mutated GAAGAAG) (lanes 14–17) reacted in vitro (60%). This gain of function is attributable exclusively to the rat COT exon 2 downstream ESE, which appears to promote trans-splicing.
Figure 6.
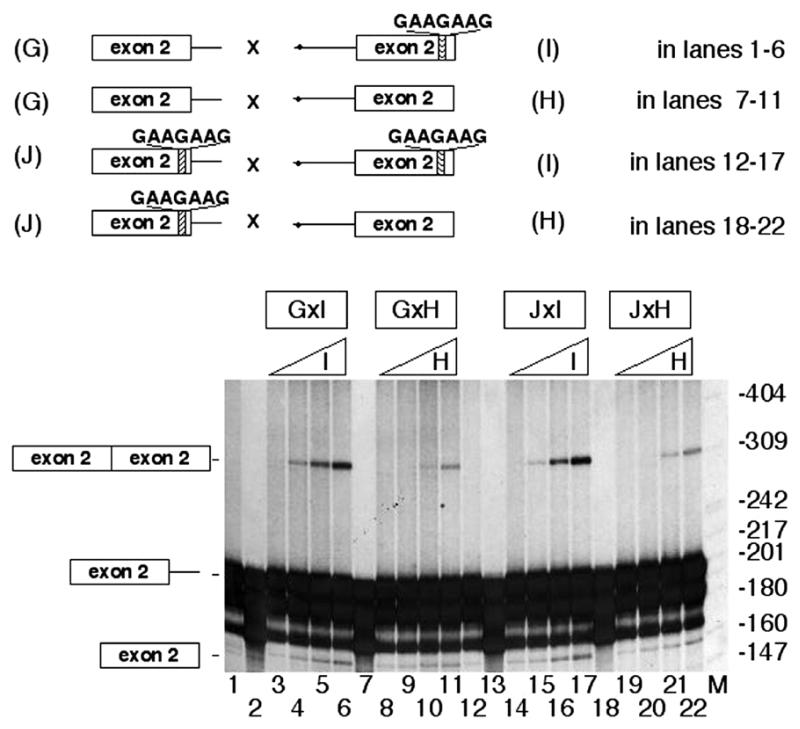
Mutation on the downstream box of human exon 2 from AAAAAAA to GAAGAAG increased trans-splicing in vitro. Trans-splicing assays in vitro contained 0.9 ng (16 fmol) of 32P-radiolabeled G pre-mRNA (lanes 1–11) or 32P-radiolabeled J pre-mRNA (lanes 12–22) and increasing amounts, 25, 50, 100 and 200 ng (280, 555, 1108 and 2220 fmol), of either acceptor I (lanes 3–6 and 14–17) or acceptor H (lanes 8–11 and 19–22). The letters GAAGAAG in the I acceptor and J donor indicate that the human wild-type downstream box AAAAAAA was mutated to GAAGAAG. Lane M contains the molecular weight markers. Lanes 1 and 12 correspond to incubations in the absence of any acceptor but in the presence of ATP. Lanes 2, 7, 13 and 18 represent controls without ATP but with 200 ng of I or H pre-mRNAs. Bands in lanes 3–6 and 14–17 represent an average of 160 and 170%, respectively, of bands seen in lanes 8–11. Bands 19–22 are similar to those of 8–11.
Relevance of the rat downstream ESE in heterologous trans-splicing in vitro
To test the relevance of the rat COT exon 2 ESE sequence in heterologous trans-splicing, wild-type rat donor COT exon 2 (construct A1) was reacted with human acceptor COT exon 2 (construct H) in the same conditions. No trans-splicing product was observed (Fig. 7, lanes 1–3). When the human acceptor COT exon 2 was mutated in the downstream box AAAAAAA to the rat ESE GAAGAAG (construct I), trans-splicing occurred (compare lanes 4–6 with 1–3). This experiment showed that, on the one hand, rat COT exon 2 as a donor does not by itself produce trans-splicing when it reacts with a human acceptor (lacking the ESE box). On the other hand, when the human construct is mutated in the corresponding box AAAAAAA with the rat ESE sequence (GAAGAAG) trans-splicing is produced.
Figure 7.
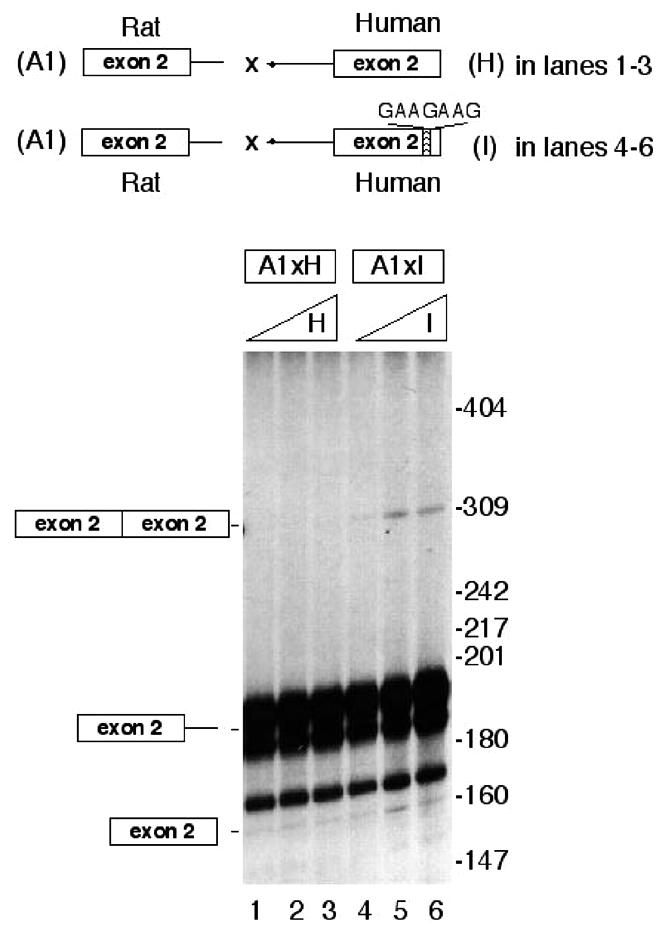
Mutation of the downstream box of human COT exon 2 produces a gain of function in heterologous trans-splicing in vitro. Trans-splicing assays in vitro contained 0.9 ng (16 fmol) of 32P-radiolabeled rat A1 pre-mRNA (lanes 1–6) and increasing amounts, 10, 25 and 50 ng (111, 280 and 555 fmol), of human acceptor H (lanes 1–3) or human acceptor I (lanes 4–6).
Mutation of rat COT exon 2 ESE sequence to the sequence observed in COT exon 2 from other mammals strongly decreases trans-splicing in vivo
To test the role of the rat COT exon 2 ESE sequence in trans-splicing in vivo, HeLa cells were transfected with two rat COT minigenes (Fig. 8A). The first of these minigenes contained the rat COT exon 2 ESE sequence (GAAGAAG) intact and the second was identical to the first but the ESE sequence had been mutated to AAAAAAA. After 24 h, total RNA was isolated from transfected cells and RT–PCR amplification was performed with specific primers to detect either the formation of trans-spliced mRNA (Fig. 8B, left) or the simultaneous formation of cis- and trans-spliced mRNAs (Fig. 8B, right).
Transfected HeLa cells produced a trans-spliced band (Fig. 8B, wt, left) composed of exon2–exon2–exon3, which was not produced when the ESE sequence GAAGAAG was mutated to AAAAAAA (Fig. 8B, Mut, left). The RT–PCR amplified bands using a different set of primers that involve exon 2 and exon 3 show that in addition to trans-splicing, cis-spliced bands exon2–exon3 were also produced. In the conditions of the experiment, cis-spliced bands were stronger than trans-spliced bands, showing that cis-splicing is preferred over trans-splicing. When the rat COT ESE sequence was mutated, both the cis- and the trans-splicing were affected, but while the cis-splicing was slightly decreased, trans-splicing disappeared (Fig. 8B, right). Any of the sets of primers used could amplify endogenous HeLa COT. These experiments show that the splicing produced in vitro was also reproduced in vivo, and that the ESE sequence GAAGAAG plays a role in trans-splicing both in vitro and in vivo, since its mutation to the sequence observed in other mammals decreases trans-splicing.
DISCUSSION
The occurrence of natural trans-splicing in the rat COT pre-mRNAs, producing two different transcripts involving repetition of exons 2 and 3, had been reported previously by our group (5). The question was then to determine the molecular basis of this phenomenon. One possibility was that metabolic, sexual or developmental factors might be responsible for trans-splicing. However, the present study clearly shows that all conditions tested determine the generation of natural trans-splicing. These results emphasize that the characteristics of the gene are responsible for such phenomena.
The occurrence of in vivo trans-splicing in the rat COT gene and its absence in other mammals is surprising. The simplest explanation was that some structural feature was present in the rat COT gene and not present in other mammals. In one type of trans-splicing an intron of one pre-mRNA interacts with an intron of a second pre-mRNA, enhancing the recombination of splice sites between two pre-mRNAs (23). This is not the case in the COT pre-mRNA constructs (5), which do not require intronic RNA–RNA interactions among substrates. Another explanation is that the COT gene requires specialized elements such as ESEs for trans-splicing to occur.
The examination of exon 2 of the rat COT gene revealed two purine-rich sequences, which may facilitate the trans-splicing reaction as ESEs. Accordingly, we independently mutated these two putative ESE sequences and observed the effects on trans-splicing in vitro. The upstream sequence, which was nearly identical in the COT exon 2 of all mammals sequenced, did not produce any quantitative change in trans-splicing after mutation, from which we concluded that, by itself, it was not responsible for the trans-splicing observed in the rat.
Different behavior was observed in downstream rat exon 2 ESE sequence GAAGAAG, which was different from that seen in the rest of the mammals studied, which is invariably AAAAAAA. Experiments in vitro with the randomly mutated downstream ESE produced a marked decrease in trans-splicing (residual capacity 40%) with respect to control. These results were confirmed by the mutation of the rat downstream ESE sequence to AAAAAAA. Once more, this mutation decreased trans-splicing by 50%. From this study it is clear that sequence GAAGAAG in exon 2 of the rat COT gene strongly increases trans-splicing.
The experiments of trans-splicing with human COT exon 2 illustrate the relevance of the ESE GAAGAAG sequence in trans-splicing. The inclusion of the putative rat downstream ESE sequence, instead of that naturally found in human, restored the ability of human exon 2 to produce trans-splicing, since there was a marked increase in trans-splicing compared to control. This was confirmed in heterologous trans-splicing in vitro, in which the inclusion of the rat ESE sequence GAAGAAG instead of the human AAAAAAA sequence induced to human exon 2 to produce trans-splicing when interacting with the rat exon 2.
To confirm these results in vitro, a set of experiments was performed in vivo. Rat COT minigenes containing regions of intron 1, exon 2, intron 2 and exon 3 produced trans-splicing in transfected HeLa cells. These constructs also produced cis-spliced products in a much higher amount than trans-spliced products. This could be shown because under competitive conditions PCR amplifications showed stronger cis-spliced bands. Importantly, mutation in the ESE box abolished trans-splicing bands in vivo. Moreover, the mutation of the ESE band also decreased cis-splicing, although to a lesser extent than trans-splicing. This result is not surprising since the influence of ESE boxes on cis-splicing is well documented.
Previous experiments with artificial genes had shown that ESE enhancers activate trans-splicing (2,3). Moreover, ESE enhancers are also necessary for many cis-splicing reactions (20–22), which they trigger by binding to SR proteins, a family of modular splicing factors bearing one or more RNA recognition motifs and an arginine/serine (RS)-rich region (24). Curiously, previous cis-splicing experiments showed that if an SR-dependent enhancer is changed either to (AAAAAAGGGGGG)2 or poly(A) the cis-splicing reaction is inhibited (25). Recently, it was also shown that when another enhancer, activated by SR proteins, from the the human β-globin pre-mRNA, is changed to AAAAAAA, the cis-splicing reaction is also inhibited (21). These observations indicate that the function of SR proteins is compromised if the enhancer sequence is changed to AAAAAAA. From an evolutionary point of view, we can postulate that there was a mutation in the COT gene in rat as a result of which it produced trans-splicing and thus generated two peroxisomal proteins. It is important to stress that no change is produced in the encoded protein when GAAGAAG is mutated to AAAAAAA (in both cases the predicted sequence is Leu–Lys–Lys). Thus, overlapping protein coding in rat and RNA recognition elements to facilitate trans-splicing may be co-selected during evolution to acquire the encoding of a new protein with unknown catalytic properties or cellular localization. This is the first report of the occurrence of an ESE box that enhances trans-splicing in mammals.
Acknowledgments
ACKNOWLEDGEMENTS
The editorial help of Robin Rycroft is greatly acknowledged. This work was supported by Grant PB95-0012 from the Dirección General de Investigación Científica y Técnica, Grant 1999SGR0075 of the Generalitat de Catalunya (given to F.G.H.) and by a grant from Fundación Ramón Areces (given to M.B.-E.). C.C. is the recipient of a fellowship from the Dirección General de Investigación Científica y Técnica, Spain.
DDBJ/EMBL/GenBank accession nos AF144397–AF144399
References
- 1.Bonen L. (1993) Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J., 7, 40–46. [DOI] [PubMed] [Google Scholar]
- 2.Bruzik J.P. and Maniatis,T. (1995) Enhancer-dependent interaction between 5′ and 3′ splice sites in trans. Proc. Natl Acad. Sci. USA, 92, 7056–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiara M.D. and Reed,R. (1995) A two-step mechanism for 5′ and 3′ splice-site pairing. Nature, 375, 510–513. [DOI] [PubMed] [Google Scholar]
- 4.Chew S.L., Liu,H.X., Mayeda,A. and Krainer,A.R. (1999) Evidence for the function of an exonic splicing enhancer after the first catalytic step of pre-mRNA splicing. Proc. Natl Acad. Sci. USA, 96, 10655–10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caudevilla C., Serra,D., Miliar,A., Codony,C., Asins,G., Bach,M. and Hegardt,F.G. (1998) Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc. Natl Acad. Sci. USA, 95, 12185–12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimizu A., Nussenzweig,M.C., Han,H., Sanchez,M. and Honjo,T. (1991) Trans-splicing as a possible molecular mechanism for the multiple isotype expression of the immunoglobulin gene. J. Exp. Med., 173, 1385–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan P.M., Petrusz,P., Szpirer,C. and Joseph,D.R. (1991) Alternative processing of androgen-binding protein RNA transcripts in fetal rat liver. Identification of a transcript formed by trans splicing. J. Biol. Chem., 266, 143–154. [PubMed] [Google Scholar]
- 8.Vellard M., Sureau,A., Soret,J., Martinerie,C. and Perbal,B. (1992) A potential splicing factor is encoded by the opposite strand of the trans-spliced c-myb exon. Proc. Natl Acad. Sci. USA, 89, 2511–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breen M.A. and Ashcroft,S.J.H. (1997) A truncated isoform of Ca2+/calmodulin-dependent protein kinase II expressed in human islets of Langerhans may result from trans-splicing. FEBS Lett., 409, 375–379. [DOI] [PubMed] [Google Scholar]
- 10.Akopian A.N., Okuse,K., Souslova,V., England,S., Ogata,N. and Wood,J.N. (1999) Trans-splicing of a voltage-gated sodium channel is regulated by nerve growth factor. FEBS Lett., 445, 177–182. [DOI] [PubMed] [Google Scholar]
- 11.Li B.-L., Li,X.-L. Duan,Z.-J., Lee,O., Lin,S., Ma,Z.M., Chang,C.C.Y., Yang,X.-Y., Park,J.P., Mohandas,T.K., Noll,W., Chan,L. and Chang,T.-Y. (1999) Human acyl-CoA cholesterol acyltransferase-1 (ACAT-1) gene organization and evidence that the 4.3-kilobase ACAT-1 mRNA is produced from two different chromosomes. J. Biol. Chem., 274, 11060–11071. [DOI] [PubMed] [Google Scholar]
- 12.Frantz S.A., Thiara,A.S., Lodwick,D., Ng,L.L., Eperon,I.C. and Samani,N.J. (1999) Exon repetition in mRNA. Proc. Natl Acad. Sci. USA, 96, 5400–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asins G., Serra,D. and Hegardt,F.G. (1994) The effect of etomoxir on the mRNA levels of enzymes involved in ketogenesis and cholesterogenesis in rat liver. Biochem. Pharmacol., 47, 1373–1379. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 15.Choi S.J., Oh,D.H., Song,C.S., Roy,A.K. and Chatterjee,B. (1995) Molecular cloning and sequence analysis of the rat liver carnitine octanoyltransferase cDNA, its natural gene and the gene promoter. Biochim. Biophys. Acta, 1264, 215–222. [DOI] [PubMed] [Google Scholar]
- 16.Datta A.K. (1995) Efficient amplification using ‘megaprimer’ by asymmetric polymerase chain reaction. Nucleic Acids Res., 23, 4530–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferdinandusse S., Mulders,J., Ijist,L., Denis,S., Dacremont,G., Waterham,H.R. and Wanders,R.J. (1999) Molecular cloning and expression of human carnitine octanoyltransferase: evidence for its role in the peroxisomal beta-oxidation of branched-chain fatty acids. Biochem. Biophys. Res. Commun., 263, 213–218. [DOI] [PubMed] [Google Scholar]
- 18.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1987) Current Protocols in Molecular Biology. Green Publishing Associates/Wiley-Interscience, New York, NY.
- 19.Miliar A., Serra,D., Casaroli,R., Vilaró,S., Asins,G. and Hegardt,F.G. (2001) Developmental changes in carnitine octanoyltransferase gene expression in intestine and liver of suckling rats. Arch. Biochem. Biophys., 385, 283–289. [DOI] [PubMed] [Google Scholar]
- 20.Sun Q., Mayeda,A., Hampson,R.K., Krainer,A.R. and Rottman,F.M. (1993) General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev., 7, 2598–2608. [DOI] [PubMed] [Google Scholar]
- 21.Schaal T.D. and Maniatis,T. (1999) Multiple distinct splice enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol., 19, 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watakabe A., Tanaka,K. and Shimura,Y. (1993) The role of exon sequences in splice site selection. Genes Dev., 7, 407–418. [DOI] [PubMed] [Google Scholar]
- 23.Puttaraju M., Jamison,S.F., Mansfield,S.G., Garcia-Blanco,M.A. and Mitchell,L.G. (1999) Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat. Biotechnol., 17, 246–252. [DOI] [PubMed] [Google Scholar]
- 24.Zahler A.M., Lane,W.S., Stolk,J.A. and Roth,M.B. (1992) SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev., 6, 837–847. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka K., Watakabe,A. and Shimura,Y. (1994) Polypurine sequences within a downstream exon function as a splicing enhancer. Mol. Cell. Biol., 14, 1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]