Abstract
Hoxa1 has diverse functional roles in differentiation and development. We identify and characterize properties of regions bound by HOXA1 on a genome-wide basis in differentiating mouse ES cells. HOXA1-bound regions are enriched for clusters of consensus binding motifs for HOX, PBX, and MEIS, and many display co-occupancy of PBX and MEIS. PBX and MEIS are members of the TALE family and genome-wide analysis of multiple TALE members (PBX, MEIS, TGIF, PREP1, and PREP2) shows that nearly all HOXA1 targets display occupancy of one or more TALE members. The combinatorial binding patterns of TALE proteins define distinct classes of HOXA1 targets, which may create functional diversity. Transgenic reporter assays in zebrafish confirm enhancer activities for many HOXA1-bound regions and the importance of HOX-PBX and TGIF motifs for their regulation. Proteomic analyses show that HOXA1 physically interacts on chromatin with PBX, MEIS, and PREP family members, but not with TGIF, suggesting that TGIF may have an independent input into HOXA1-bound regions. Therefore, TALE proteins appear to represent a wide repertoire of HOX cofactors, which may coregulate enhancers through distinct mechanisms. We also discover extensive auto- and cross-regulatory interactions among the Hoxa1 and TALE genes, indicating that the specificity of HOXA1 during development may be regulated though a complex cross-regulatory network of HOXA1 and TALE proteins. This study provides new insight into a regulatory network involving combinatorial interactions between HOXA1 and TALE proteins.
Hoxa1 displays the earliest expression during mouse embryogenesis of the clustered Hox genes (Hunt et al. 1991; Murphy and Hill 1991) and is one of the most rapidly induced genes during retinoid-induced differentiation of murine ES cells (Lin et al. 2011; De Kumar et al. 2015). Hox genes are subject to extensive auto- and cross-regulatory interactions (Tumpel et al. 2009; Parker et al. 2016). The characterization of Hox-response elements associated with these regulatory processes in vertebrate and invertebrate species has uncovered important roles for PBX/EXD and MEIS/HTH as HOX cofactors (Chan et al. 1994; Pöpperl et al. 1995; Rieckhof et al. 1997; Ryoo et al. 1999; Ferretti et al. 2000; Merabet et al. 2007). PBX and MEIS are members of the TALE (three amino-acid loop extension) family of homeodomain proteins, which consists of six different classes: IRX, MKX, MEIS, PBX, PREP, and TGIF (Burglin 1997). Beyond PBX and MEIS, very little is known about whether there are similar roles for other TALE proteins as cofactors contributing to HOX binding properties. PBX and MEIS have more general roles as cofactors for a variety of homeodomain and nonhomeodomain classes of transcription factors (e.g., PDX1, EN, PAX, ZFPIP [Zn finger], PTF1 [bHLH], and THRA). Genetic and regulatory studies have shown that the general cofactor roles of PBX and MEIS have diverse functional inputs in regulating cell and developmental processes (Moens and Selleri 2006; Laurent et al. 2008; Schulte and Frank 2014).
Physical interactions between PBX and HOX proteins have been shown to enhance their binding specificity and in vivo site selection (for review, see Mann and Chan 1996; Merabet and Mann 2016). This is achieved in part through formation of a PBX-HOX heterodimer that binds on an overlapping bipartite HOX-PBX site (Jabet et al. 1999; Piper et al. 1999). PBX also forms a heterodimer with MEIS or PREP using a different domain, and this complex binds to a distinct PBX-MEIS/PREP site (Abu-Shaar et al. 1999; Berthelsen et al. 1999; Penkov et al. 2013). The HOX-PBX and PBX-MEIS/PREP sites are often in close proximity, which facilitates the formation of a ternary complex that helps in fine tuning binding specificity and potentiating regulatory activity (Berthelsen et al. 1998; Ryoo et al. 1999; Ferretti et al. 2000, 2005). MEIS/HTH proteins have also been shown to be important for controlling the nuclear localization of PBX/EXD and stability of PBX-mediated complexes (Rieckhof et al. 1997; Abu-Shaar et al. 1999; Berthelsen et al. 1999; Waskiewicz et al. 2001). PBX/EXD-HOX interactions are involved in repression as well as activation of target genes (Rauskolb and Wieschaus 1994). Hence, TALE proteins as HOX cofactors dictate transcriptional regulatory outputs in a context-dependent manner (Galant et al. 2002; Merabet et al. 2003). These studies illustrate the diverse and important roles of cofactors in modulating the binding specificity, selectivity, and functional outcomes of HOX proteins.
Previous studies investigating the conserved roles of PBX/EXD and MEIS/HTH as cofactors for HOX proteins on selected vertebrate and invertebrate loci have not addressed the contribution of other TALE proteins to HOX specificity. Similarly, very few genome-wide studies have been performed in mammalian systems to investigate the binding of HOX proteins and their relationships to TALE family members in a tissue-specific or developmental context (Jung et al. 2010; Donaldson et al. 2012; Huang et al. 2012; Sorge et al. 2012; De Kumar et al. 2017). Therefore, we use programmed differentiation of mouse ES cells in combination with genomic approaches to characterize the genome-wide binding properties and regulatory interactions of HOXA1 and a set of TALE (PBX, MEIS, PREP1, PREP2, TGIF) proteins.
Results
Characterization of ES cell line carrying an inducible epitope-tagged variant of HOXA1
In this study, we sought to characterize the genome-wide occupancy of HOXA1 and to understand the nature of its downstream targets. Due to the transient expression of Hoxa1 in development and the limiting amounts of appropriate embryonic neural tissue, we used the programmed differentiation of ES cells into neuroectoderm as a model system. Human and mouse ES cells are differentiated into neuronal fates using retinoic acid (RA), and this leads to sequential activation of Hox genes in a manner mimicking their induction during embryonic development (Simeone et al. 1990, 1991; Papalopulu et al. 1991; Mazzoni et al. 2013; Sheikh et al. 2014; De Kumar et al. 2015). Previously, we have extensively characterized RA-induced differentiation of KH2 ES cells and provided insight into the mechanistic basis for rapid and early induction of Hoxa1 in ES cells through paused polymerase and control of transcriptional elongation (Lin et al. 2011; Gaertner et al. 2012; De Kumar et al. 2015).
We generated a variant of the KH2 ES cell line (Beard et al. 2006) with a locus specific insertion (Col1A1) of a doxycycline (Dox)-inducible version of HOXA1 tagged with 3×-Flag and Myc epitopes at the C terminus (Fig. 1A). We optimized the expression of the epitope-tagged version of Hoxa1 in response to RA so that it was comparable to that of the endogenous gene. Three independent methods, single-molecule RNA fluorescent in situ hybridization (FISH), qPCR, and RNA-seq, demonstrated that the epitope-tagged version was expressed at a ratio of 1:1.5 compared to endogenous Hoxa1 (Supplemental Fig. S1B–D). Western blot hybridization showed that tagged HOXA1 protein is detected at 6 h following induction and that levels increase for 48 h (Supplemental Fig. S1A). Previous analyses indicated that a 24-h time point closely reflects gene expression profiles of early mouse neural ectoderm (De Kumar et al. 2015).
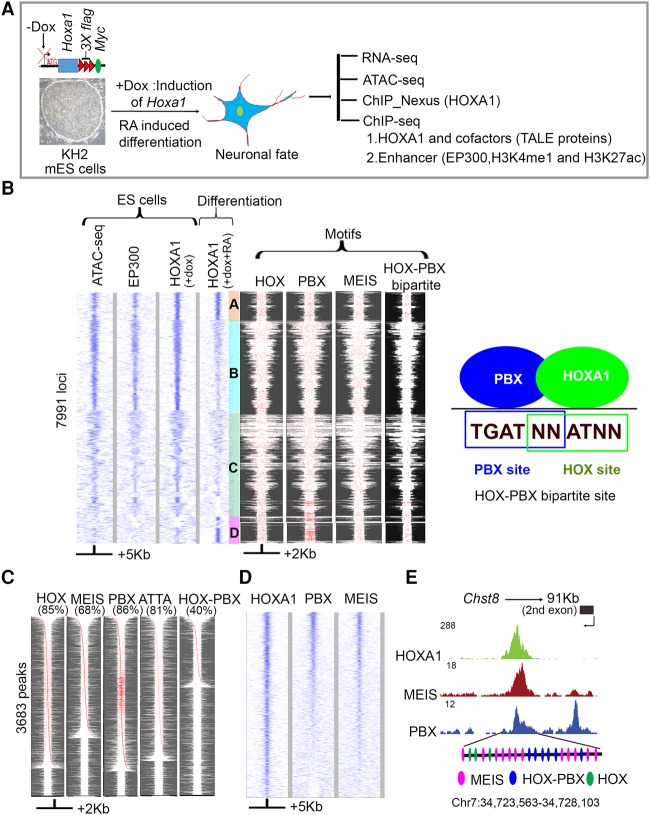
Figure 1.
Genome-wide identification and analysis of HOXA1-bound regions reveal overlaps with PBX and MEIS binding. (A) Experimental design for differentiation and analysis where Dox (+Dox) induces epitope-tagged Hoxa1 and retinoic acid (+RA) induces programmed differentiation of ES cells into neural fates. (B) Heat maps of genome-wide occupancy of HOXA1 in ES cells (+dox) and differentiated cells (+dox +RA) defined by ChIP-seq. The occupancy of EP300 and chromatin state (ATAC-seq) in ES cells is also shown. Analyses of ES and differentiated cells reveal four distinct groups (A–D) of bound regions. The four columns on the right show the spatial distribution of consensus motifs for HOX, PBX, MEIS, and HOX-PBX bipartite binding sites in the corresponding genomic regions. Best FIMO matches (P ≤ 1 × 10−4) for all variants of the HOX, PBX, and MEIS, and HOX-PBX bipartite motifs are highlighted in red. A diagram illustrates the relationship of PBX and HOX binding sites in HOX-PBX bipartite motifs (far right panel). Groups A and D, representing HOXA1-bound regions in differentiated cells, show increased presence of HOX-PBX bipartite sites and tandem clusters of PBX motifs. (C) HOXA1-bound regions in differentiated cells are enriched for consensus HOX, PBX, and MEIS motifs. For each TransFac motif, FIMO matches (P ≤ 1 × 10−4) are shown in red. The respective motif and % of peaks with the motif are indicated (top of each column). Each row is independently sorted by the distance of the central-most motif to the peak center. Many rows of HOXA1-bound regions show multiple binding motifs in a peak (red dots). (D) Heat map shows extensive co-occupancy of HOX, PBX, and MEIS on HOXA1 targets in differentiated cells. The plots use IP coverage of z-score matrix for PBX and MEIS binding at HOXA1 peaks. Rows are sorted by the intensity of the PBX signal within the central 1-kb region. In B–D, the size of regions flanking the center of the peak of HOXA1 binding are shown at the bottom. (E) UCSC Genome Browser shots showing PBX, MEIS, and HOXA1 in exon 2 of Chst8. Clustering of multiple HOX, HOX-PBX, and MEIS binding sites are also shown at the bottom for the HOXA1-bound region.
Clustered binding sites for HOX, PBX, and MEIS
We performed HOXA1 ChIP-seq experiments in undifferentiated and RA-induced ES cells. ES cells normally do not express Hoxa1, but since they represent a well-characterized cellular state, we were interested in the potential of HOXA1 to bind in this cell type. Therefore, we induced HOXA1 in ES cells by Dox treatment. ChIP-seq experiments identified HOXA1 binding in 8836 genomic regions near 6029 genes (Fig. 1B; Supplemental Table S1). These peaks are found in regions of open chromatin, as assayed using ATAC-seq (assay for transposase-accessible chromatin with high throughput sequencing) and show occupancy of coactivators (EP300) (Fig. 1B). In contrast, there is a marked change in the binding profile upon RA-induced differentiation of the ES cells. Only a subset of the HOXA1-bound regions in ES cells retain occupancy upon differentiation (group A), and many other regions show a reduction or loss of binding (groups B and C) (Fig. 1B). In differentiated cells, a total of 3682 HOXA1-bound regions near 2595 genes are found. This includes those that retain occupancy (group A) (Supplemental Table S2) and a new binding group (group D), which was not found in the ATAC assay for open chromatin in ES (Fig. 1B). These sites appear to become newly open during differentiation.
In ES and differentiated cells, while the number of peaks per gene was similar in both samples, the peaks in undifferentiated ES cells showed a relatively low enrichment over input, on average around twofold. In differentiated cells, most HOXA1-bound peaks showed greater enrichment (three- to sixfold) (Supplemental Fig. S2A). Nearly all top-ranking peaks correspond to bound regions in differentiated cells (Supplemental Fig. S2B). This suggests that the major proportion of HOXA1-bound regions in ES cells, have a low level of enrichment, presumably due to lower binding affinity compared to sites bound after differentiation.
To explore this difference in binding, we analyzed the motif content by searching for enriched consensus binding sites using HOMER (Heinz et al. 2010). Both HOX and HOX-PBX bipartite sites are significantly enriched in regions occupied by HOXA1 in differentiated cells (Supplemental Fig. S2C; Supplemental Tables S3, S4), suggesting that the increased enrichment in differentiated cells may be mediated in part by the association of HOXA1 with PBX and MEIS on HOX-PBX bipartite sites.
Beyond HOX-PBX bipartite sites, PBX, MEIS, and PBX/MEIS heterodimers also bind to a variety of unique and overlapping consensus motifs (Penkov et al. 2013). Hence, we extended our analysis to search for the presence and spatial distribution of these motifs in HOXA1-bound regions using FIMO (Matys et al. 2006) and motif definitions in the TransFac database (Fig. 1B,C). The subset of HOXA1-bound regions that retained occupancy upon differentiation (group A) and the set of new binding sites that appeared in differentiated cells (group D) both showed increased presence of HOX-PBX bipartite sites and tandem clusters of PBX motifs (Fig. 1B). Further analysis of the peaks in differentiated cells reveals that they contain known consensus motifs for HOX (85%), PBX (86%), MEIS (68%), and HOX-PBX bipartite sites (40%) which are often present in multiple copies per peak (Fig. 1C). The high fraction of motifs among HOXA1-bound regions suggests that the increased binding of HOXA1 in differentiated cells may be related to the cis-regulatory motif composition.
The presence of PBX and MEIS motifs raises the question of whether these cofactors physically occupy these genomic loci along with HOXA1. Hence, we performed ChIP-seq experiments in differentiated cells using α-PBX1/2/3 and α-MEIS1/2 antibodies. We identified 5761 PBX and 1410 MEIS binding regions (1% IDR), many of which display co-occupancy of PBX and MEIS (Fig. 1D). The regions bound by each factor were enriched for the corresponding consensus motifs, similar to those previously identified in 11.5-d mouse embryos (Supplemental Tables S5, S6; Penkov et al. 2013), confirming the specificity of these antibodies. The larger number of PBX-bound regions compared to MEIS is consistent with its more general role as a cofactor for a variety of TFs (Laurent et al. 2008; Schulte and Frank 2014). In comparing these genome-wide profiles with HOXA1-bound regions, a large number also display co-occupancy of PBX and MEIS (Fig. 1D). Strong occupancy of HOXA1, MEIS, and PBX was particularly observed at loci with multiple motifs. For example, all three transcription factors occupy a region at the Chst8 locus where multiple MEIS, HOX, and HOX-PBX bipartite sites are present (Fig. 1E).
To examine HOXA1 binding at high-resolution, we used the lambda exonuclease-based ChIP-nexus approach (He et al. 2015). ChIP-nexus revealed that single peaks identified by ChIP-seq often represent clusters of binding events. HOMER analysis of the ChIP-nexus binding profiles showed that HOXA1 was not only bound to HOX and HOX-PBX bipartite motifs but also to PBX and MEIS consensus motifs (Supplemental Table S7). HOXA1 physically co-occupies at least some of the sites with PBX and MEIS, indicating that HOXA1 may bind to DNA directly or indirectly through association with PBX and MEIS as cofactors.
In addition to PBX and MEIS motifs, our de novo motif analysis from ChIP-seq and ChIP-nexus revealed a series of enriched motifs for other transcription factors coassociated with HOXA1 binding (Supplemental Tables S4, S7). Some of these transcription factors, including KROX20, SOX1, TCF3, and MAFB, are known to genetically interact with Hox genes in developmental contexts and may contribute to binding of HOXA1. Together, these data indicate that, on a genome-wide basis, HOXA1 binding is frequently associated with PBX and MEIS proteins and that a wide variety of motif combinations underlie and are coassociated with the binding of HOXA1.
Distinct combinations of TALE proteins on HOXA1-bound regions
HOXA1-bound regions are frequently co-occupied by PBX and MEIS, hence, we investigated whether other TALE proteins may be associated with HOXA1-bound regions. Pbx1-3, Tgif1-2, and Prep1 are expressed in ES cells, and upon RA treatment these and other TALE genes show dynamic changes in their expression profiles (Supplemental Fig. S3A–C). There is a significant increase in the levels of Pbx1-2, Meis1-3, and Prep2, while Tgif1-2 and Pbx4 are down-regulated, generating a changed repertoire of TALE cofactors. Therefore, we performed additional ChIP-seq experiments with α-TGIF, α-PREP 1, and α-PREP2 antibodies to compare the genome-wide binding profiles of the TALE family before and after differentiation.
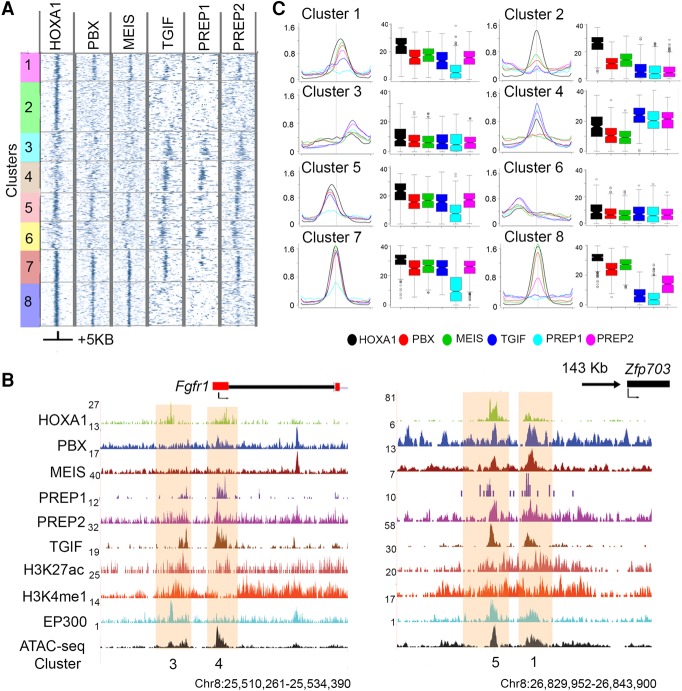
Our analysis revealed that nearly all HOXA1-bound regions display occupancy with one or more TALE proteins, yet each TALE protein has a unique binding pattern (Fig. 2). Eight classes were identified through k-means clustering (Cluster 1–8), based on their distinct relative levels of TALE proteins and their spatial relationship to the center of HOXA1 binding (Fig. 2A,B). The Fgfr1 and Zfp703 loci are shown as examples of four of these binding classes (Fig. 2C,D).
Figure 2.
Patterns of co-occupancy of TALE proteins on HOXA1 targets define eight distinct clusters. (A) Heat maps for occupancy of individual TALE factors at HOXA1 peaks using IP coverage of z-score matrix after k-means-clustering (k = 8). Only positions of positive z-score are shown. A ±5-kb region flanking the center of the peaks of HOXA1 binding is shown. (B) Average binding levels, shown as line and box plots, illustrate the distinct properties with respect to relative binding of each protein and its spatial distribution of each cluster in A. Box plots are row sums from the central 1 kb of each column. (C) UCSC Genome Browser shots (coordinates from version mm10) of Fgfr1 and Zfp703 loci, showing HOXA1-bound regions along with occupancy of TALE cofactors, activator protein EP300, enhancer marks, and chromatin state. The clusters to which these bound regions map are noted at the bottom.
The characteristics of these classes uncover distinct interactions of TALE family members on HOXA1-bound regions. Cluster 8 represents the classical group where PBX and MEIS are the primary cofactors for HOXA1, and there is little or no binding of other TALE members. In contrast, Cluster 4 contains a group of targets with little or no occupancy of PBX and MEIS but high levels of binding with other TALE proteins. Cluster 7 illustrates a class where nearly all TALE proteins co-occur (Fig. 2).
To explore whether the clusters of target genes are potentially associated with different functions, we performed a KEGG pathway analysis on the genes associated with each cluster. There is an enrichment of distinct and overlapping pathways for each cluster (Supplemental Fig. S4). For example, Clusters 3, 4, 6, and 8 are all enriched for components of the Hippo signaling pathway. Clusters 4 and 6 are also enriched for Hedgehog signaling, while Clusters 6 and 8 are enriched for Wnt signaling components. This suggests that the binding patterns observed in different clusters may be functionally relevant for context-dependent regulation of diverse biological processes by HOXA1. Together, these findings indicate that the TALE family provides a wide repertoire of HOX cofactors beyond PBX and MEIS.
TALE binding preferences are associated with distinct classes of cis-motifs
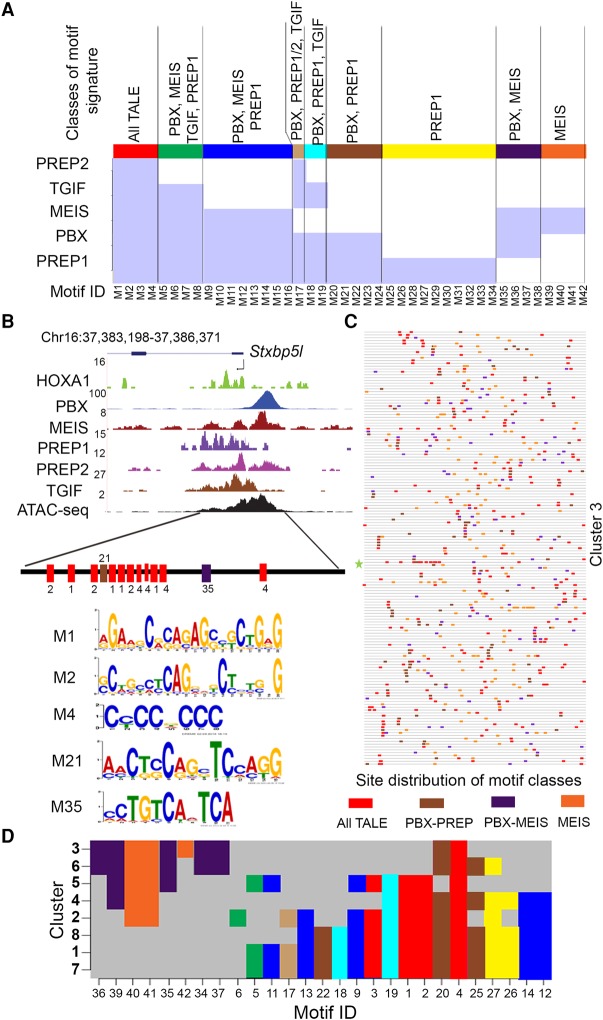
The clusters of differential occupancy of TALE proteins raise the possibility that in each cluster there is a distinct combination of motif signatures at the level of cis-elements (combinatorial code). This generates permutations that mediate differential recruitment of TALE proteins to the HOXA1-bound regions. To explore this, analysis of the general motif composition of the genome-wide binding preferences of individual TALE proteins generates a series of known and novel enriched consensus motifs for each protein. We then compared the relative enrichment of these motifs in regions bound by other TALE proteins (Supplemental Tables S6, S9). We define nine distinct classes of motif signatures that reflect unique and overlapping combinations of binding preferences (Fig. 3A; Supplemental Table S9). For example, motifs M1–M4 are enriched for all TALE proteins, while M25–M34 are specifically enriched in PREP1-bound regions.
Figure 3.
Unique and overlapping genome-wide binding preferences of individual TALE proteins and their distribution in HOXA1-bound regions. (A) MEME and AME (analysis of motif enrichment) identify motifs in genome-wide binding preferences for members of the TALE family (PBX, MEIS, TGIF, PREP1, and PREP2). There are 42 enriched motif signatures (M1–M42) comprised of both novel and known consensus TALE binding sites. These 42 cis-motifs are divided into nine distinct classes based on their presence or absence within regions bound by one or more of the TALE family members (shaded light blue areas in the matrix). Each class is identified by a separate color (top) along with the respective TALE family members showing occupancy on these motifs. (B) UCSC Genome Browser shot (mm10) showing occupancy of HOXA1 and TALE factors near the Stxbp5l locus (a representative binding region in Cluster 3). The diagram at the bottom illustrates the five different motifs (M1, M2, M4, M21, and M35) and represents three classes of motif signatures defined in A, above. There are multiple copies of three of the cis-motifs (M1, M2, and M4). (C) A block diagram shows the occurrence of four distinct motif classes with multiple copies in all the loci present in Cluster 3. Each respective motif class is colored as indicated in A. Length of peaks are not drawn to scale. (D) A matrix indicating the presence or absence of the individual classes of cis-motif signatures in the respective clusters defined by combinatorial TALE binding on HOXA1-bound regions. Distribution of various motif signature classes in each cluster are colored as defined in A. This plots the unique combinations of motif signatures which underlie the differential binding of TALE proteins in each cluster.
We then looked at the presence or absence of these individual classes of cis-motif signatures in the respective clusters defined by TALE binding and found that, apart from Clusters 1 and 7, each cluster had a distinct and unique combination of motif signatures (Fig. 3D). Hence, the binding patterns found in Clusters 1–8 correlate with the underlying specific patterns of motif enrichment. For example, in Cluster 5, PREP1 displays a low level of occupancy compared to the other TALE proteins, and there is a complete absence of PREP1-specific motifs (Figs. 2, 3D). Cluster 2–6 are enriched for MEIS/PBX and MEIS classes and have a reduced repertoire of other classes of motifs when compared to the other clusters. Together, these data suggest a combinatorial cis-binding code provides a mechanistic basis for differential recruitment of TALE proteins to the HOXA1-bound regions.
In general, the clusters of differential TALE occupancy in HOXA1-bound regions consist of multiple cis-motifs from these classes. Stxbp5l is an example of a binding profile of a locus from Cluster 3 containing five different motifs, representing four classes of motif signatures (Fig. 3B). There are multiple copies of three of the cis-motifs (M1, M2, and M4). Similar patterns with multiple motifs and classes are observed for all loci in Cluster 3 (Fig. 3C). These findings reinforce the observation that in HOXA1-bound regions, there are distinct combinations of multiple binding motifs for both HOXA1 and TALE proteins. Enriched motifs for other transcription factors in each cluster may also contribute to differential patterns of binding (Supplemental Table S10).
Functional roles of HOX-PBX and TGIF motifs in HOXA1-bound regions
The computational analysis above identified various motif combinations enriched for TALE proteins, including HOX-PBX bipartite sites, among HOXA1-bound regions (Supplemental Tables S4, S6). Many of these sites also showed specific footprints in ChIP-nexus data confirming their occupancy at the motifs (Supplemental Table S7). To functionally validate the importance of HOX-PBX and TALE sites, we assayed HOXA1-bound regions for cis-regulatory activity in zebrafish (Fig. 4; Supplemental Fig. S5). We focused on regions that display enhancer-related histone modifications (H3K27Ac and H3K4me1), occupancy of coactivator EP300, and an open chromatin confirmation based on ATAC-seq. In examining the putative target genes of these regions, we noticed that multiple candidate enhancers are located near the TALE genes themselves. Since these enhancers could mediate auto- and cross-regulation among Hox and TALE genes, we specifically tested these regions for enhancer activity in vivo.
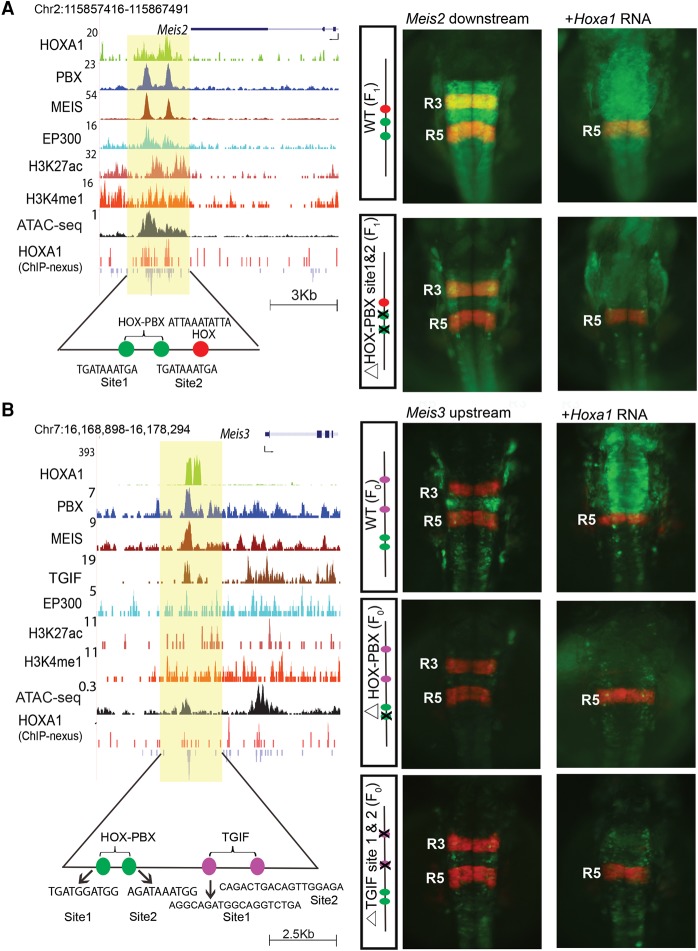
Figure 4.
The Meis2 and Meis3 genes are downstream targets of HOXA1. UCSC Genome Browser shots of Meis2 (A) and Meis3 (B) show HOXA1-bound regions along with occupancy of PBX and MEIS, activator protein EP300, and open chromatin. For Meis3, occupancy of TGIF is also shown. ChIP-nexus data reveal the presence of multiple binding motifs for HOXA1 and TALE cofactors in the protected regions as noted at the bottom. The shaded area indicates bound regions. Panels to the right are transgenic zebrafish reporter images demonstrating the regulatory potential of the binding regions ±250 bp. For both Meis2 and Meis3, transgenic embryos show restricted reporter expression in the hindbrain and a response to ectopic expression of Hoxa1, as indicated by the loss of RFP (red)-specific expression in R3 and expansion of GFP (green) expression. In both cases, deletion of the HOX-PBX sites abolishes or reduces GFP reporter activity and reduces response to ectopic Hoxa1 mRNA. For Meis3 (B), mutation of the two TGIF motifs in the enhancer result in low expression of reporter and weak anterior expansion upon coinjection with Hoxa1 mRNA. HOX-PBX bipartite motifs and TGIF motifs in each of the enhancers are shown at the bottom. All embryos are shown in dorsal views. (F0) Founder embryos, (F1) stable lines carrying the reporter.
Transgenic GFP-reporter assays demonstrate that the regions from Pbx1, Meis1-3, and Tgif2 all mediate restricted expression in the developing hindbrain and other neural regions in zebrafish, implying that they can function as enhancers (Fig. 4; Supplemental Fig. S5). The enhancer for Meis2 mediates expression in r2-r8 in the hindbrain, while that of Meis3 directs expression primarily in r4 and r7-spinal cord (Fig. 4). To test whether these regions are responsive to Hoxa1 dosage, we assayed reporter expression after injection of Hoxa1 mRNA. Indeed, the reporter expression domains directed by the wild type (WT) Meis2 and Meis3 enhancers expanded into more anterior regions of the hindbrain, midbrain, and forebrain, showing they are responsive to ectopic Hoxa1 in vivo (Fig. 4A,B).
Both the Meis2 and the Meis3 enhancers contain two HOX-PBX sites (sites 1 and 2), and ChIP-seq data reveal occupancy of HOXA1, PBX, and MEIS. However, ChIP-nexus data indicate occupancy of HOXA1 only on site 1 in both cases (Fig. 4). When we specifically mutated the HOX-PBX site 2 of the Meis2 enhancer, which lacked binding of HOXA1, no effect on transgene activity was observed (data not shown). However, deletion of the HOX-PBX motifs that were bound by HOXA1 led to a marked reduction of transgene expression for both the Meis2 and Meis3 enhancers, and they weakly respond to ectopic Hoxa1 expression. This shows that the HOX-PBX bipartite sites bound by HOXA1 are required for regulatory activity and respond to Hoxa1 in vivo, implying that these HOX-PBX bipartite sites play important roles in potentiating Hoxa1-mediated regulation of these TALE genes.
In the Meis3 enhancer, we also identified two putative TGIF binding motifs adjacent to the HOX-PBX motifs and observed co-occupancy of TGIF (Fig. 4B). Mutation of the TGIF motifs from this Meis3 enhancer resulted in considerable reduction in reporter expression and a failure to respond to ectopic Hoxa1 (Fig. 4B). Together, these regulatory analyses show that both the TGIF and HOX-PBX motifs contribute to regulatory activity and are required for the HOXA1-dependent function of these enhancers.
Physical interaction of HOXA1 with TALE proteins
The clustering of diverse HOX, HOX-PBX, and TALE motifs within HOXA1-bound regions and evidence for co-occupancy of TALE proteins raises the question of whether these factors physically interact on chromatin. Previous evidence suggests cooperative binding among HOX, PBX, MEIS, and PREP proteins and the formation of ternary complexes involving HOX, PBX, MEIS, and PREP, bound to HOX-PBX and MEIS-PREP sites (Ferretti et al. 2000, 2005; Penkov et al. 2000, 2013). However, whether all TALE proteins act through direct physical interactions with HOXA1 is not clear. Since our analysis of the Meis3 enhancer indicates the importance of regulatory inputs from both TGIF and HOX-PBX motifs, we wondered whether TGIF proteins can also participate in physical interactions with HOX or HOX-PBX complexes.
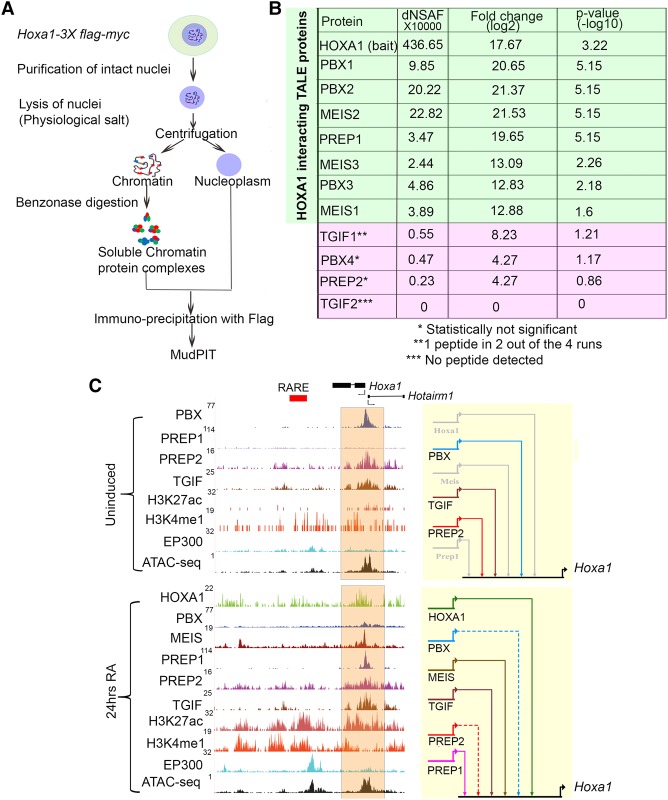
To address this question, we performed proteomic analysis on the epitope-tagged HOXA1 ES line to search globally for HOXA1 interacting partners in RA-differentiated cells (Fig. 5A). We immunoprecipitated chromatin-bound protein complexes containing HOXA1 (Aygun et al. 2008) and identified the interacting proteins with mass spectrometry-based multi-dimensional protein identification technology (MudPIT). Four independent differentiated cell samples were analyzed and compared with a control cell line lacking the epitope-tagged HOXA1 (Supplemental Table S8). Quantitative proteomic analysis revealed that, among all interacting proteins detected, MEIS2 and PBX2 are among the top 20 enriched proteins. Furthermore, other TALE proteins such as PBX1, PBX3, MEIS1, MEIS3, and PREP1 were significantly enriched with HOXA1-bound proteins on chromatin (Fig. 5B). These data for HOXA1 are consistent with published studies and add further in vivo support for the ability of HOX proteins to interact with PBX, MEIS, and PREP on DNA.
Figure 5.
Interactions between HOXA1 and TALE proteins on chromatin and binding of TALE proteins to the Hoxa1 locus. (A) Strategy for isolation and identification of chromatin-bound complexes interacting with HOXA1 by immunoprecipitation and MudPIT. (B) MudPIT analyses averaged from four replicates showing dNSAF(X10000), fold enrichment, and P-values of TALE proteins associated with HOXA1 on chromatin. (C) UCSC Genome Browser shots (mm10) for the Hoxa1 locus. Binding of HOXA1 and occupancy of the TALE family members, modified histone marks (H3K4me1 and H3K27Ac) characteristic of enhancers, occupancy of the EP300 activator protein, and accessibility of chromatin (ATAC-seq) are shown in uninduced (top) and differentiated (bottom) cells. The orange shaded area indicates Hoxa1-bound regions near the promoter. In the BioTapestry plots (right panels), solid colored lines indicate binding of each specific TALE factor, while gray lines indicate lack of binding. Dotted lines represent decreased binding.
Interestingly, no unique peptides were detected for TGIF2 and the levels of detection for TGIF1, PBX4, and PREP2 were not statistically significant (Fig. 5B; Supplemental Table S8). We suggest that HOXA1 globally interacts with or is part of chromatin-bound complexes containing the PBX, MEIS, and PREP family of proteins, but there is no evidence for similar physical interactions with TGIF family members. Since we show that TGIF motifs are bound by TGIF and are necessary for enhancer activity in the case of MEIS3, TGIF proteins may provide a parallel regulatory input independent of interactions with HOXA1.
Auto- and cross-regulatory interactions between Hoxa1 and TALE genes
Extensive occupancy of HOXA1 and TALE proteins on the TALE genes themselves and our validation of many of these putative enhancers in transgenic reporter assays (Fig. 4; Supplemental Fig. S5) suggest frequent auto- and cross-regulatory interactions between HOXA1 and TALE regulators. Since Pbx1-3, Tgif1-2, and Prep1 are expressed in ES cells, we performed ChIP-seq experiments and found that all three factors bind to a region upstream of the Hoxa1 promoter. This is illustrated by a browser shot of the Hoxa1 locus and the combined regulatory inputs depicted as a BioTapestry model (Fig. 5C; Longabaugh et al. 2005). The putative Hoxa1 enhancer is accessible based on ATAC-seq, but epigenetic enhancer marks are absent. This is consistent with the absence of Hoxa1 expression in ES cells but the presence of a paused polymerase on the Hoxa1 promoter and the rapid induction of Hoxa1 upon RA-induced differentiation (Lin et al. 2011; De Kumar et al. 2015). Upon RA-induced differentiation, the occupancy of TALE proteins and the epigenetic state undergo a dynamic change (Fig. 5C). Binding of PBX and PREP2 is reduced, while MEIS and PREP1 are recruited to this site along with the appearance of histone modifications associated with active enhancers. We observe increased occupancy of HOXA1 over this region, suggesting that it may provide auto-regulatory input into its own expression (Fig. 5C). Similarly, we find that the genes encoding TALE proteins are themselves downstream targets of HOXA1 and TALE cofactors (Fig. 4; Supplemental Fig. S5), suggesting the presence of extensive auto- and cross-regulatory interactions.
To test whether these auto- and cross-regulatory elements are functional in vivo, we used reporter assays in zebrafish to monitor the activity of 13 of these regions. We found that 11 of them mediate reporter expression in neural tissues (Figs. 4, 6, 7; Supplemental Fig. S5; Supplemental Table S11). For example, TALE- and HOXA1-bound regions flanking Meis1, Pbx1, and Tgif2 mediate expression of a GFP reporter in the hindbrain and some other areas of the nervous system (Supplemental Fig. S5). Thus, we confirm that a large fraction of these putative auto- and cross-regulatory elements function as enhancers in vivo contributing to regulation of TALE genes.
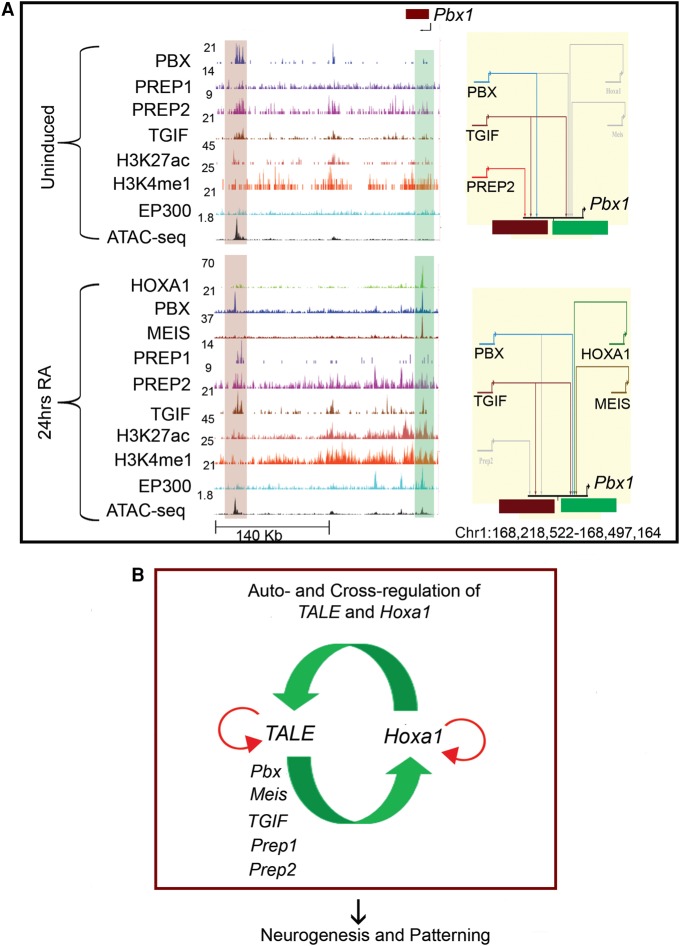
Figure 6.
Auto- and cross-regulatory interactions between HOXA1 and TALE proteins. (A) UCSC Genome Browser shots (left) for the Pbx1 locus showing binding of HOXA1 and occupancy of the TALE family members in ES cells (top) and differentiated cells (bottom). The presence of modified histone marks (H3K4me1 and H3K27Ac), occupancy of the EP300 activator protein and ATAC-seq at these genomic loci are also shown. The shaded areas indicate bound regions. In the BioTapestry plots (right panels), solid colored lines indicate binding of TALE factors, gray lines indicate lack of binding, and dotted lines represent decreased binding. (B) Model summarizes auto- and cross-regulatory interactions among Hoxa1 and TALE genes and their output on downstream targets.
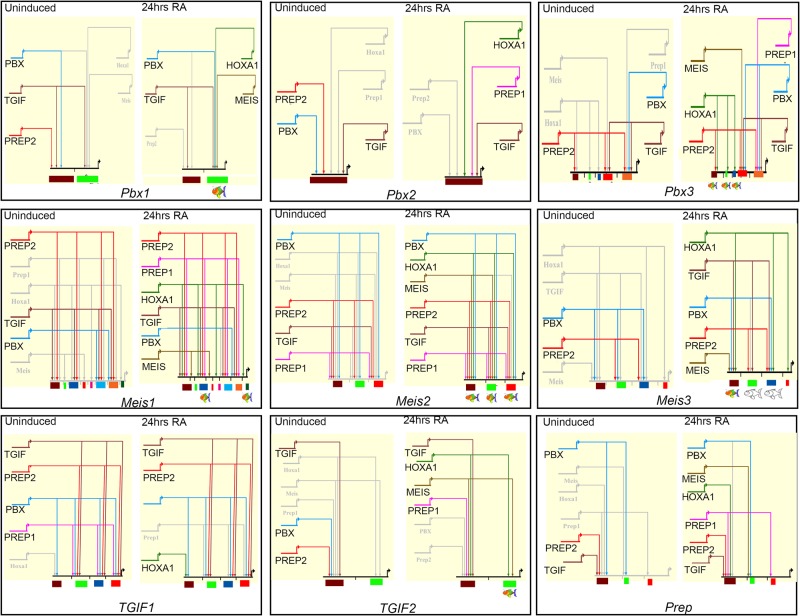
Figure 7.
BioTapestry models depicting the auto- and cross-regulatory interactions between HOXA1 and TALE members. ES cells are depicted on the left and differentiated cells on the right. Solid colored lines indicate binding of each specific TALE factor or HOXA1, gray lines indicate lack of binding, and dotted lines represent decreased binding. At the bottom, the colored boxes indicate each TALE locus and the respective cis-regulatory regions that integrate the specific binding inputs. Colored cartoon fish under cis-regulatory elements indicate those with spatially-restricted regulatory activity, while the uncolored fish indicate a lack of reporter activity.
By analyzing the occupancy of HOXA1 and TALE proteins on these cis-regulatory regions before and after differentiation, we observe a complex and dynamic interplay among these factors. There are diverse patterns of binding on multiple sites spread in and around these loci and we observed changes in occupancy during differentiation (Figs. 6A, 7). For example, a single 5′ cis-regulatory element of Pbx2 is occupied by PREP2, TGIF, and PBX in ES cells, but upon differentiation, the occupancy of PREP2 and PBX is replaced by that of HOXA1 and PREP1. Likewise, two regions flanking the Pbx1 gene are bound by PBX, TGIF, and PREP2 in ES cells. However, this binding pattern changes upon RA treatment, when HOXA1, MEIS, and PBX are recruited to a second distinct downstream region, while TGIF remains bound to both enhancers (Figs. 6A, 7). These examples suggest that an array of cis-elements integrates dynamic inputs by HOXA1 and TALE proteins to mediate specific expression during differentiation.
To functionally test a Hoxa1 role in regulating TALE genes predicted by this interaction network, we used CRISPR/Cas9 technology to generate three independent ES cell lines with one copy of Hoxa1 deleted (Hoxa1+/−) (Supplemental Methods). In these lines, there was an ∼50% reduction in Hoxa1 expression levels compared to wild-type ES cells (Supplemental Fig. S3D; Supplemental Table S12). To examine the impact of this reduction in Hoxa1 expression, we quantified levels of TALE mRNA in these lines using qPCR and compared it to levels in nonmanipulated wild-type ES cells (Supplemental Fig. S3E; Supplemental Table S12). We detected changes in the levels of expression of several TALE genes. For example, expression of Pbx1 and Meis3 are decreased upon loss of one copy of Hoxa1, while Pbx2, Pbx3, Meis1, Prep1, and Prep2 display increased expression. This implies that, through cross-regulatory interactions, variations in the levels of HOXA1 contribute to the relative levels and availability of TALE proteins, which in turn could impact its binding to downstream targets. From a gene regulatory network perspective, these interactions imply that Hoxa1 and TALE genes utilize an extensive auto- and cross-regulatory circuitry to control their expression, which may be important for their functions during patterning and development (Fig. 6B).
Discussion
We have identified and characterized regions bound by HOXA1 on a genome-wide basis in differentiating mouse ES cells. These analyses show that HOXA1 binding is frequently associated with a wide variety of motif combinations, including consensus sites for PBX and MEIS. High resolution mapping (ChIP-nexus) of individual binding peaks revealed evidence for multiple sites of HOXA1 binding on a series of distinct consensus motifs for HOX, HOX-PBX, PBX, and MEIS (Figs. 1E, 5). PBX and MEIS also show physical occupancy on these HOXA1-bound regions (Fig. 1D), and transgenic enhancer reporter assays in zebrafish demonstrate the importance of selected HOX-PBX sites. Finally, proteomic analysis of HOXA1-interacting complexes on chromatin indicates physical interactions between HOXA1 and PBX and MEIS family members (Fig. 5B). These data strongly support that HOXA1 is recruited to many of its target sites through physical interactions with PBX and MEIS as cofactors, as previously reported based on individual examples (Berthelsen et al. 1998; Ryoo et al. 1999; Ferretti et al. 2000, 2005).
In addition to the strong association of HOXA1 with PBX and MEIS, we also uncovered associations with other members of the TALE family. PREP1, PREP2, and TGIF also occupy HOXA1-bound regions and their binding motifs are enriched (Fig. 2). Transgenic enhancer reporter assays demonstrate that TGIF-bound motifs are required for regulatory activity (Fig. 4). This suggests that TALE proteins represent a wider repertoire of HOX cofactors and coregulators on HOX target genes than previously appreciated. On the other hand, proteomic analysis of HOXA1-interacting complexes on chromatin indicates that there are physical interactions of HOXA1 with PREP family members but not with TGIF (Fig. 5B). Thus, HOX proteins may not universally interact with all TALE proteins, which may reflect cell type specificity, differences between individual HOX proteins, and/or variations in the binding ability of individual subclasses of TALE proteins. The exact nature by which additional TALE proteins contribute to HOXA1 binding and enhancer activity remains to be elucidated.
The extent to which nearly all HOXA1-bound regions are associated with the occupancy of diverse combinations of TALE proteins is remarkable. These binding patterns fall into eight distinct clusters based on differential occupancy of TALE proteins. Motif enrichment analysis reveals that these clusters are composed of a unique collection of cis-elements. These cis-elements include distinct classes of TALE motif signatures, which match the occupancy of the corresponding proteins. Furthermore, these distinct binding clusters may be biologically meaningful because KEGG analysis shows that the target genes associated with the different clusters are involved in different processes (Supplemental Fig. S4). This underscores the important general role that the TALE family proteins have in broadly modulating the specificity and contributing to the function of HOX proteins. It suggests that a unique combinatorial cis-binding code provides a mechanistic basis that determines HOXA1 binding specificity through the differential recruitment of various permutations of TALE proteins. The combination of multiple binding preferences and clustering of motifs for both HOXA1 and TALE proteins may add robust recruitment and contribute to overall specificity to these regions as HOX response elements.
An interesting common feature of the cis-element composition is the presence of multiple sites for HOXA1 and TALE cofactors. Comparative analysis of differential binding properties of HOXA1 in ES cells and differentiated cells suggests that the presence of clustered motifs results in more binding enrichment in differentiated cells (Fig. 1B; Supplemental Fig. S2). This is consistent with the idea that HOXA1 binding is stabilized through cooperative interactions with TALE proteins, and thus multiple sites could add robustness to HOXA1 binding (Berman et al. 2002; Crocker et al. 2015; Farley et al. 2015). However, in the Meis2 and Meis3 enhancers, some HOX-PBX sites within HOXA1-bound regions were more strongly occupied by HOXA1, and these sites were functionally more important (Fig. 4). This suggests that HOX-PBX sites may not always be sufficient for binding even when the enhancer region is accessible. Efficient binding of Hox1 to HOX-PBX sites may require additional factors, which in turn vary in a tissue-specific fashion. Indeed, while we have focused on the clustering of HOX and TALE motifs in HOXA1-bound regions in this study, we have also observed enrichment for a variety of other novel and known motifs for factors such as, SOX1, KROX20, TCF3, and MAFB (Supplemental Table S4). Furthermore, individual TALE proteins might themselves occupy a variety of different motifs (Supplemental Tables S6, S10). Together, this opens the possibility that the HOX binding code is highly complex and tissue-specific, with multiple additional proteins contributing to its specificity and robustness.
From a gene regulatory network perspective, our genome-wide binding analyses point to an extensive series of auto- and cross-regulatory interactions among Hoxa1 and TALE genes in ES cells and their differentiated derivatives (Figs. 5–7). HOXA1 and TALE proteins bind to several cis-regulatory regions around Hoxa1 and TALE genes, and many putative enhancers for TALE genes indeed mediate restricted expression in the zebrafish hindbrain and function as Hoxa1-responsive elements (Fig. 4; Supplemental Fig. S5). This is consistent with the broad expression of TALE proteins during hindbrain patterning and the phenotype of Pbx mutants in mice and zebrafish, which recapitulate some of the Hox loss-of-function phenotypes in the hindbrain and other tissues (Pöpperl et al. 2000; Waskiewicz et al. 2001, 2002; Moens and Selleri 2006; Vitobello et al. 2011). It is also consistent with the conserved role for auto-and cross-regulatory interactions between Hox genes in hindbrain patterning (Parker et al. 2016). Therefore, the auto- and cross-regulatory interactions we characterized using the ES cell model are likely to be relevant for patterning of the hindbrain and other neural tissues during embryonic development.
Taken together, our data suggest a model in which auto- and cross-regulatory feedback interactions among Hoxa1 and TALE genes result in the tissue-specific regulation of Hoxa1 and cofactors (summarized in Fig. 6B). The specific combination of factors in each tissue then specifies and potentiates the downstream roles of HOXA1 in patterning and development.
Methods
Experimental animals
All experiments involving zebrafish (Protocol ID: 2015-0149) and mice (Protocol ID: 2016-0164) were done under approved protocols issued to R.K. as the P.I. by the Institutional Animal Care and Use Committee of the Stowers Institute for Medical Research.
ChIP and ChIP-nexus
Epitope-tagged Hoxa1 KH2 ES cells were differentiated with doxycycline and retinoic acid for 24 h. ChIP was performed per the Upstate protocol described in Smith et al. (2010) and peaks were selected based on IDR (≤0.01). ChIP-nexus performed and analyzed as described by He and coworkers (He et al. 2015).
Coverage heat maps
Signal across peak coordinates were visualized with the CoverageView package in R (https://rdrr.io/bioc/CoverageView/; R Core Team 2016) using windows ±5 kb from peak midpoints. To focus on binding trend and not magnitude, IP z-score values were used for transcription factor samples. For ATAC-seq, we transformed the raw IP counts matrix to percent-of-maximum-value, so that values would be within the same general range as z-scores. Heat maps were clustered by k-means (k = 8).
Zebrafish reporter assay
Zebrafish transgenesis was performed as previously described (Fisher et al. 2006). The following pre-existing zebrafish lines were used for experiments: Slusarski AB, wild type; egr2b:KalTA4BI-1xUASkCherry, mCherry inserted in the endogenous egr2b locus and expressed in r3 and r5 (Distel et al. 2009).
Immunoprecipitation
Protein complexes associated with HOXA1 on chromatin were identified as described (Aygun et al. 2008). KH2 line bearing Hoxa1 with a C-terminal 3×Flag-Myc epitope tag were treated with 3.3 µM retinoic acid and 1 mg/mL doxycycline for 24 h and immunoprecipitated using Flag agarose beads (Sigma Aldrich, #A2220) and were analyzed using an LTQ linear ion trap mass spectrometer. Spectra were interpreted using SEQUEST, and statistical analyses were performed using the QSPEC/QPROT software (version 1.2.2).
Data access
All raw sequencing data from this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA341679. Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://odr.stowers.org/websimr/.
Supplementary Material
Acknowledgments
We thank Stowers Institute (SIMR) Tissue Culture, Molecular Biology, Aquatics Facility, members of the Krumlauf and Zeitlinger labs, and Leanne Wiedemann for valuable discussions and feedback. This work was performed to fulfill, in part, requirements for I.P.’s PhD thesis research as a student registered in the Stowers Institute Graduate Program. This research was supported by funds from the Stowers Institute for Medical Research to R.K. (Grant 1001).
Author contributions: B.D.K., H.J.P., M.E.P., B.D.S., J.R.U., and R.K. contributed the experimental conception and design; B.D.K., H.J.P., M.E.P., I.P., B.D.S., and J.R.U. performed the acquisition of data; B.D.K., H.J.P., A.P., I.P., B.D.S., J.R.U., J.Z., and R.K. provided the analysis and interpretation of data; M.E.P. and N.P.S. conducted immunoprecipitation; L.F. and Y.Z. carried out MuPIT runs and analysis; B.D.K., H.J.P., M.E.P., A.P., I.P., B.D.S., J.R.U., J.Z., and R.K. drafted and revised the article; M.E.P. and J.Z. contributed unpublished data, protocols, and reagents.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.219386.116.
References
- Abu-Shaar M, Ryoo HD, Mann RS. 1999. Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev 13: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygun O, Svejstrup J, Liu Y. 2008. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci 105: 8580–8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. 2006. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44: 23–28. [DOI] [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci 99: 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F. 1998. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J 17: 1434–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelsen J, Kilstrup-Nielsen C, Blasi F, Mavilio F, Zappavigna V. 1999. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev 13: 946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin TR. 1997. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res 25: 4173–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SK, Jaffe L, Capovilla M, Botas J, Mann RS. 1994. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 78: 603–615. [DOI] [PubMed] [Google Scholar]
- Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, Alsawadi A, Valenti P, Plaza S, Payre F, et al. 2015. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 160: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kumar B, Parrish ME, Slaughter BD, Unruh JR, Gogol M, Seidel C, Paulson A, Li H, Gaudenz K, Peak A, et al. 2015. Analysis of dynamic changes in retinoid-induced transcription and epigenetic profiles of murine Hox clusters in ES cells. Genome Res 25: 1229–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kumar B, Parker HJ, Parrish ME, Lange JJ, Slaughter BD, Unruh JR, Paulson A, Krumlauf R. 2017. Dynamic regulation of Nanog and stem cell-signaling pathways by Hoxa1 during early neuro-ectodermal differentiation of ES cells. Proc Natl Acad Sci 114: 5838–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel M, Wullimann MF, Koster RW. 2009. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc Natl Acad Sci 106: 13365–13370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson IJ, Amin S, Hensman JJ, Kutejova E, Rattray M, Lawrence N, Hayes A, Ward CM, Bobola N. 2012. Genome-wide occupancy links Hoxa2 to Wnt-β-catenin signaling in mouse embryonic development. Nucleic Acids Res 40: 3990–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS. 2015. Suboptimization of developmental enhancers. Science 350: 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, Marshall H, Pöpperl H, Maconochie M, Krumlauf R, Blasi F. 2000. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development 127: 155–166. [DOI] [PubMed] [Google Scholar]
- Ferretti E, Cambronero F, Tümpel S, Longobardi E, Wiedemann LM, Blasi F, Krumlauf R. 2005. Hoxb1 enhancer and control of rhombomere 4 expression: complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol Cell Biol 25: 8541–8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. 2006. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc 1: 1297–1305. [DOI] [PubMed] [Google Scholar]
- Gaertner B, Johnston J, Chen K, Wallaschek N, Paulson A, Garruss AS, Gaudenz K, De Kumar B, Krumlauf R, Zeitlinger J. 2012. Poised RNA polymerase II changes over developmental time and prepares genes for future expression. Cell Rep 2: 1670–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galant R, Walsh CM, Carroll SB. 2002. Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites. Development 129: 3115–3126. [DOI] [PubMed] [Google Scholar]
- He Q, Johnston J, Zeitlinger J. 2015. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotechnol 33: 395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Sitwala K, Bronstein J, Sanders D, Dandekar M, Collins C, Robertson G, MacDonald J, Cezard T, Bilenky M, et al. 2012. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood 119: 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt P, Gulisano M, Cook M, Sham MH, Faiella A, Wilkinson D, Boncinelli E, Krumlauf R. 1991. A distinct Hox code for the branchial region of the vertebrate head. Nature 353: 861–864. [DOI] [PubMed] [Google Scholar]
- Jabet C, Gitti R, Summers MF, Wolberger C. 1999. NMR studies of the pbx1 TALE homeodomain protein free in solution and bound to DNA: proposal for a mechanism of HoxB1-Pbx1-DNA complex assembly. J Mol Biol 291: 521–530. [DOI] [PubMed] [Google Scholar]
- Jung H, Lacombe J, Mazzoni EO, Liem KF Jr, Grinstein J, Mahony S, Mukhopadhyay D, Gifford DK, Young RA, Anderson KV, et al. 2010. Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron 67: 781–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent A, Bihan R, Omilli F, Deschamps S, Pellerin I. 2008. PBX proteins: much more than Hox cofactors. Int J Dev Biol 52: 9–20. [DOI] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. 2011. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev 25: 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longabaugh WJ, Davidson EH, Bolouri H. 2005. Computational representation of developmental genetic regulatory networks. Dev Biol 283: 1–16. [DOI] [PubMed] [Google Scholar]
- Mann R, Chan S-K. 1996. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. TIG 12: 258–262. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni EO, Mahony S, Peljto M, Patel T, Thornton SR, McCuine S, Reeder C, Boyer LA, Young RA, Gifford DK, et al. 2013. Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat Neurosci 16: 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabet S, Mann RS. 2016. To be specific or not: the critical relationship between Hox and TALE proteins. Trends Genet 32: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabet S, Kambris Z, Capovilla M, Berenger H, Pradel J, Graba Y. 2003. The hexapeptide and linker regions of the AbdA Hox protein regulate its activating and repressive functions. Dev Cell 4: 761–768. [DOI] [PubMed] [Google Scholar]
- Merabet S, Saadaoui M, Sambrani N, Hudry B, Pradel J, Affolter M, Graba Y. 2007. A unique Extradenticle recruitment mode in the Drosophila Hox protein Ultrabithorax. Proc Natl Acad Sci 104: 16946–16951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Selleri L. 2006. Hox cofactors in vertebrate development. Dev Biol 291: 193–206. [DOI] [PubMed] [Google Scholar]
- Murphy P, Hill RE. 1991. Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development 111: 61–74. [DOI] [PubMed] [Google Scholar]
- Papalopulu N, Lovell-Badge R, Krumlauf R. 1991. The expression of murine Hox-2 genes is dependent on the differentiation pathway and displays a collinear sensitivity to retinoic acid in F9 cells and Xenopus embryos. Nucleic Acids Res 19: 5497–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HJ, Bronner ME, Krumlauf R. 2016. The vertebrate Hox gene regulatory network for hindbrain segmentation: evolution and diversification: coupling of a Hox gene regulatory network to hindbrain segmentation is an ancient trait originating at the base of vertebrates. Bioessays 38: 526–538. [DOI] [PubMed] [Google Scholar]
- Penkov D, Tanaka S, Di Rocco G, Berthelsen J, Blasi F, Ramirez F. 2000. Cooperative interactions between PBX, PREP, and HOX proteins modulate the activity of the α 2(V) collagen (COL5A2) promoter. J Biol Chem 275: 16681–16689. [DOI] [PubMed] [Google Scholar]
- Penkov D, Mateos San Martin D, Fernandez-Diaz LC, Rossello CA, Torroja C, Sanchez-Cabo F, Warnatz HJ, Sultan M, Yaspo ML, Gabrieli A, et al. 2013. Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Rep 3: 1321–1333. [DOI] [PubMed] [Google Scholar]
- Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C. 1999. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96: 587–597. [DOI] [PubMed] [Google Scholar]
- Pöpperl H, Bienz M, Studer M, Chan S, Aparicio S, Brenner S, Mann R, Krumlauf R. 1995. Segmental expression of Hoxb1 is controlled by a highly conserved autoregulatory loop dependent upon exd/Pbx. Cell 81: 1031–1042. [DOI] [PubMed] [Google Scholar]
- Pöpperl H, Rikhof H, Chang H, Haffter P, Kimmel CB, Moens CB. 2000. lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol Cell 6: 255–267. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Rauskolb C, Wieschaus E. 1994. Coordinate regulation of downstream genes by extradenticle and the homeotic selector proteins. EMBO J 13: 3561–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. 1997. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91: 171–183. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Marty T, Casares F, Affolter M, Mann RS. 1999. Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development 126: 5137–5148. [DOI] [PubMed] [Google Scholar]
- Schulte D, Frank D. 2014. TALE transcription factors during early development of the vertebrate brain and eye. Dev Dyn 243: 99–116. [DOI] [PubMed] [Google Scholar]
- Sheikh BN, Downer NL, Kueh AJ, Thomas T, Voss AK. 2014. Excessive versus physiologically relevant levels of retinoic acid in embryonic stem cell differentiation. Stem Cells 32: 1451–1458. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Arcioni L, Andrews PW, Boncinelli E, Mavilio F. 1990. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature 346: 763–766. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Nigro V, Faiella A, D'Esposito M, Stornaiuolo A, Mavilio F, Boncinelli E. 1991. Differential regulation by retinoic acid of the homeobox genes of the four HOX loci in human embryonal carcinoma cells. Mech Dev 33: 215–227. [DOI] [PubMed] [Google Scholar]
- Smith KT, Martin-Brown SA, Florens L, Washburn MP, Workman JL. 2010. Deacetylase inhibitors dissociate the histone-targeting ING2 subunit from the Sin3 complex. Chem Biol 17: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge S, Ha N, Polychronidou M, Friedrich J, Bezdan D, Kaspar P, Schaefer MH, Ossowski S, Henz SR, Mundorf J, et al. 2012. The cis-regulatory code of Hox function in Drosophila. EMBO J 31: 3323–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpel S, Wiedemann LM, Krumlauf R. 2009. Hox genes and segmentation of the vertebrate hindbrain. Curr Top Dev Biol 88: 103–137. [DOI] [PubMed] [Google Scholar]
- Vitobello A, Ferretti E, Lampe X, Vilain N, Ducret S, Ori M, Spetz J, Selleri L, Rijli FM. 2011. Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev Cell 20: 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Hernandez RE, Moens CB. 2001. Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development 128: 4139–4151. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Moens CB. 2002. Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev Cell 3: 723–733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.