During carbon starvation, autophagy is associated with protein degradation and impact energy status by regulating alternative respiration via the ETF/ETFQO through a yet unclear mechanism.
Abstract
Under heterotrophic conditions, carbohydrate oxidation inside the mitochondrion is the primary energy source for cellular metabolism. However, during energy-limited conditions, alternative substrates are required to support respiration. Amino acid oxidation in plant cells plays a key role in this by generating electrons that can be transferred to the mitochondrial electron transport chain via the electron transfer flavoprotein/ubiquinone oxidoreductase system. Autophagy, a catabolic mechanism for macromolecule and protein recycling, allows the maintenance of amino acid pools and nutrient remobilization. Although the association between autophagy and alternative respiratory substrates has been suggested, the extent to which autophagy and primary metabolism interact to support plant respiration remains unclear. To investigate the metabolic importance of autophagy during development and under extended darkness, Arabidopsis (Arabidopsis thaliana) mutants with disruption of autophagy (atg mutants) were used. Under normal growth conditions, atg mutants showed lower growth and seed production with no impact on photosynthesis. Following extended darkness, atg mutants were characterized by signatures of early senescence, including decreased chlorophyll content and maximum photochemical efficiency of photosystem II coupled with increases in dark respiration. Transcript levels of genes involved in alternative pathways of respiration and amino acid catabolism were up-regulated in atg mutants. The metabolite profiles of dark-treated leaves revealed an extensive metabolic reprogramming in which increases in amino acid levels were partially compromised in atg mutants. Although an enhanced respiration in atg mutants was observed during extended darkness, autophagy deficiency compromises protein degradation and the generation of amino acids used as alternative substrates to the respiration.
Energy availability is a key factor influencing both plant growth and development. Accordingly, plants obtain their energy both by harvesting light energy during photosynthesis and by oxidation of organic substrates via mitochondrial respiration. The latter process is mostly dependent on carbohydrates through the oxidation of organic acids by the tricarboxylic acid cycle (Plaxton and Podesta, 2006; Sweetlove et al., 2010; Araújo et al., 2011). In situations where carbohydrates become limiting, including environmental stress conditions and natural plant senescence, plant cells are forced to use alternative substrates, such as lipids and amino acids, to provide energy and maintain active mitochondrial metabolism (Buchanan-Wollaston et al., 2005; Araújo et al., 2011; Kirma et al., 2012).
Compelling evidence has demonstrated that amino acids produced from protein degradation can be an important source of alternative substrates for plant respiration, supporting ATP synthesis through a route that differs from the classical respiratory pathway (Araújo et al., 2010; Engqvist et al., 2011; Peng et al., 2015). The electron transfer flavoprotein (ETF)/ETF:ubiquinone oxidoreductase (ETF/ETFQO) system has been characterized substantially in mammals for the catabolism of fatty acids, amino acids, and choline (Watmough and Frerman, 2010). Although the entire pathway for the degradation of branched-chain amino acids (BCAAs) has been found in plant mitochondria (Salvato et al., 2014), only two dehydrogenases have been identified to date in plants as able to donate electrons to the ETF/ETFQO complex. These two enzymes are (1) isovaleryl-CoA dehydrogenase (IVDH), which is involved in the degradation of BCAAs, and (2) 2-hydroxyglutarate dehydrogenase (D2HGDH), which uses Lys as an alternative substrate (Engqvist et al., 2009, 2011; Araújo et al., 2010). These alternative pathways can provide electrons from amino acid oxidation directly to the mitochondrial electron transport chain via the ETF complex as well as by the direct feeding of catabolic products into the tricarboxylic acid cycle (Ishizaki et al., 2005, 2006; Araújo et al., 2010, 2011; Kirma et al., 2012). Notably, not only the dehydrogenases related to the ETF/ETFQO complex but also enzymes associated with amino acid catabolism in general (Hildebrandt et al., 2015), including the oxidation of sulfur-containing amino acids such as Cys and Met by the Ethylmalonic Encephalopathy Protein1 (Krüßel et al., 2014), have been associated with mitochondrial metabolism.
The physiological role of the ETF/ETFQO system during plant stress responses has been demonstrated unequivocally (for review, see Araújo et al., 2011). Additionally, several studies have demonstrated the induction of transcripts of these proteins during dark-induced senescence (Buchanan-Wollaston et al., 2005), oxidative stress (Lehmann et al., 2009), and under conditions in which free amino acids are plentiful (Weigelt et al., 2008). Furthermore, the function of this alternative pathway, and by corollary of BCAA catabolism in stress tolerance mechanisms including drought and carbon starvation, have been demonstrated (Ishizaki et al., 2005, 2006; Araújo et al., 2010; Engqvist et al., 2011; Peng et al., 2015; Pires et al., 2016). Although these studies have clearly enhanced our understanding concerning the use of amino acids as respiratory substrates, the functional linkage between protein degradation, amino acid turnover, and the alternative pathways of respiration remains to be fully elucidated.
Macroautophagy (referred to hereafter as autophagy) is a highly conserved and regulated catabolic process involved in the degradation of cytoplasmic constituents including soluble proteins, protein aggregates, or even entire organelles, allowing the recycling of cell components into primary molecules (Li and Vierstra, 2012; Liu and Bassham, 2012; Zientara-Rytter and Sirko, 2016). Briefly, during this process, cell components are sequestered by the autophagosomes and delivered into the vacuoles, where this material is then degraded and macromolecules are released back into the cytosol for reuse (Feng et al., 2014). The knowledge of autophagic mechanisms has been expanded primarily through genetic analyses in Saccharomyces cerevisiae, leading to the identification of autophagy-related (ATG) genes that encode the central autophagy machinery. Homologs of ATG genes also have been identified in plants, and their functions are similar to those in yeast cells (Liu and Bassham, 2012; Lv et al., 2014; Michaeli et al., 2016). The physiological importance of autophagy has been demonstrated extensively through the characterization of several Arabidopsis (Arabidopsis thaliana) loss-of-function mutants (atg mutants). Early senescence and hypersensitivity to carbon starvation are commonly observed phenotypes in atg mutants, providing evidence of a significant role of the autophagic process in nutrient recycling, particularly under stress conditions (Doelling et al., 2002; Bassham, 2009; Wada et al., 2009; Li and Vierstra, 2012; Liu and Bassham, 2012; Yoshimoto et al., 2014). Notably, the involvement of autophagy in providing respiratory substrates following stress conditions was demonstrated only recently. Growth impairments under both long- and short-day conditions have been observed in the Arabidopsis starchless atg double mutant, which was associated with reduced soluble sugar availability during the night (Izumi et al., 2013). Since amino acids can be used as alternative substrates for energy supply following carbon starvation (Araújo et al., 2010), autophagy seems to have a potential role in energetic maintenance under this condition. Decreased levels of free amino acids also have been reported in etiolated Arabidopsis seedlings following carbon starvation (Avin-Wittenberg et al., 2015). The reduced levels of BCAA and Lys coupled with the increased redistribution of labeled Lys to malate indicate a potential role for amino acids in supporting the respiratory flux observed in atg mutants (Avin-Wittenberg et al., 2015).
Although the importance of autophagy under nutrient-starved and other stressful conditions has been demonstrated, the exact linkage between autophagy and alternative pathways of respiration remains unclear. Here, we investigated how primary metabolism and physiological aspects are impaired in three independent atg T-DNA insertion mutant lines during dark-induced senescence. We used mutants for ATG5 and ATG7 genes that have a full inhibition of autophagy (Thompson et al., 2005; Phillips et al., 2008; Shin et al., 2014) and the atg9-1 mutant that displayed a milder reduction of the autophagic process (Shin et al., 2014; Zhuang et al., 2017). Our results demonstrate an early-senescence phenotype coupled with an accumulation of organic acids following extended darkness in all atg mutants. Reduced levels of several amino acids in the atg mutants were associated with an induction at the transcriptional level of enzymes of the ETF/ETFQO pathway following extended darkness. Collectively, the data obtained indicate that metabolite recycling during autophagy and alternative pathways of respiration are both required to provide correct respiratory function under conditions of carbon starvation. The results are discussed in the context of the importance of the autophagic process and the current models of reserve mobilization and alternative pathways of respiration during extended dark-induced senescence in leaves.
RESULTS
Characterization of T-DNA Insertional Mutants of ATG Genes
To examine the involvement of autophagy in metabolic responses during plant development and carbon limitation, we analyzed three previously described loss-of-function mutants of the autophagy pathway, namely (1) ATG5 (atg5-1; Thompson et al., 2005), (2) ATG7 (atg-7-2; Hofius et al., 2009), and (3) ATG9 (atg-9-1; Hanaoka et al., 2002). To this end, the homozygosity of each mutant line was confirmed using primer pairs designed to span the T-DNA insertion sites of each locus. The Arabidopsis glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used as a control to demonstrate the integrity and quantity of the RNA preparation (Supplemental Fig. S1D). ATG5, ATG7, and ATG9 mRNAs were detected in the wild-type control (Columbia-0) using the primer sets L1/R1, L2/R2, and L3/R3, respectively (Supplemental Fig. S1, A–C). However, no amplification products for the gene corresponding to the mutation were observed in the atg mutants (Supplemental Fig. S1D), confirming that transcripts spanning the T-DNA insertion site are absent in these mutant lines.
Importance of Autophagy for Both Growth and Seed Yield
In order to obtain further insight into the consequences of the lack of ATG genes under optimal short-day conditions, mutant plants were grown side by side with their respective wild-type controls in order to evaluate their morphological and physiological characteristics. Morphological analysis revealed a reduction of rosette area and a decrease in both fresh and dry weight matter but only in atg7-2 mutants (Table I). The specific leaf area, however, was invariant between wild-type plants and all atg mutant lines (Table I).
Table I. Growth parameters of atg mutants (4-week-old plants).
Values presented are means ± se of at least six independent biological replicates per genotype. Values in boldface were determined by Student’s t test to be significantly different (P < 0.05) from the wild type.
| Parameter | Genotype |
|||
|---|---|---|---|---|
| Wild Type | atg5-1 | atg7-2 | atg9-1 | |
| Fresh weight (mg) | 88.4 ± 9.10 | 77.8 ± 3.94 | 62.3 ± 4.33 | 91.7 ± 10.91 |
| Dry weight (mg) | 9.9 ± 1.19 | 7.6 ± 0.44 | 5.9 ± 0.30 | 8.62 ± 0.92 |
| Rosette area (cm2) | 30.0 ± 2.15 | 29.0 ± 1.28 | 24.6 ± 0.30 | 30.5 ± 1.88 |
| Specific leaf area (cm2 g−1) | 426 ± 15.4 | 460 ± 19.9 | 482 ± 20.8 | 449 ± 34.7 |
Given that the disruption of autophagy seems to result in minor growth impairments, we next evaluated a range of physiological parameters in order to assess whether changes in growth may be associated with an alteration in those parameters. In 4-week-old plants, no differences were observed in net assimilation rate (Supplemental Fig. S2A), stomatal conductance (Supplemental Fig. S2B), or internal CO2 concentration (Supplemental Fig. S2C). Thus, although autophagy seems to be a limiting factor for normal growth in Arabidopsis plants, this is unlikely to be related to the photosynthetic efficiency of the lines.
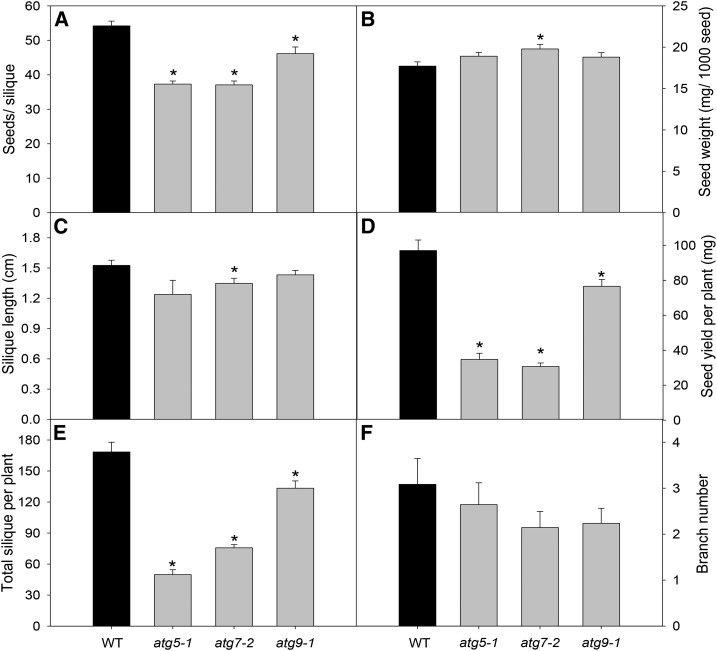
We next investigated the impact of the autophagic process during the reproductive stage. Although a reduction in the number of seeds per silique was observed (Fig. 1A), no major changes in 1,000 seeds weight (Fig. 1B) or in silique length (Fig. 1C) were found in atg mutants. The total number of siliques per plant was reduced in atg mutants (Fig. 1E) with no changes in branch numbers (Fig. 1F), leading to a reduction in seed yield per plant in atg mutants (Fig. 1D), in agreement with a previous observation of lower total seed weight in Arabidopsis plants lacking core components of the ATG system (Guiboileau et al., 2012).
Figure 1.
Seed and silique phenotypes observed in Arabidopsis atg mutants. A, Number of seeds per silique. B, Seed weight. C, Silique length. D, Seed yield. E, Total siliques per plant. F, Branch number. Seed weight was obtained by measuring 500 seeds (n = 10). Silique length (C) was determined in images taken with a digital camera (Canon Powershot A650 IS) attached to a stereomicroscope (Zeiss Stemi 2000-C). The measurements were performed on the images using ImageJ software. Values presented are means ± se of at least 10 biological replicates per genotype. Asterisks designate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild type (WT).
atg Mutants Are More Susceptible to Energy Deprivation
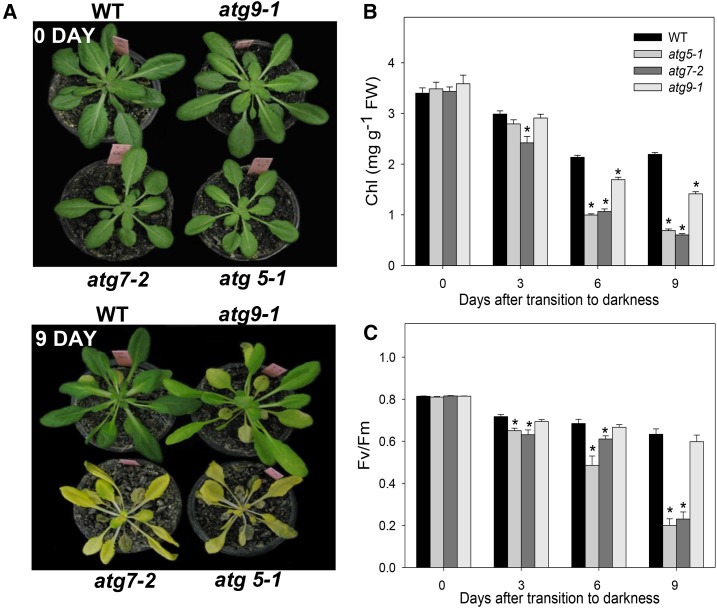
Following the confirmation of the molecular identity of the T-DNA insertional mutants and given that previous studies have implied the function of autophagy under stress conditions (Izumi et al., 2013; Avin-Wittenberg et al., 2015), we next transferred 4-week-old mutant plants, alongside wild-type plants, to carbon starvation induced by extended dark conditions. Under these conditions, a range of phenotypes became apparent (Fig. 2). The atg5-1 and atg7-2 mutants started to wilt and show signs of senescence after only 6 d of continuous darkness, and both mutants were apparently dead after 12 d of continuous darkness (Supplemental Fig. S3). It should be pointed out that wild-type plants were still alive and exhibited only limited signs of senescence and no visible abnormalities after 12 d of continuous darkness. The atg9-1 mutant also showed signs of senescence after 9 d of continuous darkness, but with a less severe phenotype compared with atg5-1 and atg7-2 mutants, thus showing an intermediate senescence phenotype between wild-type plants and the other two atg mutant lines (Fig. 2A).
Figure 2.
Phenotypes of Arabidopsis atg mutants under extended dark treatment. A, Images of 4-week-old, short-day-grown Arabidopsis plants immediately light was turned off (0 d) and after further treatment for 9 d in darkness conditions. B and C, Chlorophyll (Chl) content (B) and Fv/Fm (C) of leaves of 4-week-old, short-day-grown Arabidopsis plants after further treatment for 0, 3, 6, and 9 d in extended darkness. Values are means ± se of five independent samplings. Asterisks indicate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild type (WT) at each time point analyzed. FW, Fresh weight.
In order to further investigate the accelerated senescence symptoms, two parameters related to chloroplast function, chlorophyll content and maximum photochemical efficiency of PSII (maximum variable fluorescence/maximum yield of fluorescence [Fv/Fm]), were analyzed (Fig. 2, B and C). During the extended dark treatment, chlorophyll content declined more rapidly in the mutants than in the wild type (Fig. 2B). Accordingly, these results were associated with a more rapid decline in Fv/Fm in atg5-1 and atg7-2 mutants after 6 d of darkness (Fig. 2C). By contrast, Fv/Fm values in wild-type and atg9-1 lines remained similar throughout the entire time period of the experiment (Fig. 2C). Thus, these parameters are in good agreement with an early-senescence phenotype observed in both atg5-1 and atg7-2 mutants in comparison with the wild type and atg9-1.
The Respiratory Response of atg Mutants Under Dark-Induced Starvation
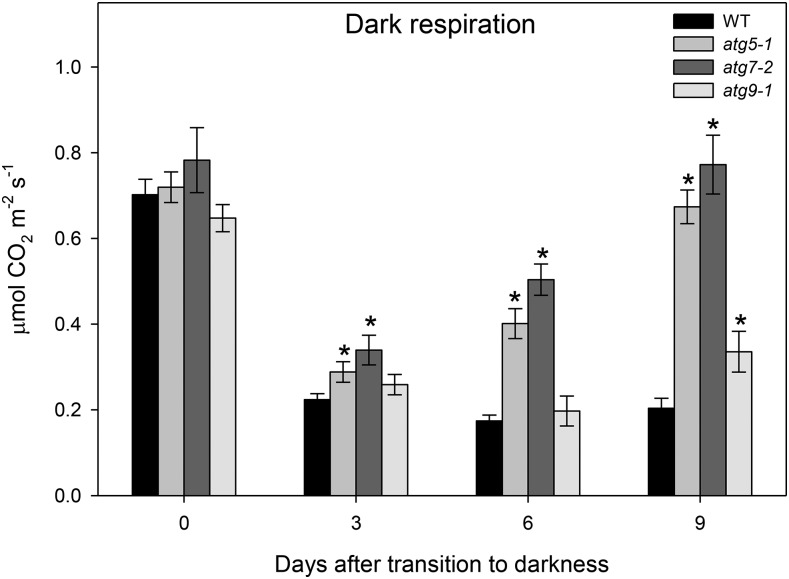
Given the severe phenotype of atg mutants under carbon starvation and that alternative pathways of respiration are important for plant survival during darkness (Ishizaki et al., 2005, 2006; Araújo et al., 2010), we next investigated the connection between autophagy and plant respiration. To this end, we evaluated the CO2 gas-exchange rate, since leaf respiration represents a major source of CO2 release in plants (Atkin et al., 2000). Dark respiration rates measured immediately at the start of the dark treatment (day 0) were virtually invariant between the genotypes, indicating that autophagy deficiency has a minor impact on plant respiration under nonstressed conditions prior to bolting (Fig. 3). Interestingly, dark respiration rates were reduced after 3 d under darkness in all genotypes, and such low levels of dark respiration were maintained in wild-type plants during extended darkness. Following darkness, the atg mutants also presented a reduction of dark respiration, but this reduction seemed to be lower than the one observed in wild-type plants. In addition, we observed that, after 6 d of treatment, dark respiration had increased again in atg5-1 and atg7-2 mutants, reaching higher levels after 9 d under darkness. It was also observed that dark respiration rates remained similar to those of the wild type in the atg9-1 mutants, with a significant increase starting only from 9 d after darkness (Fig. 3).
Figure 3.
Dark respiration during extended dark treatment. The CO2 efflux rates of 4-week-old Arabidopsis plants immediately after light was turned off (0 d) and during further treatment for 9 d in darkness were analyzed. Gas-exchange measurements were performed with an open-flow infrared gas-exchange analyzer system with a portable photosynthesis system to fit a whole-plant cuvette. Values presented are means ± se of seven biological replicates per genotype. Asterisks designate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild type (WT).
Deficiency of Autophagy Leads to a Differential Metabolic Response following Carbon Starvation
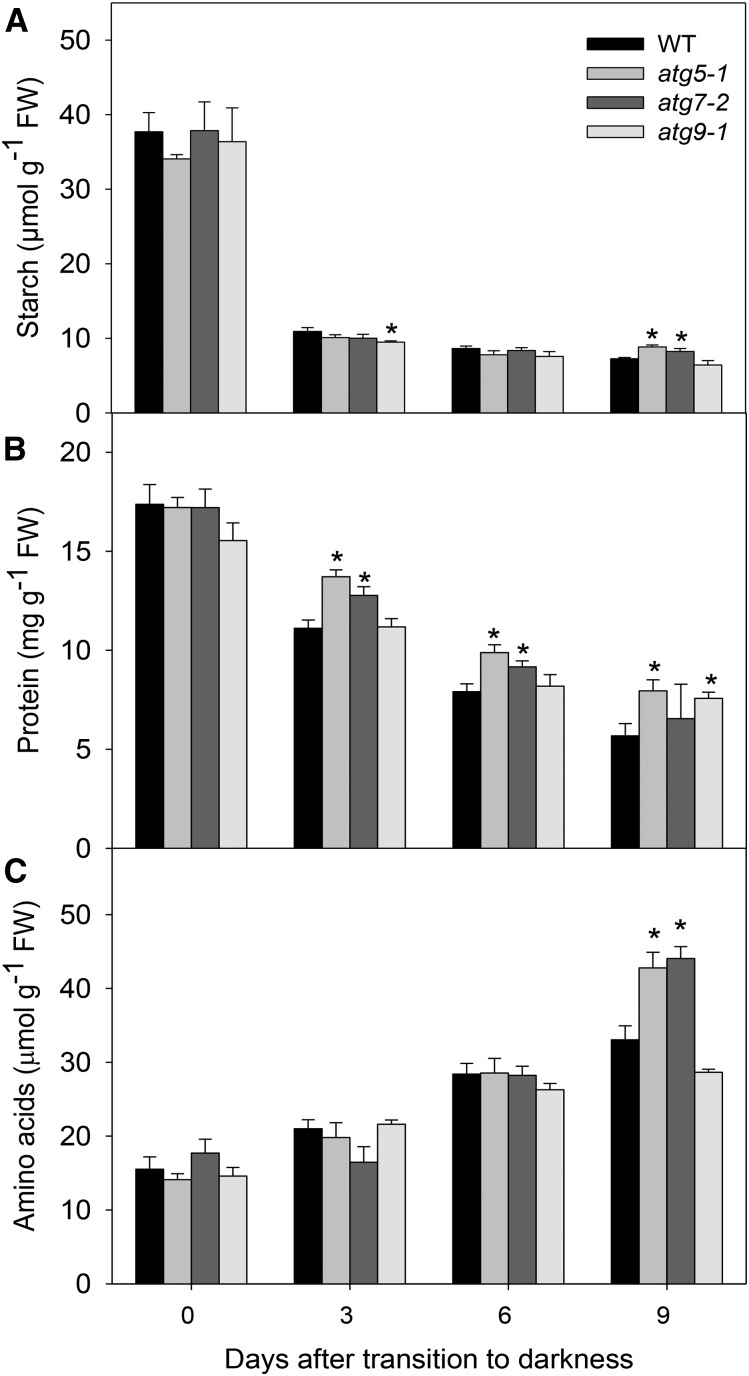
To elucidate the connection between autophagy and nutrient recycling following carbon starvation, we further conducted a detailed metabolic analysis in leaves during the extended dark treatment. It is important to mention that all genotypes used in this study showed similar levels of total soluble proteins, total amino acids, organic acids, soluble sugars, and starch in samples harvested immediately prior to the start of the dark treatment (day 0; Fig. 4; Supplemental Fig. S4).
Figure 4.
Metabolite levels in Arabidopsis atg mutants. Starch (A), protein (B), and amino acids (C) were measured using whole rosettes of 4-week-old short-day-grown Arabidopsis plants after further treatment for 0, 3, 6, and 9 d in extended darkness. Values presented are means ± se of five biological replicates per genotype. Asterisks designate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild type (WT) at each time point analyzed. FW, Fresh weight.
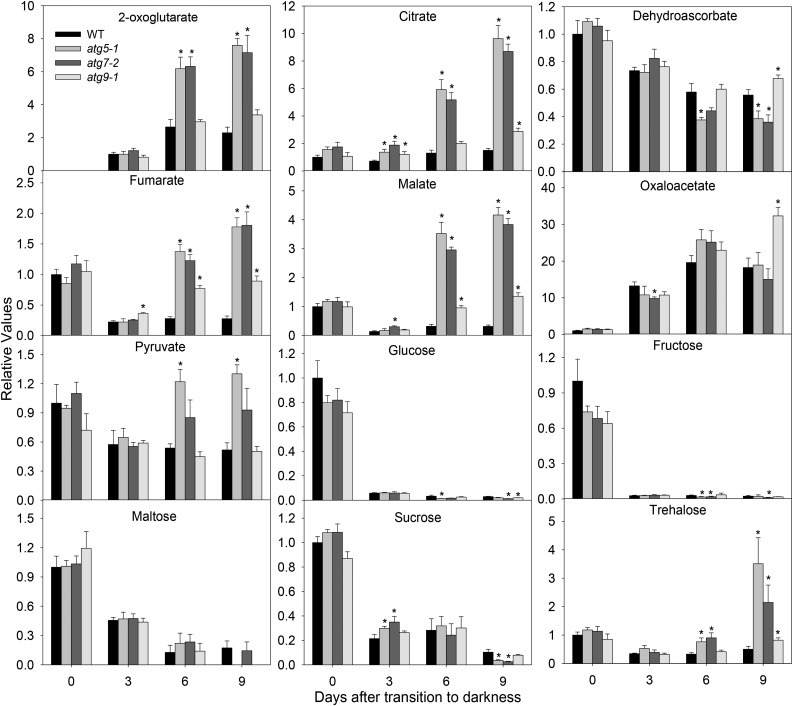
As might be expected, starch levels declined rapidly from 3 d of dark treatment onward (Fig. 4A). Interestingly, after 9 d of darkness, higher starch content was observed in atg5-1 and atg7-2 mutants coupled with lower levels of Suc and Glc in atg mutants compared with wild-type plants (Supplemental Fig. S4). While these changes are striking, it was suggested previously that autophagy contributes to leaf starch degradation (Wang et al., 2013).
Given that proteins are degradation targets of the autophagy machinery (Li and Vierstra, 2012), we next decided to examine the protein content during the extended dark treatment. Thus, while total protein content decreased during dark treatment in all genotypes (Fig. 4B), the levels were reduced to a lesser extent in the mutants, especially in atg5-1 and atg9-1 lines, in comparison with wild-type plants. Increases in the levels of total amino acids were observed throughout the dark treatment, most likely as a result of enhanced protein degradation prompted by the carbon starvation conditions (Fig. 4C). Accordingly, after 9 d of darkness, the amino acid content was significantly higher in atg5-1 and atg7-2 mutants, while no change was observed in the atg9-1 mutant.
In order to obtain a more detailed characterization of changes in the metabolism of atg mutants, we next decided to extend this study to encompass the major pathways of metabolism by metabolite profiling, which was able to provide information on 30 primary metabolites across the experiment. We observed considerable changes in the levels of a wide range of organic acids, amino acids, and sugars in atg mutants in response to dark treatment (Figs. 5 and 6). The tricarboxylic acid cycle intermediates citrate, fumarate, malate, and pyruvate were increased in atg mutants at the end of the dark treatment, mainly in atg5-1 and atg7-2 genotypes (Fig. 5). By contrast, in wild-type plants, the levels of these organic acids were constant or tended to be reduced, indicating an altered operation of the tricarboxylic acid cycle in the atg mutants during darkness. Although not different from wild-type plants, the levels of oxaloacetate clearly increased whereas the levels of dehydroascorbate were reduced dramatically at the end of the dark treatment, declining to as low as 35% of the levels measured at the start of treatment. In agreement with our spectrometric assays (Supplemental Fig. S4), reduced levels of sugars were observed in all genotypes starting after 3 d of darkness. It is important to mention that minor differences, including significantly reduced levels of Suc, Glc, and Fru in atg mutants, were observed after 3 d of darkness. Additionally, increased trehalose levels starting from 6 d of darkness were observed in all mutant lines coupled with the absence of changes in maltose levels (Fig. 5).
Figure 5.
Relative levels of sugars and organic acids in Arabidopsis atg mutants during extended dark conditions as measured by gas chromatography-mass spectrometry (GC-MS). The y axis values represent the metabolite level relative to the wild type (WT). Data were normalized relative to the mean content calculated for the 0-d dark-treated leaves of the WT (in case no response was detected at 0 d, normalization was performed against 3-d dark-treated leaves of the wild type). Values presented are means ± se of five biological replicates per genotype. Asterisks designate values that were determined by Student’s t test to be significantly different (P < 0.05) from the WT at each time point analyzed.
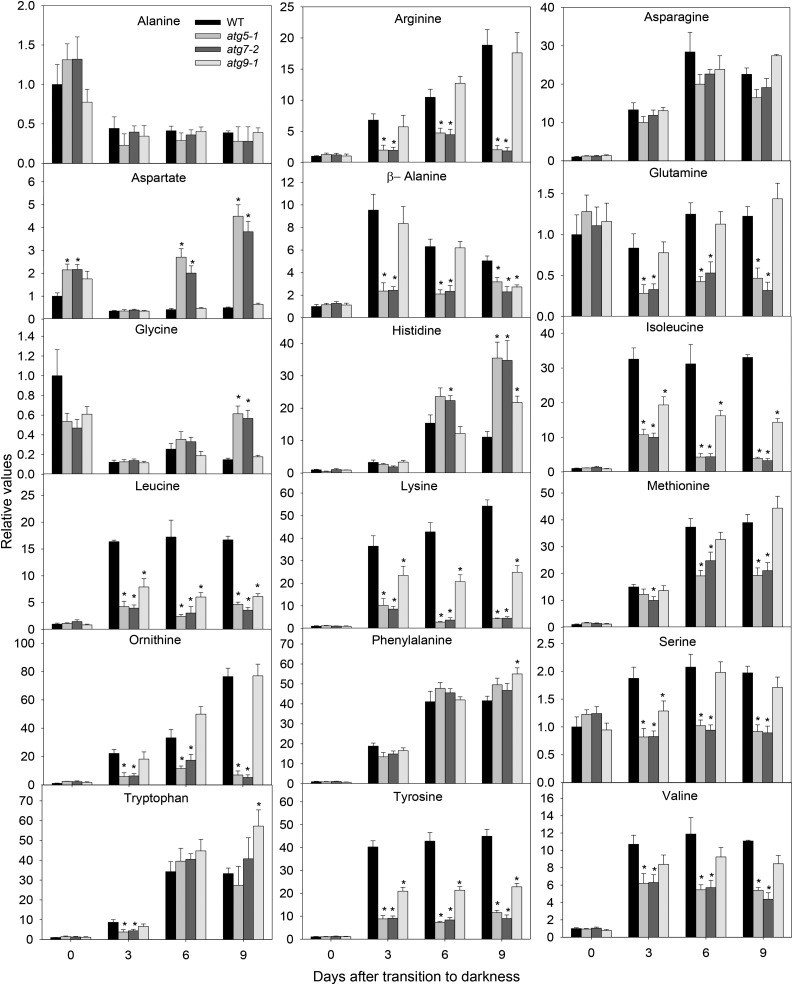
Figure 6.
Relative levels of amino acids in Arabidopsis atg mutants during extended dark conditions as measured by GC-MS. The y axis values represent the metabolite level relative to the wild type (WT). Data were normalized to the mean response calculated for the 0-d dark-treated leaves of the WT (in case no response was detected at 0 d, normalization was performed against 3-d dark-treated leaves of the wild type). Values presented are means ± se of five biological replicates per genotype. Asterisks designate values that were determined by Student’s t test to be significantly different (P < 0.05) from the WT at each time point analyzed.
Analyses of the levels of individual amino acids revealed that Asn, Ile, Leu, Lys, Met, Phe, Trp, Tyr, and Val increased significantly in all genotypes following dark treatment, while the levels of Ala were reduced (Fig. 6). Intriguingly, some metabolites showed distinct direction of changes with respect to the wild type, as in the case of β-Ala, Orn, and Ser, which only increased in the wild type and atg9-1, whereas Asp, His, and Gly increased more in atg5-1 and atg7-2 mutants (Fig. 6). Although the Gln levels were virtually constant in wild-type plants following dark treatment, reductions in the levels of this amino acid were observed for both atg5-1 and atg7-2 mutant lines. Interestingly, increases in BCAAs (Leu, Ile, and Val), Lys, and Tyr observed in wild-type plants following dark treatment were less pronounced in the atg mutants.
Carbon Starvation Leads to the Induction of Alternative Pathways in atg Mutants
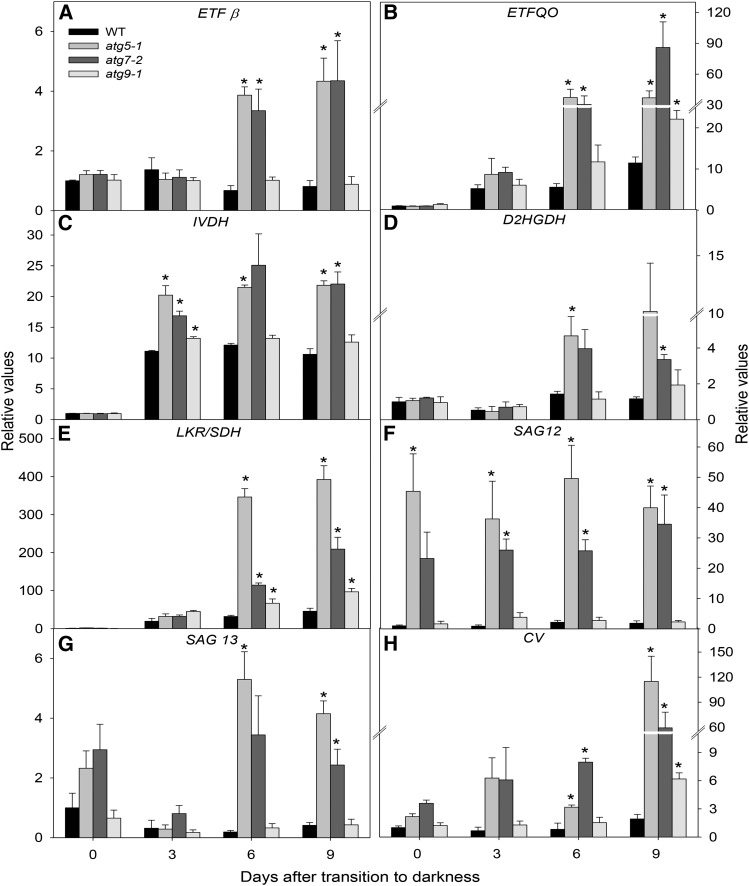
In order to investigate whether autophagic impairment coupled with amino acid degradation has an influence on alternative respiratory pathways, expression analysis of genes related to the ETF/ETFQO system was performed by quantitative reverse transcription (RT)-PCR (Fig. 7). During carbon starvation, the importance of BCAAs, aromatic amino acids, and Lys as alternative respiratory substrates was demonstrated via the characterization of loss-of-function mutants for IVDH, D2HGDH, ETF, and ETFQO (Ishizaki et al., 2005, 2006; Araújo et al., 2010). Here, we demonstrated that the transcript levels of IVDH, ETFQO, ETFβ, and D2HGDH were generally induced in atg5-1 and atg7-2 in comparison with the levels observed in wild-type plants, while a mild induction was observed in atg9-1 mutants when compared with the other atg mutants under extended dark treatment (Fig. 7, A–D). More specifically, ETFβ was only up-regulated in atg5-1 and atg7-2 plants after 6 d of darkness, with no changes observed in either wild-type or atg9-1 mutant plants (Fig. 7A). In addition, ETFQO was up-regulated following dark treatment in all genotypes, but with higher expression levels in atg mutants after 6 d of darkness (Fig. 7B). Also, there was an early induction of IVDH transcripts in both the wild type and atg mutants after 3 d of darkness (Fig. 7C). Such strong induction of IVDH, reaching increments higher than 20-fold after 3 d of dark transition, reinforces claims of its pivotal role in amino acid degradation (Araújo et al., 2010; Peng et al., 2015). Given that Lys catabolism can occur by either D2HGDH or Lys-ketoglutarate reductase/saccharopine dehydrogenase (LKR/SDH; Engqvist et al., 2009, 2011; Galili, 2011; Kirma et al., 2012), we next decided to investigate the changes in the expression of these genes. The expression of D2HGDH was only up-regulated in atg5-1 and atg7-2 plants after 6 d of darkness, with no changes in the other genotypes studied here (Fig. 7D). Interestingly, the expression of LKR/SDH was induced strongly in all genotypes following dark treatment, with higher induction being observed in atg5-1 and atg7-2 following 6 and 9 d of darkness (Fig. 7E). Our data showed a higher induction of LKR/SDH (about 200-fold) than of D2HGDH (10-fold; Fig. 7, D and E).
Figure 7.
Changes in transcript levels in 4-week-old, short-day-grown Arabidopsis plants after further treatment for 0, 3, 6, and 9 d in extended darkness. Transcript abundance is shown for genes associated with the alternative pathways of respiration, including ETFβ (A), ETFQO (B), IVDH (C), D2HGDH (D), LKR/SDH (E), SAG12 (F), SAG13 (G), and CV (H). The y axis values represent the metabolite level relative to the wild type (WT). Data were normalized with respect to the mean response calculated for the 0-d dark-treated leaves of the wild type. Values presented are means ± se of three independent biological replicates. Asterisks indicate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild type at each time point analyzed.
Impairment of Autophagy Induces Senescence and Chloroplast Degradation Events
Given that several senescence parameters were induced in response to darkness, we next investigated the expression of the commonly known senescence-associated genes SAG12 and SAG13 during dark-induced senescence. Interestingly, although no changes in the transcript levels of SAG12 or SAG13 were observed in either wild-type or atg9-1 mutant plants following darkness, it was observed that, in atg5-1 and atg7-2 mutants, the transcript levels of SAG12 were highly induced at all time points coupled with an up-regulation of SAG13 after 6 d of darkness (Fig. 7, F and G). Taken together with chlorophyll content and Fv/Fm values, these results are in good agreement with an early-senescence phenotype observed in those genotypes.
The degradation of chloroplasts is a hallmark of both natural and stress-induced plant senescence (Ishida et al., 2014), and autophagy is an established cellular pathway involved in targeting chloroplast proteins for degradation (Ishida et al., 2008, 2014; Michaeli et al., 2014; Izumi et al., 2017). Recently, an autophagy-independent pathway for chloroplast degradation, the chloroplast vesiculation (CV) pathway, which is associated with thylakoid and stroma protein degradation, was demonstrated unequivocally (Wang and Blumwald, 2014). Thus, we next investigated the expression of the CV gene during our experimental conditions. Interestingly, a higher induction of CV gene expression was observed in atg5-1 and atg7-2 mutants under dark-induced senescence (110- and 60-fold at 9 d of darkness, respectively), while the transcript levels were essentially invariant in wild-type plants and showed only a minor induction in atg9-1 mutants (Fig. 7H).
DISCUSSION
During the last decade, a growing body of evidence has emerged showing the function of autophagy in nutrient recycling under energy-limited conditions (Thompson et al., 2005; Phillips et al., 2008; Chung et al., 2010; Izumi et al., 2010). Although the connection between autophagy, protein degradation, and amino acid availability during energetic limitation has been demonstrated (Izumi et al., 2013; Avin-Wittenberg et al., 2015), our current understanding of the metabolic process associated with energy supply following carbon starvation remains fragmentary. Here, we provide further evidence of the importance of autophagy in governing an exquisite metabolic reprogramming during both dark-induced carbon starvation and developmental transitions in the plant life cycle.
The Significance of Autophagy during Plant Development
By using well-characterized autophagy-deficient mutants, we provide further evidence that this process impacts both vegetative and reproductive development (Fig. 1; Table I). A reduction of growth was observed for the atg7-2 mutant (Table I). Growth inhibition also has been observed in atg mutants grown under both short-day conditions and mineral-rich medium without Suc, providing a mechanism where the autophagic process participates in nighttime energy availability and sustains growth (Izumi et al., 2013). Furthermore, it was observed that the lack of an autophagic process culminates in a negative impact on seed production (Fig. 1D). Lower seed yield already has been demonstrated in atg mutants (Avila-Ospina et al., 2014), and impairments of nutrient remobilization also have been associated with the lack or reduction of autophagy (Avila-Ospina et al., 2014; Li et al., 2015). Therefore, it is highly tempting to suggest that the impaired reproductive growth phenotype of atg mutants can be at least partly related to the impairment of protein degradation and remobilization processes during seed formation (Guiboileau et al., 2012; Li et al., 2015). That said, the exact mechanistic relationship between energetic metabolism, seed production, and autophagy itself will be examined in detail in future studies.
Autophagy Plays a Pivotal Role in Plant Survival and Respiration during Extended Darkness
In addition to the function of autophagy during developmental processes, autophagy seems to be strictly necessary for plant cell survival following carbon starvation. The first evidence was the early onset of dark-induced senescence observed in atg mutants accompanied by the loss of chlorophyll and photosynthetic competence (Fig. 2). In agreement with the observed phenotype, senescence-associated genes such as SAG12 and SAG13 were up-regulated mainly in atg5-1 and atg7-2 mutants during dark treatment (Fig. 7, F and G). Since a large number of reporters showed that SAG12 is usually up-regulated in natural senescence but not in dark-induced senescence (Noh and Amasino, 1999; Weaver and Amasino, 2001; Grbić, 2003), it is reasonable to assume that the mildly induced senescence observed in wild-type plants was not sufficient to induce the transcription of SAG12 and SAG13. However, transcript induction of SAG12 and SAG13 in atg5-1 and atg7-2 mutants can be at least partially associated with changes in the salicylic acid signaling and accumulation usually observed in atg mutants (Morris et al., 2000; Yoshimoto et al., 2009; Zhao et al., 2016).
To decipher the autophagy function in the maintenance of cellular energy status, we paid particular attention to the respiration of atg mutants. The respiratory rates were reduced in all genotypes as a consequence of carbon depletion with extended treatment (Fig. 3), and it was kept low in wild-type plants during darkness. In agreement, a lower respiratory rate was observed previously in dark-treated Arabidopsis plants (Keech et al., 2007) as well as in cell culture under Suc starvation (Contento et al., 2004). The atg mutants also showed reduction of total dark respiration after 3 d in darkness; however, increases were observed during extended dark treatment (after 6 and 9 d) in atg 5-1 and atg7-2 and relatively later and to a lesser extent in atg9-1 plants (Fig. 3). It seems reasonable to assume that wild-type plants are waiting in a so-called standby mode for better suboptimal environmental conditions (Keech et al., 2007). In sharp contrast, atg mutants are not able to fine-tune this basal metabolism.
Interestingly, we also observed higher levels of tricarboxylic acid cycle intermediates from 6 d of darkness in atg 5-1, atg7-2, and, to a lesser extent, atg9-1 plants (Fig. 5). When considered together with higher CO2 rates, these results suggest a misregulation of respiratory metabolism characterized by a higher flux through the tricarboxylic acid cycle as a consequence of the respiratory activity in atg mutants. This conclusion is consistent with previous observations of the flow of labeled carbon showing that (1) a higher proportion of carbohydrate oxidation occurs via the tricarboxylic acid cycle and (2) increased label distribution in malate pools from [13C]Lys occurs in atg5-1 etiolated seedlings, indicating a possible higher flux for respiration under carbon depletion (Avin-Wittenberg et al., 2015).
Autophagy Deficiency Compromises Amino Acid Release and Impacts Alternative Pathways of Respiration
The levels of the majority of the amino acids generally increased within the first 3 d of darkness, albeit to a lesser extent in atg mutants (Fig. 6). BCAAs, aromatic amino acids, and Lys have been characterized previously as alternative substrates able to sustain respiration during carbon starvation (Ishizaki et al., 2005, 2006; Araújo et al., 2010). Our findings demonstrate that, following dark treatment, increases in the amounts of these amino acids were partially compromised in atg mutants in comparison with wild-type plants (Fig. 6). It should be mentioned that a minor impact on the levels of BCAAs and Lys (Fig. 6) as well as a milder sensitivity phenotype under darkness were observed in atg9-1 in relation to atg5-1 and atg7-2 mutants (Fig. 2). Accordingly, both ATG5 and ATG7 participate in autophagosome formation by ubiquitin-like conjugating systems, and as such, correspondent Arabidopsis mutants fail to form autophagic structures (Li and Vierstra, 2012; Liu and Bassham, 2012; Lv et al., 2014). On the other hand, although ATG9 operates in lipid delivery for phagophore growth, it seems not to be strictly necessary for the whole autophagic flux (Shin et al., 2014; Zhuang et al., 2017). Thus, autophagic activity is not fully blocked in the atg9 mutants, which can explain the relatively minor phenotype observed in this mutant line. Nonetheless, our data indicate that functional autophagy is probably required to allow the precise provision of energetic substrates, since the same pattern of reduced levels of most amino acids also was observed in other reports that also used atg mutants following carbon starvation (Izumi et al., 2013; Avin-Wittenberg et al., 2015).
The use of amino acids under energy-limited conditions is seemingly related to the operation of alternative pathways of respiration. In this scenario, we observed a significant transcript induction of genes involved with alternative respiration (ETFβ, ETFQO, IVDH, and D2HGDH) and Lys catabolism (LKR/SDH) mainly in atg5-1 and atg7-2 mutants following extended darkness (Fig. 7, A–E). The induction of amino acid degradation and alternative pathways of respiration has been observed during prolonged darkness and developmental leaf senescence (Ishizaki et al., 2005, 2006; Peng et al., 2015; Chrobok et al., 2016). The transcriptional regulation seems to be dependent on the energy status of plants and might contribute to the reduced amino acid levels of atg mutants. Thus, when there is autophagy deficiency, the generation of amino acids following protein degradation is compromised but the alternative pathways of respiration are induced, leading to an even higher reduction in the levels of amino acids that is likely associated with a complex regulation of respiratory metabolism in plants.
Although the pathways of essential amino acids and their interactions with the regulatory networks in plants have long been established, certain gaps remain unfilled, such as how amino acid metabolism is associated with the autophagic process. We cannot formally exclude that the decreased levels in amino acids (Fig. 6) can be, at least partially, related to their increased utilization as carbon skeletons, allowing the synthesis of specific compounds required under carbon limitation in atg mutants. In fact, amino acids have a plethora of functions in addition to their role as a catabolic substrate. The dynamic of amino acid levels depends on both catabolic and biosynthetic reactions, and as such, several amino acids represent precursors of nucleotides, phytohormones, or secondary metabolites (for review, see Hildebrandt et al., 2015). Therefore, the accurate recycling of amino acids, lipids, carbohydrates, or micronutrients and macronutrients available in the plant cell becomes a critical factor that ensures plant survival and growth. This is particularly true given that, in the functional deficiency of autophagy, plants are still likely able to have a complete and normal development (Figs. 1 and 2A).
The Chloroplast Degradation Pathway Is Induced during Autophagy Deficiency
The differential respiratory response observed between wild-type and atg plants is seemingly able to explain partially the plant survival following carbon starvation conditions. Generally speaking, low cellular metabolic activity provides longer maintenance of cell viability during extended dark conditions. This response may be related directly to the inhibition of energy consumption that allows preservation of the photosynthetic machinery (Keech et al., 2007). While wild-type plants started to show a few signs of senescence from 12 d of darkness onward (Supplemental Fig. S3), atg mutants presented a markedly early-senescence phenotype, with faster loss of photochemical efficiency (Fig. 2) and slower reduction in protein levels (Fig. 4). Whereas all our data point to a dominant role of autophagy in reducing protein levels during carbon deprivation, we cannot yet rule out a simultaneous induction of other catabolic pathways. In this scenario, we observed an up-regulation of the CV gene only in atg mutants following carbon starvation (Fig. 7H). The role of autophagy in chloroplast degradation is well known (Ishida et al., 2008; Liu and Bassham, 2012; Xie et al., 2015; Izumi et al., 2017). However, recently, an autophagy-independent process of chloroplast degradation associated with the CV pathway was reported (Wang and Blumwald, 2014). Our findings suggest that CV is highly induced in the absence of autophagy, contributing to the early-senescence phenotype observed in atg mutants.
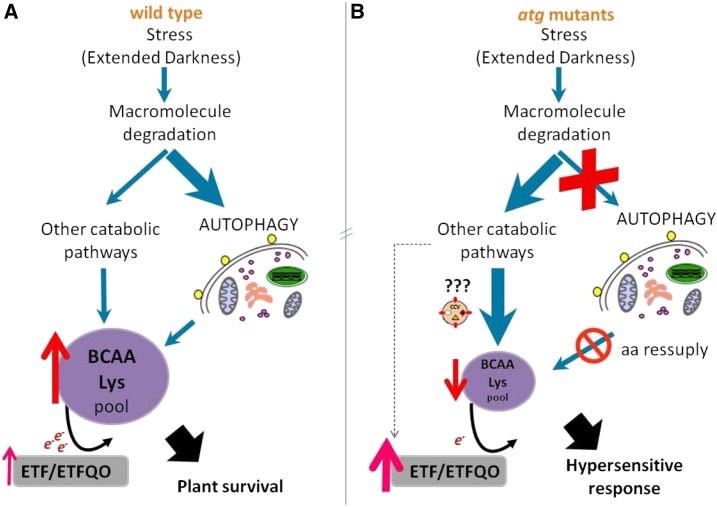
Although our results provide circumstantial evidence, we are not able to ascertain whether autophagy is linked directly with the provision of amino acids or the extent to which the absence of autophagy is able to induce other catabolic pathways. It seems tempting to suggest that the increased stress response observed in atg mutants is likely due to multiple feedback mechanisms able not only to regulate autophagy but also to impact amino acid and protein catabolism as well as plant respiration under carbon limitation (Fig. 8). The molecular mechanisms involved in the regulation of amino acid catabolism are largely unknown (Hildebrandt et al., 2015; Galili et al., 2016), and as such, the exact mechanism connecting autophagy and dark respiration in plants seems to be unclear from this study. It will be interesting to determine this linkage in a manner that does not affect any other connected pathway, potentially by the use of conditional inhibition (inducible artificial microRNA lines) of the autophagic pathway.
Figure 8.
Schematic model showing the catabolic process involved in macromolecule degradation leading to electron donation to the ETF-ETFQO pathway during dark-induced senescence in wild-type plants (A) and atg mutants (B). Carbon starvation conditions promoted by extended darkness are associated with macromolecule degradation via several catabolic pathways (e.g. autophagy), releasing amino acids to be oxidized. The electrons generated are transferred, via the ETF/ETFQO system, to the respiratory chain through the ubiquinol pool, promoting plant survival. In atg mutants, there is a compromised amino acid (aa) supply, particularly BCAAs and Lys, recognized previously to be able to feed electrons to the ETF/ETFQO system. Simultaneously, there is a higher induction of genes associated with the ETF/ETFQO pathways and autophagy independent of chloroplast degradation, CV, which leads to a hypersensitivity response to energetic limitations in atg mutants.
CONCLUSION
We have presented compelling evidence that autophagy has an important role during various stages of Arabidopsis development or during carbon deprivation conditions. Although the impairment of growth observed is related to neither changes in photosynthesis nor dark respiration under optimal conditions, it is noteworthy that the reduction in seed yield in atg mutants strongly reinforces the significant contribution of autophagy to metabolic processes affecting plant developmental fitness. Furthermore, during conditions of prolonged darkness, the impairment of protein degradation and thereby amino acid remobilization experienced by atg mutants, coupled with an increased use of amino acids as alternative substrates to mitochondrial respiration, culminate in a hypersensitive phenotype. Thus, despite the higher up-regulation of genes related to alternative pathways of respiration, the supply of amino acids seems to be insufficient to match the enhanced respiration rates of atg mutants (Fig. 8). Collectively, this energetic depletion may favor an induction of other catabolic pathways, including the degradation of chloroplastic proteins via autophagy-independent routes such as the CV. Taken together, the results presented here highlight the complexity and specificity of plant metabolism in response to carbon limitation and suggest that myriad interplays are involved between autophagy and plant respiration. Dissecting the intertwined mechanisms involved in their coregulation will be required to fully understand the implications of autophagy for the regulation of plant metabolism.
MATERIALS AND METHODS
Plant Material and Dark Treatment
All Arabidopsis (Arabidopsis thaliana) plants used in this study were of the Columbia-0 ecotype. The T-DNA mutant lines atg9-1 (Hanaoka et al., 2002), atg5-1 (SAIL_129B079; Yoshimoto et al., 2009), and atg7-2 (GK-655B06; Hofius et al., 2009) were used in this study. Seeds were surface sterilized and imbibed for 4 d at 4°C in the dark on 0.7% (w/v) agar plates containing one-half-strength Murashige and Skoog medium (Sigma-Aldrich; pH 5.7). Seeds were subsequently germinated and grown at 22°C under short-day conditions (8 h of light/16 h of dark), 60% relative humidity, with 150 μmol photons m−2 s−1. For dark treatments, 10- to 14-d-old seedlings were transferred to soil and then grown at 22°C under short-day conditions (8 h of light/16 h of dark). Four-week-old plants were transferred to darkness in the same growth cabinet. Whole rosettes of two different plants of six independent samples by genotype were harvested at intervals of 0, 3, 6, and 9 d after the transition to darkness and immediately frozen in liquid nitrogen and then stored at ‒80°C until further analysis.
T-DNA Insertion Mutants and Genotype Characterization
Homozygous mutant lines were confirmed by PCR using ATG7-specific primers (forward, 5′-GACTGTACCTAACTCAGTGGGATG-3′, and reverse, 5′-GCTCCTGCAATAGGAGCTAGAC-3′) in combination with the T-DNA left border primer (GABI-08474, forward, 5′-ATAATAACGCTGCGGACATCTACATTTT-3′) for the atg7-2 mutant; ATG5-specific primers (forward, 5′-TTAGCACCAAGAATAGGATATTTGC-3′, and reverse, 5′-TGCAATTTCCATTGATGATATATTG-3′) in combination with the T-DNA left border primer (LB1, forward, 5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′) for the atg5-1 mutant; and ATG9- specific primers (forward, 5′-CTAAGAGATGGCGTGGAAAGG-3′, and reverse, 5′-CTTGAGGTTTGAGGCATTTCA-3′) with the T-DNA left border primer (LB1, 5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTC-3′) for the atg9-1 mutant. Glyceraldehyde-3-phosphate dehydrogenase (forward, 5′-TGGTTGATCTCGTTGTGCAGGTCTC-3′, and reverse, 5′-GTCAGCCAAGTCAACAACTCTCTG-3′) was used for the normalization of gene transcript levels.
Evaluation of Biometric Parameters of Seeds
To phenotype reproductive tissues, seeds were submitted to the procedure described above, and the seedlings were transferred to commercial substrate and kept in a growth chamber at 22°C ± 2°C, 60% relative humidity, and irradiance of 150 µmol photons m−2 s−1 with a photoperiod of 12 h of light and 12 h of dark for seed production. Siliques were harvested and cleared with 0.2 m NaOH and 1% SDS solution to remove pigments. For length determination, images of at least 50 Arabidopsis siliques were taken with a digital camera (Canon Powershot A650 IS) attached to a stereomicroscope (Zeiss Stemi 2000-C). The measurement of silique length was performed on the images using ImageJ software. Seed weight was determined by weighing aliquots of a known number of seeds (500 seeds per aliquot). Total seed yield was determined by weighing seeds collected from at least 10 individual plants.
Biochemical Assays
Leaf samples were harvested at the time points indicated, immediately frozen in liquid nitrogen, and stored at −80°C until further analysis. Extraction was performed by rapid grinding of tissue in liquid nitrogen and immediate addition of ethanol as described by Gibon et al. (2004). Photosynthetic pigments were determined exactly as described by Porra et al. (1989). The levels of starch, Suc, Glc, and Fru were determined exactly as described previously (Fernie et al., 2001). Malate and fumarate contents were determined as described before (Nunes-Nesi et al., 2007). Protein and total amino acid levels were determined as described previously (Cross et al., 2006).
Measurements of Photosynthetic Parameters
Gas-exchange measurements were performed with an open-flow infrared gas-exchange analyzer system (Li-Cor 6400XT) with a portable photosynthesis system. Light was supplied from a series of light-emitting diodes located above the cuvette, providing an irradiance of 150 µmol photons m−2 s−1, exactly as in our growth conditions. The reference CO2 concentration was set at 400 μmol CO2 mol−1 air. All measurements were performed at 25°C, and vapor pressure deficit was maintained at 2 ± 0.2 kPa, while the amount of blue light was set to 10% of photon flux density to optimize stomatal aperture. The determination of the photosynthetic parameters was performed in 4-week-old plants. Fv/Fm, which corresponds to the potential quantum yield of the photochemical reactions of PSII and represents a measure of photochemical efficiency, was measured as described previously (Oh et al., 1996) during the dark treatment. Dark respiration was measured using the same gas exchange described above in full rosettes of plants maintained in darkness.
Metabolite Profiling
Metabolite profiling was performed using approximately 50 mg of whole rosette material. The extraction, derivatization, standard addition, and sample injection were performed based on the GC-MS-based metabolite profiling protocol of Lisec et al. (2006). Peak detection, retention time alignment, and library matching were performed using the Target Search R package (Cuadros-Inostroza et al., 2009). Metabolites were identified in comparison with database entries of authentic standards (Kopka et al., 2005; Schauer et al., 2005). Identification and annotation of detected peaks followed the recommendations for reporting metabolite data described by Fernie et al. (2011). The full data set from the metabolite profiling study is additionally available as Supplemental Table S1.
Expression Analysis by qRT-PCR
Total RNA was isolated using TRIzol reagent (Ambion, Life Technology) according to the manufacturer’s recommendations. Total RNA was treated with DNase I (RQ1 RNase free DNase I; Promega). The integrity of the RNA was checked on 1% (w/v) agarose gels, and the concentration was measured using a Nanodrop spectrophotometer. Finally, 2 μg of total RNA were reverse transcribed with SuperScript II RNase H2 reverse transcriptase (Invitrogen) and oligo(dT) primer according to the manufacturer’s recommendations. Real-time PCR was performed on a 96-well microtiter plate with an ABI PRISM 7900 HT sequence detection system (Applied Biosystems Applera) using Power SYBR Green PCR Master Mix according to Piques et al. (2009). The primers used here were designed using the open-source program QuantPrime-qPCR primer design tool (Arvidsson et al., 2008) and are described in Supplemental Table S2. The transcript abundance was calculated by the standard curves of each selected gene and normalized using the constitutively expressed gene ACTIN (AT2G37620). Data analyses were performed as described by Caldana et al. (2007). Three biological replicates were processed for each experimental condition.
Statistical Analyses
The experiments were conducted in a completely randomized design with three to seven replicates of each genotype. Data were statistically examined using ANOVA and tested for significant (P < 0.05) differences using Student’s t test. All statistical analyses were performed using the algorithm embedded into Microsoft Excel.
Accession Numbers
The Arabidopsis Genome Initiative locus numbers for the major genes discussed in this article are as follows: ATG5 (At5g17290), ATG7 (At5g45900), and ATG9 (At2g31260).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Schematic representation of the sites of T-DNA insertion in atg mutants.
Supplemental Figure S2. Gas-exchange parameters are not affected in atg mutants.
Supplemental Figure S3. Phenotypes of Arabidopsis mutants under extended dark treatment.
Supplemental Figure S4. Metabolite levels in atg Arabidopsis mutants.
Supplemental Table S1. Relative metabolite contents of leaves of Arabidopsis knockout mutants atg5-1, atg7-2, and atg9-1 and wild-type plants after further treatment for 0, 3, 6, and 9 d in darkness.
Supplemental Table S2. Primers utilized for quantitative RT-PCR.
Acknowledgments
Discussions with Dimas M. Ribeiro and Agustin Zsögön (Universidade Federal de Viçosa) were highly valuable in the development of this work. We also thank Gad Galili (Weizmann Institute of Science) for contributing the seeds used in this study.
Glossary
- BCAA
branched-chain amino acid
- RT
reverse transcription
- GC-MS
gas chromatography-mass spectrometry
Footnotes
This work was supported by funding from the Max Planck Society, the National Council for Scientific and Technological Development (CNPq-Brazil grant no. 402511/2016-6), and the Foundation for Research Assistance of the Minas Gerais State (FAPEMIG-Brazil grant no. APQ-01078-15) to W.L.A. Scholarships granted by the Agency for the Support and Evaluation of Graduate Education (CAPES-Brazil) to J.A.S.B., FAPEMIG to J.H.F.C (grant no BDP-00018-16) and D.B.M., and research fellowships granted by CNPq to A.N.-N. and W.L.A. are gratefully acknowledged. The work performed by T.A.-W. was supported by Minerva, Alexander von Humboldt, and EMBO postdoctoral fellowships.
References
- Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, et al. (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR (2011) Protein degradation: an alternative respiratory substrate for stressed plants. Trends Plant Sci 16: 489–498 [DOI] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B (2008) QuantPrime: a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin OK, Evans JR, Ball MC, Lambers H, Pons TL (2000) Leaf respiration of snow gum in the light and dark: interactions between temperature and irradiance. Plant Physiol 122: 915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C (2014) Autophagy, plant senescence, and nutrient recycling. J Exp Bot 65: 3799–3811 [DOI] [PubMed] [Google Scholar]
- Avin-Wittenberg T, Bajdzienko K, Wittenberg G, Alseekh S, Tohge T, Bock R, Giavalisco P, Fernie AR (2015) Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 27: 306–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC. (2009) Function and regulation of macroautophagy in plants. Biochim Biophys Acta 1793: 1397–1403 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Chrobok D, Law SR, Brouwer B, Lindén P, Ziolkowska A, Liebsch D, Narsai R, Szal B, Moritz T, Rouhier N, et al. (2016) Dissecting the metabolic role of mitochondria during developmental leaf senescence. Plant Physiol 172: 2132–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Contento AL, Kim SJ, Bassham DC (2004) Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol 135: 2330–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross JM, von Korff M, Altmann T, Bartzetko L, Sulpice R, Gibon Y, Palacios N, Stitt M (2006) Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol 142: 1574–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros-Inostroza A, Caldana C, Redestig H, Kusano M, Lisec J, Peña-Cortés H, Willmitzer L, Hannah MA (2009) TargetSearch: a Bioconductor package for the efficient preprocessing of GC-MS metabolite profiling data. BMC Bioinformatics 10: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Engqvist M, Drincovich MF, Flügge UI, Maurino VG (2009) Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and β-oxidation pathways. J Biol Chem 284: 25026–25037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist MK, Kuhn A, Wienstroer J, Weber K, Jansen EEW, Jakobs C, Weber APM, Maurino VG (2011) Plant D-2-hydroxyglutarate dehydrogenase participates in the catabolism of lysine especially during senescence. J Biol Chem 286: 11382–11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, Klionsky DJ (2014) The machinery of macroautophagy. Cell Res 24: 24–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Aharoni A, Willmitzer L, Stitt M, Tohge T, Kopka J, Carroll AJ, Saito K, Fraser PD, DeLuca V (2011) Recommendations for reporting metabolite data. Plant Cell 23: 2477–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Roscher A, Ratcliffe RG, Kruger NJ (2001) Fructose 2,6-bisphosphate activates pyrophosphate:fructose-6-phosphate 1-phosphotransferase and increases triose phosphate to hexose phosphate cycling in heterotrophic cells. Planta 212: 250–263 [DOI] [PubMed] [Google Scholar]
- Galili G. (2011) The aspartate-family pathway of plants: linking production of essential amino acids with energy and stress regulation. Plant Signal Behav 6: 192–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G, Amir R, Fernie AR (2016) The regulation of essential amino acid synthesis and accumulation in plants. Annu Rev Plant Biol 67: 153–178 [DOI] [PubMed] [Google Scholar]
- Galili G, Avin-Wittenberg T, Angelovici R, Fernie AR (2014) The role of photosynthesis and amino acid metabolism in the energy status during seed development. Front Plant Sci 5: 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M (2004) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić V. (2003) SAG2 and SAG12 protein expression in senescing Arabidopsis plants. Physiol Plant 119: 263–269 [Google Scholar]
- Guiboileau A, Yoshimoto K, Soulay F, Bataillé MP, Avice JC, Masclaux-Daubresse C (2012) Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol 194: 732–740 [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt TM, Nunes Nesi A, Araújo WL, Braun HP (2015) Amino acid catabolism in plants. Mol Plant 8: 1563–1579 [DOI] [PubMed] [Google Scholar]
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jørgensen LB, Jones JD, Mundy J, Petersen M (2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Ishida H, Izumi M, Wada S, Makino A (2014) Roles of autophagy in chloroplast recycling. Biochim Biophys Acta 1837: 512–521 [DOI] [PubMed] [Google Scholar]
- Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T (2008) Mobilization of Rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, Leaver CJ (2005) The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Schauer N, Larson TR, Graham IA, Fernie AR, Leaver CJ (2006) The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J 47: 751–760 [DOI] [PubMed] [Google Scholar]
- Izumi M, Hidema J, Makino A, Ishida H (2013) Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol 161: 1682–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Ishida H, Nakamura S, Hidema J (2017) Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell 29: 377–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Wada S, Makino A, Ishida H (2010) The autophagic degradation of chloroplasts via Rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol 154: 1196–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O, Pesquet E, Ahad A, Askne A, Nordvall D, Vodnala SM, Tuominen H, Hurry V, Dizengremel P, Gardeström P (2007) The different fates of mitochondria and chloroplasts during dark-induced senescence in Arabidopsis leaves. Plant Cell Environ 30: 1523–1534 [DOI] [PubMed] [Google Scholar]
- Kirma M, Araújo WL, Fernie AR, Galili G (2012) The multifaceted role of aspartate-family amino acids in plant metabolism. J Exp Bot 63: 4995–5001 [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: the Golm metabolome database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Krüßel L, Junemann J, Wirtz M, Birke H, Thornton JD, Browning LW, Poschet G, Hell R, Balk J, Braun HP, et al. (2014) The mitochondrial sulfur dioxygenase ETHYLMALONIC ENCEPHALOPATHY PROTEIN1 is required for amino acid catabolism during carbohydrate starvation and embryo development in Arabidopsis. Plant Physiol 165: 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Schwarzländer M, Obata T, Sirikantaramas S, Burow M, Olsen CE, Tohge T, Fricker MD, Møller BL, Fernie AR, et al. (2009) The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol Plant 2: 390–406 [DOI] [PubMed] [Google Scholar]
- Li F, Chung T, Pennington JG, Federico ML, Kaeppler HF, Kaeppler SM, Otegui MS, Vierstra RD (2015) Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27: 1389–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vierstra RD (2012) Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Liu Y, Bassham DC (2010) TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bassham DC (2012) Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Lv X, Pu X, Qin G, Zhu T, Lin H (2014) The roles of autophagy in development and stress responses in Arabidopsis thaliana. Apoptosis 19: 905–921 [DOI] [PubMed] [Google Scholar]
- Michaeli S, Galili G, Genschik P, Fernie AR, Avin-Wittenberg T (2016) Autophagy in plants: what’s new on the menu? Trends Plant Sci 21: 134–144 [DOI] [PubMed] [Google Scholar]
- Michaeli S, Honig A, Levanony H, Peled-Zehavi H, Galili G (2014) Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 26: 4084–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K, MacKerness SA, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23: 677–685 [DOI] [PubMed] [Google Scholar]
- Noh YS, Amasino RM (1999) Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol Biol 41: 181–194 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Oh SA, Lee SY, Chung IK, Lee CH, Nam HG (1996) A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol Biol 30: 739–754 [DOI] [PubMed] [Google Scholar]
- Peng C, Uygun S, Shiu SH, Last RL (2015) The impact of the branched-chain ketoacid dehydrogenase complex on amino acid homeostasis in Arabidopsis. Plant Physiol 169: 1807–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD (2008) The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Höhne M, Usadel B, Gibon Y, Rohwer J, Stitt M (2009) Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires MV, Pereira Júnior AA, Medeiros DB, Daloso DM, Pham PA, Barros KA, Engqvist MK, Florian A, Krahnert I, Maurino VG, et al. (2016) The influence of alternative pathways of respiration that utilize branched-chain amino acids following water shortage in Arabidopsis. Plant Cell Environ 39: 1304–1319 [DOI] [PubMed] [Google Scholar]
- Plaxton WC, Podesta FE (2006) The functional organization and control of plant respiration. Crit Rev Plant Sci 25: 159–198 [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Salvato F, Havelund JF, Chen M, Rao RSP, Rogowska-Wrzesinska A, Jensen ON, Gang DR, Thelen JJ, Møller IM (2014) The potato tuber mitochondrial proteome. Plant Physiol 164: 637–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Shin KD, Lee HN, Chung T (2014) A revised assay for monitoring autophagic flux in Arabidopsis thaliana reveals involvement of AUTOPHAGY-RELATED9 in autophagy. Mol Cells 37: 399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Beard KF, Nunes-Nesi A, Fernie AR, Ratcliffe RG (2010) Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 15: 462–470 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A (2009) Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 149: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Blumwald E (2014) Stress-induced chloroplast degradation in Arabidopsis is regulated via a process independent of autophagy and senescence-associated vacuoles. Plant Cell 26: 4875–4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu B, Zhao J, Guo J, Li Y, Han S, Huang L, Du Y, Hong Y, Tang D, et al. (2013) Autophagy contributes to leaf starch degradation. Plant Cell 25: 1383–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watmough NJ, Frerman FE (2010) The electron transfer flavoprotein:ubiquinone oxidoreductases. Biochim Biophys Acta 1797: 1910–1916 [DOI] [PubMed] [Google Scholar]
- Weaver LM, Amasino RM (2001) Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol 127: 876–886 [PMC free article] [PubMed] [Google Scholar]
- Weigelt K, Küster H, Radchuk R, Müller M, Weichert H, Fait A, Fernie AR, Saalbach I, Weber H (2008) Increasing amino acid supply in pea embryos reveals specific interactions of N and C metabolism, and highlights the importance of mitochondrial metabolism. Plant J 55: 909–926 [DOI] [PubMed] [Google Scholar]
- Xie Q, Michaeli S, Peled-Zehavi H, Galili G (2015) Chloroplast degradation: one organelle, multiple degradation pathways. Trends Plant Sci 20: 264–265 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Bassham DC (2005) AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Shibata M, Kondo M, Oikawa K, Sato M, Toyooka K, Shirasu K, Nishimura M, Ohsumi Y (2014) Organ-specific quality control of plant peroxisomes is mediated by autophagy. J Cell Sci 127: 1161–1168 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, Hu Y, You J, Shi H, Zhu Y, et al. (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA 113: 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Chung KP, Cui Y, Lin W, Gao C, Kang BH, Jiang L (2017) ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci USA 114: E426–E435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientara-Rytter K, Sirko A (2016) To deliver or to degrade: an interplay of the ubiquitin-proteasome system, autophagy and vesicular transport in plants. FEBS J 283: 3534–3555 [DOI] [PubMed] [Google Scholar]