A large-scale survey of photosynthetic parameters and biomass accumulation in a global rice diversity panel reveals critical leaf photosynthetic parameters controlling biomass accumulation.
Abstract
Mining natural variations is a major approach to identify new options to improve crop light use efficiency. So far, successes in identifying photosynthetic parameters positively related to crop biomass accumulation through this approach are scarce, possibly due to the earlier emphasis on properties related to leaf instead of canopy photosynthetic efficiency. This study aims to uncover rice (Oryza sativa) natural variations to identify leaf physiological parameters that are highly correlated with biomass accumulation, a surrogate of canopy photosynthesis. To do this, we systematically investigated 14 photosynthetic parameters and four morphological traits in a rice population, which consists of 204 U.S. Department of Agriculture-curated minicore accessions collected globally and 11 elite Chinese rice cultivars in both Beijing and Shanghai. To identify key components responsible for the variance of biomass accumulation, we applied a stepwise feature-selection approach based on linear regression models. Although there are large variations in photosynthetic parameters measured in different environments, we observed that photosynthetic rate under low light (Alow) was highly related to biomass accumulation and also exhibited high genomic inheritability in both environments, suggesting its great potential to be used as a target for future rice breeding programs. Large variations in Alow among modern rice cultivars further suggest the great potential of using this parameter in contemporary rice breeding for the improvement of biomass and, hence, yield potential.
Improving photosynthetic efficiency is regarded as a major target to improve crop biomass production and yield potential (Long et al., 2006; for review, see Zhu et al., 2010). The canopy photosynthetic efficiency, which is determined by leaf area index, canopy architecture, and leaf photosynthetic properties, plays an important role in determining biomass accumulation (Long et al., 2006; Zhu et al., 2012). Historically, the improvement of canopy architecture (i.e. creating cultivars with semidrawf architecture, more erect leaves, and higher leaf area index) has played an important role in traditional crop breeding (Hedden, 2003; Peng et al., 2008); in contrast, the improvement of leaf photosynthetic properties has played a minor or no role during this process. Broadly, there are two major approaches to improve photosynthetic efficiency: by genetically engineering photosynthetic efficiency if an engineering target is well defined and by conventional crop breeding (i.e. identifying those lines with superior photosynthetic efficiency and then crossing this superior photosynthetic property into desired target cultivars; Gepts, 2002; Long et al., 2006). In either case, the major challenge now is to define effective photosynthetic traits that can lead to enhanced biomass production. We earlier demonstrated that a systems approach can be used to identify new targets to improve photosynthesis by combining systems modeling and an evolutionary algorithm (Zhu et al., 2008). The identified targets to improve photosynthesis have been tested transgenically both in the laboratory and in the field (Rosenthal et al., 2011; Simkin et al., 2015). Similarly, increasing the speed of recovery from photoprotection has been demonstrated to be a major approach to increase canopy photosynthesis and crop yield potential (Zhu et al., 2004), which was validated recently in the model crop species tobacco (Nicotiana tabacum) in the field (Kromdijk et al., 2016). This success of enhancing biomass production through the manipulation of photosynthesis clearly demonstrates that there is huge potential to improve photosynthetic efficiency for greater biomass and yield production.
Besides using a systems approach, another potentially rewarding approach to identify parameters related to biomass production is through mining natural variations (Flood et al., 2011; Lawson et al., 2012). The systems approach, to a certain degree, increases the potential range of physiological parameters that can be explored and then observed in existing cultivars. However, the success of this approach relies on the availability of highly sophisticated and accurate systems models for the process under study. The advantage of mining natural variation is that we can collect biomass data and many physiological parameters for a large number of germplasms simultaneously, which facilitates the identification of parameters before the availability of highly mechanistic models for the involved processes.
So far, however, large-scale systematic studies of natural variations of photosynthetic parameters in major crops are scarce. Driever et al. (2014) reported natural variations of photosynthetic parameters in 64 elite wheat (Triticum aestivum) cultivars and found that, although there are significant variations in photosynthetic capacity, biomass, and yield, no correlation exists between grain yield and photosynthetic capacity. They suggested that, during the breeding process, some traits might have been unintentionally selected out; hence, photosynthetic efficiency should be a major target to utilize during wheat breeding in the future (Driever et al., 2014; Carmo-Silva et al., 2017). Similar experiments have been conducted in rice (Oryza sativa) that reached similar conclusions (i.e. leaf photosynthetic rates measured under saturating light levels do not show positive correlation with biomass accumulation; Jahn et al., 2011). At first sight, this is rather contradictory to the current theory of photosynthesis. However, if we consider the canopy, then the overall crop light use efficiency, where biomass accumulation can be used as a surrogate, is determined by the total canopy photosynthesis instead of leaf photosynthesis. Indeed, our earlier modeling work showed that light-limited photosynthesis can contribute up to 70% of the total canopy photosynthetic CO2 uptake rates, even at a moderate leaf area index of 4.8 (Song et al., 2013). The proportion of light-limited photosynthesis will be even higher under either high leaf area index or future elevated CO2 conditions (Zhu et al., 2012; Song et al., 2013). Large-scale surveys of rice grain yield, harvest index, and biomass accumulation for rice cultivars released since 1966 have shown clearly that the grain yield of cultivars released after 1980 was highly correlated with biomass accumulation, suggesting improved canopy photosynthesis during recent rice breeding (Peng et al., 2001; Hubbart et al., 2007). The potential factors contributing to canopy photosynthesis in rice remain unknown.
In this study, we aim to identity leaf photosynthetic parameters that are highly correlated with biomass accumulation, a surrogate of canopy photosynthesis. To do this, we surveyed a large number of leaf photosynthetic parameters and crop architectural parameters at two different locations in China (i.e. Shanghai [SH] and Beijing [BJ]). In this study, to enable a comprehensive survey of parameters relevant to canopy photosynthesis, we measured photosynthetic parameters not only under high light but also under limiting light conditions, with the intention to examine whether photosynthetic rates under low light are positively correlated with biomass accumulation. Finally, to minimize the potential complexity of source-sink interaction during the grain-filling stage, we used biomass accumulation before flowering to avoid the complexity of source-sink interaction (Chang et al., 2017). To maximize the genetic diversity utilized in this study, we used both a global rice diversity population consisting of 204 minicore accessions and 11 elite Chinese rice cultivars. Our results revealed that photosynthetic rate under low light (Alow) is highly correlated with biomass accumulation in this diverse rice germplasm population under both Beijing and Shanghai environments. Genetic analysis further shows that Alow is under strong genetic control and, hence, is amenable for breeding or genetic manipulations. The large variations of Alow in modern rice variations and the high genetic inheritance suggest that Alow can be used as a promising target in rice marker-assisted breeding.
RESULTS
Variability of the Parameters in the Global Rice Diversity Panel
As shown from Table I, natural variations for both 14 photosynthetic traits (PTs) and four morphological traits (MTs) in BJ and SH conditions showed different levels of heterogeneity. The PGV is used to represent the levels of natural variation of traits. The PGV is calculated by the differences between extreme values over mean values in the population (for details, see “Materials and Methods”). The values of PGV in BJ ranged from 3.6 to 197.6 for PTs and from 77.2 to 227.9 for MTs; while the values of PGV in SH ranged from 21.5 to 280.1 for PTs and from 85.9 to 170 for MTs. The trait with minimum natural variation is the maximal quantum yield of PSII (Fv/Fm), and its PGV under BJ/SH environmental conditions is only 12%. For gas exchange-related parameters under full light, the PGV values across the two conditions decreased as follows: stomatal conductance under normal light (gs) > stomatal limitation (Ls) > water use efficiency (WUE) > photosynthetic CO2 uptake rate (A) > internal CO2 concentration under normal light (Ci), while the PGV values of these parameters under low light decreased as follows: water use efficiency under low light (Wlow) > stomatal limitation under low light (Lslow) > photosynthetic CO2 uptake rate under low light (Alow) > stomatal conductance under low light (gslow) > internal CO2 concentration under low light (Cilow; Table I). The PGV of both dark respiration (Adark) and stomatal conductance under dark (gsdark) were at least 120%, which was 2 times higher than the PGV of the SPAD value, a surrogate of chlorophyll concentration. For morphological traits (Table I), PGV values showed drastic differences between experiments in BJ/SH environmental conditions. The ranking of PGVs for biomass, tiller number, and leaf thickness decreased gradually under BJ/SH environmental conditions. Most of the MTs showed higher variations in PGV values under the BJ environment than under SH, except for the PGV of plant height (Table I).
Table I. Natural variation and SNP-based heritability of PTs in the global minicore panel and elite rice lines grown in BJ and SH environments.
Percentage genetic variation (PGV) was calculated as described in “Materials and Methods.” Asterisks represent P < 0.05 (*) and P < 0.01 (**). Abbreviations for PTs are as follows: A, photosynthetic rates under high light; Alow: photosynthetic rates under low light; Biomass, aboveground biomass; Ci, internal CO2 under high light; Cilow, internal CO2 under low light; Fv/Fm, maximum PSII efficiency; gs, stomatal conductance under high light; gslow, stomatal conductance under low light; Ls, stomatal limitation under high light; Lslow, stomatal limitation under low light; SPAD, SPAD values; WUE, water use efficiency under high light; Wlow, water use efficiency under low light. h2SNP represents SNP-based heritability. The two shaded rows mean that the data are available only for the BJ site. N represents the number of the accession.
| Traits | Sites | N | Range | Mean ± sd | PGV | h2SNP ± se |
|---|---|---|---|---|---|---|
| A (μmol m−2 s−1) | BJ | 214 | 13.65–28.19 | 21.02 ± 3.00 | 69.17 | 0.13 ± 0.07 |
| SH | 186 | 12.44–39.76 | 24.35 ± 5.02 | 112.20 | 0.15 ± 0.03 | |
| gs (mmol m−2 s−1) | BJ | 214 | 0.18–0.99 | 0.41 ± 0.12 | 197.56 | 0.34 ± 0.14* |
| SH | 186 | 0.14–1.16 | 0.56 ± 0.18 | 182.14 | 0.25 ± 0.08* | |
| WUE (mmol mol−1) | BJ | 214 | 28.77–67.51 | 44.51 ± 0.66 | 87.04 | 0.73 ± 0.20** |
| SH | 187 | 20.50–77.71 | 43.44 ± 10.03 | 131.70 | 0.61 ± 0.11** | |
| Ci (μmol mol−1) | BJ | 215 | 204.4–316.4 | 276.53 ± 17.75 | 40.50 | 0.71 ± 0.20** |
| SH | 187 | 245.91–347.75 | 308.66 ± 21.58 | 32.99 | <0.001 | |
| Ls | BJ | 215 | 0.18–0.47 | 0.28 ± 0.05 | 103.57 | 0.72 ± 0.20** |
| SH | 188 | 0.15–0.51 | 0.26 ± 007 | 138.46 | 0.48 ± 0.21** | |
| Alow (μmol m−2 s−1) | BJ | 214 | 2.56–6.62 | 4.87 ± 0.65 | 83.37 | 0.37 ± 0.12* |
| SH | 187 | 2.26–6.42 | 3.76 ± 0.67 | 110.64 | 0.36 ± 0.12* | |
| gslow (mmol m−2 s−1) | BJ | 214 | 0.17–0.26 | 0.12 ± 0.04 | 75.00 | 0.08 ± 0.13 |
| Cilow (μmol mol−1) | BJ | 214 | 242.10–342.60 | 299.76 ± 17.19 | 33.53 | 0.29 ± 0.13* |
| SH | 188 | 265.12–380.37 | 353.63 ± 15.56 | 32.59 | <0.001 | |
| Wlow (mmol mol−1) | BJ | 214 | 20.91–73.25 | 39.18 ± 0.83 | 133.59 | <0.001 |
| SH | 187 | 8.62–72.07 | 22.63 ± 9.55 | 280.38 | <0.001 | |
| Lslow | BJ | 214 | 0.27–0.79 | 0.61 ± 0.83 | 85.25 | 0.08 ± 0.03 |
| SH | 188 | 0.13–0.95 | 0.44 ± 0.17 | 186.36 | 0.15 ± 0.04 | |
| Adark (μmol mol-2s−1) | BJ | 214 | −0.32–−0.87 | −0.47 ± 0.09 | 117.02 | 0.40 ± 0.16* |
| SH | 183 | −0.21–−1.25 | −0.53 ± 0.22 | 233.96 | 0.26 ± 0.06* | |
| gsdark (mol mol−1) | BJ | 214 | 0.01–0.05 | 0.02 ± 0.01 | 200.00 | 0.08 ± 0.12 |
| Fv/Fm | BJ | 214 | 0.82–0.85 | 0.84 ± 0.01 | 3.57 | 0.34 ± 0.13* |
| SH | 182 | 0.66–0.83 | 0.79 ± 0.03 | 21.52 | 0.12 ± 0.03 | |
| SPAD | BJ | 213 | 27.13–65.2 | 37.28 ± 4.54 | 102.12 | 0.60 ± 0.16* |
| SH | 180 | 23.53–47.70 | 36.47 ± 4.77 | 66.27 | 0.53 ± 0.17* | |
| Biomass (g) | BJ | 214 | 4.60–63.40 | 25.8 ± 9.36 | 227.9 | 0.35 ± 0.22* |
| SH | 184 | 6.69–64.35 | 22.74 ± 1.04 | 97.40 | 0.25 ± 0.03* | |
| Plant height (cm) | BJ | 214 | 58.33–138.0 | 103.26 ± 14.65 | 77.2 | 0.07 ± 0.02 |
| SH | 188 | 55.0–130.75 | 91.11 ± 13.78 | 170.04 | <0.001 | |
| Tiller number | BJ | 214 | 5.67–30.67 | 12.93 ± 4.36 | 193.3 | 0.25 ± 0.12* |
| SH | 188 | 6.25–25.67 | 23.27 ± 2.99 | 85.93 | 0.20 ± 0.05* | |
| Leaf thickness (μm) | BJ | 214 | 105.67–420.0 | 210.85 ± 49.86 | 149.1 | <0.001 |
| SH | 183 | 175.07–355.01 | 209.43 ± 32.30 | 95.22 | <0.001 |
The estimation of single-nucleotide polymorphism (SNP)-based heritability (h2SNP) on a functional trait provides information about whether any particular trait is under strong genetic control and, hence, can be used as a potential parameter during crop breeding. h2SNP of PTs was in the range of less than 0.001 to 0.72 in BJ/SH environmental conditions (Table I). Among these PTs, only four PTs exhibited significant h2SNP under BJ/SH environmental conditions: WUE, Alow, Adark, and SPAD (Table I). For the MTs, biomass accumulation and tiller number showed high h2SNP under BJ/SH environmental conditions (Table I).
We further employed two-way ANOVA to analyze genotype × environment interaction with regard to PTs and MTs. The results show that all PTs and MTs were significantly different between BJ and SH environments. On the one hand, we found strong environmental effects on most of the collected PTs and MTs (Table II); in contrast, 12 out of 18 collected traits were affected significantly by a genotype factor, except Ls, gslow, Cilow, Lslow, gsdark, and leaf thickness (Table II). Only Cilow was not significantly affected by environment and genotype interactions (Table II).
Table II. ANOVA (F values and significance) for the effects of environment, genotype, and environment × genotype interaction for each photosynthetic trait.
Stomatal conductance measured under low light (gslow) and at night (gsdark) were not determined under the SH experimental condition. ND, No determination. Asterisks represent P < 0.05 (*) and P < 0.001 (***).
| Traits | Genotype | Environment | Interactions |
|---|---|---|---|
| A | 5.494*** | 165.50*** | 8.931*** |
| gs | 5.607*** | 358.80*** | 11.100*** |
| WUE | 6.795*** | 16.40*** | 10.360*** |
| Ci | 3.350*** | 376.50*** | 2.488*** |
| Ls | 0.296 | 135.80*** | 4.681*** |
| Alow | 3.617*** | 708.80*** | 6.958*** |
| gslow | 1.155 | ND | ND |
| Wlow | 1.197* | 333.40*** | 17.340*** |
| Cilow | 1.907 | 109.10*** | 1.127 |
| Lslow | 0.397 | 114.72*** | 15.110*** |
| Adark | 4.338*** | 55.84*** | 21.120*** |
| gsdark | 0.980 | ND | ND |
| SPAD | 7.68*** | 10.18*** | 5.438*** |
| Fv/Fm | 3.695*** | 118.20*** | 0.328*** |
| Biomass | 6.549*** | 45.40*** | 16.860*** |
| Plant height | 2.554*** | 176.80*** | 5.650*** |
| Tiller number | 1.696*** | 2022.00*** | 17.910*** |
| Leaf thickness | 0.463 | 986.00*** | 15.440*** |
Correlation between Biomass and Other Biological Parameters
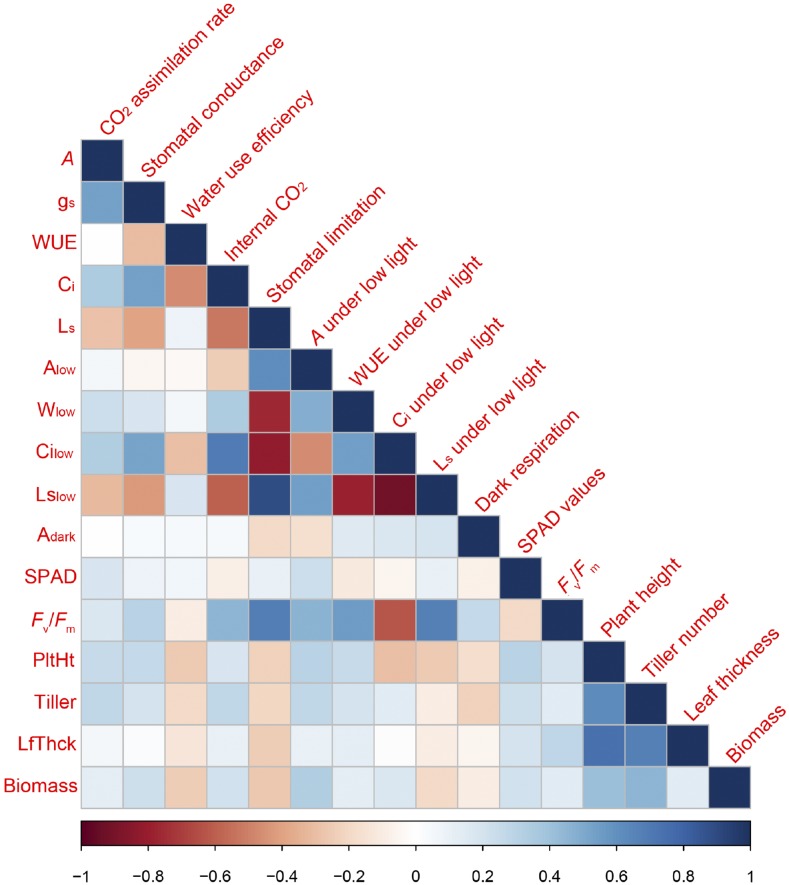
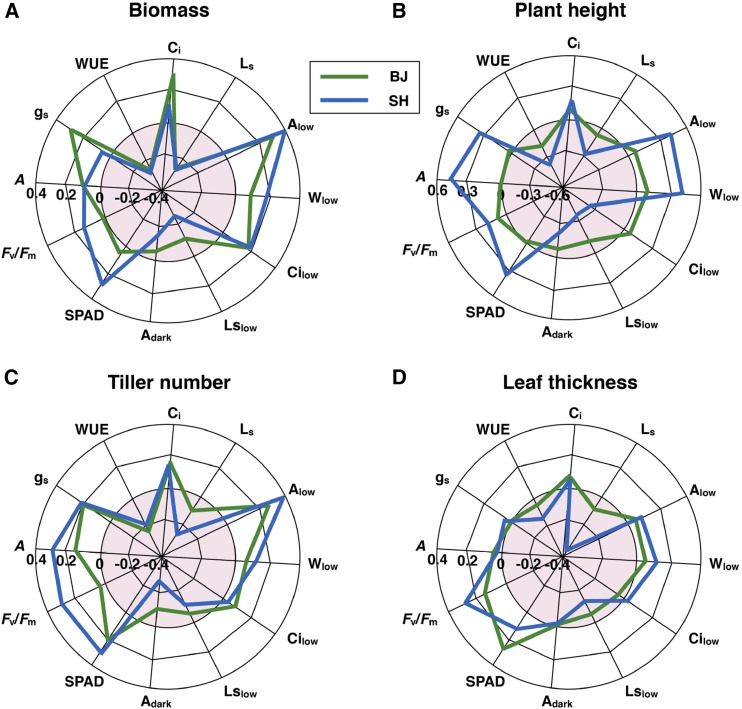
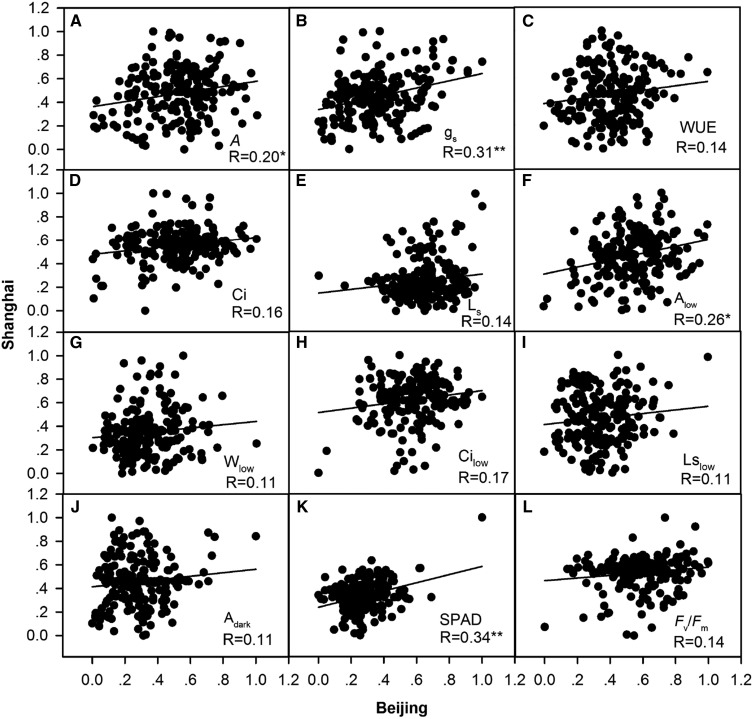
The Pearson correlation coefficient was determined to evaluate the relatedness of biomass with different PTs and MTs under BJ/SH environmental conditions (Fig. 1; Supplemental Table S1; Supplemental Figs. S2 and S3). As shown in Figure 1 and Supplemental Table S1, strong correlations were observed between PTs under both normal light and low light conditions when data sets measured under BJ/SH environmental conditions were combined. Figure 2 shows the correlation between MTs and PTs in BJ/SH environmental conditions (Supplemental Table S2). The results reveal that A, gs, Ci, Alow, Wlow, Cilow, Fv/Fm, and SPAD show positive correlation with plant height, tiller number, and biomass. On the other hand, WUE, Ls, Lslow, and Adark showed negative correlation with plant height, tiller number, leaf thickness, and biomass. As expected, there is huge variation in the measured PTs between BJ and SH environmental conditions, suggesting strong environment impacts on most of the PTs (Fig. 3), as shown earlier by the strong environment effect on these parameters (Table II). Certain photosynthetic parameters, including A, gs, Alow, and SPAD, showed high correlation index (R2) between both BJ and SH sites (Fig. 3).
Figure 1.
Correlation of PTs and MTs in the global minicore panel and elite rice lines. Data were combined from BJ and SH experiments. Abbreviations are defined in the figure.
Figure 2.
Graphic representation of the correlations of different PTs with MTs in the global minicore panel and elite rice lines under BJ and SH environments. The shaded area at the center of each circle represents negative correlation. Trait abbreviations are given in Table I.
Figure 3.
Self-correlation of each photosynthetic trait in the global minicore panel and elite rice lines grown in BJ and SH environments. The Pearson correlation coefficient (R) values were calculated. Asterisks represent P < 0.05 (*) and P < 0.01 (**). The PT abbreviations are defined in Figure 1.
Linear Regression Model and Stepwise Feature Selection
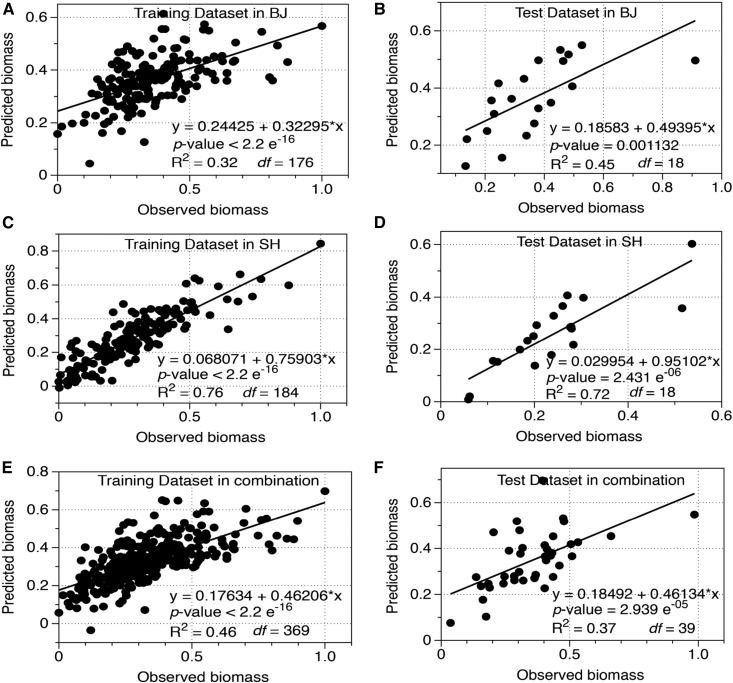
To identify the key parameters that dominate biomass variation under BJ/SH environmental conditions, we employed a linear regression model (LRM) with a stepwise optimization method based on the Akaike information criterion. We first evaluated the prediction accuracy of the derived LRMs under BJ, SH, and combined data sets (Fig. 4). Our approach used a training data set consisting of 90% of the original data and a test data set of the remaining 10% of the data (for details, see “Materials and Methods”). As shown in Figure 4, the models predicted the values of biomass under BJ, SH, and combined environments (P < 0.001), with R2 between the predicted and measured biomass ranging from 0.32 to 0.76 in the training data set (Fig. 4, A, C, and E). Furthermore, the model predicted the test data set, with R2 ranging from 0.37 to 0.72 across the three models (Fig. 4, B, D, and F). These results suggest that the derived LRMs can predict the biomass accumulation with a high level of confidence.
Figure 4.
Model construction and cross-validation in the global minicore panel and elite rice lines under BJ (A and B), SH (C and D), and the combined (E and F) data sets. The training data set consists of 90% of the whole data set, and the remaining 10% of items were used as a test data set (for details, see “Materials and Methods”). Predicted values of biomass versus observed values of biomass were used during the cross-validation. The determination index (R2) reflects the accuracy of regression between predicted and observed values.
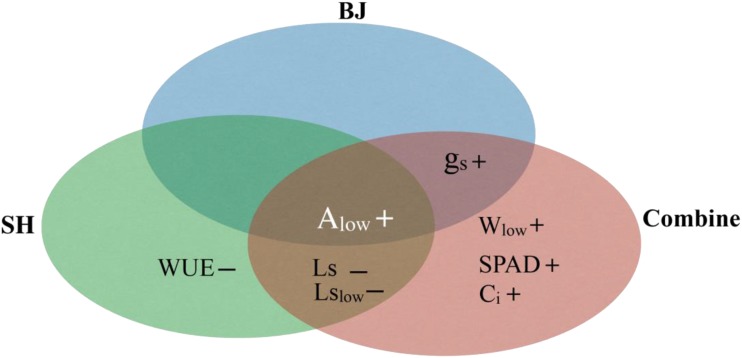
The PTs identified in these three LRMs were used for further analysis. The PTs identified as highly correlated with biomass accumulation from different data sets are shown in Figure 5 and Supplemental Table S3. The fitting equations under BJ/SH environmental conditions are as follows: biomass (BJ) = 0.096 gs + 0.115 Alow + 0.311 × plant height + 0.355 × tiller number; biomass (SH) = 0.053 Ls − 0.081 WUE + 0.152 Alow + 0.077 Lslow + 0.127 × leaf thickness + 0.341 × plant height + 0.671 × tiller number; biomass (combined) = 0.072 gs + 0.169 Ci − 0.089 Ls + 0.107 Alow + 0.192 Wlow − 0.145 Lslow + 0.139 SPAD + 0.448 × plant height + 0.556 × tiller number. Two PTs (gs and Alow) were identified in models for BJ; four PTs (WUE, Ls, Lslow, and Alow) were identified for SH; and seven PTs (gs, Wlow, SPAD, Ci, Ls, Lslow, and Alow) were identified for the combined environments. These large variations in the identified PTs responsible for biomass accumulation in different locations reflect the strong environment impacts on many photosynthetic parameters. Surprisingly, even under such great impacts of environments on photosynthetic parameters, Alow was identified consistently to be closely associated with biomass accumulation in BJ/SH environmental conditions (Fig. 5). gs also was shown to be a major variable associated with biomass accumulation in both the BJ and combined data sets. Ls and Lslow were identified in both SH and combined data sets (Fig. 5). Based on these obtained LRMs, the PTs expected to be increased to improve biomass accumulation are gs, Alow, Wlow, and SPAD, while those expected to be decreased are WUE, Ls, and Lslow (Supplemental Table S3).
Figure 5.
Feature selection analysis of PTs using LRMs. Key PTs identified by the models were represented under BJ, SH, and combined environments (Combine). The equations under BJ, SH, and combined environments are as follows: biomass (BJ) = 0.096 gs + 0.115 Alow + 0.311 × plant height + 0.355 × tiller number; biomass (SH) = 0.053 Ls − 0.081 WUE + 0.152 Alow + 0.077 Lslow + 0.127 × leaf thickness + 0.341 × plant height + 0.671 × tiller number; biomass (combined) = 0.072 gs + 0.169 Ci − 0.089 Ls + 0.107 Alow + 0.192 Wlow − 0.145 Lslow + 0.139 SPAD + 0.448 × plant height + 0.556 × tiller number.
Ranking of Elite Cultivars within the Minicore Collection
To further evaluate the scope to manipulate Alow for improved biomass production, we examined the distribution of Alow among the minicore panel and the distribution of Alow in 11 current elite rice lines (Fig. 6). Under the BJ environment, Alow exhibits a normal distribution in the minicore population (Fig. 6A) and there is a huge variation of Alow among the 11 elite rice lines (Fig. 6B). The distribution pattern and ranking of Alow in the SH environment are shown in Supplemental Figure S1. We further evaluated the potential improvements of Alow by calculating the percentage difference between Alow of the elite lines and the highest Alow observed in the minicore population. There are potentially 76.77% and 85.49% improvement in DHX-Z and ZH11, respectively, if their corresponding Alow can reach the maximal Alow in the minicore (i.e. that for P4140).
Figure 6.
Trait distribution of elite cultivars and the minicore accessions under the BJ environment. A, Histogram representing the distribution of Alow in the minicore diversity panel. B, Phenotypic distribution of Alow of elite cultivars within the minicore accessions. The circled numbers are as follows: 1, WCC1; 2, WCC2; 3, DHX-Z; 4, HE19; 5, KY131; 6, XS134; 7, ZH11; 8, MH63; 9, KALS; 10, 9311; and 11, WY-4.
DISCUSSION
Natural variation in PTs is a largely unexploited resource that can be used to identify new targets to breed or engineer higher photosynthetic efficiency (Flood et al., 2011; Driever et al., 2014). Comparing the relatively long-term perspective of engineering photosynthesis for greater yield (Long et al., 2015), mining natural variations of photosynthesis using natural populations can lead to reasonably short-term (less than 5 years) crop improvements (Parry et al., 2011). In this study, we explored natural variations in photosynthetic parameters in rice that might be related to biomass accumulation, a surrogate of canopy photosynthesis. Using LRMs constructed under different environments, we identified Alow as a major photosynthetic parameter with high correlation with biomass accumulation under two drastically different environments. Here, we briefly discuss the major findings of this study and their implications for rice breeding.
Natural Variations and Heritability of All Photosynthetic Parameters
Since the 1960s, researchers started working on improving photosynthesis through introgression (e.g. in soybean [Glycine max]; Ojima, 1974). However, the progress was rather limited because, on the one hand, it remained unclear what PTs should be the targets, and on the other hand, there were no effective molecular marks related to PTs defined well enough to be used in breeding programs (Flood et al., 2011). The aim of this study was to identify highly heritable PTs relevant to biomass production under different environments. Since screening PTs is labor intensive and time consuming, instead of using the global rice core collection of 1,794 accessions, we used a minicore diversity panel consisting of 204 global rice accessions, which is sufficiently diverse to effectively represent the original core collection (Agrama et al., 2009) and also is manageable, especially for detailed photosynthesis phenotyping.
As expected, our data suggest that there are substantial variations among photosynthetic parameters under BJ/SH environmental conditions (Table I), suggesting that there is genetic diversity in PTs in rice that can be potentially exploited. Furthermore, our heritability analysis shows that h2SNP values in many PTs, including SPAD, Ls, and WUE, under BJ/SH environmental conditions were around 0.6 to 0.7, which are close to some earlier reports (Schuster et al., 1992; Geber and Dawson, 1997; McKown et al., 2014; Table I), suggesting that these parameters are under strong genetic control in different species. It is worth emphasizing that, in this study, our estimate of h2SNP utilizes not only causal genes, as in the traditional variance method (for review, see Zaitlen and Kraft, 2012), but also considers other SNP markers (Yang et al., 2011). The observed high levels of heterogeneity and relatively high h2SNP for many PTs suggest that these traits can be used as potential candidates in marker-assisted breeding for rice (Ackerly et al., 2000).
Alow Is a Photosynthetic Trait That Is Highly Correlated with Biomass Accumulation under Different Environments
In this study, a stepwise feature selection approach was applied to the data collected under either the BJ or SH environment. With this method, we identified two PTs (i.e. gs and Alow) in both the BJ and combined data sets (Fig. 5). Both gs and Alow exhibited high correlation with biomass (Fig. 2; Supplemental Figs. S2 and S3). The values of both parameters show strong correlation between BJ and SH environments (Fig. 3). gs showed a high h2SNP and substantial natural variation among rice cultivars under BJ/SH environmental conditions (Tables I and II), suggesting that gs is a good parameter to be used in rice breeding. In fact, gs screening based on thermal imaging (Takai et al., 2010b) has already been used in some breeding programs (e.g. the wheat yield potential breeding in the physiology breeding program of International Maize and Wheat Improvement Center; Rajaram et al., 1994). It is worth noting that, in addition to gs itself being a potentially important parameter for breeding, the faster response of gs to fluctuating light can be an adaptive trait for rice under severe drought conditions (Qu et al., 2016).
Remarkably, Alow instead of A under normal light was identified to be a photosynthetic parameter highly correlated with biomass accumulation in the BJ, SH, and combined data sets (Fig. 5; Supplemental Table S3). Alow also is under strong genetic control, as shown by its high h2SNP (Table I). Therefore Alow is a promising target for future rice breeding improvements. This finding is remarkable since, although it has long been recognized that canopy, instead of leaf, photosynthesis is a major determinant of biomass accumulation, so far, direct experimental evidence supporting the importance of photosynthetic efficiency under low light is lacking. The strong correlation between Alow and biomass accumulation reported here strongly supports the notation that photosynthetic CO2 uptake of the lower layer leaves, which usually experience low light levels, contributes substantially to the overall canopy photosynthesis and, hence, biomass production. This finding increases the repertoire of parameters known so far that can potentially improve canopy photosynthesis, which includes faster speed of recovery from photoprotective status (Zhu et al., 2004), rapid recovery of stomata conductance under fluctuating light (Lawson and Blatt, 2014; Qu et al., 2016), and Rubisco with optimized kinetic properties (Zhu et al., 2004). Large-scale genetic screening of these different parameters and gene identification in this global minicore are under way in our laboratory.
Potential Value of the Identified Traits in Current Rice Breeding
The natural distribution of Alow across the minicore panel yielded a normal distribution (Fig. 6B); furthermore, there is substantial variation of Alow in the modern elite rice cultivars (Table III; Fig. 6), suggesting large space to improve Alow to enhance biomass production in contemporary elite rice cultivars. By comparing the value of Alow in the modern elite cultivars with the extreme values observed in the minicore diversity panel under the BJ environment, we identified candidate donors that can be used as genetic resources for Alow. For example, 76.77% and 85.49% improvement of Alow can be achieved in DHX-Z and ZH11 (elite cultivar), respectively (Table III), if the causal genes (or quantitative trait loci) controlling Alow in P4140 can be transferred into these two cultivars.
Table III. Comparison of Alow between elite cultivars and the minicore accessions showing highest Alow.
Percentage difference was expressed as (Alow of extreme accession − Alow of elite cultivar)/(Alow of extreme accession) × 100%. P4140 is an accession in the minicore population that showed the highest values in Alow in the BJ environment.
| Elite Accessions | Mean ± sd | Percentage Difference |
|---|---|---|
| WCC1 | 5.39 ± 0.45 | 22.88 |
| WCC2 | 5.82 ± 0.83 | 13.65 |
| DHX-Z | 3.74 ± 1.03 | 76.77 |
| HE19 | 5.93 ± 0.29 | 11.68 |
| KY131 | 5.66 ± 0.88 | 17.02 |
| XS134 | 4.87 ± 0.51 | 35.91 |
| ZH11 | 3.57 ± 0.60 | 85.49 |
| MH63 | 4.66 ± 0.15 | 41.97 |
| KALS | 4.98 ± 0.56 | 32.97 |
| 9311 | 5.12 ± 0.51 | 29.21 |
| WY-4 | 4.57 ± 0.72 | 44.83 |
| Target accession | P4140 | |
CONCLUSION
By mining natural variations of photosynthesis-related traits in a natural rice diversity panel, we found that, among many photosynthetic parameters, Alow is highly correlated with biomass accumulation under different environments. Furthermore, Alow shows a high level of variability among contemporary elite rice lines, and it has high inheritability. All these findings suggest that Alow is a promising target for future rice breeding programs.
MATERIALS AND METHODS
Plant Material
The accessions from the U.S. Department of Agriculture collected minicore rice (Oryza sativa) diversity panel are from 76 countries covering 15 geographic regions, which consists of six groups: indica (35.4%), aus (18.7%), tropical japonica (18.2%), temperate japonica (15.2%), aromatic (3%), and their admixtures (9.6%; Agrama et al., 2010; Li et al., 2010). The population accounts for 12.1% of the global rice core accessions and displayed 100% coverage in genetic variation (Agrama et al., 2010). In this study, we used 204 out of 217 accessions in the minicore population, since the remaining 13 accessions have extremely long growing seasons. In addition, we used 11 Chinese elite rice cultivars (WCC1, WCC2, DHX-Z, HE19, KY131, XS134, ZH11, MH63, KALS, 9311, and WY-4; Hamdani et al., 2015).
Measurements of Leaf Gas Exchange
The 204 minicore panel and 11 elite rice lines were transplanted under two environments, BJ (116.3943°E, 39.9820°N) in May 2013 and SH (121.4530°E, 31.0428°N) in May 2015. Average atmosphere temperature under BJ/SH environmental conditions, during the growth periods from transplanting to booting stage until large-scale measurements started, which spanned around 60 d, were around 24.8°C ± 3.1°C and 25.5°C ± 4.4°C, respectively. Experiments were conducted in pots, and detailed experimental procedures were described by Hamdani et al. (2015). Briefly, plants were sown in 12-L pots filled with commercial peat soil (Pindstrup Substrate no. 4; Pindstrup Horticulture). For each accession, six plants were planted in two pots with three plants per pot. Two pots for the same accession were arranged close together to ensure the formation of a canopy. Pots from different accessions were separated to avoid shading from a different accession. During the growth period, plants were exposed to natural sunlight and were irrigated daily. Fertilizers were applied twice per month. Experiments of leaf gas exchange were conducted at 60 d after emergence (DAE). For each accession, we used four replicates during the measurements of gas exchange-related parameters. All the photosynthesis measurements were finished within 10 d. To minimize the potential errors introduced by potential growth stage differences, we measured photosynthetic parameters from accession 1 through 215 sequentially for the first and third replicates and then from accession 215 through 1 sequentially for the second and fourth replicates.
Plants were acclimated in a controlled room with a temperature around 27°C and a photosynthetic photon flux density (PPFD) around 600 μmol m−2 s−1 for at least 60 min before gas-exchange measurements. During the measurements, two levels of PPFD, 1,200 μmol mol−1 s−1 (normal light) and 100 μmol mol−1 s−1 (low light), were used. Four portable infrared gas-exchange systems (Li-6400XT; LI-COR) were used simultaneously. An automatic program was applied to measure gas-exchange traits under two light levels. Traits under normal light include A, gs, Ci, WUE, and Ls; traits under low light include Alow, Gslow, Cilow, Wlow, and Lslow. The process of the program was as follows: a leaf was first maintained under a PPFD of 1,200 μmol m−2 s−1 for at least 5 min or until gs reached a steady state, then PPFD was changed to 100 μmol m−2 s−1 for 25 min, allowing gs to approach steady state, as described by Qu et al. (2016). During the measurements, the leaf temperature was maintained at 25°C and relative humidity was maintained at ∼75%, the reference CO2 concentration was set as 400 μmol mol−1, and we used the top fully expanded leaves for this measurement. Data were recorded automatically, and average values within the last 1 min before light switch were used for data analysis.
Measurements of Dark Respiration and Maximal Quantum Yield
Experiments for dark respiration were conducted at 60 DAE. Respiration rates were determined as net rates of CO2 efflux in darkness during the night after 8 pm according to Bunce (2007). Leaf temperatures were set to 25°C, reference CO2 concentration was set to 400 μmol mol−1, and light level was set to 0 μmol mol−1 s−1.
A Multi-Function Plant Efficiency Analyzer chlorophyll fluorometer (Hansatech) was used to measure Fv/Fm, following Hamdani et al. (2015). Fm represents the maximum chlorophyll fluorescence, Fo is the minimum chlorophyll fluorescence, and Fv = Fm − Fo (Oxborough and Baker, 1997; Huang et al., 2016; Essemine et al., 2017).
Measurements of SPAD and Leaf Thickness
Experiments for SPAD and leaf thickness were conducted at 60 DAE. To estimate leaf total chlorophyll content and leaf thickness, a SPAD 502 Plus Chlorophyll Meter (Spectrum Technologies; Takai et al., 2010a) and a Micrometer Screw (Mitutoyo) were used, respectively. For each leaf, the chlorophyll content was estimated as the mean of five chlorophyll content measurements at different positions in the middle section of the leaf. Four replicates from four different plants were determined for both leaf chlorophyll concentration and leaf thickness.
Measurements of Plant Morphological Traits
Aboveground biomass accumulation, plant height, and tiller number were determined at 60 DAE according to Qu et al. (2016). At least four replicates were measured for each parameter. Samples for biomass determinations were kept at 120°C for 1 h and then under 70°C for at least 24 h in a baking oven until constant weight was reached before the weights of biomass or leaf segments were measured.
Regression Model between Biomass and Morphological and Photosynthetic Traits
We used an LRM to capture the correlation of biomass with PTs and MTs. The model is defined as follows:
where y is a vector representing the biomass values of each rice accession, x is a vector of independent variables, β is a weighted coefficient corresponding to x, and ε is an error vector. The model was constructed with a stepwise manner, which can identify highly relevant parameters and remove low-relevance parameters based on the Akaike information criterion according to Jin et al. (2014). In practice, a training data set including 90% of items of the whole data set was randomly extracted from the original data set, and the remaining 10% of data was used as a test data set (Kawamura et al., 2010; Iwasaki et al., 2013). The training data set was first defined to build the regression model, and then an independent validation was conducted on the test data set to check the performance of the model.
Estimation of h2SNP
GCTA software (version 1.25.2; Yang et al., 2011) was employed to estimate the h2SNP of 23 functional traits using 2.3 million filtered SNPs of the minicore population (Wang et al., 2016). GCTA implements the method in two steps: generating a high-dimensional genetic relatedness matrix between individuals and then estimating the variance explained by all SNPs by a restricted maximum likelihood analysis of the phenotypes with the genetic relatedness matrix (Yang et al., 2011). The significance of h2SNP is assessed by a likelihood ratio test, which is the ratio of likelihood under the alternative hypothesis (H1, h2SNP ≠ 0) to that under the null hypothesis (H0, h2SNP = 0). The likelihood ratio test and its corresponding P value were reported in the GCTA output file.
Data Analysis
In order to quantitatively evaluate the genetic variation of biological traits in the combined population, PGV was calculated as (Xmax − Xmin)/X × 100 (%), where Xmax, Xmin, and X stand for maximum, minimum, and mean values in the population, respectively (Gu et al., 2014). The Pearson correlation coefficient was calculated using the R package (Corrplot; version 3.2.1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Trait distribution of elite cultivars and the minicore accessions under the SH environment.
Supplemental Figure S2. Correlation of photosynthetic traits with biomass under the BJ environment.
Supplemental Figure S3. Correlation of photosynthetic traits with biomass under the SH environment.
Supplemental Table S1. Correlation of photosynthetic traits with morphological traits in the global minicore panel and elite rice cultivars across the experiments of BJ and SH sites.
Supplemental Table S2. Correlation of photosynthetic traits with morphological traits under BJ and SH experiments.
Supplemental Table S3. Feature selection across BJ, SH, and combined sites.
Acknowledgments
We thank the anonymous reviewers for constructive comments, which helped us improve our revision.
Glossary
- PT
photosynthetic trait
- MTs
morphological traits
- BJ
Beijing
- SH
Shanghai
- PGV
percentage genetic variation
- WUE
water use efficiency
- SNP
single-nucleotide polymorphism
- LRM
linear regression model
- DAE
days after emergence
- PPFD
photosynthetic photon flux density
Footnotes
This work was supported by the CAS Strategic Research Project (grant no. XDA08020301), the Shanghai Municipal Natural Science Foundation (grant nos. 17YF1421800 and 14ZR1446700), and the Bill & Melinda Gates Foundation (grant no. OPP1014417).
Articles can be viewed without a subscription.
References
- Ackerly DD, Dudley SA, Sultan SE, Schmitt H, Coleman JS, Linder CR, Sandquist DR, Geber MA, Evans AS, Dawson TE, et al. (2000) The evolution of plant ecophysiological traits: recent advances and future directions. Bioscience 50: 979–995 [Google Scholar]
- Agrama HA, Yan W, Lee F, Robert F, Chen MH, Jia M, McClung A (2009) Genetic assessment of a mini-core subset developed from the USDA rice genebank. Crop Sci 49: 1336–1346 [Google Scholar]
- Agrama HA, Yan WG, Jia M, Fjellstrom R, McClung A (2010) Genetic structure associated with diversity and geographic distribution in the USDA rice world collection. Nat Sci 2: 247–291 [Google Scholar]
- Bunce JA. (2007) Direct and acclimatory responses of dark respiration and translocation to temperature. Ann Bot (Lond) 100: 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Silva E, Andralojc PJ, Scales JC, Driever SM, Mead A, Lawson T, Raines CA, Parry MAJ (2017) Phenotyping of field-grown wheat in the UK highlights contribution of light response of photosynthesis and flag leaf longevity to grain yield. J Exp Bot 61: 235–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TG, Xin CP, Qu MN, Zhu XG (2017) Evaluation of protocols for measuring leaf photosynthetic properties of field-grown rice. Rice Sci 24: 1–9 [Google Scholar]
- Driever SM, Lawson T, Andralojc PJ, Raines CA, Parry M A J (2014) Natural variation in photosynthetic capacity, growth, and yield in 64 field-grown wheat genotypes. J Exp Bot 65: 4959–4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essemine J, Xiao Y, Qu M, Mi H, Zhu XG (2017) Cyclic electron flow may provide some protection against PSII photoinhibition in rice (Oryza sativa L.) leaves under heat stress. J Plant Physiol 211: 138–146 [DOI] [PubMed] [Google Scholar]
- Flood PJ, Harbinson J, Aarts MGM (2011) Natural genetic variation in plant photosynthesis. Trends Plant Sci 16: 327–335 [DOI] [PubMed] [Google Scholar]
- Geber MA, Dawson TE (1997) Genetic variation in stomatal and biochemical limitations to photosynthesis in the annual plant, Polygonum arenastrum. Oecologia 109: 535–546 [DOI] [PubMed] [Google Scholar]
- Gepts P. (2002) A comparison between crop domestication, classical plant breeding, and genetic engineering. Crop Sci 42: 1780–1790 [Google Scholar]
- Gu J, Yin X, Stomph TJ, Struik PC (2014) Can exploiting natural genetic variation in leaf photosynthesis contribute to increasing rice productivity? A simulation analysis. Plant Cell Environ 37: 22–34 [DOI] [PubMed] [Google Scholar]
- Hamdani S, Qu M, Xin CP, Li M, Chu C, Govindjee, Zhu XG (2015) Variations between the photosynthetic properties of elite and landrace Chinese rice cultivars revealed by simultaneous measurements of 820 nm transmission signal and chlorophyll a fluorescence induction. J Plant Physiol 177: 128–138 [DOI] [PubMed] [Google Scholar]
- Hedden P. (2003) Constructing dwarf rice. Nat Biotechnol 21: 873–874 [DOI] [PubMed] [Google Scholar]
- Huang W, Yang YJ, Hu H, Cao KF, Zhang SB (2016) Sustained diurnal stimulation of cyclic electron flow in two tropical tree species Erythrophleum guineense and Khaya ivorensis. Front Plant Sci 7: 1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbart S, Peng S, Horton P, Chen Y, Murchie EH (2007) Trends in leaf photosynthesis in historical rice varieties developed in the Philippines since 1966. J Exp Bot 58: 3429–3438 [DOI] [PubMed] [Google Scholar]
- Iwasaki T, Takeda Y, Maruyama K, Yokosaki Y, Tsujino K, Tetsumoto S, Kuhara H, Nakanishi K, Otani Y, Jin Y, et al. (2013) Deletion of tetraspanin CD9 diminishes lymphangiogenesis in vivo and in vitro. J Biol Chem 288: 2118–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn CE, Mckay JK, Mauleon R, Stephens J, McNally KL, Bush DR, Leung H, Leach JE (2011) Genetic variation in biomass traits among 20 diverse rice varieties. Plant Physiol 155: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Qiao F, Chen L, Lu C, Xu L, Gao X (2014) Serum metabolomic signatures of lymph node metastasis of esophageal squamous cell carcinoma. J Proteome Res 13: 4091–4103 [DOI] [PubMed] [Google Scholar]
- Kawamura K, Watanabe N, Sakanoue S, Lee HJ, Inoue Y, Odagawa S (2010) Testing genetic algorithm as a tool to select relevant wavebands from field hyperspectral data for estimating pasture mass and quality in a mixed sown pasture using partial least squares regression. Grassl Sci 56: 205–216 [Google Scholar]
- Kromdijk J, Głowacka K, Leonelli L, Gabilly ST, Iwai M, Niyogi KK, Long SP (2016) Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354: 857–861 [DOI] [PubMed] [Google Scholar]
- Lawson T, Blatt MR (2014) Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol 164: 1556–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T, Kramer DM, Raines CA (2012) Improving yield by exploiting mechanisms underlying natural variation of photosynthesis. Curr Opin Biotechnol 23: 215–220 [DOI] [PubMed] [Google Scholar]
- Li X, Yan W, Agrama H, Hu B, Jia L, Jia M, Jackson A, Moldenhauer K, McClung A, Wu D (2010) Genotypic and phenotypic characterization of genetic differentiation and diversity in the USDA rice mini-core collection. Genetica 138: 1221–1230 [DOI] [PubMed] [Google Scholar]
- Long SP, Marshall-Colon A, Zhu XG (2015) Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161: 56–66 [DOI] [PubMed] [Google Scholar]
- Long SP, Zhu XG, Naidu SL, Ort DR (2006) Can improvement in photosynthesis increase crop yields? Plant Cell Environ 29: 315–330 [DOI] [PubMed] [Google Scholar]
- McKown AD, Guy RD, Quamme L, Klápště J, La Mantia J, Constabel CP, El-Kassaby YA, Hamelin RC, Zifkin M, Azam MS (2014) Association genetics, geography and ecophysiology link stomatal patterning in Populus trichocarpa with carbon gain and disease resistance trade-offs. Mol Ecol 23: 5771–5790 [DOI] [PubMed] [Google Scholar]
- Ojima M. (1974) Improvement of photosynthetic capacity in soybean variety. Jpn Agric Res Q 8: 6–12 [Google Scholar]
- Oxborough K, Baker NR (1997) Resolving chlorophyll a fluorescence images of photosynthetic efficiency into photochemical and non-photochemical components: calculation of qP and Fv’/Fm’ without measuring Fo’. Photosynth Res 54: 135–142 [Google Scholar]
- Parry MAJ, Reynolds M, Salvucci ME, Raines C, Andralojc PJ, Zhu XG, Price GD, Condon AG, Furbank RT (2011) Raising yield potential of wheat. II. Increasing photosynthetic capacity and efficiency. J Exp Bot 62: 453–467 [DOI] [PubMed] [Google Scholar]
- Peng S, Khush GS, Virk P, Tang Q, Zou Y (2008) Progress in ideotype breeding to increase rice yield. Field Crops Res 108: 32–38 [Google Scholar]
- Peng S, Laza RC, Visperas RM, Sanico AL, Cassman KGKG (2001) Grain yield of rice cultivars and lines developed in the Philippines since 1966. Crop Sci 40: 307–314 [Google Scholar]
- Qu M, Hamdani S, Li W, Wang S, Tang J, Chen Z, Song Q, Li M, Zhao H, Chang T, et al. (2016) Rapid stomatal response to fluctuating light: an under-explored mechanism to improve drought tolerance in rice. Funct Plant Biol 43: 727–738 [DOI] [PubMed] [Google Scholar]
- Rajaram S, van Ginkel M, Fischer RA (1994) CIMMYT’s wheat breeding mega-environments (ME). In Li ZS, Xin ZY, eds, Proceedings of the 8th International Wheat Genetic Symposium, July 19–24, Beijing, China. China Agric Scientech Press, Beijing, pp 1101–1106 [Google Scholar]
- Rosenthal DM, Locke AM, Khozaei M, Raines CA, Long SP, Ort DR (2011) Over-expressing the C3 photosynthesis cycle enzyme Sedoheptulose-1-7 Bisphosphatase improves photosynthetic carbon gain and yield under fully open air CO2 fumigation (FACE). BMC Plant Biol 11: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster WSF, Phillips SL, Sandquist DR, Ehleringer JR (1992) Heritability of carbon isotope discrimination in Gutierrezia-microcephala (Asteraceae). Am J Bot 79: 216–221 [Google Scholar]
- Simkin AJ, McAusland L, Headland LR, Lawson T, Raines CA (2015) Multigene manipulation of photosynthetic carbon assimilation increases CO2 fixation and biomass yield in tobacco. J Exp Bot 66: 4075–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Zhang G, Zhu XG (2013) Optimal crop canopy architecture to maximise canopy photosynthetic CO2 uptake under elevated CO2: a theoretical study using a mechanistic model of canopy photosynthesis. Funct Plant Biol 40: 109–124 [DOI] [PubMed] [Google Scholar]
- Takai T, Kondo M, Yano M, Yamamoto T (2010a) A quantitative trait locus for chlorophyll content and its association with leaf photosynthesis in rice. Rice (N Y) 3: 172–180 [Google Scholar]
- Takai T, Yano M, Yamamoto T (2010b) Canopy temperature on clear and cloudy days can be used to estimate varietal differences in stomatal conductance in rice. Crop Res 115: 165–170 [Google Scholar]
- Wang H, Xu X, Vieira FG, Xiao Y, Li Z, Wang J, Nielsen R, Chu C (2016) The power of inbreeding: NGS-based GWAS of rice reveals convergent evolution during rice domestication. Mol Plant 9: 975–985 [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88: 76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N, Kraft P (2012) Heritability in the genome-wide association era. Hum Genet 131: 1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR (2008) What is the maximum efficiency with which photosynthesis can convert solar energy into biomass? Curr Opin Biotechnol 19: 153–159 [DOI] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR (2010) Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol 61: 235–261 [DOI] [PubMed] [Google Scholar]
- Zhu XG, Ort DR, Whitmarsh J, Long SP (2004) The slow reversibility of photosystem II thermal energy dissipation on transfer from high to low light may cause large losses in carbon gain by crop canopies: a theoretical analysis. J Exp Bot 55: 1167–1175 [DOI] [PubMed] [Google Scholar]
- Zhu XG, Song Q, Ort DR (2012) Elements of a dynamic systems model of canopy photosynthesis. Curr Opin Plant Biol 15: 237–244 [DOI] [PubMed] [Google Scholar]