The 2-oxoglutarate-dependent dioxygenase 16DOX catalyzes steroid 16α-hydroxylation in the steroidal glycoalkaloid (SGA) pathway and is a suitable target for controlling toxic SGA levels in potato.
Abstract
Steroidal glycoalkaloids (SGAs) are toxic specialized metabolites that are found in the Solanaceae. Potato (Solanum tuberosum) contains the SGAs α-solanine and α-chaconine, while tomato (Solanum lycopersicum) contains α-tomatine, all of which are biosynthesized from cholesterol. However, although two cytochrome P450 monooxygenases that catalyze the 22- and 26-hydroxylation of cholesterol have been identified, the 16-hydroxylase remains unknown. Feeding with deuterium-labeled cholesterol indicated that the 16α- and 16β-hydrogen atoms of cholesterol were eliminated to form α-solanine and α-chaconine in potato, while only the 16α-hydrogen atom was eliminated in α-tomatine biosynthesis, suggesting that a single oxidation at C-16 takes place during tomato SGA biosynthesis while a two-step oxidation occurs in potato. Here, we show that a 2-oxoglutarate-dependent dioxygenase, designated as 16DOX, is involved in SGA biosynthesis. We found that the transcript of potato 16DOX (St16DOX) was expressed at high levels in the tuber sprouts, where large amounts of SGAs are accumulated. Biochemical analysis of the recombinant St16DOX protein revealed that St16DOX catalyzes the 16α-hydroxylation of hydroxycholesterols and that (22S)-22,26-dihydroxycholesterol was the best substrate among the nine compounds tested. St16DOX-silenced potato plants contained significantly lower levels of SGAs, and a detailed metabolite analysis revealed that they accumulated the glycosides of (22S)-22,26-dihydroxycholesterol. Analysis of the tomato 16DOX (Sl16DOX) gene gave essentially the same results. These findings clearly indicate that 16DOX is a steroid 16α-hydroxylase that functions in the SGA biosynthetic pathway. Furthermore, St16DOX silencing did not affect potato tuber yield, indicating that 16DOX may be a suitable target for controlling toxic SGA levels in potato.
Steroidal glycoalkaloids (SGAs) are typically found in plants in the family Solanaceae (Harrison, 1990; Petersen et al., 1993; Helmut, 1998). In particular, potato (Solanum tuberosum) is known to contain the SGAs α-solanine and α-chaconine, while tomato (Solanum lycopersicum) contains α-tomatine, which are toxic to fungi, bacteria, insects, animals, and humans (Friedman, 2002, 2006). The mechanisms of toxicity include the disruption of membranes and the inhibition of acetylcholine esterase activity (Roddick, 1989). α-Solanine and α-chaconine occur in most tissues of potato plants, but they are present at particularly high levels in floral and tuber sprout tissues (Kozukue and Mizuno, 1985, 1989; Ginzberg et al., 2009). Therefore, since these potato SGAs are toxic and cause a bitter taste, controlling their content in potato tubers is an important focus of potato breeding (Friedman, 2006). Similarly, α-tomatine exists in all tomato plant tissues, but it is richly accumulated in the leaves and immature fruit, with its content decreasing during fruit ripening (Friedman, 2002).
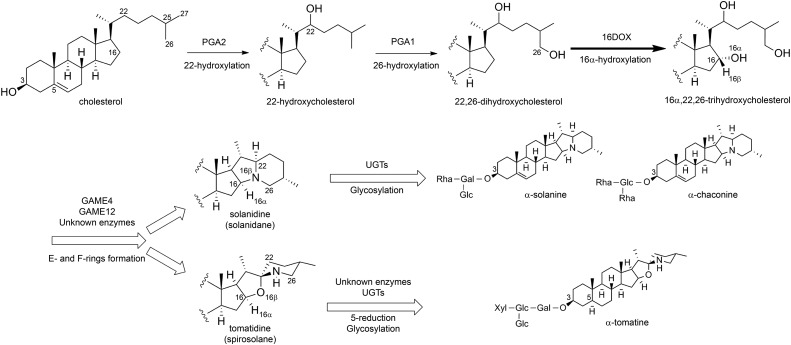
SGAs are composed of C27 steroids with an oligosaccharide at the hydroxy group at the C-3 position (Friedman, 2002, 2006). In α-solanine and α-chaconine in potato, the oligosaccharides solatriose and chacotriose are attached to the C-3 hydroxy group of solanidine, which is a solanidane-type aglycone; similarly, in α-tomatine in tomato, lycotetraose is linked to the C-3 hydroxy group of tomatidine, which is spirosolane-type aglycone. These SGAs are biosynthesized from cholesterol (Sawai et al., 2014), which is modified subsequently via oxidation at the C-16, C-22, and C-26 positions, amination at the C-26 position, and glycosylation at the C-3 hydroxy group (Petersen et al., 1993; Friedman, 2002; Ginzberg et al., 2009; Ohyama et al., 2013). Cytochrome P450 monooxygenases (CYPs) are likely involved in the oxidative modification of cholesterol, while a transaminase catalyzes the amination at C-26, and UDP-dependent glycosyltransferases (UGTs) also function in the glycosylation at the C-3 hydroxy group (Fig. 1). Some UGTs have now been identified as the enzymes that are involved in the glycosylation steps for SGA biosynthesis (Moehs et al., 1997; McCue et al., 2005, 2006, 2007; Itkin et al., 2011). Itkin et al. (2013) reported that several genes that are involved in the SGA biosynthetic pathways exist as gene clusters on chromosomes 7 and 12 in the genomes of tomato and potato. In addition, sterol side chain reductase2 (SSR2) has been identified as a gene that is committed to cholesterol biosynthesis and involved in SGA production (Sawai et al., 2014). However, the genes that are involved in the later steps of the SGA biosynthetic pathways remain to be completely elucidated.
Figure 1.
Putative biosynthetic pathway for SGAs in potato and tomato. The thick solid arrow indicates the reaction step characterized in this work. Thin solid arrows indicate the reaction steps reported by Umemoto et al. (2016). White arrows represent multiple reaction stages.
CYPs are heme-thiolate membrane proteins that are generally bound to the cytoplasmic surface of the endoplasmic reticulum and are known to be involved in the oxidation steps during the metabolism of various plant natural products. A number of CYPs have been reported to catalyze the oxidation of triterpene skeletons, such as oleanane, lupine, ursane, and dammarane types (Moses et al., 2014; Seki et al., 2015). In addition, several CYP genes have been identified as being responsible for the oxidation of the steroid backbone in the biosynthesis and catabolism of brassinosteroids, which are plant steroid hormones (Ohnishi et al., 2009). Therefore, we previously investigated SGA biosynthesis in potato and tomato by selecting several CYP genes that show high expression in SGA-rich organs as candidate oxidases of cholesterol using the public EST databases of potato and tomato. This showed that two CYPs (PGA1/CYP72A208 and PGA2/CYP72A188) are involved in the hydroxylation of cholesterol at the C-26 and C-22 positions (Umemoto et al., 2016). In addition, another CYP (PGA3/GAME4/CYP88B1) also was identified as being involved in SGA biosynthesis, but its catalytic function remains unknown (Itkin et al., 2013; Umemoto and Sasaki, 2013). We were unable to find a C-16 hydroxylase among the candidate CYPs, however.
The 2-oxoglutarate-dependent dioxygenase (2OGD) superfamily is the second largest enzyme family, following the CYP superfamily, in the plant genome (Kawai et al., 2014). 2OGDs are nonheme iron-containing proteins that localize in the cytosol as soluble proteins. Many 2OGDs play crucial roles in the oxidation steps in the biosynthesis of diverse specialized metabolites such as flavonoids and phytohormones (Kawai et al., 2014). However, unlike CYPs, no 2OGDs have been identified as being involved in the biosynthesis of triterpenoids and steroids to date. Here, we characterized the 2OGD gene named 16DOX and found that it is involved in SGA biosynthesis. The 16DOX gene was coexpressed with the previously identified SGA biosynthetic genes in potato and tomato, and the 16DOX protein was found to catalyze the hydroxylation of cholesterol at the C-16α position. Furthermore, 16DOX silencing in transgenic potato plants led to significantly reduced endogenous SGA levels and the suppression of potato tuber sprouting, which are similar to the phenotypes observed in PGA1- and PGA2-silenced plants (Umemoto et al., 2016). Thus, to our knowledge, 16DOX is the first 2OGD to be identified as functioning as a steroid 16α-hydroxylase involved in SGA biosynthesis.
RESULTS
SGA Analysis of Potato Shoots and Tomato Seedlings Fed with Stable Isotope-Labeled Compounds
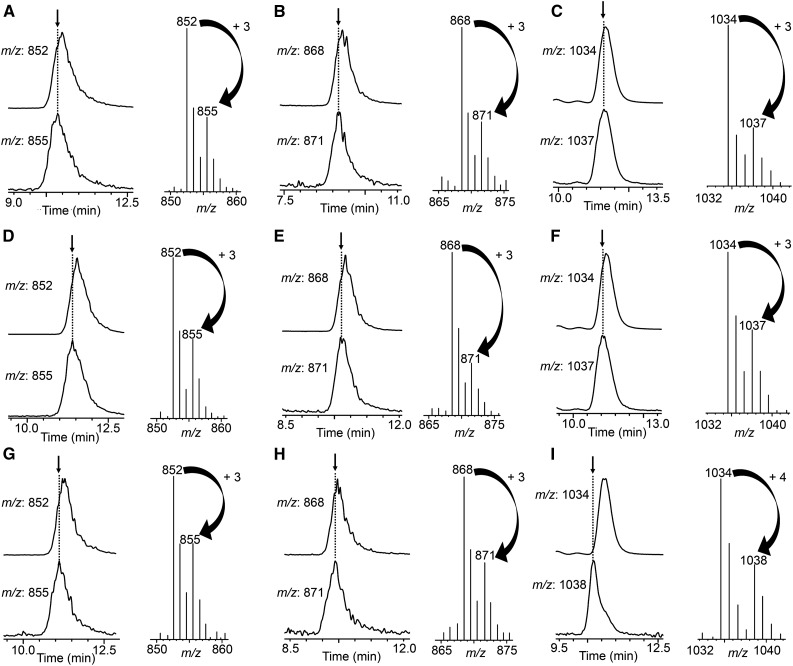
To examine the fates of hydrogen atoms at the C-16 position during SGA biosynthesis in potato shoots and tomato seedlings, we conducted feeding experiments using three kinds of stable isotope-labeled compounds: [15,15,16α,17α-2H4]cholesterol, [15,15,16β,17α-2H4]cholesterol, and [15,15,17α-2H3]cholesterol. The accumulated SGAs in the harvested shoots and seedlings were extracted and analyzed by liquid chromatography-mass spectrometry (LC-MS; Fig. 2). In the case of potato shoots, labeled α-solanine and α-chaconine were detected by monitoring the mass-to-charge ratio (m/z) 871 and 855 ions [M+H]+, respectively (Fig. 2, A–F), while nonlabeled α-solanine and α-chaconine were detected by monitoring the m/z 868 and 852 ions [M+H]+, respectively, in shoots that were fed with the three labeled cholesterols. These results indicate that the 15- and 17-hydrogens of cholesterol were retained but both hydrogens at the 16α and 16β positions were eliminated in the process of potato SGA biosynthesis. In the case of tomato seedlings, labeled α-tomatine was detected by monitoring the m/z 1,037 ion [M+H]+ (Fig. 2, G and H), while nonlabeled α-tomatine was detected by monitoring the m/z 1,034 ion [M+H]+ in seedlings that were fed with [15,15,16α,17α-2H4]cholesterol and [15,15,17α-2H3]cholesterol. By contrast, when [15,15,16β,17α-2H4]cholesterol was administered, labeled α-tomatine was detected by monitoring the m/z 1,038 ion [M+H]+ (Fig. 2I), which is one mass higher than the results obtained with [15,15,16β,17α-2H4]cholesterol and [15,15,17α-2H3]cholesterol. These results indicate that 16β-hydrogen was retained but 16α-hydrogen was eliminated in tomato SGA biosynthesis. These results are consistent with the previous observation reported by Canonica et al. (1977). Taken together, these findings suggest that oxidation (dehydrogenation) occurs at both the C-16α and C-16β positions in potato and at the C-16α position in tomato during SGA biosynthesis.
Figure 2.
LC-MS analysis of SGAs upon feeding stable isotope-labeled compounds to in vitro-grown potato shoots and tomato seedlings. The two traces on the left are mass chromatograms for the indicated m/z ions. The mass spectrum on the right was obtained at the retention time indicated by the arrow. Plants were fed with [15,15,17α-2H3]cholesterol (A–C), [15,15,16α, 17α-2H4]cholesterol (D–F), or [15,15,16β,17α-2H4]cholesterol (G–I). A, D, and G, LC-MS analysis of α-chaconine in potato shoots. B, E, and H, LC-MS analysis of α-solanine in potato shoots. C, F, and I, LC-MS analysis of α-tomatine in tomato seedlings.
Identification of the Candidate 16DOX Gene in Potato and Tomato
To identify the candidate genes that are involved in the oxidative modification of cholesterol for SGA biosynthesis, we surveyed several enzyme superfamilies that are included in the EST databases of potato from the DFCI Plant Gene Indices (http://compbio.dfci.harvard.edu/tgi/) and the tomato databases from MiBASE (http://www.pgb.kazusa.or.jp/mibase/) and Sol Genomics (http://solgenomics.net). We initially selected CYP genes based on the correlation between the read numbers of the EST contigs and the tissue-specific accumulation of SGAs in potato and tomato. This led to the identification of the CYPs PGA1 (Sotub06g021140, CYP72A208) and PGA2 (Sotub07g016580, CYP72A188) as a steroid 26-hydroxylase and a steroid 22-hydroxylase, respectively, for potato SGA biosynthesis (Umemoto et al., 2016). However, we were unable to identify a 16-hydroxylase among the candidate CYPs. The 2OGD superfamily also is associated with the oxygenation reactions of various specialized metabolites (Kawai et al., 2014). Therefore, we surveyed the unigenes encoding 2OGDs from the potato database as a second focus.
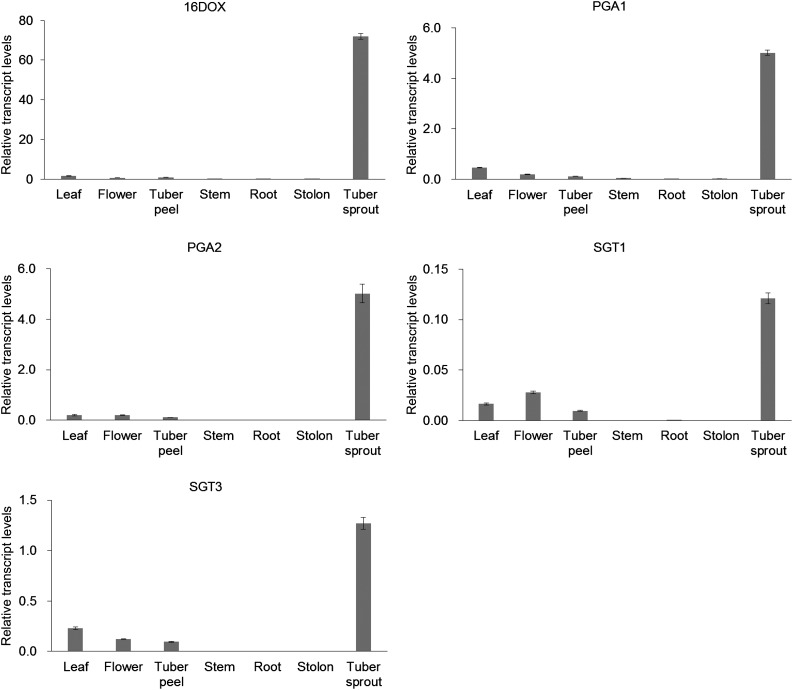
A total of 256 2OGD transcripts were extracted by BLASTX search of nucleotide sequences of all transcript sequences obtained from the Spud DB potato genomics resource (http://solanaceae.plantbiology.msu.edu/) against protein sequences of 130 2OGDs from Arabidopsis (Arabidopsis thaliana; Kawai et al., 2014). One of the 2OGD transcripts (PGSC0003DMT400030676) showed the highest fragments per kilobase of exon per million mapped fragments (FPKM) value in tuber sprout among the 256 2OGD transcripts in RNA-Seq Gene Expression Data obtained from Spud DB (Supplemental Table S1), and we selected the transcript as a candidate gene encoding a 16-hydroxylase, which was designated as St16DOX. Quantitative reverse transcription (RT)-PCR analysis showed that potato St16DOX was highly expressed in the tuber sprouts (Fig. 3), where large amounts of SGAs are accumulated (Friedman and Dao, 1992; Smith et al., 1996; Milner et al., 2011). BLAST searches against the genome databases of Solanum spp. (http://solgenomics.net) showed that St16DOX is identical to Sotub07g016570 in the Potato ITAG protein database and shows 93% amino acid identity to tomato Solyc07g043420 (designated as Sl16DOX) in the Tomato ITAG protein database. Tomato Sl16DOX was highly expressed in flowers, which accumulate large amounts of α-tomatine (Supplemental Fig. S1). These results suggest that 16DOX genes are a candidate for 16-hydroxylase in SGA biosynthesis (Fig. 3; Supplemental Fig. S1).
Figure 3.
Quantitative RT-PCR analysis of the expression patterns of SGA biosynthetic genes in various organs of potato plants. Transcript levels of SGA biosynthetic genes are shown relative to that of EF1α as an internal reference gene. Error bars indicate sd (n = 3).
In a recent phylogenetic classification of the plant 2OGD superfamily (Kawai et al., 2014), 16DOXs were found to belong to clade DOXC41, which includes hyoscyamine 6β-hydroxylase (H6H), which is involved in the biosynthesis of the tropane alkaloid scopolamine (Matsuda et al., 1991). The 16DOXs share around 44% amino acid sequence identity with H6H in Hyoscyamus niger and contain several sequence motifs that are highly conserved among 2OGDs (Supplemental Fig. S2). The St16DOX and Sl16DOX proteins contain the Fe(II)-binding motif His-X-Asp-Xn-His (His-217, Asp-219, and His-272 in 16DOX), which is conserved in the 2OGD superfamily (Lukačin and Britsch, 1997; Wilmouth et al., 2002; Bugg, 2003). The Arg-X-Ser motif (Arg-282 and Ser-284), which binds to the C-5 carboxy group of 2-oxoglutarate, also is conserved in the 16DOX proteins (Lukačin et al., 2000; Wilmouth et al., 2002).
In Vitro Functional Analysis of the Recombinant 16DOX Protein
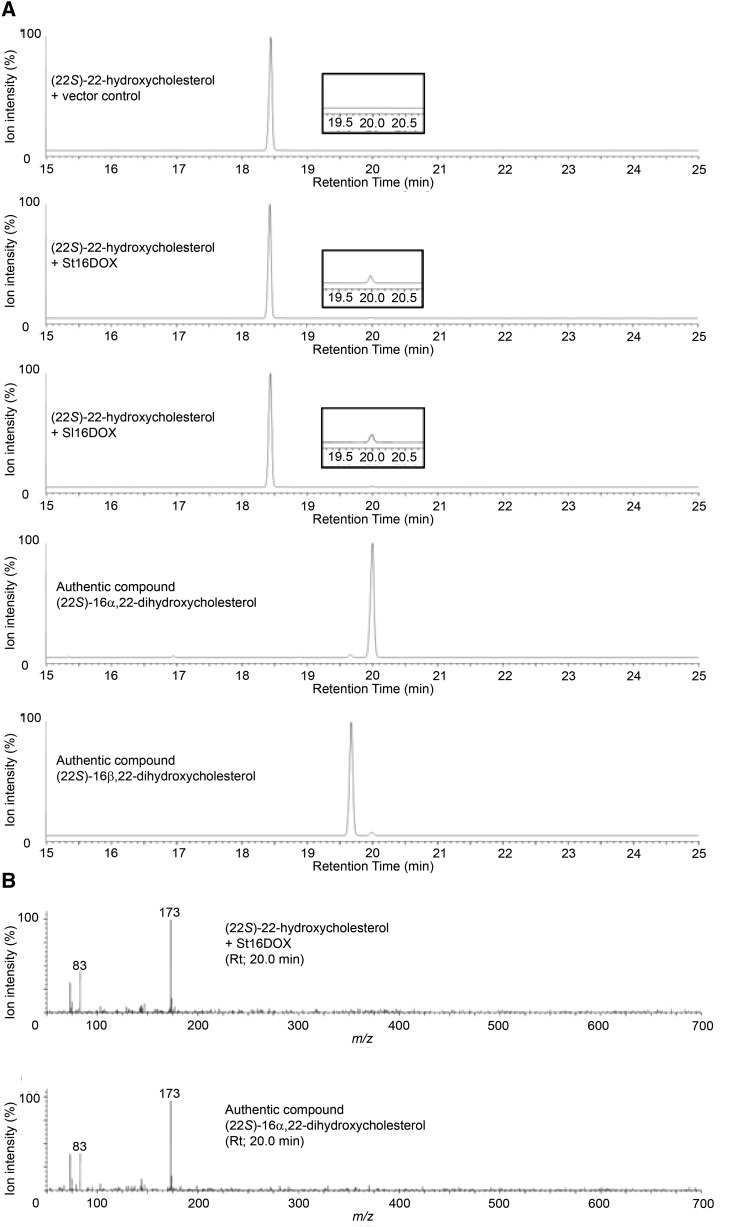
To investigate the catalytic functions of 16DOX, recombinant St16DOX protein was prepared with a bacterial expression system in Escherichia coli and an in vitro enzyme assay was performed with (22S)-22-hydroxycholesterol as a substrate. The reaction products were analyzed using gas chromatography-mass spectrometry (GC-MS). St16DOX metabolized (22S)-22-hydroxycholesterol to a product with a retention time of 20 min and a major mass fragment ion at m/z 173 (Fig. 4). This product was identical to the authentic compound (22S)-16α,22-dihydroxycholesterol in terms of both the retention time and the mass spectrum but different from the authentic compound (22S)-16β,22-dihydroxycholesterol, indicating that St16DOX catalyzes the 16-hydroxylation of (22S)-22-hydroxycholesterol specifically in the 16α configuration. The catalytic activity of tomato Sl16DOX was consistent with that of St16DOX (Fig. 4).
Figure 4.
GC-MS analysis of the reaction products from the recombinant St16DOX and Sl16DOX proteins with (22S)-22-hydroxycholesterol as a substrate. A, Extracted ion chromatograms targeting ion m/z 173 of the reaction products and the authentic compounds. B, Mass spectra of the peaks shown in A at a retention time of 20 min.
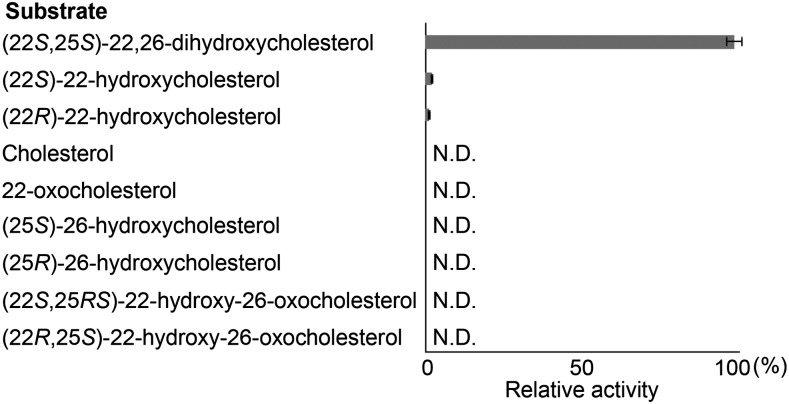
We next determined the substrate specificity of St16DOX toward cholesterol and several oxygenated cholesterols (Fig. 5). St16DOX showed the highest activity toward (22S,25S)-22,26-dihydroxycholesterol, reaching a rate that was approximately 50 times higher than that with (22S)-22-hydroxycholesterol (Fig. 5; Supplemental Fig. S3). LC-MS analysis showed a fragment ion at m/z 435 in the product, which corresponds with the deduced parent ion of 16,22,26-trihydroxycholesterol (Supplemental Fig. S4). St16DOX weakly metabolized (22R)-22-hydroxycholesterol to a new product (Supplemental Fig. S5), but the structure of the metabolite was unknown. By contrast, the assays with the other substrates did not give any product peaks. In terms of kinetic parameters, the Km value for St16DOX toward (22S,25S)-22,26-dihydroxycholesterol was determined to be 4.19 ± 0.17 μm (Supplemental Fig. S6). These results strongly suggest that St16DOX functions as a 16α-hydroxylase of (22S,25S)-22,26-dihydroxycholesterol in SGA biosynthesis.
Figure 5.
Relative activities of recombinant St16DOX toward cholesterol and several oxygenated cholesterols. N.D., Not detected.
SGA Analysis of St16DOX-Silenced Transgenic Potato Plants
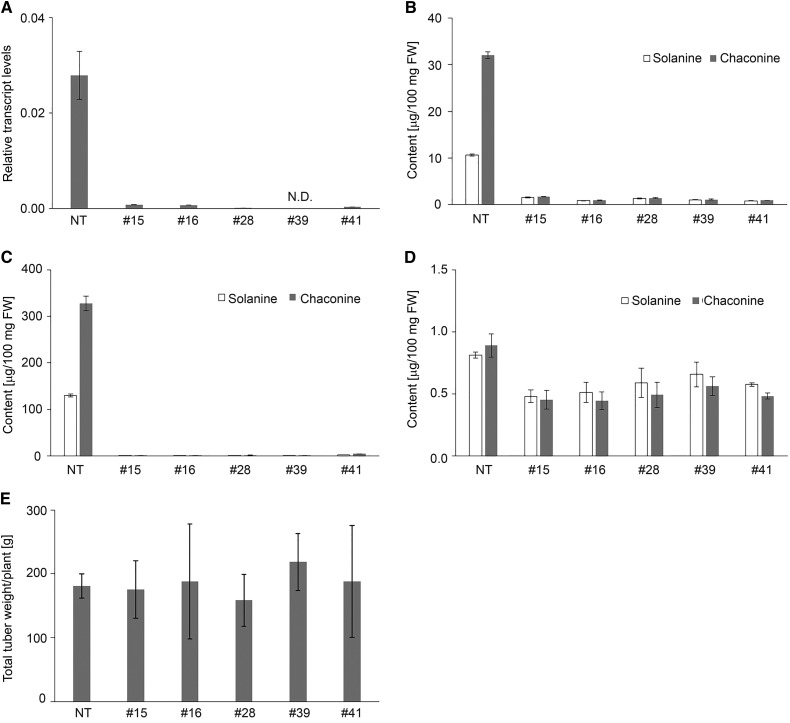
To confirm the contribution of St16DOX to SGA biosynthesis in potato, we transformed potato plants with an RNA interference vector to create St16DOX-silenced transgenic potato plants. Among 41 lines of St16DOX-silenced transgenic plants, the in vitro-grown shoots of five independent lines (#15, #16, #28, #39, and #41; Fig. 6) had significantly lower St16DOX transcript levels than the control (Fig. 6A) and consistently had a much lower SGA content (Fig. 6B). All of the silenced lines grew normally in a greenhouse, and the potato tubers were harvested. The SGA contents in the tuber peel and cortex of the five silenced lines were significantly lower than in the control (Fig. 6, C and D), indicating that St16DOX is involved in SGA biosynthesis in potato. Similarly, the SGA contents in Sl16DOX-silenced transgenic tomato plants also were severely reduced (Supplemental Fig. S7).
Figure 6.
SGA contents and yields of tubers from St16DOX-silenced transgenic potato plants. A, Quantitative RT-PCR analysis of St16DOX transcript levels in the in vitro-grown shoots of St16DOX-silenced plants. B, LC-MS analysis of SGA levels in the in vitro-grown shoots of St16DOX-silenced plants. C and D, LC-MS analysis of SGA levels in the peel (C) and cortex (D) of harvested tubers from St16DOX-silenced plants. E, Yields of tubers from St16DOX-silenced plants. Error bars indicate sd (n = 3). FW, Fresh weight; NT, nontransgenic control plants; #15, #16, #28, #39, and #41, independent transgenic lines.
Phenotype of St16DOX-Silenced Transgenic Potato Plants
The St16DOX-silenced transgenic potato plants had similar tuber yields to the control (Fig. 6E); however, the tuber sprouts of these plants stopped elongation under dark and light conditions. Although sprout initiation seemed to be normal, the sprouts did not grow even after more than 3 months at 20°C or for a 1 year at 4°C after the cessation of usual dormancy in the control plants (Supplemental Fig. S8A). Interestingly, tiny sprouts could start to grow after the tubers were planted in soil (Supplemental Fig. S8B) but not in water under light or dark conditions. In addition, after placing separated tops of the tiny sprouts on tissue culture medium, the sprouts could grow (Supplemental Fig. S8C). This phenotype is completely the same as that of PGA1- and PGA2-silenced potato plants (Umemoto et al., 2016). PGA1- and PGA2-silenced potato plants do not bloom and are sterile. Unlike PGA1- and PGA2-silenced potato plants, the St16DOX-silenced transgenic potato plants bloomed normally. The Sl16DOX-silenced transgenic tomato plants are confirmed to be fertile and did not demonstrate any outward difference compared with the control.
Endogenous Metabolite Analysis in St16DOX-Silenced Transgenic Potato Plants
To examine the effects of St16DOX gene silencing on the endogenous metabolites, we analyzed changes in steroidal compounds in the St16DOX-silenced plants using LC-MS. This analysis detected seven distinctive peaks that were presumed to be steroidal saponins, which had retention times of 17.44, 17.94, 20.02, 20.56, 23.36, 24.05, and 28.45 min, and gave putative masses of parental ions at m/z 1,236, 1,220, 1,074, 1,058.0, 911.8, 895.8, and 603.7, respectively (Supplemental Fig. S9). These peaks had a major mass fragment ion at m/z 383.6, which corresponds to [dihydroxycholesterol – 2H2O + H+]+, and therefore were assumed to be the glycosides (1-5 saccharides) of dihydroxycholesterol.
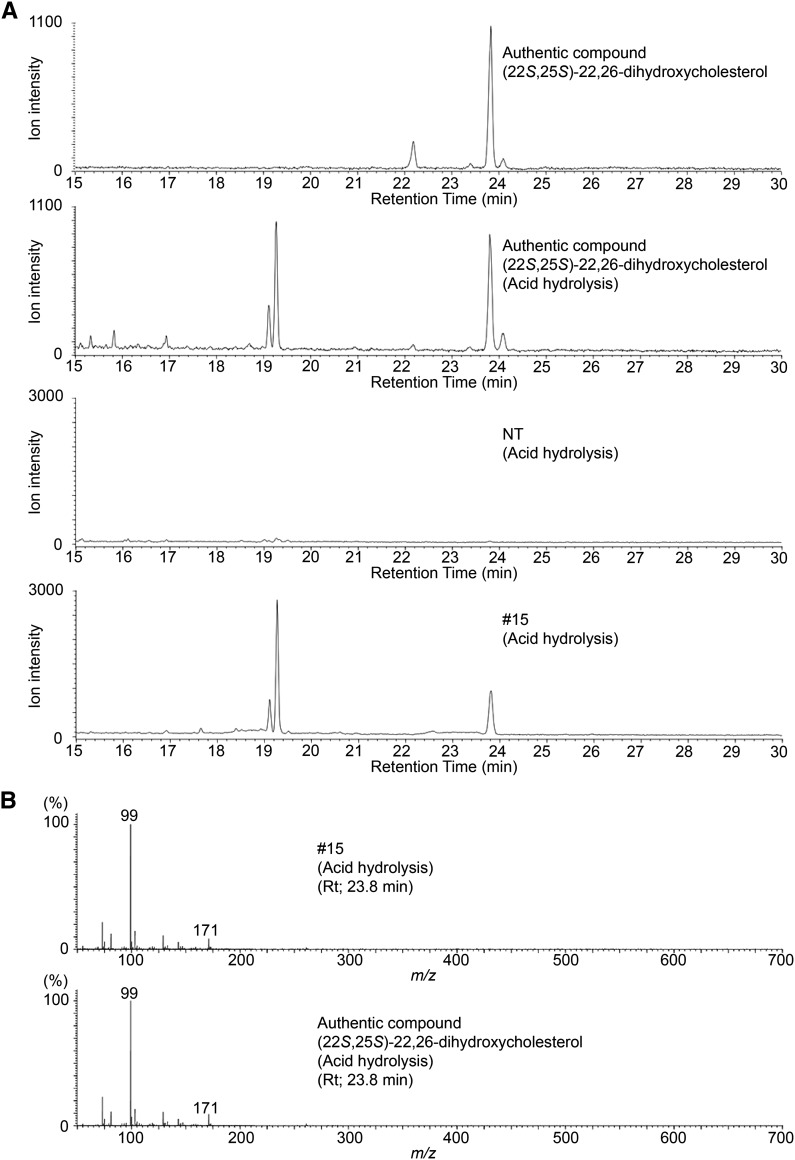
To determine the aglycone structure of these steroidal glycosides that were accumulated in the St16DOX-silenced plants, we extracted the endogenous metabolites and hydrolyzed them with 1 n HCl and then analyzed the trimethylsilylated metabolites using GC-MS. Compared with nontransgenic plants, the St16DOX-silenced plants had three new peaks in the extracted ion chromatogram targeting the ion m/z 171 (Fig. 7A). One of these peaks had a retention time of 23.8 min and was identical to the authentic (22S,25S)-22,26-dihydroxycholesterol in terms of both the retention time and the mass spectrum (Fig. 7). The other two peaks, which had retention times of 19.1 and 19.3 min, were identical to the artifacts that occur during the processes of acid hydrolysis and trimethylsilylated derivatization of authentic (22S,25S)-22,26-dihydroxycholesterol (Fig. 7A). These results indicate that St16DOX gene silencing results in the accumulation of the glycosides of (22S,25S)-22,26-dihydroxycholesterol, which was determined to be the best substrate for St16DOX in the in vitro assay. Similarly, Sl16DOX-silenced transgenic tomato plants also accumulated the glycosides of (22S,25S)-22,26-dihydroxycholesterol (Supplemental Fig. S10).
Figure 7.
GC-MS analysis of the accumulated compounds in leaves of St16DOX-silenced transgenic potato plants. A, Extracted ion chromatograms targeting ion m/z 171 of the accumulated compounds and the authentic compound. B, Mass spectra of the peaks shown in A at a retention time of 23.8 min. NT, Nontransgenic control line; #15, a transgenic line.
DISCUSSION
16DOX Is a Dioxygenase That Shows Steroid 16α-Hydroxylase Activity in SGA Biosynthesis
SGA biosynthesis in potato and tomato is thought to require the oxidation of cholesterol at the C-16, C-22, and C-26 positions, followed by transamination and glycosylation (Petersen et al., 1993; Friedman, 2002; Ginzberg et al., 2009). We recently identified two CYPs (PGA1 and PGA2) that are involved in SGA biosynthesis, which catalyze the hydroxylation of cholesterol at C-26 and C-22, respectively (Umemoto et al., 2016). In this study, we investigated the role of a 2OGD gene named 16DOX, which belongs to clade DOXC41. An in vitro functional analysis of the recombinant 16DOX protein confirmed that 16DOX catalyzes the hydroxylation of steroids at C-16α (Fig. 4). In addition, 16DOX-silenced transgenic plants contained significantly lower amounts of SGAs (Fig. 6, B–D). These results clearly demonstrate that 16DOX is a steroid 16α-hydroxylase that is involved in SGA biosynthesis in Solanum spp. Although many CYPs from various organisms have been characterized as catalyzing the oxidative modification of steroid compounds, to our knowledge, this is the first 2OGD to be identified as functioning in the oxidation of steroids.
Reaction Orders during Steroidal Alkaloid Biosynthesis in Solanum and Veratrum spp.
In this study, we found that 16DOX-silenced transgenic plants accumulated the glycosides of dihydroxycholesterol (Supplemental Fig. S8). Similarly, Itkin et al. (2013) also reported that transgenic tomato plants in which GAME11, a gene coding a putative dioxygenase, was silenced by virus-induced gene silencing accumulated dihydroxycholesterol saponins of an unknown structure. This GAME11 corresponds to Sl16DOX, and their results were consistent with our observations. In this study, we determined the aglycone structure of the glycoside product as (22S,25S)-22,26-dihydroxycholesterol (Fig. 7); furthermore, functional characterization revealed that this was the best substrate for the recombinant St16DOX (Fig. 5). Therefore, we conclude that 16DOX catalyzes the C-16 hydroxylation of (22S,25S)-22,26-dihydroxycholesterol, which is biosynthesized via the C-22 and C-26 hydroxylation of cholesterol by PGA2 and PGA1, respectively.
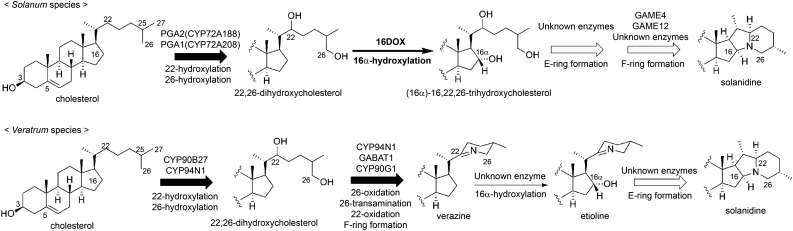
Veratrum spp. are known to contain the steroidal alkaloid cyclopamine, which exhibits potent pharmacological activities, and Veratrum spp. alkaloids are proposed to be biosynthesized from cholesterol via the predicted intermediates verazine and solanidine (Kaneko et al., 1976, 1977). Therefore, since solanidine is a common precursor for the biosynthesis of SGA as well as Veratrum spp. alkaloids, verazine also has been proposed as a precursor in potato SGA biosynthesis (Friedman, 2006). Recently, Augustin et al. (2015) reported the functional characteristics of the four genes (CYP90B27, CYP94N1, GABAT1, and CYP90G1) that catalyze the first six reactions in the production of verazine from cholesterol in Veratrum californicum. In this pathway, oxidation and transamination at C-26, subsequent oxidation at C-22, and F-ring closure occurred in the absence of the hydroxylation at C-16 (Augustin et al., 2015), and previous phytochemical studies also have supported this reaction order (Kaneko et al., 1970, 1975, 1976, 1977). By contrast, our study demonstrated that 16DOX hydroxylated (22S,25S)-22,26-dihydroxycholesterol at C-16 (Fig. 5), and this was supported by the observation that the glycosides of 22,26-dihydroxycholesterol accumulated in 16DOX-silenced transgenic plants (Supplemental Fig. S9), whereas verazine-type compounds were not detected. These different reaction orders may be explained by the independent origins of the biosynthetic genes in Solanum and Veratrum spp. (Fig. 8). Potato PGA2 (CYP72A188) and PGA1 (CYP72A208) belong to the CYP72 family and differ from Veratrum spp. CYP90B27 and CYP94N1, which catalyze C-22 hydroxylation and two steps of C-26 oxidation to form C-26 aldehyde, which can be transaminated by GABAT1 (Augustin et al., 2015). The identification of a C-22 oxidase in Solanum spp. and a C-16 hydroxylase in Veratrum spp. will provide further support for the different reaction orders and origins of the steroidal alkaloid pathways in these genera.
Figure 8.
Putative reaction orders in the biosynthetic pathways of steroidal alkaloids in Solanum and Veratrum spp. Thick arrows indicate the reaction steps that were characterized in this work. The black-filled arrow indicates the multiple reaction stages reported by Umemoto et al. (2016) and Augustin et al. (2015). White arrows represent unclear multiple reaction stages.
16DOX Catalyzes C-16 Hydroxylation Specifically in the 16α Configuration
Feeding with the three labeled cholesterols indicated that the 16α-hydrogen of cholesterol was eliminated but the 16β-hydrogen was retained in tomato SGA biosynthesis (Fig. 2, G–I), which supported the finding that 16DOX catalyzes C-16 hydroxylation specifically in the 16α configuration. However, the structure of the final product α-tomatine is not consistent with this, as the oxygen atom in the E-ring of α-tomatine is in the 16β configuration and 16α-hydrogen also is present. Curiously, our tracer experiments suggested that the 16α-hydrogen of α-tomatine is derived from the 16β-hydrogen of cholesterol. In the proposed biosynthetic pathway of α-tomatine (Itkin et al., 2013), 16,22,26-trihydroxycholesterol is likely reoxidized at C-22, followed by E-ring closure to form furostanol-type aglycone, in which the oxygen atom is in the 16β configuration. Therefore, this apparent discrepancy may be explained by the process of E-ring closure, during which the inversion of 16β-hydrogen to the C-16α position occurs, although a second step of C-22 oxidase and an E-ring cyclase that are involved in tomato SGA biosynthesis have not yet been characterized.
Our tracer experiments also suggested that both the 16α- and 16β-hydrogen atoms of cholesterol were eliminated during potato SGA biosynthesis while St16DOX catalyzes only 16α-hydroxylation (Fig. 2, A–F). These results suggest that loss of the 16β-hydrogen atom is catalyzed by an additional unknown C-16 oxidase and that this oxidation step occurred specifically in potato SGA biosynthesis. In addition, 16α-hydrogen was added subsequently to form solanidine in potato SGA biosynthesis (Fig. 2, A–F), suggesting that an unknown C-16 reductase that introduces 16α-hydrogen may be involved in the final stage of solanidine biosynthesis. Taken together, these findings indicate that further investigation is required to elucidate the mechanism by which the E- and F-rings of spirosolane and solanidane are formed during SGA biosynthesis.
Coexpression of 16DOX with SGA Biosynthetic Genes
Quantitative RT-PCR analysis in various tissues of potato showed that the 16DOX gene was coexpressed with the PGA1 and PGA2 genes as well as the UGT genes that are responsible for the glycosylation steps in SGA biosynthesis (Fig. 3). Itkin et al. (2013) reported that SGA biosynthetic genes including GAME11 (which corresponds to 16DOX) exist as gene clusters on chromosomes 7 and 12 of tomato and potato. Such physical clustering of functionally related genes may enable the coordinated regulation of gene expression at the chromatin level (Field et al., 2011). Previously, Sl16DOX was reported as the pistil-expressed dioxygenase that has been variously named TPP1, GAD2, SPP2, and H6H-like protein in tomato (Milligan and Gasser, 1995; Jacobsen and Olszewski, 1996; Lantin et al., 1999; Nakane et al., 2003). The findings of these studies were consistent with our results that Sl16DOX showed high expression levels in flowers (Fig. 3). Recently, Thagun et al. (2016) reported that the overexpression and suppression of Jasmonate-Responsive ERF (JRE) genes (JRE3, JRE4, and JRE5) in tomato significantly influenced SGA accumulation and the expression of the genes encoding the SGA biosynthetic enzymes, including those involved in the upstream mevalonate pathway. JRE4 was found to bind a GCC box-like element that is enriched in the promoters of JRE-regulated genes and also controlled the coordinated expression of the SGA biosynthetic genes. Similarly, Cárdenas et al. (2016) revealed that GAME9 (which is identical to JRE4) in tomato and potato regulates the expression of the SGA biosynthetic genes by directly or indirectly acting on the promoters of downstream target genes. Thus, coexpression analysis of the SGA biosynthetic genes that are regulated by JRE4/GAME9 will help to identify novel genes that are involved in the formation of the E- and F-rings of spirosolane and solanidane, including a C-16 oxidase in potato.
Phenotypes of 16DOX-Silenced Transgenic Plants
16DOX-silenced transgenic potato plants contained significantly lower amounts of α-solanine and α-chaconine, and the silenced lines grew normally in a greenhouse with comparable yields of tubers to the wild type. Interestingly, 16DOX-silenced transgenic potato plants did not sprout (Supplemental Fig. S8A), but the sprouts could grow by planting the tubers in soil or placing the excised sprout tips on tissue culture medium (Supplemental Fig. S8, B and C). These phenotypes are similar to those of PGA1- and PGA2-silenced plants, except that the latter also exhibited abnormal flowers (Umemoto et al., 2016). By contrast, GAME4/PGA3- or SSR2-silenced transgenic potato plants were phenotypically identical to control plants, despite containing much lower SGA contents (Itkin et al., 2013; Umemoto and Sasaki, 2013; Sawai et al., 2014). These findings demonstrate that the amounts of SGA are unrelated to sprouting morphogenesis, so 16DOX-silenced plants may accumulate unknown compounds that inhibit sprouting and can be removed by planting the tubers in soil (Umemoto et al., 2016). We found that 16DOX-silenced transgenic plants accumulated dihydroxycholesterol and/or its glycosides (Supplemental Fig. S9), and PGA1- and PGA2-silenced plants also were found to accumulate 22- or 26-hydroxycholesterols and/or their glycosides (Umemoto et al., 2016). Therefore, it is possible that the suppression of sprouting morphogenesis is caused by the accumulation of hydroxylated cholesterol or its glycosides in the sprout clusters of the silenced potatoes.
Our observations demonstrate that it may be possible to breed SGA-free potato, as reported by Umemoto et al. (2016). 16DOX is a valuable target for the development of a loss-of-function mutant because it occurs as a single-copy gene in the genomes of potato and tomato. Such a loss-of-function mutant for 16DOX could be obtained through mutation breeding using targeting induced local lesions in genomes (Elias et al., 2009) and genome-editing techniques (Sawai et al., 2014; Nicolia et al., 2015).
MATERIALS AND METHODS
Chemicals
Authentic samples of α-solanine, α-chaconine, α-tomatine, (22R)-22-hydroxycholesterol, and (22S)-22-hydroxycholesterol were purchased from Sigma-Aldrich. Cholesterol was purchased from Tama Biochemical. Authentic compounds of (25R)-26-hydroxycholesterol, (25S)-26-hydroxycholesterol, (22S)-16α,22-dihydroxycholesterol, (22S)-16β,22-dihydroxycholesterol, (22S,25S)-22,26-dihydroxycholesterol, (22R,25S)-22-hydroxy-26-oxocholesterol, (22S,25RS)-22-hydroxy-26-oxocholesterol, [15,15,16α,17-2H4]cholesterol, [15,15,16β,17-2H4]cholesterol, and [15,15,17-2H3]cholesterol were synthesized as described in Supplemental Materials and Methods S1.
RNA Extraction and RT
Total RNA extraction was performed using the RNeasy Plant Mini Kit (Qiagen) and the RNase-Free DNase Set (Qiagen). Total RNAs of potato (Solanum tuberosum) and tomato (Solanum lycopersicum) were prepared from the leaves, flowers, tuber peels, stems, roots, stolons, and tuber sprouts of potato cv Sassy and the leaves, flowers, mature green fruits, yellow fruits, orange fruits, and red fruits of tomato cv Micro-Tom, respectively. The extracted total RNAs of potato and tomato were used to synthesize the first-strand cDNAs using the SuperScript First-Strand Synthesis System for RT-PCR (Life Technologies) and the Transcriptor First-Strand cDNA Synthesis Kit (Toyobo), respectively.
Cloning of 16DOX cDNAs
The cDNA fragments that contained the open reading frame of each 16DOX gene were amplified by RT-PCR with primers 1 and 2 for St16DOX and primers 3 and 4 for Sl16DOX (Supplemental Table S2), which were designed from the potato and tomato unigene sequences (Sotub07g016570 in the Potato ITAG protein database and Solyc07g043420 in the Tomato ITAG protein database, respectively). The PCR products were cloned into the pENTR/D-TOPO plasmid (Life Technologies).
Expression of the Recombinant 16DOX Protein in Escherichia coli
The coding sequence for each 16DOX gene was amplified from the pENTR/D-TOPO plasmid using primers 5 and 6 for St16DOX and primers 7 and 8 for Sl16DOX (Supplemental Table S2), which contained restriction sites. The amplified DNA fragments were ligated into the pMD19 vector (TaKaRa) and digested with BamHI and SalI. The DNA fragments were then ligated into the BamHI-SalI sites of pGEX4T-1. E. coli strain BLR (DE3) (Clontech) transformed with the constructed plasmid was grown at 37°C in lysogeny broth with 50 μg mL−1 ampicillin until its OD600 reached 0.5. Recombinant protein expression was induced by adding 0.1 mm isopropyl β-d-1-thiogalactopyranoside and was continued for 20 h at 18°C. The culture was then centrifuged at 3,500 rpm for 30 min at 4°C, and the cell pellets were resuspended in 5 mL of cold sonication buffer containing 50 mm sodium phosphate (pH 7.4), 300 mm NaCl, and 20% (v/v) glycerol. The solution was then sonicated three times for 30 s each on ice using a Bandelin Sonopuls HD 2070 ultrasonic homogenizer type MS73 (Sigma-Aldrich) at a sound intensity of 200 W cm−2 and centrifuged at 15,000 rpm for 10 min at 4°C. The GST-tagged proteins present in the supernatant were purified using GST Spin Trap columns (GE Healthcare) according to the manufacturer’s instructions. After two column washes, the adsorbed proteins were eluted twice in 200 μL of a solution containing 50 mm Tris-HCl (pH 8), 20 mm reduced glutathione, and 20% (v/v) glycerol, and the elution was mixed. The concentration of the purified proteins was determined by the Bradford system. The purified recombinant proteins were visualized by SDS-PAGE. The proteins were revealed by staining the gel with Coomassie Brilliant Blue R-250 and then were used for further analyses.
In Vitro Enzyme Activity Assay
An in vitro enzyme activity assay was performed using 100 μL of reaction mixture that consisted of 100 mm Bis-Tris-HCl (pH 7.2), 5 mm 2-ketoglutaric acid, 10 mm sodium ascorbate, 0.2 mm FeSO4, 25 μm (22S)-22-hydroxycholesterol as a substrate, and each of the purified recombinant 16DOX proteins of potato and tomato as an enzyme. The reaction was initiated by the addition of the enzyme being tested and was carried out at 30°C for 3 h. The reaction was then stopped by the addition of 100 μL of ethyl acetate, followed by the addition of 0.2 μg of 25-hydroxycholesterol in 100% ethanol as an internal standard. The reaction products were extracted three times with an equal volume of ethyl acetate, and the organic phase was collected and evaporated. The residue was trimethylsilylated with the TMS-HT Kit (Tokyo Chemical Industry) at 80°C for 30 min.
GC-MS analysis of the reaction products was performed as described previously by Seki et al. (2008) with minor modifications. GC-MS was conducted using a GC-MS-QP2010 Ultra (Shimadzu) with a DB-5MS (30 m × 0.25 mm, 0.25 μm film thickness; J&W Scientific) capillary column. The injection temperature was 250°C, and the following column temperature program was used: 80°C for 1 min, followed by a rise to 300°C at a rate of 20°C min−1 and a hold at 300°C for 20 min. The carrier gas was He, and the flow rate was 1 mL min−1. The interface temperature was 300°C, with a splitless injection.
Biochemical Analysis of Recombinant St16DOX
To determine the substrate specificity of St16DOX, its activity was assayed using 25 μm cholesterol, (22R)- and (22S)-22-hydroxycholesterols, 22-oxocholesterol, (25R)- and (25S)-26-hydroxycholesterols, (22S,25S)-22,26-dihydroxycholesterol, and (22R,25S)- and (22S,25RS)-22-hydroxy-26-oxocholesterols. Each reaction was carried out at 30°C for 3 h. Extraction and GC-MS analysis of the reaction product were performed as described above. In addition, the reaction product for (22S,25S)-22,26-dihydroxycholesterol also was analyzed by LC-MS by extracting and evaporating the product as described above and dissolving the residue in 200 μL of ethanol. LC-MS analysis was performed using a system consisting of an ACQUITY UPLC H-Class System (Waters) and an SQ Detector 2 (Waters), and data acquisition and analysis were performed using MassLynx 4.1 software (Waters). Each sample (5 μL) was injected into an ACQUITY UPLC BEH C-18 chromatographic column (50 × 2.1 mm, 1.7 μm; Waters), in which the column temperature was set at 40°C and the flow rate was set at 0.2 mL min−1. The mobile phases were water with 0.1% (v/v) formic acid (A) and acetonitrile (B), using a gradient elution of 10% to 90% B at 0 to 30 min, 90% to 100% B at 30 to 40 min, and 100% B at 40 to 46 min (0–30 min and 30–40 min, linear gradient). The mass spectra were obtained in positive electrospray ionization, with a capillary voltage of 3 kV and a sample cone voltage of 60 V. Mass spectrometry scan mode with a mass range of m/z 250 to 1,400 was used.
Next, we determined the kinetic parameters of recombinant St16DOX in triplicate assays. The activity was assayed using (22S,25S)-22,26-dihydroxycholesterol at a concentration ranging from 1 to 50 μm. The reaction was carried out at 30°C for 30 min. Extraction and GC-MS analysis of the reaction product were performed as described above. Kinetic parameters were determined by nonlinear regression using the ANEMONA program (Hernández and Ruiz, 1998).
Generation of Transformation Vectors, Plant Transformation, and Growth Conditions
A 370-bp fragment of St16DOX cDNA was PCR amplified using primers 9 and 10 (Supplemental Table S2), which contained restriction sites. An RNA interference binary vector that targeted the St16DOX gene (pKT258) was constructed from the binary vector pKT11 (Umemoto et al., 2001) by locating two 370-bp fragments of St16DOX in opposite directions that interposed the third intron of the Arabidopsis (Arabidopsis thaliana) At4g14210 gene under the control of the cauliflower mosaic virus 35S promotor in the T-DNA region, as described previously (Umemoto et al., 2016). pKT258 was then electroporated into Agrobacterium tumefaciens GV3110 mp90. Potato (cv Sassy) plants were transformed using A. tumefaciens GV3110 mp90 cells with pKT258, as reported previously (Momma, 1990). In vitro-grown plants were cultured at 20°C under a 16-h-light/8-h-dark cycle, following which 41 transformants were individually selected by genomic PCR of in vitro-grown shoots using primers 11 and 12 (Supplemental Table S2), which targeted the kanamycin resistance gene in the T-DNA region that was integrated into the potato genome. Quantitative RT-PCR analysis of St16DOX was performed using primers 13 and 14 (Supplemental Table S2) as described in “Real-Time Quantitative RT-PCR Analysis.” Total RNA was then prepared from the stems of five independent lines of in vitro-cultured plants: #5, #16, #28, #39, and #41. Primers 15 and 16 (Supplemental Table S2), which targeted the potato Elongation Factor1α gene (EF1α; Nicot et al., 2005), were used as a control. Tomato (cv Micro-Tom) plants also were transformed using A. tumefaciens GV3110 mp90 cells with pKT258, as reported previously (Sun et al., 2006)
LC-MS Analysis of SGAs in 16DOX-Silenced Transgenic Plants
The SGAs that were contained in 16DOX-silenced transgenic plants were extracted and quantified as described previously (Umemoto et al., 2016). LC-MS analysis of the steroidal compounds that accumulated in the 16DOX-silenced plants was performed as described in “Biochemical Analysis of Recombinant St16DOX” with minor modifications: LC-MS was conducted with an ACQUITY UPLC HSS T3 chromatographic column (100 × 2.1 mm, 1.8 μm; Waters); water with 0.1% (v/v) formic acid (A) and acetonitrile (B) were used as the mobile phases, using a gradient elution of 10% B at 0 to 2 min and 10% to 55% B at 2 to 32 min (linear gradient); and mass spectrometry scan mode was used with a mass range of m/z 300 to 1,250.
GC-MS Analysis of Steroids in 16DOX-Silenced Transgenic Plants
The steroid compounds that accumulated in the in vitro-grown shoots of 16DOX-silenced transgenic potato plants and the leaves of 16DOX-silenced transgenic tomato plants were extracted and analyzed using a similar method to that described previously (Ohyama et al., 2013). The extracted residue was trimethylsilylated with N-methyl-N-trimethylsilyltrifluoroacetamide (Sigma-Aldrich) at 80°C for 30 min. GC-MS analysis was performed as described above with minor modifications: a DB-1MS (30 m × 0.25 mm, 0.25 μm film thickness; J&W Scientific) capillary column was used.
Real-Time Quantitative RT-PCR Analysis
Quantitative RT-PCR was performed with a LightCyclerNano (Roche) using THUNDERBIRD SYBR qPCR Mix (Toyobo) with the following primer sets: 17 and 18 for St16DOX, 19 and 20 for PGA1, 21 and 22 for PGA2, 23 and 24 for SGT1, 25 and 26 for SGT3, and 15 and 16 for EF1α (Nicot et al., 2005), using the cDNA of various potato tissues as templates; and 17 and 18 for Sl16DOX, 27 and 28 for PGA1-homolog, 29 and 30 for PGA2-homolog, 31 and 32 for GAME1, and 33 and 34 for ubiquitin, using the cDNA of various tomato tissues as templates (Supplemental Table S2). Cycling was carried out at 95°C for 10 min, 45 cycles at 95°C for 10 s, 60°C for 10 s, and 72°C for 15 s for amplification, followed by holding at 95°C for 30 s and ramping up from 60°C to 95°C at 0.1°C s−1 to perform a melting curve analysis. Three biological replicates were analyzed in duplicate. The gene expression levels were normalized against the values obtained for the EF1α and Ubiquitin genes, which were used as internal references in potato and tomato, respectively. Data acquisition and analysis were performed using LightCyclerNano software (Roche).
Tracer Experiments Using Stable Isotope-Labeled Compounds
Tracer experiments were performed according to a previously reported method (Ohyama et al., 2013) with minor modifications: each stable isotope-labeled compound (1 mg) was dissolved in acetone (25 μL)-Tween 80 (25 μL). In vitro-grown potato (cv Sassy) shoots and tomato (cv Micro-Tom) seedlings were prepared for tracer experiments and fed with the stable isotope-labeled compounds [15,15,16α,17-2H4]cholesterol, [15,15,16β,17-2H4]cholesterol, and [15,15,17-2H3]cholesterol. After 7 d, the SGAs that accumulated in the harvested potato shoots and tomato seedlings were extracted and analyzed by LC-MS as described above.
Accession Numbers
Accession numbers are as follows: St16DOX, LC222743; and Sl16DOX, LC222744.
Supplemental Data
The following supplemental materials are available.
Supplemental Materials and Methods S1. Chemical synthesis of steroidal compounds.
Supplemental Figure S1. Quantitative RT-PCR analysis of the expression pattern of SGA biosynthetic genes in various organs of tomato.
Supplemental Figure S2. Amino acid sequence alignment of St16DOX, Sl16DOX, and hyoscyamine 6β-hydroxylase from H. niger (HnH6H).
Supplemental Figure S3. GC-MS analysis of the reaction products obtained from the recombinant St16DOX protein with (22S,25S)-22,26-dihydroxycholesterol as a substrate.
Supplemental Figure S4. LC-MS analysis of the reaction products from the recombinant St16DOX protein with (22S,25S)-22,26-dihydroxycholesterol as a substrate.
Supplemental Figure S5. GC-MS analysis of the reaction products from the recombinant 16DOX proteins with (22R)-22-hydroxycholesterol as a substrate.
Supplemental Figure S6. Kinetic analysis of the recombinant St16DOX with (22S,25S)-22,26-dihydroxycholesterol.
Supplemental Figure S7. LC-MS analysis of α-tomatine levels in the leaves of Sl16DOX-silenced transgenic tomato plants.
Supplemental Figure S8. Phenotypes of St16DOX-silenced transgenic potato plants.
Supplemental Figure S9. LC-MS analysis of the accumulated compounds in the leaves of St16DOX-silenced transgenic potato plants.
Supplemental Figure S10. GC-MS analysis of the accumulated compounds in leaves of Sl16DOX-silenced transgenic tomato plants.
Supplemental Table S1. List of the 256 2OGD transcripts extracted from the Spud DB potato genomics resource.
Supplemental Table S2. Oligonucleotides that were used in this study.
Supplemental Materials and Methods S1. Supplemental materials and method.
Acknowledgments
We thank Masako Otsuka, Noriko Yasuno, and Ryo Negoya for technical assistance. Parts of the experimental measurements were carried out using the Bruker and JEOL 600 MHz NMR spectrometers, as well as the JEOL JMS-700 mass spectrometer, in the Joint Usage/Research Center at the Institute for Chemical Research, Kyoto University.
Glossary
- SGA
steroidal glycoalkaloid
- LC-MS
liquid chromatography-mass spectrometry
- RT
reverse transcription
- GC-MS
gas chromatography-mass spectrometry
Footnotes
This work was supported by the Program for the Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN), Japan, by the Scientific Technique Research Promotion Program for Agriculture, Forestry, Fisheries, and Food Industry, Japan, and by the Cross-Ministerial Strategic Innovation Promotion Program (SIP), Japan. The synthetic research was supported by 2nd Seiken Academic Incentive Fund provided by Research Instituted for Production Development, Kyoto, Japan.
References
- Augustin MM, Ruzicka DR, Shukla AK, Augustin JM, Starks CM, O’Neil-Johnson M, McKain MR, Evans BS, Barrett MD, Smithson A, et al. (2015) Elucidating steroid alkaloid biosynthesis in Veratrum californicum: production of verazine in Sf9 cells. Plant J 82: 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg TDH. (2003) Dioxygenase enzymes: catalytic mechanisms and chemical models. Tetrahedron 59: 7075–7101 [Google Scholar]
- Canonica L, Ronchetti F, Russo G (1977) Fate of the 16β-hydrogen atom of cholesterol in the biosynthesis of tomatidine and solanidine. J Chem Soc Chem Commun 286–287 [Google Scholar]
- Cárdenas PD, Sonawane PD, Pollier J, Vanden Bossche R, Dewangan V, Weithorn E, Tal L, Meir S, Rogachev I, Malitsky S, et al. (2016) GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat Commun 7: 10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias R, Till BJ, Mba C, Al-Safadi B (2009) Optimizing TILLING and Ecotilling techniques for potato (Solanum tuberosum L). BMC Res Notes 2: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Fiston-Lavier AS, Kemen A, Geisler K, Quesneville H, Osbourn AE (2011) Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc Natl Acad Sci USA 108: 16116–16121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman M. (2002) Tomato glycoalkaloids: role in the plant and in the diet. J Agric Food Chem 50: 5751–5780 [DOI] [PubMed] [Google Scholar]
- Friedman M. (2006) Potato glycoalkaloids and metabolites: roles in the plant and in the diet. J Agric Food Chem 54: 8655–8681 [DOI] [PubMed] [Google Scholar]
- Friedman M, Dao L (1992) Distribution of glycoalkaloids in potato plants and commercial potato products. J Agric Food Chem 40: 419–423 [Google Scholar]
- Ginzberg I, Tokuhisa J, Veilleux R (2009) Potato steroidal glycoalkaloids: biosynthesis and genetic manipulation. Potato Res 52: 1–15 [Google Scholar]
- Harrison DM. (1990) Steroidal alkaloids. Nat Prod Rep 7: 139–147 [Google Scholar]
- Helmut R. (1998) Solanum steroid alkaloids: an update. Alkaloids: Chem Biol Perspect 12: 103–185 [Google Scholar]
- Hernández A, Ruiz MT (1998) An EXCEL template for calculation of enzyme kinetic parameters by non-linear regression. Bioinformatics 14: 227–228 [DOI] [PubMed] [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, et al. (2013) Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341: 175–179 [DOI] [PubMed] [Google Scholar]
- Itkin M, Rogachev I, Alkan N, Rosenberg T, Malitsky S, Masini L, Meir S, Iijima Y, Aoki K, de Vos R, et al. (2011) GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell 23: 4507–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE (1996) Gibberellins regulate the abundance of RNAs with sequence similarity to proteinase inhibitors, dioxygenases and dehydrogenases. Planta 198: 78–86 [DOI] [PubMed] [Google Scholar]
- Kaneko K, Mitsuhashi H, Hirayama K, Yoshida N (1970) Dormantinol, a possible precursor in solanidine biosynthesis, from budding Veratrum grandiflorum. Phytochemistry 9: 2489–2495 [Google Scholar]
- Kaneko K, Seto H, Motoki C, Mitsuhashi H (1975) Biosynthesis of rubijervine in Veratrum grandiflorum. Phytochemistry 14: 1295–1301 [Google Scholar]
- Kaneko K, Watanabe M, Mitsuhashi H (1976) Origin of nitrogen in the biosynthesis of solanidine by Veratrum grandiflorum. Phytochemistry 15: 1391–1393 [Google Scholar]
- Kaneko K, Watanabe M, Mitsuhashi H (1977) Dormantinol, a possible precursor in solanidine biosynthesis, from budding Veratrum grandiflorum. Phytochemistry 16: 1247–1251 [Google Scholar]
- Kawai Y, Ono E, Mizutani M (2014) Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J 78: 328–343 [DOI] [PubMed] [Google Scholar]
- Kozukue N, Mizuno S (1985) Studies on glycoalkaloids of potatoes. Part II. Analysis of glycoalkaloid content in potato tissues and tubers. J Jpn Soc Hortic Sci (Suppl) 54: 496–497 [Google Scholar]
- Kozukue N, Mizuno S (1989) Studies on glycoalkaloids of potatoes. Part IV. Changes of glycoalkaloid content in four parts of a sprouted potato tuber and in potato tubers during storage. J Jpn Soc Hortic Sci 58: 231–235 [Google Scholar]
- Lantin S, O’Brien M, Matton DP (1999) Pollination, wounding and jasmonate treatments induce the expression of a developmentally regulated pistil dioxygenase at a distance, in the ovary, in the wild potato Solanum chacoense Bitt. Plant Mol Biol 41: 371–386 [DOI] [PubMed] [Google Scholar]
- Lukačin R, Britsch L (1997) Identification of strictly conserved histidine and arginine residues as part of the active site in Petunia hybrida flavanone 3β-hydroxylase. Eur J Biochem 249: 748–757 [DOI] [PubMed] [Google Scholar]
- Lukačin R, Gröning I, Pieper U, Matern U (2000) Site-directed mutagenesis of the active site serine290 in flavanone 3β-hydroxylase from Petunia hybrida. Eur J Biochem 267: 853–860 [DOI] [PubMed] [Google Scholar]
- Matsuda J, Okabe S, Hashimoto T, Yamada Y (1991) Molecular cloning of hyoscyamine 6 β-hydroxylase, a 2-oxoglutarate-dependent dioxygenase, from cultured roots of Hyoscyamus niger. J Biol Chem 266: 9460–9464 [PubMed] [Google Scholar]
- McCue KF, Allen PV, Shepherd LV, Blake A, Maccree MM, Rockhold DR, Novy RG, Stewart D, Davies HV, Belknap WR (2007) Potato glycosterol rhamnosyltransferase, the terminal step in triose side-chain biosynthesis. Phytochemistry 68: 327–334 [DOI] [PubMed] [Google Scholar]
- McCue KF, Allen PV, Shepherd LV, Blake A, Whitworth J, Maccree MM, Rockhold DR, Stewart D, Davies HV, Belknap WR (2006) The primary in vivo steroidal alkaloid glucosyltransferase from potato. Phytochemistry 67: 1590–1597 [DOI] [PubMed] [Google Scholar]
- McCue KF, Shepherd LVT, Allen PV, Maccree MM, Rockhold DR, Corsini DL, Davies HV, Belknap WR (2005) Metabolic compensation of steroidal glycoalkaloid biosynthesis in transgenic potato tubers: using reverse genetics to confirm the in vivo enzyme function of a steroidal alkaloid galactosyltransferase. Plant Sci 168: 267–273 [Google Scholar]
- Milligan SB, Gasser CS (1995) Nature and regulation of pistil-expressed genes in tomato. Plant Mol Biol 28: 691–711 [DOI] [PubMed] [Google Scholar]
- Milner SE, Brunton NP, Jones PW, O’Brien NM, Collins SG, Maguire AR (2011) Bioactivities of glycoalkaloids and their aglycones from Solanum species. J Agric Food Chem 59: 3454–3484 [DOI] [PubMed] [Google Scholar]
- Moehs CP, Allen PV, Friedman M, Belknap WR (1997) Cloning and expression of solanidine UDP-glucose glucosyltransferase from potato. Plant J 11: 227–236 [DOI] [PubMed] [Google Scholar]
- Momma T. (1990) Recent study for genetic engineering of soybean glycinin gene. Plant Tissue Cult Lett 7: 57–63 [Google Scholar]
- Moses T, Papadopoulou KK, Osbourn A (2014) Metabolic and functional diversity of saponins, biosynthetic intermediates and semi-synthetic derivatives. Crit Rev Biochem Mol Biol 49: 439–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane E, Kawakita K, Doke N, Yoshioka H (2003) Elicitation of primary and secondary metabolism during defense in the potato. J Gen Plant Pathol 69: 378–384 [Google Scholar]
- Nicolia A, Proux-Wéra E, Åhman I, Onkokesung N, Andersson M, Andreasson E, Zhu LH (2015) Targeted gene mutation in tetraploid potato through transient TALEN expression in protoplasts. J Biotechnol 204: 17–24 [DOI] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56: 2907–2914 [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Yokota T, Mizutani M (2009) Insights into the function and evolution of P450s in plant steroid metabolism. Phytochemistry 70: 1918–1929 [DOI] [PubMed] [Google Scholar]
- Ohyama K, Okawa A, Moriuchi Y, Fujimoto Y (2013) Biosynthesis of steroidal alkaloids in Solanaceae plants: involvement of an aldehyde intermediate during C-26 amination. Phytochemistry 89: 26–31 [DOI] [PubMed] [Google Scholar]
- Petersen HW, Mølgaard P, Nyman U, Olesen CE (1993) Chemotaxonomy of the tuber-bearing Solanum species, subsection Potatoe (Solanaceae). Biochem Syst Ecol 21: 629–644 [Google Scholar]
- Roddick JG. (1989) The acetylcholinesterase-inhibitory activity of steroidal glycoalkaloids and their aglycones. Phytochemistry 28: 2631–2634 [Google Scholar]
- Sawai S, Ohyama K, Yasumoto S, Seki H, Sakuma T, Yamamoto T, Takebayashi Y, Kojima M, Sakakibara H, Aoki T, et al. (2014) Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell 26: 3763–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H, Ohyama K, Sawai S, Mizutani M, Ohnishi T, Sudo H, Akashi T, Aoki T, Saito K, Muranaka T (2008) Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci USA 105: 14204–14209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H, Tamura K, Muranaka T (2015) P450s and UGTs: key players in the structural diversity of triterpenoid saponins. Plant Cell Physiol 56: 1463–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Roddick JG, Jones JL (1996) Potato glycoalkaloids: some unanswered questions. Trends Food Sci Technol 7: 126–131 [Google Scholar]
- Sun HJ, Uchii S, Watanabe S, Ezura H (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47: 426–431 [DOI] [PubMed] [Google Scholar]
- Thagun C, Imanishi S, Kudo T, Nakabayashi R, Ohyama K, Mori T, Kawamoto K, Nakamura Y, Katayama M, Nonaka S, et al. (2016) Jasmonate-responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant Cell Physiol 57: 961–975 [DOI] [PubMed] [Google Scholar]
- Umemoto N, Nakayasu M, Ohyama K, Yotsu-Yamashita M, Mizutani M, Seki H, Saito K, Muranaka T (2016) Two cytochrome P450 monooxygenases catalyze early hydroxylation steps in the potato steroid glycoalkaloid biosynthetic pathway. Plant Physiol 171: 2458–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto N, Sasaki K June 27, 2013. Protein having glycoalkaloid biosynthetic enzyme activity and gene encoding the same. US Patent Application No. 20130167271 A1
- Umemoto N, Tsukahara M, Yoshioka M June 19, 2001. JP Patent Publication (Kokai) 2001-161373 A
- Wilmouth RC, Turnbull JJ, Welford RWD, Clifton IJ, Prescott AG, Schofield CJ (2002) Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure 10: 93–103 [DOI] [PubMed] [Google Scholar]