Auxin influences maximum leaf photosynthetic rate through leaf venation and thus water transport capacity.
Abstract
Land plants lose vast quantities of water to the atmosphere during photosynthetic gas exchange. In angiosperms, a complex network of veins irrigates the leaf, and it is widely held that the density and placement of these veins determines maximum leaf hydraulic capacity and thus maximum photosynthetic rate. This theory is largely based on interspecific comparisons and has never been tested using vein mutants to examine the specific impact of leaf vein morphology on plant water relations. Here we characterize mutants at the Crispoid (Crd) locus in pea (Pisum sativum), which have altered auxin homeostasis and activity in developing leaves, as well as reduced leaf vein density and aberrant placement of free-ending veinlets. This altered vein phenotype in crd mutant plants results in a significant reduction in leaf hydraulic conductance and leaf gas exchange. We find Crispoid to be a member of the YUCCA family of auxin biosynthetic genes. Our results link auxin biosynthesis with maximum photosynthetic rate through leaf venation and substantiate the theory that an increase in the density of leaf veins coupled with their efficient placement can drive increases in leaf photosynthetic capacity.
A central requirement for photosynthesis on land is the uptake of CO2 from the atmosphere. To assimilate just one molecule of atmospheric CO2, plants on average lose 150 molecules of water (Wong et al., 1979). To supply this high demand for water an internal transport system comprised of xylem conduits irrigates the leaf. Higher rates of photosynthesis (A) are linked to higher rates of transpiration, thus demanding greater efficiency in the transport and delivery of water to evaporating leaf tissue (Brodribb and Feild, 2000; Tyree and Zimmermann, 2002; Sack and Scoffoni, 2013). The xylem transports water efficiently through nonliving tubular cells in the leaf, yet the last part of the pathway involves a significantly slower transfer through living mesophyll cells to the stomata. The length of this final nonxylem transfer has a major influence on the efficiency of the entire water transport system (Brodribb et al., 2007). Current theory posits that as hydraulic supply is essential for A, a reduction in the mean path length for water flow through the mesophyll will increase leaf hydraulic conductance (Kleaf), which will increase stomatal conductance (gs) and consequently theoretical maximum A (Sack and Frole, 2006; Brodribb et al., 2007, 2010; Scoffoni et al., 2011, 2016).
A core body of literature has identified a number of adaptations that reduce the terminal path length for water flow in the leaf and thus increase Kleaf. These adaptations range from increasing the density of the veins per unit area of leaf (Brodribb et al., 2007; Sack et al., 2013; Caringella et al., 2015), modifying leaf thickness and bundle sheath extensions (Sack et al., 2003; Zsögön et al., 2015) through to the formation of accessory transfusion tissue (Brodribb and Holbrook, 2005), and the formation and placement of free-ending veinlets (FEVs) within areoles (the small area of leaf mesophyll bound on all sides by veins; Scoffoni et al., 2011; Fiorin et al., 2016). Of these adaptations, vein density has been a major focus due to the high plasticity of this trait both within species (Carins Murphy et al., 2012; Scoffoni et al., 2015) and across vascular plant taxa (Boyce et al., 2009). Using this variation in vein density, strong correlations have been established between the terminal path length for water flow through the mesophyll and maximum Kleaf (Sack and Frole, 2006; Brodribb et al., 2007, 2010; Sack et al., 2013; Scoffoni et al., 2016). These correlations are supported by single-gene vein density mutants in Arabidopsis (Arabidopsis thaliana) and Solanum lycopersicum spanning a spectrum of vein modifications that influence Kleaf (Caringella et al., 2015; Zsögön et al., 2015).
The importance of Kleaf lies in its positive correlation with A. Selection for greater A and productivity is believed to have been the primary driver behind the >10-fold increase in vein density that is observed in the fossil record over the past 400 million years, from the single-veined leaves of lycophytes to the highly complex, hierarchical networks of leaf veins in angiosperms (Brodribb et al., 2005; Boyce et al., 2009; Zhang et al., 2014). This increase in vein density is correlated with an equally substantial increase in maximum A across extant lineages that diverged during this major transition in leaf anatomy. Similar correlations between vein density, Kleaf, and A can be observed within angiosperms (Brodribb and Feild, 2010; Carins Murphy et al., 2012; Scoffoni et al., 2015, 2016).
However, this correlative evidence does not prove that changes in venation are responsible for changes in A, especially considering that stomatal and/or epidermal traits, which can also strongly influence maximum leaf gas exchange, are highly coordinated with vein density (Brodribb and Jordan, 2011; Carins Murphy et al., 2012, 2016). Here we use the powerful tool of physiological genetics to address the relationships between venation, water transport, and A. A number of mutants, particularly those related to auxin biosynthesis or signaling, have reduced vein densities, aberrant vein topologies, or defective vein formation (Tobeña-Santamaria et al., 2002; Scarpella and Meijer, 2004; Cheng et al., 2006, 2007; Verna et al., 2015). Indeed, the auxin canalization theory, based on observations of a self-organizing flux of auxin that initiates a vascular cambium, effectively predicts the formation and development of leaf vein networks (Sachs 1981; Lee et al., 2014; Rolland-Lagan and Prusinkiewicz, 2005). While the vein-patterning mutant resource has been invaluable for studying both auxin biosynthesis and the role of the auxin stream in leaf vascular formation, no study to date has examined whether these mutations affect vein networks independently of changes in stomatal density or anatomy, and if so, whether these vein defects influence Kleaf, maximum leaf gas exchange, and most importantly, A.
In this study, we characterize mutations at the Crispoid (Crd) locus of pea (Pisum sativum; Swiecicki, 1989; Berdnikov et al., 2000). We show that Crd corresponds to the PsYUC1 gene, a member of the YUCCA gene family known to encode key auxin biosynthesis enzymes (Zhao et al., 2001; Mashiguchi et al., 2011). We comprehensively describe aberrations in the vein morphology, topology, maximum leaf gas exchange, and Kleaf in the crd-4 mutant. We use this mutant to test the hypothesis that maximum A is determined by the distance water must travel through the mesophyll. We show that maximum A is regulated by auxin through leaf venation.
RESULTS
crd Mutants Carry Lesions in the Pea Homolog of AtYUC1/AtYUC4/PhFLOOZY
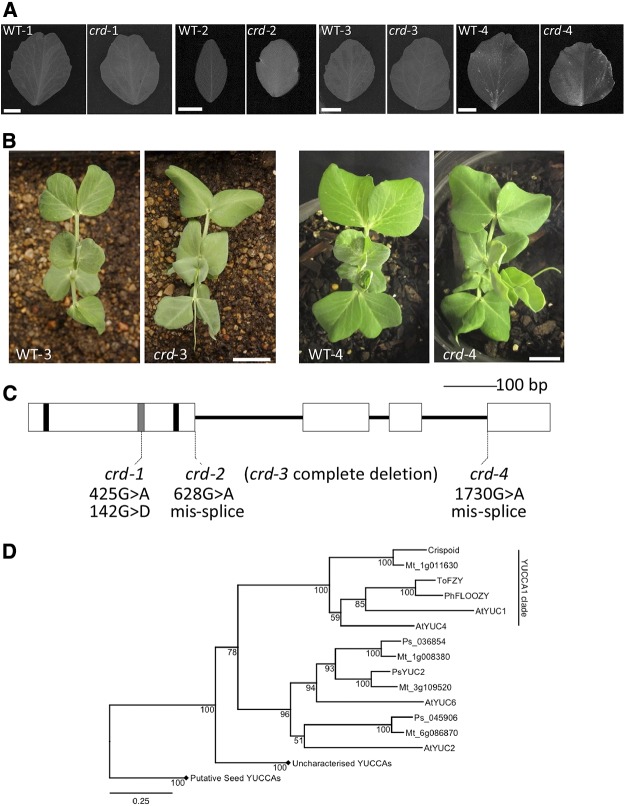
Homozygous recessive crd mutants are easily distinguished from wild-type (Swiecicki, 1989; Berdnikov et al., 2000) plants due to a noticeable reduction in vein density (Fig. 1, A and B; Supplemental Table S1). Two mutants with similar defective venation phenotypes were identified and shown to be additional alleles of the Crd locus by complementation testing (Supplemental Fig. S1). The recessive mutant alleles were named crd-1 (Swiecicki, 1989), crd-2 (Berdnikov et al., 2000), crd-3 (FN 1522/1), and crd-4 (UTILLdb L905). To identify the gene affected in the crd mutants, we used a next-generation RNA sequencing approach, focusing on the identification of any transcripts missing from the crd-3 allele, as most lesions characterized so far from the JI 2822-derived fast-neutron mutant population have been large, gene-sized deletions (Domoney et al., 2013). We assembled a reference transcriptome from the corresponding wild-type line (WT-3) comprising 79,693 contigs (N50 = 1,191 bp), and used this reference transcriptome to map reads from triplicate crd-3 samples. We identified two contigs that showed significant differences in expression between the crd-3 and WT-3 genotypes (false-discovery rate [FDR] corrected P value < 0.05). Only one contig, 435 bp in length, was expressed in WT-3 samples but showed complete absence of expression in crd-3 mutant samples, consistent with a fast neutron-generated deletion. This contig corresponded to the entire last exon and portion of the 3′ UTR of PsYUC1, a previously identified pea homolog of AtYUC1/AtYUC4/PhFLOOZY, encoding a 411-amino acid flavin-containing mono-oxygenase (FMO) protein (Tivendale et al., 2010).
Figure 1.
A, Representative leaflets from node 6 taken from each crd mutant allele and respective wild type. A 1-cm scale marker for each pair is shown in the wild-type image of each pairing. B, Aerial view of seedlings of WT-3 and crd-3 (left) and WT-4 and crd-4 (right) showing node 4 leaves uppermost in each panel. A 2-cm scale marker for each pair of images is shown on the mutant image of each pairing. Similar images or comprehensive phenotypic details of crd-1 and crd-2 seedlings are provided by Swiecicki (1989) and Berdnikov et al. (2000), respectively. C, Diagram of Crd gene showing regions encoding conserved motifs and the positions of crd mutations. Exons are shown as boxes, nucleotides encoding FAD- and NADPH-binding motifs are shown as black bars, ATG-containing motif 1 is shown as a gray bar. D, Phylogenetic analysis of YUCCA protein sequences from M. truncatula (Mt), pea (Ps), and Arabidopsis (At), as well as ToFZY (Solanum lycopersicum) and PhFLOOZY (Petunia hybrida). Bootstrap values (from 1,000 iterations) for this BIONJ tree are displayed adjacent to nodes, and the scale bar indicates genetic distance (0.25 changes/amino acid). Diamonds indicate collapsed clades belonging to the putative but uncharacterized seed YUCCA clades. Accession numbers for all genes are given in Supplemental Table S2.
To test whether PsYUC1 corresponds to the Crd locus, the four known crd mutant alleles were characterized further. We identified a 425G > A SNP in the crd-1 allele (Fig. 1C), which would result in a 142G > D amino acid substitution within the ATG-containing motif 1 conserved between YUCCA proteins (Liu et al., 2012). A single 628G > A SNP was identified in the coding sequence of the crd-2 allele, which is predicted to cause an intron 1 splicing defect (Fig. 1C). Accordingly, PCR analysis of crd-2 cDNA showed several transcripts of different sizes (Supplemental Fig. S2). Sequencing confirmed that the largest of these transcripts contained the unspliced intron 1, which would encode a protein product with a frameshift at position 210 and premature termination after 244 amino acids (Supplemental Fig. S3). A smaller misspliced crd-2 transcript was found to contain 4 bp of intron 1, which would also result in a frameshift at position 210 and premature termination of the protein product after 262 amino acids. No portion of the PsYUC1 gene could be amplified from crd-3 genomic DNA template using PCR primer pairs spanning the full length of the gene, suggesting that crd-3 is a null allele corresponding to complete deletion of PsYUC1 (Supplemental Fig. S4). The crd-4 allele was characterized as a 1730G > A SNP, which would shift the intron 3 splice acceptor site 1 bp downstream relative to WT-4. This would cause a frameshift at position 334 of the protein product, with 27 nonsense amino acids preceding premature termination of the protein.
Crd and PsYUC1 cosegregated in an F3 population of 96 individuals, which is consistent with these representing the same locus. Furthermore, genetic mapping located PsYUC1 on pea LG II, at a position consistent with the Crd locus. On the basis of the strong evidence provided by these RNA-seq data, cosegregation analysis and the significant lesions characterized in four independent alleles, we concluded that the Crd locus corresponds to the PsYUC1 gene. Phylogenetic analysis revealed that of the 12 FMO genes most closely related to AtYUC1/AtYUC4/PhFLOOZY identified in the pea gene atlas (Alves-Carvalho et al., 2015), Crd is the only gene that groups with members of the AtYUC1/AtYUC4/PhFLOOZY/SlFZY clade (Fig. 1D), indicating that Crd is the pea ortholog. This conclusion is further supported by the finding that there is also only a single Medicago truncatula gene, Medtr1g011630, that groups within this clade present in the genome of M. truncatula (Fig. 1D). M. truncatula is a sequenced legume closely related to pea, and their genomes are collinear in this region (Tayeh et al., 2015). Reciprocal BLASTn searches of the M. truncatula genome (v4) and pea transcriptome databases showed that Crd is most similar to Med1g011630; they likely represent an orthologous pair.
Altered Vein Topology in crd Mutants
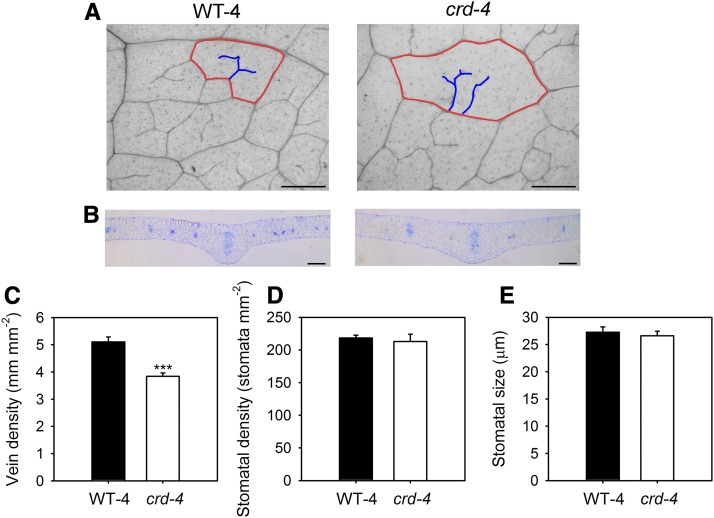
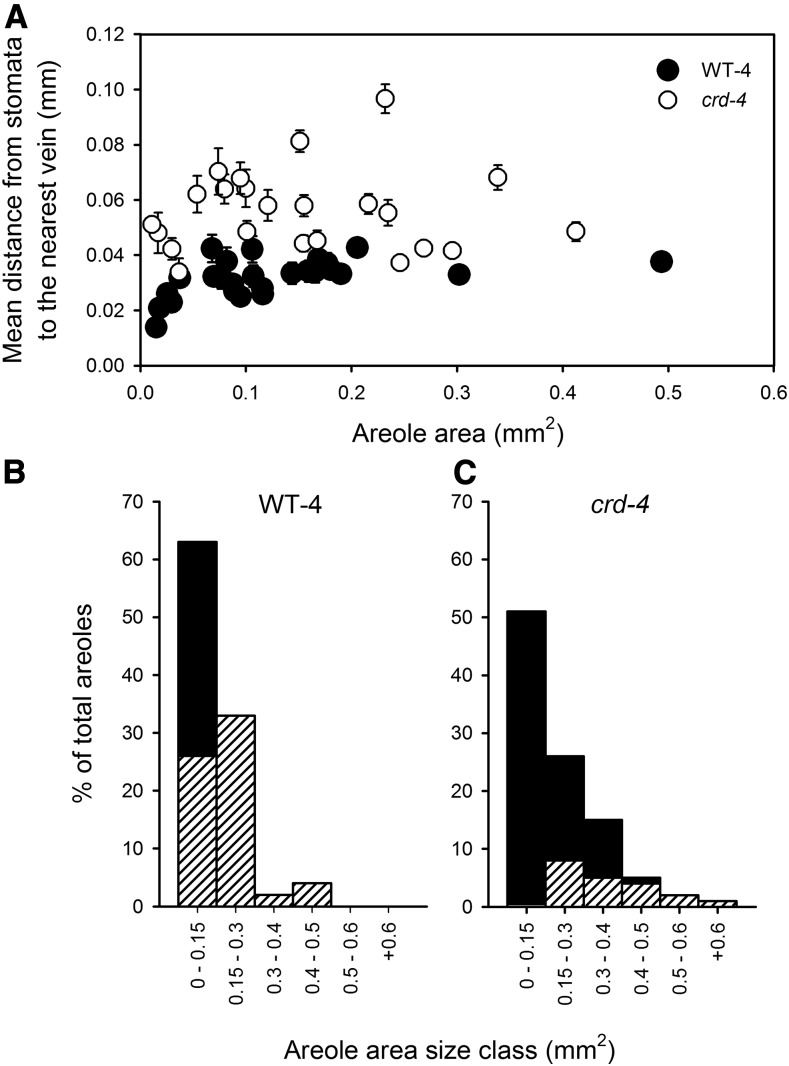
A common feature of all crd mutant alleles is a reduction in vein density at both a macroscopic and microscopic level (Figs. 1, A and B and 2, A and B; Supplemental Table S1). Major vein density is reduced in the mutant alleles, with a significant reduction in the density of either or both secondary and tertiary orders of veins in mutants compared to wild-type leaves (Fig. 1A; Supplemental Table S1). A more comprehensive microscopic examination of crd-4 mutant leaves revealed a 20% reduction in minor vein density compared to WT-4 leaves (Fig. 2C). This reduction is not associated with changes in stomatal density or stomatal size, with both of these traits being the same as in WT-4 plants (Fig. 2, D and E). The placement and development of FEVs in areoles was also severely altered in the crd-4 mutant (Fig. 3). In WT-4 plants, the mean distance from the stomata to the nearest vein showed little variability across the spectrum of areole sizes. In contrast, and consistent with observations of lower vein density, crd-4 mutants had a significantly greater mean distance from the stomata to the nearest vein compared to WT-4 across areole sizes (t test, P < 0.001), as well as a significantly greater variance in the mean distance from stomata to the nearest vein (F-test, P < 0.001; Fig. 3). The development of FEVs was substantially reduced in crd-4 mutant plants compared to WT-4 plants (Fig. 3, B and C), which in addition to a lower vein density contributed to the greater mean distance from stomata to the nearest vein. In WT-4 plants, only 60% of the smallest size class of areoles lacked FEVs, with all areoles larger than 0.15 mm2 having these structures (Fig. 3B). In crd-4 plants, all areoles <0.15 mm2 lacked FEVs, and only areoles >0.5 mm2 consistently developed FEVs in this mutant (Fig. 3C).
Figure 2.
A, A representative longitudinal image of leaf venation in WT-4 and crd-4 leaves (scale bar = 200 µm, red lines outline an areole, blue lines highlight FEVs within the areole). B, A representative cross-sectional image of WT-4 and crd-4 mutant leaves through the midrib (scale bar = 200 µm). C, Mean vein density (n = 7 leaves, ±se), D, stomatal density (n = 7 leaves, ±se), and E, stomatal size (n = 50 stomata, ±se) in WT-4 and crd-4 mutant plants. Asterisks denote significant difference in means (P < 0.001).
Figure 3.
A, The relationship between the mean distance from stomata to the nearest vein (including FEV) in an areole (±se) and areole area for WT-4 (white circles) and crd-4 mutant (black circles) plants. FEV occurrence across size classes of areoles in WT-4 (B) and crd-4 (C) mutant plants; bars represent the percentage of areoles that fall within each size class, with hatching depicting the areoles in that size class that had FEVs.
Leaf Hydraulic Capacity and Gas Exchange in crd Mutants
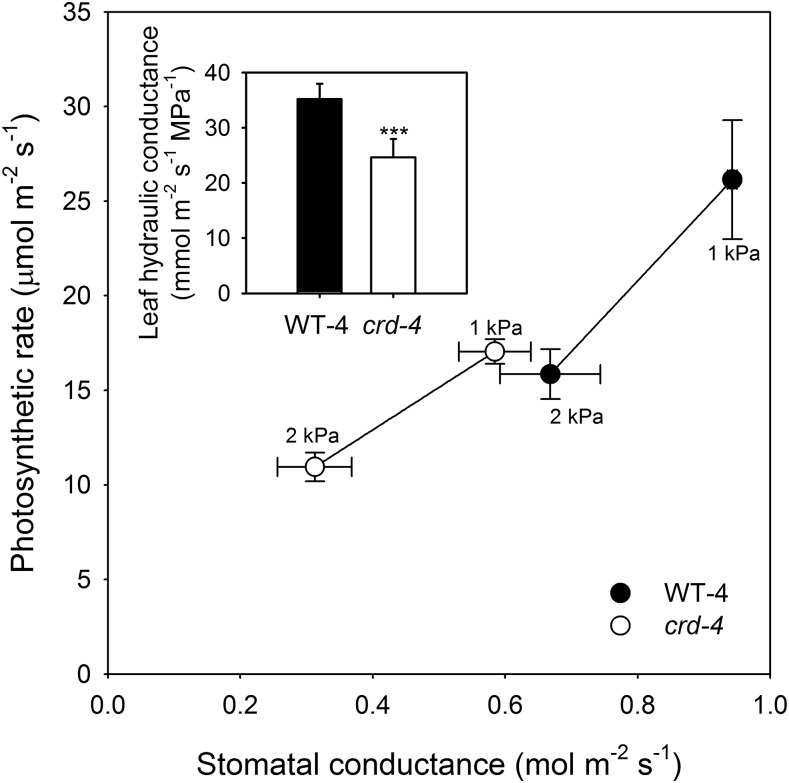
Maximum Kleaf was reduced in crd-4 mutant plants, being 30% lower than in WT-4 plants (Fig. 4). Leaf gas exchange was also significantly reduced in the crd-4 mutant plants compared to WT-4 plants at both high and low vapor pressure deficit (VPD; Fig. 4). Leaf gas exchange parameters including A and gs in WT-4 plants were double that of crd-4 plants at low VPD and significantly higher at high VPD (Fig. 4). Photosynthetic apparatus was not compromised by the crd-4 mutation, with both WT-4 and crd-4 plants having similar A-internal CO2 concentration (Ci) relationships (Supplemental Fig. S5). The lower maximum A observed in crd-4 mutant plants is likely to have contributed to a reduction in shoot biomass observed in these plants compared to WT-4 plants (Supplemental Fig. S6). Other factors may contribute to reduced growth in the crd mutants, which, in addition to altered vein anatomy, have an occasional reduction or modification in tendril number as well as, rarely, leaflet/pinna number (Supplemental Figs. S7 and S8).
Figure 4.
Mean photosynthetic rate and stomatal conductance (n = 3; ±se) measured in leaves of whole plants exposed to a VPD of 1 or 2 kPa in WT-4 (black circles) and crd-4 mutants (white circles). Insert depicts mean maximum leaf hydraulic conductance (n = 6; ±se) in WT-4 and crd-4 mutant plants. Asterisks denote a significant difference in means (P < 0.001).
Mutation in Crd Reduces Auxin Activity and Levels in Expanding Leaves
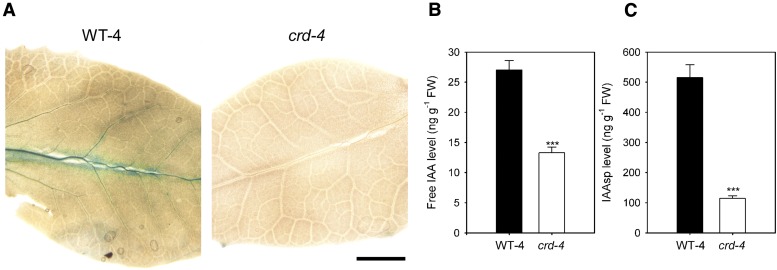
FMO enzymes are believed to catalyze the conversion of indole-3-pyruvic acid to auxin (indole-3-acetic acid [IAA]; Mashiguchi et al., 2011). To visualize potential differences in auxin activity in the developing leaves of crd-4 and WT-4 plants, we utilized a DR5::GUS reporter construct, for which GUS expression is driven by the promoter of a highly active synthetic auxin response element (DeMason and Polowick, 2009). A substantial reduction in auxin activity was noted in the veins of developing leaves of crd-4 DR::GUS plants compared with those of WT-4 DR5::GUS plants, by minimal GUS staining (Fig. 5A). In contrast to WT-4 leaves, no GUS staining was observed in young leaves (<4 mm in length) of crd-4 DR5::GUS plants, indicating reduced auxin activity (Supplemental Fig. S9). We further investigated this difference in auxin activity in developing leaves by quantifying the levels of both free IAA and the auxin conjugate, IAA-Asp (IAAsp), in apical tissue containing developing leaves and meristematic tissue. Free IAA and IAAsp levels were reduced by approximately 50% and 80%, respectively, in the apical tissue of crd-4 plants compared to WT-4 plants (Fig. 5, B and C). In contrast, a reduction in free IAA levels was not observed in whole shoots (Supplemental Fig. S10).
Figure 5.
A, GUS expression in expanding pinnae of representative F2 segregant plants (<8 mm in length) taken from WT-4, DR5::GUS, and crd-4, DR5::GUS plants (scale bar = 2 mm). B, Mean levels of IAA and IAAsp (C) from apical tissue of WT-4 and crd-4 mutant plants (n = 4, ±se). Asterisks denote a significant difference in means (P < 0.001).
DISCUSSION
Auxin via Vein Anatomy Drives Maximum Leaf Gas Exchange
Current theory indicates that the terminal path length for water flow through the mesophyll should govern maximum Kleaf (Buckley et al., 2015), gs, and thus maximum A (Brodribb et al., 2010). However, both parts of this proposition are supported largely by correlative evidence, derived from natural variation within and between species (Sack and Frole, 2006; Brodribb et al., 2007; Brodribb and Feild, 2010; Scoffoni et al., 2015, 2016). Recently, the influence of the distance through the mesophyll for water flow on Kleaf was demonstrated experimentally using a genetic approach (Caringella et al., 2015), and here we present genetic evidence that auxin, through leaf venation, acts as a regulator of both maximum leaf hydraulic supply and maximum A, and likely whole plant productivity. Our physiological analyses of the auxin-biosynthetic crd mutants indicate that altered vein density and topology results in an increase in the hydraulic path length for water flow through the mesophyll, reduced maximum Kleaf, and as a consequence maximum A. Our data support the conclusions derived from correlations that have highlighted the importance of an efficient internal leaf water transport system for maximizing both gs and A (Sack and Frole, 2006; Brodribb et al., 2007; Carins Murphy et al., 2012).
The effects of auxin deficiency on water transport, and consequently A, observed in the crd-4 mutant occurred independently of any changes in stomatal anatomy, photosynthetic apparatus, or intrinsic photosynthetic capacity. This breaking of coordination between vein and stomatal development is surprising, given that angiosperms are extremely efficient at coordinating hydraulic supply (the formation of veins) with demand (the density of stomata; Brodribb and Jordan, 2011; Carins Murphy et al., 2016). The absence of a distinctive stomatal phenotype in the crd-4 mutant plants is at odds with the suggestion that auxin transport influences stomatal development (Le et al., 2014). Interestingly, there have been no reports of aberrant vein development in the collection of stomatal patterning mutants (Berger and Altmann, 2000; Kanaoka et al., 2008), which suggests independent pools of auxin must be involved in the developmental regulation of these two morphological features. The endogenous signal that may be responsible for coordinating vein and stomatal density in a developing leaf thus remains unknown.
The Formation of FEVs
In addition to providing experimental evidence for the link between maximum Kleaf and A, our observations of the crd-4 mutant revealed substantial rearrangements in the topology of the leaf vein network. We found that, unlike in WT-4 plants, both major and minor vein density were reduced in mutant plants and, in contrast to a wide diversity of angiosperms species (Fiorin et al., 2016), the development of FEVs in both large and small areoles in crd-4 plants was affected, as evidenced by a greater distance from stomata to the nearest vein in all size classes of areoles. Recent work has suggested that a strong selective pressure is placed on plants to develop leaves with a uniform distance from the nearest vein to the stomata (Fiorin et al., 2016). The correct positioning of FEVs minimizes the mean distance from the minor veins to the nearest stomata. The mechanism regulating this key anatomical feature of reticulate veined angiosperm leaves is unknown; however, auxin clearly plays a major role.
Auxin Biosynthesis as a Central Regulator in the Formation of Veins
Auxin biosynthesis in plants occurs through a two-step pathway from the amino acid Trp to IAA via the intermediate indole-3-pyruvic acid (Zhao, 2012). These two steps are sequentially catalyzed by Trp aminotransferases and FMOs from the YUCCA family (Tao et al., 2008; Mashiguchi et al., 2011). We show here that Crd is the only transcript recovered from pea database searches that is embedded within the clade of FMO auxin biosynthetic genes, including YUCCA1 and YUCCA4 from Arabidopsis and FLOOZY from Petunia (Petunia hybrida; Fig. 1D). Consistent with this, we found reduced free IAA levels in crd-4 apical tissue when compared with WT-4 plants (Fig. 5B). This finding demonstrates the importance of local auxin synthesis in the early stages of leaf and vascular development. In addition to reduced IAA levels, our observation of substantially lower levels of IAAsp in the apical tissue of crd-4, compared to WT-4 plants, is indicative of impaired IAA biosynthesis in the apical bud. Interestingly, we observed no difference in free IAA levels in whole shoot tissue of seedlings of the crd-4 mutant plants compared to WT-4 plants (Supplemental Fig. S10), suggesting the reduction in auxin levels in these mutants is localized to the developing leaves. Our DR5::GUS reporter observations further support this, indicating that a reduction in IAA levels may be localized to the developing vascular tissue in expanding leaves of mutant plants (Fig. 5A). In wild-type plants, high auxin activity in vascular tissue of developing leaves has been reported previously in pea (Demason et al., 2013) as well as Arabidopsis (Mattsson et al., 2003); in crd-4 leaves we found no GUS staining apparent in the developing veins (Fig. 5A). Similar reductions in GUS staining have been observed in the veins of multiple yuc mutants in Arabidopsis lines carrying a DR5::GUS reporter (Cheng et al., 2006). Furthermore, systemic reductions in auxin activity were also noted in very young developing leaves of crd-4 mutant plants (Supplemental Fig. S9).
The development of the leaf vascular network can be explained by the auxin canalization theory, which describes a self-forming auxin transport stream initiating the formation of a vascular cambium (Sachs 1981). The profound and highly dysfunctional vein phenotypes of mutants in PIN-FORMED proteins strongly implicate auxin efflux as a major regulator of this auxin transport stream and thus the formation of leaf veins (Mattsson et al., 1999). As a result, modifications to the auxin transport stream by auxin efflux dominates models seeking to simulate vein patterning (Bennett et al., 2014; Feugier et al., 2005; Lee et al., 2014; Mitchison, 1980; Rolland-Lagan and Prusinkiewicz, 2005). Our results, however, add to previous evidence suggesting that, in addition to auxin efflux rates, the biosynthesis of this hormone also plays an important role in establishing a normal leaf vascular network (Tobeña-Santamaria et al., 2002; Cheng et al., 2006, 2007). Indeed, Cheng et al. (2007) found that auxin deficiency on a background of reduced auxin efflux had an additive effect on compromised leaf vascular development. Whether current models for leaf venation patterning, based primarily on auxin efflux dynamics, can mimic venation patterns observed in auxin biosynthetic mutants or require substantial modification will be an interesting avenue for future research.
Auxin as a Driver of Maximum Photosynthetic Rate
The importance of Crd, an FMO, in normal vein formation and thus maximum A suggests that this clade of auxin biosynthetic genes has been central to the formation of complex, high-density vein networks in the leaves of derived angiosperms, a prerequisite for the high productivity of these species (Scoffoni et al., 2016). Phylogenetic analysis of closely related FMOs suggests that duplication and functional specialization in this family occurred prior to and during the diversification of angiosperms, allowing for the evolution of a clade of genes that play key roles in the development and topology of veins. This clade includes Crd, PhFLOOZY, AtYUCCA1, and AtYUCCA4, which all result in compromised leaf vascular development when mutated (Tobeña-Santamaria et al., 2002; Cheng et al., 2006, 2007). Given the importance of auxin in the development of efficient reticulate venation patterns in angiosperms, it is an intriguing possibility that this hormone may have played an instrumental role in the Cretaceous radiation and ecological rise of the angiosperms. Indeed, as the hydraulic pathway through the leaf accounts for the greatest limitation to maximum realized leaf gas exchange, whether enhancing auxin levels in a developing leaf will improve maximum productivity remains to be tested. However, our new mechanistic understanding of auxin as a key developmental determinant of maximum leaf gas exchange provides exciting potential for future increases in plant productivity.
MATERIALS AND METHODS
Plant Material
Previously described crd mutants include the first described allele (crd-1; JI 2460) selected after N-ethyl-N-nitrosourea mutagenesis of the line Paloma (WT-1; Swiecicki, 1989) and the crispoid-whip allele (crd-2; JI 3160), found after ethyl methane sulfonate mutagenesis of the line SG (WT-2; Berdnikov et al., 2000). These mutants were obtained from the John Innes Pisum Germplasm collection. In this paper, two novel crd mutant alleles were identified. The crd-3 mutant was identified as line FN 1522/1 in a fast neutron mutagenesis population of the JI 2822 line (WT-3; Domoney et al., 2013). The crd-4 mutant was observed incidentally in line 905, derived from an ethyl methane sulfonate mutagenesis population of the Caméor line in the UTILLdb program (Dalmais et al., 2008). Initially this mutant was named pipes in short supply (pss). Allelism was confirmed by noncomplementation of the crd phenotype in F1 progeny from the crosses of FN1522/1 × crd1, FN1522/1 × crd2, and pss × crd1 (Supplemental Fig. S1). At least six generations of backcross selection were undertaken to provide near-isogenic wild-type (WT-4) and crd-4 lines for morphological and physiological experiments. Three generations of backcross selection were undertaken to provide crd-3 lines for RNA sequencing.
crd-1, crd-2, crd-3, and respective wild-type plants (as well as plants from allelism tests between these lines) were sown in a glasshouse in Aberystwyth, UK, under natural light between June and September (5.2–9.5/18.2–22.4°C day/night temperatures) in peat-free Jiffy pellets 7C, 45 × 40 mm (Jiffy products S.L. (pvt) Ltd) and after 2 weeks transferred into 12-cm-long and 12-cm-diameter pots with John Innes potting mix number 3 (http://www.johninnes.info/about.htm) supplemented with 30% chick grit. Jiffy pellets and pots were placed on a carpet that was watered every day, allowing the plants to absorb water through the carpet. WT-4 and crd-4 plants (and plants for the allelism test with this line, as well as DR5::GUS reporter lines) were grown under the controlled glasshouse conditions in Hobart, Australia, described by McAdam et al. (2016). All morphological and physiological observations were made on fully expanded node 4 leaves, unless specified otherwise.
RNA-Sequencing and Sequence Data Analysis to Identify the crd-3 Mutation
Total RNA from 20-d-old crd-3 and WT-3 shoot tips (three replicates each) was extracted using the RNeasy Plant Mini Kit (Qiagen), and RNA concentration was determined using a Qubit fluorescence spectrophotometer (Thermo Fischer). Each amplified library for sequencing was prepared from 2 μg of total RNA (Illumina TruSeq RNA v2 Sample Preparation Kit) and uniquely indexed to enable multiplex sequencing. Libraries were analyzed by gel electrophoresis and quantified using Qubit and then adjusted to 10 nm in 0.1 m Tris-HCl/0.01% (v/v) Tween 20 buffer and pooled equally prior to denaturation/dilution in a final loading concentration of 8 pm. Uniquely indexed, amplified libraries prepared for RNA-seq were typically 300 bp in size. These were multiplexed and sequenced in 2 × 126-bp format in a single lane of a HiSeq v4 high-output flowcell on an Illumina HiSeq2500 platform. Data in FASTQ format were analyzed using the Genomics Workbench v6.5 software package (CLC Bio). RNA-seq data obtained ranged from 68.3 to 79.5 million reads per sample for crd-3 (average 73 million) and from 73 to 90 million reads for WT-3 (average 80.4 million). Following trimming to remove low-quality sequence and reads <50 bp, these were reduced to an average of 71.4 million reads for crd-3 and 78.9 million reads for WT-3, with an average read length of 113 bp for all samples. Overlapping WT-3 read-pairs were merged (approximately 54% of each wild-type dataset), and these data were used along with unmerged WT-3 read-pairs (21.85 Gbp in total) for de novo assembly of a reference transcriptome using the de Bruijn graph method with a k-mer value of 25. Maximum bubble size for conflict resolution within the graph was set at 50. Repeat regions within the graph were resolved using scaffolding based on paired-end sequences. Following initial contig assembly, reads were mapped back to contigs, requiring 50% identity and 80% similarity across the read. Ambiguous reads mapping to more than one contig were discarded. Insertion and deletion penalties were set at 3 and mismatch penalty at 2. Contigs from the initial assembly were removed if no reads mapped. Reads from all six individual samples were subsequently mapped to these reference contigs and used to calculate the number of reads mapped per kb of exon, per million mapped reads (RPKM; Mortazavi et al., 2008). On average, 85% of reads from each sample could be mapped to reference contigs and used to generate RPKM expression values. RPKM values were analyzed as a two-group experiment, crd-3 versus WT-3, to identify significantly different expression values between the two groups, with P values adjusted using a FDR (Benjamini and Hochberg, 1995). One significantly different (FDR-corrected P value < 0.05) contig sequence, showing complete absence of expression in crd-3 mutants, was used to query NCBI nucleotide and protein databases, using BLASTn and BLASTx, respectively, to identify the corresponding gene.
Sanger Sequencing for Mutant Characterization
For cDNA template preparation, total RNA was first extracted as for RNA sequencing and treated with DNase (Ambion; WT-1 to -3, crd-1 to -3), or with the SV total RNA isolation system (Promega) including a DNase step (WT-4, crd-4), and quality-checked by gel electrophoresis. RNA was reverse-transcribed using oligo(dT) primers and the Superscript III First-Strand Synthesis System (Invitrogen) in a total volume of 50 μL containing 0.4 μg RNA (WT-1 and crd-1) or 0.2 μg RNA (WT-2 and crd-2; WT-3 and crd-3), or using the Tetro cDNA synthesis kit (Bioline) in 20 μL containing 1 μg RNA (WT-4 and crd-4). Genomic DNA template was extracted from leaves following the DNeasy Plant Mini Kit protocol (Qiagen; WT-1 to -3, crd-1 to -3) or using a CTAB-based method (WT-4, crd-4; Allen et al., 2006). All primers are listed in Supplemental Table S3. Sanger sequencing of purified PCR products was performed at Aberystwyth University or by Macrogen Inc. (Korea).
Genetic Analysis
Plants grown in the glasshouse were scored for crd or wild-type phenotype. A WT-1 × crd-1 cross yielded 96 F3 progeny segregating 74:22 (3:1, χ2 = 0.2, P > 0.6, n.s.). To test for cosegregation of the crd phenotype and the crd-1 mutation, phenotype and genotype scores of the F3 progeny were compared. Genomic DNA prepared from each plant was genotyped using a cleaved-amplified polymorphic sequence marker for the Crd locus, amplified by PCR using primers 5F and 7R in Supplemental Table S1 and digested with DdeI. Digestion products were distinguished after gel electrophoresis (WT-1: 385 bp, 147 bp and 110 bp; crd-1: 385 bp and 257 bp). All plants homozygous for the crd-1 allele had the crd phenotype, and no heterozygous plant or any plants homozygous for the WT-1 allele had the crd phenotype (χ2= 93.2, P = 6 × 10−21). The same cleaved-amplified polymorphic sequence marker was used to place the Crd locus on the genetic map of the JI 281 × JI 399 recombinant inbred population (Laucou et al., 1998) at position 130 cm on LG II, between markers cDNA39 and SNP190.
Phylogenetic Analysis
The phylogenetic relationships between YUCCA-related genes in Arabidopsis (Arabidopsis thaliana), Medicago truncatula, and pea (Pisum sativum) were assessed in conjunction with selected characterized genes from other species using amino acid sequences obtained from GenBank (www.ncbi.nlm.nih.gov) and the pea gene atlas (Alves-Carvalho et al., 2015; Supplemental Table S2). Sequences were aligned using the Clustal Omega algorithm (Sievers et al., 2011), and phylogenies were reconstructed using an LG substitution model (Le and Gascuel, 2008) based on a BIONJ tree using the statistical analysis software R. The packages APE (Paradis et al., 2004) and phangorn (Schliep, 2011) were used for sequence management and modeling, and ggtree (Yu et al., 2017) was used for tree visualization.
Leaf Anatomy
Leaves of crd-1, crd-2, and crd-3 mutant lines and respective wild types were prepared for vein density measurements by incubating overnight in a solution of 3:1 ethanol:acetic acid (v/v) at room temperature. Leaves were then incubated for 1 h in a solution of 70% ethanol in water (v/v) and later transferred to 50% ethanol in water (v/v) until scanned at 600 to 1200 DPI (HP Scanjet 8200).
Paradermal sections of WT-4 and crd-4 leaves were cleared in household bleach (50 g/L sodium hypochlorite and 13 g/L sodium hydroxide) and stained in 1% toluidine blue. Five fields of view (FOV) at 4× magnification (for vein density measurements, FOV: 3.47 mm2), five FOV at 20× magnification (for stomatal density measurements), sufficient images at 40× magnification to capture 50 stomata (for measuring stomatal size), and images at 20× magnification focusing on the abaxial stomatal surface and covering the entire areole (for calculating mean stomatal distance to the nearest vein) were taken using a Nikon Digital Sight DS-L1 camera mounted on a Leica DM 1000 microscope. Vein density and stomatal density were measured using ImageJ, as the total length of leaf vascular tissue and total number of stomata per mm2 of leaf area. Stomatal size was determined by measuring the longest distance parallel to the stomatal pore from the edges of the guard cells using ImageJ. The mean distance from stomata to the nearest vein for 24 areoles, randomly selected to span the full-size spectrum of areoles across the leaf, was determined by making a composite image of each areole using the 20× objective FOVs, which were then assembled together in Adobe Photoshop. This composite image of each areole was analyzed using specifically designed software that calculated the mean distance to the nearest vein for each stomata in the areole once veins and stomata were manually marked. Leaf cross-sections were made from leaf segments spanning the midrib that were fixed in 2.5% buffered glutaraldehyde under vacuum then dehydrated in acetone and embedded in Spurr’s resin. Semithick sections (5–7 µm) were stained with toluidine blue O.
Leaf Gas Exchange and Kleaf
One leaf from three individuals of both WT-4 and crd-4 were used to determine the response of leaf gas exchange to variation in VPD. A portable infrared gas analyzer (Li-6400; Li-Cor Biosciences) was used to measure leaf gas exchange between 1130 and 1300 h when gas exchange was expected to be maximal. Conditions within the leaf cuvette were maintained as close to external conditions as possible (temperature at 22°C and light intensity at 1,000 µmol m−2 s−1, VPD was regulated relative to external conditions by controlling intake air flow through a desiccant column). On the first day of measurement, VPD in the glasshouse was held at 1 kPa from dawn by a dehumidifier with integrated humidity sensors (ADH-1000, Airrex Portable dehumidifier, Hephzibah Co. Ltd.); on the second day of measurement, VPD in the glasshouse was held at 2 kPa. On each day, midday leaf water potential was measured in all plants using a pressure chamber. Kleaf was measured in three leaves of WT-4 and crd-4 plants using the evaporative flux method and flowmeter according to Carins Murphy et al. (2012).
Shoot Biomass and Growth
Whole shoots of WT-4 and crd-4 plants (n = 3), grown under the same glasshouse conditions, were harvested 28 d after sowing and dried at 70°C for at least 62 h before being weighed (± 0.0001 g). Plant height in WT-1, crd-1, WT-3, and crd-3 plants (n = 13–14), grown under the same glasshouse conditions, was measured every 10 d (from 20 d after sowing) until plants had dried at 60 d after sowing.
Quantification of Free IAA and IAAsp Levels
Growing apical tissue, including all tissue immediately proximal to the uppermost fully expanded leaf of WT-4 and crd-4 plants, was harvested. Tissue was weighed (± 0.0001 g; with approximately 25 mg harvested per replicate for both genotypes) and placed in 1 mL of sodium phosphate buffer (50 nm, pH7). Samples were homogenized with a bead mill for 2 min. To quantify the endogenous compounds, stable isotope-labeled internal standards were added; these were [13C6]-IAA (Cambridge Isotope Laboratories) and [15N, 2H5]-IAAsp (OlChemim). Compounds were extracted according to Novák et al. (2012). In addition, free auxin was quantified in two-week-old whole seedlings of WT-4 and crd-4, which were weighed and harvested into 3 mL of 80% methanol in water (v/v) containing 250 mg/L butylated hydroxytoluene. All samples were analyzed by ultra-performance liquid chromatography tandem mass spectrometry as described by Cook et al. (2016).
DR5::GUS Staining for Auxin Activity
Transgenic DR5::GUS lines carrying the WT-4 and crd-4 alleles were constructed by crossing the RTP9 line (DeMason and Polowick, 2009) with crd-4 mutant pollen. The segregating F2 generation consisting of 42 Crd GUS+: 15 crd GUS+: 14 Crd GUS-: 7 crd GUS- (9:3:3:1, χ2 = 1.0, P = 0.8, n.s.) was genotyped by PCR for DR5::GUS using the primers and conditions described by DeMason and Polowick (2009), and distinctive leaf morphology phenotypes were used to identify crd-4 plants. Young developing leaflets <8 mm in length (one per individual, harvested from eight randomly selected individuals per genotype) were removed from within stipules enclosing the apical meristem and immediately immersed in GUS staining buffer containing 2 mm 5-bromo- 4-chloro-3-indolyl b-d-glucuronide, 100 mm sodium phosphate (pH 7.5), 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, 10 mm EDTA, and 0.1% (v/v) Triton X-100. Samples were vacuum-infiltrated on ice at 200 mbar for 45 min then incubated for 48 h at 37°C. The staining buffer was refreshed after 24 h. The samples were then cleared in 70% ethanol, dissected, and photographed as described above. To ensure an unbiased methodological approach and morphological assessment of GUS staining in the leaves, the samples were stained, cleared, and photographed without prior knowledge of the genotype using a double-blind methodology. After decoding the genotypes, staining was compared in three randomly selected leaflets of <4 mm in length from crd-4 and wild-type plants carrying the DR5::GUS reporter in addition to a randomly selected leaflet <8 mm in length from each of the two genotypes.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers shown in Supplementary Table S2.
Supplemental Materials
The following supplemental materials are available.
Supplemental Figure S1. Representative images of leaves or leaflets from F1 plants of crosses between putative mutant lines carrying the crd phenotype.
Supplemental Figure S2. The G628A SNP in crd-2 affects PsYUC1 splicing.
Supplemental Figure S3. Alignment of Crispoid protein from three wild-type and mutant lines.
Supplemental Figure S4. No portion of Crispoid/PsYUC1 could be amplified from crd-3 mutant genomic DNA template.
Supplemental Figure S5. Photosynthetic rate/internal CO2 concentration curves for WT-4 and crd-4 mutant plants.
Supplemental Figure S6. Mean dry shoot biomass and plant height in wild-type and crd plants.
Supplemental Figure S7. Missing and underdeveloped tendrils in the crd-1 mutant leaves.
Supplemental Figure S8. Missing and underdeveloped tendrils and pinnae in leaves of the crd-3 mutant.
Supplemental Figure S9. GUS expression in F2 segregant plants.
Supplemental Figure S10. Mean levels of free IAA from whole aerial potions of 14-d-old seedlings of WT-4 and crd-4 mutant plants.
Supplemental Table S1. Mean secondary and tertiary vein densities in leaves of allelic crd mutants and respective wild-types.
Supplemental Table S2. Accession numbers of proteins used for phylogenetic analysis.
Supplemental Table S3. Details of Crd primers used in this study.
Acknowledgments
The authors thank Charly Potter for preparation of libraries for RNA-seq, Caron James for Sanger sequencing, Vincent Vadez for helpful discussions, Patricia Polowick for generously providing the RTP9 line carrying the DR5::GUS insert, Noel Ellis for statistical advice and comments on the manuscript, and two reviewers for astute recommendations on auxin quantifications.
Glossary
- FEV
free-ending veinlet
- FOV
fields of view
- IAA
indole-3-acetic acid
- IAAsp
indole-3-acetic acid-Asp
- RPKM
reads mapped per kb of exon, per million mapped reads
- VPD
vapor pressure deficit
Footnotes
The TILLING platform was supported by the ERC Advanced Grant SEXYPARTH (M.D.). This work was supported by the Australian Research Council (grant nos. DE140100946 [S.A.M.M.], DP140100666 [T.J.B.], and DP130103357 [J.J.R.]).
Articles can be viewed without a subscription.
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1: 2320–2325 [DOI] [PubMed] [Google Scholar]
- Alves-Carvalho S, Aubert G, Carrère S, Cruaud C, Brochot A-L, Jacquin F, Klein A, Martin C, Boucherot K, Kreplak J, et al. (2015) Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant J 84: 1–19 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bennett T, Hines G, Leyser O (2014) Canalization: what the flux? Trends Genet 30: 41–48 [DOI] [PubMed] [Google Scholar]
- Berdnikov VA, Gorel’ FL, Kosterin OE (2000) A new allele at Crd distrubs development of the compound leaf. Pisum Genet 32: 6–8 [Google Scholar]
- Berger D, Altmann T (2000) A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev 14: 1119–1131 [PMC free article] [PubMed] [Google Scholar]
- Boyce CK, Brodribb TJ, Feild TS, Zwieniecki MA (2009) Angiosperm leaf vein evolution was physiologically and environmentally transformative. Proc Biol Sci 276: 1771–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodribb TJ, Feild TS (2000) Stem hydraulic supply is linked to leaf photosynthetic capacity: evidence from New Caledonian and Tasmanian rainforests. Plant Cell Environ 23: 1381–1388 [Google Scholar]
- Brodribb TJ, Feild TS (2010) Leaf hydraulic evolution led a surge in leaf photosynthetic capacity during early angiosperm diversification. Ecol Lett 13: 175–183 [DOI] [PubMed] [Google Scholar]
- Brodribb TJ, Feild TS, Jordan GJ (2007) Leaf maximum photosynthetic rate and venation are linked by hydraulics. Plant Physiol 144: 1890–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodribb TJ, Feild TS, Sack L (2010) Viewing leaf structure and evolution from a hydraulic perspective. Funct Plant Biol 37: 488–498 [Google Scholar]
- Brodribb TJ, Holbrook NM (2005) Water stress deforms tracheids peripheral to the leaf vein of a tropical conifer. Plant Physiol 137: 1139–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodribb TJ, Holbrook NM, Zwieniecki MA, Palma B (2005) Leaf hydraulic capacity in ferns, conifers and angiosperms: impacts on photosynthetic maxima. New Phytol 165: 839–846 [DOI] [PubMed] [Google Scholar]
- Brodribb TJ, Jordan GJ (2011) Water supply and demand remain balanced during leaf acclimation of Nothofagus cunninghamii trees. New Phytol 192: 437–448 [DOI] [PubMed] [Google Scholar]
- Buckley TN, John GP, Scoffoni C, Sack L (2015) How does leaf anatomy influence water transport outside the xylem? Plant Physiol 168: 1616–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caringella MA, Bongers FJ, Sack L (2015) Leaf hydraulic conductance varies with vein anatomy across Arabidopsis thaliana wild-type and leaf vein mutants. Plant Cell Environ 38: 2735–2746 [DOI] [PubMed] [Google Scholar]
- Carins Murphy MR, Jordan GJ, Brodribb TJ (2012) Differential leaf expansion can enable hydraulic acclimation to sun and shade. Plant Cell Environ 35: 1407–1418 [DOI] [PubMed] [Google Scholar]
- Carins Murphy MR, Jordan GJ, Brodribb TJ (2016) Cell expansion not cell differentiation predominantly co-ordinates veins and stomata within and among herbs and woody angiosperms grown under sun and shade. Ann Bot (Lond) 118: 1127–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SD, Nichols DS, Smith J, Chourey PS, McAdam EL, Quittenden L, Ross JJ (2016) Auxin biosynthesis: are the indole-3-acetic acid and phenylacetic acid biosynthesis pathways mirror images? Plant Physiol 171: 1230–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmais M, Schmidt J, Le Signor C, Moussy F, Burstin J, Savois V, Aubert G, Brunaud V, de Oliveira Y, Guichard C, et al. (2008) UTILLdb, a Pisum sativum in silico forward and reverse genetics tool. Genome Biol 9: R43–R43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demason DA, Chetty V, Barkawi LS, Liu X, Cohen JD (2013) Unifoliata-Afila interactions in pea leaf morphogenesis. Am J Bot 100: 478–495 [DOI] [PubMed] [Google Scholar]
- DeMason Darleen A, Polowick Patricia L (2009) Patterns of DR5:GUS expression in organs of pea (Pisum sativum). Int J Plant Sci 170: 1–11 [Google Scholar]
- Domoney C, Knox M, Moreau C, Ambrose M, Palmer S, Smith P, Christodoulou V, Isaac PG, Hegarty M, Blackmore T, et al. (2013) Exploiting a fast neutron mutant genetic resource in Pisum sativum (pea) for functional genomics. Funct Plant Biol 40: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Feugier FG, Mochizuki A, Iwasa Y (2005) Self-organization of the vascular system in plant leaves: inter-dependent dynamics of auxin flux and carrier proteins. J Theor Biol 236: 366–375 [DOI] [PubMed] [Google Scholar]
- Fiorin L, Brodribb TJ, Anfodillo T (2016) Transport efficiency through uniformity: organization of veins and stomata in angiosperm leaves. New Phytol 209: 216–227 [DOI] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu J-K, Torii KU (2008) SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laucou V, Haurogné K, Ellis N, Rameau C (1998) Genetic mapping in pea. 1. RAPD-based genetic linkage map of Pisum satium. Theor Appl Genet 97: 905–915 [Google Scholar]
- Le J, Liu X-G, Yang K-Z, Chen X-L, Zou J-J, Wang H-Z, Wang M, Vanneste S, Morita M, Tasaka M, et al. (2014) Auxin transport and activity regulate stomatal patterning and development. Nat Commun 5: 3090. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Lee S-W, Feugier FG, Morishita Y (2014) Canalization-based vein formation in a growing leaf. J Theor Biol 353: 104–120 [DOI] [PubMed] [Google Scholar]
- Liu H, Ying Y-Y, Zhang L, Gao Q-H, Li J, Zhang Z, Fang J-G, Duan K (2012) Isolation and characterization of two YUCCA flavin monooxygenase genes from cultivated strawberry (Fragaria × ananassa Duch.). Plant Cell Rep 31: 1425–1435 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J, Ckurshumova W, Berleth T (2003) Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol 131: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J, Sung ZR, Berleth T (1999) Responses of plant vascular systems to auxin transport inhibition. Development 126: 2979–2991 [DOI] [PubMed] [Google Scholar]
- McAdam SAM, Brodribb TJ, Ross JJ (2016) Shoot-derived abscisic acid promotes root growth. Plant Cell Environ 39: 652–659 [DOI] [PubMed] [Google Scholar]
- Mitchison GJ. (1980) Model for vein formation in higher-plants. Proc R Soc Lond B Biol Sci 207: 79–109 [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K (2012) Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J 72: 523–536 [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290 [DOI] [PubMed] [Google Scholar]
- Rolland-Lagan AG, Prusinkiewicz P (2005) Reviewing models of auxin canalization in the context of leaf vein pattern formation in Arabidopsis. Plant J 44: 854–865 [DOI] [PubMed] [Google Scholar]
- Sachs T. (1981) The control of the patterned differentiation of vascular tissue. Adv Bot Res 9: 151–262 [Google Scholar]
- Sack L, Cowan PD, Jaikumar N, Holbrook NM (2003) The ‘hydrology’ of leaves: co-ordination of structure and function in temperate woody species. Plant Cell Environ 26: 1343–1356 [Google Scholar]
- Sack L, Frole K (2006) Leaf structural diversity is related to hydraulic capacity in tropical rain forest trees. Ecology 87: 483–491 [DOI] [PubMed] [Google Scholar]
- Sack L, Scoffoni C (2013) Leaf venation: structure, function, development, evolution, ecology and applications in the past, present and future. New Phytol 198: 983–1000 [DOI] [PubMed] [Google Scholar]
- Sack L, Scoffoni C, John GP, Poorter H, Mason CM, Mendez-Alonzo R, Donovan LA (2013) How do leaf veins influence the worldwide leaf economic spectrum? Review and synthesis. J Exp Bot 64: 4053–4080 [DOI] [PubMed] [Google Scholar]
- Scarpella E, Meijer AH (2004) Pattern formation in the vascular system of monocot and dicot plant species. New Phytol 164: 209–242 [DOI] [PubMed] [Google Scholar]
- Schliep KP. (2011) phangorn: phylogenetic analysis in R. Bioinformatics 27: 592–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoffoni C, Chatelet DS, Pasquet-Kok J, Rawls M, Donoghue MJ, Edwards EJ, Sack L (2016) Hydraulic basis for the evolution of photosynthetic productivity. Nat Plants 2: 16072. [DOI] [PubMed] [Google Scholar]
- Scoffoni C, Kunkle J, Pasquet-Kok J, Vuong C, Patel AJ, Montgomery RA, Givnish TJ, Sack L (2015) Light-induced plasticity in leaf hydraulics, venation, anatomy, and gas exchange in ecologically diverse Hawaiian lobeliads. New Phytol 207: 43–58 [DOI] [PubMed] [Google Scholar]
- Scoffoni C, Rawls M, McKown A, Cochard H, Sack L (2011) Decline of leaf hydraulic conductance with dehydration: relationship to leaf size and venation architecture. Plant Physiol 156: 832–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Sys Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecicki WK. (1989) A new gene CRISPOID (crd) on chromosome 1. Pisum Newsletter 21: 73–74 [Google Scholar]
- Tao Y, Ferrer J-L, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayeh N, Aluome C, Falque M, Jacquin F, Klein A, Chauveau A, Bérard A, Houtin H, Rond C, Kreplak J, et al. (2015) Development of two major resources for pea genomics: the GenoPea 13.2K SNP Array and a high-density, high-resolution consensus genetic map. Plant J 84: 1257–1273 [DOI] [PubMed] [Google Scholar]
- Tivendale ND, Davies NW, Molesworth PP, Davidson SE, Smith JA, Lowe EK, Reid JB, Ross JJ (2010) Reassessing the role of N-hydroxytryptamine in auxin biosynthesis. Plant Physiol 154: 1957–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobeña-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JNM, Souer E, Koes R (2002) FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev 16: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyree MT, Zimmermann MH (2002) Xylem structure and the ascent of sap. Springer, Berlin [Google Scholar]
- Verna C, Sawchuk MG, Linh NM, Scarpella E (2015) Control of vein network topology by auxin transport. BMC Biol 13: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SC, Cowan IR, Farquhar GD (1979) Stomatal conductance correlates with photosynthetic capacity. Nature 282: 424–426 [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y (2017) ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8: 28–36 [Google Scholar]
- Zhang S-B, Sun M, Cao K-F, Hu H, Zhang J-L (2014) Leaf photosynthetic rate of tropical ferns is evolutionarily linked to water transport capacity. PLoS One 9: e84682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2012) Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 5: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zsögön A, Negrini AC, Peres LEP, Nguyen HT, Ball MC (2015) A mutation that eliminates bundle sheath extensions reduces leaf hydraulic conductance, stomatal conductance and assimilation rates in tomato (Solanum lycopersicum). New Phytol 205: 618–626 [DOI] [PubMed] [Google Scholar]