The assembly of plant defense-related acylsugars in Petunia axillaris trichomes requires four acyltransferase activities that are distinct from those catalyzing acylsugar biosynthesis in tomato.
Abstract
Acylsugars are synthesized in the glandular trichomes of the Solanaceae family and are implicated in protection against abiotic and biotic stress. Acylsugars are composed of either sucrose or glucose esterified with varying numbers of acyl chains of differing length. In tomato (Solanum lycopersicum), acylsugar assembly requires four acylsugar acyltransferases (ASATs) of the BAHD superfamily. Tomato ASATs catalyze the sequential esterification of acyl-coenzyme A thioesters to the R4, R3, R3ʹ, and R2 positions of sucrose, yielding a tetra-acylsucrose. Petunia spp. synthesize acylsugars that are structurally distinct from those of tomato. To explore the mechanisms underlying this chemical diversity, a Petunia axillaris transcriptome was mined for trichome preferentially expressed BAHDs. A combination of phylogenetic analyses, gene silencing, and biochemical analyses coupled with structural elucidation of metabolites revealed that acylsugar assembly is not conserved between tomato and petunia. In P. axillaris, tetra-acylsucrose assembly occurs through the action of four ASATs, which catalyze sequential addition of acyl groups to the R2, R4, R3, and R6 positions. Notably, in P. axillaris, PaxASAT1 and PaxASAT4 catalyze the acylation of the R2 and R6 positions of sucrose, respectively, and no clear orthologs exist in tomato. Similarly, petunia acylsugars lack an acyl group at the R3ʹ position, and congruently, an ortholog of SlASAT3, which catalyzes acylation at the R3ʹ position in tomato, is absent in P. axillaris. Furthermore, where putative orthologous relationships of ASATs are predicted between tomato and petunia, these are not supported by biochemical assays. Overall, these data demonstrate the considerable evolutionary plasticity of acylsugar biosynthesis.
Plants synthesize an array of specialized metabolites that contribute to fitness through their roles in defense against pathogens and herbivory or the promotion of reproduction through the attraction of pollinators and seed-dispersing fauna (Tewksbury and Nabhan, 2001; Klahre et al., 2011; Mithöfer and Boland, 2012; Dudareva et al., 2013). Specialized metabolites are often synthesized at a specific stage of development or in response to environmental stimuli, and their biosynthesis may be restricted to a single tissue or cell type (Bird et al., 2003; Murata et al., 2008; Arimura et al., 2009; Bedewitz et al., 2014; Rambla et al., 2014). Glandular trichomes are epidermal cell appendages present on the surface of many plant species that serve as the location for the synthesis, storage, and secretion of diverse classes of specialized metabolites, including phenylpropanoids, terpenoids, and acylsugars (Schilmiller et al., 2008; Tissier, 2012). These compounds act directly as toxins or repellants to deter herbivory or serve as signaling molecules in tritrophic interactions to attract predators and also have been coopted by humans for use as medicines, narcotics, aromatics, and flavorings (Schilmiller et al., 2008; Weinhold and Baldwin, 2011; Bleeker et al., 2012). The glandular trichomes of members of the Solanaceae family are structurally and chemically diverse and synthesize specialized metabolites that are often restricted to individual species or specific accessions (Luckwill, 1943; Shapiro et al., 1994; Fridman et al., 2005; Gonzales-Vigil et al., 2012; Kim et al., 2012, 2014a). This chemical diversity is manifest through tremendous genetic variation that includes gene duplication and loss, expression variation, and amino acid substitutions that alter enzyme activity (Schilmiller et al., 2010b; Kim et al., 2012, 2014a; Matsuba et al., 2013; Kang et al., 2014). Together, these studies illustrate the plasticity and rapid evolution of glandular trichome-derived specialized metabolites, supporting their role as primary defense mechanisms that likely facilitate plant adaptation to differing environments.
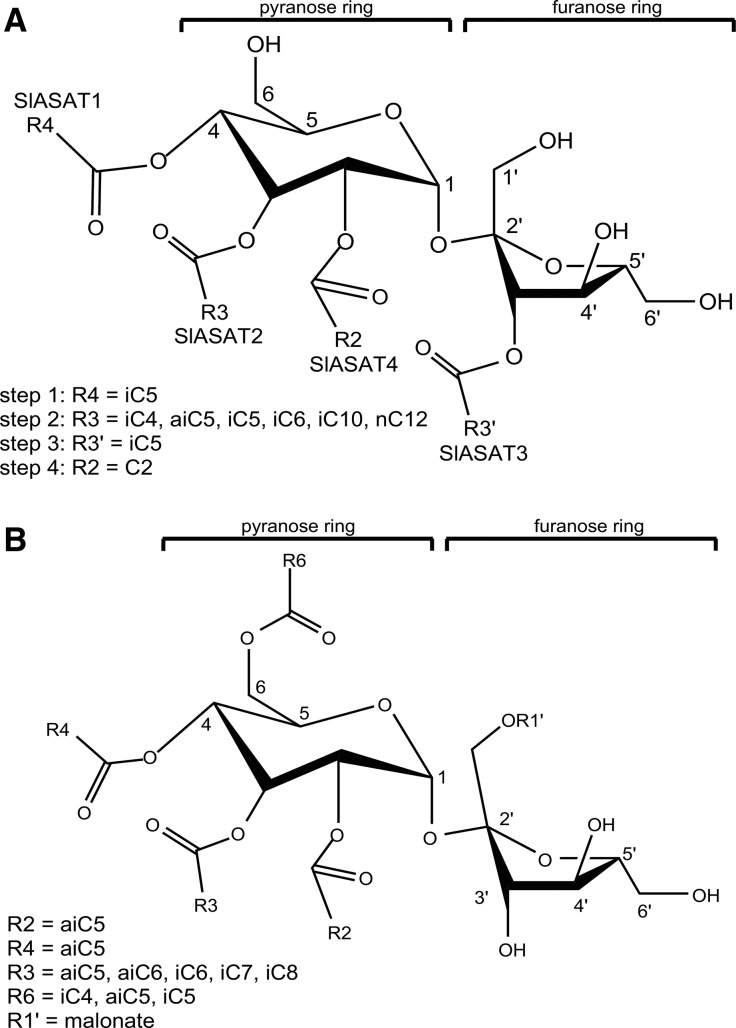
Acylsugars are synthesized in the glandular trichomes of several solanaceous species and are particularly abundant in Nicotiana spp., Petunia spp., and the wild tomato species Solanum pennellii, which can synthesize acylsugars up to 20% of leaf dry weight (Fobes et al., 1985; Severson et al., 1985; Arrendale et al., 1990; Chortyk et al., 1997). Secretion of acylsugars creates a sticky exudate that covers the aerial surfaces of plants that is implicated in direct defense against herbivores, tritrophic interactions, and possesses antimicrobial activity (Goffreda et al., 1989; Chortyk et al., 1993; Hare, 2005; Weinhold and Baldwin, 2011; Luu et al., 2017). Acylsugars consist of either a Suc or Glc backbone onto which acyl groups of differing lengths, typically C2 to C12, are esterified at various positions (King et al., 1986, 1988; Halinski and Stepnowski, 2013; Ghosh et al., 2014). Acyl groups can be straight chain (n) or branched chain, with the latter present in the iso (i) and anteiso (ai) configurations (Ghosh et al., 2014; Liu et al., 2017). A combination of ultra-high-performance liquid chromatography-mass spectrometry (MS) followed by purification and structural characterization by NMR spectroscopy revealed that trichomes of cultivated tomato (Solanum lycopersicum) synthesize a relatively simple mixture of approximately 15 tri- and tetra-acylsucroses (Ghosh et al., 2014). This acylsugar profile is dominated by the compound S4:17 (2,5,5,5), which consists of a Suc (S) molecule esterified with four (S4) acyl groups that includes a C2 chain and three C5 chains, to give a total number of carbons in the acyl groups of 17 (Schilmiller et al., 2010a; Ghosh et al., 2014; Fig. 1A). In addition to the short-chain acyl groups, several acylsugars from S. lycopersicum contain a single acyl group ranging from C10 to C12 in length that is esterified at either the R3 or R3ʹ position of Suc (Fig. 1A). Additional chemical complexity is manifest through isomers, which have the same elemental formulae and mass but varied positions of esterification (Ghosh et al., 2014). In contrast to the relatively simple acylsugar profile of S. lycopersicum, diverse accessions of the wild tomato species Solanum habrochaites collectively synthesize at least 34 acylsucroses and two acylglucoses, although these numbers are likely underestimates once isomers are considered (Kim et al., 2012; Ghosh et al., 2014). Diversity also exists between accessions of S. pennellii, some of which preferentially accumulate acylglucoses rather than acylsucroses (Shapiro et al., 1994). Overall, these data illustrate that, although acylsugars are fairly simple molecules, considerable diversity can occur as a result of acylation pattern and the length and structure of the acyl donors.
Figure 1.
Acylsucroses from S. lycopersicum and P. axillaris. A, Structures of characteristic tetra-acylsucroses from S. lycopersicum. Details of the length of the acyl chains typically found at each position in the most abundant metabolites are provided together with the identity of the ASAT enzymes responsible for acylsucrose assembly and the order in which they catalyze acylation reactions. Data represented are summarized from previous reports (Schilmiller et al., 2012, 2015; Ghosh et al., 2014; Fan et al., 2016). B, Structures of characteristic acylsucroses from P. axillaris. Details of the length of the acyl chains typically found at each position are derived from NMR-based structural characterization (Liu et al., 2017). The order of assembly of P. axillaris acylsucroses and the identification of the corresponding ASAT enzymes is the subject of this study.
Until recently, the genes required for the assembly of acylsugars remained elusive. However, a combination of metabolite profiling of introgression lines of tomato, coupled with positional cloning and RNA interference screens of trichome-expressed genes, identified four acylsugar acyltransferases (SlASAT1–SlASAT4) that belong to the BAHD acyltransferase superfamily (Schilmiller et al., 2012, 2015; Fan et al., 2016). Biochemical characterization revealed that these ASATs sequentially catalyze the esterification of acyl chains at the R4, R3, R3′, and R2 positions of Suc to generate a tetra-acylsucrose, and their activities with different substrates are sufficient to explain the structures of S. lycopersicum acylsugars (Fig. 1A; Ghosh et al., 2014; Fan et al., 2016). Furthermore, a combination of gene duplication, gene loss, and allelic variation at the asat2, asat3, and asat4 loci determines much of the chemical diversity observed in the wild tomato species S. habrochaites and S. pennellii (Kim et al., 2012; Schilmiller et al., 2012, 2015; Fan et al., 2016).
The trichomes of Petunia spp. synthesize large quantities of acylsugars, and ultra-high-performance liquid chromatography-MS analysis identified more than 100 acylsucroses in Petunia axillaris, Petunia exserta, and Petunia integrifolia (Liu et al., 2017). Twenty-nine of these acylsugars were purified and their structures determined by NMR. Together, these approaches revealed that petunia synthesizes a mixture of neutral and malonated acylsugars. Each acylsugar contains four acyl chains esterified to the pyranose ring at the R2, R3, R4, and R6 positions and, depending on the species, a single malonate ester can be formed on the furanose ring at either the R1′ or R6′ position (Fig. 1B; Liu et al., 2017). In petunia, the length of the acyl groups ranges from C4 to C8, and NMR analyses revealed that the structures of these groups were a mixture of iC4, iC5, iC6, iC7, and iC8 together with aiC5 and aiC6 (Fig. 1B). However, these acyl groups are not equally represented across the acylsugars, as aiC5 predominates, whereas iC4, iC5, and aiC6 are relatively minor components. NMR analyses also revealed that the medium chain length acyl groups (C6–C8) are almost exclusively esterified to the R3 position of Suc, whereas the shorter chain acyl groups (C4 and C5) are located on the R2, R4, and R6 positions. Thus, petunia synthesizes a suite of acylsugars that are completely distinct from those found in tomato, and these differences are manifest through acyl chains of different lengths and conformation that are esterified to different positions of Suc.
With the exception of tomato and closely related wild relatives, the enzymes required for acylsugar assembly remain unknown. As petunia synthesizes a completely different suite of acylsugars than tomato, with acyl chains of different lengths that are esterified at alternative positions on the Suc molecule, it was hypothesized that the mechanisms of acylsugar assembly would differ between the two species. This hypothesis was tested and confirmed through the identification and characterization of four ASATs that are required for the assembly of tetra-acylsucroses in P. axillaris. Notably, the order of acylsugar assembly differs between tomato and petunia, and the orthologous relations and biochemical activities of tomato and petunia ASATs are not conserved, suggesting that acylsugar biosynthesis has evolved differentially within diverse members of the Solanaceae family.
RESULTS
Identification of Putative ASATs from P. axillaris
The four ASATs from S. lycopersicum, SlASAT1 (Solyc12g006330), SlASAT2 (Solyc04g012020), SlASAT3 (Solyc11g067270), and SlASAT4 (Solyc01g105580), were used as query sequences in tBLASTN searches against a de novo transcriptome assembly of P. axillaris that includes transcripts generated from trichomes (Guo et al., 2015). Two P. axillaris unigenes, Pax36474 and Pax31629, were recovered that share 68% and 61% amino acid identity with SlASAT1 and SlASAT2, respectively. Examination of their expression patterns derived from the transcriptome data, which included callus, whole seedlings, shoot apices, flowers of mixed developmental stages, and trichomes, revealed that both Pax36474 and Pax31629, like SlASAT1 and SlASAT2 (Fan et al., 2016), are expressed preferentially in trichomes (Supplemental Table S1). Confirmation of the trichome-enriched expression of Pax36474 and Pax31629 was obtained by real-time quantitative reverse transcription (qRT)-PCR in whole petioles, shaved petioles, and trichomes, which indicated ∼100-fold enrichment in trichomes for both genes when compared with shaved petioles (Fig. 2). A phylogeny of ASAT-related BAHDs constructed using available sequences from across the Solanaceae family confirmed the close relationship between both SlASAT1 and Pax36474 and SlASAT2 and Pax31629 (Fig. 3). Furthermore, comparison of ∼120 kb of genomic sequence surrounding the SlASAT1 and Pax36474 loci (Tomato Genome Consortium, 2012; Bombarely et al., 2016) revealed considerable synteny between tomato and P. axillaris within this region, suggesting that the genes are orthologous (Supplemental Fig. S1). However, no syntenic relationship was observed between the genomic regions of SlASAT2 and Pax31629. Together, these data suggest that the Pax36474 and Pax31629 unigenes represent candidate ASATs. Two additional P. axillaris unigenes, Pax21699 and Pax39119, also were recovered in tBLASTN searches using SlASAT1 as the query sequence (Fig. 3). These unigenes are more divergent to SlASAT1 than Pax36474, each sharing approximately 38% amino acid identity with the query sequence. In particular, Pax21699 does not appear to possess a putative ortholog in any of the Solanaceae genomes investigated (tomato, potato [Solanum tuberosum], and pepper [Capsicum annuum]), and while a small amount of synteny exists between the corresponding genomic region of petunia and two regions of tomato chromosome 2, the latter do not contain any predicted BAHD acyltransferases (Supplemental Fig. S2). Similarly, although there is synteny between tomato and petunia in a genomic region that flanks the Pax39119 locus, tomato does not contain a putative ortholog of this petunia BAHD acyltransferase (Supplemental Fig. S3). Analyses of expression by qRT-PCR revealed that both Pax21699 and Pax39119 are strongly preferentially expressed in trichomes relative to shaved petioles (Fig. 2).
Figure 2.
Expression analysis of putative P. axillaris ASATs. The expression of candidate ASATs was determined in whole petioles, shaved petioles with trichomes removed, and trichomes of P. axillaris. Three biological and three technical replicates for each sample were analyzed. Data are presented as mean expression values ± se relative to those observed in shaved stems. Values labeled with different letters are significantly different (least square means, P < 0.05).
Figure 3.
Phylogenetic analysis of putative ASATs. An unrooted phylogenetic tree of known S. lycopersicum (tomato) ASATs together with related amino acid sequences from S. lycopersicum, Solanum tuberosum (potato), Capsicum annuum (pepper), and P. axillaris was constructed using the maximum likelihood method from a multiple sequence alignment of deduced full-length amino acid sequences. The tree was constructed using MEGA version 5, and bootstrap values of 50 or greater are shown from 2,000 replicates. Known tomato ASATs are shown in red, and putative trichome preferentially expressed ASATs from P. axillaris are in blue. Subsequent functional characterization of the putative P. axillaris ASATs reported herein led to the identification of four enzymes involved in acylsugar assembly, PaxASAT1 to PaxASAT4. The positions on the Suc molecule where the addition of acyl chains occurs following ASAT-mediated catalysis are provided in parentheses.
In tomato, SlASAT3 catalyzes the addition of acyl groups to the 3′ position of the furanose ring, a position that is not acylated in petunia acylsugars (Schilmiller et al., 2015; Liu et al., 2017). Reciprocal BLASTP searches and phylogenetic analysis indicated that SlASAT3 is 53% identical to, and most closely related to, the P. axillaris unigene, Pax16120. However, Pax16120 is not expressed in trichomes (Supplemental Table S1), suggesting that this unigene is unlikely to be involved in acylsugar assembly. Tandem gene duplications are a feature of many plant genes and are particularly prevalent in genes involved in specialized metabolism (Matsuba et al., 2013; Chae et al., 2014; Boutanaev et al., 2015). In tomato, SlASAT4 resides at a locus on chromosome 1 that contains two additional BAHD acyltransferases (Schilmiller et al., 2012), and similar clustering of tandem duplicates of SlASAT4-related BAHDs occurs in the potato and pepper genomes (Potato Genome Sequencing Consortium, 2011; Kim et al., 2014b). In P. axillaris, several unigenes were identified that share approximately 50% amino acid identity to SlASAT4 (Fig. 3), and three of these, Pax24741, Pax28156, and Pax35115, are expressed preferentially in trichomes (Supplemental Table S1). However, qRT-PCR analyses revealed that the transcript abundance of Pax24741, Pax28156, and Pax35115 is enriched between 5- and 18-fold in trichomes compared with shaved petioles, which is in contrast to the at least 100-fold enriched expression of Pax21699, Pax31629, Pax36474, and Pax39119 in P. axillaris trichomes (Fig. 2). Phylogenetic analyses revealed that the SlASAT4-related proteins form a separate clade from those more closely related to SlASAT1, SlASAT2, and SlASAT3 (Fig. 3).
Silencing of Putative ASATs in P. axillaris
Based on phylogenetic analyses and their highly enriched (100-fold or greater) expression in trichomes (Figs. 2 and 3), the role of four putative ASATs, Pax21699, Pax31629, Pax36474, and Pax39119, in acylsugar biosynthesis was investigated using virus-induced gene silencing (VIGS) in P. axillaris. The four candidate P. axillaris ASATs are highly divergent at the nucleotide level, and it was possible to identify fragments of each gene ranging from 247 to 375 nucleotides in length that are highly specific for each target. For example, BLASTN searches against P. axillaris transcriptome and genome assemblies (Guo et al., 2015; Bombarely et al., 2016) failed to identify any additional nucleotide sequences longer than 19 bp that share 100% identity with each VIGS fragment, indicating that there is very limited potential for off-target silencing. Upon silencing, the abundance of each target transcript was reduced by approximately 80% to 90% in silenced lines when compared with those of TRV2 empty vector controls (Supplemental Fig. S4).
Prior analyses using MS and NMR spectroscopy revealed that P. axillaris acylsugars contain four acyl groups on the pyranose ring and often possess a malonate ester on the furanose ring (Fig. 1B; Liu et al., 2017). In agreement with these prior findings, the acylsugar profile of petioles of P. axillaris TRV2 empty vector control plants is composed of approximately 10 compounds and is dominated by the S5 ester S(m)5:26 (m,5,5,5,8) (Fig. 4, A and B). Furthermore, the molecular formulae of the acylsugars identified in TRV2 empty vector control plants (Fig. 4A) match those of a number of structurally resolved acylsugars from P. axillaris (Supplemental Table S2).
Figure 4.
The P. axillaris acylsugar profile is altered by the silencing of Pax21699 and Pax31629. A, Comparison of the acylsugar profile of P. axillaris expressing TRV2 empty vector, TRV2::Pax21699, or TRV2::Pax31629 constructs. Negative ion mode total ion current liquid chromatography-mass spectrometry (LC-MS) chromatograms are shown, indicating both reduced [e.g. S(m)5:26 (m,5,5,5,8)] and increased [e.g. S3:18 (5,5,8), S(m)4:21 (m,5,5,8), S4:19 (4,5,5,5), and S4:20 (5,5,5,5)] abundance of individual acylsugars in TRV2::Pax21699- and TRV2::Pax31629-silenced lines. The peak corresponding to a 5 µm telmisartan internal standard is labeled IS. Details of individual acylsugars are provided in Supplemental Table S2 and Supplemental Figure S5. B and C, Quantification of acylsugar levels in TRV2::Pax21699- and TRV2::Pax31629-silenced lines. The abundances of individual and total (insets) acylsugars in TRV2::Pax21699- and TRV2::Pax31629-silenced lines relative to those in TRV2 empty vector controls are shown. Data are presented as peak area normalized to dry weight and internal standard peak area and represent means ± se of 12 biological replicates. Asterisks denote significant differences (**, P < 0.01; ***, P < 0.001) as determined by Student's t tests.
Silencing of Pax21699 resulted in qualitative and quantitative changes to the acylsugar profile (Fig. 4, A and B). Total acylsugars were reduced in Pax21699 silenced lines by approximately 60%, which was manifest by a reduction in all of the acylsugars detected in TRV2 empty vector lines. In addition, several acylsugars were apparent in the Pax21699 silenced lines that were undetectable in the empty vector lines, and each lacks a single C5 acyl group. For example, S(m)5:26 (m,5,5,5,8) is reduced in the silenced lines while S(m)4:21 (m,5,5,8) and S3:18 (5,5,8) increase (Fig. 4, A and B; Supplemental Fig. S5). These data suggest that Pax21699 possesses ASAT activity and may be responsible for adding a C5 acyl group to the pyranose ring of P. axillaris acylsugars (Fig. 1B). Similarly, silencing of Pax31629 also resulted in an overall reduction in acylsugar levels of approximately 50%, and this was accompanied by an increased abundance of minor acylsugars containing only C4 or C5 acyl groups, including S4:20 (5,5,5,5) and S4:19 (4,5,5,5) (Fig. 4, A and C). These data indicate a shift from the C6 to C8 acyl groups in favor of shorter acyl chains. Notably, in structurally resolved acylsugars of P. axillaris, the C6 to C8 acyl groups are esterified predominantly to the R3 position of the pyranose ring (Liu et al., 2017). In contrast to silencing Pax21699 and Pax31629, which resulted in both qualitative and quantitative changes to the acylsugar profile of P. axillaris trichomes, only quantitative changes were observed following the silencing of both Pax36474 and Pax39119, with an overall reduction in acylsugar levels of between 50% and 60% (Fig. 5). Overall, these data suggest that the four highly trichome-enriched unigenes, Pax21699, Pax31629, Pax36474, and Pax39119, each encoding predicted BAHD acyltransferases, are involved in acylsugar assembly.
Figure 5.
P. axillaris acylsugar levels are reduced in Pax36474- and Pax39119-silenced lines. The abundances of individual and total (insets) acylsugars in TRV2::Pax36474-silenced (A) and TRV2::Pax39119-silenced (B) lines relative to those in TRV2 empty vector controls are shown. Data are presented as peak area normalized to dry weight and internal standard peak area and represent means ± se of 12 biological replicates. Asterisks denote significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) as determined by Student's t tests.
Biochemical Characterization of P. axillaris ASATs
In tomato, SlASAT1 (Solyc12g006330) catalyzes the first step in acylsucrose assembly through the acylation of Suc at the R4 position, using several acyl-CoA donors (Fan et al., 2016). As the P. axillaris unigene, Pax36474, is the putative ortholog of SlASAT1 (Fig. 3; Supplemental Fig. S1), it was hypothesized that this enzyme also would catalyze the initial acylation of Suc. However, Pax36474 failed to catalyze the acylation of Suc using several acyl-CoA donors (C4–C8 in length), including aiC5, which represents the most frequent acyl group esterified to the R4 position of P. axillaris acylsugars (Fig. 1B; Supplemental Table S3; Liu et al., 2017). The failure of Pax36474 to utilize Suc as a substrate indicated that this enzyme does not possess ASAT1 activity, suggesting that the assembly of acylsugars differs between tomato and petunia and prompting investigation of the activity of the additional putative P. axillaris ASATs, Pax21699, Pax31629, and Pax39119. Only Pax39119 was active with Suc as an acyl acceptor and led to the synthesis of various mono-acylsucroses when supplied with acyl-CoA donors ranging from C5 to C8 in length but not with iC4-CoA (Supplemental Fig. S6; Supplemental Table S3). These data suggest that Pax39119 catalyzes a reaction consistent with ASAT1 activity in P. axillaris; therefore, we renamed this protein PaxASAT1. Notably, different retention times were observed for the S1:5 products generated by SlASAT1 and PaxASAT1, using either iC5 or aiC5 as acyl donor, suggesting that the acyl groups are not esterified to the same position of Suc (Fig. 6). To determine the position at which PaxASAT1 acylates Suc, a reaction using Suc and aiC5 was scaled up to generate sufficient material for purification and structural determination by NMR spectroscopy. Subsequent heteronuclear single-quantum correlation (HSQC) spectroscopy together with correlation spectroscopy NMR spectra revealed that PaxASAT1 catalyzes the addition of the acyl group to the R2 position of Suc, which is in contrast to SlASAT1, which catalyzes addition at the R4 position (Fan et al., 2016; Fig. 6B; Supplemental Table S4).
Figure 6.
ASAT1 activity is not conserved in P. axillaris and S. lycopersicum. A, In vitro formation of S1:5 by recombinant S. lycopersicum ASAT1 (SlASAT1) and P. axillaris ASAT1 (PaxASAT1) using Suc as the acyl acceptor and either iC5-CoA or aiC5-CoA as the acyl donor. Negative ion mode LC-MS extracted ion chromatograms for the reaction product mass-to-charge ratio (m/z) 461.1 (S1:5; [M+Cl]−) are shown. The vertical dashed line highlights a retention time shift of S1:5 products formed by SlASAT1 and PaxASAT1, suggesting that different S1:5 isomers are formed by these enzymes. B, Structures of the S1:5 reaction products formed by SlASAT1 and PaxASAT1 using iC5-CoA and aiC5-CoA, respectively. Data for S1:5(iC5R4) formed by SlASAT1 are based on prior structural characterization (Schilmiller et al., 2015; Fan et al., 2016), while the structure of S1:5(aiC5R2) was determined by NMR (Supplemental Table S4).
To define the second step in acylsucrose assembly, the S1:5 (aiC5R2) product generated with PaxASAT1 was utilized as a substrate together with acyl-CoA donors, ranging from C4 to C8 in length, in sequential reactions with one of the additional putative P. axillaris ASATs, Pax21699, Pax31629, or Pax36474. Only Pax36474 acylated S1:5 (aiC5R2), and the enzyme was active with iC4-CoA, iC5-CoA, aiC5-CoA, and iC6-CoA acyl donors, indicating that it possesses PaxASAT2 activity (Fig. 7A; Supplemental Table S3). The synthesis of S2:10 (aiC5R2, aiC5), formed through sequential reactions of Suc and aiC5-CoA catalyzed by PaxASAT1 and PaxASAT2, was scaled up, and the product was purified by preparative HPLC and its structure was determined by NMR spectroscopy. The resulting NMR spectra (Supplemental Table S5) revealed that the two acyl groups of S2:10 are esterified to the R2 and R4 positions of Suc, yielding S2:10 (aiC5R2, aiC5R4). This is consistent with the structural characterization of acylsucroses from P. axillaris, indicating that while aiC5 is the predominant acyl group at the R4 position, iC4-CoA also can be incorporated (Liu et al., 2017). However, enzyme activity assays (Supplemental Table S3) suggest that, at least in vitro, PaxASAT2 possesses greater catalytic promiscuity with respect to the acyl donor substrate than is observed in the acylsugar profile in planta.
Figure 7.
Identification of ASAT2, ASAT3, and ASAT4 activities of P. axillaris. A, In vitro formation of S2:10 by recombinant P. axillaris ASAT2 (PaxASAT2) using S1:5 (aiC5R2) as the acyl acceptor and aiC5-CoA as the acyl donor. A negative ion mode LC-MS extracted ion chromatogram for the reaction product m/z 555.2 is shown. B, In vitro formation of S3:17 by recombinant P. axillaris ASAT3 (PaxASAT3) using S2:10 (aiC5R2, aiC5R4) as the acyl acceptor and iC7-CoA as the acyl donor. A negative ion mode LC-MS extracted ion chromatogram for the reaction product m/z 667.3 is shown. C, In vitro formation of S4:22 by recombinant P. axillaris ASAT4 (PaxASAT4) using S3:17 (aiC5R2, iC7R3, aiC5R4) as the acyl acceptor and aiC5-CoA as the acyl donor. A negative ion mode LC-MS extracted ion chromatogram for the reaction product m/z 751.4 is shown. D to F, Tandem mass spectra of the acylsucroses formed by PaxASAT2, PaxASAT3, and PaxASAT4. The fragmentation of specific reaction products of m/z 555.2, 667.3, and 751.4 presented in A to C is shown. Fragmentation in negative mode reveals the length of the acyl chains esterified to the Suc core due to the loss of acyl groups as ketenes.
Sequential reactions were performed subsequently with PaxASAT1 and PaxASAT2 and either Pax21699 or Pax31629. These reactions revealed that tri-acylsucroses were formed only in the presence of Pax31629 using iC6, iC7, and iC8 as acyl-CoA donors but not iC4- or aiC5-CoAs, suggesting that this enzyme corresponds to PaxASAT3 (Fig. 7B; Supplemental Table S3). In P. axillaris, these medium chain length acyl groups (C6–C8) are esterified predominantly to the R3 position of acylsucroses and are not present at the R6 position (Liu et al., 2017), the remaining open position on the pyranose ring that can serve as an acyl acceptor once the R2 and R4 positions are occupied in S2:10 (aiC5R2, aiC5R4). These data suggest that PaxASAT3 catalyzes the addition of acyl groups to the R3 position of a diacylsucrose, a hypothesis supported by reduced levels of acylsugars containing C6 to C8 groups in Pax31629 VIGS lines (Fig. 4). Furthermore, SlASAT2 (Solyc04g012020), which is phylogenetically closely related to PaxASAT3 (Fig. 3), also catalyzes acylation at the R3 position in tomato (Fan et al., 2016).
P. axillaris acylsugars possess four acyl groups on the pyranose ring (Fig. 1B; Liu et al., 2017), and the current biochemical analysis indicates that the R2, R4, and R3 positions are substituted by the activities of Pax39119, Pax36474, and Pax31629, respectively. Thus, the R6 position is the final position of the pyranose ring to be acylated. Silencing of Pax21699 led to an overall reduction in acylsugars that was also associated with the accumulation of acylsucroses that lack a C5 chain, suggesting that Pax21699 possesses ASAT activity (Fig. 4; Supplemental Fig. S5). Although Pax21699 was inactive with Suc, S1:5 (aiC5R2), and S2:10 (aiC5R2, aiC5R4) as substrates, tetra-acylsucroses were formed following incubation of Pax21699 with tri-acylsucroses, generated by the sequential action of PaxASAT1, PaxASAT2, and PaxASAT3 and either iC4- or aiC5-CoAs (Fig. 7C; Supplemental Table S3). These data are congruent with the structural characterization of P. axillaris acylsucroses (Fig. 1B; Liu et al., 2017). Pax21699 was also active with S3:16 and S3:17 to yield S4:21 and S4:22 when supplied with aiC5 as an acyl donor (Supplemental Table S3). Together, these data indicate that Pax21699 possesses PaxASAT4 activity, and overall, all four of the trichome-expressed candidate BAHD enzymes demonstrate ASAT activity.
DISCUSSION
The tremendous chemical diversity observed in plant-specialized metabolites is mediated in part by the formation of ester linkages between starter molecules that result in the formation of increasingly complex phytochemicals (D’Auria, 2006; Mugford et al., 2009; Nishizaki et al., 2013). Acylsugars are plant-specialized metabolites that are synthesized in the glandular trichomes of members of the Solanaceae family and are formed through the direct coupling of primary metabolites or their immediate breakdown products. Glc or Suc serves as the backbone of the acylsugar molecule, and ester linkages are formed at the various hydroxyl groups of these sugars through the addition of acyl groups derived from the metabolism of branched-chain amino acids (Kandra et al., 1990; Walters and Steffens, 1990; Slocombe et al., 2008; Ning et al., 2015) and, in the case of Petunia spp., the presence of a malonate ester derived from malonyl-CoA, a precursor of fatty acid biosynthesis (Liu et al., 2017). Hence, relative to more structurally complex specialized metabolites, including many alkaloids and terpenoids that are synthesized by multistep pathways involving several enzyme classes (Winzer et al., 2012; Lange and Turner, 2013; Bedewitz et al., 2014; Dugé de Bernonville et al., 2015), acylsucrose assembly is seemingly simple, as only four acylation steps catalyzed by the same class of enzyme, BAHD acyltransferases, are required in S. lycopersicum trichomes to synthesize the characteristic tetra-acylsucroses (Fan et al., 2016). However despite this apparent simplicity and the metabolic proximity to primary metabolism, the acylsugars with structures depicted in Figure 1 are not widely distributed across the plant kingdom and, thus far, are found only in the glandular trichomes of the Solanaceae, where expression data indicate that their synthesis is restricted to the single glandular cell at the tip of type I and IV trichomes (Schilmiller et al., 2012, 2015; Ning et al., 2015; Fan et al., 2016).
Although acylsugars are structurally simple, chemical complexity is apparent between and within species, and this is mediated by the varied lengths and conformations of acyl chains (C2–C12) that are esterified to different positions of the sugar backbone together with the catalytic promiscuity of the BAHD acyltransferases that possess the ability to utilize multiple acyl-CoA donors (Ghosh et al., 2014; Schilmiller et al., 2015; Fan et al., 2016; Liu et al., 2017). Plant-specialized metabolism is highly variable and evolves rapidly, a phenomenon attributed to the reduced evolutionary constraints compared with those imposed upon primary metabolism (Milo and Last, 2012). This rapid evolution frequently involves gene duplication, gene loss, and allelic variation that subsequently alter enzyme activity, and these phenomena are apparent within the Solanaceae and contribute to the acylsugar phenotype. For example, deletion at the asat4 locus (formerly asat2) in S. pennellii and select accessions of S. habrochaites alters the acylsugar phenotype (Kim et al., 2012; Schilmiller et al., 2012). Similarly, variation at the asat3 locus in the tomato clade also contributes to acylsugar diversity. For example, while the allele from S. lycopersicum acylates a di-acylsucrose on the furanose ring to form a tri-acylsucrose, the allele from S. pennellii acylates a mono-acylsucrose on the pyranose ring to form a di-acylsucrose (Schilmiller et al., 2015). Furthermore, duplication of the asat3 locus in multiple accessions of S. habrochaites coupled with neofunctionalization creates ASATs with altered acyl donor and acyl acceptor specificities that explain much of the acylsugar diversity observed in the species (Kim et al., 2012; Schilmiller et al., 2015).
Petunia and S. lycopersicum diverged from a common ancestor ∼30 million years ago (Särkinen et al., 2013), and the acylsugar profile of Petunia spp. is completely different from that of Solanum spp. due to the structure of the acyl donors and their locations on the sugar backbone (Ghosh et al., 2014; Liu et al., 2017). This chemical diversity prompted the examination of whether acylsugar assembly is conserved across the Solanaceae and involves putative orthologs of the tomato ASATs or whether additional enzymes have evolved ASAT function in P. axillaris. Utilizing a combination of transcriptome-guided candidate gene identification, gene silencing, and biochemical analyses, four trichome-expressed BAHD enzymes designated PaxASAT1 to PaxASAT4 were identified from P. axillaris that are required for acylsugar assembly and whose activities are congruent with the acylsugar profile of the species. For example, Pax39119/PaxASAT1 can utilize C5, C6, C7, and C8 acyl donors to acylate the R2 position of Suc (Supplemental Fig. S6; Supplemental Table S4), and NMR-based structural characterization of P. axillaris acylsugars indicates that, while aiC5 is the most frequent acyl group at this position, iC6 and iC8 acyl groups are present in S4:24[1] (iC6R2, iC8R3, aiC5R4, aiC5R6) and S4:24[2] (iC8R2, aiC6R3, aiC5R4, aiC5R6) (Liu et al., 2017). In all structurally resolved P. axillaris acylsugars characterized to date, either an iC4 or aiC5 group is present at the R4 position (Liu et al., 2017), and PaxASAT2, which catalyzes addition to the R4 position, is active with acyl donors up to C6 in length. Silencing of Pax31629/PaxASAT3 leads to a reduction in acylsugars with C6 to C8 acyl chains, which are esterified primarily at the R3 position of the pyranose ring (Fig. 4; Liu et al., 2017). This is accompanied by a small increase in very minor acylsugars that contain four short acyl chains, including S4:18 (4,4,5,5), S4:19 (4,5,5,5), and S4:20 (5,5,5,5) (Fig. 4C). In agreement with the hypothesis that PaxASAT3 adds C6 to C8 acyl groups to the R3 position of the pyranose ring, recombinant PaxASAT3 was active with iC6, iC7, and iC8 acyl donors but inactive with C4 and C5 acyl donors (Fig. 7B; Supplemental Table S3). Similarly, silencing of Pax21699/PaxASAT4 leads to a reduction in total acylsugars together with the accumulation of several tri-acylsucroses that lack a C5 acyl group, and this enzyme is active with C4 and C5 acyl donors (Fig. 4; Supplemental Table S3). Together, these data indicate a general consensus between the known structures of P. axillaris acylsugars and the observed silencing and biochemical phenotypes. However, the assays performed using recombinant enzymes illustrate broader catalytic potential among the P. axillaris ASATs than is currently observed among the structurally resolved acylsugars from this species. For example, recombinant PaxASAT2 possesses activity with acyl donors up to C6 in length (Supplemental Table S3), whereas only iC4 and aiC5 acyl groups are present at the R4 position in structurally resolved P. axillaris acylsugars (Liu et al., 2017). Such discrepancies may simply reflect acylsugar abundance in the species, which typically drives the selection of targets for purification and NMR structural elucidation. Alternatively, a restricted in planta acylsugar profile compared with that obtained from the in vitro biochemical activity of a recombinant enzyme may reflect reduced availability of an acyl donor or lower levels of ASAT activity with specific substrates.
Silencing of each petunia ASAT transcript led to a quantitative reduction in total acylsugar abundance (Figs. 4 and 5). This reduction would be expected, particularly for early steps in the pathway, such as that catalyzed by Pax39119/PaxASAT1, and indeed was observed previously in SlASAT1-silenced lines (Fan et al., 2016). However, in general, metabolic precursors did not accumulate to levels that might be expected based on the extent of reduction of some of the individual acylsugars. For example, while the most abundant petunia acylsucrose, S(m)5:26 (m,5,5,5,8), is reduced by approximately 70% in Pax21699/PaxASAT4-silenced lines (Fig. 4B), there is only a slight increase in abundance of the predicted precursors S3:18 (5,5,8) and S(m)4:21 (m,5,5,8). Similarly, when Pax36474/PaxASAT2 and Pax31629/PaxASAT3 were silenced, accumulation of either mono-acylsucroses or di-acylsucroses would be predicted, respectively. However, these intermediary metabolites were not detected in any of the silenced lines. While the underlying reasons for this remain unknown, it is possible that when ASAT activity is reduced by silencing, precursors may be turned over by acylsugar hydrolases or further metabolized to compounds undetected by our metabolite analyses. Together, these processes may contribute to the overall reduction in acylsugars observed in VIGS lines.
Notable differences exist between the P. axillaris and S. lycopersicum ASATs, and their orthologous relationships and acyl acceptor preferences are not fully conserved, leading to an altered order of tetra-acylsucrose assembly between the two species (Fig. 8). For example, based on phylogeny (Fig. 3), it was hypothesized that Pax36474, like SlASAT1, would encode an enzyme with ASAT1 activity that utilizes Suc as the acyl acceptor and catalyzes the addition of an acyl group to the R4 position of the pyranose ring (Fig. 1A). However, Pax36474 was inactive with Suc as an acyl acceptor and every acyl donor tested (C4–C8), whereas subsequent biochemical analyses and NMR-based structural characterization of reaction products identified Pax39119 as PaxASAT1 that catalyzes the addition of a range of acyl groups to the R2 position of the pyranose ring of Suc (Supplemental Fig. S6; Supplemental Tables S3 and S4). Thus, rather than initiation at the R4 position, as in tomato, acylsugar assembly in P. axillaris begins with acyl chain addition at the R2 position, and subsequent acylations at the R4, R3, and R6 positions complete tetra-acylsucrose assembly. This is in contrast to the sequence of acylsugar assembly in S. lycopersicum, which occurs through sequential acylations at the R4, R3, R3ʹ, and R2 positions (Fan et al., 2016; Fig. 8). These data indicate that primary sequence is not a good predictor of ASAT substrate preference, as SlASAT1/Solyc12g006330 of tomato utilizes Suc as an acyl acceptor whereas Pax36474 has ASAT2 activity and utilizes a mono-acylsucrose substrate. This shift in substrate preference also is observed between SlASAT2/Solyc04g012020 and Pax31629/PaxASAT3, which are phylogenetically related and share approximately 61% amino acid identity (Fig. 3). While SlASAT2 utilizes a mono-acylsucrose as an acyl acceptor (Fan et al., 2016), PaxASAT3 utilizes a di-acylsucrose (Fig. 7B). Despite these changes in acyl acceptor specificity between related P. axillaris and tomato ASATs, SlASAT1 and PaxASAT2 both acylate at the R4 position of the pyranose ring, while SlASAT2 and PaxASAT3 each catalyze additions to the R3 position. These data suggest that sequence conservation between ASATs of diverse species may be a useful predictor of acylation position. Indeed, structure-function analyses of ASATs from tomato and closely related wild species has proven to be a powerful approach to identify amino acid residues that specify substrate preferences (Fan et al., 2016). However, given the evolutionary distance between petunia and tomato and the fairly low sequence identity of their ASATs, additional comparisons between putative ASATs of diverse species from across the Solanaceae phylogeny will likely be required to enhance the power of this approach. Alternatively, resolving the structures of ASATs bound to their substrates may reveal the underlying biochemical changes that have led to the ability of seemingly orthologous enzymes to evolve new substrate preferences.
Figure 8.
Comparison of tetra-acylsucrose assembly in tomato and P. axillaris. A, Structure of a generic tetra-acylsucrose from tomato. B, Structure of a generic acylsucrose from P. axillaris. The four steps required for the formation of tetra-acylsucroses in each species are shown. Orthologous enzymes predicted by sequence analysis are indicated by the same color. Note that the predicted orthologous relationships between SlASAT1 and PaxASAT2 and between SlASAT2 and PaxASAT3 are not conserved at the biochemical level, and this, together with the presence-absence variation of other ASATs between tomato and petunia, results in differential acylsucrose assembly in these two species.
While sequence conservation is observed between SlASAT1 and PaxASAT2 and between SlASAT2 and PaxASAT3, additional PaxASATs are less well conserved. For example, P. axillaris does not appear to contain a putative ortholog of SlASAT3, which is in agreement with the lack of a branched acyl group on the furanose ring of acylsugars from this species (Liu et al., 2017). Similarly, additional ASATs are not fully conserved across the Solanaceae. For example, Pax39119/PaxASAT1 is 72% identical to Ca06g15270, a predicted BAHD from pepper, although no closely related proteins are found in tomato or potato (Fig. 3). In addition, while putative orthologs of Pax31629/PaxASAT3 are readily identifiable in tomato, potato, and pepper, putative orthologs of Pax36474/PaxASAT2 are present in tomato and potato but not in pepper (Fig. 3). In contrast, Pax21699/PaxASAT4 is more divergent, sharing less than 40% identity with the most closely related sequences from tomato, potato, and pepper, suggesting that a putative ortholog of this gene is likely absent from these species. While the functions of the putative ASATs from potato and pepper need to be characterized to determine whether they are involved in acylsugar biosynthesis, the lack of complete conservation of the ASATs and related BAHDs across the Solanaceae (Fig. 3) reflects the plasticity of specialized metabolism, which frequently involves gene duplication and loss (Pichersky and Lewinsohn, 2011; Chae et al., 2014).
Although four P. axillaris ASATs have been identified that can explain much of the acylsugar diversity within this species, there are ASAT activities in P. axillaris that remain to be identified. For example, many P. axillaris acylsugars possess an unusual malonate ester on the furanose ring at the R1ʹ position that has thus far not been detected in acylsugars of other members of the Solanaceae (Liu et al., 2017). However, despite our efforts to identify the ASAT activity responsible for adding this malonate group, the enzyme remains unknown. Thus, it is unclear at what step in acylsugar assembly the malonate group is added and whether this may affect ASAT activity. However, our favored hypothesis is that malonylation is the final step in acylsugar assembly in Petunia spp., as neutral and malonate ester forms of multiple acylsugars coexist. For example, S(m)5:25 (m,5,5,5,7) is present with S4:22 (5,5,5,7) in P. axillaris trichomes and both S(m)5:26 (m,5,5,5,8) and S4:23 (5,5,5,8) occur in the trichomes of P. exserta (Liu et al., 2017). In addition, the enzyme responsible for catalyzing the esterification of either a C4 or C5 acyl group to the R3 position of the low-abundance acylsucroses that accumulate in Pax31629/PaxASAT3-silenced lines, S4:18 (4,4,5,5), S4:19 (4,5,5,5), and S4:20 (5,5,5,5), is uncharacterized, and further experimentation will be required to confirm its identity.
Although there has been a longstanding interest in the role of acylsugars in plant defense (Goffreda et al., 1989; Buta et al., 1993; Chortyk et al., 1997), until relatively recently, the enzymes involved in trichome-derived acylsucrose assembly remained unknown. A combination of genetics- and genomics-enabled biochemistry led to the identification of several BAHDs from tomato and closely related wild species that are involved in acylsugar assembly and chemical diversity (Kim et al., 2012; Schilmiller et al., 2012, 2015; Fan et al., 2016). This study utilized the tomato ASATs as a framework to explore the potential conservation of acylsugar assembly. Acylsugar composition differs between tomato and Petunia spp., and although four trichome-expressed ASATs were identified from P. axillaris that can explain much of the acylsugar diversity within this species, the data indicate that the genes encoding ASATs are not fully conserved across the Solanaceae and that considerable variation exists in the substrate preference of tomato and P. axillaris ASATs as well as in the mechanism of acylsucrose assembly between these two species. These data expand the existing knowledge of acylsugar biosynthesis in plants, highlighting the evolutionary complexity that contributes to the chemical diversity of these structurally simple specialized metabolites. Additional studies are required using species from across the Solanaceae to determine the extent of acylsugar chemical diversity, their assembly, and the biochemical evolution of ASATs.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of Petunia axillaris (PI667515) were obtained from the Germplasm Resources Information Network. Plants were grown in 4-inch pots in peat-based compost supplemented with one-half-strength Hoagland solution in a growth room under a 16-h photoperiod (145 μmol m−2 s−1) at a constant temperature of 24°C.
Multiple Sequence Alignments and Phylogenetic Analyses
Sequence analysis was performed using MEGA version 5 (Tamura et al., 2011). Amino acid alignments were generated using MUSCLE (Edgar, 2004), and phylogenetic trees were constructed using the maximum likelihood method and the Jones-Taylor-Thornton model. A bootstrap test of 2,000 replicates was used to assess the reliability of the phylogeny.
VIGS
Constructs were assembled in TRV2-LIC vector as described previously (Dong et al., 2007) using the primers listed in Supplemental Table S6. VIGS experiments were performed as described (Velásquez et al., 2009). The P. axillaris first true leaf was inoculated with Agrobacterium tumefaciens strain GV3101 cultures. Plants inoculated with A. tumefaciens carrying the TRV2-LIC empty vector were used as controls, and TRV2::PHYTOENE DESATURASE plants were used to monitor the progression of gene silencing. Plants were grown as described above. Petiole segments of 10 mm were harvested from the fifth true leaf of 5-week-old plants for acylsugar profiling, and trichomes for RNA extraction were harvested from frozen petioles by brushing and collection directly into liquid nitrogen.
RNA Extraction, cDNA Synthesis, and qRT-PCR
Total RNA was extracted from tissues of P. axillaris using the RNeasy Plant Mini Kit and subjected to on-column DNase treatment (Qiagen). cDNA was synthesized from 800 ng of total RNA using the SuperScript III first-strand synthesis kit (Invitrogen). Following synthesis, cDNAs were column purified, and their quantity and quality were determined by UV absorbance. Gene-specific primers (Supplemental Table S6) were designed using Primer Express 3.0 software (Applied Biosystems), and primers from petunia EF1α (Mallona et al., 2010) were utilized for normalization. Each primer pair was validated to ensure that the amplification efficiency of each target gene was similar to the EF1α endogenous control using serial dilutions of cDNA synthesized from trichome RNA. Only primer pairs that had an absolute value of the slope of ΔCT versus log of the input cDNA concentration of 0.1 or less were utilized. Experiments to determine the relative expression of target genes in trichomes versus shaved stems were performed using 20 ng of cDNA template, while those investigating the efficiency of gene silencing were performed with 30 ng of cDNA template. PCRs (10 µL) containing 2× FAST SYBR Master Mix (Applied Biosystems), cDNA template, and 300 nm of each primer were assembled using a Biomek 3000 liquid handler and amplified using an Applied Biosystems ABI Prism 7900HT Real-Time PCR System. Amplification conditions were as follows: 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Data were analyzed using the ΔΔCT method to generate relative expression values.
Acylsugar Profiling by LC-MS
For acylsugar profiling, 10-mm segments of petioles were placed in 1 mL of extraction solvent (acetonitrile:isopropanol:water, 3:3:2, v/v) containing 0.1% formic acid and 5 μm telmisartan as an internal standard. Samples were gently agitated for 1 min, and the solvent was transferred to HPLC vials. The acylsugar extracts were analyzed using a Shimadzu LC-20AD HPLC system connected to a Waters LCT Premier ToF-MS device. Ten microliters of sample was injected onto a fused-core Ascentis Express C18 column (2.1 mm × 10 cm, 2.7 μm particle size) for reverse-phase separation. Column temperature was set to 50°C. The flow rate was set at 0.3 mL min−1, and start conditions were 99% solvent A (10 mm ammonium formate, pH 2.8) and 1% solvent B (acetonitrile). The 20-min elution gradient was as follows: ramp to 60% B at 1.25 min, 90% B at 12.5 min, and 100% B at 15 min. Hold at 100% B up to 19 min, return to 1% B at 19.01 min, and hold to 20 min. The MS parameters employed electrospray ionization and were as described (Schilmiller et al., 2015). Five separate acquisition functions were utilized to generate spectra at different collision potentials (10, 25, 40, 55, and 80 V), providing both nonfragmenting and a range of fragmenting conditions. Acylsugar abundances were determined using the Waters QuanLynx analysis tool to integrate extracted ion chromatogram peak areas relative to the peak area of the internal standard. Representative samples (Fig. 4A; Supplemental Fig. S5) were analyzed using a Waters Acquity UPLC system connected to a Waters Xevo G2-S QToF LC-MS device. Chromatography conditions were identical to those described above, except that the 20-min elution gradient was modified slightly such that 100% acetonitrile was maintained from 12.5 to 18 min followed by 1% acetonitrile from 18.01 to 20 min.
Synthesis of Acyl-CoA Thioesters
Isobutyryl-CoA (iC4-CoA) and isovaleryl-CoA (iC5-CoA) were purchased from Santa Cruz Biotechnology and Sigma-Aldrich, respectively. Additional acyl-CoA thioesters were synthesized from their cognate fatty acids using a carbonyldiimidazole method (Kawaguchi et al., 1981) with modifications outlined by Schilmiller et al. (2015). 2-Methylbutyric acid (aiC5), 4-methylpentanoic acid (iC6), and 5-methylhexanoic acid (iC7) were purchased from Sigma-Aldrich, and 6-methylheptanoic acid (iC8) and CoA trilithium salt (CoA) were obtained from BOC Sciences and Crystal Chem, respectively. The concentrations of the resulting acyl-CoAs were obtained by UV spectroscopy using the CoA extinction coefficient of 16,400 at 260 nm.
Recombinant Expression of BAHD Acyltransferases and Enzyme Assays
Full-length open reading frames of candidate ASAT enzymes were amplified from P. axillaris trichome cDNA using the primers described in Supplemental Table S6. Linkers containing either NheI or NdeI together with BamHI or SalI were added to permit subcloning of the fragments into the pET28b vector (EMD Millipore). Constructs were transformed into BL21 Rosetta Cells (EMD Millipore) for recombinant protein production. His-tagged protein production was induced in Escherichia coli cultures by 0.05 mm isopropyl β-d-1-thiogalactopyranoside for 16 h at 16°C. Soluble protein was partially purified by nickel-affinity chromatography as described previously (Schilmiller et al., 2012). Enzyme assays were performed for 30 min at 30°C in a 30-μL volume containing 50 mm ammonium acetate (pH 6) with 100 µm of an appropriate acyl-CoA donor and 1 mm Suc as the acyl acceptor. Assays were terminated by the addition of 60 μL of acetonitrile:isopropanol:formic acid (1:1:0.001) containing 10 μm propyl-4-hydroxy benzoate internal standard and centrifugation at 13,000g for 10 min.
For sequential enzyme assays, heat inactivation of previous enzyme activity was achieved by incubation at 65°C for 5 min followed by cooling on ice and the subsequent addition of an appropriate acyl-CoA donor (100 µm) and a candidate ASAT enzyme. Enzyme reactions were terminated as described previously (Schilmiller et al., 2015). Acylsugars produced in vitro together with representative samples from plant extracts were analyzed using a Waters Xevo G2-S QToF LC-MS device interfaced to a Waters Acquity UPLC system. Ten microliters of leaf dip extract was injected onto a fused-core Ascentis Express C18 column (5 cm × 2.1 mm, 2.7 µm particle size) interfaced to a Waters Acquity UPLC system. The column temperature was maintained at 40°C. The start conditions were 95% A and 5% solvent B with flow rate set to 0.3 mL min−1. The 7-min linear gradient was as follows: ramp to 60% B at 1 min, then to 100% B at 5 min, hold at 100% B to 6 min, return to 95% A at 6.01 min, and hold until 7 min. The MS settings are identical to those of Schilmiller et al. (2015). Three acquisition functions were set up to generate spectra at different collision potentials (5, 25, and 60 V). The method and MS parameters for electrospray ionization in both positive and negative ion modes are similar except that, in positive ion mode, the capillary voltage was 2.61 kV. Liquid chromatography-tandem MS in negative ion mode was used to determine the fragmentation of selected acylsucrose products from enzyme assays detected as m/z 555.2, 667.3, and 751.4 under the same conditions as above but with a single acquisition function at the collision potential of 6 V and acquisition mass range from m/z 50 to 600, 700, and 800, respectively.
Collision-induced dissociation MS in negative ion mode was used to obtain acylsugar structure information about the number and length of acyl chains according to the carboxylate fragment ion masses, whereas MS in positive ion mode was used to cleave the glycosidic bond linking the Suc pyranose and furanose rings and obtain information on the number and length of acyl chains present on each ring as described previously (Schilmiller et al., 2010a; Ghosh et al., 2014).
Analysis of Mono-Acylsucroses
To analyze the shift in retention time of mono-acylsucrose (S1:5) products formed by PaxASAT1 and SlASAT1, reaction products were analyzed using a Waters Acquity UPLC system connected to a Waters Xevo G2-S QToF LC-MS system. Ten microliters of reaction product was injected into a fused-core ACQUITY UPLC Ethylene Bridged Hybrid amide column (2.1 mm × 10 cm, 2.7 μm particle size; Waters) with column oven set to 40°C. The flow rate was 0.4 mL min−1, with initial conditions at 5% solvent A (0.15% formic acid) and 95% B (acetonitrile). The 9-min elution gradient was as follows: hold at 5% A to 1 min, then ramp to 40% A at 6 min, ramp to 95% A at 7 min, return to 5% A at 7.01 min, and hold until 9 min. The MS parameters were as described (Schilmiller et al., 2015), except that the mass range was m/z 50 to 1,000 and five separate acquisition functions were utilized to generate spectra at different collision potentials (10, 25, 40, 55, and 80 V).
Purification of Acylsugars and Structural Elucidation by NMR
One-liter cultures of E. coli expressing either PaxASAT1 or PaxASAT2 were induced, and protein was extracted and purified as described above. Enzyme assays with PaxASAT1 to form S1:5 (aiC5) were performed in a 30-mL reaction volume containing 50 mm ammonium acetate (pH 6), 0.66 mm Suc, and 50 μm aiC5-CoA at 30°C for 3 h. For the sequential assay with PaxASAT2 to form S2:10 (aiC5, aiC5), PaxASAT1 was inactivated at 65°C for 1 h, the reaction was cooled on ice and centrifuged, 30 μm aiC5-CoA and PaxASAT2 were added, and the reaction was incubated for 3 h at 30°C. Reactions were terminated by the addition of 60 mL of acetonitrile:isopropanol:formic acid (1:1:0.001) followed by centrifugation at 4,500 rpm for 10 min. Solvent was evaporated to dryness, and the residue was dissolved in 1 mL of water.
S1:5 (aiC5) and S2:10 (aiC5, aiC5) purification was performed using a Waters 2795 HPLC device and a Thermo Scientific Acclaim 120 C18 semipreparative HPLC column (4.6 × 150 mm, 5 μm particle size). The mobile phase consisted of 0.15% formic acid in water (solvent A) and acetonitrile (solvent B) with flow rate of 1.5 mL min−1. For the purification of S1:5, a linear gradient consisted of 1% solvent B at 0 min, 8% solvent B at 3 min, 9% solvent B at 11 min, and 100% solvent B at 12 min. Solvent composition was held at 100% solvent B for 12 to 14 min, brought back to 1% solvent B at 15 min, and held at 1% solvent B for 15 to 16 min. For the purification of S2:10, a 40-min method with a linear gradient consisted of 1% solvent B at 0 min, 8% solvent B at 3 min, 9% solvent B at 11 min, 17% solvent B from 12 to 35 min, ramped to 18% solvent B at 35 min and then to 100% solvent B at 36 min, held at 100% solvent B until 38 min, returned to 1% solvent B at 39 min, and held until 40 min. Eluted fractions of S1:5 were collected in 1-min fractions for five injections and eight injections for S2:10, using an injection volume of 200 μL for each injection. The desired compound was collected in 7 to 10 min for S1:5 and 23 min for S2:10 and evaporated to dryness. The residue was dissolved in 1 mL of water with sonication for 10 min, followed by transfer to a polypropylene centrifuge tube and centrifugation at 3,000g for 2 min at 25°C. Supernatants were collected in HPLC vials with glass low-volume inserts. Purified acylsugars were dissolved in 250 μL of deuterium oxide and transferred into Shigemi solvent-matched NMR tubes. NMR spectra were recorded using a Bruker Avance 900 NMR spectrometer equipped with a TCI triple-resonance inverse detection cryoprobe at the Michigan State University Max T. Rogers NMR facility. NMR experiments measured 1H (899.13 MHz), correlation spectroscopy, and HSQC spectroscopy (F1, 226.09 MHz; F2, 899.13 MHz). Acylsugar substitution patterns were determined by chemical shifts of H based on 1H and HSQC spectroscopy.
Accession Numbers
A P. axillaris transcriptome assembly and associated sequences are available under GenBank/EMBL/DDBJ BioProject PRJNA261953 (Guo et al., 2015). Sequences of P. axillaris ASATs are deposited in GenBank under the following accession numbers: PaxASAT1 (KT716258), PaxASAT2 (KT716259), PaxASAT3 (KT716260), and PaxASAT4 (KT716261).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Synteny between the SlASAT1 and Pax36474 genomic regions.
Supplemental Figure S2. Synteny between the Pax21699 genomic region of P. axillaris and two separate regions of tomato chromosome 2.
Supplemental Figure S3. Synteny between tomato and P. axillaris within a region that flanks the Pax39119 locus.
Supplemental Figure S4. Transcript abundance in candidate ASAT-silenced lines.
Supplemental Figure S5. Mass spectra of acylsugars that accumulate in TRV2::Pax21699-silenced lines.
Supplemental Figure S6. Activity of PaxASAT1 with multiple acyl-CoA donors.
Supplemental Table S1. Putative ASATs identified from P. axillaris transcriptome data.
Supplemental Table S2. P. axillaris acylsugars detected in petioles of P. axillaris.
Supplemental Table S3. In vitro activities of candidate ASAT enzymes of P. axillaris.
Supplemental Table S4. NMR chemical shifts of S1:5 (aiC5) produced in vitro by PaxASAT1.
Supplemental Table S5. NMR chemical shifts of S2:10 (aiC5) produced in vitro by sequential action of PaxASAT1 and PaxASAT2.
Supplemental Table S6. Oligonucleotide primers utilized in this study.
Acknowledgments
We thank members of the Solanum Trichome Project (www.trichome.msu.edu), particularly Matthew Bedewitz, Tony Schilmiller, and Pengxiang Fan, for sharing materials and providing advice and Krystle Wiegert-Rininger for assistance with RNA isolation; we also thank the Michigan State University RTSF Mass Spectrometry and Metabolomics Core Facility and Kermit Johnson and Dan Holmes of the Michigan State University Max T. Rogers NMR facility for assistance with collecting NMR spectra.
Glossary
- qRT
quantitative reverse transcription
- VIGS
virus-induced gene silencing
- HSQC
heteronuclear single-quantum correlation
- MS
mass spectrometry
- LC-MS
liquid chromatography-mass spectrometry
Footnotes
This research was supported by NSF IOS-1025636 and NSF IOS-1546617. C.S.B. and A.D.J. are supported in part by Michigan AgBioResearch and through the USDA National Institute of Food and Agriculture, Hatch project numbers MICL02265 and MICL02143.
Articles can be viewed without a subscription.
References
- Arimura G, Matsui K, Takabayashi J (2009) Chemical and molecular ecology of herbivore-induced plant volatiles: proximate factors and their ultimate functions. Plant Cell Physiol 50: 911–923 [DOI] [PubMed] [Google Scholar]
- Arrendale RF, Severson RF, Sisson VA, Costello CE, Leary JA, Himmelsbach DS, Vanhalbeek H (1990) Characterization of the sucrose ester fraction from Nicotiana glutinosa. J Agric Food Chem 38: 75–85 [Google Scholar]
- Bedewitz MA, Góngora-Castillo E, Uebler JB, Gonzales-Vigil E, Wiegert-Rininger KE, Childs KL, Hamilton JP, Vaillancourt B, Yeo YS, Chappell J, et al. (2014) A root-expressed L-phenylalanine:4-hydroxyphenylpyruvate aminotransferase is required for tropane alkaloid biosynthesis in Atropa belladonna. Plant Cell 26: 3745–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird DA, Franceschi VR, Facchini PJ (2003) A tale of three cell types: alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell 15: 2626–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker PM, Mirabella R, Diergaarde PJ, VanDoorn A, Tissier A, Kant MR, Prins M, de Vos M, Haring MA, Schuurink RC (2012) Improved herbivore resistance in cultivated tomato with the sesquiterpene biosynthetic pathway from a wild relative. Proc Natl Acad Sci USA 109: 20124–20129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A, Moser M, Amrad A, Bao M, Bapaume L, Barry CS, Bliek M, Boersma MR, Borghi L, Bruggmann R, et al. (2016) Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat Plants 2: 16074. [DOI] [PubMed] [Google Scholar]
- Boutanaev AM, Moses T, Zi J, Nelson DR, Mugford ST, Peters RJ, Osbourn A (2015) Investigation of terpene diversification across multiple sequenced plant genomes. Proc Natl Acad Sci USA 112: E81–E88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buta JG, Lusby WR, Neal JW, Waters RM, Pittarelli GW (1993) Sucrose esters from Nicotiana gossei active against the greenhouse whitefly Trialeuroides vaporariorum. Phytochemistry 32: 859–864 [Google Scholar]
- Chae L, Kim T, Nilo-Poyanco R, Rhee SY (2014) Genomic signatures of specialized metabolism in plants. Science 344: 510–513 [DOI] [PubMed] [Google Scholar]
- Chortyk OT, Kays SJ, Teng Q (1997) Characterization of insecticidal sugar esters of Petunia. J Agric Food Chem 45: 270–275 [Google Scholar]
- Chortyk OT, Severson RF, Cutler HC, Sisson VA (1993) Antibiotic activities of sugar esters isolated from selected Nicotiana species. Biosci Biotechnol Biochem 57: 1355–1356 [DOI] [PubMed] [Google Scholar]
- D’Auria JC. (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Dong Y, Burch-Smith TM, Liu Y, Mamillapalli P, Dinesh-Kumar SP (2007) A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol 145: 1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Klempien A, Muhlemann JK, Kaplan I (2013) Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol 198: 16–32 [DOI] [PubMed] [Google Scholar]
- Dugé de Bernonville T, Clastre M, Besseau S, Oudin A, Burlat V, Glévarec G, Lanoue A, Papon N, Giglioli-Guivarc’h N, St-Pierre B, et al. (2015) Phytochemical genomics of the Madagascar periwinkle: unravelling the last twists of the alkaloid engine. Phytochemistry 113: 9–23 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan P, Miller AM, Schilmiller AL, Liu X, Ofner I, Jones AD, Zamir D, Last RL (2016) In vitro reconstruction and analysis of evolutionary variation of the tomato acylsucrose metabolic network. Proc Natl Acad Sci USA 113: E239–E248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobes JF, Mudd JB, Marsden MPF (1985) Epicuticular lipid accumulation on the leaves of Lycopersicon pennellii (Corr.) D’Arcy and Lycopersicon esculentum Mill. Plant Physiol 77: 567–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman E, Wang J, Iijima Y, Froehlich JE, Gang DR, Ohlrogge J, Pichersky E (2005) Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell 17: 1252–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh B, Westbrook TC, Jones AD (2014) Comparative structural profiling of trichome specialized metabolites in tomato (Solanum lycopersicum) and S. habrochaites: acylsugar profiles revealed by UHPLC/MS and NMR. Metabolomics 10: 496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffreda JC, Mutschler MA, Avé DA, Tingey WM, Steffens JC (1989) Aphid deterrence by glucose esters in glandular trichome exudate of the wild tomato, Lycopersicon pennellii. J Chem Ecol 15: 2135–2147 [DOI] [PubMed] [Google Scholar]
- Gonzales-Vigil E, Hufnagel DE, Kim J, Last RL, Barry CS (2012) Evolution of TPS20-related terpene synthases influences chemical diversity in the glandular trichomes of the wild tomato relative Solanum habrochaites. Plant J 71: 921–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Wiegert-Rininger KE, Vallejo VA, Barry CS, Warner RM (2015) Transcriptome-enabled marker discovery and mapping of plastochron-related genes in Petunia spp. BMC Genomics 16: 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halinski LP, Stepnowski P (2013) GC-MS and MALDI-TOF MS profiling of sucrose esters from Nicotiana tabacum and N. rustica. Z Naturforsch C J Biosci 68: 210–222 [DOI] [PubMed] [Google Scholar]
- Hare JD. (2005) Biological activity of acyl glucose esters from Datura wrightii glandular trichomes against three native insect herbivores. J Chem Ecol 31: 1475–1491 [DOI] [PubMed] [Google Scholar]
- Kandra G, Severson R, Wagner GJ (1990) Modified branched-chain amino acid pathways give rise to acyl acids of sucrose esters exuded from tobacco leaf trichomes. Eur J Biochem 188: 385–391 [DOI] [PubMed] [Google Scholar]
- Kang JH, Gonzales-Vigil E, Matsuba Y, Pichersky E, Barry CS (2014) Determination of residues responsible for substrate and product specificity of Solanum habrochaites short-chain cis-prenyltransferases. Plant Physiol 164: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Yoshimura T, Okuda S (1981) A new method for the preparation of acyl-CoA thioesters. J Biochem 89: 337–339 [DOI] [PubMed] [Google Scholar]
- Kim J, Kang K, Gonzales-Vigil E, Shi F, Jones AD, Barry CS, Last RL (2012) Striking natural diversity in glandular trichome acylsugar composition is shaped by variation at the Acyltransferase2 locus in the wild tomato Solanum habrochaites. Plant Physiol 160: 1854–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Matsuba Y, Ning J, Schilmiller AL, Hammar D, Jones AD, Pichersky E, Last RL (2014a) Analysis of natural and induced variation in tomato glandular trichome flavonoids identifies a gene not present in the reference genome. Plant Cell 26: 3272–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Park M, Yeom SI, Kim YM, Lee JM, Lee HA, Seo E, Choi J, Cheong K, Kim KT, et al. (2014b) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46: 270–278 [DOI] [PubMed] [Google Scholar]
- King RR, Calhoun LA, Singh RP (1988) 3,4-Di-O-acylated and 2,3,4-tri-O-acylated glucose esters from the glandular trichomes of nontuberous Solanum species. Phytochemistry 27: 3765–3768 [Google Scholar]
- King RR, Pelletier Y, Singh RP, Calhoun LA (1986) 3,4-Di-o-isobutyryl-6-o-caprylsucrose: the major component of a novel sucrose ester complex from type B glandular trichomes of Solanum berthaultii Hawkes (PI 473340). J Chem Soc Chem Commun 1078–1079 [Google Scholar]
- Klahre U, Gurba A, Hermann K, Saxenhofer M, Bossolini E, Guerin PM, Kuhlemeier C (2011) Pollinator choice in Petunia depends on two major genetic loci for floral scent production. Curr Biol 21: 730–739 [DOI] [PubMed] [Google Scholar]
- Lange BM, Turner GW (2013) Terpenoid biosynthesis in trichomes: current status and future opportunities. Plant Biotechnol J 11: 2–22 [DOI] [PubMed] [Google Scholar]
- Liu X, Enright M, Barry CS, Jones AD (2017) Profiling, isolation and structure elucidation of specialized metabolites accumulating in trichomes of Petunia species. Metabolomics 13: 85 [Google Scholar]
- Luckwill L. (1943) The Genus Lycopersicon: A Historical, Biological, and Taxonomic Survey of the Wild and Cultivated Tomatoes. Aberdeen University Press, Aberdeen, Scotland [Google Scholar]
- Luu VT, Weinhold A, Ullah C, Dressel S, Schoettner M, Gase K, Gaquerel E, Xu S, Baldwin IT (2017) O-Acyl sugars protect a wild tobacco from both native fungal pathogens and a specialist herbivore. Plant Physiol 174: 370–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol 10: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuba Y, Nguyen TTH, Wiegert K, Falara V, Gonzales-Vigil E, Leong B, Schäfer P, Kudrna D, Wing RA, Bolger AM, et al. (2013) Evolution of a complex locus for terpene biosynthesis in Solanum. Plant Cell 25: 2022–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Last RL (2012) Achieving diversity in the face of constraints: lessons from metabolism. Science 336: 1663–1667 [DOI] [PubMed] [Google Scholar]
- Mithöfer A, Boland W (2012) Plant defense against herbivores: chemical aspects. Annu Rev Plant Biol 63: 431–450 [DOI] [PubMed] [Google Scholar]
- Mugford ST, Qi X, Bakht S, Hill L, Wegel E, Hughes RK, Papadopoulou K, Melton R, Philo M, Sainsbury F, et al. (2009) A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell 21: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata J, Roepke J, Gordon H, De Luca V (2008) The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 20: 524–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J, Moghe GD, Leong B, Kim J, Ofner I, Wang Z, Adams C, Jones AD, Zamir D, Last RL (2015) A feedback-insensitive isopropylmalate synthase affects acylsugar composition in cultivated and wild tomato. Plant Physiol 169: 1821–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizaki Y, Yasunaga M, Okamoto E, Okamoto M, Hirose Y, Yamaguchi M, Ozeki Y, Sasaki N (2013) p-Hydroxybenzoyl-glucose is a zwitter donor for the biosynthesis of 7-polyacylated anthocyanin in Delphinium. Plant Cell 25: 4150–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Lewinsohn E (2011) Convergent evolution in plant specialized metabolism. Annu Rev Plant Biol 62: 549–566 [DOI] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195 [DOI] [PubMed] [Google Scholar]
- Rambla JL, Tikunov YM, Monforte AJ, Bovy AG, Granell A (2014) The expanded tomato fruit volatile landscape. J Exp Bot 65: 4613–4623 [DOI] [PubMed] [Google Scholar]
- Särkinen T, Bohs L, Olmstead RG, Knapp S (2013) A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evol Biol 13: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A, Shi F, Kim J, Charbonneau AL, Holmes D, Jones AD, Last RL (2010a) Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J 62: 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Charbonneau AL, Last RL (2012) Identification of a BAHD acetyltransferase that produces protective acyl sugars in tomato trichomes. Proc Natl Acad Sci USA 109: 16377–16382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Last RL, Pichersky E (2008) Harnessing plant trichome biochemistry for the production of useful compounds. Plant J 54: 702–711 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Miner DP, Larson M, McDowell E, Gang DR, Wilkerson C, Last RL (2010b) Studies of a biochemical factory: tomato trichome deep expressed sequence tag sequencing and proteomics. Plant Physiol 153: 1212–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Moghe GD, Fan P, Ghosh B, Ning J, Jones AD, Last RL (2015) Functionally divergent alleles and duplicated loci encoding an acyltransferase contribute to acylsugar metabolite diversity in Solanum trichomes. Plant Cell 27: 1002–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson RF, Arrendale RF, Chortyk OT, Green CR, Thome FA, Stewart JL, Johnson AW (1985) Isolation and characterization of the sucrose esters of the cuticular waxes of green tobacco leaf. J Agric Food Chem 33: 870–875 [Google Scholar]
- Shapiro JA, Steffens JC, Mutschler MA (1994) Acylsugars of the wild tomato Lycopersicon pennellii in relation to geographic distribution of the species. Biochem Syst Ecol 22: 545–561 [Google Scholar]
- Slocombe SP, Schauvinhold I, McQuinn RP, Besser K, Welsby NA, Harper A, Aziz N, Li Y, Larson TR, Giovannoni J, et al. (2008) Transcriptomic and reverse genetic analyses of branched-chain fatty acid and acyl sugar production in Solanum pennellii and Nicotiana benthamiana. Plant Physiol 148: 1830–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewksbury JJ, Nabhan GP (2001) Seed dispersal: directed deterrence by capsaicin in chilies. Nature 412: 403–404 [DOI] [PubMed] [Google Scholar]
- Tissier A. (2012) Glandular trichomes: what comes after expressed sequence tags? Plant J 70: 51–68 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velásquez AC, Chakravarthy S, Martin GB (2009) Virus-induced gene silencing (VIGS) in Nicotiana benthamiana and tomato. J Vis Exp 28: e1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters DS, Steffens JC (1990) Branched-chain amino acid metabolism in the biosynthesis of Lycopersicon pennellii glucose esters. Plant Physiol 93: 1544–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold A, Baldwin IT (2011) Trichome-derived O-acyl sugars are a first meal for caterpillars that tags them for predation. Proc Natl Acad Sci USA 108: 7855–7859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer T, Gazda V, He Z, Kaminski F, Kern M, Larson TR, Li Y, Meade F, Teodor R, Vaistij FE, et al. (2012) A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336: 1704–1708 [DOI] [PubMed] [Google Scholar]