Pea plants with altered amino acid transport processes produce higher seed yields and use nitrogen (N) more efficiently than wild-type plants in N-poor and N-rich soils.
Abstract
Improving the efficiency of nitrogen (N) uptake and utilization in plants could potentially increase crop yields while reducing N fertilization and, subsequently, environmental pollution. Within most plants, N is transported primarily as amino acids. In this study, pea (Pisum sativum) plants overexpressing AMINO ACID PERMEASE1 (AAP1) were used to determine if and how genetic manipulation of amino acid transport from source to sink affects plant N use efficiency. The modified plants were grown under low, moderate, or high N fertilization regimes. The results showed that, independent of the N nutrition, the engineered plants allocate more N via the vasculature to the shoot and seeds and produce more biomass and higher seed yields than wild-type plants. Dependent on the amount of N supplied, the AAP1-overexpressing plants displayed improved N uptake or utilization efficiency, or a combination of the two. They also showed significantly increased N use efficiency in N-deficient as well as in N-rich soils and, impressively, required half the amount of N to produce as many fruits and seeds as control plants. Together, these data support that engineering N allocation from source to sink presents an effective strategy to produce crop plants with improved productivity as well as N use efficiency in a range of N environments.
Nitrogen (N) is an essential nutrient that plants require in large amounts for growth and development. In industrial countries, high N fertilization enables maximum crop yields, and in the last 50 years, the use of synthetic N fertilizers has increased dramatically to meet the agricultural demands of a growing population (FAO, 2012; Conant et al., 2013). However, depending on the crop species and soil conditions, plants take up less than half of the applied N fertilizer (Raun and Johnson, 1999; Hodge et al., 2000; Kumar and Goh, 2002; Yang et al., 2015; Zhu et al., 2016). The remaining soil N may contaminate aquatic systems through runoffs or leached water (Crews and Peoples, 2005; Gruber and Galloway, 2008) or it may undergo denitrification and be released into the atmosphere as nitrous oxide, a powerful greenhouse gas (Bouwman, 1996; Bouwman et al., 2002), altogether resulting in negative effects on the environment and human health. Oppositely, in developing countries, access to N fertilizer is limited, and insufficient N nutrition results in low crop productivity and, ultimately, in reduced food supply (Brown et al., 2009; Lal, 2009).
One solution to these issues is the production of crop varieties that are highly efficient in using N and produce high yields with reduced N input (Masclaux-Daubresse et al., 2010; McAllister et al., 2012; Garnett et al., 2015). Plant nitrogen use efficiency (NUE) is determined by plant seed yield relative to the amount of N applied and is generally composed of both N uptake and N utilization efficiency (Moll et al., 1982). Nitrogen uptake efficiency (NUpE) is defined as total shoot N relative to the amount of N supplied to the soil. The uptake of N by the root is influenced by the availability of soil N and controlled by plant N assimilation processes and N metabolite levels as well as the N demand of the plant (Ruffel et al., 2011; Nacry et al., 2013; Stahl et al., 2016). In addition, recent work has resolved that amino acid transport processes in root and shoot strongly influence N uptake and metabolism, most probably through a feedback regulatory mechanism and shoot-to-root signaling (Miller et al., 2008; Sanders et al., 2009; Tan et al., 2010; Zhang et al., 2010, 2015; Forde, 2014; Santiago and Tegeder, 2016). Nitrogen utilization efficiency (NUtE) is defined as total seed yield relative to total shoot N content (Moll et al., 1982) and is affected by several physiological factors, including N uptake, metabolism, allocation, and remobilization (Habash et al., 2001; Tsay et al., 2011; Girondé et al., 2015). In addition, regulated transport of N within the plant strongly influences the amount of N allocated to seeds and final seed yields (Rolletschek et al., 2005; Schmidt et al., 2007; Weigelt et al., 2008; Tan et al., 2010; Zhang et al., 2010, 2015; Carter and Tegeder, 2016; Santiago and Tegeder, 2016).
Approaches to improve plant NUE have focused mainly on the genetic manipulation of N uptake (Fang et al., 2013; Araus et al., 2016; Chen et al., 2017), nitrate allocation (Tsay et al., 2011), N metabolism (Ameziane et al., 2000; Chichkova et al., 2001; Habash et al., 2001; Yamaya et al., 2002; Seiffert et al., 2004; Good et al., 2007; Shrawat et al., 2008; Peña et al., 2017), and its regulation (Yanagisawa et al., 2004). While generally successful, these studies often did not address the different components of NUE (i.e. NUtE and NUpE) and/or the effects of different N environments on NUE, or they only demonstrated improved NUE when transgenic plants were grown under a specific N condition. Furthermore, N partitioning processes within the plant and associated amino acid transporters have been discussed as potentially important factors influencing NUE (Masclaux-Daubresse et al., 2010; Xu et al., 2012; Gaju et al., 2014; Mu et al., 2015; see above), but their role in efficient plant N use has not yet been investigated. The goal of this study was to tackle this lack of knowledge.
Plants generally take up nitrate and ammonium from the soil, which are assimilated into amino acids in either roots or shoots, depending on the plant species (von Wirén et al., 2000; Miller and Cramer, 2005; Miller et al., 2007; Xu et al., 2012; Krapp et al., 2014). In legumes like pea (Pisum sativum), amino acid synthesis occurs mainly in the roots followed by its transport via the xylem to mature source leaves (Atkins et al., 1983). Within leaves, the amino-N is utilized for a variety of metabolic processes, transiently stored as amino acids or protein, or loaded into the phloem in order to supply N to developing sinks, such as fruit and seeds (Urquhart and Joy, 1982; Tegeder and Rentsch, 2010; Zhang et al., 2010; Tegeder, 2014; Santiago and Tegeder, 2016). Some of the root-derived amino acids also might be transferred from the xylem to the phloem for direct N supply of sinks (Urquhart and Joy, 1982; Zhang et al., 2010; Tegeder, 2014) or exit the xylem along the transport pathway for transient storage in the stem (McNeil et al., 1979). During the reproductive stage, N stored in stem and leaves is remobilized and transported in the phloem to developing flowers, pods, and seeds (Pate and Flinn, 1973; Schiltz et al., 2005). Within the seed, amino acids are taken up by the embryo for metabolism and the accumulation of storage compounds, such as proteins and starch (Lanfermeijer et al., 1990; Jenner et al., 1991; Hirner et al., 1998; Sanders et al., 2009; Zhang et al., 2015). Both loading of amino acids into the phloem and uptake into the embryo are key regulatory steps in N allocation from source to sink and require the activity of transport proteins (Schmidt et al., 2007; Tan et al., 2010; Zhang et al., 2010, 2015; Tegeder, 2014; Santiago and Tegeder, 2016). The function of these amino acid transporters impacts the number of pods and seeds that develop as well as the amount of seed storage compounds (Atkins et al., 1975; Hirel et al., 2007; Sanders et al., 2009; Tan et al., 2010; Zhang et al., 2015; Santiago and Tegeder, 2016). We hypothesized that these organic N transporters in source and sink also contribute to plant NUE and that their activities affect plant growth and development in N-poor as well as N-rich environments.
In recent work, we found that simultaneous overexpression of AMINO ACID PERMEASE1 (AAP1) in the phloem and embryos of pea plants improved source-to-sink allocation of amino acids and led to increased seed yield and seed storage protein levels when the plants were grown with highly abundant N nutrition (Zhang et al., 2015). The results further indicated positive feedback regulation of N uptake and assimilation in roots and increased shoot N supply. Using AAP1-overexpressing (AAP1-OE) plants, this study aimed to resolve the role of source-to-sink N partitioning processes in NUE and to determine if increased amino acid transporter expression in source and sink results in improved plant performance, even in N-deficient environments. The AAP1-OE and wild-type pea plants were grown in low, moderate, and high N-fertilized soils, and the effects on the efficiency of N uptake and utilization were evaluated. The results demonstrate that AAP1-OE plants outperform the control plants and achieve higher seed yields in all N fertilization regimes. While the effects on NUpE and NUtE varied with the different N treatments, NUE was improved significantly under all N conditions tested. Remarkably, with half the amount of N fertilizer supplied, the modified plants performed as well as wild-type plants grown with twice as much N. Overall, our work demonstrates that the manipulation of source-to-sink N transport provides a promising approach for increasing plant productivity while optimizing N use.
RESULTS
Shoot Biomass and Root-to-Shoot N Partitioning Are Increased in AAP1-OE Plants Grown under Low, Moderate, and High N Nutrition
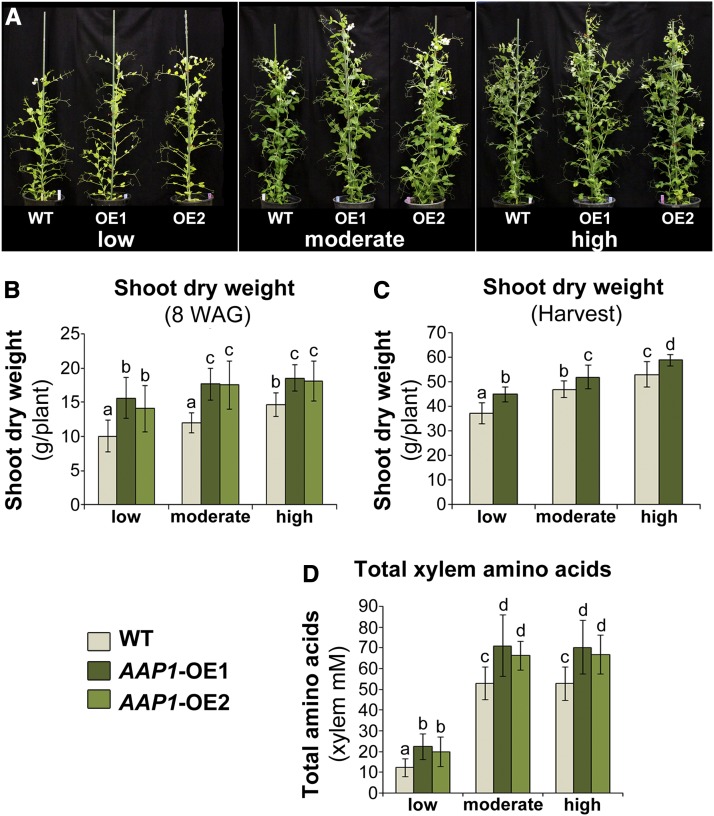
Previous studies in our laboratory demonstrated that, when grown in very high N environments, pea plants overexpressing AAP1 in the leaf phloem and embryos produce more shoot biomass and seed yield, as well as higher seed protein levels, relative to the wild type (Zhang et al., 2015). In this study, we examined if the improved performance of the AAP1-OE plants is due in part to high N nutrition or if the genetic manipulation provides the AAP1-OE plants with an essential trait resulting in improved performance even in N-deficient environments. Non-nodulated transgenic and wild-type plants were grown in nutrient-deficient soil with 2 g (low), 4 g (moderate), or 8 g (high) of N per week and a modified Hoagland nutrient solution. These N amounts were established in preliminary experiments with wild-type pea plants. High N fertilization resulted in maximum shoot growth, while moderate N nutrition caused a significant decrease in shoot biomass (Fig. 1, A to C). Wild-type plants grown under low N supply showed severe N deficiency symptoms, including yellow leaves and reduced shoot growth (Fig. 1, A to C). To analyze the growth of AAP1-OE compared with wild-type plants in the different N soils, shoot dry weights of 8-week-old plants (Fig. 1, A and B) and of plants at harvest (Fig. 1C) were examined. The results resolved that AAP1-OE plants produced significantly more biomass than the wild type for all N treatments.
Figure 1.
Shoot biomass and root-to-shoot amino acid transport in AAP1-OE and wild-type (WT) pea plants grown in low (2 g of N per week), moderate (4 g of N per week), or high N (8 g of N per week) environments. A, Image of two AAP1-OE lines (OE1 and OE2) and wild-type plants after 8 weeks of growth. B, Shoot dry weight of plants 8 weeks after germination (WAG; n ≥ 6). C, Shoot dry weight of desiccated plants at harvest (16-week-old plants; n ≥ 6). D, Total free amino acids in xylem (n ≥ 7). Data are means ± sd. Significant differences are indicated by letters (ANOVA; P ≤ 0.05).
Non-nodulated pea plants mainly assimilate N in the roots and transport the resulting amino acids to the shoot via the xylem. To analyze if and how N delivery from root to shoot is affected in the non-nodulated AAP1-OE and wild-type plants, xylem amino acid content was analyzed. Generally, the total xylem amino acid content was similar in pea plants grown under moderate and high N, while it was strongly reduced in plants with low N supply (Fig. 1D). This corroborates other studies showing that plants display severe reductions in N uptake and assimilation in extremely low N environments (Black and Wight, 1979; Delogu et al., 1998; Tillard et al., 1998). When comparing AAP1-OE lines and the wild type, the results showed a significant increase in total xylem amino acids for the transgenic plants under all N conditions tested (Fig. 1D). Together, the results support that, independent of the amount of N supplied, AAP1-OE plants perform better than wild-type plants with respect to root-to-shoot N allocation and shoot biomass production.
AAP1-OE Plants Display Increased Source-to-Sink Transport of Amino Acids under Low, Moderate, and High N Nutrition
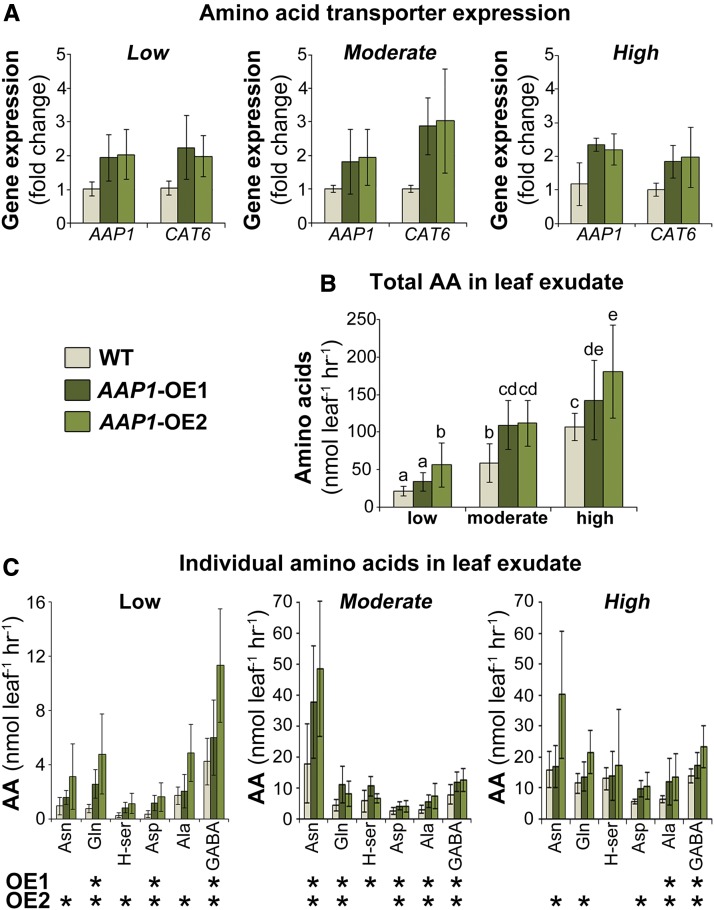
To analyze the effects of N nutrition on N partitioning to sinks, the expression of phloem amino acid transporters as well as amino acid levels in leaf exudate were examined (Fig. 2). The expression of AAP1 as well as CAT6 (CATIONIC AMINO ACID TRANSPORTER6), which also may contribute to increased amino acid phloem loading in AAP1-OE leaves (Zhang et al., 2015), was analyzed (Fig. 2A). The results showed an up-regulation of both transporters in AAP1-OE plants exposed to low, moderate, and high soil N levels (Fig. 2A). Generally, exudates released from leaf petioles consist mainly of phloem sap but also may contain fluids from other leaf cells, the leaf apoplast, and xylem (Fiehn, 2003; Zhang et al., 2012). Analysis of leaf exudates resolved that the total amino acid concentrations were generally increased for all pea genotypes grown with higher N fertilization (Fig. 2B; Winzer et al., 1996; Tillard et al., 1998). Most importantly, when comparing transgenic versus wild-type plants, amino acid levels were significantly elevated in the AAP1-OE leaf exudates under all N treatments (Fig. 2B). Analysis of the most abundant amino acids showed that the amounts of Asn, Gln, Asp, and Ala were increased for all N fertilization regimes for one or both of the AAP1-OE lines. Homoserine levels, however, seem to have been up-regulated only under low and moderate N conditions (Fig. 2C). In addition, the amounts of γ-aminobutyric acid (GABA), a non-protein amino acid, were increased in the AAP1-OE leaf exudates under all N conditions tested. GABA levels were highest when AAP1-OE plants were grown in low N soil and contributed considerably to the total amino acid pool in leaf exudates (Fig. 2C). This observed GABA accumulation under N deficiency is in line with the proposed function of the amino acid in environmental stress responses (Kinnersley and Turano, 2000; Shelp et al., 2012). Overall, the results support that improved source-to-sink translocation of N in AAP1-OE pea plants is achieved not only under high but also under deficient N nutrition.
Figure 2.
Analysis of source-to-sink amino acid transport in two AAP1-OE lines (AAP1-OE1 and AAP1-OE2) and wild-type (WT) pea plants. Eight-week-old plants grown under low (2 g of N per week), moderate (4 g of N per week), or high N (8 g of N per week) supply were examined. A, Expression analysis of the amino acid transporters AAP1 (Zhang et al., 2010) and CAT6 (Tan et al., 2010) in source leaves using reverse transcription-quantitative PCR. Results are shown as fold change compared with the wild type (n = 5 biological replicates). A mean fold change of greater than 2 between the wild type and mutants was used to identify differentially expressed genes. B, Total amino acids in leaf exudates (n ≥ 7). C, Individual amino acids (AA) in leaf exudates (n ≥ 7). Data are means ± sd. Significant differences for each AAP1-OE line are indicated by letters (A and B) or asterisks (C; ANOVA; P ≤ 0.05).
AAP1-OE Plants Produce More Seeds Than Wild-Type Plants, Independent of the N Condition, But Their Seed N Content Depends on N Availability
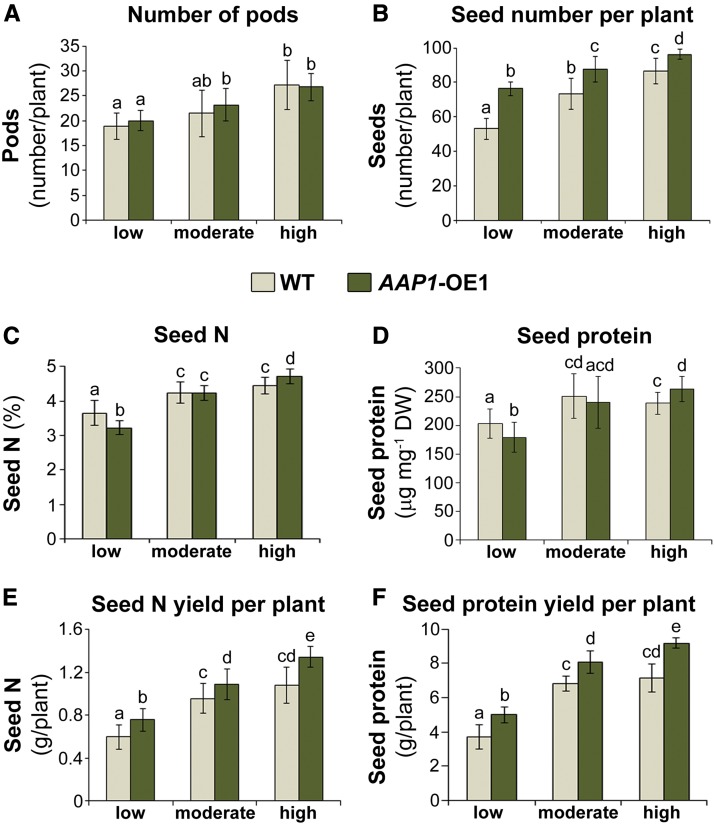
The effects of N fertilization on reproductive AAP1-OE sink development were analyzed. No differences in pod number per plant were found between AAP1-OE and wild-type plants (Fig. 3A), including under high N nutrition. This varies from the work by Zhang et al. (2015) showing that pod numbers were increased significantly in AAP1-OE versus wild-type plants. The discrepancy is most probably due to differences in the type of growth medium and the type and amount of fertilization. Our study used nutrient-deficient soil and a modified Hoagland solution. In the previous work, plants were grown on nutritious soil and with plentiful nutrition using a commercial fertilizer at high volume to maximize pod and seed development (see “Materials and Methods”; Zhang et al., 2015). Nevertheless, and most importantly, in this work, the seed number per pod was higher in the transgenic plants for all N treatments, resulting in a significant increase in the number of seeds per AAP1-OE plant compared with wild-type plants independent of the N nutrition (Fig. 3B). Notably, a comparison between moderate and high N treatment showed that AAP1-OE plants fed with half the amount of N produced as many seeds as wild-type plants grown under high N supply (Fig. 3B).
Figure 3.
Analyses of sink number and seed N levels of AAP1-OE1 and wild-type (WT) plants grown with low (2 g of N per week), moderate (4 g of N per week), or high N (8 g of N per week) supply. Plants were grown for 16 weeks and analyzed after desiccation. A, Total number of pods per plant (n ≥ 6). B, Total seed number per plant (n ≥ 6). C, Seed elemental N content (n ≥ 5 plants). D, Seed protein content (n ≥ 5). E, Total seed N yield per plant (n ≥ 5). F, Total seed protein yield per plant (n ≥ 5). Data are means ± sd. Significant differences are indicated by letters (ANOVA; P ≤ 0.05). DW, Dry weight.
In order to evaluate the effects of N nutrition on seed quality of AAP1-OE plants, seed N and protein levels were determined (Fig. 3, C and D). Both N and protein content were increased in AAP1-OE versus wild-type seeds under high N supply, unchanged under moderate N, and reduced under low N fertilization. These data support that, independent of the amount of N fed to the plant, the increased N allocation to sinks primarily improves seed number rather than protein amount per seed (Zhang et al., 2015). Only when N supply is plentiful, as under high N, are both maximum sink development and seed N/protein accumulation achieved (Fig. 3, C–F; Zhang et al., 2015). Nevertheless, when calculating total seed N and protein per plant, the results showed that AAP1-OE plants perform significantly better than wild-type plants for all N treatments. They allocate more N to seed sinks and produce higher protein yields (Fig. 3, E and F).
AAP1-OE Plants Contain Less N in Stubble Tissue under Low and Moderate N Supply
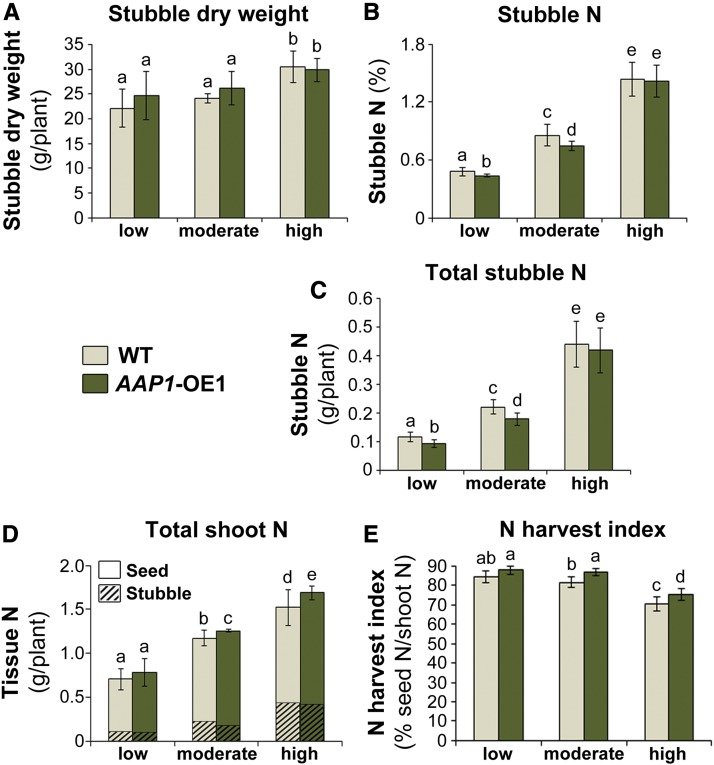
In order to determine the N uptake and utilization efficiencies of AAP1-OE and wild-type plants, total shoot N needed to be examined at harvest. Shoot N is composed of seed N (Fig. 3E) as well as stubble N, which includes N present in stem, leaf, and pod wall tissues (all shoot tissue except seeds). First, the effects of the different N treatments on stubble mass and N content were evaluated (Fig. 4, A–C). The results showed that high versus moderate and low N fertilization generally led to significantly higher stubble dry weight in both transgenic and wild-type pea plants (Fig. 4A). But, within each N treatment, no difference in stubble dry weight was observed between AAP1-OE lines and the wild type. Furthermore, when grown with high N, no differences were found between AAP1-OE and wild-type plants with respect to the percentage of N in stubble tissue and total stubble N content (Fig. 4, B and C). However, under low and moderate N conditions, the stubble matter of AAP1-OE plants contained significantly less N than in the wild type.
Figure 4.
Analyses of stubble biomass and shoot N content of AAP1-OE1 and wild-type (WT) plants grown with low (2 g of N per week), moderate (4 g of N per week), or high N (8 g of N per week) supply. Stubble biomass includes stem, leaves, and pod walls and excludes seeds. A, Stubble dry weight (n ≥ 5 plants). B, Percentage of N in stubble (n ≥ 5). C, Total stubble N (n ≥ 5). D, Total shoot N as a sum of the total amount of N in stubble and seeds per plant (n ≥ 5). E, N harvest index, which presents the percentage of shoot N in seeds (n ≥ 5). Data are means ± sd. Significant differences are indicated by letters (ANOVA; P ≤ 0.05).
Dependent on the N application, stubble N contributed between 12% and 29% to the total shoot N of the pea plants analyzed, while seed N represented the major N pool (Fig. 4D). Although stubble N was reduced in AAP1-OE plants grown in environments with low and moderate N supply relative to the wild type, the transgenic plants showed no change or an increase in total shoot N, respectively, due to significant increases in total seed N per plant (Figs. 3E and 4D). Together, these results suggest that AAP1-OE plants supplied with limited N amounts store less N in stubble than wild-type plants and allocate the N to sinks. However, when plenty of N is available, as in the high N treatment, AAP1-OE plants contain similar N amounts to the wild type in stubble tissues while increasing N delivery to seed sinks, together resulting in an increased total shoot N (Figs. 3E and 4D). Furthermore, increased shoot N levels in AAP1-OE plants grown with moderate and high N amounts indicate that, under those N conditions, the transgenic plants take up and distribute more N to the shoot than in the wild type. Overall, the AAP1-OE plants displayed a higher N harvest index (i.e. the percentage of shoot N present in seeds; Fig. 4E), suggesting that they preferentially allocate N to seeds, which results in an overall increase in seed production as well as in higher seed N and protein yields (Fig. 3, B, E, and F).
NUE Is Greatly Improved in AAP1-OE Plants
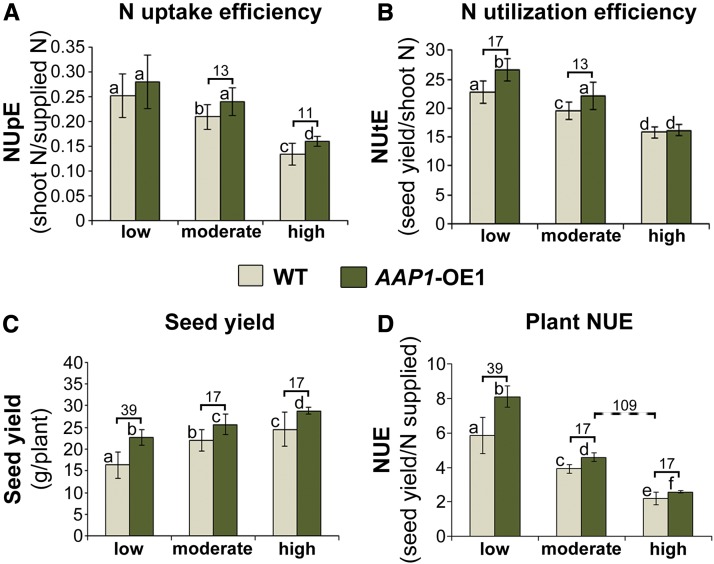
Plant NUE is generally composed of both NUpE and NUtE (Moll et al., 1982). NUpE and NUtE of AAP1-OE and wild-type plants grown under low, moderate, and high N conditions were calculated as described in “Materials and Methods.” Overall, NUpE (and NUtE) decreased in all pea plants analyzed with increasing N supply, as generally expected (Fig. 5, A and B; Beatty et al., 2010). Under low N conditions, no change in NUpE was detected for AAP1-OE plants compared with wild-type plants (Fig. 5A). However, when the supply of N was moderate or high, AAP1-OE plants displayed higher NUpE and contained 13% and 11%, respectively, more N in the shoot than control plants (Fig. 5A).
Figure 5.
Analysis of NUE in AAP1-OE1 and wild-type (WT) plants grown with low (2 g of N per week), moderate (4 g of N per week), or high N (8 g of N per week) supply. NUE is determined by plant seed yield relative to the amount of N applied and is generally composed of both NUpE and NUtE (see “Materials and Methods”). A, Plant NUpE (n ≥ 5). B, Plant NUtE (n ≥ 5). C, Seed yield per plant (n ≥ 5). D, Plant NUE (n ≥ 5). Data are means ± sd. Significant differences are indicated by letters (ANOVA; P ≤ 0.05). Numbers above the columns indicate percentage changes between AAP1-OE and wild-type plants grown under the same (solid brackets) or different (dashed brackets) N conditions.
To be able to determine NUtE, total seed yield per plant was measured. The results showed an increase in seed yields with increasing N supply for transgenic and wild-type plants (Fig. 5C). More importantly, under each N condition tested, AAP1-OE plants outperformed wild-type plants. Dependent on the N treatment, seed yield increases of 17% to 39% were obtained. Equally impressive, when comparing moderate versus high N treatments, the AAP1-OE plants achieved similar yields to the wild type with half the amount of N fertilizer (Fig. 5C). Seed weight was unchanged in each treatment (data not shown), indicating that the increase in yield is due to an increased number of seeds (Fig. 3B).
Analysis of N utilization demonstrated that AAP1-OE plants grown under low and moderate N conditions show a significant increase in NUtE by 17% and 13%, respectively (Fig. 5B). This further supports that the transgenic plants allocate more shoot N to seeds compared with the wild type (Figs. 3E and 4, D and E), resulting in higher seed production (Fig. 5C). No differences in NUtE were detected between AAP1-OE and wild-type plants grown with a high N supply (Fig. 5B).
Finally, NUE was determined for AAP1-OE and wild-type plants grown under low, moderate, and high N conditions. Overall, NUE of the analyzed pea plants increased with decreasing N nutrition, as expected (Fig. 5D; Beatty et al., 2010). Remarkably, compared with the wild type, AAP1-OE plants displayed significant improvements in NUE under all N conditions (Fig. 5D). Dependent on the N treatment, NUE was enhanced between 17% and 39%. Furthermore, AAP1-OE plants with a moderate N supply, which produced similar yields to wild-type plants grown under high N, were 109% more efficient in using N (Fig. 5D). Overall, the results demonstrate that enhancing source-to-sink amino acid partitioning provides a successful strategy for achieving high crop yields in non-fixing or low N-fixing legumes and non-legume seed crops while supplying less N fertilizer.
DISCUSSION
Source-to-Sink Partitioning of Amino Acids Is a Bottleneck for Shoot Biomass and Seed Production in Both N-Deficient and N-Sufficient Environments
This work examined the importance of N source-to-sink partitioning for plant performance and NUE in a range of N environments. Recent research has demonstrated that amino acid transport processes that occur downstream of N uptake and assimilation play essential roles in plant development, seed yield, and seed protein accumulation (Rolletschek et al., 2005; Sanders et al., 2009; Tan et al., 2010; Zhang et al., 2010, 2015; Carter and Tegeder, 2016; Santiago and Tegeder, 2016). Even more important, transporter function in movement of amino acids from source (i.e. mature leaf) to sink (e.g. fruits and seeds) seems to exert a regulatory control over N uptake, amino acid synthesis, and amino acid allocation (Rolletschek et al., 2005; Weigelt et al., 2008; Tan et al., 2010; Zhang et al., 2015; Santiago and Tegeder, 2016). Essential for this work, positive feedback regulation of N assimilation and root-to-shoot partitioning, as well as an increase in seed productivity, were observed in pea plants overexpressing AAP1 in the phloem and embryo and that were grown under a very high N supply (Zhang et al., 2015). In this study, we determined if the observed increases in seed yield and seed protein require high N availability and if the modified pea plants use N more efficiently than the wild type in a range of N conditions. When AAP1-OE plants were grown under low, moderate, and high N nutrition, the expression of transporters involved in phloem loading as well as amino acid levels in leaf exudates were increased (Fig. 2), indicating that more N was moved from sources to sinks under all N conditions. In addition, seed number (Fig. 3B) and seed yield (Fig. 5C) were increased in every N environment tested, supporting that the transgenic pea plants outperform the wild type independent of N supply. This further demonstrates that transporters functioning in source-to-sink partitioning of amino acids strongly influence seed production in all N fertilization regimes and that altering transporter expression presents a successful approach to increase seed yields, even when N supply is suboptimal.
While AAP1-OE plants produce higher seed yields compared with the wild type in each N treatment, including in low N environments, seed N and protein levels were influenced by the N fertilization level (Fig. 3, C and D). The importance of amino acid partitioning for seed development and quality was recently elucidated through studies in Arabidopsis (Arabidopsis thaliana) and pea. Knocking out an Arabidopsis transporter involved in amino acid phloem loading led to a decreased number of seeds but no change in seed protein levels (Santiago and Tegeder, 2016), whereas a mutation in an amino acid seed loader negatively affected seed protein content (Sanders et al., 2009). Furthermore, studies overexpressing amino acid transporters in the parenchyma of legume cotyledons resulted in improved seed N and protein content (Weigelt et al., 2008; Rolletschek et al., 2005). Together, these findings suggest that seed protein levels are controlled by amino acid transporter function in the embryo, while amino acid phloem loading affects sink/seed number (Tan et al., 2010; Santiago and Tegeder, 2016). As demonstrated in this and previous studies, concurrent up-regulation of both amino acid phloem and seed loading leads to increased seed yield as well as seed storage protein levels when N is plentiful (Figs. 3D and 5C; Zhang et al., 2015). However, our work also demonstrates that, under low and moderate N supply, AAP1-OE seed N/protein levels were decreased or unchanged, respectively, compared with the wild type (Fig. 3, C and D), while seed number/yield was increased (Figs. 3B and 5C). The amount of N allocated to many seed sinks versus the N taken up by the individual seed might depend not only on the N available to the plant but also on the total number of sinks that develop at the early reproductive phase. Once seeds are established, and especially during the seed-filling phase when N demand is high, competing dynamics might exist between seed number and protein content per seed (Seiffert et al., 2004; Drechsler et al., 2015). In AAP1-OE plants, the increased amounts of phloem amino acids (Fig. 2, B and C) seem to accommodate both the increased number of seeds as well as seed loading, but at some cost for seed protein accumulation when N availability is low (Fig. 3, B and D). This contrasts with Arabidopsis, which produces fewer seeds but with sufficient N/storage protein when N phloem loading, and subsequently, delivery to seeds is reduced (Schmidt et al., 2007; Bennett et al., 2012; Guan et al., 2015; Santiago and Tegeder, 2016). These variations in seed number or seed protein levels under N stress may point to species-specific survival strategies (Fageria and Baligar, 2005; Hirel et al., 2007). Nevertheless, the transgenic pea plants produced higher total seed N and protein yields per plant, due to increased seed number, and displayed a higher N harvest index compared with wild-type plants in each N treatment (Figs. 3E, 4E, and 5C). This underpins that the AAP1-OE plants are more effective at using available N to produce harvestable seed protein, no matter the N condition.
Alteration of Source-to-Sink N Partitioning Is an Effective Strategy to Improve NUE
The observed increase in seed yield in AAP1-OE plants in each N treatment suggests that the transgenic plants are generally better equipped in using N compared with control plants. In fact, the AAP1-OE plants display increased NUE independent of the soil N status (Fig. 5D). However, the engineered pea plants exhibit improved NUpE, NUtE, or both, depending on N availability (Fig. 5, A and B). NUpE has been discussed as one of the most critical components of plant NUE in both high and deficient N conditions (Le Gouis et al., 2000; López-Bellido and López-Bellido, 2001; Fageria and Baligar, 2005; Coque and Gallais, 2007). However, efforts to increase N uptake and NUpE through the manipulation of nitrate and ammonium transporters or associated transcription factors have had only limited success (Fraisier et al., 2000; Yuan et al., 2007; Tsay et al., 2011; Ranathunge et al., 2014; Qu et al., 2015; Araus et al., 2016; Chen et al., 2017). Furthermore, varying outcomes have been reported from the ectopic expression of genes involved in amino acid synthesis and N metabolism (Ameziane et al., 2000; Chichkova et al., 2001; Habash et al., 2001; Yamaya et al., 2002; Seiffert et al., 2004; Good et al., 2007; Shrawat et al., 2008; McAllister et al., 2012; Peña et al., 2017). However, these studies rarely addressed the different components of NUE (i.e. NUtE and NUpE) and the effects of different N environments or only demonstrated improved NUE under specific N conditions (Shrawat et al., 2008; McAllister et al., 2012).
Some plant species take up N primarily during the vegetative growth phase (Guindo et al., 1992; Rossato et al., 2001; Malagoli et al., 2005; Coque and Gallais, 2007; Gallais et al., 2007), but pea plants generally acquire N throughout the life cycle (Salon et al., 2001; Schiltz et al., 2005). When grown under high or moderate N fertilization, AAP1-OE plants displayed significantly higher xylem amino acid levels (Fig. 1D) and N accumulation in the shoot (Fig. 4D), supporting increased root-to-shoot allocation and potentially increased root N acquisition and assimilation. This is also in line with our previous studies showing the up-regulation of genes involved in nitrate uptake and amino acid synthesis under high N (Zhang et al., 2015) and agrees with the observed significant increases in NUpE for AAP1-OE plants under moderate and high N supply (Fig. 5A). The regulation of N uptake and assimilation is multifaceted and may involve N metabolite sensing and signaling mechanisms in shoot and root as well as shoot-to-root communication (Fig. 6; Ruffel et al., 2008; Thum et al., 2008; Vidal and Gutiérrez, 2008; Ho et al., 2009; Nunes-Nesi et al., 2010; Wang et al., 2012). Amino acids like Glu, Gln, or Asn have been shown to regulate the expression of genes involved in N uptake and assimilation (Cooper and Clarkson, 1989; Imsande and Touraine, 1994; Lam et al., 1998; Gutiérrez et al., 2008). While increased shoot N levels could result from increased N remobilization from root to shoot, one might also predict that, in AAP1-OE plants, increased source-to-sink amino acid transport and associated changes in the levels of amino acids or related compounds result in positive feedback on root N uptake and assimilation when N availability is sufficient (Fig. 6; Zhang et al., 2015).
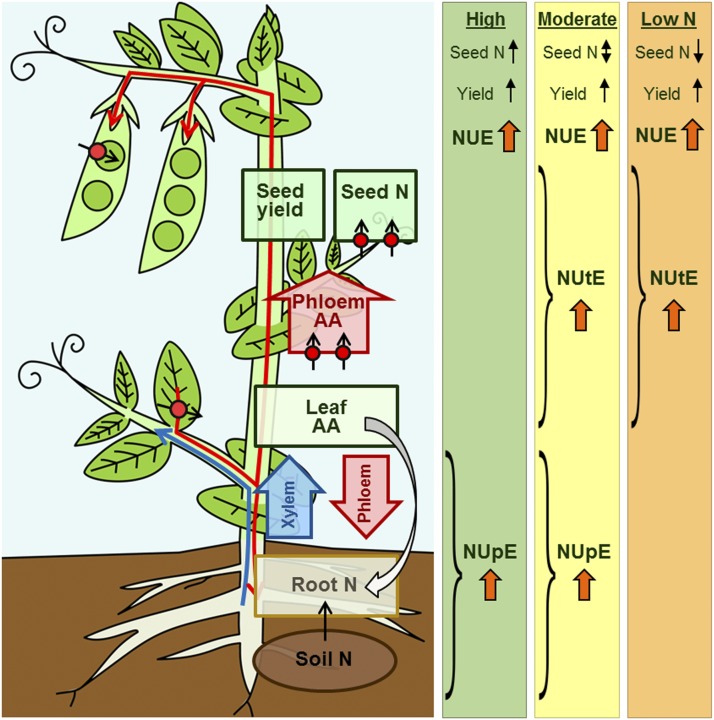
Figure 6.
Overview model of AAP1 overexpression in pea and the effects on N uptake, allocation, and use under high N and N-deficient growth conditions. Left, In previous work, it was found that increased AAP1 transporter expression in the phloem and seeds of pea plants grown under very high N supply results in (1) increased N partitioning from source leaves to sinks, affecting seed development, and (2) improved N import into seed cotyledons, influencing seed N/protein level (Zhang et al., 2015). Enhanced leaf-to-seed N transport positively affected root-to-shoot N allocation in the xylem, probably due to feedback regulation through root-shoot signaling via the phloem (large white arrow). Arrows with red circles point to increased expression of AAP1 transporters in the phloem and seeds. Right, AAP1-OE and wild-type plants were grown with low (2 g of N per week), moderate (4 g of N per week), or high N (8 g of N per week) supply. Arrows indicate relative change in AAP1-OE plants compared with the wild type. Under high N nutrition, shoot N supply, seed yield, and seed N content were increased in AAP1-OE pea plants compared with the wild type. The transgenic plants displayed improved NUE due to increased NUpE, but NUtE was unchanged. When N fertilization was reduced by half (moderate N), AAP1-OE plants continued to outperform wild-type plants with respect to N allocation to sinks and seed yield, but seed N/protein levels were unchanged. Under these strongly reduced N conditions, NUE of AAP1-OE plants was increased significantly due to increases in both NUpE as well as NUtE. Under extreme N deficiency (low N), AAP1-OE plants still produced higher seed yields, although with lower N content. NUE was increased significantly in the N-starved plant due to more efficient N utilization for seed production, but NUpE was not changed. Overall, this model supports that manipulation of source-to-sink N transport is an effective strategy to improve plant NUE and plant productivity independent of the N treatment.
Increased shoot biomass as well as xylem amino acid levels in 8-week-old AAP1-OE plants grown under low N (Fig. 1) support that these plants take up more N and move larger amounts of amino acids from the root to the shoot than wild-type plants, at least until the early reproductive phase. However, total N acquisition for the whole growing period was not changed in the transgenic compared with control plants, as total shoot N content at harvest (Fig. 4D) and NUpE (Fig. 5A) were unchanged. This suggests that N allocation from root to shoot in AAP1-OE plants is lower or at best similar to that in the wild type during the later reproductive growth phase, when plant N demand for seed development and protein accumulation is high and soil N is depleted. While future studies might address how extremely N-deficient soils affect N acquisition throughout all phases of AAP1-OE development, overall, our data demonstrate that the overexpression of AAP1 provides the transgenic plants with an advantage with respect to NUpE in N-limiting environments (i.e. moderate N) but not under severe N stress (i.e. low N).
A crop with optimum N use would not only take up N more effectively from the soil but also utilize the acquired N more efficiently for biomass and seed production (Lhuillier-Soundélé et al., 1999; Salon et al., 2001; Schiltz et al., 2005; Burstin et al., 2007). Following N uptake and assimilation, the amino acids may be transported to developing sinks (Lewis and Pate, 1973; Pate and Flinn, 1973; Urquhart and Joy, 1982; Schiltz et al., 2005), or, during the vegetative phase, some N also might be transiently stored in leaves or stem as amino acids or proteins (Pate and Flinn, 1973; Egle et al., 2015; Girondé et al., 2015). At the reproductive phase, the stored N is remobilized and redistributed in support of fruit growth and seed filling (Masclaux-Daubresse and Chardon, 2011; Girondé et al., 2015). In addition, the remobilization of N occurs from senescing tissues (Feller and Fischer, 1994; Guiboileau et al., 2010; Avila-Ospina et al., 2014, 2015). AAP1-OE plants grown under a low or moderate amount of N produced similar amounts of stubble to wild-type plants (Fig. 4A); however, the tissue contained less N (Fig. 4, B and C). On the other hand, total seed N per transgenic plant was significantly higher (Figs. 3E and 4D). This suggests that, under insufficient N nutrition, the transgenic plants remobilize more N, probably triggered by improved sink strength (Fig. 5C; Crafts-Brandner and Egli, 1987; Schulze et al., 1994; Lemaître et al., 2008; Masclaux-Daubresse et al., 2008; Masclaux-Daubresse and Chardon, 2011). Indeed, NUtE was improved significantly in the AAP1-OE plants under low and moderate N conditions (Fig. 5B). On the other hand, no change in NUtE between transgenic and wild-type plants was detected under high N fertilization (Fig. 5B). Obviously, when soil N is abundant, shoot adjustments in N usage are not required for AAP1-OE plants and improved N allocation to the shoot is sufficient to accommodate the increased N demand of sinks (Fig. 5, A and B). These results are in line with studies in Arabidopsis and different maize (Zea mays) cultivars showing that NUpE contributes to NUE in N-rich soils while NUtE influences NUE under N deficiency (Moll et al., 1982; Chardon et al., 2010; Mu et al., 2015). However, in barley (Hordeum vulgare) cultivars and wheat (Triticum aestivum) plants that overexpressed a Gln synthetase gene, observed increases in seed yield under low N were due to both improved NUpE and NUtE (Habash et al., 2001; Beatty et al., 2010). These discrepancies might imply species-dependent differences in physiological responses to varying N environments (Xu et al., 2012).
CONCLUSION
In conclusion, this study established that improving amino acid partitioning from source to sink presents an effective strategy to produce high seed yields while reducing N fertilization (Fig. 6). It is demonstrated that the engineered pea plants outperform wild-type plants in low, moderate, and high N soils. NUE is generally higher in the transgenic plants, and they produce seed yields that are similar to the controls when grown with half the amount of fertilizer. The components contributing to the improved NUE, however, vary, depending on the amount of N supplied. Specifically, the transgenic plants display improved (1) NUpE in high N soils, (2) NUtE in low N environments, and (3) both NUpE and NUtE under moderate N supply (Fig. 6). Clearly, the AAP1-OE plants demonstrate important physiological plasticity through a flexible response to changing N environments. Such a flexible adjustment of NUpE and NUtE requires serious consideration in breeding programs. In the past, selection for optimal crop productivity has often been performed in high N environments probably favoring effective N uptake and with little selection pressure for improved NUtE. However, to avoid poor plant performance on marginal land, and/or to allow for reduced fertilization to diminish costly energy inputs and to protect the environment from N pollution, modern and conventional breeding approaches need to be evaluated under varying N nutrition and to select for both NUpE and NUtE to achieve reliable improvements in NUE. Nevertheless, future studies with AAP1-OE plants will have to address if they grow as impressively under field conditions and on marginal soils and if they continue to outperform wild-type plants. Ultimately, this research has the strong potential to be transferred to other crop plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Pea (Pisum sativum ‘Bohatyr’) wild-type plants and transgenic pea plants overexpressing PsAAP1 (AY956395) under the control of the Arabidopsis (Arabidopsis thaliana) AAP1 (At1g58360) promoter (AAP1-OE; Zhang et al., 2015) were grown in 2-gallon pots in nitrogen- and nutrient-deficient soil (Sun Gro growing mix LB2) consisting of a mix of peat, perlite, and gypsum (70%–80%) and domestic limestone (20%–30%). In addition to using fresh nutrient-deficient soil, growth chambers, greenhouse benches, and pots were cleaned thoroughly with bleach to prevent rhizobial infection. Furthermore, roots were inspected postharvest for the presence of N-fixing nodules. Generally, nonnodulated, non-fixing AAP1-OE lines and wild-type plants were grown at 14-h daylength with photosynthetically active radiation between 400 and 500 µmol photons m−2 s−1 and at day and night temperatures of 18°C and 15°C, respectively. Fertilization was performed weekly using 500 mL of a modified Hoagland solution containing 1 mm MgSO4, 1 mm CaCl2, 0.25 mm KH2PO4, 0.25 mm K2SO4, 50 μm CoCl2, 50 μm H3BO4, 25 μm MnCl2, 2 μm Fe-EDTA, 0.5 μm ZnSO4, 0.5 μm CuSO4, and 0.5 μm Na2MoO4. In addition, N was supplied at high and low concentrations that were defined as follows: low N (2 g of N once per week using 5 mm NH4NO3), moderate N (4 g of N once per week using 10 mm NH4NO3), or high N (4 g of N twice per week using 10 mm NH4NO3). Source leaves, xylem sap, and leaf exudates were collected from 8-week-old pea plants grown in growth chambers. Seed yield and total N of shoots, stubble, and dry seeds were determined from greenhouse-grown, desiccated plants and after a 4-month growth period. Initial experiments were performed with two AAP1-OE lines (OE1 and OE2), but since the results were generally similar for both lines (Figs. 1 and 2; Zhang et al., 2015) and greenhouse growth space was restricted, later experiments focused on AAP1-OE1.
Gene Expression Analyses
Total RNA was extracted from mature source leaves using TRIzol reagent (Invitrogen; Chomczynski, 1993). Reverse transcription was performed according to Santiago and Tegeder (2016). Quantitative PCR was done following Sanders et al. (2009) using source leaf RNA from two independently grown sets of plants and primer sets targeting genes associated with N transport (Zhang et al., 2015). For each experimental group and treatment, five independent biological replicates were analyzed. Fold changes in gene expression were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001) by comparing the expression of a specific gene relative to the expression of actin (X90378) and ubiquitin (PUB1–PUB4; L81139, L81140, L81141, and L81142; Zhang et al., 2015). A mean fold change of greater than 2 or less than 0.5 between the wild type and overexpressors was used to identify differentially expressed genes (Rajeevan et al., 2001; Guether et al., 2009).
Collection of Leaf Exudates and Xylem Sap
Leaf exudates were collected according to Urquhart and Joy (1981) and by using the fifth and sixth source leaves from the plant base. Leaf petioles were placed into a tube containing 360 μL of 20 mm Na2EDTA at pH 7 and covered with plastic wrap to avoid transpiration. Exudates were collected for 2 h in a dark humid chamber. The EDTA was precipitated by adding 40 μL of 1 m HCl to each tube and subsequent centrifugation at 13,500g for 30 min. The supernatant was transferred to a new tube and stored at −80°C or used for analysis. Xylem sap was collected during the reproductive stage from 8-week-old, non-nodulated plants by using positive root pressure following a standard procedure. Roots were flooded with water 2 h prior to decapitation of the shoot at the stem base with a razor blade. The cut surface was rinsed with deionized water. Xylem sap that exudated during the first 5 min was discarded to avoid contamination with fluid from phloem or other cells in exudate samples. Subsequent exudate was then collected every 5 min for 20 min with a pipette and stored at −80°C until analysis (Pélissier and Tegeder, 2007). Total xylem sap volume from plants grown with low N was around 50 µL and up to 120 µL for plants grown with moderate and high N. Undiluted xylem sap was used for HPLC analysis as described below.
Amino Acid, Protein, and Elemental N Analyses
For amino acid analyses, 22 μL of leaf exudate samples and 4.5 μL of undiluted xylem sap were used. Derivatization was performed as described previously (Aoyama et al., 2004; Zhang et al., 2015), and HPLC analyses were done according to Tan et al. (2010). Total amounts of soluble seed proteins were determined according to Zhang et al. (2015). Protein was extracted from 1 mg of ground dry seed tissue and quantified using the NanoOrange reagent (Invitrogen). Analysis of elemental N was performed with dry seeds (2 mg) or stubble tissue (100 mg) according to Sanders et al. (2009).
Determining NUE
NUpE, NUtE, and NUE were calculated according to Moll et al. (1982). NUpE was defined as the ratio of total N in the aboveground shoot mass to total N supplied. NUtE is the ratio of seed yield to total shoot N. NUE is described as the combination of these two parameters, or the ratio of seed yield to the total N supplied:
 |
Statistical Analyses
The results presented are from one growth set but are representative of at least two independently grown sets of plants. Data are generally shown as means ± sd of at least four biological repetitions. One-way ANOVA followed by a Holm-Sidak test was used to determine statistical significance with SigmaPlot 11.0 software. Statistical significance was determined by P values less than or equal to 0.05.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PsAAP1 (AY956395), AtAAP1 (At1g58360), PsActin (X90378), and PsUbiquitin (PUB1-PUB4; L81139, L81140, L81141, and L81142).
Acknowledgments
We thank Dr. Raymond Lee and Margaret Davies for the isotope analyses. We are greatly appreciative of the support from the School of Biological Sciences greenhouse manager Chuck Cody.
Glossary
- NUE
nitrogen use efficiency
- NUpE
nitrogen uptake efficiency
- NUtE
nitrogen utilization efficiency
- GABA
γ-aminobutyric acid
Footnotes
This work was funded by grants from the U.S. National Science Foundation (IOS 1021286 and IOS 1457183).
Articles can be viewed without a subscription.
References
- Ameziane R, Bernhard K, Lightfoot D (2000) Expression of the bacterial gdhA gene encoding a NADPH glutamate dehydrogenase in tobacco affects plant growth and development. Plant Soil 22: 147–157 [Google Scholar]
- Aoyama C, Santa T, Tsunoda M, Fukushima T, Kitada C, Imai K (2004) A fully automated amino acid analyzer using NBD-F as a fluorescent derivatization reagent. Biomed Chromatogr 18: 630–636 [DOI] [PubMed] [Google Scholar]
- Araus V, Vidal EA, Puelma T, Alamos S, Mieulet D, Guiderdoni E, Gutiérrez RA (2016) Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiol 171: 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins CA, Pate JS, Peoples MB, Joy KW (1983) Amino acid transport and metabolism in relation to the nitrogen economy of a legume leaf. Plant Physiol 71: 841–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins CA, Pate JS, Sharkey PJ (1975) Asparagine metabolism: key to the nitrogen nutrition of developing legume seeds. Plant Physiol 56: 807–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Ospina L, Marmagne A, Talbotec J, Krupinska K, Masclaux-Daubresse C (2015) The identification of new cytosolic glutamine synthetase and asparagine synthetase genes in barley (Hordeum vulgare L.), and their expression during leaf senescence. J Exp Bot 66: 2013–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C (2014) Autophagy, plant senescence, and nutrient recycling. J Exp Bot 65: 3799–3811 [DOI] [PubMed] [Google Scholar]
- Beatty PH, Anbessa Y, Juskiw P, Carroll RT, Wang J, Good AG (2010) Nitrogen use efficiencies of spring barley grown under varying nitrogen conditions in the field and growth chamber. Ann Bot (Lond) 105: 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett E, Roberts JA, Wagstaff C (2012) Manipulating resource allocation in plants. J Exp Bot 63: 3391–3400 [DOI] [PubMed] [Google Scholar]
- Black A, Wight J (1979) Range fertilization: nitrogen and phosphorous uptake and recovery over time. J Range Manage 32: 349–353 [Google Scholar]
- Bouwman A. (1996) Direct emission of nitrous oxide from agricultural soils. Nutr Cycl Agroecosyst 46: 53–70 [Google Scholar]
- Bouwman A, Boumans L, Batjes N (2002) Emissions of N2O and NO from fertilized fields: summary of available measurement data. Global Biogeochem Cycles 16: 1058 [Google Scholar]
- Brown ME, Hintermann B, Higgins N (2009) Markets, climate change, and food security in West Africa. Environ Sci Technol 43: 8016–8020 [DOI] [PubMed] [Google Scholar]
- Burstin J, Marget P, Huart M, Moessner A, Mangin B, Duchene C, Desprez B, Munier-Jolain N, Duc G (2007) Developmental genes have pleiotropic effects on plant morphology and source capacity, eventually impacting on seed protein content and productivity in pea. Plant Physiol 144: 768–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AM, Tegeder M (2016) Increasing nitrogen fixation and seed development in soybean requires complex adjustments of nodule nitrogen metabolism and partitioning processes. Curr Biol 26: 2044–2051 [DOI] [PubMed] [Google Scholar]
- Chardon F, Barthélémy J, Daniel-Vedele F, Masclaux-Daubresse C (2010) Natural variation of nitrate uptake and nitrogen use efficiency in Arabidopsis thaliana cultivated with limiting and ample nitrogen supply. J Exp Bot 61: 2293–2302 [DOI] [PubMed] [Google Scholar]
- Chen J, Fan X, Qian K, Zhang Y, Song M, Liu Y, Xu G, Fan X (2017) pOsNAR2.1:OsNAR2.1 expression enhances nitrogen uptake efficiency and grain yield in transgenic rice plants. Plant Biotechnol J (in press) http://doi.org/10.1111/pbi.12714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichkova S, Arellano J, Vance CP, Hernández G (2001) Transgenic tobacco plants that overexpress alfalfa NADH-glutamate synthase have higher carbon and nitrogen content. J Exp Bot 52: 2079–2087 [DOI] [PubMed] [Google Scholar]
- Chomczynski P. (1993) A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15: 532–537 [PubMed] [Google Scholar]
- Conant R, Berdanier A, Grace P (2013) Patterns and trends in nitrogen use and nitrogen recovery efficiency in world agriculture. Global Biogeochem Cycles 27: 558–566 [Google Scholar]
- Cooper HD, Clarkson DT (1989) Cycling of amino-nitrogen and other nutrients between shoots and roots in cereals: a possible mechanism integrating shoot and root in the regulation of nutrient uptake. J Exp Bot 40: 753–762 [Google Scholar]
- Coque M, Gallais A (2007) Genetic variation for nitrogen remobilization and postsilking nitrogen uptake in maize recombinant inbred lines: heritabilities and correlations among traits. Crop Sci 47: 1787–1796 [Google Scholar]
- Crafts-Brandner SJ, Egli DB (1987) Sink removal and leaf senescence in soybean: cultivar effects. Plant Physiol 85: 662–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews T, Peoples M (2005) Can the synchrony of nitrogen supply and crop demand be improved in legume and fertilizer-based agroecosystems? A review. Nutr Cycl Agroecosyst 72: 101–120 [Google Scholar]
- Delogu G, Cattivelli L, Pecchioni N, De Falcis D, Maggiore T, Stanca AM (1998) Uptake and agronomic efficiency of nitrogen in winter barley and winter wheat. Eur J Agron 9: 11–20 [Google Scholar]
- Drechsler N, Zheng Y, Bohner A, Nobmann B, von Wirén N, Kunze R, Rausch C (2015) Nitrate-dependent control of shoot K homeostasis by the nitrate transporter1/peptide transporter family member NPF7.3/NRT1.5 and the stelar K+ outward rectifier SKOR in Arabidopsis. Plant Physiol 169: 2832–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egle K, Beschow H, Merbach W (2015) Nitrogen allocation in barley: relationships between amino acid transport and storage protein synthesis during grain filling. Can J Plant Sci 95: 451–459 [Google Scholar]
- Fageria NK, Baligar VC (2005) Enhancing nitrogen use efficiency in crop plants. Adv Agron 88: 97–185 [Google Scholar]
- Fang Z, Xia K, Yang X, Grotemeyer MS, Meier S, Rentsch D, Xu X, Zhang M (2013) Altered expression of the PTR/NRT1 homologue OsPTR9 affects nitrogen utilization efficiency, growth and grain yield in rice. Plant Biotechnol J 11: 446–458 [DOI] [PubMed] [Google Scholar]
- Feller U, Fischer A (1994) Nitrogen metabolism in senescing leaves. Crit Rev Plant Sci 13: 241–273 [Google Scholar]
- Fiehn O. (2003) Metabolic networks of Cucurbita maxima phloem. Phytochemistry 62: 875–886 [DOI] [PubMed] [Google Scholar]
- FAO (2012) Current World Fertilizer Trends and Outlook to 2016. Food and Agriculture Organization of the United Nations, Rome [Google Scholar]
- Forde BG. (2014) Glutamate signalling in roots. J Exp Bot 65: 779–787 [DOI] [PubMed] [Google Scholar]
- Fraisier V, Gojon A, Tillard P, Daniel-Vedele F (2000) Constitutive expression of a putative high-affinity nitrate transporter in Nicotiana plumbaginifolia: evidence for post-transcriptional regulation by a reduced nitrogen source. Plant J 23: 489–496 [DOI] [PubMed] [Google Scholar]
- Gaju O, Allard V, Martre P, Le Gouis J, Moreau D, Bogard M, Hubbart S, Foulkes MJ (2014) Nitrogen partitioning and remobilization in relation to leaf senescence, grain yield and grain nitrogen concentration in wheat cultivars. Field Crops Res 155: 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallais A, Coque M, Le Gouis J, Prioul JL, Hirel B, Quillere I (2007) Estimating the proportion of nitrogen remobilization and of postsilking nitrogen uptake allocated to maize kernels by nitrogen-15 labeling. Crop Sci 47: 685–691 [Google Scholar]
- Garnett T, Plett D, Heuer S, Okamoto M (2015) Genetic approaches to enhancing nitrogen-use efficiency (NUE) in cereals: challenges and future directions. Funct Plant Biol 42: 921–941 [DOI] [PubMed] [Google Scholar]
- Girondé A, Etienne P, Trouverie J, Bouchereau A, Le Cahérec F, Leport L, Orsel M, Niogret MF, Nesi N, Carole D, et al. (2015) The contrasting N management of two oilseed rape genotypes reveals the mechanisms of proteolysis associated with leaf N remobilization and the respective contributions of leaves and stems to N storage and remobilization during seed filling. BMC Plant Biol 15: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good A, Johnson S, De Pauw M, Carroll R, Savidov N, Vidmar J, Lu Z, Taylor G, Stroeher V (2007) Engineering nitrogen use efficiency with alanine aminotransferase. Can J Bot 85: 252–262 [Google Scholar]
- Gruber N, Galloway JN (2008) An Earth-system perspective of the global nitrogen cycle. Nature 451: 293–296 [DOI] [PubMed] [Google Scholar]
- Guan M, Møller IS, Schjoerring JK (2015) Two cytosolic glutamine synthetase isoforms play specific roles for seed germination and seed yield structure in Arabidopsis. J Exp Bot 66: 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P (2009) Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182: 200–212 [DOI] [PubMed] [Google Scholar]
- Guiboileau A, Sormani R, Meyer C, Masclaux-Daubresse C (2010) Senescence and death of plant organs: nutrient recycling and developmental regulation. C R Biol 333: 382–391 [DOI] [PubMed] [Google Scholar]
- Guindo D, Wells BR, Wilson CE, Norman RJ (1992) Seasonal accumulation and partitioning of nitrogen-15 in rice. Soil Sci Soc Am J 56: 1521–1527 [Google Scholar]
- Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habash DZ, Massiah AJ, Rong HL, Wallsgrove RM, Leigh RA (2001) The role of cytosolic glutamine synthetase in wheat. Ann Appl Biol 138: 83–89 [Google Scholar]
- Hirel B, Le Gouis J, Ney B, Gallais A (2007) The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J Exp Bot 58: 2369–2387 [DOI] [PubMed] [Google Scholar]
- Hirner B, Fischer WN, Rentsch D, Kwart M, Frommer WB (1998) Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J 14: 535–544 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hodge A, Robinson D, Fitter A (2000) Are microorganisms more effective than plants at competing for nitrogen? Trends Plant Sci 5: 304–308 [DOI] [PubMed] [Google Scholar]
- Imsande J, Touraine B (1994) N demand and the regulation of nitrate uptake. Plant Physiol 105: 3–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner CF, Ugalde TD, Aspinall D (1991) The physiology of starch and protein deposition in the endosperm of wheat. Funct Plant Biol 18: 211–226 [Google Scholar]
- Kinnersley AM, Turano FJ (2000) Gamma aminobutyric acid (GABA) and plant responses to stress. Crit Rev Plant Sci 19: 479–509 [Google Scholar]
- Krapp A, David LC, Chardin C, Girin T, Marmagne A, Leprince AS, Chaillou S, Ferrario-Méry S, Meyer C, Daniel-Vedele F (2014) Nitrate transport and signalling in Arabidopsis. J Exp Bot 65: 789–798 [DOI] [PubMed] [Google Scholar]
- Kumar K, Goh K (2002) Recovery of 15N-labelled fertilizer applied to winter wheat and perennial ryegrass crops and residual 15N recovery by succeeding wheat crops under different crop residue management practices. Nutr Cycl Agroecosyst 62: 123–130 [Google Scholar]
- Lal R. (2009) Soils and food sufficiency: a review. Agron Sustain Dev 29: 113–133 [Google Scholar]
- Lam HM, Hsieh MH, Coruzzi G (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J 16: 345–353 [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Koerselman-Kooij JW, Borstlap AC (1990) Changing kinetics of L-valine uptake by immature pea cotyledons during development: an unsaturable pathway is supplemented by a saturable system. Planta 181: 576–582 [DOI] [PubMed] [Google Scholar]
- Le Gouis J, Béghin D, Heumez E, Pluchard P (2000) Genetic differences for nitrogen uptake and nitrogen utilisation efficiencies in winter wheat. Eur J Agron 12: 163–173 [Google Scholar]
- Lemaître T, Gaufichon L, Boutet-Mercey S, Christ A, Masclaux-Daubresse C (2008) Enzymatic and metabolic diagnostic of nitrogen deficiency in Arabidopsis thaliana Wassileskija accession. Plant Cell Physiol 49: 1056–1065 [DOI] [PubMed] [Google Scholar]
- Lewis OAM, Pate JS (1973) The significance of transpirationally derived nitrogen in protein synthesis in fruiting plants of pea (Pisum sativum L.). J Exp Bot 24: 596–606 [Google Scholar]
- Lhuillier-Soundélé A, Munier-Jolain NG, Ney B (1999) Dependence of seed nitrogen concentration on plant nitrogen availability during the seed filling in pea. Eur J Agron 11: 157–166 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- López-Bellido RJ, López-Bellido L (2001) Efficiency of nitrogen in wheat under Mediterranean conditions: effect of tillage, crop rotation and N fertilization. Field Crops Res 71: 31–46 [Google Scholar]
- Malagoli P, Laine P, Rossato L, Ourry A (2005) Dynamics of nitrogen uptake and mobilization in field-grown winter oilseed rape (Brassica napus) from stem extension to harvest. I. Global N flows between vegetative and reproductive tissues in relation to leaf fall and their residual N. Ann Bot (Lond) 95: 853–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Chardon F (2011) Exploring nitrogen remobilization for seed filling using natural variation in Arabidopsis thaliana. J Exp Bot 62: 2131–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot (Lond) 105: 1141–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Orsel M (2008) Leaf nitrogen remobilisation for plant development and grain filling. Plant Biol (Stuttg) (Suppl 1) 10: 23–36 [DOI] [PubMed] [Google Scholar]
- McAllister CH, Beatty PH, Good AG (2012) Engineering nitrogen use efficient crop plants: the current status. Plant Biotechnol J 10: 1011–1025 [DOI] [PubMed] [Google Scholar]
- McNeil DL, Atkins CA, Pate JS (1979) Uptake and utilization of xylem-borne amino compounds by shoot organs of a legume. Plant Physiol 63: 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Cramer MD (2005) Root nitrogen acquisition and assimilation. Plant Soil 274: 1–36 [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM (2007) Nitrate transport and signalling. J Exp Bot 58: 2297–2306 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Fan X, Shen Q, Smith SJ (2008) Amino acids and nitrate as signals for the regulation of nitrogen acquisition. J Exp Bot 59: 111–119 [DOI] [PubMed] [Google Scholar]
- Moll R, Kamprath E, Jackson W (1982) Analysis and interpretation of factors which contribute to efficiency of nitrogen utilization. Agron J 74: 562–564 [Google Scholar]
- Mu X, Chen F, Wu Q, Chen Q, Wang J, Yuan L, Mi G (2015) Genetic improvement of root growth increases maize yield via enhanced post-silking nitrogen uptake. Eur J Agron 63: 55–61 [Google Scholar]
- Nacry P, Bouguyon E, Gojon A (2013) Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 370: 1–29 [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Pate JS, Flinn AM (1973) Carbon and nitrogen transfer from vegetative organs to ripening seeds of field pea (Pisum arvense L.). J Exp Bot 24: 1090–1099 [Google Scholar]
- Pélissier H, Tegeder M (2007) PvUPS1 plays a role in source-sink transport of allantoin in French bean (Phaseolus vulgaris). Funct Plant Biol 18: 282–291 [DOI] [PubMed] [Google Scholar]
- Peña PA, Quach T, Sato S, Ge Z, Nersesian N, Changa T, Dweikat I, Soundararajan M, Clemente TE (2017) Expression of the maize Dof1 transcription factor in wheat and sorghum. Front Plant Sci 8: 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu B, He X, Wang J, Zhao Y, Teng W, Shao A, Zhao X, Ma W, Wang J, Li B, et al. (2015) A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol 167: 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeevan MS, Ranamukhaarachchi DG, Vernon SD, Unger ER (2001) Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods 25: 443–451 [DOI] [PubMed] [Google Scholar]
- Ranathunge K, El-Kereamy A, Gidda S, Bi YM, Rothstein SJ (2014) AMT1;1 transgenic rice plants with enhanced NH4+ permeability show superior growth and higher yield under optimal and suboptimal NH4+ conditions. J Exp Bot 65: 965–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raun W, Johnson G (1999) Improving nitrogen use efficiency for cereal production. Agron J 91: 357–363 [Google Scholar]
- Rolletschek H, Hosein F, Miranda M, Heim U, Götz KP, Schlereth A, Borisjuk L, Saalbach I, Wobus U, Weber H (2005) Ectopic expression of an amino acid transporter (VfAAP1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiol 137: 1236–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossato L, Lainé P, Ourry A (2001) Nitrogen storage and remobilization in Brassica napus L. during the growth cycle: nitrogen fluxes within the plant and changes in soluble protein patterns. J Exp Bot 52: 1655–1663 [PubMed] [Google Scholar]
- Ruffel S, Freixes S, Balzergue S, Tillard P, Jeudy C, Martin-Magniette ML, van der Merwe MJ, Kakar K, Gouzy J, Fernie AR, et al. (2008) Systemic signaling of the plant nitrogen status triggers specific transcriptome responses depending on the nitrogen source in Medicago truncatula. Plant Physiol 146: 2020–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM (2011) Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci USA 108: 18524–18529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salon C, Munier-Jolain N, Duc G, Voisin AS, Grandgirard D, Larmure A, Emery R, Ney B (2001) Grain legume seed filling in relation to nitrogen acquisition: a review and prospects with particular reference to pea. Agronomie 21: 539–552 [Google Scholar]
- Sanders A, Collier R, Trethewy A, Gould G, Sieker R, Tegeder M (2009) AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J 59: 540–552 [DOI] [PubMed] [Google Scholar]
- Santiago JP, Tegeder M (2016) Connecting source with sink: the role of Arabidopsis AAP8 in phloem loading of amino acids. Plant Physiol 171: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiltz S, Munier-Jolain N, Jeudy C, Burstin J, Salon C (2005) Dynamics of exogenous nitrogen partitioning and nitrogen remobilization from vegetative organs in pea revealed by 15N in vivo labeling throughout seed filling. Plant Physiol 137: 1463–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Stransky H, Koch W (2007) The amino acid permease AAP8 is important for early seed development in Arabidopsis thaliana. Planta 226: 805–813 [DOI] [PubMed] [Google Scholar]
- Schulze W, Schulze ED, Stadler J, Heilmeier H, Stitt M, Mooney HA (1994) Growth and reproduction of Arabidopsis thaliana in relation to storage of starch and nitrate in the wild-type and in starch-deficient and nitrate-uptake-deficient mutants. Plant Cell Environ 17: 795–809 [Google Scholar]
- Seiffert B, Zhou Z, Wallbraun M, Lohaus G, Möllers C (2004) Expression of a bacterial asparagine synthetase gene in oilseed rape (Brassica napus) and its effect on traits related to nitrogen efficiency. Physiol Plant 121: 656–665 [Google Scholar]
- Shelp BJ, Bozzo GG, Trobacher CP, Zarei A, Deyman KL, Brikis CJ (2012) Hypothesis/review: contribution of putrescine to 4-aminobutyrate (GABA) production in response to abiotic stress. Plant Sci 193-194: 130–135 [DOI] [PubMed] [Google Scholar]
- Shrawat AK, Carroll RT, DePauw M, Taylor GJ, Good AG (2008) Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol J 6: 722–732 [DOI] [PubMed] [Google Scholar]
- Stahl A, Friedt W, Wittkop B, Snowdon RJ (2016) Complementary diversity for nitrogen uptake and utilisation efficiency reveals broad potential for increased sustainability of oilseed rape production. Plant Soil 400: 245–262 [Google Scholar]
- Tan Q, Zhang L, Grant J, Cooper P, Tegeder M (2010) Increased phloem transport of S-methylmethionine positively affects sulfur and nitrogen metabolism and seed development in pea plants. Plant Physiol 154: 1886–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M. (2014) Transporters involved in source to sink partitioning of amino acids and ureides: opportunities for crop improvement. J Exp Bot 65: 1865–1878 [DOI] [PubMed] [Google Scholar]
- Tegeder M, Rentsch D (2010) Uptake and partitioning of amino acids and peptides. Mol Plant 3: 997–1011 [DOI] [PubMed] [Google Scholar]
- Thum KE, Shin MJ, Gutiérrez RA, Mukherjee I, Katari MS, Nero D, Shasha D, Coruzzi GM (2008) An integrated genetic, genomic and systems approach defines gene networks regulated by the interaction of light and carbon signaling pathways in Arabidopsis. BMC Syst Biol 2: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillard P, Passama L, Gojon A (1998) Are amino acids involved in the shoot to root control of nitrate uptake in Ricinus communis plants? J Exp Bot 49: 1371–1379 [Google Scholar]
- Tsay YF, Fan SC, Chen HY, Chen KE (2011) Method for changing nitrogen utilization efficiency in plants. US Patent Application No. 12/832, 234
- Urquhart AA, Joy KW (1981) Use of phloem exudate technique in the study of amino acid transport in pea plants. Plant Physiol 68: 750–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart AA, Joy KW (1982) Transport, metabolism, and redistribution of xylem-borne amino acids in developing pea shoots. Plant Physiol 69: 1226–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Gutiérrez RA (2008) A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol 11: 521–529 [DOI] [PubMed] [Google Scholar]
- von Wirén N, Gazzarrini S, Gojon A, Frommer WB (2000) The molecular physiology of ammonium uptake and retrieval. Curr Opin Plant Biol 3: 254–261 [PubMed] [Google Scholar]
- Wang YY, Hsu PK, Tsay YF (2012) Uptake, allocation and signaling of nitrate. Trends Plant Sci 17: 458–467 [DOI] [PubMed] [Google Scholar]
- Weigelt K, Küster H, Radchuk R, Müller M, Weichert H, Fait A, Fernie AR, Saalbach I, Weber H (2008) Increasing amino acid supply in pea embryos reveals specific interactions of N and C metabolism, and highlights the importance of mitochondrial metabolism. Plant J 55: 909–926 [DOI] [PubMed] [Google Scholar]
- Winzer T, Lohaus G, Heldt HW (1996) Influence of phloem transport, N‐fertilization and ion accumulation on sucrose storage in the taproots of fodder beet and sugar beet. J Exp Bot 47: 863–870 [Google Scholar]
- Xu G, Fan X, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63: 153–182 [DOI] [PubMed] [Google Scholar]
- Yamaya T, Obara M, Nakajima H, Sasaki S, Hayakawa T, Sato T (2002) Genetic manipulation and quantitative-trait loci mapping for nitrogen recycling in rice. J Exp Bot 53: 917–925 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T (2004) Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA 101: 7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Cao W, Thorup-Kristensen K, Bai J, Gao S, Chang D (2015) Effect of Orychophragmus violaceus incorporation on nitrogen uptake in succeeding maize. Plant Soil Environ 61: 260–265 [Google Scholar]
- Yuan L, Loqué D, Ye F, Frommer WB, von Wirén N (2007) Nitrogen-dependent posttranscriptional regulation of the ammonium transporter AtAMT1;1. Plant Physiol 143: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yu X, Ayre BG, Turgeon R (2012) The origin and composition of cucurbit “phloem” exudate. Plant Physiol 158: 1873–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Garneau MG, Majumdar R, Grant J, Tegeder M (2015) Improvement of pea biomass and seed productivity by simultaneous increase of phloem and embryo loading with amino acids. Plant J 81: 134–146 [DOI] [PubMed] [Google Scholar]
- Zhang L, Tan Q, Lee R, Trethewy A, Lee YH, Tegeder M (2010) Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 22: 3603–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Vivanco JM, Manter DK (2016) Nitrogen fertilizer rate affects root exudation, the rhizosphere microbiome and nitrogen-use-efficiency of maize. Appl Soil Ecol 107: 324–333 [Google Scholar]