Liposome-reconstituted, heterologously expressed cellulose synthases contribute to primary/secondary plant cell walls synthesized the glucan chains that assembled into cellulose microfibrils.
Abstract
Cellulose, the major component of plant cell walls, can be converted to bioethanol and is thus highly studied. In plants, cellulose is produced by cellulose synthase, a processive family-2 glycosyltransferase. In plant cell walls, individual β-1,4-glucan chains polymerized by CesA are assembled into microfibrils that are frequently bundled into macrofibrils. An in vitro system in which cellulose is synthesized and assembled into fibrils would facilitate detailed study of this process. Here, we report the heterologous expression and partial purification of His-tagged CesA5 from Physcomitrella patens. Immunoblot analysis and mass spectrometry confirmed enrichment of PpCesA5. The recombinant protein was functional when reconstituted into liposomes made from yeast total lipid extract. The functional studies included incorporation of radiolabeled Glc, linkage analysis, and imaging of cellulose microfibril formation using transmission electron microscopy. Several microfibrils were observed either inside or on the outer surface of proteoliposomes, and strikingly, several thinner fibrils formed ordered bundles that either covered the surfaces of proteoliposomes or were spawned from liposome surfaces. We also report this arrangement of fibrils made by proteoliposomes bearing CesA8 from hybrid aspen. These observations describe minimal systems of membrane-reconstituted CesAs that polymerize β-1,4-glucan chains that coalesce to form microfibrils and higher-ordered macrofibrils. How these micro- and macrofibrils relate to those found in primary and secondary plant cell walls is uncertain, but their presence enables further study of the mechanisms that govern the formation and assembly of fibrillar cellulosic structures and cell wall composites during or after the polymerization process controlled by CesA proteins.
Cellulose is the most abundant biopolymer on earth (Pauly and Keegstra, 2008; McNamara et al., 2015). It consists of bundles of β-1,4-glucan chains typically organized as microfibrillar structures. It is mostly found in plants, bacteria, oomycetes, and green algae (Fugelstad et al., 2009; John et al., 2011; Augimeri et al., 2015). Cellulose is the main structural component of plant cell walls, and is used for several technological applications including manufacturing of paper, textiles, and furniture (McFarlane et al., 2014). In recent years cellulose has become a focus of those pursuing the production of sustainable biofuels due to its potential to be converted to ethanol and other compounds (Pauly and Keegstra, 2008). Detailed knowledge regarding the structure and function of cellulose synthases (CesAs) and the mechanisms of cellulose polymerization and microfibril assembly is likely to facilitate downstream processing of cellulose (Cantarel et al., 2009; McNamara et al., 2015).
A simplified in vitro system that includes levels of CesA assembly sufficient to produce cellulose microfibrils would greatly facilitate the acquisition of such knowledge. There are many CesA genes in plants whose products exhibit complex interactions (Popper et al., 2011). For example, Arabidopsis (Arabidopsis thaliana) has 10 CesA genes that are divided into two groups by the type of cell wall that they make: CesA1, -2, -3, -5, -6, and -9 are involved in primary cell wall synthesis (Arioli et al., 1998; Caño-Delgado et al., 2003; Desprez et al., 2007; Persson et al., 2007) and CesA4, -7, and -8 are involved in secondary cell wall synthesis (Taylor et al., 2000; Brown et al., 2005; Bosca et al., 2006; Mendu et al., 2011; McFarlane et al., 2014). These CesAs form a hexameric rosette structure in the plasma membrane, called Cellulose Synthase Complex (CSC), and it was believed until recently that each of the six lobes have six subunit CesA proteins (Kimura et al., 1999; Doblin et al., 2002; Cosgrove, 2005; Somerville, 2006). The latest developments based on imaging, structural, and computational approaches suggest that each CSC lobe possesses three CesA molecules (Newman et al., 2013; Hill et al., 2014; Nixon et al., 2016; Vandavasi et al., 2016). A single CSC should thus produce up to 18 single glucan chains, presuming that all individual CesA proteins are active synthases; these chains subsequently crystallize to form cellulose microfibrils that are 3 nm to 5 nm in diameter (Ha et al., 1998; Cosgrove, 2005; Thomas et al., 2013). Higher-order bundles of microfibrils are also seen in cell walls (Marga et al., 2005; Somerville, 2006; Zhang et al., 2016).
Although assembly of CesAs and assembly of glucan chains involve different macromolecules, they are likely to be linked processes, but the extent of linkage remains unclear. One scenario is that the CesA enzymes assemble into trimeric lobes, and these into rosette CSCs. Subsequent glucan synthesis and extrusion from the organized rosette provides chains to form fibrillar cellulose. Alternatively, CesA might assemble into trimeric lobes that each synthesize three adjacent glucan chains that self-assemble into a submicrofibril. The latter further assemble into complete microfibrils providing feedback onto individual lobes, moving them into a rosette distribution. The microfibrils could then further assemble with each other and the wall matrix materials.
Some clues have been garnered regarding assembly of CesA proteins. The secondary structure of most plant CesA proteins consists of at least six transmembrane helices (TMHs) that TMH-anchor and separate a cytoplasmic glycosyltransferase (GT) domain from an N-terminal, intrinsically unfolded domain (Sethaphong et al., 2013). In addition to possessing the catalytic function, the GT domain includes a plant conserved region (P-CR) and a class-specific region (CSR; Slabaugh et al., 2014). The catalytic function is provided in part by three broadly conserved Asp-containing motifs designated as DDG, DCD, and TED, followed by a conserved QXXRW motif (Saxena and Brown, 1997; Saxena et al., 2001; Sethaphong et al., 2013). In vitro studies have shown that the catalytic domain of rice CesA1 forms redox-sensitive dimers (Olek et al., 2014), and that the same region of Arabidopsis CesA1 forms redox-insensitive trimers (Vandavasi et al., 2016). Small-angle x-ray scattering and transmission electron microscopy (TEM)-based low-resolution volumes of the trimer fit well to averaged lobe volumes from Physcomitrella patens rosettes imaged by freeze-fracture TEM (Nixon et al., 2016). A Zn2+-binding RING domain is encoded in the N-terminal regions of CesAs, which may be responsible for dimerization of CesA proteins (Kurek et al., 2002).
Superimposed on this as yet poorly defined tendency for CesA subunits to assemble is an intrinsic propensity for nascent β-1,4-glucan chains to form microfibrils with varied packing of the chains (Ha et al., 1998; Cosgrove, 2005; Thomas et al., 2013). Cellulose microfibrils are deposited in a transverse direction with respect to the cell growth (Szymanski and Cosgrove, 2009), and reoriented during cell wall expansion (Baskin, 2005; Anderson et al., 2010). Within a microfibril, the cellulose chains are held together by hydrogen bonds and van der Waals forces (Nishiyama et al., 2002, 2003). In nature, parallel chains are packed with low order, hence called “amorphous”, or with high order, as found in crystalline cellulose. Because individual β-glucan chains are synthesized as a flat ribbon with 180° flipping of the orientation of successive Glc monomers, it is possible to pack them in crystalline sheets with the lateral register of the “up” or “down” Glc moieties in adjacent chains in different ways, giving rise to the Iα and Iβ crystalline forms. The Iα form is mostly found in bacterial and algal cellulose, and the Iβ form in higher plants and tunicate animals (Atalla and Vanderhart, 1984; Belton et al., 1989). A single microfibril may contain a mixture of these forms, and interactions between microfibrils contribute significantly to the mechanical properties of cellulose. The parallel alignment of glucan chains is consistent with simultaneous synthesis of cellulose chains in CSCs (Somerville, 2006). It is not known if CesA facilitates the close placement of cellulose chains to enable hydrogen bonding and hydrophobic interactions; if the close distance between nascent glucan chains is sufficient to induce the formation of the higher-order cellulose microfibrils; or if other protein(s) is (are) required for this assembly (Somerville, 2006).
Recent work demonstrated that a single CesA isoform (CesA8 from hybrid aspen, PttCesA8) synthesizes cellulose in vitro and packs it into fibrils observable by TEM (Purushotham et al., 2016). PttCesA8 contributes to the formation of secondary cell walls. Here we report similar synthesis of cellulose microfibrils by reconstituted CesA5 of P. patens (PpCesA5), which is involved in primary cell wall formation (Goss et al., 2012). Previously, we had shown that overexpression of PpCesA5 in P. patens itself yielded partially purified protein that synthesized cellulose microfibrils, but the presence of other CesA isoforms and accessory proteins could not be ruled out (Cho et al., 2015). Now we eliminate the possible presence of additional isoforms and other proteins by purifying heterologously expressed PpCesA5 from Pichia pastoris, and reconstituting it into proteoliposomes. Reconstituted PpCesA5 produced cellulose microfibrils, which was demonstrated by incorporation of radioactive Glc from UDP-[3H]Glc, cellulase-specific degradation, TEM analysis, and linkage analysis. Furthermore, TEM analysis identified cellulose microfibrils within and on the surface of proteoliposomes bearing PpCesA5 or PttCesA8. These microfibrils coalesced into higher-order macrofibrils. These results confirm that single isoforms of CesAs for primary and secondary cell wall synthesis are sufficient to produce cellulose microfibrils, and suggest that glucan chains can form higher-order cellulose structures by self-assembly without the need for accessory proteins.
RESULTS
Heterologous Expression and Purification of PpCesA5
PpCesA5 is predicted to have seven transmembrane domains, an N-terminal Zn-binding domain, and a long cytosolic region between the second and the third transmembrane helices (Supplemental Fig. S1). The cytosolic region contains P-CR and CSR regions as well as a TED motif, the latter believed to deprotonate the acceptor hydroxyl via its Asp residue (Morgan et al., 2014). PpCesA5 conjugated with 12× His-tag at the C terminus was heterologously expressed in P. pastoris. Expression in P. pastoris was directed by using methanol to induce transcription from the alcohol oxidase1 promoter. Membrane proteins were isolated and further purified with TALON (cobalt) resin. PpCesA5 proteins were copurified with other membrane proteins, as shown by immunoblot using anti-PpCesA5 antibody no. 1 (Lanes 1 to 3; Fig. 1). The final elution fraction contained a highly enriched PpCesA5 protein of approximately 125 kD (Lane 9; Fig. 1). Two other PpCesA5 specific antibodies no. 2 and no. 3 also detected similar band patterns, whereas anti-His antibody identified an additional band at around 55 kD (Supplemental Fig. S2). Whereas the epitopes of the PpCesA5 specific antibodies were in the cytosolic domain (Supplemental Fig. S1), the His-tag was at the C terminus, suggesting that the N-terminal region is more prone to degradation. Similarly, heterologously expressed PttCesA8 also showed N-terminal degradation that was speculated to be due to conformational flexibility or loose association of the N-terminal RING finger region (Purushotham et al., 2016).
Figure 1.
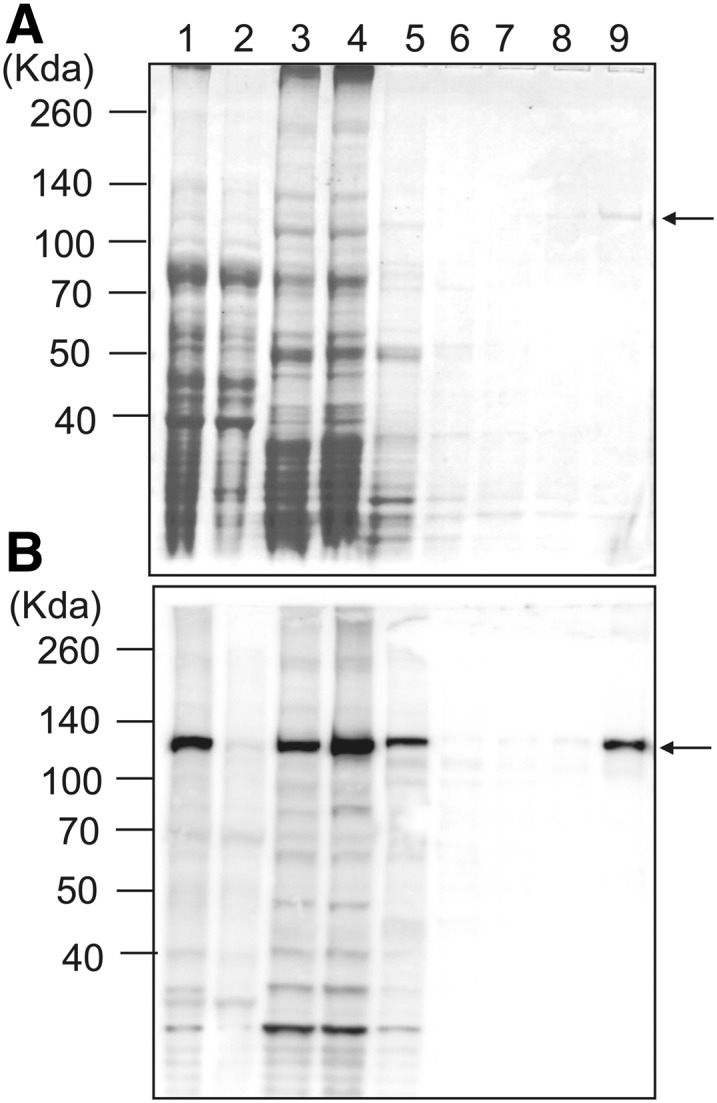
Partial purification of heterologously expressed PpCesA5 from P. pastoris. Membrane proteins were isolated from P. pastoris expressing PpCesA5::12×His and purified using TALON (cobalt) resin. A, SDS-PAGE gel stained with Coomassie Blue. Lane 1, total protein extract; Lane 2, nonmembrane proteins; Lane 3, membrane proteins; Lane 4, flow through; Lane 5, washing fraction 1 (20 mM imidazole); Lane 6, washing fraction 2 (40 mM imidazole); Lane 7, washing fraction 3 (60 mM imidazole); Lane 8, washing fraction 4 (80 mM imidazole); Lane 9, elution (350 mM imidazole). B, Immunoblot of gel as in (A) using PpCesA5-specific antibody no. 1. The arrows point to bands of mass expected for full-length PpCesA5. (See Supplemental Fig. S1 for epitope location.)
To further confirm that PpCesA5 was enriched, proteins in the region between 110 kD and 140 kD were excised from an SDS-PAGE gel and analyzed by mass spectrometry (MS). Three peptides of PpCesA5 including 444VQPTFVK450, 538AGAMNALVR546, and 808GSAPINlsdR817 were identified, together with fragments of 18 proteins from P. pastoris, including the catalytic subunit of β-1,3-glucan synthase (Supplemental Table S1).
PpCesA5 Is Functional When Reconstituted in Proteoliposomes
Because CesA proteins are integral membrane proteins, the enriched PpCesA5 was reconstituted into proteoliposomes using total lipid extract from yeast. These proteoliposomes were then subjected to biochemical characterization using radiolabeled UDP-[3H]Glc as a tracer, followed by detergent disruption and descending paper chromatography (Fig. 2), as described in Omadjela et al. (2013) and Purushotham et al. (2016). Radiolabeled Glc was incorporated into insoluble material as shown by radioactivity that failed to leave the origin, indicating that PpCesA5s in proteoliposomes were functional.
Figure 2.
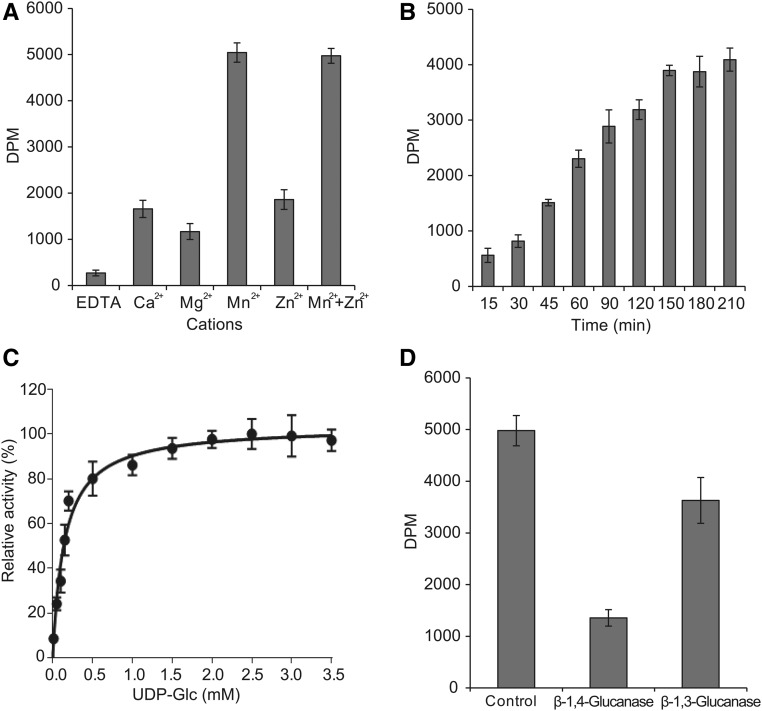
Functional characterization of PpCesA5 reconstituted into liposomes. A, Partially purified PpCesA5 protein was reconstituted into liposomes using yeast total lipids and detergent removal by BioBeads (Bio-Rad), and then assayed for function by incubation with UDP-Glc and UDP-[3H]Glc in the presence of the indicated cations. After stopping reactions by the addition of Triton X-100, the product was applied to descending paper chromatography, from which insoluble material at the origin was evaluated in a scintillation counter. Error bars represent sd (n = 3). B, Time course. C, Kinetic analysis. Michaelis-Menten parameter estimates and 95% confidence intervals: KM = 137 (102,179) μM; Vmax = 103 (96,111) relative units. D, Treatments with β-1,4-glucanase (cellulase) or β-1,3-glucanase (callase).
CesA proteins require cations as cofactors for function. Activity of PpCesA5 proteoliposomes was tested with different cations including Ca2+, Mg2+, Mn2+, and Zn2+. We found that Mn2+ was most effective, and that Zn2+ in combination with Mn2+ appeared to have no synergistic effect (Fig. 2A). These results were consistent with those from PttCesA8 (Purushotham et al., 2016). Mn2+ alone was used in all subsequent tests.
The incorporation of radiolabeled Glc from UDP-[3H]Glc into synthesized product was monitored over time (Fig. 2B). The radioactive signal reached saturation at 150 min, which might be due to the depletion of substrate, loss of protein activity, or product inhibition. In contrast, it took 90 min for PttCesA8 to reach a plateau (Purushotham et al., 2016). Enzyme kinetics assays were conducted using a constant level of UDP-[3H]Glc with a variation of UDP-Glc concentration. These assays show monophasic Michaelis-Menten kinetics with KM = 137 (102 to 179) μM (Fig. 2C; 95% confidence interval in parentheses).
To confirm that the in vitro product contained cellulose, it was treated with β-1,4-glucanse, solubilizing 73% of the radiolabeled product (Fig. 2D). The enriched preparation of PpCesA5 contained other proteins from P. pastoris that were present in the 110-kD to 140-kD region of an SDS-PAGE gel (Supplemental Table S1), including the catalytic subunit of β-1,3-d-glucan synthase (callose synthase). To examine potential contribution of callose synthase to the incorporated radioactive product signal, in vitro products were subjected to treatment with β-1,3-glucanase, solubilizing 27% of the radiolabeled product (Fig. 2D).
To further confirm that the observed signal resulted from PpCesA5 activity, we introduced amino acid substitution D782N of the catalytically crucial TED motif of PpCesA5 (Supplemental Fig. S3). The analogous substitution inactivated PttCesA8 (Purushotham et al., 2016). As evident in immuno-blots, PpCesA5(D782N) was sensitive to proteolysis. Proteoliposomes prepared as for the wild-type PpCesA5 showed no capacity to incorporate [3H]Glc (Supplemental Fig. S3).
In Vitro-Produced Glucan Chains Assemble into Cellulose Microfibrils
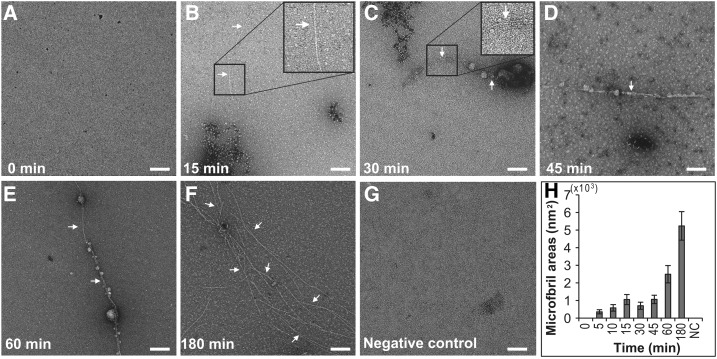
β-1,4-d-glucan chains produced from CesAs form microfibrils and these are sometimes bundled into larger arrays. To determine if β-1,4-glucan chains produced by PpCesA5 proteoliposomes form microfibrils or higher-order bundles, proteoliposomes that had been incubated with UDP-Glc were examined over time by TEM (Fig. 3). Individual glucan chains are too small to be seen with this method, but microfibrils are large enough and they were first observed within 5 min of incubation with UDP-Glc. The initial amount of microfibrils increased upon incubation for 60 min and 180 min. There were no microfibrils formed in a negative control reaction lacking UDP-Glc, nor any jagged-edge fibrils as reported for callose (Him et al., 2001). Repeated experiments with the TED mutant protein PpCesA5(D782N) yielded no observable microfibrils (Supplemental Fig. S5). Pretreatment of proteoliposomes bearing wild-type PpCesA5 with Triton X-100 caused them to fail to yield microfibrils (Supplemental Fig. S6). Also, the addition of Zn2+ to PpCesA5-proteoliposomes had no effect on the level of microfibril formation (Supplemental Fig. S7). The thickness of microfibrils made by PpCesA5 was estimated by cryo-TEM and was found to be 4.3 ± 0.8 nm (Supplemental Fig. S8). Most microfibrils disappeared after 15 min of incubation with cellulase, whereas a negative control without cellulase showed no loss of microfibrils (Fig. 4).
Figure 3.
PpCesA5 in proteoliposomes produces microfibrils. A to F, Proteoliposomes bearing PpCesA5 were incubated with UDP-Glc, during which samples were taken at designated time points for imaging by negative stain TEM. Representative random images are shown. Arrows indicate microfibrils, magnified in insets of (B) and (C). G, Negative control, representative image for a reaction incubated for 180 min without UDP-Glc. Scale bars, 100 nm. H, Plots of the mean and se for the areas occupied by microfibrils in 40 random images. NC, negative control.
Figure 4.
Microfibrils are sensitive to cellulose-specific glucosidases. To examine if the microfibrils were made of cellulose, in vitro products were subjected to cellulase treatment and sampled at the designated time points by negative staining. Randomly taken TEM images were analyzed for areas occupied by fibrils. A to F, Representative images for reaction times of 0, 5, 10, 30, 45, and 60 min. G, Negative control, representative image for a reaction incubated for 60 min without cellulase. Arrows indicate microfibrils. Arrowheads in (A) and (G) indicate bundled microfibrils. Scale bars,100 nm. H, Plots of the mean values of the areas occupied by microfibrils. Gray bars and white bars represent total microfibrils and bundled microfibrils, respectively. Error bars indicate ses (n = 40). NC, negative control.
To further confirm that the fibrils were cellulose microfibrils with 1,4-Glc linkage, a glycosidic linkage analysis was performed by permethylation followed by gas chromatography coupled to electron-impact mass spectrometry (GC/EI-MS). In vitro products from a large-scale reaction were pretreated with α-amylase and amyloglucosidase to remove P. pastoris-derived glycogen-like α-1,4-glucan chains. Thereafter, the sample was permethylated to alditol acetates, and then subjected to GC/EI-MS analysis. Comparison with reference peaks confirmed that the sample mainly contained 1,4-d-Glc, with no 1,3-d-Glc being observed (Fig. 5; Supplemental Fig. S9A). The sample also contained a small level of terminal Glc, as further identified by electron-impact mass spectrometry (Supplemental Fig. S9B). Analogously treated material that was produced by proteoliposomes bearing the inactive PpCesA5(D782N) showed no 1,4-d-Glc (Supplemental Fig. S10A). However, 1,4-d-Glc was present before eliminating α-1,4-glucans with amylase (Supplemental Fig. S10B).
Figure 5.
Microfibrils contain 1 to 4 linked Glc. In vitro products were pretreated with 2% SDS and proteinase K, followed by α-amylase and amyloglucosidase to remove P. pastoris-derived α-1,4-glucan chains. Thereafter, the sample was permethylated to alditol acetates, and then subjected to GC/MS analysis. The resulting spectra are compared with reference peaks. Identities of each peak are marked. In particular, terminal-Glc and 4-linked Glc were detected at 9.348 min and 19.059 min, respectively. t-Glc, terminal-Glc; 4-Glc, 4-linked Glc.
Higher-Order Self-Assembly of Cellulose Microfibrils
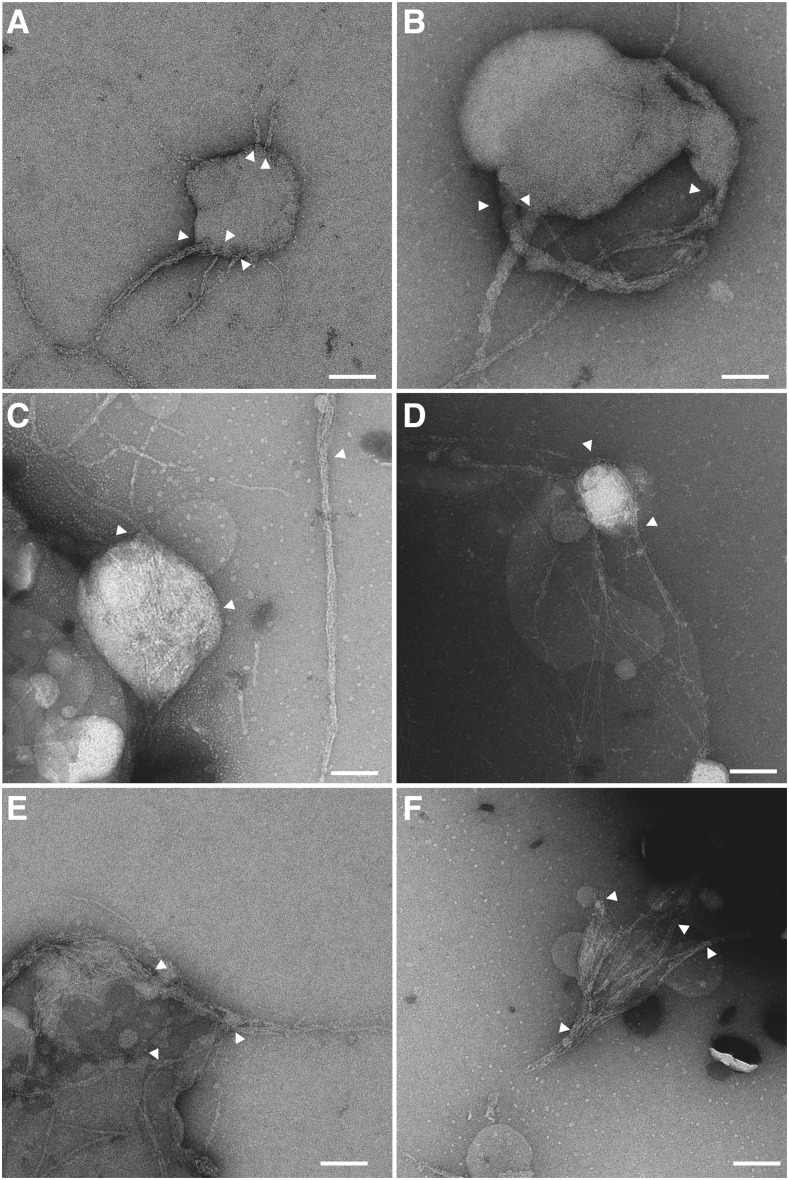
It is known that cellulose microfibrils are often bundled into larger fibrils in plant cell walls. To observe possible higher-order assembly that might reflect a bundling process in the PpCesA5 and PttCesA8/proteoliposome systems, we thoroughly explored negatively stained TEM grids with in vitro products to find unbroken or partially broken proteoliposomes. This revealed cellulose microfibrils near and apparently attached to the surface of proteoliposomes with PpCesA5, as if they were spawned from the proteoliposomes (Fig. 6, A and B). The thicknesses of such fibrils varied, with thinner fibrils merging to form thicker fibrils. We also found PpCesA5-proteoliposomes that contained numerous cellulose microfibrils inside and a few microfibrils exiting out through the membranes, which also formed higher-order assemblies outside (Fig. 6C). Some cellulose microfibrils wrapped around the proteoliposomes (Fig. 6D). In many images, thin cellulose fibrils coalesced into higher-order micro- or macrofibrils (Fig. 6, C, E, and F) that in some cases might be considered bundles. This coalescence was also observed in cellulose microfibrils produced by proteoliposomes bearing PttCesA8 (Fig. 7). Such bundled microfibrils were also observed in other grid areas (Fig. 4, A and G, marked by arrowheads). In total, the area of bundled microfibrils accounted for 36% to 43% of the microfibrillar area in each image. These bundled microfibrils were also eliminated by cellulase digestion before imaging (Fig. 4H), indicating that they were cellulose microfibrils.
Figure 6.
Cellulose microfibrils are produced from PpCesA5 in proteoliposomes and coalesce into higher-order macrofibrils. Proteoliposomes bearing PpCesA5 were incubated with UDP-Glc, followed by negative staining and TEM analysis. Arrows indicate where cellulose microfibrils were attached to proteoliposomes (A and B). Scale bars, 100 nm.
Figure 7.
Cellulose microfibrils from PttCesA8-proteoliposomes coalesce into higher-order macrofibrils. Proteoliposomes bearing PttCesA8 were incubated with UDP-Glc, followed by negative staining and TEM analysis. Arrows indicate where cellulose microfibrils coalesced into macrofibrils. Scale bars, 100 nm.
DISCUSSION
In this study, proteoliposomes bearing single isoforms of CesAs known to synthesize primary cell walls were able to synthesize β-1,4-glucan chains that were assembled into cellulose microfibrils. The cellulose microfibrils were often seen to merge into higher-order forms. It is not yet clear if these in vitro products relate to the microfibrils, macrofibrils, or bundles seen in plant cell walls, so in the following discussion we will explicitly denote the in vitro cellulose assemblies with superscript iv (thus, microfibrilsiv, macrofibrilsiv, or bundlesiv).
Both biochemical and TEM data showed the synthesis of β-1,4-glucan chains that assembled into microfibrilsiv. For incorporation of [3H]Glc from UDP-[3H]Glc into insoluble material that was sensitive to cellulase, the presence of cellulose-like fibers with diameters of 4.3 nm in negative stain and cryo-TEM images and the presence of β-1,4-d-Glc in linkage analysis in the in vitro product confirm the synthesis of cellulose microfibrilsiv. In addition to the synthesis of cellulose, there was some evidence for the production of β-1,3-glucan chains. The presence of a 110-kD to 140-kD fragment of the 220 kD β-1,3-glucan synthase of P. pastoris and observed partial digestion of in vitro synthesized insoluble polymers by β-1,3-glucanase are consistent with callose synthesis. However, we observed no fibers in negative stain images that had jagged edges such as reported for callose, and the linkage analysis showed no evidence of β-1,3-d-Glc. These contradictory observations could be explained by variable presence of functional P. pastoris β-1,3-glucan synthase in the PpCesA5 preparations of this study. Such variation was seen for preparations of PttCesA8 of hybrid aspen (Purushotham et al., 2016).
A key aspect of our previous report on PttCesA8 and this report on PpCesA5 is that a single isoform of plant CesA can alone produce β-1,4-glucan chains of cellulose. These observations force, to our knowledge, new considerations regarding the prior models of CSC composition. We note that the CSC is a complex of CesA proteins, arranged in tightly bound lobes that are more loosely contained within mostly hexamers (sometimes pentamers) of lobes (Kimura et al., 1999; Desprez et al., 2007; Nixon et al., 2016). Recent evidence strongly suggests that each lobe is a trimer of CesA, and until recently these trimers were assumed to be heterotrimers of different CesA isoforms, the exact isoforms depending on whether the CSC is involved in making cellulose for primary versus secondary cell walls (Cosgrove, 2005). The genetic redundancy has been interpreted as a way that plants provide differential regulation of cellulose synthesis in specific tissues and developmental events (McFarlane et al., 2014). The results presented here for CesA5 from the nonvascular plant P. patens and previously for CesA8 from the vascular plant hybrid aspen undeniably show that a single isoform is capable of forming cellulose microfibrilsiv in in vitro systems. This is thus true for CesAs known for contributing in vivo to the production of primary (PpCesA5) or secondary (PttCesA8) cell walls. These observations of single isoforms making cellulose microfibrilsiv are consistent with reported 1:1:1 stoichiometric presence of CesA isoforms needed for normal primary and secondary cell wall formation (Gonneau et al., 2014; Hill et al., 2014), but they do raise the possibility of homomeric complexes.
If one only considers the moss P. patens, it could be argued that homomeric lobes may be restricted to nonvascular plants. This would be reasonable because the P. patens’ genome has seven nearly identical CesA genes (Roberts and Bushoven, 2007) and also has CSCs in the plasma membranes similar to the rosettes seen in vascular plants (Roberts et al., 2012; Nixon et al., 2016). Based at least in part on such observations, it was predicted that a single CesA isoform might be able to substitute for other CesAs in the CSCs of P. patens. Consistent with that prediction, single knockout of PpCesA6 or PpCesA7 showed no phenotype; only the double knockout resulted in reduction of stem lengths (Wise et al., 2011). However, our concurrent study of hybrid aspen PttCesA8 that was also heterologously expressed in P. pastoris and reconstituted into proteoliposomes makes it clear that only one isoform of CesA from vascular plants is sufficient to yield cellulose microfibrilsiv (Purushotham et al., 2016). In both the moss and poplar studies, CesA in its detergent-solubilized form failed to incorporate Glc from UDP-Glc into glucan chains, regaining this function only when incorporated into proteoliposomes. The amino acid sequences of CesA proteins in hybrid aspen are diverse like those in Arabidopsis (Djerbi et al., 2004). Thus, we propose that CesA proteins will generally need to be in a lipid bilayer to adopt functional conformation(s), and that they can assemble into homotrimer as well as heterotrimer lobes (the latter not yet having been demonstrated in vitro).
In plants, the individual β-1,4 glucan chains made by CesA proteins are assembled into microfibrils, and these into macrofibrils or higher-order bundles, such as those noted in atomic force microscopic images of onion epidermal cell walls (Zhang et al., 2016). The relationship between trimeric lobes in rosettes and assembly of crystalline cellulose microfibrils is not yet understood. It is very intriguing that we observe micro- and macrofibrilsiv in both the CesA studies, including the one reported here for CesA5 and in the CesA8 studies presented earlier (Purushotham et al., 2016). Such microfibrils are not made by the bacterial cellulose synthase BcsA-BcsB, even when present at high concentrations (Purushotham et al., 2016). However, when BscA-BcsB was immobilized on a nickel-film, followed by reaction with UDP-Glc, fibrillar cellulose was produced (Basu et al., 2016, 2017). This suggests that close proximity of cellulose synthase proteins is required and sufficient for formation of microfibrilsiv, and we infer that such close proximity is present in the membrane-reconstituted moss and hybrid aspen synthases CesA5 and CesA8.
The lateral dimension of an individual cellulose glucan chain is lower than 0.5 nm, which makes isolated chains invisible in negative stain TEM images. As shown in Figures 6 and 7, variably thick fibers are seen, and these tend to coalesce into larger fibrils. Although negative stain images do not give reliable size estimates, these relative differences in size are clearly present. We think it possible that a homotrimer of CesA5 or CesA8 is capable of making an elemental fibril of three glucan chains, and that these can further assemble to form more typical microfibrils of approximately 4.3 nm to 4.8 nm in diameter. The estimated diameter presented here from cryo-TEM images is reliable, given the absence of any staining, and such sizes are consistent with diameters reported for plant cell walls (Ha et al., 1998; Cosgrove, 2005; Thomas et al., 2013; Zhang et al., 2016). This raises the possibility that homotrimer lobes could be organized into higher-order rosettes as the microfibrilsiv are formed. It is simultaneously possible that the dimerization propensity of N-terminal domains of CesA could help organize rosette formation by bringing together lobes (Kurek et al., 2002). Indeed, truncation of N-terminal Zn2+-binding domains from PttCesA8 resulted in the formation of few or no microfibrils despite significant production of β-1,4-glucan chains (Purushotham et al., 2016). Future use of these in vitro systems may reveal the membrane distributions of wild-type and mutant CesAs before, during, and after catalysis with UDP-Glc and thus allow testing these hypotheses.
CONCLUSION
PpCesA5 of the moss P. patens was successfully expressed heterologously in P. pastoris and partially purified, followed by reconstitution into proteoliposomes. Proteoliposomes bearing PpCesA5 synthesize glucan chains that assembled into higher-order cellulose microfibrilsiv and macrofibrilsiv or bundlesiv comparable to what is seen for a vascular plant CesA. We discuss how formation of cellulose microfibrils from single isoforms of CesA could be attributable to close proximity of CesA proteins in lipid bilayers, formation of higher-order complexes among CesAs, or a combination of that and feedback from glucan chain crystallization—these assembly processes remain to be investigated. These systems for reconstitution of functional cellulose synthases in vitro serve as foundations for further studies on the synthesis of cellulose by CesA proteins and the assembly of nascent glucan chains into elementary fibrils, microfibrils, and microfibril bundles in the absence or presence of matrix polysaccharides. Access to purified, membrane-embedded, functional CesAs may also yield structures of CesA from nonvascular and vascular plants.
MATERIALS AND METHODS
Cloning and Transformation of PpCesA5
The PpCesA5 gene was amplified by PCR using a primer set designed to add 12×His tag at the C terminus: 5′-ACTAATCAAGGTACCATGGAGGCTAATGCAGGCCTTATT-3′/5′-ACAATAGCGGCCGCTCAGTGATGGTGATGGTGATGGTGATGGTGATGGTGATGACAGCTAAGCCCGCACTCGAC-3′. Once synthesized, the PCR product was digested with KpnI and NotI restriction enzymes. The resulting fragment was ligated into KpnI and NotI-linearized pPICZ A vector. Five µg of pPICZ A plasmid containing PpCesA5 was used to transform into the Pichia pastoris strain SMD1168H. For preparation of the SMD1168H cells for transformation, a colony grown on YPDS plate containing 1% yeast extract, 2% peptone, and 2% Glc was inoculated into 50 mL YPDS liquid medium, followed by culture at 30°C by shaking until growth reached early stationary phase (approximately 0.6 to 2 × 108 cells/mL). Cells were harvested by centrifugation at 3000 rpm for 5 min at 4°C, followed by washing with 40 mL ice-cold sterile water and centrifugation at 2500 rpm for 5 min at 4°C. After washing with 20 mL ice-cold water, cells were resuspended in 5 mL ice-cold sorbitol solution (2% sorbitol in water) and pelleted at 2000 rpm for 5 min at 4°C. The cells were resuspended in 150 μL ice-cold sorbitol solution, from which 40 μL of cells were mixed with 5 μg PmeI-linearized DNA in a prechilled electroporation cuvette. Electroporation was performed at 1.5 kV, 25 μF, and 200 Ohms for 5 s, which was immediately followed by addition of 1 mL 1 m ice-cold sorbitol solution. Transformed cells were screened on YPDS medium containing 100 μg/mL of Zeocin.
The catalytically inactive PpCesA5 construct was generated using the QuikChange Mutagenesis Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. Primers used for mutagenesis are: 5′-CTGTCACCGAGAATATTCTGACGG-3′ and 5′-CCGTCAGAATATTCTCGGT GACAG-3′.
Enrichment of PpCesA5 and PttCesA8
PpCesA5 and PttCesA8 were partially purified and reconstituted to proteoliposomes following the method previously described in Purushotham et al. (2016). The P. pastoris strain expressing PpCesA5 or PttCesA8 was grown on YPDS medium at 30°C overnight. A colony was inoculated into a 500 mL preculture medium containing 1% yeast extract, 2% peptone, 1% glycerol, 0.34% yeast nitrogen base, 1% ammonium sulfate, and 0.00004% biotin, followed by shaking incubation at 30°C overnight. Cells were collected by centrifugation at 2600g at 4°C for 15 min and resuspended in 1 L of induction medium (1% yeast extract, 2% peptone, 0.7% methanol, 0.34% yeast nitrogen base, 1% ammonium sulfate, and 0.00004% biotin) to an OD600 of 0.5, followed by shaking incubation at 30°C for 24 h. Grown cells were collected by centrifugation and frozen at −80°C until use. Frozen cells were thawed in a Cell Resuspension buffer containing 20 mm Tris-HCl pH 7.5, 1 m sorbitol, 1× EDTA-free protease inhibitor tablet (Thermo Fisher Scientific) and 1 mm PMSF and lysed by three passes using a microfluidizer at 20,000 psi. The lysate was centrifuged at 10,000g at 4°C for 10 min and its supernatant was subjected to ultracentrifugation at 200,000g at 4°C for 2 h. The pellet corresponding to vesicle fraction was resuspended in 50 mL Membrane Resuspension buffer containing 150 mm sodium P pH 7.5, 100 mm sodium chloride, 40 mm n-dodecyl-β-d-maltoside, 10% glycerol, 1× EDTA-free Protease inhibitor tablet (Thermo Fisher Scientific), and 1 mm PMSF, followed by gentle agitation at 4°C for 1 h. Insoluble materials were removed by ultracentrifugation at 200,000g at 4°C for 40 min. The obtained membrane protein fraction was incubated with 5 mL preequilibrated TALON Superflow Resin (GE Healthcare) with gentle agitation at 4°C overnight. The mix was applied to an empty column and washed with a series of 15 mL washing buffer (150 mm sodium P pH 7.5, 100 mm sodium chloride, and 1 mm LysoFos Choline Ether 14) containing 20, 40, 60, or 80 mm imidazole. Protein was eluted by 15 mL washing buffer containing 350 mm imidazole. Imidazole was reduced to less than 0.5 mm by repeated concentration/dilution cycles through a Vivaspin 20 desalting column (30 kD cutoff; GE Healthcare). All fractions were examined by Western-blot analysis.
Reconstitution of Proteoliposomes
Chloroform was evaporated from a predissolved yeast total lipid extract (Avanti Polar Lipids) by a stream of N gas, which was then completely dried by overnight incubation in a vacuum chamber. The lipid (100 mg) was resuspended in 1 mL of 120 mm lauryldimethylamine oxide, solubilized by heating at 60°C for 1 h, and stored at −20°C until use. Bio-Beads SM-2 Adsorbent Media (Bio-Rad) was washed in deionized water at room temperature for 10 min and dried on a filter paper. Six-hundred μL of partially purified PpCesA5 or PttCesA8 was mixed with 400 μL yeast lipid, to which dried Bio-Beads SM-2 Adsorbent Media was added to 75% of the total volume. This mix was incubated overnight with gentle rotation at 4°C. Reconstitution was determined by visual turbidity.
Immunoblot Analysis
Protein fractions were separated on a 4% to 12% gradient PAGE gel (GenScript) and transferred to a nitrocellulose membrane (GE Healthcare), which was hybridized with one of three monoclonal PpCesA5-specific antibodies raised against the synthetic peptides 681CKFSRKKTAPTRSDS694, 506CGHSGGHDTDGNELP519, or 473CAQKVPEEGWTMQDG486 (GenScript). The hybridized blot was incubated with secondary antibody conjugated with horseradish peroxidase. Chemiluminescent signals generated by Super Signal West Dura Extended Duration Substrate (Thermo Fisher Scientific) were detected by a GelLogic 4000 Pro System (Carestream Health).
Activity Assay
In general, reconstituted proteoliposome was mixed with a reaction buffer containing 20 mm Tris-HCl, pH7.5, 100 mm sodium chloride, 10% glycerol, 20 mm manganese chloride, 3 mm UDP-Glc, and 0.25 μCi UDP-[3H]Glc, followed by incubation at 37°C for 3 h. However, assays with alternate components and conditions were performed as indicated in each experiment. Reactions were stopped by adding Triton X-100 to a final concentration of 0.1%. The reaction mix was then centrifuged at 15,000 rpm for 20 min to collect the product, which was resuspended in 20 μL of cellulose resuspension buffer containing 20 mm Tris-HCl, pH7.5, 100 mm sodium chloride, and 10% glycerol and applied to a descending chromatography paper (2 mm; Whatman) using 60% ethanol. After air-drying, the chromatography paper was submerged in 5 mL ScintiVerse BD Cocktail (Thermo Fisher Scientific), and radioactivity was measured in a model no. LS6500 Scintillation Counter (Beckman Coulter). To prepare samples for electron microscopy, proteoliposomes were mixed with a reaction buffer containing 20 mm Tris-HCl, pH 7.5, 100 mm sodium chloride, 10% glycerol, 20 mm manganese chloride, and 3 mm UDP-Glc and incubated at 37°C for 3 h, which was followed by treatment with 0.1% Triton X-100.
Kinetic Analysis
Reactions were performed with 20 mm MnCl2 and 0 to 3.5 mm UDP-Glc. A 4-fold concentrated stock solution of 20 mm UDP-Glc and 1.0 μCi UDP-[3H]Glc was diluted to the final substrate concentration required in each reaction. After incubation for 30 min, the reactions were treated as described above. Untransformed data were fit to the Michaelis-Menton equation to extract parameter values.
TEM
Four-hundred-mesh Glider Copper grids (Ted Pella) were coated by evaporation of carbon using a model no. 502B Vacuum Carbon Coater (Denton). A quantity of 3.5 μL of prepared sample was loaded onto a glow-discharged grid, incubated for 1 min, and negatively stained with 10 serial drops of 0.75% uranyl formate on a parafilm. TEM images were taken using a Tecnai 12 Spirit BioTWIN TEM [120 kV, 6.3 mm spherical aberration (Cs), 56 K magnification, 4 K × 4 K Eagle CCD camera; FEI; located at the Huck Institutes of the Life Sciences, Pennsylvania State University].
Cryo-EM Analysis
To measure the width of cellulose microfibrils, in vitro cellulose microfibril products were applied to cryo-EM analysis as described in Grassucci et al. (2008). A never-dried sample was loaded on a QUANTIFOIL holey carbon grid (300-mesh; Ted Pella), blotted for 3 s, and vitrified with a Vitrobot (FEI; at the Huck Institute of the Life Sciences, Pennsylvania State University).
Cellulase Sensitivity Assay
For assays of incorporation of radioactive UDP Glc, the in vitro products were incubated with 10 U of β-1,4-glucanases or β-1,3-glucanases (Megazyme) at 37°C for 1 h. For TEM observation, in vitro produced cellulose microfibrils were incubated with a total of 150 ng highly purified cellulase mix containing Cel6A, Cel7A, and Cel7B proteins (kindly provided by Giridhar Poosarla, Pennsylvania State University), which was incubated at 50°C. Sample was taken for negative staining.
Linkage Analysis
In vitro products were pretreated for linkage analysis as described in Purushotham et al. (2016). Briefly, 2% SDS and 1 mg/mL proteinase K were used to eliminate proteins, followed by washing twice with water, treatment with hexane, and then freeze-drying. Dried samples were treated with chloroform/methanol followed by washing with 70% ethanol, and then a solution containing 50 mm Tris-HCl, 2% SDS, 10 mm Na-EDTA, 40 mm β-mercaptoethanol was used to remove residual proteins, followed by washing three times with 70% ethanol. To eliminate P. pastoris-derived glycogen-like polysaccharide, the samples were subjected to α-amylase and amyloglucosidase, and then washed with 70% ethanol. Subsequently, samples were freeze-dried. These treatments greatly reduced the mass [from 10 to 2 mg for wild type and 33 to 4.2 mg for PpCesA5(D782N)]. The prepared samples were permethylated to alditol acetates and analyzed by GC/EI-MS following the method previously described in Omadjela et al. (2013).
Image Analysis
All image analyses were performed using the software ImageJ (National Institutes of Health). To measure areas occupied by microfibrils, the Set Scale option was used to set the real length, the Polygon selection tool was used to define areas along the microfibrils, and the “Measure” tool was used to measure area. Measured areas were analyzed by mean and se. For measurement of thickness of individual microfibrils, the “Straight Line” tool was used to define widths and the “Measure” tool was used for measurement of thickness.
MS
For MS analysis, partially purified protein samples were separated by SDS-PAGE, followed by Coomassie Blue staining. The gel region corresponding to 110 kD to 140 kD was cut out and incubated with 5 mm Tris(2-carboxyethyl)phosphine hydrochloride/0.8% (w/v) ammonium bicarbonate at 60°C for 10 min, followed by washing with 100 mm iodoacetamide/0.8% (w/v) ammonium bicarbonate at 37°C for 15 min. MS spectra were taken from 60 min gradient from a NanoLC-Ultra-2D Plus and LC through a 200 μm × 0.5 mm ChromXP C18-CL 3 μm 120 Å Trap Column, and elution was performed through a 75 μm × 15 cm NanoLCMS C18-CL 2.6 μm 120 Å Nano LC Column (all by Eksigent). Model no. 5600 TripleTOF settings (Sciex) were as follows: parent scan was acquired for 250 ms, and then up to 50 MS/MS spectra were acquired over 2.5 s for a total cycle time of 2.8 s Gas 1 (N) = 7 and Gas 3 (N) = 25. MS analysis was performed by the Proteomics and Mass Spectrometry Core Facility of Pennsylvania State University.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Diagram of PpCesA5
Supplemental Figure S2. Purification of heterologously expressed PpCesA5 from P. pastoris
Supplemental Figure S3. Purification of PpCesA5(D782N)
Supplemental Figure S4. Biochemical activity of PpCesA5(D782N) protein
Supplemental Figure S5. Proteoliposomes bearing PpCesA5(D782N) produce no microfibrils
Supplemental Figure S6. TEM analysis of in vitro products from disrupted proteoliposomes bearing PpCesA5
Supplemental Figure S7. TEM analysis of in vitro products from proteoliposomes bearing PpCesA5
Supplemental Figure S8. Cryo-EM image of cellulose microfibrils
Supplemental Figure S9. Fragmentation spectra from electron-impact mass spectrometry of the permethylated alditol acetates of the in vitro generated polysaccharide
Supplemental Figure S10. Linkage analysis of in vitro products of proteoliposomes bearing PpCesA5(D782N)
Supplemental Table S1. List of proteins identified by mass spectrometry
Acknowledgments
S.H.C., M.K., and B.T.N. designed the experiments, interpreted the data, and wrote the paper; P.P. and J.Z. constructed Pichia pastoris strains and performed radiolabeling assays; S.H.C. was involved in most experiments; S.H.C., C.F., and C.M. conducted TEM analysis; S.M.D. and V.B. performed linkage analysis.
Acknowledgments
We thank Missy Hazen and John Cantolina (Huck Institutes of the Life Sciences, Penn State University) for technical help with TEM and Giridhar Poosarla (Penn State University) for providing Cel6A, Cel7A, and Cel7B proteins.
Footnotes
This material is based upon work supported as part of The Center for LignoCellulose Structure and Formation, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, and Office of Basic Energy Sciences under Award no. DE-SC0001090.
Articles can be viewed without a subscription.
References
- Anderson CT, Carroll A, Akhmetova L, Somerville C (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, Cork A, Glover J, et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Atalla RH, Vanderhart DL (1984) Native cellulose: a composite of two distinct crystalline forms. Science 223: 283–285 [DOI] [PubMed] [Google Scholar]
- Augimeri RV, Varley AJ, Strap JL (2015) Establishing a role for bacterial cellulose in environmental interactions: lessons learned from diverse biofilm-producing proteobacteria. Front Microbiol 6: 1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI. (2005) Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Basu S, Omadjela O, Gaddes D, Tadigadapa S, Zimmer J, Catchmark JM (2016) Cellulose microfibril formation by surface-tethered cellulose synthase enzymes. ACS Nano 10: 1896–1907 [DOI] [PubMed] [Google Scholar]
- Basu S, Omadjela O, Zimmer J, Catchmark JM (2017) Impact of plant matrix polysaccharides on cellulose produced by surface-tethered cellulose synthases. Carbohydr Polym 162: 93–99 [DOI] [PubMed] [Google Scholar]
- Belton CT, Tanner SF, Cartier N, Chanzy H (1989) High-resolution solid-state carbon-13 nuclear magnetic resonance spectroscopy of tunicin, an animal cellulose. Macromolecules 22: 1615–1617 [Google Scholar]
- Bosca S, Barton CJ, Taylor NG, Ryden P, Neumetzler L, Pauly M, Roberts K, Seifert GJ (2006) Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiol 142: 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34: 351–362 [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37: D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Du J, Sines I, Poosarla VG, Vepachedu V, Kafle K, Park YB, Kim SH, Kumar M, Nixon BT (2015) In vitro synthesis of cellulose microfibrils by a membrane protein from protoplasts of the non-vascular plant Physcomitrella patens. Biochem J 470: 195–205 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djerbi S, Aspeborg H, Nilsson P, Sundberg B, Mellerowicz E, Blomqvist K, Teeri TT (2004) Identification and expression analysis of genes encoding putative cellulose synthases (CesA) in the hybrid aspen, Populus tremula (L.) × P. tremuloides (Michx.). Cellulose 11: 301–312 [Google Scholar]
- Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP (2002) Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 43: 1407–1420 [DOI] [PubMed] [Google Scholar]
- Fugelstad J, Bouzenzana J, Djerbi S, Guerriero G, Ezcurra I, Teeri TT, Arvestad L, Bulone V (2009) Identification of the cellulose synthase genes from the Oomycete Saprolegnia monoica and effect of cellulose synthesis inhibitors on gene expression and enzyme activity. Fungal Genet Biol 46: 759–767 [DOI] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Guillot A, Vernhettes S, Höfte H (2014) Catalytic subunit stoichiometry within the cellulose synthase complex. Plant Physiol 166: 1709–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss CA, Brockmann DJ, Bushoven JT, Roberts AW (2012) A CELLULOSE SYNTHASE (CESA) gene essential for gametophore morphogenesis in the moss Physcomitrella patens. Planta 235: 1355–1367 [DOI] [PubMed] [Google Scholar]
- Grassucci RA, Taylor D, Frank J (2008) Visualization of macromolecular complexes using cryo-electron microscopy with FEI Tecnai transmission electron microscopes. Nat Protoc 3: 330–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha MA, Apperley DC, Evans BW, Huxham IM, Jardine WG, Viëtor RJ, Reis D, Vian B, Jarvis MC (1998) Fine structure in cellulose microfibrils: NMR evidence from onion and quince. Plant J 16: 183–190 [DOI] [PubMed] [Google Scholar]
- Hill JL Jr., Hammudi MB, Tien M (2014) The Arabidopsis cellulose synthase complex: a proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 26: 4834–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Him JL, Pelosi L, Chanzy H, Putaux JL, Bulone V (2001) Biosynthesis of (1→3)-β-D-glucan (callose) by detergent extracts of a microsomal fraction from Arabidopsis thaliana. Eur J Biochem 268: 4628–4638 [DOI] [PubMed] [Google Scholar]
- John RP, Anisha GS, Nampoothiri KM, Pandey A (2011) Micro and macroalgal biomass: a renewable source for bioethanol. Bioresour Technol 102: 186–193 [DOI] [PubMed] [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM Jr (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D (2002) Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci USA 99: 11109–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marga F, Grandbois M, Cosgrove DJ, Baskin TI (2005) Cell wall extension results in the coordinate separation of parallel microfibrils: evidence from scanning electron microscopy and atomic force microscopy. Plant J 43: 181–190 [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Döring A, Persson S (2014) The cell biology of cellulose synthesis. Annu Rev Plant Biol 65: 69–94 [DOI] [PubMed] [Google Scholar]
- McNamara JT, Morgan JL, Zimmer J (2015) A molecular description of cellulose biosynthesis. Annu Rev Biochem 84: 895–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendu V, Stork J, Harris D, DeBolt S (2011) Cellulose synthesis in two secondary cell wall processes in a single cell type. Plant Signal Behav 6: 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, McNamara JT, Zimmer J (2014) Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21: 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman RH, Hill SJ, Harris PJ (2013) Wide-angle x-ray scattering and solid-state nuclear magnetic resonance data combined to test models for cellulose microfibrils in mung bean cell walls. Plant Physiol 163: 1558–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Langan P, Chanzy H (2002) Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron x-ray and neutron fiber diffraction. J Am Chem Soc 124: 9074–9082 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Sugiyama J, Chanzy H, Langan P (2003) Crystal structure and hydrogen bonding system in cellulose I(α) from synchrotron x-ray and neutron fiber diffraction. J Am Chem Soc 125: 14300–14306 [DOI] [PubMed] [Google Scholar]
- Nixon BT, Mansouri K, Singh A, Du J, Davis JK, Lee JG, Slabaugh E, Vandavasi VG, O’Neill H, Roberts EM, Roberts AW, Yingling YG, et al. (2016) Comparative structural and computational analysis supports eighteen cellulose synthases in the plant cellulose synthesis complex. Sci Rep 6: 28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olek AT, Rayon C, Makowski L, Kim HR, Ciesielski P, Badger J, Paul LN, Ghosh S, Kihara D, Crowley M, Himmel ME, Bolin JT, et al. (2014) The structure of the catalytic domain of a plant cellulose synthase and its assembly into dimers. Plant Cell 26: 2996–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omadjela O, Narahari A, Strumillo J, Mélida H, Mazur O, Bulone V, Zimmer J (2013) BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc Natl Acad Sci USA 110: 17856–17861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Keegstra K (2008) Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J 54: 559–568 [DOI] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Hervé C, Domozych DS, Willats WG, Tuohy MG, Kloareg B, Stengel DB (2011) Evolution and diversity of plant cell walls: from algae to flowering plants. Annu Rev Plant Biol 62: 567–590 [DOI] [PubMed] [Google Scholar]
- Purushotham P, Cho SH, Díaz-Moreno SM, Kumar M, Nixon BT, Bulone V, Zimmer J (2016) A single heterologously expressed plant cellulose synthase isoform is sufficient for cellulose microfibril formation in vitro. Proc Natl Acad Sci USA 113: 11360–11365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AW, Bushoven JT (2007) The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Mol Biol 63: 207–219 [DOI] [PubMed] [Google Scholar]
- Roberts AW, Roberts EM, Haigler CH (2012) Moss cell walls: structure and biosynthesis. Front Plant Sci 3: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Brown RM Jr (1997) Identification of cellulose synthase(s) in higher plants: sequence analysis of processive β-glycosyltransferases with the common motif “D, D, D35Q(R,Q)XRW”. Cellulose 4: 33–49 [Google Scholar]
- Saxena IM, Brown RM Jr., Dandekar T (2001) Structure-function characterization of cellulose synthase: relationship to other glycosyltransferases. Phytochemistry 57: 1135–1148 [DOI] [PubMed] [Google Scholar]
- Sethaphong L, Haigler CH, Kubicki JD, Zimmer J, Bonetta D, DeBolt S, Yingling YG (2013) Tertiary model of a plant cellulose synthase. Proc Natl Acad Sci USA 110: 7512–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabaugh E, Davis JK, Haigler CH, Yingling YG, Zimmer J (2014) Cellulose synthases: new insights from crystallography and modeling. Trends Plant Sci 19: 99–106 [DOI] [PubMed] [Google Scholar]
- Somerville C. (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Cosgrove DJ (2009) Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr Biol 19: R800–R811 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LH, Forsyth VT, Sturcová A, Kennedy CJ, May RP, Altaner CM, Apperley DC, Wess TJ, Jarvis MC (2013) Structure of cellulose microfibrils in primary cell walls from collenchyma. Plant Physiol 161: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandavasi VG, Putnam DK, Zhang Q, Petridis L, Heller WT, Nixon BT, Haigler CH, Kalluri U, Coates L, Langan P, Smith JC, Meiler J, et al. (2016) A structural study of CESA1 catalytic domain of Arabidopsis cellulose synthesis complex: evidence for CESA trimers. Plant Physiol 170: 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise HZ, Saxena IM, Brown RM (2011) Isolation and characterization of the cellulose synthase genes PpCesA6 and PpCesA7 in Physcomitrella patens. Cellulose 18: 371–384 [Google Scholar]
- Zhang T, Zheng Y, Cosgrove DJ (2016) Spatial organization of cellulose microfibrils and matrix polysaccharides in primary plant cell walls as imaged by multichannel atomic force microscopy. Plant J 85: 179–192 [DOI] [PubMed] [Google Scholar]