Adventitious root cultures provide insights to elucidating the biosynthesis of pharmaceutically relevant diterpenoids in the model genus Tripterygium.
Abstract
Adventitious root cultures were developed from Tripterygium regelii, and growth conditions were optimized for the abundant production of diterpenoids, which can be collected directly from the medium. An analysis of publicly available transcriptome data sets collected with T. regelii roots and root cultures indicated the presence of a large gene family (with 20 members) for terpene synthases (TPSs). Nine candidate diterpene synthase genes were selected for follow-up functional evaluation, of which two belonged to the TPS-c, three to the TPS-e/f, and four to the TPS-b subfamilies. These genes were characterized by heterologous expression in a modular metabolic engineering system in Escherichia coli. Members of the TPS-c subfamily were characterized as copalyl diphosphate (diterpene) synthases, and those belonging to the TPS-e/f subfamily catalyzed the formation of precursors of kaurane diterpenoids. The TPS-b subfamily encompassed genes coding for enzymes involved in abietane diterpenoid biosynthesis and others with activities as monoterpene synthases. The structural characterization of diterpenoids accumulating in the medium of T. regelii adventitious root cultures, facilitated by searching the Spektraris online spectral database, enabled us to formulate a biosynthetic pathway for the biosynthesis of triptolide, a diterpenoid with pharmaceutical potential. Considering the significant enrichment of diterpenoids in the culture medium, fast-growing adventitious root cultures may hold promise as a sustainable resource for the large-scale production of triptolide.
Tripterygium wilfordii, also known as léi gōng téng in Mandarin Chinese (generally translated as “thunder god vine”), has a long history of use in traditional Chinese medicine for the treatment of fever, chills, edema, and carbuncles (Helmstädter, 2013). The genus Tripterygium (Celastraceae) is known to be a rich source of specialized metabolites, of which more than 400 have been isolated, structurally characterized, and assessed in cell-based assays (Brinker et al., 2007). Root extracts have been evaluated as a medication for rheumatoid arthritis, cancer, hepatitis, nephritis, ankylosing spondylitis, polycystic kidney disease, and obesity; more than a dozen clinical trials with such extracts (often referred to as Tripterygium glycoside) have been completed, but, in part due to shortcomings in study designs, the efficacy has remained a matter of debate (Chen et al., 2010; Liu et al., 2013; Zhu et al., 2013). More promising results have been obtained with purified constituents, which are usually extracted from Tripterygium roots. Semisynthetic chemical derivatives of triptolide, a diterpenoid epoxide, have been evaluated in phase I clinical trials (Zhou et al., 2012; Meng et al., 2014). Minnelide, a water-soluble prodrug analog of triptolide, has shown very promising activity in multiple animal models of pancreatic cancer (Chugh et al., 2012) and, in 2013, was advanced to phase I clinical trials in the United States (clinicaltrials.gov identifier NCT01927965).
One of the critical challenges for clinical evaluations is a supply shortage for triptolide and other diterpenoids. The most abundant specialized metabolites in Tripterygium roots are quinone methide triterpenoids and sesquiterpene pyridine alkaloid macrolides, whereas triptolide and other diterpenoids occur only at very low concentrations ranging from 0.0001% to 0.002% of dry weight biomass (Zhou et al., 2012; Zeng et al., 2013; Guo et al., 2014). Accordingly, the extraction yields of diterpenoids from Tripterygium roots are poor and alternative, sustainable production methods need to be developed. Tissue cultures represent a promising alternative for the production of high-value plant metabolites. Early studies with T. wilfordii suspension cultures were designed to unravel the structures of small-molecule constituents (Kutney et al., 1981, 1992, 1993; Kutney and Han, 1996; Nakano et al., 1997, 1998). Similarly, hairy root cultures were initially employed for phytochemical investigations (Nakano et al., 1998). It was recognized only recently that triptolide concentrations produced by tissue cultures (up to 0.15% of dry weight; Miao et al., 2013, 2014; Zhu et al., 2014) far exceed those reported for roots.
Surprisingly, despite considerable pharmaceutical interest, the biosynthesis of triptolide has only recently attracted the deserved attention. Several diterpene synthases, some of which are relevant to triptolide formation, have now been characterized in T. wilfordii (Zerbe et al., 2013; Andersen-Ranberg et al., 2016; Hansen et al., 2017). However, the remaining genes involved in the biosynthesis of the highly functionalized triptolide structure have remained enigmatic. In this article, we describe the development of Tripterygium adventitious root cultures in which diterpenoids are produced as principal metabolites that can be harvested sustainably from the medium. Based on the structures of these metabolites, we can now postulate the biochemical steps leading to the main diterpenoid metabolites. In addition, analysis of transcriptome data sets, acquired with diterpenoid-producing adventitious root cultures, enabled the selection and subsequent functional characterization of genes involved in the early steps of diterpenoid biosynthesis. Our results indicate that Tripterygium tissue cultures offer opportunities to both unravel diterpenoid biosynthetic pathways and produce target diterpenoids, such as triptolide, at larger scale.
RESULTS
Development of Diterpenoid-Secreting Adventitious Root Cultures
Tripterygium regelii adventitious root cultures were initiated based on standard protocols. Over a 1-year period, during which medium was extracted with ethyl acetate and hydrophobic metabolites were analyzed by HPLC-quadrupole time of flight-mass spectrometry (QTOF-MS), the highest triptolide producers were selected for further propagation. In contrast to roots of mature Tripterygium plants, where triterpenoids occur at low concentrations (less than 1%) and diterpenoids are trace constituents (less than 0.01%), our adventitious root cultures accumulated diterpenoids abundantly in the medium and triterpenoids (in particular celastrol) in roots (Fig. 1, A and B). By comparing adjusted peak areas from HPLC-QTOF-MS runs, we estimated that diterpenoids constitute approximately 77% of all detected metabolites in an organic extract of the culture medium (Supplemental Fig. S1).
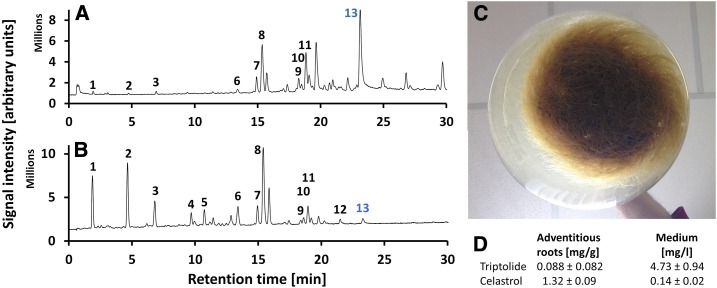
Figure 1.
Accumulation of abietane diterpenoids in T. regelii adventitious root cultures. A and B, HPLC-QTOF-MS traces (total ion current) obtained with ethanolic extracts of cultured roots (A) and culture medium (B). The identities of the metabolite peaks 1 to 13 are given in Table I (peak 13 [celastrol] is highlighted in blue because it is the only metabolite that does not belong to the abietane diterpenoids). C, Fernbach flask with root culture photographed from below. The arm of the person holding the 4-L flask is visible at the bottom right and serves as a size comparison. D, Concentrations of triptolide and celastrol in cultured roots and culture medium.
An accurate quantitation of all diterpenoids was not possible because we did not have a complete set of authentic standards, but we performed absolute quantitations for signature metabolites (triptolide and celastrol). Various common reagents and elicitors were tested (methyl jasmonate, methyl salicylate, chitosan, yeast extract, and cold exposure), but only the methyl salicylate treatment resulted in a significant increase in triptolide accumulation (2.2-fold; P value of 0.01) and a concomitant decrease in celastrol concentration (Supplemental Table S1). The disadvantage of methyl salicylate treatments was that root color darkened and growth ceased, often for months. Therefore, we focused our efforts on further selecting cultures that were high diterpenoid producers under noninducing conditions and could be maintained for extended periods of time. These cultures were gradually scaled up to 2.8-L flasks (500 mL of medium; Fig. 1C). Triptolide accumulated at 4.7 mg L−1 (Fig. 1D) and was readily extractable from the culture medium every 2 weeks (when adventitious roots were transferred to fresh medium).
Hundreds of structurally diverse metabolites have been isolated and characterized from various Tripterygium organs and tissues (Lange et al., 2017). Our adventitious root cultures, in contrast, secreted primarily diterpenoids into the culture medium and, therefore, are an excellent experimental model system in which to study the biosynthesis of these clinically relevant metabolites. As a first step to further assess the potential of these tissue cultures, it was important to identify the major constituents of medium extracts. Following HPLC fractionation of medium extracts of T. regelii adventitious root cultures, fractions containing metabolites that corresponded to prominent peaks in HPLC-QTOF-MS chromatograms were characterized by mass spectrometry (MS), tandem mass spectrometry (MS/MS), and 1H-NMR spectroscopy (Supplemental Fig. S2; Supplemental Methods and Data File S1), and metabolites were identified by searches against the Spektraris database (Fischedick et al., 2015). We ascertained the identities of five diterpenoids, were able to annotate three additional metabolites with high confidence, and acquired tentative identifications for an additional two metabolites (Table I and Fig. 2). Three metabolites showed the mass fragmentation patterns typical of diterpenoids, but their identities remained unknown (Table I).
Table I. Metabolites detected by HPLC-QTOF-MS in the medium of T. regelii adventitious root cultures.
MS/MS spectra were acquired with a fragmentation voltage of 30 eV.
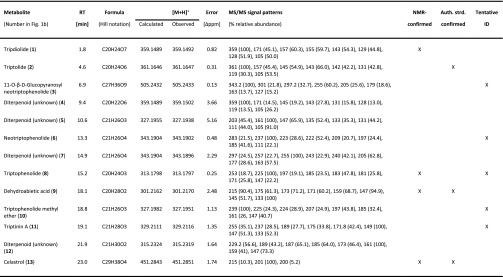 |
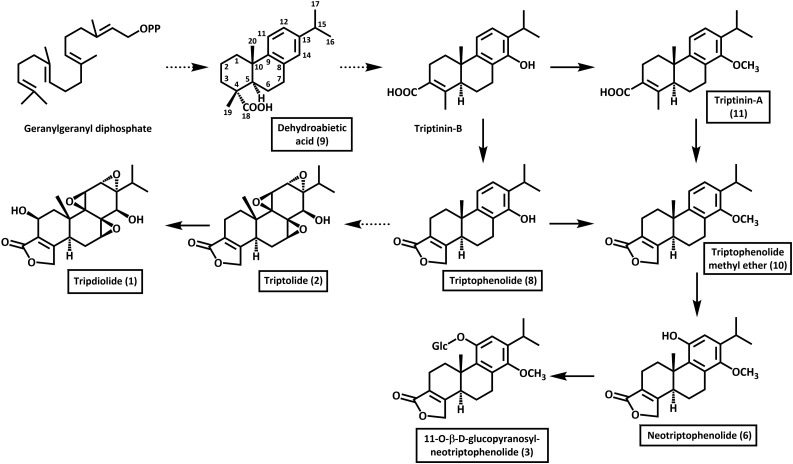
Figure 2.
Putative biosynthetic pathway toward abietane diterpenoids in Tripterygium. Metabolites found in this study are boxed. The numbering of the carbons forming the abietane skeleton is indicated using dehydroabietic acid as an example. Reactions requiring multiple steps are indicated by dotted arrows. The numbering of metabolites is the same as in Figure 1.
Functional Characterization of Candidate Diterpene Synthases
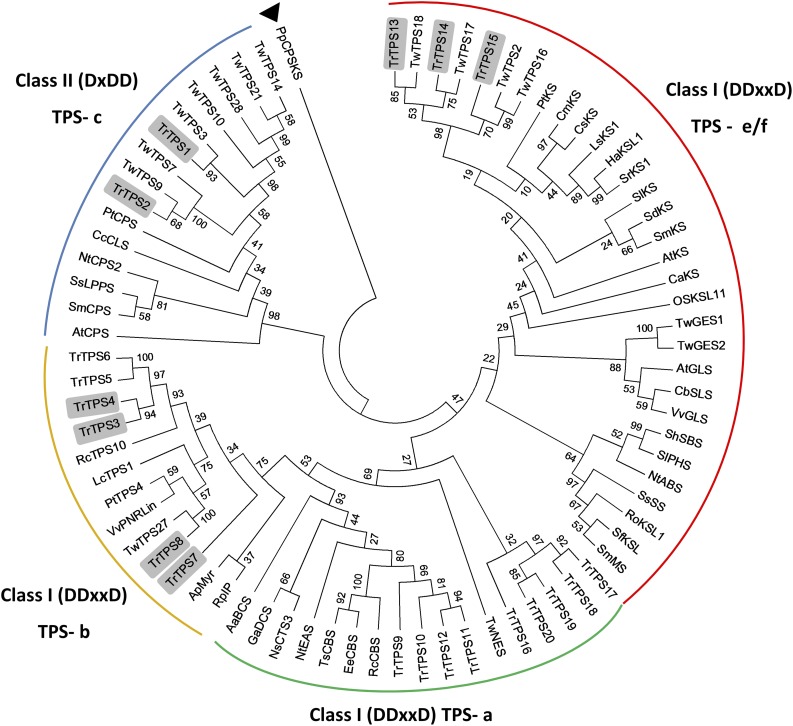
To obtain full-length candidate genes, we deemed it advantageous to employ all available T. regelii sequences. Therefore, we downloaded several publicly available transcriptome data sets obtained with roots and root cultures and then generated a consensus assembly. tBLASTn searches with peptide sequences of diterpene synthases of dicotyledons were performed against our assembly data. A phylogenetic analysis indicated that translated peptide sequences of two T. regelii genes (TrTPS2 and TrTPS1) clustered with functionally characterized class II diterpene synthases of the TPS-c family (copalyl diphosphate synthases; Fig. 3; Supplemental Table S2). Full-length cDNAs were cloned (KX533964 and KX533965, respectively; for primer sequences, see Supplemental Table S3), and their biochemical functions were evaluated (see below). Translated peptide sequences of three genes (TrTPS13, TrTPS14, and TrTPS15) clustered with functionally characterized class I ent-kaurene synthases of the TPS-e/f family (Fig. 3). Full-length clones (KX533966, KX533967, and KX533968, respectively) were obtained by PCR and subjected to functional characterization (see below). Five additional genes (TrTPS19, TrTPS17, TrTPS20, TrTPS18, and TrTPS16) clustered with linear-type diterpene synthases of the TPS-a family but, because of the significant separation from sequences of previously characterized diterpene synthases, also might catalyze the formation of nonlinear diterpenes of as yet unknown structure. While this was an interesting finding, it was not of direct interest to this investigation (involvement in the biosynthesis of kauranes or abietanes unlikely); therefore, we did not proceed with further characterizations. The translated sequences of two cDNAs (TrTPS8 [KY856995] and TrTPS7 [KY856996]) clustered with a recently discovered class I diterpene synthase of the TPS-b family from T. wilfordii (Hansen et al., 2017; Fig. 3) and were selected for functional characterization (see below). Other members from the TPS-b family were TrTPS3 (KY856993) and TrTPS4 (KY856994), which were also functionally characterized (see below), and two additional transcripts (TrTPS5 and TrTPS6) that were quite short (less than 600 bp) and could not be extended to a length that would have allowed functional characterization. Finally, nine members of the TPS-a family (TrTPS9, TrTPS10, TrTPS11, TrTPS12, TrTPS16, TrTPS17, TrTPS18, TrTPS19, and TrTPS20) clustered with known genes that encode linear diterpene cyclases and macrocyclases, which were not of direct interest to this study and, therefore, were not further characterized.
Figure 3.
Molecular phylogeny of dicot (di)terpene synthases, with an emphasis on those from the genus Tripterygium. Functionally characterized terpene synthases from T. regelii are highlighted by gray boxes. The analysis was carried out using the maximum likelihood method based on a matrix-based model (Jones et al., 1992). The bootstrap consensus tree inferred from 1,000 replications (Felsenstein, 1985) is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% of bootstrap replicates are collapsed. Initial tree(s) for the heuristic search were obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances and then selecting the topology with superior log likelihood value. The analysis involved 77 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 94 positions in the final data set. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016). The filled triangle indicates the position of the sequence chosen as an outgroup for rooting the phylogenetic tree.
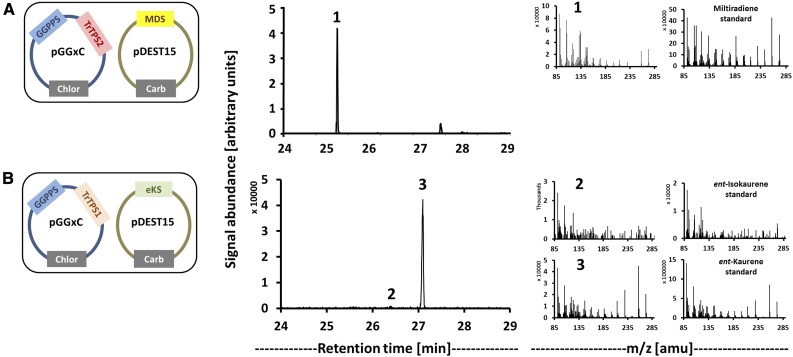
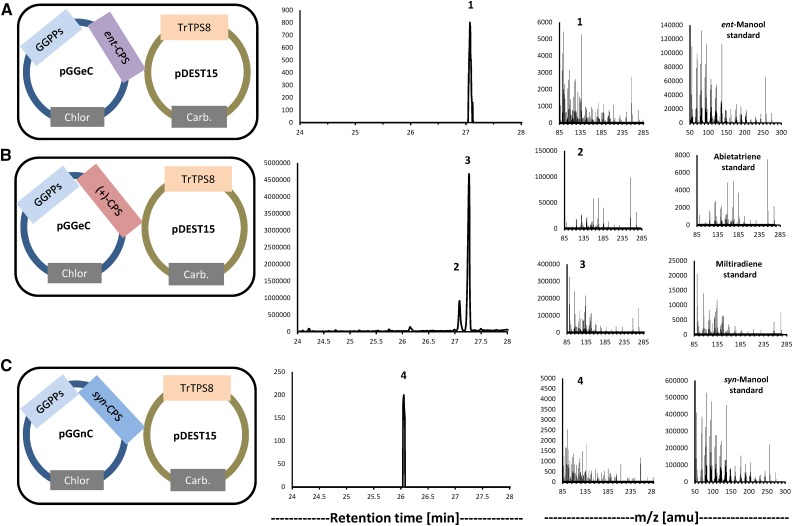
Diterpene synthase candidate genes were characterized by functional expression in engineered Escherichia coli strains that provide appropriate precursors (Cyr et al., 2007). Appropriately truncated cDNAs of class II diterpene synthase candidates from T. regelii (lacking the plastidial targeting sequence; Supplemental Fig. S3) were introduced into E. coli harboring a recombinant geranylgeranyl diphosphate synthase (GGPPS) gene from Abies grandis (Burke and Croteau, 2002), in combination with either the ent-kaurene synthase (eKS) gene from Arabidopsis (Arabidopsis thaliana; Yamaguchi et al., 1998) or the miltiradiene synthase (MDS) gene from Saliva miltiorrhiza (Gao et al., 2009). The expression of TrTPS2 (a candidate diterpene synthase gene from the TPS-c family) led to the production of miltiradiene when combined with MDS but did not generate detectable products in other combinations, indicating that the gene encodes a (+)-copalyl diphosphate synthase [(+)-CPS; Fig. 4A]. The combination of TrTPS1 (also a TPS-c family member) with eKS yielded ent-kaurene and small amounts of ent-isokaurene, but no products were detected when it was expressed in other gene combinations, indicating that this gene encodes an ent-copalyl diphosphate synthase (ent-CPS; Fig. 4B).
Figure 4.
Modular in vivo assay results obtained with T. regelii class II diterpene synthase candidates of the TPS-c subfamily. From left to right, constructs (selectable markers in gray boxes; Chlor, chloramphenicol; Carb, carbenicillin; MDS [from Salvia miltiorrhiza]; eKS [from Arabidopsis]), gas chromatography (GC)-MS chromatograms, and mass spectra of assay products and authentic standards. TrTPS2 was identified as a (+)-CPS (A), while TrTPS1 showed activity as an ent-CPS (B).
A similar modular approach was taken for the characterization of class I diterpene synthase candidates (once again lacking the plastidial targeting sequence; Supplemental Fig. S4), involving coexpression with GGPPS (from A. grandis) and either ent-CPS (from Arabidopsis), (+)-CPS [a mutant version of abietadiene synthase from A. grandis that produces (+)-copalyl diphosphate], or syn-CPS (from rice [Oryza sativa]; Cyr et al., 2007). ent-Kaurene was the most prominent reaction product (with ent-isokaurene as a minor side product) when TrTPS13 or TrTPS14 (TPS-e/f family members) were expressed in combination with ent-CPS (Fig. 5, A and B), indicating ent-kaurene synthase functions for these isoforms. When combined with (+)-CPS, TrTPS13 expression produced small amounts of sandaracopimaradiene and isopimaradiene, while coexpression of TrTPS13 with syn-CPS led to the formation of syn-pimara-7,15-diene and syn-stemod-13(17)-ene (Fig. 5, C and D). The combination of both class II and class I diterpene synthases from T. regelii (TrTPS2 and TrTPS13) in one modular construct resulted in the production of ent-kaurene (with smaller quantities of ent-isokaurene; Supplemental Fig. S5), thus confirming the function of these genes in ent-kaurane diterpenoid biosynthesis. TrTPS15 (also a member of the TPS-e/f family) produced small quantities of ent-manool, syn-manool, or (+)-manool when coupled with ent-CPS, syn-CPS, or (+)-CPS, respectively (Fig. 5, E–G). When coupled with (+)-CPS, TrTPS8 (from the TPS-b family; Supplemental Fig. S6) produced primarily miltiradiene (Fig. 6A) along with small amounts of abietatriene, presumably arising from a previously noted autoxidation (Zi and Peters, 2013). Both products also were found to be present in the medium of T. regelii adventitious root cultures (Supplemental Fig. S1). When combined with ent-CPS or syn-CPS, TrTPS8 generated small quantities of ent-manool or syn-manool, respectively (Fig. 6, B and C).
Figure 5.
Modular in vivo assay results obtained with T. regelii class I diterpene synthase candidates of the TPS-e/f family. From left to right: constructs (selectable markers in gray boxes; Chlor, chloramphenicol; Carb, carbenicillin), GC-MS chromatograms, and mass spectra of assay products and authentic standards. The following gene combinations were tested: ent-CPS/TrTPS13 (A), ent-CPS/TrTPS14 (B), (+)-CPS/TrTPS13 (C), syn-CPS/TrTPS13 (D), ent-CPS/TrTPS15 (E), (+)-CPS/TrTPS15 (F), and syn-CPS/TrTPS15 (G).
Figure 6.
Modular in vivo assay results obtained with TrTPS8, a class I diterpene synthase candidate of the TPS-a subfamily. From left to right: constructs (selectable markers in gray boxes; Chlor, chloramphenicol; Carb, carbenicillin), GC-MS chromatograms, and mass spectra of assay products and authentic standards. The following gene combinations were tested: ent-CPS/TrTPS8 (A), (+)-CPS/TrTPS8 (B), and syn-CPS/TrTPS8 (C).
TrTPS3 and TrTPS4, members of the TPS-b subfamily (Supplemental Fig. S6), did not form products in any combination with CPSs. Instead, both TrTPS3 and TrTPS4 produced mixtures of monoterpenes upon reaction with geranyl diphosphate (substrate for monoterpene synthases), with (−)-linalool and (+)-linalool as major products (Table II; Supplemental Fig. S7). When reacted with linalyl diphosphate (a reaction intermediate of monoterpene synthases), TrTPS3 and TrTPS4 both formed (−)-terpineol and (+)-terpineol. In assays with linalyl diphosphate, TrTPS3, but not TrTPS4, also generated appreciable amounts (−)-limonene (∼20% of total products; Table II). TrTPS7 was expressed in the recombinant E. coli strains but was not active in any combination with copalyl diphosphate synthases. It also did not generate products from geranyl diphosphate as a substrate in in vitro assays.
Table II. Product distribution in in vitro assays with TrTPS3 and TrTPS4.
Dashes indicate quantities lower than 0.1% (v/v) of total detected volatiles.
| Monoterpene Product | Retention Time | Geranyl Diphosphate as Substrate |
Linalyl Diphosphate as Substrate |
||
|---|---|---|---|---|---|
| TrTPS3 | TrTPS4 | TrTPS3 | TrTPS4 | ||
| min | % of total | ||||
| Myrcene | 17.135 | – | 4.39 | 1.45 | 1.57 |
| (−)-Limonene | 20.567 | 7.77 | – | 2.05 | 2.06 |
| (+)-Limonene | 20.823 | – | – | 1.87 | 1.85 |
| Terpinolene | 23.734 | – | – | 2.18 | 2.22 |
| (−)-Linalool | 32.171 | 35.60 | 40.38 | 21.88 | 23.81 |
| (+)-Linalool | 32.327 | 44.77 | 46.04 | 28.32 | 29.93 |
| (−)-Terpineol | 40.083 | 7.07 | 4.83 | 20.63 | 20.22 |
| (+)-Terpineol | 40.359 | 4.78 | 4.35 | 21.62 | 18.33 |
DISCUSSION
Proposing a Diterpenoid Biosynthetic Pathway Based on Metabolites That Accumulate in Tripterygium Adventitious Root Cultures
In contrast to roots of mature Tripterygium plants, where diterpenoids are only very minor constituents (less than 0.01%), our adventitious root cultures secreted these metabolites at very high levels (approximately 77% of the area under all peaks detected by HPLC-QTOF-MS in organic extracts of the culture medium). The structure and abundance of the principal diterpenoids found in our tissue cultures can thus be employed to develop hypotheses regarding the biosynthesis of abietane-type diterpenoids of Tripterygium (Fig. 2). It has been proposed previously that the two-step cyclization of the universal diterpenoid precursor geranylgeranyl diphosphate, via (+)-copalyl diphosphate, yields miltiradiene as an abietane hydrocarbon intermediate (Hansen et al., 2017), and our data agree with this suggestion. Oxidation at C18, like that described for CYP720 in conifers (Ro et al., 2005; Hamberger et al., 2011; Geisler et al., 2016), would produce dehydroabietic acid (which we detected in T. regelii root cultures; Fig. 2). Demethylation at C4, carboxylation at C3, and hydroxylation at C14 would result in the formation of triptinin B. This metabolite was not detected in our root cultures, but its methyl ether, triptinin A, was identified tentatively as a significant constituent. The tentatively identified neotriptophenolide and its glycoside, both likely derived from triptinin B and/or triptinin A, accumulated as significant by-products in our root cultures. The lactonization of triptinin B would generate triptophenolide, which accumulated as the most abundant abietane diterpenoid in our root cultures (Fig. 2). The functionalization of the aromatic ring would then produce triptolide and, following an additional hydroxylation, tripdiolide, as the primary end products of the biosynthetic conversions detected in our adventitious root cultures.
Schemes for abietane diterpenoid biosynthesis in the genus Tripterygium have been proposed before (Kutney et al., 1981; Kutney and Han, 1996), but these were based on the available phytochemical evidence at the time and did not incorporate information about relative metabolite abundance (Fig. 1; Supplemental Fig. S1). By adding this new dimension, we are increasing the confidence in predictions regarding the design principles of the pathways leading to structurally unusual diterpene triepoxides. Because of the high abundance of diterpenoids in the medium of T. regelii adventitious root cultures, we have been able to employ simple purification protocols for intermediates (triptophenolide) and end products (triptolide and tripdiolide). This provides commercially unavailable substrates and products for in vitro enzyme assays and, therefore, enables the functional characterization of candidate genes.
Identifying Genes with Roles in Generating Diterpenoid Structural Diversity
One T. regelii clone (TrTPS1) was characterized as encoding an ent-CPS that, in combination with eKSs (TrTPS13 and TrTPS14), generates ent-kaurene (and very small amounts of ent-isokaurene; Fig. 5). These reactions are common to all vascular plants, which require endogenous production of the derived GA hormones (Zi et al., 2014). However, ent-kaurane diterpenoids also occur as specialized metabolites in some plant families, including the Celastraceae (which contains the genus Tripterygium). The previously reported metabolites can be grouped into two major classes: four-ring ent-kauranoic acids/alcohols and five-ring oxygen bridge-containing ent-kauranes (Duan et al., 1999, 2001; Tanaka et al., 2004). In the closely related species T. wilfordii, recent studies also identified an ent-CPS (TwTPS3) and several isoforms of ent-kaurene/ent-isokaurene synthase (TwTPS2, TwTPS16, TwTPS17, and TwTPS18; Zerbe et al., 2013; Hansen et al., 2017). Various biological activities of Tripterygium ent-kauranes have been documented in in vitro assays (for review, see Brinker et al., 2007), but the in vivo functions remain to be elucidated. When combined with a (+)-CPS, TrTPS13 formed sandaracopimaradiene and isopimaradiene; in combination with a syn-CPS, TrTPS13 showed activity for the production of syn-pimara-7,15-diene and syn-stemod-13(17)-ene (Fig. 5). We did not find a syn-CPS in T. regelii, nor does there appear to be such an activity among the class II diterpene synthases of T. wilfordii (Hansen et al., 2017); therefore, the biological relevance of this finding is unknown at this time.
Genes from T. wilfordii were characterized previously as coding for ent-copal-8-ol diphosphate synthase (termed TwTPS21) or kolavenyl diphosphate synthase (TwTPS10, TwTPS14, and TwTPS28; Andersen-Ranberg et al., 2016; Hansen et al., 2017). However, the stereochemistry at C8 and C9 (8S, 9S) is the opposite of that of all labdane-type diterpenoids characterized from Tripterygium thus far (8R, 9R; Duan et al., 1999, 2001). In our functional assays, TrTPS15 produced ent-manool, (+)-manool, and syn-manool when combined with ent-CPS, (+)-CPS, or syn-CPS, respectively. Interestingly, (+)-manool has the correct stereochemistry (8R, 9R) to serve as a precursor for the known labdane diterpenoids of Tripterygium (Duan et al., 1999, 2001). The other manools and manoyl oxides, generated by diterpene synthases of T. regelii and T. wilfordii, respectively, may only be produced in vivo under specific environmental conditions or may simply reflect enzymatic substrate promiscuity. However, the generation of structural diversity by combinations of class II and class I diterpene synthases, whether with recognizable in vivo relevance, does provide biosynthetic access to these and derived diterpenoids.
The gene coding for T. regelii TrTPS8 [converts (+)-copalyl diphosphate to miltiradiene, a likely intermediate in triptolide and tripdiolide biosynthesis] is an ortholog of the recently characterized TwTPS27 gene from T. wilfordii (Hansen et al., 2017). Based on phylogenetic analyses, Hansen et al. (2017) concluded that TwTPS27 diversified from members of the TPS-b subfamily, more specifically from terpene synthases that catalyze the formation of acyclic monoterpenes. The location of TwTPS27 on the phylogenetic tree of angiosperm terpene synthases was a surprise because all angiosperm diterpene synthases involved in abietane/labdane biosynthesis characterized until then belonged to the TPS-e/f subfamily (Chen et al., 2011). This evolutionary history also applies to TrTPS8, which is very closely related to TwTPS27.
Hansen et al. (2017) expressed TwTPS23, TwTPS24, and TwTPS26 transiently in Nicotiana benthamiana and then subjected volatiles to a head-space analysis. The authors did not have authentic standards for monoterpenes and, therefore, could only use spectral comparisons for tentative identification. We are in the fortunate position to host a sizable repository of monoterpenes and, therefore, were able to perform in vitro assays with unequivocal results. TrTPS3 and TrTPS4 catalyzed the formation of a mixture of (−)-linalool and (+)-linalool from geranyl diphosphate as a substrate. These results from early termination (by water capture) immediately following the formation of a linalyl cation (Supplemental Fig. S6). To test if these enzymes would be capable of catalyzing cyclization if initial water capture was avoided, we also performed assays with linalyl diphosphate as a substrate. Indeed, in these assays, TrTPS3 and TrTPS4 formed (−)-terpineol and (+)-terpineol as products [with TrTPS3 also releasing (−)-limonene], indicating that, although water capture is still the main means of terminating the catalyzed reaction, these TPSs can catalyze cyclization. To the best of our knowledge, these are the first monoterpene synthases to be unequivocally identified from Tripterygium.
In summary, the high abundance of diterpene synthase transcripts in T. regelii adventitious root cultures facilitated the rapid cloning of candidate genes. These were functionally characterized using a suitable modular expression system (Cyr et al., 2007). Thus, our tissue cultures are an excellent experimental system for further pathway elucidation.
Can Adventitious Root Cultures Serve as Sustainable Resources for the Production of Pharmaceutically Relevant Diterpenoids?
Cultures of Tripterygium hairy roots, adventitious roots, or cell suspensions all have the advantage of accumulating abietane diterpenoids at high concentrations (Miao et al., 2013; Zhu et al., 2014) and, in comparison with the unsustainable harvest of roots (where the concentrations of these specialized metabolites are very low), should be considered as commercial sources. We demonstrate here that diterpenoids are secreted into the culture medium of T. regelii adventitious root cultures, which significantly reduces the complexity of the matrix for extraction. Triptolide is detected as the peak with the second highest intensity (16% of the total peak area) in our HPLC-QTOF-MS chromatograms of culture medium extracts. Furthermore, it is well separated from other medium constituents, which enabled a one-step HPLC processing to greater than 95% purity (as judged by HPLC-QTOF-MS and 1H-NMR). In our hands, the production of triptolide in adventitious root cultures also has been reliable (grown continuously for more than 2 years) and readily scalable (from 50- to 500-mL volumes within weeks). Others have scaled up Tripterygium cultures to 10- to 20-L bioreactors (Kutney et al., 1992; Miao et al., 2014); therefore, it appears that typical shortcomings of tissue culture (reliability and scalability) have already been addressed.
It is probable that triptolide levels could be enhanced further if flux through the abietane diterpenoid pathway was increased in transgenic tissue cultures, where genes involved in diterpenoid biosynthesis are already expressed at fairly high levels (Supplemental Fig. S8). One approach would be the overexpression of genes involved in providing diterpenoid precursors. A different and potentially complementary approach would be the down-regulation of genes involved in competing pathways (e.g. reducing triterpenoid formation). We are currently investigating methods for the transformation of Tripterygium to enable such endeavors. Alternatively, the genes required for triptolide biosynthesis, once discovered, could be transferred to an engineered microbial strain, akin to the successful efforts to produce another diterpenoid, forskohlin (Pateraki et al., 2017). At this point in time, yields of highly functionalized plant diterpenoids in synthetic hosts have been relatively low; therefore, plant tissue cultures would seem to be a competitive option for triptolide production.
MATERIALS AND METHODS
Chemicals
Triptolide and celastrol were purchased from Cayman Chemical. Dehydroabietic acid was synthesized according to a literature protocol (González et al., 2010). Acetone was of HPLC grade (Fisher Scientific), ethyl acetate was ACS quality (Avantor Performance Materials), ethanol was Omnisolv (EMD Serono), and acetonitrile and water were liquid chromatography-mass spectrometry grade (Sigma-Aldrich). CDCl3 was obtained from Cambridge Isotope Laboratories. All other reagents were purchased from Sigma-Aldrich.
Adventitious Root Cultures
Young Tripterygium regelii plants were purchased from Woodlanders and maintained under greenhouse conditions (illumination, 16-h day/8-h night [250-500 µE]; temperature, 24°C day/20°C night; relative humidity, 45%–55%). To initiate tissue cultures, leaf material was harvested, rinsed with sterile water, and surface sterilized by soaking in 20% (v/v) commercial bleach. Sterilized leaves were cut into 1-cm2 pieces and placed onto Murashige and Skoog medium with macronutrients and micronutrients plus Gamborg’s vitamins (Caisson), Suc (20 g L−1; Sigma-Aldrich), 1-naphthaleneacetic acid (0.2 mg L−1; Caisson), 6-benzylaminopurine (1 mg L−1; Sigma-Aldrich), and phytagel (2.4 g L−1; Sigma-Aldrich), with pH adjusted to 5.8. After 4 to 6 weeks, callus that developed along the leaf square edges was moved to plates containing root induction medium (Murashige and Skoog medium with macronutrients and micronutrients plus Gamborg’s vitamins, Suc [20 g L−1], 1-naphthaleneacetic acid [1 mg L−1], 6-benzylaminopurine [0.2 mg L−1], and phytagel [2.4 g L−1], with pH adjusted to 5.8). Developing adventitious roots were transferred to fresh plates every 4 to 6 weeks until they appeared strong enough for transfer to liquid medium (Murashige and Skoog medium with macronutrients and micronutrients plus Gamborg’s vitamins, Suc [20 g L−1], and 1-naphthaleneacetic acid [1 mg L−1], pH 5.8). Adventitious roots were partially submerged in medium (50 mL in a 250-mL Erlenmeyer flask) and maintained at 25°C, by shaking at 80 rpm, in the dark, with transfer to fresh medium every 2 weeks. As adventitious roots grew, the size of the flask and the volume of liquid medium were adjusted up to 500 mL in a 2-L Fernbach flask.
Metabolite Extraction from Adventitious Root Cultures
Solid materials from adventitious root cultures were washed with water (volume equivalent to the wet weight of material) and freeze dried for 3 d (Lyph-Lock 12L; LabConco), and the resulting dry matter was stored at −80°C until further use. Aliquots of 50 ± 0.5 mg were transferred to receptacles of an MM01 Dry Mill (Retsch) and further homogenized by ball shaking for 45 s at a rate of 20 shakes per second. The homogenate was then transferred to glass test tubes with Teflon-lined caps. Each sample was extracted four times at 23°C with 5 mL of acetone for 30 min (Ultrasound Bath FS30H; Fisher Scientific). After each extraction step, samples were centrifuged at 3,000g for 5 min, and the combined supernatants were collected in a separate glass test tube. The solvent was removed under reduced pressure (EZ Bio; GeneVac), and the remaining residue was redissolved in 1 mL of 90% acetonitrile containing 10 μg mL−1 9-anthracene carboxylic acid as an internal standard. Prior to further analysis, samples were passed through syringe filters (polytetrafluoroethylene; 0.22-μm pore size) and stored at 4°C for a maximum of 2 d.
A medium aliquot of adventitious root cultures (generally 5 mL) was extracted twice with ethyl acetate (2 mL each time) by thorough mixing for 1 min at 23°C. The combined organic extracts (4 mL) were extracted against water (2 mL) and transferred to a new glass vial, and the solvent was removed under reduced pressure (EZ Bio; GeneVac). The residue was redissolved in 200 µL of 90% aqueous acetonitrile containing 10 μg mL−1 9-anthracene carboxylic acid as an internal standard. Prior to further analysis, samples were passed through syringe filters (polypropylene; 0.22-μm pore size) and stored at 4°C for a maximum of 2 d.
HPLC-QTOF-MS and MS/MS Data Acquisition and Method Validation
The separation of metabolites was almost identical to that described previously (Fischedick et al., 2015). The conditions for metabolite separation were modified slightly: the initial conditions were 70% solvent A (water with 0.1% [v/v] formic acid) and 30% solvent B (acetonitrile with 0.1% [v/v] formic acid). A linear gradient (flow rate, 0.6 mL min−1) was used to increase solvent B to 80% over 35 min, followed by a more rapid gradient to 95% solvent B at 40 min. The diode array detector was set to record at 219, 254, and 424 nm, with UV/visible spectra being recorded from 200 to 500 nm. Mass spectral data were obtained based on the protocols outlined previously (Fischedick et al., 2015) with the following differences: the electrospray ionization source was operated in positive polarity and MS/MS data were acquired with a collision energy of 30 eV. Data analysis was performed using the MassHunter software version B.03.01 (Agilent Technologies). The approach for validating the quantitation of triptolide and celastrol was reported previously for various analytes in plant matrices (Cuthbertson et al., 2013), and only the relevant values are given here: recovery from adventitious roots (n = 3): 85.6% ± 5.6% for triptolide, 96.8% ± 2.8% for celastrol; recovery from culture medium (n = 3): 106.6% ± 3.9% for triptolide, 86.1% ± 2.9% for celastrol; reproducibility of extraction/detection (as relative sd; n = 3): 6.5% (interday) and 0.9% (intraday) for triptolide, 0.5% (interday) and 0.6% (intraday) for celastrol; limit of detection at signal-to-noise ratio of 1: 5 (n = 3): 0.01 ng for triptolide (MS detection) and 1 ng for celastrol (diode array detection at 424 nm); limit of quantitation at signal-to-noise ratio of 1: 10 (n = 3): 0.05 for triptolide (MS detection) and 10 ng for celastrol (diode array detection at 424 nm); regression equation for calibration curve (n = 3): y = 25,938x + 16,475x (R2 = 0.989) for triptolide (corrected for matrix effects) and y = 0.9077x (R2 = 0.999) for celastrol (matrix effects irrelevant for diode array detection); and linear range, 0.05 to 50 ng for triptolide and 1 to 2,500 ng for celastrol.
Cloning and Functional Characterization of Terpene Synthases
Raw data from several transcriptome sequencing projects performed previously with T. regelii tissues (National Center for Biotechnology Information Short Read Archive accession no. SRP075639 [adventitious root cultures]) and data sets available at http://www.medplantrnaseq.org/ (roots and root cultures) were downloaded, and a consensus assembly was generated using the Trinity (Haas et al., 2013) and TransABySS (Robertson et al., 2010) assemblers. tBLASTn searches with peptide sequences of characterized class I and class II diterpene synthases (Supplemental Table S2) were performed against the T. regelii consensus assembly. RNA was isolated from T. regelii adventitious root cultures (harvested at 10 d after transfer to fresh medium) using the Plant RNA Purification reagent (Invitrogen) and the RNEasy Mini Kit (Qiagen) according to each manufacturer’s instructions, converted to cDNA using Maxima Reverse Transcriptase (Thermo Fisher Scientific), and PCR with gene-specific primers was then employed to clone candidate diterpene synthase sequences (details in Supplemental Table S3). RACE-PCR was performed according to the manufacturer’s instructions (SMARTer-RACE 5′/3′ kit; Clontech/TaKaRa). The Gateway cloning system (Invitrogen) was used to insert TrTPS1 and TrTPS2 into a previously described pGG-DEST vector, and the other putative diterpene synthases into pDEST15, for functional expression in the Escherichia coli C41 (DE3) (Cyr et al., 2007). Diterpenoids were extracted directly from 50-mL cultures with an equal volume of n-hexanes (Fisher Scientific). The extract was then run through silica gel 60 and magnesium sulfate columns as described elsewhere (Cyr et al., 2007). The eluents were dried under a flow of nitrogen, and the residue was dissolved in 200 µL of n-hexanes. Aliquots (1 µL) were injected onto an HP-5MS column (30-m length × 0.25-mm diameter, 0.25-µm film thickness; J&W Scientific, distributed through Agilent) of a 6890N gas chromatograph (operated in splitless mode) coupled to a 5973 inert mass selective detector (Agilent). Settings were as follows: helium as carrier gas at a flow rate of 1 mL min−1; inlet temperature set to 250°C; oven program with 50°C for 1 min, first linear gradient to 300°C at 7°C min−1, second linear gradient to 330°C at 20°C min−1, and final hold of 5 min; transfer line set to 180°C; electron impact spectra recorded at 70 eV with MS data collection from mass-to-charge ratio of 50 to 450. Peaks were identified based on comparisons of retention times and MS fragmentation patterns with those of authentic standards. The functional evaluation of monoterpene synthases was performed according to Srividya et al. (2015).
Accession Numbers
All genes characterized as part of this study have been deposited in GenBank with the following accession numbers: KX533964 (TrTPS2), KX533965 (TrTPS1), KX533966 (TrTPS13), KX533967 (TrTPS14), KX533968 (TrTPS15), KY856993 (TrTPS3), KY856994 (TrTPS4), KY856995 (TrTPS8), and KY856996 (TrTPS7).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Accumulation of abietane diterpenoids in the medium of T. regelii adventitious root cultures.
Supplemental Figure S2. 1H-NMR spectra of abietane diterpenoids.
Supplemental Figure S3. Sequences of T. regelii diterpene synthases of the TPS-c family.
Supplemental Figure S4. Sequences of T. regelii diterpene synthases of the TPS-e/f family.
Supplemental Figure S5. Sequences of T. regelii diterpene synthases of the TPS-b family.
Supplemental Figure S6. Modular in vivo combination of T. regelii class II (TrTPS2) and class I (TrTPS13) diterpene synthases.
Supplemental Figure S7. Proposed mechanism for T. regelii monoterpene synthases.
Supplemental Figure S8. Comparison of expression levels of candidate diterpene synthase genes in T. regelii adventitious root cultures and roots based on RNA sequence by expectation-maximization analysis.
Supplemental Figure S9. Alignment of diterpene synthase sequences included in the phylogenetic analysis.
Supplemental Table S1. Changes in triptolide and celastrol concentrations in T. regelii adventitious root cultures following various elicitor treatments.
Supplemental Table S2. List of dicot diterpene synthase protein sequences used in the phylogenetic analysis.
Supplemental Table S3. List of primers used in T. regelii terpene synthase gene cloning.
Supplemental Methods and Data File S1. Purification and characterization of diterpenoids from T. regelii adventitious root cultures.
Acknowledgments
We thank Sean R. Johnson for generating sequence assemblies with publicly available transcriptome data sets, Washington State University’s NMR Core Facility for access to instruments, and Dr. Greg Helms for expert support.
Glossary
- QTOF-MS
quadrupole time of flight-mass spectrometry
- MS/MS
tandem mass spectrometry
- MS
mass spectrometry
- GC
gas chromatography
Footnotes
This work was supported by the National Institutes of Health (award nos. RC2GM092561 to B.M.L. and GM076324 to R.J.P.) and McIntire-Stennis formula funds from the Agricultural Research Center at Washington State University (to B.M.L.).
Articles can be viewed without a subscription.
References
- Andersen-Ranberg J, Kongstad KT, Nielsen MT, Jensen NB, Pateraki I, Bach SS, Hamberger B, Zerbe P, Staerk D, Bohlmann J, et al. (2016) Expanding the landscape of diterpene structural diversity through stereochemically controlled combinatorial biosynthesis. Angew Chem Int Ed Engl 55: 2142–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinker AM, Ma J, Lipsky PE, Raskin I (2007) Medicinal chemistry and pharmacology of genus Tripterygium (Celastraceae). Phytochemistry 68: 732–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke C, Croteau R (2002) Interaction with the small subunit of geranyl diphosphate synthase modifies the chain length specificity of geranylgeranyl diphosphate synthase to produce geranyl diphosphate. J Biol Chem 277: 3141–3149 [DOI] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66: 212–229 [DOI] [PubMed] [Google Scholar]
- Chen YZ, Gao Q, Zhao XZ, Chen XM, Zhang F, Chen J, Xu CG, Sun LL, Mei CL (2010) Meta-analysis of Tripterygium wilfordii Hook F in the immunosuppressive treatment of IgA nephropathy. Intern Med 49: 2049–2055 [DOI] [PubMed] [Google Scholar]
- Chugh R, Sangwan V, Patil SP, Dudeja V, Dawra RK, Banerjee S, Schumacher RJ, Blazar BR, Georg GI, Vickers SM, et al. (2012) A preclinical evaluation of Minnelide as a therapeutic agent against pancreatic cancer. Sci Transl Med 4: 156ra139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson DJ, Johnson SR, Piljac-Žegarac J, Kappel J, Schäfer S, Wüst M, Ketchum RE, Croteau RB, Marques JV, Davin LB, et al. (2013) Accurate mass-time tag library for LC/MS-based metabolite profiling of medicinal plants. Phytochemistry 91: 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr A, Wilderman PR, Determan M, Peters RJ (2007) A modular approach for facile biosynthesis of labdane-related diterpenes. J Am Chem Soc 129: 6684–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Takaishi Y, Momota H, Ohmoto Y, Taki T, Jia Y, Li D (1999) Immunosuppressive diterpenoids from Tripterygium wilfordii. J Nat Prod 62: 1522–1525 [DOI] [PubMed] [Google Scholar]
- Duan HQ, Takaishi Y, Momota H, Ohmoto Y, Taki T, Tori M, Takaoka S, Jia YF, Li D (2001) Immunosuppressive terpenoids from extracts of Tripterygium wilfordii. Tetrahedron 57: 8413–8424 [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Fischedick JT, Johnson SR, Ketchum REB, Croteau RB, Lange BM (2015) NMR spectroscopic search module for Spektraris, an online resource for plant natural product identification: taxane diterpenoids from Taxus × media cell suspension cultures as a case study. Phytochemistry 113: 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B, Peters RJ (2009) A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett 11: 5170–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler K, Jensen NB, Yuen MM, Madilao L, Bohlmann J (2016) Modularity of conifer diterpene resin acid biosynthesis: P450 enzymes of different CYP720B clades use alternative substrates and converge on the same products. Plant Physiol 171: 152–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González MA, Pérez-Guaita D, Correa-Royero J, Zapata B, Agudelo L, Mesa-Arango A, Betancur-Galvis L (2010) Synthesis and biological evaluation of dehydroabietic acid derivatives. Eur J Med Chem 45: 811–816 [DOI] [PubMed] [Google Scholar]
- Guo L, Duan L, Liu K, Liu EH, Li P (2014) Chemical comparison of Tripterygium wilfordii and Tripterygium hypoglaucum based on quantitative analysis and chemometrics methods. J Pharm Biomed Anal 95: 220–228 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8: 1494–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Ohnishi T, Hamberger B, Séguin A, Bohlmann J (2011) Evolution of diterpene metabolism: Sitka spruce CYP720B4 catalyzes multiple oxidations in resin acid biosynthesis of conifer defense against insects. Plant Physiol 157: 1677–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen NL, Heskes AM, Hamberger B, Olsen CE, Hallström BM, Andersen-Ranberg J, Hamberger B (2017) The terpene synthase gene family in Tripterygium wilfordii harbors a labdane-type diterpene synthase among the monoterpene synthase TPS-b subfamily. Plant J 89: 429–441 [DOI] [PubMed] [Google Scholar]
- Helmstädter A. (2013) Tripterygium wilfordii Hook. f.: how a traditional Taiwanese medicinal plant found its way to the West. Pharmazie 68: 643–646 [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutney JP, Han K (1996) Studies with plant-cell cultures of the Chinese herbal plant, Tripterygium wilfordii: isolation and characterization of diterpenes. Recl Trav Chim Pays Bas 115: 77–93 [Google Scholar]
- Kutney JP, Hewitt GM, Kurihara T, Salisbury PJ, Sindelar RD, Stuart KL, Townsley PM, Chalmers WT, Jacoli GG (1981) Cyto-toxic diterpenes triptolide, tripdiolide, and cyto-toxic triterpenes from tissue-cultures of Tripterygium wilfordii. Can J Chem 59: 2677–2683 [Google Scholar]
- Kutney JP, Hewitt GM, Lee G, Piotrowska K, Roberts M, Rettig SJ (1992) Studies with tissue-cultures of the Chinese herbal plant, Tripterygium-wilfordii: isolation of metabolites of interest in rheumatoid-arthritis, immunosuppression, and male contraceptive activity. Can J Chem 70: 1455–1480 [Google Scholar]
- Kutney JP, Samija MD, Hewitt GM, Bugante EC, Gu H (1993) Anti-inflammatory oleanane triterpenes from Tripterygium wilfordii cell suspension cultures by fungal elicitation. Plant Cell Rep 12: 356–359 [DOI] [PubMed] [Google Scholar]
- Lange BM, Fischedick JT, Lange MF, Srividya N, Šamec D, Poirier BC (2017) Integrative approaches for the identification and localization of specialized metabolites in Tripterygium roots. Plant Physiol 173: 456–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YF, Tu SH, Gao WN, Wang Y, Liu PL, Hu YH, Dong H (2013) Extracts of Tripterygium wilfordii Hook F in the treatment of rheumatoid arthritis: a systemic review and meta-analysis of randomised controlled trials. Evid Based Complement Alter Med 2013: 410793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C, Zhu H, Song H, Wang Z, Huang G, Li D, Ma Z, Ma J, Qin Q, Sun X, et al. (2014) Targets and molecular mechanisms of triptolide in cancer therapy. Chin J Cancer Res 26: 622–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao GP, Zhu CS, Feng JT, Han J, Song XW, Zhang X (2013) Aggregate cell suspension cultures of Tripterygium wilfordii Hook f for triptolide, wilforgine, and wilforine production. Plant Cell Tissue Organ Cult 112: 109–116 [Google Scholar]
- Miao GP, Zhu CS, Yang YQ, Feng MX, Ma ZQ, Feng JT, Zhang X (2014) Elicitation and in situ adsorption enhanced secondary metabolites production of Tripterygium wilfordii Hook. f. adventitious root fragment liquid cultures in shake flask and a modified bubble column bioreactor. Bioprocess Biosyst Eng 37: 641–650 [DOI] [PubMed] [Google Scholar]
- Nakano K, Oose Y, Takaishi Y (1997) A novel epoxy-triterpene and nortriterpene from callus cultures of Tripterygium wilfordii. Phytochemistry 46: 1179–1182 [Google Scholar]
- Nakano K, Yoshida C, Furukawa W, Takaishi Y, Shishido K (1998) Terpenoids in transformed root culture of Tripterygium wilfordii. Phytochemistry 49: 1821–1824 [DOI] [PubMed] [Google Scholar]
- Pateraki I, Andersen-Ranberg J, Jensen NB, Wubshet SG, Heskes AM, Forman V, Hallström B, Hamberger B, Motawia MS, Olsen CE, Staerk D, Hansen J, et al. (2017) Total biosynthesis of the cyclic AMP booster forskolin from Coleus forskohlii. eLife 6: e23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro DK, Arimura G, Lau SYW, Piers E, Bohlmann J (2005) Loblolly pine abietadienol/abietadienal oxidase PtAO (CYP720B1) is a multifunctional, multisubstrate cytochrome P450 monooxygenase. Proc Natl Acad Sci USA 102: 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, et al. (2010) De novo assembly and analysis of RNA-seq data. Nat Methods 7: 909–912 [DOI] [PubMed] [Google Scholar]
- Srividya N, Davis EM, Croteau RB, Lange BM (2015) Functional analysis of (4S)-limonene synthase mutants reveals determinants of catalytic outcome in a model monoterpene synthase. Proc Natl Acad Sci USA 112: 3332–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Ooba N, Duan H, Takaishi Y, Nakanishi Y, Bastow K, Lee KH (2004) Kaurane and abietane diterpenoids from Tripterygium doianum (Celastraceae). Phytochemistry 65: 2071–2076 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Sun Tp, Kawaide H, Kamiya Y (1998) The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol 116: 1271–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Wang W, Guan S, Cheng C, Yang M, Avula B, Khan IA, Guo DA (2013) Simultaneous quantification of 18 bioactive constituents in Tripterygium wilfordii using liquid chromatography-electrospray ionization-mass spectrometry. Planta Med 79: 797–805 [DOI] [PubMed] [Google Scholar]
- Zerbe P, Hamberger B, Yuen MM, Chiang A, Sandhu HK, Madilao LL, Nguyen A, Hamberger B, Bach SS, Bohlmann J (2013) Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol 162: 1073–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZL, Yang YX, Ding J, Li YC, Miao ZH (2012) Triptolide: structural modifications, structure-activity relationships, bioactivities, clinical development and mechanisms. Nat Prod Rep 29: 457–475 [DOI] [PubMed] [Google Scholar]
- Zhu B, Wang Y, Jardine M, Jun M, Lv JC, Cass A, Liyanage T, Chen HY, Wang YJ, Perkovic V (2013) Tripterygium preparations for the treatment of CKD: a systematic review and meta-analysis. Am J Kidney Dis 62: 515–530 [DOI] [PubMed] [Google Scholar]
- Zhu C, Miao G, Guo J, Huo Y, Zhang X, Xie J, Feng J (2014) Establishment of Tripterygium wilfordii Hook. f. hairy root culture and optimization of its culture conditions for the production of triptolide and wilforine. J Microbiol Biotechnol 24: 823–834 [DOI] [PubMed] [Google Scholar]
- Zi J, Mafu S, Peters RJ (2014) To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu Rev Plant Biol 65: 259–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J, Peters RJ (2013) Characterization of CYP76AH4 clarifies phenolic diterpenoid biosynthesis in the Lamiaceae. Org Biomol Chem 11: 7650–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]