The Arabidopsis RopGEF1 is important for the localization of auxin influx carrier AUX1 and accumulation of efflux carrier PIN proteins, thereby affecting auxin-dependent growth and development.
Abstract
Polar auxin transport, facilitated by the combined activities of auxin influx and efflux carriers to maintain asymmetric auxin distribution, is essential for plant growth and development. Here, we show that Arabidopsis (Arabidopsis thaliana) RopGEF1, a guanine nucleotide exchange factor and activator of Rho GTPases of plants (ROPs), is critically involved in polar distribution of auxin influx carrier AUX1 and differential accumulation of efflux carriers PIN7 and PIN2 and is important for embryo and early seedling development when RopGEF1 is prevalently expressed. Knockdown or knockout of RopGEF1 induces embryo defects, cotyledon vein breaks, and delayed root gravity responses. Altered expression from the auxin response reporter DR5rev:GFP in the root pole of RopGEF1-deficient embryos and loss of asymmetric distribution of DR5rev:GFP in their gravistimulated root tips suggest that auxin distribution is affected in ropgef1 mutants. This is reflected by the polarity of AUX1 being altered in ropgef1 embryos and roots, shifting from the normal apical membrane location to a basal location in embryo central vascular and root protophloem cells and also reduced PIN7 accumulation at embryos and altered PIN2 distribution in gravistimulated roots of mutant seedlings. In establishing that RopGEF1 is critical for AUX1 localization and PIN differential accumulation, our results reveal a role for RopGEF1 in cell polarity and polar auxin transport whereby it imapcts auxin-mediated plant growth and development.
Auxin is a unique plant hormone that displays polar transport. It is transported from the site of biosynthesis to distal target tissues by an intercellular transport system. Polar auxin transport (PAT) and local auxin metabolism lead to its asymmetric distribution and generation of auxin gradients and auxin maxima within plant cells, tissues, and organs. Auxin gradients and maxima are essential for plant growth and development, including establishment of the embryonic axis, formation and maintenance of the root stem cell niche, and mediating tropic response and organogenesis (Vanneste and Friml, 2009). Polar auxin movement is facilitated by the combined activities of auxin influx and efflux carrier proteins. The AUX1/LIKE-AUX1 (AUX/LAX) family of auxin transporters comprises major influx carriers, whereas PIN-FORMED (PIN) and B subfamily of ABC transporters are major auxin efflux carriers. AUX1 has a cell-type-dependent polar plasma membrane (PM) localization and accumulates on the apical face of protophloem cells in root meristem (Swarup et al., 2001; Kleine-Vehn et al., 2006) facilitating auxin uptake. PIN proteins also display polar localization at the PM and regulate the direction of auxin flow (Wisniewska et al., 2006). For example, PIN1, PIN3, and PIN7 are localized at the basal membrane of root stele cells, where they mediate the downward flow of auxin to the root tip. PIN2, on the other hand, localizes at the apical membrane of root epidermal cells and mediates the upward flow of auxin to the root elongation zone (Petrásek and Friml, 2009). Thus, PIN efflux carriers together with AUX/LAX influx carriers act concomitantly in the directionality of intercellular auxin movement (Swarup and Péret, 2012).
AUX1/LAX family contains four members, AUX1, LAX1, LAX2, and LAX3. AUX1 is the founding member of the family and has been confirmed as a high-affinity auxin transporter in Xenopus laevis oocytes (Yang et al., 2006) and baculovirus-infected insect cells (Carrier et al., 2008). Functional studies showed that AUX/LAX genes play critical roles in auxin-regulated development. For example, aux1lax1 and aux1lax1lax2 mutations affect embryogenesis including cotyledon and root patterning (Robert et al., 2015). Mutations in AUX1 result in root agravitropic response, reduced lateral roots, and short root hairs (Bennett et al., 1996; Marchant et al., 1999; Swarup et al., 2001). Loss of function in LAX3 reduces lateral root emergence (Swarup et al., 2008). lax2 mutant displays vascular vein discontinuity in the cotyledons (Péret et al., 2012).
AUX1 polar localization is cell-type-specific in the root as it resides at the apical PM of protophloem cells but evenly distributes around the cell in root cap (Swarup et al., 2001; Kleine-Vehn et al., 2006). Auxin-Resistant4, an endoplasmic reticulum-localized protein is required for AUX1 localization by regulating AUX1 trafficking, loss of function in Auxin-Resistant4 causes the accumulation of AUX1 in the endoplasmic reticulum of root epidermis cells (Dharmasiri et al., 2006). AUX1 polarity is also dependent on the actin cytoskeleton and sterol composition of the membrane (Kleine-Vehn et al., 2006). Brefeldin A inhibits vesicle trafficking and induces intercellular accumulation of constitutively cycling PM proteins (Geldner et al., 2001). Brefeldin A-sensitive as well as insensitive ARF guanine nucleotide exchange factors (GEFs) might be involved in AUX1 subcellular trafficking (Grebe et al., 2002; Kleine-Vehn et al., 2006).
Asymmetric distribution of PIN and AUX1 in a cell is important for mediating auxin into and out of the cell. Multiple factors for regulating PIN polarity have been identified. Differential distribution of PIN proteins requires regulated endocytosis, ARF-GEF GNOM-dependent recycling to the PM and retromer-dependent vascular targeting for degradation (Steinmann et al., 1999; Geldner et al., 2003; Jaillais et al., 2007). The phosphorylation status of PIN proteins is also critical for determining PIN polarity (Adamowski and Friml, 2015). The dynamic cycles of PIN phosphorylation and dephosphorylation were mediated by the AGC kinase or PINOID-related AGC3 and phosphatase PP2C (Friml et al., 2004; Michniewicz et al., 2007). Additionally, cellulose-based cell wall connections to PM are important for the maintenance of PIN polarity (Kleine-Vehn et al., 2006). While differential distribution of PIN proteins has been extensively studied (Adamowski and Friml, 2015), molecular components that maintain the AUX1 polarity remain largely unknown. Here, we report that RopGEF1, which activate ROP GTPases (Berken et al., 2005), plays an important role in maintaining the polarity of AUX1 in phrotophloem cells.
ROPGTPases are important regulators of many biological processes including polar growth of pollen tubes and root hairs, hormone responses, PAT, and defense responses (Yalovsky et al., 2008; Wu et al., 2011; Yang and Lavagi, 2012). They function as molecular switches and cycle between GTP-bound active and GDP-bound inactive forms. Activation of ROPs is predominantly controlled by RopGEFs, a plant-specific family of GEFs with a conserved central domain (called PRONE) for catalytic activity. The Arabidopsis genome encodes 14 RopGEFs. Transient overexpression of a subset of RopGEFs, including RopGEF1, in tobacco (Nicotiana benthamiana) and Arabidopsis led to depolarized growth of pollen tubes (Gu et al., 2006; Zhang and McCormick, 2007; Cheung et al., 2008). Hypersensitive responses to abscisic acid (ABA)-mediated stomata closure and ABA-inhibited root growth have been reported for ropgef1ropgef4 and ropgef1ropgef4ropgef10 mutant plants (Li and Liu, 2012; Yu et al., 2012). A recent study found that RopGEF1 was targeted for degradation by ABA and functioned in ABA-mediated inhibition of lateral root growth (Li et al., 2016). RopGEF11 has been shown to be involved in root development by interacting with phytochrome (Shin et al., 2010). We have previously reported that RopGEF7 is important for auxin-dependent maintenance of root stem cell niche (Chen et al., 2011) and showed that the accumulation of auxin efflux carriers PINs in the PM was reduced in knockdown plants with reduced RopGEF7 expression (Chen et al., 2011; Huang et al., 2014). A subset of RopGEFs are specific and highly expressed in pollen, while other members of the RopGEF family are expressed in a variety of developmental stages and tissues, such as embryos, seedlings, roots, leaves, and flowers (Zhang and McCormick, 2007). As activators of ROPs, RopGEFs are inevitably critical throughout plant development. However, except for their biochemical activity to activate ROPs, how they might impact molecular events that underlie growth and developmental processes are largely unknown.
In this work, we have characterized the function of Arabidopsis RopGEF1 during embryo development and seedling growth. Phenotypic analyses show that RopGEF1 is important for auxin-regulated processes including embryo development and root gravitropism. Our data indicate that RopGEF1 is asymmetrically localized in embryonic and root meristem cells. It is required for proper AUX1 localization in central vascular embryonic cells and root protophloem cells and impacts the distribution of two PIN proteins, PIN7 and PIN2. Our results therefore reveal an important role for RopGEF1 in maintaining polar auxin transport via impacting the properties of auxin influx and efflux carriers, thereby affecting auxin-dependent plant growth and development by modulating polar auxin transport.
RESULTS
RopGEF1 Is Expressed Broadly and RopGEF1 Protein Shows Polar Localization in the PM
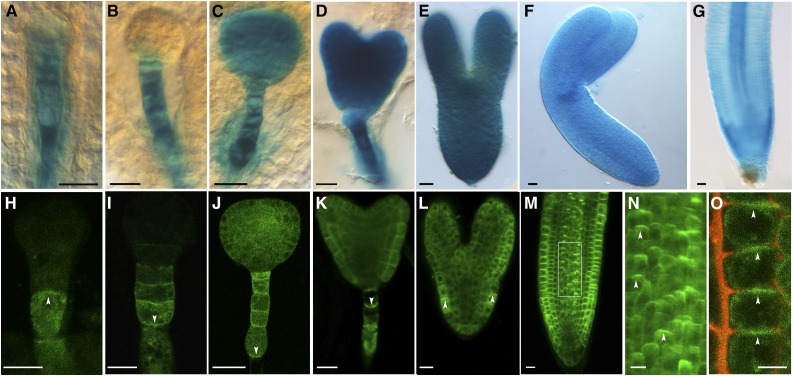
To study RopGEF1 expression and subcellular localization in detail, the GEF1 promoter (RopGEF1pro):GUS and RopGEF1pro:GFP-RopGEF1 cDNA fusions were constructed. Analysis of eight RopGEF1pro:GUS transformed lines showed that GUS activity was detected broadly throughout embryogenesis. From the four-cell stage to the globular stage, GUS activity was predominantly detected in the cells of the suspensor (Fig. 1, A and B). Subsequently, GUS expression was detected in all the cells of the embryo proper and the suspensor at the transition and heart stages (Fig. 1, C and D) and remained strongly expressed in the embryo by the torpedo and bent cotyledon stages (Fig. 1, E and F). In seedlings, RopGEF1pro:GUS was detected in the root meristem, showing strong expression in the epidermis, lateral root caps, and stele and relatively weak expression in the cortex, endodermis, quiescent center, and columellum cells (Fig. 1G). In seedlings, RopGEF1pro:GUS was also expressed in the shoot apical meristem (Supplemental Fig. S1, A and B), the vascular tissue (Supplemental Fig. S1, A and C), cotyledons (Supplemental Fig. S1A), and lateral root primordia (Supplemental Fig. S1D), whereas in open flowers GUS was detected in petals and pistils but not in the anther and pollen (Supplemental Fig. S1, E and F), similar to that previously reported (Li and Liu, 2012).
Figure 1.
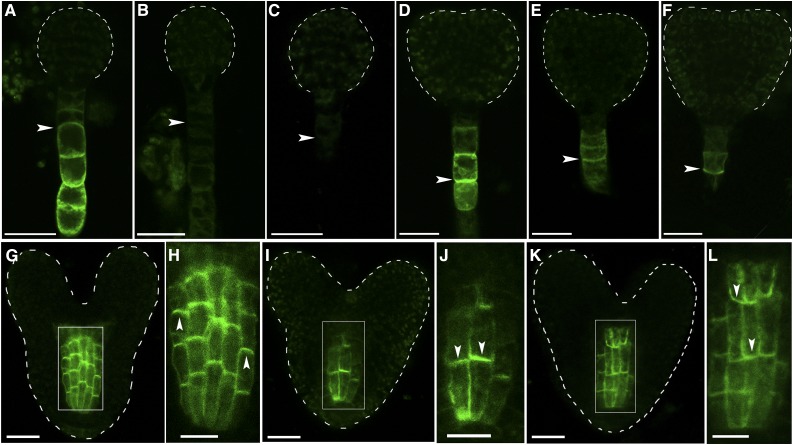
Expression patterns and polar plasma membrane localization of RopGEF1 in Arabidopsis. A to G, RopGEF1pro:GUS expression in embryos at four-cell (A), globular (B), transition (C), heart (D), torpedo (E), and bent cotyledon (F) stages and the root apical meristem (RAM) of a 4-d-old seedling (G). GUS staining of embryos and seedlings was for 16 and 2 h, respectively. H to N, RopGEF1pro:GFP-RopGEF1 expression at four-cell (H), globular (I), transition (J), heart (K), and torpedo (L) embryos and the RAM of a 3-d-old seedling (M). The white dotted boxed region of M is shown magnified in N. O, Root epidermal cells were plasmolyzed by 10% d-Mannitol for 5 min. The arrowheads indicate the localization of GFP-labeled RopGEF1 in root epidermal cells. The propidiumiodide dye outlines the position of the cell walls, GFP-labeled RopGEF1 is associated with apical PM. Bars = 10 μm in A, H, and O; 20 μm in B to G and I to M. Upward and downward white arrowheads indicate the polarity of GFP-RopGEF1.
To study RopGEF1 subcellular localization, we used GFP-RopGEF1 translational fusion expressed under the control of RopGEF1pro. The pattern of RopGEF1pro:GFP-RopGEF1 expression in embryogenesis and seedling roots was identical to that seen for RopGEF1pro:GUS (Fig. 1). GFP-RopGEF1 was localized in both the PM and cytoplasm (Fig. 1, H–M). Interestingly, RopGEF1 PM localization underwent a stage-specific reversal in polarity (Fig. 1, H–K). At the four-cell stage, it was localized in the apical side of the suspensor (Fig. 1H). From the globular stage onward, the asymmetric localization of RopGEF1 reversed and was detected at the basal side of suspensor cells (Fig. 1, I–K). The asymmetric subcellular localization of RopGEF1 in the suspensor cells (Fig. 1, H–K) was very similar to that of the auxin efflux carrier PIN7 (Friml et al., 2003), suggesting a potential correlation between RopGEF1 and directional auxin flow during embryo development. At the torpedo stage, increased GFP-RopGEF1 signal was detected in embryos, and its localization was polarized and again became more strongly associated with the apical PM of the protoderm cells (Fig. 1L). In the root meristem, GFP-RopGEF1 was predominantly detected at the apical PM of the stele and epidermal cells (Fig. 1, M–O). GFP-RopGEF1 was colocalized with PM marker PIP2-mCherry in the PM of root cells (Supplemental Fig. S2, A–C).
Knockdown and Knockout of RopGEF1 Perturb Distinct Auxin-Regulated Developmental Processes
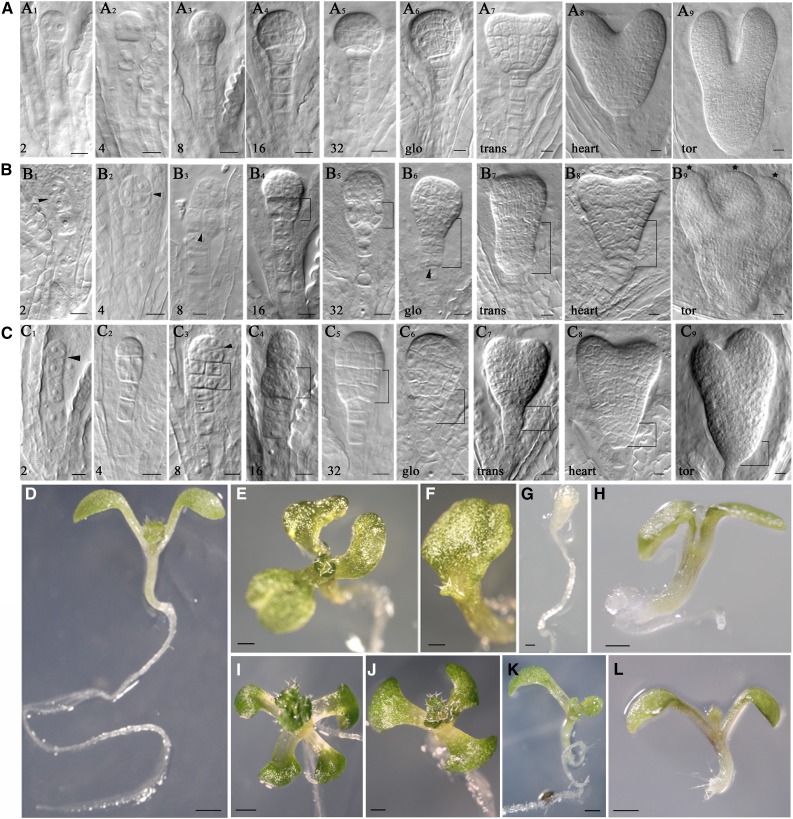
To assess the functional role of RopGEF1, we identified two T-DNA insertion lines and named them as ropgef1-3 and ropgef1-4 (Supplemental Fig. S3, A and B). Quantitative reverse transcription (qRT)-PCR analysis showed that ropgef1-3 is a knockdown mutant with reduced expression of RopGEF1 (Supplemental Fig. S3C), whereas ropgef1-4 with its T-DNA being inserted into the PRONE domain is a null mutant (Supplemental Fig. S3, A and C). As RopGEF1 was expressed during embryogenesis (Fig. 1), we analyzed the embryo phenotypes of ropgef1-3 and ropgef1-4. In comparison with wild-type control (Fig. 2A), the stereotypical pattern of cell division during embryogenesis was affected in ropgef1 mutants (Figs. 2, B and C). The earliest defect was observed in the 2-cell stage where the apical cell underwent an abnormal anticlinal cell division instead of periclinal (Fig. 2, B1 and C1, compared with Fig. 2A1). At the four- to eight-cell stage, abnormal cell divisions in the embryo proper and suspensor had also been observed (Fig. 2, B2, B3, C2, and C3, compared with Fig. 2, A1–A3). From the 16-cell to the heart stage, mutant embryos displayed abnormal shapes such as apical-basally enlarged embryos (Fig. 2, B4, B5, B7, C4 and C5), extra cell divisions in the suspensor (Fig. 2, B6 and C7), and disorganized hypophyseal derivatives (Fig. 2, B6, B8, C6, and C8). At the torpedo stage, occasionally, ropgef1 embryos showed three abnormal leaf primordia or asymmetric size of two embryonic leaves (Fig. 2, B9 and C9). The embryo defects in ropgef1 strongly resembled auxin transport mutants pin1pin3pin4pin7 and aux1lax1lax2, auxin biosynthesis mutant yuc1yuc4yuc10yuc11, and auxin signaling mutants mp and bdl (Friml et al., 2003; Blilou et al., 2005; Cheng et al., 2007; Robert et al., 2015). Similar embryo phenotypes were also found in ROP3 dominant-negative and loss-of-function mutants (Huang et al., 2014). In addition to these more severe cell division defects, ropgef1 embryos also displayed weak phenotypes derived from abnormal cell divisions in the protoderm layer (Supplemental Fig. S4, D–I, compared with Supplemental Fig. S4, A–C). A significantly elevated level of ropgef1 showed relatively strong embryo defects at the globular stage relative to wild-type embryos (see Supplemental Table S2; ropgef1-3 = 19%, n = 101; ropgef1-4 = 27.2%, n = 132; wild type = 1.8%, n = 57). However, when examined at the heart stage, the level of defective embryos dropped in the ropgef1 mutants, albeit remaining significantly higher than that observed in a wild-type population (Supplemental Table S2; ropgef1-3 = 14.1%, n = 92; ropgef1-4 = 13.2%, n = 98; wild type = 0%, n = 51). Overall and through the entire embryogenesis, we found a higher frequency of ropgef1-4 embryos (17.7%, n = 503) with division defects than ropgef1-3 embryos (10.9%, n = 650; Supplemental Table S2), as expected since ropgef1-4 was a null mutant. The declining levels of ropgef1 mutant embryos as development progressed suggest that some of the defective globular embryos had recovered and managed to develop past the heart stage. The recovery of ropgef1-defective embryos at heart stage coincided with RopGEF7 expression (Chen et al., 2011), which probably mitigated the reduction or loss of RopGEF1 function. Moreover, mutations in RopGEF1 did not induce embryo lethality or disruption of seed sets (Supplemental Table S4). Two other RopGEFs (RopGEF5 and RopGEF2) are also highly expressed in embryos (Winter et al., 2007) and likely contribute to embryo development.
Figure 2.
Mutations of RopGEF1 affect embryogenesis. A to C, Embryogenesis from two-cell to the torpedo stage in wild type (A1–A9), ropgef1-3 (B1–B9), and ropgef1-4 (C1–C9). Figures at bottom left indicate the number of embryo cells. Glo, trans, tor = globular, transit, torpedo, respectively. D to L, 7-d-old seedlings from wild type (D), ropgef1-3 (E–H), and ropgef1-4 (I–L). Scale bars = 10 μm in A to C; 1 mm in D to L.
ropgef1 seeds have similar a germination rate to the wild type (Supplemental Figure S5). Some ropgef1 seedlings (Supplemental Table S3) displayed notable cotyledon phenotypes, including four (Fig. 2I), three (Fig. 2, E and J, compared with Fig. 2D), single (Fig. 2, F and K), and no cotyledons (Fig. 2G). Occasionally, ropgef1 seedlings without or with a very short root were observed (Fig. 2, H and L). The total percentage of seedlings with phenotypes was 10.7% (n = 542) for ropgef1-3 and 11.7% (n = 587) for ropgef1-4, whereas only 1.87% (n = 482) of wild-type seedlings developed defects (Supplemental Table S3).
The strong RopGEF1pro:GUS expression in embryonic leaves (Fig. 1, E and F) led to our observing in detail the cotyledon vascular pattern in ropgef1 mutants. We found that the development of vascular strands in the mutant cotyledons showed frequent discontinuity compared to wild-type cotyledons (Supplemental Fig. S6, B and C, compared with Supplemental Fig. S6A). The frequency of seedlings with vascular defects was approximately 51.22% (n = 82) for ropgef1-3 and 55.05% (n = 109) for ropgef1-4, while only 14.8% of wild-type seedlings (n = 108) displayed defects. PIN1pro:PIN1-GFP expression in cotyledons of ropgef1 was not altered relative to the wild type (Supplemental Fig. S6, D–F).
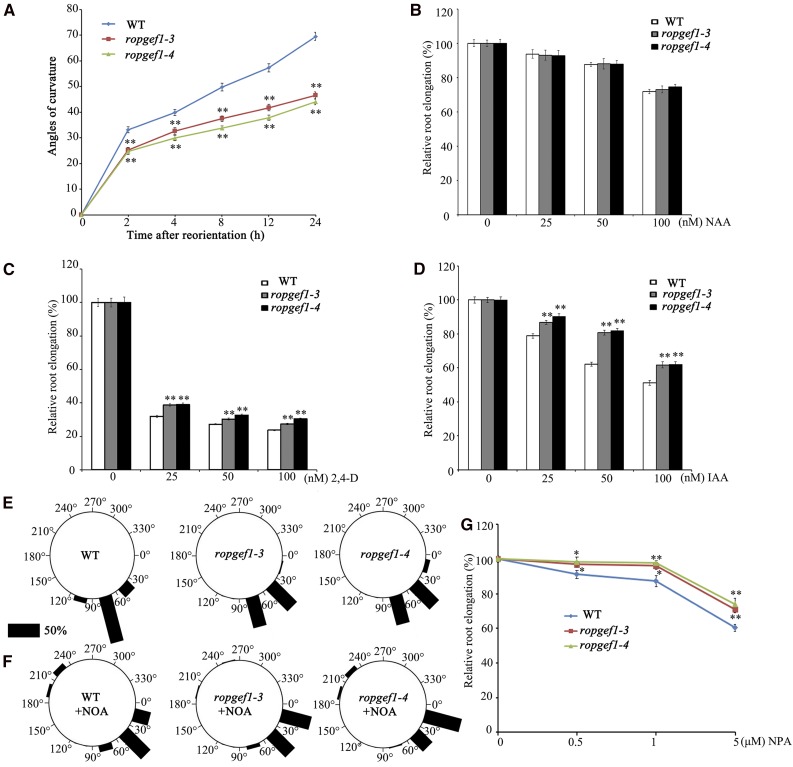
We also analyzed the effects of RopGEF1 on root growth. Under the standard growth condition, the majority of ropgef1-3 and ropgef1-4 seedlings (Supplemental Fig. S7A) had relatively normal root length and lateral root number phenotypes (Supplemental Fig. S7B). However, the root gravitropic response in ropgef1 mutants was noticeably retarded relative to the wild-type response (Fig. 3A). Measurements of root curvature in response to gravity change showed that ropgef1 bent more slowly than the wild type (Fig. 3A). The mutant phenotypes of ropgef1 were complemented when the RopGEF1pro:GFP-RopGEF1 line was crossed into ropgef1 mutants or RopGEF1pro:GFP-RopGEF1 was directly transformed to ropgef1 background (Supplemental Fig. S8, A–E; Supplemental Table S2). These results indicate that RopGEF1 plays an important role in embryo patterning, vascular development, and root gravitropism, properties known to be regulated by auxin.
Figure 3.
The ropgef1 mutants exhibit retarded root gravitropic response and altered sensitivities to auxin transport inhibitors. A, ropgef1 mutants show delayed gravity responses compared with the wild-type (WT) control. Four-day-old WT or ropgef1 seedlings were vertically grown on 1/2 MS medium and reoriented by 90°. After reorientation, root curvatures were measured at indicated time points. Data are means ± sd (n > 30). Asterisks highlight significant differences from the wild type by Student’s t test (*P < 0.05; **P < 0.01). B to D, WT, ropgef1-3, and ropgef1-4 were germinated on hormone-free medium and grown for 4 d then were transferred into medium containing different concentrations of NAA (B), 2,4-d (C), and IAA (D), and grown for another 5 d. Relative root elongation was calculated with the primary root length of untreated WT seedlings set as 100%. Data are means ± sd (n > 30) from one representative experiment that had been repeated at least three times with comparable results. Asterisks highlight significant differences from the wild type by Student’s t test (**P < 0.01). E and F, ropgef1 seedlings were more sensitive to the treatment of the influx carrier inhibitor 1-NOA. Four-day-old WT or ropgef1 seedlings were vertically grown on 1/2 MS medium supplemented without (E) or without 10 μm 1-NOA (F). After 90° for 24 h reorientation, curvatures of the root tip growth were measured. Roots in each of the 12 30°-sectors were counted. Vertical position (90°) represents normal gravitropic response. n > 80 seedlings per experiment. The length of each bar represents the percentage of seedlings showing direction of root growth within that sector. G, WT, ropgef1-3 and ropgef1-4 were germinated and grown on different concentrations of NPA for 9 d. Relative root length was calculated. Data are means ± sd (n > 30). Asterisks highlight significant differences from the wild type by Student’s t test (*P < 0.05; **P < 0.01).
RopGEF1 Mutations Affect Polar Auxin Transport-Dependent Root Growth
The ropgef1 phenotypes in embryos and seedlings (Figs. 2 and 3) strongly suggest that the ropgef1 mutants might have altered auxin sensitivity or auxin transport. To investigate these possibilities, we examined the effect of exogenous auxin including the synthetic auxin 1-naphthaleneacetic acid (NAA), 2,4-dichlorophenoxyacetic acid (2,4-d), and natural auxin inodle-3-acetic acid (IAA) on ropgef1 root growth. As shown in Figure 3, in the plant medium, root elongation in wild type, ropgef1-3, and ropgef1-4 was inhibited in the presence of various concentrations of auxin (Fig. 3, B–D). Compared with wild type, both ropgef1-3 and ropgef1-4 had almost normal sensitivity to NAA, but both mutants exhibited significantly reduced sensitivity to the indicated concentrations of 2,4-d and IAA (Fig. 3, C and D). The differential auxin sensitivity of ropgef1 mutants suggests that RopGEF1 might play a role in the regulation of auxin influx, given that IAA and 2,4-d represent substrates of the auxin influx carrier but not NAA (Delbarre et al., 1996; Swarup et al., 2001).
Moreover, auxin influx inhibitor 1-naphtoxyacetic acid (1-NOA) was applied to investigate whether polar auxin transport is impaired in ropgef1 mutants. Seedlings were grown vertically on the medium with or without 10 μm 1-NOA for 5 d and followed by 24 h gravistimulation. In the absence of NOA, 100% of wild-type seedlings (n = 119) showed more than a bent of 30°, whereas the gravitropic response (i.e. more than a 30° bent) decreased to 73.9% (n = 92) when grown in the presence of the inhibitor (compare Fig. 3, E and F). For ropgef1 mutants, more than 90% of seedlings (97.96%, n = 98 for ropgef1-3; 92.5%, n = 80 for ropgef1-4) showed a gravitropic response in the absence of 1-NOA (Fig. 3E). When grown in the presence of 1-NOA, their gravitropic response decreased significantly relative to that of wild type, with 52.31% of ropgef1-3 (n = 130) and 42.88% of ropgef1-4 (n = 116) seedlings showing a bent of >30° (Fig. 3F). These results indicate that root gravitropism was more severely inhibited in ropgef1 mutants than that in wild type in the presence of the inhibitor (Fig. 3, E and F).Auxin efflux inhibitor naphthylphthalamic acid (NPA) was also used to treat ropgef1 mutants. As shown in Figure 3G, root growth of ropgef1 mutants displayed a clear resistance to NPA compared with the wild type. Taken together, our data suggest that polar auxin transport, both auxin influx and efflux, might be defective in ropgef1 mutants.
Auxin Distribution Is Altered in ropgef1 Embryos and Roots
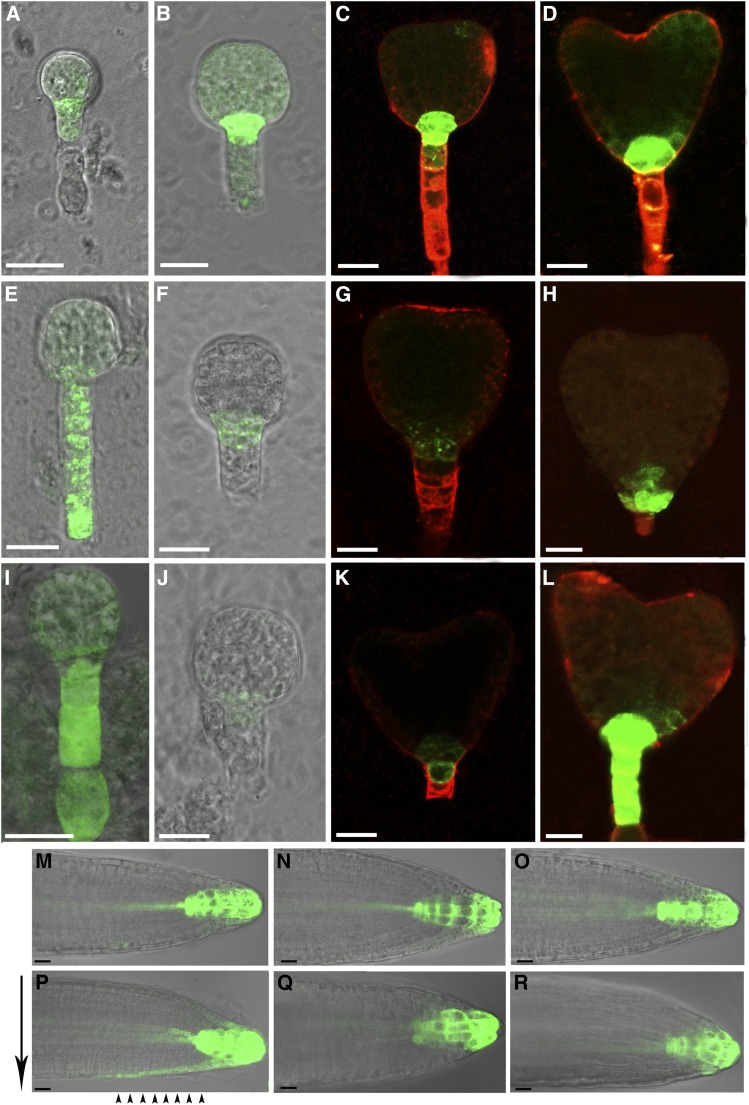
Since ropgef1 mutants displayed auxin-related phenotypes, we examined the auxin reporter DR5rev:GFP in ropgef1 mutants. As shown in Figure 4, strong GFP signals were detected in the root pole of wild type early globular, globular, and heart embryos (Fig. 4, A–D; 96.7%, n = 30) but substantially suppressed in ropgef1-3 (Fig. 4, F and G; 40%, n = 35) and ropgef1-4 (Fig. 4, J and K; 46.7%, n = 30) embryos. Some embryos of ropgef1 displayed abnormal DR5 activity pattern in distal suspensor cells (Fig. 4, E, I, and L; 11.4%, n = 35 for ropgef1-3; 16.7%, n = 30 for ropgef1-4) or altered auxin maximum in the root pole (Fig. 4H). These data are consistent with auxin distribution being perturbed in ropgef1 embryos.
Figure 4.
The expression of auxin reporter gene is altered in ropgef1 mutants. A to L, The expression of DR5rev:GFP in early globular (A and I), globular (B, E, F, and J), and heart embryos (C, D, G, H, K, and L) in wild type (A–D), ropgef1-3 (E–H), and ropgef1-4 (I–L). M to O, DR5rev:GFP activity in the root apex in 4-d-old wild type (M), ropgef1-3 (N), and ropgef1-4 (O). P to R, After gravistimulation for 3.5 h, auxin translocation was visualized by DR5rev:GFP at the lower side of roots in 4-d-old wide type (P), but not in ropgef1-3 (Q) and ropgef1-4 (R). Black arrows indicate the direction of gravity and black arrowheads show the lateral DR5rev:GFP signal on lower side of RAM. Scale bars = 20 μm.
We also examined DR5rev:GFP in ropgef1 roots. In vertically grown roots, the level of DR5rev:GFP expression was slightly lower or similar in the quiescent center and columellum tiers of ropgef1-3 and ropgef1-4 mutants relative to the wild type (Fig. 4, M–O). Upon gravistimulation by 90° rotation, asymmetric activation of DR5rev:GFP in the lower side of lateral root cap and epidermal cells was clearly observed by 3.5 h after stimulation in the wild type root (Fig. 4P). Similar pattern of GFP redistribution was not observed in ropgef1-3 and ropgef1-4, consistent with the lack of gravistimulation induced auxin flow at the lower side of these mutant roots (Fig. 4, Q and R). This observation indicates mutations in RopGEF1 impair auxin distribution, thereby affecting auxin-dependent root gravitropic responses.
PIN7 Accumulation and AUX1 Polarity Were Altered during Embryo Development in ropgef1 Mutants
Mutations in ropgef1 displayed auxin-related embryo phenotypes (Fig. 2) and affected the expression of DR5rev:GFP (Fig. 4), suggesting that PIN-or AUX1/LAX-dependent auxin transport might be perturbed. Moreover, RopGEF1 was strongly expressed in the suspensor throughout embryogenesis (Fig. 1, H–K), which overlapped with the expression pattern of PIN7 (Friml et al., 2003). To determine whether RopGEF1 affects polar localization of PIN proteins, we observed the localization of PIN1pro:PIN1-GFP and PIN7pro:PIN7-GFP in wild type and ropgef1 mutants. PIN1-GFP displayed identical localization in the embryos of wild type and ropgef1 mutants (Supplemental Fig. S9, A–C). However, the level of PIN7-GFP was markedly reduced in the suspensor cells of the globular and early heart embryos of ropgef1 mutants (Fig. 5, B, C, E, and F; 81.3%, n = 48 for ropgef1-3; 90.9%, n = 44 for ropgef1-4) compared with strong GFP signal in wild-type embryos (Fig. 5, A and D; 94%, n = 45). Next, we investigated if RopGEF1 was involved in the regulation of auxin influx carrier AUX1. We compared polar localization of AUX1 in wild type and ropgef1 mutants by using a AUX1pro:AUX1-YFP reporter line. AUX1 displayed apical membrane localization in cells of the central vasculature in heart-stage embryos of wild type (100%, n = 44; Fig. 5, G and H; Supplemental Fig. S11A). Strikingly, an apical to basal shift in AUX1 localization was observed in ropgef1-3 (53.2%, n = 94) and ropgef1-4 (81%, n = 42) embryos (Fig. 5, I–L; Supplemental Fig. S11, B and C). These data suggest that RopGEF1 is involved in the regulation of PIN7 and AUX1 during embryo development, and the diminished PIN7 accumulation together with altered AUX1 polarity could be responsible for the embryo patterning defects in ropgef1 mutants.
Figure 5.
RopGEF1 regulates the expression of PIN7 and the polarity of AUX1 during embryo development. A to C, PIN7pro:PIN7-GFP in the suspensor cells of wild type (A), ropgef1-3 (B), and ropgef1-4 (C) at the stage of globular embryos. D to F, PIN7pro:PIN7-GFP in the suspensor cells of wild type (D), ropgef1-3 (E), and ropgef1-4 (F) at the stage of early heart embryos. G to L, The localization of AUX1pro:AUX1-YFP in the procambium cells of wild type (G and H), ropgef1-3 (I and J), and ropgef1-4 (K and L) embryos. Boxed regions in G, I, and K are shown magnified in H, J, and L, respectively. Arrowheads indicate the localization of PIN7-GFP at the basal PM of suspensor cells (A–F) and the polarity of AUX1 in embryos (H, J, and L). Scale bars = 20 μm.
RopGEF1 Modulates the Localization of Auxin Influx Protein AUX1 and Gravity-Stimulated Distribution of PIN2 Protein in sSeedlings
The auxin influx carrier AUX1 and efflux carrier PIN2 mediate basipetal auxin flux and are required for the root gravity response (Bennett et al., 1996; Luschnig et al., 1998). To investigate if the delayed root gravitropic response in ropgef1 depends on AUX1 and PIN2, we first examined the expression and localization of AUX1pro:AUX1-YFP in ropgef1 primary roots of seedlings. An apical-to-basal reversal also occurred in AUX1 polarity in developing seedlings. AUX1-YFP showed apical membrane localization in root protophloem cells of wild type (100%, n = 38; Fig. 6, A and B; Supplemental Fig. S12A), similar to that reported by Swarup et al. (2001), whereas an apical-to-basal switch in AUX1 polarity appeared in ropgef1 mutants (ropgef1-3: 53.8%, n = 52; ropgef1-4: 63.7%, n = 52; Fig. 6, C–F; Supplemental Fig. S12, B and C). On the other hand, auxin efflux carriers PIN1-GFP and PIN7-GFP localization in the basal PM of stele cells did not differ between wild type (Supplemental Figs. S9, D and E, and S10A) and ropgef1 mutants (Supplemental Figs. S9, F–I, and S10, B and C).
Figure 6.
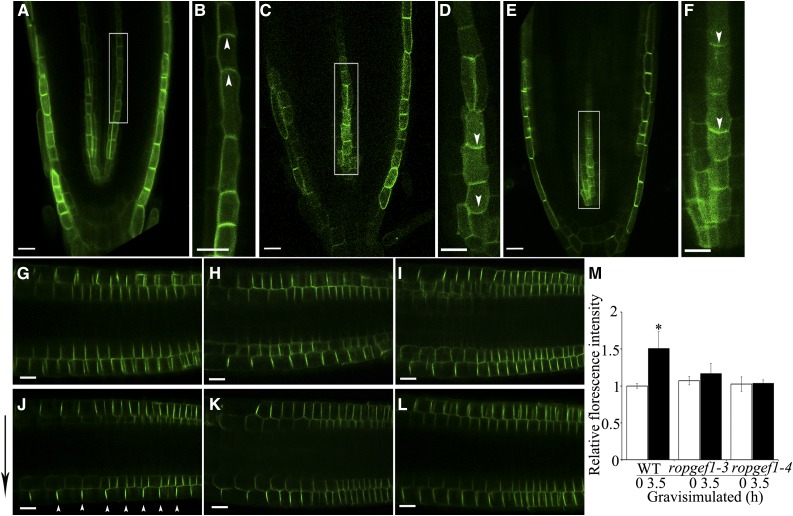
Mutations in RopGEF1 affect AUX1 polarity and PIN2 distribution in the root tips of seedlings. A to F, The localization of AUX1pro:AUX1-YFP in the root protophloem cells of wild type (A and B), ropgef1-3 (C and D), and ropgef1-4 (E and F). Boxed regions in A, C, and E are shown magnified in B, D, and F, respectively. G to I, The localization of PIN2pro:PIN2-GFP in the root epidermis and cortical cells of wild type (G), ropgef1-3 (H), and ropgef1-4 (I). J to L, After gravistimulation for 3.5 h, the localization of PIN2-GFP in 4-d-old seedlings of wild type (WT; J), ropgef1-3 (K), and ropgef1-4 (L). M, Comparions of the ratios of PIN2-GFP signal in the lower epidermal cell file versus in the upper epidermal cell file between the start (0 h) and end (3.5 h) of gravistimulation in WT, ropgef1-3, and ropgef1-4. Data present means ± sd (n > 30). Asterisks highlight significant differences from the wild type by Student’s t test (*P < 0.05). Scale bars = 20 μm. White arrowheads indicate the polarity of AUX1-YFP, and black arrow indicates the direction of gravity, respectively.
We next assessed whether RopGEF1 affected PIN2 property. Confocal microscopy observations showed that the polar localization and accumulation of PIN2-GFP was not affected in ropgef1 roots (Fig. 6, G–I). When wild-type control was positioned at horizontal reorientation for 3.5 h, PIN2-GFP displayed higher expression at the lower side than upper side (Fig. 6, J and M), whereas asymmetric distribution of PIN2-GFP at the lower side of root epidermal cells did not occur in ropgef1 mutants (Fig. 6, K–M). These results indicate that RopGEF1 function as regulators of AUX1 polarity and PIN2 dynamic distribution, affecting polar auxin distribution. The changed AUX1 polarity in the protophloem cells might interfere auxin flux into cells and affect the acropetal auxin transport in the root tip, as suggested earlier by Robert et al. (2015). A previous study showed that the expression of AUX1 in the stele cells was not required for root gravitropic response (Swarup et al., 2005), consistent with the notion that the observed altered root gravitropic response in ropgef1 mutants (Fig. 3A) could be linked to changed abnormal PIN2 distribution.
RopGEF1 Is Required for the Regulation of Actin Network in Roots
It is well-established that ROP GTPases function as the master regulators of actin cytoskeleton (Yalovsky et al., 2008). To test whether RopGEF1 affects the actin cytoskeleton in root development, we crossed the actin marker 35Spro:ABD2-GFP (Wang et al., 2008) into ropgef1 mutants to visualize the actin cytoskeleton configuration in vivo. Long, intensely fluorescent filaments were observed in the epidermal cells (95.9%, n = 49) of elongation and maturation zones in wild-type roots (Fig. 7A), whereas the accumulation of actin filaments was dramatically reduced in ropgef1-3 mutant with 64.5% (n = 121) of cells showing weak ABD2-GFP signal (Fig. 7B). Similarly, cells in ropgef1-4 (62.2%, n = 172) displayed an actin network that was detectable but considerably less dense than in wild-type cells (Fig. 7C). To quantify the effect of RopGEF1 on F-actin network, filament density (occupancy) in the epidermis cells from the root maturation region was measured. As shown in Figure 7G, ropgef1-3 and ropgef1-4 had significantly lower F-actin density values than that in the wild type, indicating that RopGEF1 is important for the maintenance of F-actin cytoskeleton in the root cells.
Figure 7.
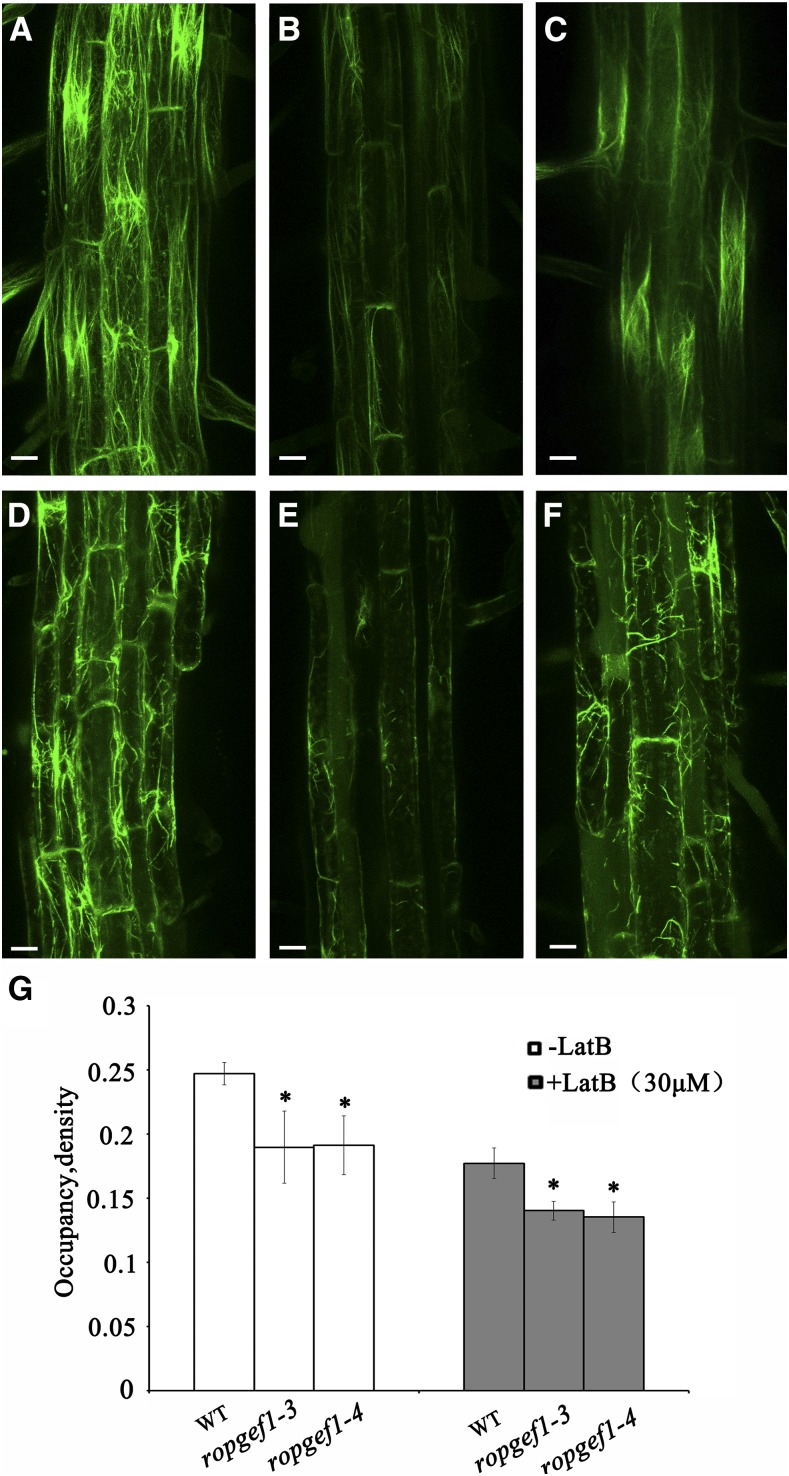
RopGEF1 affects the stability of actin cytoskeleton. A to C, Visualization of F-actin using 35Spro:ABD2-GFP in root epidermal cells of wild type (A), ropgef1-3 (B), and ropgef1-4 (C). D to F, Treatment with 30 μm LatB for 2h in root epidermal cells of wild type (D), ropgef1-3 (E), and ropgef1-4 (F). G, Quantitative analysis of F-actin density (occupancy) in epidermal cells from the root maturation zone. Data are means ± sd (n > 50). Asterisks indicate significant differences from the wild type by Student’s t test (*P < 0.05). Images are projections of 7 to 9 confocal optical sections. Scale bars = 20 μm.
We next assessed the effect of Latrunculin (LatB), a drug that disassembles actin. In wild-type cells, the actin filaments were degraded into short fragments after 2 h of LatB treatment (Fig. 7D). Strikingly, actin filaments in ropgef1-3 and ropgef1-4 root cells became highly fragmented and often appeared as punctuate structures (Fig. 7, E and F), resulting in a highly disrupted actin filamentous network. Therefore, compared with wild type, ropgef1 mutants were more sensitive to LatB treatment. The quantitative analyses on F-actin also showed that LatB-treated ropgef1 mutants had reduced occupancy values compared to the wild type (Fig. 7G). Together, these data suggest that RopGEF1 is required for the integrity of actin cytoskeleton.
DISCUSSION
ROPs function as important regulators of hormone responses and PAT (Wu et al., 2011; Yang and Lavagi, 2012), and auxin is known to rapidly activate these small GTPases (Tao et al., 2002; Xu et al., 2010). As activators of ROPs, RopGEFs are inevitable links in auxin-signaled and ROP-mediated pathways, and they have been implicated in several auxin-regulated developmental processes (Chen et al., 2011; Duan et al., 2010). ROPGEF1 is the most broadly expressed ROP activator. In addition to elucidating a biological role for RopGEF1 in several auxin-transport-dependent processes, results reported here also provide evidence for RopGEF1 playing a critical role in maintaining PAT by acting on the polarity of the auxin influx carrier AUX1 and differential accumulation of PIN7 and PIN2 efflux carriers.
RopGEF1 Is Important for Auxin-Regulated Plant Development
RopGEF1pro:GUS expression in embryos and roots was robust but highly cell- and developmental-stage-specific with a pattern resembling those of genes encoding auxin transport carriers (Friml et al., 2003; Robert et al., 2015). For instance, RopGEF1 expression (Fig. 1) overlapped with those of AUX1 and LAX1 in the embryo proper and was similar to LAX2 and PIN7 in the suspensor (Robert et al., 2015; Friml et al., 2003). Auxin is known to be key for embryogenesis (Lau et al., 2012; Smit and Weijers, 2015). Our results demonstrate that disruption of ropgef1 led to defects (Fig. 2) similar to those observed in various auxin mutants, such as pin1pin3pin4pin7, aux1lax1lax2, mp, bdl, and yuc1yuc4yuc10yuc11 (Friml et al., 2003; Blilou et al., 2005; Cheng et al., 2007; Robert et al., 2015), therefore strongly support an important role for RopGEF1 in embryogenesis. Mutations in RopGEF1 induced embryo phenotypes (Fig. 2), and our observations showed that the embryo phenotypes were associated with altered AUX1 polarity and PIN7 level (Fig. 5; Supplemental Fig. S11). Previous genetic analysis revealed that pin7 knockout mutant exhibited early embryo defects in establishing apical-basal axis during the 1- to 32-cell stages (Fig. 4 in Friml. et al., 2003). The ropgef1 phenotypes at 2 to 32 cells (Fig. 2) are very similar to that of pin7 (Fig. 4D in Friml et al., 2003). AUX1 is expressed at the 32-cell stage (Robert, et al., 2015), although aux1 single mutant has no embryo phenotype, a recent genetic study showed that AUX1 and closely related genes LAX1 and LAX2 played redundant roles in embryo development (Robert, et al., 2015). AUX1/LAX auxin influx transporters and PIN auxin efflux carriers function cooperatively to regulate embryo development, because combining mutations in PIN and AUX1/LAX genes had stronger embryo defects (Robert, et al., 2015). That ropgef1 phenotypes at the globular stage are stronger at the root pole than pin7 and that ropgef1 seedlings also showed cotyledon defects not observed in pin7 (Friml et al., 2003) at late stages of development (Fig. 2) suggests that altered PIN7 level and AUX1 polarity may together result in the observed ropgef1 phenotypes. However, we cannot exclude the possibility that RopGEF1 may also regulate other embryo-expressed auxin influx carriers LAX1 and LAX2.
Auxin is also known to be critical for root gravitropism. The delayed root gravitropic response in ropgef1 mutants (Fig. 3A) could be attributable to perturbed auxin transport and thus altered auxin distribution. This is supported by the observation that PIN2 redistribution during gravistimulation was affected in ropgef1 mutants (Fig. 6). It has been reported that the AUX1 expression domain in protophloem cells may not contribute to root gravitropic response (Swarup et al., 2005), but it is likely that apically localized AUX1 may play a role in acropetal auxin transport toward the root tip. A recent study showed that aux1lax1lax2 triple mutant had reduced auxin maxima in the root pole and defects in the formation of the root pole during embryogenesis (Robert, et al., 2015), suggesting a role of auxin influx carriers in the shoot-pole-to-root-pole auxin transport. Interestingly, both auxin efflux and influx carriers are required for establishing the auxin gradient at the root tip by simulation and experimental analyses (Band et al., 2014). In showing that knockout or severely knocked-down ropgef1 mutants displayed perturbed AUX1 polarity and gravity-stimulated PIN2 redistribution and delayed root gravitropic response (Figs. 6 and 3A; Supplemental Fig. S12), our results are consistent with RopGEF1 mediating interplay between AUX1 and PIN2, which could be an underlying factor important for root gravitropic response. It is possible that polarized AUX1-mediated influx works together with PIN-dependent efflux and provides auxin to the stele for PIN-dependent auxin flow toward the root tip, where PIN2 controls shootward auxin transport to root elongation zone to result in root gravitropic response, as previously speculated (Robert, et al., 2015).
RopGEF1 Impacts the AUX1 Polarity and PIN Distribution and Is Important for Polar Auxin Transport
Uptake of natural auxin IAA and synthetic auxin 2,4-d is known to be dependent on auxin influx carriers, whereas NAA uptake is independent of the auxin influx carriers (Delbarre et al., 1996; Swarup et al., 2001). Interestingly, ropgef1 mutants displayed differential sensitivities to NAA, 2,4-d, and IAA (Fig. 3, B–D), therefore strongly implicating RopGEF1 involvement in the regulation of auxin influx-dependent PAT. 1-NOA had been used to disrupt auxin influx. A previous study showed that 1-NOA acted on both auxin influx and efflux proteins in tobacco BY-2 cells (Lanková et al., 2010). Our observation that the hypersensitivity to the auxin influx inhibitor NOA of ropgef1 seedlings relative to wild type in root agravitropic response (Fig. 3, E and F) suggests that RopGEF1 plays a crucial role in overall auxin transport, including auxin influx and efflux.
The polar localization pattern of GFP-RopGEF1 in root stele cells (Fig. 1N) is identical to the apical PM localization exhibited by AUX1 (Fig. 6, A and B; Kleine-Vehn et al., 2006). This and the observation that knockdown or knockout of RopGEF1 resulted in the reversal of AUX1 polarity (Fig. 6, A–F; Supplemental Fig. S12, B and C) strongly correlated polar localization of GFP-RopGEF1 in the cell with AUX1 polarization. These data further support the idea that RopGEF1 plays an important role in controlling AUX1 polarity, thereby impacting PAT.
RopGEF1 also appears to play a role in the accumulation of PIN7 (Fig. 5, A–F) and gravity-stimulated redistribution of PIN2 (Fig. 6, J–L). Nonetheless, it is unclear by which mechanism RopGEF1 affects PIN proteins. Previous observations showed that ROP signaling was linked to the regulation of PIN localization (Lin et al., 2012; Huang et al., 2014). ROP6 interacts with ROP-interactive CRIB motif-containing protein1 to inhibit PIN2 endocytosis by the stabilization of actin filaments in roots (Lin et al., 2012). ROP3 affects PIN1 and PIN3 abundance in the PM of the root stele cells through regulating their recycling (Huang et al., 2014). A ROP effecter protein, interactor of constitutive active ROP1, regulated PIN recycling and polarity of PIN1 and PIN2 and was required for embryo development and root meristem maintenance (Hazak et al., 2010). Therefore, in controlling ROP activities, RopGEF1 could affect endocytic or exocytic trafficking of PIN proteins, rendering this upstream activator of ROPs an important regulator for polar auxin transport. RopGEF1 had been identified as the activator of ROP1, and its PRONE domain is critical for its function in mediating pollen tube growth when examined in a transient expression system (Gu et al., 2006). To determine whether RopGEF1-mediated ROP activation underlies its activity to impact PAT, e.g. by mutating the conserved residues within the PRONE domain that binds to ROP (Thomas et al., 2007), will need to be explored in the future. It remains to be determined which one among eleven ROPs is part of RopGEF1-mediated PAT pathway.
RopGEFs are important for polarized pollen tube growth, which strongly depends on a dynamic and elaborately organized actin cytoskeleton (Cheung and Wu, 2008). It was reported that AUX1 polarity in root protophloem cells was dependent on the actin cytoskeleton (Kleine-Vehn et al., 2006). Depolymerization of actin by LatB led to the failure of AUX1 localization in the PM or a more random distribution of AUX1 in root protophloem cells (Kleine-Vehn et al., 2006). Another report showed that ROP6 and its effecter ROP-interactive CRIB motif-containing protein1 inhibited the PIN2 endocytosis through actin regulation in root cells (Lin et al., 2012). PIN2 internalization was blocked in rop6 and ric1 roots by treatment with Jasplakinolide, a drug that stabilizes F-actin (Lin et al., 2012). Results reported here (Fig. 7, B, C, and G) indicate that accumulation of actin filaments were reduced in the root cells of ropgef1 mutants, which inevitably would disrupt vesicle trafficking and in turn AUX1 and PIN protein polar targeting to the PM. Our data therefore support that RopGEF1 regulates the polarity of AUX1 and PIN distribution to participate in PAT through actin modulation. In elucidating the role of RopGEF1 in auxin transport, our finding further consolidated the crucial role played by the broader ROP signaling network in plant development.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used in this study. Some of the marker lines used were previously described: AUX1pro:AUX1-YFP (Swarup et al., 2004); PIN1pro:PIN1-GFP (Benkova et al., 2003); PIN7pro:PIN7-GFP (Blilou et al., 2005); DR5rev:GFP (Benkova et al., 2003); 35Spro:ABD2-GFP (Wang et al., 2008).
Seeds were surface sterilized and germinated on half-strength Murashige and Skoog (MS) medium supplemented with 1.5% Suc and solidified by 0.7% agar. After being cold treated at 4°C for 2 d for synchronization, plates were transferred to 22°C for germination with a 16 h light/8 h dark cycle in a growth chamber. Seedlings were transferred to soil about 10 d after germination and grown under the same conditions.
Chimeric Gene Construction and Plant Transformation
RopGEF1pro:GUS was generated by insertion of a 1270-bp genomic fragment containing RopGEF1 promoter sequence and 5′ untranslated region into pBI101.1 at the HindIII and SalI sites. RopGEF1pro:GFP-RopGEF1 contains a GFP coding sequence fused in-frame with a RopGEF1 coding region under the control of the same 1270-bp RopGEF1 promoter region, and it was cloned into Agrobacterium Ti plasmid pAC1352 (Chen et al., 2011).
Arabidopsis was transformed by floral dip (Clough and Bent, 1998), and transgenic plants were selected on kanamycin- or hygromycin-containing (25μg/mL for kanamycin, 15 μg/mL for hygromycin) medium.
The sequences of the primers used in chimeric gene constructions are listed in Supplemental Table S1.
Identification of ropgef1 Mutants and Complementation of ropgef1
The T-DNA insertion mutants of RopGEF1, ropgef1-3 (GABI_GK524D01), and ropgef1-4 (GABI_GK586B11) were obtained from GABI-Kat (https://www.gabi-kat.de/). T-DNA insertions were confirmed by PCR analysis. qRT-PCR assays were used to determine RopGEF1 expression in homozygous mutant lines. ropgef1-3 and ropgef1-4 were crossed with RopGEFpro:GFP-RopGEF1 plants for complementation studies. In the F2 population, ropgef1 homozygotes were screened among F2 progeny by PCR, then hybrid plants harboring RopGEF1pro:GFP-RopGEF1 were identified by GFP fluorescence, followed by PCR to confirm presence of the complementing transgene. Seeds (F3 generation) collected from individual plants were germinated on medium containing both antibiotics sulfadiazine (ropgef1 mutants carry a sulfadiazine-resistant gene) and kanamycin (RopGEF1pro:GFP-GEF1 plants carry a kanamycin-resistant gene) to reconfirm homozygosity of both the ropgef1 alleles and the complementing transgene. F3 seeds producing progeny plants resistant to both antibiotics without any further segregation are homozygotes; they (and subsequent generations) were used for further analysis. In parallel experiments, the RopGEF1pro:GFP-RopGEF1 construct was also transformed into ropgef1 mutants for complementation studies.
The sequences of the gene-specific primers used are listed in Supplemental Table S1.
Root-Growth Assay for Auxin Sensitivity
To test the effect of various auxin on root growth, seedlings of wild type and ropgef1 mutants were germinated and grown on 1/2 MS medium containing 1.5% Suc and 0.8% agar for 4 d, then transferred to medium supplemented with different concentrations (25, 50, and 100 nm) of NAA, 2,4-d, or IAA for another 5 d. Then the primary root length was measured. Approximately 30 seedlings were used for measurement, and data presented are the averages of 30 seedlings with sd. The statistical significance was evaluated by Student’s t test analysis. Comparable results were observed in at least three independent experiments.
Phenotypic Analysis of ropgef1 Mutants and Gravity Response Measurements
Samples including ovules and seedlings were cleared as previously described (Chen et al., 2011) before viewing under Nomarski optics for analysis of embryonic phenotypes. Root gravitropic response experiments were performed using 4 d after germination seedlings grown vertically and under 16/8 h light/dark condition. The angles of root curvatures were measured at 2, 4, 8, 12, and 24 h after 90° reorientation of the seedling plates, with the aid of the image-analysis program Image J.
NOA, NPA, and LatB Treatments
Seeds were germinated on medium containing 10 μm 1-NOA (Sigma-Aldrich), 4-d-old vertically grown seedlings were reorientated by 90° for 24 h, and curvature of root tip growth was measured. NPA treatment experiments were performed on 9-d-old seedlings on vertically positioned plates containing different concentrations of NPA (Sigma-Aldrich), and primary root lengths were measured. For LatrunculinB (LatB) treatment, 3-d-old seedlings were incubated in 1/2 MS liquid medium containing 30 μm LatB (Sigma-Aldrich) for 2 h; actin cytoskeleton in root cells was observed under the confocal microscope (Zeiss LSM710 META).
GUS Histochemical Assay
GUS staining of ovules and seedlings followed the standard procedure (Chen et al., 2011) in 0.25 mg/mL 5-bromo-4-chloro-3-indolyl-glucronide at 37°C for 2 to 16 h.
Marker Gene Analysis
Reporter lines PIN1pro:PIN1-GFP, PIN7pro:PIN7-GFP, AUX1pro:AUX1-YFP, 35Spro:ABD2-GFP, and DR5rev:GFP were individually crossed to ropgef1-3 and ropgef1-4. Homozygous plants were obtained in the F2 population, and F3 seeds (and later generations) were used for further study with progeny seedlings from parental plants serving as controls.
Microscopy
DIC microscopy was carried out on an Olympus BX51 microscope connected to a Ritiga 2000R digital camera. Seedling phenotypes were photographed by using a Nikon SMZ1000 dissection microscope equipped with Nikon Digital sight DS-Fi1 camera. Confocal images were taken using a Zeiss LSM710 META laser scanning microscope.
Quantification of Seedling F-actin Density
To quantify percent occupancy (density) of F-actin, we used images of epidermal cells in root maturation zone from at least nine serial optical sections (1-μm-step) following the methods of Higaki et al. (2010) and Dyachok et al. (2014). The original confocal images were converted into binary images and skeletonized images for the measurement of F-actin pixels. Filament occupancy was calculated with the total pixel numbers of all marked filaments. Density was defined as the ratio of the filament occupancy and cell area as described by Dyachok et al. (2014). The above measurements were performed by using the software Image J (version 1.4.3.67). At least 50 cells from 10 to 15 seedlings were used for measurements. The statistical significance was evaluated by Student’s t test analysis. Signal asterisks indicate significant differences from control at the level of P < 0.05.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Information Resource or GenBank/EMBL databases under the following accession numbers: RopGEF1 (At4g38430), ACTIN2 (At3g18780), PIN1 (At1g73590), PIN2 (At5g57090), PIN7 (At1g23080), AUX1 (At2g38120).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression pattern of RopGEF1 during postembryonic development.
Supplemental Figure S2. Colocalization of GFP-RopGEF1 and PM marker PIP2-mCherry in root epidermis cells.
Supplemental Figure S3. Identification of the ropgef1 T-DNA insertion mutants.
Supplemental Figure S4. Cell division defects in the protoderm layer of ropgef1 embryos.
Supplemental Figure S5. Germination of ropgef1 seeds.
Supplemental Figure S6. Cotyledon venation defects in ropgef1 mutants.
Supplemental Figure S7. Comparison of the root length and lateral root density of ropgef1 mutants and wild type.
Supplemental Figure S8. RopGEF1pro:GFP-RopGEF1 can rescue ropgef1 phenotypes.
Supplemental Figure S9. Localization of PIN1pro:PIN1-GFP is not altered in ropgef1 mutants.
Supplemental Figure S10. Expression of PIN7pro:PIN7-GFP is not altered in the seedlings of ropgef1 mutants.
Supplemental Figure S11. Mutations in RopGEF1 affect the polarity of AUX1-YFP in Arabidopsis embryos.
Supplemental Figure S12. Mutations in RopGEF1 affect the polarity of AUX1-YFP in Arabidopsis roots.
Supplemental Table S1. Primers used in this study.
Supplemental Table S2. Quantitative analysis of ropgef1 embryonic phenotypes.
Supplemental Table S3. Quantitative analysis of ropgef1 seedling phenotypes.
Supplemental Table S4. Mutations in RopGEF1 do not affect seed sets.
Acknowledgments
We thank Drs. Malcolm Bennett and Chuanyou Li for providing seeds for this study, respectively. We thank Guitao Zhong from Dr. Hao Wang’s laboratory for technical assistance on particle bombardment experiment. We thank W.N. Zhang (Center for Agrobiological Gene Research, Guangdong Academy of Agricultural Sciences, Guangdong, Guangzhou) for technical assistance with confocal microscopy.
Glossary
- PAT
polar auxin transport
- PM
plasma membrane
Footnotes
This work was supported by grants from the National Natural Science Foundation of China (91117007, 31570274, and 31370312) to L.-z.T. and the U.S. National Science Foundation (IOS no. 1146941) to H.-M.W.
Articles can be viewed without a subscription.
References
- Adamowski M, Friml J (2015) PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 27: 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Wells DM, Fozard JA, Ghetiu T, French AP, Pound MP, Wilson MH, Yu L, Li W, Hijazi HI, et al. (2014) Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 26: 862–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Berken A, Thomas C, Wittinghofer A (2005) A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436: 1176–1180 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Carrier DJ, Bakar NT, Swarup R, Callaghan R, Napier RM, Bennett MJ, Kerr ID (2008) The binding of auxin to the Arabidopsis auxin influx transporter AUX1. Plant Physiol 148: 529–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Liu H, Kong J, Yang Y, Zhang N, Li R, Yue J, Huang J, Li C, Cheung AY, et al. (2011) RopGEF7 regulates PLETHORA-dependent maintenance of the root stem cell niche in Arabidopsis. Plant Cell 23: 2880–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wu HM (2008) Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu Rev Plant Biol 59: 547–572 [DOI] [PubMed] [Google Scholar]
- Cheung AY, Duan QH, Costa SS, de Graaf BH, Di Stilio VS, Feijo J, Wu HM (2008) The dynamic pollen tube cytoskeleton: Live cell studies using actin-binding and microtubule-binding reporter proteins. Mol Plant 1: 686–702 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delbarre A, Müller P, Imhoff V, Guern J (1996) Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198: 532–541 [DOI] [PubMed] [Google Scholar]
- Dharmasiri S, Swarup R, Mockaitis K, Dharmasiri N, Singh SK, Kowalchyk M, Marchant A, Mills S, Sandberg G, Bennett MJ, et al. (2006) AXR4 is required for localization of the auxin influx facilitator AUX1. Science 312: 1218–1220 [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM (2010) FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyachok J, Sparks JA, Liao F, Wang YS, Blancaflor EB (2014) Fluorescent protein-based reporters of the actin cytoskeleton in living plant cells: Fluorophore variant, actin binding domain, and promoter considerations. Cytoskeleton 71: 311–327 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, Hooykaas PJ, Palme K, Offringa R (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Grebe M, Friml J, Swarup R, Ljung K, Sandberg G, Terlou M, Palme K, Bennett MJ, Scheres B (2002). Cell polarity signaling in Arabidopsis involves a BFA-sensitive auxin influx pathway. Curr Biol 12: 329–334. [DOI] [PubMed] [Google Scholar]
- Gu Y, Li S, Lord EM, Yang Z (2006) Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell 18: 366–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazak O, Bloch D, Poraty L, Sternberg H, Zhang J, Friml J, Yalovsky S (2010) A rho scaffold integrates the secretory system with feedback mechanisms in regulation of auxin distribution. PLoS Biol 8: e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T, Kutsuna N, Sano T, Kondo N, Hasezawa S (2010) Quantification and cluster analysis of actin cytoskeletal structures in plant cells: Role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J 61: 156–165 [DOI] [PubMed] [Google Scholar]
- Huang JB, Liu H, Chen M, Li X, Wang M, Yang Y, Wang C, Huang J, Liu G, Liu Y, et al. (2014) ROP3 GTPase contributes to polar auxin transport and auxin responses and is important for embryogenesis and seedling growth in Arabidopsis. Plant Cell 26: 3501–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Santambrogio M, Rozier F, Fobis-Loisy I, Miège C, Gaude T (2007) The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell 130: 1057–1070 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Dhonukshe P, Swarup R, Bennett M, Friml J (2006) Subcellular trafficking of the Arabidopsis auxin influx carrier AUX1 uses a novel pathway distinct from PIN1. Plant Cell 18: 3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanková M, Smith RS, Pesek B, Kubes M, Zazímalová E, Petrásek J, Hoyerová K (2010) Auxin influx inhibitors 1-NOA, 2-NOA, and CHPAA interfere with membrane dynamics in tobacco cells. J Exp Bot 61: 3589–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S, Slane D, Herud O, Kong J, Jürgens G (2012) Early embryogenesis in flowering plants: Setting up the basic body pattern. Annu Rev Plant Biol 63: 483–506 [DOI] [PubMed] [Google Scholar]
- Li Z, Liu D (2012) ROPGEF1 and ROPGEF4 are functional regulators of ROP11 GTPase in ABA-mediated stomatal closure in Arabidopsis. FEBS Lett 586: 1253–1258 [DOI] [PubMed] [Google Scholar]
- Li Z, Waadt R, Schroeder JI (2016) Release of GTP exchange factor mediated down-regulation of abscisic acid signal transduction through ABA-induced rapid degradation of RopGEFs. PLoS Biol 14: e1002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Nagawa S, Chen J, Cao L, Chen X, Xu T, Li H, Dhonukshe P, Yamamuro C, Friml J, et al. (2012). A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr Biol 22: 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12: 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18: 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Péret B, Swarup K, Ferguson A, Seth M, Yang Y, Dhondt S, James N, Casimiro I, Perry P, Syed A, et al. (2012) AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Friml J (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Robert HS, Grunewald W, Sauer M, Cannoot B, Soriano M, Swarup R, Weijers D, Bennett M, Boutilier K, Friml J (2015) Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 142: 702–711 [DOI] [PubMed] [Google Scholar]
- Shin DH, Cho MH, Kim TL, Yoo J, Kim JI, Han YJ, Song PS, Jeon JS, Bhoo SH, Hahn TR (2010) A small GTPase activator protein interacts with cytoplasmic phytochromes in regulating root development. J Biol Chem 285: 32151–32159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit ME, Weijers D (2015) The role of auxin signaling in early embryo pattern formation. Curr Opin Plant Biol 28: 99–105 [DOI] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Gälweiler L, Palme K, Jürgens G (1999) Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286: 316–318 [DOI] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, Palme K, Bennett M (2001) Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev 15: 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kargul J, Marchant A, Zadik D, Rahman A, Mills R, Yemm A, May S, Williams L, Millner P, et al. (2004) Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16: 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HM, Haseloff J, Beemster GT, Bhalerao R, Bennett MJ (2005) Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol 7: 1057–1065 [DOI] [PubMed] [Google Scholar]
- Swarup R, Péret B (2012) AUX/LAX family of auxin influx carriers-an overview. Front Plant Sci 3: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao LZ, Cheung AY, Wu HM (2002) Plant Rac-like GTPases are activated by auxin and mediate auxin-responsive gene expression. Plant Cell 14: 2745–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Fricke I, Scrima A, Berken A, Wittinghofer A (2007) Structural evidence for a common intermediate in small G protein-GEF reactions. Mol Cell 25: 141–149 [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J (2009) Auxin: A trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Wang YS, Yoo CM, Blancaflor EB (2008) Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol 177: 525–536 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertová D, Brewer PB, Ruzicka K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Wu HM, Hazak O, Cheung AY, Yalovsky S (2011) RAC/ROP GTPases and auxin signaling. Plant Cell 23: 1208–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z (2010) Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Bloch D, Sorek N, Kost B (2008) Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol 147: 1527–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E (2006) High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16: 1123–1127 [DOI] [PubMed] [Google Scholar]
- Yang Z, Lavagi I (2012) Spatial control of plasma membrane domains: ROP GTPase-based symmetry breaking. Curr Opin Plant Biol 15: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Qian L, Nibau C, Duan Q, Kita D, Levasseur K, Li X, Lu C, Li H, Hou C, et al. (2012) FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc Natl Acad Sci USA 109: 14693–14698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, McCormick S (2007) A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 18830–18835 [DOI] [PMC free article] [PubMed] [Google Scholar]