MYB26 regulates anther secondary thickening via NST1 and NST2 specifically in the endothecium through a series of regulatory controls.
Abstract
Successful fertilization relies on the production and effective release of viable pollen. Failure of anther opening (dehiscence), results in male sterility, although the pollen may be fully functional. MYB26 regulates the formation of secondary thickening in the anther endothecium, which is critical for anther dehiscence and fertility. Here, we show that although the MYB26 transcript shows expression in multiple floral organs, the MYB26 protein is localized specifically to the anther endothecium nuclei and that it directly regulates two NAC domain genes, NST1 and NST2, which are critical for the induction of secondary thickening biosynthesis genes. However, there is a complex relationship of regulation between these genes and MYB26. Using DEX-inducible MYB26 lines and overexpression in the various mutant backgrounds, we have shown that MYB26 up-regulates both NST1 and NST2 expression. Surprisingly normal thickening and fertility rescue does not occur in the absence of MYB26, even with constitutively induced NST1 and NST2, suggesting an additional essential role for MYB26 in this regulation. Combined overexpression of NST1 and NST2 in myb26 facilitates limited ectopic thickening in the anther epidermis, but not in the endothecium, and thus fails to rescue dehiscence. Therefore, by a series of regulatory controls through MYB26, NST1, NST2, secondary thickening is formed specifically within the endothecium; this specificity is essential for anther opening.
Fertilization is important for seed production; a number of factors are required for successful fertilization, such as the production of viable pollen and then its efficient release at the optimal time to allow for pollination. Failure of pollen release results in male sterility even if the pollen itself is fully functional. Pollen is formed within anthers, a specialized structure that is supported on a filament, which provides vascular connections to the developing anther. The filament also enables the anther to extend and protrude away from the petals to facilitate effective pollen dispersal. The anther is comprised of four cell layers, which encase the microspores as they develop into mature pollen grains: the tapetum, middle cell layer, endothecium, and the outer epidermal layer. Defects in these cell layers, particularly the tapetum, frequently result in a failure of pollen development, with the degeneration of the pollen, empty anther locules, and male sterility (Scott et al., 2004; Ma, 2005; Ariizumi and Toriyama, 2011). The endothecium, however, plays a principal role in anther dehiscence by providing the force required for opening due to localized secondary thickening and anther dehydration.
After microspore release, the endothecium layer starts to undergo selective expansion followed by secondary thickening, and specific epidermal cells differentiate to form the stomium region. This region subsequently defines the position of anther opening and does not develop the secondary thickening seen in the endothecium and connective tissues. Dehiscence is a two-phase process involving initial enzymatic degradation of the septum separating the two locules, followed by retraction of the locules resulting in a split at the stomium (Wilson et al., 2011). By a combination of molecular genetic analysis and mathematical modeling, we have shown that the mechanical control of opening is mediated by the bilayer structure of the mature anther wall (Nelson et al., 2012). This is comprised of an outer epidermal cell layer, whose turgor pressure is related to its hydration, and the endothecial layer, whose walls contain helical secondary thickening that resist stretching and bending. This model predicts that epidermal dehydration, in association with the thickened endothecial layer, creates forces in the anther wall, causing it to bend outwards, which results in splitting of the stomium, locule retraction, anther opening, and pollen release (Nelson et al., 2012). The requirement for endothecium thickening for dehiscence has been demonstrated genetically by mutants of MYB26/MALE STERILE35 (Dawson et al., 1993; Steiner-Lange et al., 2003) and NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1) and NST2 (Mitsuda et al., 2005). Both the myb26 and nst1nst2 mutants produce viable pollen but lack endothecium secondary thickening and are male sterile because the pollen is not released.
We previously showed that the MYB26 gene is able to induce ectopic secondary thickening when expressed under the control of the CaMV35S promoter (Yang et al., 2007). Similar phenotypes to those seen with MYB26 are also generated by overexpression of the NST1 and NST2 genes (Mitsuda et al., 2005). Cecchetti et al. (2013) demonstrated that the timing of anther dehiscence was negatively regulated by auxin inhibiting MYB26 expression and thus endothecium lignification, but also stomium opening via the control of jasmonic acid (JA) biosynthesis. It has recently been shown that an auxin maxima is formed due to transport of auxin from the tapetum into the middle cell layer, and this is important for the regulation of pollen development and dehiscence (Cecchetti et al., 2017).
NST1 and NST2 belong to the large NAC-domain family, which is made up of plant-specific transcription factors associated with a variety of developmental processes (Olsen et al., 2005). A subgroup of these has been identified as master regulators of secondary thickening. These appear to function redundantly in groups exhibiting differential expression throughout the plant. NST3/SECONDARY WALL ASSOCIATED NAC DOMAIN PROTEIN1 (SND1; At1g32770) are specifically expressed in fibers (Zhong et al., 2006; Mitsuda et al., 2007; Mitsuda and Ohme-Takagi, 2008), while VASCULAR RELATED NAC-DOMAIN1–7 (VND1–VND7) are expressed in vessels with VND6 and VND7 important for the formation of proto and metaxylem, and VND1–5 for parenchyma cells (Kubo et al., 2005; Yamaguchi et al., 2008; Zhong et al., 2008; Zhou et al., 2014). NST1 and NST2 act redundantly to facilitate secondary thickening in the anther (Mitsuda et al., 2005). Previous work has shown that NST2 is expressed predominantly in the anther with some expression in the interfascicular fibers and xylary fibers (Zhong and Ye, 2015), whereas NST1 is expressed in the anther and other tissues where secondary thickening is observed, where it acts alongside NST3/SND1 to regulate secondary wall biosynthesis in these tissues (Mitsuda et al., 2005). The double mutant nst1snd1 only has limited thickening within these tissues suggesting that NST2 plays a minor role in the regulation of secondary wall biosynthesis in fibers (Zhong and Ye, 2015).
The anther endothecium thickening forms as striated bands that resemble tracheary elements and are formed of lignocellulose, as indicated by phloroglucinol and ethidium acridine-orange staining (Dawson et al., 1999; Yang et al., 2007). The composition of the thickening appears to be critical for dehiscence, since the triple ccc mutant, which is defective for cinnamoyl CoA reductase1, cinnamyl alcohol dehydrogenase c and cinnamyl alcohol dehydrogenase d, has hypolignified stems and accumulates higher amounts of flavonol glycosides, sinapoyl malate. and feruloyl malate, has abnormal endothecium thickening, and is male sterile (Thévenin et al., 2011).
Previous studies have demonstrated the roles that the NAC domain genes play in regulating secondary thickening biosynthesis genes; however, little is known regarding the relationship between the NAC domain genes and the upstream transcription factors that regulate the tissue specificity of secondary thickening formation. Here, we have conducted a molecular genetic analysis of the interactions between the MYB26 and NST1/NST2 genes, which has shown that MYB26 is an initial switch required for subsequent secondary thickening formation in the anther, acting directly via regulating NST1 and NST2. Using a functional inducible translational fusion, we have shown that the MYB26 protein is nuclear localized specifically within the anther endothecium, despite the transcript being detected in multiple cell layers in the anther. We have also shown that expression of NST1/NST2 cannot rescue dehiscence and fertility in the myb26 mutant, thus demonstrating that the downstream NST1/NST2 factors are insufficient for secondary thickening and that expression of MYB26 and presumably the equivalent regulator in the vegetative tissues is essential for correctly localized secondary thickening formation. This series of controls ensures the specificity of location of secondary thickening, which is essential for anther dehiscence.
RESULTS
Dexamethasone-Inducible Expression of MYB26 Rescues Fertility in the myb26 Mutant
A translational MYB26 fusion protein was constructed (MYB26pro:MYB26-GR-YFP) under the control of the native MYB26 promoter, with the MYB26 genomic sequence fused to the glucocorticoid receptor (GR) ligand-binding domain and YFP reporter gene; thus, the translated protein was localized in the cytoplasm and inactive until treated with dexamethasone (DEX), allowing it to become nuclear localized. The construct was transformed into heterozygous myb26MYB26 mutant plants, and the T1 generation was screened by Basta and PCR for the transgene. Confirmed transgenic plants were genotyped to identify myb26 homozygous plants; these mutants carrying the MYB26-GR-YFP transgene were male sterile as expected (Fig. 1, A, C, and F), due to a failure of dehiscence because of a lack of secondary thickening in the endothecium, as seen in the myb26 mutant (Fig. 1I). However, a single spray of 25 µm DEX solution on the young flower buds was able to produce flowers with normal dehiscent anthers and rescued fertility (Fig. 1, B, D, G, and J).
Figure 1.
Rescue of fertility by DEX induction of MYB26. A, myb26 mutant carrying the MYB26pro:MYB26-GR-YFP transgene line before DEX treatment showing short, sterile siliques due to a lack of self-fertilization as a result of a failure of anther dehiscence. B, myb26 mutant carrying the MYB26pro:MYB26-GR-YFP transgene line after DEX treatment, showing rescued fertility and elongated, filled siliques on the upper region of the inflorescences (arrows); below the rescued fertile siliques were male sterile short, seedless siliques which developed before the DEX treatment. C, Close-up of the inflorescence from the transgene line before DEX treatment, showing short siliques (arrow), which do not contain seeds. D, Close-up of the inflorescence from the transgene line showing rescue of fertility and elongated, filled siliques (arrows) after DEX treatment. E, Wild-type flower showing anther dehiscence and pollen release. F, Flower from the myb26 mutant carrying the MYB26pro:MYB26-GR-YFP transgene before DEX treatment, showing a lack of anther dehiscence and pollen release. G, Flower from the myb26 mutant line carrying the MYB26pro:MYB26-GR-YFP transgene line after DEX treatment, showing rescue of anther dehiscence. H to J, Confocal images of anthers after ethidium bromide/acridine orange staining for secondary thickening. H, Wild-type anther showing lignified endothecium layer (arrow). I, myb26 mutant carrying the MYB26pro::MYB26-YFP-GR transgene before DEX treatment, which lacks endothecium secondary thickening (arrow). J, myb26 MYB26pro:MYB26-GR-YFP transgene line showing restoration of endothecium thickening after DEX treatment (arrow). Scale bars: 100 µm.
The response of transgenic myb26 mutant plants to the DEX was dependent on the stage of anther development during the treatment. Old unopened flower buds containing postpollen mitosis I stage anthers were not affected, and these formed short siliques without seeds. However in younger buds, prior to pollen mitosis I stage, that developed in the 4 to 7 d after the DEX treatment, full fertility was restored with anther dehiscence occurring normally and silique elongation as seen in wild type (Fig. 1D). Pollen development stage was confirmed using DAPI staining of the pollen, and this corresponded to when endothecium expansion and deposition of secondary thickening normally occurred (Sanders et al., 1999). The effect of a single DEX treatment lasted approximately 7 d; after this point, the plants reverted to sterility, unless the DEX treatment was repeated. The flowers from lines carrying the transgene appeared normal, with no ectopic thickening or abnormalities in the DEX-treated inducible lines regardless of whether the transgene was in the mutant or wild-type background (Fig. 1, E–G). Fertility was not affected in the wild-type transgenic lines by DEX treatment. The lignification of the endothecium in the complemented myb26 mutant buds was variable, with some anthers forming a fully developed endothecial layer while others showed only a partially lignified endothecium layer (Fig. 1J). This did not appear to correspond to bud age and was possibly due to the uneven distribution of the DEX and nuclear-localized MYB26 within the anthers.
MYB26 Protein Is Specifically Localized to the Anther Endothecium
The myb26 mutant lines carrying the functional MYB26-GR-YFP fusion protein were analyzed for localization of the MYB26-YFP protein. After DEX treatment the MYB26-YFP protein was observed in the nuclei of endothecium cells during the pollen mitosis I stage (Fig. 2, A and B). Prior to this, during pollen mother cell meiosis and microspore release and after pollen mitosis II, no MYB26-YFP expression was seen. No MYB26-YFP expression was seen in other tissues in the flowers or vegetative tissues despite detection of GUS expression using the same length promoter in a transcriptional fusion (MYB26pro:GUS) in the nectaries, style, filaments, and anthers (Fig. 2C).
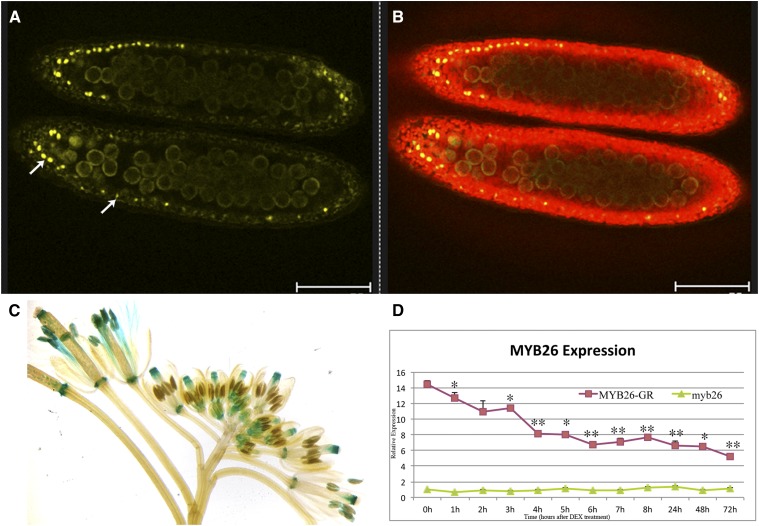
Figure 2.
Localization of MYB26 after DEX-induced expression. A and B, Confocal imaging of expression of the functional MYB26pro:MYB26-YFP fusion protein in anthers; expression is only seen in the nuclei of the anther endothecium cells during pollen mitosis I. A, MYB26-YFP fusion protein localized in endothecium nuclei (arrows; excitation 514 nm). B, Overlay of anther chlorophyll autofluorescence (excitation 488 nm) and MYB26-YFP fusion protein. Scale bar represents 75 µm. C, MYB26Pro:GUS expression is seen in many floral tissues, including nectaries, style, filaments, and anthers. D, Time course of MYB26 expression by qRT-PCR in myb26 mutant buds and myb26 mutant carrying the MYB26pro:MYB26-GR-YFP transgene after DEX treatment. Expression levels of the transgene fluctuated slightly but were reduced 1 h post-DEX treatment and strongly reduced by 4 h post-DEX treatment with all samples being at least P < 0.05 after 3 h compared to 0 h control (t test statistical analysis; *P ≤ 0.05; **P ≤ 0.01).
MYB26 expression was determined by time course quantitative reverse transcription (qRT)-PCR analysis in buds from the inducible line; expression showed an initial fluctuation immediately post-DEX treatment (and thus nuclear localization of the MYB26 protein); however, approximately 3 h post-DEX treatment reduced MYB26 expression was seen, which was subsequently maintained throughout the analysis (72 h; Fig. 2D). This suggests that the functional MYB26 protein may directly or indirectly inhibit its own (MYB26) expression.
MYB26 Can Induce Expression of NST1 and NST2, But Cannot Rescue Secondary Thickening in the nst1nst2 Mutant Background
Previous work suggested that MYB26 may act upstream of the two NAC domain genes NST1 and NST2, with a reduction of the expression of both these genes in the myb26 mutant, and up-regulation in MYB26 overexpression line (Yang et al., 2007); however, the genetic relationship between these genes has not been fully established. The ability of MYB26 to induce NST1 and NST2 expression and regulate secondary thickening in the absence of NST1NST2 expression was therefore investigated. Expression of NST1 and NST2 was analyzed in lines overexpressing MYB26 (regulated by the CaMV35S promoter), and in our DEX-inducible MYB26 line (MYB26pro:MYB26-GR-YFP in the myb26 mutant background). Increased expression of NST1pro:GUS was seen in the lines overexpressing MYB26 (35Spro:MYB26) with intense NST1pro:GUS staining visible, particularly in the peduncle, sepals, and anthers (Fig. 3, B and C). Expression of NST1 and NST2 was analyzed by qRT-PCR over a 72 h period after MYB26 induction by DEX treatment; induction of NST1 and NST2 occurring approximately 4 to 6 h post-DEX treatment (Fig. 3, D and E). These data suggest that NST1 and NST2 are induced by and act downstream of MYB26. To confirm this and to check whether overexpression of MYB26 was able to rescue fertility in the double nst1nst2 mutant, the nst1nst1NST2nst2 heterozygous mutant was transformed with the 35Spro:MYB26 construct (see “Materials and Methods”). Transgenic lines were selected on hygromycin plates and PCR screened for presence of the MYB26 transgene and segregation of the nst2 mutation. T1 and T2 transgenic lines were analyzed for male fertility, anther development, and secondary thickening in anther and vegetative tissues. qRT-PCR was also conducted to establish the levels of MYB26 and NST1/2 gene expression.
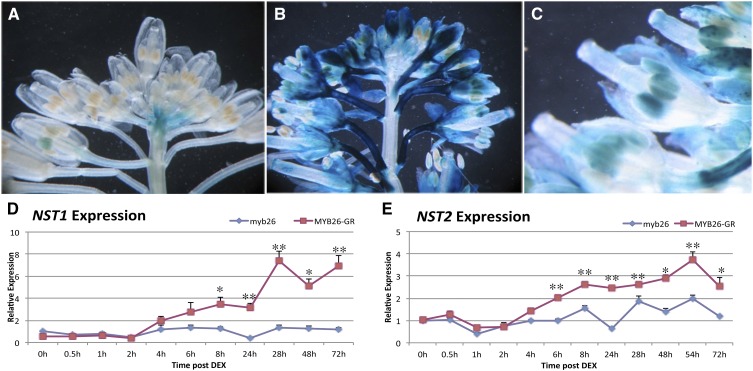
Figure 3.
Induction of NST1 and NST2 expression by MYB26. A, NST1Pro:GUS expression in wild type and (B) increased NST1pro:GUS expression in MYB26 overexpression line, particularly in the peduncle, sepals. and anthers. C, Increased magnification of NST1pro:GUS expression in MYB26 overexpression line showing expression in anthers. D and E, Time course analysis of expression of NST1 and NST2 by qRT-PCR after DEX activation of MYB26 in the transgenic (MYB26pro:MYB26-GR-YFP) myb26 mutant line and in the myb26 mutant control lacking the transgene. D, Induction of NST1 occurred 4 to 6 h after DEX treatment. E, Induction of NST2 was seen 4 to 6 h after DEX treatment. Error bars represent sd (t test statistical analysis compared to 0 h in each line; *P ≤ 0.05; **P ≤ 0.01).
NST1 and NST2 have been previously shown to act redundantly, with male sterility in the double mutant but normal fertility and vegetative growth seen if one functional NST1 or NST2 copy is present (Mitsuda et al., 2005). We also observed that the nst1nst2 double mutant was male sterile as previously reported (Mitsuda et al., 2005), with viable pollen but indehiscent anthers due to a lack of secondary thickening in the anther endothecium (Fig. 4, B and F). Secondary thickening was still present in the inflorescence stem and other tissues in the nst1nst2 mutant, if slightly reduced compared to wild type (Fig. 4, I and J), presumably due to the normal expression of NST3/SND1, which acts redundantly with NST1 in the stem (Mitsuda et al., 2007). The nst1nst2 double mutant tended to be bushier than the wild type (Fig. 4, A and B), probably due to the lack of NST1 and NST2 expression throughout the plant, as well as the reduced levels of fertilization and seed set in the double mutant. MYB26 expression levels varied in buds from the whole inflorescence, between individual nst1nst2 lines. In some instances, MYB26 expression was slightly increased in the nst1nst2 double mutant (Fig. 4M); however the minor changes observed suggests that the absence of NST1 and NST2 did not have a significant regulatory role on MYB26 expression.
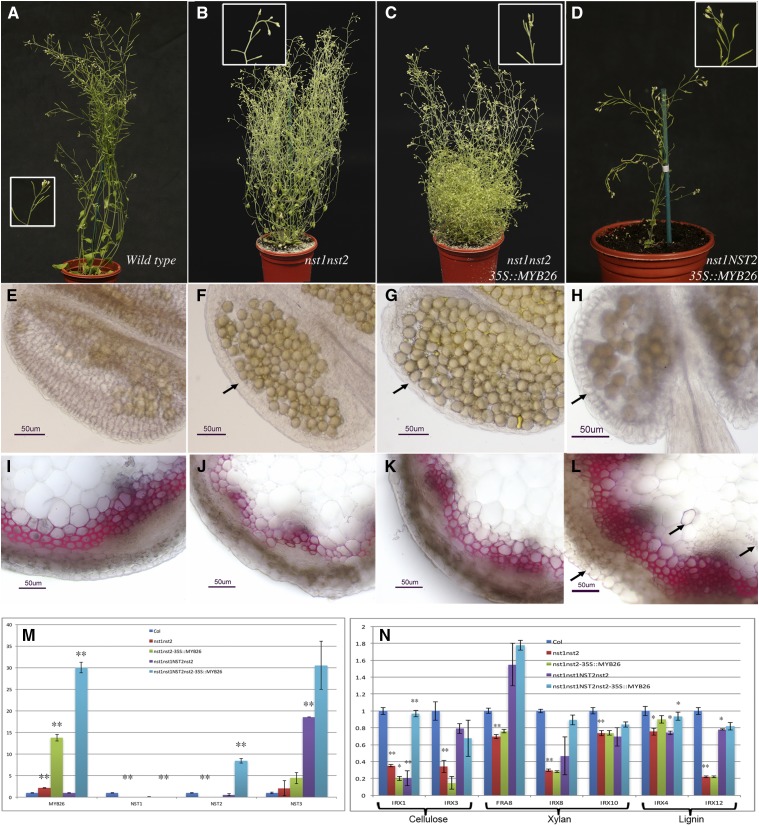
Figure 4.
Ectopic expression of MYB26 is unable to induce secondary thickening in the anther in the absence of NST1 or NST2 expression. A, Wild-type plant showing full fertility as evidenced by silique elongation and full seed set. B, nst1nst2 double mutant showing sterility as indicated by a lack of silique elongation; plants also showed increased bushy growth. C, Expression of 35Spro:MYB26 in nst1nst2 background does not rescue male fertility or bushy growth. D, Expression of 35Spro:MYB26 in nst1nst1NST2nst2 background, which is heterozygous for and thus expressing NST2, is fertile, and growth resembles wild type. NST2 acts redundantly with NST1, and the phenotypes of heterozygous lines are equivalent to wild type, with full fertility and normal growth habit (Mitsuda et al., 2005). Boxed regions show increased magnification of the same lines. E to L, Sections stained with phloroglucinol to detect lignin; scale bar represents 50 µm. E Wild-type anther showing secondary thickening in the endothecium. F The nst1nst2 double mutant fails to develop endothecium secondary thickening (arrow). G, Endothecium secondary thickening is not rescued by expression of 35Spro:MYB26 in the nst1nst2 background (arrow). H, Increased levels of anther endothecium thickening were, however, seen with the 35Spro:MYB26 in the nst1nst1Nst2nst2 heterozygous background (arrow). I to L, Secondary thickening in the inflorescence stems (I) wild type, (J) nst1nst2 double mutant (thickening is slightly reduced), and (K) the nst1nst2 double mutant expressing 35Spro:MYB26. L, Ectopic secondary thickening is seen in the inflorescence stem (arrows) when MYB26 is overexpressed in presence of NST2 (35Spro:MYB26 in the nst1nst1NST2nst2 heterozygous background). M, qRT-PCR expression analysis of MYB26, NST1, NST2, and NST3 in the whole inflorescence of wild type, nst1nst2 mutant, nst1nst2 mutant expressing 35Spro:MYB26, and in the nst1 single mutant (nst1nst1NST2nst2 heterozygous line), and nst1nst1NST2nst2 heterozygous line expressing 35Spro:MYB26. N, qRT-PCR expression of genes involved in secondary thickening pathways in the whole inflorescence of various backgrounds shown in M. Error bars represent sd in M and N (t test statistical analysis compared to its relevant background for each line; *P ≤ 0.05; **P ≤ 0.01).
As expected, ectopic expression of MYB26 under control of the CaMV35S promoter was unable to complement the male sterile phenotype of the nst1nst2 double mutant (Fig. 4C), indicating it acts upstream of NST1NST2. These lines failed to produce endothecium secondary thickening (Fig. 4G) and therefore did not undergo anther dehiscence and pollen release. The heterozygous mutant nst1nst1NST2nst2 carrying 35Spro:MYB26 was fertile due to the NST2 expression alongside the MYB26 expression but showed enhanced secondary thickening (Fig. 4, D and H). In this line, the expression of NST2 was enhanced compared to wild type, presumably as a consequence of induction by the high levels of MYB26 (Fig. 4M). This heightened expression of NST2 therefore resulted in the increased anther secondary thickening observed in these lines (Fig. 4H). However, enhanced thickening in the anther was observed only in the endothecium, allowing normal anther dehiscence and fertility. No ectopic expression was seen in the other anther cell layers, suggesting that strong spatial regulation limiting secondary thickening deposition occurs in the anther and that NST2 is principally acting in the endothecium. Previously, when MYB26 was overexpressed in the wild-type background, which is expressing both NST1 and NST2, ectopic epidermal thickening was seen alongside increased endothecium thickening (Yang et al., 2007). This suggests that MYB26 acts with NST1/NST2, and that NST2 and MYB26 are principally acting in the endothecium, while NST1 is present in both cell layers. Therefore, the expression of NST1 in both endothecium and epidermal tissue allowing for the ectopic epidermal thickening when NST1 is up-regulated in this cell layer by constitutive MYB26 expression (CaMV35S promoter). The growth pattern of the 35Spro:MYB26 in the nst1nst1NST2nst2 background appeared as wild type and did not show the bushiness seen in the nst1nst2 mutant. This is likely to be a consequence of redundancy between NST1 and NST2 (Mitsuda et al., 2005) and the expression of NST2, which has been recently reported in stem tissues (Zhong and Ye, 2015), and rescue of sterility. The level of MYB26 overexpression was also strongly increased in the presence of functional NST2 (Fig. 4M), suggesting that NST2 may also up-regulate or stabilize MYB26 expression.
NST1 and NST2 have been shown to act redundantly with NST3/SND1, which is expressed in the inflorescence stems, in the regulation of secondary wall thickenings in interfasicular fibers and secondary xylem (Mitsuda et al., 2007; Zhong and Ye, 2015). NST3 expression was therefore also analyzed by qRT-PCR in buds from the MYB26-overexpressing lines. No significant native expression of NST3 was seen in the floral tissues, although a slight increase in NST3 was observed in the nst1nst2 mutant samples (Fig. 4M). This may reflect a compensatory increase in NST3 expression in the peduncle due to the absence of NST1. Although NST3 is still expressed in the nst1nst2 double mutant, the lack of significant ectopic thickening when 35Spro:MYB26 was expressed in the absence of NST1 or NST2 suggests that MYB26 is principally acting via NST1 and NST2, rather than NST3. Nevertheless, NST3 expression was greatly increased in the nst1nst1NST2nst2 line and by high levels of MYB26 in the NST2nst2 background (Fig. 4M); this increase was not seen in the nst1nst2 lines overexpressing MYB26, suggesting that this up-regulation may be mediated by NST2 (and also potentially NST1), in combination with MYB26. Ectopic lignification of the stem tissues (Fig. 4L) and also other tissues, e.g. sepals and petals, was seen in the nst1nst1NST2nst2 lines carrying the 35Spro:MYB26 gene, which may be due to the MYB26 expression in the presence of NST2 or increased NST3 expression in the stem tissues, as this lignification was not seen in the double nst1nst2 mutant lines overexpressing MYB26. qRT-PCR was also used to determine the effect of MYB26 and NST1NST2 on the expression of key genes linked to secondary thickening deposition. In the nst1nst2 mutant inflorescences, there was a significant down-regulation of IRREGULAR XYLEM1 (IRX1), IRX3, IRX8, and IRX12. Expression of NST2 in nst1nst1NST2nst2 rescued IRX3 and IRX12 expression, suggesting that NST2 directly or indirectly regulates these genes (Fig. 4N), while presence of NST2 and overexpression of MYB26 led to rescue of IRX1 and IRX8, suggesting that these genes may also require the presence of MYB26 or are primarily regulated by NST2 and require increased NST2 expression to reach normal levels. IRX1/Ces8 and IRX3/Ces7 have been shown to be coordinately expressed alongside IRX5 and to interact to form the cellulose synthase complex (Taylor et al., 2003), whereas IRX8/Galacturonosyltransferase 12 (GAUT12) and IRX12/LACCASE are involved in xylan (Persson et al., 2005; Caffall et al., 2009) and lignin biosynthesis (Zhao et al., 2013), respectively. Other genes (IRX4, IRX10) associated with secondary thickening showed a slight reduction of expression in nst1nst2 mutant and MYB26 overexpression in this double mutant background. IRX4 expression was increased with the presence of NST2 and overexpression of MYB26 in the nst1nst1NST2nst2 background, suggesting that the presence of NST2 and MYB26 is important for IRX4 expression (Fig. 4N). While FRA8 showed a slight increase in expression in the presence of NST2 (Fig. 4, M and N). This agrees with the observed development of endothecium secondary thickening and rescue of fertility in the 35Spro:MYB26 nst1nst1NST2nst2 lines, suggesting that NST2 and NST1 are acting downstream of MYB26 to regulate the biosynthesis of secondary thickening, including cellulose, hemicelluloses, and lignin biosynthesis.
Chromatin Immunoprecipitation (ChIP)-PCR Enrichment Supports MYB26 as Directly Regulating NST1 and NST2
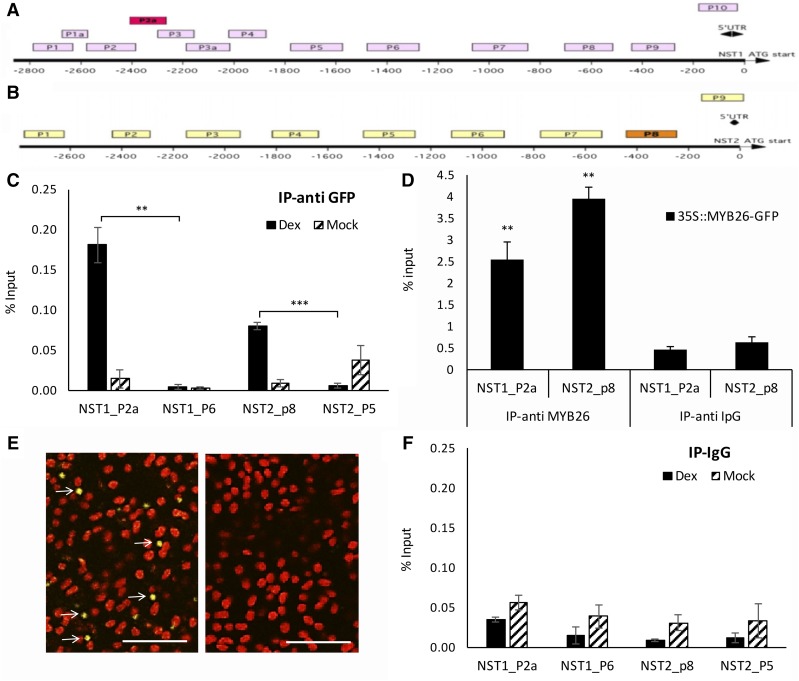
ChIP-PCR analysis was conducted to establish if the interaction between MYB26 and NST1/2 was via direct binding using a number of upstream regions of the NST1 and NST2 genes (Fig. 5, A and B) and a peptide-derived anti-MYB26 antibody with chromatin isolated from 35Spro:MYB26-GFP buds (Fig. 5D). An independent experiment using an anti-GFP antibody with buds collected from the MYB26pro:MYB26-GR-YFP line, which had been DEX-induced with the noninduced line as a control (mock), was also conducted (Fig. 5, C and F). MYB26-YFP within the nucleus of the endothecium was detected in the DEX-induced MYB26pro:MYB26-GR-YFP line (Fig. 5E). In both experiments, enrichment was seen in selected regions of the NST1 and NST2 promoter compared to negative controls of negative promoter fragments or nonspecific antibodies (IpG; Fig. 5, C, D and F). EMSA was subsequently conducted to further confirm this result; however, no retardation was observed (data not shown). ChIP therefore indicates that direct binding is occurring between MYB26 and NST1 and -2, but the lack of gel retardation implies that another factor/modification is needed for this regulation or that the conditions for in vitro binding were not suitable for complex formation.
Figure 5.
ChIP indicates that MYB26 directly binds to upstream regions of NST1 and NST2. A and B, Diagram of upstream region of (A) NST1 (B) NST2; boxes P1 to P10 indicate regions used for ChIP analysis; red/orange boxes are regions that showed positive binding. C and D, ChIP qPCR showing enrichment for (C) P2a in NST1 and P8 for NST2 using anti-GFP, and (D) anti-MYB26 antibodies. E, MYB26-YFP within the nucleus of the endothecium (left; arrows) was detected in the DEX-induced MYB26pro:MYB26-GR-YFP line; no nuclear localized expression was seen in the non-DEX-induced line (right). F, No ChIP qPCR enrichment was seen in the IP-ipG controls. Error bars represent sd (t test statistical analysis compared to control primer [C and F] or anti IpG [D] controls; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Overexpression of NST1 and NST2 Cannot Complement the myb26 Mutation
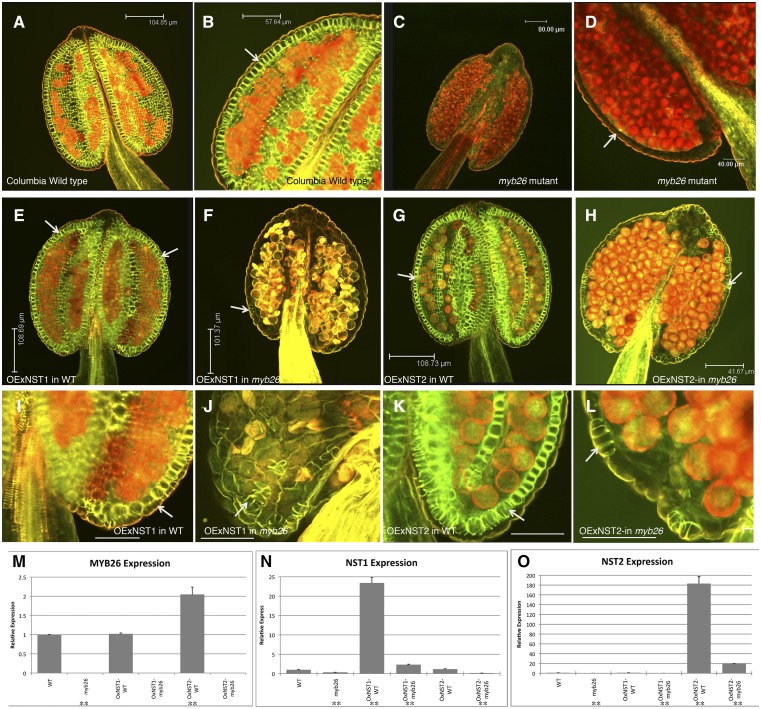
Previously it has been shown that individually the MYB26, NST1, and NST2 gene under control of the CaMV35S promoter induced ectopic secondary thickening (Mitsuda et al., 2005; Yang et al., 2007). Given that the NST1 and NST2 genes are responsible for induction of secondary thickening biosynthesis genes and appear to be downstream and regulated by MYB26, we tried to complement the myb26 mutation by overexpression of NST1 and NST2 using the CaMV35S promoter. Secondary thickening in anthers was observed using a combined stain of ethidium bromide, which indicates lignified cells (red fluorescence) and acridine orange, which stains lignified walls with a drop in fluorescence for nonlignified walls (green fluorescence; Stockert et al., 1984; Yang et al., 2007; Thévenin et al., 2011). As previously reported, we observed that the myb26 mutant failed to develop endothecium thickening (Fig. 6, C and D). As expected in the wild-type background, both the 35Spro:NST1 and 35Spro:NST2 lines showed increased secondary thickening in the flowers and leaves. In the wild-type anthers when NST2 was overexpressed, this was limited to the endothecium cell layer (Fig. 6, G and K), whereas when NST1 was expressed, using the same CaMV35S promoter, thickening was seen in the epidermis as well and in the endothecium (Fig. 6, E and I). However, when either NST1 or NST2 was overexpressed in the myb26 mutant background, the levels of secondary thickening in the anthers were not significantly increased (Fig. 6, F, J, H, and L), with no significant secondary thickening forming except limited secondary thickening in a very few isolated epidermal cells. This analysis was initially conducted using myb26 SALK_112372 insertional mutant but was subsequently repeated using the ms35 x-ray mutant ms35gl, in case gene silencing of the transgene was occurring as both constructs contained the CaMV35S promoter. Similar results were seen with these lines: increased secondary thickening in the endothecium (NST1 and NST2) and epidermis (NST1) in the heterozygous ms35MS35 and a lack of ectopic thickening without MYB26 expression except for the occasional isolated epidermal cell (Supplemental Fig. S1). This suggests that NST1 or NST2 singularly in the absence of MYB26 are not able to induce secondary thickening and that the presence of MYB26 in the anther is required to initiate normal endothecium thickening; nevertheless, MYB26 is acting through NST1/2. This lack of complementation by NST1/2 expression may be a reflection that MYB26 is controlling the expression of an additional factor that is required for accumulation, or potentially activation, of the NST1/2 transcripts; for example, this could be acting by the removal of a repressor that serves to limit the level of NST1 and NST2 transcript.
Figure 6.
Expression of NST1 or NST2 under the control of the CaMV35S promoter is unable to rescue anther secondary thickening in the myb26 mutant. A to L, Anthers stained for secondary thickening with acridine orange/ethidium bromide and visualized by confocal microscopy. A and B, Wild-type anther showing endothecium thickening (arrow). C and D, myb26 mutant lacking endothecium thickening (arrow). E and I, Overexpression of NST1 (35Spro:NST1) in wild-type background; increased levels of secondary thickening are seen in both the endothecium and epidermal tissues (arrows). F and J, Overexpression of NST1 (35Spro:NST1) in the myb26 mutant background; occasional patches of secondary thickening are seen in the epidermal tissues (arrow), but these are extremely limited, and no endothecium thickening is seen. G and K Overexpression of NST2 (35Spro:NST2) in wild-type background; increased levels of secondary thickening are seen in the endothecium (arrow), but not in the epidermal tissues as seen with NST1 overexpression in wild type. H and L, Overexpression of NST2 (35Spro:NST2) in the myb26 mutant background; occasional patches of secondary thickening are seen in the epidermal cells (arrow); however, these are extremely limited, and the endothecium cells are abnormal and lack the usual expansion seen in these cells prior to secondary thickening deposition. I to L are higher magnifications of the same anther shown in E to H. Scale bars represent 104.85 µm in A, 57.64 µm in B, 80 µm in C, 50 µm in D, 108.69 µm in E, 101.37 µm in F, 108.73 µm in G, 41.67 µm in H, and 50 µm in I to L. M to O, Expression by qRT-PCR analysis in the wild type, myb26 mutant, and overexpression lines of (M) MYB26, (N) NST1, and (O) NST2. Error bars represent sd (t test statistical analysis compared to its relevant background for each line; **P ≤ 0.01).
qRT-PCR indicated that the levels of NST1 and NST2 expression (35Spro:NST1 or 35Spro:NST2) were greatly enhanced in the wild-type background (Fig. 6, N and O), confirming that the observed phenotypic changes correlated with levels of NST1/2 expression, whereas lines carrying the 35Spro:NST1 or 35Spro:NST2 constructs in the myb26 or ms35gl mutant background showed a much-reduced level of NST1 or NST2 expression, as appropriate to the transgene (Fig. 6, N and O; Supplemental Fig. S1, R and S). This was observed in multiple lines and with both NST1 and NST2 constructs and therefore is unlikely to reflect position effects in the different overexpression lines. Given that in these lines the expression of the NST1 and NST2 genes is under regulation of the CaMV35S promoter, the low level of NST1 and NST2 observed may be the consequence of posttranscriptional regulation or direct or indirect action of the MYB26 protein on the stabilization of the NST1/NST2 RNA.
NST1 and NST2 Cannot Induce High-Level Expression of Genes Involved in the Biosynthesis of Secondary Thickening in the myb26 Background
In wild-type lines carrying the 35Spro:NST1 or 35Spro:NST2 construct, an up-regulation of genes involved in wall biosynthesis was observed. This was particularly evident for cellulose (IRX1 and IRX3) and hemicellulose (FRAGILE FIBER8 [FRA8], IRX8, and IRX10) biosynthesis genes. Genes associated with lignin formation, IRX4, and COMT however, did not show a major change, although IRX12 showed slight up-regulation (Supplemental Fig. S2). This up-regulation was more pronounced in lines carrying the 35Spro:NST1 transgenes than those with 35Spro:NST2. It was observed that NST1 was more effective in initiating ectopic secondary thickening than NST2; with high levels of NST1 showing extensive secondary thickening in epidermis and endothecium, while high levels of NST2 caused enhanced thickening in the endothecium (Fig. 6, I and K; Supplemental Fig. S1, J and N, and L and P); however, this was only seen when there was expression of MYB26. In the wild-type background, there was a direct correlation between the levels of NST1/2 gene expression, enhanced expression of the secondary thickening biosynthesis genes (Supplemental Figs. S1 and 2), and the formation of increased secondary thickening (Fig. 6; Supplemental Fig. S1); however, this induction appears to be dependent on the presence of MYB26. In the wild-type background, ectopic thickening by NST1 overexpression was linked to high levels of NST1 expression, with a cut off point of expression (∼8× normal expression) not having ectopic thickening (Supplemental Fig. S3). This suggests that although NST1 and NST2 are both able to induce the expression of genes associated with secondary wall biosynthesis, NST1 is more effective, agreeing with previous observations made by Mitsuda et al. (2005). However, in the myb26 mutant background, no enhancement of expression of these secondary wall biosynthesis genes was observed, regardless of whether NST1 or NST2 was expressed, and despite the fact the respective overexpression lines have higher expression compared to wild type (Supplemental Fig. S1, R and S).
Overexpression of Both NST1 and NST2 Together Can Induce Ectopic Thickening in the Anther Epidermis in the myb26 Background
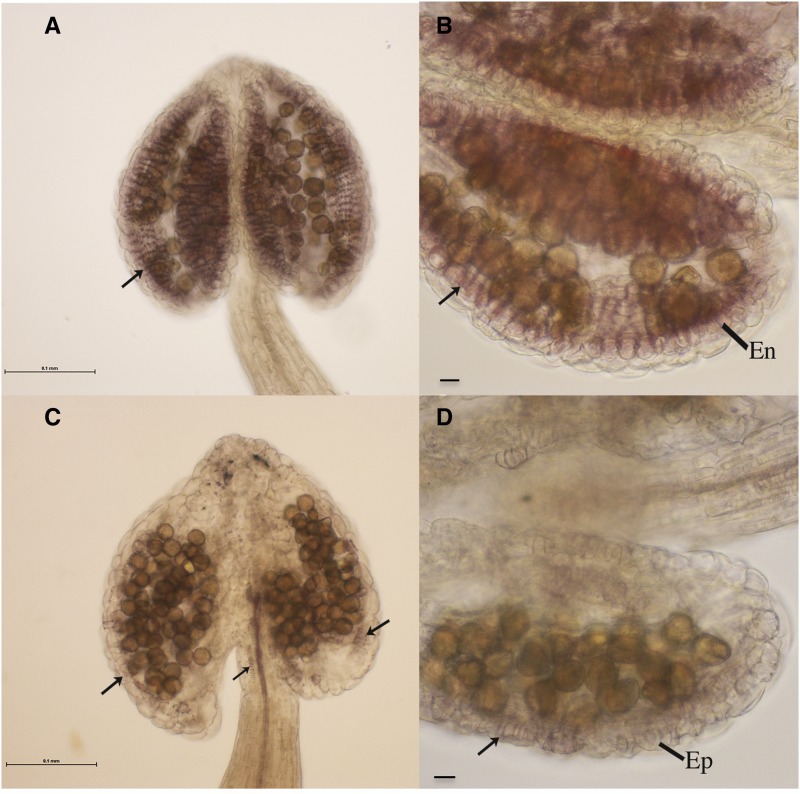
The NST1 and NST2 genes appear to act redundantly in the anther to regulate secondary thickening (Mitsuda et al., 2005), with expression of either NST1 or NST2 sufficient to induce secondary thickening. However, expression of either individually under the CaMV35S promoter was not able to complement the myb26 mutation or induce significant secondary thickening. Despite the fact that NST1 and NST2 are able to function independently, we also tested both transgenes in combination to determine whether together they were able to affect anther secondary thickening. Overexpression of both NST1 and NST2 together in the MYB26myb26 background resulted in increased secondary thickening in the anther endothecium and also ectopic anther filament and some epidermal thickening (Fig. 7, A and B). These lines were fertile since endothecium thickening was present and the ectopic epidermal thickening was at a low level, such that it did not prevent dehiscence. However, expression of both transgenes in the myb26 mutant background had a surprising effect—anther endothecium thickening failed to develop, but extensive ectopic thickening in the anther epidermis occurred (Fig. 7, C and D). These lines failed to dehisce and were male sterile due to the lack of endothecium thickening, but also because of the ectopic anther epidermal thickening. The native endothecium thickening and the ectopic epidermal thickening forms across the cell length of the cells; however, the anther epidermal cells are arranged in a different orientation (along the anther length opposed to those of the endothecium, which form along the anther width; Kelliher and Walbot, 2011). Therefore, the thickening forms in the alternate (crossed) orientation to that of the endothecium. This means that as the anther dehydrates, it is still unable to open. This effect of indehiscence as a consequence of alternate thickening due to the orientation of the epidermal cells preventing dehiscence was also previously observed in wild-type lines overexpressing MYB26 (Yang et al., 2007). Mutants of MYB26 were previously observed to have changes in the cell expansion of the endothecium layer (Dawson et al., 1999; Yang et al., 2007); this may be as a consequence of MYB26 acting on other factors to induce cell differentiation or by it repressing a repressor to allow endothecium expansion and development. NST1/2 do not appear to play a role in this, since when overexpressed, either individually or in combination, the endothecium and, in the case of ectopic expression, the other cell layers appeared contorted and failed to expand.
Figure 7.
Anthers from MYB26myb26 heterozygotes and myb26 mutants that are expressing both NST1 and NST2 under the control of the CaMV35S promoter. A to D, Anthers isolated and stained with phloroglucinol HCl to detect lignified thickening from lines overexpressing both NST1 and NST2. A and B, High levels of native secondary thickening are seen in the endothecium (En) layer (arrows) in the MYB26myb26 heterozygote background with both NST1 and NST2 transgenes. C and D, In the myb26 mutant the anthers appear contorted with ectopic thickening in epidermal tissues (arrows shows ectopic thickening in anther epidermis [Ep] and also in the filament); normal secondary thickening is not seen in the endothecium in the myb26 background, regardless of expression of both NST1 and NST2. Scale bar represents 0.1 mm.
qRT-PCR expression analysis indicated that high levels of NST1 and NST2 expression were seen in lines expressing both transgenes and that this was effective in inducing IRX1 and IRX3, downstream genes linked to cellulose biosynthesis, and also FRA8, associated with hemicellulose formation (Supplemental Fig. S4). NST2 expression was high in both the myb26 mutant and heterozygous myb26MYB26 lines; however, this did not equate to secondary thickening formation, except where MYB26 was present or when high levels of NST1 were also present. The levels of downstream gene expression did not appear to directly correlate with levels of secondary thickening. The low changes in gene expression observed are likely to be a consequence of the very limited numbers of cells forming thickening in these lines, and therefore cell-by-cell changes may be masked.
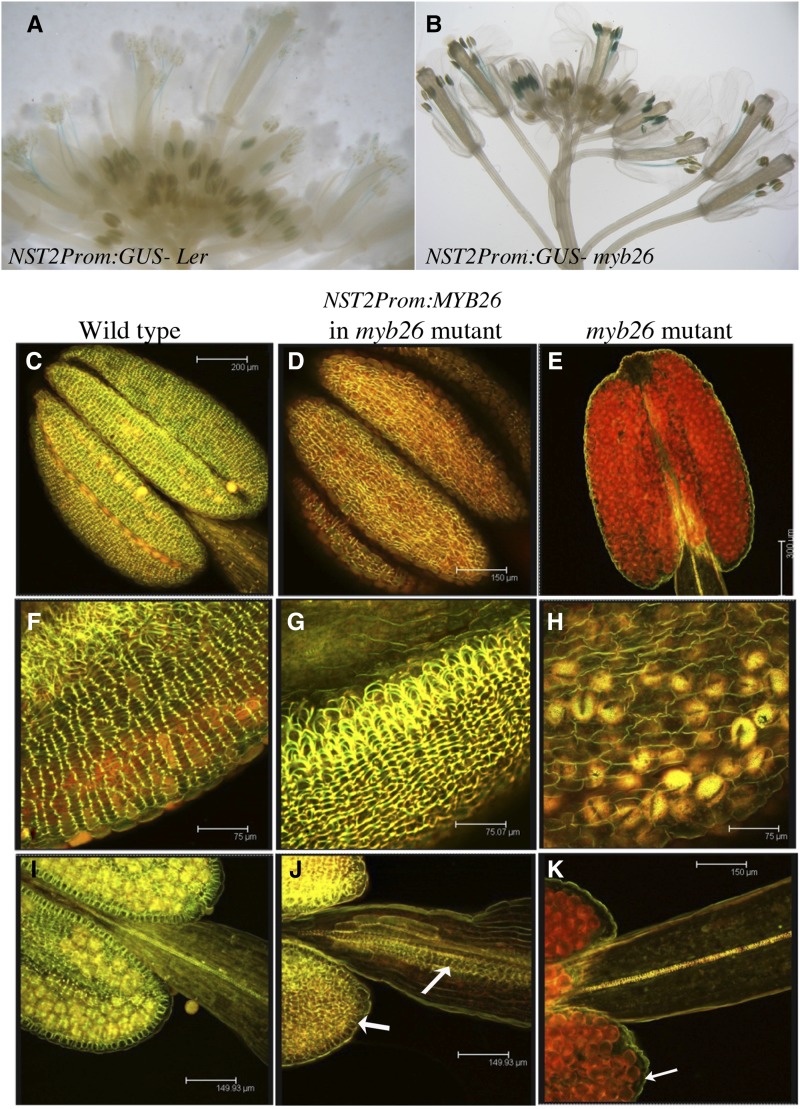
Expression of MYB26 under the Regulation of the NST2 Promoter Rescues Fertility in the myb26 Mutant
The NST2 promoter has been shown to be expressed in the floral tissues (Mitsuda et al., 2005); we confirmed this by using an NST2pro:GUS transgenic line (Fig. 8A). Expression was observed in the young postmeiotic anthers, older filaments, and pollen around the time of filament extension, prior to dehiscence. This construct was introgressed into the myb26 mutant; the expression pattern was as seen for the wild type (Fig. 8B), indicating that NST2 is induced by some other factor in addition to MYB26. We subsequently expressed MYB26 under the control of the NST2 promoter; the transgenic line showed complete rescue and full fertility in the myb26 mutant. This confirms that a factor additional to MYB26 is switching on expression of NST2, since expression is still observed in the myb26 mutant. Analysis of secondary thickening in these anthers showed that ectopic thickening formed in the endothecium and also in the filament (Fig. 8, D, G, and J). The thickening in these transgenic lines was increased as compared to the wild-type lines; this was expected since expression of MYB26 would result in a feedback loop that enhanced expression of both NST2 and NST2pro:MYB26, thus resulting in enhanced expression and secondary thickening in areas where NST2 expression was initially occurring. The deposition of thickening in the anther and filament confirms the GUS expression pattern of NST2; the lack of epidermal thickening indicates that functional NST2 is not present in the anther epidermis.
Figure 8.
Rescue of fertility in the myb26 mutant by expression of MYB26 regulated by the NST2 promoter. A, NST2pro:GUS expression in wild type, showing expression in extending filaments prior to dehiscence, and in postmeiotic anthers. B, NST2pro:GUS expression in the myb26 mutant, showing expression in extending filaments prior to dehiscence, and in postmeiotic anthers. C to K, Stamen stained for secondary thickening with acridine orange/ethidium bromide and visualized by confocal microscopy; (C, F, and I) wild-type anthers and filaments; (D, G, and J) NST2pro:MYB26 expression in the myb26 mutant showing high levels of secondary thickening in the endothecium and increased secondary thickening in the filament; (E, H, and K) myb26 mutant with no thickening in the anther endothecium. Scale bars represent 200 µm in C, 150 µm in D, 300 µm in E, 75 µm in F to H, and 150 µm in I to K.
Neither NST1 nor NST2 Interacts with MYB26 in Yeast
The full-length cDNA of MYB26, NST1 and NST2 were cloned into the Yeast two-hybrid pDEST22 Activation domain (AD) vector and pDEST32 DNA Binding domain (DB) vector (Invitrogen). These were used in pairwise combinations in yeast strain MaV203 and analyzed for activation of the expression of the three reporter genes (HIS3, URA3, and lacZ). A low level of autoactivation was seen with MYB26 fused to the DB domain, which could be overcome using at least 50 mm 3-amino-1,2,4-triazole; however, relatively strong autoactivation was also observed from the NST2 equivalent clone. Nevertheless, combinations of MYB26 as bait (DB) with NST1, or NST2 as prey (AD), suggest that there is no interaction occurring between the NST1 or NST2 proteins and MYB26 (Supplemental Fig. S5); however, MYB26-MYB26 may form as a homodimer, and homo- and heterodimerization of NST1 and -2 is also likely to occur, as predicted from the NAC domain structure (Olsen et al., 2005).
DISCUSSION
MYB26 Expression Regulates Tissue-Specific Localization of Secondary Thickening in the Anther Endothecium
MYB26, NST1, and NST2 initiate secondary thickening in the anther by a complex pathway that involves multiple regulatory points. The specific cellular localization of this thickening is critical for efficient anther opening. Our data indicate that the expression of MYB26 is essential to the formation and spatial arrangement of secondary thickening in the anther and that it acts via induction of NST1 and NST2. Nevertheless, it is clear that although the NAC domain genes are required for induction of secondary thickening biosynthesis, they are only able to do this if MYB26 is present, implying an additional regulatory step that is controlled by MYB26 that is required for progression of the tissue-specific secondary thickening in the anther.
Using a functional, inducible MYB26-YFP fusion protein, we have shown the MYB26 protein shows specific targeted localization that is different from the MYB26 transcript (Fig. 2). Previously, we reported that MYB26 expression, determined using a MYB26pro:GUS construct, was observed in many floral tissues, including the nectaries, style, filaments, and anthers (Yang et al., 2007). However, the MYB26 protein shows specific localization to the anther endothecium (Fig. 2), which agrees with the phenotype seen in the myb26/ms35 mutants, with defects in the anther endothecium, rather than alterations in the style and other floral tissue (Dawson et al., 1999; Steiner-Lange et al., 2003; Yang et al., 2007). This suggests that posttranscriptional or translational regulation of MYB26 is occurring, which confines MYB26 protein to the endothecium layer. In addition, activation by nuclear localization of the functional MYB26-GR-YFP protein after DEX treatment resulted in a decrease of MYB26 transcript (Fig. 2D), suggesting that the MYB26 protein may down-regulate its own expression. The presence of the MYB26-YFP protein was also only seen for a limited period after DEX treatment, implying rapid turnover of the MYB26 protein (data not shown). An F-box gene, Secondary wall thickening-Associated F-box1 (SAF1) has recently been reported to negatively regulate endothecium secondary thickening, which, when overexpressed, results in defective endothecium thickening and indehiscence (Kim et al., 2012). It may be that SAF1, or another factor, may act by targeting the breakdown of MYB26, or NST1/2, and preventing accumulation of these proteins and thus secondary thickening gene expression.
Gene Expression Network Associated with Secondary Thickening in the Anther
The genetic evidence suggests that MYB26 acts upstream of NST1/2, since MYB26 overexpression was unable to rescue the nst1nst2 double mutant (Fig. 4), and MYB26-GR was able to induce expression of NST1 and NST2 (Fig. 3). This appears to be via direct regulation, with MYB26 binding to both promoters by ChIP-PCR (Fig. 5) and rapid induction (within 4–6 h) of NST1/2 seen after DEX activation of MYB26. However, NST2 also appears regulated by an additional factor(s), since the NST2 promoter can drive gene expression in the myb26 background, as demonstrated by the NST2pro:GUS and NST2pro:MYB26 constructs (Fig. 8). In the absence of myb26, NST2 appears to show similar expression within the endothecium, as indicated by the rescue of fertility and endothecium thickening by NST2:MYB26; nevertheless, MYB26 is essential for induction of endothecium secondary thickening.
In the wild-type or MYB26myb26 heterozygous background, overexpression of NST1 led to increased secondary thickening within the endothecium and ectopic secondary thickening in the epidermis. However, in the myb26/ms35 mutant, overexpression of NST1/2 singularly or combined did not result in secondary thickening within the endothecium and therefore was unable to rescue the myb26/ms35 mutants (Figs. 6 and 7; Supplemental Fig. S1). This is unlikely to be a consequence of the promoter since 35Spro:MYB26 was previously able to rescue fertility in the myb26 mutant (Yang et al., 2007). Nevertheless, combined overexpression of NST1 and NST2 resulted in ectopic thickening in the anther epidermis in the myb26 mutant, but endothecium thickening still did not occur. It therefore appears that it is easier for the epidermis to form ectopic thickening than other cell layers in the anther. Epidermal tissues have been reported as highly metabolically active (Mahroug et al., 2006). The ability of the epidermis to develop thickening if NST1 expression is sufficiently high may be a reflection of the enhanced competency of this tissue for such metabolic activity. NST1 is more effective at inducing secondary thickening biosynthesis (Mitsuda et al., 2005); therefore, this may explain why overexpression of NST2 in the wild-type background is unable to induce epidermal thickening.
This lack of rescue appears to be at least partly due to the insufficient expression of NST1 and NST2 in the absence of MYB26, as 35Spro:NST1 and 35Spro:NST2 expression was reduced in the myb26/ms35 mutants in comparison to overexpression within the wild-type background (Supplemental Fig. S3). The relationship among MYB26 and NST1 and NST2 is therefore more complex than a linear network. It appears that an additional factor(s) controlled by MYB26 enables an increase of the NST1 and NST2 transcripts and thus induction of secondary thickening genes. This could be a consequence of altered stability of the NST1/2 transcripts/proteins, or of the removal of an additional repressor facilitating transcript increase, which facilitates secondary thickening formation (Fig. 9). This additional role of MYB26 does not appear to be a consequence of direct interactions at the protein level, since NST1/NST2 and MYB26 do not appear to interact in a yeast two-hybrid analysis (Supplemental Fig. S5).
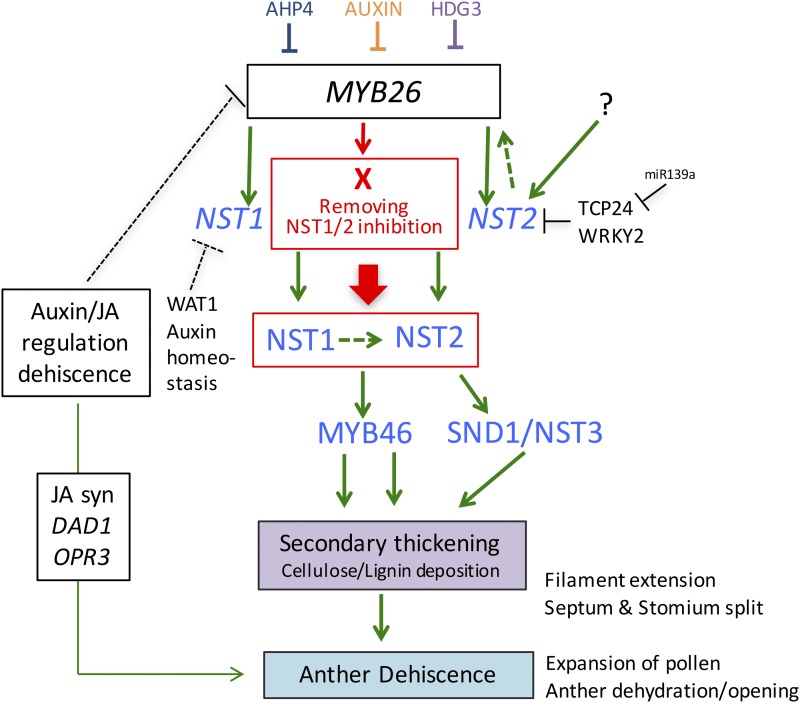
Figure 9.
Model of MYB26 regulation of anther secondary thickening pathway. MYB26 regulation of secondary thickening through downstream the redundant transcription factors NST1/NST2. Arrows represent direct regulation, while bar represents repression, and dotted lines represent predicted regulation/repression. X = unknown factor that enables NST1/2 to initiate secondary thickening. This could be via NST1/2 protein activation/stabilization or removal of an inhibitor involved in NST1/2 degradation/turnover. AHP4, ARABIDOPSIS HISTIDINE-CONTAINING PHOSPHOTRANSFER FACTOR 4; HDG3, HOMEODOMAIN GLABRA 2-LIKE PROTEIN 3; TCP24 and WRKY2, transcription factor; JA, Jasmonic Acid; DAD1, DEFECTIVE IN ANTHER DEHISCIENCE1; OPR3, 12-OXOPHYTODIENOATE REDUCTASE 3; WAT1, WALLS ARE THIN1.
In the absence of MYB26, secondary thickening can only be achieved in the anther if both NST1 and NST2 are expressed at high levels and then only ectopically in the wrong cell layer, the epidermis. This suggests that there is a highly cell-specific, spatial regulation of thickening involving MYB26, which is easier to overcome in the epidermis than in other cell layers in the anther, in particularly the endothecium. NAC domain genes are a large group of plant-specific transcription factors that show specific regulation, by various mechanisms, including miRNA cleavage and ubiquitin-mediated proteolysis (Olsen et al., 2005). For example NAM, CUC1, and CUC2, which function in shoot meristem formation and boundary specification, are regulated by miRNAs (Aida et al., 1997). It can be speculated that similar regulation of NST1 and NST2 may be occurring via miRNAs, which may be repressed by MYB26. The F-box protein Secondary wall thickening-Associated F-box 1 (SAF1) could also potentially be regulating the protein turnover of NST1/2, since when this is overexpressed it negatively regulates endothecial secondary wall thickening (Kim et al., 2012). This also agrees with the observation that SAF1 is up-regulated in the myb26 mutant (https://www.cpib.ac.uk/anther; Pearce et al., 2015). The WRKY12 transcription factor has also been shown to negatively regulate NST2 (Wang et al., 2010), and Homeobox-leucine zipper protein 15 (AtHB15) negatively regulates NST3 and NST2 within pith parenchyma cells (Du et al., 2015). WRKY13, however, positively regulates NST1–3 within the stem and has been shown to bind directly to the NST2 promoter (Li et al., 2015). It is therefore possible that there is a similar transcription factor regulating expression or turnover of NST1 and NST2 within the anther cell layers. In wild-type plants transcription factor (TCP24) is strongly expressed in the early stages of endothecium formation, and this expression reduces and eventually disappears by the time secondary wall thickening occurs (Wang et al., 2015). TCP24, which is regulated by miR139, has been shown to repress endothecium secondary thickening and NST1/2 expression, but not MYB26 (Wang et al., 2015); however, it does not appear to show significant expression changes in the myb26 mutant (https://www.cpib.ac.uk/anther; Pearce et al., 2015).
Lack of NST1/NST2 Alters Plant Stature alongside Regulating Secondary Thickening
The nst1nst2 double mutant shows altered stature, with a very bushy appearance, which is rescued by the presence of a single copy of either the NST1 or NST2 gene (Mitsuda et al., 2005). This phenotype is not seen with the myb26 mutant (Dawson et al., 1999; Steiner-Lange et al., 2003), suggesting that this is not associated with reduced fertility but may reflect the lack of NST1 and NST2 expression throughout the plant. A similar phenotype was reported for the saf1 mutant, and it was suggested that this may be a consequence of altered auxin levels, which is also seen when flavonoid balance is altered (Kim et al., 2012). This phenotype is not seen in the nst1nst1NST2nst2 lines, suggesting that a single copy of NST2 is able to compensate for the lack of NST1 in the plant. Recently, it has been shown that NST2 together with NST1 and NST3 regulate secondary cell wall synthesis in fibers of stems (Zhong and Ye, 2015). qRT-PCR expression analysis in the different transgenic mutant lines indicated that when expressed at very high levels NST2 may alter the level of expression or enhance the stability/reduce the turnover of the MYB26 and NST3 transcripts (Figs. 4 and 6M). However, NST1 does not appear to affect the expression levels of either MYB26 or NST2 (Figs. 6 and 9).
Expression of Secondary Thickening Biosynthesis Genes Is Regulated by NST1/2
Overexpression of MYB26 in the wild-type background resulted in increased thickening in the endothecium, epidermis, and ectopically throughout the plant; however, it was unable to induce lignin and cellulose biosynthesis genes in the absence of NST1/2 (Fig. 4N) and appears to act by directly up-regulating expression of both NST1/2, which in turn regulates cellulose biosynthesis (particularly IRX1 and -3) and lignin biosynthetic genes. NST1/2 act redundantly, and presence of one of them was sufficient for secondary thickening induction. Nevertheless, it appears that secondary thickening biosynthesis is principally mediated via NST1, with NST1 more effective in the induction of secondary thickening biosynthesis genes, as previously reported (Mitsuda et al., 2005). However qRT-PCR data suggests that NST2 may also indirectly cause up-regulation of NST1 via a feedback loop of up-regulation of MYB26 (Fig. 9).
Studies of the NST2-NST3/SND1 and VND1–VND7 genes suggest that secondary cell wall regulating NAC-domain genes are all able to directly bind targets associated with cellulose, lignin, and hemicellulose biosynthesis, through a 19-bp consensus sequence secondary wall NAC-binding element (Zhong et al., 2010; Yamaguchi et al., 2011; Taylor-Teeples et al., 2015). Complementation studies have shown that by misexpression of one NAC-domain gene is able to rescue the mutant phenotype, indicating that these genes are functional paralogs (Zhong et al., 2010; Yamaguchi et al., 2011; Zhong and Ye, 2014). Almost all of these transcription factors contain at least one secondary wall NAC-binding element site in their own promoter (except VASCULAR-RELATED NAC-DOMAIN 6 [VND6]; Zhong and Ye, 2014), and NST3 has been shown to up-regulate its own expression (Wang et al., 2011). Given the observed similarities between the NAC domain genes that regulate secondary thickening in different plant tissues, it seems likely that NST1 may also be able to up-regulate its own expression.
CONCLUSION
Overall, it appears that there is tight regulation of secondary thickening in the anther, which is controlled by localization of the MYB26 protein to the endothecium cell layer and direct induction of NST1 and NST2 expression by MYB26 (Fig. 9). However, there is an additional mechanism involving MYB26 that enables the accumulation of the NAC domain transcripts that is essential for thickening. This may be needed to maintain cell specificity, since other factors are also involved in the activation of these NAC domain genes, e.g. NST2, thereby facilitating strict temporal and boundary control to thickening. Such high-level cell-specific control is a prerequisite to effective regulation of dehiscence at optimal developmental stages.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Two Arabidopsis (Arabidopsis thaliana) MYB26 mutant lines were used as previously described by Yang et al. (2007); the x-ray line ms35gl (Z.A.W. lab, University of Nottingham) and the myb26 T-DNA SALK line (SALK_112372) (SIGnal; Alonso et al., 2003), as well as the T-DNA SALK lines nst1, nst2, and nst1nst2 double mutants previously described by Mitsuda et al. (2005). T1 seeds of NST2pro:GUS myb26 (SALK 112372; CR684), NST1pro:GUS myb26 (DR0561), NST2pro:MYB26 myb26 (DR0562), and 35Spro:MYB26 NST1pro:GUS (DR0816) were generated by Dr. Nobutaka Mitsuda (National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan). Plants were selected on hygromycin/kanamycin plates, as appropriate, then transferred into Levington M3 (The Scotts Company) compost supplemented with 0.2 g l-1 of Intercept 70 WG (Scotts, Monro South) and grown in a glasshouse at 21°C/17°C (day/night) and 22/2 h photoperiod as previously described (Dawson et al., 1999), along with their appropriate wild-type controls (ecotype Heynh. var Landsberg erecta [Ler] for ms35gl; and ecotype Columbia [Col-0] for myb26 SALK line).
DEX-Inducible MYB26 Construct
A 5-kb region of MYB26 including a 3-kb upstream region was amplified from genomic DNA of Ler using primers MS35prom-KpnI and MS35cDNA-R-SpeI (Supplemental Table S1) and then cloned into TOPO PCR Blunt II (Invitrogen). The fragment was then digested with KpnI/SpeI and cloned into pGREEN0229-GR-YFP (kindly provided by the Bennett Lab, University of Nottingham) upstream of GR-YFP to produce the construct pGREEN0229MYB26pro:MYB26-GR-YFP. The construct was confirmed by PCR and sequencing, then transferred into Agrobacterium (GV3101 + pSOUP) by electroporation (Sambrook et al., 1989). Arabidopsis heterozygous myb26 SALK mutant and ms35gl plants were transformed by floral dipping (Clough and Bent, 1998). The T1 generation were screened for Basta resistance and PCR tested for the transgene. These plants grew to flowering stage; the sterile plants with flower buds showing myb26 mutant phenotype were sprayed with, or dipped into 25 µm DEX + 0.02% (v/v) Silwet l-77 solution. YFP was observed using confocal microscope (TCS SP2, Leica) with 514-nm excitation.
Overexpression Lines
The coding region of NST1 and NST2 with stop codons was amplified by PCR (Supplemental Table S1), cloned into pDONR211 (Invitrogen), and then transferred by Gateway cloning into the PGWB5 (Invitrogen) destination vector to form 35Spro:NST1 and 35Spro:NST2 overexpression constructs. The constructs were then transferred into Agrobacterium (C58) by electroporation (Sambrook et al., 1989) and transformed into Arabidopsis heterozygous myb26 SALK line and heterozygous ms35glMS35 plants by floral dipping (Clough and Bent, 1998). The T1 generation were screened for hygromycin resistance and PCR tested for the transgene (Supplemental Table S1). The selected homozygous lines of 35Spro:NST1 and 35Spro:NST2 in the heterozygous myb26MYB26 and ms35glMS35 background were then subsequently crossed to produce an overexpression of both NST1 and NST2 lines in the homozygous myb26 and ms35 background.
Expression Analysis
RNA was isolated from buds and leaves (RNeasy, Qiagen) and cDNA prepared using 5 µg total RNA in a 20 µL reaction (Superscript II reverse transcriptase, Invitrogen). qRT-PCR was carried out using a Light Cycler (Roche) in a 384 plate using the Maxima SYBRR Green QPCR Master Mix in a final volume of 9 μL containing 0.2 μL of cDNA and 0.2 μL of the appropriate primers (Supplemental Table S1). PCR cycling conditions for amplification were 95°C for 10 min, then 40 cycles of 95°C for 30 s, 58°C for 1 min, and 72°C for 1 min. All samples were run at least in duplicate. Data acquisition and analyses were performed using the Light Cycler software. Relative expression levels were determined in comparison to actin or PP2A expression using the 2-ΔΔCT analysis method (Livak and Schmittgen, 2001).
Microscopy
For analysis of lignin, fresh samples were stained with phloroglucinol-HCl (Ruzin, 1999) and were observed under a light microscope (Nikon); for confocal microscopy (TCS SP2, Leica) observation a modified ethidium bromide/acridine orange stain was used (Yang et al., 2007). The ethidium bromide stains lignified cells (red fluorescence; 514 nm excitation; emission collection 590 nm [570–620 nm]) and the acridine orange stains lignified walls with a drop of fluorescence for nonlignified walls (green fluorescence; 488 nm excitation; emission collection 520 nm [510–530 nm]). A minimum of ten independent transformants were analyzed.
Yeast Two-Hybrid Analysis
A yeast two-hybrid screen was conducted using the Gateway yeast two-hybrid system (Invitrogen) according to the manufacturer’s instructions. The full-length MYB26, NST1, and NST2 coding regions were cloned into pDEST32 (DNA DB) and pDEST22 (AD) vectors and used to check pairwise interactions in yeast strain MaV203 carrying three reporter genes (HIS3, URA3, and lacZ). Interactions and autoactivation were tested by His selection supplied with 30, 60, and 80 mm of 3-Amino-1,2,4-triazole and X-Gal assay, following the manufacturer’s instructions. Control assays were used as positive and negative controls for the analysis; these consisted of empty pDEST22 and pDEST32 (A—negative control for growth); pEXPTM22/RalGDS-m2 and pEXPTM32/Krev1 (B—negative control for interaction); pEXPTM22/RalGDS-m1 and pEXPTM32/Krev1 (C—weak positive control for interaction); pEXPTM22/RalGDS-wt and pEXPTM32/Krev1 (D—strong positive control for interaction). They were used as described by the manufacturer’s instructions.
ChIP Analysis
ChIP analysis was conducted on MYB26-DNA complexes in the 35Spro:MYB26-GFP, and DEX-inducible MYB26pro:MYB26-GR-YFP line using both a peptide-derived anti-MYB26 antibody and ChIP grade anti-GFP (Abcam; ab290, 3%–5% [v/v] final concentration), respectively. Following a modified protocol from Ferguson et al., (2017), chromatin was isolated from 5 g bud tissue. All samples were run in triplicate with at least two biological replicates. Negative controls were as follows, noninduced MYB26pro:MYB26 GR YFP line (treated with water rather than DEX), nonspecific antibody (anti-HA or anti-HIS IgG), and negative promoter primers (NST1-P6 and NST2-P5) were used. Primers for qChIP-PCR are shown in Supplemental Table S1. Data are presented as %input to test whether there was enrichment of the NST1 and NST2 promoters in comparison to all the controls used.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: MYB26 (At3g13890, Q9SPG3); NST1 (At2g46770 Q84WP6); NST2 (At3g61910, Q9M274); SND1 (AT1G32770, Q9LPI7); VND7 (AT1G71930, Q9C8W9); IRX1 (At4g18780, Q8LPK5); IRX3 (At5g17420, Q9SWW6); FRA8 (AT2G28110, Q9ZUV3); IRX8 (At5g54690, Q9FH36); IRX10 (At1g27440, Q9FZJ1); IRX4 (At1g15950, Q9S9N9); IRX12 (At2g38080, O80434); COMT (At1g67980, Q9C9W3).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression of NST1 or NST2 under the control of the CaMV35S promoter is unable to rescue anther secondary thickening in the ms35 mutant.
Supplemental Figure S2. qRT-PCR expression and secondary thickening analysis of NST1 in wild-type and ms35 mutant buds overexpressing NST1.
Supplemental Figure S3. Ectopic expression of NST1 under the control of CaMV35S promoter is proportional to the level of NST1 expression.
Supplemental Figure S4. qRT-PCR expression analysis in wild type and myb26 mutant, and in MYB26myb26 and myb26myb26 lines overexpressing both NST1 and NST2.
Supplemental Figure S5. No interactions are detected by yeast-two hybrid analysis between MYB26 and NST1, or MYB26 and NST2.
Supplemental Table S1. Primers used.
Acknowledgments
Seed stocks were provided by NASC.
Footnotes
This work was funded by the Biotechnology and Biological Science Research Council (BB/F021062/1 and BB/J001295/1); R.M. was supported by a PhD Scholarship from the China Scholarship Council.
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Caffall KH, Pattathil S, Phillips SE, Hahn MG, Mohnen D (2009) Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol Plant 2: 1000–1014 [DOI] [PubMed] [Google Scholar]
- Cecchetti V, Altamura MM, Brunetti P, Petrocelli V, Falasca G, Ljung K, Costantino P, Cardarelli M (2013) Auxin controls Arabidopsis anther dehiscence by regulating endothecium lignification and jasmonic acid biosynthesis. Plant J 74: 411–422 [DOI] [PubMed] [Google Scholar]
- Cecchetti V, Celebrin D, Napoli N, Ghelli R, Brunetti P, Costantino P, Cardarelli M (2017) An auxin maximum in the middle layer controls stamen development and pollen maturation in Arabidopsis. New Phytol 213: 1194–1207 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dawson J, Sözen E, Vizir I, Van Waeyenberge S, Wilson ZA, Mulligan BJ (1999) Characterization and genetic mapping of a mutation (ms35) which prevents anther dehiscence in Arabidopsis thaliana by affecting secondary wall thickening in the endothecium. New Phytol 144: 213–222 [Google Scholar]
- Dawson J, Wilson ZA, Aarts MGM, Braithwaite AF, Briarty LG, Mulligan BJ (1993) Microspore and pollen development in six male-sterile mutants of Arabidopsis thaliana. Can J Bot 71: 629–638 [Google Scholar]
- Du Q, Avci U, Li S, Gallego-Giraldo L, Pattathil S, Qi L, Hahn MG, Wang H (2015) Activation of miR165b represses AtHB15 expression and induces pith secondary wall development in Arabidopsis. Plant J 83: 388–400 [DOI] [PubMed] [Google Scholar]
- Ferguson AC, Pearce S, Band LR, Yang C, Ferjentsikova I, King J, Yuan Z, Zhang D, Wilson ZA (2017) Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis. New Phytol 213: 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Walbot V (2011) Emergence and patterning of the five cell types of the Zea mays anther locule. Dev Biol 350: 32–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YY, Jung KW, Jeung JU, Shin JS (2012) A novel F-box protein represses endothecial secondary wall thickening for anther dehiscence in Arabidopsis thaliana. J Plant Physiol 169: 212–216 [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tian Z, Yu D (2015) WRKY13 acts in stem development in Arabidopsis thaliana. Plant Sci 236: 205–213 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Mahroug S, Courdavault V, Thiersault M, St-Pierre B, Burlat V (2006) Epidermis is a pivotal site of at least four secondary metabolic pathways in Catharanthus roseus aerial organs. Planta 223: 1191–1200 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M (2008) NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J 56: 768–778 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Band LR, Dyson RJ, Lessinnes T, Wells DM, Yang C, Everitt NM, Jensen OE, Wilson ZA (2012) A biomechanical model of anther opening reveals the roles of dehydration and secondary thickening. New Phytol 196: 1030–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Pearce S, Ferguson A, King J, Wilson ZA (2015) FlowerNet: A gene expression correlation network for anther and pollen development. Plant Physiol 167: 1717–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin SE. (1999) Plant Microtechnique and Microscopy. Oxford University Press, New York. [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning: A laboratory manual, 2nd Edition. Cold Spring Harbour [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell 16(Suppl): S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner-Lange S, Unte US, Eckstein L, Yang C, Wilson ZA, Schmelzer E, Dekker K, Saedler H (2003) Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J 34: 519–528 [DOI] [PubMed] [Google Scholar]
- Stockert JC, Cañete M, Colman OD (1984) Histochemical mechanism for the orthochromatic staining and fluorescence reaction of lignified tissues. Cell Mol Biol 30: 503–508 [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Teeples M, Lin L, de Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R, et al. (2015) An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517: 571–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenin J, Pollet B, Letarnec B, Saulnier L, Gissot L, Maia-Grondard A, Lapierre C, Jouanin L (2011) The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol Plant 4: 70–82 [DOI] [PubMed] [Google Scholar]
- Wang H, Avci U, Nakashima J, Hahn MG, Chen F, Dixon RA (2010) Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc Natl Acad Sci USA 107: 22338–22343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Mao Y, Yang J, He Y (2015) TCP24 modulates secondary cell wall thickening and anther endothecium development. Front Plant Sci 6: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhao Q, Chen F, Wang M, Dixon RA (2011) NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J 68: 1104–1114 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Song J, Taylor B, Yang C (2011) The final split: The regulation of anther dehiscence. J Exp Bot 62: 1633–1649 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Kubo M, Fukuda H, Demura T (2008) Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55: 652–664 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T (2011) VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J 66: 579–590 [DOI] [PubMed] [Google Scholar]
- Yang C, Xu Z, Song J, Conner K, Vizcay Barrena G, Wilson ZA (2007) Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19: 534–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang ZY, Dixon RA (2013) Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18: 3158–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye ZH (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 3: 1087–1103 [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2014) Complexity of the transcriptional network controlling secondary wall biosynthesis. Plant Sci 229: 193–207 [DOI] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2015) The Arabidopsis NAC transcription factor NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of inflorescence stems. Plant Signal Behav 10: e989746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Zhong R, Ye ZH (2014) Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS One 9: e105726. [DOI] [PMC free article] [PubMed] [Google Scholar]