The zygotic wall program is executed by the heterodimeric homeobox transcription factors GSM1/GSP1 via transcriptional activation and post-transcriptional suppression in Chlamydomonas.
Abstract
The sexual cycle of the unicellular Chlamydomonas reinhardtii culminates in the formation of diploid zygotes that differentiate into dormant spores that eventually undergo meiosis. Mating between gametes induces rapid cell wall shedding via the enzyme g-lysin; cell fusion is followed by heterodimerization of sex-specific homeobox transcription factors, GSM1 and GSP1, and initiation of zygote-specific gene expression. To investigate the genetic underpinnings of the zygote developmental pathway, we performed comparative transcriptome analysis of both pre- and post-fertilization samples. We identified 253 transcripts specifically enriched in early zygotes, 82% of which were not up-regulated in gsp1 null zygotes. We also found that the GSM1/GSP1 heterodimer negatively regulates the vegetative wall program at the posttranscriptional level, enabling prompt transition from vegetative wall to zygotic wall assembly. Annotation of the g-lysin-induced and early zygote genes reveals distinct vegetative and zygotic wall programs, supported by concerted up-regulation of genes encoding cell wall-modifying enzymes and proteins involved in nucleotide-sugar metabolism. The haploid-to-diploid transition in Chlamydomonas is masterfully controlled by the GSM1/GSP1 heterodimer, translating fertilization and gamete coalescence into a bona fide differentiation program. The fertilization-triggered integration of genes required to make related, but structurally and functionally distinct organelles—the vegetative versus zygote cell wall—presents a likely scenario for the evolution of complex developmental gene regulatory networks.
Metazoa, Embryophyta, and Fungi, the so-called crown groups, independently evolved developmental programs that give rise to complex multicellular organisms. Their explosive morphological diversity is largely driven via modification or reuse of existing gene regulatory networks (GRNs) consisting of transcriptional regulators and signaling pathways (Carroll, 2008; Davidson and Erwin, 2006; Pires and Dolan, 2012; Rudel and Sommer, 2003). Therefore, inquiry about the origins of the crown groups has been focused on analyzing the evolutionary precursors of the GRNs that underlie their key developmental strategies.
Large-scale comparative genomics studies and more focused analyses of ancestral lineages closely related to the Metazoa (e.g. choanoflagellates) and Embryophyta (e.g. charophytes) have indeed reported that many GRN components predate the origins of the crown groups (Fairclough et al., 2013; Hori et al., 2014; King et al., 2008; de Mendoza et al., 2013; Wickett et al., 2014; Worden et al., 2009). These findings encourage inquiry into the roles of pan-eukaryotic GRN modules in the differentiation repertoires of the unicellular eukaryotes that share common ancestry with crown group organisms. Research on the unicellular green alga Chlamydomonas reinhardtii has been particularly useful for gaining insights into the origins of basal GRNs of the Embryophytes, since large numbers of gene families that are not found in fungi or animals are shared within Viridiplantae (Merchant et al., 2007; Worden et al., 2009).
Given that the most recent eukaryotic common ancestor engaged in sexual reproduction (Goodenough and Heitman, 2014), an obvious focus for these inquiries has been the strategies and proteins involved with gametic differentiation, mate recognition, zygote/spore formation, and meiosis. Particularly fruitful have been studies of these processes in unicellular yeasts, where transcriptional and signaling cascades are found to be conserved in the sexual reproduction of mushrooms and beyond (for review, see Honigberg and Purnapatre, 2003; Mata et al., 2002; Neiman, 2011). Sexual development of Chlamydomonas, detailed below, uses several such strategies and proteins. As an example, both Chlamydomonas and yeast gametes contain a transcription factor that forms a heterodimer in the diploid zygote that is involved in diploid development (Goutte and Johnson, 1988; Lee et al., 2008). Furthermore, the Chlamydomonas heterodimeric transcription factors GSM1/GSP1 in the zygote are structural and functional homologs to the KNOX/BELL homeobox heterodimers that are also involved in the diploid development of land plants (Horst et al., 2016; Sakakibara et al., 2008, 2013). Such deeply rooted conservation of sexual GRNs suggests their fundamental importance for complex multicellular evolution.
The proteins involved in Chlamydomonas sexual development also exemplify the deep ancestry of sexual processes (Speijer et al., 2015). GEX1, for example, has been identified as a gamete-expressed nuclear envelope fusion protein that is required for nuclear fusion in protists, fungi, plants, and many vertebrates (except those, including humans and mice, whose pronuclei do not fuse in the zygote; Ning et al., 2013). And the primordial gamete fusogen HAP2 likely was present at the origins of sexual reproduction in eukaryotes (Liu et al., 2008). Recently, HAP2 was shown to be a class II fusion protein, in the same family as dengue and Zika virus fusion proteins (Fedry et al., 2017).
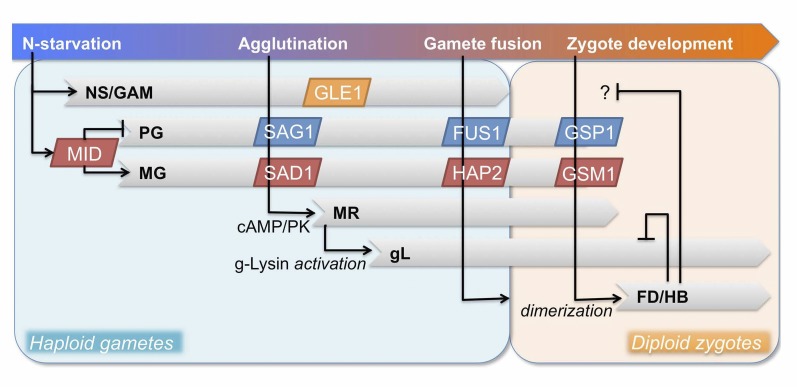
Molecular understanding of sexual development of Chlamydomonas has been driven by molecular genetics studies (for review, see Goodenough et al., 2007). The Chlamydomonas sexual cycle is invoked by nitrogen starvation and entails expression of sex-specific proteins involved in pre- and post-fertilization events during gamete interactions (mating) and zygote formation (Fig. 1). Sex-specific (plus and minus) gametogenesis programs are governed by the biallelic mating type loci called MTL+ and MTL− (De Hoff et al., 2013; Ferris et al., 2002). The FUS1 gene, exclusive to MTL+, encodes a glycoprotein required for the plus gametes to bind and fuse to minus gametes (Ferris et al., 1996), and the MID gene, exclusive to MTL−, encodes a transcription factor in the RWP-RK family that induces the minus sex program and suppresses the plus sex program (Lin and Goodenough 2007; Ferris and Goodenough 1997). Therefore, both MTL+ and MTL− are necessary for a successful progression through the sexual cycle.
Figure 1.
A gene-regulatory network model of sexual development in Chlamydomonas reinhardtii consisting of at least six regulatory programs. Gray bars depict the duration of each program. The upper bar shows physiological/cellular events providing cues for the programs. Arrows denote the consequence of specific events/factors as induction or repression. Known molecular players are shown as boxes, blue for plus-specific and red for minus-specific. NS/GAM, N-starvation induced/gametogenesis program; PG/MG, plus- and minus-specific gamete programs; MR, mating reaction-induced program (dibutyryl-cAMP-inducible); gL, g-lysin-induced program; FD/HB, fusion-dependent/homeobox transcription factor-dependent program.
Minus and plus sexual programs employ a series of key players acting in pairs that are expressed in a sex-specific manner: agglutinins (SAD1 and SAG1) on the flagellar membrane (Ferris et al., 2005; Goodenough et al., 1985), fusion-enabling factors (HAP2 and FUS1) on the plasma membrane (Liu et al., 2015, 2008; Misamore et al., 2003), and heterodimeric transcription factors in the cytosol, GSM1 and GSP1 (Lee et al., 2008; Zhao et al., 2001). SAD1 and SAG1, large Hyp-rich glycoproteins (HRGPs), serve as both ligands and receptors for the initial gamete-recognition event (Ferris et al., 2005), which triggers downstream events collectively called the mating reaction that involves cAMP-mediated signal transduction via a cyclic GMP-activated protein kinase (for review, see Snell and Goodenough, 2009). Responses to cAMP elevation include activation of a periplasmic proenzyme called gametolysin (g-lysin; Buchanan et al., 1989) that removes cell walls, promotes translocation of additional agglutinins to flagella (Cao et al., 2015), and activates the sex-specific mating structures where HAP2 and FUS1 are localized (Goodenough and Jurivich, 1978; Liu et al., 2008; Misamore et al., 2003). During gamete differentiation, the homeobox transcription factors GSM1 and GSP1 are synthesized and stored in the minus and plus cytoplasms; upon gametic cell fusion they heterodimerize and translocate to the nuclei, turning on the zygote developmental pathway and preparing fused cells for eventual meiotic germination (Lee et al., 2008). Therefore, the MID-controlled minus and plus programs prepare minus and plus gametes for a series of events, occurring within a few minutes in a step-wise manner, that initiates the diploid phase of the life cycle (Fig. 1).
The transition from gametes to zygotes (the haploid-to-diploid transition) represents a key step in the Chlamydomonas life cycle, analogous to the embryogenesis following fertilization in plants without entailing mitotic growth. Zygote development entails signature events largely completed within the first 12 h, such as zygote-specific wall assembly (Minami and Goodenough, 1978; Suzuki et al., 2000; Woessner and Goodenough, 1989), selective destruction of chloroplast nucleoids derived from minus gametes and of mitochondrial DNAs from plus gametes (Aoyama et al., 2006; Kuroiwa et al., 1982), flagellar resorption and basal-body disassembly (Cavalier-Smith, 1974; Pan and Snell, 2005), and nuclear and chloroplastic fusion (Cavalier-Smith, 1976). Such a rapid differentiation process would require robust regulatory networks to prevent promiscuous execution, networks that combine intra- and perhaps intercellular cues and coordinate a multitude of events.
The walls of vegetative and gametic cells serve two core functions. (1) They must protect cells from mechanical and osmotic stresses, and (2) they must be easily removed. Vegetative cells shed the so-called mother cell wall after their mitotic progeny have formed their own cell walls, and upon encountering a gamete of the opposite mating type, gametes must be able to rapidly shed their walls to allow cell fusion. In both vegetative cells and gametes, transcripts for cell wall genes are up-regulated when their walls are experimentally released by incubating cells in g-lysin (Hoffmann and Beck, 2005; Kurvari, 1997). One notable difference is that only the vegetative wall needs to support large increases in cell volume, which requires continuous remodeling of the existing wall layers.
In contrast, the zygote wall has only a single function, which is to provide physical and chemical enclosure that allows Chlamydomonas to survive loss of nutrient sources and extreme environmental conditions, including dessication. The zygote cell wall is unaffected by g-lysin (Schlosser, 1976) but is removed by the zygote hatching enzyme (z-lysin) whose biochemical nature is unknown.
The zygote wall-specific features include secretion and assembly of zygote-specific extracellular materials, such as (1-3)β-d-glucans into 4 to 8 complex layers. These are distinct in structure and composition from the vegetative wall (Catt, 1979; Cavalier-Smith, 1976; Grief et al., 1987), which is organized into seven layers entirely made of HRGPs (Goodenough and Heuser, 1985; Roberts et al., 1972). In liquid cultures, zygote walls connect neighboring zygotes into extensive sheets called pellicle (Minami and Goodenough, 1978; Suzuki et al., 2000).
Two previous reports document that the haploid-to-diploid transition of C. reinhardtii is initiated by the GSM1/GSP1 heterodimer: (1) ectopic expression of GSM1/GSP1 heterodimers in vegetative cells can induce early zygote genes, pellicle formation, and ultimately commitment to meiosis (Lee et al., 2008); and (2) zygotes of gsp1 null (biparental31) plus gametes and wild-type minus gametes cannot activate early zygotic genes nor degrade minus-derived chloroplast DNA, and they undergo extra fusions (Nishimura et al., 2012).
The molecular details of the zygote differentiation program have been explored by a transcriptome analysis (Lopez et al., 2015), which reported ∼600 zygote-specific genes. However, further details of the regulatory networks for the early-zygote developmental pathway, and the functional contexts of the zygote-specific genes for zygote differentiation, are largely unexplored.
The current study analyzes the regulatory network controlling the majority of the early zygote (EZ) transcriptome by considering the mating-induced signaling events and contribution of the GSM1/GSP1 heterodimer. Our results define the predominant role of the GSM1/GSP1 heterodimer, and provide molecular insights concerning how various cellular machineries participate in zygote differentiation.
RESULTS
Experimental Design for Identifying EZ-Specific Genes
To identify which mRNAs derive from bona fide EZ-specific genes, we considered the following in the sample choice for transcriptome sequencing. First, we included haploid and diploid strains in N-replete samples, called TAP1/2 in order to minimize mating-type-dependent or -limited or ploidy-dependent expression biases. Second, genetically diverse strains were used in the two EZ samples combining 45 min and 2 h sets after mating as EZ1/2 in order to minimize strain-dependent biases. We compared our samples with the following published samples representing gene regulatory programs (given in parentheses) that function prior to zygote development: N-replete and N-starved samples (program NS; Miller et al., 2010); plus and minus gametes (program GAM), some being mating-type specific (programs PG and MG; Ning et al., 2013); and gamete samples following dibutyryl cAMP-treatment (program MR, > 350 genes) or subjected to g-lysin treatment (program gL, > 100 genes; Ning et al., 2013). In the end, the zygote-enriched transcripts that are induced by the NS, GAM, MR, or gL programs were excluded to collect bona fide transcripts controlled by the fusion-dependent program (program FD).
To assess the contribution of the GSM1/GSP1 heterodimer to the FD program, we compared zygote samples in a gsp1 null background (bp31) and its complemented strain (bp31C) (Nishimura et al., 2012). Finally, gamete samples ectopically expressing both GSM1 and GSP1 were included in the above multiway comparison as pseudo-zygotes (PZ condition); these cells exhibit zygotic traits such as pellicle formation without undergoing gamete fusion (Lee et al., 2008). Hence, the entire dataset included 15 conditions in 11 genetically diverse strains (Table I; Supplemental Table S1).
Table I. Details of the RNA-seq samples and conditions used in this study. Note that samples are of different genetic backgrounds as well as prepared in four laboratories. Additional details in Supplemental Table S1.
| Name | Description | Strain | Reference |
|---|---|---|---|
| TAP | Midlog phase, liquid culture | I2-152 (iso2, mt-), R9-53 (mt+/mt-[mid-1]) | This paper |
| EZ | Zygote mix, 45 min and 2 hr after mating | CC-125 (mt+) X CC-2342 (mt-), | This paper |
| CC-2937 (mt+) X CC-124 (mt-) | |||
| PZ | Pseudo zygotes induced by ectopic expression of GSM1 and/or GSP1 | N7RM-110 (T-GSM1, mt+/mt-[mid-1]), | This paper |
| M2MP-10 (T-GSM1, T-GSP1, mt-[mid-2]) | |||
| B-TAP | Nitrogen-replete, 48 hr | CC-4691 (cw15, mt+) | Miller et al. (2010) |
| B-NF | Nitrogen-deplete, 48 hr | CC-4691 (cw15, mt+) | Miller et al. (2010) |
| bp31_30 | Zygote mix, GSP1 deletion, 0.5 hr | bp31 (mt+) X CC-124 (mt-) | This paper |
| bp31C_30 | Zygote mix, GSP1 deletion complemented, 0.5 hr | bp31C (bp31, T-GSP1,T-INM1, mt+) X CC-124 (mt-) | This paper |
| bp31_90 | Zygote mix, GSP1 deletion, 1.5 hr | bp31 (mt+) X CC-124 (mt-) | This paper |
| bp31C_90 | Zygote mix, GSP1 deletion complemented, 1.5 hr | bp31C (bp31, T-GSP1,T-INM1, mt+) X CC-124 (mt-) | This paper |
| VNS | Nonsynchronized, midlog, liquid | CC-1690 (mt+) + CC-1690 (mt-) | Ning et al. (2013) |
| VS | Synchronized, L4, liquid | CC-1690 (mt+) + CC-1690 (mt-) | Ning et al. (2013) |
| MA | Minus gametes, db-cAMP treatment, 1 hr | CC-1691 (mt-) | Ning et al. (2013) |
| MG | Minus gametes | CC-1691 (mt-) | Ning et al. (2013) |
| ML | Minus gametes, g-lysin treatment, 1 hr | CC-1691 (mt-) | Ning et al. (2013) |
| PA | Plus gametes, db-cAMP treatment, 1 hr | CC-1690 (mt+) | Ning et al. (2013) |
| PG | Plus gametes | CC-1690 (mt+) | Ning et al. (2013) |
| PL | Plus gametes, g-lysin treatment, 1 hr | CC-1690 (mt+) | Ning et al. (2013) |
Our dataset includes field isolates that are highly polymorphic. Therefore, during the RNA-seq mapping, we allowed three mismatches after considering average synonymous polymorphism among the field isolates (nucleotide diversity at synonymous sites, dS = 0.0317, Flowers et al., 2015). As a result, mapping rates range between 82.1% and 98.2% of the total reads (Supplemental Table S2). Further details of data processing and analysis are given in Supplemental Note S1. Following read mapping, gene expression values normalized as FPKM (fragments per kilobase transcript per million reads) were calculated using the V5.3 genome annotation available at Phytozome (http://www.phytozome.net/, Supplemental Table S3).
Clustering Analysis Identifies EZ-Specific Genes in Two Distinct Pools
Since defining zygote-specific genes relies on the cross-referencing of all other conditions, we performed unguided cluster analysis with 13 conditions, excluding two zygote samples of the gsp1 null strain, averaged from 33 independent samples following FPKM calculations. Prior to the analysis, 5656 (out of 18823) gene models were discarded as Cluster 0 by low expression (having less than 4 FPKM in more than 31 samples), likely representing either pseudo-genes or genes specifically expressed under conditions not included in our dataset. The rest were normalized per model by converting FPKM into percentage values of expression over the sum of average FPKM values under 13 conditions, so that condition-specific expression patterns, rather than absolute expression or fold-change, could be highlighted. The resulting percentage values were used to generate 50 clusters of similar expression patterns by Ward’s hierarchical clustering algorithm implemented in the Genomics workbench software v4.
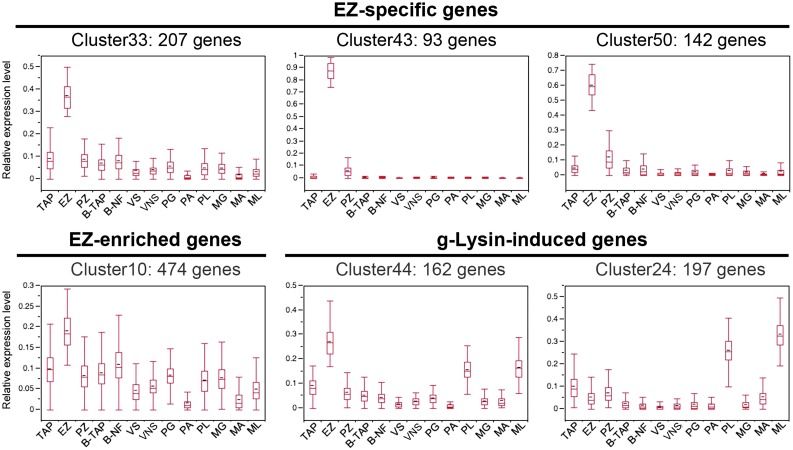
Among the 50 clusters, C33, C43, and C50 show highly up-regulated or nearly exclusive expression in the EZ condition (box plots, Fig. 2, A–C), and these are designated EZ-specific genes. C10 also shows significant up-regulation in EZ, but relatively strong basal expression in all the other conditions as well, and was therefore excluded from the EZ-specific set (Fig. 2D). Of special interest is C44, where similar enrichment is found between EZ and g-lysin-treated gametes (PL and ML conditions), in contrast to C24 where g-lysin-induction is strong but EZ-enrichment is absent (Fig. 2, E–F). Hereafter, these two gene clusters are referred to as gL+EZ (C44) and gL-EZ (C24). Detailed FPKM statistics and description of the 50 clusters are available in Supplemental Tables S4 and S5.
Figure 2.
Identification of coregulated genes in the early zygote transcriptome by hierarchical clustering analysis. Relative expression patterns among 13 conditions are shown for selected gene clusters representing the early zygote-specific clusters (C33, C43, C50, and C10) and the g-lysin-induced clusters (C44 and C24). Box plots show distribution of percentage expression values per condition, calculated relative to the sum of FPKM values in all 13 conditions per gene. Boxes show the median and the 25th and 75th percentile. Whiskers indicate the 10th and 90th percentiles. Details pertaining to the remaining clusters are found in Supplemental Tables S4 and S5.
Out of 444 candidate genes from the three EZ-specific clusters (after discarding redundant gene models), we further refined the EZ set by removing genes with less than a 4-fold induction between EZ and the vegetative condition (TAP; 98 genes) or with maximum expression of less than 12 FPKM in EZ (54 genes). We further eliminated 39 genes whose zygote-specific expression is only evident in one of the two biological duplicates (EZ1 and EZ2 in Supplemental Table S1). The remaining 253 genes are defined as the core EZ genes (EZ-core, Supplemental Table S6).
We cross-examined the EZ-core with previously reported EZ genes (Supplemental Table S7 for EZ-core annotation). EZY1, ZYS1-4, and ZSP1-2 are found in the EZ-core, while EZY2 is absent due to its low expression. Genes EZY3-23 were identified in a microarray study by Kubo et al. (2008), where EZY genes were divided into two classes based on g-lysin-inducibility. Most of their non-g-lysin class genes (EZY4, -7, -8, -9, -15, -17, -18, -19, -20, -22) are found in our EZ-core, while their g-lysin-induced genes are mostly found in gL+EZ (EZY11, -12, -13, -14) and in gL-EZ (EZY5,23). EZY3 and 10 in their non-g-lysin class are found in gL+EZ, with clear g-lysin-inducibility both in our data and in Figure 1 of Kubo et al. (2008). EZY6, -16, and -21 in their g-lysin class show strong expression elsewhere in our dataset and therefore fall in different clusters (EZY6, -16 in C38, and EZY21 in C13). This comparison confirms that our clustering approach collected the majority of EZY genes and precisely distinguished those that are g-lysin-induced.
Another EZ transcriptome study by Lopez et al. (2015) reported a total of 627 zygote-specific gene based on 4-fold up-regulation by RNA-seq, which is more than twice as large as our EZ-core. We found that the difference is mainly due to the inclusion of low-expressed genes (144 models with < 2 FPKM), g-lysin induced genes (60 models), and multiple models for the same gene (30 cases). Of the remaining 393 genes, 82.7% (325 genes) are found among our EZ-specific clusters and C10 (showing EZ up-regulation with strong basal expression). Overall, the comparison to the Lopez dataset shows that our clustering method can sort the complex EZ transcriptome into subclasses and is sensitive enough to collect the majority of differentially expressed genes.
Expression of EZ-Core Genes Is >94% Controlled by the GSP1/GSM1 Heterodimer
To learn which genes in the EZ-core are under the control of GSM1/GSP1 heterodimers, we analyzed differential gene expression between the zygotes generated from the gsp1 null and its complemented strain (bp31 and bp31C zygotes; Nishimura et al., 2012) using a bioconductor package, DESeq2 (Love et al., 2014). A significant increase in bp31C zygotes compared to bp31 zygotes is interpreted to indicate GSM1/GSP1 dependency. In total, 186 genes show >4-fold increased expression (484 genes at >2-fold increase) and only 11 genes show >4-fold decreased expression (115 genes at >2-fold decrease) in bp31C zygotes over bp31 zygotes (Supplemental Tables S8, S9, and S10). Those up-regulated genes are mostly from the EZ-specific clusters (166/186). Of the 253 EZ-core genes, 207 (81.8%) show significant >2-fold up-regulation in bp31C zygotes over bp31 zygotes (FDR < 0.05, Supplemental Table S6). In contrast, a >2-fold enrichment of bp31C zygotes was found for only 158 out of 13,185 genes (1.2%) from the non-EZ clusters (excluding C10/33/43/50, Supplemental Table S11). Of the remaining 46 EZ-core genes showing no significant up-regulation in bp31C zygotes, 34 genes are either poorly expressed in bp31C zygotes or found to be g-lysin-inducible, meaning that nearly all (207/219, 94.5%) of the EZ-core genes are dependent on GSP1 for EZ-expression.
We also examined the differential expression of EZ-core in the pseudozygotes (PZ condition) where the zygote program is activated by ectopic expression of GSM1/GSP1 without gamete fusion. We found that 146 of the 253 EZ-core genes show >2-fold induction (Supplemental Table S6, column Z) over the TAP condition, supporting the dominant role of the GSM1/GSP1 heterodimer in activating EZ-specific genes. The level of up-regulation in the PZ condition is consistently lower than in the EZ condition, likely due to the fact that the zygotic program is nonsynchronously induced in the pseudozygotes following N-starvation (data not shown).
To check the reproducibility of our analysis, 10 of the EZ-core genes at high (>200 FPKM), medium (100–200), and low (<100) expression levels (Supplemental Table S12A) were analyzed by quantitative RT-PCR (qRT-PCR) in three biological replicates of 30 min, 1 h, and 2 h zygote samples and compared to plus and minus gametes (Fig. 3A and Supplemental Fig. S1). All show early induction at 30 min and continued expression until 2 h following gametic fusion in a GSP1-dependent manner.
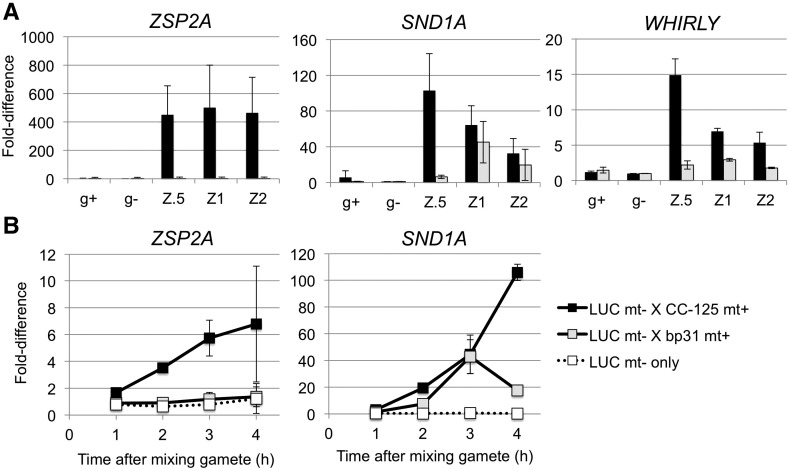
Figure 3.
GSM1/GSP1-dependent activation of EZ-core genes. A, Relative abundance of transcripts in early stage zygotes to minus gametes was analyzed using qRT-PCR. Error bars: sd of three biological replicates. g+: plus gametes; g−: minus gametes; Z.5, Z1, and Z2 represent 0.5 h, 1 h, and 2 h after mixing gametes. Black bar, CC-621 (mt−) X CC-125 (mt+); Gray bar: CC-621 (mt−) X bp31 (mt+). B, Promoter-luciferase reporter assay of two EZ-core promoters. Fold-difference was calculated from cumulative luciferase-based promoter activity during the early zygote development relative to samples without mating. Error bars, sd of three biological replicates.
The EZ-Core Is Regulated at the Transcriptional Level via TGAC Motif
We next asked whether the GSM1/GSP1-dependent gene activation is regulated at the level of transcription given that RNA-seq cannot distinguish between transcriptional and posttranscriptional controls. We constructed reporter transgenes with luciferase coupled to the promoter sequences of two EZ-core genes—g15252 (ZSP2A) with very clean EZ-specific expression and Cre06.g256800 (SND1A) with additional up-regulation in the activated gametes (MA/PA conditions)—and analyzed their expression in wild type and gsp1 null zygotes (Fig. 3B). The ZSP2A promoter drove luciferase activity from 1 h up to 4 h after zygote formation (∼6 fold increase), whereas no activation was found in gsp1 null zygotes. The SND1A promoter showed >100-fold increase in luciferase activity early in wild-type zygotes, whereas a delayed but significant >40-fold increase was found in gsp1 null zygotes. A similar delayed up-regulation of SND1A in gsp1 null zygotes was observed in qRT-PCR analysis (Fig. 3A), indicating that the SND1A promoter is controlled by the mating reaction in addition to the GSM1/GSP1 heterodimer. In the EZ-core, we have found 11/253 additional mating reaction-induced genes based on the average FPKM ratio of the MA/PA conditions over the MG/PG conditions (Supplemental Table S6). However, they all show greater expression in the EZ condition, suggesting that their functions are mainly focused on zygote development.
With the promoter-reporter assay results demonstrating the action of GSM1/GSP1 heterodimer at promoters, we performed in silico analysis to identify putative cis-acting elements in the pool of EZ-specific genes by Amadeus software (Linhart et al., 2008). The analysis identified three 10-base motifs (Supplemental Fig. S2A). The most significant hit, “gtGACacGAC,” shares the same GACnnGAC consensus with the previously reported ZYgote Responsive cis-acting Element candidates (ZYRE; Hamaji et al., 2016; Lee et al., 2008). The second and third motifs also contain in common a single TGAC sequence, possibly representing the core motif recognized by the GSM1/GSP1 heterodimer, which is also the motif repeatedly identified for TALE-class homeobox transcription factors with “WFI50N” (also shared by GSM1) (Knoepfler et al., 1997; Krusell et al., 1997).
Distribution of the three ZYRE motifs was examined to find additional genes under the direct regulation of GSM1/GSP1 heterodimer. Overall, the three motifs are found 6,024 times in 4,468 out of 18,458 genes (24%, Supplemental Table S13). Per-cluster distribution shows significant increase in the three EZ-specific clusters—C33, C43, and C50—as expected (the highest occurrence being 57% in C43), but also in C10 and C44 (gL+EZ) (χ2 test, P < 0.001, Supplemental Fig. S2B, Supplemental Table S14).
We also asked independently whether C10 (showing strong basal expression throughout the examined samples) and C44 (gL+EZ) genes are directly regulated by the GSM1/GSP1 heterodimer using the differential expression analysis between bp31C and bp31 zygotes. C10 includes 74/475 genes that are significantly up-regulated in bp31C zygotes, and C44 includes 9/162 such up-regulated genes (>2-fold, FDR < 0.05, Supplemental Table S11). Comparing bp31C/bp31 ratio distribution per our coexpressed gene cluster indicates that both C10 and C44 are indeed significantly higher in the bp31C/bp31 ratio than the average ratio distribution of all the other clusters combined (Wilcoxon test, P < 0.001, Supplemental Table S11). In conclusion, the two statistical analyses suggest that at least some of the C10 and C44 genes are directly up-regulated by the GSM1/GSP1 heterodimer.
A Subset of g-lysin-Inducible Genes Is Selectively Suppressed by the Zygote Program
g-lysin is released during the mating reaction to remove cell walls and allow gametes to fuse. It has long been known that if the supernatant from mating cells is harvested and applied to vegetative cells or unmated gametes, their walls are also removed and the cells proceed to assemble new walls via activating genes encoding wall proteins and proteins of unknown functions (Hoffmann and Beck, 2005; Kubo et al., 2008; Ning et al., 2013). The walls of vegetative and gametic cells are apparently identical (defined as the VG-wall in contrast to the zygotic wall [Z-wall]; Monk, 1988). Therefore, g-lysin-treated unmated gametes, and newly fused gametes exposed to g-lysin during a natural mating, are a priori expected to up-regulate the same VG-wall transcripts that are up-regulated in g-lysin-treated vegetative cells.
As noted earlier, two clusters containing the g-lysin-induced genes present an interesting anomaly: the C44 genes (gL+EZ) exhibit EZ-enrichment, whereas the C24 genes (gL-EZ) do not show any sign of EZ-induction. Moreover, the bp31C/bp31 zygote expression ratio is significantly less than one for the gL-EZ set (Supplemental Fig. S3; Supplemental Table S11). This suggests that the gL+EZ subset of g-lysin-induced transcripts persists in early zygotes, whereas the gL-EZ subset does not, either because the transcription of the gL-EZ set is switched off and/or because their transcripts are actively targeted for degradation or decay.
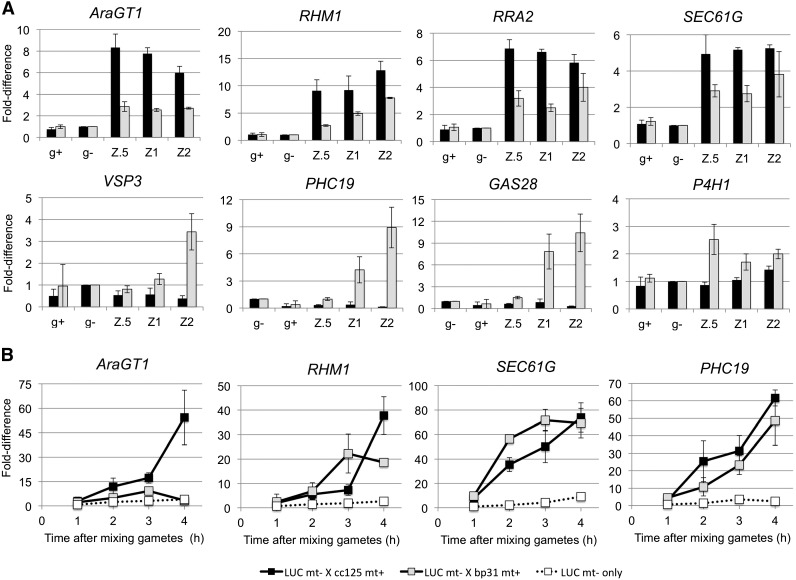
To explore the differential regulation of gL+EZ and gL-EZ genes, we analyzed several g-lysin-inducible genes in wild-type and bp31 (gsp1 null) zygotes by qRT-PCR and the promoter-reporter assay used for the EZ-core analysis. Three gL+EZ genes (RHM1, AraGT1, RRA2) and five gL-EZ genes (SEC61G, VSP3, PHC19, GAS28, P4H1) of various expression levels were selected for the analysis (Supplemental Table S12).
qRT-PCR results show that all three gL+EZ genes, and also SEC61G, were up-regulated 3 to 6-fold in gsp1 null zygotes and 5 to 8-fold in wild-type zygotes (Fig. 4A). In contrast, the other four gL-EZ genes show significant up-regulation in gsp1 null zygotes but almost no changes or decrease in expression in wild-type zygotes (Fig. 4A). Hence our qRT-PCR results document that the differential regulation of the two gene sets is GSM1/GSP1-dependent.
Figure 4.
Differential activation of g-lysin-induced genes during the early zygote development. A, Relative abundance of transcripts in early stage zygotes to minus gametes was analyzed using qRT-PCR. Error bars, sd of three biological replicates. g+: plus gametes; g−: minus gametes; Z.5, Z1, and Z2 represent 0.5 h, 1 h, and 2 h after mixing gametes. Black bar, CC-621 (mt−) X CC-125 (mt+); Gray bar, CC-621 (mt−) X bp31 (mt+). B, Promoter-luciferase reporter assay of two EZ-core promoters. Fold-difference was calculated from cumulative luciferase-based promoter activity during the early zygote development relative to samples without mating. Error bars, sd of three biological replicates.
The exceptional SEC61G gene actually shows comparable RNA-seq data to our qRT-PCR results, where its expression in bp31C zygotes is 1.5-fold higher than in bp31 zygotes, similar to other gL+EZ genes (Supplemental Table S12). We manually inspected RNA-seq data of the gL-EZ set (172 genes) and identified 16 genes showing comparable expression between the EZ and ML/PL conditions (less than 2-fold difference) as potential gL+EZ genes (noted in the column K of Supplemental Table S17), which contains five out of seven ER-residing secretion-related genes including SEC61G in the original gL-EZ set.
We next sought to learn more about the g-lysin program. We first used our promoter-reporter assay to test if the g-lysin program operates at the transcriptional level. We constructed transgenic strains harboring promoter-reporter constructs for AraGT1, RHM1, and SEC61G (representing gL+EZ) and for PHC19 (representing gL-EZ); all showed consistent inducible reporter expression when subjected to exogenous g-lysin exposure, indicating that the g-lysin program regulates both sets via transcriptional activation (Supplemental Fig. S4 for the promoter cloning schemes and Supplemental Fig. S5 for g-lysin inducibility test). We then asked how the g-lysin program is affected during zygote development by examining those g-lysin inducible promoters. The RHM1, SEC61G, and PHC19 promoters showed comparable up-regulation during the first four hours after gametic fusion in both wild-type and bp31 (gsp1-null) zygotes (10-15-fold in 3 h zygotes, Fig. 4B). The AraGT1 promoter showed stronger up-regulation in wild-type zygotes, although weak but clear up-regulation occurred in gsp1 null zygotes (3-fold in 3 h zygotes, Fig. 4B).
The promoter assay results document that these test genes, and by extension the g-lysin program, operates normally during zygote development and hence is not inhibited by the GSM1/GSP1-dependent program. Therefore, the observed paucity of transcripts produced by gL-EZ genes in wild-type zygotes suggests the existence of a posttranscriptional mechanism for their down-regulation, one that does not affect gL+EZ genes.
Gamete-Specific Genes Are Not Apparently Suppressed by the GSM1/GSP1 Heterodimer during Early Zygote Development
The zygote developmental program is expected to be accompanied by a negative control of gametic features, such as agglutinins and mating structures, to prevent additional gametic fusion. We examined expression of known gamete/sex-specific genes in our RNA-seq dataset (Supplemental Table S15). Most known minus-specific genes in C18, including SAD1, GSM1, HAP2, and MTD1, are expressed at comparable levels in gametes and early zygotes. The same trend is true for the plus-specific genes residing in MTL+ such as FUS1 and MTA1. This trend indicates that the gamete-specific programs are not apparently suppressed by the zygote program. In contrast, the plus-specific genes residing outside MTL+, such as GSP1 and SAG1, showed rapid down-regulation as early as 30 min zygotes. Earlier examination of GSP1 and SAG1 expressions by qRT-PCR also showed rapid turn-off in early zygotes (Nishimura et al., 2012). Interestingly, SAG1 down-regulation was found equally in both bp31C and bp31 zygotes, suggesting that the down-regulation of the plus-specific genes may be triggered by cellular fusion rather than the GSM1/GSP1 heterodimer.
Prediction of Functional Contexts of the Early Zygote Transcriptome
Our analysis has revealed that EZ-core genes and the gL+EZ set of g-lysin-induced genes constitute the main body of the early-stage zygote transcriptome dependent on the GSM1/GSP1 heterodimer. Although 203/253 members of the EZ-core were previously identified as zygote-specific genes by Lopez et al. (2015) and 30/159 of the gL+EZ as g-lysin induced genes by Ning et al. (2013), these earlier studies offered annotations but not detailed analyses. Therefore, we have gone on to analyze these coexpressed gene clusters, focusing on which functional systems are enriched in the EZ transcriptome and assessing the differences among EZ-core/gL+EZ/gL-EZ gene contents.
To analyze molecular details, we performed exhaustive manual annotation as described in “Materials and Methods,” and categorized the genes into seven major functional groups relevant to the zygote differentiation processes: cell wall synthesis and assembly, organelle-targeted, miscellaneous metabolism including transporters, putative gene regulation, signal transduction, secretion/endomembrane/cytoskeleton system, and ubiquitin system (detailed annotation results in Supplemental Tables S7, S16, and S17). Figure 5 and Supplemental Table S18 summarize the functional categories among the three coregulated gene sets. In the following, we focus on Z-wall assembly where our transcriptome has provided new insights and putative molecular players.
Figure 5.
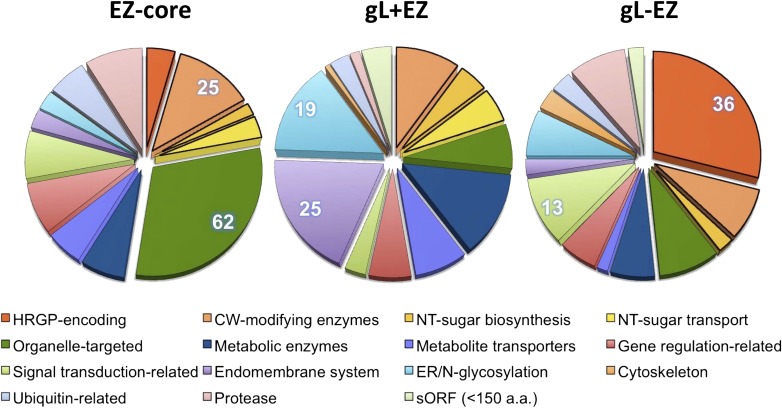
Distribution of annotated EZ-core/gL+EZ/gL-EZ genes in functional categories related to cellular differentiation (excluding the unknowns). The categories listed in the legend represent, in order, the categories in the pie charts starting from midnight and proceeding clockwise. Cell wall-related categories are marked by thick outlines. Left, EZ-core (n = 204); middle, gL+EZ (C44) (n = 129); right, gL-EZ (C24) (n = 113). Number of genes in the top two categories are shown in pie charts. Annotation details are found in Supplemental Tables S18, S19, and S20.
Molecular Components of the Zygotic Cell Wall Assembly
The largest annotated functional category associated with EZ-core/gL+EZ/gL-EZ is cell wall-related genes (46/253, 25/159, and 49/172, respectively), including (1) HRGP-encoding, (2) HRGP-modifying enzymes, and (3) nucleotide-sugar metabolism-related, including sugar-converting enzymes and nucleotide-sugar transporters (Fig. 5).
Cell Wall Constituents: HRGPs and Products of Glycosyl Transferases
Previous studies (Grief et al., 1987; Minami and Goodenough, 1978) have documented that the zygotic wall and vegetative wall consist of distinct sets of glycoproteins, primarily HRGPs, recognized by domains dominated by Pro (P) and Ser (S), often in repeated motifs. While the gL+EZ contains no HRGP-encoding genes, the EZ-core and gL-EZ include unique sets of HRGP-encoding genes. Of particular interest is the presence of a C-terminal hydrophobic domain in six out of the nine HRGPs in the EZ-core (Supplemental Table S7). Such a hydrophobic tail serves as either a transmembrane domain or a signal for GPI-like lipid-modification. In silico tools predicting GPI-like lipid modification signals (PredGPI and BIG-PI; Borner et al., 2003; Pierleoni et al., 2008) showed that five of the six candidates likely function as GPI-anchor signal (Supplemental Table S19). In contrast, none of the 36 HRGPs in gL-EZ displays a hydrophobic C-terminal tail (Supplemental Table S17).
A GPI-anchor motif is often associated with plant AGP (arabinogalactan protein)-type HRGPs but has not yet been experimentally characterized in Chlamydomonas. GPI-type lipid-modification was predicted for a fasciclin-like HRGP, Algal-CAM, in the Volvox vegetative wall (Huber and Sumper, 1994), introducing the idea that GPI-anchors or similar structures are likely present in algal cell walls. In support of this idea, most of the GPI-anchor biosynthesis genes are found in the Chlamydomonas genome, of which two enzymes functioning at rate-limiting steps, PIG-O and PGAP1, are up-regulated in early zygotes (C33; Supplemental Table S20).
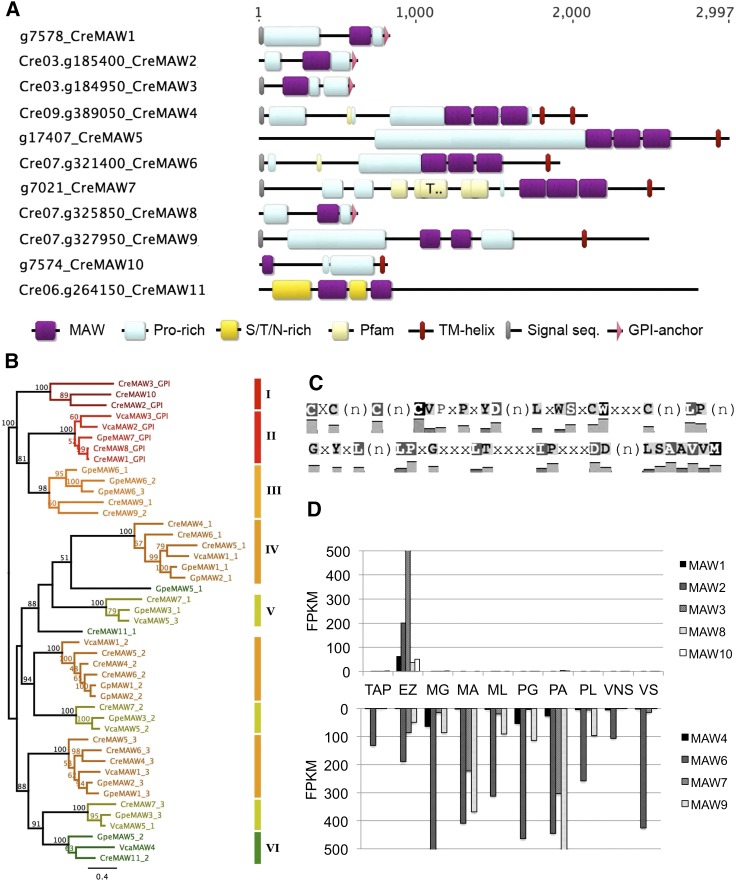
We found a novel six-Cys-containing homology domain among all the EZ-core HRGPs with C-terminal anchors except Zsp1, which we named MAW (membrane-associated wall protein) (Fig. 6C for the conserved amino acids; Supplemental Fig. S6 for the domain sequence alignment). We searched for additional MAW family members by HMMER3 (Mistry et al., 2013), and identified 11 Chlamydomonas members, five members in Volvox carteri, and seven members in Gonium pectorale (Supplemental Table S19). We could not find MAW homologs in any other published green algal genome sequences. Chlamydomonas MAW proteins have up to three MAW domains, and the majority (10/11) contain Pro-rich domains of PPSPX, PPX, and/or PS repeats, characteristics of HRGP shafts (Fig. 6A; Lee et al., 2007). Intriguingly, all MAW members identified are found to possess either one or two transmembrane domains or a putative GPI-anchor signal at the C-terminus (Supplemental Table S19), indicating that the MAW domain associates with plasma membrane-anchored cell wall proteins in the Volvocales.
Figure 6.
Identification of the MAW (membrane-anchored cell wall protein) family encoding HRGPs with a C-terminal hydrophobic domain. A, Domain structure of MAW family proteins in Chlamydomonas. B, Bayesian phylogeny of MAW domains among Volvocacean MAW proteins. Predicted GPI-anchored MAW proteins are found in clades I and II. C, Cys-rich MAW domain consensus sequence. Domain sequence alignment is found in Supplemental Figure S6. D, Differential expression of MAW family genes in Chlamydomonas based on average FPKM values among nine conditions. Upper graph, EZ-specific genes. Lower graph, Non-EZ-specific genes. Due to very low expression, CreMAW5 and CreMAW11 are not included in the graph.
Phylogenetic analysis of the MAW family reveals six well-supported clades among Volvocalean species (Fig. 6B). Clades I and II are distinguished as having a putative GPI-anchor signal at the C terminus (except CreMAW10). Since all Chlamydomonas genes in Clade I and II are exclusively expressed in early zygotes, we propose that they may serve as key organizers of Z-wall development in Volvocacean algae. Clades IV and V share triple MAW domain configurations and have diverse expression patterns in Chlamydomonas (Fig. 6D).
The absence of HRGP-encoding genes in the gL+EZ set that contribute both to the VG- and Z-walls suggests that VG-wall and Z-wall may be assembled with exclusive sets of materials. We therefore asked what features are unique to the VG-wall, taking the gL-EZ set as its representative. The gL-EZ includes 36 putative HRGP-encoding genes, 20 of which are pherophorins, the largest known HRGP family in the Volvocales. The Chlamydomonas genome encodes 73 pherophorin genes, 28 of which are up-regulated in g-lysin-enriched clusters (C24/8/34), while 10 are found in gamete- and mating reaction-induced clusters (C22/26/41) but none are in EZ-specific clusters (C10/33/43/44/50) (Supplemental Table S21). Hence, we propose that pherophorins are hallmark components of the VG-wall.
HRGP-Modifying Enzymes
HRGPs are made from nascent polypeptides that undergo extensive O-glycosylation on serines and hydroxylated prolines (HYPs) within Pro-rich segments during ER-Golgi trafficking. Therefore, production of HRGPs requires prolyl 4-hydroxylases (P4Hs) and various glycosyltransferases. In land plants, distinct glycosyltransferase families are known to build either unbranched O-glycans found in extensins or complex O-glycans found in AGPs (for review, see Hijazi et al., 2014).
Both Ser- and HYP-glycosyltransferases have been recently identified in plants from studies using plant extensins as substrates: SGT (Peptidyl Ser alpha-Galactosyltransferase) and HPAT (Hyp O-arabinosyltransferases) belong to the GT8 family and catalyze initial O-glycosylation on Ser or HYP (Ogawa-Ohnishi et al., 2013; Saito et al., 2014), and subsequent arabinosylation is catalyzed by members of two GT77 subfamilies, RRA and xyloglucanase-113 (Egelund et al., 2007; Gille et al., 2009). To learn whether HRGP modification capacity or specificity is regulated during wall assembly, we prepared an extensive catalog of the Chlamydomonas homologs for the known HRGP modifying enzymes (Supplemental Table S22). A majority of the HRGP-modification-related genes turned out to be up-regulated by the g-lysin- or EZ-program (6/17 P4Hs, and 15/16 HRGP-O-glycosylation enzymes), of which four P4Hs and four O-glycosylation genes are specifically up-regulated during zygote development. In contrast, we did not find a concerted up-regulation of N-glycosylation-related genes by the g-lysin- or EZ-program (6/31 are up-regulated, Supplemental Table S23).
Chlamydomonas HRGPs are also decorated by branched glycans (ex. GP1, Ferris et al., 2001), whose core structure consists of Hyp1-Ara1 - 3 that are mono- or di-galactosylated. By contrast, the complex glycans associated with AGPs in land plants consist of a Hyp1-Gal1-3 core, perhaps explaining the absence of any Chlamydomonas homologs to currently identified glycosyltransferases associated with AGP-glycosylation that belong to the GT14, GT29 and GT31 families (Geshi et al., 2013; Liang et al., 2013; Nguema-Ona, 2014; Ogawa-Ohnishi and Matsubayashi, 2015).
EZ-specific and g-lysin-induced clusters include many putative glycosyltransferases, mainly of GT47 and GT90 members. Characterized GT47 members of Arabidopsis are reported to add sugars to glycans during pectin and hemicellulose biosynthesis (Geshi et al., 2011); however, pectin or hemicellulose has not been found in the Chlamydomonas vegetative wall. Phylogenetic analysis of GT47 family including 47 Chlamydomonas and 39 Arabidopsis members shows a peculiar dichotomy wherein all Chlamydomonas members are related to one Arabidopsis member, At3g57630, which carries an EGF domain, and the remaining Arabidopsis members form separate clades (Supplemental Fig. S7). Six GT47 genes are exclusively expressed in early zygotes, and three are found in g-lysin-inducible clusters (C8 and C24).
GT90 proteins are known as multifunctional xylosyltransferases on mannans when involved in capsule formation in fungi, and as Ser-O-glucosyltransferases when involved in Notch signaling in Metazoa (Acar et al., 2008; Reilly et al., 2009), whereas the role of GT90 in plants has not been elucidated. GT90 phylogeny shows great diversity and Chlamydomonas-specific expansion in a number of clades in comparison to Volvox proteins (Supplemental Fig. S8). Both EZ- and g-lysin-induced programs include at least one member of each clade (in red and in blue), suggesting the involvement of GT90 members in both VG- and Z-wall assemblies (Supplemental Fig. S8).
Pellicle Formation
Pellicle formation in liquid culture begins with wall-to-wall adherence by early-stage zygotes, followed by extensive build-up of HRGP fibers (Minami and Goodenough, 1978; Suzuki et al., 2000). Sulfated polysaccharides or glycoproteins are known to be involved in cell-cell adhesion and to be critical for ECM structural integrity in Metazoans and multicellular Chlorophytes, including Volvox carteri (Domozych and Domozych, 2014; Misevic et al., 2004). The EZ-core contains three genes encoding sulfotransferases with distant homology to Metazoan proteins involved in chondroitin and heparan sulfate biosynthesis; we therefore named them CSR (carbohydrate sulfotransferase-related; Supplemental Table S22E). The CSR phylogeny shows seven clades conserved in Volvocacean algae, and two of the three zygote-specific clades are specifically expanded in the multicellular Volvox, possibly participating in cell-cell adhesion (Supplemental Fig. S9).
The zygote wall components become cross-linked into a matrix that resists most solubilizing agents (e.g. SDS), and evidence for H2O2-mediated crosslinking and isodityrosine (IdT)-linkages in the Chlamydomonas vegetative and zygote wall has been presented in Waffenschmidt et al. (1993), where Tyr-Gly-Gly (YGG) was proposed as a IdT motif. In the EZ-core, we found many YGG motifs in MAW10 with a C-terminal transmembrane domain. A member of the candidate protein family involved in H2O2-mediated cross-linking, glyoxal/Gal oxidase (GOX), is among the most highly expressed genes in the EZ-core. This family has been implicated in cross-linking of plant cell wall constituents such as pectin (K. Šola, E. Gilchrist, M.-C. Ralet, C.S., Mansfield, and G. Haughn, personal communication) and in direct defense against pathogens via its expected catalysis of H2O2 generation (Zhao et al., 2013). GOX genes are present in Chlamydomonas as a large gene family of 19 members, five of which are up-regulated by the g-lysin- or EZ-program, suggesting their general involvement in cell wall assembly. An interesting feature unique to the Volvocacean GOX is the presence of N-terminal Pro-rich domains (15/19) with repeat patterns such as PPSPX, PS, or PX, indicating that these GOX proteins are likely part of the cell wall network (Supplemental Table S22F). Phylogenetic analysis of the GOX family shows a dichotomy similar to the GT47 phylogeny, where Arabidopsis and Chlamydomonas have undergone independent expansion (Supplemental Fig. S10).
Support of Metabolic Demands of Glycosylation Precursors
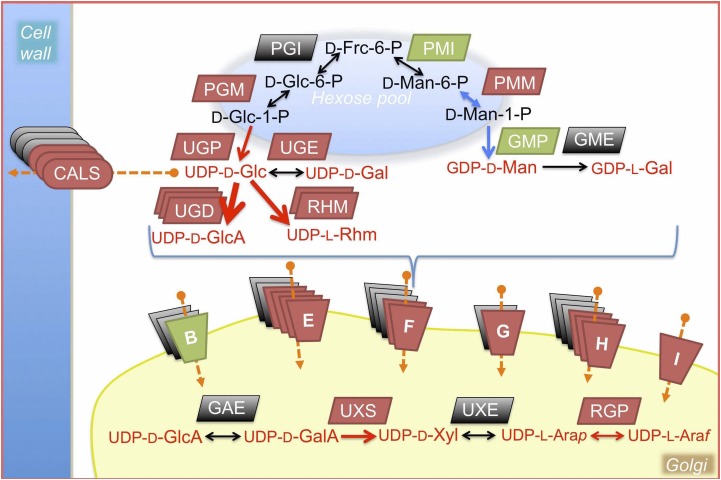
To support increased HRGP-secretion for zygote wall formation and reported extracellular callose accumulation (Catt, 1979; Grief et al., 1987), the cytosolic hexose pool would be in great demand during zygote wall assembly. Hexoses must be activated via UDP or GMP nucleotidylation, and the resultant nucleotide sugars then feed glycosyltransferases (Seifert, 2004). A total of 20 genes involved in nucleotide-sugar biosynthesis and 38 putative sugar transporters, mostly in triose-phosphate transporter/nucleotide-sugar transporter (TPT/NST) family, are found in the Chlamydomonas genome (Supplemental Tables S24 and S25), allowing us to propose a model for the flow of sugars for glycosylation reactions (Fig. 7). A notable difference between the Chlamydomonas model and its plant counterpart is the absence of GMD/GER and AXS to produce GDP-l-Fuc and UDP-d-Api, since homologs of these genes have not been identified (Supplemental Table S24). According to our model, the sugar flow via UDP-d-Glc up to UDP-l-Ara would become highly activated during EZ development by up-regulation of both nucleotide-sugar transporters (13/34) at nonplastidic locations and sugar-converting enzymes for UDP-l-Ara (red arrows in Fig. 7). Of the NST clades, we noticed size expansion in the clades D, E, and H in both Chlamydomonas and Arabidopsis, where 12 genes (out of 22) are found to be up-regulated in the EZ condition and four genes are up-regulated by g-lysin treatment, suggesting that high demands of glycosylation for zygote wall formation may have driven the expansion of NST family in Chlamydomonas (Supplemental Table S25, Supplemental Fig. S11).
Figure 7.
Predicted flow of cytosolic hexoses into nucleotide-sugar metabolism in support of glycosyl transferases up-regulated during the early zygote development. Cytosolic hexose pool is supplied by gluconeogenesis and export from the chloroplast. Arrows show directional or bidirectional conversions. Red arrows, predicted sugar flow changes in early zygotes, resulting in a large increase in available nucleotide-sugars both in the cytosol and in Golgi via NST-family transporters. Blue arrows, predicted sugar flow increase into GDP-d-Man and GDP-l-Gal in the cytosol. Red metabolites, forms usable by glycosyl transferases. Shaded symbols represent genes. Brown, early zygote-specific or up-regulated (EZ or gL+EZ). Green, g-lysin induced (gL-EZ). Gray, found in other clusters. Details of these genes are found in Supplemental Tables S24 and S25. Putative nucleotide-sugar transporters in ER/Golgi are found in six classes (B-I excluding C and D classes known to be localized to the plasma membrane; Supplemental Table S26). Frc, Fructose; Man, Mannose; Glc, Glucose; Gal, Galactose; Rhm, Rhamnose; GlcA, Glucuronic acid; GalA, Galacturonic acid; Xyl, Xylose; Arap, Arabinose-pyranose; Araf, Arabinose-furanose.
In addition to the flow of hexose into HRGPs via glycosylation in the Golgi, Chlamydomonas zygotes have another major sink for the hexose pool, callose deposition, contributing up to 30% of all extractable sugars in the zygotic wall (Catt, 1979). Two callose synthases and one callose synthase-like gene (GT48) that are exclusively expressed in early zygotes are likely enzymes for such a huge extracellular build-up (Supplemental Table S26). So far, only trace amounts of glucose have been detected in the Chlamydomonas VG-wall (Catt, 1979).
DISCUSSION
Our transcriptome analysis comparing wild-type and gsp1 null zygotes shows that the EZ transcriptome is mostly, if not entirely, under the regulation of the GSM1/GSP1 homeobox heterodimer, positively at the transcriptional level for the zygote-specific genes, and negatively at the posttranscriptional level for the VG-wall specific genes. The combination of the two regulatory modules achieves differential regulation of the EZ-core/gL+EZ/gL-EZ gene sets, whose annotation points to molecular differences of the cell-type specific wall assemblies as a hallmark of cellular differentiation.
Nonguided Clustering Reveals Crosstalk between the EZ and g-lysin-Induced Programs
Our study design explicitly considers strain variability and multiway comparison by hierarchical clustering of 13 different conditions and diverse genetic backgrounds. Since zygotes are naturally of mixed genetic make-ups, their study needs to consider genetic variation that may bias the analysis. Therefore, we aimed to identify reliable and robust sets of genes (such as the EZ-core) that warrant more thorough examination. Nonguided clustering provided opportunities to discover expected and unexpected crosstalk among the five transcriptional programs: plus- and minus-specific gametogenesis, mating reaction-induced, g-lysin-induced, and EZ-specific. Such a blinded approach has documented that the EZ transcriptome includes a major contribution from the g-lysin-induced program, but not the other mating-associated programs.
Regulatory Network for the Assembly of Two Wall Types
The VG- and Z-wall assembly programs expectedly share factors involved in HRGP modification and secretory routes for HRGPs, since both walls consist mainly of HRGPs. Such shared factors are clearly represented by the gL+EZ genes that are expressed equally well during the VG- and Z-wall assembly. Following our prediction, the gL+EZ cluster includes 42 ER/secretion-related genes and 26 HRGP-modifying- and nucleotide-sugar metabolic enzyme-encoding genes, but no obvious HRGP-encoding genes (Fig. 5, Supplemental Table S16).
Our study defines the EZ-core and gL-EZ genes that are exclusively expressed during the Z-wall or VG-wall assembly, respectively, reflecting the fact that the VG wall and the Z wall are very different in their organization and properties. Accordingly, the EZ-core and gL-EZ genes include distinct sets of HRGP-encoding genes—for example, GPI-anchored HRGPs for the EZ-core and pherophorins for the gL-EZ—indicating that they are indeed the Z- and VG-wall-specific cohorts.
Interaction between EZ- and g-lysin programs is highly selective, affecting about half of the g-lysin-induced genes. Follow up analysis of three g-lysin-inducible promoters provided compelling evidence for persistent expression from these promoters in early zygotes; we also document a subsequent posttranscriptional repression of > 150 genes that is GSM1/GSP1-dependent. Hence, the early zygote appears to be programmed to discard the transcripts encoding VG-wall structural proteins that might interfere with the Z-wall assembly, but to maintain production of enzymes for glycosylation and other modifications for cell wall proteins employed in both VG-wall and Z-wall construction.
The posttranscriptional mechanisms employed by the early zygote to recognize and discard VG-wall-specific transcripts remain to be investigated. One possibility is suggested by the presence of Tudor Staphylococcal Nuclease (TSN) in the gL+EZ set (Supplemental Table S16). The TSN homolog in Arabidopsis has been reported to be involved in regulating the stability of the mRNAs that are targeted to the ER for cotranslational translocation (Sundström et al., 2009); more recently, its regulatory action has been found to depend on its nuclease domain and on selective decapping of mRNAs at the stress granule (Gutierrez-Beltran et al., 2015), where TSN operates together with a decapping enzyme like Dcp1 and a 5′-3′ exonuclease like Xrn1, proteins that are also conserved in Chlamydomonas.
Departure from Unicellular Contexts
In flowering plants, the haploid-to-diploid transition is followed by sporophyte development, giving rise to multiple tissue and organ types, whereas the haploid phase is dramatically shortened, entailing only a few mitotic divisions. In the moss Physcomitrella patens, key players involved in the haploid-to-diploid transition include KNOX-family and their putative binding partners BELL-family transcription factors, necessary for spore maturation and suppression of the haploid program, and Clf /Fie as a component of the PRC2 that suppresses the diploid program (Horst et al., 2016; Mosquna et al., 2009; Okano et al., 2009; Sakakibara et al., 2008; 2013). Since GSM1 is the only KNOX-family transcription factor in Chlamydomonas (Lee et al., 2008), it is of great interest to understand how the GSM1/GSP1-dependent program evolved into the KNOX-dependent regulation of land-plant haploid-to-diploid transition. A recent study has shown that the two divergent KNOX subfamilies in land plants function in antagonistic roles: type II promotes differentiation programs, whereas type I counteracts type II by promoting the undifferentiated state of stem cells (Furumizu et al., 2015). A plausible scenario, therefore, is that the type II KNOX retains the ancestral differentiation-triggering function of GSM1 and that type I evolved to delay the differentiation program leading to meiosis, thereby allowing the emergence of a multicellular diploid phase. Recently, Frangedakis et al. (2016) reported that the type I and II KNOX already diversified in charophyte algae before the evolution of multicellular diploid generation. The dual-action mechanisms of the GSM1/GSP1 heterodimer described in this study, controlling both positive gene expression and posttranscriptional mRNA degradation, may have opened up an evolutionary path for the KNOX-based GRN to become bifurcated by subfunctionalization following gene duplication.
MATERIALS AND METHODS
Strains and Culture Conditions
Details of strains used in RNA-seq are found in Supplemental Table S1. CC-numbered strains are available in the Chlamydomonas Resource Center at University of MN (www.chlamycollection.org). JL28 (nic7; mt−) was generated by mating between CC-125 and CC-2663 (nic7; mt−). Pseudo-zygote strains are described in Lee et al. (2008). bp31/bp31C strains are described in Nishimura et al. (2012). Plate cultures were maintained in continuous light on Tris-acetate-phosphate (TAP) medium (Harris, 1989) solidified with 1.5% Bacto agar. Liquid cultures were inoculated at 5 × 104 cells/mL concentration and maintained in continuous light on TAP medium until harvest at 5 × 106 cells/mL.
Sample Preparation and RNA-Seq Library Construction
Vegetative cells in logarithmic growth were obtained in TAP-liquid culture at 22°C in constant light under 40 μmol/m2/sec and harvested at 1 × 106 cells/mL. Early zygotes were obtained from mating mixtures of fully differentiated gametes prepared from 7-d-old TAP-plate culture and incubated in NF-TAP for 5 h until more than 90% of cells became active gametes. Once plus and minus gametes were mixed in equal number (1E+7 cells), samples were harvested at 45 min and 2 h after mixing and combined during the RNA-seq library preparation considering slightly low mating efficiencies (50-80% at 1 h). Pseudozygotes from JL-N7RM-110 (T-GSM1, mt+/[mt−;imp11]) and JL-M2MP-10 (T-GSM1, T-GSP1, mt−;mid-2) were obtained from 7-d-old TAP-plate culture and incubated in NF-TAP for 24 h until the majority of cells have formed large aggregates (pellicle) in the medium.
RNA Extraction and Sequencing
Total and poly(A)+ RNAs were extracted by methods described earlier (Lee et al., 2008). Poly(A)+ RNAs were reverse-transcribed using random primers and dT(15) primers. Resulting cDNAs were sheared into 300- to 400-bp fragments and used to prepare six mRNA sequencing libraries according to the manufacturer’s protocol (cat no. RS-930-1001, Illumina). Libraries for TAP/EZ/PZ samples were sequenced on the Illumina GAIIx for 76 bp single-end cycle except TAP duplicate no. 2 for 36 bp single reads, following the manufacturer's standard cluster generation and sequencing protocol. bp31/bp31C zygote samples were sequenced for 36 bp single reads.
RNA-Seq Analysis
All the new and published RNA-seq reads (Miller et al., 2010, Ning et al., 2013) were mapped with Tophat2 (Kim et al., 2013) to the v5 genome combined with the mating-type minus sequence obtained from CC-2290 (GenBank no. GU814015, Ferris et al., 2010) with the allowance of three mismatches. The mapping details are given in the Supplemental Table S2. Normalized FPKM values were calculated accordingly to our modified V5.3 annotation that contains extra mating-type minus genes defined in GU814015 by cufflinks ver. 2.11 (cufflinks -I 2000 -no-faux -G, Trapnell et al., 2012), which performs correction for multihits by dividing a pool of multihits into corresponding genes according to their unique hit distribution. Following our analysis, an updated annotation (V5.5) was released and also used to calculate FPKMs of our mapped reads; however, we found no significant effect in our zygotic transcriptome analysis and decided to utilize V5.3 information in this study (see Supplemental Note S2 for details.)
Hierarchical Clustering Analysis
Following FPKM calculation, we combined 33 samples of FPKM data (excluding bp31/bp31C zygote samples) representing 13 conditions in triplicates or duplicates (for 6 conditions) and performed clustering analysis following the method described in Ning et al. (2013). Prior to the analysis, 5656/18823 (as Cluster 0) gene models were discarded by the low-expression criterion (32/33 with less than 4 FPKM). The rest with 33 data points were averaged per 13 conditions and normalized by converting the average FPKM values into percentage values of expression over the sum of 13 average FPKM values. The resulting percentage values were used to generate 50 clusters by Ward's hierarchical clustering algorithm applied to Euclidean distances using Genomics Workbench v4 (see Supplemental Table S3 for clustering results, Supplemental Table S4 for clustering statistics, and Supplemental Table S5 for cluster descriptions based on their bias patterns).
In Silico Promoter Motif Analysis
Sequences combining 1500 bp upstream and 500 bp downstream to the transcription start site were compiled using a perl script (http://github.com/minillinim/gff2fasta) and assigned to 51 bins matching their coexpression clusters. Statistical enrichment of 10-base motifs was analyzed by AmadeusPBM 1.0 (Linhart et al., 2008) with default parameters.
Gametogenesis and Mating
Gametogenesis was induced as described (Ferris and Goodenough, 1997). Unless otherwise stated, cultures were grown on TAP plates for 6 d, resuspended in TAP, and aliquoted into two tubes, one for gametogenesis and the other for control. Gamete samples were washed once and resuspended in nitrogen free-TAP (NF-TAP) to a final 5 X 107 cells/mL, while control samples were washed and resuspended in TAP to the same concentration. The resulting liquid cultures were incubated under continuous light in 24-well plates. Four hr later, the cells in NF-TAP displayed vibrant agglutinating activity when mixed with tester gametes of opposite mating type, the sign of gametic differentiation, but not the cells in TAP medium. Mating efficiency was calculated based on QFC formation as previously described (Goodenough et al., 1993). Zygotes were produced by mating equal numbers of plus and minus gametes.
Transformation by the glass bead method
To introduce promoter-reporter constructs, nic7 mutant cells (mt-) were grown in liquid TAP containing 4 μg/mL nicotinamide until they reached a density of 5 X 106 cells/mL. Approximately 1 X 108 cells were harvested and incubated with 2.5 mL g-lysin for 1-2 h until > 90% had lost cell walls, as monitored by the sensitivity of cells to lysis in 0.02% NP-40. Protoplasts were harvested in a 10-mL glass tube and resuspended in 300 μL TAP, to which were added 0.3 g glass beads (0.5 mm) and 100 μL 20% PEG-8000 containing 0.5 - 2 μg linearized transforming DNA derived from pNic7.9 (Ferris, 1995). Cells were vortexed for 25 s at the top setting of a Vortex-Genie mixer, washed twice with 10 mL TAP, resuspended in 300 μL TAP, mixed with 2.5 mL 0.5% Top Agar in TAP (melted at 45°C), and poured onto TAP supplemented with 15 ppm of 3-acetylpyridine (Sigma), which is toxic to nicotinamide requiring cells (Harris, 1989). Colonies of Nic+ transformants became visible after 3-5 d. g-lysin, which removes Chlamydomonas cell walls (Kinoshita et al., 1992), was prepared as described in Lee at el. (2008).
Real-Time qRT-PCR
To analyze the relative abundance of transcripts using qRT-PCR, DNase-treated 5 μg of total RNA was reverse-transcribed with Superscript II RT-PCR kit (Invitrogen). Primers were designed to produce PCR amplicon lengths of 100–150 bp. qRT–PCR was performed in optical 96-well plates using Bio-Rad CFX96 Real-Time PCR systems. Reactions were performed in a final volume of 20 μL containing 10 μL of 23 SYBR Green Master Mix, 0.5 μm each primer, and 10 ng of cDNA. PCR conditions were as follows: 30 s at 95°C, followed by 45 cycles of 5 s at 95°C and 30 s at 60°C. Fluorescence threshold data (Ct) were analyzed using Bio-Rad CFX Manager software (version 2.0). Quantitative reactions were done in duplicate and averaged. Relative expression levels in each cDNA sample were normalized to the RACK1 reference gene under the same conditions. Three biological replicates were averaged for quantitative analysis. Primers used in Real-time qPCR are found in the Supplemental Table S12.
Cloning of Promoters
To isolate promoters of the genes of interests, genomic DNAs of CC-125 strain were used in PCR amplification. 5′ primers are tagged by XbaI and 3′ primers are tagged by EcoRI. PCR amplicons were cloned into pGEM-T Easy vector and fully sequenced for verification. Resulting XbaI and EcoRI fragments were ligated into XbaI and EcoRI of linearized pNIC7-GL that contains a Gaussian Luciferase ORF derived from pHsp70A/RbcS2-cgLuc (Ruecker et al., 2008) in pNic7.9 (Ferris, 1995). The resulting constructs were used for transformation into nic7 mutant strains.
Promoter-Luciferase Assays
Promoter constructs were transformed into mt− Chlamydomonas cells (nic7, mt−, JL28). Randomly picked 24 colonies were screened for secreted luciferase activity with or without g-lysin treatment for C24 and C44 promoters and with or without mating to plus gametes for C43 and C50 promoters. Two representatives were chosen for further studies. Luciferase assays were done under the following conditions to test their effects on promoter activation. Luciferase-containing transformants were prepared as gametes and mixed with CC-125 or bp31 as mating partners. Hourly accumulation of luciferase activity in fresh culture media was measured along with unmated gametes.
Annotation of EZ-Specific and g-lysin-Inducible Genes
Collected protein sequences were initially analyzed by three approaches: (1) reciprocal best hits with Arabidopsis proteins for retrieving available functional annotation from Arabidopsis orthologs (TAIR v10); (2) Interpro (http://www.ebi.ac.uk/interpro/, Finn et al., 2017) search for collecting functional/homology domain information; and (3) PredAlgo (https://giavap-genomes.ibpc.fr/cgi-bin/predalgodb.perl?page=main, Tardif et al., 2012) and/or TargetP (http://www.cbs.dtu.dk/services/TargetP/, Emanuelsson et al., 2007) search for N-terminal leader sequences. This initial analysis allowed us to choose five major categories for further investigation: Nucleotide-sugar biosynthesis/transport (Seifert 2004), cell wall protein-modifying enzymes (Kim and Brandizzi, 2016; Hijazi et al., 2014; Nguema-Ona et al., 2014; Atmodjo et al., 2013; McCarthy et al., 2014), ER-translocon/ERAD/N-glycosylation (Ruiz-May et al., 2012; Mathieu-Rivet et al., 2013), organellar genome replication/recombination/repair (Day and Madesis, 2007), and secretion/endomembrane system (Vernoud et al., 2003; Richter et al., 2007; Sanderfoot 2007; Fujimoto and Ueda, 2012; Fendrych et al., 2010). For each category, we compiled the lists of known molecular players from recent primary research papers and reviews, mostly focused on Arabidopsis/plant systems. The known Arabidopsis proteins were used to identify Chlamydomonas homologs by reciprocal best hits. If a best-hit Chlamydomonas protein showed an E-value of larger than 1E-10, or many proteins showed similar homology, phylogenetic analysis of all the homologs with similar domain structure was performed to define evolutionarily conserved clades. In case of multigene families, homologous sequences that belong to a family were collected by Hmmer3 search of the key homology domain against the nonredundant proteomes in relevant species. The final genome-wide collection of Chlamydomonas genes related to cell wall is given in the Supplemental Tables S19, S20, S21, S22, S24, S25, and S26. Supplemental Table S31 is made available for cross-referencing of EZ-core, g-lysin-induced genes, and gamete-specific genes with the annotation and the compiled transcriptome data.
Phylogenetic Tree Construction
For each target family, family member protein sequences were collected by a genome-wide search by HMMer3 using homology domain motifs specific to the target families retrieved from the Pfam 3.0 database or generated from our own domain alignments. Family searches were done in nonredundant predicted protein sets of the selected species listed in Supplemental Table S27. Collected protein sequences were aligned using MAFFT v7.017 (Katoh et al., 2002). Resulting alignments were manually inspected for refinement. A consensus Neighbor-Joining (N-J) tree was initially constructed with 500 bootstrapping. If clade structure was not satisfactory in the N-J tree, MrBayes V3.2.6 (Huelsenbeck and Ronquist, 2001) was used for constructing a second phylogeny with a best-fit model searched by Prottest v.3.4.2 (Darriba et al., 2011) or with the WAG model of protein evolution. The BMCMC was run mostly for 106 generations, sampling every 100 generations and discarding the first 2,000 samples as burn-in. The remaining 8,000 samples were used to construct the 25% majority-rule consensus phylogeny.
Accession Numbers
RNA-seq reads were submitted to NCBI with accession no. GEO91400 except bp31/bp31C zygotes data available at http://dandelion.liveholonics.com/pothos/Nishimura_GSP1/ for download. Additional data accession numbers for the published datasets and sequencing statistics are shown in Supplemental Table S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Note S1. Gene model consideration between V5.3 and V5.5
Supplemental Note S2. Cautionary tales in FPKM counting.
Supplemental Table S1. Details of the samples used in the transcriptome analysis.
Supplemental Table S2. RNA-seq mapping statistics by Tophat2.
Supplemental Table S3. Average FPKM values and cluster ID of 13167 gene models analyzed by hierarchical clustering.
Supplemental Table S4. Statistics of FPKM estimates among the 50 hierarchical clusters collected in this study.
Supplemental Table S5. Description of the coexpression patterns of the 50 final clusters.
Supplemental Table S6. FPKM estimates and differential expression analysis of the 253 EZ-core genes.
Supplemental Table S7. Detailed annotation of the EZ-core genes.
Supplemental Table S8. DESeq2 results for differential expression analysis between the bp31C- and bp31-zygotes.
Supplemental Table S9. Significantly up-regulated genes (>2-fold) in bp31C- over bp31-zygotes by DESeq2 analysis (cutoff FDR < 0.05).
Supplemental Table S10. Significantly down-regulated genes (>2-fold) in bp31C- over bp31-zygotes by DESeq2 analysis (cutoff FDR < 0.05).
Supplemental Table S11. Nonparametric Wilcoxon test for the difference in the bp31C/bp31 ratio distribution among clusters.
Supplemental Table S12. Primers used for qPCR and promoter cloning.
Supplemental Table S13. List of genes having at least one of the three ZYRE motifs identified in this study.
Supplemental Table S14. Statistics of TGAC motif occurrence among the clusters.
Supplemental Table S15. FPKM estimates of the known gamete-specific genes.
Supplemental Table S16. Detailed annotation of the gL+EZ (C44) genes.
Supplemental Table S17. Detailed annotation of the gL-EZ (C24) genes.
Supplemental Table S18. Distribution of functional categories among EZ-core, gL+EZ, and gL-EZ genes.
Supplemental Table S19. Details of the MAW gene family.
Supplemental Table S20. Chlamydomonas genes involved in GPI-anchor biosynthesis.
Supplemental Table S21. Pherophorin-encoding genes in Chlamydomonas.
Supplemental Table S22. Chlamydomonas genes involved in cell wall protein maturation.
Supplemental Table S23. Chlamydomonas genes involved in N-glycosylation.
Supplemental Table S24. Chlamydomonas genes involved in nucleotide-sugar metabolism for cell wall biosynthesis.
Supplemental Table S25. Chlamydomonas genes encoding nucleotide-sugar transporters.
Supplemental Table S26. Chlamydomonas genes encoding plasma membrane-localized glycosyl transferase.
Supplemental Table S27. Genomic resources used for the phylogenetic analysis of the target gene families.
Supplemental Table S28. Comparison of the gene models, version 5.3 and version 5.5.
Supplemental Table S29. Analysis of correlation in FPKM estimates between V5.3 model- and V5.5 model-based analysis.
Supplemental Table S30. Comparison of the > 4 fold up-regulated genes in EZ/TAP using V5.5 models-based FPKM estimates to the results using V5.3 models.
Supplemental Table S31. The Chlamydomonas haploid-to-diploid transition (HDT) transcriptome
Supplemental Figure S1. qRT-PCR analysis of EZ-core gene expression.
Supplemental Figure S2. TGAC motifs are most significantly enriched among the early zygote-specific genes.
Supplemental Figure S3. Distribution of bp31C/bp31 FPKM ratio among the EZ-specific (C33, C43, and C50) and g-lysin-induced (C24 and C44) gene clusters.
Supplemental Figure S4. Structure of the genes that were analyzed by the promoter-reporter assay.
Supplemental Figure S5. Analysis of g-lysin inducible luciferase expression of the selected transgenic strains harboring a g-lysin-inducible promoter-luciferase construct.
Supplemental Figure S6. MAW domain sequence alignment.
Supplemental Figure S7. A phylogenetic analysis of the Exostosin (GT47) family in Arabidopsis and Chlamydomonas.
Supplemental Figure S8. A phylogenetic analysis of the GT90 family in green plants.
Supplemental Figure S9. A phylogenetic analysis of the CSR (Carbohydrate Sulfurtransferase Related) family.
Supplemental Figure S10. A phylogenetic analysis of the GOX (glyoxal oxidase) family by the Neighbor-Joining method.
Supplemental Figure S11. A phylogenetic analysis of the TPT/NST family using Arabidopsis and Chlamydomonas members.
Supplemental Figure S12. RNA-seq results based on V5.3 models show little difference from the results based on V5.5 models.
Acknowledgments
We are grateful to Sean Gallaher and Sabeeha Merchant for supporting RNA-seq reads and data-handling in her laboratory. We thank David Casero and Matteo Pellegrini for their support for next-gen sequencing and sequence analysis. We thank William Snell for sharing his RNA-seq reads prior to the publication. We thank Masaki Odahara for technical assistance in cDNA sequencing library preparation. Also, tremendous thanks to JGI for providing V5.3/V5.5 assembly and annotation. We also thank George Haughn for reading manuscripts and sharing unpublished results.
Footnotes
This work was supported by the Korea CCS R&D Center (KCRC), Korean Ministry of Science, grant no. 2016M1A8A1925345, and by the Natural Sciences and Engineering Research Council of Canada, Discovery Grant 418471-12, from to J.-H. Lee. Y. Nishimura was supported by the NEXT program of Japan: GS015 and Ministry of Education, Culture, Sports, Science, and Technology of Japan grants to Y.N. (17H05840). Sequencing was supported by the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research program under Award No. DE-FC02-02ER63421 to Sabeeha Merchant.
Articles can be viewed without a subscription.
References
- Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ (2008) Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell 132: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama H, Hagiwara Y, Misumi O, Kuroiwa T, Nakamura S (2006) Complete elimination of maternal mitochondrial DNA during meiosis resulting in the paternal inheritance of the mitochondrial genome in Chlamydomonas species. Protoplasma 228: 231–242 [DOI] [PubMed] [Google Scholar]
- Atmodjo MA, Hao Z, and Mohnen D (2013) Evolving Views of Pectin Biosynthesis. Annu Rev Plant Biol 64: 747–779 [DOI] [PubMed] [Google Scholar]
- Borner GHH, Lilley KS, Stevens TJ, Dupree P (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol 132: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan MJ, Imam SH, Eskue WA, Snell WJ (1989) Activation of the cell wall degrading protease, lysin, during sexual signalling in Chlamydomonas: the enzyme is stored as an inactive, higher relative molecular mass precursor in the periplasm. J Cell Biol 108: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Ning J, Hernandez-Lara CI, Belzile O, Wang Q, Dutcher SK, Liu Y, Snell WJ (2015) Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. eLife 4: e05242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. (2008) Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Catt JW. (1979) The isolation and chemical composition of the zygospore cell wall of Chlamydomonas reinhardii. Plant Sci Lett 15: 69–74 [Google Scholar]
- Cavalier-Smith T. (1974) Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J Cell Sci 16: 529–556 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. (1976) Eelectron microscopy of zygospore formation in Chlamydomonas reinhardii. Protoplasma 87: 297–315 [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH (2006) Gene regulatory networks and the evolution of animal body plans. Science 311: 796–800 [DOI] [PubMed] [Google Scholar]
- De Hoff PL, Ferris P, Olson BJSC, Miyagi A, Geng S, Umen JG (2013) Species and population level molecular profiling reveals cryptic recombination and emergent asymmetry in the dimorphic mating locus of C. reinhardtii. PLoS Genet 9: e1003724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendoza A, Sebé-Pedrós A, Šestak MS, Matejčić M, Torruella G, Domazet-Lošo T, Ruiz-Trillo I (2013) Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc Natl Acad Sci USA 110: E4858–E4866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domozych DS, Domozych CE (2014) Multicellularity in green algae: upsizing in a walled complex. Front Plant Sci 5: 64910.3389/fpls.2014.00649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund J, Obel N, Ulvskov P, Geshi N, Pauly M, Bacic A, Petersen BL (2007) Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol Biol 64: 439–451 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, Heijne von G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protocols 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Fairclough SR, Chen Z, Kramer E, Zeng Q, Young S, Robertson HM, Begovic E, Richter DJ, Russ C, Westbrook MJ, Manning G, Lang BF, et al. (2013) Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol 14: R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fédry J, Liu Y, Péhau-Arnaudet G, Pei J, Li W, Tortorici MA, Traincard F, Meola A, Bricogne G, Grishin NV, Snell WJ, Rey FA, et al. (2017) The ancient gamete fusogen HAP2 Is a eukaryotic class II fusion protein. Cell 168: 904–915.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Synek L, Pecenkova T, Toupalova H, Cole R, Drdova E, Nebesarova J, Sedinova M, Hala M, Fowler JE, Zarsky V (2010) The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. Plant Cell 22: 3053–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P, Olson BJSC, De Hoff PL, Douglass S, Casero D, Prochnik S, Geng S, Rai R, Grimwood J, Schmutz J, Nishii I, Hamaji T, et al. (2010) Evolution of an expanded sex-determining locus in Volvox. Science 328: 351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ. (1995) Localization of the nic-7, ac-29 and thi-10 genes within the mating-type locus of Chlamydomonas reinhardtii. Genetics 141: 543–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Armbrust EV, Goodenough UW (2002) Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160: 181–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Goodenough UW (1997) Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics 146: 859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Waffenschmidt S, Umen JG, Lin H, Lee J-H, Ishida K, Kubo T, Lau J, Goodenough UW (2005) Plus and minus sexual agglutinins from Chlamydomonas reinhardtii. Plant Cell 17: 597–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Woessner JP, Goodenough UW (1996) A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol Biol Cell 7: 1235–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Woessner JP, Waffenschmidt S, Kilz S, Drees J, Goodenough UW (2001) Glycosylated polyproline II rods with kinks as a structural motif in plant hydroxyproline-rich glycoproteins. Biochemistry 40: 2978–2987 [DOI] [PubMed] [Google Scholar]
- Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang H-Y, Dosztanyi Z, El-Gebali S, Fraser M, et al. (2017) InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res 45: D190–D199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JM, Hazzouri KM, Pham GM, Rosas U, Bahmani T, Khraiwesh B, Nelson DR, Jijakli K, Abdrabu R, Harris EH, Lefebvre PA, Hom EF, et al. (2015) Whole-Genome Resequencing Reveals Extensive Natural Variation in the Model Green Alga Chlamydomonas reinhardtii. Plant Cell 27: 2353–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangedakis E, Saint-Marcoux D, Moody LA, Rabbinowitsch E, and Langdale JA (2016) Nonreciprocal complementation of KNOX gene function in land plants. The New phytologist 341: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Ueda T (2012) Conserved and plant-unique mechanisms regulating plant post-Golgi traffic. Front Plant Sci 3: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumizu C, Alvarez JP, Sakakibara K, Bowman JL (2015) Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PLoS Genet 11: e1004980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geshi N, Harholt J, Sakuragi Y, Krüger Jensen J, Scheller HV (2011). Glycosyltransferases of the GT47 Family. In Plant Polysaccharides, Biosynthesis and Bioengineering, Plant Polysaccharides, Biosynthesis and Bioengineering, Wiley-Blackwell, Oxford, UK, pp. 265–283. [Google Scholar]
- Geshi N, Johansen JN, Dilokpimol A, Rolland A, Belcram K, Verger S, Kotake T, Tsumuraya Y, Kaneko S, Tryfona T, Dupree P, Scheller HV, et al. (2013) A galactosyltransferase acting on arabinogalactan protein glycans is essential for embryo development in Arabidopsis. Plant J 76: 128–137 [DOI] [PubMed] [Google Scholar]
- Gille S, Hänsel U, Ziemann M, Pauly M (2009) Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc Natl Acad Sci USA 106: 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Heitman J (2014) Origins of eukaryotic sexual reproduction. Cold Spring Harb Perspect Biol 6: a016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Lin H, Lee J-H (2007) Sex determination in Chlamydomonas. Semin Cell Dev Biol 18: 350–361 [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Adair WS, Collin-Osdoby P, Heuser JE (1985) Structure of the Chlamydomonas agglutinin and related flagellar surface proteins in vitro and in situ. J Cell Biol 101: 924–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE (1985) The Chlamydomonas cell wall and its constituent glycoproteins analyzed by the quick-freeze, deep-etch technique. J Cell Biol 101: 1550–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Jurivich D (1978) Tipping and mating-structure activation induced in Chlamydomonas gametes by flagellar membrane antisera. J Cell Biol 79: 680–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Shames B, Small L, Saito T, Crain RC, Sanders MA, Salisbury JL (1993) The role of calcium in the Chlamydomonas reinhardtii mating reaction. J Cell Biol 121: 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutte C, Johnson AD (1988) a1 protein alters the DNA binding specificity of α 2 repressor. Cell 52: 875–882 [DOI] [PubMed] [Google Scholar]
- Grief C, O’Neill MA, Shaw PJ (1987) The zygote cell wall of Chlamydomonas reinhardii: a structural, chemical and immunological approach. Planta 170: 433–445 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Moschou PN, Smertenko AP, Bozhkov PV (2015) Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 27: 926–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaji T, Lopez D, Pellegrini M, Umen J (2016) Identification and Characterization of a cis-Regulatory Element for Zygotic Gene Expression in Chlamydomonas reinhardtii. G3 (Bethesda) 6: 1541–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook, Academic Press, San Diego, CA, [Google Scholar]
- Hijazi M, Velasquez SM, Jamet E, Estevez JM, Albenne C (2014) An update on post-translational modifications of hydroxyproline-rich glycoproteins: toward a model highlighting their contribution to plant cell wall architecture. Front Plant Sci 5: 39510.3389/fpls.2014.00395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann XK, Beck CF (2005) Mating-induced shedding of cell walls, removal of walls from vegetative cells, and osmotic stress induce presumed cell wall genes in Chlamydomonas. Plant Physiol 139: 999–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg SM, Purnapatre K (2003) Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J Cell Sci 116: 2137–2147 [DOI] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, Moriyama T, Ikeuchi M, et al. (2014) Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst NA, Katz A, Pereman I, Decker EL, Ohad N, Reski R (2016) A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nat Plants 2: 15209. [DOI] [PubMed] [Google Scholar]
- Huber O, Sumper M (1994) Algal-CAMs: isoforms of a cell adhesion molecule in embryos of the alga Volvox with homology to Drosophila fasciclin I. EMBO J 13: 4212–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-I, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Gen Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Brandizzi F (2016) The plant secretory pathway for the trafficking of cell wall polysaccharides and glycoproteins. Glycobiology 26: 940–949 [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, et al. (2008) The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451: 783–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Fukuzawa H, Shimada T, Saito T, Matsuda Y (1992) Primary structure and expression of a gamete lytic enzyme in Chlamydomonas reinhardtii: similarity of functional domains to matrix metalloproteases. Proc Natl Acad Sci USA 89: 4693–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Calvo KR, Chen H, Antonarakis SE, Kamps MP (1997) Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc Natl Acad Sci USA 94: 14553–14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Rasmussen I, Gausing K (1997) DNA binding sites recognised in vitro by a knotted class 1 homeodomain protein encoded by the hooded gene, k, in barley (Hordeum vulgare). FEBS Lett 408: 25–29 [DOI] [PubMed] [Google Scholar]
- Kubo T, Abe J, Oyamada T, Ohnishi M, Fukuzawa H, Matsuda Y, Saito T (2008) Characterization of novel genes induced by sexual adhesion and gamete fusion and of their transcriptional regulation in Chlamydomonas reinhardtii. Plant Cell Physiol 49: 981–993 [DOI] [PubMed] [Google Scholar]
- Kuroiwa T, Kawano S, Nishibayashi S, Sato C (1982) Epifluorescent microscopic evidence for maternal inheritance of chloroplast DNA. Nature 298: 481–483 [DOI] [PubMed] [Google Scholar]
- Kurvari V. (1997) Cell wall biogenesis in Chlamydomonas: molecular characterization of a novel protein whose expression is up-regulated during matrix formation. Mol Gen Genet 256: 572–580 [DOI] [PubMed] [Google Scholar]
- Lee J-H, Lin H, Joo S, Goodenough U (2008) Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell 133: 829–840 [DOI] [PubMed] [Google Scholar]
- Lee J-H, Waffenschmidt S, Small L, Goodenough U (2007) Between-species analysis of short-repeat modules in cell wall and sex-related hydroxyproline-rich glycoproteins of Chlamydomonas. Plant Physiol 144: 1813–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Basu D, Pattathil S, Xu W-L, Venetos A, Martin SL, Faik A, Hahn MG, Showalter AM (2013) Biochemical and physiological characterization of fut4 and fut6 mutants defective in arabinogalactan-protein fucosylation in Arabidopsis. J Exp Bot 64: 5537–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Goodenough UW (2007) Gametogenesis in the Chlamydomonas reinhardtii minus mating type is controlled by two genes, MID and MTD1. Genetics 176: 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart C, Halperin Y, Shamir R (2008) Transcription factor and microRNA motif discovery: the Amadeus platform and a compendium of metazoan target sets. Genome Res 18: 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Pei J, Grishin N, Snell WJ (2015) The cytoplasmic domain of the gamete membrane fusion protein HAP2 targets the protein to the fusion site in Chlamydomonas and regulates the fusion reaction. Development 142: 962–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tewari R, Ning J, Blagborough AM, Garbom S, Pei J, Grishin NV, Steele RE, Sinden RE, Snell WJ, Billker O (2008) The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev 22: 1051–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Hamaji T, Kropat J, De Hoff P, Morselli M, Rubbi L, Fitz-Gibbon S, Gallaher SD, Merchant SS, Umen J, Pellegrini M (2015) Dynamic Changes in the Transcriptome and Methylome of Chlamydomonas reinhardtii throughout Its Life Cycle. Plant Physiol 169: 2730–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bähler J (2002) The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32: 143–147 [DOI] [PubMed] [Google Scholar]
- Mathieu-Rivet E, Scholz M, Arias C, Dardelle F, Schulze S, Le Mauff F, Teo G, Hochmal AK, Blanco-Rivero A, Loutelier-Bourhis C, et al. (2013) Exploring the N-glycosylation pathway in Chlamydomonas reinhardtii unravels novel complex structures. Mol Cell Proteomics 12: 3160–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy TW, Der JP, Honaas LA, dePamphilis CW, Anderson CT (2014) Phylogenetic analysis of pectin-related gene families in Physcomitrella patens and nine other plant species yields evolutionary insights into cell walls. BMC Plant Biol 14: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF, Qu LH, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R, Wu G, Deshpande RR, Vieler A, Gärtner K, Li X, Moellering ER, Zäuner S, Cornish AJ, Liu B, Bullard B, Sears BB, et al. (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol 154: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami SA, Goodenough UW (1978) Novel glycopolypeptide synthesis induced by gametic cell fusion in Chlamydomonas reinhardtii. J Cell Biol 77: 165–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misamore MJ, Gupta S, Snell WJ (2003) The Chlamydomonas Fus1 protein is present on the mating type plus fusion organelle and required for a critical membrane adhesion event during fusion with minus gametes. Mol Biol Cell 14: 2530–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misevic GN, Guerardel Y, Sumanovski LT, Slomianny MC, Demarty M, Ripoll C, Karamanos Y, Maes E, Popescu O, Strecker G (2004) Molecular recognition between glyconectins as an adhesion self-assembly pathway to multicellularity. J Biol Chem 279: 15579–15590 [DOI] [PubMed] [Google Scholar]
- Mistry J, Finn RD, Eddy SR, Bateman A, Punta M (2013) Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res 41: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk BC. (1988) The cell wall of Chlamydomonas reinhardtii gametes: composition, structure, and autolysin-mediated shedding and dissolution. Planta 176: 441–450 [DOI] [PubMed] [Google Scholar]
- Mosquna A, Katz A, Decker EL, Rensing SA, Reski R, Ohad N (2009) Regulation of stem cell maintenance by the Polycomb protein FIE has been conserved during land plant evolution. Development 136: 2433–2444 [DOI] [PubMed] [Google Scholar]
- Neiman AM. (2011) Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189: 737–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema-Ona E, Vicré-Gibouin M, Gotté M, Plancot B, Lerouge P, Bardor M, Driouich A (2014) Cell wall O-glycoproteins and N-glycoproteins: aspects of biosynthesis and function. Front Plant Sci 5: 49910.3389/fpls.2014.00499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J, Otto TD, Pfander C, Schwach F, Brochet M, Bushell E, Goulding D, Sanders M, Lefebvre PA, Pei J, Grishin NV, Vanderlaan G, et al. (2013) Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev 27: 1198–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Shikanai T, Nakamura S, Kawai-Yamada M, Uchimiya H (2012) Gsp1 triggers the sexual developmental program including inheritance of chloroplast DNA and mitochondrial DNA in Chlamydomonas reinhardtii. Plant Cell 24: 2401–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsushita W, Matsubayashi Y (2013) Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat Chem Biol 9: 726–730 [DOI] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsubayashi Y (2015) Identification of three potent hydroxyproline O-galactosyltransferases in Arabidopsis. Plant J 81: 736–746 [DOI] [PubMed] [Google Scholar]
- Okano Y, Aono N, Hiwatashi Y, Murata T, Nishiyama T, Ishikawa T, Kubo M, Hasebe M (2009) A polycomb repressive complex 2 gene regulates apogamy and gives evolutionary insights into early land plant evolution. Proc Natl Acad Sci USA 106: 16321–16326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Snell WJ (2005) Chlamydomonas shortens its flagella by activating axonemal disassembly, stimulating IFT particle trafficking, and blocking anterograde cargo loading. Dev Cell 9: 431–438 [DOI] [PubMed] [Google Scholar]
- Pierleoni A, Martelli PL, Casadio R (2008) PredGPI: a GPI-anchor predictor. BMC Bioinformatics 9: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires ND, Dolan L (2012) Morphological evolution in land plants: new designs with old genes. Philos Trans R Soc Lond B Biol Sci 367: 508–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MC, Levery SB, Castle SA, Klutts JS, Doering TL (2009) A novel xylosylphosphotransferase activity discovered in Cryptococcus neoformans. J Biol Chem 284: 36118–36127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Geldner N, Schrader J, Wolters H, Stierhof Y-D, Rios G, Koncz C, Robinson DG, Jürgens G (2007) Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448: 488–492 [DOI] [PubMed] [Google Scholar]
- Roberts K, Gurney-Smith M, Hills GJ (1972) Structure, composition and morphogenesis of the cell wall of Chlamydomonas reinhardi. I. Ultrastructure and preliminary chemical analysis. J Ultrastruct Res 40: 599–613 [DOI] [PubMed] [Google Scholar]
- Rudel D, Sommer RJ (2003) The evolution of developmental mechanisms. Dev Biol 264: 15–37 [DOI] [PubMed] [Google Scholar]
- Ruecker O, Zillner K, Groebner-Ferreira R, Heitzer M (2008) Gaussia-luciferase as a sensitive reporter gene for monitoring promoter activity in the nucleus of the green alga Chlamydomonas reinhardtii. Mol Genet Genomics 280: 153–162 [DOI] [PubMed] [Google Scholar]
- Ruiz-May E, Kim S-J, Brandizzi F, Rose JKC (2012) The secreted plant N-glycoproteome and associated secretory pathways. Front Plant Sci 3: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito F, Suyama A, Oka T, Yoko-O T, Matsuoka K, Jigami Y, Shimma Y-I (2014) Identification of Novel Peptidyl Serine α-Galactosyltransferase Gene Family in Plants. J Biol Chem 289: 20405–20420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara K, Ando S, Yip HK, Tamada Y, Hiwatashi Y, Murata T, Deguchi H, Hasebe M, Bowman JL (2013) KNOX2 genes regulate the haploid-to-diploid morphological transition in land plants. Science 339: 1067–1070 [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Nishiyama T, Deguchi H, Hasebe M (2008) Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evol Dev 10: 555–566 [DOI] [PubMed] [Google Scholar]
- Sanderfoot A. (2007) Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol 144: 6–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ. (2004) Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Curr Opin Plant Biol 7: 277–284 [DOI] [PubMed] [Google Scholar]
- Schlosser UG. (1976) Entwicklungsstadien- und sippenspezifische Zellwand-Lysine bei der Freisetzung von Fortpflanzungszellen in der Gattung Chlamydomonas. Ber Dtsch Bot Ges 89: 1–56 [Google Scholar]
- Snell WJ, Goodenough UW (2009). Chapter 12 - Flagellar Adhesion, Flagellar-Generated Signaling, and Gamete Fusion during Mating. In EH. Harris, DB Stern, GB Witman, eds, The Chlamydomonas Sourcebook (Second Edition), Academic Press, London. pp. 369–394. [Google Scholar]
- Speijer D, Lukeš J, Eliáš M (2015) Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc Natl Acad Sci USA 112: 8827–8834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E, Tiwari BS, Rodriguez-Nieto S, Zamyatnin AA Jr., Välineva T, Saarikettu J, Frilander MJ, et al. (2009) Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat Cell Biol 11: 1347–1354 [DOI] [PubMed] [Google Scholar]
- Suzuki L, Woessner J, Uchida H (2000) A zygote-specific protein with hydroxyproline-rich glycoprotein domains and lectin-like domains involved in the assembly of the cell wall of Chlamydomonas reinhardtii. J Phycol 36: 571–583 [DOI] [PubMed] [Google Scholar]
- Tardif M, Atteia A, Specht M, Cogne G, Rolland N, Brugiere S, Hippler M, Ferro M, Bruley C, Peltier G, Vallon O, Cournac L (2012) PredAlgo: A new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol 29: 3625–3639 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131: 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waffenschmidt S. (1993) Isodityrosine cross-linking mediates insolubilization of cell walls in Chlamydomonas. Plant Cell 5: 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, Ruhfel BR, Wafula E, et al. (2014) Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA 111: E4859–E4868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner JP, Goodenough UW (1989). Molecular characterization of a zygote wall protein: an extensin-like molecule in Chlamydomonas reinhardtii. Plant Cell 1: 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, Foulon E, Grimwood J, et al. (2009) Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324: 268–272 [DOI] [PubMed] [Google Scholar]
- Zhao H, Guan X, Xu Y, Wang Y (2013) Characterization of novel gene expression related to glyoxal oxidase by agro-infiltration of the leaves of accession Baihe-35-1 of Vitis pseudoreticulata involved in production of H2O2 for resistance to Erysiphe necator. Protoplasma 250: 765–777 [DOI] [PubMed] [Google Scholar]
- Zhao H, Lu M, Singh R, Snell WJ (2001) Ectopic expression of a Chlamydomonas mt+-specific homeodomain protein in mt- gametes initiates zygote development without gamete fusion. Genes Dev 15: 2767–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]