The plant hormone strigolactone promotes infection thread formation but does not appear to influence other stages of nodulation, including nitrogen fixation.
Abstract
Strigolactones (SLs) influence the ability of legumes to associate with nitrogen-fixing bacteria. In this study, we determine the precise stage at which SLs influence nodulation. We show that SLs promote infection thread formation, as a null SL-deficient pea (Pisum sativum) mutant forms significantly fewer infection threads than wild-type plants, and this reduction can be overcome by the application of the synthetic SL GR24. We found no evidence that SLs influence physical events in the plant before or after infection thread formation, since SL-deficient plants displayed a similar ability to induce root hair curling in response to rhizobia or Nod lipochitooligosaccharides (LCOs) and SL-deficient nodules appear to fix nitrogen at a similar rate to those of wild-type plants. In contrast, an SL receptor mutant displayed no decrease in infection thread formation or nodule number, suggesting that SL deficiency may influence the bacterial partner. We found that this influence of SL deficiency was not due to altered flavonoid exudation or the ability of root exudates to stimulate bacterial growth. The influence of SL deficiency on infection thread formation was accompanied by reduced expression of some early nodulation genes. Importantly, SL synthesis is down-regulated by mutations in genes of the Nod LCO signaling pathway, and this requires the downstream transcription factor NSP2 but not NIN. This, together with the fact that the expression of certain SL biosynthesis genes can be elevated in response to rhizobia/Nod LCOs, suggests that Nod LCOs may induce SL biosynthesis. SLs appear to influence nodulation independently of ethylene action, as SL-deficient and ethylene-insensitive double mutant plants display essentially additive phenotypes, and we found no evidence that SLs influence ethylene synthesis or vice versa.
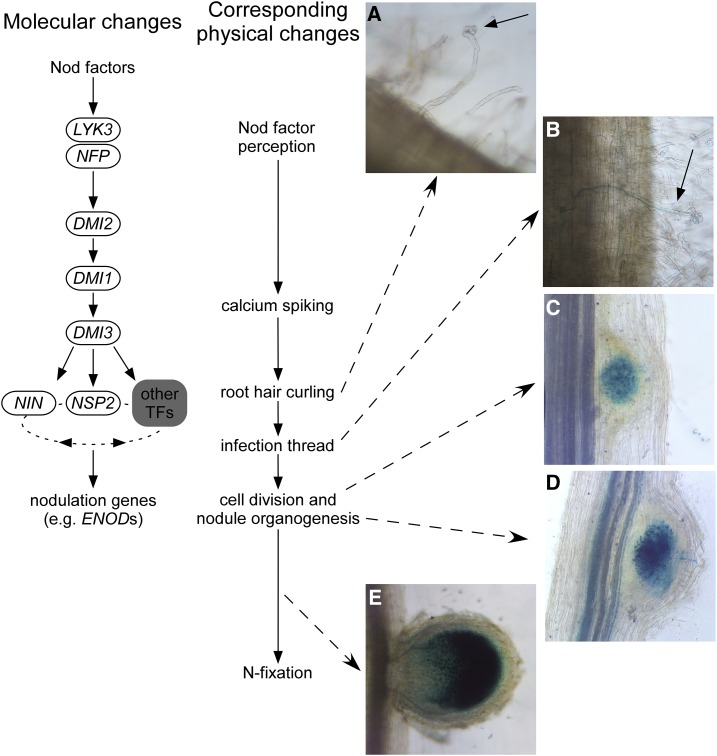
Nodulation results from the intimate relationship of nitrogen (N)-fixing rhizobacteria and leguminous plants. The uptake of rhizobia and the organogenesis of the nodule in which the bacteria are hosted only occur after the exchange of specific chemical signals through the rhizosphere between the rhizobia and plant host. The exudate from roots of the host includes flavonoids that induce the rhizobia to produce signals including specific Nod lipochitooligosaccharides (LCOs; Peters et al., 1986). Following the recognition of these Nod LCOs by receptor-like kinases, including the Nod LCO receptors NFP and LYK3, there is an induction of oscillations in nucleus-associated calcium levels via the action of DMI2 and DMI1. This is sensed by DMI3, which, along with parallel pathways, influence a range of transcription factors (e.g. NIN, NSP1, NSP2, and IPD3) that, in turn, coordinate the expression of nodulation-associated genes such as ENODs (for early nodulation; Yano et al., 2008; Venkateshwaran et al., 2013; Singh et al., 2014; Genre and Russo, 2016; Fig. 1). Corresponding physical changes induced by this perception pathway include root hair curling, infection thread formation, and concomitant cell division in the inner cortical and pericycle cells that leads to the formation of the nodule meristem and, ultimately, colonized nodules (Fig. 1). In some species, the meristem is maintained in the mature nodule (indeterminant nodulators), while in other species, the nodule meristem is lost at maturity (determinant nodulators; Ferguson et al., 2010).
Figure 1.
Scheme of the physical and molecular changes during nodule development in pea, including the proposed positions of genes in the Nod LCO signaling pathway that influence a complex nonlinear network of transcription factors (TFs). A, Root hair curling. B, Infection thread formation. C, Developing nodule not visible to naked eye. D, Maturing nodules. E, Young mature nodule. All photographs show pea roots colonized with blue-stained lacZ-labeled rhizobia.
Plant hormones play critical roles in a number of these processes. One of these is strigolactones (SLs), a group of plant hormones synthesized from a carotenoid precursor via the sequential action of enzymes including D27, CCD7, CCD8, MAX1, and LBO and perceived via a complex of the D14 receptor, MAX2 F-box protein, and D53 (Smith and Li, 2014; Brewer et al., 2016). SLs play important roles in shoot and root development and during the interaction of plants with microorganisms (Foo and Reid, 2013; Smith and Li, 2014; López-Ráez et al., 2017). For example, SLs are exuded from plant roots and act as a rhizosphere signal to promote the symbioses of plants with phosphorus-acquiring arbuscular mycorrhizal (AM) fungi by promoting spore germination and hyphal branching (Akiyama et al., 2005). This AM signaling pathway shares functional elements with nodulation, including elements of the Nod LCO perception pathway outlined above (Venkateshwaran et al., 2013). SLs appear to be a common signal in AM and nodulation, as reports in several species, using both genetic and application approaches, indicate that SLs also exert a primarily positive role during nodulation (Soto et al., 2010; Foo and Davies, 2011; Liu et al., 2013; De Cuyper et al., 2015).
The specific stage at which SLs influence the interaction with rhizobia is not yet clear. A significant reduction in SL biosynthesis, due to lesions in the CCD7 or CCD8 gene, leads to significant reductions in the number of nodules in species that form both indeterminate (pea [Pisum sativum]) and determinate (Lotus japonicus) nodules (Foo and Davies, 2011; Liu et al., 2013). Consistent with this positive role of SLs in nodulation is the observation that nodule number is elevated by the application of the synthetic SL, GR24, in pea and Medicago sativa (Soto et al., 2010; Foo and Davies, 2011). In contrast, GR24 application has been reported to lead to a small decrease in nodule number in Medicago truncatula (De Cuyper et al., 2015), and D27 RNAi lines in this species did not have altered nodulation (van Zeijl et al., 2015). However, gene expression studies in M. truncatula from two independent groups show that the expression of several key SL biosynthesis genes (D27, CCD7, and CCD8) is elevated following challenge with rhizobia (Breakspear et al., 2014; van Zeijl et al., 2015). In the case of CCD8, promoter fusion studies revealed that this expression occurred specifically in infected root hairs and developing nodule primordia (Breakspear et al., 2014), and in mature nodules, the expression of D27, CCD7, and CCD8 was restricted to the meristem and distal infection zone (van Zeijl et al., 2015). Overall, these mutant and gene expression studies suggest that SLs may act at several stages to promote rhizobial infection and nodule formation.
As outlined above, one of the first steps in the interaction of legumes and N-fixing rhizobia is signal exchange through the rhizosphere. However, although they are well known for their role as a rhizosphere signal in AM symbioses, there is no direct evidence that SLs influence rhizobia in an analogous way. Application of the synthetic SL GR24 directly to rhizobia does not promote bacterial growth or signaling (Moscatiello et al., 2010; Soto et al., 2010). The recent report that GR24 induces increased swarming behavior in some rhizobia is intriguing (Peláez-Vico et al., 2016). However, there is currently no known connection between swarming and nodulation, and indeed, plant root exudates from nonlegumes and some, but not all, legume hosts were shown to induce swarming behavior (Tambalo et al., 2014). One possibility yet to be explored is that SL status may influence other root exudates, including flavonoids.
Outputs of the Nod LCO signaling cascade include changes to hormone biosynthesis and/or perception. There is some indication that the Nod LCO signaling pathway may influence SL biosynthesis. M. truncatula mutants with lesions in NSP1 and/or NSP2 (transcription factors downstream of Nod LCO signaling) have reduced levels of SLs in the absence of rhizobia, and this is reflected in reduced expression of the SL biosynthesis gene D27 (Liu et al., 2011). The gene expression studies outlined above by van Zeijl et al. (2015) found that application of Nod LCOs induced the expression of several SL biosynthesis genes, and this was disrupted in plant mutants with lesions in the Nod LCO response pathway (dmi1, dmi2, dmi3, nsp1, and nsp2), although SL levels were not quantified in that study. Like many developmental processes, hormones often interact to control nodulation. Ethylene is known to be a negative regulator of nodulation (Penmetsa and Cook, 1997; Guinel, 2015; Foo et al., 2016c). Recent work in M. truncatula investigated the potential interaction between ethylene and SL during nodulation and found that ethylene-insensitive skl/ein2 mutants did not display altered nodulation when treated with GR24 (De Cuyper et al., 2015). This suggests that SLs may act upstream of ethylene action during nodulation, but further studies are required to fully test this hypothesis.
In this study, we pinpoint the stage at which SLs appear to influence nodulation and examine interactions between SLs, the Nod LCO signaling pathway, and ethylene during nodulation in pea. Studies with severe SL biosynthesis ccd8 mutants and application studies with the synthetic SL GR24 suggest that SLs act specifically to promote infection thread formation. We found no evidence that SL deficiency influences physical events before or after this crucial stage, including flavonoid production, root hair curling, or nitrogen fixation in mature nodules. We also report studies with an SL receptor mutant, d14, which suggest that CCD8 products may promote infection thread formation by influencing the bacterial partner rather than via the SL response system in the plant. This influence of SL deficiency on infection thread formation is accompanied by a reduction in the expression of certain ENOD genes that are well-known markers of the early events during the formation of symbioses. SL levels are up-regulated by elements of the Nod LCO signaling pathway, and SLs appear to influence nodulation largely independently of ethylene action.
RESULTS
Stages of Nodulation in an SL-Deficient Mutant
SLs have been reported to have a generally positive role in promoting nodule formation (Foo and Davies, 2011), although it is not clear at what stage SLs influence the interaction between legumes and rhizobia. We used null SL-deficient ccd8 mutants, disrupted in a key enzyme of the SL biosynthesis pathway (Sorefan et al., 2003; Gomez-Roldan et al., 2008), in a series of studies to pinpoint the stage at which nodulation was disrupted in this mutant.
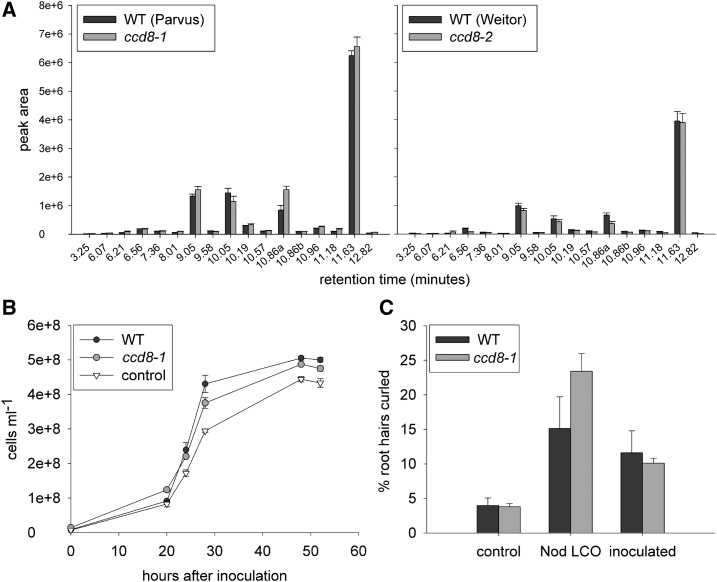
One of the earliest events in the interaction between a legume host and rhizobia is flavonoid exudation by the host root. This, in turn, induces the production of Nod LCOs from rhizobia. We examined the flavonoid profile of several alleles of ccd8 SL-deficient mutants and compared them with their wild-type progenitor lines. We found no difference in the identity or significant reduction in the levels of flavonoids between two null SL-deficient mutants, ccd8-1 and ccd8-2, and their respective progenitor lines when analyzed by ANOVA (Fig. 2A). We also examined the influence of root exudates from wild-type and SL-deficient ccd8-1 plants on bacterial growth in vitro and found that both had a positive influence on bacterial growth and that there was no significant difference overall between the genotypes (Fig. 2B). These studies suggest that SLs do not influence nodulation by influencing the production of flavonoids or rhizobial population growth.
Figure 2.
Influence of the wild type (WT) and SL-deficient ccd8 mutants on early events in nodulation. A, Flavonoid profile in root exudate of ccd8-1 and ccd8-2 mutants and their respective wild types, cv Parvus and cv Weitor (n = 4). B, Growth of Rhizobium leguminosarum bv viciae (RLV248) over time after treatment with wild-type cv Parvus or ccd8-1 root exudates compared with a solvent control (n = 4). C, Percentage of root hairs curled in wild-type cv Parvus and ccd8-1 mutant plants 5 d after treatment with 0.1 μm Nod LCO or 10% solution of R. leguminosarum bv viciae culture (inoculated) compared with a solvent control (n = 4–5). Values are means ± se, and analysis of each experiment by ANOVA indicated no significant differences between ccd8 and its wild-type progenitor.
In many legume species, including pea, root hairs are the site of rhizobial infection, and one of the first physical changes during nodulation is root hair curling. Previous studies have reported that pea ccd8 mutants do not display altered root hair number or length (Foo and Davies, 2011). We found that there also was no significant difference in the number of curled root hairs produced by the SL-deficient mutant ccd8-1 compared with wild-type plants in response to pure Nod LCO or application of the compatible rhizobia (Fig. 2C).
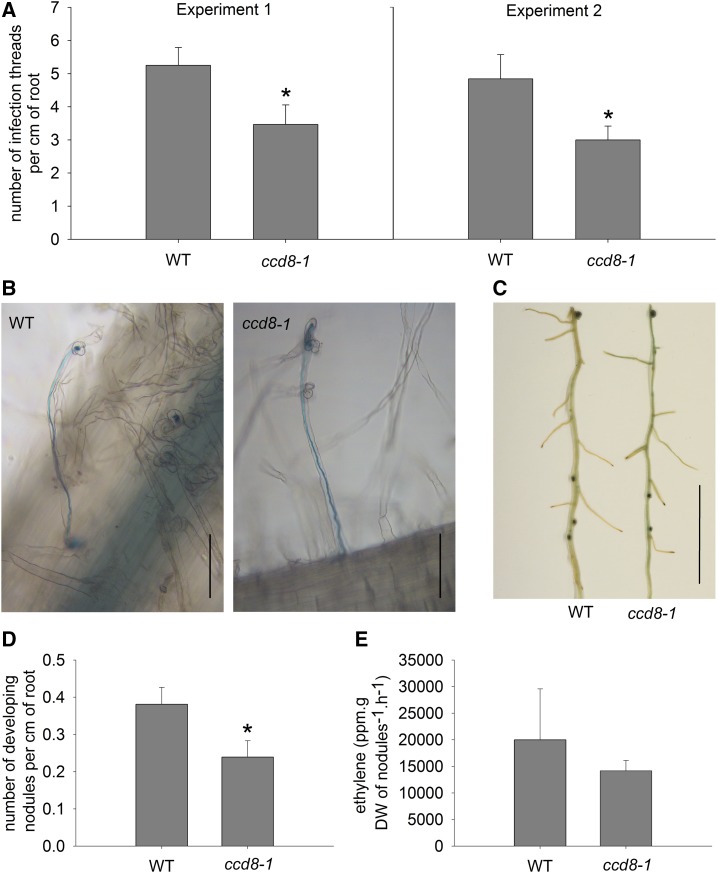
Following root hair curling, uptake of the rhizobial bacteria occurs through infection thread structures that channel rhizobia into dividing cells that will form the nodule. Using lacZ-labeled rhizobia, we were able to investigate the formation of infection threads and developing nodules (nodules not yet visible to the naked eye but visible and stained blue under magnification) in an SL-deficient ccd8-1 line (Fig. 3, A–D). We found that ccd8-1 mutants formed significantly fewer infection threads compared with wild-type plants in two independent experiments (Fig. 3A; P < 0.05), and a two-way ANOVA across experiments found a significant genotype effect (P < 0.01). The infection threads that did form in ccd8-1 mutants were similar in morphology to those observed in the wild type (Fig. 3B), ranging from immature infection threads to mature infection threads connected to developing nodules. There did not appear to be a reduction in the frequency of infection threads leading to nodules (i.e. a higher abortion rate) in SL-deficient mutants, as the approximate 30% to 40% reduction in infection thread formation in ccd8-1 mutants was mirrored by a similar significant reduction in the number of developing nodules compared with the wild type (Fig. 3, C and D; P < 0.05). Both reductions in infection thread formation and developing nodules in ccd8-1 are consistent with the 30% to 40% decrease in mature nodules reported previously in ccd8-1 mutant plants (Foo and Davies, 2011). No other changes were observed in the roots (or root hairs) of ccd8-1 plants as a consequence of the reduced infection thread formation.
Figure 3.
Nodule development in wild-type cv Parvus (WT) and SL-deficient ccd8-1 plants infected with lacZ-labeled R. leguminosarum bv viciae. A, Number of infection threads per cm of root (four to 10 root segments from n = 6–11 plants) in two independent experiments. B, Photograph of stained roots showing infection threads in wild-type and ccd8-1 plants. Bars = 0.1 mm. C, Photograph of stained root showing nodules in wild-type and ccd8-1 plants. Bar = 5 mm. D, Number of developing nodules per cm of root (10 root segments from n = 15 plants). E, Ethylene evolution per gram nodule dry weight (DW) from the acetylene reductase assay (n = 4). Values are means ± se, and values statistically different from the wild type are indicated by asterisks: *, P < 0.05. In B and C, R. leguminosarum is stained blue.
To examine whether the influence of CCD8 products (presumably SLs) on infection thread formation is via the SL receptor D14, we compared infection thread formation in SL-deficient ccd8 mutant plants with that in d14 mutant plants (de Saint Germain et al., 2016). As observed above, ccd8-2 mutants formed significantly fewer infection threads and nodules than wild-type plants (Supplemental Fig. S1, A and B). In contrast, we found no decrease in the number of infection threads or nodules formed on d14 mutants compared with the wild type, indicating that the D14 SL receptor is not required for the effect of the CCD8 product on nodule development. As observed previously (Urquhart et al., 2015), both ccd8-2 and d14 mutants have somewhat shorter lateral roots than the wild type, consistent with the reported SL deficiency and insensitivity of these mutants (Supplemental Fig. S1C).
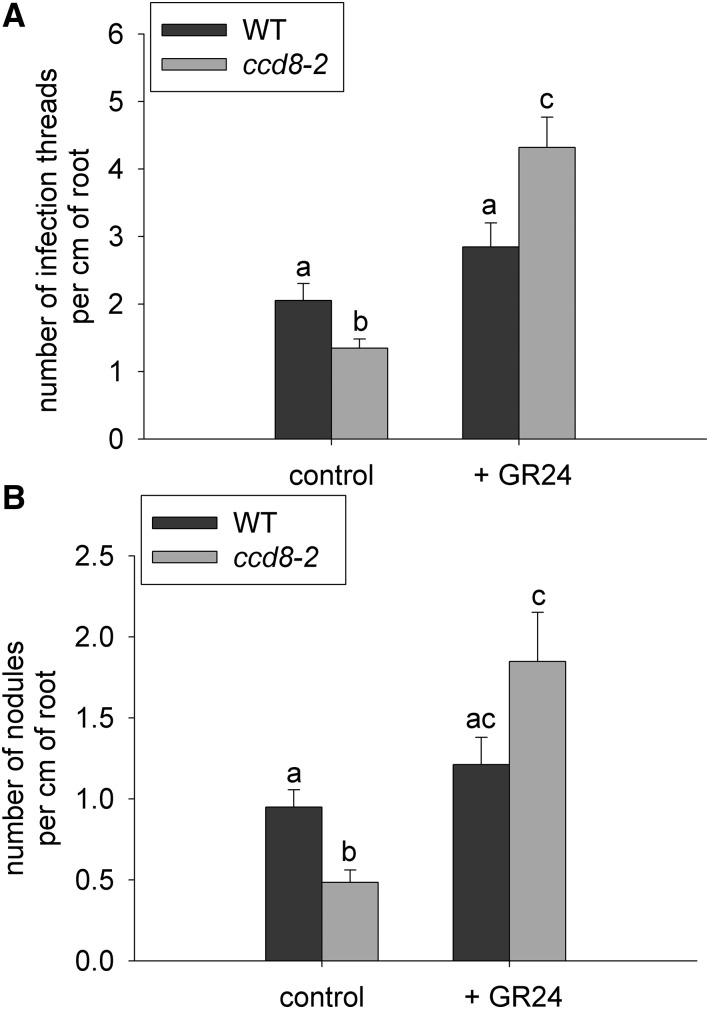
To check whether the reduced infection thread formation in ccd8 plants was due to the SL deficiency in this mutant, the response to applied GR24 was examined in wild-type and ccd8 plants (Fig. 4). A two-way ANOVA showed a significant effect of GR24 application (P < 0.01) and also a significant interaction between genotype and GR24 treatment (P < 0.01). As observed previously (Fig. 2A; Supplemental Fig. S1A), infection thread formation was reduced significantly in ccd8 mutants compared with the wild type. The fact that GR24 significantly increased infection thread formation in ccd8 mutants (Fig. 4A) provides strong support for the view that the action of CCD8 on infection thread formation is via its known effect on SL biosynthesis. Similarly, a two-way ANOVA also showed a strong treatment effect of GR24 on the total number of nodules (P < 0.001) and a significant interaction between treatment and genotype (P < 0.05). The alterations seen in infection thread formation across both control and GR24-treated genotypes were reflected in similar changes in total nodule number (Fig. 4B). The approximately 50% reduction in the total number of nodules seen in ccd8 mutants compared with wild-type plants was fully restored by GR24 treatment, consistent with the previously reported positive effects of SLs on the number of visible nodules seen in pea (Foo and Davies, 2011).
Figure 4.
Nodule development in wild type cv Torsdag (WT) and SL-deficient ccd8-2 plants infected with lacZ-labeled R. leguminosarum bv viciae treated with synthetic SL (+)-GR24 or solvent control. A, Number of infection threads per cm of root. B, Total number of nodules per cm of root. Values are means ± se (one to two root segments from n = 12–13 plants), and values with different letters are significantly different (P < 0.05).
Some legumes, such as Lupinus spp., undergo infection not through root hairs but via cracks in the epidermis (Tang et al., 1993; González‐Sama et al., 2004). Interestingly, we found that application of the synthetic SL (+)-GR24 did not enhance nodule formation in blue lupin (Lupinus angustifolius; Supplemental Fig. S2), while similar doses of GR24 have been shown to alter nodulation in pea, M. truncatula, and M. sativa (Soto et al., 2010; Foo and Davies, 2011; De Cuyper et al., 2015). The fact that SLs only appear to influence nodule formation in species that use root hair infection is consistent with an important role for SLs during the infection thread stage of infection.
Maturation of the nodule ultimately results in a functional nodule in which rhizobia are able to fix nitrogen. We found that, like other root tissue, mature nodules also contain SLs, although the levels were approximately 4 times lower than in surrounding mature root (the level of fabacyl acetate in nodule tissue was 0.05 ng g−1 fresh weight, compared with 0.19 ng g−1 fresh weight in root tissue). To determine whether SLs influence nodule function, we examined the ability of nodules that do form on SL-deficient mutants to fix nitrogen using the acetylene reductase assay (Fig. 3E). The SL-deficient ccd8-1 nodules were clearly functional and, for a given mass of nodules, did not have a significantly different acetylene reductase rate from comparable wild-type nodules, indicating that SL-deficient nodules can fix nitrogen.
Gene Expression during Early Nodulation in an SL-Deficient Mutant
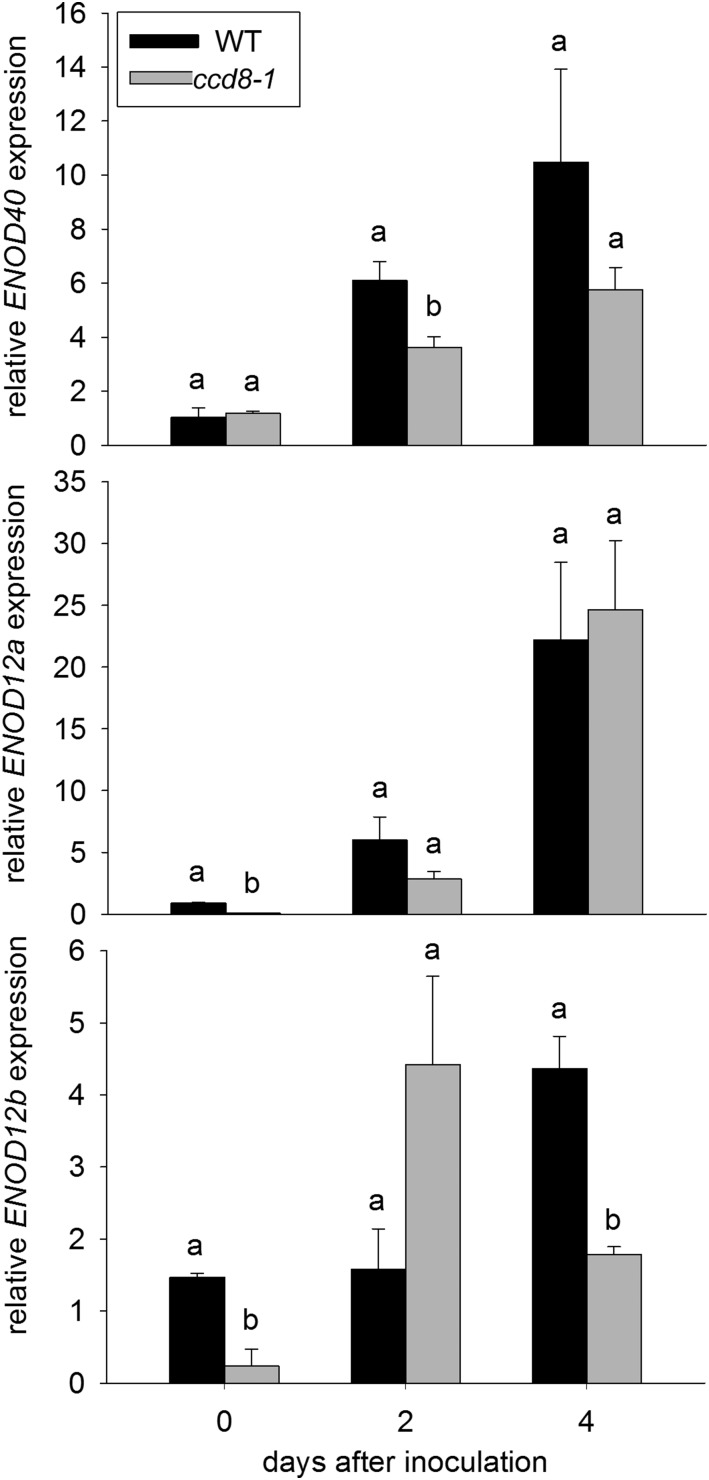
During early interactions with rhizobia, the expression of a suite of ENODs is induced via the action of the Nod LCO signal transduction pathway. In pea, this includes ENOD12a, ENOD12b, and ENOD40 (Govers et al., 1991; Schneider et al., 1999). Induction of these genes by compatible rhizobia is severely impaired, at least at some time points, in mutants disrupted in elements of the Nod LCO signal transduction pathway, such as dmi2 (Schneider et al., 1999). We found that, in the days following inoculation with rhizobia, the induction of some of these genes was reduced significantly in SL-deficient ccd8-1 mutant plants compared with wild-type cv Parvus plants (Fig. 5). For example, 4 d after inoculation, the expression of ENOD12b was significantly lower in roots of ccd8-1 plants compared with the wild type (Fig. 5; P < 0.01). We also found that, in wild-type plants, the expression of the SL biosynthesis gene D27 was elevated significantly following challenge with the rhizobia (Supplemental Fig. S3; P < 0.05), consistent with similar reports in M. truncatula (van Zeijl et al., 2015). Recent reports suggest that SLs may also influence disease development in some systems (López-Ráez et al., 2017), although studies with a pea SL-deficient ccd8 mutant have not revealed any influence of this mutation on pea disease caused by Pythium irregulare or Fusarium oxysporum (Blake et al., 2016; Foo et al., 2016a). To examine if a ccd8 mutation may influence disease response after challenge with rhizobium, the expression of three key disease marker genes (Supplemental Fig. S4) as well as callose and lignin deposition were examined in ccd8 and wild-type plants exposed to rhizobia. No trends of consistently elevated or reduced gene expression (Supplemental Fig. S4) or callose or lignin deposition were observed between ccd8 and wild-type roots.
Figure 5.
Expression of ENOD genes 0, 2, and 4 d following inoculation with R. leguminosarum bv viciae in wild type (WT) cv Parvus and the SL-deficient ccd8-1 mutant. Values are means ± se (n = 3). For days 0, 2 and 4, values with different letters are significantly different (P < 0.05).
In addition to inducing nodulation-specific molecular and physical changes, rhizobia (via Nod LCO signaling) also induce changes in root architecture (Oláh et al., 2005). Given that SLs have been implicated in the control of root architecture (Rasmussen et al., 2013), it was interesting to examine whether CCD8 plays a role in modifying root architecture in response to Nod LCO. Two-way ANOVAs showed that Nod LCO treatment significantly affected lateral root number and length (Supplemental Fig. S5; P < 0.01). However, there was no significant interaction between genotype and treatment, indicating that ccd8-1 plants responded to Nod LCO in a similar way to wild-type plants.
Common SYM Mutants Have Impaired SL Biosynthesis
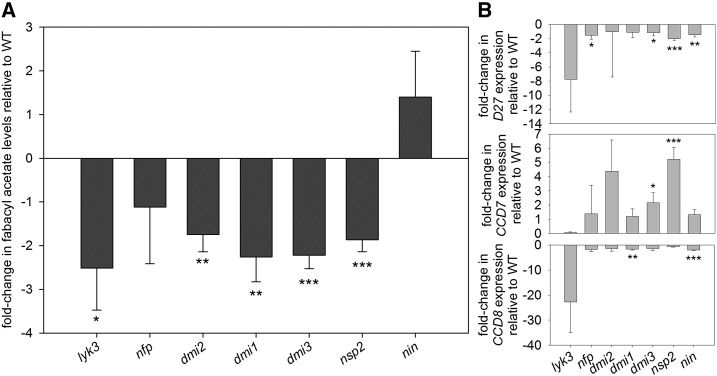
Previous studies in M. truncatula have identified SL biosynthesis and/or the expression of SL biosynthesis genes as dependent in part on some elements of the LCO signaling pathway (Liu et al., 2011; Breakspear et al., 2014; van Zeijl et al., 2015), but it is not clear from these studies if this is due to indirect effects of individual genes in this pathway or if the LCO signaling pathway as a whole influences SL biosynthesis. To address this question, we systematically examined SL levels in nonnodulated roots of pea mutants disrupted in Nod LCO receptors (nfp and lyk3), LCO signaling elements (dmi1, dmi2, and dmi3), and downstream transcription factors (nsp2 and nin). We found that the major canonical SL present in pea tissue, fabacyl acetate, was reduced significantly in mutants disrupted in one Nod LCO receptor (lyk3), signaling elements (dmi1, dmi2, and dmi3), and one downstream transcription factor (nsp2) compared with their respective wild-type progenitor lines (Fig. 6A; P < 0.05–0.001). The SLs orobanchol and orobanchyl acetate also were detected in some experiments, and these also were reduced in dmi3 (Supplemental Fig. S6). In contrast, there was no significant reduction in fabacyl acetate levels in mutants disrupted in the NIN transcription factor (Fig. 6A). This indicates that SL levels are influenced by the Nod LCO signaling pathway but not via the transcription factor encoded by the NIN gene. This relatively small (approximately 2-fold) reduction in SL levels in the roots is not sufficient to significantly promote shoot branching in these lines (data not shown), a phenotype observed in severely SL-deficient lines (Beveridge et al., 1997).
Figure 6.
SL levels and expression of SL biosynthesis genes in root tissue of various symbiosis mutants. A, Fold change relative to respective wild types (WT) of the major canonical SL in pea, fabacyl acetate, in root tissue of various pea symbiosis mutants. B, Fold change relative to the respective wild type of the expression of SL biosynthesis genes CCD7, CCD8, and D27 in root tissue of pea symbiosis mutants. Values statistically different from the wild type are indicated by asterisks: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Values are means ± 95% confidence interval (n = 4).
It is interesting that, in some cases, this reduction in SL levels was accompanied by significant reductions in the expression of the SL biosynthesis genes D27 and CCD8, but in many cases, the expression of these genes did not reflect SL level, and for CCD7, the expression was elevated in several cases (Fig. 6B). This may be due to the well-established feedback regulation of the SL biosynthesis pathway (Foo et al., 2005: Umehara et al., 2008) and is consistent with other hormones, where the expression of hormone biosynthesis genes often is not an accurate proxy for hormone levels (Symons and Reid, 2008).
SLs Act Largely Independently of Ethylene to Influence Nodulation
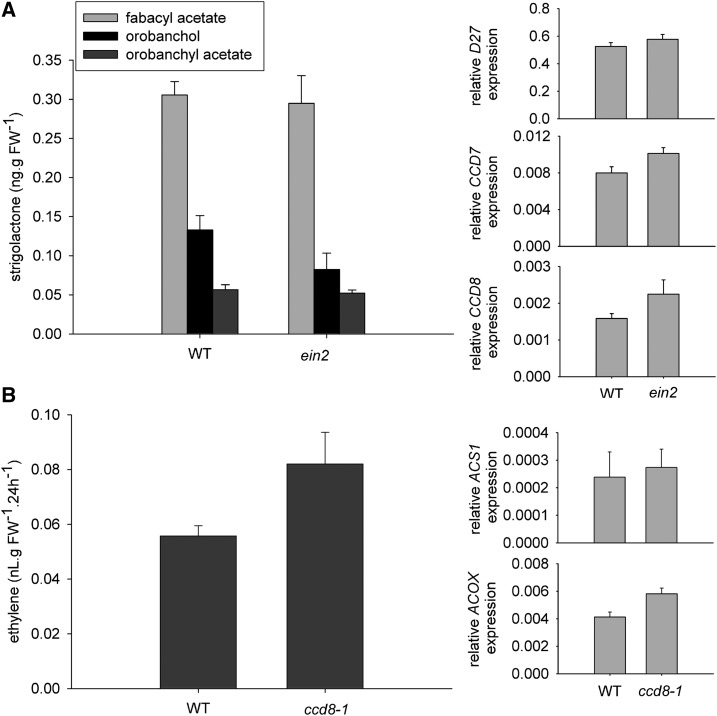
It was suggested recently that ethylene may act downstream of SLs during nodulation (De Cuyper et al., 2015), and we used several approaches to examine ethylene-SL interactions during nodulation. We examined whether endogenous SLs influence endogenous ethylene and visa versa in pea using the ethylene-insensitive ein2 mutant (Weller et al., 2015; Foo et al., 2016c) and the SL-deficient ccd8-1 mutant. We found no indication that the ein2 mutant significantly altered the expression of the SL biosynthesis genes CCD7, CCD8, and D27 or the level of the major SLs produced by pea when compared with wild-type cv Torsdag plants (Fig. 7A). Similarly, we found no significant difference in ethylene production or in the expression of the key ethylene metabolism genes ACC synthase (ACS1) or ACC oxidase (ACOX) in ccd8-1 mutants compared with wild-type cv Parvus plants (Fig. 7B). An independent experiment with both SL-deficient ccd8-1 and ccd7-3 mutants confirmed that SL deficiency does not substantially alter ethylene production (data not shown).
Figure 7.
SL and ethylene levels and expression of biosynthesis genes in SL-deficient ccd8-1 or ethylene-insensitive ein2 plants compared with their respective wild types (WT). A, Levels of three SLs, fabacyl acetate, orobanchol, and orobanchyl acetate, and relative expression of the SL biosynthesis genes CCD7, CCD8, and D27 in root tissue of wild-type (cv Torsdag) and ein2 mutant plants. B, Ethylene evolution from whole plants of the wild type (cv Parvus) and ccd8-1 and expression of the ethylene metabolism genes ACS1 and ACOX in roots of these plants. Values are means ± se (n = 4), and analysis of each parameter by Student’s t test indicated no significant differences between the mutant and the wild type. FW, Fresh weight.
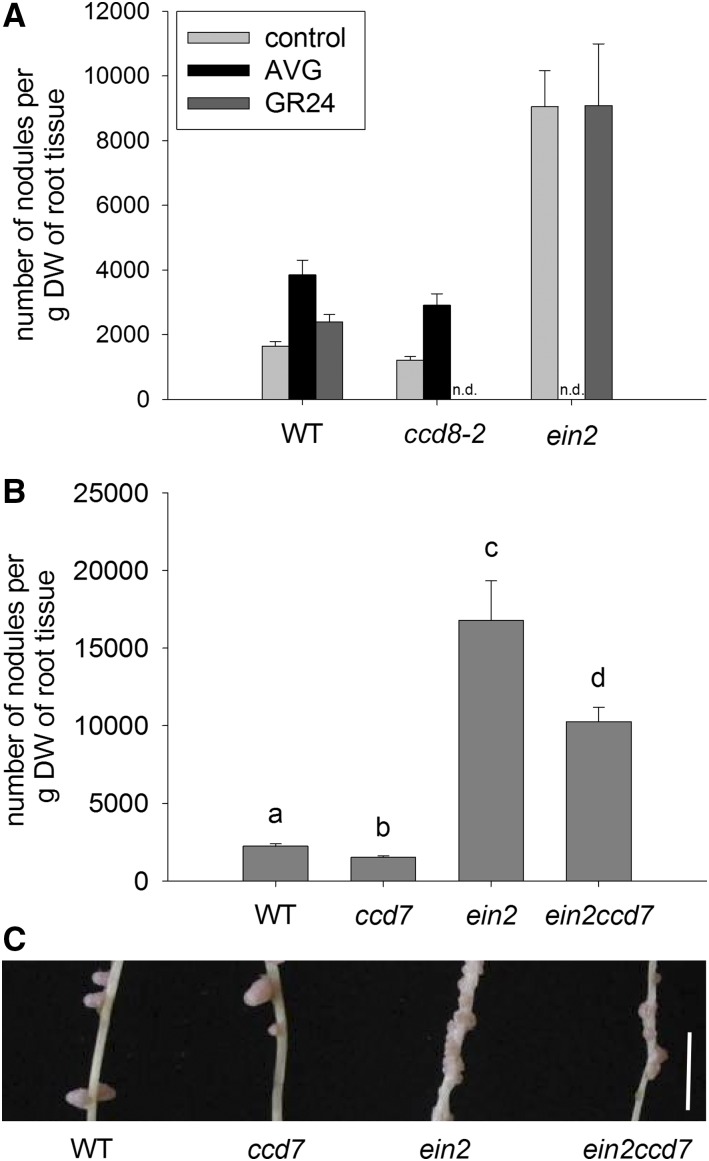
During nodulation, SLs do not appear to act downstream of ethylene, as, like wild-type plants, the SL-deficient ccd8-2 mutant could significantly elevate visible nodule number in response to the ethylene synthesis inhibitor aminoethoxyvinylglycine (AVG; Fig. 8A). However, GR24 could not elevate nodulation in the ethylene-insensitive ein2 mutant even though a small but significant increase occurred in wild-type cv Torsdag plants (Fig. 8A) and ccd8 mutants (Foo and Davies, 2011). We further investigated the relationship between SL and ethylene during nodulation by examining the nodulation phenotype of a double mutant disrupted in the SL biosynthesis gene CCD7 and also the ethylene signaling component EIN2 and comparing these double mutant plants with wild-type, ein2, and ccd7 segregants from the cross between ein2 and ccd7 (Fig. 8, B and C). We found that, like ein2 single mutant plants, the ein2 ccd7 double mutants formed significantly more visible nodules than the wild type (P < 0.01). However, visible nodule number in the double mutant plants was significantly lower than in the ein2 single mutant plants (P < 0.05). Indeed, the phenotype of ein2 ccd7 double mutants appears to be essentially additive, as the approximate 40% reduction in nodule number in ein2 ccd7 compared with the ein2 single mutant is consistent with an approximate 40% significant reduction in nodule number of the ccd7 single mutant compared with the wild type (P < 0.05). Taken together with the application studies, this finding suggests that SL and ethylene are likely to act largely independently to control nodule number.
Figure 8.
Interactions between ethylene and SL during nodulation. A, Nodule number in wild-type cv Torsdag (WT), SL-deficient ccd8-2, or ethylene-insensitive ein2 pea mutants treated with the synthetic SL (+)-GR24, the ethylene synthesis inhibitor AVG, or solvent control (n = 6–11). n.d., Not done. B, Nodule number in wild-type, SL-deficient ccd7, ein2, and double mutant ccd7 ein2 segregants (n = 5–7). Values are means ± se. For A, separate two-way ANOVAs were performed for the ccd8 and ein2 comparisons, and for B, a one-way ANOVA was performed, and values with different letters are significantly different (P < 0.05). DW, Dry weight. C, Photograph of nodules on secondary roots (tertiary roots have been removed). Bar = 5 mm.
DISCUSSION
Over the last decade, SLs have been implicated in a wide range of developmental processes (Foo and Reid, 2013; Smith and Li, 2014). However, the exact mechanism by which SLs influence each process is still emerging. This has been the case for the role of SLs in the interaction between legumes and N-fixing rhizobia, with a number of studies indicating that SLs influence the ultimate number of mature nodules in several species, but the specific point during nodulation at which this occurs has not been established (for review, see López-Ráez et al., 2017). In this study, we show that infection thread formation is reduced specifically in SL-deficient ccd8 mutants during nodulation and that this effect may occur due to the action of the CCD8 product on the bacterial partner. This is presumably due to the reduced SL levels in this mutant, as exogenous GR24 can overcome this reduction in infection thread formation. Application and genetic studies indicate that this influence of SLs early in nodulation appears to be independent of ethylene signaling. The disruption of infection thread formation in an SL-deficient mutant is accompanied by reduced expression of some early nodulation genes, indicating that SLs result in a modification of the Nod LCO signaling pathway. Furthermore, we show that SL levels are down-regulated by mutations in the Nod LCO signaling pathway, and this appears to be dependent on NSP2 but independent of the NIN transcription factor.
In species that undergo root hair infection, root hair curling is one of the first physical events observed following challenge with rhizobia or specific Nod LCOs. We found that SLs do not appear to influence this very early event, as SL-deficient plants had a similar number of curled root hairs when challenged with compatible rhizobia or Nod LCOs as wild-type plants (Fig. 2C). However, SL-deficient ccd8 mutants did have reduced formation of infection threads (Figs. 3A and 4A; Supplemental Fig. S1), the next physical event in nodulation that allows the uptake of rhizobia from the soil and the ultimate delivery of the rhizobia into the dividing cells that will form the nodule. This reduction could be overcome by the application of GR24 (Fig. 4). These results suggest a positive influence of SLs on infection thread formation in pea, which is consistent with studies in M. truncatula that found that the expression of the SL biosynthesis gene CCD8 was specifically up-regulated in infected root hairs and developing nodule primordia (Breakspear et al., 2014). Other studies in M. truncatula also found that, later in development, the SL biosynthesis genes D27, CCD7, and CCD8 were expressed in the meristem and distal infection zone of mature nodules, leading to speculation that SLs may play a role during the later stages of nodule organogenesis or function (van Zeijl et al., 2015). However, although we found that nodules do contain low levels of SLs, the nodules that do develop on SL-deficient mutants of pea are of a similar size to those on wild-type plants (Foo and Davies, 2011) and also can fix atmospheric nitrogen (Fig. 3E).
In addition to infection via root hairs, many other legume-rhizobia interactions involve the uptake of rhizobia through cracks in the epidermis. Interestingly, when we examined the influence of SLs on the nodulation of lupin, a legume that uses crack-entry uptake of rhizobia, we found that the synthetic SL (+)-GR24 had no influence on nodule number (Supplemental Fig. S2). This is in contrast to reports of the effects of GR24 on nodule number in a range of root hair-entry species (pea, M. truncatula, M. sativa, and L. japonicus; Soto et al., 2010; Foo and Davies, 2011; Liu et al., 2013; De Cuyper et al., 2015). This observed lack of effect of SL on a species that does not form infection threads is consistent with our hypothesis that SLs appear to influence infection thread formation specifically.
While SL-deficient ccd8 mutants form fewer infection threads and ultimately nodules than wild-type plants, this is not observed in d14 SL receptor mutants (Supplemental Fig. S1). This is consistent with previous studies that showed that pea max2 mutants, disrupted in another key SL signaling component, also did not display reduced nodulation (Foo et al., 2013). This suggests that, like its role in AM development, SLs may influence nodulation by affecting the microbial partner rather than the plant partner. One possibility is that SL-deficient ccd8 plants produce altered levels of compounds known to be important in nodulation, such as flavonoids. However, we found no major difference in the profile or level of flavonoids in root exudates from SL-deficient ccd8 and wild-type roots (Fig. 2A). We also found that SL-deficient ccd8 root exudates had a similar positive influence on bacterial growth to wild-type root exudates (Fig. 2B), suggesting that products resulting from CCD8 action do not influence nodulation by directly affecting bacterial growth. This is consistent with previous studies showing that application of the synthetic SL GR24 does not influence the growth or Nod LCO production of rhizobia, including the pea-compatible R. leguminosarum (Moscatiello et al., 2010; Soto et al., 2010). Recent reports suggest that SLs such as GR24 may influence rhizobial motility by affecting swarming behavior, although the specific function of swarming in nodulation per se is still unclear (Tambalo et al., 2014; Peláez-Vico et al., 2016). Given the discrete window in which we have shown that CCD8 influences nodulation, after root hair curling but before infection thread formation, future work could examine the role of products of CCD8 action, presumably SLs, in the dialogue between the plant and its bacterial partner during this crucial stage of nodule development.
The physical and molecular events that occur during nodulation are regulated in part via the Nod LCO signaling pathway (Fig. 1). Indeed, previous studies have indicated that the Nod LCO signaling pathway may influence SL biosynthesis, with some mutants in this pathway reported to have lower SL levels and/or expression of SL biosynthesis genes (Liu et al., 2011; van Zeijl et al., 2015). We examined this systematically, quantifying the level of SLs produced in pea mutants with lesions in each step of the signaling pathway (Fig. 6; Supplemental Fig. S6). We found that SL levels are reduced in mutants in core elements of the Nod LCO pathway, including a Nod LCO receptor (LYK3), and signaling elements (DMI1, DMI2, and DMI3). This positive influence on SL levels in wild-type plants requires the downstream transcription factor NSP2 but not NIN. Nonmycorrhizal species such as Arabidopsis (Arabidopsis thaliana) have lost several of these key SYM genes (DMI2 and DMI3; Delaux et al., 2014) but retained others. It would be interesting to examine the influence of these genes on noncanonical SL levels, since recent reports indicate that Arabidopsis does not appear to produce canonical SLs (Xie et al., 2015; Brewer et al., 2016). Given the role of CCD8 in influencing infection thread number, it is interesting that the nsp2 mutation blocks bacterial colonization of the curled root hairs while the nin mutation (also known as sym35) blocks the next stage, which is the initiation of infection threads (Tsyganov et al., 2002; Borisov et al., 2003). Recent progress has been made in our understanding of infection thread formation (Fournier et al., 2015), and it will be interesting to explore specific nonplant roles for SLs during this process in the future.
The promotion of SL levels by the Nod LCO signaling pathway, together with the fact that the Nod LCO pathway is required for rhizobia and/or Nod LCOs to elevate the expression of some SL biosynthesis genes (van Zeijl et al., 2015), suggest that Nod LCOs may induce SL biosynthesis. However, when the SL levels in the root infection zone (1–2 cm behind the root tip) of wild-type pea were measured 1, 2, 3, and 7 d after inoculation, we did not find a consistent increase in SL levels (data not shown). If SL levels are elevated specifically in root hairs, as suggested by the root hair expression of CCD8 in M. truncatula (Breakspear et al., 2014), such localized increases in levels may be difficult to detect in extracts from root sections.
Overall, we found that SLs act largely independently of ethylene in nodulation. We found no evidence that SL deficiency influences ethylene levels, as ethylene levels and the expression of ethylene metabolism genes were not altered significantly in ccd8-1 compared with wild-type plants (Fig. 7B). Importantly, we found a 7- to 8-fold increase in nodule number of ein2 mutants on either a wild-type or SL-deficient ccd7 background (Fig. 8B). This additive phenotype suggests that ethylene and SLs control nodule number largely independently. The fact that the nodulation of ethylene-insensitive ein2 pea mutants was not responsive to GR24 (Fig. 8A) but that nodule number could be elevated in SL-deficient pea mutants treated with the ethylene synthesis inhibitor AVG (Fig. 8A) suggests that the ethylene response of the plant has a much greater influence on nodulation than SL exudation. SL-ethylene interactions also have been investigated in root hair elongation and leaf senescence in Arabidopsis, and there is evidence for somewhat different interactions and independent effects of each hormone (Kapulnik et al., 2011; Ueda and Kusaba, 2015), reflecting the common observation that plant-hormone interactions are often specific to a given developmental process (Vanstraelen and Benková, 2012).
In conclusion, we have highlighted a specific role for SLs during nodule development. SLs appear to promote infection thread formation and may do so by influencing the bacterial partner. We have revealed that many elements of the Nod LCO signaling pathway are required to up-regulate SL levels, and this and other studies support the idea that Nod LCOs may elevate SL biosynthesis during nodulation. Although important roles for SLs during the nutrient regulation of development have been established (Brewer et al., 2013), it is important to note that SLs are not required to modulate nodulation in response to nitrogen or phosphorus (Foo et al., 2013). It appears that SLs may play a small but significant role during nodulation, and future studies may explore the role of SL action, either directly or indirectly, on the rhizobial partner, including in other plant-rhizobium symbioses with different developmental features, such as actinorhizal interactions that form via intracellular infection and root hair penetration.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The pea (Pisum sativum) lines used were the SL-deficient line ccd8-1 (also known as rms1-1; Beveridge et al., 1997) derived from wild-type cv Parvus; the SL receptor mutant d14 (de Saint Germain et al., 2016) derived from cv Torsdag (also known as rms3-1 and K487; Arumingtyas et al., 1992); the ethylene-insensitive ein2 mutant (Weller et al., 2015) derived from cv Torsdag; and the symbiosis mutants nfp (sym10; Madsen et al., 2003) and dmi2 (sym19; Stracke et al., 2002) derived from cv Frisson, dmi1 (sym8; Edwards et al., 2007) and nin (sym35; Borisov et al., 2003) derived from cv Finale, and dmi3 (sym9; Lévy et al., 2004) and nsp2 (sym7; Kaló et al., 2005) derived from cv Sparkle. The ccd8-2 line (also known as rms1-2) mutant was derived from cv Weitor (Beveridge et al., 1997), and this line was used in Figure 2. The ccd8-2 line also has been backcrossed to cv Torsdag (also known as rms1-2T; Foo et al., 2013), and this line was employed in Figures 4 and 8 to allow direct comparison with ein2 and in Supplemental Figure S1 to allow direct comparison with d14. The double mutant ein2 ccd7 was derived from a cross between ccd7-3 (also known as rms5-3T; Foo et al., 2013) and ein2. In all cases, comparisons were made between the mutant line and its wild-type progenitor line. Seeds were surface sterilized with 70% (v/v) ethanol and grown in growth cabinets (18-h photoperiod, 20°C day, 15°C night, under cool-white fluorescent tubes [100 µmol m−2 s−1]), two per pot in vermiculite, under conditions to exclude rhizobial bacteria, unless stated otherwise.
Flavonoid Analysis and Bacterial Growth Assays
Plants were grown for 21 d as described by Foo et al. (2016c), removed from pots, placed on damp paper, and five 10- × 10-mm filter papers were placed 1 cm from root tips. After 2 h, the papers were pooled from 20 plants per genotype, and flavonoids were extracted in 1:1 ethyl acetate:methanol overnight at 4°C. The sample was dried and resuspended in 100 µL of 0.4% acetic acid. A total of 10 μL was injected onto an Acquity UPLC BEH C18 column using an Acquity H-series UPLC device coupled to an Acquity Photodiode Array detector (PDA) (Waters) in series with a Xevo triple quadrupole mass spectrometer. The column was held at 35°C, flow rate was 0.35 mL min−1, with 100% A (1% acetic acid):0% B (acetonitrile) increasing to 90% A:10% B (v/v) at 0.5 min and 70% A:30% B (v/v) at 20 min, and the PDA was monitored continuously over the range 230 to 500 nm. The initial selection of targeted flavonoids was based on peaks with UV λmax between 325 and 380 nm, supported by subsequent positive and negative ion tandem mass spectrometry spectra and, where possible, by characteristic flavonoid full UV spectra.
The mass spectrometer was operated in several different modes in separate injections. Initially, negative ion full-scan survey scans were acquired over the mass-to-charge ratio (m/z) range 200 to 1,700 every 0.3 s, with a cone voltage of 40 V, after which ScanWave daughter scans at 32 and 45 V collision energy at 2,000 m/z per second were acquired automatically from the strongest ions. Subsequent runs targeted some of the stronger putative flavonoids (e.g. m/z 771, 787, and 1,023) to acquire higher quality ScanWave daughters with a collision energy of 40 V. The ion source temperature was 130°C, the desolvation gas was nitrogen at 950 L h−1, the desolvation temperature was 450°C, and the capillary voltage was 2.7 kV in all cases. Positive ion tandem mass spectrometry data also were acquired in precursor scan mode using m/z 287 and 303 as precursor ions (targeting kaempferol- and quercetin-containing flavonoids, respectively). This indicated that the majority of flavonoids present were highly glycosylated and acylated, consistent with the finding of kaempferol and quercetin sophorotriosides and several acylated versions of these in pea shoots (Ferreres et al., 1995). While the molecular masses of five of the compounds found in pea roots through this process (772, 788, 934, 950, and 964 D) were the same as those reported for pea shoot flavonoid sophorotriosides, there was insufficient sample to determine their structures. Peaks identified as probable flavonoids through this method were then targeted by selected ion monitoring, using a dwell time of 25 ms per ion (Supplemental Table S1).
For bacterial growth assays, root exudates were collected and extracted as described above and resuspended in 1 mL of water. Four replicate cultures of Rhizobium leguminosarum bv viciae (RLV248) were grown in yeast-mannitol broth with 250 µL of the extracted root exudate (or 250 µL of water in control samples) at 25°C and 120 rpm. Bacterial growth was measured 0, 20, 24, 28, 48, and 52 h after inoculation by measuring the A600 of 1 mL of culture on an Spectrostar Nano spectrophotometer (BMG Labtech).
Root Hair Curling, Infection Thread, and Developing Nodule Studies
Plants grown for root hair curling studies were grown for 7 d and inoculated with 75 mL of sterile water (control), 0.1 × 10−6 m Nod LCO (CO-IV [C18:1 Δ11Z, Ac]), or a 10% solution (v/v) of a 3-d-old culture of R. leguminosarum bv viciae (RLV248) grown in yeast-mannitol broth (an equal volume of sterile yeast-mannitol broth and/or solvent [50% acetonitrile (v/v)] also was included in all treatments/controls). Nod LCO was prepared by a chemoenzymatic approach combining biotechnological synthesis of the saccharidic backbone in transgenic Escherichia coli followed by chemical acylation with cis-vaccenic acid (Samain et al., 1999; Ohsten Rasmussen et al., 2004; Chambon et al., 2015). Five days after treatment, six to 10 roots per plant were stained briefly with Toluidine Blue and examined with a light microscope, and the percentage of curled root hairs was recorded.
For infection thread and developing nodule studies, seedlings were inoculated 10 d after planting with 75 mL of a 10% solution (v/v) of a 3-d-old culture of R. leguminosarum bv viciae (RLV3841) carrying pXLGD4 (carrying the lacZ reporter gene; supplied by John Innes Centre) grown in Tryptone Yeast (TY) medium with streptomycin (200 µg mL−1) and tetracycline (5 µg mL−1). Plants received a weekly dose of modified Long Ashton nutrient solution with no N and 5 mm NaH2PO4, as described by Foo et al. (2013). Nine days after inoculation, root segments from the root tip to the first visible nodule (approximately 3–6 cm) or entire lateral roots were harvested and fixed in 25% glutaraldehyde (v/v) in wash buffer (100 mm sodium phosphate at pH 7, 10 mm KCl, and 1 mm MgCl2·6H2O), then twice washed in wash buffer, and stained overnight in the dark in wash buffer containing 5 mm K3[Fe(CN)6], 5 mm K4[Fe(CN)6]·3H2O, and 800 µg mL−1 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid. Root segments were viewed with a Zeiss Axiolab light microscope with a 20× objective, and images were taken with a Nikon Digital Sight DS-Fi2 camera. The root length, number of blue-stained infection threads, developing nodules (nodules not visible with the naked eye but visible under a 20× objective; Fig. 1), and, for Figure 4 and Supplemental Figure S1, the number of nodules visible to naked eye were counted in one to 10 roots of six to 15 plants per genotype. Total nodule number is the sum of developing and visible nodules. Root samples were scored blind, meaning that roots were scored without the scorer knowing the genotype.
Nodule Function Studies
For the acetylene reduction assay, plants were grown and inoculated with R. leguminosarum bv viciae (RLV248) as described previously (Foo and Davies, 2011). A whole-root system of mature nodulated plants was placed in 100-mL bottles sealed with a gas-tight lid fitted with a septum, with four replicate bottles per genotype. Acetylene was added to each bottle to make a final concentration of 1% (v/v), and the roots were incubated for 4 h at room temperature. The amount of ethylene generated from the reduction of acetylene via nitrogenase was measured by gas chromatography-mass spectrometry as described by Foo et al. (2006), except that no cryotrap was used. Injections were split 20:1, and the oven temperature was 50°C. A standard of mixed ethylene and acetylene (1% [v/v] of each) was analyzed between each replicate so that the concentration of ethylene in the samples could be calculated. After analysis, the dry weight of nodules was measured, and ethylene evolved was expressed on a per gram dry weight of nodules basis.
Hormone and Nod LCO Application Studies
For hormone application studies with blue lupin (Lupinus angustifolius), the plants were grown and inoculated as described previously (Foo et al., 2016b). Pea plants were inoculated on day 7 as described previously (Foo and Davies, 2011). For both pea and lupin, pots were then treated on days 9, 13, 15, and 18 with 75 mL of control (water), 2 × 10−5 m (+)-GR24 (also referred to as GR245DS; Scaffidi et al., 2014), or 1.5 × 10−5 m AVG (Sigma-Aldrich). Chemicals were dissolved in dimethyl sulfoxide, and control plants received an equal concentration of dimethyl sulfoxide in water only. On day 28, plants were harvested, and the number of nodules and root dry weight were recorded. The effects of GR24 application on infection thread formation and nodule development were scored as described above.
To study the effects of Nod LCO on root architecture, 4-d-old sterile pea seedlings were transplanted to slants (20 mL of one-half-strength modified Long Ashton nutrient solution with 0.8 mm KNO3 and 0.25 mm NaH2PO4 solidified with 5 g L−1 Phytagel [Sigma-Aldrich]). While the medium was molten, Nod LCO (CO-IV [C18:1 Δ11Z, Ac]) was added to a final concentration of 1 × 10−8 m. Nod LCO was dissolved in a minimal volume of 50% acetonitrile (v/v), and control slants received an equal concentration of acetonitrile. On day 18, plants were harvested, and tap root length, lateral root number, and the length of the longest lateral root were recorded.
Hormone Analysis
For SL quantification, approximately 2 g (fresh weight) of whole-root tissue (three to four plants per replicate) was harvested from 3- to 4-week-old plants, and SLs were purified and measured as described by Foo et al. (2013) with the inclusion of labeled SL standards ([6′-2H1]fabacyl acetate, [6′-2H1]orobanchol, and [6′-2H1]orobanchyl acetate). For ethylene quantification, plants were grown in sterile vermiculite, three per 250-mL glass jar in a growth cabinet (as described in “Plant Material and Growth Conditions”) for 12 d, and ethylene evolution from whole plants was performed from four replicate jars, as described by Foo et al. (2016c).
Gene Expression
For the gene expression studies in Figure 5 and Supplemental Figures S3 and S4, plants were grown for 10 d and root tip tissue was harvested (day 0). The remaining plants were inoculated with a 10% solution (v/v) of a 3-d-old culture of RLV248, and 2 and 4 d later, root tip tissue was harvested. For the gene expression studies in Figures 6 and 7A, whole-root tissue (three to four plants per replicate) was harvested from 3- to 4-week-old plants, and after grinding, a subset was taken for gene expression analysis and the remainder was processed for hormone analysis (see above). For the gene expression studies in Figure 7B, after ethylene levels were measured, plants were removed from jars and the whole-root tissue (three plants per replicate) was harvested.
Tissue was ground and RNA was extracted from approximately 100 mg of tissue using the ISOLATE II RNA Mini Kit (Bioline). cDNA was synthesized from 1 µg of RNA using the SensiFAST cDNA Synthesis Kit (Bioline). cDNA was diluted, and duplicate, real-time PCRs were performed in a Rotor Gene 2000 (Corbett) using the SensiFAST SYBR Hi-ROX Kit (Bioline) and 100 to 200 pmol of a primer pair. Primer pairs for genes analyzed in this study were as follows: PsD27 (forward, 5′-CAAGCAGCAACAGGAATCAG-3′, and reverse, 5′-TTGATGGTGGCATCACTCTC-3′), PsCCD7 (RMS5 forward and reverse; Johnson et al., 2006), PsCCD8 (RMS1 forward and reverse; Johnson et al., 2006), PsENOD12a (Foo et al., 2016b), PsENOD12b (forward, 5′-TGAACCACCAGTGAATGAGC-3′, reverse, 5′-TGGATGTTATGTTCCGCTGT-3′), PsENOD40 (Foo et al., 2016b), PsACS1 (Foo et al., 2006), PsACOX (Foo et al., 2006), the defense markers PsPAL1, PsHmm6 and PsB-Gluc (Blake et al., 2016) and the housekeeping gene ACTIN (Foo et al., 2005). Standard curves were created for each gene using serially diluted plasmids containing cloned fragments of each amplicon. The average concentration of technical replicates was calculated. The relative gene expression of four biological replicates was determined by comparing the concentration of the gene of interest with the ACTIN concentration of that sample.
Statistical Analysis
For pairwise comparisons, Student’s t tests were performed in Excel. For other experiments, one- or two-way ANOVAs were performed in R version 3.2.2 (R Core Team), followed by Tukey’s honest significant difference posttests where appropriate. When appropriate, the data were log transformed prior to analysis.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Nodule development in wild-type, SL-deficient ccd8-2, and SL-insensitive d14-1 plants infected with lacZ-labeled R. leguminosarum bv viciae.
Supplemental Figure S2. Nodule number in blue lupin following treatment with (+)-GR24 or solvent control.
Supplemental Figure S3. Expression of the SL biosynthesis genes CCD7, CCD8, and D27 at 0, 2, and 4 d following inoculation with R. leguminosarum bv viciae in wild-type cv Parvus.
Supplemental Figure S4. Expression of the disease marker genes at 0, 2, and 4 d following inoculation with R. leguminosarum bv viciae in wild-type cv Parvus and ccd8-1 plants.
Supplemental Figure S5. Root development in wild-type cv Parvus and SL-deficient ccd8-1 mutant plants following treatment with 0.1 μm Nod LCO or solvent control.
Supplemental Figure S6. Fold change relative to the respective wild type of the canonical SLs orobanchol and orobanchyl acetate in various pea symbiosis mutants.
Supplemental Table S1. Putative flavonoid ions monitored via selected ion monitoring compound retention times and UV absorbance data for stronger signals.
Acknowledgments
We thank Dr. David Nichols for assistance with hormone analysis; Shelley Urquhart, Tracey Winterbottom, and Michelle Lang (University of Tasmania) for assistance with experiments and plant care; and Dr. Jim Weller (University of Tasmania) for ein2 seed, Dr. Mike Ambrose (John Innes Centre) for pea sym mutant seed, Dr. Catherine Delaitre (Institut National de la Recherche Agronomique) for lyk3/sym37 seed, Sonali Roy and Dr. Phillip Poole (John Innes Centre) for the kind gift of lacZ-labeled R. leguminosarum, Dr. Chris McErlean (University of Sydney) for the kind gift of (+)-GR24, and Koichi Yoneyama (Utsunomiya University) and Dr. Kohki Akiyama (Osaka Prefecture University) for the kind gifts of measured quantities of labeled SLs.
Glossary
- LCO
lipochitooligosaccharide
- SL
strigolactone
- AM
arbuscular mycorrhizal
- AVG
aminoethoxyvinylglycine
Footnotes
This work was supported by the Australian Research Council through Discovery Project and Future Fellowship grants to J.B.R., E.F., and N.W.D. S.F., E.S., and S.C. acknowledge support from ICMG FR 2607, LabEx ARCANE (ANR-11-LABX-0003-01), PolyNat Carnot Institute.
References
- Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Arumingtyas EL, Floyd RS, Gregory MJ, Mufert IC (1992) Branching in Pisum: inheritance and allelism tests with 17 ramosus mutants. Pisum Genet 24: 17–31 [Google Scholar]
- Beveridge CA, Symons GM, Murfet IC, Ross JJ, Rameau C (1997) The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s). Plant Physiol 115: 1251 [Google Scholar]
- Blake SN, Barry KM, Gill WM, Reid JB, Foo E (2016) The role of strigolactones and ethylene in disease caused by Pythium irregulare. Mol Plant Pathol 17: 680–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N, et al. (2003) The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol 131: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear A, Liu C, Roy S, Stacey N, Rogers C, Trick M, Morieri G, Mysore KS, Wen J, Oldroyd GE, et al. (2014) The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26: 4680–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Koltai H, Beveridge CA (2013) Diverse roles of strigolactones in plant development. Mol Plant 6: 18–28 [DOI] [PubMed] [Google Scholar]
- Brewer PB, Yoneyama K, Filardo F, Meyers E, Scaffidi A, Frickey T, Akiyama K, Seto Y, Dun EA, Cremer JE, et al. (2016) LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 113: 6301–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon R, Despras G, Brossay A, Vauzeilles B, Urban D, Beau JM, Armand S, Cottaz S, Fort S (2015) Efficient chemoenzymatic synthesis of lipo-chitinoligosaccharides plant growth promotors. Green Chem 17: 3923–3930 [Google Scholar]
- De Cuyper C, Fromentin J, Yocgo RE, De Keyser A, Guillotin B, Kunert K, Boyer FD, Goormachtig S (2015) From lateral root density to nodule number, the strigolactone analogue GR24 shapes the root architecture of Medicago truncatula. J Exp Bot 66: 137–146 [DOI] [PubMed] [Google Scholar]
- Delaux PM, Varala K, Edger PP, Coruzzi GM, Pires JC, Ané JM (2014) Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet 10:e1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Clavé G, Badet-Denisot MA, Pillot JP, Cornu D, Le Caer JP, Burger M, Pelissier F, Retailleau P, Turnbull C, et al. (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol 12: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A, Heckmann AB, Yousafzai F, Duc G, Downie JA (2007) Structural implications of mutations in the pea SYM8 symbiosis gene, the DMI1 ortholog, encoding a predicted ion channel. Mol Plant Microbe Interact 20: 1183–1191 [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Indrasumunar A, Hayashi S, Lin MH, Lin YH, Reid DE, Gresshoff PM (2010) Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52: 61–76 [DOI] [PubMed] [Google Scholar]
- Ferreres F, Esteban E, Carpena-Ruiz R, Jiménez MA, Tomás-Barberán FA (1995) Acylated flavonol sophorotriosides from pea shoots. Phytochemistry 39: 1443–1446 [DOI] [PubMed] [Google Scholar]
- Foo E, Blake SN, Fisher BJ, Smith JA, Reid JB (2016a) The role of strigolactones during plant interactions with the pathogenic fungus Fusarium oxysporum. Planta 243: 1387–1396 [DOI] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA (2005) The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17: 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Davies NW (2011) Strigolactones promote nodulation in pea. Planta 234: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Foo E, Heynen EM, Reid JB (2016b) Common and divergent shoot-root signalling in legume symbioses. New Phytol 210: 643–656 [DOI] [PubMed] [Google Scholar]
- Foo E, McAdam EL, Weller JL, Reid JB (2016c) Interactions between ethylene, gibberellins, and brassinosteroids in the development of rhizobial and mycorrhizal symbioses of pea. J Exp Bot 67: 2413–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Reid JB (2013) Strigolactones: new physiological roles for an ancient signal. J Plant Growth Regul 32: 429–442 [Google Scholar]
- Foo E, Ross JJ, Davies NW, Reid JB, Weller JL (2006) A role for ethylene in the phytochrome-mediated control of vegetative development. Plant J 46: 911–921 [DOI] [PubMed] [Google Scholar]
- Foo E, Yoneyama K, Hugill CJ, Quittenden LJ, Reid JB (2013) Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol Plant 6: 76–87 [DOI] [PubMed] [Google Scholar]
- Fournier J, Teillet A, Chabaud M, Ivanov S, Genre A, Limpens E, de Carvalho-Niebel F, Barker DG (2015) Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol 167: 1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Russo G (2016) Does a common pathway transduce symbiotic signals in plant-microbe interactions? Front Plant Sci 7: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- González‐Sama A, Lucas MM, De Felipe MR, Pueyo JJ (2004) An unusual infection mechanism and nodule morphogenesis in white lupin (Lupinus albus). New Phytol 163: 371–380 [DOI] [PubMed] [Google Scholar]
- Govers F, Harmsen H, Heidstra R, Michielsen P, Prins M, van Kammen A, Bisseling T (1991) Characterization of the pea ENOD12B gene and expression analyses of the two ENOD12 genes in nodule, stem and flower tissue. Mol Gen Genet 228: 160–166 [DOI] [PubMed] [Google Scholar]
- Guinel FC. (2015) Ethylene, a hormone at the center-stage of nodulation. Front Plant Sci 6: 1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C (2006) Branching genes are conserved across species: genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol 142: 1014–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kapulnik Y, Delaux PM, Resnick N, Mayzlish-Gati E, Wininger S, Bhattacharya C, Séjalon-Delmas N, Combier JP, Bécard G, Belausov E, et al. (2011) Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233: 209–216 [DOI] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Liu J, Novero M, Charnikhova T, Ferrandino A, Schubert A, Ruyter-Spira C, Bonfante P, Lovisolo C, Bouwmeester HJ, Cardinale F (2013) Carotenoid cleavage dioxygenase 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus. J Exp Bot 64: 1967–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Kohlen W, Lillo A, Op den Camp R, Ivanov S, Hartog M, Limpens E, Jamil M, Smaczniak C, Kaufmann K, et al. (2011) Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23: 3853–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez JA, Shirasu K, Foo E (2017) Strigolactones in plant interactions with beneficial and detrimental organisms: the yin and yang. Trends Plant Sci 22: 527–537 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Moscatiello R, Squartini A, Mariani P, Navazio L (2010) Flavonoid-induced calcium signalling in Rhizobium leguminosarum bv. viciae. New Phytol 188: 814–823 [DOI] [PubMed] [Google Scholar]
- Ohsten Rasmussen M, Hogg B, Bono JJ, Samain E, Driguez H (2004) New access to lipo-chitooligosaccharide nodulation factors. Org Biomol Chem 2: 1908–1910 [DOI] [PubMed] [Google Scholar]
- Oláh B, Brière C, Bécard G, Dénarié J, Gough C (2005) Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. Plant J 44: 195–207 [DOI] [PubMed] [Google Scholar]
- Peláez-Vico MA, Bernabéu-Roda L, Kohlen W, Soto MJ, López-Ráez JA (2016) Strigolactones in the Rhizobium-legume symbiosis: stimulatory effect on bacterial surface motility and down-regulation of their levels in nodulated plants. Plant Sci 245: 119–127 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR (1997) A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275: 527–530 [DOI] [PubMed] [Google Scholar]
- Peters NK, Frost JW, Long SR (1986) A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233: 977–980 [DOI] [PubMed] [Google Scholar]
- Rasmussen A, Depuydt S, Goormachtig S, Geelen D (2013) Strigolactones fine-tune the root system. Planta 238: 615–626 [DOI] [PubMed] [Google Scholar]
- Samain E, Chazalet V, Geremia RA (1999) Production of O-acetylated and sulfated chitooligosaccharides by recombinant Escherichia coli strains harboring different combinations of nod genes. J Biotechnol 72: 33–47 [DOI] [PubMed] [Google Scholar]
- Scaffidi A, Waters MT, Sun YK, Skelton BW, Dixon KW, Ghisalberti EL, Flematti GR, Smith SM (2014) Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol 165: 1221–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Walker SA, Poyser S, Sagan M, Ellis TH, Downie JA (1999) Genetic mapping and functional analysis of a nodulation-defective mutant (sym19) of pea (Pisum sativum L.). Mol Gen Genet 262: 1–11 [DOI] [PubMed] [Google Scholar]
- Singh S, Katzer K, Lambert J, Cerri M, Parniske M (2014) CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15: 139–152 [DOI] [PubMed] [Google Scholar]
- Smith SM, Li J (2014) Signalling and responses to strigolactones and karrikins. Curr Opin Plant Biol 21: 23–29 [DOI] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Chatfield S, Haurogné K, Goussot M, Rameau C, Foo E, Beveridge CA, Leyser O (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17: 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto MJ, Fernandez-Aparicio M, Castellanos-Morales V, Garcia-Garrido JA, Delgado MJ, Vierheilig H (2010) First indications for the involvement of strigolactones on nodule formation in alfalfa (Medicago sativa). Soil Biol Biochem 42: 383–385 [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al. (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Symons GM, Reid JB (2008) Brassinosteroids, de-etiolation and the re-emerging art of plant hormone quantification. Plant Signal Behav 3: 868–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambalo DD, Vanderlinde EM, Robinson S, Halmillawewa A, Hynes MF, Yost CK (2014) Legume seed exudates and Physcomitrella patens extracts influence swarming behavior in Rhizobium leguminosarum. Can J Microbiol 60: 15–24 [DOI] [PubMed] [Google Scholar]
- Tang C, Robson AD, Dilworth MJ (1993) Anatomical and ultrastructural observations on infection of Lupinus angustifolius L. by Bradyrhizobium sp. J Comput Assist Microsc 5: 47–51 [Google Scholar]
- Tsyganov VE, Voroshilova VA, Priefer UB, Borisov AY, Tikhonovich IA (2002) Genetic dissection of the initiation of the infection process and nodule tissue development in the Rhizobium-pea (Pisum sativum L.) symbiosis. Ann Bot (Lond) 89: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Kusaba M (2015) Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol 169: 138–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Urquhart S, Foo E, Reid JB (2015) The role of strigolactones in photomorphogenesis of pea is limited to adventitious rooting. Physiol Plant 153: 392–402 [DOI] [PubMed] [Google Scholar]
- Vanstraelen M, Benková E (2012) Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol 28: 463–487 [DOI] [PubMed] [Google Scholar]
- van Zeijl A, Liu W, Xiao TT, Kohlen W, Yang WC, Bisseling T, Geurts R (2015) The strigolactone biosynthesis gene DWARF27 is co-opted in rhizobium symbiosis. BMC Plant Biol 15: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateshwaran M, Volkening JD, Sussman MR, Ané JM (2013) Symbiosis and the social network of higher plants. Curr Opin Plant Biol 16: 118–127 [DOI] [PubMed] [Google Scholar]
- Weller JL, Foo EM, Hecht V, Ridge S, Vander Schoor JK, Reid JB (2015) Ethylene signaling influences light-regulated development in pea. Plant Physiol 169: 115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Kisugi T, Nomura T, Akiyama K, Asami T, Yoneyama K (2015) Strigolactones are transported from roots to shoots, although not through the xylem. J Pestic Sci 40: 214–216 [Google Scholar]
- Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]