The low level of organic S in Sarcocornia as compared to Salicornia is the result of a higher L-cysteine degradation rate by O-acetylserine-(thiol) lyases, especially when supplemented with sulfate.
Abstract
Salicornia and Sarcocornia are almost identical halophytes whose edible succulent shoots hold promise for commercial production in saline water. Enhanced sulfur nutrition may be beneficial to crops naturally grown on high sulfate. However, little is known about sulfate nutrition in halophytes. Here we show that Salicornia europaea (ecotype RN) exhibits a significant increase in biomass and organic-S accumulation in response to supplemental sulfate, whereas Sarcocornia fruticosa (ecotype VM) does not, instead exhibiting increased sulfate accumulation. We investigated the role of two pathways on organic-S and biomass accumulation in Salicornia and Sarcoconia: the sulfate reductive pathway that generates Cys and l-Cys desulfhydrase that degrades Cys to H2S, NH3, and pyruvate. The major function of O-acetyl-Ser-(thiol) lyase (OAS-TL; EC 2.5.1.47) is the formation of l-Cys, but our study shows that the OAS-TL A and OAS-TL B of both halophytes are enzymes that also degrade l-Cys to H2S. This activity was significantly higher in Sarcocornia than in Salicornia, especially upon sulfate supplementation. The activity of the sulfate reductive pathway key enzyme, adenosine 5′-phosphosulfate reductase (APR, EC 1.8.99.2), was significantly higher in Salicornia than in Sarcocornia. These results suggest that the low organic-S level in Sarcocornia is the result of high l-Cys degradation rate by OAS-TLs, whereas the greater organic-S and biomass accumulation in Salicornia is the result of higher APR activity and low l-Cys degradation rate, resulting in higher net Cys biosynthesis. These results present an initial road map for halophyte growers to attain better growth rates and nutritional value of Salicornia and Sarcocornia.
Soil salinity is one of the oldest and most important abiotic stresses affecting agricultural productivity globally. According to the Food and Agricultural Organization of the United Nations, roughly 800 million hectares of land are affected by salt. It has further been predicted that approximately 50% of arable land will be affected by salt stress by the year 2050 (Wang et al., 2003). Therefore, there is an urgent need to develop techniques to confront the adverse effects of salinity stress and develop strategies to enhance crop production under saline conditions. To do so, it is necessary to understand the physiological processes and molecular mechanisms that have evolved in plants to tolerate salt resistance, and exploit them for sustainable crop production (Fatma et al., 2013; Iqbal et al., 2013; Khan et al., 2013).
Most crop plants are glycophytes that grow in nonsaline soils and bodies of fresh water. Glycophytes are able to adapt to moderate levels of salinity, albeit with decreased productivity. Halophytic plants, on the other hand, grow and thrive in highly saline waters and soils. Among the most promising candidates for the development of novel halophytic crops are species of the Salicornia and Sarcocornia. Both genera are phenotypically and ecologically very similar and occur naturally throughout the world, along coastal salt marshes, edges of saline lakes, and in areas where the vegetation is often subjected to daily tides that contain high sulfate concentrations (5 mM to 30 mM in interstitial water; Howarth and Giblin, 1983; Davy et al., 2001, 2006; de la Fuente et al., 2013; Steffen et al., 2015). The species of both genera are often referred to as pioneer plants on the sea coasts (Davy et al., 2001, 2006) and several Salicornia species are already used as a both fodder and a vegetable crop and can be irrigated with highly saline water, even with full seawater (Ventura et al., 2011a). The Sarcocornia genus differs from the annual Salicornia genus by its distinct perennial growth habit (Davy et al., 2006) and by differences in floral morphology (Kadereit et al., 2007). Both genera produce succulent shoots suitable for leafy vegetable production, but they differ in terms of yield and nutritional value (Ventura et al., 2011b).
Mineral nutrient levels are a major determinant of crop yield and quality; and saline environments complicate mineral nutrition and affect crop sustainability (Nazar et al., 2011a). The supply of optimal sulfur nutrition to plants is important because sulfur is an integral part of several important plant compounds, such as iron-sulfur clusters, polysaccharides, and sulfolipids, as well as a broad variety of biomolecules including vitamins such as biotin and thiamine, cofactors such as Coenzyme A and S-adenosyl-Met, peptides such as glutathione (GSH) and phytochelatins, secondary metabolites such as allyl Cys sulfoxides and glucosinolates, and the sulfur-containing amino acids Cys and Met (Kopriva 2006; Nocito et al., 2011). Cys residues (thiols) have the capacity to react with a broad spectrum of agents, ranging from free radicals, reactive oxygen species (ROSs), and cytotoxic electrophilic and organic xenobiotics, to affect the redox state of tissues and serve as signals in plant responses to stress (Mullineaux and Rausch, 2005; Koprivova et al., 2008a).
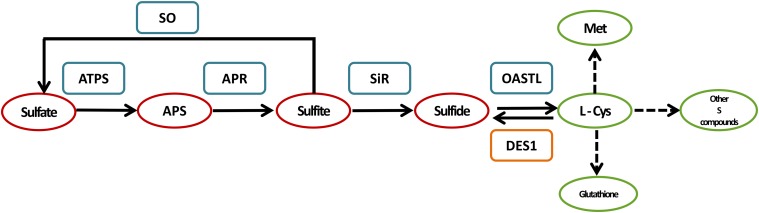
The main source of sulfur, sulfate, can either be taken up from the environment or generated within the plants from other S-containing compounds, such as sulfite (Brychkova et al., 2013, 2015). The sulfate reduction pathway (Fig. 1) is initiated in plastids (Leustek et al., 2000) and/or in the cytosol (Leustek, 2002) by the adenylation of transported sulfate by ATP sulfurylase (ATPS, EC 2.7.7.4) to generate adenosine 5′-phosphosulfate (APS). APS is then reduced to sulfite by the plastidic APS reductase (APR, EC 1.8.99.2). Further, the toxic sulfite can be oxidized to sulfate by peroxisomal sulfite oxidase (SO, E.C. 1.8.3.1.) or reduced to sulfide by the chloroplastic sulfite reductase (SiR, EC 1.8.7.1). Sulfide, together with O-acetyl-Ser (OAS) whose biosynthesis is catalyzed by Ser acetyltransferase (SAT, EC 2.3.1.30), is then incorporated into Cys in a reaction catalyzed by the O-acetyl-Ser-(thiol) lyase (OAS-TL, EC 2.5.1.47; Wirtz et al., 2004). The generated l-cys is a precursor of thiols containing metabolites (Kopriva, 2006). Cys homeostasis is controlled by the cytosolic l-Cys desulfhydrase 1 (DES1, EC 4.4.1.1), which catalyzes the breakdown of Cys to sulfide, ammonia, and pyruvate (Fig. 1; Álvarez et al., 2010).
Figure 1.
Schematic representation of the Sulfate reduction and Cys degradation pathways in Arabidopsis plants. ATPS catalyzes the adenylation of sulfate to APS using ATP as an electron donor. Then, APS is reduced by the plastidic enzyme APR to sulfite in the presence of two molecules of reduced GSH, which acts as an electron donor. The generated sulfite can be oxidized to sulfate by SO with the formation of H2O2 as a byproduct or further be reduced to sulfide by the SiR employing three molecules of reduced ferredoxin. The sulfide together with O-acetyl-l-Ser is the substrate for Cys biosynthesis catalyzed by OAS-TL. Cys homeostasis is controlled by l-Cys desulfhydrase (DES1, EC 4.4.1.1), which catalyzes the breakdown of Cys to sulfide, ammonia, and pyruvate. Red circle, inorganic S compounds; green circle, organic S compounds; blue rectangle, sulfate reduction pathway enzymes; orange rectangle, Cys degradation pathway enzyme.
The sulfur reduction pathway in glycophyte plants is modified in response to salinity stress (López-Berenguer et al., 2007; Koprivova and Kopriva, 2008b). ATPS, the first rate-limiting enzyme of the S assimilation pathway, is up-regulated in the glycophyte Brassica napus upon exposure to 150 mm NaCl (Ruiz and Blumwald, 2002). Exposure to this NaCl concentration also affects the expression of key enzymes of the sulfate reduction pathway, enhancing APR activity and increasing the abundance of the 3 APR isoforms 3-fold. Interestingly, an increase in APR activity was correlated with a higher rate of Cys biosynthesis to regulate the increased demand for GSH in response to the salinity stress as a defense response to ROSs (Koprivova and Kopriva, 2008b). Additionally, it has been shown that both the rate of S assimilation and the biosynthesis of thiols were greatly increased in B. napus (Ruiz and Blumwald, 2002) and barley (Hordeum vulgare L.; Astolfi and Zuchi, 2013) exposed to saline conditions.
The limited investigation of sulfur metabolism in halophytes has mainly focused on the role of S-containing metabolites such as reduced GSH and dimethylsulfonioproprionate (Nguyen et al., 2014; Colmer et al., 1996; Mulholland and Otte, 2000). Thus, it has been reported that increasing the sulfate concentration in the growth medium of 2% seawater-grown marsh cordgrass Spartina alterniflora resulted in a positive growth response, but no such growth response was seen in Spartina cynosuroides and in Spartina anglica grown with 0 mm to 1.6 mm sulfate supply (Stribling, 1997; Mulholland and Otte, 2000). Interestingly, Salicornia europaea has been determined to be extremely tolerant to sulfide ion accumulation (Ingold and Havill, 1984; Havill et al., 1985), although the tolerance mechanism is not understood. In contrast, Martin and Maricle (2015) examined 17 estuarine species, reporting that those with higher levels of cytochrome c oxidase activity were more sulfide-tolerant than those with lower levels.
Sulfate assimilation in glycophytes such as Arabidopsis (Arabidopsis thaliana), Brassica, and tobacco (Nicotiana tabacum) has been studied mainly from the perspective of S deprivation (Lappartient and Touraine, 1996; Lappartient et al., 1999; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003; Lewandowska and Sirko, 2008; Rouached et al., 2011; Király et al., 2012; Lee et al., 2012; Wipf et al., 2014). There is little information on sulfate assimilation in the presence of excessive S in glycophytes and even less in halophytes (Nazar et al., 2011b).
In contrast, sulfate was widely studied as a source of salinity and the response of halophytes to sodium sulfate was compared to that of sodium chlorides in various halophytes. Employing sodium sulfate at levels of 38 mm to 500 mm in comparison to sodium chloride resulted in toxic effects and a significant decrease in biomass accumulation in halophytes such as Prosopis strombulifera (Reginato et al., 2012, 2014; Llanes et al., 2013, 2014). Interestingly, Salicornia and Sarcocornia followed this inhibitory notion, exhibiting a significant decrease in biomass accumulation when grown with 100 mm sodium sulfate compared to 100 mm or even 200 mm sodium chloride (Supplementary Fig. S1).
Previously we showed the feasibility of cultivating Salicornia and Sarcocornia by applying a multiple harvest regime and irrigating with 100% seawater, generating economic yields with high nutritional value (Ventura et al., 2011a). The essentiality of supplementing artificial lighting to the natural day-length in Salicornia and successive harvesting regime in Sarcocornia for all-year-round cultivation was demonstrated, as well as the importance of molybdenum for improving total biomass accumulation in Salicornia grown in seawater (Ventura et al., 2010, 2011b, 2015; Ventura and Sagi, 2013).
Employing RNA and protein sequences of Salicornia (being highly similar to Sarcocornia) allows us to explore new avenues for enhancing yield and quality of this crop. Here we show that biomass and organic-S accumulation were significantly increased in S. europaea (ecotype RN) in response to sulfate supplementation, whereas S. fruticosa (ecotype VM) accumulated higher sulfate, but showed no increase in biomass. The sulfate-reductive pathway and the l-Cys desulfhydrase (DES) activities were explored for factors affecting sulfate and organic-S levels in the two halophytes. The major function of OAS-TLs is known to be the formation of l-Cys, but we found that OAS-TL also functions as a DES, degrading l-Cys to H2S. We attribute the higher organic-S and greater biomass accumulation in Salicornia to the significantly lower l-Cys DES activity of OAS-TL A and OAS-TL B, especially in the presence of sulfate supplementation, as well as to the higher APR activity, both of which should lead to higher net l-Cys. By contrast, Sarcocornia exhibited significantly higher DES and lower APR activity levels and did not accumulate biomass in response to sulfate supplementation. These results will hold great promise for sustainable agriculture, and will help halophyte growers to improve the nutritional value and productivity of edible halophytes, such as Salicornia and Sarcocornia.
RESULTS
High Sulfate Increased Biomass in Salicornia But Not in Sarcocornia, whereas Salinity Enhancement Increased Biomass Accumulation in Both Genera Grown in Low and High Sulfate
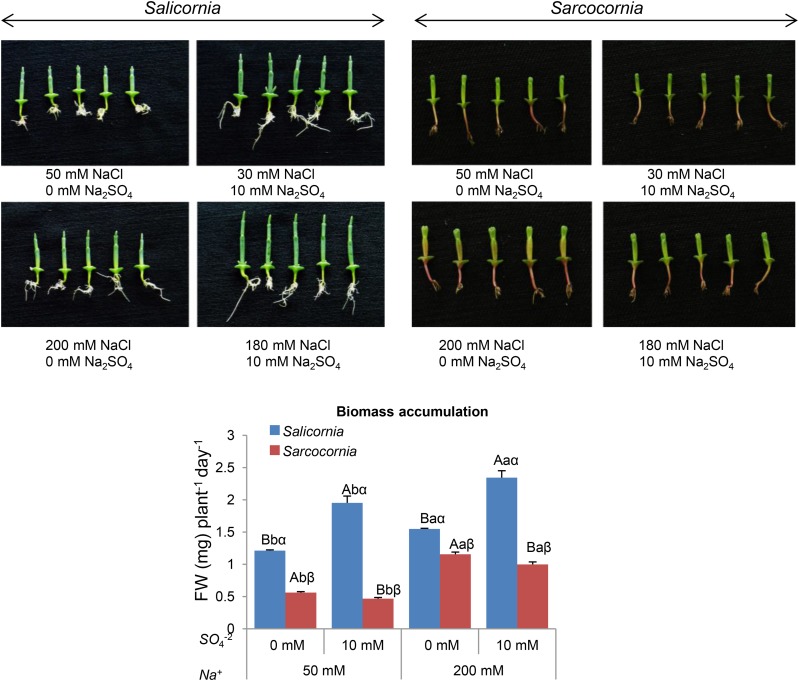
Enhanced demand for sulfur nutrition may be expected among halophyte plants such as Salicornia and Sarcocornia that are adapted to growth in saltmarshes and sea shores exposed to frequent seawater tides containing high concentrations of sulfate, ranging between 5 mm and 30 mM (Howarth and Giblin, 1983). Interestingly, irrigation with a solution containing 50% seawater improved biomass accumulation in both genera, as compared with the absence of seawater (Ventura et al., 2011a). These results led us to examine the effect of supplementation of high sulfate levels such as 10 mM, because optimization of sulfur nutrition may not only affect biomass but also the organic sulfur accumulation in plants. Because both Salicornia and Sarcocornia exhibit poor growth in the absence of NaCl in the growth medium (Ventura et al., 2011a), treatment conditions without NaCl were not compared to those with NaCl. Assessment of the effect of the 10 mm Na2SO4 supplementation to the 1/2 MS (containing 0.87 mm sulfate) growth medium was carried out in the presence of either 30 mm or 180 mm NaCl, so that both levels include 50 mm and 200 mm total Na+. As expected for halophytes, the biomass accumulation of both Salicornia and Sarcocornia was greater at the higher than at the lower salt concentration (Fig. 2). Although both Salicornia and Sarcocornia belong to the Amaranthaceae family, they differed in the effect on biomass accumulation of enhanced sulfate in the growth medium. Salicornia responded positively to the 10-mM sulfate supplementation, exhibiting a significant increase in the rate of biomass accumulation even when grown without NaCl supplementation (Fig. 2, left; Supplemental Fig. S2). In contrast to Salicornia, Sarcocornia exhibited a reduction in biomass accumulation in response to the addition of 10 mm sulfate to the growth mediums (Supplemental Fig. S2; Fig. 2, right). In summary, these results indicate that high sulfate is essential for optimal growth of Salicornia, but has a negative effect in Sarcocornia.
Figure 2.
Effect of sodium (50, 200 mM) and sulfate (0, 10 mM) concentrations on biomass accumulation of Salicornia (left) and Sarcocornia (right). Sulfate is applied as sodium sulfate and the remainder sodium as sodium chloride. The lower and higher salinity treatments are shown in the top and bottom photos, respectively. The values are means ± se (n = 30). Growth of the plants was measured as increase in biomass per day. Values denoted with different letters are significantly different according to the Tukey-Kramer HSD test, P < 0.05 (JMP 8.0 software). Different upper-case letters indicate significant differences between sulfate treatments in Salicornia or Sarcocornia. Different lower-case letters indicate significant differences between salinity treatments in Salicornia or Sarcocornia. Different Greek letters indicate significant difference between Salicornia and Sarcocornia subjected to the same treatment. The data are representative of one of 15 different experiments that yielded similar results.
Interestingly, a similar response to sulfate was evident with Salicornia (RN) and Sarcocornia (VM) grown in pots filled with the highly air-permeable and high-water-capacity perlite, irrigated with 100 mm and 200 mm NaCl solution supplemented with 1/2 Hoagland nutrient solution, containing 0 mm and 10 mm sulfate (compare Fig. 2 to Supplemental Fig. S3), i.e. biomass accumulation was improved in Salicornia in response to excess sulfate, but not in Sarcocornia. The results indicate that growth conditions, either in plates containing 1% plant agar mixed with 1/2 MS localized in a growth-room or grown in perlite supplied with 1/2 Hoagland nutrient solution and localized in a controlled greenhouse (see the “Materials and Methods”), did not affect the biomass accumulation response to high sulfate in Salicornia and Sarcocornia.
Effect of Sulfate and Salinity on Anthocyanin, Hydrogen Peroxide, and Superoxide Levels in Salicornia and Sarcocornia
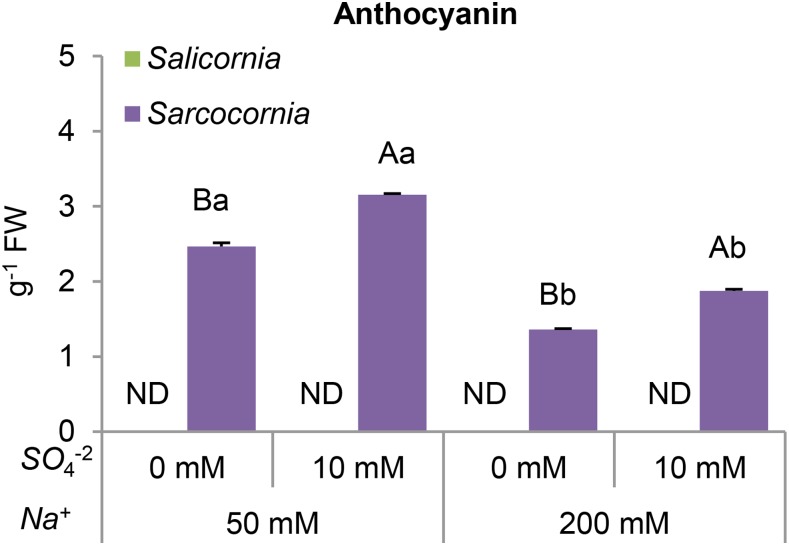
Enhanced anthocyanin biosynthesis is a characteristic response of flowering plants to unfavorable environmental conditions (Chalker-Scott, 1999). Anthocyanin was not detectable in Salicornia plants (Fig. 3), whereas anthocyanin was produced in Sarcocornia and its level was enhanced by the addition of 10 mm sulfate. The increase in anthocyanin content was greater in plants grown in the lower salinity medium (Fig. 3), suggesting that both low salinity conditions and sulfate supplementation are stressful for Sarcocornia.
Figure 3.
Effect of sodium (50, 200 mM) and sulfate (0, 10 mM) concentrations on the anthocyanin content of Salicornia and Sarcocornia. Sulfate is applied as sodium sulfate and the remainder sodium as sodium chloride. The values are means ± se (n = 5). Values denoted with different letters are significantly different according to the Tukey-Kramer HSD test, P < 0.05 (JMP 8.0 software). Different upper-case letters indicate significant differences between sulfate treatments in Salicornia or Sarcocornia. Different lower-case letters indicate significant differences between salinity treatments in Salicornia or Sarcocornia. ND, not detectable.
The generation of ROS, such as superoxide (O2−) and hydrogen peroxide (H2O2), are well-known components of the oxidative stress response and constitute one of the earliest responses of plant cells to nutrient imbalances. Increased ROS levels may result in accelerated catabolism leading to premature senescence (Yarmolinsky et al., 2014; Brychkova et al., 2015) and reduction in plant growth and loss of crop yield (You and Chan, 2015). In light of the high anthocyanin levels detected in Sarcocornia shoots, we investigated the effect of supplemental sulfate on ROS production in Salicornia and Sarcocornia shoots.
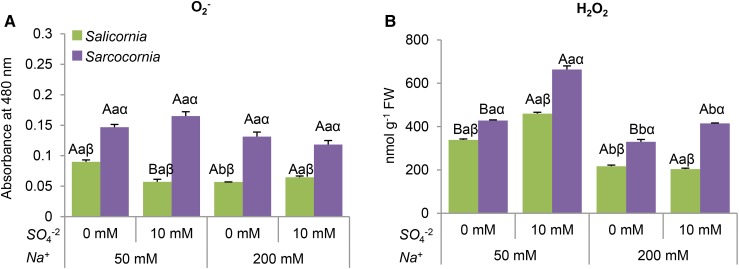
Superoxide levels in Salicornia were significantly higher in plants grown in the low salinity medium without sulfate supplementation, when compared to the other treatments (Fig. 4A). By contrast, superoxide production was at a similar level in all the treatments, being significantly higher in Sarcocornia than in Salicornia (Fig. 4A).
Figure 4.
Effect of sodium (50, 200 mM) and sulfate (0, 10 mM) concentrations on superoxide and hydrogen peroxide content of Salicornia and Sarcocornia. Sulfate is applied as sodium sulfate and the remainder sodium as sodium chloride. A, Superoxide (O2−) content in Salicornia and Sarcocornia. The values are means ± se (n = 4). B, Hydrogen peroxide (H2O2) content in Salicornia and Sarcocornia. The values are means ± se (n = 4). Values denoted with different letters are significantly different according to the Tukey-Kramer HSD test, P < 0.05 (JMP 8.0 software). Different upper-case letters indicate significant differences between sulfate treatments in Salicornia or Sarcocornia. Different lower-case letters indicate significant differences between salinity treatments in Salicornia or Sarcocornia. Different Greek letters indicate significant difference between Salicornia and Sarcocornia subjected to the same treatment.
Lower hydrogen peroxide levels were detected in both halophytic plants at the higher salinity, suggesting that high salinity conditions are preferable for both halophytes (Fig. 4B). The sulfate supplementation increased H2O2 in Sarcocornia at both salinities (Fig. 4B), whereas in Salicornia, the enhancement was evident only at the lower salinity (Fig. 4B). These results indicate that sulfate supplementation is more stressful for Sarcocornia than for Salicornia.
The Effect of Sulfate and Salinity on S-related Metabolites in Salicornia and Sarcocornia
The Effect on Sulfate, Sulfite, and Sulfide Levels
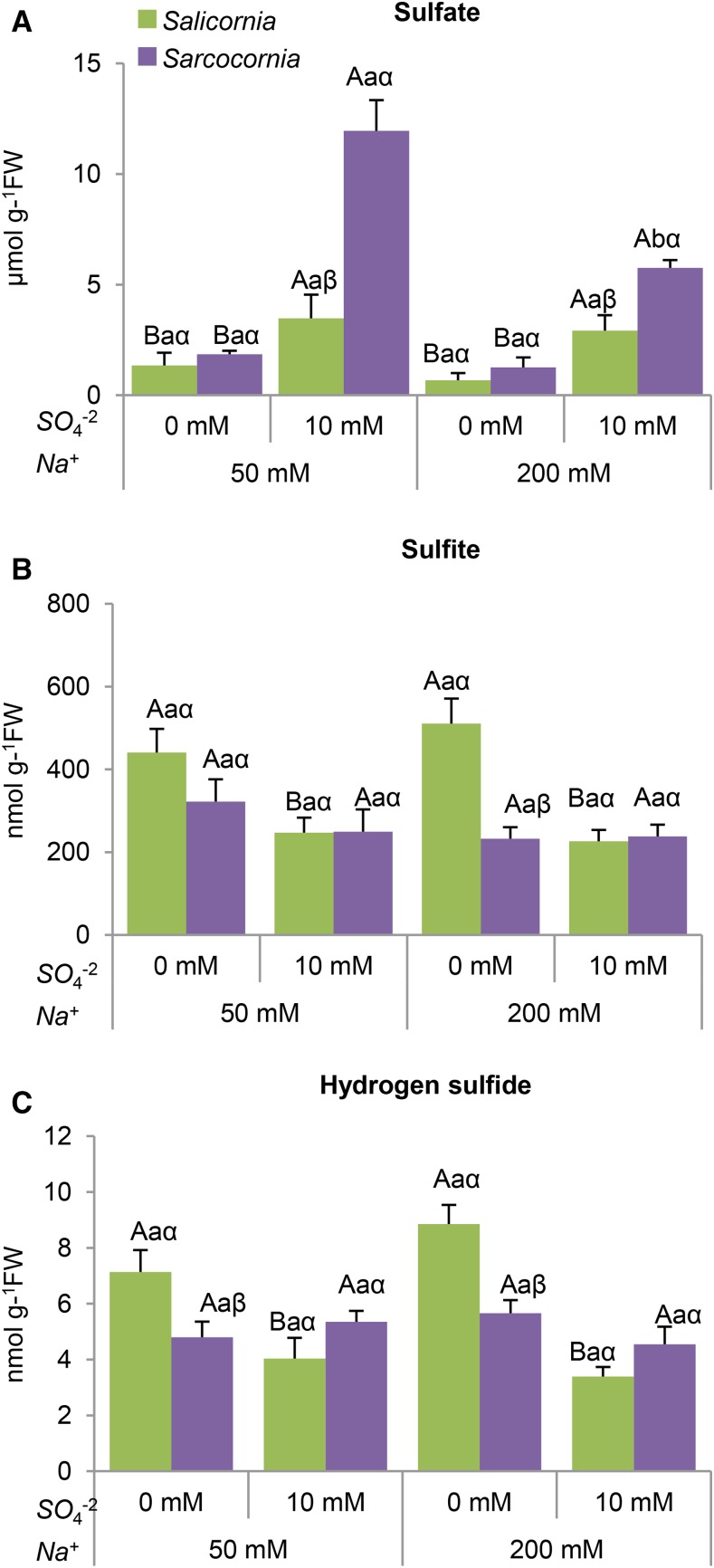
Sulfate is taken up and reduced via the sulfate-reductive pathway (Fig. 1), hence S is mostly available to plants in its fully oxidized form, the sulfate anion (Brychkova et al., 2013). As shown in Figure 5A, sulfate supplementation significantly increased sulfate levels in both plants. The effect was greater in Sarcocornia, especially when grown under the lower salinity (Fig. 5A), indicating that Sarcocornia accumulates more sulfate than Salicornia. Additionally, the results indicate that increasing salinity reduces sulfate accumulation. The higher sulfate accumulation in Sarcocornia as compared with Salicornia grown on plates, as shown here (Fig. 5A), is in agreement with the significantly enhanced sulfate shown in Sarcocornia when both halophytes were grown in pots filled with perlites and irrigated with 50% to 100% seawater supplemented with 200 ppm commercial N–P–K fertilizer (20–20–20 + microelements; Haifa Chemicals; Ventura et al., 2011a). The results further indicate that growth conditions in the plates did not affect the response of Salicornia and Sarcocornia to high sulfate.
Figure 5.
Effect of sodium (50, 200 mM) and sulfate (0, 10 mM) concentrations on sulfate, sulfite, and hydrogen sulfide contents in Salicornia and Sarcocornia. Sulfate is applied as sodium sulfate and the remainder sodium as sodium chloride. A, Sulfate levels in Salicornia and Sarcocornia. The values are means ± se (n = 3). B, Sulfite levels in Salicornia and in Sarcocornia. The values are means ± se (n = 3). The data are from one of three different experiments that yielded similar results. C, Hydrogen sulfide levels in Salicornia and in Sarcocornia. The values are means ± se (n = 4). Values denoted with different letters are significantly different according to the Tukey-Kramer HSD test, P < 0.05 (JMP 8.0 software). Different upper-case letters indicate significant differences between sulfate treatments in Salicornia or Sarcocornia. Different lower-case letters indicate significant differences between salinity treatments in Salicornia or Sarcocornia. Different Greek letters indicate significant difference between Salicornia and Sarcocornia subjected to the same treatment.
Sulfite is generated in the chloroplast by the GSH-dependent APR, but excess accumulation of sulfite is toxic to plants. To maintain sulfite homeostasis in the chloroplast, sulfite is further reduced to sulfide by the ferredoxin-dependent SiR in the sulfate-reductive pathway. Sulfite can also be detoxified to the less toxic thiosulfate by the sulfurtransferases or be oxidized to sulfate by the molybdenum cofactor enzyme, the peroxisomal SO, or can enter the sulfolipid reductive pathway in the chloroplast to generate sulfolipid (Nakamura et al., 2000; Papenbrock and Schmidt, 2000; Tsakraklides et al., 2002).
Salinity had no effect on the sulfite level in either plant (Fig. 5B). Sulfite levels in Salicornia were negatively affected by sulfate supplementation (Fig. 5B), whereas in Sarcocornia, sulfate supplementation had no significant effect (Fig. 5B).
Sulfide, the substrate for Cys biosynthesis, is the product of sulfite reduction by sulfite reductase, and/or a product of sulfur-containing metabolite degradation (Yarmolinsky et al., 2014; Brychkova et al., 2015). Sulfide levels were unaffected by salinity increase in both plants (Fig. 5C). A significant reduction in H2S content was seen in Salicornia, but not in Sarcocornia plants supplemented with 10 mm sulfate at the low salinity (Fig. 5C).
The Effect on Cys and GSH
Cys is the final product of the S assimilation pathway, and is the rate-limiting factor for GSH and Met biosynthesis. GSH is a storage form of reduced S in plants, playing an important role in controlling the redox status of plant tissue, protection against biotic and abiotic stresses, precursor of phytochelatins, detoxification of xenobiotics, and more (Rao and Reddy, 2008; Zechmann et al., 2008).
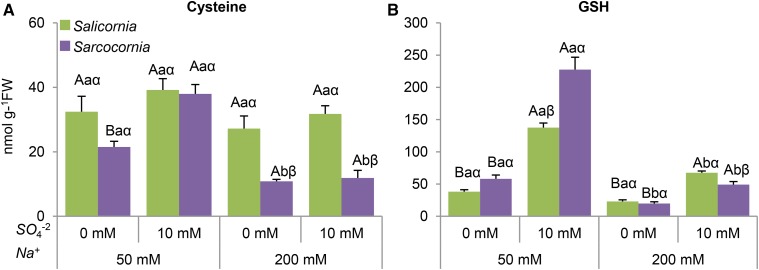
The level of free Cys in Salicornia was unaffected by salinity and sulfate treatments, whereas in Sarcocornia, Cys level decreased with salinity, being significantly lower than in Salicornia. Sulfate supplementation resulted in Cys enhancement at the lowest salinity level tested (Fig. 6A). Sulfate supplementation increased total GSH in both Salicornia and Sarcocornia, but was lower at the higher salt concentration in both plants (Fig. 6B).
Figure 6.
The effect of sodium (50, 200 mM) and sulfate (0, 10 mM) concentrations on Cys and GSH content in the shoots of Salicornia and Sarcocornia. Sulfate is applied as sodium sulfate and the remainder sodium as sodium chloride. A, Cys levels in Salicornia and Sarcocornia. Error bars indicate SE (n = 4). The data are from one of four different experiments that yielded similar results. B, Glutathione levels in Salicornia and Sarcocornia. The values are means ± se (n = 4). The data are from one of four different experiments that yielded similar results. Values denoted with different letters are significantly different according to the Tukey-Kramer HSD test, P < 0.05 (JMP 8.0 software). Different upper-case letters indicate significant differences between sulfate treatments in Salicornia or Sarcocornia. Different lower-case letters indicate significant differences between salinity treatments in Salicornia or Sarcocornia. Different Greek letters indicate significant difference between Salicornia and Sarcocornia subjected to the same treatment.
Total Sulfur and Organic Sulfur
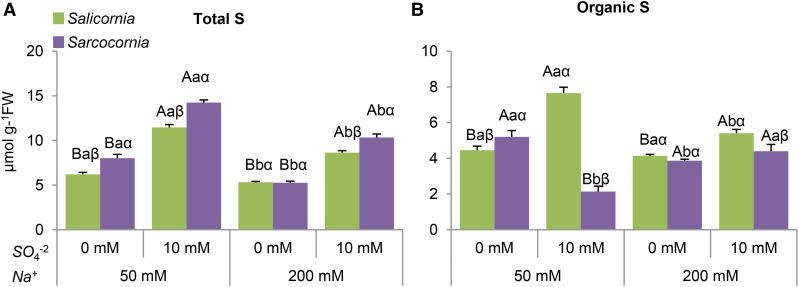
The effect of salinity and sulfate supplementation on total and organic-S was measured. Remarkably, except for plants grown at the highest salinity without supplementation of sulfate, the total sulfur level in Sarcocornia was significantly higher than in Salicornia plants (Fig. 7A). In both halophytes, the total sulfur level increased with increasing sulfate and decreased with increasing salinity in the growth medium (Fig. 7A).
Figure 7.
The effect of sodium (50, 200 mM) and sulfate (0, 10 mM) concentrations on total and organic sulfur content in the shoots of Salicornia and Sarcocornia. Sulfate is applied as sodium sulfate and the remainder sodium as sodium chloride. A, Total S content in Salicornia and Sarcocornia. The values are means ± se (n = 3). B, Total organic S-compounds were calculated by the subtraction of total sulfur content from the known inorganic S-metabolites content in Salicornia and Sarcocornia. The values are means ± se (n = 3). Values denoted with different letters are significantly different according to the Tukey-Kramer HSD test, P < 0.05 (JMP 8.0 software). Different upper-case letters indicate significant differences between sulfate treatments in Salicornia or Sarcocornia. Different lower-case letters indicate significant differences between salinity treatments in Salicornia or Sarcocornia. Different Greek letters indicate significant difference between Salicornia and Sarcocornia subjected to the same treatment.
Importantly, the organic sulfur level followed biomass accumulation in Salicornia but not in Sarcocornia when both halophytes were supplemented with high sulfate (compare Fig. 2, lowest insets, to Fig. 7B). Salinity negatively affected organic sulfur level in Salicornia, and in the higher salinity treatment without supplementation of sulfate in Sarcocornia (Fig. 7B). Impressively, at the lower salinity treatments without sulfate supplementation, the organic sulfur level in Sarcocornia was higher as compared to Salicornia, yet in the other treatments, organic sulfur was significantly higher in Salicornia (Fig. 7A).
Effect of Sulfate and Salinity Levels on Sulfate-Reduction Pathway Components in Salicornia and Sarcocornia
The differences in organic sulfur between Sarcocornia and Salicornia can be the result of differences in the sulfate assimilation pathways and/or in organic-S catabolism expressed as Cys degradation. These possibilities were further examined to uncover the factor/s responsible for these differences.
APR Expression
Chloroplast-localized APR is known to be a key regulatory point in the sulfate assimilation pathway (Vauclare et al., 2002; Kopriva, 2006; Khan et al., 2010) and may shed light on the cause/s for the different response of Salicornia as compared to Sarcocornia under sulfate supplementation at different salinity levels described above (Figs. 2, 3, and 4).
A reduction in APR transcripts was evident in response to 10 mm sulfate treatment in both Salicornia and Sarcocornia, whereas APR transcript abundance was significantly higher in Salicornia grown in 200 mm NaCl without sulfate supplementation than in either halophyte exposed to other treatments (Supplemental Fig. S4A).
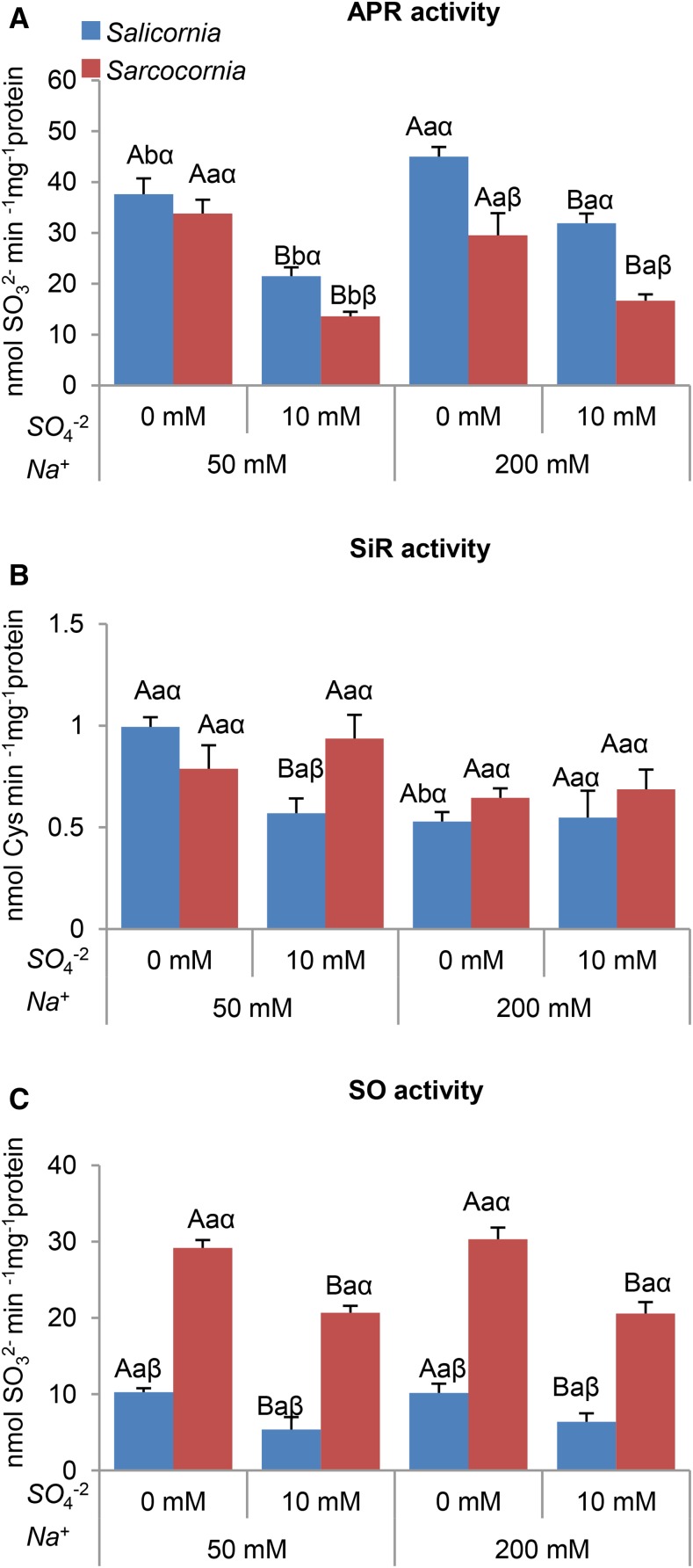
Salinity positively affected APR activity in Salicornia. In Sarcocornia a positive response was evident only when high sulfate was supplemented. A significant decline in APR activity was evident in both types of plants when supplemented with 10 mm sulfate at either salinity level. Importantly, APR activity was higher in Salicornia than Sarcocornia under all growth conditions, being insignificantly higher only in plants grown with the low salinity medium without sulfate supplementation (Fig. 8A).
Figure 8.
Effect of sodium (50, 200 mM) and sulfate (0, 10 mM) concentrations on APR, SiR, and SO activities in Salicornia and Sarcocornia. Sulfate is applied as sodium sulfate and the remainder sodium as sodium chloride. A, APR activity was detected by sulfite appearance in Salicornia and Sarcoconia. The values are means ± se (n = 4). B, SiR activity was detected by Cys appearance in Salicornia and Sarcocornia. The values are means ± se (n = 3). C, SO activity was detected as sulfite disappearance in Salicornia and Sarcoconia. The values are means ± se (n = 4). Values denoted with different letters are significantly different according to the Tukey-Kramer HSD test, P < 0.05 (JMP 8.0 software). Different upper-case letters indicate significant differences between sulfate treatments in Salicornia or Sarcocornia. Different lower-case letters indicate significant differences between salinity treatments in Salicornia or Sarcocornia. Different Greek letters indicate significant difference between Salicornia and Sarcocornia subjected to the same treatment.
These results indicate that in the halophytes Salicornia and Sarcocornia, APR expression is reduced by sulfate supplementation. The results also show that APR activity is higher in Salicornia, especially in sulfate-supplemented plants.
SiR Expression
Chloroplast-localized SiR catalyzes the reduction of sulfite to sulfide in the sulfate-reductive pathway (Yarmolinsky et al., 2014; Brychkova et al., 2015).
SiR transcript abundance was not affected by salinity and sulfate treatments in Salicornia. In contrast, sulfate supplementation to Sarcocornia caused a decrease in SiR transcript levels at low salinity, whereas in the absence of sulfate supply, the enhanced salinity resulted in decreased SiR transcript. (Supplemental Fig. S4B).
SiR activity in both plants decreased with increasing salinity, being significant only without sulfate supply in Salicornia (Fig. 8B). The supplementation with 10 mm sulfate of the low salinity medium resulted in a 2-fold decrease in SiR activity in Salicornia, but there was no difference in SiR activity in Sarcocornia at either salinity level (Fig. 8B). SiR activity in Salicornia plants grown in high salinity was unaffected by supplementation with 10 mm sulfate.
SO Expression
The internal sulfite generated during the sulfate reduction pathway or as a result of S amino acids catabolism (Brychkova et al., 2013) can be oxidized to sulfate by the molybdenum cofactor-containing peroxisomal SO. The abundance of SO transcripts decreased with sulfate addition under low salinity conditions in Salicornia, but not in Sarcocornia. At the higher salinity, sulfate supplementation had little effect on SO transcript abundance in Salicornia, but increased it in Sarcoconia (Supplemental Fig. S4C). Importantly, salinity level had little effect on SO activity, whereas sulfate addition decreased SO activity in either plant (Fig. 8C). Impressively, more than 2-fold higher SO activity was noticed in Sarcocornia in any of the treatments applied, suggesting a requirement for high SO activity to oxidize excess sulfite to sulfate.
OAS-TL Expression
OAS-TL catalyzes the biosynthesis of Cys using OAS and sulfide generated by the sulfate-reduction pathway and/or as a result of the degradation of thiol-containing metabolites (Álvarez et al., 2010, 2012).
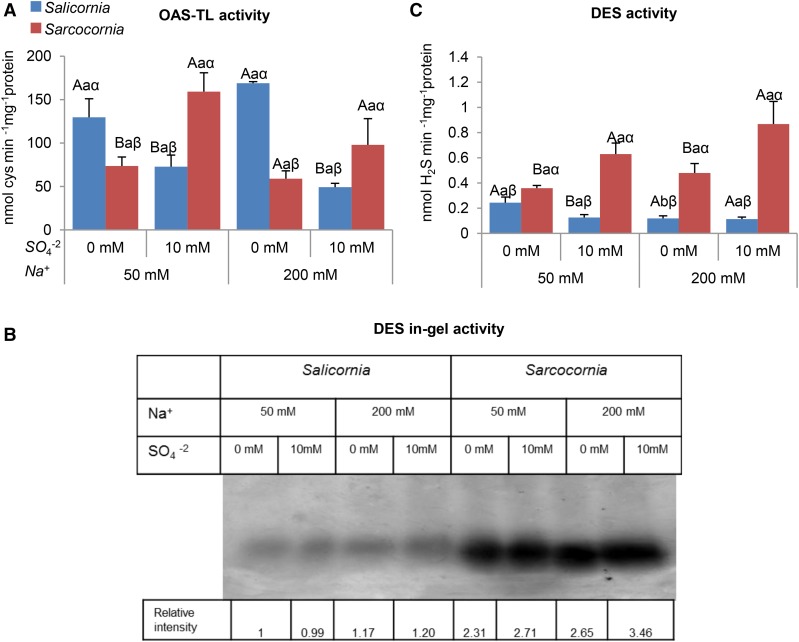
The genes encoding OAS-TL A and OAS-TL B, which are localized to the cytosol and chloroplast, respectively, were examined for changes in transcript abundance in response to sulfate supplementation. At the lower salinity, we observed a decrease in the abundance of Salicornia OAS-TL A, but not OAS-TL B transcripts in response to sulfate supplementation. By contrast, Sarcocornia showed a significant increase in the abundance of both OAS-TL A and OAS-TL B transcripts in response to sulfate supplementation. We observed an increase in both OAS-TL A and OAS-TL B transcript abundance in Sarcocornia at the higher salinity level, whereas in Salicornia, the increase was observed mainly in the abundance of OAS-TL B transcripts (Supplemental Fig.S4, D and E). Interestingly, whereas salinity levels in the tested range had no effect on OAS-TL activity in either halophyte, the OAS-TL activities detected as Cys generation followed transcript expression, exhibiting enhanced activity in Sarcocornia supplemented with high sulfate. The results were significant at the lowest salinity. In contrast, a higher OAS-TL activity rate was noticed in Salicornia at the low compared to high sulfate supplementation (Fig. 9A) conditions. This may indicate a response to sulfur starvation (Barroso et al., 1998; Ravina et al., 1999; Carfagna et al., 2011, 2016), as the organic-S and biomass accumulation in Salicornia were indeed significantly increased at the highest level of supplemental sulfate (Figs. 7B and 2). The results indicate also a higher capacity of Cys biosynthesis by OAS-TL activity in Sarcocornia as compared to Salicornia supplied with the highest sulfate concentration. Yet, considering the organic-S content, one would expect the opposite, unless higher organic-S degradation activities exist in Sarcocornia.
Figure 9.
Effect of sodium (50, 200 mM) and sulfate (0, 10 mM) concentrations on OAS-TL and Cys degradation (DES) activities in Salicornia and Sarcocornia. Sulfate is applied as sodium sulfate and the remainder sodium as sodium chloride. A, OAS-TL activity was detected as Cys appearance in Salicornia and Sarcocornia. The values are means ± se (n = 3). B, l-Cys DES was detected in the gel assay as a brown precipitate resulting from the reaction of hydrogen sulfide, generated by DES enzymatic activity, with lead acetate. The data are from three different experiments that yielded similar results. C, DES activity was also quantified as the release of sulfide from l-Cys. The values are means ± se (n = 4). Values denoted with different letters are significantly different according to the Tukey-Kramer HSD test, P < 0.05 (JMP 8.0 software). Different upper-case letters indicate significant differences between sulfate treatments in Salicornia or Sarcocornia. Different lower-case letters indicate significant differences between salinity treatments in Salicornia or Sarcocornia. Different Greek letters indicate significant difference between Salicornia and Sarcocornia subjected to the same treatment.
A Higher l-Cys DES Activity Was Evident in Sarcocornia as Compared with Salicornia
The differences in organic-S levels between the halophytes might result from either anabolic or catabolic processes. We therefore investigated the levels of molecular and biochemical factors playing a role in the catabolism of sulfur-containing compounds. l-Cys degradation activity was monitored by sulfide production using both in gel and kinetic assays (Fig. 9, B and C, respectively). Significantly, Sarcocornia exhibited a higher l-Cys-degrading activity level (l-Cys desulfhydrase) than did Salicornia and exhibited a significant increase when supplemented with high sulfate, whereas in Salicornia l-Cys desulfhydrase activity was decreased with the low salinity and did not change much with high salinity when supplemented with enhanced sulfate (Fig. 9, B and C). The results of both in-gel and kinetic assays that detected sulfide production show that the l-Cys-degrading activity is higher in Sarcocornia than in Salicornia and that it shows a more marked increase in response to sulfate supplementation in Sarcocornia.
Identification of the Cys Desulfhydrase Activity Source by Trypsinization of the Activity Bands in Salicornia and Sarcocornia
Specific unique peptides trypsinized from the l-Cys desulfhydrase activity bands of Salicornia and Sarcocornia (sliced from the bands shown in Figure 9B) were identified by their similarity to a OAS-TL A (Q00834) of Spinacia oleracea, being 93% identical to Salicornia sequence (Supplemental Fig. S5A). The number of identified unique trypsinized peptides were able to overlap 67% of the Salicornia’s OAS-TL A protein full sequence and 55% of the Sarcocornia protein sequence (Supplemental Table S2). Further, additional unique peptides were identified by their similarity to the chloroplast-localized OAS-TL B, exhibiting 87% sequence identity when AAA16973 (S. oleracea) was compared to Salicornia’s OAS-TL B protein sequence (Supplemental Fig. S5B). The obtained number of trypsinized peptides overlapped 91% of Salicornia’s OAS-TL B protein whereas in Sarcocornia the amount of identified unique peptides overlapped 57.5% of OAS-TL B (Supplemental Table S3). Importantly, no peptide was found when the search for unique peptides was based on the similarity to Arabidopsis l-Cys desulfhydrase 1 (DES1, AT5G28030) or Pyridoxal P-dependent transferases DES1 (AT3G62130). Considering the absence of proteins with sequence similarity to l-Cys desulfhydrase 1 activity among the proteins identified in the sliced activity bands, these results indicate that OAS-TL A and OAS-TL B proteins may participate in l-Cys degradation in Salicornia and Sarcocornia (Fig. 9).
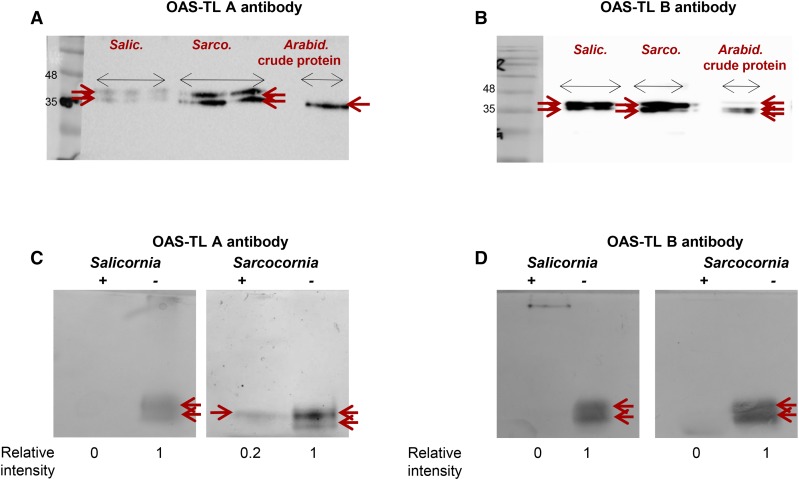
Identification of the Cys DES Activity Source by Immunodetection and Immunoprecipitation
The Western blot analysis of the DES activity bands (sliced from the band shown in Fig. 9B and fractionated by SDS-PAGE), employing antibody raised against Arabidopsis OAS-TL A, revealed two cross-reacting bands; the lower is likely OAS-TL A, as it shows identical gel mobility as shown for Arabidopsis OAS-TL A protein. Both protein bands exhibited significantly higher intensity in Sarcocornia as compared to Salicornia (Fig. 10A). Using OAS-TL B antibody, which cross reacted with Arabidopsis OAS-TL A, OAS-TL B, and OAS-TL, also cross reacted with two bands in Sarcocornia and Salicornia (Fig. 10B). The suitability of the antibodies raised against the Arabidopsis recombinant OAS-TL (kindly provided by Prof. Dr. S. Kopriva, University of Cologne, Köln) with Salicornia’s (and Sarcocornia’s) OAS-TL A and OAS-TL B proteins can be explained by their 81% and 78% sequence identity, respectively (Supplemental Fig. S6, A and B).
Figure 10.
Immunodetection and immunoprecipitation of Salicornia and Sarcocornia OAS-TL proteins. A and B, Immunodetection of OAS-TL A and OAS-TL B, respectively. Fifty µg proteins of Salicornia and Sarcocornia were subjected to in-gel Cys desulfhydrase activity as shown in Fig. 9B. The activity bands were then sliced and were fractionated by 12.5% SDS PAGE together with Arabidopsis wild-type (Col) crude extract and analyzed by Western blot with antibodies raised against Arabidopsis OAS-TL A or OAS-TL B. C and D, Immunoprecipitation of 30 µg Salicornia and Sarcocornia desulfhydrase activity employing antibodies raised against Arabidopsis OAS-TL A and OAS-TL B, respectively. Plus sign (+) indicates “with antibody”; (−) minus sign indicates “without antibody”; red pointer indicates activity band.
Immunoprecipitation analysis of Salicornia and Sarcocornia protein extract was performed employing OAS-TL A and OAS-TL B antibodies (see “Materials and Methods”). The l-Cys DES activity bands were fully, or almost fully, abrogated when proteins were extracted from Salicornia or Sarcocornia, respectively, were incubated with Arabidopsis OAS-TL A antibody as compared to the normal activity with proteins incubated without the antibody (Fig. 10C). Immunoprecipitation with OAS-TL B antibody in both plants revealed a complete disappearance of the l-Cys DES activity bands whereas the proteins that were incubated at the absence of the antibody in the pull-down assay exhibited desulfhydrase activity bands (Fig. 10D). Considering the relatively close identity of Salicornia OAS-TL A and OAS-TL B to the Arabidopsis proteins (Supplemental Fig. S6, A and B; GenBank/EMBL data libraries, accession nos. P47998 and P47999, respectively), as well as the absence of l-Cys DES1 protein (accession no. NP-001330588) among the proteins that generated the l-Cys desulfhydrase activity bands, these results indicate that the DES activity by the halophyte protein extracts resulted from OAS-TL A and OAS-TL B.
DISCUSSION
Salicornia Is Better Adapted to High Sulfate than Sarcocornia
Studies on S nutrition of Salicornia and Sarcocornia are few, and those that exist were carried out with sulfur sources other than sulfate. The positive response of Salicornia europaea to sulfide as compared with other tested halophyte species, including Aster tripolium, Halimione portulacoides, Suaeda maritima, and Puccinellia maritima, was attributed to its habitat (Ingold and Havill, 1984).
Here we show that the addition of 10 mm sulfate to the 1/2 MS growth medium resulted in a significant increase in biomass accumulation in Salicornia, whereas in Sarcocornia, it gave the opposite response (Fig. 2). Salicornia therefore appears to be better adapted to high sulfate conditions, likely because Salicornia’s natural habitat is seawater that contains relatively high sulfate levels (Howarth and Giblin, 1983). Whereas S. europaea is found in areas exposed to frequent seawater tidal flooding, S. fruticosa is normally found in environments exposed to high soil salinities, high vegetation coverage, and less frequent flooding (Rogel et al., 2000).
At typical soil sulfate concentrations, most of the sulfate enters glycophytic plants, is reduced in the leaves and is allocated to the various sinks, yet 10% to 20% of the sulfur generally accumulates as sulfate (Cram, 1990). In coastal halophytes, at high environmental sulfate levels, up to 93% of total sulfur is normally expected to appear as sulfate (Ernst, 1990). Interestingly, at the lowest sulfate concentration used in this study, both Salicornia and Sarcocornia exhibited similar sulfate accumulation ratios of approximately 25% to 15% and 22.5% to 24% of the total sulfur content, respectively. However, when supplemented with high sulfate, Sarcocornia behaved more like a coastal halophyte, with 85% and 55% of the total sulfur being sulfate, whereas Salicornia exhibited more efficient use of the applied sulfate for growth and organic-S biosynthesis, resulting in 35% to 29% sulfate to the total sulfur ratio (calculated from Figs. 5A and 7A). The results here are in agreement with the previously reported observation that Sarcocornia accumulated twice as much sulfate as Salicornia when irrigated with Red Sea water (Ventura et al., 2011a), indicating that Salicornia is very well adapted to its natural habitat in areas exposed to frequent seawater tidal flooding.
High Salinity Is Favorable for Both Salicornia and Sarcocornia, Whereas High Sulfate Is a Stressor for Sarcocornia
Above a certain threshold, specifically for glycophytes as well as halophytes, salinity may generate ionic imbalance, which results in ionic toxicity, osmotic stress, and the generation of ROS (Allakhverdiev et al., 2000; Hasegawa et al., 2000; Chaparzadeh et al., 2004; Parida and Jha, 2010; Chawla et al., 2013). Salicornia and Sarcocornia thrived with 200 mm sodium rather than with 50 mm (Fig. 2), exhibiting lower anthocyanins in Sarcocornia and H2O2 in both Salicornia and Sarcocornia treated with the highest salinity (Figs. 3 and 4B). Interestingly, the enhanced sulfate supplementation resulted in an increase in anthocyanins and H2O2 in Sarcocornia (Figs. 3 and 4B). The H2O2 enhancement is most likely the cause for anthocyanin production, serving as a ROS scavenger in Sarcocornia (Takahama, 1992; Yamasaki et al., 1997; Chalker-Scott, 1999; Gould et al., 2002; Schüssler et al., 2008). The results indicate that high salinity is favorable for both halophytes, whereas the high sulfate is a stressor for Sarcocornia.
The high sulfate level detected in Sarcocornia shoots (Fig. 5A) could be the result of massive sulfate uptake that did not enter the sulfate reduction pathway. It also could be the result of the oxidation by SO of excess endogenous sulfite generated by APR and/or the result of S-amino acids degradation as demonstrated recently (Brychkova et al., 2013; Yarmolinsky et al., 2014). Excess sulfate uptake is thought to be energetically wasteful, employed to avoid osmotic potential imbalances (Hawkesford and De Kok, 2006). Yet, the capacities for osmotic adjustment can be reduced when relatively high sulfate is present in the growth medium, resulting in toxicity symptoms, as was shown in halophytes such as P. strombulifera (Llanes et al., 2013). Interestingly, estimation of the osmotic potential in both halophyte plants studied revealed a significantly higher osmolality in Sarcocornia than in Salicornia extracts when sulfate was added to the growth medium (Supplemental Fig. S7). Because biomass accumulation in Sarcocornia is generally decreased in the presence of high sulfate, whereas in Salicornia biomass accumulation is enhanced (Fig. 2), the sulfate accumulation (Fig. 5A) can be seen also as a cause of energetic waste. Whether the accumulated sulfate is a result of endogenous sulfite oxidation and/or avoidance of osmotic potential imbalances, both are energetically wasteful.
OAS-TL A and OAS-TL B Exhibit Significant l-Cys DES Activity in Sarcocornia, Especially in the Presence of High Sulfate
The synthesis of Cys and its degradation should be well coordinated. l-Cys desulfhydrase activity results in the release of sulfide, whereas OAS-TL consumes sulfide for Cys biosynthesis. It has been claimed that the kinetic properties of the OAS-TLs A, OAS-TL B, and OAS-TL C in Arabidopsis most likely do not allow a significant reverse reaction (Wirtz et al., 2004). Yet, free H2S was shown to be partially released by OAS-TLs (Papenbrock et al., 2007), acting not only in l-Cys de novo biosynthesis but also in its homeostasis (Riemenschneider et al., 2005). Interestingly, both OAS-TL and DES activities showed a positive correlation in response to the Brassicaceae-infecting pathogen, Pyrenopeziza brassicae (Bloem et al., 2004). Similar results have been reported in Vitis vinifera, consequent from chilling stress (Fu et al., 2013). These indicate the feasibility of certain conditions where l-Cys desulfhydrase activities of OAS-TL can be affected at least to a certain level.
Sulfide for Cys biosynthesis can be derived from Cys degradation, cyanide detoxification, and iron-sulfur cluster degradation, but the bulk of sulfide is believed to be generated by the assimilatory sulfate reduction pathway (Birke et al., 2015a, 2015b). Yet, this appears to be in doubt because the rate of sulfide generation by sulfite reductase activity in Arabidopsis leaves has been reported to be 2.5 to 4 nmol mg−1 protein min−1 (Khan et al., 2010; Yarmolinsky et al., 2013), whereas the activity of Arabidopsis l-Cys DES1 that degrades Cys to sulfide, pyruvate, and ammonia has been reported to be 7 to 10 nmol mg−1 protein min−1 (Álvarez et al., 2010). This indicates that the DES activity is a possible source of sulfide, in addition to sulfite reductase activity. Intriguingly, the DES1 activity in Arabidopsis leaves accounted for only 13% to 19% of the total desulfhydrase activity (calculated from Álvarez et al., 2010), indicating the existence of additional unidentified enzymes with DES activity.
In view of the significantly lower organic-S content of Sarcocornia compared to Salicornia (Fig. 7B), it seems possible that sulfate supplementation stimulates l-Cys degradation by the Sarcocornia OAS-TL (Fig. 9, A and B). Several lines of evidence support this notion. First, we showed a significantly higher DES activity in Sarcocornia as compared to Salicornia in the presence of supplemental sulfate (Fig. 9B). The enhancement of DES activity in sulfate-supplemented Sarcocornia was also detected using the in-gel activity, as well as by employing the alternative kinetic DES assay, both, based on the detection of H2S (Fig. 9, B and C). Finally, the DES activity was shown to be attributable directly to OAS-TL A and OAS-TL B by peptide identification in the l-Cys DES activity bands of Salicornia and Sarcocornia (Tables S2 and S3), as well as by identification of the activity bands and their abrogation by immunoprecipitation, with Arabidopsis OAS-TL A or OAS-TL B antibodies (Fig. 10). The complete pull-down of the l-Cys desulfhydrase activity bands by OAS-TL B antibody in both Salicornia and Sarcocornia (Fig. 10D), and the complete abrogation in Salicornia and almost full pull-down in Sarcocornia by OAS-TL A antibody (Fig. 10C), indicate that even if DES1 comigrated with the OAS-TLs, the vast majority of DES activity is still that of OAS-TL A and OAS-TL B.
The Higher APR Activity in Salicornia than in Sarcocornia Suggests Enhanced Sulfate Reduction Activity in Salicornia
The higher total-S but lower organic-S accumulated in Sarcocornia as compared with Salicornia supplemented with high sulfate, in either salinities levels (Fig. 7), indicates that the lower sulfate level in Salicornia (Fig. 5A) is more the result of a higher sulfate assimilation rate by the sulfate reduction pathway, than only the result of lower sulfate uptake. The key step in the pathway is the reduction of APS to sulfite catalyzed by APR (Vauclare et al., 2002) considered to be the key control point in the sulfate assimilation pathway (Vauclare et al., 2002; Kopriva, 2006; Khan et al., 2010). APR activity and expression increase upon sulfur starvation and decrease with sulfate availability (Hirai et al., 2003; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003). In accordance with the concept of demand-driven regulation of sulfate assimilation, APR activity was down-regulated when 10 mm sulfate was supplied to either Salicornia or Sarcocornia (Fig. 8A). Yet, APR activity was significantly higher in Salicornia than in Sarcocornia (Fig. 8A), which may explain the lower sulfate and higher organic-S levels in Salicornia (Figs. 5A and 7B). Enhanced APR activity indicates a higher need for S-containing metabolites and is followed by higher synthesis levels of thiols such as Cys (Romero et al., 2001; Koprivova et al., 2008a). Similarly, a significantly higher Cys level was detected in Salicornia than in Sarcocornia (Fig. 6A), indicating a higher demand for reduced sulfur for the synthesis of essential S-containing metabolites. Moreover, sulfur assimilation is a highly regulated process that controls responses to developmental cues. In plants, the organic sulfur (such as Cys and Met) is extremely important for growth, especially when plant organs are developing rapidly (Martin et al., 2005). Accordingly, the higher organic sulfur content appears to be important in achieving the higher growth rate observed in Salicornia as compared with Sarcocornia (Fig. 2). In summary, the higher organic-S level in Salicornia (as compared with Sarcocornia) is likely indicative of an enhanced APR response to the demand for enhanced biomass production and is likely to be the result of reduced degradation of organic-S compounds such as l-Cys.
High Sulfate Accumulation and SO Activity Rates in Sarcocornia But Not in Salicornia Are Indicative of Enhanced Organic S Degradation Activity in Sarcocornia
The high sulfate level detected in Sarcocornia (Fig. 5A) is first of all a consequence of the higher uptake of sulfate, as inferred from its generally higher total sulfur content as compared with Salicornia (Fig. 7A). It also could be the result of the high oxidation rate of sulfite generated as the result of plant metabolism, as part of the sulfate reduction pathway, and/or degradation of sulfur-containing metabolites. Turnover of S-containing amino acids, a massive component of plant organic S (Stulen and De Kok, 1993), has recently been reported (Brychkova et al., 2013). The absence of active plant SO resulted in the accumulation of sulfite as a result of dark-induced accelerated catabolism of protein-bound Cys and Met, whereas a significant enhancement of sulfate was evident in the presence of active SO in wild-type plants (Brychkova et al., 2013, 2015). This notion is supported by the high SO activity rate in Sarcocornia, because high expression of the constitutively expressed SO protein (0.1% of total crude leaf protein; Lang et al., 2007) can be explained mainly by the need to detoxify excess sulfite, most likely the result of degraded organic sulfur. Although SO activity decreased when Sarcocornia plants were supplied with sulfate, the greater sulfate accumulation can be attributed to the generally high SO activity (Fig. 8C), which efficiently oxidizes excess sulfite resulting in decreased nontoxic sulfite levels (Fig. 5B). In this context, the significantly higher ROS levels in Sarcocornia than in Salicornia plants (Fig. 4) are likely indicative of the higher ROS generation by SO (Hänsch et al., 2006; Brychkova et al., 2007, 2012).
CONCLUSIONS
By exploring the sulfate reductive pathway that generates l-Cys and the l-Cys DES activities to identify factors affecting organic-S and sulfate levels, we identified a major role for OAS-TLs, enzymes known to catalyze l-Cys as the final step of the sulfate reductive pathway. We showed that OAS-TL A and OAS-TL B exhibit significant l-Cys DES activity and that this activity is significantly higher in Sarcocornia than in Salicornia, especially upon sulfate supplementation. In addition, the activity of APR, the key enzyme in sulfate assimilation, was significantly higher in Salicornia than in Sarcocornia, whereas the activity of SO, which regulates sulfite levels in the sulfate reductive pathway, was significantly higher in Sarcocornia. These results indicate that the low organic-S in Sarcocornia is the result of enhanced organic-S degradation by the Cys degrading activity of OAS-TL A and OAS-TL B, likely followed by SO oxidation of sulfite originated from protein-bound sulfur amino acid degradation. The low Cys generation by the reduced APR activity observed in Sarcocornia further contributed to the lower organic-S in this plant. The significantly higher APR activity rate and the very low l-Cys DES activities in Salicornia is suggestive for its higher net Cys generation, resulting in higher organic-S levels for biomass accumulation. The results of this study provide evidence that Salicornia thrives at a high sulfate concentration, indicating its adaptation not only to high salinity, but also to high sulfate that exists in its natural habitat. In contrast, Sarcocornia is sensitive to high sulfate, as evident not only by the lower biomass and organic-S accumulation but also by the high anthocyanin and ROS production. These results present an initial road map for halophyte growers to gain better growth and nutritional value of Salicornia and Sarcocornia.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Biomass Determination
Salicornia europaea (ecotype RN, collected in the Dead Sea area, Israel) and Sarcocornia fruticosa (ecotype VM, collected in the Ramat HaNegev district, Israel) were used in the experiments (Ventura et al., 2010; Ventura and Sagi, 2013). Experiments were carried out in the growth room at Ben-Gurion University of the Negev, under the following conditions: 14-h light/10-h dark, 25°C, 75% to 85% relative humidity, and under light intensity of 150 μmol m−2 s−1.
Seeds were germinated and grown in standard 90 mm petri dishes on 1/2 MS medium supplemented 1% plant agar and 1% Suc for 10 d. The seedlings were transferred to large petri dishes of 155-mm diameter × 30-mm height, supplied with 1/2 MS medium (Murashige and Skoog, 1962; containing 0.87 mm sulfate level) supplemented with either 50 mm NaCl or 200 mm NaCl without the addition of Na2SO4. The NaCl concentration was 30 mm NaCl or 180 mm NaCl when 10 mm Na2SO4 was supplemented. All treatments were performed in three replicates.
The weight of shoot biomass accumulation was determined 14 d after the treatment onset for S. europaea (RN) and 21 d for S. fruticosa (VM). Results were expressed as average plant growth rate in mg d−1 plant−1. Samples were immediately frozen in liquid N and stored at −80°C for further use.
To estimate biomass accumulation in the preliminary experiments, Salicornia and Sarcocornia seeds were germinated in pots (0.4 L) filled with the highly air-permeable and high-water-capacity perlite (up to 2 mm particles size; Agrekal Habonim Industries). After germination, seedlings were thinned out to similar numbers of 40 seedlings per pot in four replicas and subjected to 100 mm NaCl and 200 mm NaCl in half-strength Hoagland’s nutrient solution (Hoagland and Arnon, 1950) supplemented with 0 mm or 10 mm sulfate. Plants were grown in controlled greenhouses at Ben Gurion University of the Negev, Israel, under approximately 13.5-h sunlight (May 2012), day temperature 25 to 31°C, night temperature approximately 20°C, relative humidity approximately 60%, and light intensity ranged between 400 and 600 μmol m−2 s−1. The biomass accumulation data from this experiment is presented in Supplemental Fig. S3.
For RNA sequencing, S. europaea (RN) seeds were germinated in plastic trays on Metromix-360 garden soil in a greenhouse at King Abdullah University of Science and Technology (Biological and Environmental Science and Engineering Division) and the seedlings were maintained under natural daylight conditions until sampled.
RNA Sequencing and Transcriptome De Novo Assembly
Four-month-old S. europaea (RN) plants were sampled for RNA sequencing and transcriptome de novo assembly. Total RNA was extracted using Trizol and used for preparation of RNAseq libraries. Paired-end libraries were prepared from S. europaea RNA using a TruSeq RNA Library Prep Kit v2 (Illumina) to prepare RNASeq libraries using manufacturers low sample protocol and libraries were sequenced on a HiSeq2000 platform (Illumina). A total of approximately 550 million paired-end reads of insert size 100 bp were produced. Low-quality reads and adapter sequences were filtered using a Trimmomatic (Bolger et al., 2014), leaving approximately 500 million high quality reads for downstream analyses. Transcriptome de novo assembly was performed using the software Trinity (Grabherr et al., 2011) and annotated following the Trinotate pipeline (Haas et al., 2013). A total of 56,107 protein sequences were predicted using TransDecoder (Haas et al., 2013). Predicted protein sequences were annotated by performing BLASTp against Swiss-Prot, protein domains were searched for through Pfam, and annotated using Cluster of Orthologous Genes and Gene Ontology annotations.
RNA Extraction, cDNA Synthesis, and Quantitative Real-Time Reverse Transcription-PCR in Salicornia and Sarcocornia
Total RNA extraction, cDNA synthesis, and the quantitative analysis of transcripts of Salicornia and Sarcocornia shoots grown in 1/2 MS medium was performed as described in Brychkova et al. (2007) employing the primers shown in Supplemental Table S1 as described below. To employ at least one of the two primers as splice junction overlapping primer, Actin (Act), SiR, Transcription initiation factor (TFIID), APR, OAS-TL A, and OAS-TL B primers were designed based on similarity analysis to Beta vulgaris, whereas SO primers for Salicornia were designed based on similarity to Populus trichocarpa. All the primers designed for Salicornia were suitable for Sarcocornia transcripts analysis as well. The quantitative PCR products were separated on a 1% agarose gel, excised from the gel, and sequenced for identity verification (see alignment results with each of the related transcripts in Supplemental Fig. S8). ACT (88% identity to B. vulgaris subsp. vulgaris actin-related protein 7-like, Accession XR_789363.1| PREDICTED) and TFIID (85% identity to B. vulgaris subsp. vulgaris transcription initiation factor TFIID subunit 6, Accession XM_010690649) were unaffected by the treatments and thus were employed as reference housekeeping genes. The transcript levels in Salicornia or Sarcocornia were compared to 50 mm NaCl and 0 mm Na2SO4 treatment after normalization to TFIID or ACT and presented as relative expression (means ± se, n = 4). Only results normalized with TFIID are presented.
Protein Extraction, Determination of Soluble Protein, and Kinetic Assays for APR, SO, SiR, and OAT-TL Enzymes
Protein extraction, desalting, concentration determination, and kinetic assays for APR, SO, SiR, and OAS-TL in Salicornia and Sarcocornia shoots grown in 1/2 MS medium were carried out as previously described for tomato leaves Solanum lycopersicum “Rheinlands Ruhm” (Brychkova et al., 2013). 1 U of 124-fold purified Arabidopsis (Arabidopsis thaliana) OAS-TL was added to the reaction mix of the SiR assay as a sulfide trap. SiR and OAS-TL activities were expressed in nmol Cys mg–1 protein min–1 (Brychkova et al., 2012) and APR and SO in nmol sulfite mg–1 protein min–1.
Protein Extraction, Determination of Soluble Protein, and Kinetic Assays for DES Activity
Protein extraction of Salicornia and Sarcocornia shoots grown in 1/2 MS medium and desalting for DES was performed as described before for sulfurtransferases (Brychkova et al., 2013). DES activity was detected based on direct detection of H2S formation in the presence of l-Cys as described in Riemenschneider et al. (2005). The modified assay solution contained 0.1 m Tris-HCl, pH 9.0, 2.5 mM DTT, 0.8 mM l-Cys, and 10 μg of desalted protein in a total volume of 0.2 mL. After incubation at 37°C for 30 min, H2S was detected according to Bloem et al. (2004) with 30 mm FeCl3 dissolved in 1.2 n HCl and 20 mm N,N-dimethyl-p-phenylenediamine dihydrochloride dissolved in 7.2 n HCl. The formation of methylene blue was measured at 670 nm with Epoch Microplate Spectrophotometer supported by Gen5 1.10 software (BioTek). Na2S*9H2O was used as a standard.
Measurement of Sulfur-containing Metabolites
To determine total sulfur content, approximately 10 mg dried and powdered Salicornia and Sarcocornia shoots grown in 1/2 MS medium were placed in tin containers in an elemental analyzer (EA1108 CHNS/O; Fisons Instruments) and the amounts of total S in each sample were quantified according to a standard calibration curve prepared with reduced GSH.
Sulfate and sulfite from Salicornia and Sarcocornia shoots grown in 1/2 MS medium were determined as described in Brychkova et al. (2012). Briefly, for sulfate determination, the leaf samples were extracted in 24 mm formaldehyde in 2 mm Na2CO3/0.75 mm NaHCO3 solution (1:100, w/v) and determined employing a DIONEX LC20 ion chromatograph with an ED 50 Electrochemical Detector and an AS9-SC Analytical IonPak (4 mm × 250 mm). Sulfite was determined by employing chicken SO using NADH-peroxidase-dependent determination of H2O2 generated as a byproduct of sulfite oxidation to sulfate, catalyzed by chicken SO. For sulfide detection, samples were extracted in a solution containing 50 mm citric acid, 100 mm Na2HPO4, and 1 mm salicylic acid, pH 5.0 in 1:40 (w:v) ratio and detected by employing the ISO-H2S-100 microsensor for H2S according to Yarmolinsky et al. (2014). Cys and total GSH were separated and quantified by HPLC according to Ohkama-Ohtsu et al. (2007). Organic S was calculated by subtracting the sum of sulfate, sulfite, thiosulfate, and sulfide from the total sulfur content.
Protein Extraction for an In-Gel DES Assay
Shoots samples of Salicornia and Sarcocornia plants grown in 1/2 MS medium were ground using a mortar and pestle in extraction buffer (ratio 1:4) containing 0.25 m Suc, 50 mm Tris-HCl (pH 8.5), 3 mm EDTA, 1 mm sodium molybdate, 4 mm DTT, 15 mm GSH, 0.025% of Triton X-100, and a cocktail of protease inhibitors including aprotinin (10 μg mL−1), leupeptin (10 μg mL−1), and pepstatin (10 μg mL−1). The homogenate was centrifuged at 18,000g for 20 min and the total soluble protein content was determined by the Bradford assay (Bradford, 1976).
DES In-Gel Activity Assay
Fifty μg of extracted protein was loaded into each lane and fractionated by 7.5% Native PAGE. DES activity was detected using a modification of an in-gel visualization protocol for H2S (Manchenko, 2002). Lead acetate was employed to detect the generated H2S, producing dark brown lead sulfide bands. The reaction solution contained 0.15 m Tris–HCl, pH 8.5, 1 mm DTT, 50 mm β-mercaptoethanol, 20 mm l-Cys, 0.1 mm pyridoxal 5-P, 0.4 mm lead acetate. The reaction was stopped by immersion of the gel in double-distilled water.
Protein Sequencing
To identify the proteins participating in the l-Cys desulfhydrase activity the activity bands from the in-gel activity of DES (Fig. 9B) were sliced from the native gel, and fractionated with 12% SDS-PAGE. Thereafter the proteins were stained by Coomassie Brilliant Blue, and the stained bands were excised from the gel, trypsinized, and the resulting peptides were separated by HPLC and analyzed by LC-MS/MS on Q-Exactive (Thermo Fisher Scientific) at The Smoler Protein Research Center (Technion University, Haifa, Israel).
All the identified peptides were filtered with high confidence, top rank, mass accuracy, and a minimum of two peptides. High confidence peptides passed the 1% false discovery rate (FDR) threshold (*FDR = the estimated fraction of false positives in a list of peptides). Semiquantitation was done by calculating the peak area of each peptide. The area of the protein is the average of the three most intense peptides from each protein. Analysis of peptide sequences was performed by employing the Proteome Discoverer software, ver. 1.4.1.14 (Thermo Fisher Scientific) against the Salicornia sequence (kindly supplied by Prof. Nina Fedoroff and performed as described in the paragraph titled “RNA Sequencing and Transcriptome De Novo Assembly”).
Immunodetection and Immunoprecipitation of l-Cys DES
For immunodetection analysis, the DES activity bands (sliced from the band shown in Fig. 9B) together with Arabidopsis wild-type bands were fractionated by 12.5% SDS PAGE as described in Brychkova et al. (2013) and blotted onto polyvinylidene difluoride membranes (Immuno-Blot membranes; Bio-Rad). The blotted proteins were subjected to immuno-detection with antibodies raised against Arabidopsis cytosolic OAS-TL A (diluted 1:2000) and chloroplastic OAS-TL B. The latter (diluted 1:5000; kindly provided by Prof. Dr. S. Kopriva, University of Cologne, Köln) could identify the three OAS-TL (A, B, and C). PBS-diluted (1:5000) secondary antibodies (antirabbit IgG; Sigma-Aldrich) was followed. Protein bands were visualized with the ChemiDoc Touch Imaging system (Bio-Rad) after staining with an ECL detection system, using the SuperSignal West Pico Chemiluminescent Substrate (Pierce) according to the manufacturer’s instructions. Band intensities were quantified by using ImageJ software (National Institutes of Health).
For the immunoprecipitation assay, 50 μg of protein from Salicornia and Sarcocornia were incubated with 30 μL of the OAS-TL A or OAS-TL B antibodies in Tris-buffer saline for 30 min at room temperature and then kept at 4°C for overnight. Protein extract that had not been mixed with antibody was employed as control. The mixture and the control solutions were then incubated with 50 μL of Protein G Agarose at 4°C for 2 h with continuous shaking and then centrifuged at 10,000g for 5 min, followed by removal of supernatant for analysis by the in-gel DES assay.
Determination of Anthocyanin Content and Osmolality
The anthocyanin content was determined based on a modification of protocols described by Laby et al. (2000) and Kant et al. (2006). Approximately 100 mg of fresh shoot tissue of Salicornia and Sarcocornia plants grown in 1/2 MS medium was crushed in 600 μL methanol acidified with 1% HCl. The extract was centrifuged for 10 s at 4000g. Five-hundred μL of double distilled water was added to the collected sand, mixture was gently vortexed, and then 700 μL chloroform was added and mixed for 20 s followed by centrifugation at 4000g for 2 min. The total anthocyanin in the aqueous phase was determined by detecting the optical density at A530 nm and A657 nm. The amount of anthocyanin was calculated by subtracting the A657 from the A530 (Laby et al., 2000).
The osmolality was measured according to Thalmann et al. (2016). The fresh shoot tissues of Salicornia and Sarcocornia grown in 1/2 MS medium were crushed with iron beads, diluted 1/3 with water, and centrifuged for 10 min at 18,000g at 4°C. The supernatant was used to determine the osmolality employing a Micro-Osmometer (Advance Instruments).
ROS Determination
For detecting O2− and H2O2, frozen shoots of Salicornia and Sarcocornia were extracted in 50 mm P buffer (pH 7.5) at a ratio of 1:8 (w/v) and centrifuged (Eppendorf 5417R) twice at 18,000g for 20 min. The reaction mixture for detecting O2− consisted of 4 mm epinephrine as an electron acceptor in 100 mm Tris-HCl buffer (pH 7.8) in the presence or absence of 2100 U/mL CuZn-SOD as described in Yesbergenova et al. (2005). Absorbance was measured at 480 nm employing an Epoch Microplate Spectrophotometer supported by Gen5 1.10 software (BioTek).
The reaction mixture for detecting H2O2 consisted of 0.85 mm 4-aminoantipyrine, 3.4 mm 3,5-dichloro-2-hydroxobenzene sulfonate, 4.5 U mL−1 HRP in 2 mL of 50 mm P buffer (pH 7.5) in the presence or absence of 2 mm tungstic acid and 100 μm DPI as described in Yesbergenova et al. (2005). Absorbance was measured after 5 min at 515 nm as described above.
Accession Numbers
Protein sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: NP_001330588 (DES1, Arabidopsis), P47998 (OAS-TL A, Arabidopsis), P47999 (OAS-TL B, Arabidopsis), Q00834 (OAS-TL A, Spinach oleracea), and AAA16973 (OAS-TL B, S. oleracea).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effect of NaCl (100 and 200 mM) and Na2SO4 (100 mM) on biomass accumulation of Salicornia (RN) and Sarcocornia (VM).
Supplemental Figure S2. Effect of sodium (0 mM) and sulfate (0 and 10 mM) levels on biomass accumulation of Salicornia (upper panels) and Sarcocornia (lower panels).
Supplemental Figure S3. Effect of sodium (100 and 200 mM) and sulfate (0 and 10 mM) levels on biomass accumulation of Salicornia and Sarcocornia.
Supplemental Figure S4. Effect of sodium (50 and 200 mM) and sulfate (0, 10 mM) application on selected enzymes’ expression in Salicornia and Sarcocornia.
Supplemental Figure S5. Multiple sequence alignment of Salicornia’s OAS-TL A and OAS-TL B protein sequence against S. oleracea’s OAS-TL A and OAS-TL B protein sequence.
Supplemental Figure S6. Multiple sequence alignment of Salicornia’s OAS-TL A and OAS-TL B protein sequence against Arabidopsis’s OAS-TL A and OAS-TL B protein sequence.
Supplemental Figure S7. Effect of sodium (50 and 200 mM) and sulfate (0 and 10 mM) application on osmolality level in Salicornia and Sarcocornia.
Supplemental Figure S8. Salicornia selected gene sequences and quantitative real-time PCR product verification results in multiple sequence alignment.
Supplemental Table S1. List of gene primers used for quantitative real-time PCR with Salicornia and Sarcocornia.
Supplemental Table S2. List of identified and overlapped unique peptides of OAS-TL A in Salicornia and Sarcocornia.
Supplemental Table S3. List of identified and overlapped unique peptides of OAS-TL B in Salicornia and Sarcocornia.
Acknowledgments
We thank Dr. Dominic Standing and Ms. Talya Samani for their technical support and Prof. S. Kopriva (University of Cologne, Köln) for providing OAS-TL A and OAS-TL B antibodies.
Footnotes
This research was supported by the Israel Science Foundation grant no. 212/13, by the I-CORE Program of The Israel Science Foundation grant no. 152/11 and by the Chief Scientist, Ministry of Agriculture and Rural Development, Israel grant no. 857-0691-13. We also thank COST Action FA0901 “Putting Halophytes to Work—From Genes to Ecosystems” for their support.
Articles can be viewed without a subscription.
References
- Allakhverdiev SI, Sakamoto A, Nishiyama Y, Inaba M, Murata N (2000) Ionic and osmotic effects of NaCl-induced inactivation of photosystems I and II in Synechococcus sp. Plant Physiol 123: 1047–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez C, Bermúdez MÁ, Romero LC, Gotor C, García I (2012) Cysteine homeostasis plays an essential role in plant immunity. New Phytol 193: 165–177 [DOI] [PubMed] [Google Scholar]
- Álvarez C, Calo L, Romero LC, García I, Gotor C (2010) An O-acetylserine(thiol)lyase homolog with L-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol 152: 656–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astolfi S, Zuchi S (2013) Adequate S supply protects barley plants from adverse effects of salinity stress by increasing thiol contents. Acta Physiol Plant 35: 175–181 [Google Scholar]
- Barroso C, Vega JM, Gotor C (1998) The role of roots in cysteine biosynthesis by Arabidopsis thaliana. J Physiol Biochem 54: 189–194 [PubMed] [Google Scholar]
- Birke H, De Kok LJ, Wirtz M, Hell R (2015b) The role of compartment-specific cysteine synthesis for sulfur homeostasis during H2S exposure in Arabidopsis. Plant Cell Physiol 56: 358–367 [DOI] [PubMed] [Google Scholar]
- Birke H, Hildebrandt TM, Wirtz M, Hell R (2015a) Sulfide detoxification in plant mitochondria. Methods Enzymol 555: 271–286 [DOI] [PubMed] [Google Scholar]
- Bloem E, Riemenschneider A, Volker J, Papenbrock J, Schmidt A, Salac I, Haneklaus S, Schnug E (2004) Sulphur supply and infection with Pyrenopeziza brassicae influence L-cysteine desulphydrase activity in Brassica napus L. J Exp Bot 55: 2305–2312 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brychkova G, Grishkevich V, Fluhr R, Sagi M (2013) An essential role for tomato sulfite oxidase and enzymes of the sulfite network in maintaining leaf sulfite homeostasis. Plant Physiol 161: 148–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brychkova G, Xia Z, Yang G, Yesbergenova Z, Zhang Z, Davydov O, Fluhr R, Sagi M (2007) Sulfite oxidase protects plants against sulfur dioxide toxicity. Plant J 50: 696–709 [DOI] [PubMed] [Google Scholar]
- Brychkova G, Yarmolinsky D, Batushansky A, Grishkevich V, Khozin-Goldberg I, Fait A, Amir R, Fluhr R, Sagi M (2015) Sulfite oxidase activity is essential for normal sulfur, nitrogen and carbon metabolism in tomato leaves. Plants (Basel) 4: 573–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brychkova G, Yarmolinsky D, Fluhr R, Sagi M (2012) The determination of sulfite levels and its oxidation in plant leaves. Plant Sci 190: 123–130 [DOI] [PubMed] [Google Scholar]
- Carfagna S, Bottone C, Cataletto PR, Petriccione M, Pinto G, Salbitani G, Vona V, Pollio A, Ciniglia C (2016) Impact of sulfur starvation in autotrophic and heterotrophic cultures of the extremophilic microalga Galdieria phlegrea (Cyanidiophyceae). Plant Cell Physiol 57: 1890–1898 [DOI] [PubMed] [Google Scholar]
- Carfagna S, Vona V, Di Martino V, Esposito S, Rigano C (2011) Nitrogen assimilation and cysteine biosynthesis in barley: evidence for root sulphur assimilation upon recovery from N deprivation. Environ Exp Bot 71: 18–24 [Google Scholar]
- Chalker-Scott L. (1999) Environmental significance of anthocyanins in plant stress responses. Photochem Photobiol 70: 1–9 [Google Scholar]
- Chaparzadeh N, D’Amico ML, Khavari-Nejad R-A, Izzo R, Navari-Izzo F (2004) Antioxidative responses of Calendula officinalis under salinity conditions. Plant Physiol Biochem 42: 695–701 [DOI] [PubMed] [Google Scholar]
- Chawla S, Jain S, Jain V (2013) Salinity induced oxidative stress and antioxidant system in salt-tolerant and salt-sensitive cultivars of rice (Oryza sativa L.). J Plant Biochem Biotechnol 22: 27–34 [Google Scholar]
- Colmer TD, Fan TWM, Läuchli A, Higashi RM (1996) Interactive effects of salinity, nitrogen and sulphur on the organic solutes in Spartina alterniflora leaf blades. J Exp Bot 47: 369–375 [Google Scholar]
- Cram W. (1990) Uptake and transport of sulfate. Rennenberg H, Brunold C, De Kok LJ, Stulen I, eds. In Sulfur Nutrition and Sulfur Assimilation in Higher Plants: Fundamental, Environmental, and Agricultural Aspects. SPB Academic Publishing, The Hague, The Netherlands: pp 3–11 [Google Scholar]
- Davy AJ, Bishop GF, Costa CSB (2001) Salicornia L. (Salicornia pusilla J. Woods, S. ramosissima J. Woods, S. europaea L., S. obscura P.W. Ball & Tutin, S. nitens P.W. Ball & Tutin, S. fragilis P.W. Ball & Tutin and S. dolichostachya Moss). J Ecol 89: 681–707 [Google Scholar]
- Davy AJ, Bishop GF, Mossman H, Redondo-Gómez S, Castillo JM, Castellanos EM, Luque T, Figueroa ME (2006) Biological flora of the British Isles: Sarcocornia perennis (Miller). A.J. Scott. Journal of Ecology 94: 1035–1048 [Google Scholar]
- de la Fuente V, Oggerin M, Rufo L, Rodríguez N, Ortuñez E, Sánchez-Mata D, Amils R (2013) A micromorphological and phylogenetic study of Sarcocornia AJ Scott (Chenopodiaceae) on the Iberian Peninsula. Plant Biosyst 147: 158–173 [Google Scholar]
- Ernst WHO. (1990) Ecological aspects of sulfur metabolism. Rennenberg H, Brunold C, De Kok LJ, Stulen I, eds. In Sulfur Nutrition and Sulfur Assimilation in Higher Plants: Fundamental, Environmental, and Agricultural Aspects. SPB Academic Publishing, The Hague, The Netherlands: pp 131–144 [Google Scholar]
- Fatma M, Khan MIR, Masood A, Khan NA (2013) Coordinate changes in assimilatory sulfate reduction are correlated to salt tolerance: involvement of phytohormones. Annu Rev Res Biol 3: 267–295 [Google Scholar]
- Fu P, Wang W, Hou L, Liu X (2013) Hydrogen sulfide is involved in the chilling stress response in Vitis vinifera L. Acta Soc Bot Pol 82: 295–302 [Google Scholar]
- Gould K, McKelvie J, Markham K (2002) Do anthocyanins function as antioxidants in leaves? Imaging of H2O2 in red and green leaves after mechanical injury. Plant Cell Environ 25: 1261–1269 [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8: 1494–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänsch R, Lang C, Riebeseel E, Lindigkeit R, Gessler A, Rennenberg H, Mendel RR (2006) Plant sulfite oxidase as novel producer of H2O2: combination of enzyme catalysis with a subsequent non-enzymatic reaction step. J Biol Chem 281: 6884–6888 [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499 [DOI] [PubMed] [Google Scholar]
- Havill DC, Ingold A, Pearson J (1985) 6. Sulphide tolerance in coastal halophytes. Beeftink WG, Rozema J, Huiskes AHL, eds. In Ecology of Coastal Vegetation. Springer, New York: pp 279–285 [Google Scholar]
- Hawkesford MJ, De Kok LJ (2006) Managing sulphur metabolism in plants. Plant Cell Environ 29: 382–395 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33: 651–663 [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI (1950) The Water-Culture Method for Growing Plants without Soil. College of Agriculture, University of California, Berkeley, CA, C347 [Google Scholar]
- Howarth RW, Giblin A (1983) Sulfate reduction in the salt marshes at Sapelo Island, Georgia. Limnol Oceanogr 28: 70–82 [Google Scholar]
- Ingold A, Havill D (1984) The influence of sulphide on the distribution of higher plants in salt marshes. J Ecol 72: 1043–1054 [Google Scholar]
- Iqbal N, Masood A, Khan MIR, Asgher M, Fatma M, Khan NA (2013) Cross-talk between sulfur assimilation and ethylene signaling in plants. Plant Signal Behav 8: e22478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadereit G, Ball P, Beer S, Mucina L, Sokoloff D, Teege P, Yaprak AE, Freitag H (2007) A taxonomic nightmare comes true: Phylogeny and biogeography of glassworts (Salicornia L., Chenopodiaceae). Taxon 56: 1143–1170 [Google Scholar]
- Kant S, Kant P, Raveh E, Barak S (2006) Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ 29: 1220–1234 [DOI] [PubMed] [Google Scholar]
- Khan MIR, Asgher M, Iqbal N, Khan NA (2013) Potentiality of sulphur-containing compounds in salt stress tolerance. In Ecophysiology and Responses of Plants under Salt Stress. Springer, New York: pp 443–472 [Google Scholar]
- Khan MS, Haas FH, Samami AA, Gholami AM, Bauer A, Fellenberg K, Reichelt M, Hänsch R, Mendel RR, Meyer AJ, Wirtz M, Hell R (2010) Sulfite reductase defines a newly discovered bottleneck for assimilatory sulfate reduction and is essential for growth and development in Arabidopsis thaliana. Plant Cell 22: 1216–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Király L, Künstler A, Höller K, Fattinger M, Juhász C, Müller M, Gullner G, Zechmann B (2012) Sulfate supply influences compartment specific glutathione metabolism and confers enhanced resistance to Tobacco mosaic virus during a hypersensitive response. Plant Physiol Biochem 59: 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S. (2006) Regulation of sulfate assimilation in Arabidopsis and beyond. Ann Bot (Lond) 97: 479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Kopriva S (2008b) Lessons from investigation of regulation of APS reductase by salt stress. Plant Signal Behav 3: 567–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, North KA, Kopriva S (2008a) Complex signaling network in regulation of adenosine 5′-phosphosulfate reductase by salt stress in Arabidopsis roots. Plant Physiol 146: 1408–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI (2000) The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23: 587–596 [DOI] [PubMed] [Google Scholar]
- Lang C, Popko J, Wirtz M, Hell R, Herschbach C, Kreuzwieser J, Rennenberg H, Mendel RR, Hänsch R (2007) Sulphite oxidase as key enzyme for protecting plants against sulphur dioxide. Plant Cell Environ 30: 447–455 [DOI] [PubMed] [Google Scholar]
- Lappartient AG, Touraine B (1996) Demand-driven control of root ATP sulfurylase activity and SO42- uptake in intact canola (the role of phloem-translocated glutathione). Plant Physiol 111: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappartient AG, Vidmar JJ, Leustek T, Glass ADM, Touraine B (1999) Inter-organ signaling in plants: regulation of ATP sulfurylase and sulfate transporter genes expression in roots mediated by phloem-translocated compound. Plant J 18: 89–95 [DOI] [PubMed] [Google Scholar]
- Lee BR, Huseby S, Koprivova A, Chételat A, Wirtz M, Mugford ST, Navid E, Brearley C, Saha S, Mithen R, Hell R, Farmer EE, et al. (2012) Effects of fou8/fry1 mutation on sulfur metabolism: is decreased internal sulfate the trigger of sulfate starvation response? PLoS One 7: e39425 10.1371/journal.pone.0039425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T. (2002) Sulfate metabolism. Arabidopsis Book 1: e0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick JA, Davies JP (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol 51: 141–165 [DOI] [PubMed] [Google Scholar]
- Lewandowska M, Sirko A (2008) Recent advances in understanding plant response to sulfur-deficiency stress. Acta Biochim Pol 55: 457–471 [PubMed] [Google Scholar]
- Llanes A, Bertazza G, Palacio G, Luna V (2013) Different sodium salts cause different solute accumulation in the halophyte Prosopis strombulifera. Plant Biol (Stuttg) 15(Suppl 1): 118–125 [DOI] [PubMed] [Google Scholar]
- Llanes A, Masciarelli O, Ordoñez R, Isla M, Luna V (2014) Differential growth responses to sodium salts involve different abscisic acid metabolism and transport in Prosopis strombulifera. Biol Plant 58: 80–88 [Google Scholar]
- López-Berenguer C, Carvajal M, Garcéa-Viguera C, Alcaraz CF (2007) Nitrogen, phosphorus, and sulfur nutrition in broccoli plants grown under salinity. J Plant Nutr 30: 1855–1870 [Google Scholar]
- Manchenko GP. (2002) Handbook of Detection of Enzymes on Electrophoretic Gels. CRC Press, Boca Raton, FL [Google Scholar]
- Martin NM, Maricle BR (2015) Species-specific enzymatic tolerance of sulfide toxicity in plant roots. Plant Physiol Biochem 88: 36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MN, Tarczynski MC, Shen B, Leustek T (2005) The role of 5′-adenylylsulfate reductase in controlling sulfate reduction in plants. Photosynth Res 86: 309–323 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H (2003) Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol 132: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland M, Otte M (2000) Effects of varying sulphate and nitrogen supply on DMSP and glycine betaine levels in Spartina anglica. J Sea Res 43: 199–207 [Google Scholar]
- Mullineaux PM, Rausch T (2005) Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynth Res 86: 459–474 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakamura T, Yamaguchi Y, Sano H (2000) Plant mercaptopyruvate sulfurtransferases: molecular cloning, subcellular localization and enzymatic activities. Eur J Biochem 267: 5621–5630 [DOI] [PubMed] [Google Scholar]
- Nazar R, Iqbal N, Masood A, Syeed S, Khan NA (2011b) Understanding the significance of sulfur in improving salinity tolerance in plants. Environ Exp Bot 70: 80–87 [Google Scholar]
- Nazar R, Iqbal N, Syeed S, Khan NA (2011a) Salicylic acid alleviates decreases in photosynthesis under salt stress by enhancing nitrogen and sulfur assimilation and antioxidant metabolism differentially in two mungbean cultivars. J Plant Physiol 168: 807–815 [DOI] [PubMed] [Google Scholar]
- Nguyen X-V, Klein M, Riemenschneider A, Papenbrock J (2014) Distinctive features and role of sulfur-containing compounds in marine plants, seaweeds, seagrasses and halophytes, from an evolutionary point of view. In Sabkha Ecosystems. IV: Cash Crop Halophyte and Biodiversity Conservation. Springer, New York, pp 299–312 [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33: 633–650 [DOI] [PubMed] [Google Scholar]
- Nocito FF, Lancilli C, Dendena B, Lucchini G, Sacchi GA (2011) Cadmium retention in rice roots is influenced by cadmium availability, chelation and translocation. Plant Cell Environ 34: 994–1008 [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Zhao P, Xiang C, Oliver DJ (2007) Glutathione conjugates in the vacuole are degraded by γ-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J 49: 878–888 [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Riemenschneider A, Kamp A, Schulz-Vogt HN, Schmidt A (2007) Characterization of cysteine-degrading and H2S-releasing enzymes of higher plants—from the field to the test tube and back. Plant Biol (Stuttg) 9: 582–588 [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Schmidt A (2000) Characterization of a sulfurtransferase from Arabidopsis thaliana. Eur J Biochem 267: 145–154 [DOI] [PubMed] [Google Scholar]
- Parida AK, Jha B (2010) Antioxidative defense potential to salinity in the euhalophyte Salicornia brachiata. J Plant Growth Regul 29: 137–148 [Google Scholar]
- Rao AC, Reddy AR (2008) Glutathione reductase: a putative redox regulatory system in plant cells. In Sulfur Assimilation and Abiotic Stress in Plants. Springer, New York: pp 111–147 [Google Scholar]
- Ravina CG, Barroso C, Vega JM, Gotor C (1999) Cysteine biosynthesis in Chlamydomonas reinhardtii. Molecular cloning and regulation of O-acetylserine(thiol)lyase. Eur J Biochem 264: 848–853 [DOI] [PubMed] [Google Scholar]
- Reginato M, Sgroy V, Llanes A, Cassán F, Luna V (2012) The American halophyte Prosopis strombulifera, a new potential source to confer salt tolerance to crops. In Crop Production for Agricultural Improvement. Springer, New York, pp 115–143 [Google Scholar]
- Reginato M, Sosa L, Llanes A, Hampp E, Vettorazzi N, Reinoso H, Luna V (2014) Growth responses and ion accumulation in the halophytic legume Prosopis strombulifera are determined by Na2SO4 and NaCl. Plant Biol (Stuttg) 16: 97–106 [DOI] [PubMed] [Google Scholar]
- Riemenschneider A, Riedel K, Hoefgen R, Papenbrock J, Hesse H (2005) Impact of reduced O-acetylserine(thiol)lyase isoform contents on potato plant metabolism. Plant Physiol 137: 892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogel JA, Ariza FA, Silla RO (2000) Soil salinity and moisture gradients and plant zonation in Mediterranean salt marshes of southeast Spain. Wetlands 20: 357–372 [Google Scholar]
- Romero LC, Domínguez-Solís JR, Gutiérrez-Alcalá G, Gotor C (2001) Salt regulation of O-acetylserine(thiol)lyase in Arabidopsis thaliana and increased tolerance in yeast. Plant Physiol Biochem 39: 643–647 [Google Scholar]
- Rouached H, Secco D, Arpat B, Poirier Y (2011) The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JM, Blumwald E (2002) Salinity-induced glutathione synthesis in Brassica napus. Planta 214: 965–969 [DOI] [PubMed] [Google Scholar]
- Schüssler MD, Alexandersson E, Bienert GP, Kichey T, Laursen KH, Johanson U, Kjellbom P, Schjoerring JK, Jahn TP (2008) The effects of the loss of TIP1;1 and TIP1;2 aquaporins in Arabidopsis thaliana. Plant J 56: 756–767 [DOI] [PubMed] [Google Scholar]
- Steffen S, Ball P, Mucina L, Kadereit G (2015) Phylogeny, biogeography and ecological diversification of Sarcocornia (Salicornioideae, Amaranthaceae). Ann Bot (Lond) 115: 353–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stribling JM. (1997) The relative importance of sulfate availability in the growth of Spartina alterniflora and Spartina cynosuroides. Aquat Bot 56: 131–143 [Google Scholar]
- Stulen I, De Kok LJ (1993) Whole-plant regulation of sulfur metabolism, a theoretical approach and comparison with current ideas on regulation of nitrogen-metabolism. De Kok LJ, Stulen I, Rennenberg H, Brunold C, Rauser WE, eds. In 2nd Workshop on Sulfur Metabolism in Higher Plants. SPB Academic Publishing, The Hague, The Netherlands. [Google Scholar]
- Takahama U. (1992) Hydrogen peroxide scavenging systems in vacuoles of mesophyll cells of Vicia faba. Phytochemistry 31: 1127–1133 [Google Scholar]
- Thalmann MR, Pazmino D, Seung D, Horrer D, Nigro A, Meier T, Kölling K, Pfeifhofer HW, Zeeman SC, Santelia D (2016) Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell tpc.00143.2016 [DOI] [PMC free article] [PubMed]
- Tsakraklides G, Martin M, Chalam R, Tarczynski MC, Schmidt A, Leustek T (2002) Sulfate reduction is increased in transgenic Arabidopsis thaliana expressing 5′-adenylylsulfate reductase from Pseudomonas aeruginosa. Plant J 32: 879–889 [DOI] [PubMed] [Google Scholar]
- Vauclare P, Kopriva S, Fell D, Suter M, Sticher L, von Ballmoos P, Krähenbühl U, den Camp RO, Brunold C (2002) Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J 31: 729–740 [DOI] [PubMed] [Google Scholar]
- Ventura Y, Eshel A, Pasternak D, Sagi M (2015) The development of halophyte-based agriculture: past and present. Ann Bot (Lond) 115: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura Y, Sagi M (2013) Halophyte crop cultivation: the case for Salicornia and Sarcocornia. Environ Exp Bot 92: 144–153 [Google Scholar]
- Ventura Y, Wuddineh WA, Ephrath Y, Shpigel M, Sagi M (2010) Molybdenum as an essential element for improving total yield in seawater-grown Salicornia europaea L. Sci Hortic (Amsterdam) 126: 395–401 [Google Scholar]
- Ventura Y, Wuddineh WA, Myrzabayeva M, Alikulov Z, Khozin-Goldberg I, Shpigel M, Samocha TM, Sagi M (2011a) Effect of seawater concentration on the productivity and nutritional value of annual Salicornia and perennial Sarcocornia halophytes as leafy vegetable crops. Sci Hortic (Amsterdam) 128: 189–196 [Google Scholar]
- Ventura Y, Wuddineh WA, Shpigel M, Samocha TM, Klim BC, Cohen S, Shemer Z, Santos R, Sagi M (2011b) Effects of day length on flowering and yield production of Salicornia and Sarcocornia species. Sci Hortic (Amsterdam) 130: 510–516 [Google Scholar]
- Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218: 1–14 [DOI] [PubMed] [Google Scholar]
- Wipf D, Mongelard G, van Tuinen D, Gutierrez L, Casieri L (2014) Transcriptional responses of Medicago truncatula upon sulfur deficiency stress and arbuscular mycorrhizal symbiosis. Front Plant Sci 5: 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M, Droux M, Hell R (2004) O-acetylserine (thiol) lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J Exp Bot 55: 1785–1798 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Sakihama Y, Ikehara N (1997) Flavonoid-peroxidase reaction as a detoxification mechanism of plant cells against H2O2. Plant Physiol 115: 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmolinsky D, Brychkova G, Fluhr R, Sagi M (2013) Sulfite reductase protects plants against sulfite toxicity. Plant Physiol 161: 725–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmolinsky D, Brychkova G, Kurmanbayeva A, Bekturova A, Ventura Y, Khozin-Goldberg I, Eppel A, Fluhr R, Sagi M (2014) Impairment in sulfite reductase leads to early leaf senescence in tomato plants. Plant Physiol 165: 1505–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesbergenova Z, Yang G, Oron E, Soffer D, Fluhr R, Sagi M (2005) The plant Mo-hydroxylases aldehyde oxidase and xanthine dehydrogenase have distinct reactive oxygen species signatures and are induced by drought and abscisic acid. Plant J 42: 862–876 [DOI] [PubMed] [Google Scholar]
- You J, Chan Z (2015) ROS regulation during abiotic stress responses in crop plants. Front Plant Sci 6: 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechmann B, Müller M, Zellnig G (2008) Modified levels of cysteine affect glutathione metabolism in plant cells. In Sulfur Assimilation and Abiotic Stress in Plants. Springer, New York, pp 193–206 [Google Scholar]