Phenotypic characterization of Anaphase Promoting Complex/Cyclosome Inhibitors reveals tissue-dependent roles for the APC/CCCS52A1 and APC/CCCS52A2 complexes.
Abstract
The endocycle represents a modified mitotic cell cycle that in plants is often coupled to cell enlargement and differentiation. Endocycle onset is controlled by activity of the Anaphase Promoting Complex/Cyclosome (APC/C), a multisubunit E3 ubiquitin ligase targeting cell-cycle factors for destruction. CELL CYCLE SWITCH52 (CCS52) proteins represent rate-limiting activator subunits of the APC/C. In Arabidopsis (Arabidopsis thaliana), mutations in either CCS52A1 or CCS52A2 activators result in a delayed endocycle onset, whereas their overexpression triggers increased DNA ploidy levels. Here, the relative contribution of the APC/CCCS52A1 and APC/CCCS52A2 complexes to different developmental processes was studied through analysis of their negative regulators, being the ULTRAVIOLET-B-INSENSITIVE4 protein and the DP-E2F-Like1 transcriptional repressor, respectively. Our data illustrate cooperative activity of the APC/CCCS52A1 and APC/CCCS52A2 complexes during root and trichome development, but functional interdependency during leaf development. Furthermore, we found APC/CCCS52A1 activity to control CCS52A2 expression. We conclude that interdependency of CCS52A-controlled APC/C activity is controlled in a tissue-specific manner.
The endocycle represents a modified version of the mitotic cell cycle during which the genome is replicated in the absence of mitosis and cytokinesis, resulting in a doubling of the nuclear DNA content (De Veylder et al., 2011; Edgar at al., 2014). Endoreplication is a common feature among eukaryotes, frequently observed in cell types with a high metabolic activity (Larkins et al., 2001). In Drosophila melanogaster, endoreplication is predominantly seen in larval tissues and the salivary glands (Lilly and Duronio, 2005), whereas in Caenorhabditis elegans, it is observed in the syncytium (Flemming et al., 2000). In higher plants, like Arabidopsis (Arabidopsis thaliana), endoreplication is observed in most tissues and is coupled to cell differentiation (Breuer et al., 2010), such as in developing leaves, where the onset of the endocycle marks the exit from cell division (Beemster et al., 2006). Furthermore, endoreplication is believed to be an important trigger for cell and organ growth, because cell size is frequently correlated with the DNA ploidy levels (Melaragno et al., 1993; De Veylder et al., 2011), although such a relationship is not always observed (Beemster et al., 2002). In tomato (Solanum lycopersicum), increased ploidy levels are correlated with increased levels of rRNA and protein synthesis per-nucleus, indicating increased metabolism to support cell growth (Bourdon et al., 2012). Endoreplication has also been demonstrated to instruct cell fate, as observed for Arabidopsis trichomes (Bramsiepe et al., 2010). More recently, endocycle modulators have been implicated in the control of innate immunity in Arabidopsis (Hamdoun et al., 2016), suggestive for the importance of the endocycle in the plant immunity.
In all eukaryotes, control of the mitotic cell cycle, cell differentiation, and endocycle onset is achieved by the Anaphase Promoting Complex/Cyclosome (APC/C). The APC/C is a conserved E3 ubiquitin ligase that ubiquitinates key cell cycle proteins containing APC/C recognition motifs known as the Destruction or KEN/GxEN-boxes, resulting in their destruction by the proteasome, thereby ensuring the unidirectional cell cycle progression (Heyman and De Veylder, 2012). During the late G2 and early M phase, the APC/C is activated by the CDC20/Fizzy activator subunit, which itself is targeted for destruction during the anaphase and substituted by the CDH1/FZR type activator subunit, known in plants as CELL CYCLE SWITCH52 (CCS52) proteins (Peters, 2002; Baker et al., 2007). The Arabidopsis genome encodes two types of CCS52 proteins, namely the A-type, consisting of CCS52A1 and CCS52A2, and B-type CCS52B (Cebolla et al., 1999). Whereas the function of CCS52B in the cell cycle is still unclear, both CCS52A1 and CCS52A2 activator subunits have been demonstrated to control endocycle onset (Cebolla et al., 1999; Lammens et al., 2008; Narbonne-Reveau et al., 2008; Mathieu-Rivet et al., 2010). In the Arabidopsis root, APC/CCCS52A1 activity controls the timing of endocycle onset by marking the A-type cyclin CYCA2;3 for destruction (Imai et al., 2006; Boudolf et al., 2009). Correspondingly, ccs52a1 mutants display roots with an expanded meristem size owing to an increased number of meristematic cells (Vanstraelen et al., 2009). CCS52A1 also drives the trichome endocycle and trichome branching, as ccs52a1 loss-of-function mutant trichomes display two branches in contrast to wild-type trichomes that predominantly contain three branches. Correspondingly, CCS52A1 overexpression results in trichome overbranching (Imai et al., 2006; Kasili et al., 2010). In contrast to CCS52A1, the CCS52A2 activator subunit appears not to control root meristem size, but is instead required for stem cell maintenance by suppressing cell division of the Quiescent Center (QC) stem cells, as ccs52a2 mutants display increased QC cell division rates being correlated with a disorganization of the root meristem (Vanstraelen et al., 2009). CCS52A2 might play a similar role in the shoot, as its absence results in a disrupted cell organization of the L1 and L2 layers (Liu et al., 2012). In leaves, both CCS52A1- and CCS52A2-activated APC/C complexes control endocycle onset, as mutation of either results in reduced DNA ploidy levels (Lammens et al., 2008). Functional redundancy between CCS52A1 and CCS52A2 is additionally suggested by the observation that no viable double mutant plants can be obtained (Baloban et al., 2013).
Due to its importance during development, APC/CCCS52A activity is tightly controlled at both the transcriptional and posttranslational levels. On the transcriptional level, expression of both CCS52A1 and CCS52A2 is negatively regulated by E2Fa in complex with RETINOBLASTOMA-RELATED PROTEIN1 (RBR1). Overexpression of a mutated E2Fa allele, lacking the RBR1 interaction domain, results in increased CCS52A expression, indicating that recruitment of RBR1 is required to suppress gene expression (Magyar et al., 2012). CCS52A1 expression is additionally negatively regulated by the GT2-LIKE1 trihelix transcription factor (Breuer et al., 2012), whereas its transcription is activated by the cytokinin-activated ARABIDOPSIS RESPONSE REGULATOR2 (Takahashi et al., 2013). CCS52A2 expression rather appears to be specifically repressed by the atypical E2F transcription factor DEL1, which acts as a negative regulator of endocycle onset (Vlieghe et al., 2005; Lammens et al., 2008). Mutation of DEL1 results in increased CCS52A2 expression and APC/CCCS52A2 activity, resulting in a premature endocycle onset and increased DNA ploidy levels. At the posttranslational level, the UVI4 protein, in association with the UBIQUITIN-SPECIFIC PROTEASE14, has been found to be an inhibitor of the APC/CCCS52A1 complex (Heyman et al., 2011; Xu et al., 2016). Double mutant analysis of plants lacking a functional CCS52A1 and UVI4 gene revealed that CCS52A1 functions epistatically over UVI4 in controlling endocycle onset. Correspondingly, UVI4 regulates endocycle onset in leaves and root meristem size maintenance by inhibiting APC/CCCS52A1 activity, as observed by the increased DNA ploidy levels in leaves and trichomes, and a reduced number of meristematic cells in the root tip of uvi4 mutant plants (Hase et al., 2006; Heyman et al., 2011). Here we aimed to study the interplay and specificity of APC/CCCS52A1 and APC/CCCS52A2 during plant development. For this purpose, we performed a comparative phenotypic analysis of their negative regulators, being UVI4 and DEL1, respectively, as such circumventing the artificial and unspecific effects that might result from constitutive CCS52A1 and CCS52A2 overexpression.
RESULTS
CCS52A Activators Are Indispensable for Plant Development
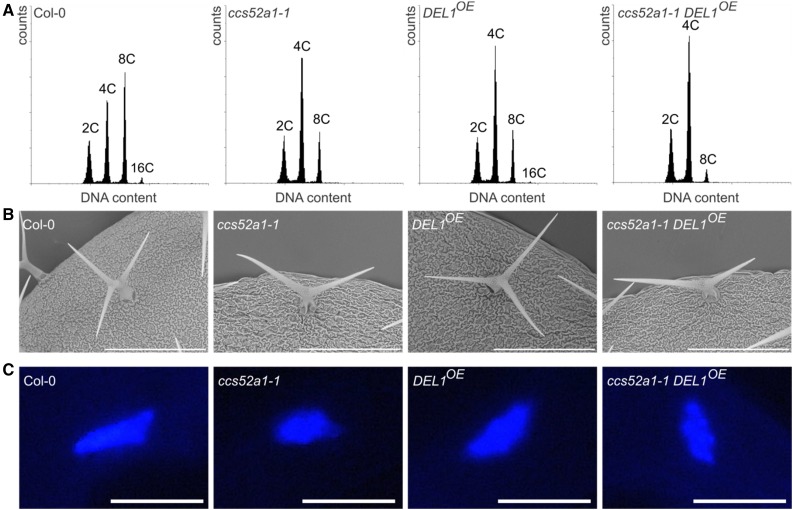
Because of the importance of the CCS52A proteins in cell cycle exit and endocycle onset, we aimed to test the effects of deficiency in both CCS52A1 and CCS52A2 during plant development. Because ccs52a1-1 ccs52a2-1 double mutants are not viable (Vanstraelen et al., 2009; Baloban et al., 2013), we generated a ccs52a1-1 DEL1OE double mutant, which lacks a functional CCS52A1 and displays reduced CCS52A2 expression, owing to increased activity of the DEL1 transcriptional repressor (Lammens et al., 2008). Using flow cytometry, we confirmed the previously observed reduced DNA ploidy levels of the ccs52a1-1 and DEL1OE single mutant leaves compared to the wild type (Vlieghe et al., 2005; Vanstraelen et al., 2009; Baloban et al., 2013; Fig. 1A; Supplemental Table S1). In ccs52a1-1 DEL1OE double mutant leaves, an additional decrease in the DNA ploidy level compared to those of the single mutants could be observed (Fig. 1A; Supplemental Table S1), confirming that expression of both CCS52A genes contribute to endocycle onset in the leaf.
Figure 1.
Endoreplication phenotypes of the ccs52a1-1 DEL1OE double mutant. A, Flow cytometric analysis of wild-type (Col-0), ccs52a1-1, DEL1OE, and ccs52a1-1 DEL1OE three-week-old first true leaves. Data are representative for the mean (n = 3). B, Scanning electron microscope images of wild-type (Col-0), ccs52a1-1, DEL1OE, and ccs52a1-1 DEL1OE trichomes. Images are representative for the mean (n = 3). Bars = 300 μm. C, Epifluorescence images of DAPI-stained wild-type, ccs52a1-1, DEL1OE, and ccs52a1-1 DEL1OE trichome nuclei. Images are representative for the mean (n > 13). Bars = 10 μm.
Independently, the trichome branch number was quantified, which frequently correlates with the DNA content (Perazza et al., 1999a). Whereas the ccs52a1-1 mutant plants display trichomes with a reduced number of branches, the DEL1OE trichome branch number did not differ from that of the wild type, but the ccs52a1-1 DEL1OE double mutant showed a trichome branch number similar to that of the ccs52a1-1 mutant (Fig. 1B; Supplemental Fig. S1A). In addition, the trichome nuclear size was investigated. Corresponding to the reduced trichome branch number, a reduction in trichome nuclear size of the ccs52a1-1 mutant was observed (Fig. 1C; Supplemental Fig. S1B), confirming previous findings (Heyman et al., 2011). Whereas no difference in trichome nuclear size of the DEL1OE mutant could be detected, the ccs52a1-1 DEL1OE double mutant displayed a reduced nuclear size, comparable to that of the ccs52a1-1 single mutant (Fig. 1C; Supplemental Fig. S1B), confirming previous data that CCS52A1 is the main APC/C activator in trichomes.
UVI4 Is a Specific Inhibitor of CCS52A1
Previously, we demonstrated that UVI4 acts as an inhibitor of APC/CCCS52A1 (Heyman et al., 2011). Using leaf ploidy levels, and trichome nuclear size and branching phenotype as a readout, the ccs52a1-1 mutation was found to be epistatic over uvi4 (Heyman et al., 2011). To test whether a similar genetic relationship exists between CCS52A2 and UVI4, the leaf ploidy level and trichome nuclear size of the uvi4 ccs52a2-1 double mutant was compared with that of the single mutants. Whereas the uvi4 and ccs52a2-1 single mutants showed an increase and decrease in DNA ploidy levels compared to the wild type, respectively, the uvi4 ccs52a2-1 double mutant contained DNA ploidy levels intermediate to those of the single mutants (Fig. 2A; Supplemental Table S2). Additionally, the trichome branch number was quantified. As demonstrated previously, uvi4 mutants display increased trichome branching (Perazza et al., 1999b; Hase et al., 2006; Heyman et al., 2011), whereas the ccs52a2-1 mutant showed a mild reduction in the number of trichomes containing four branches compared to the wild type (Fig. 2B; Supplemental Fig. S2A), correlating with trichome nuclear size (Fig. 2C and Supplemental Fig. S2B). The uvi4 ccs52a2-1 double mutant displayed a trichome branch number intermediate to that of the uvi4 and ccs52a2-1 single mutants, again corresponding to the observed nuclear size (Fig. 2, B and C; Supplemental Fig. S2B). Combined with the previously observed protein-protein interaction of UVI4 with CCS52A1 but not CCS52A2 (Heyman et al., 2011), these data suggest that UVI4 is a specific inhibitor of APC/CCCS52A1.
Figure 2.
Endoreplication phenotypes of the uvi4 ccs52a2-1 double mutant. A, Flow cytometric analysis of wild-type (Col-0), uvi4, ccs52a2-1, and uvi4 ccs52a2-1 three-week-old first true leaves. Data are representative for the mean (n = 3). B, Scanning electron microscope images of wild-type (Col-0), uvi4, ccs52a2-1, and uvi4 ccs52a2-1 trichomes. Images are representative for the mean. Bars = 300 μm. C, Epifluorescence images of DAPI-stained wild-type, uvi4, ccs52a2-1, and uvi4 ccs52a2-1 trichome nuclei. Images are representative for the mean (n > 8). Bars = 10 μm.
UVI4 and DEL1 Are Coexpressed in Arabidopsis Seedlings
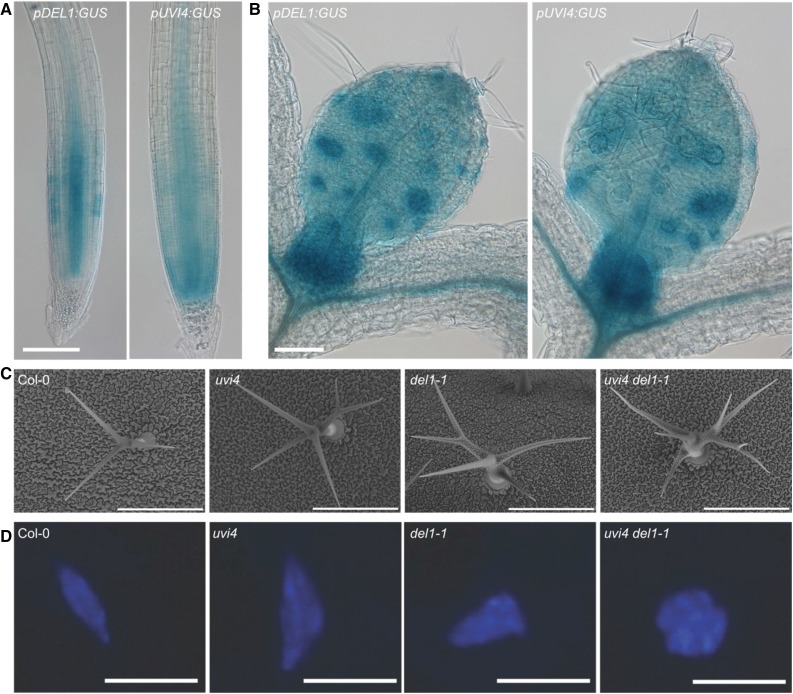
To investigate the putative interplay between UVI4 and DEL1 during plant development, we compared their expression patterns using transcriptional GUS reporter lines. Expression of both UVI4 and DEL1 could be detected in tissues showing a high cell division activity, such as the root meristem (Fig. 3A) and shoot meristem, and young leaves (Fig. 3B). Thus it appears that both APC/C regulators are tightly coexpressed, suggesting a role for both APC/CCCS52A1 and APC/CCCS52A2 in the development of different tissues.
Figure 3.
UVI4 and DEL1 independently control trichome ploidy levels. A and B, Expression pattern of UVI4 and DEL1 in the root meristem (A) and leaves (B). Bars = 0.1 mm. C, Scanning electron microscope images of wild-type (Col-0), uvi4, del1-1, and uvi4 del-1 trichomes. Images are representative for the mean. Bars = 300 μm. D, Epifluorescence images of DAPI-stained wild-type, uvi4, del-1, and uvi4 del-1 trichome nuclei. Images are representative for the mean (n > 8). Bars = 10 μm.
UVI4 and DEL1 Contribute Independently to Trichome Development
To study the effects of a lack of both a functional UVI4 and DEL1, we generated a uvi4 del1-1 double mutant, which is anticipated to result in an increased activity of both APC/CCCS52A1 and APC/CCCS52A2 complexes. Similar to the uvi4 mutant, del1-1 mutants were found to display an increased trichome branching phenotype (Fig. 3C; Supplemental Fig. S3S). Correspondingly, quantification of the trichome nuclear size revealed an increase in the DNA content in del1-1 mutant trichomes, similar to the uvi4 mutant, indicative for a role of DEL1 in suppressing endoreplication in trichomes (Fig. 3D; Supplemental Fig. S3B). In the uvi4 del1-1 double mutant, a clearly enhanced effect on trichome branching could be observed (Fig. 3C; Supplemental Fig. S3A), with a correspondingly increased trichome nuclear size compared to the single mutants (Fig. 3D; Supplemental Fig. S3B). These data suggest that both UVI4 and DEL1, and hence APC/CCCS52A1 and APC/CCCS52A2, control trichome branching.
UVI4 and DEL1 Independently Regulate Root Meristem Size Maintenance
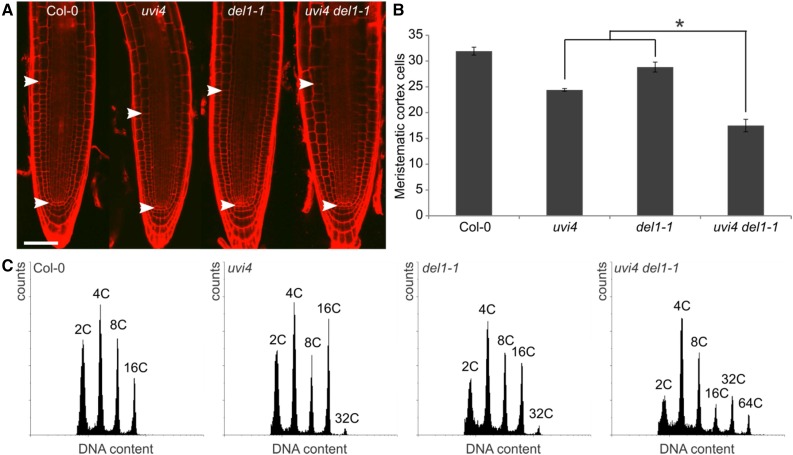
In the Arabidopsis root, CCS52A1 expression can be found in the root elongation zone, where it controls the timing of cell cycle exit. Accordingly, mutation of UVI4 results in a decreased root meristem size, reflected by a decrease in the number of meristematic cortex cells, probably owing to an increased APC/CCCS52A1 activity (Heyman et al., 2011). On the other hand, CCS52A2 expression can be found predominantly in the QC cells, where it is required to keep these cells from dividing and differentiating (Vanstraelen et al., 2009). Because the DEL1 transcriptional repressor of CCS52A2 is expressed throughout the root meristem, it was tested whether DEL1 mutation affects root meristem size maintenance. No significant decrease in the number of meristematic cortex cells could be observed in the del1-1 mutant compared to wild-type roots (Fig. 4, A and B). In contrast, when determining the root meristem size of the uvi4 del1-1 double mutant, a significant reduction in the number of meristematic cortex cells could be observed compared to both the uvi4 and del1-1 single mutants (Fig. 4, A and B). This reduction was offset by an increased cortex cell size, resulting in a meristem size similar to that of the wild type.
Figure 4.
UVI4 and DEL1 independently control cell cycle exit in the root. A, Representative confocal microscopy images of wild-type (Col-0), uvi4, del1-1, and uvi4 del1-1 one-week-old root meristems stained with propidium iodide. Arrowheads indicate the meristem size based on the cortical cell length. Bar = 50 μm. B, Number of meristematic cortex cells for lines presented in (A). Data represent mean ± se (n > 8, *P < 0.05, Student’s t test). C, Flow cytometric analysis of wild-type (Col-0), uvi4, del1-1, and uvi4 del1-1 one-week-old mutant roots. Data are representative for the mean.
When determining the DNA ploidy levels of uvi4 and del1-1 mutant roots, an increase could be observed for both compared to those of wild-type roots (Fig. 4C; Supplemental Table S3). When determining the DNA ploidy levels of uvi4 del1-1 double mutant roots, an increase could be detected compared to the single uvi4 and del1-1 mutants (Fig. 4C; Supplemental Table S3). These observations suggest that both APC/CCCS52A1 and APC/CCCS52A2 contribute to the DNA ploidy levels of roots.
UVI4 and DEL1 Control the Onset of Endoreplication in Leaves through a Common Mechanism
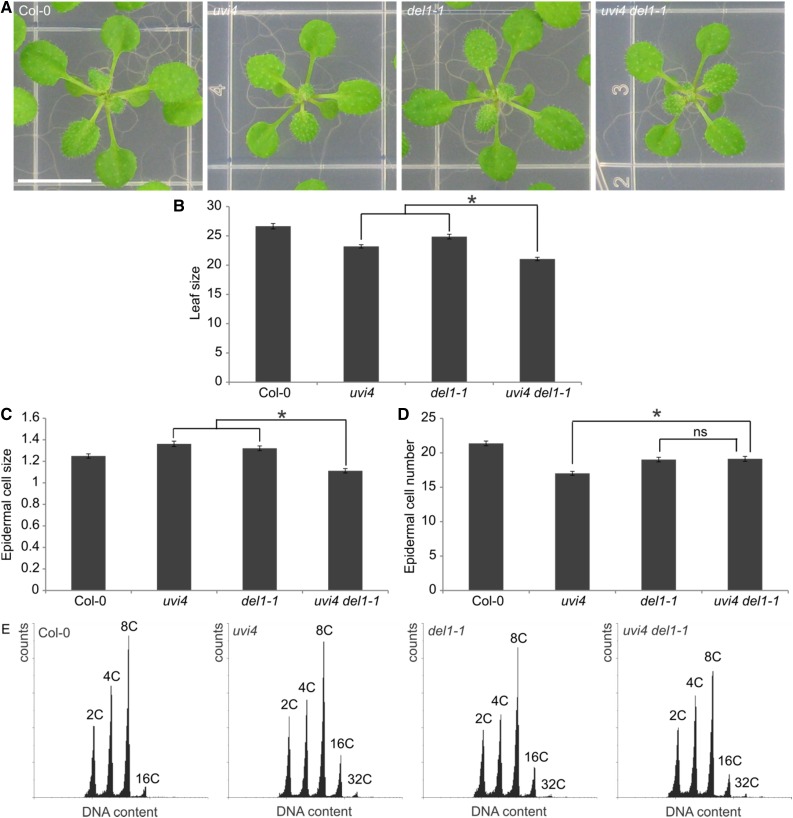
Next to the effect of the uvi4 de11-1 double mutation in trichomes and roots, we tested its effect on leaf development by analyzing the size of mature first true leaves. Both the uvi4 and del1-1 single mutants displayed a reduced leaf size compared to wild-type leaves (Fig. 5, A and B). The uvi4 del1-1 double mutant displayed an additive effect on the significantly reduced leaf size compared to both uvi4 and del1-1 single mutants (Fig. 5, A and B). To determine the cause of the reduced leaf size, the average abaxial epidermal cell number and size were investigated. The uvi4 and del1-1 single mutants’ reduced leaf size was caused by a reduction in cell number, despite the small increase in cell size, compared to wild-type plants (Fig. 5, C and D). The uvi4 del1-1 double mutant displayed a decrease in epidermal cell number, being equal to that seen for the single mutants, but without compensatory increase in cell size (Fig. 5, C and D). These data show that the apparent additive effect of uvi4 and del1 mutations on leaf size in the double mutant is not caused by the additive effects of the single mutations on epidermal cell size and cell number.
Figure 5.
UVI4 and DEL1 control leaf ploidy levels through a common mechanism. A, Images of three-week-old wild-type (Col-0), uvi4, del1-1, and uvi4 del1-1 rosettes. Bar = 1 cm. B, Average first true leaf sizes of three-week-old wild-type (Col-0), uvi4, del1-1, and uvi4 del1-1 plants (in mm2). Data represent mean ± se (n = 45, *P < 0.05, Student’s t test). C and D, Average abaxial epidermal cell size (× 1.000 μm2) and average cell number (× 1.000), respectively, of first leaves of three-week-old lines presented in (B). Data represent mean ± se (n = 18, *P < 0.05, Student’s t test). E, Flow cytometric analysis of wild-type (Col-0), uvi4, del1-1, and uvi4 del1-1 three-week-old first true leaves. Data are representative for the mean (n = 3).
Seeing how double mutation of UVI4 and DEL1 has strong additive effects on the DNA ploidy levels in trichomes and roots, we tested whether a similar additive effect could hold true for leaves. When determining the DNA ploidy levels of mature leaves in the uvi4 and del1-1 single mutants, an increase could be confirmed compared to wild-type plants (Vlieghe et al., 2005; Heyman et al., 2011; Fig. 5E; Supplemental Table S4). However, the DNA ploidy distributions of the uvi4 del1-1 double mutant leaves did not differ significantly from the uvi4 and del1-1 single mutants (Fig. 5E; Supplemental Fig. S4; Supplemental Table S4), which is in contrast to the results obtained in trichomes and roots, suggesting that APC/CCCS52A1 and APC/CCCS52A2 are interdependent in the leaf.
CCS52A2 Expression Is Affected by CCS52A1 Activity
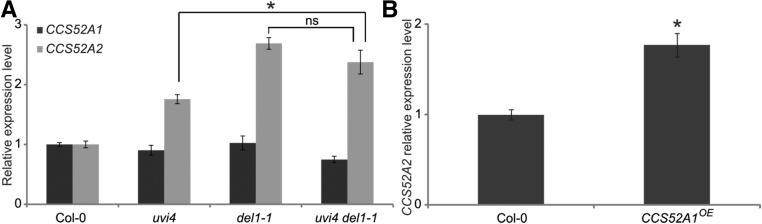
The observation that the DNA ploidy levels of the uvi4 del1-1 double mutant leaves are similar to those of the single mutants, is contradicting the gene dosage effect observed in ccs52a1-1 DEL1OE mutants. To investigate this apparent paradox, we analyzed the CCS52A expression levels in developing first true leaves of the uvi4, del1-1, and uvi4 del1-1 mutants using RT-qPCR. To ensure the seedlings were at a similar developmental age, leaves were harvested at stage 1.04 (Boyes et al., 2001). For CCS52A1, no major differences in expression could be observed in the mutants compared to the wild type (Fig. 6A). By contrast, when determining the CCS52A2 transcript levels in the del1-1 mutant, an up-regulation could be observed, confirming previous data (Lammens et al., 2008; Fig. 6A). Surprisingly, mutation of UVI4 also resulted in an increased CCS52A2 expression in first true leaves compared to wild-type leaves (Fig. 6A), suggesting that activation of APC/CCCS52A1 results in increased CCS52A2 expression. In the uvi4 del1-1 double mutant stage 1.04 leaves, however, no additive effect on CCS52A2 expression could be observed, as CCS52A2 transcript levels were found to be increased, identical to those observed in the del1-1 mutant (Fig. 6A).
Figure 6.
CCS52A1 activity affects CCS52A2 expression. A, Relative expression levels of CCS52A1 and CCS52A2 in first true leaves from wild-type (Col-0), uvi4, del1-1, and uvi4 del1-1. First true leaves were harvested at stage 1.04. The relative expression levels for wild-type leaves were arbitrarily set to 1. Data represent mean ± se (n = 3, *P < 0.05, Student’s t test). B, Increased CCS52A2 transcript levels in CCS52A1OE first true leaves compared to the wild type. First true leaves were harvested at stage 1.04. The relative expression levels for wild-type leaves were arbitrarily set to 1. Data represent mean ± se (n = 3, *P < 0.05, Student’s t test).
To confirm that an increase in APC/CCCS52A1 activity results in transcriptional activation of CCS52A2, CCS52A2 transcript levels were analyzed in wild-type versus CCS52A1OE leaves harvested at stage 1.04. Indeed, CCS52A2 transcript levels were increased in CCS52A1OE leaves, similar to uvi4 mutant leaves (Fig. 6B), suggesting that increased APC/CCCS52A1 activity results in increased CCS52A2 expression.
DISCUSSION
(Non)Redundant Functions of CCS52A1 and CCS52A2
Both APC/C activity-controlling proteins CCS52A1 and CCS52A2 play important functions during plant development. CCS52A1 has been predominantly implicated in controlling trichome branching and root ploidy levels (Vanstraelen et al., 2009; Kasili et al., 2010), whereas CCS52A2 has been demonstrated to control the proliferation status of the root QC stem cells (Vanstraelen et al., 2009) and shoot apical meristem maintenance (Liu et al., 2012). Although CCS52A1 and CCS52A2 control different developmental processes, functional redundancy is expected, as plants being deficient for both CCS52A genes are not viable (Baloban et al., 2013). Indeed, endoreplication onset in leaves is ensured by both CCS52A-type proteins. Furthermore, during plant growth, a decreased expression of CCS52A1 appears to be compensated by an increased expression of CCS52A2 (Baloban et al., 2013). To overcome the problem of lethality, we generated a partial loss-of-function mutant using the ccs52a1-1 DEL1OE double mutant in which the absence of CCS52A1 is accompanied with reduced CCS52A2 transcription owing to its increased repression by DEL1 (Lammens et al., 2008). Whereas no clear obvious additive effects could be observed in the development of tissues that are predominantly controlled by CCS52A1, being the trichomes or the root meristem, the leaf DNA ploidy levels were further decreased in the ccs52a1-1 DEL1OE double mutant compared to the single mutants. Therefore, it could be stated that some tissues are more dependent on the CCS52A gene redundancy compared to others.
UVI4 and DEL1 Specifically Fine-Tune APC/CCCS52A Activity
The uvi4 ccs52a2-1 double mutant displays a trichome branching and DNA ploidy phenotype being intermediate to that observed for the single uvi4 and ccs52a2-1 mutants, which is in contrast to the observation that the ccs52a1-1 mutation is epistatic over the uvi4 mutation (Heyman et al., 2011). Together with the previously observed lack of protein-protein interactions between UVI4 and CCS52A2, these observations support the idea that UVI4 is a specific inhibitor of the APC/CCCS52A1 complex. How such specificity might be achieved at the protein level is unclear. The CCS52A1 and CCS52A2 proteins share a high sequence homology, including the C-box, Cdh1 specific motif, cyclin-binding, and C-terminal IR motif, together with predicted CDK phosphorylation sites (Fülöp et al., 2005). Extending our knowledge of CCS52A structural domains might help shed light on the preference of the UVI4 protein inhibitor.
The specificity of DEL1 toward regulating CCS52A2 expression was demonstrated previously by specific binding of DEL1 to the CCS52A2 promoter and exclusive changes in CCS52A2 transcription levels in DEL1OE and del1-1 knockout lines (Lammens et al., 2008). Whereas these observations were made for complete seedling and leaf tissue, respectively, a control of CCS52A2 expression by DEL1 can be observed as well for root tissue (Supplemental Fig. S6, A and B), strongly suggesting that DEL1 is a specific repressor of CCS52A2 across all DEL1 expressing cells.
Additive Effects of UVI4 and DEL1
The specificity of UVI4 and DEL1 toward APC/CCCS52A1 and APC/CCCS52A2, respectively, offers a unique tool to study the relative contribution of both APC/C complexes during development, complementing available knockout data. This strategy has as benefit that the APC/C activity levels obtained remain within a physiological range, in contrast to constitutive CCS52A1 or CCS52A2 overexpression, where it can be expected that complexes lose substrate specificity when being highly abundant. Using this strategy, we revealed an additive effect of uvi4 and del1 knockout on trichome branching and ploidy level, indicating that DEL1 functions to actively repress APC/CCCS52A2 activity in trichome cells. Accordingly, ectopic expression of CCS52A2 triggers trichome hyperbranching (Baloban et al., 2013). A similar situation likely holds true for the root, as uvi4 del1-1 double knockouts display a phenotypic enhancement of the uvi4 and del1-1 single knockouts, again suggesting that DEL1 plays an active role in suppressing APC/CCCS52A2 activity in the meristematic cells, supported by the DEL1 expression pattern. Apparently contrasting with an APC/CCCS52A2-repressing role for DEL1, del1-1 single mutant roots are phenotypically indistinguishable from wild-type roots in terms of meristem size. These data suggest that APC/CCCS52A1 is the primary APC/C complex controlling cell cycle exit in the root elongation zone (Vanstraelen et al., 2009). Strikingly, the aggravated meristematic cell number phenotype of del1-1 in the uvi4 mutant background was accompanied by an increased meristematic cortex cell size, suggesting an increased cell cycle duration. Knowing that APC/CCCS52A2 activity represses cell division activity of the QC cells, it is appealing to speculate that transcriptional activation of CCS52A2 throughout the root meristem here also delays cell cycle progression, resulting in the observed increased cell size.
Cross Talk between APC/CCCS52A1 and APC/CCCS52A2 Activity
Contrary to trichomes and root meristems, where APC/CCCS52A1 activity predominantly controls endoreplication onset, in the leaf, both CCS52A1 and CCS52A2 control the DNA ploidy level (Lammens et al., 2008; Baloban et al., 2013). Correspondingly, mutation of UVI4 or DEL1 results in leaves containing increased DNA ploidy levels, likely due to increased APC/CCCS52A1 or APC/CCCS52A2 activity, respectively. Surprisingly, no additive effect on the leaf DNA ploidy levels could be detected upon UVI4 and DEL1 double mutation. Rather, the double mutant results suggest a linear pathway. Strikingly, increased APC/CCCS52A1 activity was found to boost CCS52A2 expression. Here, the effect of CCS52A1 on CCS52A2 expression might be DEL1-dependent, because the increased CCS52A2 transcript level in the uvi4 del1-1 double mutant leaves appeared to be similar to that of del1-1 single mutant leaves. How DEL1 activity could be controlled by APC/CCCS52A1 remains unknown. In mammalian systems, atypical E2F proteins are recognized by APC/CCDH1 through a KEN-box motif and are subsequently marked for proteolytic degradation (Boekhout et al., 2016). However, no obvious APC/C recognition degron can be found in the DEL1 protein sequence. Another possibility is that DEL1 activity is regulated through its phosphorylation, because a predicted CDK phosphorylation motif is present in the N terminus of the DEL1 protein sequence (Supplemental Fig. S5; Chang et al., 2007). Increased APC/CCCS52A1-mediated destruction of cyclins could reduce putative CDK-mediated phosphorylation of DEL1, possibly rendering it unable to repress CCS52A2 expression. Although nothing is known about the regulation of DEL proteins through phosphorylation, DEL1 has been shown to interact with CYCB2;3 and CYCD1;1 in a yeast two-hybrid screen, hinting to a connection between DEL1 and the cyclin-CDK machinery (Boruc et al., 2010).
Another possible explanation might be that, in leaves, activation of APC/CCCS52A1 or APC/CCCS25A2 alone is sufficient to surpass a specific activity threshold to engage into the endocycle, e.g. through destruction of a factor being rate limiting for endocycle onset. This hypothesis would imply that CCS52A1 and CCS52A2 only control leaf endocycle onset, and not endocycle progression itself. Correspondingly, in trichomes, APC/CCCS52 activity has been postulated to mediate endoreplication onset, whereas following endocycles are thought to be maintained by the CULLIN4-RING FINGER-LIGASE ubiquitin ligase (Roodbarkelari et al., 2010). A similar endocycle onset/maintenance mechanism might hold true for leaves as well. In this scenario, the additive effects of UVI4 and DEL1 mutations observed in root meristems and trichomes would be suggestive for CCS52A1- and CCS52A2-independent functions, e.g. through different substrate specificity.
In conclusion, gaining more insight into the tissue-specific substrates and substrate specificity of APC/CCCS52A1 and APC/CCCS52A2 will strongly contribute to the understanding of how APC/C-dependent endocycle onset is fine-tuned during development. In the root, CYCA2;3 has been reported to be a specific target of APC/CCCS52A1 in the root elongation zone (Boudolf et al., 2009), whereas the ERF115 transcription factor was proposed to be a APC/CCCS52A2-specific target in root stem cells (Heyman et al., 2013). A possible method to uncover specific APC/C substrates is by using a biochemical approach. A proteomics screen to identify differentially ubiquitinated proteins (Walton et al., 2016) in the ccs52a1 and ccs52a2 mutants might prove a major step forward. Alternatively, mutant suppressor screens could be adverted to search for APC/CCCS52A1 or APC/CCCS52A2 specific targets. Finding the answer to these questions might shed light on why certain tissues predominantly advert only one of the two CCS52A proteins, whereas other organs depend on both CCS52A-type isoforms.
MATERIALS AND METHODS
Plant Medium and Growth Conditions
Arabidopsis thaliana wild type and mutants are in the Columbia-0 (Col-0) background. Plants were grown under a long-day/short-night regime (16-h light/8-h darkness) at 21°C on agar-solidified culture medium (Murashige & Skoog medium, 10 g/L saccharose, 0.43 g/L MES, and 0.8% [w/v] plant tissue culture agar).
Mutant Lines
The uvi4 (Hase et al., 2006), pUVI4:GUS/GFP (Heyman et al., 2011), pDEL1:GUS/GFP, ccs52a1-1 and ccs52a2-1 (Lammens et al., 2008), del1-1 and DEL1OE (Vlieghe et al., 2005), and CCS52A1OE (Vanstraelen et al., 2009) mutants and reporter lines used have been described previously. Double mutants were generated by crossing and validated by genotyping.
Flow Cytometry Analysis
Leaves were chopped with a razor blade in 200 μL CyStain UV Precise nuclei extraction buffer (Partec) and DNA was stained by adding 800 μL staining buffer (Partec). Nuclei were measured with CyFlow Flow Cytometer (Partec) and analyzed with the CXP Analysis software (Partec). Three or more leaves originating from different plants were analyzed for each technical repeat. The endoreduplication index was calculated as follows: EI = [(0 × %2C nuclei) + (1 × %4C nuclei) + (2 × %8C nuclei) + (3 × %16C nuclei) + (4 × %32C nuclei)].
Leaf and Cellular Parameter Determination
Mature first true leaves were harvested and cleared using a 75:25 (v/v) ethanol/acetic acid solution. Next, leaves were fixed and mounted on a slide using lactic acid. Cells were drawn using a DF microscope (Leica). Analysis of the leaf area was performed using the software ImageJ 1.41 (National Institutes of Health).
Quantification of Trichome Nuclear DNA Content
For 4′,6-diamidino-2-phenylindole (DAPI) staining, 3-week-old mature leaves were fixed using acetic acid (75:25 [v/v] acetic acid/ethanol) for at least 2 h and washed for at least 1 h with 70% (v/v) ethanol. Leaves were briefly submerged in 0.5 m EDTA and trichomes were removed using forceps. DNA was stained using 20 μg/mL DAPI in McIlvaine’s buffer (60 mm citric acid, 80 mm Na2HPO4, pH 4.1). Trichomes were mounted in Vectashield mounting medium (No. CA94010; Vector Laboratories) for fluorescence H1000 (Vector Laboratories) and observed via epifluorescence on an Axioscope Imager microscope (Zeiss). Nuclear size and epifluorescence signal were analyzed using the software ImageJ 1.41. The integrated density was calculated by multiplication of nuclear size and fluorescence intensity, normalized against the integrated density of wild-type Col-0 trichome nuclei, of which the size was arbitrarily set to 32C.
Confocal and Scanning Electron Microscopy
Root meristems were analyzed with Axiovert 100M confocal laser scanning microscopy (Zeiss). Plant material was incubated for 3 min in a 10-μM propidium iodide solution to stain the cell walls and observed after excitation using a 543-nm laser and detected using the 650-nm long-pass emission filter. Images of leaf trichomes were acquired with a TM-1000 Tabletop electron microscope (Hitachi).
RT-qPCR Analysis
RNA was extracted from the respective tissues with the RNeasy Kit (Qiagen). After treatment with the RQ1 RNase-Free DNase (Promega), cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad). Relative expression levels were determined with the LightCycler 480 Real-Time SYBR green PCR System (Roche). The ACT and CAK2 reference genes were used for normalization. Primer sequences can be found in Supplemental Table S5.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CCS52A1 (At4G22910); CCS52A2 (At4G11920); DEL1 (At3G48160); UVI4 (At2G42260)
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Quantification of wild-type (Col-0), ccs52a1-1, DEL1OE, and ccs52a1-1 DEL1OE trichome branch number and nuclear DNA content.
Supplemental Figure S2. Quantification of wild-type (Col-0) and uvi4, ccs52a2-1, and uvi4 ccs52a2-1 trichome branch number and nuclear DNA content.
Supplemental Figure S3. Quantification of wild-type (Col-0) and uvi4, del1-1, and uvi4 del1-1 mutant trichome branch number and nuclear DNA content.
Supplemental Figure S4. Endoreduplication index of wild-type (Col-0), uvi4, del1-1, and uvi4 del1-1 mature leaves reveals no significant difference in leaf ploidy distributions of the uvi4 del1-1 double mutant compared to the uvi4 and del1-1 single mutants.
Supplemental Figure S5. Putative CDK phosphorylation site in the DEL1 N-terminal sequence.
Supplemental Figure S6. CCS52A1 and CCS52A2 expression levels in del1-1 and DEL1OE mutant leaves and roots.
Supplemental Table S1. DNA ploidy distribution of three-week-old first true leaves of wild-type (Col-0), ccs52a1-1, DEL1OE, and ccs52a1-1 DEL1OE plants.
Supplemental Table S2. DNA ploidy distribution of three-week-old mature first true leaves of the wild-type (Col-0), uvi4, ccs52a2-1, and uvi4 ccs52a2-1 plants.
Supplemental Table S3. DNA ploidy distribution of one-week-old roots of the wild-type (Col-0), uvi4, del1-1, and uvi4 del1-1 plants.
Supplemental Table S4. DNA ploidy distribution of three-week-old first true leaves of wild-type (Col-0), uvi4, del1-1, and uvi4 del1-1 plants.
Supplemental Table S5. List of primers used for RT-qPCR analysis.
Acknowledgments
The authors thank Annick Bleys for help in preparing the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Baker DJ, Dawlaty MM, Galardy P, Van Deursen JM (2007) Mitotic regulation of the anaphase-promoting complex. Cell Mol Life Sci 64: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloban M, Vanstraelen M, Tarayre S, Reuzeau C, Cultrone A, Mergaert P, Kondorosi E (2013) Complementary and dose-dependent action of AtCCS52A isoforms in endoreduplication and plant size control. New Phytol 198: 1049–1059 [DOI] [PubMed] [Google Scholar]
- Beemster GT, De Vusser K, de Tavernier E, De Bock K, Inzé D (2002) Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol 129: 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, Vercruysse S, De Veylder L, Kuiper M, Inzé D (2006) The Arabidopsis leaf as a model system for investigating the role of cell cycle regulation in organ growth. J Plant Res 119: 43–50 [DOI] [PubMed] [Google Scholar]
- Boekhout M, Yuan R, Wondergem AP, Segeren HA, Van Liere EA, Awol N, Jansen I, Wolthuis RMF, De Bruin A, Westendorp B (2016) Feedback regulation between atypical E2Fs and APC/CCdh1 coordinates cell cycle progression. EMBO Rep 17: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Van Den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, Inzé D, De Veylder L, Russinova E (2010) Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22: 1264–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Lammens T, Boruc J, Van Leene J, Van Den Daele H, Maes S, Van Isterdael G, Russinova E, Kondorosi E, Witters E, De Jaeger G, Inzé D, et al. (2009) CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol 150: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon M, Pirrello J, Cheniclet C, Coriton O, Bourge M, Brown S, Moïse A, Peypelut M, Rouyère V, Renaudin J-P, Chevalier C, Frangne N (2012) Evidence for karyoplasmic homeostasis during endoreduplication and a ploidy-dependent increase in gene transcription during tomato fruit growth. Development 139: 3817–3826 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsiepe J, Wester K, Weinl C, Roodbarkelari F, Kasili R, Larkin JC, Hülskamp M, Schnittger A (2010) Endoreplication controls cell fate maintenance. PLoS Genet 6: e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Ishida T, Sugimoto K (2010) Developmental control of endocycles and cell growth in plants. Curr Opin Plant Biol 13: 654–660 [DOI] [PubMed] [Google Scholar]
- Breuer C, Morohashi K, Kawamura A, Takahashi N, Ishida T, Umeda M, Grotewold E, Sugimoto K (2012) Transcriptional repression of the APC/C activator CCS52A1 promotes active termination of cell growth. EMBO J 31: 4488–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla A, Vinardell JM, Kiss E, Oláh B, Roudier F, Kondorosi A, Kondorosi E (1999) The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J 18: 4476–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EJ, Begum R, Chait BT, Gaasterland T (2007) Prediction of cyclin-dependent kinase phosphorylation substrates. PLoS One 2: e656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A (2011) Molecular control and function of endoreplication in development and physiology. Trends Plant Sci 16: 624–634 [DOI] [PubMed] [Google Scholar]
- Edgar BA, Zielke N, Gutierrez C (2014) Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat Rev Mol Cell Biol 15: 197–210 [DOI] [PubMed] [Google Scholar]
- Flemming AJ, Shen Z-Z, Cunha A, Emmons SW, Leroi AM (2000) Somatic polyploidization and cellular proliferation drive body size evolution in nematodes. Proc Natl Acad Sci USA 97: 5285–5290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fülöp K, Tarayre S, Kelemen Z, Horváth G, Kevei Z, Nikovics K, Bakó L, Brown S, Kondorosi A, Kondorosi E (2005) Arabidopsis anaphase-promoting complexes: multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle 4: 1084–1092 [PubMed] [Google Scholar]
- Hamdoun S, Zhang C, Gill M, Kumar N, Churchman M, Larkin JC, Kwon A, Lu H (2016) Differential roles of two homologous cyclin-dependent kinase inhibitor genes in regulating cell cycle and innate immunity in Arabidopsis. Plant Physiol 170: 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase Y, Trung KH, Matsunaga T, Tanaka A (2006) A mutation in the uvi4 gene promotes progression of endo-reduplication and confers increased tolerance towards ultraviolet B light. Plant J 46: 317–326 [DOI] [PubMed] [Google Scholar]
- Heyman J, Cools T, Vandenbussche F, Heyndrickx KS, Van Leene J, Vercauteren I, Vanderauwera S, Vandepoele K, De Jaeger G, Van Der Straeten D, De Veylder L (2013) ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342: 860–863 [DOI] [PubMed] [Google Scholar]
- Heyman J, De Veylder L (2012) The anaphase-promoting complex/cyclosome in control of plant development. Mol Plant 5: 1182–1194 [DOI] [PubMed] [Google Scholar]
- Heyman J, Van Den Daele H, De Wit K, Boudolf V, Berckmans B, Verkest A, Alvim Kamei CL, De Jaeger G, Koncz C, De Veylder L (2011) Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome. Plant Cell 23: 4394–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai KK, Ohashi Y, Tsuge T, Yoshizumi T, Matsui M, Oka A, Aoyama T (2006) The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18: 382–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasili R, Walker JD, Simmons LA, Zhou J, De Veylder L, Larkin JC (2010) SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics 185: 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens T, Boudolf V, Kheibarshekan L, Zalmas LP, Gaamouche T, Maes S, Vanstraelen M, Kondorosi E, La Thangue NB, Govaerts W, Inzé D, De Veylder L (2008) Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc Natl Acad Sci USA 105: 14721–14726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins BA, Dilkes BP, Dante RA, Coelho CM, Woo YM, Liu Y (2001) Investigating the hows and whys of DNA endoreduplication. J Exp Bot 52: 183–192 [PubMed] [Google Scholar]
- Lilly MA, Duronio RJ (2005) New insights into cell cycle control from the Drosophila endocycle. Oncogene 24: 2765–2775 [DOI] [PubMed] [Google Scholar]
- Liu Y, Ye W, Li B, Zhou X, Cui Y, Running MP, Liu K (2012) CCS52A2/FZR1, a cell cycle regulator, is an essential factor for shoot apical meristem maintenance in Arabidopsis thaliana. BMC Plant Biol 12: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, Horváth B, Khan S, Mohammed B, Henriques R, De Veylder L, Bakó L, Scheres B, Bögre L (2012) Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J 31: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu-Rivet E, Gévaudant F, Sicard A, Salar S, Do PT, Mouras A, Fernie AR, Gibon Y, Rothan C, Chevalier C, Hernould M (2010) Functional analysis of the anaphase promoting complex activator CCS52A highlights the crucial role of endo-reduplication for fruit growth in tomato. Plant J 62: 727–741 [DOI] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW (1993) Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne-Reveau K, Senger S, Pal M, Herr A, Richardson HE, Asano M, Deak P, Lilly MA (2008) APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development 135: 1451–1461 [DOI] [PubMed] [Google Scholar]
- Perazza D, Herzog M, Hülskamp M, Brown S, Dorne A-M, Bonneville J-M (1999) Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152: 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J-M. (2002) The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell 9: 931–943 [DOI] [PubMed] [Google Scholar]
- Roodbarkelari F, Bramsiepe J, Weinl C, Marquardt S, Novák B, Jakoby MJ, Lechner E, Genschik P, Schnittger A (2010) Cullin 4-ring finger-ligase plays a key role in the control of endoreplication cycles in Arabidopsis trichomes. Proc Natl Acad Sci USA 107: 15275–15280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Kajihara T, Okamura C, Kim Y, Katagiri Y, Okushima Y, Matsunaga S, Hwang I, Umeda M (2013) Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr Biol 23: 1812–1817 [DOI] [PubMed] [Google Scholar]
- Vanstraelen M, Baloban M, Da Ines O, Cultrone A, Lammens T, Boudolf V, Brown SC, De Veylder L, Mergaert P, Kondorosi E (2009) APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc Natl Acad Sci USA 106: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GTS, Maes S, Magyar Z, Atanassova A, De Almeida Engler J, De Groodt R, Inzé D, De Veylder L (2005) The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol 15: 59–63 [DOI] [PubMed] [Google Scholar]
- Walton A, Stes E, Cybulski N, Van Bel M, Iñigo S, Durand AN, Timmerman E, Heyman J, Pauwels L, De Veylder L, Goossens A, De Smet I, et al. (2016) It’s time for some “site”-seeing: novel tools to monitor the ubiquitin landscape in Arabidopsis thaliana. Plant Cell 28: 6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jin W, Li N, Zhang W, Liu C, Li C, Li Y (2016) UBIQUITIN-SPECIFIC PROTEASE14 interacts with ULTRAVIOLET-B INSENSITIVE4 to regulate endoreduplication and cell and organ growth in Arabidopsis. Plant Cell 28: 1200–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]