Stress-inducible non-coding antisense RNAs generated by RNA-dependent RNA polymerases function in a novel abiotic stress response mechanism different from the known endogenous small RNA pathways.
Abstract
Our previous study identified approximately 6,000 abiotic stress-responsive noncoding transcripts existing on the antisense strand of protein-coding genes and implied that a type of antisense RNA was synthesized from a sense RNA template by RNA-dependent RNA polymerase (RDR). Expression analyses revealed that the expression of novel abiotic stress-induced antisense RNA on 1,136 gene loci was reduced in the rdr1/2/6 mutants. RNase protection indicated that the RD29A antisense RNA and other RDR1/2/6-dependent antisense RNAs are involved in the formation of dsRNA. The accumulation of stress-inducible antisense RNA was decreased and increased in dcp5 and xrn4, respectively, but not changed in dcl2/3/4, nrpd1a and nrpd1b. RNA-seq analyses revealed that the majority of the RDR1/2/6-dependent antisense RNA loci did not overlap with RDR1/2/6-dependent 20–30 nt RNA loci. Additionally, rdr1/2/6 mutants decreased the degradation rate of the sense RNA and exhibited arrested root growth during the recovery stage following a drought stress, whereas dcl2/3/4 mutants did not. Collectively, these results indicate that RDRs have stress-inducible antisense RNA synthesis activity and a novel biological function that is different from the known endogenous small RNA pathways from protein-coding genes. These data reveal a novel mechanism of RNA regulation during abiotic stress response that involves complex RNA degradation pathways.
When growing under natural conditions, plants are exposed to a variety of environmental stresses that negatively affect their growth and productivity (Boyer, 1982; Chevin et al., 2010). Drought stress is one of the major limiting environmental factors affecting more than 10% of arable lands, resulting in a greater than 50% decline in the average yield of major crops worldwide (Bray et al., 2000). Therefore, it is important to understand the molecular mechanisms associated with abiotic stress responses in order to improve the molecular breeding of stress-tolerant plants.
Recent genome-wide transcriptome technologies, such as tiling arrays and next generation sequencing, have revealed a large number of stress-responsive ncRNAs that were not translated into proteins. Emerging evidence indicates that ncRNAs are major products of the plant transcriptome (Matsui et al., 2013; Liu et al., 2012). Previously, several transcriptome analyses identifying antisense RNAs and their associated regulatory mechanisms have been reported (Yamada et al., 2003; Osato et al., 2003; Jin et al., 2008; Matsui et al., 2008; Hazen et al., 2009). Current evidence suggests that natural cis-antisense RNAs have various functional roles in transcriptional and posttranscriptional regulation in order to achieve different biological responses (Terryn and Rouzé, 2000; Prescott and Proudfoot, 2002; Yamada et al., 2003; Faghihi et al., 2010; Zubko et al., 2011). These roles include gene silencing (Aravin et al., 2001; Tufarelli et al., 2003; Katiyar-Agarwal et al., 2006), poly (A) signal modification (Gu et al., 2009), inhibition of miRNA function (Faghihi et al., 2010), and RNA editing (Peters et al., 2003). Using a tiling array approach, our previous study of the Arabidopsis transcriptome revealed a unique transcriptomic phenomenon where more than 6,000 non-protein-coding antisense transcripts were expressed in response to abiotic stress (Matsui et al., 2008). These antisense transcripts were fully overlapping sense-antisense transcripts (fSATs), in which the sequence of one transcript covered more than 80% of the other antisense transcript. Pairs of the sense- and antisense transcripts exhibited a positive correlation in their expression in response to abiotic stress. These results suggested that there is novel regulation of RNA metabolism that functions under abiotic stress conditions.
At the present time, there is still limited understanding pertaining to the biogenesis mechanisms and biological function of antisense RNAs that are induced in response to abiotic stress. This is primarily due to the fact that various types of antisense RNAs are synthesized on protein-coding loci. The most intensively studied antisense RNAs have been mRNA-like transcripts that possess a cap structure, intron gaps, and a poly (A) tail (Osato et al., 2003; Jen et al., 2005; Wang et al., 2005). In many cases, their existence can be estimated from the alignment of full-length cDNAs and ESTs to the genome sequence. In general, it is believed that they are transcribed from their 5′-upstream promoters by RNA polymerase II and that most of them partially overlap with sense-stranded protein-coding genes. However, our subcloning analysis revealed that stress-induced antisense RNAs did not have cap-structures or a poly (A) tail and have a sequence complementary to the mRNA mapped onto the opposite strand (Matsui et al., 2008). The stress-inducible antisense RNAs are coexpressed with the sense mRNA under abiotic stresses (Matsui et al., 2008). These findings support the hypothesis that RNA-dependent RNA polymerases (RDRs) are involved in the biosynthesis of antisense RNAs, although another possibility for stress-induced antisense RNA synthesis may also exist. Recent findings have indicated that uncapped sense-stranded RNAs are linked to post-transcriptional gene silencing (PTGS)-related small RNA synthesis, synthesizing dsRNA during the process of small RNA biosynthesis. Additionally, recent studies have also reported that disruption of several genes, including the decapping enzyme complex component (DCP1, DCP2, VCS), a 5′-3′ exoribonuclease (XRN4(EIN5)), and a DExH-box RNA helicase functioning in 3′-5′ RNA degradation (SKI2), induced the expression of endogenous siRNAs (Gregory et al., 2008; Martínez de Alba et al., 2015; Zhang et al., 2015). The increased levels of uncapped sense-stranded RNAs in these mutants were used as templates to produce dsRNAs during the PTGS process. The dsRNAs were synthesized by RNA-dependent RNA polymerase 6 (RDR6), one of six Arabidopsis RDRs (Rajeswaran et al., 2012; Martínez de Alba et al., 2015; Zhang et al., 2015), and processed into 21-22 nt siRNA by DCL4 (Zhang et al., 2015). Another possible mechanism of antisense RNA synthesis is transcriptional gene silencing (TGS) of related RNAs that are synthesized by a DNA-directed RNA polymerase IV (PolIV) and RDR2 (Xie et al., 2004; Swiezewski et al., 2009). PolIV and RDR2 generate dsRNAs from heterochromatic regions (Xie et al., 2004). The generation of siRNAs from overlapping regions of protein-coding gene pairs has been previously reported (Borsani et al., 2005; Nobuta et al., 2007; Zhou et al., 2009; Ron et al., 2010).
In order to elucidate the mechanism responsible for the biosynthesis of abiotic stress-induced antisense RNA, we tried to identify the antisense RNA biosynthesis-defective mutants based upon the information from a previous study where the sequences of antisense RNAs complementary to their sense RNAs on protein-coding gene loci were reported (Matsui et al., 2008). The current study indicated that RDRs function redundantly during antisense RNA synthesis. Surprisingly, most of the RDR1/2/6-dependent antisense RNA loci did not overlap with regions of 20–30 nt RDR1/2/6-dependent small RNAs, suggesting that RDR synthesized dsRNA in endogenous loci where small RNAs were limited at very low level in wild-type plants. The rdr1/2/6 mutants also exhibited arrested root growth during the recovery stage following a drought stress, while dcl2/3/4 mutants did not show this phenotype. These results indicate that RDRs have a novel biological function that is different from the known small RNA pathway involved in abiotic stress response. Our study reveals the importance of a novel RNA metabolism and that RDR1/2/6-mediated biosynthesis of antisense RNAs is involved in maintaining the integrity of the transcriptome network in abiotic stress response.
RESULTS
Accumulation of a Fully Overlapping Antisense Transcript, fAsRD29A1, from the RD29A Locus Is Reduced in the rdr1/2/6 Mutant
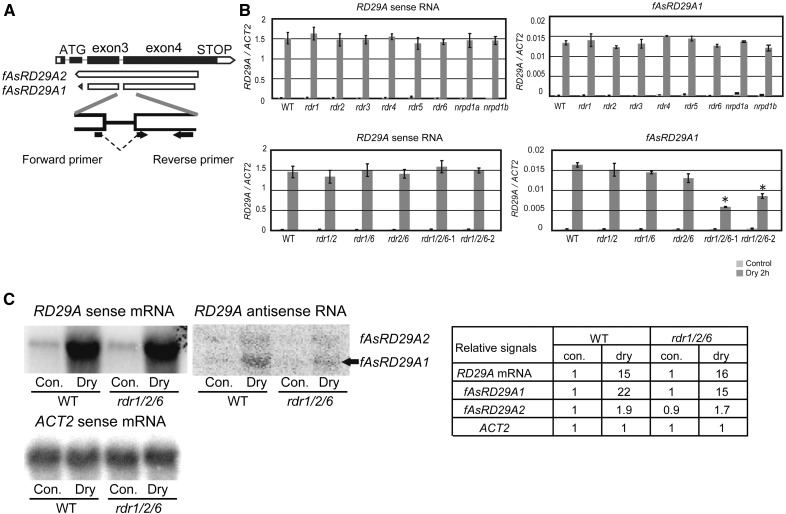
We previously found that two types of antisense transcripts are generated at the RD29A locus in Arabidopsis plants subjected to abiotic stress (Matsui et al., 2008). The sequence of a fully overlapping antisense RNA (fAsRD29A1, named antisense TU2 in Matsui et al. [2008]) on the RD29A locus is complementary to the sense mRNA sequence containing an intron gap, and the fAsRD29A2 (named antisense TU1 in Matsui et al. [2008]) sequence is complementary to the genomic sequence (Fig. 1A). In order to investigate the biosynthesis of the fully overlapping antisense RNA, the accumulation of fAsRD29A1 was measured in single gene knockout lines of RDRs subjected to drought stress. To measure the accumulation of fAsRD29A1, qRT-PCR primers were designed with forward primers containing part of exon 3 and exon 4 (Fig. 1A). The RNA samples were used for the reverse transcription with the forward and reverse primers to generate the strand-specific cDNA at 55°C for preventing miss-annealing (Yassour et al., 2010; Feng et al., 2012), and the accumulation of sense and antisense RNAs was measured by qRT-PCR. Accumulation of fAsRD29A1 increased in wild-type plants in response to a drought stress treatment in the same manner as the poly (A+) sense RNA. However, the accumulation of fAsRD29A1 was approximately 1/100 to the accumulation of the poly (A+) sense RNA. qRT-PCR results also indicated that the accumulation of fAsRD29A1 was not affected in the single rdr, nrpd1a, and nrpd1b mutants (Fig. 1B). This result was consistent with our previous finding that the accumulation of CYP707A1 antisense RNA was unaffected in six single rdr mutant lines (Matsui et al., 2008). The accumulation of fAsRD29A1 in the double and two independent triple knockout lines, rdr1/2, rdr1/6, rdr2/6, rdr1/2/6-1, and rdr1/2/6-2 was subsequently measured. The results indicated that disruption of RDR1/2/6 reduced the expression of fAsRD29A1 transcripts to half (the P values for t test < 0.01) (Fig. 1B). Accumulation of the stress-inducible antisense RNA of RD20 (AT2G33380) and PP2CA (AT3G11410), where the antisense RNAs were identified in our previous study (Matsui et al., 2008), were reduced in rdr1/2/6-1 mutant (Supplemental Fig. S1A). The linker RT-PCR method (Lepere et al., 2008) also showed that the accumulation of the antisense poly (A−) RNAs was reduced in rdr1/2/6-1 and in rdr1/2/6-2 (Supplemental Fig. S2). Northern analysis using the strand-specific probe confirmed that the signal of fAsRD29A1 fragment was lower in rdr1/2/6-1 as compared to wild type (Fig. 1C). The expression analyses suggest that the loss of RDR1/2/6 reduces the expression of stress-inducible antisense RNAs. Loss of NRPD1a and NRPD1b, which resulted in impaired PolIV and PolV, respectively, did not affect accumulation of fAsRD29A1 (Supplemental Fig. S1A), suggesting that abiotic stress-induced fAsRD29A1 is not generated by TGS-related transcription. In contrast, the accumulation of fAsRD29A2 was not affected in the rdr1/2/6 mutant (Supplemental Fig. S3).
Figure 1.
Analysis of the accumulation of sense and antisense RNA at the RD29A locus in rdr and nrpd1 mutants. A, Schematic diagram of two types of antisense RNAs, fAsRD29A1 and fAsRD29A2, that exist at the RD29A locus. In order to detect the accumulation of fAsRD29A1 complementary to the sense RNA, an RD29A forward primer containing part of exon 3 and 4, but lacking the intron, was used for qRT-PCR. B, The accumulation level of RD29A sense RNA and antisense RNA (fAsRD29A1) in wild type, single mutants of six RDRs, NRPD1a, and NRPD1b, and double and triple mutants of RDR1/2/6 was measured by qRT-PCR. One μg of total RNA was used. Data represent the mean ± SD (n = 3). Asterisk indicates the P values for t test < 0.01, when mutants and wild type were compared under the same condition. C, northern analysis of RD29A sense RNA and antisense RNAs in wild-type and rdr1/2/6-1 plants. Ten μg of total RNA was loaded into each lane and hybridized with RI-labeled strand-specific RNA probes of RD29A and the ACT2 control (that shows the similar expression level between nontreatment and 2 h drought treatment). The exposure times for detecting the sense- and antisense- RNAs were 1 h and 24 h, respectively. The fAsRD29A1 fragment is indicated by an arrow. The table shows relative RI-intensities of each band at the northern blots using Image-J program.
Mutation of XRN4 and DCP5 Affects Accumulation of fAsRD29A1, Indicating that Antisense RNA Is Generated from Uncapped Sense RNA
Mutation of xrn4 increases the accumulation of uncapped and unpolyadenylated RNAs that serve as substrates for RDRs, leading to the formation of dsRNA precursors. Indeed, it has been reported that the xrn4 mutation induced a class of small RNAs that are processed from the antisense strand of endogenous genes (Gregory et al., 2008). The mutant lines xrn4 and dcp5 were selected in order to test whether degraded RNAs affect the biosynthesis of fully overlapping antisense RNAs. DCP5 is involved in the de-capping of mRNAs by interacting with DCP1 or DCP2 (Xu and Chua, 2012). We examined the circular RT-PCR, which connects the 5′-end of uncapped RNAs to its 3′ end by self-ligation and amplifies the self-ligated molecules and synthesizes cDNA using the random RT primer to analyze the uncapped RD29A sense RNA. The circular RT-PCR revealed that the accumulation of RD29A uncapped sense RNA increased in xrn4 and decreased in dcp5 (Supplemental Fig. S1B). qRT-PCR analysis using a random primer or oligo dT primer showed that poly (A−) RD29A sense RNA accumulated in xrn4 and poly (A+) RD29A sense RNA accumulated in dcp5 with and without drought stress treatment (Supplemental Fig. S1C). Expression of fAsRD29A1 was increased in xrn4 but was reduced in dcp5 (Supplemental Fig. S1C). Especially, these results indicate the accumulation of uncapped RD29A sense RNA in xrn4 and the reduction of fAsRD29A1 in dcp5, suggesting that fAsRD29A1 is synthesized by using the degraded fragment of RD29A sense mRNA as a template. The same expression profile in rdr1/2/6-1, dcp5, and xrn4 was observed in RD20 and PP2CA antisense RNAs under drought stress (Supplemental Fig. S1A). RD29A poly (A−) sense RNA might be specially targeted by XRN4-dependent RNA degradation under normal condition and drought stress (Supplemental Fig. S1C). RDRs have dsRNA synthesis activity under both control and drought conditions, but the generation of the stress-inducible antisense RNAs is induced with the amount of its substrate by changes in RNA degradation metabolism under drought stress. Collectively, the results indicate that antisense RNAs of RD29A and several other loci whose expression is drought-inducible are synthesized using endogenous uncapped sense RNA as a template.
Custom Microarray Analysis Identified 1,136 RDR1/2/6-Dependent Antisense RNA Loci and Suggested that RDR1/2/6 Activities Synthesizing Antisense RNAs Increase under Drought Stress
Sequence analysis of subcloned cDNAs showed that fAsRD29A1 did not have a cap structure and a long poly (A) tail (Matsui et al., 2008). Although recent developments of sequencing technology have made more sequencing reads possible, it was difficult to compare the expression of antisense RNA in wild type with those in rdr1/2/6-1 using SOLiD 3 sequencing technology (GSE39033). This was primarily due to very low expression of stress-induced antisense RNAs, and no methods were available to separate the stress-inducible antisense RNAs from other poly (A−) RNAs such as tRNAs, snoRNAs, 5S rRNAs, and uncapped sense RNAs. Therefore, a custom microarray analysis was selected so that it could be used to detect the probe signal of the target antisense RNAs by overloading a high amount of labeled cRNAs onto the array. The custom array was designed for detecting novel transcriptional regions, including 7,918 antisense transcripts on 7,138 TAIR10-annotated gene loci (Supplemental Fig. S4A) based on previous transcriptome data obtained using a tiling array and RNA sequencing (Matsui et al., 2008; Okamoto et al., 2010; Kawaguchi et al., 2012). Poly (A+) and poly (A−) RNA fractions were separated from total RNA extracted from rdr1/2/6-1 and wild-type plants with and without a drought stress treatment. The poly (A+) and poly (A−) RNA fractions were then used for custom microarray analysis (Supplemental Fig. S4B). Signals of tRNAs, U2 RNAs, and U6 RNAs that do not have poly (A) tails had a higher intensity in the poly (A−) RNA microarray data as compared to the poly (A+) RNA microarray data (Supplemental Fig. S4C). The custom microarray analysis successfully detected poly (A−) and poly (A+) RNA (Supplemental Fig. S4C). We focused on 7,138 loci of the sense and antisense transcripts (SAT loci) that are detectable in wild-type or rdr1/2/6-1 plants on the custom array (Supplemental Table S1).
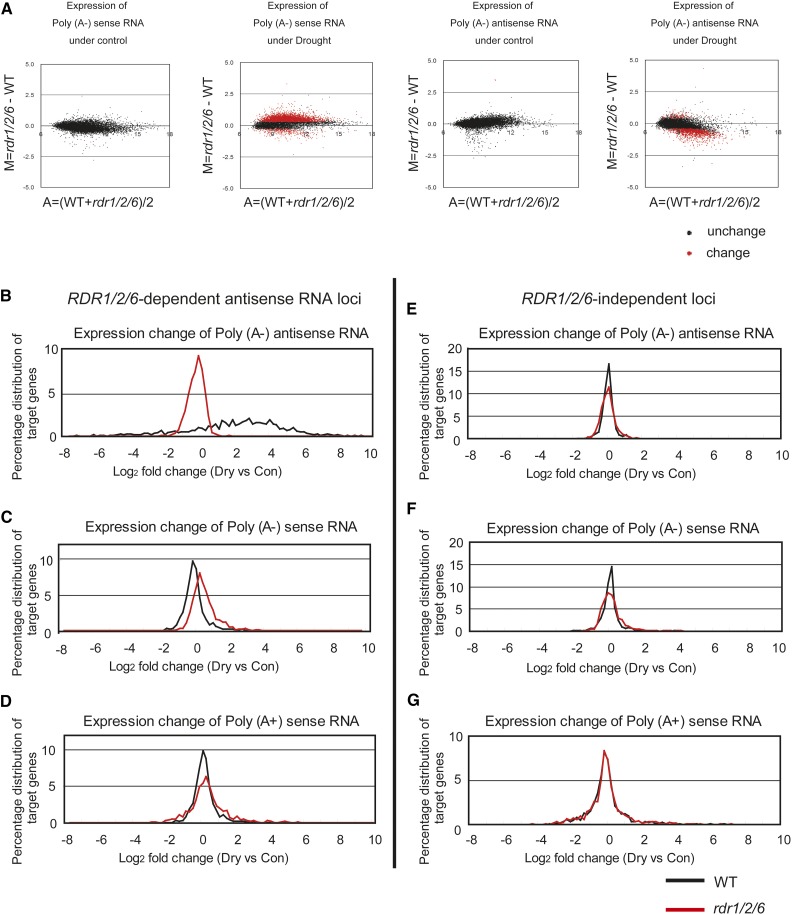
Differential expression of poly (A−) RNAs in wild-type and rdr1/2/6-1 plants was evaluated in response to drought stress (Fig. 2A, Supplemental Fig. S4D). Loss of RDR1/2/6 not only decreased poly (A−) antisense RNAs, but also increased poly (A−) sense RNAs (Fig. 2A, Supplemental Fig. S5, B and D). Poly (A−) RNAs on 7,138 SAT loci were compared between control and drought stress conditions. When rdr1/2/6-1 plants were subjected to drought stress, poly (A−) antisense RNA was reduced (Supplemental Figure S5D), whereas poly (A−) sense transcripts accumulated (Supplemental Figure S5B). On the other hand, the accumulation of poly (A−) antisense and sense RNAs was stable in wild-type plants (Supplemental Fig. S5, B and D). The trend seen in the poly (A−) RNA microarray data were not observed in the poly (A+) RNA data (Supplemental Fig. S5, A and C). When subjected to drought stress, the overall trend in the ratio of poly (A−) antisense RNA/poly (A−) sense RNA per each gene was more greatly reduced in rdr1/2/6 plants (Supplemental Fig. S5, E and F).
Figure 2.
Custom microarray analysis of the accumulation of antisense RNAs in wild-type and rdr1/2/6 plants subjected to drought stress. A, MA plots illustrating the accumulation level of poly (A-) sense RNA and poly (A-) antisense RNA at 7,138 gene loci in which sense and antisense RNAs are detectable in wild-type and rdr1/2/6-1 plants. Red and black dots indicate differentially expressed genes and genes with similar expression levels, respectively, in WT and rdr1/2/6-1 plants. Differentially expressed genes were selected with an FDR < 0.1 using linear models and empirical Bayes methods (Ritchie et al., 2015) and Benjamini-Hochberg p-adjustment. B, Histograms illustrating the distribution of log2 fold changes (Dry/Control) in RDR1/2/6-dependent antisense RNA loci- and RDR1/2/6-independent-loci. Black and red lines indicate wild-type and rdr1/2/6-1 samples, respectively.
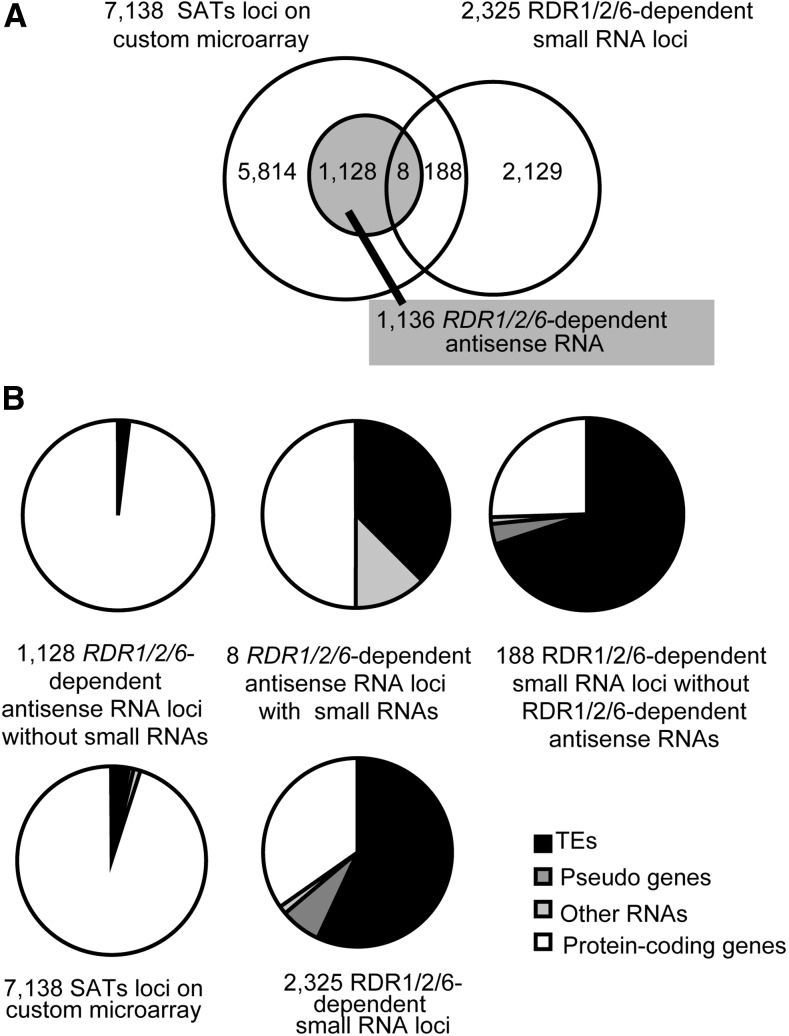
RDR1/2/6-dependent antisense RNA loci were selected by the criteria that the ratio of antisense RNA to sense RNA is lower in rdr1/2/6-1 as compared with the wild-type plants under control or drought stress conditions (FDR < 0.075). The 1,136 RDR1/2/6-dependent antisense RNA loci were identified only under drought stress (Fig. 3A; Supplemental Table S1). Twenty-six percent of the RDR1/2/6-dependent antisense RNA loci were drought-inducible genes, including RD29A (Supplemental Table S1). Our previous study, which compared the gene expression between control and drought stress, revealed the presence of the antisense RNAs on the stress-responsive genes (Matsui et al., 2008). In the current study, comparisons of rdr1/2/6-1 and wild type revealed a novel finding that RDR1/2/6-dependent antisense RNA was synthesized from the genes with various expression patterns. Microarray analyses revealed that the efficiency of RDR1/2/6-dependent antisense RNA synthesis was gradually regulated in a gene dependent manner.
Figure 3.
Comparative analysis of RDR1/2/6-dependent antisense RNA loci and RDR1/2/6-dependent small RNA loci. A, Venn diagram comparing 1,136 RDR1/2/6-dependent antisense RNA loci within 7,138 sense-antisense transcripts (SATs) loci with custom array probes and 2,325 RDR1/2/6-dependent small RNA loci. B, Classification of five groups based on functional categories in the TAIR10 dataset.
In order to address their expression trend of sense and antisense RNAs in RDR1/2/6-dependent antisense RNA loci, we therefore needed to compare the RDR1/2/6-dependent antisense RNA loci and RDR1/2/6-independent RNA loci using a relatively weak threshold. When the threshold of FDR > 0.25 was used in this study based upon the data that the change in log2 ratio of poly (A−) antisense RNA/poly (A−) sense RNA between rdr1/2/6-1 and wild type was less than 1/2, one thousand and fifteen RDR1/2/6-independent loci were also selected (Supplemental Table S1). Changes in the accumulation of poly (A−) sense and poly (A−) antisense RNA in the presence and absence of drought stress were examined in these gene sets (Fig. 2, B–G). A distinct trend was observed with an increased signal intensity of poly (A−) antisense RNAs in wild-type plants in response to the drought stress treatment (Fig. 2B). In contrast, the signal intensity of poly (A−) sense RNAs did not exhibit a large change in wild type in response to drought stress at RDR1/2/6-dependent antisense RNA loci (Fig. 2C). The expression trends of poly (A−) antisense and sense RNAs for the response to drought stress in RDR1/2/6-dependent antisense RNA loci were not observed in the RDR1/2/6-independent RNA loci (Fig. 2, E and F).
qRT-PCR analysis confirmed the decrease in the expression of poly (A−) antisense RNAs in rdr1/2/6-1 after drought stress in several RDR1/2/6-dependent antisense RNA loci (Supplemental Fig. S6). Collectively, these data indicate that RDR1/2/6-dependent antisense RNAs exist on the annotated protein-coding gene loci throughout the genome and that their accumulation increases in response to drought stress. In addition, qRT-PCR analyses also confirmed the increase of poly (A−) sense RNA in some RDR1/2/6-dependent antisense RNA loci in rdr1/2/6-1 plants (Supplemental Fig. S6). The microarray analysis showed that the loss of RDR1/2/6 distinctly affected antisense RNA accumulation and slightly affected poly (A−) sense RNA under drought stress.
RD29A and fAsRD29A1 Form dsRNA by the Action of RDRs
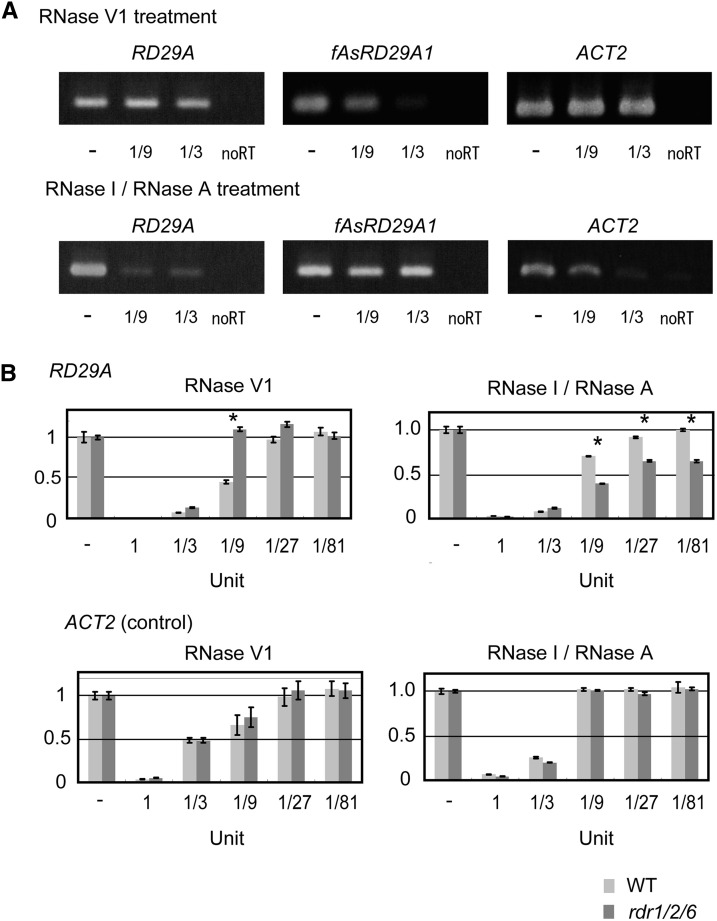
The custom microarray analysis indicated the possibility that RDR1/2/6 affects the accumulation of poly (A−) sense RNA in RDR1/2/6-dependent antisense RNA loci. It is known that RDRs generate dsRNAs by transcribing antisense RNA from its sense RNA (Yoshikawa et al., 2005; Curaba and Chen, 2008; Willmann et al., 2011). In order to determine whether dsRNAs are formed from sense and antisense RNAs, a RNase protection assay was utilized in RD29A, a RDR1/2/6-dependent antisense RNA locus, and in ACT2, which does not have antisense RNAs, thus serving as a negative control (Fig. 4A). A fAsRD29A1-derived fragment could be amplified after using single-stranded RNA-specific RNases (RNase I/RNase A). In contrast, after using a dsRNA-specific RNase (RNase V1), the fAsRD29A1-derived fragment could not be amplified. The amount of the RD29A sense RNA-derived fragment decreased slightly after RNase V1 treatment and decreased significantly after RNase I/RNase A Mix treatment. The RNA structure in wild type and rdr1/2/6-1 was compared. qRT-PCR of samples after being treated with the RNase I/RNase A Mix indicated that dsRNA structure decreased in rdr1/2/6-1 (Fig. 4B). These results imply that a part of RD29A sense RNA forms dsRNA with fAsRD29A1 by the action of RDRs and that most of the RD29A sense RNA exists in the form of single-stranded RNA. Similar results were obtained by a RNA protection assay in other RDR1/2/6-dependent antisense RNA loci (Supplemental Fig. S7).
Figure 4.
RNase protection assay of sense and antisense RNA at the RD29A locus. A, One μg of total RNA extracted from 2 h drought-stress-treated wild-type plants was treated with 1/900 U of RNase V1 or 1/9 U of RNase I/RNase A Mix for 15 min at room temperature. RNase-treated RNAs were converted to cDNAs using RD29A strand-specific primers. Then, 1/10 of the cDNAs were amplified using 35 cycles of PCR. ACT2 sense RNA, which does not have antisense RNAs, was used as a negative control. For ACT2 sense RNA, 1/300 U of RNase V1 and 1/3 U of RNase I/RNase A Mix were used. “-” indicates no treatment of RNases. “no RT” indicates no reverse transcription of RNA to cDNA. B, Accumulation of RD29A sense RNA in wild-type and rdr1/2/6-1 plants was measured by qRT-PCR analysis after RNase V1 and RNase I/RNase A Mix treatment. Then, 1/10 of the cDNAs, which were converted by a random primer, were used for qRT-PCR. Asterisk indicates the P values for t test < 0.01.
Most of the RDR1/2/6-Dependent Antisense RNA Loci Do Not Overlap with RDR1/2/6-Dependent small RNA Loci
Recent studies have shown that various RDR-dependent noncoding RNAs are produced on protein-coding loci and TEs (Gregory et al., 2008; Cao et al., 2014, Martínez de Alba et al., 2015; Zhang et al., 2015) and that small RNAs are generated for mRNA degradation on protein-coding gene loci under specific conditions, such as virus-infection or in RNA degradation-related mutants. These pathways are similar to that of the accumulation of RDR1/2/6-dependent antisense RNA under abiotic stress. Therefore, we examined whether RDR1/2/6-dependent antisense RNAs were processed into small RNAs by comparing the small RNAs between rdr1/2/6-1 and wild type by the use of HiSeq2000 deep sequencing. These efforts identified 2,325 RDR1/2/6-dependent small RNA loci in drought stress or control conditions (Fig. 3A; Supplemental Table S2). Only eight small RNA loci overlapped with RDR1/2/6-dependent antisense RNA loci (Fig. 3, A and B). Specifically, they were a transacting siRNA locus 1a (TAS1a), three transposable elements, and four protein-coding genes in which DNA methylation was observed (Zhang et al., 2006). Two RDR1/2/6-dependent antisense RNAs that were mapped on TAS1B existed upstream of a major small RNA-generated region (Supplemental Fig. S8A, Rajeswaran et al., 2012). In addition, we tested whether accumulation of sense and antisense RNAs was reduced through the small RNA production process on RDR-dependent antisense RNA loci (Supplemental Fig. S9). The mutation of DCL2/3/4 increased accumulation of sense and antisense RNA in TAS2 locus where transacting siRNA is produced by RDR6 and DCL4 (Supplemental Fig. S9). On the other hand, the dcl2/3/4 mutation did not change the accumulation sense and antisense RNAs on stress-inducible RDR-dependent antisense RNA loci (Supplemental Fig. S9).
Recently, it was reported that DCL1/2/3/4-independent and Pol IV-dependent small RNAs can also function in RdDM pathway (Yang et al., 2016). The expression of antisense RNAs was not changed in nrpd1a and nrpd1b (Fig. 1A; Supplemental Fig. S1A). The possibility of generating a small amount of small RNAs was investigated. A histogram of small RNAs revealed that the average number of small RNA on RDR1/2/6-dependent antisense RNA loci was less than 2−4-fold compared with the average number on RDR1/2/6-dependent small RNA loci (Supplemental Fig. S8B). These data indicate that small RNAs at RDR1/2/6-dependent antisense RNA loci were at a background level (Supplemental Fig. S8B; Supplemental Table S1), and that they represent 20–30 nt small RNAs corresponding to RNA degradation fragments generated from sense RNA. The result of the custom microarray showed that the signal intensity of poly (A−) sense and poly (A−) antisense RNA at RDR1/2/6-dependent small RNA loci was lower than the signal at RDR1/2/6-dependent antisense RNA loci (Supplemental Fig. S8C). These observations suggest that most of the dsRNAs did not accumulate as intermediates of small RNA synthesis, because they were quickly processed into small RNAs (Rajeswaran et al., 2012). The RDR1/2/6-dependent antisense RNA loci were also compared with the specific small RNA loci reported previously (Martínez de Alba et al., 2015; Zhang et al., 2015). Only 24% of RDR1/2/6-dependent antisense RNA loci overlapped with the specific small RNA loci (Supplemental Fig. S10A). MA plots showed that the overlapped 277 loci did not change siRNA accumulation in rdr1/2/6-1 compared with wild type (Supplemental Fig. S10B). The results suggested that these loci produced siRNAs only under some specific conditions (Martínez de Alba et al., 2015; Zhang et al., 2015) and did not accumulate siRNAs under our control and drought stress conditions. These results suggest that RDRs synthesize double-strand RNAs on the loci that do not generate small RNAs in addition to the small RNA-generating loci in wild type under drought stress.
In order to study the linkage between RDR1/2/6-dependent antisense RNA synthesis and siRNA production, the accumulation of antisense RNA was measured in rdr6 at a transacting siRNA locus, TAS2. The rdr6 mutant showed a 2-fold increase in the accumulation of antisense RNAs on the 3′ fragment of TAS2 after the miR173 cleavage site where small RNAs are synthesized (Supplemental Fig. S11A), although RDR6 is necessary for generating antisense RNA during an intermediate step in transacting siRNA synthesis (Allen et al., 2005; Yoshikawa et al., 2005; Curaba and Chen, 2008). The strand-specific circular RT-PCR for cloning for TAS antisense RNAs, using the strand-specific primers during RT, identified various lengths of TAS2 antisense RNAs in rdr6 (Supplemental Fig. S11B). Relative to the levels in rdr6, TAS2 antisense RNAs were lower in rdr1/6, rdr2/6, and rdr1/2/6. TAS2 transcripts were a common target substrate of dsRNA synthesis. When RDR6 was missing, however, the increase of antisense RNAs could be induced by other redundant RDRs, and they did not serve as substrates for processing into siRNAs (Allen et al., 2005; Yoshikawa et al., 2005; Curaba and Chen, 2008). These results indicate that siRNA synthesis and antisense RNA synthesis are not always cooperated and that the recognition of target RNAs by each synthesis-related component occurs prior to the synthesis of dsRNA.
RNA Decay Rate of RD29A Poly (A+) Sense RNA Is Reduced in rdr1/2/6, dcp5, and Is Not Affected in dcl2/3/4
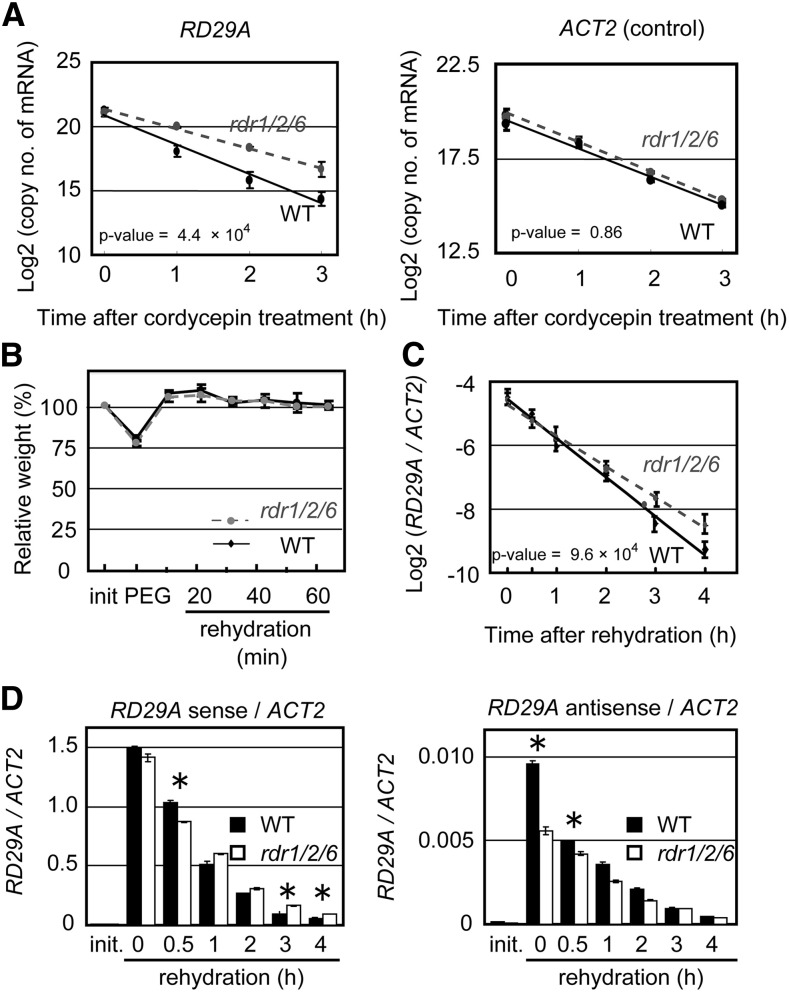
Results of the current study indicate that antisense RNAs are involved in poly (A+) sense RNA degradation. Therefore, the RNA decay rate of RD29A poly (A+) sense RNA in rdr1/2/6-1 and wild-type plants were compared as described previously (Lidder et al., 2005). Plants were pretreated with 30% polyethyleneglycol (PEG) solution for 2 h and then transferred to a PEG solution containing a transcriptional inhibitor, cordycepin. The reduction in the RNA decay rate of RD29A mRNA in rdr1/2/6-1, relative to the wild type, was obvious (Fig. 5A). The decay rate of RD29A poly (A+) sense RNA was 1.4-fold greater in wild type than in rdr1/2/6-1. In contrast, the decay rate of ACT2 poly (A+) sense RNA was similar between rdr1/2/6-1 and wild type (Fig. 5A). It was also confirmed that the RNA decay rate of other RDR1/2/6-dependent antisense RNA loci decreased in rdr1/2/6 subjected to osmotic stress (Supplemental Fig. S12). In addition, we addressed the RNA decay rate of RD29A and other RDR1/2/6-dependent antisense RNA loci in dcp5 and dcl2/3/4 (a mutant that cannot synthesize canonical siRNA after dsRNA synthesis) plants that affected and did not affect RDR1/2/6-dependent antisense RNA synthesis (Supplemental Fig. S13). The RNA decay rates of targets were reduced in dcp5 but were not affected in dcl2/3/4 (Supplemental Fig. S13). It was apparent that DCP5 was also required for RNA degradation that is the initial step of RDR1/2/6-dependent antisense RNA- mediating RNA degradation on RDR1/2/6-dependent antisense RNA loci. In addition, RDR1/2/6-dependent antisense RNA may also assist with poly (A+) RNA degradation without small RNA-dependent RNA degradation in these loci. On the other hand, this result caused a discrepancy because the similar levels of poly (A+) RNA expression for a part of the RDR1/2/6-dependent antisense RNA loci between rdr1/2/6-1 and wild type could not be explained by the RNA decay rate between rdr1/2/6-1 and wild type (Supplemental Figs. S6 and S12). The short-term drought stress drastically changes the transcriptional activity, and the transcriptional regulation has a strong effect on gene expression as compared to degradation. Therefore, it might not yet have reached at the state of static equilibrium between transcription and degradation. It might be also due to that mRNA synthesis is robustly controlled by feedback signal from RNA degradation (Haimovich et al., 2013; Skalska et al., 2017).
Figure 5.
RNA Decay Rate of RD29A Poly (A+) Sense RNA in wild-type and rdr1/2/6 plants subjected to an osmotic stress. A, Two-week-old wild-type and rdr1/2/6-1 plants were pretreated with a 30% PEG solution for 2 h and then treated with 0.6 mm cordycepin, a transcription inhibitor. The RNA decay rate of RD29A (a RDR1/2/6-dependent antisense RNA locus) and ACT2 (a locus without antisense RNA) mRNAs was measured by qRT-PCR. The vertical axis indicates the log2 copy numbers of mRNA per 100 ng of total RNA. The horizontal axis indicates the time-course following cordycepin treatment. Covariance analysis (ANCOVA) was used to determine whether there are different RNA decay rates between wild-type and rdr1/2/6-1 plants. B, rdr1/2/6-1 and wild-type plants were treated with 30% PEG6000 for 2 h and returned to a well-watered condition (rehydration). Relative fresh weight of rdr1/2/6-1 and wild-type plants were then measured. Covariance analysis (ANCOVA) was used to determine whether there are different on the RNA decay rates between wild type and rdr1/2/6-1 plants. C, Accumulation of RD29A mRNA during the rehydration phase was measured by qRT-PCR and normalized to ACT2 accumulation. D, Accumulation of RD29A mRNA and antisense RNA (fAsRD29A1) during the rehydration phase was measured by qRT-PCR and normalized to ACT2 accumulation. Asterisk indicates the P values for t test < 0.05.
rdr1/2/6 Exhibits Reduced Recovery of Root Growth after a Temporal Osmotic Stress as Compared to WT and dcl2/3/4
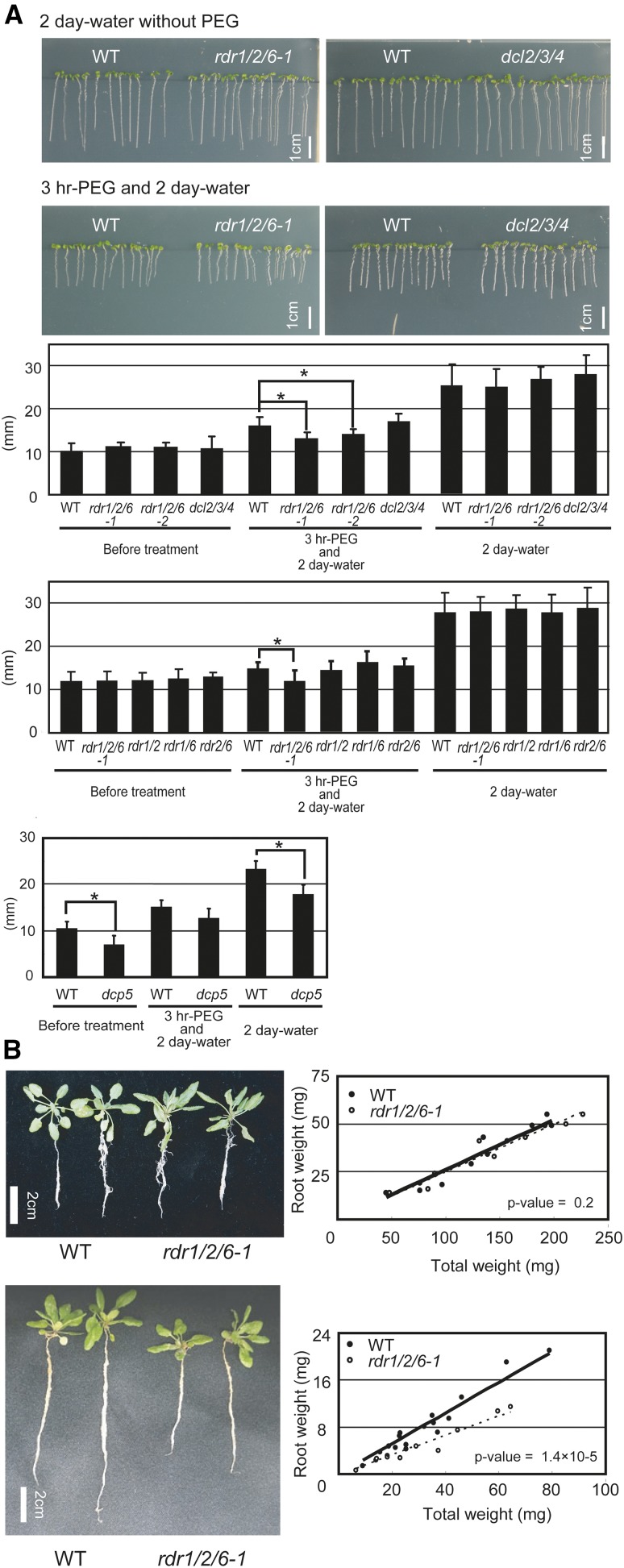
The expression of sense RNA was observed to be regulated by antisense RNAs after a reduction of the transcriptional activity (Fig. 5A). Specifically, qRT-PCR detected a partial decrease in the degradation of the sense mRNAs on RD29A and RDR1/2/6-dependent antisense RNA loci in rdr1/2/6-1 during the recovery stage when water was applied after a temporal osmotic stress (Figs. 5B, 5C, and 5D; Supplemental Fig. S14). We hypothesized that RDR1/2/6-dependent antisense RNA loci functioned in the RNA degradation of abiotic-response-related genes after transcription levels were reduced when plants were re-watered. In order to investigate biological function of RDR1/2/6-dependent antisense RNA, the phenotype of rdr1/2/6-1, rdr1/2/6-2, dcl2/3/4, and wild-type plants were characterized and compared during the recovery phase following an osmotic stress treatment (Fig. 6A). All of the genotypes exhibited similar root lengths on the initial day and after two days of well-watered conditions (control). In contrast, a temporal osmotic stress treatment (three-hour osmotic stress treatment) followed by two days of a well-watered condition inhibited root growth in rdr1/2/6-1, rdr1/2/6-2, dcl2/3/4, and wild-type plants. In particular, root growth in rdr1/2/6 plants was severely inhibited, relative to dcl2/3/4 and wild-type plants. On the other hand, rdr double mutants did not exhibit the severe inhibition of the root growth phenotype as compared to rdr1/2/6 plants after a temporal osmotic stress was administered (Fig. 6A). Additionally, the repetition of a temporal drought stress treatment inhibited root growth more greatly in rdr1/2/6-1 and rdr1/2/6-2 plants as compared to wild-type plants (Fig. 6B). Collectively, these results indicate that the redundant RDRs had a novel biological function in drought stress response.
Figure 6.
Inhibition of root growth in rdr1/2/6 plants in response to a temporal drought stress treatment. A, Six day-old wild-type, rdr1/2/6-1, rdr1/2/6-2, and dcl2/3/4 plants were treated with 30% PEG6000 for three hours and were then grown under well-watered conditions for two days. The length of ten primary roots was measured and averaged and the experiment was repeated three times (n = 3). The bar graph shows the primary root length for each of the plant-types before treatment, in PEG (osmotic stress)-treated plants, and after two days of rehydration (recovery). Scale bars = 1 cm. Asterisks indicate significantly different values (P value < 0.05). B, Seven-day-old wild-type and rdr1/2/6-1 plants were subjected to three cycles of three-day drought stress and recovery treatments (by adding 20 mL of water) (total 9 d treatment). Scale bars = 2 cm. Scatter plots illustrate root fresh weight per total fresh weight of each plant. Covariance analysis (ANCOVA) was used to determine whether there are different on the rate of root weight to total weight between wild-type and rdr1/2/6-1 plants.
We also addressed the root growth phenotype of dcp5 that showed a resistance of RNA decay and a reduction of antisense RNA accumulation in some RDR1/2/6-dependent antisense RNA loci. Phenotypic analyses of the dcp5 mutant revealed a short root phenotype as compared to wild type under normal and temporal osmotic stress conditions (Fig. 6A). These data suggest that DCP5-mediated RNA decay and RDR1/2/6-dependent antisense RNA are also required for normal root growth.
RDR1/2/6-Dependent Antisense RNA Synthesis Functions in Enhanced Down-Regulation of its Poly (A+) Sense RNA during the Recovery Stage after a Temporal Osmotic Stress
In order to better understand the regulation of gene expression by RDR1/2/6-dependent antisense RNA, we compared gene expression patterns between wild type and rdr1/2/6-1 under control conditions, 2 h drought stress and 2 h rehydration, in which antisense RNAs were gradually down-regulated (Fig. 5D). The microarray analysis showed that the change in the expression of sense poly (A+) RNA between rdr1/2/6-1 and wild type was slightly affected under drought stress conditions. In addition, more than 1,200 genes showed a change in expression of the sense poly (A+) RNA between rdr1/2/6 and wild-type plants under the recovery stage (Supplemental Fig. S15A). It is important to note that RDR1/2/6-dependent antisense RNA loci, such as RD29A, were among the significantly differentially expressed genes under the recovery stage (Supplemental Table S3). These data indicate that RDR1/2/6-dependent antisense RNAs triggered the gene expression changes under the recovery stage. In order to clarify the relationship between the regulation of gene expression and RDR1/2/6-dependent antisense RNA, we selected the drought-stress-up-regulated and rehydration-down-regulated genes, whose transcriptional activities were estimated to be greatly reduced during recovery stage, from RDR1/2/6-dependent antisense RNA loci (Supplemental Fig. S15B). In comparison to wild-type plants, rdr1/2/6-1 plants showed a reduced down-regulation of the expression ratio (2 h rehydration/2 h dry) during the recovery stage on a portion of RDR1/2/6-dependent antisense RNA loci (Supplemental Fig. S15C). Such a decrease in the down-regulation of gene expression during the recovery stage was not observed in the drought-stress-up-regulated and rehydration-down-regulated genes from RDR1/2/6-independent loci and RDR1/2/6-dependent small RNA loci (Supplemental Fig. S15C). These results indicate that RDR1/2/6-dependent antisense RNA synthesis functions in enhanced degradation of its poly (A+) sense RNA during the recovery stage after a temporal osmotic stress. It is important to note that the expression in RDR1/2/6-dependent small RNA loci was increased under drought stress in rdr1/2/6-1 as compared with wild type (Supplemental Fig. S15C).
DISCUSSION
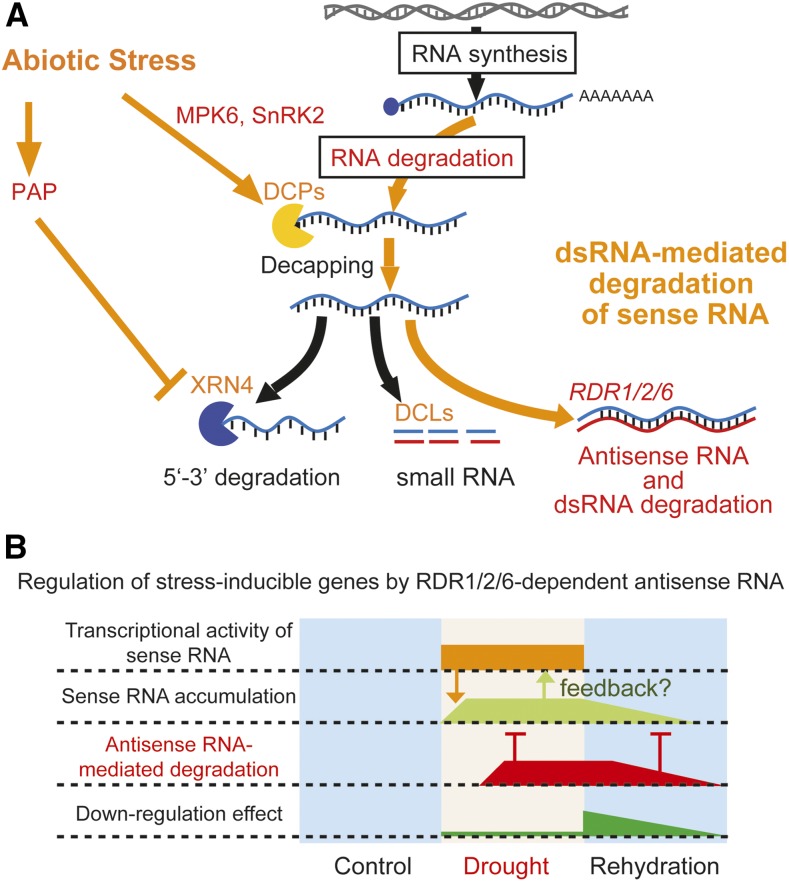
The present studies demonstrate that poly (A−) antisense RNAs are synthesized by RDR1/2/6 on protein-coding regions where 20–30 nt small RNAs have not been generated under drought stress conditions (Figs. 3A and 7A). The poly (A−) antisense RNAs form dsRNAs with the poly (A−) sense RNAs and are involved in the degradation of poly (A+) sense RNAs (Fig. 5, A and C). Our data indicate that antisense RNA-mediated degradation of sense RNA functions in a part of RNA degradation during drought stress and the recovery stage after the drought stress (Fig. 7B).
Figure 7.
Schematic model for antisense RNA-mediated degradation of sense mRNAs under abiotic stress. A, The hypothetical model is constructed from the current study and previous reports. Antisense RNAs are generated from uncapped sense RNAs by RDRs. The antisense RNAs and sense RNAs form double-stranded RNAs that function in the degradation of sense mRNAs. DCPs and XRNs are involved in this mechanism. B, The model of RNA regulation by RDR1/2/6-dependent antisense RNAs. RNA regulation is robustly controlled by transcription during the increasing phase prior to static equivalence and/or a feedback mechanism. RDR1/2/6-dependent antisense RNAs reinforce the down-regulation of drought stress-increased mRNAs after their transcriptions are strongly decreased during the recovery stage.
When compared to control conditions, microarray analyses of rdr1/2/6 plants showed a trend of poly (A−) sense RNA induction and a reduction of poly (A−) antisense RNA under drought stress conditions (Fig. 2, B and C; Supplemental Fig. S5, B and D). Linker RT-PCR also did not detect antisense RNA in poly (A+) RNA fraction (Supplemental Fig. S2). RDR6 has been reported to synthesize antisense RNAs from noncanonical sense RNAs of transgenes with aberrant features, such as the noncapped structure of the 5′ end (Yoshikawa et al., 2005; Willmann et al., 2011). These data suggest that antisense RNA forms dsRNAs with poly A(−) sense RNA, resulting in dsRNA degradation. rdr1/2/6 plants also showed a decreased decay rate of poly (A+) sense RNA in RDR1/2/6-dependent antisense RNA loci (Fig. 5; Supplemental Fig. S12). We speculate that poly (A−) dsRNA degradation is associated with a degradation pathway of poly A (+) sense RNA (Fig. 7A).
However, microarray and qRT-PCR data of poly (A+) sense RNA also suggest that gene expression is robustly controlled under control and drought stress conditions (Fig. 2; Supplemental Figs. S5 and S6; Supplemental Table S1). RNA degradation by RDR1/2/6-dependent antisense RNA was observed under the conditions in which transcription was stopped by cordycepin treatment or during the recovery stage. The reason why sense RNA degradation by RDR1/2/6-dependent antisense RNA was not observed under drought stress might be due to lack of time to reach static equilibrium between transcription and RNA degradation or feedback regulation mechanism of transcription (Fig. 7B). Under drought stress condition, as poly A(+) RNAs of drought stress-inducible genes are generated by the transcription by RNA polymerase II, it is difficult to see the decrease of poly A(+) sense RNA. Recently, it has been also reported that mRNA synthesis was robustly controlled by feedback signal from RNA degradation. For example, “degradation factor” of RNA decay complex shuttles between the cytoplasm and the nucleus and increased mRNA level (Haimovich et al., 2013). The nascent pre-mRNA and nascent ncRNA change chromatin activating transcription as regulatory feedback system (Skalska et al., 2017).
RDR seemed to keep their activity of antisense RNA synthesis under control and drought stress, because the accumulation of RD29A antisense RNA was increased due to increase of poly (A−) sense RNA by loss of XRN4-dependent degradation under normal condition and drought stress (Supplemental Fig. S1, B and C). The accumulation of antisense RNA was affected by the amount of the substrate that is generated by changes of RNA degradation metabolism under drought stress. Recently, it has been reported that mRNA decay pathway plays an important role in plant stress and hormone responses (Abler and Green, 1996, Kuhn and Schroeder, 2003, Riehs-Kearnan et al., 2012, Xu and Chua, 2012; Soma et al., 2017). DCP1 and VARICOSE were phosphorylated by MPK6 and SnRK2, respectively, when plants were subjected to drought stress (Xu and Chua, 2012; Soma et al., 2017). Phosphorylated DCP1 preferentially bound to DCP5 and promoted the decapping of direct mRNA targets, whereas dcp5 mutants were hypersensitive to osmotic stress (Xu and Chua, 2012). The increase of a phosphonucleotide (3′-phosphoadenosine 5′-phosphate [PAP]) in response to drought inhibits XRN activity (Estavillo et al., 2011), which degrades RNA substrates after de-capping process. It seems that RNA metabolism is maintained in a balanced state under drought stress by an increase of poly (A−) sense RNA, antisense RNA synthesis, and dsRNA degradation. It is plausible that the activation of a specific RNA metabolism in response to abiotic stress is required for the removal of uncapped sense RNAs by RDR1/2/6-dependent antisense RNA. For the biological function, it is possible that the effects of RDR1/2/6-dependent antisense RNA may reinforce the degradation of sense RNA of critical developmental-related genes for the adaptation to drought stress.
Recent studies have shown that various RDR-dependent noncoding RNA were produced on protein-coding loci and TEs. (Gregory et al., 2008; Cao et al., 2014, Martínez de Alba et al., 2015; Zhang et al., 2015). The small RNA production occurs through the activity of RDRs and DCLs on protein-coding genes and is involved in mRNA degradation under specific conditions, such as virus-infection or loss of functions of decapping enzyme and ribonucleases (Gregory et al., 2008; Cao et al., 2014, Martínez de Alba et al., 2015; Zhang et al., 2015). Twenty-four percent of RDR1/2/6-dependent antisense RNA loci were overlapped with virus-derived siRNAs or RNA degradation-related siRNAs, suggesting that these loci had potential to make dsRNA by RDRs. However, it was reported that accumulation of known siRNAs was significantly changed by disruption of key component of siRNA synthesis. For example, the virus-derived siRNAs were affected by RDR1, DCL4, and AGO2 (Cao et al., 2014), the decapping-related siRNAs were affected by RDR6 (Martínez de Alba et al., 2015), and cytoplasmic RNA decay-related siRNAs were affected by RDR6, SGS3 and DCL2/4 (Zhang et al., 2015). The present study showed an 20–30 nt small RNAs were not accumulated on the antisense RNA loci under our control or drought stress conditions (Fig. 3A, Supplemental Figs. S8B and S10). The antisense RNA was accumulated redundantly by RDRs and the degradation of sense RNA was reduced in rdr1/2/6, but not in dcl2/3/4 (Fig. 1A, Supplemental Figs. S1, S12, and S13). rdr1/2/6 plants exhibited severe inhibition of root growth after the application of a temporal drought stress, whereas the phenotype was not observed in dcl2/3/4 and rdr double mutants (Fig. 6A). The dsRNA accumulation and small RNA accumulation can be subjected depending on the situation. It is thought that siRNA biosynthesis is required for the recognition and construction of a protein-RNA complex, including other components of siRNA synthesis (Yoshikawa et al., 2005; Curaba and Chen, 2008; Willmann et al., 2011). It was also reported that another small RNAs over 30 nt DCL1/2/3/4-independent and pol IV-dependent small RNAs can also function in RdDM pathway, that mostly targeted TEs (Yang et al., 2016). Our present analysis showed that the synthesis of stress-inducible antisense RNAs requires RDR1/2/6, but not NRPD1a and NRPD1b (Fig. 1A; Supplemental Fig. S1A) and antisense RNA loci contains a lot of protein-conding genes (Fig. 3). We speculate that dsRNAs formed from poly(A−) sense and poly(A−) antisense RNAs are degraded by other dsRNases independently of siRNA generation.
Interestingly, RDRs are conserved in RNA viruses, plants, fungi, protists, and worms, but are absent in flies, mice, and humans (Willmann et al., 2011). In humans, a telomerase reverse transcriptase catalytic subunit (TERT) and an RNA component of mitochondrial RNA processing endoribonuclease (RMRP) form a distinct ribonucleoprotein complex that has RDR activity and produces double-stranded RNAs that can be processed into siRNAs (Maida et al., 2009). In plants, among a total of six RDRs, RDR1, RDR2, and RDR6 are known to synthesize dsRNA to generate siRNA in utilizing TGS and PTGS (Willmann et al., 2011). When compared to wild-type plants, the accumulation of antisense RNAs was reduced in rdr1/2/6, but not in single or double mutants of RDRs (Fig. 1; Supplemental Fig. S1A; Supplemental Table S1). The cloning of TAS2 antisense RNA using rdr mutants also showed an independent recognition to select the genes that are targeted by each RDR (Supplemental Fig. S11). These results suggest that RDRs with redundant functions are necessary for the synthesis of stress-inducible antisense RNAs. Among the six RDR genes in Arabidopsis, RDR3/4/5 may also function in the synthesis of stress-inducible antisense RNAs because the loss of RDR1/2/6 only affected half of the antisense RNA synthesis. RDR1, -2, and -6 are thought to convert aberrant single-stranded RNAs into double-stranded RNAs (Voinnet, 2008; Willmann et al., 2011). Each RDR has specific substrates that are processed into various forms of siRNAs. It is also known that RDRs function in a coordinated manner. Analysis of female gametophyte development suggests that RDR2 and RDR6 function redundantly in gene silencing (Autran et al., 2011). RDR1 and RDR6 have redundant functions in the biosynthesis or amplification of viral dsRNAs for TuMV and CMV (Garcia-Ruiz et al., 2010, Wang et al., 2010). RDR2 and RDR6 compete for RNA substrates produced by transgenes that are subjected to sense transgene-mediated posttranscriptional gene silencing (S-PTGS) (Jauvion et al., 2012). Data from the previous and present studies indicates that RDR1, RDR2, and RDR6 have redundant functions in the biosynthesis of antisense RNAs from sense transcripts of protein-coding genes.
Sequence characteristics of the 1,136 RDR1/2/6-dependent antisense RNA loci were revealed by comparing them with the 1,015 RDR1/2/6-independent loci and 7,138 SAT loci (Supplemental Figure S16A). The multi exon transcripts (>5 exons; χ2 test P value = 4.5*10−24) and long 5′ UTR transcripts (>200 nt; P value = 2.4*10−3) were enriched in the RDR1/2/6-dependent antisense RNA loci. Their sequence motifs at the (+) strand of the 50–150 nt length 5′UTR category and the 200–300 nt length 5′UTR category were searched by comparing RDR1/2/6-dependent antisense RNA loci to RDR1/2/6-independent loci, using Multiple Em for Motif Elicitation (MEME) software (Bailey et al., 2006). The analysis identified a CT-rich motif (5′-TCTNNNTCT-3) and a AG-rich motif (5′-AGANNNAGA-3′) in both 5′ UTR categories (Supplemental Figure S16B). About 40% of the transcripts with a 200–300 nt 5′ UTR had both motifs, while most of the 50–150 nt 5′UTR transcripts only had one of the pair. When the search of sequences of the 50–150 nt 5′ UTR mRNAs was expanded to UTRs that were 250 nt in length, including the exon, the number of loci with both motifs increased. It was thought that transcripts with the 50–150 nt UTR might have their secondary structures unraveled by translation and reduced the possibility to produce antisense RNA. Since antisense RNA was transcribed from the 3′ end of mRNA, it is plausible that RNA secondary structures with the CT-rich and AG-rich motifs are formed and that motifs with the 5′ UTR perhaps indirectly regulate antisense RNA generation via inhibition of a 5′-3′ exoribonuclease or by recruitment of RDRs.
The current study indicates that, during the RNA decay response to abiotic stress, the selectivity of the target RNA might affect various biological processes in plants. It also suggests that RDR1/2/6-dependent antisense RNA may be linked to other forms of RNA regulation and together regulate stress response and adaptation. The current study illuminates a novel mechanism that RDR1/2/6-mediated biosynthesis of antisense RNAs is involved in RNA turnover in response to specific environmental conditions.
MATERIALS AND METHODS
Linker RT-PCR and qRT-PCR Analysis
Two-week-old mutant and wild-type plants were subjected to a drought stress as previously described (Matsui et a., 2008). Total RNAs were extracted with a Plant RNA Isolation Reagent (Life Technologies) and treated with DNase I (Life Technologies). For linker RT-PCR, RNAs were reverse transcribed with the target gene primers with linker sequence (5′-CGACTGGAGCACGAGGACACTGA-3′), and the strand-specific PCR was used with one primer specific to the linker and the other primer specific for the target genes (Lepere et al., 2008)(Supplemental Table S4). For strand-specific qRT-PCR, cDNAs of sense and antisense RNAs were reverse transcribed with the strand-specific primers (Supplemental Table S4) using Superscript III (Life Technologies) at 55°C for preventing miss-annealing (Yassour et al., 2010; Feng et al., 2012). Accumulation of the transcripts was measured in an ABI Prism 3100 or Step One Plus (Life Technologies) using SYBR Premix Ex Taq II (Takara) or SYBR Green Master Mix (Life Technologies), respectively. Three independent biological replicates were used for each RT-PCR analysis.
Northern Analysis
Northern analysis was performed as previously described (Matsui et al., 2008).
RNase Protection Assay
One μg of total RNA and 1 μg of yeast tRNA were combined. Enzymatic reactions were used for the single-stranded RNA degradation assay according to a previously described approach (Dodds et al., 1984). RNA was dissolved in 0.3 m NaCl, 50 mm Tris-HCl (pH 6.5) and the RNA solution was added to a dilution series of a mixture of 1U of RNase I (Life Technologies) and 1 U of RNase A (Life Technologies) and incubated for 15 min at room temperature. For the dsRNA degradation assay, RNase V1 (Life Technologies) was used. The RNA was prepared in 0.3 m NaCl, 50 mm Tris-HCl (pH 6.5) and the RNA solution was added to a dilution series of 0.01 U of RNase V1 and incubated for 15 min at room temperature. RNase-treated RNA was purified by proteinase K treatment, phenol extraction, and ethanol precipitation. The quantity of recovered RNA was measured using the qRT-PCR protocol as described previously.
Circular RT-PCR
Circular RT-PCR was carried out as previously described (Zakrzewska-Placzek et al., 2010). Four μg of total RNA was self-ligated for 8 h using T4 RNA ligase (Takara). The ligated RNA was then converted to cDNA using a random primer for RD29A. 1/20 dilution of cDNA was amplified by 30 cycles of PCR using circular PCR primers (Supplemental Table S4). For the cloning of TAS2 antisense RNA, the circular RNA after self-ligation is converted to cDNA using the strand-specific primer “TAS2_circular_RT-PCR_RT” in Supplemental Table S4, and PCR is performed with the primers “TAS2_circular_RT-PCR_F” and “TAS2_circular_RT-PCR_R”.
Measurement of RNA Decay Rate
In order to determine whether antisense RNA synthesis affects RNA stability, the RNA decay rate of RD29A poly (A+) sense RNA in rdr1/2/6 and wild-type plants was measured using a half-life analysis as described by Lidder et al. (2005). Two-week-old plants were transferred to a 30% PEG solution and incubated for two hours. The plants were then transferred to a 30% PEG solution with and without 0.6 mm 3′-deoxyadenosine (cordycepin). Total RNA was isolated from untreated and 1, 2, and 3 h-treated plants using a Plant RNA Purification reagent (Invitrogen). cDNA was reverse-transcribed from samples containing 2 μg of total RNA using SuperScript III (Life Technologies) and an oligo dT primer. The remaining RD29A and ACT2 poly (A+) sense RNAs were measured by qRT-PCR analysis.
Custom Microarray Analysis
A custom microarray was used for gene expression analysis during the drought (Matsui et al., 2008) and rehydration (Oono et al., 2003) treatments. Two-week-old rdr1/2/6-1 and wild-type plants were transferred from Murashige and Skoog (MS) medium to plastic Petri dishes and kept for 2 h. Then, water was added to the petri dishes essentially as described previously (Oono et al., 2003). Total RNA was extracted from whole plants using Plant RNA Isolation Reagent (Life Technologies). Then, 100 μg of total RNA was separated into poly (A+) RNA and poly (A−) fractions using Ambion Poly A purist MAG. Poly (A−) RNA samples were prepared by depleting rRNAs using an Invitrogen RiboMinus Plant Kit. Five ng of the poly (A+) RNA fraction and 1400 ng of poly (A−) from each sample was used to prepare for Cy3-labeled cRNA using a Low Input Quick Amp Labeling Kit (Agilent). In a subsequent step, 600 ng of cRNAs from each poly (A+) RNA sample and 3 mg of cRNAs from each poly (A-) RNA sample were hybridized to the microarray (GPL19830) at 65°C for 17 h using a Gene Expression Hybridization kit. After hybridization, the microarray was washed with Gene Expression Wash buffer 1 (Agilent) and Gene Expression Wash buffer (Agilent), and then dried immediately with a brief centrifugation step. The microarray was scanned using an Agilent DNA Microarray Scanner G2539A ver. C and raw data were extracted using the Feature Extraction program ver 9.1. RMA normalization was performed for signals of microarray probes using the limma package (Ritchie et al., 2015) in the R 2.12.1 program (R Core Team). The statistical significance of differences in gene expression was determined using a Student’s t test and Benjamini Hochberg FDR (Benjamini and Hochberg, 1995) < 0.075.
Deep Sequencing of Small RNAs
Small RNA fractions were extracted from plants using a mirVana miRNA Isolation Kit (Life Technologies). Ten μg from the small RNA fractions was separated and purified into 20–30 nt RNAs using 8M urea and 7.5% acrylamide gel electrophoresis, followed by gel extraction. Then, the 20–30 nt small RNAs were used to construct RNA libraries using a TruSeq Small RNA Library Preparation Kits (Illumina). The small RNA libraries were sequenced on a HiSequation 2000 (Illumina). The library preparation and sequencing were performed by the Genome Network Analysis Support Facility, RIKEN CLST. Small RNA libraries for SOLiD were constructed according to the protocol provided in the SOLiD Total RNA-Seq Kit (Life Technologies). First, small RNAs were ligated with a 5′ and a 3′ adapter to generate cDNAs. The cDNAs were separated in a 7.5% acrylamide gel containing 8M urea and the 60–80 nt cDNAs were recovered. The cDNAs were amplified for 12–15 cycles of PCR using barcode primers (Life Technologies) and were subjected to SOLiD sequencing from the 5′-end.
Data Analysis of Small RNAs
The sequence of the 3′-end (AATTCTCGGGTGCCAAGGAACT) for the Hi-Seq2000 sequence data or the first 26-nt barcode for the SOLiD sequence data were first removed. The sequence data were then mapped to the Arabidopsis genome using bowtie software (Langmead et al., 2009). The following two-step normalization was used for data analysis of small RNAs. (1) The small RNAs mapped to annotated genes were counted and normalized independently for rdr1/2/6 and wild type by adjusting the slope of the linear approximate equation between the control and drought stress condition. The top 10% of highly expressed small RNA loci was used for the normalization. (2) Tag numbers that mapped to miRNA loci were used by adjusting the slope of the linear approximate equation between rdr1/2/6 and wild-type samples because the accumulation of miRNAs is thought to be less affected by the loss of RDR1/2/6. Then, the P values of AGI-annotated genes with mapped small RNAs were calculated using the DESeq package (Anders and Huber, 2010) and a FDR method (Benjamini and Hochberg, 1995) in R 2.12.1 software.
Accession Numbers
Arabidopsis microarray expression profiling data are available in GEO under the accession numbers, GSE37137, GSE72309 and GSE83846. Small RNA sequence data are available under the accession number GSE39024, GSE39033 and GSE72982.
Supplemental Data
The following supplemental materials are available:
Supplemental Figure S1. Accumulation of sense and antisense RNAs of drought-inducible RD29A, RD20, PP2CA in rdr1/2/6, xrn4, dcp5 and wild-type plants.
Supplemental Figure S2. Linker RT-PCR of sense RNAs and antisense RNAs in poly (A+) RNA fraction and poly (A-) RNA fraction.
Supplemental Figure S3. Accumulation of fAsRD29A2 in rdr1/2/6 and wild-type plants.
Supplemental Figure S4. Preparation scheme for the custom microarray for the analysis of poly (A+) RNA and poly (A-) RNA.
Supplemental Figure S5. Custom microarray analysis of sense- and antisense RNAs in wild-type and rdr1/2/6 plants.
Supplemental Figure S6. Confirmation of RDR1/2/6-dependent antisense RNA loci by qRT-PCR analysis.
Supplemental Figure S7. RNase protection assay of RDR1/2/6-dependent antisense RNA loci.
Supplemental Figure S8. Number of small RNAs at the TAS1B locus and the distribution of the expression level of small RNAs, poly (A-) sense RNA, and poly (A-) antisense RNA.
Supplemental Figure S9. Accumulation of RDR1/2/6-dependent antisense RNA loci in dcl2/3/4 by qRT-PCR analysis.
Supplemental Figure S10. Comparison between RDR1/2/6-dependent antisense RNA loci and previous reported endogenous loci with small RNA accumulation associated with RNA degradation.
Supplemental Figure S11. Accumulation of antisense RNAs on TAS2 loci in wild-type and rdr mutants.
Supplemental Figure S12. Analysis of the RNA decay rate at RDR1/2/6-dependent antisense RNA loci in wild-type and rdr1/2/6 plants subjected to an osmotic stress treatment.
Supplemental Figure S13. Analysis of the RNA decay rate at RDR1/2/6-dependent antisense RNA loci in plants subjected to an osmotic stress.
Supplemental Figure S14. Accumulation level of sense RNA at RDR1/2/6-dependent antisense RNA loci in wild-type and rdr1/2/6 plants during the rehydration phase following a temporal osmotic stress treatment.
Supplemental Figure S15. Comparative expression analysis of drought stress-upregulated- and rehydration- downregulated genes in RDR1/2/6-dependent antisense RNA loci, RDR1/2/6-independent antisense RNA loci and RDR1/2/6-dependent small RNA loci during the rehydration stage.
Supplemental Figure S16. Sequence characteristics of RDR1/2/6-dependent antisense RNA loci.
Supplemental Table S1. Expression data from a custom microarray analysis of antisense RNAs and sense RNAs at 7,138 SAT loci using poly (A+) RNA and poly (A-) RNA.
Supplemental Table S2. List of 2,325 RDR1/2/6-dependent small RNA loci.
Supplemental Table S3. Expression data from a custom microarray analysis of sense RNAs on 1,136 RDR1/2/6-dependent antisense RNA loci during recovery process after drought stress treatment.
Supplemental Table S4. Primer sequences.
Acknowledgments
The authors would like to thank Shou-Wei Ding for providing the seeds of rdr1/2, rdr1/6, rdr2/6, and rdr1/2/6-1 (rdr1-1/rdr2-1/6-15); Taku Sasaki for providing seed of rdr1/2/6-2 (SALK_112300/rdr2-2/rdr6-11); Nam-Hai Chua for providing dcp5-1 seeds; Craig S. Pikaard for providing nrpd1a-3; and David Baulcombe for providing nrpd1b-1. The seeds of the xrn4 (SALK_014209) mutant were obtained from ABRC. We would like to thank Tetsuya Kurata for providing kind advice and technical expertise for SOLiD sequencing. We would also like to thank Ri-ichiroh Manabe for small RNA sequencing.
Footnotes
This work was supported by a grant from the RIKEN Center for Sustainable Resource Science (to M.S.), the Japan Science and Technology Agency (JST) [Core Research for Evolutionary Science and Technology (CREST)] (JPMJCR13B4) (to M.S.), Grants-in-Aid for Scientific Research on Kiban (C) (no. 21570056), Innovative Areas (no. 16H01476) and Challenging Exploratory Research (no. 24657041) of the Ministry of Education Culture, Sports and Technology of Japan (to M.S.), NIBB Cooperative Research Program (Next-generation DNA Sequencing Initiative: 11-725) and the Japan Advanced Plant Science Network.
Articles can be viewed without a subscription.
References
- Abler ML, Green PJ (1996) Control of mRNA stability in higher plants. Plant Mol Biol 32: 63–78 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA (2001) Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M, Grob S, Steimer A, Barann M, Klostermeier UC, Leblanc O, Vielle-Calzada JP, Rosenstiel P, et al. (2011) Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34 (Web Server issue): W369-373 [DOI] [PMC free article] [PubMed]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JS. (1982) Plant productivity and environment. Science 218: 443–448 [DOI] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G (2004) GO:TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA, Bailey-Serres J, Weretilnyk E (2000) Responses to abiotic stresses. In Biochemistry and Molecular Biology of Plants, Buchanan BB, Gruissem W, Jones RL eds (American Society of Plant Physiologists, Rockville, MD: ), pp1158-1203 [Google Scholar]
- Cao M, Du P, Wang X, Yu YQ, Qiu YH, Li W, Gal-On A, Zhou C, Li Y, Ding SW (2014) Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc Natl Acad Sci USA 111: 14613–14618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevin LM, Lande R, Mace GM (2010) Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol 8: e1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J, Chen X (2008) Biochemical activities of Arabidopsis RNA-dependent RNA polymerase 6. J Biol Chem 283: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds JA, Morris TJ, Jordan RL (1984) Plant viral double-stranded RNA. Annu Rev Phytopathol 22: 151–168 [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, Brearley C, Hell R, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G III, Wahlestedt C (2010) Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol 11: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Lintula S, Ho TH, Anastasina M, Paju A, Haglund C, Stenman UH, Hotakainen K, Orpana A, Kainov D, Stenman J (2012) Technique for strand-specific gene-expression analysis and monitoring of primer-independent cDNA synthesis in reverse transcription. Biotechniques 52: 263–270 [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC (2010) Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22: 481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR (2008) A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Gu R, Zhang Z, DeCerbo JN, Carmichael GG (2009) Gene regulation by sense-antisense overlap of polyadenylation signals. RNA 15: 1154–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín JE, Darzacq X, Choder M (2013) Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 153: 1000–1011 [DOI] [PubMed] [Google Scholar]
- Hazen SP, Naef F, Quisel T, Gendron JM, Chen H, Ecker JR, Borevitz JO, Kay SA (2009) Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol 10: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauvion V, Rivard M, Bouteiller N, Elmayan T, Vaucheret H (2012) RDR2 partially antagonizes the production of RDR6-dependent siRNA in sense transgene-mediated PTGS. PLoS One 7: e29785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen CH, Michalopoulos I, Westhead DR, Meyer P (2005) Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol 6: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Vacic V, Girke T, Lonardi S, Zhu JK (2008) Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol Biol 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A Jr., Zhu JK, Staskawicz BJ, Jin H (2006) A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA 103: 18002–18007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi S, Iida K, Harada E, Hanada K, Matsui A, Okamoto M, Shinozaki K, Seki M, Toyoda T (2012) Positional correlation analysis improves reconstruction of full-length transcripts and alternative isoforms from noisy array signals or short reads. Bioinformatics 28: 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JM, Schroeder JI (2003) Impacts of altered RNA metabolism on abscisic acid signaling. Curr Opin Plant Biol 6: 463–469 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepère G, Bétermier M, Meyer E, Duharcourt S (2008) Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev 22: 1501–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidder P, Gutiérrez RA, Salomé PA, McClung CR, Green PJ (2005) Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol 138: 2374–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, Kasim V, Hayashizaki Y, Hahn WC, Masutomi K (2009) An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 461: 230–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez de Alba AE, Moreno AB, Gabriel M, Mallory AC, Christ A, Bounon R, Balzergue S, Aubourg S, Gautheret D, Crespi MD, Vaucheret H, Maizel A (2015) In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res 43: 2902–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, Satou M, Kim JM, et al. (2008) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49: 1135–1149 [DOI] [PubMed] [Google Scholar]
- Matsui A, Nguyen AH, Nakaminami K, Seki M (2013) Arabidopsis non-coding RNA regulation in abiotic stress responses. Int J Mol Sci 14: 22642–22654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K, Venu RC, Lu C, Beló A, Vemaraju K, Kulkarni K, Wang W, Pillay M, Green PJ, Wang GL, Meyers BC (2007) An expression atlas of rice mRNAs and small RNAs. Nat Biotechnol 25: 473–477 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Tatematsu K, Matsui A, Morosawa T, Ishida J, Tanaka M, Endo TA, Mochizuki Y, Toyoda T, Kamiya Y, Shinozaki K, Nambara E, et al. (2010) Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. Plant J 62: 39–51 [DOI] [PubMed] [Google Scholar]
- Oono Y, Seki M, Nanjo T, Narusaka M, Fujita M, Satoh R, Satou M, Sakurai T, Ishida J, Akiyama K, Iida K, Maruyama K, et al. (2003) Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca 7000 full-length cDNA microarray. Plant J 34: 868–887 [DOI] [PubMed] [Google Scholar]
- Osato N, Yamada H, Satoh K, Ooka H, Yamamoto M, Suzuki K, Kawai J, Carninci P, Ohtomo Y, Murakami K, Matsubara K, Kikuchi S, et al. (2003) Antisense transcripts with rice full-length cDNAs. Genome Biol 5: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NT, Rohrbach JA, Zalewski BA, Byrkett CM, Vaughn JC (2003) RNA editing and regulation of Drosophila 4f-rnp expression by sas-10 antisense readthrough mRNA transcripts. RNA 9: 698–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott EM, Proudfoot NJ (2002) Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci USA 99: 8796–8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeswaran R, Aregger M, Zvereva AS, Borah BK, Gubaeva EG, Pooggin MM (2012) Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res 40: 6241–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehs-Kearnan N, Gloggnitzer J, Dekrout B, Jonak C, Riha K (2012) Aberrant growth and lethality of Arabidopsis deficient in nonsense-mediated RNA decay factors is caused by autoimmune-like response. Nucleic Acids Res 40: 5615–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Alandete Saez M, Eshed Williams L, Fletcher JC, McCormick S (2010) Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev 24: 1010–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalska L, Beltran-Nebot M, Ule J, Jenner RG (2017) Regulatory feedback from nascent RNA to chromatin and transcription. Nat Rev Mol Cell Biol 18: 331–337 [DOI] [PubMed] [Google Scholar]
- Soma F, Mogami J, Yoshida T, Abekura M, Takahashi F, Kidokoro S, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2017) ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat Plants 3: 16204. [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Terryn N, Rouzé P (2000) The sense of naturally transcribed antisense RNAs in plants. Trends Plant Sci 5: 394–396 [DOI] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR (2003) Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 34: 157–165 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2008) Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci 13: 317–328 [DOI] [PubMed] [Google Scholar]
- Wang XJ, Gaasterland T, Chua NH (2005) Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol 6: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Wu Q, Ito T, Cillo F, Li WX, Chen X, Yu JL, Ding SW (2010) RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann MR, Endres MW, Cook RT, Gregory BD (2011) The functions of RNA-dependent RNA polymerases in Arabidopsis. Arabidopsis Book 9: e0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH (2012) Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J 31: 1975–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, Pham P, Cheuk R, et al. (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yang DL, Zhang G, Tang K, Li J, Yang L, Huang H, Zhang H, Zhu JK (2016) Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res 26: 66–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Pfiffner J, Levin JZ, Adiconis X, Gnirke A, Nusbaum C, Thompson DA, Friedman N, Regev A (2010) Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol 11: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska-Placzek M, Souret FF, Sobczyk GJ, Green PJ, Kufel J (2010) Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res 38: 4487–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhu Y, Liu X, Hong X, Xu Y, Zhu P, Shen Y, Wu H, Ji Y, Wen X, Zhang C, Zhao Q, et al. (2015) Plant biology. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science 348: 120–123 [DOI] [PubMed] [Google Scholar]
- Zhou X, Sunkar R, Jin H, Zhu JK, Zhang W (2009) Genome-wide identification and analysis of small RNAs originated from natural antisense transcripts in Oryza sativa. Genome Res 19: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko E, Kunova A, Meyer P (2011) Sense and antisense transcripts of convergent gene pairs in Arabidopsis thaliana can share a common polyadenylation region. PLoS One 6: e16769. [DOI] [PMC free article] [PubMed] [Google Scholar]