OsPRI1 functions downstream of the Fe-binding sensor OsHRZ1 and directly controls the expression of OsIRO2 and OsIRO3 for iron homeostasis in rice.
Abstract
Oryza sativa HEMERYTHRIN MOTIF-CONTAINING REALLY INTERESTING NEW GENE AND ZINC-FINGER PROTEIN1 (OsHRZ1) is a putative iron-binding sensor. However, it is unclear how OsHRZ1 transmits signals. In this study, we reveal that POSITIVE REGULATOR OF IRON HOMEOSTASIS1 (OsPRI1) interacts with OsHRZ1. A loss-of-function mutation to OsPRI1 increased the sensitivity of plants to Fe-deficient conditions and down-regulated the expression of Fe-deficiency-responsive genes. Yeast one-hybrid and electrophoretic mobility shift assay results suggested that OsPRI1 binds to the OsIRO2 and OsIRO3 promoters. In vitro ubiquitination experiments indicated that OsPRI1 is ubiquitinated by OsHRZ1. Cell-free degradation assays revealed that the stability of OsPRI1 decreased in wild-type roots but increased in the hrz1-2 mutant, suggesting OsHRZ1 is responsible for the instability of OsPRI1. The hrz1-2 seedlings were insensitive to Fe-deficient conditions. When the pri1-1 mutation was introduced into hrz1-2 mutants, the pri1hrz1 double mutant was more sensitive to Fe deficiency than the hrz1-2 mutant. Additionally, the expression levels of Fe-deficiency-responsive genes were lower in the hrz1pri1 double mutant than in the hrz1-2 mutant. Collectively, these results imply that OsPRI1, which is ubiquitinated by OsHRZ1, mediates rice responses to Fe deficiency by positively regulating OsIRO2 and OsIRO3 expression as part of the OsHRZ1-OsPRI1-OsIRO2/3 signal transduction cascade.
Iron (Fe) is an essential mineral for plant growth and development. It affects many biochemical processes, including photosynthesis and respiration (Balk and Schaedler, 2014). In humans, Fe deficiency is one of the most prevalent nutritional disorders worldwide (Mayer et al., 2008). Fruits and vegetables are the major Fe sources for humans. Thus, characterizing the mechanism underlying Fe homeostasis in plants may have important consequences for human health. Plants take up inorganic Fe from soils. Although Fe is the second most abundant metal in the earth, in many natural and agricultural ecosystems, plants are often exposed to insufficient availability of Fe due to the insolubility of inorganic Fe, particularly in alkaline soils (Kobayashi and Nishizawa, 2012).
To cope with Fe-deficient conditions, plants have evolved sophisticated regulatory mechanisms to optimize the acquisition of Fe from soils. Higher plants obtain Fe from the rhizosphere using two major strategies, namely the reduction strategy (Strategy I) and chelation strategy (Strategy II). Strategy I consists of the following three steps: rhizosphere acidification, reduction of Fe3+ to Fe2+, and uptake of Fe2+ (Hell and Stephan, 2003; Walker and Connolly, 2008; Hindt and Guerinot, 2012). The Arabidopsis (Arabidopsis thaliana) Strategy I components (i.e., H+-ATPase 2 [Santi and Schmidt, 2009], ferric reductase [Robinson et al., 1999], and ferrous transporter [IRT1; Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002]) have been well characterized. In contrast, plants that use Strategy II secrete phytosiderophores, such as mugineic acids that bind Fe3+ with high affinity, to acquire Fe from the rhizosphere under Fe-limited conditions (Walker and Connolly, 2008; Hindt and Guerinot, 2012). Phytosiderophores are synthesized from nicotianamine, which is a nonproteinogenic amino acid. Nongraminaceous plants usually employ Strategy I, while graminaceous plants use Strategy II. As a graminaceous species, rice (Oryza sativa) employs Strategy II, but it also uses a partial Strategy I system. Rice plants secrete mugineic acid (MA) family phytosiderophores to chelate ferric Fe, but they also directly absorb ferrous Fe with OsIRT1 (IRON TRANSPORTER1) and OsIRT2 (Ishimaru et al., 2006).
In rice, Strategy II-associated genes Oryza sativa NICOTIANAMINE SYNTHASE (OsNAS1/2), NICOTIANAMINE AMINOTRANSFERASE, and DEOXYMUGINEIC ACID SYNTHASE are up-regulated during Fe limitation and participate in the synthesis of MAs (Inoue et al., 2003, Cheng et al., 2007; Bashir et al., 2017). Another Strategy II-associated gene Oryza sativa YS-Like15 (OsYSL15) is required for the transport of Fe3+-DMA from soils to roots (Inoue et al., 2009; Lee et al., 2009). It is noteworthy that the expression of all these Strategy II-associated genes is controlled by OsIRO2 (Ogo et al., 2007). However, the expression of the Strategy I-associated gene OsIRT1 is not affected by OsIRO2. In contrast, OsIRO3 negatively regulates the expression of OsNAS1/2, NICOTIANAMINE AMINOTRANSFERASE1, OsYSL15, as well as OsIRT1, in rice (Zheng et al., 2010). IRON DEFICIENCY-RESPONSIVE CIS-ACTING ELEMENT-BINDING FACTOR1 and 2 (IDEF1 and IDEF2), which belong to the ABA INSENSITIVE 3/VIVIPAROUS 1 (ABI3/VP1) and NAC (NAC DOMAIN CONTAINING PROTEIN (NAC) transcription factor families, respectively, were identified as positive regulators of rice responses to Fe deficiency (Kobayashi et al., 2007; Ogo et al., 2008). Unlike OsIRO2 and OsIRO3 expression levels, which are up-regulated by Fe deficiency, the expression levels of IDEF1 and IDEF2 are unaffected by plant Fe status. Additionally, OsbHLH133 expression is inducible under Fe-deficient conditions, and the encoded protein is responsible for regulating the distribution of Fe between the roots and shoots (Wang et al., 2013).
It is unclear how rice senses external and internal Fe contents and regulates the expression of Fe-homeostasis-associated genes. Recently, HEMERYTHRIN MOTIF-CONTAINING REALLY INTERESTING NEW GENE (RING) AND ZINC-FINGER PROTEIN1 (OsHRZ1) and OsHRZ2 were characterized as putative iron-binding sensors that negatively regulate Fe acquisition under Fe-sufficient conditions (Kobayashi et al., 2013). Similarly, its Arabidopsis homolog BRUTUS (BTS) also binds Fe and possesses ubiquitination function (Selote et al., 2015) However, how OsHRZ1 and OsHRZ2 affect the expression of Fe-deficiency-responsive genes has not been determined. Both OsHRZ1 and OsHRZ2 exhibit E3 ligase activity, implying that they function by ubiquitinating and degrading substrates. Therefore, identifying and characterizing the substrates of OsHRZ1 and OsHRZ2 may provide novel insights into rice responses to Fe deficiency.
In this study, a bHLH transcription factor, which was identified as an interacting partner of OsHRZ1, was named POSITIVE REGULATOR OF IRON HOMEOSTASIS1 (OsPRI1). A loss-of-function mutation led to hypersensitivity to Fe deficiency. Additional analyses confirmed that OsPRI1 bound to the promoters of OsIRO2 and OsIRO3 to activate expression. Moreover, the stability of OsPRI1 was negatively correlated with OsHRZ1 production. Our data revealed that OsPRI1 is a missing link between the iron-binding sensor OsHRZ1 and the Fe-deficiency-responsive genes OsIRO2 and OsIRO3 in rice.
RESULTS
OsPRI1 Is an Interacting Partner of OsHRZ1
OsHRZ1 was characterized as a putative iron-binding sensor that exhibits E3 ligase activity. However, the substrate of OsHRZ1 is unknown. Yeast two-hybrid assays were used to identify the interacting partner of OsHRZ1. The N- and C-terminals of OsHRZ1 contain the iron-binding and RING domains, respectively. The C-terminal of OsHRZ1 (containing the CHY-type zinc-finger domain, RING zinc-finger domain, and Rubredoxin fold) was used as the bait to screen an iron-depleted rice cDNA library. Sequencing analyses suggested that four of the 75 positive clones (Supplemental Table S1) contained the same insert sequence encoding the LOC_Os08g04390 bHLH transcription factor (also known as OsbHLH60), which was named POSITIVE REGULATOR OF IRON HOMEOSTASIS1 (OsPRI1).
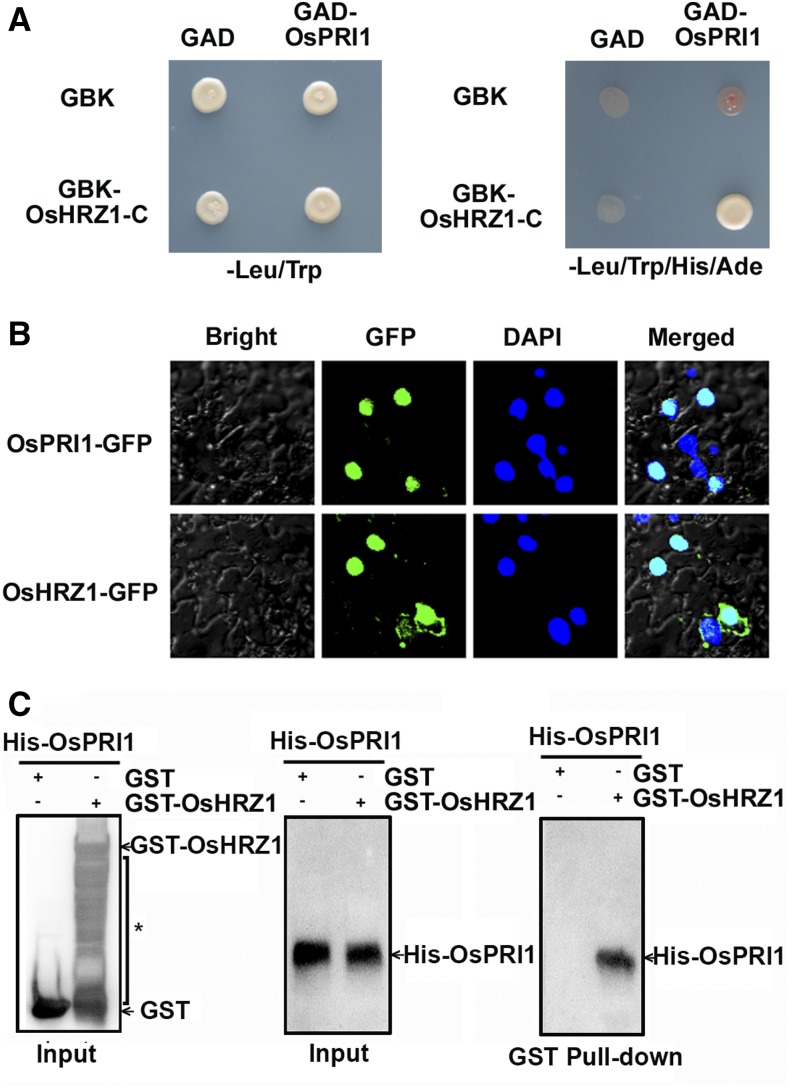
The full-length OsPRI1 coding region was cloned, after which the interaction between the encoded protein and the C-terminal of OsHRZ1 was confirmed in yeast cells (Fig. 1A). Considering that OsHRZ1 is a nucleus-localized protein (Kobayashi et al., 2013), we examined whether OsPRI1 is also located in the nucleus. To determine the subcellular localization of OsPRI1, the GFP was fused to the C-terminal of OsPRI1. Similarly, GFP was also fused to the C-terminal of OsHRZ1. Transient expression of OsPRI1-GFP or OsHRZ1-GFP in tobacco leaves revealed that OsPRI1 and OsHRZ1 localized to nuclei (Fig. 1B), further supporting the likelihood of an interaction between OsPRI1 and OsHRZ1. Pull-down assays were conducted to verify this interaction. Glutathione S-transferase (GST)-fused OsHRZ1 and poly-His-fused OsPRI1 were produced in Escherichia coli cells. In vitro pull-down assays revealed that GST-OsHRZ1, but not GST alone, could pull down His-OsPRI1 (Fig. 1C). These observations suggest that OsPRI1 interacts with OsHRZ1.
Figure 1.
OsPRI1 interacts with OsHRZ1. A, Yeast-two-hybrid assay. Interaction was indicated by the ability of cells to grow on synthetic dropout medium lacking Leu/Trp/His/Ade. The C-terminal truncated OsHRZ1 and full-length OsPRI1 were cloned into pGBKT7 (GBK) and pGADT7 (GAD), respectively. B, Subcellular localization. OsPRI1 and OsHRZ1 were respectively fused to the N-terminal of GFP protein. Fluorescence was observed in nuclear compartments of N. benthamiana leaf epidermal cells. The 4,6-diamidino-2-phenylindole (DAPI) staining indicates the localization of nuclei. Merged images show colocalization of GFP and DAPI signals. C, Pull-down assay. OsHRZ1 was fused with the GST tag, and OsPRI1 was fused with the His tag. Recombinant proteins were expressed in E. coli. Proteins were pulled down by glutathione Sepharose 4B and detected using the anti-His or anti-GST antibody. The asterisk indicates nonspecific signals.
OsPRI1 Is Ubiquitously Expressed
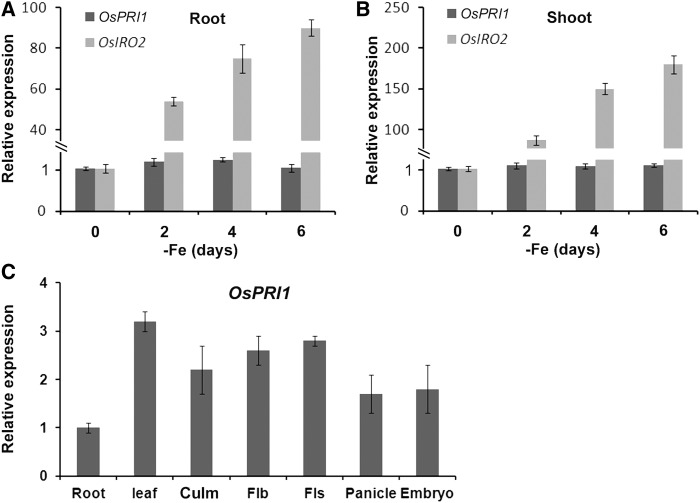
The OsPRI1 expression patterns were analyzed using a quantitative real-time PCR (qRT-PCR) to elucidate the OsPRI1 functions. Given that OsHRZ1 expression is induced by Fe deficiency (Kobayashi et al., 2013), we determined whether OsPRI1 transcript abundance is also affected by insufficient Fe. A time-course analysis of the effects of a lack of Fe was conducted. The qRT-PCR data indicated that OsPRI1 expression in the roots and shoots was not responsive to changes in external Fe concentrations. In contrast, the expression of OsIRO2 (i.e. positive control) was considerably induced by Fe deficiency (Fig. 2, A and B).
Figure 2.
Expression of OsPRI1. A, Expression of root OsPRI1 in response to Fe deficiency. B, Expression of shoot OsPRI1 in response to Fe deficiency. A and B, 7-d-old seedlings grown in Fe-sufficient media were shifted to Fe-deficient media. Roots and shoots were sampled respectively at the indicated time and used for RNA extraction. OsIRO2 was used as a positive control. Error bars represent the sd; n = 3. C, Tissue-specific expression pattern of OsPRI1. Root, 7-d-old roots; Leaf, 7-d-old shoots; Culm, 80-d-old culm; Flb, flag leaf blades; Fls, flag leaf sheaths; Panicle, panicles at 1 to 2 cm length; Embryo, embryos at 10 to 15 d after pollination. Error bars represent the sd; n = 3.
We subsequently generated transgenic plants in which a 3-kb sequence upstream from the OsPRI1 translation start site was used to induce the expression of the GUS reporter gene. Unfortunately, no GUS staining was observed in plants regardless of Fe availability. Therefore, we examined the OsPRI1 expression patterns by qRT-PCR (Fig. 2C), which revealed that OsPRI1 was expressed in various tissues and organs. Except for the slightly lower expression level in the roots, OsPRI1 was similarly expressed in the leaves, culms, flag leaf blades and sheaths, panicles, and embryos.
Loss-of-Function Mutation to OsPRI1 Results in Enhanced Sensitivity to Iron Deficiency
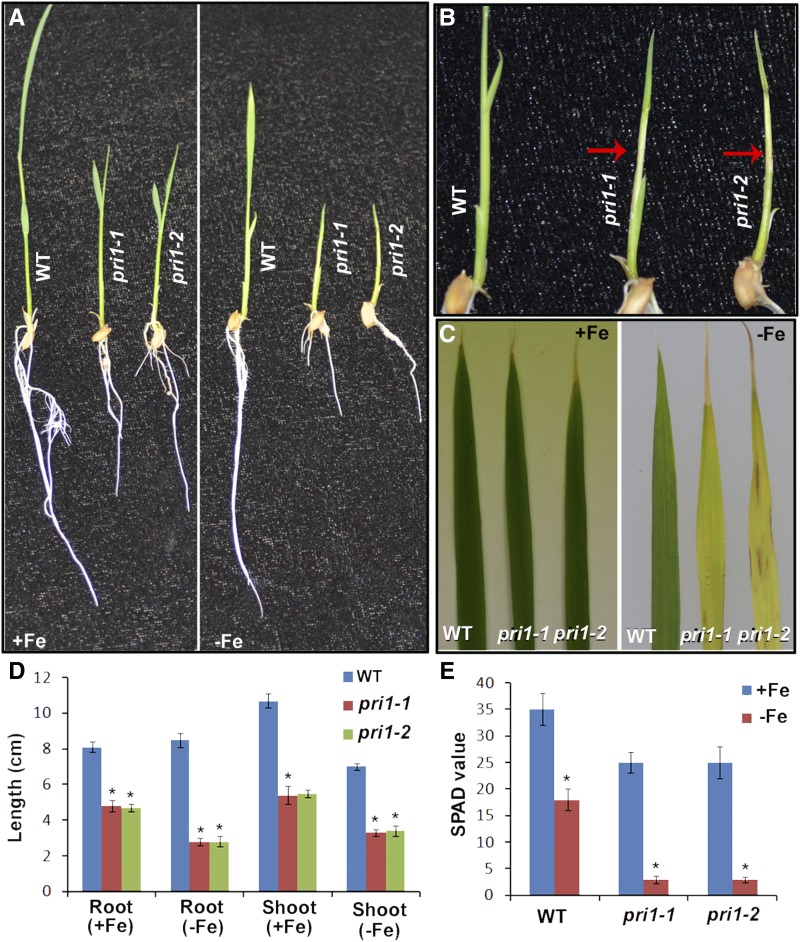
Having confirmed that OsPRI1 interacts with OsHRZ1, we investigated whether OsPRI1 influences Fe homeostasis in rice plants. We used CRISPR/Cas9 technology to construct OsPRI1 loss-of-function mutants. Two OsPRI1 gene sequences were targeted as mutation sites and respectively used in two independent rice transformations (Supplemental Fig. S1A). The resulting homozygous pri1 mutants (i.e. pri1-1 and pri1-2) were identified by sequencing. Both mutants were affected by a frame shift due to base deletion (Supplemental Fig. S1B), which resulted in the N-terminal truncated PRI1 proteins lacking partial basic helix-loop-helix (bHLH) domain (Supplemental Fig. S1C). To assess whether OsPRI1 affects Fe homeostasis in rice, plants were exposed to Fe-deficient conditions in a hydroponic system (Fig. 3A). The mutants developed severe chlorotic culm lesions (Fig. 3B) and yellow leaves with brown spots (Fig. 3C). The roots and shoots of the pri1 mutants were shorter than those of the wild-type control plants regardless of Fe availability. The wild-type shoots were shorter under Fe-deficient conditions than under Fe-sufficient conditions, whereas the wild-type roots grew similarly under both conditions. In contrast, the roots and shoots of both mutants were shorter under Fe-deficient conditions than under Fe-sufficient conditions (Fig. 3D). Consistent with these results, the leaf soil and plant analyzer development (SPAD) values of the pri1-1 and pri1-2 mutants were significantly lower than that of wild-type plants under Fe-deficient conditions (Fig. 3E). By simultaneously editing in two target sites of OsPRI1, we generated a pri1-3 mutant with a 223-bp deletion in the OsPRI1 genome and a 46-amino-acid deletion in the bHLH domain of OsPRI1 protein, and this mutant displayed phenotypes similar to those of pri1-1 and pri1-2 (Supplemental Fig. S1).
Figure 3.
Phenotypes of OsPRI1 loss-of-function mutants. A, Phenotypes of 10-d-old seedlings. 3-d-old seedlings germinated in wet paper were shifted Fe-sufficient or Fe-deficient hydroponic media for 7 d. B, Severe chlorotic culms. Magnification of the right side of A. The red arrows indicate the chlorotic culms. C, Phenotypes of the third leaf. 7-d-old seedlings grown in Fe-sufficient media were shifted to Fe-sufficient or Fe-deficient media for 7 d. The third leaf was presented. D, Length of roots and shoots. 3-d-old seedlings germinated in wet paper were shifted Fe-sufficient or Fe-deficient hydroponic media for 7 d. Error bars represent the sd; n = 3. The value which is significantly different from the corresponding wild-type (WT) value was indicated by * (P < 0.05), as determined by Student’s t test. E, SPAD value. 7-d-old seedlings grown in Fe-sufficient media were shifted to Fe-sufficient or Fe-deficient media for 7 d. The newest leaf was used for determination of SPAD value. Error bars represent the SD; n = 3. The value which is significantly different from the corresponding wild-type value was indicated by * (P < 0.05), as determined by Student’s t test.
Loss-of-Function Mutation to OsPRI1 Impairs Iron Translocation from the Roots to the Shoots
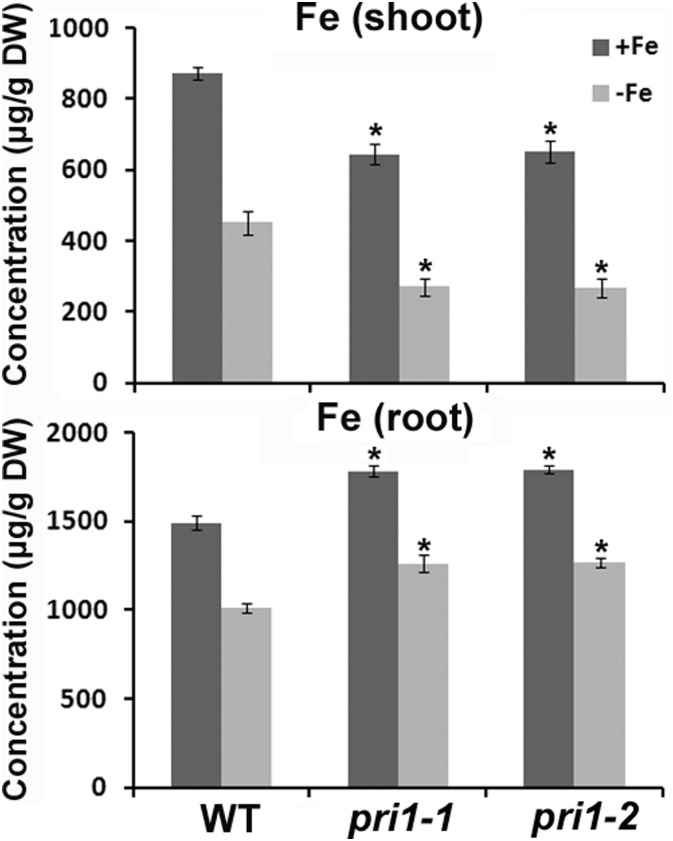
To investigate the mechanism underlying the enhanced sensitivity of OsPRI1 loss-of-function mutants to Fe deficiency, we measured the metal concentrations in pri1 mutants. Two-week-old seedlings grown in Fe-sufficient hydroponic culture medium were transferred to Fe-sufficient or Fe-deficient medium and incubated for 1 week. The shoots and roots were harvested separately for a subsequent analysis of metal concentrations. The shoot Fe concentrations were lower in the mutants than in the wild-type plants regardless of external Fe availability. In contrast, the root Fe concentrations were higher in the mutants than in the wild-type controls (Fig. 4). These data suggest that Fe translocation from the roots to the shoots was impaired in the pri1 mutants.
Figure 4.
Fe accumulation in the pri1 mutants. Two-week-old seedlings grown in Fe-sufficient media were transferred to Fe-sufficient or Fe-deficient media for 1 week. Shoots and roots were separately sampled and used for metal measurement. Error bars represent the sd; n = 3. The value which is significantly different from the corresponding wild-type (WT) value was indicated by * (P < 0.05), as determined by Student’s t test. DW, Dry weight.
Loss-of-Function Mutation to OsPRI1 Down-Regulates the Expression of Fe-Deficiency-Responsive Genes
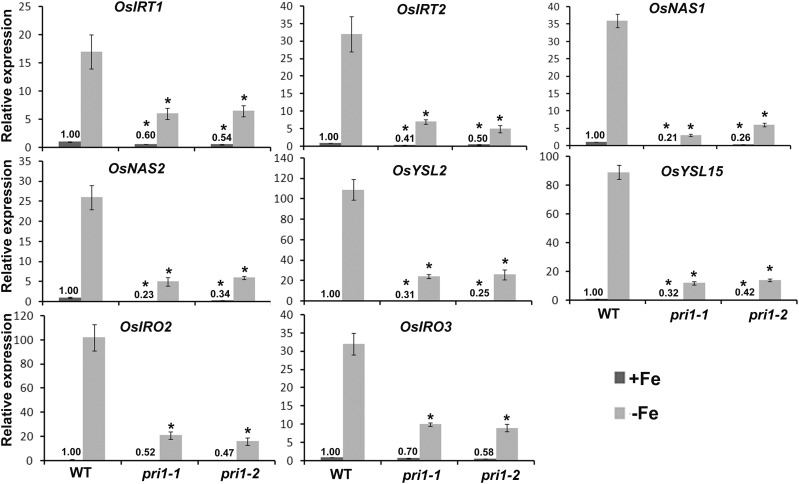
Plants can sense fluctuations in external and internal Fe contents and adjust the expression of Fe-uptake-associated genes accordingly. Considering that a loss-of-function mutation to OsPRI1 affected the sensitivity of plants to Fe deficiency and the Fe translocation from the roots to the shoots, we investigated whether the expression of Fe-deficiency-responsive genes was also affected in the pri1 mutants. To examine the effect of OsPRI1 on the regulation of Fe-deficiency-responsive genes, several representative genes were analyzed by qRT-PCR (Fig. 5). The expression of two Fe-uptake-associated genes (i.e. OsIRT1 and OsIRT2), whose transcript abundance is up-regulated by Fe deficiency (Bughio et al., 2002; Ishimaru et al., 2006), was suppressed in the pri1 mutants. Additionally, the up-regulated expression of two nicotianamine synthase genes (i.e. OsNAS1 and OsNAS2) induced by Fe deficiency (Inoue et al., 2003) was considerably inhibited by the loss-of-function mutation to OsPRI1. OsYSL2 and OsYSL15, which are responsible for the distribution of Fe in rice plants (Koike et al., 2004; Inoue et al., 2009; Lee et al., 2009; Ishimaru et al., 2010), were expressed at lower levels in the pri1 mutants than in the wild-type controls. The expression levels of OsIRO2 and OsIRO3, which encode two crucial regulators of Fe homeostasis in rice, are up-regulated by Fe deficiency (Ogo et al., 2006, 2007; Zheng et al., 2010). The expression levels of these two genes were down-regulated in the pri1 mutants. Additionally, we examined the expression of other Fe-deficiency-responsive genes, namely MITOCHONDRIAL IRON-REGULATED, NATURAL RESISTANCE-ASSOCIATED MACROPHAGE PROTEINS1 (OsNRAMP1), and TRANSPORTER OF MUGINEIC ACID FAMILY PHYTOSIDEROPHORES1 (OsTOM1; Ishimaru et al., 2009; Nozoye et al., 2011; Takahashi et al., 2011). The expression of these genes was suppressed in the pri1 mutants (Supplemental Fig. S2). These data suggest that OsPRI1 affects the transcriptional regulatory network associated with rice responses to Fe deficiency.
Figure 5.
Expression of Fe-deficiency-responsive genes in the pri1 mutants. Seven-day-old seedlings grown in Fe-sufficient media were transferred to Fe-sufficient or Fe-deficient media for 7 d. Roots were sampled and used for RNA extraction. The numbers above the bars indicate the corresponding mean values. Error bars represent the sd; n = 3. The value which is significantly different from the corresponding wild-type (WT) value was indicated by * (P < 0.05), as determined by Student’s t test.
OsPRI1 Binds to the Promoters of OsIRO2 and OsIRO3
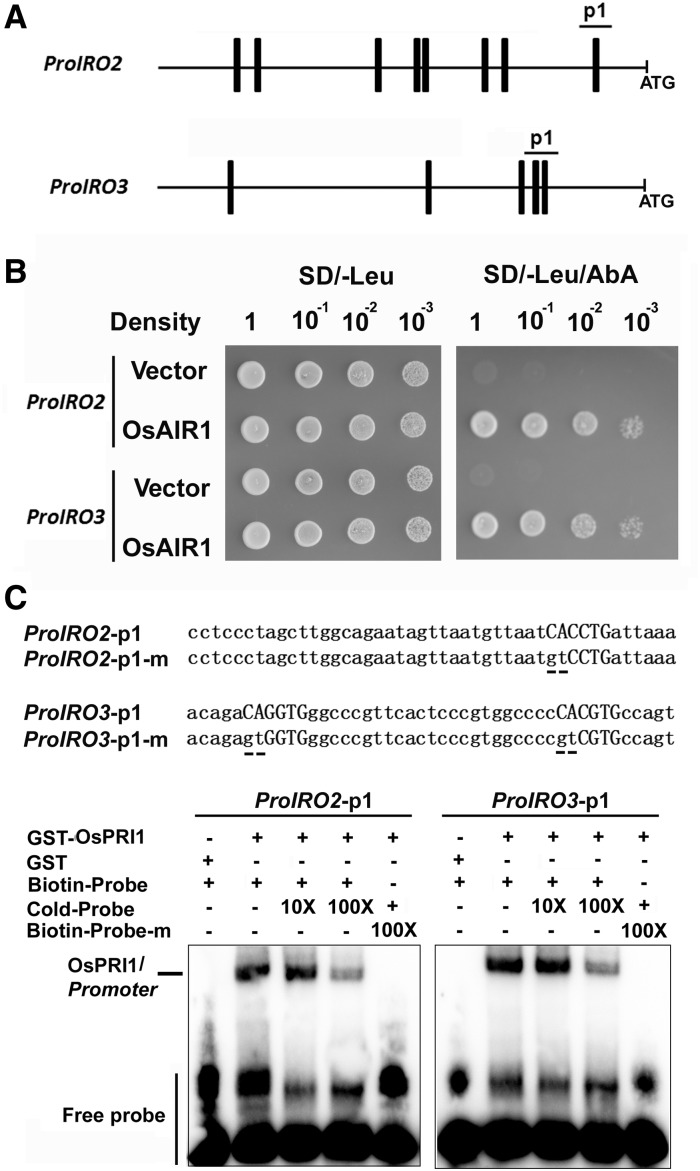
Previous reports confirmed that OsIRO2 and OsIRO3 function upstream of the Fe-deficiency-responsive genes OsIRT1, OsNRAMP1, OsNAS1, OsNAS2, and OsYSL15 to mediate Fe homeostasis in rice (Ogo et al., 2007; Zheng et al., 2010). Given that OsPRI1 positively regulates the expression of OsIRO2 and OsIRO3, we tested whether OsPRI1 directly binds and activates the promoters of OsIRO2 and OsIRO3. OsPRI1 belongs to the bHLH transcription factor family whose members often bind to the E-box in the promoters of their target genes (Fisher and Goding, 1992). We analyzed the 1.5-kb sequences upstream of the OsIRO2 and OsIRO3 translation start sites and detected several E-box motifs (Fig. 6A). Subsequent yeast one-hybrid assays were used to investigate whether OsPRI1 can bind to the promoters of OsIRO2 and OsIRO3 (Fig. 6B). We observed that OsPRI1 was able to bind to the p1 fragment in the OsIRO2 and OsIRO3 promoters. We conducted an EMSA with the GST-OsPRI1 recombinant protein purified from E. coli to verify the direct interaction between OsPRI1 and the OsIRO2 and OsIRO3 promoters. When the OsIRO2 promoter p1 fragment was used as a probe, a DNA-protein complex was detected. The abundance of this complex decreased following the addition of unlabeled wild-type competitors. In contrast, the GST-OsPRI1 protein did not bind to the mutant probe carrying a mutated E-box (Fig. 6C). The GST protein alone (i.e. negative control) also did not bind to the OsIRO2 promoter. A similar result was obtained when the OsIRO3 promoter p1 fragment was used as the probe. These data suggest that OsPRI1 binds to the promoters of OsIRO2 and OsIRO3.
Figure 6.
OsPRI1 binds to the promoters of OsIRO2 and OsIRO3. A, E-boxes in the promoter. The bar indicates the position of E-box in the 1.5-kb sequence from the translation start site of OsIRO2 and OsIRO3. B, Yeast one-hybrid assays. The p1 sequence indicated in A was used as bait and OsPRI1 as prey. The representative growth status of yeast cells is shown on synthetic dextrose medium agar plates without Leu (SD/−Leu) with or without AbA. The AbA resistance was activated by prey proteins that specifically interact with the bait sequence. C, EMSA assays. The oligonucleotides (OsIRO2/3-1p and OsIRO2/3-1p-m) were used as the probes. The underlined base indicates the mutated base. Each biotin-labeled DNA probe was incubated with the recombinant GST-OsPRI1 protein. An excess of unlabeled probe (Cold-Probe) or labeled mutated probe (Biotin-mProbe) was added to compete with labeled probe (Biotin-Probe). Biotin-probe incubated with GST protein served as the negative control.
OsPRI1 Is Ubiquitinated and Degraded by OsHRZ1
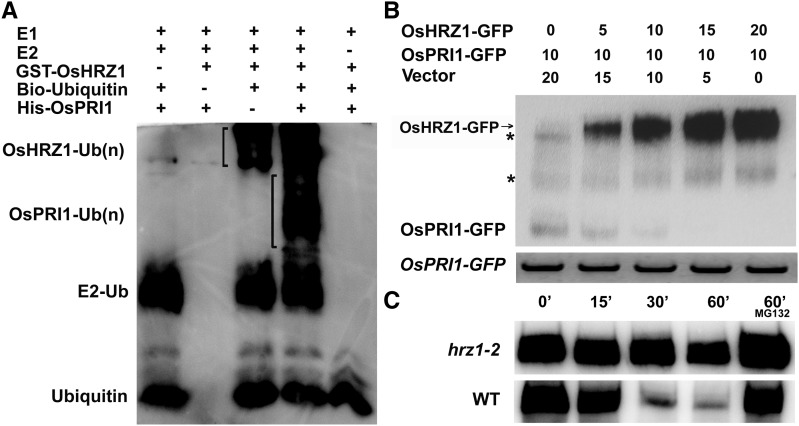
Considering OsHRZ1 exhibits E3 ligase activity and interacts with OsPRI1, we speculated that OsPRI1 is ubiquitinated by OsHRZ1. We produced GST-OsHRZ1 and His-OsPRI1 in E. coli cells and purified the fusion proteins. A subsequent in vitro ubiquitination assay indicated that the ubiquitination of OsPRI1 occurred in the presence of E1, E2, OsHRZ1, and Ubiquitin (Ub), but not if E2, OsHRZ1, or Ub were missing (Fig. 7A). These results suggest that OsHRZ1 can regulate the ubiquitination of OsPRI1.
Figure 7.
OsHRZ1 is required for ubiquitination and degradation of OsPRI1. A, Ubiquitination of OsPRI1 by OsHRZ1. E. coli-purified GST-tagged OsHRZ1 and His-tagged OsPRI1 proteins were subjected to in vitro ubiquitination assay containing ATP, biotin-tagged ubiquitin (Ub), and human E1 and E2 (UbcH5c). Proteins were immunodetected using the HRP-Streptavidin detection system. B, Degradation of OsPRI1 by OsHRZ1. Degradation of OsPRI1 was carried out by detecting the OsPRI1-GFP protein level in coinfiltration experiments with increasing amounts of OsHRZ1-GFP. GFP antibody was used. Numbers indicate the ratio of the concentrations of agrobacteria used in coinfiltration. OsPRI1-GFP mRNA expression levels were analyzed by RT-PCR. The asterisk indicates a nonspecific signal. C, Cell-free degradation. 7-d-old roots grown in Fe-sufficient media were harvested and used for protein extraction. Incubation with or without MG132 was performed over the indicated time course. WT, Wild type.
Transient expression assays were conducted to investigate whether OsHRZ1 can mediate the degradation of OsPRI1. We coinfiltrated Nicotiana benthamiana leaves with agrobacterial host constructs expressing OsHRZ1-GFP and OsPRI1-GFP together with controls. Samples were harvested to analyze the protein and RNA levels for the transfected constructs. The qRT-PCR data indicated that OsPRI1-GFP was expressed at consistent levels regardless of the coinfiltration treatment. A western blot revealed that the OsPRI1 protein abundance decreased with increasing OsHRZ1-GFP contents (Fig. 7B), suggesting that OsPRI1 protein abundance is negatively correlated with OsHRZ1.
We then assessed whether OsPRI1 is stable in an OsHRZ1 loss-of-function mutant. The hrz1-2 mutant was produced using CRISPR/Cas9 technology (Supplemental Fig. S3, A and B). The insertion of a nucleotide resulted in the deletion of CHY-type zinc-finger domain, RING zinc-finger domain, and Rubredoxin fold in the hrz1-2 mutant (Supplemental Fig. S3C). Similar to the hrz1-1 mutant (Kobayashi et al., 2013), hrz1-2 mutant also displayed low fertility. We found that hrz1-2 mutant was more tolerant to Fe deficiency than the wild-type control (Supplemental Fig. S3D). The GST-OsPRI1 fusion protein was incubated in the root extracts from the hrz1-2 mutant and wild-type plants. In contrast to the considerable decrease in the wild-type extracts, the abundance of GST-OsPRI1 only slightly decreased in the hrz1-2 mutant extracts (Fig. 7C). Collectively, these data suggest that OsHRZ1 mediates the ubiquitination and degradation of OsPRI1.
Loss-of-Function Mutation to OsPRI1 Partially Rescues the hrz1-2 Mutant Phenotype
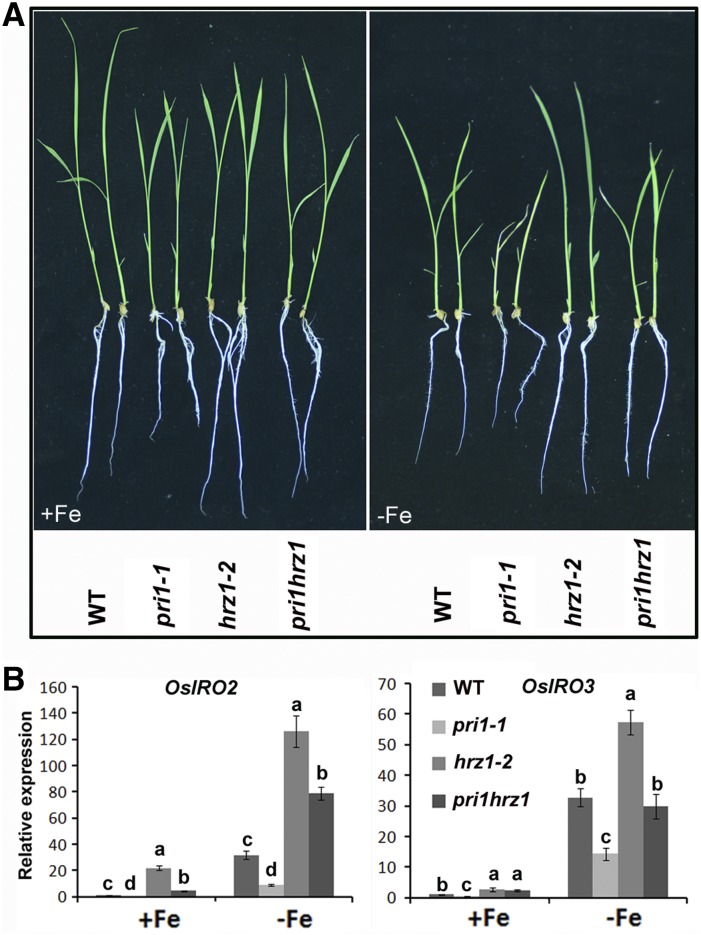
Considering OsPRI1 is negatively regulated by OsHRZ1, we speculated that the enhanced tolerance of the hrz1-2 mutant to Fe deficiency results from the increased stability of OsPRI1. Therefore, it is likely that a loss-of-function mutation to OsPRI1 would rescue the hrz1-2 mutant phenotype. To confirm this hypothesis, double mutants for both loss-of-function of OsHRZ1 and OsPRI1 were generated using CRISPR/Cas9 technology. A double mutant pri1hrz1 (Supplemental Fig. S4, A and B) in which the genotype of OsPRI1 and OsHRZ1 are identical to that of pri1-1 and hrz1-2, respectively, was selected for Fe-deficiency tolerance assays under hydroponic conditions. Seven-day-old seedlings grown under Fe-sufficient conditions were exposed to Fe-deficient or Fe-sufficient conditions for 7 d (Fig. 8A). Based on root lengths and leaf SPAD values (Supplemental Fig. S4, C and D), the pri1hrz1 double mutant was more tolerant to Fe deficiency than the pri1-1 mutant. However, it was more sensitive to Fe-deficient conditions than the hrz1-2 mutant, suggesting that a loss-of-function mutation to OsPRI1 rescues the hrz1-2 mutant phenotype.
Figure 8.
Genetic relationship between OsPRI1 and OsHRZ1. A, Two-week-old plants. Seven-day-old seedlings grown in Fe-sufficient media were transferred to Fe-sufficient or Fe-deficient media for 7 d. B, Relative expression of OsIRO2 and OsIRO3. Seven-day-old seedlings grown in Fe-sufficient media were transferred to Fe-sufficient or Fe-deficient media for 7 d. Roots were sampled and used for RNA extraction. Error bars represent the sd; n = 3. Different letters above columns indicate significant differences based on Tukey’s test (P < 0.05). WT, Wild type.
We assessed whether the difference in the susceptibilities of the pri1hrz1 double mutant and the hrz1-2 mutant to Fe deficiency resulted from altered expression of Fe-deficiency-responsive genes. The gene expression levels were analyzed in plants exposed to Fe-sufficient or Fe-deficient conditions for 7 d (Fig. 8B). Under Fe-sufficient conditions, OsIRO2 expression was up-regulated more in the hrz1-2 mutant than in the pri1hrz1 double mutant. Similar results were obtained under Fe-deficient conditions. In contrast, OsIRO3 expression was up-regulated more in the hrz1-2 mutant than in the pri1hrz1 double mutant only under Fe-deficient conditions. We also determined that OsIRT1, OsIRT2, OsNAS1, OsNAS2, OsYSL2, and OsYSL15 expression levels were lower in the pri1hrz1 double mutant than in the hrz1-2 mutant regardless of Fe availability (Supplemental Fig. S5). Therefore, a loss-of-function mutation to OsPRI1 suppressed the up-regulated expression of Fe-deficiency-responsive genes caused by a loss-of-function mutation to OsHRZ1. Collectively, these data indicate that OsPRI1 functions downstream of OsHRZ1 to maintain Fe homeostasis in rice plants.
DISCUSSION
OsPRI1 Induces Rice Responses to Fe Deficiency
The OsIRO2 and OsIRO3 bHLH transcription factors are important for regulating Fe homeostasis (Ogo et al., 2007; Zheng et al., 2010). In this study, we observed that a novel bHLH transcription factor is the interacting partner of the putative iron-binding sensor OsHRZ1. In contrast to the significant induction of OsIRO2 and OsIRO3 expression under Fe-deficient conditions, OsPRI1 was unaffected by changes in external Fe contents (Fig. 2A). Expression analyses revealed that the up-regulated expression of Fe-deficiency-responsive genes induced by limited Fe abundance was strongly suppressed by a loss-of-function mutation to OsPRI1 (Fig. 5). Under Fe-sufficient conditions, the loss-of-function of OsPRI1 resulted in the production of short plants (Fig. 3A). Additionally, under Fe-deficient conditions, the pri1 mutants exhibited more severe Fe-deficiency symptoms compared with the wild-type plants (Fig. 3, B, C, and E). These results imply that OsPRI1 is required for Fe homeostasis under both Fe-sufficient and Fe-deficient conditions. The Fe concentration of pri1 shoots was always lower than that of wild-type shoots (Fig. 4), which may explain the short shoot phenotype of pri1 mutants. Although the shoot Fe concentrations are low in the pri1 mutants, the root Fe concentrations were high, suggesting the Fe translocation from the roots to the shoots was disrupted. Therefore, OsPRI1 is a positive regulator of Fe homeostasis and is required for the translocation of Fe from rice roots to the shoots.
It is noteworthy that the expression of Strategy I and Strategy II genes was significantly inhibited in the roots of pri1 mutants regardless of Fe availability (Fig. 5). This observation appears to be inconsistent with the increased Fe concentrations in the roots of pri1 mutants (Fig. 4). OsYSL2 transports Fe from the roots to the shoots, and its loss-of-function resulted in high root Fe contents and low shoot Fe levels (Ishimaru et al., 2010). The expression of OsYSL2 was considerably down-regulated in the pri1 mutants (Fig. 5), which may have contributed to the impaired Fe translocation in these mutants. Additionally, the production of OsYSL15, which is responsible for Fe uptake and distribution (Inoue et al., 2009; Lee et al., 2009), was also repressed in the pri1 mutants. This partially explains the inhibited Fe translocation from the roots to the shoots. We noted that the Fe concentrations of pri1 mutants were significantly higher under Fe-sufficient conditions than under Fe-deficient conditions (Fig. 4), implying that a loss-of-function mutation to OsPRI1 does not completely block the Fe-uptake system in the roots. We speculated that the Fe-deficiency signals originating from the shoots of the pri1 mutants caused the roots to absorb more Fe, which was retained in the roots because of the inefficient Fe translocation from the roots to the shoots. This may be the reason for the elevated Fe concentration in the roots of pri1 mutants.
OsIRO2 and OsIRO3 Are the Target Genes of OsPRI1
OsIRO2 encodes a bHLH protein that positively regulates Fe homeostasis (Ogo et al., 2007). In contrast, OsIRO3 is a bHLH protein that negatively regulates Fe homeostasis (Zheng et al., 2010). The expression levels of OsIRO2 and OsIRO3 are strongly up-regulated by Fe deficiency in rice (Ogo et al., 2006; Zheng et al., 2010). In this study, a loss-of-function mutation to OsPRI1 down-regulated the expression of OsIRO2 and OsIRO3 (Fig. 5), suggesting that OsPRI1 is a positive regulator of OsIRO2 and OsIRO3 expression. The bHLH transcription factors are known to bind to the E-box of their target promoters. The yeast one-hybrid and EMSA experiments (Fig. 6, B and C) confirmed that OsPRI1 was able to bind to the E-box sequences in the OsIRO2 and OsIRO3 promoters, suggesting that they are the direct targets of OsPRI1. The expression of Fe-acquisition-associated genes (i.e. OsNAS1/2 and OsYSL15) was positively regulated by OsIRO2. Additionally, their expression levels decreased in the pri1 mutants (Fig. 5). These results provide evidence that OsPRI1 activates the expression of Fe-acquisition-associated genes under Fe-deficient conditions by directly up-regulating OsIRO2 expression. It is noteworthy that OsIRO3 negatively regulates OsIRO2 expression (Zheng et al., 2010). Moreover, our findings confirm that OsIRO2 and OsIRO3 expression levels are positively regulated by OsPRI1 (Fig. 5), which suggests that rice has evolved a sophisticated mechanism to maintain Fe homeostasis. The antagonistic effects of OsIRO2 and OsIRO3 may be necessary for rice to avoid the adverse effects of Fe deficiency or toxicity and to maintain Fe homeostasis. In addition, we also observed that the expression of OsYSL2, MITOCHONDRIAL IRON-REGULATED, OsIRT2, TRANSPORTER OF MUGINEIC ACID FAMILY PHYTOSIDEROPHORES1, and OsNRAMP1 was down-regulated in the pri1 mutants (Fig. 5; Supplemental Fig. S2). However, no evidence suggests that these genes are regulated by OsIRO2 or OsIRO3. Therefore, it is possible that they are directly regulated by OsPRI1 or other transcription factors targeted by OsPRI1.
Recently, CmbHLH1 and MdbHLH104 were characterized as positive regulators of Fe homeostasis in chrysanthemum and apple, respectively (Zhao et al., 2014, 2016b). In Arabidopsis, four genes (i.e. bHLH34/104/105/115) belonging to the bHLH IVc group encode positive regulators of bHLH38/39/100/101 and POPEYE expression (Zhang et al., 2015; Li et al., 2016; Liang et al., 2017). Comparative analyses suggested that the OsPRI1 amino acid sequence is similar to that of CmbHLH1, MdbHLH104, and bHLH34/104/105/115 (Supplemental Fig. S6), implying these proteins are functionally conserved across plants species. Rice contains three close homologs of OsPRI1 (Zheng et al., 2010), and we also observed that the expression of OsIRO2 and OsIRO3 was still induced by Fe deficiency in the pri1 mutants despite their low transcript abundance. Therefore, functional redundancy may exist among these homologous genes. The CmbHLH1 and MdbHLH104 transcript levels reportedly increase under Fe-deficient conditions, which is in contrast to the unaffected OsPRI1 and bHLH34/104/105/115 transcript abundances under the same conditions. These differences suggest there is some diversity in the associated gene expression regulatory activities. It is important to note that CmbHLH11 and MdbHLH104 enhance plant tolerance to Fe-deficient conditions by activating the expression of AHA genes, while MdbHLH104 also directly targets the MdAHA8 gene (Zhao et al., 2014; Zhao et al., 2016b). Additionally, OsPRI1 positively regulates the expression of many Fe-deficiency-responsive genes. Therefore, determining whether OsPRI1 also directly targets these genes may provide insights into the complex Fe-deficiency-responsive mechanism.
OsPRI1 Is Ubiquitinated by OsHRZ1
Although the OsPRI1 transcript abundance was unaffected by Fe deficiency, the downstream target genes were up-regulated by Fe-deficient conditions. Moreover, the up-regulated expression of Fe-deficiency-responsive genes induced by Fe deficiency was dependent on OsPRI1. These results imply that OsPRI1 might be regulated at the posttranscriptional level under Fe-deficient conditions (e.g. altered OsPRI1 protein abundance or activity). Our data suggested that OsHRZ1 interacts with and ubiquitinates OsPRI1 (Figs. 1 and 7A), that OsPRI1 abundance is negatively correlated with OsHRZ1 content (Fig. 7B), and that OsPRI1 is more stable in the hrz1-2 mutants than in the wild-type controls (Fig. 7C). These data suggest that OsHRZ1 mediates the degradation of OsPRI1, supporting the fact that OsHRZ1 is a negative regulator of plant responses to Fe deficiency.
Considering the fact that OsHRZ1 can bind to Fe, it is likely that OsHRZ1 activity is modulated by Fe concentrations, thereby influencing the degradation of OsPRI1 in rice. In addition to the RING-Zn-finger domain, OsHRZ1 also contains CHY- and CTCHY-Zn-finger sequences that may mediate transcriptional, posttranscriptional, or posttranslational modifications (Gamsjaeger et al., 2007). Therefore, we cannot exclude the possibility that changes in OsHRZ1 activity also affect OsPRI1 functions via protein interactions.
Several iron-binding sensors have been identified in other species. For example, FBXL5, which contains an iron-binding HHE domain, promotes the Fe-dependent degradation of IRON REGULATORY PROTEIN2 under Fe-sufficient conditions in humans (Salahudeen et al., 2009; Vashisht et al., 2009). In apple, MdBT1 and MdBT2, which contain the HHE domain, mediate the ubiquitination and degradation of MdbHLH104 under Fe-sufficient conditions (Zhao et al., 2016a). Additionally, an Arabidopsis homolog of OsHRZ1 (i.e. BTS) is required for the degradation of AtbHLH105 and AtbHLH115 (Selote et al., 2015). In this study, we suggested that OsHRZ1 is responsible for the proteolysis of OsPRI1. Moreover, the OsPRI1 amino acid sequence is highly similar to the apple MdbHLH104 and Arabidopsis AtbHLH105/115 sequences (Supplemental Fig. S6). These results imply that the associated ubiquitination and degradation are part of a conserved regulatory mechanism related to Fe homeostasis. This mechanism is especially conserved in higher plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Cultivar ‘Nipponbare’ was used in the study. Plants were grown in a greenhouse with a photoperiod of 14 h light and 10 h dark at 28°C. For hydroponic culture assays, Fe-sufficient media was prepared in half-strength Murashige and Skoog media with 0.1mm Fe(III)-EDTA and Fe-deficient media in half-strength Murashige and Skoog media without Fe. SPAD values of the newest leaf were measured in the 2-week-old plants after 1-week Fe-deficient stress conditions.
Genome Editing by the CRISPR/Cas9 System
For the editing of OsPRI1 or OsHRZ1, the single target site was inserted between OsU6a and sgRNA in the pYLsgRNA-OsU6a vector (Ma et al., 2015), and then the OsU6a-sgRNA cassette was cloned into the pMH-SA vector by the restriction enzyme sites SpeI and AscI (Liang et al., 2016). For the double editing of OsPRI1, a tandem combination of OsU6a-sgRNA and OsU6b-sgRNA was cloned between AscI and SpeI in the pMH-SA vector. For the double editing of OsPRI1 and OsHRZ1, a tandem combination of OsU6a-sgRNA (for OsPRI1) and OsU6b-sgRNA (for OsHRZ1) was cloned between AscI and SpeI in the pMH-SA vector. Homozygous mutant lines were identified by sequencing.
Yeast Assays
For the yeast-one-hybrid assay, the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech) was used. The pAbAi vector harboring the p1 fragment of OsIRO2 or OsIRO3 promoter was integrated into the genome of yeast strain Y1HGold followed by selection on synthetic dextrose medium agar plates without uracil. Growth was determined on the synthetic dextrose medium without Leu supplemented with 300 ng/mL aureobasidin A (AbA). The AbA resistance was activated by prey proteins that specifically interact with the bait sequence.
For the yeast two-hybrid assay, the C-terminal truncated OsHRZ1-C was cloned into pGBKT7, and the full-length open reading frame of OsPRI1 was cloned into pGADT7. Growth was determined as described in the Yeast Two-Hybrid System User Manual (Clontech).
Transient Expression Assays in Tobacco
Agrobacterium tumefaciens strains EHA105 was used in the transient expression experiments. Infiltration assays were performed as described previously (Li et al., 2016). The empty vector pOCA30 was used for construction of 35S:OsHRZ1-GFP and 35S:OsPRI1-GFP. In brief, the strains were first plated on Luria-Bertani (LB) medium containing the appropriate selection antibiotics. After 2 to 3 d, a single colony was inoculated into 5 mL LB medium supplemented with the appropriate antibiotics and grown at 28°C in a shaker for 48 h. The culture was transferred to new 50 mL LB medium with the appropriate antibiotics. When growth reached an OD600 of approximately 3.0, the bacteria were spun down gently (3200g, 5 min), and the pellets were resuspended in infiltration buffer (10 mm MgCl2, 10 mm MES, pH 5.6) at a final OD600 of 1.5. A final concentration of 0.2 mm acetosyringone was added, and the bacteria were kept at room temperature for at least 2 h without shaking. For coinfiltration, different agrobacterium strains carrying different constructs were mixed prior to infiltration. Leaf infiltration was conducted in 3-week-old Nicotiana benthamiana by depressing a 1-mL disposable syringe to the surface of fully expanded leaves. Protein extraction and immunoblot were conducted as described previously (Liu et al., 2010). Anti-GFP antibody (Affinity Biosciences) was used.
Pull-Down Assays
Full-length OsHRZ1 and OsPRI1 were cloned into pGEX-4T-1 and pET-28a (+), respectively. All plasmids were introduced into Escherichia coli BL21 cells (TransGen Biotech). GST, GST-OsHRZ1, and His-OsPRI1 protein expression was induced by 0.1 mm isopropyl-b-thiogalactopyranoside at 16°C for 24h. Soluble GST and GST-OsHRZ1 were extracted and immobilized to glutathione affinity resin (Thermo Fisher Scientific). For pull-down assays, His-OsPRI1 fusion protein from E. coli cell lysate was incubated with the immobilized GST and GST-OsHRZ1 proteins in phosphate-buffered saline buffer (50 mm Tris-HCl, pH 7.2, 150 mm NaCl, 10% glycerol, 0.1% Triton X-100, 1× protease inhibitor cocktail) for 2 h at 4°C. Proteins were eluted in the elution buffer, and the interaction was determined by western blot using anti-His antibody and anti-GST antibody (TransGen Biotech).
EMSA Assays
The EMSA was conducted using a Chemiluminescent EMSA Kit (Beyotime) following the manufacturer’s protocol. The recombinant GST-OsPRI1 protein and GST protein were purified from E. coli. The DNA fragments of the OsIRO2 and OsIRO3 promoters (Fig. 6C) were synthesized and biotin was labeled to the 5′ terminal of DNA. Biotin-unlabeled fragments of the same sequences or mutated sequences were used as competitors, and the GST protein alone was used as the negative control.
Ubiquitination Assay
Ubiquitination assays were carried out using a ubiquitinylation kit (ENZO Life Sciences). In brief, E. coli- purified GST-OsHRZ1 (E3) and His-OsPRI1 (substrate) were incubated with 1× ubiquitinylation buffer, 5mm Mg-ATP solution, 1 mm DTT, 100 nm human E1, 2.5μm human E2 (Ubiquitin conjugating enzyme H5c), 2.5 μm Biotin-ubiquitin, and 1 unit of inorganic pyrophosphatase (New England Biolabs) at 37°C for 4 h. Proteins were separated on a 15% (w/v) sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel under nonreducing conditions. Immunodetection was carried out using the HRP-Streptavidin detection system described in the product manual.
Protein Extraction and Cell-Free Degradation
Rice roots were harvested and ground into fine powder in liquid nitrogen. Total proteins were subsequently extracted in a degradation buffer, which consists of 25 mm Tris-HCl, pH 7.5, 10 mm NaCl, 10 mm MgCl2, 4mM phenylmethanesulfonyl fluoride (PMSF), 5mM dithiothreitol, and 10 mm ATP as previously described (Wang et al., 2009). Cell debris was removed by two 10-min centrifugations at 17,000g in 4°C. The supernatant was collected, and protein concentration was determined by the Bio-Rad protein assay. 40 μm MG132 was selectively added to various in vitro degradation assays as indicated. For E. coli-purified GST-OsPRI1 protein degradation, 100 ng of recombinant GST-OsPRI1 protein was incubated in 0.1 mL extracts (containing 500 mg total proteins) for the individual assays. All the mock controls used an equal amount of solvents for each drug. The extracts were incubated at 28°C, samples were taken at indicated intervals for determination of GST-OsPRI1 protein abundance by immunoblots.
Fe Concentration Measurement
To determine Fe content, 7-d-old seedlings grown on Fe-sufficient medium were transferred to Fe-sufficient or Fe-deficient medium for 7 d. The shoots and roots were harvested separately and used for Fe measurement. About 100 mg dry weight for roots or shoots was used as one sample and three samples were used in each independent experiment. Fe content analysis was performed using inductively coupled plasma spectroscopy.
Gene Expression Analysis
One microgram of total RNA extracted using the Trizol reagent (Invitrogen) was used for oligo(dT)18-primed cDNA synthesis according to the reverse transcription protocol (Takara). The resulting cDNA was subjected to relative quantitative PCR using the SYBR Premix Ex Taq kit (TaKaRa) on a Roche LightCycler 480 real-time PCR machine, according to the manufacturer’s instructions. OsACTIN1 was amplified as an internal control, and gene copy number was normalized to that of OsACTIN1. For the quantification of each gene, at least three biological replicates were used. The analysis of statistical significance was performed by Student’s t test or Tukey’s test. The quantitative reverse transcription-PCR primers are listed in Supplemental Table S2.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers OsPRI1 (Os08g0138500), OsHRZ1 (Os01g0689451), OsIRO2 (Os01g0952800), OsIRO3 (Os03g0379300), OsIRT1 (Os03g0667500), OsIRT2 (Os03g0667300), OsNAS1 (Os03g0307300), OsNAS2 (Os03g0307200), OsYSL2 (Os02g0649900), and OsYSL15 (Os02g0650300).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Gene editing of OsPRI1.
Supplemental Figure S2. Expression of Fe-deficiency-responsive genes in the pri1 mutants.
Supplemental Figure S3. Gene editing of OsHRZ1.
Supplemental Figure S4. Phenotypes of pri1hrz1 mutant.
Supplemental Figure S5. Expression of Fe-deficiency-responsive genes in wild-type, pri1-1, hrz1-2, and pri1hrz1.
Supplemental Figure S6. Comparative analysis of OsPRI1 homologous genes in various plant species.
Supplemental Table S1. Candidate positive clones in the Y2H screening.
Supplemental Table S2. Primers used in this paper.
Glossary
- SPAD
soil and plant analyzer development
Footnotes
This work was supported by the Applied Basic Research Project of Yunnan Province (2017FB026), the Youth Innovation Promotion Association of the Chinese Academy of Sciences, Candidates of the Young and Middle Aged Academic Leaders of Yunnan Province (2015HB095), and the program for Innovative Research Team of Yunnan Province (2014HC017).
References
- Balk J, Schaedler TA (2014) Iron cofactor assembly in plants. Annu Rev Plant Biol 65: 125–153 [DOI] [PubMed] [Google Scholar]
- Bashir K, Nozoye T, Nagasaka S, Rasheed S, Miyauchi N, Seki M, Nakanishi H, Nishizawa NK (2017) Paralogs and mutants show that one DMA synthase functions in iron homeostasis in rice. J Exp Bot 68: 1785–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bughio N, Yamaguchi H, Nishizawa NK, Nakanishi H, Mori S (2002) Cloning an iron-regulated metal transporter from rice. J Exp Bot 53: 1677–1682 [DOI] [PubMed] [Google Scholar]
- Cheng L, Wang F, Shou H, Huang F, Zheng L, He F, Li J, Zhao FJ, Ueno D, Ma JF, et al. (2007) Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiol 145: 1647–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher F, Goding CR (1992) Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J 11: 4103–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP (2007) Sticky fingers: Zinc-fingers as protein-recognition motifs. Trends Biochem Sci 32: 63–70 [DOI] [PubMed] [Google Scholar]
- Hell R, Stephan UW (2003) Iron uptake, trafficking and homeostasis in plants. Planta 216: 541–551 [DOI] [PubMed] [Google Scholar]
- Henriques R, Jásik J, Klein M, Martinoia E, Feller U, Schell J, Pais MS, Koncz C (2002) Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol Biol 50: 587–597 [DOI] [PubMed] [Google Scholar]
- Hindt MN, Guerinot ML (2012) Getting a sense for signals: Regulation of the plant iron deficiency response. Biochim Biophys Acta 1823: 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Higuchi K, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2003) Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J 36: 366–381 [DOI] [PubMed] [Google Scholar]
- Inoue H, Kobayashi T, Nozoye T, Takahashi M, Kakei Y, Suzuki K, Nakazono M, Nakanishi H, Mori S, Nishizawa NK (2009) Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J Biol Chem 284: 3470–3479 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Bashir K, Fujimoto M, An G, Itai RN, Tsutsumi N, Nakanishi H, Nishizawa NK (2009) Rice-specific mitochondrial iron-regulated gene (MIR) plays an important role in iron homeostasis. Mol Plant 2: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Masuda H, Bashir K, Inoue H, Tsukamoto T, Takahashi M, Nakanishi H, Aoki N, Hirose T, Ohsugi R, et al. (2010) Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J 62: 379–390 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, et al. (2006) Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J 45: 335–346 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nagasaka S, Senoura T, Itai RN, Nakanishi H, Nishizawa NK (2013) Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat Commun 4: 2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63: 131–152 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ogo Y, Itai RN, Nakanishi H, Takahashi M, Mori S, Nishizawa NK (2007) The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Proc Natl Acad Sci USA 104: 19150–19155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2004) OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J 39: 415–424 [DOI] [PubMed] [Google Scholar]
- Lee S, Chiecko JC, Kim SA, Walker EL, Lee Y, Guerinot ML, An G (2009) Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol 150: 786–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang H, Ai Q, Liang G, Yu D (2016) Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol 170: 2478–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Zhang H, Li X, Ai Q, Yu D (2017) bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J Exp Bot 68: 1743–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Zhang H, Lou D, Yu D (2016) Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci Rep 6: 21451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Mayer JE, Pfeiffer WH, Beyer P (2008) Biofortified crops to alleviate micronutrient malnutrition. Curr Opin Plant Biol 11: 166–170 [DOI] [PubMed] [Google Scholar]
- Nozoye T, Nagasaka S, Kobayashi T, Takahashi M, Sato Y, Sato Y, Uozumi N, Nakanishi H, Nishizawa NK (2011) Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J Biol Chem 286: 5446–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Nakanishi H, Inoue H, Kobayashi T, Suzuki M, Takahashi M, Mori S, Nishizawa NK (2006) Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J Exp Bot 57: 2867–2878 [DOI] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Nakanishi H, Kobayashi T, Takahashi M, Mori S, Nishizawa NK (2007) The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J 51: 366–377 [DOI] [PubMed] [Google Scholar]
- Ogo Y, Kobayashi T, Nakanishi Itai R, Nakanishi H, Kakei Y, Takahashi M, Toki S, Mori S, Nishizawa NK (2008) A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. J Biol Chem 283: 13407–13417 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, Grishin NV, Bruick RK (2009) An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326: 722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S, Schmidt W (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Selote D, Samira R, Matthiadis A, Gillikin JW, Long TA (2015) Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol 167: 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, Nakanishi H, Nishizawa NK (2011) The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J Exp Bot 62: 4843–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J 31: 589–599 [DOI] [PubMed] [Google Scholar]
- Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, et al. (2009) Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326: 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL, Connolly EL (2008) Time to pump iron: Iron-deficiency-signaling mechanisms of higher plants. Curr Opin Plant Biol 11: 530–535 [DOI] [PubMed] [Google Scholar]
- Wang L, Ying Y, Narsai R, Ye L, Zheng L, Tian J, Whelan J, Shou H (2013) Identification of OsbHLH133 as a regulator of iron distribution between roots and shoots in Oryza sativa. Plant Cell Environ 36: 224–236 [DOI] [PubMed] [Google Scholar]
- Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan LM, Deng XW (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu B, Li M, Feng D, Jin H, Wang P, Liu J, Xiong F, Wang J, Wang HB (2015) The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27: 787–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Ren YR, Wang QJ, Wang XF, You CX, Hao YJ (2016a) Ubiquitination-related MdBT scaffold proteins target a bHLH transcription factor for iron homeostasis. Plant Physiol 172: 1973–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Ren YR, Wang QJ, Yao YX, You CX, Hao YJ (2016b) Overexpression of MdbHLH104 gene enhances the tolerance to iron deficiency in apple. Plant Biotechnol J 14: 1633–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Song A, Li P, Chen S, Jiang J, Chen F (2014) A bHLH transcription factor regulates iron intake under Fe deficiency in chrysanthemum. Sci Rep 4: 6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Ying Y, Wang L, Wang F, Whelan J, Shou H (2010) Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol 10: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]