Down-regulation of 5-aminolevulinic acid dehydratase encoding genes results in elevated levels of reactive oxygen species, salicylic acid and resistance to Verticillium dahliae infection of cotton.
Abstract
Programmed cell death (PCD) is an important form to protect plants from pathogen attack. However, plants must precisely control the PCD process under microbe attacks to avoid detrimental effects. The complexity of how plants balance the defense activation and PCD requires further clarification. Lesion mimic mutants constitute an excellent material to study the crosstalk between them. Here, we identified a Gossypium hirsutum (cotton) lesion mimic mutant (Ghlmm), which exhibits necrotic leaf damage and enhanced disease resistance. Map-based cloning demonstrated that GhLMMD, encoding 5-aminolevulinic acid dehydratase and located on chromosome D5, was responsible for the phenotype. The mutant was resulted from a nonsense mutation within the coding region of GhLMMD. It exhibited an overaccumulation of the 5-aminolevulinic acid, elevated levels of reactive oxygen species and salicylic acid, along with constitutive expression of pathogenesis-related genes and enhanced resistance to the Verticillium dahliae infection. Interestingly, GhLMM plays a dosage-dependent role in regulating PCD of cotton leaves and resistance to V. dahliae infection. This study provides a new strategy on the modulation of plant immunity, particularly in polyploidy plants.
Cell death in plants can occur as part of normal developmental processes, which is traditionally considered to be a programmed cell death (PCD) process, or ectopically when the normal development is perturbed, such as by a genetic lesion. In addition, cell death can occur at the sites of environmental stresses, such as pathogen infection or physical wounding, or in response to low concentrations of toxins. These three distinct types of cell death—in normal development, in perturbed development, or because of environmental stresses—do not necessarily utilize the same cell death mechanism; however, they do all appear to occur via a controlled disassembly of the cell (Vaux and Korsmeyer, 1999; Pennell and Lamb, 1997).
In plants, the hypersensitive response (HR), which results in cell death at the sites of infection by an avirulent pathogen, has received considerable attention in the study of PCD and its relationship to plant disease resistance (Heath, 2000). Cell death during HR is elicited by specific recognition of pathogen-derived molecules from avirulent pathogens by plant proteins encoded by resistance (R) genes. The recognition is followed by local accumulation of reactive oxygen species (ROS), salicylic acid (SA), and defense-related proteins such as pathogenesis-related (PR) proteins, and is accompanied by the production of antimicrobial molecules and the fortification of cell walls (Shirasu and Schulze-Lefert, 2000). Considerable efforts have been made to identify the signaling components involved in the HR control and their connection to disease resistance. One approach is to identify mutants that fail to develop HR and resistance in response to infection by normally avirulent pathogens. Many of these mutations are involved in R-genes and R-protein signaling components. The second approach is to identify mutants that develop HR-like lesions and activate defense responses in the absence of pathogens. These mutants are collectively designated as lesion mimic mutants or lesion mimics, based on their spontaneous development of lesion phenotypes akin to HR or disease symptoms (Brodersen et al., 2002).
Several lesion mimic mutants have been reported in model plant Arabidopsis (Arabidopsis thaliana; Ishikawa et al., 2001; Quesada et al., 2013) as well as in cultivated crops such as maize (Zea mays; Hu et al., 1998), rice (Oryza sativa; Sun et al., 2011), and wheat (Triticum aestivum; Wang et al., 2016). Among them, about 40 genes controlling the lesion mimics have been isolated (Bruggeman et al., 2015). The proteins encoded by these genes fall into various functional groups, including membrane associated protein (Büschges et al., 1997), ion channel (Balagué et al., 2003), zinc-finger protein (Dietrich et al., 1997), and components involved in the biosynthesis/metabolic pathways of fatty acid/lipids (Kachroo et al., 2001) and porphyrin (Hu et al., 1998). Porphyrin compounds such as chlorophyll and heme play vital roles in plant development. The misregulation of chlorophyll metabolism can lead to severe photooxidative stress, resulting in cell damage, cell death, or ultimately plant death. In Arabidopsis, lesion mimic mutants lin2 and rug1 were related to the loss of function of coproporphyrinogen III oxidase (CPO) and porphobilinogen deaminase, respectively (Ishikawa et al., 2001; Quesada et al., 2013). Likewise, disruption of CPO in rice lesion initiation 1 mutant leads to cell death phenotypes (Sun et al., 2011). Interestingly, enzymes in chlorophyll biosynthesis or degradation have also been linked to system acquired resistance (SAR). Down-regulation of urophorphyrinogen decarboxylase III or CPO in tobacco (Nicotiana tabacum) and protoporphyrinogen oxidase in Arabidopsis would lead to lesions on the leaves, accompanied by the constitutive expression of PR genes, high levels of SA accumulation, and increased resistance to pathogens (Mock and Grimm, 1997; Kruse et al., 1995; Molina et al., 1999). In addition, the light signaling factors FHY3 and FAR1 could regulate plant PCD and disease immunity by modulating chlorophyll biosynthesis (Tang et al., 2012; Wang et al., 2015). Despite these advances, the molecular mechanisms by which homeostatic disruption of chlorophyll metabolism could induce PCD and SAR in plant remain elusive.
The allotetraploid Upland cotton (Gossypium hirsutum) accounts for more than 90% of the world’s cotton production, providing an important source of renewable textile fibers and oilseeds. Compared with the other widely cultivated crops such as maize, rice, and wheat, no lesion mimic mutants are discovered in cotton. In the photosynthesis biological process, 5-aminolevulinic acid dehydratase (ALAD) is an enzyme that functions at an early step in chlorophyll and heme biosynthesis. To our knowledge, no ALAD mutants have been reported in any plants. Here, we report the identification and characterization of a cotton lesion mimic mutant. Map-based cloning revealed that the mutant was developed from a premature mutation within the GhLMMD gene that encodes ALAD. The mutant exhibits high levels of aminolevulinic acid (ALA), ROS, and SA, constitutive expression of PR genes, and enhanced resistance to Verticillium dahliae infection. Remarkably, reduced ALAD enzyme activity is proportional to the increased levels of PR gene expressions and the degree of PCD. Similarly, the copy number of the mutant GhLMM was positively related to the severity of PCD, the levels of ROS, SA, and PR gene expressions, and the level of resistance to V. dahliae infection. We propose that ALAD plays a dosage-dependent role in the regulation of PCD and immunity in allotetraploid cotton and potentially other polyploidy plants, which might have practical significance in breeding crops with improved resistance to pathogens.
RESULTS
Genetic and Physical Mapping of the GhLMMD Locus
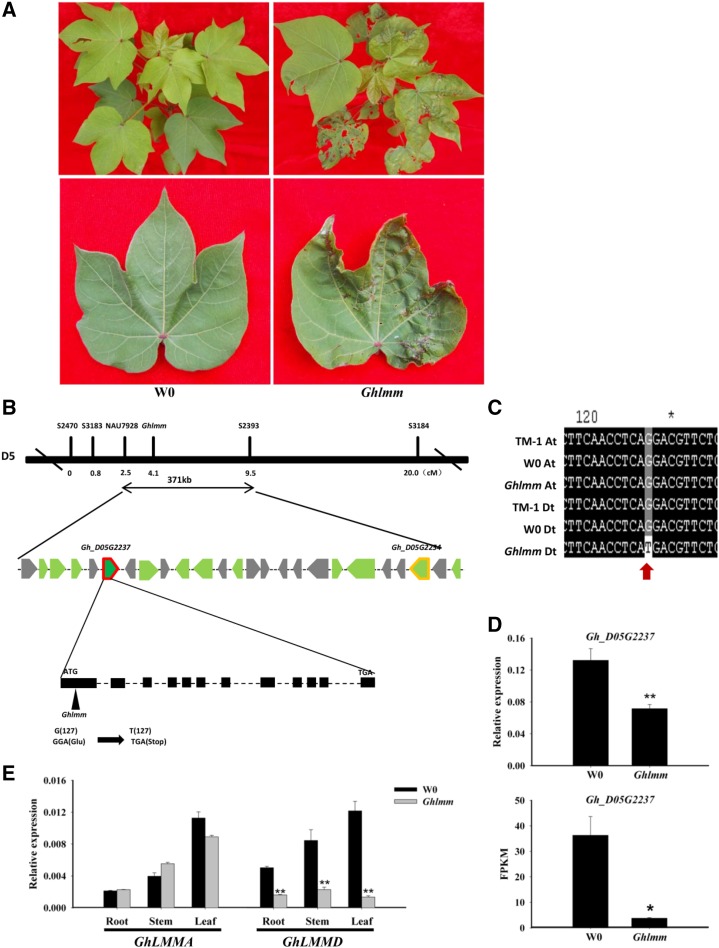
In a screen for the transgenic plants of cotton, we discovered a natural variation seedling that shows disease-like cell death phenotype during its development in the absence of pathogens. A subsequent analysis revealed that no T-DNA insertion was present in the plant, suggesting that it might originate from a mutation in the tissue culture process. We named the mutant Ghlmm (Gossypium hirsutum lesion mimic mutant). Ghlmm shows necrotic lesions only on the leaves but not in other tissues of the cotton plant (Fig. 1A). Further phenotypic investigation demonstrated that Ghlmm develops lesions in a temperature- and light-dependent manner. Under higher temperatures of 28/24°C (day/night) and long-day conditions of 16 h/8 h (light/dark), the mutant impaired leaf development and formed lesions. However, under the lower temperatures of 23/20°C (day/night) or short-day conditions of 8 h/16 h (light/dark), the leaf lesions did not appear (Supplemental Fig. S1). Though exhibiting a lesion mimic phenotype on the leaves under field conditions, other traits, such as fiber yield and quality and seed development, were almost not affected in Ghlmm compared with its corresponding wild-type accession W0 (Supplemental Table S1).
Figure 1.
Lesion phenotype of Ghlmm mutant and physical delineation of GhLMMD. A, Phenotypes of 3-month-old wild type (WT) and Ghlmm plants grown in the field. The Ghlmm mutant exhibits spontaneous disease like lesions on the leaves. B, Physical map of the GhLMMD locus. GhLMMD was delimited to a 371-kb region on Chromosome D5. Vertical lines mark the positions of DNA markers flanking GhLMMD. Twenty-five genes (denoted by gray and green thick arrows) were predicted within the region, among which 12 (green thick arrows) were verified to be active genes based on RNA-seq data and qRT-PCR analysis. The expression of two of these genes, termed Gh_D05G2237 and Gh_D05G2254, were significantly decreased in Ghlmm mutant and might be the candidates of GhLMMD. C, DNA sequence comparisons of the candidate genes, between the mutant and the wild type, identified a point mutation (G-to-T transition at the 127th position from the start codon, marked with a red thick arrow) in Gh_D05G2237 on Chromosome D5. D, Transcript abundance of Gh_D05G2237 in the mutant and the wild type analyzed by qRT-PCR and RNA-seq. Data were obtained from three independent replicates. E, Expression of GhLMMA and GhLMMD in different tissues in the mutant and the wild type, showing that transcript abundance of GhLMMA was not altered in both the mutant and the wild type, while the transcript abundance of GhLMMD was remarkably decreased in the mutant but not in wild type, further demonstrating that the mutation has taken place in the D genome. *P < 0.05 and **P < 0.01 respectively (Student’s t test). Error bars are SE of three biological replicates.
To better understand the molecular basis of the lesion mimic phenotype identified in cotton, we employed a map-based cloning strategy to isolate the target gene. As a first step to clone the gene, we initiated a genetic and physical mapping program to delimit the gene locus. The F1 and F2 mapping populations were developed by crossing Ghlmm with TM-1, a genetic standard line of G. hirsutum. All F1 plants exhibit the wild-type phenotype, and the segregation of the F2 plants meets the Mendelian ratio of 3:1 (554 individuals with wild-type phenotype and 209 individuals with lesion mimic phenotype, χ2 = 2.202 < χ20.05, 1 = 3.84), suggesting that the lesion mimic phenotype was controlled by a single nuclear recessive gene. Linkage analysis with 763 F2 plants delimited the target gene to a 371-kb DNA segment by markers NAU7928 (microsatellite) and S2393 (single nucleotide polymorphism [SNP]) on Chr. D5 (Fig. 1B), we therefore named the target gene GhLMMD. According to the high-quality reference genome sequence of the tetraploid cotton G. hirsutum acc. TM-1 (Zhang et al., 2015), the 371-kb region contains 25 open reading frames. The transcriptome analysis in TM-1 revealed that there are 12 genes whose FPKM value was >1.0 (Supplemental Fig. S2A), suggesting that they are probably active genes. The expressions of these genes were further detected by quantitative RT-PCR (qRT-PCR). It was found that among them, two genes, named Gh_D05G2237 and Gh_D05G2254, were significantly decreased in transcript abundance in Ghlmm mutant compared with that in W0 (Supplemental Fig. S2B). Therefore, the two genes were considered as the candidate gene for GhLMMD.
Identification of GhLMMD
To identify GhLMMD, the DNA and cDNA of Gh_D05G2237 and Gh_D05G2254 in Ghlmm, W0, and TM-1 were respectively amplified and sequenced. Sequence comparisons showed that no nucleotide differences in Gh_D05G2254 were detected in Ghlmm. However, a point mutation (G-to-T transition at the 127th position within the coding region) was detected in Gh_D05G2237 of Ghlmm compared with that of W0 and TM-1 (Fig. 1C), which resulted in a pretermination (GGA to TGA) in the protein. Next, we tested whether silencing Gh_D05G2237 or Gh_D05G2254 would result in the lesion mimic phenotype. They are suppressed in the wild-type cotton seedlings by the TRV-based virus-induced gene silencing (VIGS) technology, respectively. Suppression of Gh_D05G2237, but not Gh_D05G2254, led to a lesion mimic phenotype on the leaves (Supplemental Figs. S3 and S4). To further verify whether Gh_D05G2237 is the candidate of GhLMMD, qRT-PCR and a transcriptome analysis were performed for Ghlmm and W0. The results showed that the transcript abundance of Gh_D05G2237 was indeed drastically decreased in Ghlmm (Fig. 1D). Based on the point mutation that occurred at the 127th position within the coding region of Gh_D05G2237, SNP primers that could specifically detect the 127th-T genotype but not the 127th-G were developed and successfully used in genotyping 209 F2 plants exhibiting the Ghlmm phenotype.
Since upland cotton is an allotetraploid plant, it consists of the A and D subgenome. To confirm that Ghlmm was caused by a mutation in GhLMMD but not in GhLMMA, the GhLMMD homologs in the A subgenome and transcript abundance of GhLMMD and GhLMMA were respectively monitored by qRT-PCR using gene-specific primers (designed based on other SNP sites between GhLMMA and GhLMMD rather than the pretermination site in GhLMMD; Supplemental Table S2). It was shown that the transcript abundance of GhLMMD was indeed decreased in Ghlmm compared with W0, whereas the GhLMMA expression was not significantly altered in Ghlmm and W0 (Fig. 1E). These experiments indicate that the mutation leading to the lesion mimic phenotype occurred in the D subgenome and that Gh_D05G2237 is the target gene of GhLMMD.
GhLMMD Encodes the ALA Dehydratase and Localizes in Chloroplasts
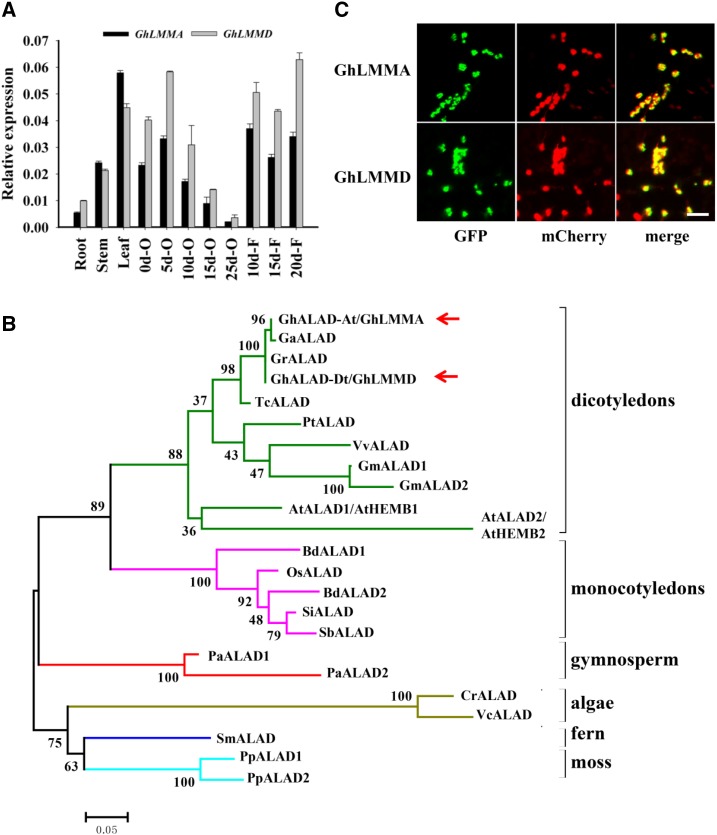
The BLAST analysis using Gh_D05G2237 as query showed that GhLMMD encodes a predicted protein of 430 amino acid residues with a conserved ALAD motif (Pfam00490) in the region from amino acid 105 to 424, with an 84% similarity to the reported AtHEMB1 protein in Arabidopsis (Supplemental Fig. S5). A nonsense mutation occurred in GhLMMD at the 43rd amino acid leads to a predicted truncated protein in Ghlmm. Wild-type GhLMMD and its corresponding gene in the A subgenome, GhLMMA, are highly conserved in the amino acid sequence (99% identity) and show similar expression patterns in all tested cotton tissues, with higher expression levels in leaves, early stage ovules, and developing fibers (Fig. 2A). The phylogenetic analysis with 23 GhLMM orthologs (temporally named ALAD genes here) derived from 17 species, including algae, fern, moss, gymnosperms, monocotyledons, and dicotyledons (Supplemental Table S3) revealed that no more than two ALAD genes was found in each tested species (Fig. 2B) and the ALAD motifs share high sequence similarities, indicating that ALAD is an ancient and conserved protein in plants (Fig. 2B). The subcellular localization showed that both GhLMMA and GhLMMD proteins localize in chloroplasts (Fig. 2C), which is consistent with their function in the tetrapyrrole biosynthesis pathway.
Figure 2.
Characterization of GhLMMD. A, Transcript abundance of GhLMMA (GhLMM in A subgenome) and GhLMMD (GhLMM in D subgenome) in different tissues of G. hirsutum acc. TM-1 quantified by qRT-PCR using gene-specific primers. GhLMMA and GhLMMD showed similar expressions in tested cotton tissues, with higher expression levels in leaves, early stage ovules, and developmental fibers. B, Phylogenetic analysis of the GhLMM orthologs (ALAD genes) in different species. Twenty-three ALAD proteins from 17 species, including algae, ferns, moss, gymnosperms, and angiosperms, were used to construct the phylogenetic tree. Species analyzed included algae (Cr: Chlamydomonas reinhardtii; Vc: Volvox carteri), fern (Sm: Selaginella moellendorffii), moss (Pp: Physcomitrella patens), gymnosperm (Pa: Picea abies), monocotyledons (Si: Setaria italic; Sb: Sorghum bicolor; Bd: Brachypodium distachyon; Os: Oryza sativa), and dicotyledons (Tc: Theobroma cacao; Gh: Gossypium hirsutum; Gr: Gossypium raimondii; Ga: Gossypium arboreum; At: Arabidopsis thaliana; Pt: Populus trichocarpa; Vv: Vitis vinifera; Gm: Glycine max). Red arrows indicate GhLMMA and GhLMMD. C, Chloroplast localization of GhLMM. GhLMMA and GhLMMD were respectively fused with GFP and transiently expressed in tobacco epidermal cells by Agro-infiltration. Chloroplast localized Rubisco protein fused with mCherry was used as positive control. GhLMM-GFP fusion proteins colocalize with Rubisco-mCherry fusion and show yellow signals after merging. Bars = 20 μm.
Reduced ALAD Activity Leads to Elevated Levels of ALA, ROS, and MDA
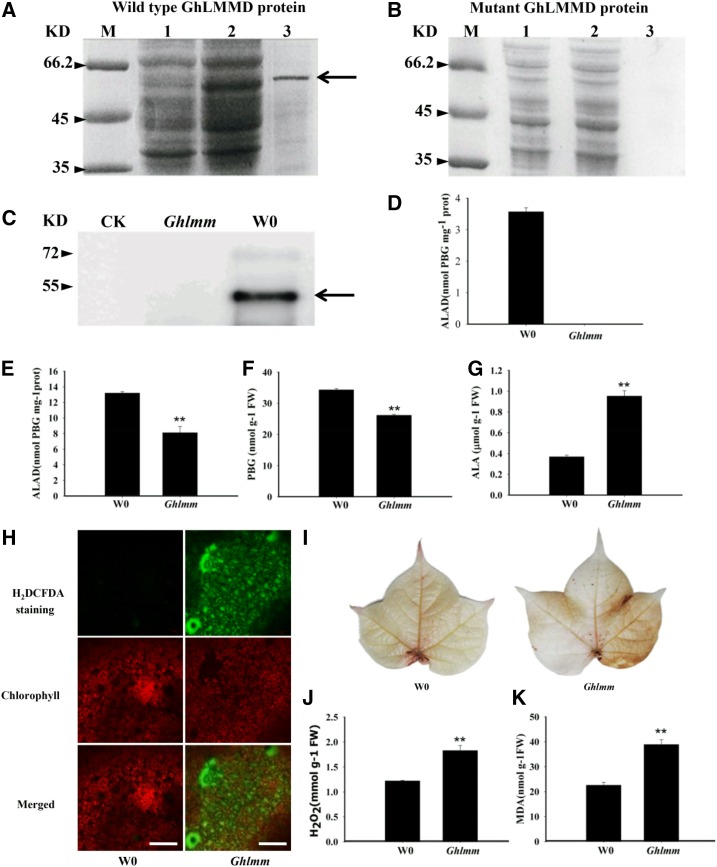
In plants, ALAD is an enzyme at an early step in the tetrapyrrole biosynthesis pathway, catalyzing the porphobilinogen (PBG) formation by condensing two molecules of the ALA. To reveal how the GhLMMD mutation affects ALAD activity, we expressed the wild-type GhLMMD protein and the corresponding mutant protein in Escherichia coli and measured their catalytic activities in vitro after purification. The wild-type GhLMMD protein was successfully expressed and purified (Fig. 3A, lanes 2 and 3), whereas the mutant protein could not (Fig. 3B, lanes 2 and 3). Western blotting using anti-His antibody further showed the purified wild-type GhLMMD protein, while no mutant protein was obtained (Fig. 3C). It is possible that the preterminated GhLMMD in Ghlmm could not be translated or the truncated protein (only 42 amino acids) was degraded in E. coli. Compared to the high enzyme activity of purified GhLMMD protein, the activity of mutant protein was undetectable (Fig. 3D).
Figure 3.
GhLMMD loss of function leads to accumulation of ALA and ROS burst. A and B, Expression and purification of wild-type and mutant GhLMMD proteins in E. coli. Lane M: protein marker; Lane 1: extracts from noninduced cells; Lane 2: extracts from IPTG-induced cells; Lane 3: GhLMMD protein purified from extracts of IPTG-induced E. coli cells. Arrow indicates the induced and purified GhLMMD protein, which is the same size with the predicted protein. Note that the wild-type GhLMMD protein could be expressed and purified from E. coli, while no mutant GhLMMD protein was expressed after induction. C, Confirmation of purified ALAD protein by western blotting. Antibodies against His-tag were used in the assay. CK, Ghlmm, and W0 indicate proteins purified from E. coli transformed with empty, mutant GhLMMD, and wild-type GhLMMD vector, respectively. D, ALAD enzyme activities of purified wild-type and mutant GhLMMD proteins. E, Analysis of ALAD enzyme activities in wild-type and mutant cotton leaves. F and G, Measurements of the contents of PBG and ALA in cotton leaves. Leaves of cotton seedlings at two-true-leaves stage were used for the analysis. H, Detection of ROS levels in cotton leaves by H2DCFDA staining. Then 10 μm 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFD), a ROS-specific fluorescent indicator, was used for the staining. Bars = 50 μm. I, H2O2 visualization in cotton leaves by staining with DAB, showing more H2O2 was produced in the mutant than the wild type. J and K, Measurements of the contents of H2O2 and MDA in cotton leaves. Leaves of cotton seedlings at two-true-leaves stage were used for the analysis. At least three biological replicates were performed in each experiment. *P < 0.05 and **P < 0.01 (Student’s t test).
In vivo, GhLMMD mutation might reduce the ALAD enzyme activity and result in overaccumulation of its substrate ALA and decreased levels of PBG. In line with our hypothesis, the enzyme activity of ALAD in the leaves of Ghlmm decreased by 28.5% (Fig. 3E), the PBG content decreased by about 29.7% (Fig. 3F), but the ALA content increased by 1.5-fold (Fig. 3G) in Ghlmm compared to W0.
Previous studies have shown that the ALA accumulation could generate reactive oxygen species from autoxidation and photochemical reactions (Ryter and Tyrrell, 2000). Thus, overaccumulation of ALA would produce excessive amounts of ROS in the Ghlmm leaves. Indeed, H2DCFDA and 3,3′-diaminobenzidine (DAB) staining demonstrated that the ROS levels were remarkably higher in mutant leaves than in wild-type leaves (Fig. 3, H and I). The measurement of the H2O2 content showed it is about 50% higher in mutant leaves than in wild-type leaves (Fig. 3J). ROS-related oxidation can cause plasma membrane damage by oxidizing the polyunsaturated fatty acids in the cells, ultimately resulting in cell death (Torres et al., 2006). The methane dicarboxylic aldehyde (MDA) production is an indication of the peroxidation of unsaturated membrane lipids in cells. Measurement of the MDA content showed that its level is about 72% higher in mutant leaves than in wild-type leaves (Fig. 3K).
Due to the high sequence similarity between GhLMMA and GhLMMD, TRV-based VIGS of GhLMM in the wild type would simultaneously suppress the expression of both of the genes and result in more severe phenotype and overaccumulation of ALA, H2O2, and MDA. To confirm this, GhLMM-silenced wild-type W0 plants were investigated at the phenotypic and molecular level. We observed that severe necrotic lesions developed in the new emerging leaves of the GhLMM-silenced cotton plants at 10 d after infiltration, and, notably, the leaf growth of the GhLMM-silenced plants was arrested and no new leaves grew out after the emergence of the first true leaf (Supplemental Fig. S4A). As expected, the expression of both GhLMMA and GhLMMD was drastically decreased (Supplemental Fig. S4B), and ALA, H2O2, and MDA were overproduced in the GhLMM-silenced cotton plants more than in the mock-treated W0 plants (Supplemental Fig. S4C). In a parallel experiment, the ALAD enzyme activity was inhibited by spraying 20 mm levulinic acid (LA), a specific inhibitor of ALAD, on the leaves of wild-type plants. The results showed that a similar necrotic lesion phenotype could appear on the leaves of the wild-type plants as on the leaves of the GhLMM-silenced wild-type plants (Supplemental Fig. S6A). Meanwhile, the levels of ALA and H2O2 had increased 2.7- and 2.5-fold, respectively, in LA-treated leaves, compared with the mock-treated leaves (Supplemental Fig. S6, B and C). This experiment demonstrated that GhLMM silencing has the same effects as the ALAD enzyme disruption.
The Lesion Mimic Mutants of Cotton Confer Resistance to V. dahliae Infection
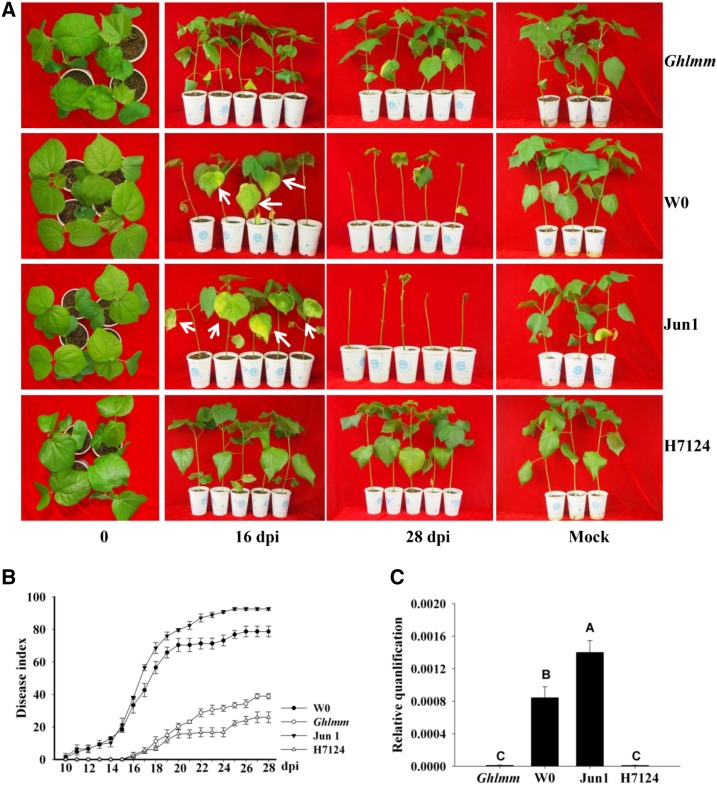
As several lesion mimic mutants exhibit resistance to pathogen attacks, particularly to biotrophs (Mock and Grimm, 1997; Kruse et al., 1995; Molina et al., 1999; Tang et al., 2012; Wang et al., 2016), we investigated whether Ghlmm has acquired an elevated resistance to the V. dahliae infection. Infection assays showed that Ghlmm reached the resistance level of H7124, a resistant control for V. dahlia infection, while W0 was as susceptible as Jun1, a susceptible control for V. dahliae infection. For instance, W0 and Jun1 showed significant wilting and etiolated leaves at 16 d postinoculation, but no obvious disease symptoms appeared in the leaves of Ghlmm and H7124 at this time point (Fig. 4A). At 28 d postinoculation, most W0 and Jun1 plants had died, the disease index of which was 79% and 92%, respectively. However, Ghlmm and H7124 showed fewer wilting and etiolated leaves, and the disease index was only 39% and 26%, respectively (Fig. 4, A and B). The resistance of Ghlmm to V. dahliae was further corroborated by the quantification of the fungal biomass in the leaves with q-PCR (Fig. 4C), microscopy of intensity of brown discoloration in stems (Supplemental Fig. S7A), and fungal recovery assays (Supplemental Fig. S7B).
Figure 4.
Ghlmm plants confers enhanced resistance to V. dahliae infection. A, Disease symptoms of Ghlmm compared with the wild-type W0, the susceptible control Jun1, and the resistant control H7124 challenged by V. dahliae. The mutant reached the resistance level of H7124. White arrows indicate disease symptoms on the leaves. B, The disease index of the mutants, W0, Jun1, and H7124. Error bars are SE calculated from three biological replicates, each containing 10 plants. C, qRT-PCR analysis of fungal biomass in leaves of the mutant and each of the controls. Different letters (A, B, C) indicate a statistically significant difference at P < 0.01 according to a randomization one-way ANOVA test. Error bars are SE of three biological replicates.
Molecular Investigation for the Increased Resistance of the Mutant
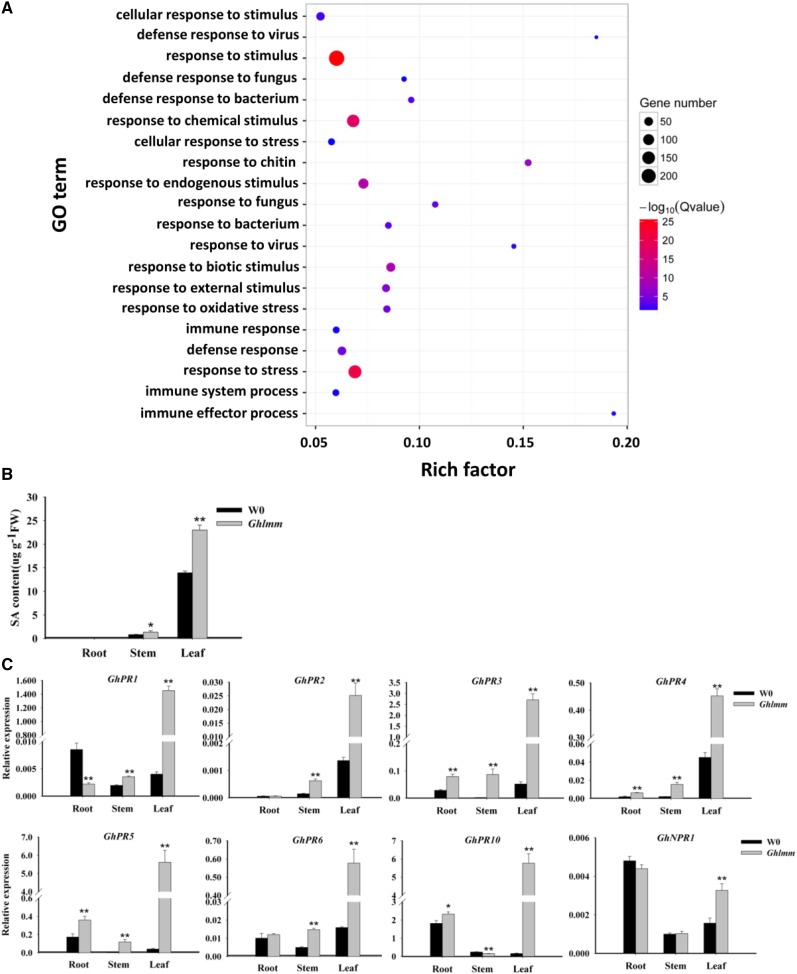
To obtain comprehensive insights into the molecular basis for the increased resistance of Ghlmm to the V. dahliae infection, an RNA-seq was performed in Ghlmm and W0. The data revealed that a total of 2,069 differential expression genes (q < 0.05 and a fold change >2), with 1,541 up-regulated and 528 down-regulated genes, were detected in Ghlmm compared with W0. Gene ontology (GO) analysis of the biological processes showed that a subset of the genes belonging to response to stimulus (FDR = 1.7E-26), response to stress (FDR = 1.2E-21), response to biotic stimulus (FDR = 5.70E-10), response to chitin (FDR = 6.60E-08), defense response (FDR = 4.70E-05), and response to oxidative stress (FDR = 5.50E-05) were overrepresented in Ghlmm, indicating the defense-related pathway initiated in Ghlmm (Fig. 5A; Supplemental Table S4). Notably, 48 PR genes were significantly up-regulated in Ghlmm, ranging from a 4- to a 449-fold increase (Supplemental Table S5).
Figure 5.
Ghlmm initiates immunity-related pathways and elevates SA accumulation and PR gene expression. A, GO enrichment analysis of the differentially expressed genes in leaves between Ghlmm and W0. The RNAs isolated from leaves of three individual plants were used for RNA-seq. P value of 0.05 adjusted by false discovery rate (FDR). Rich factor: percentage of enriched genes comparing with background in corresponding GO term. B, Measurement of SA content in different tissues of Ghlmm mutant and W0. The SA levels were significantly increased in leaves and stems of Ghlmm relative to the wild-type level. No SA could be detected in the roots. C, Transcription levels of PR and NPR1 genes in Ghlmm and W0 plants. All the tested genes were significantly up-regulated in leaves of Ghlmm plants compared with those in W0. **P < 0.01 (Student’s t test). Error bars are SE of three biological replicates.
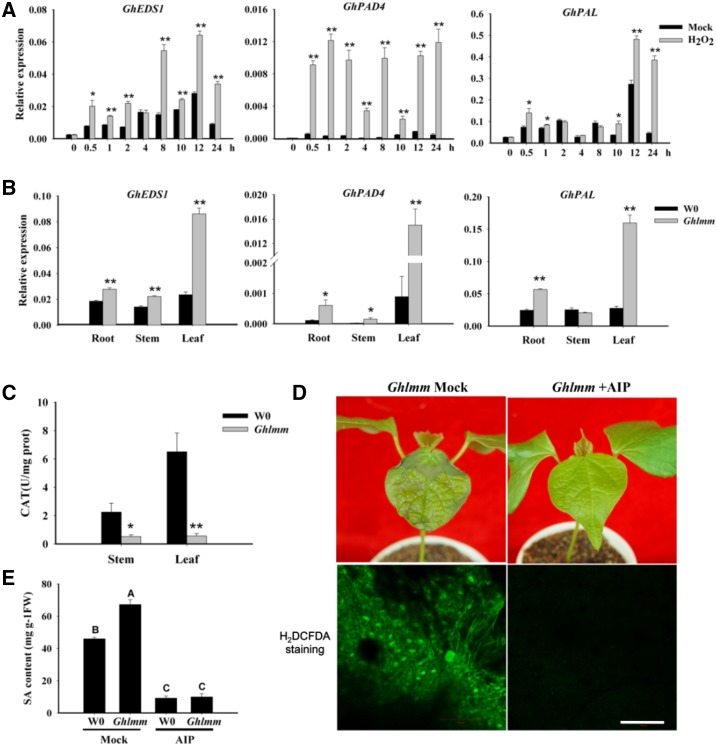
We have demonstrated that the ROS level is elevated in Ghlmm, and previous studies suggest that the ROS positively interplay with the SA. We thus measured the SA content in the roots, stems, and leaves of Ghlmm and W0. Although little was detected in roots, the SA levels significantly increased in the stems and leaves of Ghlmm compared with that in W0 plants (Fig. 5B). The SA signaling marker genes, such as the PR genes and nonexpresser of PR genes 1 (NPR1), were significantly up-regulated in Ghlmm, particularly in the leaves and stems (Fig. 5C). The genes relating to the SA biosynthesis were also investigated by qRT-PCR. Expression of EDS1 (enhanced disease susceptibility 1, encoding a lipase-like protein), PAD4 (phytoalexin deficient 4, encoding a lipase-like protein), and PAL (encoding phenylalanine ammonia-lyase) was induced by H2O2 treatment in cotton leaves (Fig. 6A). Similarly, their expression was much higher in Ghlmm than in W0 (Fig. 6B). The enzyme activities of catalase (CAT), reported to be inhibited by SA, were drastically decreased in Ghlmm (Fig. 6C). Blocking SA biosynthesis pathway by application with 2-aminoindan-2-phosphonic acid (AIP), a highly specific inhibitor of PAL, significantly decreased levels of SA and ROS in both Ghlmm and W0 and eliminated the necrotic lesions in leaves of Ghlmm (Fig. 6, D and E). These data indicate that in cotton, the ROS positively interplay with the SA, and both contribute to the development of the lesion mimic phenotype and the resistance to V. dahliae. We also analyzed the PR gene expression in response to the SA levels. All PR genes tested were induced by the SA treatment in the wild-type cotton plants (Supplemental Fig. S8), similar to the constitutive expression levels in Ghlmm. On the contrary, blocking the SA biosynthesis pathway by application of the AIP suppressed the constitutive PR genes expression (except for PR2 and PR5) in Ghlmm to a similar level as in W0 (Supplemental Fig. S9).
Figure 6.
ROS production and SA biosynthesis form a self-amplifying feedback loop in Ghlmm. A, Expression of genes regulating to SA biosynthesis were induced by H2O2 treatment. The experiments were performed with the wild-type plants. GhEDS1 and GhPAD4 are genes positively regulating SA biosynthesis; GhPAL encodes a catalytic enzyme for SA biogenesis. B, GhEDS1, GhPAD4, and GhPAL were constitutively up-regulated in Ghlmm plants relative to W0. C, CAT (catalase, inhibited by SA) enzyme activity was significantly inhibited in leaves and stems of Ghlmm plants. D, Treatment with AIP, a SA biosynthesis inhibitor, reduced the ROS content and eliminated the cell death phenotype in Ghlmm. AIP is a highly specific inhibitor of PAL enzyme. Bars = 50 μm. E, After treatment with AIP, SA content in Ghlmm decreased to the wild-type level. At least three biological replicates were performed in each experiment. *P < 0.05 and **P < 0.01 (Student’s t test). Different letters (A, B, C) indicate a statistically significant difference at P < 0.01 according to a randomization one-way ANOVA test. Error bars are SE of three biological replicates.
The GhLMM Dosage Regulates the Degree of PCD, Defense Responses, and Resistance to V. dahliae Infection
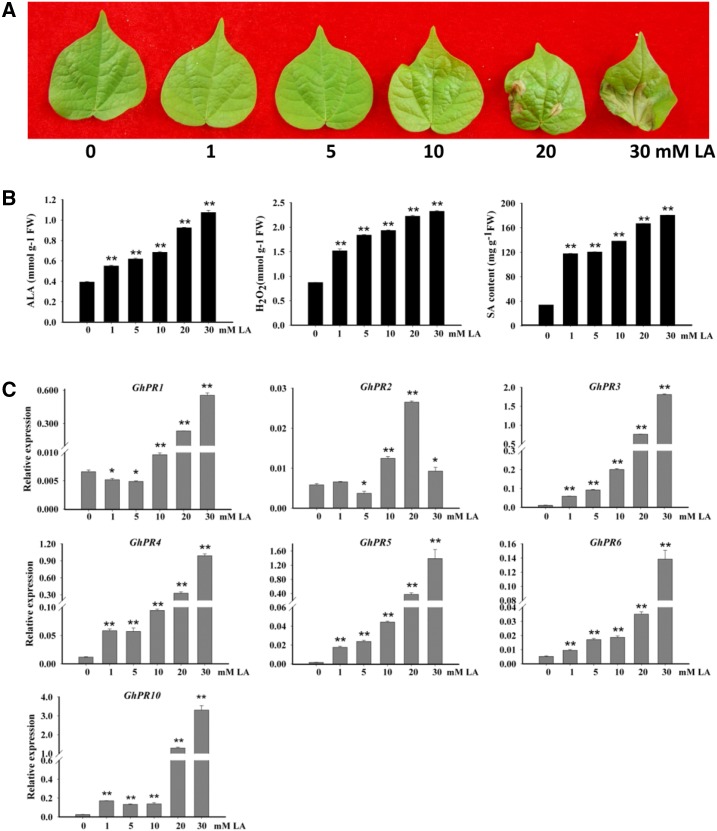
Our previous experiments demonstrated that GhLMM-silenced wild-type plants show a more severe phenotype than the Ghlmm mutant, for example, exhibiting an arrest in the leaf development (Supplemental Fig. S4). This phenomenon prompted us to hypothesize that the expression level of GhLMM should be fine-tuned in plant cells, and perturbation of GhLMM expression level or changes in the GhLMM copies may lead to the aberration of cell growth and lesion mimic phenotype. To confirm the gene dosage-dependent response, we first started the experiments by regulating the endogenous ALAD enzyme activities in cotton leaves by spraying different concentrations of LA solution (ranging from 0 to 30 mm). After 24 h of treatment, the phenotype of the leaf, the PR gene expression, and several physiological parameters were investigated. The results showed that a 20-mm LA treatment could cause an evident lesion mimic phenotype, while a 0- to 10-mm LA treatment did not lead to visible leaf lesions (Fig. 7A). The increases in the PR gene expression and levels of ALA, H2O2, and SA were parallel correlated to the increased concentrations of the LA applied during treatment (Fig. 7, B and C). These results revealed that the levels of SA and H2O2, the defense-related gene expression, and the leaf lesions in cotton could be regulated by fine modulation of ALAD enzyme activity. Furthermore, we generated F1 plants (containing one copy of the mutant gene, expressed as AADd) by crossing Ghlmm with W0. The F1 and W0 plants were exposed to different concentrations of LA solution, respectively. Interestingly, the F1 leaves exhibited evident lesions after 10 mm LA treatment, while visible cell death on W0 leaves appeared after 20 mm LA treatment (Supplemental Fig. S10A), indicating a dosage-dependent response to GhLMM copies in cotton. In addition, the phenotype of GhLMM-silenced plants in W0 and Ghlmm were characterized, respectively. The results showed that the GhLMM-silenced plants of both W0 and the mutant died, and no phenotypic difference could be observed (Supplemental Fig. S10, B and C).
Figure 7.
Mimic effects of GhLMM dosage on leaf phenotype by spraying LA. A, Leaf phenotype of W0 plants after being sprayed with different concentrations of LA solutions. Treatment with >20 mm LA solution leads to clear lesions on leaves. B, The content of ALA, H2O2, and SA was increased in W0 plants treated with different concentrations of LA solutions. C, PR gene expressions were up-regulated in W0 plants after LA treatment. At least three biological replicates were performed in each experiment. *P < 0.05 and **P < 0.01 (Student’s t test).
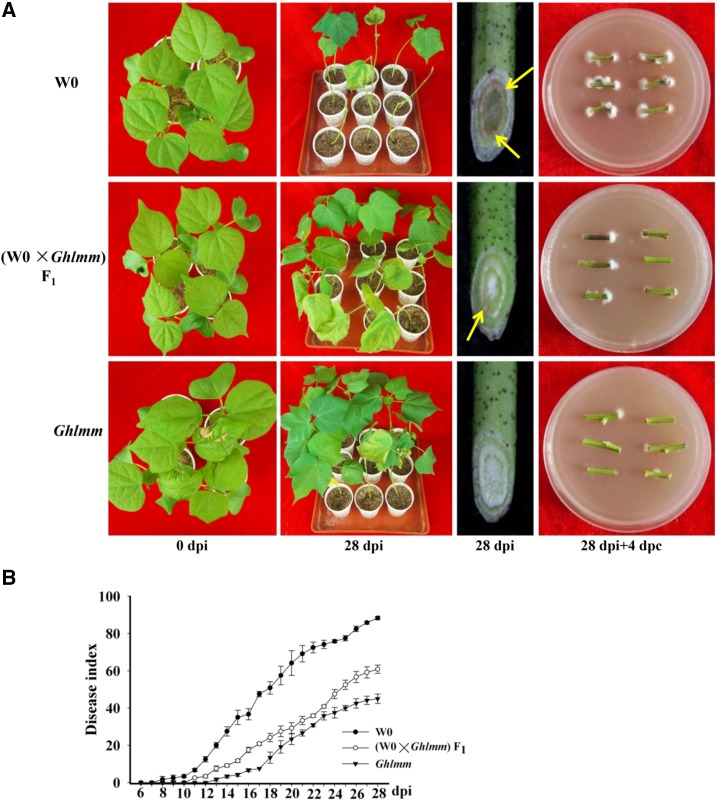
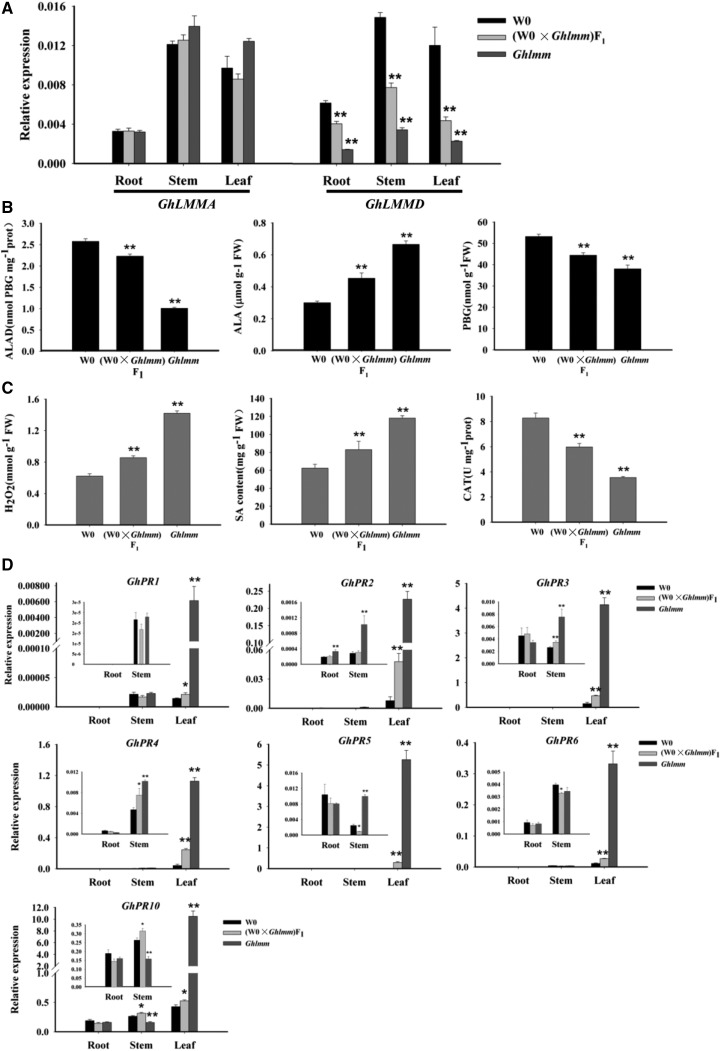
We next explored the effects of the GhLMM gene dosage on disease resistance. The resistance of the F1 plants to the V. dahliae infection was compared with Ghlmm and the wild-type cultivar W0. The results indicated that the resistance levels of these three lines were as follows: mutant > F1 > wild type (Fig. 8, A and B). To support this, the expression level of GhLMMD in these lines was detected. GhLMMD expression in the F1 plants was at midlevel between the parents, whereas no significant difference in the GhLMMA expression was detected between the parents and the F1 plants (Fig. 9A). The enzyme activity of ALAD and CAT and the contents of ALA, PBG, H2O2, and SA in the F1 plants were also at midlevel between the parents (Fig. 9, B and C). Similarly, the PR gene expression in these lines exhibited a gene dosage-dependent pattern (Fig. 9D).
Figure 8.
Disease resistance of Ghlmm plants and GhLMM dosage-dependent resistance to V. dahliae infection. A, Infection assays, visualization of fungal accumulation in stems, and fungal recovery experiments were performed to assess the disease resistance of indicated plant samples. The disease resistance of wild-type W0 (containing four copies of wild-type GhLMM, two copies of GhLMMA and GhLMMD each), the homozygote Ghlmm (containing twocopies of wild-type GhLMMA and two copies of mutant GhLMMD), and their F1 plants (containing two copies of wild-type GhLMMA, one copy of wild-type GhLMMD, and one copy of mutant GhLMMD) to V. dahliae infection was evaluated. The severity of disease symptoms was as follows: W0 > F1 > homozygote Ghlmm, exhibiting a dosage-dependent resistance to V. dahliae infection. The results from the fungal accumulation in stems and the fungal recovery experiments were similar to the infection assays. Each experiment has six biological replicates and repeated three times. Brown color indicated by arrows resulted from V. dahliae accumulation. DPC, days post culture. B, Disease index of W0, Ghlmm, and the F1 plants. Error bars are SE calculated from three biological replicates each containing 10 plants.
Figure 9.
Molecular evidence of GhLMM dosage-dependent resistance to V. dahliae infection. A, Transcript abundance of GhLMMA and GhLMMD in different tissues of W0, Ghlmm, and the F1 plants was analyzed by qRT-PCR using gene-specific primers. Expression of GhLMMA was not changed in the three lines. However, the expression of GhLMMD in the three lines was W0 > F1> Ghlmm, exhibiting a dosage-dependent expression pattern. B, ALAD enzyme activity, the content of ALA, PBG in W0, F1, and Ghlmm cotton plant leaves. C, H2O2 and SA content were positively correlated with the GhLMM gene dosage, while CAT enzyme activity was negatively correlated with it. D, PR gene expressions in different tissues of W0, F1, and Ghlmm showed a dosage-dependent expression pattern. Insets in the figures indicate the expression of corresponding genes in roots and stems. Error bars are SE calculated from five biological replicates. *P < 0.05 and **P < 0.01 (Student’s t test).
To address whether the gene dosage-dependent resistance to V. dahliae works in a different genetic background, we crossed Ghlmm with two cotton cultivars, namely Jun1 and TM-1, respectively. The infection assays were performed with the F1 plants and the parents from the two hybridization combinations. The results showed that the F1 plants derived from the two hybridizations exhibited a similar resistance level (Supplemental Fig. S11) as the F1 plants derived from the W0×Ghlmm hybridization combination. Together, our genetic and molecular evidence demonstrated that the ALAD activity or the GhLMM gene copies in cotton may well correlate with the resistance to V. dahliae infections.
DISCUSSION
Ghlmm Is Caused by a Nonsense Mutation in ALAD Encoding Gene
We have identified a lesion mimic mutant named Ghlmm in cotton. To our knowledge, it is the first lesion mimic mutant reported in cotton. Genetic and linkage analysis revealed that Ghlmm is controlled by a recessive nuclear gene located on chromosome D5. The gene responsible for the phenotype was cloned by a map-based strategy, indicating that it encodes the ALAD, a key enzyme in the tetrapyrrole biosynthesis pathway. In Ghlmm plants, a point mutation that resulted in pretermination was found in the coding region. Combined with the biochemical analysis of the wild-type and mutant GhLMMD proteins, we suppose that the mutant transcripts may be degraded more rapidly by nonsense mutation-mediated RNA surveillance system (Chang et al., 2007) and could not be translated into functional proteins. Therefore, the ALAD enzyme activity is lower in Ghlmm. We thus conclude that Ghlmm is a loss-of-function mutation derived from the wild-type gene.
Interestingly, Gh_D05G2254, another gene located in the same region of the GhLMMD locus, also showed significantly decreased expression in the mutant compared to wild-type plants (Fig S2). Gh_D05G2254 encodes DNA lyase OGG1, which functions in oxidative stress-induced DNA demethylation. It has been shown that down-regulation of OGG1 may confer cells resistant to oxidative damage (Zhou et al., 2016). Here, down-regulation of this gene might be a downstream effect of overaccumulated ROS in the mutant.
Enzymes in the Tetrapyrrole Biosynthesis Pathway Might Be Tightly Controlled in Plants
The tetrapyrrole biosynthesis pathway is important in the production of four classes of tetrapyrroles, namely chlorophyll, heme, siroheme, and phytochromobilin in plants. Among them, chlorophyll plays key roles in photosynthesis by absorbing light and transferring the light energy or the electrons to other molecules. ALA, the universal precursor for tetrapyrrole synthesis, is converted to chlorophyll via sequential enzyme activities, with the first one being the ALAD. Analysis of the ALAD gene family from lower to higher plants revealed that very few ALAD gene members are present in plants ranging from algae to angiosperms (Fig. 2B), suggesting that ALAD gene copies or their expression levels are tightly controlled for cellular demand, as either defective or enhanced activity of ALAD can have detrimental effects on plant growth. In Arabidopsis, the overexpression of HEMB1 (encoding ALAD) in the wild type displayed photobleached leaves and did not survive, and homozygotes hemb-1 mutants were also lethal (Tang et al., 2012). In our study, suppression of the ALAD in cotton leads to leaf growth arrest (Supplemental Fig. S4). In fact, most of the genes involved in tetrapyrrole biosynthesis in chloroplast might be precisely controlled in plants. Supporting information includes that (1) genes participating in tetrapyrrole biosynthesis, for example, HEMA, HEMC, HEMD, HEME, and HEMF, are only present in a few copy numbers in both Arabidopsis and O. sativa genomes (data not shown); and (2) antisense expression of HEMA1 in Arabidopsis leads to a lethal phenotype (Kumar and Söll, 2000), and disruption of HEMF1 caused plant growth alterations and the formation of necrotic lesions in leaves (Ishikawa et al., 2001; Sun et al., 2011).
ROS Derived from Chloroplast Could Enhance Plant Pathogen Resistance via SA-Dependent Pathway
In addition to the function of conducting photosynthesis in plants, chloroplast was recognized to play a central role in the defense response as the major steps of biosynthesis of defense hormones, such as SA, JA, and ABA, all occurred in chloroplast (Wildermuth et al., 2001; Wasternack and Hause, 2013; Serrano et al., 2016; Nambara and Marion-Poll, 2005). ROS has been implicated to play important roles in pathogen resistance (Shapiguzov et al., 2012). To date, most of the studies concentrate on the plant defense functions of apoplastic ROS, which was generated by plasma membrane respiratory burst oxidase homolog and cell wall peroxidase (Torres et al., 2005, 2006; Bolwell et al., 2002). Few studies provide convincing evidence that ROS produced in the chloroplast confers plant pathogen resistance. In this study, pretermination of GhLMMD decreased ALAD enzyme activity and resulted in the accumulation of ALA in chloroplast (Fig. 3, A-G). The accumulated ALA leads to relatively high levels of ROS via autoxidative decomposition (Fig. 3; Ryter and Tyrrell, 2000). ROS is proposed to act synergistically in a signal amplification loop with SA to induce the expression of PR genes and establishment of SAR (Dempsey et al., 1999). In our studies, SA biosynthesis positive regulating genes EDS1, PAD4, and PAL were significantly up-regulated in Ghlmm (Fig. 6B). On the other hand, CAT enzyme activity was inhibited in the mutant (Fig. 6C), which is consistent with previous findings that SA could inhibit CAT activity via binding its catalytic domain (Dempsey et al., 1999). Exogenous AIP (SA synthesis inhibitor) spraying on mutant leads to ROS scavenging. These results provide evidence for the self-amplifying feedback loop between ROS production and SA biosynthesis in plant. Produced ROS and induced SA could function as intercellular or intracellular second messengers to simultaneously mediate the establishment of HR, which induces PCD in plant cells, and SAR, which triggers defense-molecular production in the rest of the plant (Levine et al., 1994; Durrant and Dong, 2004).
Mutations in Essential Genes Will Not Lead to Lethality in Polyploidy Plants and Have Dosage Effect on Phenotype
To date, mutants misregulating in tetrapyrrole metabolism were discovered in diploid plants, such as Arabidopsis, maize, and rice, whereas no such research was reported in polyploidy plants including cotton. In contrast to the diploid plant species, polyploidy plants consist of more than one type of genome; they are derived from fusions between different diploid progenitors. For example, the allotetraploid cotton genome contains the A and D subgenomes, which originated from a single hybridization event between the A- and D- diploid species (Wendel, 1989). Gene redundancy and homologous genes are widely encountered in polyploidy plants, thus loss of function of a gene could be complemented by other genes with the same function and will probably not lead to visible phenotypic alterations. Thus, diploid plants could be vulnerable to genetic lesions, while polyploidy plants could be more resistant to most genetic defects. For example, mutation of Les22 gene in diploid maize caused lethal phenotype (Hu et al., 1998). However, in this study, although ALAD genes were mutated on the D subgenome, the corresponding homologs on the A subgenome are normal in the mutant, and thus the cotton plants can still survive but display necrotic leaf damage, indicating higher tolerance to the genetic defects in polyploidy plants than in diploid plants.
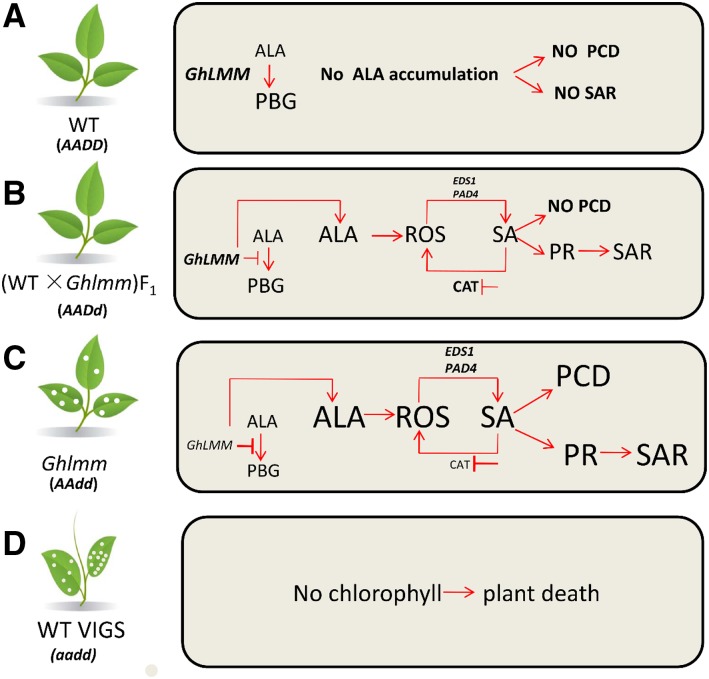
To support the idea that the fine modulation of ALAD activity can affect the intensity of the PCD and the defense response, we used an ALAD inhibitor with a series of concentrations on cotton leaves and found that both the degree of PCD and the PR gene expressions are proportional to the increasing concentration of the inhibitor (Fig. 7). Further, GhLMM gene dosage-dependent leaf cell death was demonstrated via exposing W0 and F1 plants to a range of LA solutions (Supplemental Figure S10A). These results indicated that the PCD and defense response can be tightly modulated by means of the fine regulation of ALAD activity. To investigate the effects of the ALAD gene dosage on cotton resistance to V. dahliae, we analyzed the resistance level of the homozygote mutant, the wild type, and their F1 plants to the V. dahliae infection. As expected, the resistance levels of these lines are proportional to the copies of the mutant ALAD gene in the cotton genome (Fig. 8; Supplemental Figure S11). Similarly, in transgenic Arabidopsis plants expressing antisense HEMA1 (encoding GluTR), different lines exhibiting varying degrees of chlorophyll deficiency were obtained, which displayed a range from a severe and intermediate to a negligible loss of chlorophyll (Kumar and Söll, 2000). These results reveal that the degree of mutant/lesion phenotypes could be tightly modulated by regulating the enzyme activity in steps of the tetrapyrrole biosynthesis pathway. Based on these results, we propose a model whereby the ALAD is involved in the regulation of PCD and innate immunity in allotetraploid cotton (Fig. 10). (1) Four copies of GhLMM maintain the plants in good conditions without ALA accumulation and defense activation. (2) Three copies exhibit no lesion on the leaves but lead to an increased level of ALA, ROS, and disease resistance. (3) Two copies display a significantly increased disease resistance level, but the lesion mimic phenotype occurs due to the ALA overaccumulation. (4) A simultaneous suppression of the GhLMM homeologs in tetraploid cotton blocks the tetrapyrrole and chlorophyll biosynthesis pathway and eventually leads to plant death. We suppose that a fine-tuning of ALAD activity between two and three copies might enhance the innate immunity without leaf cell death in tetraploid cotton. Moreover, as revealed by Kumar and Söll (2000), the traditional technique, the antisense method, might be superior to other techniques, such as the recently developed RNA interference and clustered regularly interspaced short palindromic repeats in modulating the ALAD gene expression due to a highly conserved sequence in the ALAD genes.
Figure 10.
Proposed model of GhLMM dosage-dependent effects on the regulation of PCD and immunity. A, Four copies of GhLMM in tetraploid cotton (wild type, expressed as AADD) maintain the plants in normal growth without ALA accumulation and SAR activation. B, Three copies of GhLMM in cotton (F1, expressed as AADd) leads to adequate ALA accumulation and proper degrees of ROS and SA levels and induction of SAR in the presence pathogen attack. Therefore, the plants exhibit no lesion on the leaves but have a high disease resistance level. C, Homozygous mutation with two copies of the mutant GhLMMD (Ghlmm mutant, expressed as AAdd) overaccumulate ALA, which might significantly increase the levels of ROS and SA, thus leading to disease lesion mimic phenotype on leaves in the absence of pathogen attack. D, Complete suppression of GhLMM by VIGS (expressed as aadd) results in no chlorophyll biosynthesis and eventually plant death. The body size in the figure denotes the content of corresponding molecules or degrees of signaling pathways involved. The bigger body size represents the more content of corresponding molecules or the higher degrees of signaling pathways.
In this study, we found that mutation in the ALAD gene located on chromosome D5 of cotton confers enhanced resistance to V. dahliae. Although the mechanism requires a deep analysis at the molecular level, we suppose that the lesion mimic mutant we identified in cotton will have great potential applications in breeding cotton hybrid seeds with improved V. dahliae resistance. Furthermore, the GhLMM gene dosage effects can serve as an example that highlights the advantages of polyploidization in plants during evolution, with improving adaptation and elasticity to the environment.
MATERIALS AND METHODS
Plant Materials
Ghlmm, a lesion mimic mutant, was found in a screening of regenerated transgenic cotton plants, with Gossypium hirsutum L. acc. W0 as the wild-type receptor. TM-1 is a standard genetic line of Upland cotton. G. hirsutum cv Jun1 is highly susceptible to Verticillium dahliae infection and was used as the negative control. H7124 is a commercial Gossypium barbadense cultivar with high resistance to V. dahliae infection and was used as the positive control. The F2 mapping population including 763 individuals was developed from a cross between TM-1 as the receptor parent and Ghlmm mutant as the donor parent. Three F1 lines were generated from crosses between Ghlmm and W0, TM-1, and Jun1, respectively. All accessions were grown in the Jiangpu Experiment Station and in the green house under 28°C/24°C (day/night) and 16-h-light/8-h-dark cycle conditions. For temperature-dependent phenotypic assay, Ghlmm plants and the wild-type controls were grown at 23°C/20°C (day/night) for low-temperature conditions and 28°C/24°C (day/night) for high-temperature conditions on a 16-h-light/8-h-dark cycle; for light-dependent phenotypic assay, Ghlmm plants and the wild-type controls were grown in a plant room under 8-h-light/16-h-dark for short-day conditions and 16-h-light/8-h-dark cycle for long-day conditions. Fresh tissues used for a series of analysis, including DNA and RNA extraction, measurement of the levels of H2O2, ALA, PBG, SA, and enzyme activity, were collected and immediately frozen in liquid nitrogen and stored at −70°C until use.
Map-Based Cloning of the GhLMMD Gene
To map the gene responsible for the lesion mimic phenotype, an F2 mapping population including 763 individuals was developed by a cross between Ghlmm and TM-1. Based on the previously published high-density tetraploid cotton linkage map (Guo et al., 2007) and the genome sequence of tetraploid cotton TM-1 (Zhang et al., 2015), the GhLMMD locus was preliminarily mapped between the molecular markers S2470 and S3184, with genetic distances of 4.1 and 15.9 cM respectively on Chr. D5. Furthermore, a microsatellite (SSR) marker NAU7928 and an SNP marker S2393 were developed, and the GhLMMD locus was narrowed to a 371-kb region. The cDNA and genomic DNA of the candidate genes within the region were amplified from W0, TM-1, and Ghlmm respectively, and the PCR products were confirmed by sequencing. All primers used in the study refer to Supplemental Table S2.
RNA Extraction and qRT-PCR
The total RNA was extracted from different tissues, including roots, stems, and leaves using the cetyltrimethyl ammonium bromide-acid phenol method (Shang et al., 2015). First-strand cDNA was generated using HiScript Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme) according to the manufacturer’s instructions. Real-time PCR was performed using a LightCycler FastStart DNA Master SYBR Green I kit (Roche) in a CFX96 Touch Real-Time PCR detection system (Bio-Rad, http://www.bio-rad.com). Three biological replicates were used for each analysis with three technical replicates each. The relative expression levels were calculated according to Livak and Schmittgen (Livak and Schmittgen, 2001). GhLMM subgenome-specific primers were designed based on the single nucleotide polymorphism between GhLMMA and GhLMMD. Other gene-specific primers were designed using Primer Premier 5. All primers are listed in Supplemental Table S2.
Subcellular Localization of GhLMM
The coding regions of GhLMMA and GhLMMD in W0 were fused with GFP in the pBinGFP4 vector to generate pBin-35S::GhLMMA-GFP4 and pBin-35S::GhLMMD-GFP4 respectively (Liu et al., 2014). The two vectors were transiently expressed in tobacco (Nicotiana benthamiana) leaf cells by Agrobacterium tumefaciens infiltration method (Liu et al., 2014). Vector (pt-rk-CD3-999)-containing chloroplast localized Rubisco protein labeled by mCherry was used as positive marker (Nelson et al., 2007). Subcellular localization of GhLMMA and GhLMMD was observed using a confocal laser scanning microscope (Zeiss LSM 780) at the specific excitation and emission wavelengths (GFP, 488 and 495–530 nm; mCherry, 587 and 600–650 nm).
Virus-Induced Gene Silencing Assay
A 412-bp fragment (corresponding to bases 585–996) of GhLMMD and a 417-bp fragment (corresponding to bases 73–489) of Gh_05D2254 were cloned into pTRV2 (Gao et al., 2011), respectively, constructing the vectors named pTRV2: GhLMMD and pTRV2: Gh_05D2254. The two vectors and pTRV1 were introduced into the Agrobacterium strain GV3101 by means of the freeze-thawing method. The Agrobacterium strains containing pTRV2: GhLMMD and pTRV2: Gh_05D2254 were respectively mixed with Agrobacterium strains containing pTRV1 at a ratio of 1:1. These mixtures were infiltrated in cotton cotyledons. More than 20 seedlings were used for each analysis. pTRV: GhCLA1 was used as a control, and the silencing of GhCLA1 produced the photobleaching phenotype. The plants were grown in pots at 23°C 20°C under a 16-h-light/8-h-dark cycle with 60% humidity. The primers used for the vector construction are listed in Supplemental Table S2. All VIGS assays were performed for three times independently.
Phylogenetic Analysis and Amino Acid Sequence Alignment of ALAD Orthologs in Plants
The ALAD genes were extracted from seventeen species, including algae, ferns, moss, gymnosperms, and angiosperms, to establish the phylogenetic tree. The genomic database of two sequenced cotton species, diploid Gossypium arboreum and tetraploid G. hirsutum acc. TM-1, was obtained from http://cgp.genomics.org.cn and http://mascotton.njau.edu.cn, respectively. The genomic database of Picea abies was obtained from http://congenie.org. The genomic database of other species was obtained from http://www.phytozome.net. The ALAD domain (Pfam00490) seed file was downloaded from http://pfam.janelia.org and used as the query to individually scan the protein databases with the HMMER software (version 3.0; Eddy, 2011). All the identified ALAD proteins were aligned with the Clustal W program. MEGA 5.0 software (www.megasoftware.net) was used for constructing the phylogenetic tree using the JTT model in the maximum-likelihood method, and the bootstrap value was set at 1,000. The Clustal X software (http://www.clustal.org/) was used for amino acid sequence alignment of ALAD proteins from rice (Oryza sativa), Arabidopsis (Arabidopsis thaliana), Theobroma cacao, and G. hirsutum, respectively. The Genedoc software (http://www.nrbsc.org/gfx/genedoc/ebinet.htm) was used for visualizing the result.
Recombinant Protein Expression and Purification
The encoding region of the wild-type GhLMMD and its corresponding mutant gene were cloned into the BamHI sites in a pET-28a vector using the ClonExpress II One Step Cloning Kit (Vazyme, C112-01), resulting in a translational fusion of the wild type and the mutant protein with six His residues at the N- and C- terminus, respectively. The plasmids were transformed into Escherichia coli BL21 (DE3). The positive clones were grown at 37°C in LB medium with 50 mg L−1 kanamycin to an OD600 of 0.8. Expression of fusion protein was induced by addition of 1 mm isopropylthio-β-galactoside (IPTG) and growth at 16°C for 16 h. Cells were suspended in buffer (0.05 m Tris-HCl, 0.1 mm dithiothreitol, 6 m urea, pH 8.2) and broken by ultrasonic method. The proteins were purified using a Ni-NTA column (Qiagen). After denaturing in urea and gradient dialysis, refolded GhLMMD protein was obtained and then used for enzyme activity assay, SDS-PAGE analysis, and western blotting. HRP-conjugated anti-His antibody (Proteintech) was used in western blotting as described (Bu et al., 2017).
Measurement of the Levels of ALA and PBG and the Enzyme Activity of ALAD
The ALA content was measured according to Mauzerall and Granick (1956). In brief, frozen samples were homogenized and suspended in a 500-μL 20 mm potassium phosphate buffer (pH 6.8). The samples were then centrifuged for 10 min at 12,000 rpm at 4°C. Then 400 μL of supernatant was transferred to a new tube and mixed with 100 μL ethyl-acetoacetate. After boiling for 10 min at 100°C, it was mixed with 500 μL of Ehrlich’s reagent for 5 min, and then the absorption was measured at 553 nm. The ALA content of the samples was calculated using the standard curve generated by the commercial ALA.
The PBG content measurement was performed as described by Kayser et al. (2005). ALAD enzyme activity was determined using the methods described by Vajpayee et al. (2000) with the buffer (0.16 mg/mL ALA, 40 mm Tris-HCl, 80 mm dithiothreitol, 9.4 mm MgCl2, pH 8.2), expressed as nmol PBG/per milligram protein per hour at 37°C.
Evaluation of the Resistance to V. dahliae Invasion
For assessment of Verticillium wilt resistance, cotton seeds were grown in pots with a mixture of peat and vermiculite (1:1, v/v). The defoliating V. dahliae isolate ‘V991’ was grown on potato dextrose agar at 25°C for 4 d. Then, mycelia were collected and cultured in liquid Czapek’s medium for 5 d. Cotton seedlings at two true-leaf stages were inoculated with V991 by soil drenching with 20 mL spore suspensions (107 conidia/mL). The disease index was calculated using the following formula: disease index = [(∑disease grades × number of infected plants)/ (total checked plants×4)] ×100. Each experiment group included at least 10 individual plants and was repeated three times. Mock inoculated plants were used as negative controls.
To quantify the fungal biomass in cotton leaves, the first true leaves of the cotton plants inoculated with V991 were collected for DNA extraction. The amounts of fungal DNA were quantified by calculating the products of the ITS1 and ITS2 regions of the rRNA genes (Z29511) of V. dahliae relative to the products of cotton histone gene (AF024716; Zhang et al., 2016). Fungal accumulation was also demonstrated by microscopy based on the intensity of the brown stem discoloration resulting from the fungi accumulation. To do this, stems were collected at the position of 1 cm above the cotyledons knot at 16 d after inoculation, free-hand sectioned, and observed using a microscope.
For fungal recovery assay, stems and leaves were surface sterilized for 1 min in 70% ethanol, rinsed with sterile water 8 times, and then sliced into sections. The stem and leaf sections were placed on the potato dextrose agar supplemented with chloramphenicol (34 mg L−1) and cultured for 4 d at 25°C, then photographed.
Detection and Measurement of the Levels of ROS, H2O2, MDA, and CAT Enzyme Activity
The ROS generation in the cotton leaf was examined with H2DCFDA staining. Cotton leaves were washed with sterile water and incubated in 10 μM H2DCFDA for 30 min in dark at 30°C. The samples were then washed with sterile water 3 times, and the staining signals were observed using a microscope.
To visualize the accumulation of H2O2, fresh cotton leaves were collected, incubated in a DAB solution (1 mg/mL, pH 3.8) for 8 h, and then decolorized in 96% ethanol. The quantification of H2O2 in leaves was performed with a commercial H2O2 detection kit (Jiancheng Bioengineering Institute).
The MDA content in leaves was measured with a commercial plant MDA detection kit (Jiancheng Bioengineering Institute). The CAT enzyme activity in leaves was determined with a CAT detection kit (Jiancheng Bioengineering Institute).
Salicylic Acid Measurement
The SA was measured according to the method described by Verberne et al. with minor modifications (Verberne et al., 2002). In brief, the cotton leaves were collected and ground in liquid nitrogen. Then, the samples were mixed with 90% methanol and centrifuged. After collecting the supernatant, the pellet was suspended in 100% methanol and centrifuged again. The supernatants were combined and added with 10 μL 0.2 m sodium hydroxide and then evaporated in a SpeedVac concentrator. The residues were resuspended in 5% trichloroacetic acid. The solutions were extracted twice with a 800-μL mixture of ethyl-acetate:cyclohexane (1:1). The upper organic phases were collected. The lower trichloroacetic acid phase was acid hydrolyzed with 300 μL 8 m hydrochloric acid (1 h, 80°C) and was extracted twice with 800 μL ethyl-acetate:cyclohexane (1:1). The organic phases were collected, combined with the above organic phase solutions, then added with 120 μL 0.2 m sodium acetate and evaporated in a SpeedVac concentrator. The residues were dissolved in 900 μL methanol, and the SA content was analyzed by HPLC and calculated according to the standard SA curve that was made from commercial SA purchased from Sigma.
Treatment of Plants with AIP and LA
AIP, the highly specific inhibitor for the PAL enzyme that catalyzes the SA biogenesis, was used to block the biosynthesis of SA. Ten days after sowing, the cotton seedlings with visible true leaves were treated with AIP at a concentration of 1 mm. The control plants were treated with sterile water. The AIP or mock treated cotton leaves were collected when the Ghlmm mutant exhibited the necrotic lesion phenotype. Fresh leaves were used for the ROS observation.
For the LA treatment, cotton plants were grown up to the two-true-leaves stage for about 16 d after sowing. Different concentrations of the LA solutions were sprayed on the leaves of the cotton seedlings. Twenty-four hours after spraying, the leaves were collected for use.
Transcriptome Sequencing
The leaves of Ghlmm and W0 were collected 15 d after sowing, and the total RNA of the leaves was extracted by means of the cetyltrimethyl ammonium bromide-sour phenol extraction method (Jiang and Zhang, 2003). Sequencing was performed on the Illumina HiSeq2000 platform. After preprocessing the RNA-seq data with an NGS QC toolkit (Patel and Jain, 2012), the reads were mapped to the G. hirsutum TM-1 genome using a Tophat spliced aligner with default parameters (Trapnell et al., 2009). The genome-matched reads from each library were assembled with Cufflinks (Trapnell et al., 2012). Cuffmerge was then used to merge the individual transcript assemblies into a single transcript set. Lastly, Cuffdiff was used to find differentially expressed genes with a cutoff of 0.05 q value and a fold change of >2. The GO analysis of the differentially expressed genes in the biological process was conducted using the AgriGO software (Du et al., 2010). The input sample list was the Arabidopsis gene ID, which was converted from the Upland cotton gene ID (http://genome.jgi.doe.gov/) and the background was constituted by the whole annotated genes in Arabidopsis. The output of enrichment needed FDR < 0.05.
Accession Numbers
All sequence files are available at DDBJ/EMBL/GenBank under the accession numbers KY652847 (GhLMMA of G. hirsutum acc. TM-1), KY652848 (GhLMMD of G. hirsutum acc. TM-1), KY652849 (GhLMMA of G. hirsutum acc. W0), and KY652850 (GhLMMD of G. hirsutum acc. W0). Transcriptome data have been deposited in GenBank under the accession number PRJNA369440.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Ghlmm mutant develops lesions on leaves in a temperature- and light-dependent manner.
Supplemental Figure S2. Expression pattern of candidate genes in leaves of TM-1, W0, and Ghlmm mutant.
Supplemental Figure S3. Gh_D05G2254 is not the causative gene responsible for Ghlmm phenotype.
Supplemental Figure S4. Functional characterization of GhLMM (Gh_D05G2237) by VIGS analysis.
Supplemental Figure S5. Amino acid sequence alignment of ALAD proteins.
Supplemental Figure S6. LA spraying leads to Ghlmm phenotype and accumulation of ALA and H2O2 in wild-type W0.
Supplemental Figure S7. V. dahliae content was significantly reduced in Ghlmm stems and leaves.
Supplemental Figure S8. The PR genes were induced by SA treatment.
Supplemental Figure S9. Expression of most PR genes in Ghlmm mutant was down-regulated to the same levels as those in W0 after AIP treatment.
Supplemental Figure S10. The effects of GhLMM dosage on leaf phenotype.
Supplemental Figure S11. Effects of GhLMM mutation on resistance to V. dahliae infection under different genetic backgrounds.
Supplemental Table S1. Fiber qualities and seed parameters in Ghlmm and W0.
Supplemental Table S2. All primers used in this study.
Supplemental Table S3. Information of ALAD genes from Gossypium, Arabidopsis, and other species.
Supplemental Table S4. GO analysis involved in biological processes.
Supplemental Table S5. PR genes significantly upregulated in Ghlmm.
Acknowledgments
This work was supported by National Key R & D Program for Crop Breeding (2016YFD0100306) and Jiangsu Collaborative Innovation Center for Modern Crop Production project (No. 10). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Glossary
- AIP
2-aminoindan-2-phosphonic acid
- ALA
5-aminolevulinic acid
- CAT
catalase
- CPO
coproporphyrinogen III oxidase
- DAB
3,3′-diaminobenzidine
- GO
gene ontology
- HR
hypersensitive response
- IPTG
isopropylthio-β-galactoside
- LA
levulinic acid
- MDA
methane dicarboxylic aldehyde
- PBG
porphobilinogen
- PCD
programmed cell death
- ROS
reactive oxygen species
- SA
salicylic acid
- SAR
system acquired resistance
Footnotes
This work was supported by National Key R & D Program for Crop Breeding (2016YFD0100306), and Jiangsu Collaborative Innovation Center for Modern Crop Production project (No.10). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Balagué C, Lin B, Alcon C, Flottes G, Malmström S, Köhler C, Neuhaus G, Pelletier G, Gaymard F, Roby D (2003) HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell GP, Bindschedler LV, Blee KA, Butt VS, Davies DR, Gardner SL, Gerrish C, Minibayeva F (2002) The apoplastic oxidative burst in response to biotic stress in plants: a three-component system. J Exp Bot 53: 1367–1376 [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Pike HM, Olszak B, Skov S, Odum N, Jørgensen LB, Brown RE, Mundy J (2002) Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev 16: 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q, Raynaud C, Benhamed M, Delarue M (2015) To die or not to die? Lessons from lesion mimic mutants. Front Plant Sci 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu W, Liang Q, Zhi L, Maciunas L, Loll PJ, Eckenhoff RG, Covarrubias M (2017) Sites and functional consequence of alkylphenol anesthetic binding to Kv1.2 channels. Mol Neurobiol 10.1007/s12035-017-0437-2 [DOI] [PubMed] [Google Scholar]
- Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, et al. (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88: 695–705 [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF (2007) The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Shah J, Klessig DF (1999) Salicylic acid and disease resistance in plants. CRC Crit Rev Plant Sci 18: 547–575 [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL (1997) A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88: 685–694 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Eddy SR. (2011) Accelerated profile HMM searches. PLOS Comput Biol 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wheeler T, Li Z, Kenerley CM, He P, Shan L (2011) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J 66: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Cai C, Wang C, Han Z, Song X, Wang K, Niu X, Wang C, Lu K, Shi B, et al. (2007) A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium. Genetics 176: 527–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath MC. (2000) Hypersensitive response-related death. Plant Mol Biol 44: 321–334 [DOI] [PubMed] [Google Scholar]
- Hu G, Yalpani N, Briggs SP, Johal GS (1998) A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10: 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A, Okamoto H, Iwasaki Y, Asahi T (2001) A deficiency of coproporphyrinogen III oxidase causes lesion formation in Arabidopsis. Plant J 27: 89–99 [DOI] [PubMed] [Google Scholar]
- Jiang J, Zhang T (2003) Extraction of Total RNA in Cotton Tissues with CTAB-acidic Phenolic Method. Cotton Sci 15: 166–167 [Google Scholar]
- Kachroo P, Shanklin J, Shah J, Whittle EJ, Klessig DF (2001) A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc Natl Acad Sci USA 98: 9448–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser H, Krull-Savage U, Rilk-van Gessel R (2005) Developmental profiles of 5-aminolevulinate, porphobilinogen and porphobilinogen synthase activity in Pieris brassicae related to the synthesis of the bilin-binding protein. Insect Biochem Mol Biol 35: 165–174 [DOI] [PubMed] [Google Scholar]
- Kruse E, Mock HP, Grimm B (1995) Reduction of coproporphyrinogen oxidase level by antisense RNA synthesis leads to deregulated gene expression of plastid proteins and affects the oxidative defense system. EMBO J 14: 3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AM, Söll D (2000) Antisense HEMA1 RNA expression inhibits heme and chlorophyll biosynthesis in arabidopsis. Plant Physiol 122: 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79: 583–593 [DOI] [PubMed] [Google Scholar]
- Liu T, Song T, Zhang X, Yuan H, Su L, Li W, Xu J, Liu S, Chen L, Chen T, et al. (2014) Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat Commun 5: 4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mauzerall D, Granick S (1956) The occurrence and determination of delta-amino-levulinic acid and porphobilinogen in urine. J Biol Chem 219: 435–446 [PubMed] [Google Scholar]
- Mock HP, Grimm B (1997) Reduction of uroporphyrinogen decarboxylase by antisense RNA expression affects activities of other enzymes involved in tetrapyrrole biosynthesis and leads to light-dependent necrosis. Plant Physiol 113: 1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina A, Volrath S, Guyer D, Maleck K, Ryals J, Ward E (1999) Inhibition of protoporphyrinogen oxidase expression in Arabidopsis causes a lesion-mimic phenotype that induces systemic acquired resistance. Plant J 17: 667–678 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7: e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell RI, Lamb C (1997) Programmed cell death in plants. Plant Cell 9: 1157–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Sarmiento-Mañús R, González-Bayón R, Hricová A, Ponce MR, Micol JL (2013) PORPHOBILINOGEN DEAMINASE deficiency alters vegetative and reproductive development and causes lesions in Arabidopsis. PLoS One 8: e53378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter SW, Tyrrell RM (2000) The heme synthesis and degradation pathways: role in oxidant sensitivity. Heme oxygenase has both pro- and antioxidant properties. Free Radic Biol Med 28: 289–309 [DOI] [PubMed] [Google Scholar]
- Serrano I, Audran C, Rivas S (2016) Chloroplasts at work during plant innate immunity. J Exp Bot 67: 3845–3854 [DOI] [PubMed] [Google Scholar]
- Shang X, Chai Q, Zhang Q, Jiang J, Zhang T, Guo W, Ruan YL (2015) Down-regulation of the cotton endo-1,4-β-glucanase gene KOR1 disrupts endosperm cellularization, delays embryo development, and reduces early seedling vigour. J Exp Bot 66: 3071–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiguzov A, Vainonen JP, Wrzaczek M, Kangasjärvi J (2012) ROS-talk - how the apoplast, the chloroplast, and the nucleus get the message through. Front Plant Sci 3: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Schulze-Lefert P (2000) Regulators of cell death in disease resistance. Plant Mol Biol 44: 371–385 [DOI] [PubMed] [Google Scholar]
- Sun C, Liu L, Tang J, Lin A, Zhang F, Fang J, Zhang G, Chu C (2011) RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J Genet Genomics 38: 29–37 [DOI] [PubMed] [Google Scholar]
- Tang W, Wang W, Chen D, Ji Q, Jing Y, Wang H, Lin R (2012) Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL (2005) Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajpayee P, Tripathi RD, Rai UN, Ali MB, Singh SN (2000) Chromium (VI) accumulation reduces chlorophyll biosynthesis, nitrate reductase activity and protein content in Nymphaea alba L. Chemosphere 41: 1075–1082 [DOI] [PubMed] [Google Scholar]
- Vaux DL, Korsmeyer SJ (1999) Cell death in development. Cell 96: 245–254 [DOI] [PubMed] [Google Scholar]
- Verberne MC, Brouwer N, Delbianco F, Linthorst HJ, Bol JF, Verpoorte R (2002) Method for the extraction of the volatile compound salicylic acid from tobacco leaf material. Phytochem Anal 13: 45–50 [DOI] [PubMed] [Google Scholar]
- Wang F, Wu W, Wang D, Yang W, Sun J, Liu D, Zhang A, Yang W, Sun J, Liu D, et al. (2016) Characterization and genetic analysis of a novel light dependent lesion mimic mutant, lm3, showing adult-plant resistance to powdery mildew in common wheat. PLoS One 11: e0155358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Tang W, Ma T, Niu D, Jin JB, Wang H, Lin R (2015) A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis. J Integr Plant Biol 58: 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot (Lond) 111: 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF. (1989) New World tetraploid cottons contain Old World cytoplasm. Proc Natl Acad Sci USA 86: 4132–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, et al. (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33: 531–537 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhao J, Ding L, Zou L, Li Y, Chen G, Zhang T (2016) Constitutive expression of a novel antimicrobial protein, Hcm1, confers resistance to both Verticillium and Fusarium wilts in cotton. Sci Rep 6: 20773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhuang Z, Wang W, He L, Wu H, Cao Y, Pan F, Zhao J, Hu Z, Sekhar C, et al. (2016) OGG1 is essential in oxidative stress induced DNA demethylation. Cell Signal 28: 1163–1171 [DOI] [PubMed] [Google Scholar]