Vesicle membrane-associated SCYL2 proteins are vital for plant development.
Abstract
Protein transport between organelles is an essential process in all eukaryotic cells and is mediated by the regulation of processes such as vesicle formation, transport, docking, and fusion. In animals, SCY1-LIKE2 (SCYL2) binds to clathrin and has been shown to play roles in trans-Golgi network-mediated clathrin-coated vesicle trafficking. Here, we demonstrate that SCYL2A and SCYL2B, which are Arabidopsis (Arabidopsis thaliana) homologs of animal SCYL2, are vital for plant cell growth and root hair development. Studies of the SCYL2 isoforms using multiple single or double loss-of-function alleles show that SCYL2B is involved in root hair development and that SCYL2A and SCYL2B are essential for plant growth and development and act redundantly in those processes. Quantitative reverse transcription-polymerase chain reaction and a β-glucuronidase-aided promoter assay show that SCYL2A and SCYL2B are differentially expressed in various tissues. We also show that SCYL2 proteins localize to the Golgi, trans-Golgi network, and prevacuolar compartment and colocalize with Clathrin Heavy Chain1 (CHC1). Furthermore, bimolecular fluorescence complementation and coimmunoprecipitation data show that SCYL2B interacts with CHC1 and two Soluble NSF Attachment Protein Receptors (SNAREs): Vesicle Transport through t-SNARE Interaction11 (VTI11) and VTI12. Finally, we present evidence that the root hair tip localization of Cellulose Synthase-Like D3 is dependent on SCYL2B. These findings suggest the role of SCYL2 genes in plant cell developmental processes via clathrin-mediated vesicle membrane trafficking.
Vesicle-mediated protein transport between organelles is a fundamental process in all eukaryotic cells. In many cases, newly synthesized proteins in the endoplasmic reticulum (ER) are targeted to specific membranes, where they function with optimal activity (Jurgens, 2004; Hwang and Robinson, 2009; Robinson and Neuhaus, 2016). Protein targeting involves processes including the formation, transport, docking, and fusion of vesicles. A number of proteins, such as coat proteins, ADP-ribosylation factor, and Soluble NSF Attachment Protein Receptors (SNAREs), also are required for the regulation of vesicle trafficking (Jurgens, 2004; Bassham and Blatt, 2008; Nielsen et al., 2008; Chen et al., 2011; Fan et al., 2015; Paez Valencia et al., 2016).
Depending on the species of coat proteins, vesicles can be classified into three major types: Coat Protein1 (COPI), COPII, and clathrin-coated vesicles (CCVs). Among them, CCVs are formed by clathrin triskelia consisting of three clathrin heavy chains (CHCs) and three clathrin light chains (CLCs). The Arabidopsis (Arabidopsis thaliana) genome contains two CHC genes (CHC1 and CHC2) and three CLC genes (CLC1, CLC2, and CLC3; Kitakura et al., 2011; Wang et al., 2013; Paez Valencia et al., 2016). In plants, CCV mediates protein trafficking from the trans-Golgi network (TGN) to the multivesicular prevacuolar compartment (PVC), which fuses with the vacuole for the degradation of proteins (Song et al., 2006; Hwang and Robinson, 2009; Park et al., 2013; Paez Valencia et al., 2016; Robinson and Neuhaus, 2016). CCVs also are known to be involved in the endocytosis of proteins such as PIN-FORMED auxin efflux carriers and Brassinosteroid Insensitive1 receptor kinase at the plasma membrane (PM; Kitakura et al., 2011; Di Rubbo et al., 2013; Wang et al., 2013). Recently, a plant-specific TPLATE adaptor protein complex was shown to be essential for clathrin-mediated endocytosis (Gadeyne et al., 2014; Wang et al., 2016). However, the CCV machinery in plants is still poorly known when compared with the CCV components identified in animals (Chen et al., 2011; Paez Valencia et al., 2016).
In animals, SCY1-LIKE2 (SCYL2), which belongs to the SCY1-like gene family, was identified as a component of the CCVs (Conner and Schmid, 2005). It was shown that animal SCYL2 binds directly to both clathrin and Adaptor Protein-2 (AP2), which is an adaptor protein (Conner and Schmid, 2005; Düwel and Ungewickell, 2006). SCYL2 has been suggested to function as a kinase (Conner and Schmid, 2005) and is found at the Golgi, TGN, and endosomes in animal systems (Düwel and Ungewickell, 2006; Borner et al., 2007). Moreover, SCYL2 selectively induces the lysosomal degradation of Frizzled5, which is a PM receptor involved in the Wnt signaling pathway (Terabayashi et al., 2009). It also has been reported that lysosomal hydrolase cathepsin D is missorted in SCYL2-depleted cells (Düwel and Ungewickell, 2006) and that knockdown of SCYL2 in Xenopus tropicalis causes severe developmental defects (Borner et al., 2007). Recently, it was shown that SCYL2 is a key regulator of neuronal function and survival in mouse brain (Gingras et al., 2015; Pelletier, 2016). These data indicate that, in animals, SCYL2 functions in clathrin-mediated vesicle trafficking between the TGN and the endosomal system with an important role in cell developmental processes.
In this study, two Arabidopsis homologs, SCYL2A and SCYL2B, are shown to play important roles in plant cell growth. SCYL2B is involved in root hair developmental processes, and both SCYL2A and SCYL2B are essential for plant growth and act redundantly in certain developmental processes. A GUS-aided promoter assay indicated that SCYL2A and SCYL2B are differentially expressed in various tissues. Moreover, we found that SCYL2B localizes at the Golgi, TGN, and PVC and interacts with CHC1 and two SNAREs: Vesicle Transport through t-SNARE Interaction11 (VTI11) and VTI12. Finally, we show that SCYL2B colocalizes with SCYL2A and CHC1 and is required for the tip localization of Cellulose Synthase-Like D3 (CSLD3) in root hairs. These data suggest that the plant SCYL2 genes, new components of CCV trafficking in plants, play vital roles in plant cell developmental processes.
RESULTS
SCYL2 Is Conserved in Many Eukaryotes
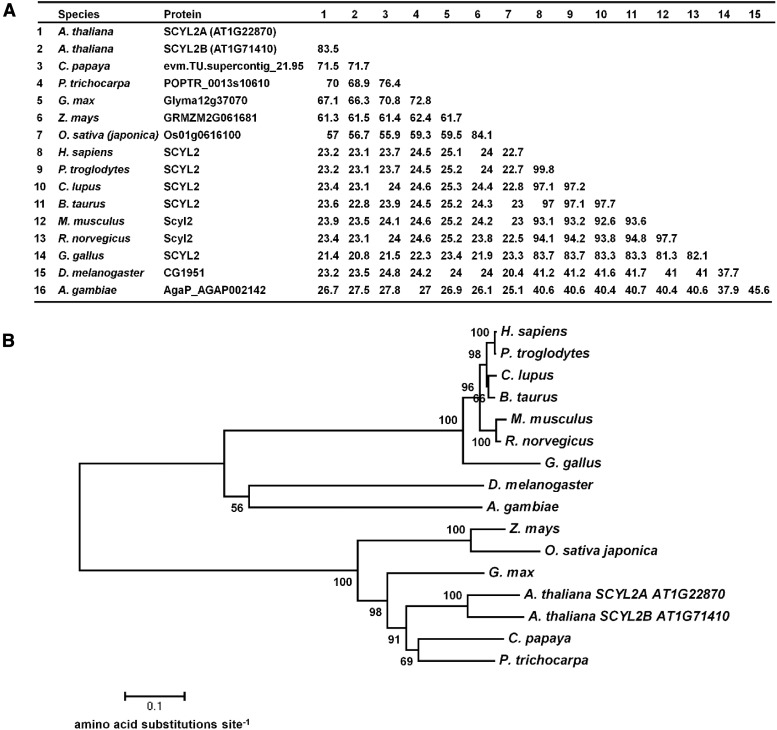
In Arabidopsis, there are two SCYL2 genes, SCYL2A and SCYL2B, which encode 913 and 909 amino acids, respectively. These two genes likely arose from a genome duplication, as the two genes fall into two syntenic chromosomal regions (Supplemental Fig. S1; Plant Genome Duplication Database [http://chibba.agtec.uga.edu/duplication/index/home]; Tang et al., 2008). Sequence alignment and phylogenetic analysis show that SCYL2 genes are conserved in the genomes of many eukaryotes (Fig. 1). Comparison of amino acid sequences of full-length Arabidopsis SCYL2 proteins revealed 57% to 83% amino acid identity to proteins in other plant species, such as papaya (Carica papaya), poplar (Populus trichocarpa), soybean (Glycine max), corn (Zea mays), and rice (Oryza sativa), and 21% to 27% amino acid identity to the SCYL proteins in animal species, such as human, chimpanzee, dog, cow, mouse, rat, chicken, fruit fly, and mosquito (Fig. 1A).
Figure 1.
SCYL2 is conserved in eukaryotes. A, A similarity matrix is shown to represent the percentage of amino acid identity between species across the SCYL2 proteins. B, Neighbor-joining phylogenetic tree showing the relationship between SCYL2 proteins from eukaryote species. The numbers above the nodes are percentages of bootstrap confidence levels from 10,000 replicates. Branch length represents the number of amino acid changes per site in accordance with the scale bar. Species of SCYL2 proteins are Arabidopsis thaliana, Carica papaya, Populus trichocarpa, Glycine max, Zea mays, Oryza sativa japonica, Homo sapiens, Pan troglodytes, Canis lupus familiaris, Bos taurus, Mus musculus, Rattus norvegicus, Gallus gallus, Drosophila melanogaster, and Anopheles gambiae.
SCYL2B Functions in Root Hair Development
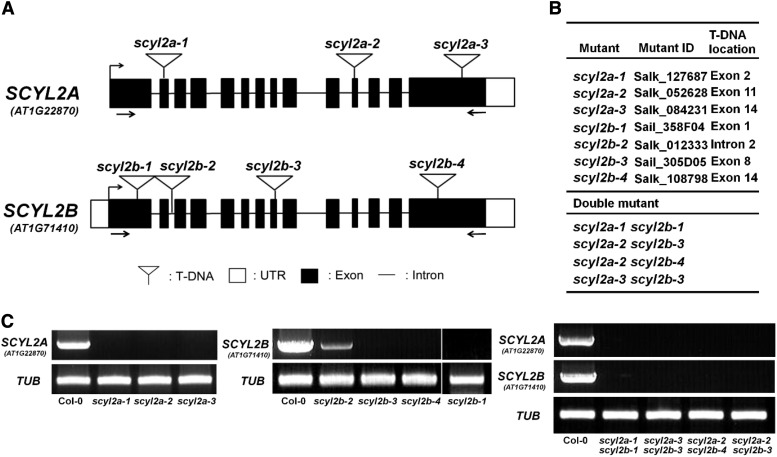
To characterize the functions of SCYL2 genes in Arabidopsis, we identified several T-DNA insertional mutant lines of SCYL2A and SCYL2B from Salk and Sail lines. Three scyl2a alleles and four scyl2b alleles were obtained (Fig. 2). Reverse transcription (RT)-PCR analysis confirmed the absence of full-length transcripts of SCYL2A or SCYL2B in scyl2a or scyl2b alleles except for scyl2b-2, which has a decreased amount of full-length transcripts due to the T-DNA insertion of the second intron in SCYL2B (Fig. 2, B and C). Four scyl2a scyl2b double mutants were obtained from the crosses between scyl2a and scyl2b alleles (Fig. 2B), and RT-PCR analysis showed that full-length transcripts of both SCYL2A and SCYL2B were absent in all scyl2a scyl2b double mutants (Fig. 2C).
Figure 2.
Gene structure of SCYL2A and SCYL2B and locations of T-DNA insertions. A, A schematic diagram shows the positions of T-DNA insertions in scyl2a and scyl2b alleles. Black boxes represent exons. Lines between black boxes represent introns. White boxes represent untranslated regions (UTR). White triangles indicate T-DNA insertion sites. The positions of primers that are used in C to check the presence of full-length transcripts of SCYL2A or SCYL2B are indicated by arrows. B, Salk and Sail lines used to isolate scyl2a and scyl2b alleles, with their T-DNA locations indicated. Four different double mutants were obtained between scyl2a and scyl2b mutants. C, RT-PCR analysis of SCYL2A and SCYL2B transcripts in wild-type Columbia-0 (Col-0), single mutants, and double mutants. The positions of the primers used are shown in A. A β-tubulin gene (TUB2) was used as a positive control for the RT-PCR. All scyl2a and scyl2b alleles are knockout mutants, except for scyl2b-2, which has a decreased level of full-length transcripts. Single PCRs were performed with cDNA from individual plants of each genotype.
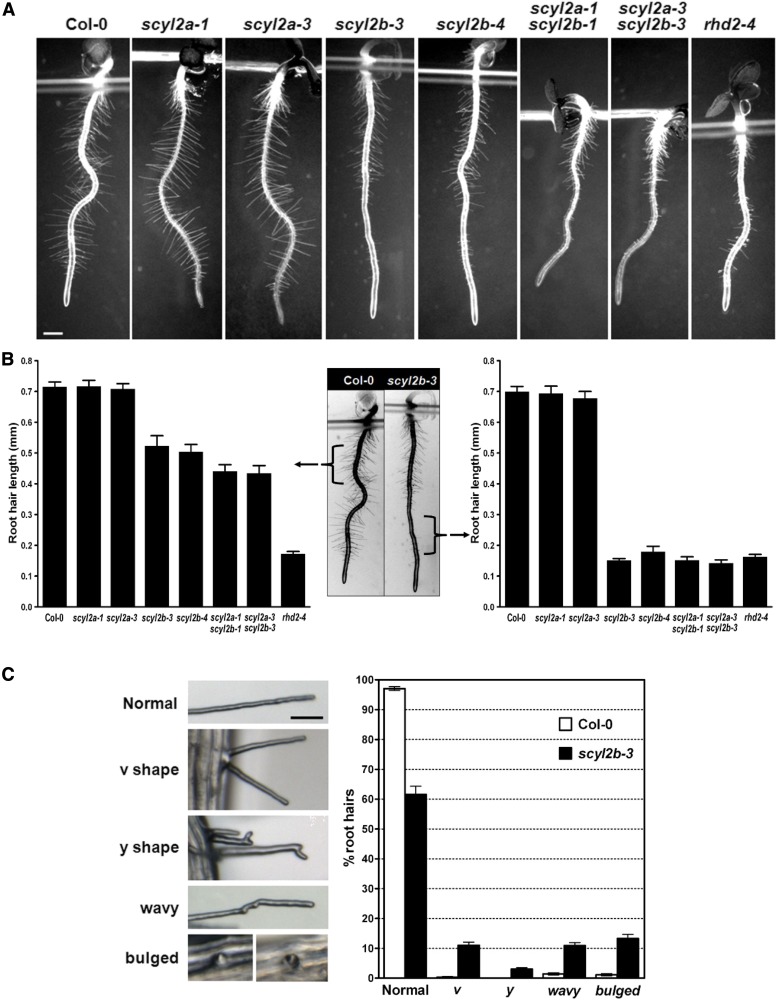
To investigate the possible role of SCYL2A and SCYL2B in plants, we analyzed the phenotypes of scyl2a, scyl2b, and scyl2a scyl2b double mutant alleles. Three days after germination, the root hairs of scyl2b alleles and scyl2a scyl2b double mutants were shorter than those of the wild type (P < 0.05; Fig. 3, A and B, left) and were as short as those of rhd2-4, which has been shown to be a root hair-defective mutant (P < 0.05; Fig. 3, A and B, right). The scyl2b alleles also had abnormal root hair phenotypes such as v shape, y shape, wavy shape, and bulged shape (Fig. 3C). The percentage of aberrant root hairs per root was much higher in scyl2b mutants than in the wild type (P < 0.05; Fig. 3C). There were no defects of root hairs in scyl2a alleles (Fig. 3, A and B). These data indicate that SCYL2B plays an important role in polarized root hair growth.
Figure 3.
SCYL2B functions in root hair development. A, Light microscopy images showing root hair length of wild-type Col-0, scyl2a, scyl2b, and scyl2a scyl2b double mutant alleles. Three-day-old roots were used for the measurement of root hair length in A and B. Bar = 0.5 mm. B, Root hair length of seedlings of wild-type Col-0, scyl2a, scyl2b, and scyl2a scyl2b double mutant alleles. Root hair length was measured in two separate regions of root: the 2-mm region below hypocotyls (left graph and middle image) and the 2-mm region starting 0.5 mm above the root hair differentiation zone (right graph and middle image). Fifteen root hairs per each seedling were measured, and 10 seedlings per genotype were used for root hair length measurements (n = 10 seedlings; means ± se). C, Abnormal root hairs in scyl2b-3. Root hairs in scyl2b-3 were classified into five classes based on their phenotype (normal, v shape, y shape, wavy, and bulged). Representative images of each class are shown (left). Percentages of each root hair class were determined in wild-type Col-0 plants and scyl2b-3 (right). About 70 to 100 root hairs per each seedling were counted in 5-mm regions starting 0.5 mm above the root hair differentiation zone (n = 10 seedlings; means ± se). At least two independent experiments were performed.
SCYL2A and SCYL2B Are Essential for Plant Growth
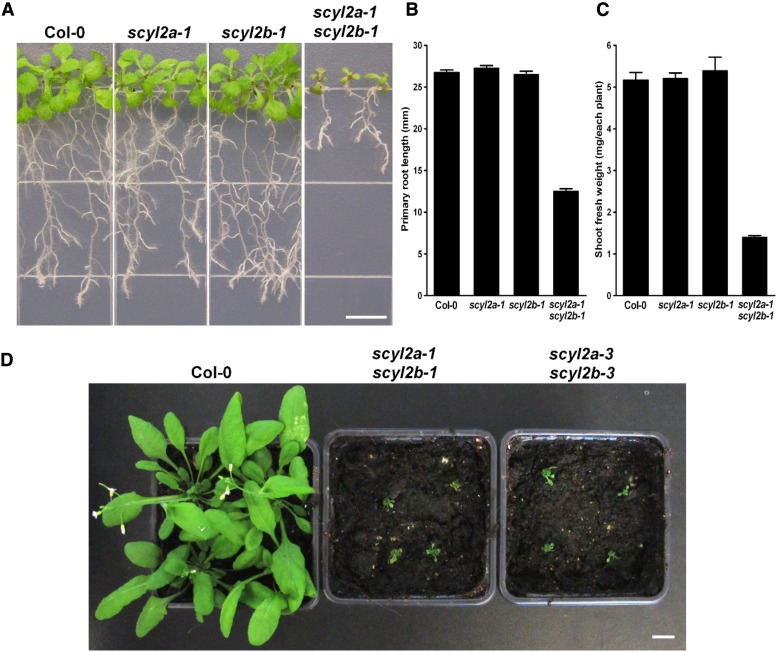
We further analyzed the phenotypes of roots and shoots in scyl2a, scyl2b, and scyl2a scyl2b alleles. Ten days after germination, scyl2a scyl2b double mutants had significantly shorter primary roots and smaller shoots (P < 0.05; Fig. 4, A–C) while scyl2a or scyl2b single mutants had no difference in roots and shoots compared with the wild type (Fig. 4, A–C). One-month-old scyl2a scyl2b double mutants grown in soil showed a striking tiny shoot phenotype (Fig. 4D). These data indicate that both SCYL2A and SCYL2B are involved in plant growth and development and redundant in certain aspects of their function.
Figure 4.
SCYL2A and SCYL2B are important in plant growth and development. A, Images of 10-d-old wild-type Col-0, scyl2a-1, scyl2b-1, and scyl2a-1 scyl2b-1 double mutants. Bar = 0.5 cm. B, Primary root length of the wild type and scyl2 knockouts. Values are means ± se from 50 roots. C, Shoot fresh weights of the wild type and scyl2 knockouts. Values are means ± se from 10 seedlings per replicate (n = 6 replicates). D, Image of 1-month-old wild-type Col-0, scyl2a-1 scyl2b-1, and scyl2a-3 scyl2b-3. Bar = 1 cm.
SCYL2A and SCYL2B Expression Levels in Various Tissues
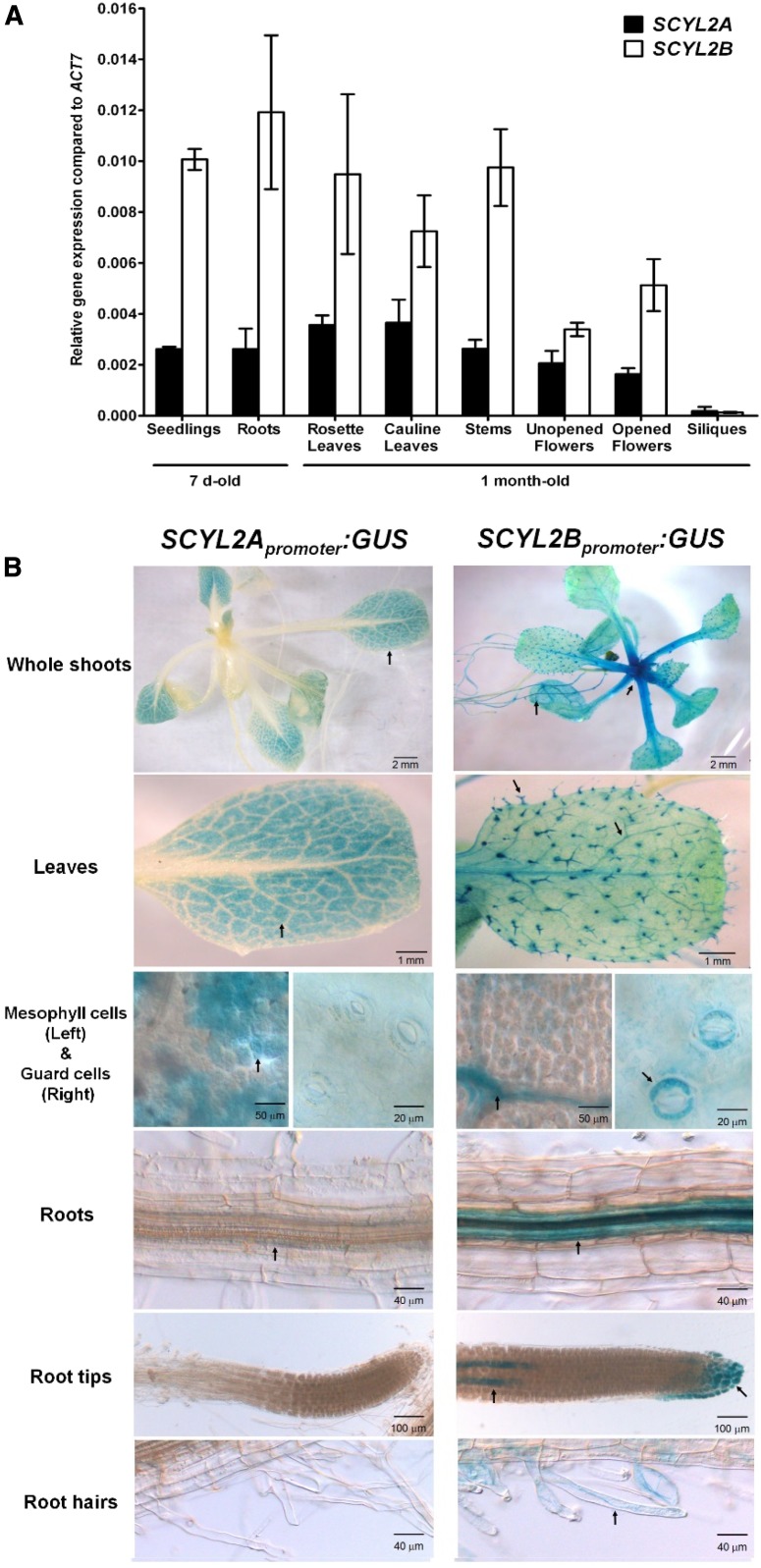
To determine in which developmental stages or tissues in Arabidopsis SCYL2A and SCYL2B are highly expressed, we analyzed gene expression levels in various tissues of 7-d-old seedlings and 1-month-old plants. Quantitative RT-PCR analysis showed that both SCYL2A and SCYL2B are coexpressed in all tissues tested, such as roots, rosette leaves, cauline leaves, stems, unopened flowers, opened flowers, and siliques (Fig. 5A), and that the expression of SCYL2B was about 2- to 3-fold higher compared with that of SCYL2A (Fig. 5A). The expression level of both SCYL2A and SCYL2B was very low in siliques (Fig. 5A).
Figure 5.
SCYL2A and SCYL2B expression levels in various tissues. A, Quantitative real-time PCR analysis of SCYL2A and SCYL2B expression in various tissues. Tissues from 7-d-old plants and 1-month-old plants were used in this analysis. The levels of SCYL2A and SCYL2B transcripts were normalized to ACTIN7. Three biological replicates were used, and error bars represent se. B, Spatial localization of SCYL2A and SCYL2B. Histochemical localization is shown for GUS activity in plants carrying SCYL2Apromoter:GUS (left) and SCYL2Bpromoter:GUS (right). About 2-week-old plants grown on plates were used for the visualization of GUS activity. Arrows indicate GUS staining.
To analyze the tissue-specific expression patterns of SCYL2A and SCYL2B in Arabidopsis, we monitored GUS expression driven by the promoters of SCYL2A and SCYL2B in transgenic plants. In plants carrying SCYL2Apromoter:GUS, GUS staining was detected mainly in leaf mesophyll cells, while very weak GUS staining was seen in root vascular tissues (Fig. 5B). In plants carrying SCYL2Bpromoter:GUS, strong GUS staining was detected in both shoot and root vascular tissues, in root cap regions including columella cells and lateral root caps, and also in some tissues of specialized cell types such as trichomes, guard cells, and root hairs (Fig. 5B).
SCYL2B Is Localized at the Golgi, TGN, and PVC
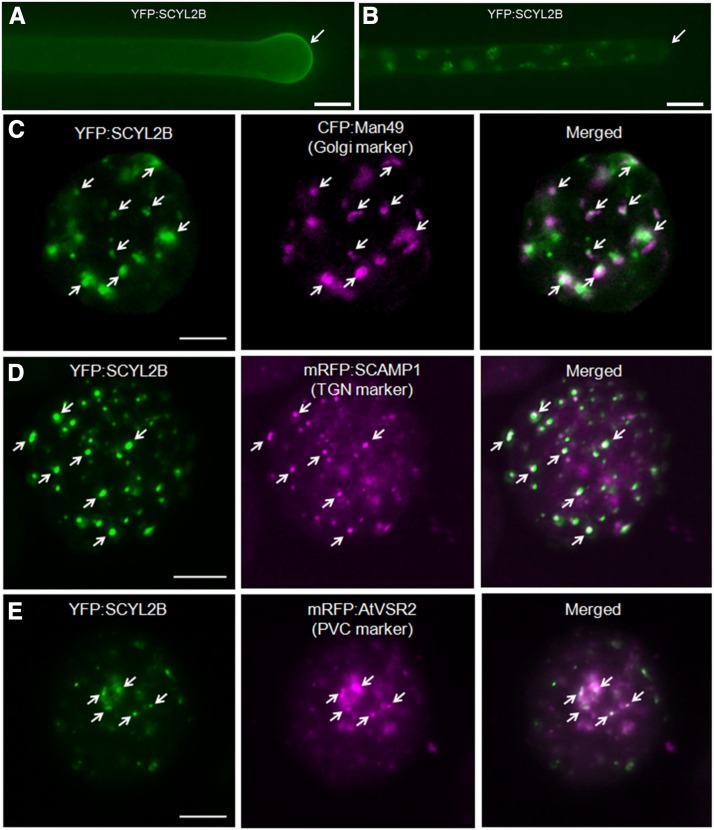
To define the subcellular localization of SCYL2B in Arabidopsis, transgenic plants carrying 35S:YFP:SCYL2B were analyzed. YFP:SCYL2B was localized to the tips of growing root hair cells (Fig. 6A; Supplemental Fig. S2). However, in mature root hairs where tip growth was finished, the tip-localization pattern of YFP:SCYL2B disappeared (Fig. 6B; Supplemental Fig. S2). In addition, YFP:SCYL2B appeared to be localized at some aggregated endosomal structures (Fig. 6B). In animals, a clathrin-coated vesicle-associated kinase of 104 kD (CVAK104), an Arabidopsis SCYL2A/B ortholog, plays an important role in vesicle trafficking and is localized at the Golgi, TGN, and endosomes (Düwel and Ungewickell, 2006; Borner et al., 2007). To determine the subcellular localization of SCYL2B, colocalization studies were performed using several fluorescent protein markers for subcellular organelles such as the Golgi, TGN, PVC, PM, mitochondria, plastids, and peroxisomes. In Arabidopsis leaf protoplasts and leaf epidermal cells, YFP:SCYL2B was colocalized with markers for the Golgi, TGN, and PVC but not with markers for mitochondria, plastids, peroxisomes, and PM (Fig. 6; Supplemental Figs. S3 and S4). The percentages of YFP:SCYL2B-positive spots overlapping with the Golgi, TGN, and PVC organelle marker proteins were about 38%, 37%, and 26%, respectively (Supplemental Table S1). The localization data of SCYL2B suggest that SCYL2B is membrane localized and may be involved in vesicle membrane trafficking in plant cells.
Figure 6.
SCYL2B is localized at the Golgi, TGN, and PVC. A, YFP:SCYL2B is tip localized in growing root hairs. B, The tip-localization pattern disappears in mature root hairs. Root hairs of transgenic Arabidopsis plants expressing YFP:SCYL2B were observed by wide-field fluorescence microscopy. Bars in A and B = 10 μm. C to E, Localization of SCYL2B in various subcellular organelles. Arabidopsis leaf protoplasts were cotransformed with YFP:SCYL2B and the indicated constructs, which encode protein markers for the Golgi (C), TGN (D), and PVC (E). The localization of SCYL2B was examined by the detection of YFP, CFP, and RFP signal by confocal microscopy. White color indicates strong colocalization. Arrows indicate spots showing colocalization. Bars = 20 μm.
SCYL2B Interacts with CHC1, VTI11, and VTI12
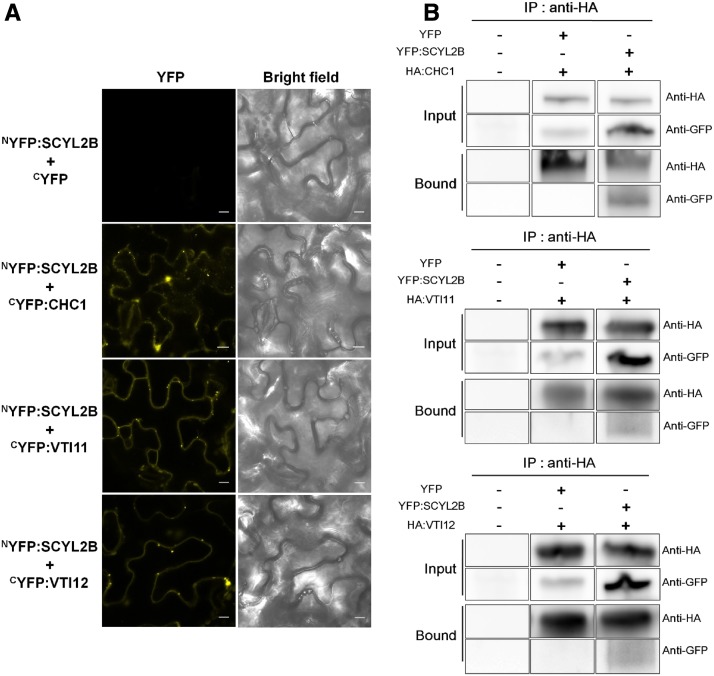
In animals, SCYL2 acts as a component of CCVs and can interact with clathrin (Conner and Schmid, 2005; Düwel and Ungewickell, 2006). To determine whether Arabidopsis SCYL2 proteins also can bind to components of CCVs, we conducted a split YFP-based bimolecular fluorescence complementation (BiFC) assay. To make constructs for BiFC, the N-terminal fragment of YFP (nYFP) was fused to the N terminus of SCYL2B and the C-terminal fragment of YFP (cYFP) was fused to the N terminus of putative SCYL2B-binding candidates such as CHC1, VTI11, and VTI12. The nYFP:SCYL2B construct was transiently coexpressed with the cYFP fusion constructs mentioned above in tobacco (Nicotiana benthamiana) leaf epidermal cells via Agrobacterium tumefaciens infiltration. Clear YFP fluorescence signal showing a punctate staining pattern was detected when nYFP:SCYL2B was transiently cotransformed with cYFP:CHC1, cYFP:VTI11, or cYFP:VTI12 (Fig. 7A). There was no YFP fluorescence signal when nYFP:SCYL2B was transiently cotransformed with cYFP (Fig. 7A). Western-blot analysis using polyclonal anti-GFP antibody showed that cYFP:CHC1, cYFP:VTI11, and cYFP:VTI12 were coexpressed well with nYFP:SCYL2B in the tobacco leaf cells (Supplemental Fig. S5). As well as BiFC experiments, we also conducted coimmunoprecipitation experiments to confirm the protein interaction of BiFC results. HA:CHC1, HA:VTI11, and HA:VTI12 were transiently coexpressed with YFP and YFP:SCYL2B in tobacco leaf cells, and then the HA-tagged proteins were pulled down by anti-HA beads and probed by anti-GFP antibody. YFP:SCYL2B was detected in the bound fraction of HA:CHC1, HA:VTI11, and HA:VTI12, whereas YFP alone was not (Fig. 7B), indicating that SCYL2B interacts with CHC1, VTI11, and VTI12. We also tested the protein interaction by pulling down YFP-tagged proteins first, followed by the detection of HA-tagged proteins, since commercial anti-GFP beads called GFP-trap also were available. When YFP and YFP:SCYL2B were immunoprecipitated by GFP-trap beads and then probed by anti-HA antibody, HA:CHC1, HA:VTI11, and HA:VTI12 were detected in the bound fraction of YFP:SCYL2B (Supplemental Figs. S6 and S7), confirming the protein interaction. However, a weak signal of HA:VTI11 and HA:VTI12 also was detected in the bound fraction of YFP alone, despite thorough washing of beads, indicating that VTI11 and VTI12 tend to bind GFP-trap beads nonspecifically (Supplemental Figs. S6 and S7). Taken together, these data indicate that Arabidopsis SCYL2B interacts with a clathrin and two v-SNAREs, VTI11 and VTI12, in vivo.
Figure 7.
SCYL2B interacts with CHC1, VTI11, and VTI12. A, BiFC assay showing in vivo interactions of SCYL2B with CHC1, VTI11, and VTI12. The nYFP:SCYL2B construct was transiently coexpressed with cYFP fusion constructs such as CHC1, VTI11, and VTI12 in tobacco leaf epidermal cells via A. tumefaciens infiltration. Bright-field images and YFP fluorescence images are shown. Bars = 10 μm. B, Coimmunoprecipitation of SCYL2B with CHC1, VTI11, and VTI12. YFP and YFP:SCYL2B constructs were transiently coexpressed with HA:CHC1, HA:VTI11, and HA:VTI12 in tobacco leaf epidermal cells. HA-tagged fusion proteins were coimmunoprecipitated by anti-HA antibody-conjugated magnetic beads and probed with anti-GFP antibody. IP, Immunoprecipitation; Input, total protein extracts before IP; Bound, bound fraction after IP.
SCYL2B Colocalizes with SCYL2A and CHC1 and Is Required for Tip Localization of CSLD3 in Root Hairs
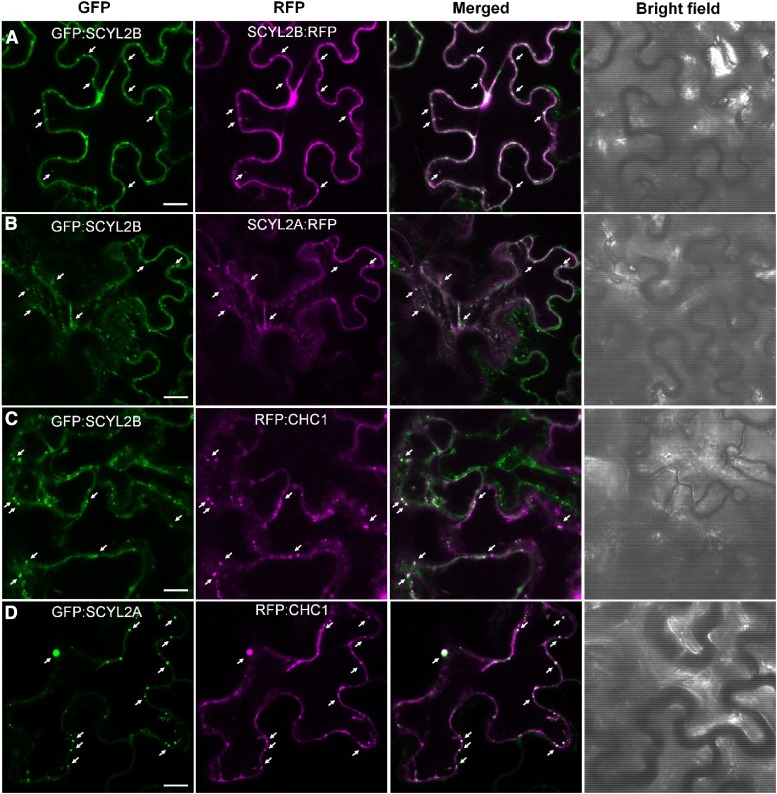
We determined the localization of SCYL2 proteins by tagging at the N or C terminus with the fluorescent proteins GFP and RFP. When N-terminal GFP-tagged SCYL2B and C-terminal RFP-tagged SCYL2B were transiently coexpressed in tobacco leaf epidermal cells, there was no difference in the localization of these proteins (Fig. 8A), suggesting that tagging fluorescent proteins to SCYL2 proteins may not alter their subcellular localization. We also compared the localization of GFP:SCYL2B with SCYL2A:RFP by coexpressing them in tobacco cells, and the results showed that there was no difference in their subcellular localization (Fig. 8B). The colocalization of SCYL2A with SCYL2B suggests that SCYL2A may act in a similar fashion to SCYL2B, probably locating at the Golgi, TGN, and PVC.
Figure 8.
SCYL2B colocalizes with SCYL2A and CHC1. Tobacco leaf epidermal cells were coinfiltrated with A. tumefaciens strains carrying the following constructs: GFP:SCYL2B and SCYL2B:RFP (A), GFP:SCYL2B and SCYL2A:RFP (B), GFP:SCYL2B and RFP:CHC1 (C), and GFP:SCYL2A and RFP:CHC1 (D). The localization of fluorescent proteins was examined by the detection of GFP and RFP signal by confocal microscopy. Representative images of each sample are shown. White color indicates strong colocalization. Arrows indicate spots showing colocalization. Bars = 10 μm.
Since BiFC and coimmunoprecipitation data showed the interaction between SCYL2B and CHC1, we also examined if they colocalize. We found that both SCYL2A and SCYL2B colocalized well with CHC1 (Fig. 8, C and D) when they were coexpressed transiently in tobacco leaf epidermal cells, supporting the notion that SCYL2 proteins are involved in clathrin-mediated vesicle trafficking.
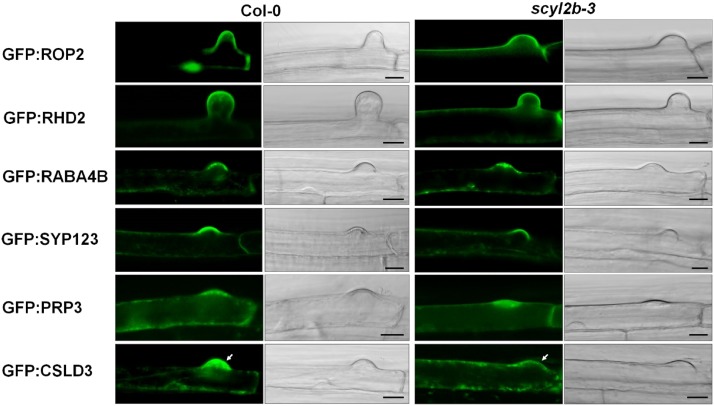
It was reported that the function of root hair tip-localized proteins, such as RHO-RELATED PROTEIN FROM PLANTS2 (ROP2), ROOT HAIR DEFECTIVE2 (RHD2), RAB GTPASE HOMOLOG A4B (RABA4B), SYNTAXIN OF PLANTS123 (SYP123), PRO-RICH PROTEIN3 (PRP3), and CSLD3, is important for proper root hair tip growth (Jones et al., 2002; Foreman et al., 2003; Preuss et al., 2004; Park et al., 2011; Ichikawa et al., 2014; Rodriguez-Furlán et al., 2016). To understand the root hair tip growth pathways in which SCYL2B functions, Col-0 and scyl2b mutants were transformed with GFP fusion constructs, including ROP2, RHD2, RABA4B, SYP123, PRP3, and CSLD3 genes, whose expression is driven by the EXPANSIN A7 (EXPA7) promoter, a root hair-specific promoter (Kim et al., 2006). Interestingly, the analysis of transgenic lines showed that only CSLD3 is mislocalized in the root hairs of scyl2b mutants (Fig. 9; Supplemental Fig. S8), indicating that SCYL2B is important for mediating the tip localization of CSLD3 proteins in root hairs.
Figure 9.
CSLD3 is mislocalized in root hairs of scyl2b mutants. Localization of proteins that are known to function at the root hair tip was observed in the background of Col-0 and scyl2b-3 mutants. The expression of N-terminal GFP fusion proteins of ROP2, RHD2, RABA4B, SYP123, PRP3, and CSLD3 was driven by the EXPA7 promoter, a root hair-specific promoter. For each gene construct, at least five independent T2 transgenic plants were used in two independent experiments that yielded similar results. Root hairs of transgenic plants were observed by confocal microscopy. White arrows indicate the difference of GFP:CSLD3 localization. Bars = 10 μm.
DISCUSSION
SCYL2 is a member of the SCY1-LIKE protein family. SCY1 is a kinase-like protein identified in Saccharomyces cerevisiae. In animals, there are three SCY1-LIKE genes: SCYL1, SCYL2, and SCYL3. SCYL1 is the gene responsible for the neurodegenerative mouse model mdf and plays important roles in COPI-mediated retrograde trafficking from the Golgi to the ER and the regulation of Golgi morphology (Burman et al., 2010). SCYL2 is involved in TGN-mediated CCV membrane trafficking. SCYL3 binds ezrin, which is a protein that mediates the interaction between transmembrane proteins and actin and is implicated in regulating cell adhesion/migration complexes in migrating cells (Sullivan et al., 2003). In plants, there are orthologs only for the animal SCYL1 and SCYL2. In Arabidopsis, the gene that encodes AT2G40730 corresponds to the ortholog of animal SCYL1, and two loci (AT1G22870 and AT1G71410) described in this study correspond to the orthologs of animal SCYL2.
We showed that Arabidopsis SCYL2B localizes at the Golgi, TGN, and PVC and colocalizes with SCYL2A and CHC1 (Figs. 6 and 8). Moreover, BiFC and coimmunoprecipitation assays showed that SCYL2B binds to CHC1 and two v-SNAREs, VTI11 and VTI12 (Fig. 7). Interestingly, the analysis of SCYL2 protein sequence alignment revealed a conserved clathrin-binding motif (SLLDLL) at the very end of the C terminus on both SCYL2A and SCYL2B (Lafer, 2002). However, based on BiFC analysis, it turned out that this binding motif is not essential for the interaction between SCYL2B and CHC1 (Supplemental Fig. S9). For the coimmunoprecipitation experiments, we used YFP alone as a negative control, and YFP alone did not bind CHC, VTI11, and VTI12. Moreover, there was no precipitation of SCYL2B in the control samples, where HA-tagged proteins are absent (Supplemental Fig. S7). We also had BiFC data showing that certain membrane proteins such as adaptor proteins, which are involved in vesicle membrane trafficking, did not interact with SCYL2B (Supplemental Fig. S10). These findings suggest that the interaction between SCYL2B and the proteins tested in this study is specific.
Our data showing SCYL2 localization and interaction indicate that Arabidopsis SCYL2 proteins play a role in CCV-mediated membrane trafficking and also suggest that plant SCYL2 may act in a similar manner to animal SCYL2. Animal SCYL2 is well known to interact with clathrin. However, there is no report showing an interaction of animal SCYL2 with SNAREs. Thus, our data showing the interaction of Arabidopsis SCYL2B with two v-SNAREs are a novel finding. In animals, SCYL2 also has been suggested to function as a SNARE adaptor (Hirst et al., 2015) due to the fact that there is a long adaptor-like structure with a predicted coiled-coil domain at the C terminus of SCYL2 (Pelletier, 2016) and that knockdowns of animal SCYL2/CVAK104 result in reduced levels of syntaxin8 and vti1b associated with CCVs (Borner et al., 2007). These findings, along with our new data showing the interaction of AtSCYL2B with two v-SNAREs, suggest that plant SCYL2 may act as a SNARE adaptor in clathrin-mediated vesicle trafficking.
In BiFC assays, a punctate staining pattern of YFP fluorescence was seen when SCYL2B was transiently coexpressed with CHC1, VTI11, and VTI12 (Fig. 7A). Moreover, a similar fluorescence pattern was observed in the colocalization studies with SCYL2 proteins and CHC1 (Fig. 8). Previously, it was shown that clathrin localizes at the TGN and PM (Blackbourn and Jackson, 1996; Dhonukshe et al., 2007; Fujimoto et al., 2010; Van Damme et al., 2011) and that VTI11 localizes primarily to the TGN, with a minor portion localized at the PVC (Zheng et al., 1999; Bassham et al., 2000; Surpin et al., 2003). VTI12 also is used as a TGN marker (Geldner et al., 2009). Likewise, the treatment of brefeldin A (BFA) and wortmannin, endomembrane trafficking inhibitors (Hicks and Raikhel, 2010; Mishev et al., 2013), induced the formation of BFA compartments and ring-like structures of SCYL2B proteins (Supplemental Fig. S11). Based on the colocalization data and chemical inhibitor assay, it is likely that SCYL2 proteins function with clathrin, VTI11, and VTI12 mainly at the TGN and/or PVC.
Since the localization of SCYL2 proteins was obtained by transient overexpression in Arabidopsis and tobacco leaf epidermal cells, we cannot completely rule out the possibility that the localization of endogenous SCYL2 could be different from that of overexpressed proteins. However, we believe that the localization of SCYL2 based on transient overexpression represents the actual localization of SCYL2 in the cell for the following reasons. First, when SCYL2 was expressed in tobacco cells using the native SCYL2B promoter or EXPA7 promoter, the localization pattern of SCYL2 was similar to that of overexpressed SCYL2 (Supplemental Fig. S12). Second, the fact that animal SCYL2 localizes to the Golgi, TGN, and endosomes also is consistent with our findings about SCYL2 localization. Lastly, the protein interaction and colocalization data showing the functional relationship of SCYL2 proteins with CHC1, VTI11, and VTI12, which mainly localize to the TGN and/or the PVC, also support our localization data.
We showed that the loss of function of SCYL2B leads to the formation of both short root hairs and morphologically abnormal root hairs such as wavy hairs and branched hairs (Fig. 3), indicating that SCYL2 is involved in both root hair elongation and the polar tip growth of root hairs. SCYL2B was located to the tips of growing root hair cells, and the tip-localization pattern of SCYL2B disappeared in mature root hairs (Fig. 6, A and B; Supplemental Fig. S2), providing further evidence for the role of SCYL2B in the polar tip growth of root hairs. Active membrane trafficking processes govern the polar tip growth of root hairs by the regulation of endocytosis and exocytosis (Ishida et al., 2008; Lee and Yang, 2008). Based on the observation that SCYL2B localizes at the Golgi, TGN, and PVC (Fig. 6) and the fact that SCYL2B binds to and colocalizes with clathrin and SNAREs (Figs. 7 and 8), it is possible that SCYL2B is involved in CCV-mediated membrane trafficking in the polar tip growth of root hairs. In plants, the Golgi and TGN/early endosomes (EE) play a critical role in the trafficking of secretory, endocytic, and vacuolar vesicles and also are important for root hair development (Hwang and Robinson, 2009). For example, RHD2, an NADPH oxidase that produces reactive oxygen species, is known to play an important role in root hair elongation. It was shown that vesicle trafficking mediated by the Golgi and TGN/EE is essential for both PM targeting and the function of RHD2, indicating that the proper function of the Golgi and TGN/EE is important for root hair elongation (Takeda et al., 2008; Lam et al., 2009). The function of PVC also is known to be important for root hair tip growth. It was shown that LY294002, a phosphatidylinositol 3-kinase-specific inhibitor, inhibits the fusion of PVC/late endosomes with vacuoles, leading to the significant reduction of root hair length (Lee et al., 2008), supporting the role of PVC in root hair elongation. Thus, the localization of SCYL2B on the Golgi, TGN, and PVC and its root hair phenotype also support the roles of these endomembrane systems in the development of root hairs.
The localization of proteins essential for root hair tip growth indicated that SCYL2B is not involved in the vesicle trafficking pathways for RHD2 and SYP123, which are membrane proteins localized at the root hair tip. However, the mislocalization of CSLD3, a membrane protein, in root hairs of scyl2b mutants showed that SCYL2B plays a role in the vesicle trafficking pathway that mediates the PM localization of CSLD3 at the root hair tip. Previously, it was reported that CSLD3 is localized at the Golgi, ER, and PM (Favery et al., 2001; Bernal et al., 2008; Zeng and Keegstra, 2008; Park et al., 2011), and it was suggested that cellulose synthases, CSLD3 homologs, may be trafficked to the plasma membrane via the Golgi and TGN (Schneider et al., 2016). Taken together, these findings suggest that SCYL2B may be involved in the Golgi/TGN-mediated exocytosis/secretion of vesicles containing CSLD3. To determine whether SCYL2B proteins are involved in endocytosis processes, we also performed an uptake assay using the endocytic tracer N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide (FM4-64) in root hair cells of the wild type and scyl2a scyl2b double mutants and found no difference in FM4-64 uptake (Supplemental Fig. S13). It was reported that FM4-64 uptake decreased only in chc2 mutants but not in chc1 mutants (Kitakura et al., 2011). Thus, it is unlikely that SCYL2 proteins, whose function may be associated with CHC1 based on protein interaction studies, are involved in endocytosis. Overall, based on both the subcellular localization and the binding characteristics of SCYL2B proteins, we suggest that SCYL2B may act as a component of clathrin-mediated vesicle membrane trafficking that regulates exocytosis/secretion mediated by TGN/EE and PVC in the process of root hair tip growth.
We also showed that plant shoot and root growth is reduced severely in scyl2a scyl2b double mutants, while the only observable phenotype in the single mutants was a root hair defect in scyl2b (Figs. 3 and 4). This indicates that SCYL2A and SCYL2B act redundantly in most tissues and play a critical role in processes other than root hair development, such as plant cell growth. However, although the phenotypic data using mutants indicate that there is some functional redundancy between SCYL2A and SCYL2B, the GUS expression pattern suggests that the function of SCYL2A and SCYL2B is somewhat specified to certain tissues. For example, based on GUS staining data, it seems that SCYL2A has a dominant role in leaf mesophyll cells whereas SCYL2B has a dominant role in vascular tissues and in certain specialized cell types, such as trichomes, guard cells, and root hairs, where lipid membrane change occurs actively. Since the GUS expression data showed that SCYL2 proteins are expressed in root columella cells, trichomes, and guard cells, we examined scyl2 mutants more carefully for the phenotype of these tissues and found an abnormal morphology of root columella cells only in scyl2a scyl2b double mutants, suggesting a role of SCYL2 proteins in the development of root columella cells (Supplemental Fig. S14). No phenotypic difference is found in the cell types of guard cells and trichomes (data not shown). We also examined the shoot and root phenotypes of several SCYL2B, HA-SCYL2B, and YFP-SCYL2B overexpression lines, and there were no phenotypic differences in these lines when compared with the wild type (data not shown).
In animals, SCYL2 also is known as CVAK104 and has a putative Ser/Thr protein kinase domain at its N terminus (Conner and Schmid, 2005). However, animal and plant SCYL2 genes lack a number of highly conserved catalytic residues within the putative kinase domain, such as Asp, which is essential in other kinases (Manning et al., 2002). It is still controversial whether SCYL2 has kinase activity. It was reported that, in the presence of poly-l-Lys, SCYL2 was able to autophosphorylate and to phosphorylate the β2-adaptin subunit of AP2 (Conner and Schmid, 2005), indicating that SCYL2 has an ability to phosphorylate some proteins under certain circumstances. However, in another report, kinase activity was not detected in immunoprecipitated or bacterially expressed SCYL2/CVAK104 (Düwel and Ungewickell, 2006). We tried to express the several recombinant forms of Arabidopsis SCYL2B with 6xHis or GST tags in Escherichia coli to find out whether it had kinase activity but failed to express the protein, possibly due to the instability of the protein. In a systematic comparison study using 1,019 Arabidopsis protein kinases, Arabidopsis SCYL2 genes were classified as unclustered kinases (Wang et al., 2003). It may be possible that Arabidopsis SCYL2 proteins may be kinases that regulate membrane trafficking by phosphorylating CCV components. More detailed studies, such as phospho-proteomic approaches using mutant alleles and mutagenesis approaches of the putative kinase domain, will be required to demonstrate that SCYL2 is a protein kinase.
In conclusion, the data presented here provide evidence that SCYL2 genes play an important role in plant cell growth and may be involved in clathrin-mediated vesicle trafficking. Additional studies will be required to pinpoint the precise function of the SCYL2 genes. The identification of other binding partners of the SCYL2 proteins and investigation about the possibility that SCYL2 proteins act as kinases will be a next step toward revealing the detailed roles of the SCYL2 genes in plant membrane trafficking and developmental processes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants used in this study were in the Columbia background. Seeds were sterilized in 70% (v/v) ethanol and 0.05% (v/v) Triton X-100 and then planted on 10-cm-diameter sterile plates containing ammonium-free complete nutrient agar medium (Jung et al., 2009). After stratification of the seeds at 4°C for 2 to 3 d, the plates were transferred to the growth chamber at 22°C with a 16-h daylength at 200 µmol m–2 s–1. Seedlings were grown vertically. Three- to 10-d-old seedlings grown on plates were used throughout the study. In addition, plants also were grown on soil under the same conditions. Unless stated otherwise, all experiments in this study were repeated at least twice with similar results.
Characterization of T-DNA Insertion Mutants and RT-PCR
T-DNA insertional Salk and Sail lines were obtained from the Arabidopsis Biological Resource Center stock center. Homozygous Salk and Sail lines for scyl2a and scyl2b alleles were isolated by PCR-based genotyping using the following primers: 5′-CAAACCACATGAAGCTCAGTATTC-3′ and 5′-GATTATCTAAAGCTGCTGAAGATGC-3′ for scyl2a-1, 5′-CTCGATGGTCAGGTAATGCTTGATAC-3′ and 5′-GTTGGTACTGATGTCCGGTTCGATGACT-3′ for scyl2a-2, 5′-CAGGAAAATAGAGGAAAAAAGAGGAGTT-3′ and 5′-TCACAATAGATCTAACAAAGATGGCTGT-3′ for scyl2a-3, 5′-GCATGTTATCCAAATTCCCAGCCTG-3′ and 5′-AACAAGTTGCAGAAAGAGAGAGAGGTG-3′ for scyl2b-1, 5′-GCGCTTGGTAATGTTGAGAATGTGG-3′ and 5′-AGTGGAACGATTGCATGTTATCCA-3′ for scyl2b-2, 5′-TCCGAAGTGACGCTAGGTTACGTGCT-3′ and 5′-AACGATCTACGGCAGTACACCGTTG-3′ for scyl2b-3, and 5′-GAATTCACAGCAGAACATGTGCT-3′ and 5′-TCATAATAGATCCAATAGAGATGGTTGTGAACCA-3′ for scyl2b-4. To check whether the transcripts of SCYL2A and SCYL2B are produced in scyl2a, scyl2b, and double mutant alleles, RNA was extracted from the mutant alleles with a previously described method (Oñate-Sánchez and Vicente-Carbajosa, 2008) and was reverse transcribed to cDNA. Then, RT-PCR was conducted with the cDNA and the following primers: 5′-TCCTGGTCTCGCTTGGAAGCTTTA-3′ and 5′-TGCTGCTTTGAACCATTTGGCTTAG-3′ for SCYL2A and 5′-CACCATGTCGATAAACATGAAAACATTTACTCAAGCTC-3′ and 5′-TCATAATAGATCCAATAGAGATGGTTGTGAACCA-3′ for SCYL2B. As a control gene, β-tubulin (AT5G62690) was amplified with the primers 5′-GCCAATCCGGTGCTGGTAACA-3′ and 5′-CATACCAGATCCAGTTCCTCCTCCC-3′. PCR amplification was performed with 37 cycles and visualized using ethidium bromide after separation on agarose gels.
Phylogenetic Analysis and Similarity Matrix
Alignment of protein sequences was performed with ClustalW in MEGA4. A phylogenetic tree was constructed using neighbor joining, Poisson correction, pairwise deletion, and bootstrap values with 10,000 replicates in MEGA4. The similarity matrix was generated using pairwise sequence comparison of 16 sequences to determine the p-distance, the number of amino acid substitutions per site. Protein sequence identity is shown as (1 − p-distance) × 100.
Root Hair Length Measurement and Root Hair Phenotype Analysis
Photographs of 3-d-old roots grown on complete nutrient plates were taken using a Nikon SMZ1500 microscope and a Q-Imaging Retiga cooled 12-bit camera. Root hair length was measured by using NIH ImageJ software program in two separate regions of roots: the 2-mm region below hypocotyls and the 2-mm region starting 0.5 mm above the root hair differentiation zone. Fifteen root hairs per each root were measured, avoiding root hairs that were growing into the medium, and 10 roots per genotype were used for root hair length measurements. Two independent experiments were performed, and similar results were obtained. The number of abnormal root hairs was counted based on five different categories: normal shape, v shape, y shape, wavy shape, and bulged shape. About 70 to 100 root hairs per each seedling were counted in the 5-mm region starting 0.5 mm above the root hair differentiation zone, and 10 seedlings were used in the analysis. Data were displayed as percentages of each root hair class in the wild type and scyl2b-3.
Phenotype Assay
Photographs of 10-d-old plants grown on complete nutrient plates were taken for primary root length measurement and subjected to shoot fresh weight measurement. For each mutant, 10 seedlings per plate were grown, and six replicate plates were used. For the measurement of primary root length, 50 roots per each mutant were used. Shoot fresh weight was measured by bulking 10 plants from each of six plates. Statistical significance was evaluated with Student’s t test. Two independent experiments were performed, and similar results were obtained.
Quantitative Real-Time PCR Analysis
Various tissues from 7-d-old seedlings or 1-month-old plants were harvested, and total RNA was isolated as described previously (Oñate-Sánchez and Vicente-Carbajosa, 2008). RNA was quantified and treated with RQ RNase-free DNase I (Promega). DNase-treated RNA was tested for genomic DNA contamination, and the quality of total RNA was determined by agarose gel electrophoresis. Two and one-half micrograms of DNA-free RNA was then reverse transcribed using the First-Stand Synthesis System (Invitrogen). Quantitative real-time RT-PCR analysis was performed using the StepOnePlus Real-Time PCR System (Applied Biosystems) and Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). To analyze the expression of SCYL2A and SCYL2B by real-time quantitative PCR, primers 5′-TCAAACCACAGTCCAGGAGGTTACAG-3′ and 5′-TCAAGCGCCTGAACAACATGAACC-3′ were used for the SCYL2A gene and primers 5′-AGTACAGGAAGTTACGGGACCAAAG-3′ and 5′-AGCGGCTCCGTAACTAAAGCCATAGC-3′ were used for the SCYL2B gene. For the normalization of SCYL2A and SCYL2B transcripts, the ACTIN7 gene (AT5G09810) was amplified with the primers 5′-ACCACTACCGCAGAAC-3′ and 5′-GCTCATACGGTCAGCA-3′ and was used as an internal control. Statistical differences in SCYL2A and SCYL2B transcript levels between samples were evaluated by Student’s t test using ΔΔCt values (Yuan et al., 2006). Three biological replicates were used to generate means and statistical significance.
Plasmid Construction and Plant Transformation
To generate the vector constructs used in this study, the open reading frames of full-length genes were amplified by PCR using NEB Phusion polymerase and the following primers: sense 5′-CACCATGTCGATTAATATGAGAACGCTAA-3′ and antisense 5′-TCACAATAGATCTAACAAAGATGGC-3′ for SCYL2A (AT1G22870), sense 5′-CACCATGTCGATTAATATGAGAACGCTAA-3′ and antisense 5′-CAATAGATCTAACAAAGATGGC-3′ for SCYL2A without a stop codon, sense 5′-CACCATGTCGATAAACATGAAAACATTTACTCAAGCTC-3′ and antisense 5′-TCATAATAGATCCAATAGAGATGGTTGTGAACCA-3′ for SCYL2B (AT1G71410), sense 5′-CACCATGTCGATAAACATGAAAACATTTACTCAAGCTC-3′ and antisense 5′-TAATAGATCCAATAGAGATGGTTGTGAACCA-3′ for SCYL2B without a stop codon, sense 5′-CACCATGGCGGCTGCTAACGCGCCCATCATTATG-3′ and antisense 5′-TTAGTAGCCGCCCATCGGTGGCATTCCA-3′ for CHC1 (AT3G11130), sense 5′-CACCATGAGTGACGTGTTTGATGGATAT-3′ and antisense 5′-TTACTTGGTGAGTTTGAAGTACAAGATGAT-3′ for VTI11 (AT5G39510), sense 5′-CACCATGAGCGACGTATTTGAAGGGTACGAGCGT-3′ and antisense 5′-TTAATGAGAAAGCTTGTATGAGATGATCAA-3′ for VTI12 (AT1G26670), sense 5′-CACCATGAATCCCTTTTCTTCT-3′ and antisense 5′-TCACAACCCGCGAGGGAA-3′ for AP-1 (AT1G23900), sense 5′-CACCATGACCGGAATGAGAGGTCTCTCCGTA-3′ and antisense 5′-TTAAAGTAAGCCAGCGAGCATAGCTCC-3′ for AP-2 (AT5G22770), and sense 5′-CACCATGTCGTCGTCTTCCACTTCTATAATG-3′ and antisense 5′-TCACAAGAGAAAATCTGGAATTATAAC-3′ for AP-3 (AT1G48760). The PCR products were inserted into pENTR/D-TOPO following the manufacturer’s instructions (Invitrogen). The inserts were sequenced to make sure that no changes were introduced by PCR. The SCYL2B gene in pENTR/D-TOPO was introduced into the destination plasmid pEARLYGATE104:N-YFP or pEARLYGATE101:N-HA to yield pEARLYGATE:YFP(or HA):SCYL2B, which allows the expression of fusion proteins with YFP or HA tag at the N-terminal position under the control of the cauliflower mosaic virus 35S promoter. For the colocalization study between SCYL2 proteins and CHC1 in tobacco (Nicotiana benthamiana) leaf epidermal cells, genes of SCYL2A, SCYL2B, and CHC1 in pENTR/D-TOPO were cloned into either pMDC43:N-GFP vector or pB7WGR2:N-RFP to generate overexpression vectors with N-terminal fluorescent protein fusion genes. The SCYL2A/2B without stop codons were cloned into pMDC83:C-GFP vector and pB7RWG2:C-RFP to yield C-terminal GFP (and RFP)-fused SCYL2A/2B overexpression vectors.
To generate the pGreenII:SCYL2A/2Bpromoter:GUS plasmids, pGreenII:GUS plasmid (Hellens et al., 2000) was first digested by NcoI and HindIII restriction enzymes. Then, DNA fragments containing SCYL2A and SCYL2B promoter regions (about 1.5 kb) were amplified by PCR using NEB Phusion polymerase and the following primers: sense 5′-ATTATAAGAACACTACCTTGATCCTTTGCAAGTATTGGA-3′ and antisense 5′-TACAGGACGTAACACCATTTTTTTTTGTCAGAATTTAACTGTTTCTCCA-3′ for the SCYL2A promoter and sense 5′-ATTATAAGAACACTAATTCATCTCTTTGGTCAGTTACGT-3′ and antisense 5′-TACAGGACGTAACACCATTTCTGCTGGATCCAATTACTCC-3′ for the SCYL2B promoter. The digested plasmid and PCR-amplified DNA fragments were recombined using a commercially available In-Fusion multiple DNA assembly cloning method (Clontech Laboratories) to yield pGreenII:SCYL2A/2Bpromoter:GUS plasmids. Transgenic Arabidopsis plants carrying pEARLYGATE:YFP:SCYL2B and pGreenII:GUS plasmids containing SCYL2A and SCYL2B promoters were generated by Agrobacterium tumefaciens-mediated transformation, and T2 or T3 transgenic lines were used in this study.
To generate the pMDC43:EXPA7promoter:N-GFP plasmids carrying various N-terminal GFP-fused genes that encode proteins with root hair tip localization, the 35S promoter of pMDC43 vector was replaced with the 1.5-kb promoter of EXPA7 by the recombination of pMDC43 DNA fragments digested with HindIII and KpnI and PCR-amplified DNA fragments using primers 5′-CGACGGCCAGTGCCAAAGCTTAATTAGGGTCCAAGGTTTGTTC-3′ and 5′-CATTTTTTCTACCGGTACCTCTAGCCTCTTTTTCTTTATTCTTA-3′. Then, the open reading frames of full-length genes of ROP2, RABA4B, RHD2, CSLD3, SYP123, and PRP3 were PCR amplified using the following primers: sense 5′-CCGGTACCGAATTCGATGGCGTCAAGGTTTATAAAGTGT-3′ and antisense 5′-ATATCTCGAGTGCGGTCACAAGAACGCGCAACGGTTCTT-3′ for ROP2 (AT1G20090), sense 5′-CCGGTACCGAATTCGATGGCCGGAGGAGGCGGATAC-3′ and antisense 5′-ATATCTCGAGTGCGGTCAAGAAGAAGTACAACAAGTGCT-3′ for RABA4B (AT4G39990), sense 5′-CCGGTACCGAATTCGATGTCTAGAGTGAGTTTTGAAGTG-3′ and antisense 5′-ATATCTCGAGTGCGGTTAGAAATTCTCTTTGTGGAAGGA-3′ for RHD2 (AT5G51060), sense 5′-CCGGTACCGAATTCGATGGCGTCTAATAATCATTTCATG-3′ and antisense 5′-ATATCTCGAGTGCGGTCATGGGAAAGTGAAAGATCCTCC-3′ for CSLD3 (AT3G03050), sense 5′-CCGGTACCGAATTCGATGAACGATCTTATCTCAAGCTCA-3′ and antisense 5′-ATATCTCGAGTGCGGTCAAGGTCGAAGTAGAGTGTTAAA-3′ for SYP123 (AT4G03330), and sense 5′-CCGGTACCGAATTCGATGGCGATCACACGCTCCTCCT-3′ and antisense 5′-ATATCTCGAGTGCGGTCAGTATTTGGGAGTGGCGGG-3′ for PRP3 (AT3G62680), and recombined with pENTR2B (Invitrogen) DNA fragments amplified by PCR using primers 5′-CCGCACTCGAGATATCTAGAC-3′ and 5′-CGAATTCGGTACCGGATC-3′. The resulting pENTR2B vectors with genes were recombined with the destination plasmid pMDC43:EXPA7promoter to yield the GFP fusion genes in pMDC43:EXPA7promoter plasmids, and T2 transgenic plants in the background of Col-0 and scyl2b mutants were generated and used for the root hair tip localization study.
GUS Histochemical Assay
Staining for GUS activity was carried out as described previously (Harrison et al., 2006). Briefly, about 2-week-old plants grown on plates were dipped in GUS solution and vacuum infiltrated for 20 min. Samples were then incubated for 2 d at 37°C, cleared in 70% (v/v) ethanol, and mounted in water prior to examination. GUS-stained tissues and plants shown in this article represent the typical results of at least two independent lines for each construct.
Protein Interaction Assay
For BiFC assay, genes in pENTR/D-TOPO were introduced into the destination plasmid pSITE-BiFC-C1nec or pSITE-BiFC-C1cec to yield nYFP or cYFP fusion constructs (Martin et al., 2009). Then, tobacco leaves were infiltrated with A. tumefaciens carrying the BiFC constructs as described previously (Sparkes et al., 2006). Three days after A. tumefaciens infiltration, YFP fluorescence images were obtained from tobacco leaf epidermal cells. The expression level of nYFP or cYFP fusion proteins in tobacco leaf epidermal cells was detected by western blotting using rabbit polyclonal anti-GFP antibody (ab290; Abcam). For coimmunoprecipitation assay, 4-week-old tobacco leaves were coinfiltrated with A. tumefaciens carrying the HA and YFP fusion genes. Three days after infiltration, leaves were frozen, ground, and protein extracted in the extraction buffer (250 mm Suc, 20 mm HEPES, pH 7.4, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1% [v/v] Nonidet P-40, 0.1% [w/v] sodium deoxycholate, 1 mm phenylmethylsulfonyl fluoride, EDTA-free protease inhibitor cocktail complete [Roche], and 10 mm NaF). The total protein extracts were centrifuged at 10,000g for 10 min to remove cell debris, and the supernatant was incubated with anti-HA magnetic beads (Pierce) or GFP-Trap beads (ChromoTek) for 2 h at 4°C. Beads were washed four times in the extraction buffer containing 0.1% Nonidet P-40 and then washed once with the same extraction buffer without detergent. The protein beads were boiled in loading buffer, and the beads were removed. One-quarter of the eluates and 10 μg of protein from total lysates (input fraction) were loaded and run on a 4-12% ExpressPlus PAGE Gel (Genscript) and then transferred with 100 V for 1 h by a wet transfer method to an Amersham Hybond-P polyvinylidene difluoride membrane (GE Healthcare). Membranes was washed briefly in PBST buffer (10 mm phosphate buffer, pH 7.2, 150 mm NaCl, and 0.2% Tween 20) and blocked in PBST buffer with 1% BSA (Sigma) for 1 h. The membrane was incubated overnight at 4°C with either rabbit polyclonal anti-GFP antibody (1:7,500 dilution; ab290; Abcam) or rat monoclonal anti-HA high-affinity antibody (1:7,500 dilution; Roche) in the PBST buffer with 1% BSA, washed three times, and incubated with either goat anti-rabbit IgG-HRP or goat anti-rat IgG-HRP (Santa Cruz Biotechnology; 1:7,500 dilution) in the PBST buffer with 1% BSA at room temperature. After washing the membrane, signal was detected using the Western Bright Sirius HRP Substrate (Advansta) and a GE Healthcare ImageQuant RT ECL Imager.
Fluorescent Organelle Markers and Microscopy
Monomeric RFP (mRFP):SCAMP1 (TGN marker) and mRFP:AtVSR2 (PVC marker) were kindly provided by Liwen Jiang (Chinese University of Hong Kong). mRFP:SCAMP1 was used as a marker for both TGN and PM, since it localizes to these two compartments (Lam et al., 2007). The other organelle markers used in this study were obtained from the Arabidopsis stock centers (http://www.arabidopsis.org) and are described on the Web site (http://www.bio.utk.edu/cellbiol/markers) and in an article published previously (Nelson et al., 2007). For the colocalization studies, Arabidopsis leaf protoplasts were generated using 3-week-old plants and transformed with plasmids by polyethylene glycol solution based on a previously described method (Yoo et al., 2007). Fluorescence signals were observed 24 to 48 h after the polyethylene glycol transformation. Images were taken using a Zeiss LSM 510 Meta confocal laser-scanning microscope equipped with a C-Apochromat 40×/1.2 numerical aperture water-immersion lens (Zeiss). For the colocalization studies using YFP and CFP signals, a 514-nm excitation wavelength with 535- to 590-nm emission filter and a 458-nm excitation wavelength with 480- to 520-nm emission filter were used, respectively. For the colocalization studies using YFP and RFP signals, a 488-nm excitation wavelength with 500- to 550-nm emission filter and a 543-nm excitation wavelength with 565- to 615-nm emission filter were used, respectively. Images obtained by confocal microscopy were processed using NIH ImageJ software to remove low-level background fluorescence and to enhance contrast for the increase of signal intensity. For the colocalization images shown in Figure 6, YFP images were rendered in green color and CFP or RFP images were rendered in magenta color. Root hairs of transgenic Arabidopsis plants expressing YFP:SCYL2B were observed with a Nikon Eclipse E800 wide-field microscope and a 40×/1.3 numerical aperture oil-immersion lens with appropriate YFP filters, and images were processed using ImageJ software.
Chemical Inhibitor and FM4-64 Uptake Assay
For the chemical inhibitor assay, 4-week-old tobacco leaves were infiltrated with A. tumefaciens carrying the 35S:GFP:SCYL2B fusion gene. Three days after infiltration, tobacco leaf discs of 1 cm diameter were incubated in water containing either 50 µm BFA or 30 µm wortmannin. Two hours after the incubation, GFP fluorescence images were taken using a Zeiss LSM 700 confocal laser-scanning microscope. Two independent experiments were performed, and about five leaf discs were used for each sample. For the FM4-64 uptake assay, about 7-d-old seedlings of Col-0 and scyl2a-3 scyl2b-3 double mutants grown on ammonium-free complete nutrient agar medium (Jung et al., 2009) were incubated with the same agar-free nutrient solution containing 2 μm FM 4-64 dye for 5, 30, and 60 min. Then, fluorescence images were taken under a Zeiss LSM 700 confocal laser-scanning microscope using a 488-nm excitation wavelength with 585- to 610-nm emission filter. Two independent experiments were performed, and about five to 10 seedlings were observed for each sample.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SCYL2A (AT1G22870), SCYL2B (AT1G71410), CHC1 (AT3G11130), VTI11 (AT5G39510), VTI12 (AT1G26670), ROP2 (AT1G20090), RABA4B (AT4G39990), RHD2 (AT5G51060), CSLD3 (AT3G03050), SYP123 (AT4G03330), PRP3 (AT3G62680), AP-1 (AT1G23900), AP-2 (AT5G22770), and AP-3 (AT1G48760).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Segmental duplication of SCYL2 genes in Arabidopsis.
Supplemental Figure S2. Tip localization of SCYL2B in growing root hairs.
Supplemental Figure S3. SCYL2B is not localized at peroxisomes, mitochondria, plastids, and PM.
Supplemental Figure S4. SCYL2B is localized at the Golgi, TGN, and PVC in Arabidopsis leaf epidermal cells.
Supplemental Figure S5. Western-blot analysis to show the expression levels of nYFP:SCYL2B with cYFP, cYFP:CHC1, cYFP:VTI11, and cYFP:VTI12 in tobacco leaf epidermal cells.
Supplemental Figure S6. Coimmunoprecipitation of SCYL2B with CHC1, VTI11, and VTI12 using GFP-trap beads.
Supplemental Figure S7. Full scans of immunoblots.
Supplemental Figure S8. CSLD3 is mislocalized in root hairs of scyl2b mutants.
Supplemental Figure S9. A clathrin-binding motif (SLLDLL) on SCYL2B does not affect the interaction between SCYL2B and CHC1.
Supplemental Figure S10. SCYL2B does not interact with adaptor proteins.
Supplemental Figure S11. Effects of BFA and wortmannin, endomembrane trafficking inhibitors, on the localization of SCYL2B in tobacco leaf epidermal cells.
Supplemental Figure S12. Comparison of the localization patterns of SCYL2B expressed by various promoters in tobacco leaf epidermal cells.
Supplemental Figure S13. FM4-64 uptake assay in Col-0 and scyl2a-3 scyl2b-3 double mutants.
Supplemental Figure S14. SCYL2 proteins function in the development of root columella cells in Arabidopsis.
Supplemental Table S1. Percentages of YFP:SCYL2B-positive spots overlapped with organelle marker proteins.
Acknowledgments
We thank Howard Berg (Donald Danforth Plant Science Center) for microscopy assistance, Liwen Jiang for providing fluorescent organelle markers, Sandeep Tata for the preparation of tobacco plants for BiFC assay, and Yves Poirier for supporting experimental work.
Glossary
- ER
endoplasmic reticulum
- CCV
clathrin-coated vesicle
- TGN
trans-Golgi network
- PVC
prevacuolar compartment
- PM
plasma membrane
- RT
reverse transcription
- BiFC
bimolecular fluorescence complementation
- Col-0
Columbia-0
- BFA
brefeldin A
- EE
early endosomes
Footnotes
D.W.L and I.H. were supported by the Cooperative Research Program for Agriculture Science and Technology Development (project no. 010953012016), Rural Development Administration, Republic of Korea, and S.B.R. was supported by the Next-Generation BioGreen 21 Program (SSAC), Korea Rural Development Administration (grant PJ01109103).
Articles can be viewed without a subscription.
References
- Bassham DC, Blatt MR (2008) SNAREs: cogs and coordinators in signaling and development. Plant Physiol 147: 1504–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Sanderfoot AA, Kovaleva V, Zheng H, Raikhel NV (2000) AtVPS45 complex formation at the trans-Golgi network. Mol Biol Cell 11: 2251–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal AJ, Yoo CM, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sørensen I, Blancaflor EB, Scheller HV, Willats WG (2008) Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol 148: 1238–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackbourn HD, Jackson AP (1996) Plant clathrin heavy chain: sequence analysis and restricted localisation in growing pollen tubes. J Cell Sci 109: 777–786 [DOI] [PubMed] [Google Scholar]
- Borner GH, Rana AA, Forster R, Harbour M, Smith JC, Robinson MS (2007) CVAK104 is a novel regulator of clathrin-mediated SNARE sorting. Traffic 8: 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman JL, Hamlin JN, McPherson PS (2010) Scyl1 regulates Golgi morphology. PLoS ONE 5: e9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Irani NG, Friml J (2011) Clathrin-mediated endocytosis: the gateway into plant cells. Curr Opin Plant Biol 14: 674–682 [DOI] [PubMed] [Google Scholar]
- Conner SD, Schmid SL (2005) CVAK104 is a novel poly-L-lysine-stimulated kinase that targets the beta2-subunit of AP2. J Biol Chem 280: 21539–21544 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J (2007) Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Di Rubbo S, Irani NG, Kim SY, Xu ZY, Gadeyne A, Dejonghe W, Vanhoutte I, Persiau G, Eeckhout D, Simon S, et al. (2013) The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 25: 2986–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düwel M, Ungewickell EJ (2006) Clathrin-dependent association of CVAK104 with endosomes and the trans-Golgi network. Mol Biol Cell 17: 4513–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Li R, Pan J, Ding Z, Lin J (2015) Endocytosis and its regulation in plants. Trends Plant Sci 20: 388–397 [DOI] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, et al. (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Arimura S, Ueda T, Takanashi H, Hayashi Y, Nakano A, Tsutsumi N (2010) Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc Natl Acad Sci USA 107: 6094–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadeyne A, Sánchez-Rodríguez C, Vanneste S, Di Rubbo S, Zauber H, Vanneste K, Van Leene J, De Winne N, Eeckhout D, Persiau G, et al. (2014) The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156: 691–704 [DOI] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras S, Earls LR, Howell S, Smeyne RJ, Zakharenko SS, Pelletier S (2015) SCYL2 protects CA3 pyramidal neurons from excitotoxicity during functional maturation of the mouse hippocampus. J Neurosci 35: 10510–10522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SJ, Mott EK, Parsley K, Aspinall S, Gray JC, Cottage A (2006) A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hicks GR, Raikhel NV (2010) Advances in dissecting endomembrane trafficking with small molecules. Curr Opin Plant Biol 13: 706–713 [DOI] [PubMed] [Google Scholar]
- Hirst J, Edgar JR, Borner GH, Li S, Sahlender DA, Antrobus R, Robinson MS (2015) Contributions of epsinR and gadkin to clathrin-mediated intracellular trafficking. Mol Biol Cell 26: 3085–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Robinson DG (2009) Transport vesicle formation in plant cells. Curr Opin Plant Biol 12: 660–669 [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Hirano T, Enami K, Fuselier T, Kato N, Kwon C, Voigt B, Schulze-Lefert P, Baluška F, Sato MH (2014) Syntaxin of plant proteins SYP123 and SYP132 mediate root hair tip growth in Arabidopsis thaliana. Plant Cell Physiol 55: 790–800 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14: 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JY, Shin R, Schachtman DP (2009) Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21: 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G. (2004) Membrane trafficking in plants. Annu Rev Cell Dev Biol 20: 481–504 [DOI] [PubMed] [Google Scholar]
- Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT (2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18: 2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitakura S, Vanneste S, Robert S, Löfke C, Teichmann T, Tanaka H, Friml J (2011) Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafer EM. (2002) Clathrin-protein interactions. Traffic 3: 513–520 [DOI] [PubMed] [Google Scholar]
- Lam SK, Cai Y, Tse YC, Wang J, Law AH, Pimpl P, Chan HY, Xia J, Jiang L (2009) BFA-induced compartments from the Golgi apparatus and trans-Golgi network/early endosome are distinct in plant cells. Plant J 60: 865–881 [DOI] [PubMed] [Google Scholar]
- Lam SK, Siu CL, Hillmer S, Jang S, An G, Robinson DG, Jiang L (2007) Rice SCAMP1 defines clathrin-coated, trans-Golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19: 296–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Bak G, Choi Y, Chuang WI, Cho HT, Lee Y (2008) Roles of phosphatidylinositol 3-kinase in root hair growth. Plant Physiol 147: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Yang Z (2008) Tip growth: signaling in the apical dome. Curr Opin Plant Biol 11: 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298: 1912–1934 [DOI] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Mishev K, Dejonghe W, Russinova E (2013) Small molecules for dissecting endomembrane trafficking: a cross-systems view. Chem Biol 20: 475–486 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nielsen E, Cheung AY, Ueda T (2008) The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol 147: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J (2008) DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 1: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez Valencia J, Goodman K, Otegui MS (2016) Endocytosis and endosomal trafficking in plants. Annu Rev Plant Biol 67: 309–335 [DOI] [PubMed] [Google Scholar]
- Park M, Song K, Reichardt I, Kim H, Mayer U, Stierhof YD, Hwang I, Jürgens G (2013) Arabidopsis μ-adaptin subunit AP1M of adaptor protein complex 1 mediates late secretory and vacuolar traffic and is required for growth. Proc Natl Acad Sci USA 110: 10318–10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Szumlanski AL, Gu F, Guo F, Nielsen E (2011) A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat Cell Biol 13: 973–980 [DOI] [PubMed] [Google Scholar]
- Pelletier S. (2016) SCYL pseudokinases in neuronal function and survival. Neural Regen Res 11: 42–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Serna J, Falbel TG, Bednarek SY, Nielsen E (2004) The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16: 1589–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG, Neuhaus JM (2016) Receptor-mediated sorting of soluble vacuolar proteins: myths, facts, and a new model. J Exp Bot 67: 4435–4449 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Furlán C, Salinas-Grenet H, Sandoval O, Recabarren C, Arraño-Salinas P, Soto-Alvear S, Orellana A, Blanco-Herrera F (2016) The root hair specific SYP123 regulates the localization of cell wall components and contributes to rizhobacterial priming of induced systemic resistance. Front Plant Sci 7: 1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Hanak T, Persson S, Voigt CA (2016) Cellulose and callose synthesis and organization in focus, what’s new? Curr Opin Plant Biol 34: 9–16 [DOI] [PubMed] [Google Scholar]
- Song J, Lee MH, Lee GJ, Yoo CM, Hwang I (2006) Arabidopsis EPSIN1 plays an important role in vacuolar trafficking of soluble cargo proteins in plant cells via interactions with clathrin, AP-1, VTI11, and VSR1. Plant Cell 18: 2258–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Sullivan A, Uff CR, Isacke CM, Thorne RF (2003) PACE-1, a novel protein that interacts with the C-terminal domain of ezrin. Exp Cell Res 284: 224–238 [DOI] [PubMed] [Google Scholar]
- Surpin M, Zheng H, Morita MT, Saito C, Avila E, Blakeslee JJ, Bandyopadhyay A, Kovaleva V, Carter D, Murphy A, et al. (2003) The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15: 2885–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Gapper C, Kaya H, Bell E, Kuchitsu K, Dolan L (2008) Local positive feedback regulation determines cell shape in root hair cells. Science 319: 1241–1244 [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH (2008) Synteny and collinearity in plant genomes. Science 320: 486–488 [DOI] [PubMed] [Google Scholar]
- Terabayashi T, Funato Y, Fukuda M, Miki H (2009) A coated vesicle-associated kinase of 104 kDa (CVAK104) induces lysosomal degradation of frizzled 5 (Fzd5). J Biol Chem 284: 26716–26724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D, Gadeyne A, Vanstraelen M, Inzé D, Van Montagu MC, De Jaeger G, Russinova E, Geelen D (2011) Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc Natl Acad Sci USA 108: 615–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Hu T, Yan X, Meng T, Wang Y, Wang Q, Zhang X, Gu Y, Sánchez-Rodríguez C, Gadeyne A, et al. (2016) Differential regulation of clathrin and its adaptor proteins during membrane recruitment for endocytosis. Plant Physiol 171: 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yan X, Chen Q, Jiang N, Fu W, Ma B, Liu J, Li C, Bednarek SY, Pan J (2013) Clathrin light chains regulate clathrin-mediated trafficking, auxin signaling, and development in Arabidopsis. Plant Cell 25: 499–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Harper JF, Gribskov M (2003) Systematic trans-genomic comparison of protein kinases between Arabidopsis and Saccharomyces cerevisiae. Plant Physiol 132: 2152–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN Jr (2006) Statistical analysis of real-time PCR data. BMC Bioinformatics 7: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Keegstra K (2008) AtCSLD2 is an integral Golgi membrane protein with its N-terminus facing the cytosol. Planta 228: 823–838 [DOI] [PubMed] [Google Scholar]
- Zheng H, von Mollard GF, Kovaleva V, Stevens TH, Raikhel NV (1999) The plant vesicle-associated SNARE AtVTI1a likely mediates vesicle transport from the trans-Golgi network to the prevacuolar compartment. Mol Biol Cell 10: 2251–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]