Regulatory uncoupling of primary and secondary cellulose synthases occurred independently in mosses and seed plants and is associated with convergent evolution of secondary wall structure.
Abstract
The secondary cell walls of tracheary elements and fibers are rich in cellulose microfibrils that are helically oriented and laterally aggregated. Support cells within the leaf midribs of mosses deposit cellulose-rich secondary cell walls, but their biosynthesis and microfibril organization have not been examined. Although the Cellulose Synthase (CESA) gene families of mosses and seed plants diversified independently, CESA knockout analysis in the moss Physcomitrella patens revealed parallels with Arabidopsis (Arabidopsis thaliana) in CESA functional specialization, with roles for both subfunctionalization and neofunctionalization. The similarities include regulatory uncoupling of the CESAs that synthesize primary and secondary cell walls, a requirement for two or more functionally distinct CESA isoforms for secondary cell wall synthesis, interchangeability of some primary and secondary CESAs, and some CESA redundancy. The cellulose-deficient midribs of ppcesa3/8 knockouts provided negative controls for the structural characterization of stereid secondary cell walls in wild type P. patens. Sum frequency generation spectra collected from midribs were consistent with cellulose microfibril aggregation, and polarization microscopy revealed helical microfibril orientation only in wild type leaves. Thus, stereid secondary walls are structurally distinct from primary cell walls, and they share structural characteristics with the secondary walls of tracheary elements and fibers. We propose a mechanism for the convergent evolution of secondary walls in which the deposition of aggregated and helically oriented microfibrils is coupled to rapid and highly localized cellulose synthesis enabled by regulatory uncoupling from primary wall synthesis.
In vascular plants, cellulose is a major component of both primary cell walls, which are deposited during cell expansion, and secondary cell walls, which are deposited after expansion has ceased (Carpita and McCann, 2000). Secondary cell walls of water-conducting tracheary elements and supportive fibers are rich in cellulose, with microfibrils arranged in helices that vary in angle according to developmental stage and environmental conditions (Barnett and Bonham, 2004). Secondary cell wall microfibrils also are more aggregated than those of primary cell walls (Donaldson, 2007; Fernandes et al., 2011; Thomas et al., 2014). Recently, sum frequency generation (SFG) spectroscopy has been used to compare the mesoscale structure of cellulose microfibrils in primary and secondary cell walls. Both high cellulose content and microfibril aggregation contribute to a strong secondary cell wall signature in SFG spectra of mature angiosperm tissues (Barnette et al., 2012; Park et al., 2013; Lee et al., 2014).
Cellulose microfibrils are synthesized by cellulose synthase (CESA) proteins that function together as cellulose synthesis complexes (CSCs) in the plasma membrane (Delmer, 1999; Kimura et al., 1999). Recent analyses of CSC and microfibril structure indicate that the rosette CSCs of land plants most likely contain 18 CESA subunits (Fernandes et al., 2011; Jarvis, 2013; Newman et al., 2013; Thomas et al., 2014; Oehme et al., 2015; Nixon et al., 2016; Vandavasi et al., 2016) in a 1:1:1 ratio (Gonneau et al., 2014; Hill et al., 2014). Seed plants have six phylogenetic and functional classes of CESA proteins, three required for primary cell wall synthesis (Desprez et al., 2007; Persson et al., 2007) and three required for the synthesis of the lignified secondary cell walls of tracheary elements and fibers (Taylor et al., 2003). Mutation of any of the secondary CESAs results in a distinctive irregular xylem phenotype characterized by collapsed xylem tracheary elements and weak stems (Taylor et al., 2004). The secondary cell wall CESAs of Arabidopsis (Arabidopsis thaliana) are regulated by master regulator NAC domain transcription factors that also activate genes required for the synthesis of other secondary cell wall components, such as xylan and lignin (Schuetz et al., 2013; Zhong and Ye, 2015; Yang and Wang, 2016).
The moss Physcomitrella patens has seven CESA genes (Roberts and Bushoven, 2007; Goss et al., 2012). Phylogenetic analysis has revealed that the P. patens CESAs do not cluster with the six CESA clades shared by seed plants (Roberts and Bushoven, 2007). Like other mosses, P. patens lacks the lignified secondary cell walls that are characteristic of vascular plant tracheary elements and fibers. However, mosses do have support cells (stereids) with thick unlignifed cell walls (Kenrick and Crane, 1997) and water-conducting cells (hydroids) that have thin cell walls and undergo programmed cell death like tracheary elements (Hebant, 1977). Although the stereid cell walls of P. patens are known to contain cellulose (Berry et al., 2016), the mesoscale structure has not been examined. Only one of the seven P. patens CESAs has been characterized functionally. When PpCESA5 was disrupted, gametophore buds failed to develop into leafy gametophores, instead forming irregular cell clumps. The associated disruptions of cell expansion and cell division are consistent with an underlying defect in primary cell wall deposition (Goss et al., 2012). Recently it was shown that PpCESA3 expression is regulated by the NAC transcription factor PpVNS7, along with the thickening of stereid cell walls (Xu et al., 2014).
Here, we show that PpCESA3 and PpCESA8 function in the deposition of stereid cell walls in the gametophore leaf midribs of P. patens and are subfunctionalized with respect to PpCESA5. We also used polarization microscopy and SFG to reveal similarities in the mesoscale organization of the microfibrils synthesized by PpCESA3 and PpCESA8 and those in the secondary cell walls of vascular plants. Finally, we propose a mechanism through which the uncoupling of primary and secondary CESA regulation played a role in the independent evolution of secondary cell walls with aggregated, helically arranged cellulose microfibrils in the moss and seed plant lineages.
RESULTS
PpCESA3 and PpCESA8 Function in Secondary Cell Wall Deposition
The CESA genes PpCESA3 and PpCESA8 were independently knocked out by homologous recombination in an effort to examine their roles in development and cell wall biosynthesis in P. patens. Stable antibiotic-resistant lines generated by transforming wild type P. patens with CESA3KO or CESA8KO vectors were tested for integration of the vector and deletion of the target gene by PCR (Supplemental Fig. S1). Integration was verified for five ppcesa8KO lines recovered from two different transformations, line 8KO-5B from a transformation of the GD06 wild type line and lines 8KO-4C, 8KO-5C, 8KO-7C, and 8KO-10C from a transformation of the GD11 wild type line (Supplemental Fig. S1). Integration was verified for three ppcesa3KO lines recovered from a single transformation of GD11 and three double ppcesa3/8KO lines recovered from a single transformation of the ppcesa8KO-5B line with the CESA3KO vector (Supplemental Fig. S1). The GD06 and GD11 lines are from independent selfings of the same haploid wild type line, as described in “Materials and Methods.”
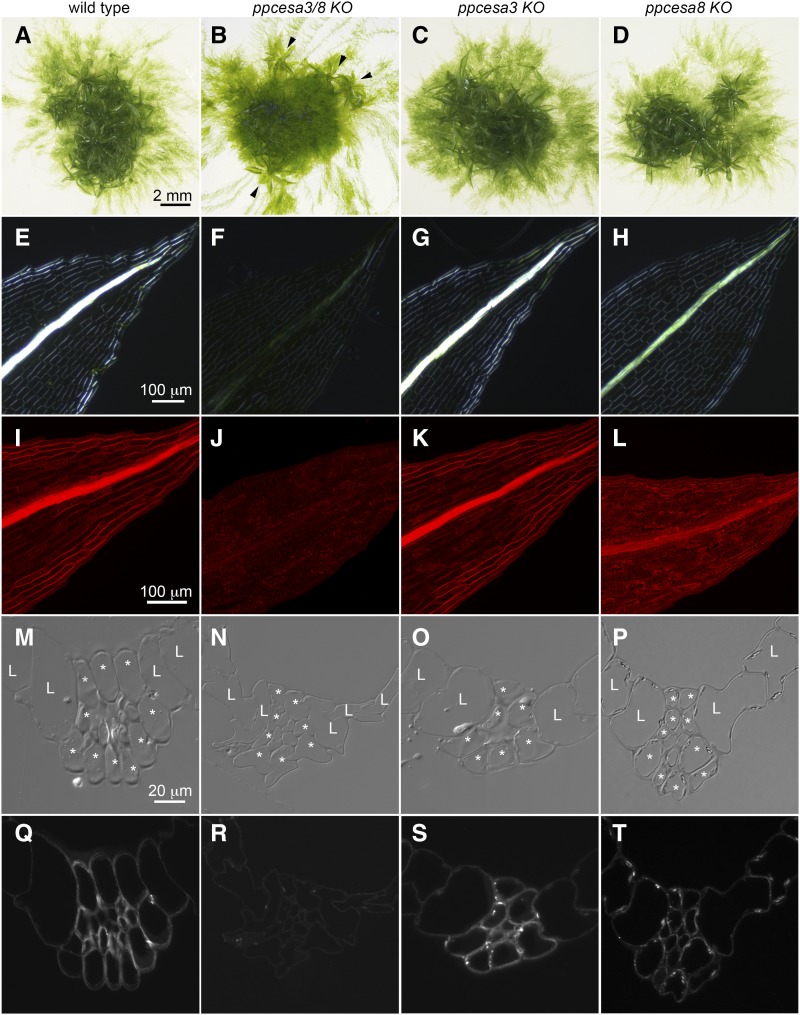
The colonies that developed from wild type and KO lines consisted of protonemal filaments and leafy gametophores (Fig. 1). Whereas wild type, ppcesa3KO, and ppcesa8KO gametophores grew vertically, the gametophores on ppcesa3/8KO colonies were unable to support themselves and adopted a horizontal orientation. Superficially, ppcesa3/8KO colonies appeared to produce fewer gametophores (Fig. 1), but dissection revealed similar numbers of horizontal gametophores that had been overgrown by protonemal filaments. Thus, PpCESA3 and PpCESA8 are not required for gametophore initiation or morphogenesis, but they appear to contribute to structural support.
Figure 1.
Phenotypes of ppcesa3/8KO, ppcesa3KO, and ppcesa8KO compared with wild type P. patens. A to D, Colony morphology is similar in the wild type, ppcesa3KOs, and ppcesa8KOs; horizontal growth is typical of gametophores produced by ppcesa3/8KO (arrowheads). E to H, Polarized light microscopy of leaves shows that the midribs of the wild type and ppcesa3KO are highly birefringent. The midribs of ppcesa3/8KO leaves have low birefringence, and ppcesa8KO midribs have moderate birefringence. I to L, Fluorescence microscopy of leaves stained with Pontamine Fast Scarlet (S4B) shows strong fluorescence in the midribs of wild type and ppcesa3KO leaves, low fluorescence in the midribs of ppcesa3/8KO leaves, and intermediate fluorescence in the midribs of ppcesa8KO leaves. M to P, Differential interference contrast microscopy of sections through the midribs of maturing leaves (L, lamina cell; *, bundle sheath cell). In the wild type and ppcesa3KO, the walls of bundle sheath cells and the stereid cells they surround show enhanced contrast due to differential refractive index. Q to T, Fluorescence microscopy of the same sections shown in M to P labeled with CBM3a. The bundle sheath and stereid cells of wild type and ppcesa3KO leaves are strongly labeled, whereas labeling is weak in ppcesa3/8KO and intermediate in ppcesa8KO leaves.
When examined with polarized light microscopy, the wild type gametophore leaves exhibited strong cell wall birefringence in the midribs and margins (Fig. 1). In contrast, the leaves produced by ppcesa3/8KOs lacked strong birefringence in these cells, consistent with the reduced crystalline cellulose content. The ppcesa3KO leaves appeared similar to wild type leaves (Fig. 1), and ppcesa8KO leaves had an intermediate phenotype. Staining with the fluorescent cellulose-binding dye S4B (Anderson et al., 2010) produced similar results, with strong fluorescence in the midribs of wild type and ppcesa3KO leaves, weak fluorescence in ppcesa3/8KO leaves, and intermediate fluorescence in ppcesa8KO leaves (Fig. 1).
Carbohydrate Binding Module 3a (CBM3a) provides a third method for detecting cellulose and can be used to probe thin sections (Blake et al., 2006). In sections from fully expanded wild type leaves, the walls of the lamina cells were labeled relatively weakly with CBM3a, whereas the thickened cell walls of the central midrib and bundle sheath cells were strongly labeled (Fig. 1). The same was true for ppcesa3KO leaves. However, midrib and bundle sheath cell labeling was nearly absent in ppcesa3/8KO and diminished in ppcesa8KO (Fig. 1) compared with the wild type and ppcesa3KO. Differential interference contrast microscopy of the same sections showed enhanced contrast in wild type and ppcesa3KO midribs (Fig. 1). Partial cell collapse occurred during embedding in ppcesa3/8KO leaves (Fig. 1).
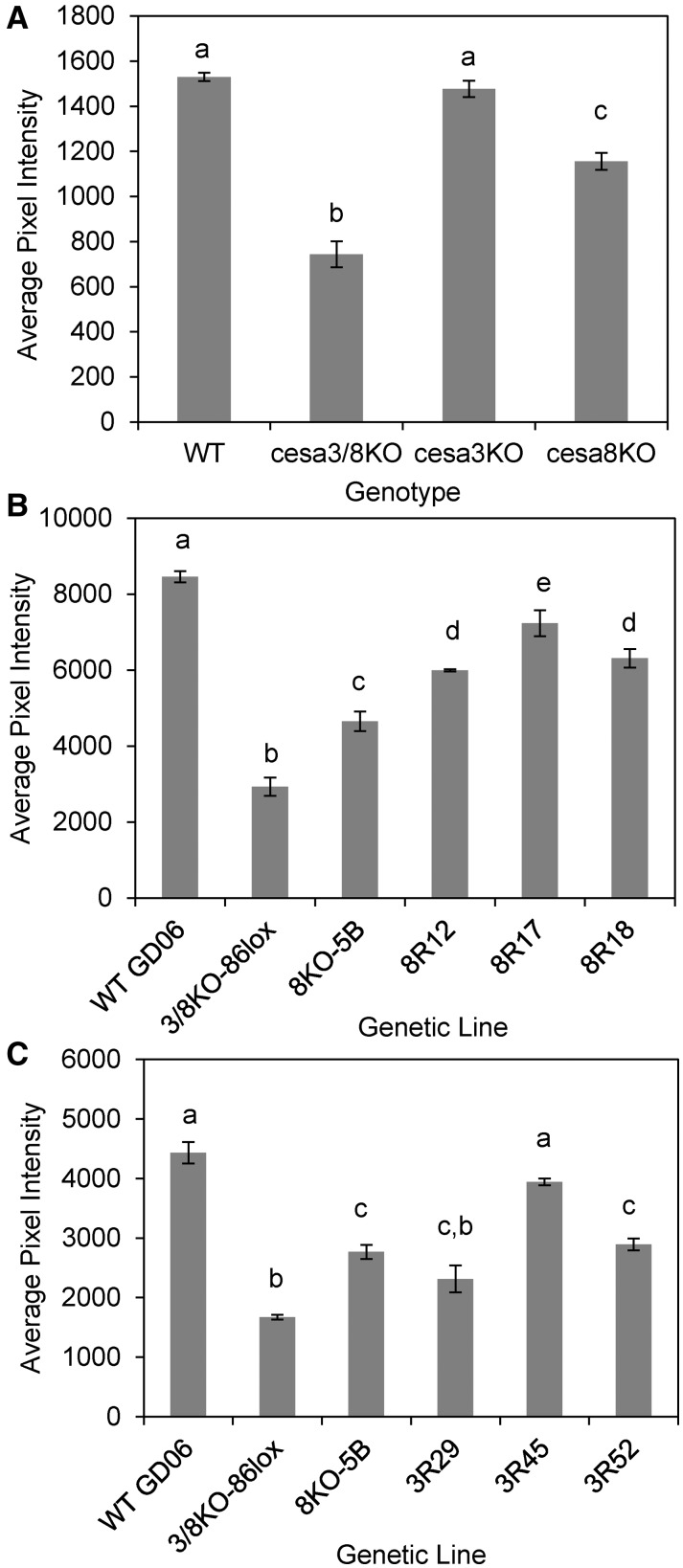
The cellulose content of the leaf midribs in the wild type and single and double ppcesaKO mutants was quantified by measuring the intensity of S4B fluorescence. Statistical analysis confirmed that the S4B fluorescence was reduced significantly in double KOs but not in ppcesa3KOs (Fig. 2). The intermediate phenotype of the ppcesa8KOs was confirmed and shown to be significantly different from both the wild type and the double KOs (Fig. 2). Updegraff analysis showed that the cellulose content of cell walls from whole ppcesa3/8KO gametophores (mean ± se of three genetic lines = 33.8% ± 0.034%) was reduced significantly (P = 0.004) compared with the wild type (GD06; mean ± se of three independent cultures = 60.1% ± 0.030%).
Figure 2.
Quantitative analysis of S4B fluorescence intensity in leaf midribs of P. patens wild type (WT), ppcesaKO, and rescue lines. Genotypes with different letters are significantly different (P < 0.05). A, Fluorescence was significantly weaker in ppcesa3/8KOs compared with the wild type. ppcesa3KOs were not significantly different from the wild type, whereas ppcesa8KOs were intermediate between the wild type and ppcesa3/8KOs and significantly different from both. For each mutant genotype, three independent genetic lines were sampled in triplicate. Two independent wild type lines (GD06 and GD11) were sampled in triplicate. Error bars indicate se for three mutant (n = 3) or two wild type (n = 2) lines. B, Lines derived from transformation of ppcesa3/8KO-86lox with proCESA8::CESA8 (8R) had significantly higher fluorescence compared with the parent double KO line and ppcesa8KO but significantly less than the wild type. C, Lines derived from transformation of ppcesa3/8-86lox with proCESA3::CESA3 (3R) had significantly higher fluorescence compared with the parent double KO line (except 3R29) and were not significantly different from either ppcesa8KO lines (3R29 and 3R52) or the wild type (3R45). For B and C, error bars indicate se for explants from the same line (n = 3 or n = 2 [WT, 3/8KO, and 8KO in C]).
To confirm that the observed ppcesa3/8KO phenotype was due to the absence of PpCESA3 and PpCESA8, the selection cassette was removed from ppcesa3/8KO-86 by Cre-mediated recombination of flanking lox-p sites (Vidali et al., 2010) to allow transformation with vectors that drive the expression of PpCESA3 or PpCESA8 with their native promoters (Supplemental Fig. S2). Stable antibiotic-resistant lines selected for the presence of numerous erect gametophores were examined with polarization microscopy (Supplemental Fig. S2). For the transformation with proCESA8::CESA8, 13 lines were examined, six of these had strong midrib birefringence, and the first three were used for further analysis. For the transformation with proCESA3::CESA3, the first three lines examined had strong midrib birefringence and were used for further analysis. S4B staining confirmed that expression of PpCESA8 or PpCESA3 rescued the defects in cellulose deposition in the leaf midribs of the double ppcesa3/8KO (Fig. 2). Lines from the transformation with proCESA8::CESA8 were expected to be restored to the wild type phenotype, because ppcesa3KO, which also expresses PpCESA8 under the control of the PpCESA8 promoter, showed no defects in cellulose deposition in the leaf midrib. All three proCESA8::CESA8 lines had significantly stronger S4B fluorescence than ppcesa8KO. This demonstrates substantial restoration of the phenotype, although fluorescence was still significantly weaker than in the wild type (Fig. 2). Two lines from a transformation with proCESA3::CESA3 (3R29 and 3R52) were not significantly different from ppcesa8KO-5B, which is expected, since they both lack PpCESA8 and express PpCESA3 under the control of the PpCESA3 promoter. In the third line (3R45), fluorescence was restored to wild type levels (Fig. 2). The y axis scales differ between experiments due to the use of different exposure time settings.
Secondary Cell Wall Microfibrils Are Helically Oriented and Laterally Aggregated
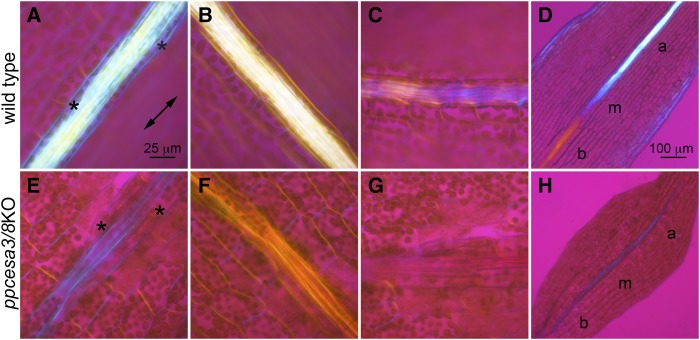
A first-order retardation plate was used with polarized light microscopy to determine the optical sign, and thus the cellulose microfibril orientation, of wild type and ppcesa3/8KO midrib cell walls (Fig. 3). In mature wild type leaves, the larger bundle sheath-like cells that surround the central stereids showed blue addition colors when oriented parallel to the major axis of the plate and yellow subtraction colors when oriented perpendicular to the major axis (Fig. 3), indicating that the net orientation of positively birefringent cellulose microfibrils is longitudinal. In contrast, the walls of the smaller central stereids were colorless when oriented parallel or perpendicular to the major axis (Fig. 3). However, when oriented at 45° to the retardation plate, these cells showed alternating bands of blue and yellow (Fig. 3), indicating that the microfibrils in their walls are helical with an angle near 45°. The central midrib cells of developing wild type leaves showed a transition from colorless to blue to yellow along the apical to basal developmental gradient when the midrib was oriented parallel to the major axis of the plate (Fig. 3). This indicates that the microfibril orientation changes from transverse to longitudinal and then to helical as the cells mature. In contrast, the central midrib stereids of mature ppcesa3/8KO leaves had blue addition colors when oriented parallel to the major axis, yellow subtraction colors when oriented perpendicular to the major axis, and no interference color when oriented at 45° to the retardation plate, indicating that microfibrils are longitudinal rather than helical. Developing ppcesa3/8KO leaves had no longitudinal gradient in interference colors (Fig. 3).
Figure 3.
Polarized light microscopy with a first-order retardation plate. The double-pointed arrow indicates the vibration direction of the major axis. A to C, Midrib of a mature wild type leaf oriented parallel, perpendicular, and at 45° to the major axis of the retardation plate. Bundle sheath cells (asterisks) flank the central midrib. D, Midrib of a developing wild type leaf oriented parallel to the major axis of the retardation plate showing change in microfibril orientations through the basal (b), medial (m), and apical (a) regions of the midrib. E to G, Midrib of a mature ppcesa3/8KO leaf oriented parallel, perpendicular, and at 45° to the major axis of the retardation plate. H, Midrib of a developing ppcesa3/8KO leaf oriented parallel to the major axis of the retardation plate showing no change in microfibril orientation through the basal, medial, and apical regions of the leaf. Bar in A is also for B, C, and E to G, and bar in D is also for H.
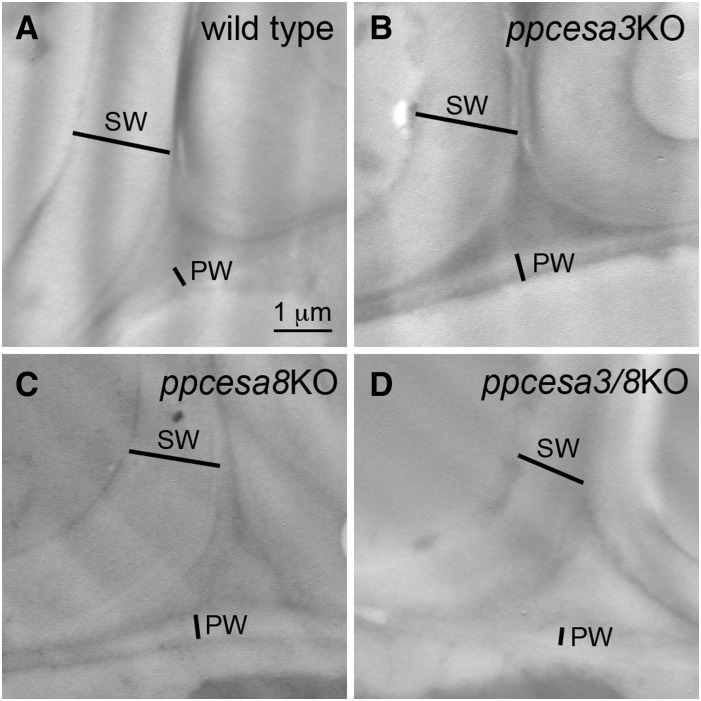
The walls of midrib cells were examined by transmission electron microscopy in ultrathin sections of chemically fixed gametophore leaves. Despite the reduced cellulose content detected by other means, the walls of midrib cells were thickened compared with the walls of adjacent lamina cells in all ppcesaKOs as well as wild type leaves (Fig. 4). When we attempted to prepare specimens by high-pressure freezing and freeze substitution, the leaves fractured in a plane parallel to the midrib. This resulted in a loss of midrib cells and precluded the examination of midrib cell walls in these specimens. We were able to examine the lamina and margin cells of freeze-substituted leaves in the wild type and two lines of each mutant. The walls of these cells appeared similar between the wild type and single and double ppcesaKOs (Supplemental Fig. S3). However, measurements revealed that lamina cell external walls (i.e. those facing the external environment) were thinner in ppcesaKOs (Supplemental Fig. S4).
Figure 4.
Transmission electron microscopy images of leaf midribs of P. patens showing adjacent cells with primary cell walls (PW) and secondary cell walls (SW) in wild type (A) and mutant (B–D) leaves.
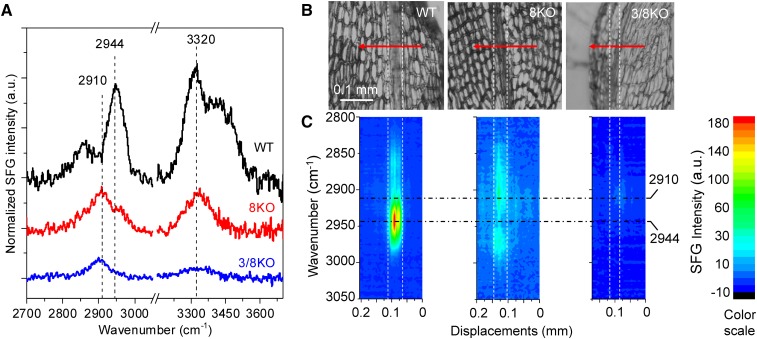
The mesoscale organization of cellulose in the midribs of wild type, ppcesa3/8KO, and ppcesa8KO leaves was examined using a broadband SFG microscope (Lee et al., 2016). Because it detects only the noncentrosymmetric ordering of functional groups, SFG provides a means of analyzing cellulose in intact cell walls with relatively little interference from matrix components (Barnette et al., 2011). For each genotype, full SFG spectra collected from three different locations along the midribs of each of three different leaves were averaged (Fig. 5). The sampling depth of the SFG microscope for cellulosic samples is 20 to 25 μm (Lee et al., 2016). Given that the thickness of turgid leaves is about 50 to 60 μm at the midrib and that they likely collapse to less than half their thickness when dried, we conclude that most of the leaf thickness contributes to the SFG signal. In spectra collected from the wild type, a strong peak at 2,944 cm−1, which is characteristic of secondary cell walls, was observed in the CH/CH2 stretch region along with a 3,320 cm−1 peak in the OH stretch region. In contrast, the spectra collected from ppcesa3/8KO and ppcesa8KO midribs had weaker peak intensity overall, with a broad CH/CH2 stretch peak centered around 2,910 cm−1. Compared with ppcesa3/8KO, the spectra from ppcesa8KO midribs had a weak signal at 2,963 cm−1 that was absent in spectra collected from ppcesa3/8KO midribs. A scan across a wild type leaf shows that the 2,944 cm−1 signal is associated with the midrib and was not observed in the cells of the lamina (Fig. 5). Equivalent scans of ppcesa3/8KO and ppcesa8KO leaves confirm the absence of a strong 2,944 cm−1 peak from the midribs of these mutants (Fig. 5).
Figure 5.
SFG spectroscopy of P. patens leaves. A, Full SFG spectra collected from leaf midribs (each is the average of nine spectra from three different positions on each of three different leaves). A strong peak in the CH stretch region (2,944 cm−1) is present in spectra from the wild type (WT), greatly diminished in spectra from ppcesa8KO (8KO), and absent in spectra from ppcesa3/8KO (3/8KO). B, P. patens wild type, ppcesa8KO, and ppcesa3/8KO leaves with SFG scan trajectories traversing the midribs. Step size was 5 μm per step. SFG spectra were collected from 2,850 to 3,150 cm−1, covering the entire CH region. C, 2D projection image of SFG spectra collected across the midribs of each leaf shown in B. Each column in each image is an entire spectrum collected from one point plotted against displacement along the scan trajectory. Colors indicate SFG intensity as shown in the scale at right.
PpCESA Proteins Are Functionally Specialized
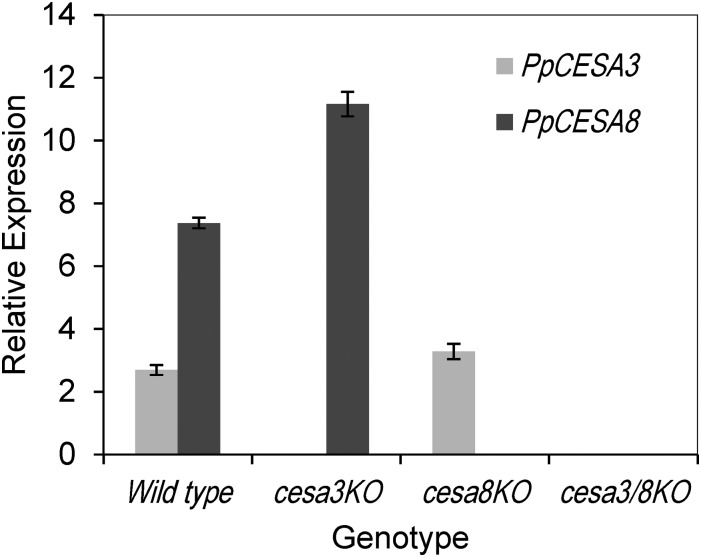
Based on the ppcesa3KO, ppcesa8KO, and ppcesa3/8KO phenotypes, PpCESA3 and PpCESA8 appear to be partially redundant. To determine whether the relative strengths of these phenotypes are related to gene expression levels, we used reverse transcription quantitative PCR to measure the expression of PpCESA3 and PpCESA8 in the wild type and mutants. In the ppcesa3KOs, PpCESA8 was up-regulated significantly compared with the wild type (Fig. 6), providing a possible explanation for the lack of a mutant phenotype in these lines. In contrast, PpCESA3 was not up-regulated significantly in ppcesa8KOs compared with the wild type, potentially explaining the intermediate phenotype in these mutants.
Figure 6.
Reverse transcription quantitative PCR analysis of PpCESA3 and PpCESA8 expression in the wild type, ppcesa3KOs, and ppcesa8KOs. Target/average reference cross-point ratios (using actin and v-type H+-translocating pyrophosphatase reference genes) were determined for three independent lines of each mutant (3KO-5, 3KO-35, 3KO-126; 8KO-5B, 8KO-4C, 8KO-10C; and 3/8KO-43, 3/8KO-57, 3/8KO-86) and two independent wild type lines (GD06 and GD11) with two technical replicates each. Error bars indicate se for the three mutant (n = 3) or two wild type (n = 2) lines.
ppcesa3KOs, ppcesa8KOs, and ppcesa3/8KOs were tested for changes in rhizoid and caulonema development to determine whether developmental defects were restricted to the gametophores. When cultured on medium containing auxin, all lines produced the expected leafless gametophores with numerous rhizoids (Supplemental Fig. S5), indicating no defects in rhizoid development in any of the KOs. Caulonema produced by colonies grown in the dark on vertically oriented plates were all negatively gravitropic (Supplemental Fig. S6). Although the appearance of the caulonema varied among experiments, those produced by KOs were always similar to the control wild type within the same experiment. Caulonemal length was not significantly different between ppcesa3/8KOs and the wild type (Table I).
Table I. Caulonema length for the wild type and ppcesa3/8KOs grown on vertical plates in the dark.
Data are from two independent experiments (n = 2). ANOVA showed no significant differences between genetic lines.
| Genetic Line | Caulonema Length | se |
|---|---|---|
| mm | ||
| Wild type GD06 | 4.69 | 0.50 |
| ppcesaA3/8KO-43 | 5.70 | 0.87 |
| ppcesaA3/8KO-57 | 4.51 | 1.14 |
| ppcesaA3/8KO-86 | 5.69 | 0.47 |
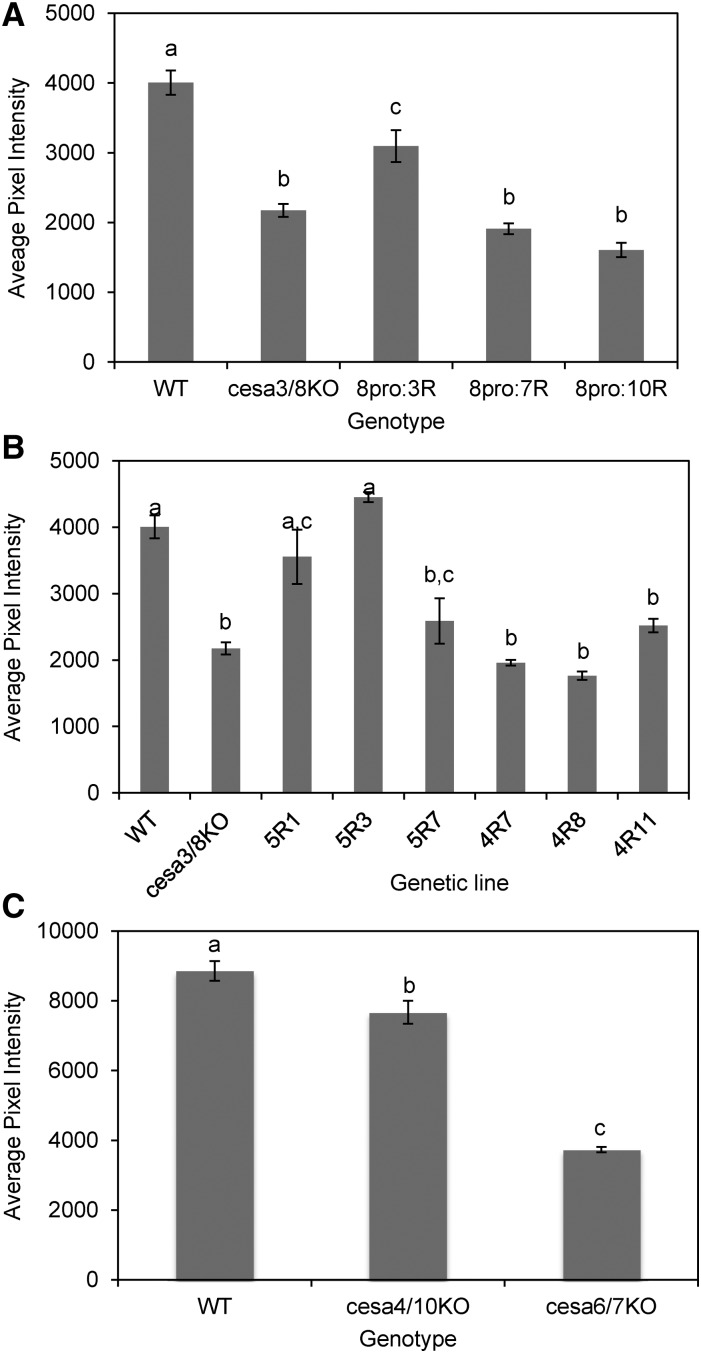
To determine whether other PpCESAs are functionally interchangeable with PpCESA3 and PpCESA8, we tested for rescue of ppcesa3/8KO-86lox by various PpCESAs driven by the PpCESA8 promoter. Polarization microscopy screening of at least 21 and up to 27 stably transformed lines for each vector revealed little or no midrib birefringence for the proCESA8::CESA4, proCESA8::CESA7, and proCESA8::CESA10 lines and moderate to strong midrib birefringence for 92% and 78% of the proCESA8::CESA3 and proCESA8::CESA5 lines, respectively. Quantitative analysis of S4B staining (Fig. 7) confirmed that the ppcesa3/8KO phenotype was partially rescued by proCESA8::CESA3 (three out of three lines) and proCESA8::CESA5 (two out of three lines), as we observed for proCESA8::CESA8 (Fig. 2). However, the proCESA8::CESA4, proCESA8::CESA7, and proCESA8::CESA10 vectors showed no rescue (Fig. 7). Western-blot analysis confirmed that PpCESA proteins were expressed in all lines except proCESA8::CESA4-11 and proCESA8::CESA5-7 (Supplemental Fig. S7). PpCESA6 differs from PpCESA7 by only two amino acids and was not tested. Although expressed with the same promoter, protein accumulation varies among the different transgenic lines (Supplemental Fig. S7). Similar differences in protein accumulation also may explain the variation in the extent of rescue by the proCESA3::CESA3 and proCESA8::CESA8 vectors (Fig. 2).
Figure 7.
Quantitative analysis of S4B fluorescence intensity in leaf midribs. A and B, Wild type (WT), ppcesa3/8KO-86lox, and ppcesa3/8KO-86lox transformed with proCESA8::CESA expression vectors. For each rescue genotype, three independent genetic lines were sampled in triplicate and measured with six samples of the wild type (GD06) and eight samples of ppcesa3/8KO-86lox. A, For lines derived from transformation of ppcesa3/8KO-86lox with proCESA8::CESA3 (8pro:3R), proCESA8::CESA7 (pro8:7R), and proCESA8::CESA10 (pro8:10R) genotypes, the three independent lines did not differ significantly and were combined. proCESA8::CESA7 and proCESA8::CESA10 lines did not differ significantly from the parent double KO line (P > 0.05), whereas proCESA8::CESA3 lines had significantly higher fluorescence compared with the parent double KO line but significantly less than the wild type (P < 0.05). Error bars indicate se for three independent lines. Genotypes with different letters are significantly different (P < 0.05). B, For lines derived from transformation of ppcesa3/8KO-86lox with proCESA8::CESA5 (pro8:5R) and proCESA8::CESA4 (pro8:4R), the three independent lines were significantly different and were analyzed separately. proCESA8::CESA5 (5R) lines were not significantly different from the wild type (P > 0.05), except for 5R7, which was not significantly different from ppcesa3/8KO-86lox (P > 0.05). proCESA8::CESA4 lines did not differ significantly from ppcesa3/8KO-86lox (P > 0.05). Error bars indicate se for three gametophores from the same line (n = 3). Lines with different letters are significantly different (P < 0.05). C, Midrib fluorescence was slightly but significantly reduced in cesa4/10KO compared with the wild type (P = 0.037). Reduction in midrib fluorescence in cesa6/7KO was substantial and highly significant (P = 0.0011). Error bars indicate se for three independent mutant lines or three replicates of the wild type (n = 3). Genotypes with different letters are significantly different (P < 0.05).
Finally, we examined ppcesa4/10KOs and ppcesa6/7KOs produced for another study to determine whether they phenocopy the ppcesa3/8KO phenotype. Genotype verification for these lines is presented in Supplemental Figures S8 and S9. The ppcesa4/10KOs showed slight but significant reduction in midrib S4B fluorescence. However, for ppcesa6/7KOs, the reduction was substantial and significant (Fig. 7), showing the PpCESA6/7 and PpCESA3/8 have nonredundant roles in secondary cell wall deposition in leaf midrib cells.
DISCUSSION
PpCESA3 and PpCESA8 Function Redundantly in Cellulose Deposition in Stereid Secondary Cell Walls
Targeted knockout of PpCESA3 and PpCESA8 blocked the deposition of cellulose in the thick walls of stereid cells, as indicated by (1) reduction of the strong birefringence associated with the midribs in ppcesa3/8KOs, (2) reduction in the midrib fluorescence of ppcesa3/8KO leaves stained with S4B, (3) lack of CBM3a labeling of sections from ppcesa3/8KO leaf midribs (Fig. 1), and (4) reduction in ppcesa3/8KO gametophore cell wall cellulose content as measured by Updegraff assay. Evidence that knockout of PpCESA3 and PpCESA8 is responsible for the observed phenotype includes the consistency of the phenotype in three independent KOs and the restoration of cellulose deposition in the midribs by the transformation of ppcesa3/8KO with vectors driving the expression of PpCESA3 or PpCESA8 (Fig. 2). Whereas we detected no reduction in midrib cellulose in ppcesa3KOs, the phenotypes of ppcesa8KOs were intermediate between the wild type and ppcesa3/8KO (Fig. 2). This, combined with the observations that only PpCESA8 is up-regulated to compensate for the loss of its paralog (Fig. 6) and the expression of PpCESA3 under the control of its native promoter only partially restores the wild type phenotype (Fig. 2), is consistent with the hypothesis that the PpCESA3 and PpCESA8 proteins are functionally interchangeable and that a dosage effect is responsible for the ppcesa8KO phenotype. The formation of morphologically normal gametophores in ppcesa3/8KOs (Fig. 1) indicates that PpCESA3 and PpCESA8 serve a different role in development than PpCESA5, which supports the normal cell division and cell expansion required for gametophore development (Goss et al., 2012). It is possible that PpCESA3 and PpCESA8 contribute to primary cell wall deposition, since ppcesa3/8KO lamina cells had thinner external walls (Supplemental Fig. S4) and tended to collapse during embedding (Fig. 1). Alternatively, PpCESA3 and PpCESA8 may contribute to the secondary thickening of lamina cell walls after they stop expanding.
CESA Evolution in Both P. patens and Arabidopsis Involves Subfunctionalization and Neofunctionalization
There are many parallels in the evolution of the P. patens and Arabidopsis CESA families. In both species, different CESAs are responsible for primary and secondary cell wall deposition. In Arabidopsis, the secondary CESAs are AtCESA4, AtCESA7, and AtCESA8 (Taylor et al., 2003) and the primary CESAs are AtCESA1, AtCESA3, and members of the 6-like group (Desprez et al., 2007; Persson et al., 2007). In P. patens, midrib secondary cell wall synthesis involves PpCESA3, PpCESA6, PpCESA7, and PpCESA8, whereas gametophore primary cell wall synthesis requires PpCESA5 (Goss et al., 2012). At least some primary CESAs can substitute for secondary CESAs and vice versa in both species. In Arabidopsis, AtCESA3pro::AtCESA7 partially rescues atcesa3 and AtCESA8pro::AtCESA1 partially rescues atcesa8 (Carroll et al., 2012). In P. patens, PpCESA8pro::PpCESA5 rescues ppcesa3/8KO. This indicates that the CESA division of labor for primary and secondary cell wall deposition in vascular plants and mosses is due at least in part to subfunctionalization. However, neofunctionalization also has occurred in both species, resulting in the requirement for two or more noninterchangeable CESA isoforms for secondary cell wall biosynthesis. In Arabidopsis, atcesa4, atcesa7, and atcesa8 null mutants share a phenotype (Taylor et al., 2000) that cannot be complemented by expressing one of the other secondary AtCESAs with the promoter for the missing isoform (Kumar et al., 2017). Likewise in P. patens, ppcesa3/8KO and ppcesa6/7KO share the same phenotype, and ppcesa3/8KO is not complemented by PpCESA8pro::PpCESA7. Studies are ongoing to determine whether the secondary PpCESAs interact physically to form a CSC, as has been shown for the secondary AtCESAs (Taylor et al., 2003; Timmers et al., 2009). Finally, the CESA families of both species show some redundancy. In Arabidopsis, the 6-like CESAs (AtCESA2, AtCESA5, AtCESA6, and AtCESA9) are partially redundant (Persson et al., 2007), as are PpCESA3 and PpCESA8 in P. patens. PpCESA6 and PpCESA7 differ by only three amino acids, and the genes that encode them appear to be redundant (Wise et al., 2011).
A recent study has shown that secondary cell wall deposition, including CESA expression, is regulated by NAC transcription factors in both P. patens and Arabidopsis (Xu et al., 2014). Three P. patens NAC genes, PpVNS1, PpVNS6, and PpVNS7, were expressed preferentially in leaf midribs, and ppvns1/ppvns6/ppvns7KOs were defective in stereid development. Overexpression of PpVNS7 activated PpCESA3 (Xu et al., 2014). Phylogenetic analyses of NACs place eight PpVNS proteins within the clade that has variously been named subfamily NAC-c (Shen et al., 2009), subfamily Ic (Zhu et al., 2012), or the VNS group (Xu et al., 2014) and also includes the Arabidopsis vascular-related NACs VND6 (ANAC101), VND7 (ANA030), NST1 (ANAC043), NST2 (ANAC066), and NST3/SND1 (ANAC012). The three PpVNS proteins that regulate stereid development form a single sister clade with five other PpVNS proteins implicated also in other processes (Xu et al., 2014). Based on this phylogenetic analysis, the common ancestor of the mosses and seed plants had a single VNS gene, and it also had a single CESA gene (Roberts and Bushoven, 2007; Yin et al., 2009; Kumar et al., 2017). Both lineages now include secondary CESAs that are regulated by VNSs and primary CESAs that are not, indicating that CESA subfunctionalization occurred independently in mosses and seed plants.
Secondary Cell Wall Microfibrillar Texture Is Similar in Mosses and Vascular Plants
In vascular plants, both water-conducting tracheary elements and supportive fibers are characterized by helical (Barnett and Bonham, 2004) and aggregated (Donaldson, 2007; Fernandes et al., 2011; Thomas et al., 2014) cellulose microfibrils. The midribs of P. patens leaves include hydroid cells that transport water and stereid cells that provide support, but only the stereids have thick cell walls (Xu et al., 2014). With highly reduced cellulose in their stereid secondary cell walls, ppcesa3/8KOs provided a negative control for the structural characterization of secondary cell walls in wild type P. patens. A sharp SFG CH/CH2 stretch peak at 2,944 cm−1 is characteristic of angiosperm secondary cell walls (Park et al., 2013), and extensive empirical testing has shown that this spectral feature is attributable to lateral microfibril aggregation (Lee et al., 2014). The 2,944 cm−1 peak also was present in SFG spectra of wild type P. patens midribs. In contrast, the spectra of ppcesa3/8KO leaf midribs lacked the 2,944 cm−1 peak and instead had a broad peak between 2,800 and 3,000 cm−1, which is characteristic of primary cell walls and other samples lacking aggregated microfibrils (Park et al., 2013; Lee et al., 2014). This suggests that lateral aggregation of microfibrils is a common feature of the secondary cell walls of moss stereids and vascular plant tracheary elements and fibers. Polarization microscopy with a first-order retardation plate revealed that the microfibrils in the stereid cell walls are deposited in a helical pattern, as observed in secondary cell walls of tracheary elements and fibers (Barnett and Bonham, 2004). Although deficient in cellulose, the stereid cell walls of ppcesa3/8KOs were thickened, indicating that secondary cell wall synthesis involves the deposition of noncellulosic components, which proceeded in the absence of cellulose deposition. This also has been observed in developing tracheary elements treated with cellulose synthesis inhibitors (Taylor et al., 1992). Thus, stereid cell walls share structural characteristics with the cell walls of tracheary elements and fibers.
Mosses and Vascular Plants Have Acquired Similar Secondary Cell Walls through Convergent Evolution
Thick, cellulose-rich secondary cell walls provide added support for the aerial organs of mosses and vascular plants alike. Within these cell walls, the lateral aggregation and helical orientation of the microfibrils contribute to their strength and resilience. Although cortical microtubules play an important role in cellulose microfibril orientation, oriented cellulose deposition can occur in the absence of cortical microtubules, and it has been suggested previously that the aggregation and helical orientation of microfibrils in secondary walls are consequences of high CSC density during rapid cellulose deposition (Emons and Mulder, 2000; Lindeboom et al., 2008). Regulation at the level of CSC secretion was emphasized in this model (Emons and Mulder, 2000), but CSC density can potentially be regulated at the level of transcription.
Rapid cellulose synthesis during secondary cell wall deposition in specific cell types requires precise temporal and spatial regulation of CESA expression that is distinct from the regulatory requirements for primary cell wall synthesis. We suggest that these distinct regulatory needs were met through the evolution of independent regulatory control of primary and secondary CESAs by subfunctionalization in both mosses and seed plants. In seed plants, phylogenetic analysis shows that the first divergence of the CESA family separated the genes that encode the primary and secondary CESAs and was followed by independent diversification within each group (Roberts et al., 2012). This, along with evidence that some primary CESAs are interchangeable with secondary CESAs (Carroll et al., 2012), indicates that subfunctionalization was an early event in the evolution of the seed plant CESA family. In P. patens, the genes that encode secondary PpCESA3 and PpCESA8 and primary PpCESA5 are also subfunctionalized and, therefore, specialized, although they encode interchangeable proteins.
Several lines of evidence indicate that the capacity to deposit a secondary cell wall evolved independently in mosses and seed plants. Structural and paleobotanical evidence suggests that the support and water-conducting cells of bryophytes and vascular plants are not homologous (Ligrone et al., 2002; Carafa et al., 2005). Phylogenetic evidence indicates that the primary and secondary CESAs diversified independently in mosses and seed plants (Roberts and Bushoven, 2007; Yin et al., 2009; Kumar et al., 2017) and, as explained above, so did the NAC transcription factors that regulate the secondary CESAs. There are even examples of convergent evolution of secondary cell walls within the angiosperm lineage. Cotton (Gossypium hirsutum) fiber secondary cell walls are synthesized by the same CESAs that are responsible for secondary cell wall deposition in tracheary elements and fibers (Haigler et al., 2012), whereas the secondary cell walls of epidermal trichomes are synthesized by the primary CESAs (Betancur et al., 2010). These observations are consistent with independent evolutionary origins for secondary cell walls in different land plant lineages and different cell types within angiosperm lineages.
Taken together, these data indicate that CESA duplication, followed by changes in the regulatory elements within the primary or secondary CESA promoters, occurred independently in mosses and vascular plants. The resulting uncoupling of the secondary CESAs from the regulatory constraints associated with primary cell wall deposition, along with a mechanistic linkage between CESA expression and microfibril texture as well as selection for strength and resilience, may have contributed to the capacity of different plants to synthesize cellulose-rich secondary cell walls with similar microfibrillar textures.
MATERIALS AND METHODS
Vector Construction
All primer pairs are shown in Supplemental Table S1, along with annealing temperatures used for PCR. Amplification programs for Taq Polymerase (New England Biolabs) consisted of a 3-min denaturation at 94°C followed by 35 cycles of 15 s at 94°C, 30 s at the annealing temperature, and 1 min kb−1 at 72°C. Amplification programs for Phusion Polymerase (New England Biolabs) consisted of a 30-s denaturation at 98°C followed by 35 cycles of 7 s at 98°C, 7 s at the annealing temperature, and 30 s kb−1 at 72°C.
To construct the CESA8KO vector, a 3′ homologous region was amplified from Physcomitrella patens genomic DNA with primers 174JB and 193JB using Taq DNA polymerase, cut with SalI and BspDI, and cloned into the SalI/BstBI site of pBHSNR (a gift of Didier Schaefer, University of Neuchâtel). The resulting plasmid was cut with KasI and NsiI to accept the KasI/NsiI fragment of a 5′ homologous region amplified from P. patens genomic DNA with primers 203JB and 185JB (Supplemental Table S1). The CESA8KO vector was cut with EcoRI and NsiI for transformation into wild type P. patens. The CESA3KO, CESA4KO, CESA6/7KO, and CESA10KO vectors were constructed using Gateway Multisite Pro cloning (Invitrogen) as described previously (Roberts et al., 2011). Flanking sequences 5′ and 3′ of the coding regions were amplified with appropriate primer pairs (Supplemental Table S1) using Phusion DNA polymerase (New England Biolabs) and cloned into pDONR 221 P1-P4 and pDONR 221 P3-P2, respectively, using BP Clonase II (Invitrogen). Similarly, an nph selection cassette was amplified from pMBL6 (a gift of Jesse Machuka, University of Leeds) and cloned into pDONR 221 P3r-P4r. All entry clones were sequence verified. For vectors conferring hygromycin resistance, entry clones with flanking sequences in pDONR 221 P1-P4 and pDONR 221 P3-P2 were inserted into BHSNRG (Roberts et al., 2011). For vectors conferring G418 resistance, entry clones with flanking sequences in pDONR 221 P1-P4 and pDONR 221 P3-P2 were linked with the entry clone containing the nph selection cassette and inserted into pGEM-gate (Vidali et al., 2009) using LR Clonase II Plus (Invitrogen). The vectors in BHSNRG or pGEM-gate were cut with BsrGI for transformation into wild type or mutant P. patens lines.
Expression vectors for hemagglutinin (HA)-tagged PpCESAs under the control of PpCESA promoters were constructed using Gateway Multisite Pro cloning (Invitrogen). The PpCESA4 (DQ902545), PpCESA5 (DQ902546), PpCESA7 (DQ160225), and PpCESA8 (DQ902549) coding sequences were amplified from cDNA clones pdp21409, pdp24095, pdp38142, and pdp39044 (RIKEN BioResource Center), respectively, using forward primers containing a single HA tag and appropriate reverse primers (Supplemental Table S1) and cloned into pDONR 221 P5-P2 using BP Clonase II (Invitrogen). The PpCESA3 (XP_001753310) and PpCESA10 (XP_001776974) coding sequences were amplified similarly from expression vectors. pDONR 221 P1-P5r entry clones containing approximately 2 kb of sequence upstream of the PpCESA3 or PpCESA8 start codon (Tran and Roberts, 2016) were linked to the sequence-verified entry clones containing the HA-PpCESA coding sequences and inserted into pSi3(TH)GW (Tran and Roberts, 2016) using LR Clonase II Plus (Invitrogen). These vectors target the expression cassettes to the intergenic 108 locus, which can be disrupted with no effect on phenotype (Schaefer and Zrÿd, 1997). Rescue vectors were cut with SwaI for transformation into a P. patens ppcesa3/8KO line from which the hph resistance cassette had been removed (see below).
Culture and Transformation of P. patens
Wild type P. patens lines (haploid) derived from the sequenced Gransden strain (Rensing et al., 2008) by selfing and propagation from a single spore in 2006 (GD06) or 2011 (GD11) were gifts of Pierre-Francois Perroud (Washington University). Wild type and transformed P. patens lines were cultured on basal medium supplemented with ammonium tartrate (BCDAT) as described previously (Roberts et al., 2011). Protoplasts were prepared and transformed as described previously (Roberts et al., 2011). Stable transformants were selected with 50 μg mL−1 G418 (CESA3KO vector) or 15 μg mL−1 hygromycin (CESA8KO and complementation vectors). The hph selection cassette was removed from ppces3/ppcesa8KO by transforming protoplasts with NLS-Cre-Zeo (Vidali et al., 2010), selecting for 7 d on BCDAT plates containing 50 μg mL−1 zeocin, replica plating zeocin-resistant colonies on BCDAT with and without 15 μg mL−1 hygromycin, and recovering hygromycin-sensitive colonies. Protein expression was tested by western-blot analysis as described previously (Scavuzzo-Duggan et al., 2015) in selected lines transformed with HA-PpCESA expression vectors.
Genotype Analysis
For PCR screening, DNA was extracted as described previously (Roberts et al., 2011) and 2.5-μL samples were subjected to 35 cycles of amplification (45 s at 94°C, 45 s at the annealing temperature shown in Supplemental Table S1, and 1 min kb−1 at 72°C) with PAQ5000 DNA polymerase (Agilent Technologies) in 25-μL reactions. Primers used to test for target integration, target-gene disruption, and selection cassette excision are listed in Supplemental Table S1.
Phenotype Analysis
The cell wall birefringence of unfixed leaves mounted in water was examined using an Olympus BHS compound microscope with D Plan-Apo UV 10×/0.4, 20×/0.7, and 40×/0.85 objectives, and polarizer and circular-polarizing analyzer, with and without a first-order retardation plate (Olympus). Images were captured with a Leica DFC310FX digital camera with Leica Application Suite software, version 4.2.0 (Leica Microsystems), with manual exposure under identical conditions.
For direct fluorescent labeling of cellulose, whole gametophores (three per line) dissected from colonies grown for 4 weeks on solid BCDAT medium were dipped in 100% acetone for 5 s to permeabilize the cuticle, rinsed in phosphate-buffered saline (PBS), incubated in PBS containing 0.01 mg mL−1 S4B (Anderson et al., 2010) for 30 min, and rinsed in PBS. All fully expanded leaves (12–20) were cut from each gametophore and mounted in PBS. Fluorescence images of each leaf, centered on the brightest part of the midrib, were captured using a Zeiss Axio Imager M2 with 43HE DsRed filter set, Plan-Neofluar 20×/0.5 objective, AxioCam MR R3 camera, and Zen Blue software, version 1.1.2.0 (Carl Zeiss Microscopy), under identical conditions using manual exposure. The midrib in each image was selected manually (Supplemental Fig. S10), and average pixel intensity was measured using ImageJ, Fiji version (Schindelin et al., 2012). For comparison of KOs with the wild type, three independent lines of each KO genotype (n = 3) and two independent wild type lines (GD06 and GD11; n = 2) were sampled in triplicate. For analysis of rescue lines, three independent explants were sampled for each genetic line (n = 3).
For affinity cytochemistry of cellulose, gametophores dissected from colonies grown for 2 weeks on BCDAT medium were fixed and embedded in LR White resin (Polysciences) as described previously (Kulkarni et al., 2012). Sections (1 μm) were mounted and labeled with CBM3a as described previously (Berry et al., 2016). Images were captured with a Zeiss Axio Imager M2 with 38 GFP filter set, EC Plan-Neofluar 40×/0.75 objective, AxioCam MR R3 camera, and Zen Blue software, version 1.1.2.0 (Carl Zeiss Microscopy), under identical conditions using manual exposure. Fluorescence and polarization images were not altered after capture. Bright-field and differential interference contrast images were captured using automatic exposure, and some images used for illustrative purposes were adjusted for uniformity using the color-balance and exposure functions in Photoshop, version CS6 (Adobe Systems).
ppcesa3KOs, ppcesa8KOs, and ppcesa3/8KOs were tested for changes in caulonema gravitropism and rhizoid development as described previously (Roberts et al., 2011). Images were captured using a Leica M165FC stereomicroscope with Leica DFC310FX camera and Leica Application Suite software, version 4.2.0 (Leica Microsystems). Caulonema length for each colony was measured as the distance from the edge of the colony to the tip of the longest caulonema filament using Leica Application Suite software.
Cell Wall Analysis
Alcohol-insoluble residue was prepared from gametophores dissected from eight to 10 4-week-old explants of P. patens wild type (three samples from independent cultures) and ppcesa3/8KO (samples from three independent lines) cultured on BCDAT medium. Tissue was ground in liquid nitrogen and extracted three times, 30 min each, with 70% (v/v) ethanol and once with 100% ethanol, and the residue was dried under vacuum. The alcohol-insoluble residue (∼1 mg) was weighed to 0.001 mg and mixed with 1 mL of acetic acid:water:nitric acid (8:2:1, v/v) in screw-cap vials, and the suspension was heated in a boiling water bath for 30 min (Updegraff, 1969). After cooling, the tubes were centrifuged at 16,900g for 5 min and the supernatant was discarded. The pellet was resuspended in 2 mL of deionized water and centrifuged, and the supernatant was discarded. The washing step was repeated at least 10 more times until the supernatant was neutralized, and the pellet was resuspended in 1 mL of water. The amount of cellulose remaining after hydrolysis was quantified by sulfuric acid assay (Albalasmeh et al., 2013) with Glc as the standard. Briefly, 100 µL of hydrolysate (six technical replicates per sample) was diluted to 1 mL with water in a glass tube, 3 mL of concentrated sulfuric acid was added, and samples were vortexed for 30 s and chilled on ice for 2 min. Reactions were measured at 315 nm against a reagent blank.
High-Pressure Freezing-Freeze Substitution and Transmission Electron Microscopy
Gametophytes of P. patens GD06 and PpCESAKOs were high-pressure frozen using a Leica EMPACT2 high-pressure freezer (Leica Microsystems) followed by freeze substitution in 0.1% uranyl acetate (w/v) in acetone for 48 h at −90°C before the temperature was ramped up slowly to −50°C (Wilson and Bacic, 2012). The samples were rinsed with acetone twice at −50°C before the acetone was replaced with ethanol, and the samples were subsequently infiltrated with LR White resin (ProSciTech) in a series of ethanol/resin dilutions. The samples were rinsed three times in 100% resin before polymerization with UV light at −20°C for 48 h. Thin sections (70 nm) were cut using a Leica Ultracut R (Leica Microsystems) and poststained with uranyl acetate and lead citrate (Wilson and Bacic, 2012). Images were taken using a Tecnai G2 Spirit transmission electron microscope (FEI). Cell wall thickness was measured using ImageJ, Fiji version (Schindelin et al., 2012).
Ultrathin sections (70 nm) also were cut from blocks prepared for affinity cytochemistry (see above), mounted on formvar-coated copper grids, and stained with uranyl acetate and lead citrate (Wilson and Bacic 2012). Sections were imaged using a FEI/Phillips CM-200 transmission electron microscope (FEI).
SFG Spectroscopy
Leaves of wild type GD06, 8KO-5B, and 3/8KO-86 lines were mounted abaxial side down in water on glass slides and allowed to air dry overnight. SFG spectra were collected in 5-µm intervals along a 200-µm line scan perpendicular to the midrib at its thickest point using an SFG microscope system described previously (Lee et al., 2016). The SFG spectra were collected with the following polarization combination: SFG signal = s-, 800 nm = s-, and broadband mid-IR = p-polarized with the laser incidence plane and the laser incidence plane aligned along the axis of midrib.
Reverse Transcription Quantitative PCR
RNA was extracted from gametophores from two independent wild type and three independent lines each of ppcesa3KO and ppcesa8KO as described previously (Tran and Roberts, 2016). cDNA samples were tested in duplicate as described previously using primer pairs for the amplification of PpCESA3 and PpCESA8. The primers were tested previously for specificity and efficiency (Tran and Roberts, 2016). Primers for actin and v-type H+-translocating pyrophosphatase reference genes were described previously (Le Bail et al., 2013). Target/average reference cross-point ratios were calculated for each sample, and se values were calculated for independent genetic lines.
Statistical Analysis
For statistical analysis, one-way ANOVA with posthoc Tukey’s Honestly Significant Difference test was performed at astatsa.com/OneWay_Anova_with_TukeyHSD/.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ902545, DQ902546, DQ902549, DQ160224, DQ160225, XP_001753310, and XP_001776974.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Genotype analysis of ppcesa8KO, ppcesa3KO, and ppcesa3/8KO lines.
Supplemental Figure S2. Phenotype analysis of a ppcesa3/8KO line transformed with vectors driving the expression of PpCESA3 or PpCESA8 with their native promoters.
Supplemental Figure S3. Transmission electron microscopy images of leaf cell walls from wild type and cesaKO lines of P. patens.
Supplemental Figure S4. Thickness of outer cell walls measured from transmission electron microscopy images.
Supplemental Figure S5. P. patens wild type and KO lines cultured on medium containing 1 μm naphthalene acetic acid to induce rhizoid initiation and inhibit leaf initiation.
Supplemental Figure S6. P. patens wild type and KO lines cultured in the dark on vertically oriented plates containing medium supplemented with 35 mm Suc to test for caulonema gravitropism.
Supplemental Figure S7. Western-blot analysis of protein expression for P. patens lines derived from the transformation of ppcesa3/8KO-86lox with vectors driving the expression of PpCESAs under the control of the PpCESA8 promoter.
Supplemental Figure S8. PCR-based genotyping of ppcesa6/7KO lines.
Supplemental Figure S9. PCR-based genotyping of ppcesa4/10KO lines.
Supplemental Figure S10. Quantification method for S4B fluorescence.
Supplemental Table S1. Primers used for vector construction and genotype analysis.
Acknowledgments
Clones pdp39044 and pdp10281 were from RIKEN BRC; we thank Chessa Goss and Virginia Lai for preliminary work on ppceas8KO, Alfred Schupp for assistance with vector construction, Evan Preisser for assistance with statistics, and Sarah Kiemle for conducting Updegraff assays.
Glossary
- SFG
sum frequency generation
- CSC
cellulose synthesis complex
- S4B
Pontamine Fast Scarlet
- PBS
phosphate-buffered saline
Footnotes
This work was supported primarily by National Science Foundation Award IOS-1257047. Analysis of mutants by SFG spectroscopy was supported as part of the Center for LignoCellulose Structure and Formation, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences Award DE-SC0001090. CBM3a affinity cytochemistry and freeze substitution transmission electron microscopy were supported by the Australian Research Council Centre for Excellence in Plant Cell Walls Grant CE1101007. High-pressure freezing and transmission electron microscopy were undertaken at the Melbourne Advanced Microscopy Facility at the Bio21 Institute and the Biosciences Microscopy Unit at the University of Melbourne. DNA sequencing and qPCR were conducted using the Rhode Island Genomics and Sequencing Center, a Rhode Island National Science Foundation EPSCoR research facility, supported in part by the National Science Foundation EPSCoR Cooperative Agreement EPS-1004057.
Articles can be viewed without a subscription.
References
- Albalasmeh AA, Berhe AA, Ghezzehei TA (2013) A new method for rapid determination of carbohydrate and total carbon concentrations using UV spectrophotometry. Carbohydr Polym 97: 253–261 [DOI] [PubMed] [Google Scholar]
- Anderson CT, Carroll A, Akhmetova L, Somerville C (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett JR, Bonham VA (2004) Cellulose microfibril angle in the cell wall of wood fibres. Biol Rev Camb Philos Soc 79: 461–472 [DOI] [PubMed] [Google Scholar]
- Barnette AL, Bradley LC, Veres BD, Schreiner EP, Park YB, Park J, Park S, Kim SH (2011) Selective detection of crystalline cellulose in plant cell walls with sum-frequency-generation (SFG) vibration spectroscopy. Biomacromolecules 12: 2434–2439 [DOI] [PubMed] [Google Scholar]
- Barnette AL, Lee C, Bradley LC, Schreiner EP, Park YB, Shin H, Cosgrove DJ, Park S, Kim SH (2012) Quantification of crystalline cellulose in lignocellulosic biomass using sum frequency generation (SFG) vibration spectroscopy and comparison with other analytical methods. Carbohydr Polym 89: 802–809 [DOI] [PubMed] [Google Scholar]
- Berry EA, Tran ML, Dimos CS, Budziszek MJ Jr, Scavuzzo-Duggan TR, Roberts AW (2016) Immuno and affinity cytochemical analysis of cell wall composition in the moss Physcomitrella patens. Front Plant Sci 7: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur L, Singh B, Rapp RA, Wendel JF, Marks MD, Roberts AW, Haigler CH (2010) Phylogenetically distinct cellulose synthase genes support secondary wall thickening in Arabidopsis shoot trichomes and cotton fiber. J Integr Plant Biol 52: 205–220 [DOI] [PubMed] [Google Scholar]
- Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, Knox JP (2006) Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem 281: 29321–29329 [DOI] [PubMed] [Google Scholar]
- Carafa A, Duckett JG, Knox JP, Ligrone R (2005) Distribution of cell-wall xylans in bryophytes and tracheophytes: new insights into basal interrelationships of land plants. New Phytol 168: 231–240 [DOI] [PubMed] [Google Scholar]
- Carpita N, McCann M (2000) The cell wall. In Buchanan B, Gruissem W, Jones R, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 52–108 [Google Scholar]
- Carroll A, Mansoori N, Li S, Lei L, Vernhettes S, Visser RG, Somerville C, Gu Y, Trindade LM (2012) Complexes with mixed primary and secondary cellulose synthases are functional in Arabidopsis plants. Plant Physiol 160: 726–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP. (1999) Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50: 245–276 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson L. (2007) Cellulose microfibril aggregates and their size variation with cell wall type. Wood Sci Technol 41: 443–460 [Google Scholar]
- Emons AMC, Mulder BM (2000) How the deposition of cellulose microfibrils builds cell wall architecture. Trends Plant Sci 5: 35–40 [DOI] [PubMed] [Google Scholar]
- Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperley DC, Kennedy CJ, Jarvis MC (2011) Nanostructure of cellulose microfibrils in spruce wood. Proc Natl Acad Sci USA 108: E1195–E1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Guillot A, Vernhettes S, Höfte H (2014) Catalytic subunit stoichiometry within the cellulose synthase complex. Plant Physiol 166: 1709–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss CA, Brockmann DJ, Bushoven JT, Roberts AW (2012) A CELLULOSE SYNTHASE (CESA) gene essential for gametophore morphogenesis in the moss Physcomitrella patens. Planta 235: 1355–1367 [DOI] [PubMed] [Google Scholar]
- Haigler CH, Betancur L, Stiff MR, Tuttle JR (2012) Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front Plant Sci 3: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebant C. (1977) The Conducting Tissues of Bryophytes. J. Cramer, Vaduz, Liechtenstein [Google Scholar]
- Hill JL Jr, Hammudi MB, Tien M (2014) The Arabidopsis cellulose synthase complex: a proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 26: 4834–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis MC. (2013) Cellulose biosynthesis: counting the chains. Plant Physiol 163: 1485–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P, Crane PR (1997) The origin and early evolution of plants on land. Nature 389: 33–39 [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM Jr (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni AR, Peña MJ, Avci U, Mazumder K, Urbanowicz BR, Pattathil S, Yin Y, O’Neill MA, Roberts AW, Hahn MG, et al. (2012) The ability of land plants to synthesize glucuronoxylans predates the evolution of tracheophytes. Glycobiology 22: 439–451 [DOI] [PubMed] [Google Scholar]
- Kumar M, Atanassov I, Turner S (2017) Functional analysis of cellulose synthase (CESA) protein class specificity. Plant Physiol 173: 970–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bail A, Scholz S, Kost B (2013) Evaluation of reference genes for RT qPCR analyses of structure-specific and hormone regulated gene expression in Physcomitrella patens gametophytes. PLoS ONE 8: e70998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Kafle K, Huang S, Kim SH (2016) Multimodal broadband vibrational sum frequency generation (MM-BB-V-SFG) spectrometer and microscope. J Phys Chem B 120: 102–116 [DOI] [PubMed] [Google Scholar]
- Lee CM, Kafle K, Park YB, Kim SH (2014) Probing crystal structure and mesoscale assembly of cellulose microfibrils in plant cell walls, tunicate tests, and bacterial films using vibrational sum frequency generation (SFG) spectroscopy. Phys Chem Chem Phys 16: 10844–10853 [DOI] [PubMed] [Google Scholar]
- Ligrone R, Vaughn KC, Renzaglia KS, Knox JP, Duckett JG (2002) Diversity in the distribution of polysaccharide and glycoprotein epitopes in the cell walls of bryophytes: new evidence for the multiple evolution of water-conducting cells. New Phytol 156: 491–508 [DOI] [PubMed] [Google Scholar]
- Lindeboom J, Mulder BM, Vos JW, Ketelaar T, Emons AM (2008) Cellulose microfibril deposition: coordinated activity at the plant plasma membrane. J Microsc 231: 192–200 [DOI] [PubMed] [Google Scholar]
- Newman RH, Hill SJ, Harris PJ (2013) Wide-angle x-ray scattering and solid-state nuclear magnetic resonance data combined to test models for cellulose microfibrils in mung bean cell walls. Plant Physiol 163: 1558–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon BT, Mansouri K, Singh A, Du J, Davis JK, Lee JG, Slabaugh E, Vandavasi VG, O’Neill H, Roberts EM, et al. (2016) Comparative structural and computational analysis supports eighteen cellulose synthases in the plant cellulose synthesis complex. Sci Rep 6: 28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehme DP, Downton MT, Doblin MS, Wagner J, Gidley MJ, Bacic A (2015) Unique aspects of the structure and dynamics of elementary Iβ cellulose microfibrils revealed by computational simulations. Plant Physiol 168: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Lee CM, Koo BW, Park S, Cosgrove DJ, Kim SH (2013) Monitoring meso-scale ordering of cellulose in intact plant cell walls using sum frequency generation spectroscopy. Plant Physiol 163: 907–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Roberts AW, Bushoven JT (2007) The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Mol Biol 63: 207–219 [DOI] [PubMed] [Google Scholar]
- Roberts AW, Dimos CS, Budziszek MJ Jr, Goss CA, Lai V (2011) Knocking out the wall: protocols for gene targeting in Physcomitrella patens. Methods Mol Biol 715: 273–290 [DOI] [PubMed] [Google Scholar]
- Roberts AW, Roberts EM, Haigler CH (2012) Moss cell walls: structure and biosynthesis. Front Plant Sci 3: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavuzzo-Duggan TR, Chaves AM, Roberts AW (2015) A complementation assay for in vivo protein structure/function analysis in Physcomitrella patens (Funariaceae). Appl Plant Sci 3: 1500023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer DG, Zrÿd JP (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11: 1195–1206 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz M, Smith R, Ellis B (2013) Xylem tissue specification, patterning, and differentiation mechanisms. J Exp Bot 64: 11–31 [DOI] [PubMed] [Google Scholar]
- Shen H, Yin Y, Chen F, Xu Y, Dixon RA (2009) A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. Bioenerg Res 2: 217–232 [Google Scholar]
- Taylor JG, Owen TP Jr, Koonce LT, Haigler CH (1992) Dispersed lignin in tracheary elements treated with cellulose synthesis inhibitors provides evidence that molecules of the secondary cell wall mediate wall patterning. Plant J 2: 959–970 [Google Scholar]
- Taylor NG, Gardiner JC, Whiteman R, Turner SR (2004) Cellulose synthesis in the Arabidopsis secondary cell wall. Cellulose 11: 329–338 [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LH, Forsyth VT, Martel A, Grillo I, Altaner CM, Jarvis MC (2014) Structure and spacing of cellulose microfibrils in woody cell walls of dicots. Cellulose 21: 3887–3895 [Google Scholar]
- Timmers J, Vernhettes S, Desprez T, Vincken JP, Visser RG, Trindade LM (2009) Interactions between membrane-bound cellulose synthases involved in the synthesis of the secondary cell wall. FEBS Lett 583: 978–982 [DOI] [PubMed] [Google Scholar]
- Tran ML, Roberts AW (2016) Cellulose synthase gene expression profiling of Physcomitrella patens. Plant Biol (Stuttg) 18: 362–368 [DOI] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Vandavasi VG, Putnam DK, Zhang Q, Petridis L, Heller WT, Nixon BT, Haigler CH, Kalluri U, Coates L, Langan P, et al. (2016) A structural study of CESA1 catalytic domain of Arabidopsis cellulose synthesis complex: evidence for CESA trimers. Plant Physiol 170: 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Burkart GM, Augustine RC, Kerdavid E, Tüzel E, Bezanilla M (2010) Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell 22: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, van Gisbergen PA, Guérin C, Franco P, Li M, Burkart GM, Augustine RC, Blanchoin L, Bezanilla M (2009) Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc Natl Acad Sci USA 106: 13341–13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SM, Bacic A (2012) Preparation of plant cells for transmission electron microscopy to optimize immunogold labeling of carbohydrate and protein epitopes. Nat Protoc 7: 1716–1727 [DOI] [PubMed] [Google Scholar]
- Wise HZ, Saxena IM, Brown RM Jr (2011) Isolation and characterization of the cellulose synthase genes PpCesA6 and PpCesA7 in Physcomitrella patens. Cellulose 18: 371–384 [Google Scholar]
- Xu B, Ohtani M, Yamaguchi M, Toyooka K, Wakazaki M, Sato M, Kubo M, Nakano Y, Sano R, Hiwatashi Y, et al. (2014) Contribution of NAC transcription factors to plant adaptation to land. Science 343: 1505–1508 [DOI] [PubMed] [Google Scholar]
- Yang JH, Wang H (2016) Molecular mechanisms for vascular development and secondary cell wall formation. Front Plant Sci 7: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Huang J, Xu Y (2009) The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol 9: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2015) Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 56: 195–214 [DOI] [PubMed] [Google Scholar]
- Zhu T, Nevo E, Sun D, Peng J (2012) Phylogenetic analyses unravel the evolutionary history of NAC proteins in plants. Evolution 66: 1833–1848 [DOI] [PubMed] [Google Scholar]