Abstract
The roundworm Caenorhabditis elegans has a haploid karyotype containing six linear chromosomes. The termini of worm chromosomes have been proposed to play an important role in meiotic prophase, either when homologs are participating in a genome-wide search for their proper partners or in the initiation of synapsis. For each chromosome one end appears to stimulate crossing-over with the correct homolog; the other end lacks this property. We have used a bioinformatics approach to identify six repetitive sequence elements in the sequenced C.elegans genome whose distribution closely parallels these putative meiotic pairing centers (MPC) or homolog recognition regions (HRR). We propose that these six DNA sequence elements, which are largely chromosome specific, may correspond to the genetically defined HRR/MPC elements.
INTRODUCTION
Meiosis in sexually reproducing eukaryotes is a process in which the number of chromosomes in a diploid cell (2n) are reduced by half to yield haploid gametes containing 1n chromosomes. In animals this event is coupled with gametogenesis, leading to the production of either mature ova or spermatozoa. A key feature that serves to minimize errors and ensure faithful segregation of only one copy of each chromosome to individual gametes during this essential event is a physical association between the two newly replicated chromosomal homologs. The classical chiasmata, formed by DNA recombination, but not visible until after synapsis, appear to hold the chromatids together and constrain univalents to form a bivalent. When microtubules from opposite meiotic spindle poles attach to discrete sites on the outside of each bivalent the resulting tension during metaphase I aligns the chromosomes at the metaphase plate. In anaphase I the connections holding the chromatids together are finally dissolved, allowing the paired chromosomes to disjoin into each daughter cell. The resulting DNA shuffling could potentially confer a selective advantage to eventual progeny that receive one of these recombinant chromosomes (1). Of course meiotic recombination can also be used to establish the relative order and distance between linked genes along a chromosome (2).
Recombination per se involves a complex series of stereotypical events that occur in early meiosis during prophase (3). Homolog recognition is likely the earliest step in this process (4,5). Each member of a pair of homologous chromosomes must sort through the other non-homologous chromosomes within the nucleus and find its one correct partner which could lead to a productive alignment. While nothing is known about the molecular events leading to proper homolog recognition, prevalent models invoke the presence of cis-acting sites located on the chromosomes which could then bind trans-acting factors (4,6,7). Once the correct homologs find and recognize each other then actual pairing, whereby the two homologs are aligned in register along the chromosome, can commence. Meiotic chromosome synapsis occurs after proper pairing when the synaptonemal complex (SC) forms a proteinaceous matrix in between the two paired homologs (8). Similarly to homolog recognition, little is known about the steps in SC formation. The precise timing of the actual recombination event is not known for most organisms, but has clearly initiated by the diplotene stage of prophase I (9,10).
Discovery and mapping of regions that appear to stimulate pairing in the roundworm Caenorhabditis elegans has been facilitated by the ease of isolating and characterizing relatively stable chromosomal aberrations (11). Specifically, the cumulative genetic behavior of not only chromosomal fragments (so-called free duplications), but also reciprocal translocations, led to the identification of a specific sub-region on each one of the six chromosomes that acts dominantly to enhance recombination of genetic markers on the chromosomal rearrangements with the wild-type copy of that homolog (12,13). These sub-regions are referred to as either homolog recognition regions (HRRs) or meiotic pairing centers (MPCs) and are located asymmetrically on one end of each chromosome (4,5,7). By examining deficiencies of chromosomes I and X it was also possible to infer that each chromosome must contain additional sites located outside the HRRs/MPCs that can aid homolog recognition or synapsis when one or both HRRs/MPCs are partially deleted (6,7).
As part of the ongoing analysis of the completed C.elegans genome sequence we sought DNA sequences that were located predominantly on only one of the six chromosomes. Among this data set we have found short (11–16 nt) repeated elements that are essentially unique to each chromosome. Examining the distribution of these sequences along a particular homolog revealed a marked asymmetry that is strongly correlated with the genetically defined HRRs/MPCs described above. Here we describe the nature of these sequence elements and propose that they may correspond to cis-acting sites mediating the initial steps of either homolog recognition or synapsis during meiotic prophase in the worm.
MATERIALS AND METHODS
The C.elegans chromosome sequence data as of 98/11/11 from the Sanger Center (www.sanger.ac.uk) were used in this study. The first step in our analysis was to identify and count all of the 2mers, 3mers, 4mers, …, 20mers contained in the DNA sequence of each one of the six C.elegans chromosomes. The second step involved dividing the number of occurrences of each nmer on one chromosome by the total length of DNA from that chromosome to arrive at the number per bp. The third step was to take each individual nmer and compare the number per bp from one chromosome to the number per bp on the other five chromosomes. For example, the number of 6mers with the sequence GAATTC found in 13 Mb of sequence from chromosome I was calculated per bp and compared to the number of 6mers per bp with this sequence found in 84 Mb of sequence comprised of chromosomes II, III, IV, V and X. Most nmers in the worm genome are present at similar frequencies on each chromosome. However, in rare instances an nmer may be enriched, or over-represented, on one chromosome in comparison to the other five chromosomes. Any DNA sequence that was at least 1.38-fold enriched on an individual chromosome was flagged for further analysis. Source code for this analysis is available upon request.
The highest scoring sequences for each chromosome were then charted on frequency plots. All the sequence elements from one chromosome were aligned manually and the flanking sequences were examined for other repeating patterns; this was repeated for each individual chromosome. Some clustering was noted and this is discussed below in the next section.
RESULTS AND DISCUSSION
Our search strategy as outlined in Materials and Methods was designed to identify relatively short sequences (up to 20 nt in length) that were over-represented on only one of the six C.elegans chromosomes. This upper limit of 20 nt was chosen in part because searches with longer sequences would have taken prohibitively long to calculate and in part because most sequences in the worm genome longer than 14 or 15 nt are likely to be unique [(1/4)14 = 1/2.68 × 108], which makes analysis of longer and longer sequences less informative. Furthermore, our methodology will also highlight longer sequences with unusual distributions in the worm genome because subsets of non-identical overlapping 20mers present in a longer sequence will be identified using this algorithm. Our method uses brute force to identify and catalog nmers contained in the sequence of each worm chromosome. By calculating how often an individual nmer occurs (e.g. once per 43 kb) along a chromosome and comparing that value to how often the same nmer occurs in the rest of the genome we discovered candidate sequences whose frequency distribution appeared skewed in the genome. At this point several additional criteria were applied. Sequences whose complement did not also show a high relative frequency were ruled out since they would be found primarily on one strand of the DNA and we thought it more likely that sites for potential sequence-specific DNA-binding proteins might be found in both orientations along the chromosome. Similarly, we focused only on sequences that were present more than twice per chromosome, reasoning that elements with multiple copies might be more relevant. Additionally, we eliminated minisatellite sequences where the same simple short element is repeated tandemly dozens or hundreds of times, since we thought that such highly clustered repeats were most likely the result of slippage and expansion during DNA replication. For five of the worm chromosomes there was one and only one sequence that met all these criteria and these were analyzed further to determine how they were distributed along their respective chromosomes. On chromosome IV two candidates were identified and the one with a strongly skewed distribution along the physical map is reported here. For all of the repeated DNA elements described in this report there was a dramatic and obvious difference between their scores and the next nmer on the list.
Table 1 shows the sequence of each chromosome-specific repeat. In accordance with the nomenclature system devised by the genome sequencing consortium we have named the repeat elements CeRep45, CeRep46, CeRep47, CeRep48, CeRep49 and CeRep50. With one noteworthy exception the six sequences we have identified are not represented in any of the previously described short repetitive elements in the worm genome (i.e. CeRep1–CeRep44). The exception is the 16 bp palindrome found predominantly on the left arm of chromosome III (CeRep47). CeRep47 is a subset of the larger palindromes CeRep25 (14,15) and CeRep25B (16). In each case we limited our analysis to sequences that are precise matches to the six defined elements. We restricted out searches to perfect repeats, in part due to computational limitations. Presumably lowering the stringency and allowing mismatches would find more sites, but we have not investigated this further.
Table 1. Incidence of chromosome-enriched repetitive elements in the C.elegans genome.
| Element name |
|
|
|
|
|
|
| CeRep45 | CeRep46 | CeRep47 | CeRep48 | CeRep49 | CeRep50 | |
| DNA sequence |
TTGGTTGAGGCT |
TTTGTAGTCTAGCA |
TGCTAAATATTTAGCA |
GTATAATCATG |
TGGGCGCTGCT |
TGGTCAGTGCA |
| Length | 12 | 14 | 16 | 11 | 11 | 11 |
| Chromosome | I | II | III | IV | V | X |
| Chromosome length (Mb) | 13.85 | 14.73 | 12.77 | 16.14 | 20.82 | 17.22 |
| Incidence on enriched chromosome, including both DNA strands (per Mb) | 611 (44.1) | 152 (10.3) | 197 (15.4) | 347 (21.5) | 713 (34.2) | 335 (19.4) |
| Incidence in the remaining genome (per Mb) | 201 (2.5) | 54 (0.7) | 1 (0.0) | 251 (3.2) | 13 (0.2) | 74 (0.9) |
| Total incidence in the C.elegans genome (per Mb) | 812 (8.5) | 206 (2.2) | 198 (2.1) | 598 (6.2) | 726 (7.6) | 409 (4.3) |
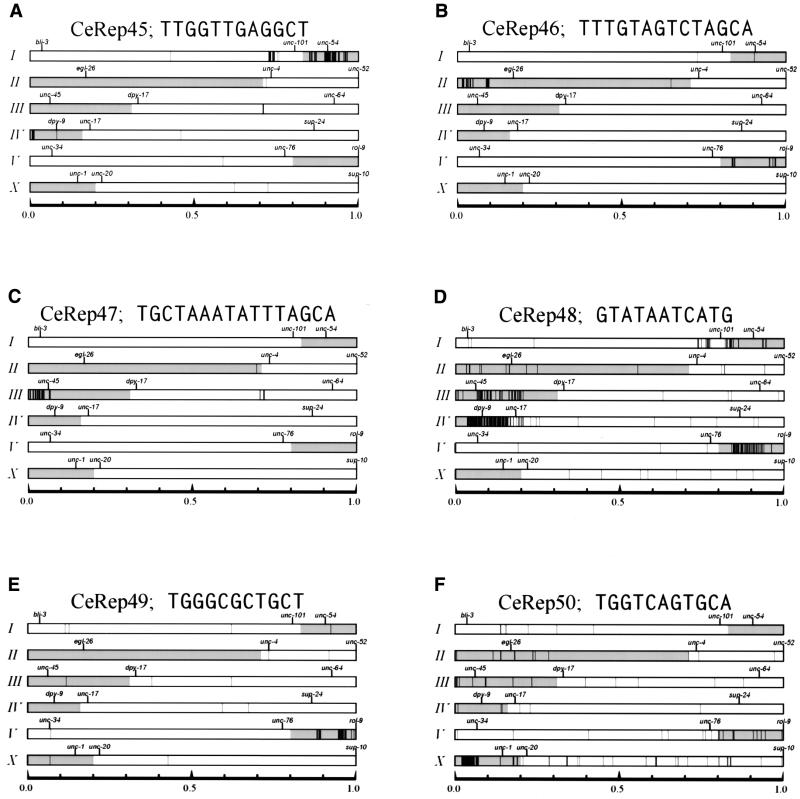
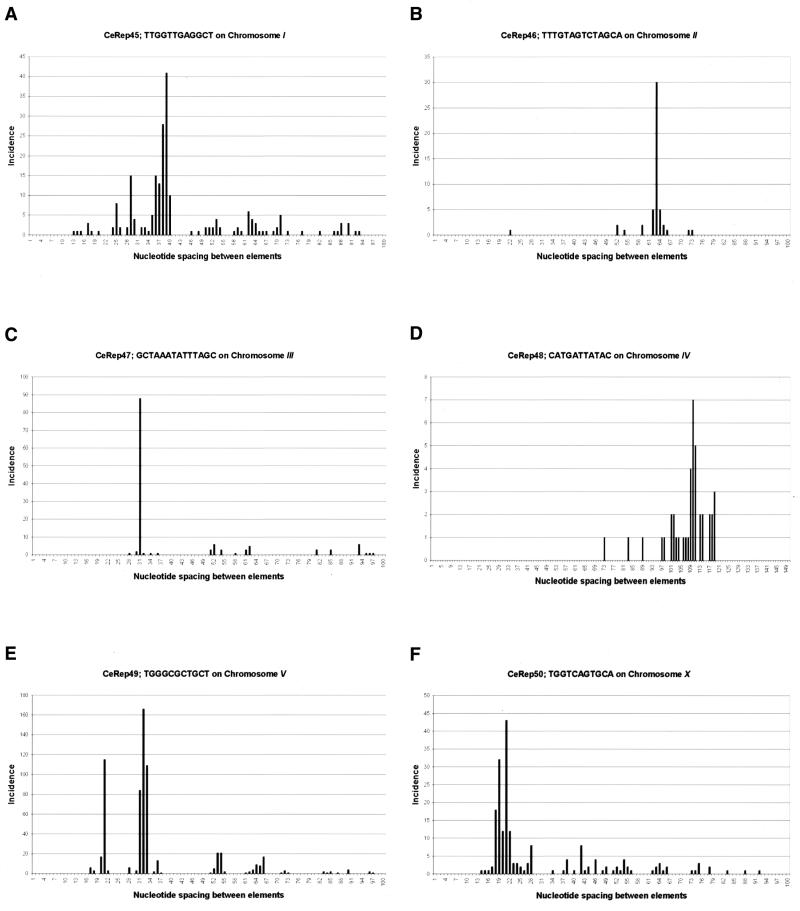
The worm genome contains 812 copies of the 12 bp sequence CeRep45 and 611 of these are found in the sequence of chromosome I. This 12 bp sequence is GC rich (50%) compared to the whole C.elegans genome, which is 36% GC overall. CeRep45 elements are found in two large clusters on the right end of chromosome I (Fig. 1A). Approximately one-third (214) of the elements are within 100 bp of another copy of CeRep45 and virtually all of these tightly linked adjacent copies of CeRep45 are present in an inverted orientation relative to each other. The largest such cluster contains 70 copies of CeRep45 distributed within 35 kb at position 12.453 (this numbering system uses the extreme left end of chromosome I as position number 0.000001). The highest density of CeRep45 is located at position 12.848, where 47 copies are repeated in a fragment of 5 kb. By plotting the distances between each occurrence of CeRep45 we noticed that certain spacings of 36, 37, 38, 39 and 40 bp are favored (Fig. 2A) (see below).
Figure 1.
Repetitive sequence elements in the C.elegans genome with asymmetrical distributions. The genome-wide distribution of each of the six repetitive elements described in the text are shown. Each chromosome is labeled with roman numerals on the left side of the panels. Horizontal rectangular boxes represent the six worm chromosomes. Caenorhabditis elegans gene names for genetic markers used to orient the physical map are shown above the chromosomes. The shaded portions of each chromosome correspond to genetically mapped HRR/MPC elements (4). Each panel shows the location of individual CeRep elements. Black vertical lines represent the locations along the chromosomes. Chromosomes vary in length from ∼13 to ∼21 Mb and are normalized in this diagram. The scale is shown along the bottom of each panel.
Figure 2.
Conserved spacings between adjacent repetitive DNA elements. For each of the six repetitive elements described in this study we tabulated the distance in nucleotides between each adjacent copy along the chromosome. The data was then plotted as bar graphs with the incidence on the ordinate and the distances on the abscissa. Elements >100 bp apart are not shown (or >120 bp apart in the case of CeRep48). (A) CeRep45; (B) CeRep46; (C) CeRep47; (D) CeRep48; (E) CeRep49; (F) CeRep50.
CeRep46 is a 14 bp long sequence of 35% GC (Table 1) and is found repeated 206 times in C.elegans. Of these copies, 152 are located on chromosome II in two small clusters at the left end (Fig. 1B). The first cluster, containing 66 copies, is 482 kb in size and begins at chromosome position 0.213 (once again using the extreme left end as position 0.000001) while the second cluster, containing 86 copies, is only 112 kb in size and is separated from the first cluster by 600 kb of genomic sequence (Fig. 1B). There are 34 sub-clusters of CeRep46 containing two (or more) copies of the element and in almost every instance they are found as direct repeats. Similarly to CeRep45 described above, we found that tightly linked copies of CeRep46 are located a characteristic distance apart (in this instance 63 or 64 bp) (Fig. 2B).
There are 197 copies of the 16 bp palindrome CeRep47 located almost exclusively on the extreme left end of chromosome III (Fig. 1C). One additional copy is located on another chromosome. At 51 of the sites there are two or more copies of CeRep47 clustered together. Some of these 197 elements correspond to CeRep25, a 31 bp repeated sequence identified by the genome sequencing consortium (14,15). Another variant of this sequence was identified previously by Pilgrim (16) as a 24 bp palindromic minisatellite DNA element highly enriched on chromosome III, which he named CeRep25B (where B denotes it is a degenerate variant). CeRep47 elements are found in three sub-clusters located at positions 0.274 (42 copies in 13 kb), 0.403 (47 copies in 35 kb) and 0.523 (42 copies in 36 kb). Interestingly, we note that in these densely covered regions the spacing between individual CeRep47s are often multiples of 31 nt (e.g. 31, 62, 93) (Fig. 2C and see below).
The 11mer sequence defining CeRep48 differs from the other five minisatellites we characterized in that there are copies of this element (251 in total) scattered along chromosomes I, II, III, V and X (Fig. 1D). While the other 347 copies of CeRep48 are clearly distributed non-randomly at the left end of chromosome IV, in contrast to the other five sequences we describe the individual elements are not clustered as tightly near each other (Fig. 2D). In addition, we do not see such a skewed distribution of inter-element spacings (Fig. 2D). Based on these three observations CeRep48 clearly seems different from the other elements (see below).
The 11 nt CeRep49 elements located almost exclusively on chromosome V are present at 733 copies on the extreme right end of this chromosome (Fig. 1E). Similarly to CeRep47 on chromosome III the spacing between individual elements is often 31, 32 or 33 nt in length and in multiples of these integers (e.g. 62, 64 and 66). Their distribution is restricted to two large clusters. One at position 18.358 is 205 kb in length. The other large cluster of repeats is located 1.1 Mb away at position 19.715 and is 528 kb in length. The vast majority of CeRep49 repeats are <60 bp away from another copy of this element. Strikingly, they are almost always in tandem arrays. Another unusual feature of this sequence is that it is uncharacteristically GC rich (72%).
The 335 copies of the 11mer CeRep50 arrayed along the X chromosome are concentrated in five sub-clusters on the left end of the chromosome (Fig. 1F). Most of the copies are located near each other in direct tandem repeats separated by multiples of 21 nt. One ramification of this spacing is that, similarly to CeRep46 and CeRep49, many CeRep50 elements will be 63 nt apart.
The only conspicuous similarity between any of these six sequences is the pentamer TAGCA, which is found on the 3′-end of CeRep46 and CeRep47 (Table 1). Since CeRep47 is a perfect palindrome, this means TAGCA is found twice in each copy of CeRep47, once on each strand. Further examination revealed that CeRep49 and CeRep50 share a degenerate version of this same sequence, YAGYR. However, CeRep45 and CeRep48 contain nothing remotely resembling this pattern. CeRep45 and CeRep48 share a different degenerate sequence, TRRTYRWG.
The literature is replete with papers reporting repetitive DNA sequence elements in the C.elegans genome. These range from classical descriptions of reassociation kinetics using C0t curves (17) and hybridization studies to detect RFLPs (18) to actual cloning and sequencing of genomic library fragments (19). With near completion of the genomic sequencing effort (15) it is now possible to apply exhaustive string searching algorithms (20) to catalog all the different types of repetitive DNA in this worm.
We set out to identify short sequences that were enriched on one of the six worm chromosomes. In each case a single oligonucleotide from each chromosome met this criterion. Since many repetitive elements in the worm genome were already known to be localized in the chromosome arms, we were not surprised to discover that CeRep45, CeRep46, CeRep47, CeRep48, CeRep49 and CeRep50 are largely absent from the gene-rich central clusters of each autosome (15,19,21). However, the distribution of each repeat element along its own individual chromosome was striking in its asymmetry. Each of these six repeated sequences is distinctly clustered at just one of the ends of their specific chromosome. While it is possible that skewed patterns in nucleotide sequences could arise by chance or that such patterns may reflect an ancient architecture for each worm chromosome (20), we were impressed by the strong correlation between the location of these six repetitive DNA elements and the genetically characterized HRRs/MPCs mapped to each chromosome (4,5,13).
The existence of HRR/MPC sequences in C.elegans was originally deduced from the observation that long (i.e. several Mb) stretches of identical DNA sequence are not always sufficient to promote wild-type levels of meiotic recombination between rearranged chromosomes (12). HRR/MPC sequences have been proposed to function as enhancers of meiotic recombination by facilitating either an early step in pairing or actual synapsis between homologous chromosomes during prophase in meiosis I in the worm. Given that low levels of recombination can occur in the absence of HRRs/MPCs, there seems to be some functional redundancy in the system that ensures proper disjunction, as if other sequences on each chromosome can also stimulate recombination (6,7). However, nothing is known about the nature of any of these important sequences.
There are two further pieces of evidence that support our hypothesis that the six sequences we have identified and analyzed in this study may play a role in either meiotic homolog recognition or synapsis. First, our tabulation and comparison of short oligonucleotides in the C.elegans genome did not turn up multiple candidates on each chromosome. This is significant because it means that there are no other sequence elements of this length present in the genome with such strongly skewed patterns of enrichment on one chromosome and asymmetry along that chromosome. Secondly, we searched for additional sequences that were more frequent closer to the end of each chromosome than our proposed HRRs. No additional candidates were found in this analysis, showing that all other short repetitive elements are less abundant.
Why do many of the elements occur with a characteristic spacing? Sequence alignment of the elements from each chromosome did not reveal any larger degenerate repeats (Materials and Methods). The explanation we currently favor is that having a high local concentration of a DNA-binding site could increase the concentration of bound proteins, perhaps cooperatively.
As noted above, there are many copies of the CeRep48 element located on chromosomes I, II, III, V and X (Fig. 1D). If CeRep48 can function as part of the HRR/MPC on chromosome IV then the cell must distinguish between elements linked to IV and elements located on other chromosomes, otherwise time and energy would be spent on non-productive pairing. Interestingly, on chromosomes I, III and V the CeRep48 elements lie predominantly near the HRR regions. Therefore, we speculate that CeRep48 may have two roles in homolog recognition: the high density of CeRep48 on chromosome IV functions as an HRR/MPC, while in the context of other HRR/MPC elements the lower density of CeRep48 on other chromosomes might act as partially redundant sequences that help to stimulate recombination (6,7).
At present the evidence for CeRep45–CeRep50 being HRR/MPC elements is entirely circumstantial. Now that we have identified candidate sequences that could fulfill this important function on each chromosome there are several experimental approaches that could be used to assess the biological function(s) (if any) of these repetitive sequence elements. It would be interesting to ask if any of the sequences we have identified in this report bind specifically to C.elegans nuclear proteins. It may also be possible to identify such proteins using the yeast one-hybrid screen with each sequence (22). Assays such as these address specificity but not function. A key test for these six CeRep will be to reintroduce them onto a chromosome fragment that lacks HRR/MPC activity and ask if they now confer recombination enhancing function. Likewise, precisely deleting clusters of these CeRep from one chromosome should lead to reduced levels of recombination. Experiments like these may be feasible in the worm since it is possible to (i) make transgenic worms (23,24), (ii) attach the arrays to genetically marked duplications (25), and (iii) screen for customized deletions from collections of mutagenized animals (26,27).
It would also be of great interest to understand at the molecular level precisely how the HRRs/MPCs function to facilitate pairing events and by eventually identifying the sequences and the proteins that control this process we hope to gain some insight into this essential process.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Diane De Abreu, Andrew Spence and an anonymous reviewer for helpful comments on the manuscript. This research was supported by a scholarship and an operating grant from the Medical Research Council of Canada (to M.D.P.)
References
- 1.Peck J.R. (1994) A ruby in the rubbish: beneficial mutations, deleterious mutations and the evolution of sex. Genetics, 137, 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sturtevant A.H. (1913) The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Zool., 14, 43–59. [Google Scholar]
- 3.Hawley R.S. and Arbel,T. (1993) Yeast genetics and the fall of the classical view of meiosis. Cell, 72, 301–303. [DOI] [PubMed] [Google Scholar]
- 4.Zetka M. and Rose,A. (1995) The genetics of meiosis in Caenorhabditis elegans. Trends Genet., 11, 27–31. [DOI] [PubMed] [Google Scholar]
- 5.Albertson D.G., Rose,A.M. and Villeneuve,A.M. (1997) Chromosome organization, mitosis and meiosis. In Riddle,D.L., Blumenthal,T., Meyer,B.J. and Preiss,J.R. (eds) C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 47–78. [PubMed]
- 6.McKim K.S., Peters,K. and Rose,A.M. (1993) Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans. Genetics, 134, 749–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villeneuve A.M. (1994) A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics, 136, 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmekel K. and Daneholt,B. (1995) The central region of the synaptonemal complex revealed in three dimensions. Trends Cell Biol., 5, 239–242. [DOI] [PubMed] [Google Scholar]
- 9.Dernburg A.F., McDonald,K., Moulder,G., Barstead,R., Dresser,M. and Villeneuve,A.M. (1998) Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell, 94, 387–398. [DOI] [PubMed] [Google Scholar]
- 10.Zalevsky J., MacQueen,A.J., Duffy,J.B., Kemphues,K.J. and Villeneuve,A.M. (1999) Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics, 153, 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herman R., Albertson,D. and Brenner,S. (1976) Chromosome rearrangements in Caenorhabditis elegans. Genetics, 83, 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rose A.M., Baillie,D.L. and Curran,J. (1984) Meiotic pairing behaviour of two free duplications of linkage group I in Caenorhabditis elegans. Mol. Gen. Genet., 195, 52–56. [DOI] [PubMed] [Google Scholar]
- 13.McKim K., Howell,A. and Rose,A. (1988) The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics, 120, 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson R., Ainscough,R., Anderson,K., Baynes,C., Berks,M., Bonfield,J., Burton,J., Connell,M., Copsey,T., Cooper,J. et al. (1994) 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature, 368, 32–38. [DOI] [PubMed] [Google Scholar]
- 15.T.C.e.S. Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 16.Pilgrim D. (1998) CeRep25B forms chromosome-specific minisatellite arrays in Caenorhabditis elegans. Genome Res., 8, 1192–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sulston S. and Brenner,S. (1974) The DNA of Caenorhabditis elegans. Genetics, 77, 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emmons S., Klass,M. and Hirsh,D. (1979) Analysis of the constancy of DNA sequences during development and evolution of the nematode Caenorhabditis elegans. Proc. Natl Acad. Sci. USA, 76, 1333–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naclerio G., Cangiano,G., Coulson,A., Levitt,A., Ruvolo,V. and La Volpe,A. (1992) Molecular and genomic organization of clusters of repetitive DNA sequences in Caenorhabditis elegans. J. Mol. Biol., 226, 159–168. [DOI] [PubMed] [Google Scholar]
- 20.Surzycki S.A. and Belknap,W.R. (2000) Repetitive-DNA elements are similarly distributed on Caenorhabditis elegans autosomes. Proc. Natl Acad. Sci. USA, 97, 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnes T.M., Kohara,Y., Coulson,A. and Hekimi,S. (1995) Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics, 141, 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M.M. and Reed,R.R. (1993) Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature, 364, 121–126. [DOI] [PubMed] [Google Scholar]
- 23.Fire A. (1986) Integrative transformation of Caenorhabditis elegans. EMBO J., 5, 2673–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mello C.C., Kramer,J.M., Stinchcomb,D. and Ambros,V. (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller L.M., Waring,D.A. and Kim,S.K. (1996) Mosaic analysis using a ncl-1(+) extrachromosomal array reveals that lin-31 acts in the Pn.p cells during Caenorhabditis elegans vulval development. Genetics, 143, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plasterk R.H.A. (1995) Reverse genetics: from gene sequence to mutant worm. In Epstein,H.F. and Shakes,D.C. (eds), Caenorhabditis elegans: Modern Biological Analysis of an Organism. Academic Press, Toronto, Canada, Vol. 48, pp. 59–80. [DOI] [PubMed]
- 27.Jansen G., Hazendonk,E., Thijssen,K.L. and Plasterk,R.H.A. (1997) Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nature Genet., 17, 119–121. [DOI] [PubMed] [Google Scholar]