Abstract
Curli are functional amyloids produced by proteobacteria like Escherichia coli, as part of the extracellular matrix that holds cells together into biofilms. The molecular events during curli nucleation and fiber extension remain largely unknown. Combining observations from curli amyloidogenesis in bulk solutions with real-time in situ nanoscopic imaging at the single fiber level, we show that curli display polar growth, and detect two kinetic regimes of fiber elongation. Single fibers exhibit stop-and-go dynamics characterized by bursts of steady-state growth alternated with periods of stagnation. At high subunit concentrations fibers show constant, unperturbed burst growth. Curli follow a one-step nucleation process, where monomers contemporaneously fold and oligomerize into minimal fiber units that have growth characteristics identical to the mature fibrils. Kinetic data and interaction studies of curli fibrillation in the presence of the natural inhibitor CsgC show the inhibitor binds curli fibers and predominantly acts at the level of fiber elongation.
Keywords: curli, functional amyloid, biofilm, nucleation, high-speed AFM
Introduction
Amyloids are aggregative protein or peptide fibrils best known for their implication in (neuro)degenerative illnesses such as type 2 diabetes and Alzheimer’s, Parkinson’s and Huntington’s diseases1,2. However, it is now well established that the structural hallmark of amyloids, the assembly into cross-β structured fibrils, is not restricted to off-pathway protein misfolding events seen in pathological amyloidosis but is also found as the native conformation of several pro- and eukaryotic proteins referred to as “functional amyloids”1,3. In bacteria, for example, functional amyloid fibrils are frequently found as part of the extracellular biopolymer matrix that ties the bacteria together into persistent multicellular communities called biofilms, or mediate bacterial attachment to the host or abiotic surfaces4–9. These functional amyloids are the product of diverse biosynthetic pathways dedicated to the controlled aggregation of pro-amyloid subunits into surface-localized fibers.
Although the repeating units of different amyloid fibers are highly diverse in primary and tertiary structure10,11, both functional and disease-associated amyloids share a number of basic characteristics including a cross-β spine quaternary structure, and a self-assembly process that involves a usually rate-limiting nucleation step and a rapid extension into linear fibrils through addition of soluble subunits to the fiber template12. In pathogenic amyloids, cytotoxicity is ascribed primarily to soluble oligomeric species that exist as intermediates or side-products during nucleation and early fiber assembly, and to a lesser extent to the mature fibers13–15. Whether this applies to functional amyloids is not known. Cells and organisms that actively form amyloids as part of their natural physiology must prevent or deal with the formation of possible cytotoxic species. From this perspective, a detailed elucidation of the molecular mechanism(s) of functional amyloid formation is needed to determine whether the adapted amyloidogenesis pathways minimize cytotoxicity, or whether control mechanisms like chaperone activity and/or controlled spatiotemporal aggregation of the amyloid subunits take priority.
Curli are one of the most widespread functional amyloids, found in many Gram-negative bacteria, where they form non-covalent polymeric protein filaments assembled as part of an extracellular matrix that encapsulates the bacteria within a protective biofilm4,16–18. Two gene clusters encode the curli subunits, as well as the accessory and channel proteins required for the controlled passage of pro-amyloid subunits across the cell envelope and subsequent deposition into amyloid fibrils on the cell surface4,18,19. The major building block of curli is CsgA, an intrinsically disordered, pseudo-repeat protein of 15,7 kDa in E. coli, which folds during self-assembly into fibers that exhibit the typical cross-β spine architecture and are resistant to denaturing agents as sodium dodecyl sulphate (SDS)20. In vitro fibrillation can be initiated by switching from denaturing to native conditions which spontaneously gives rise to a tangled mesh of fibers that appear structurally and morphologically equivalent to their in vivo counterparts14,21. The mechanism of curli nucleation and growth has been a subject of intense study over the last decade21–24. Based on various biophysical methods, a two-step precipitation pathway has been suggested that comprises an initial lag-phase characterized by a build-up of dynamic, amorphous, metastable aggregates that give rise to an amyloid-like oligomeric nucleus, followed by a growth phase that is primarily dominated by the elongation of existing fibers21. However, a direct demonstration of such a mechanism is lacking. Interestingly, the curli system also possesses a selective inhibitor, CsgC, that can prevent premature fiber formation in the periplasmic space25, presumably by targeting the level of nucleation26. The current picture of CsgA fiber formation and its inhibition has been predominantly formed based on bulk biophysical methods and ex situ transmission electron microscopy imaging, leaving a number of key questions unanswered. What are the molecular events that take place in the lag phase? What is the size and structure of the amyloid nucleus? What species precede the formation of such a nucleus, and how does CsgC prevents its formation? And how do single fibers grow?
In this contribution, we sought to answer these questions by combining established biophysical methods with real-time in situ nanoscale imaging using atomic force microscopy27,28 focusing on the earliest moments of in vitro amyloid formation. We found that curli fibers are polar, and that single fibers exhibit stop-and-go dynamics characterized by periods of steady growth, alternated with variable periods of stagnation, often associated with localized structural defects. We observed that in vitro curli nucleation does not originate from aggregate intermediates, but rather appears a direct, one-step process where monomeric species contemporaneously fold and oligomerize into minimal fiber units that have growth characteristics identical to the mature fibrils that emanate from these structures. We also found that CsgC specifically binds curli fibrils and acts at the level of fiber elongation rather than by inhibiting nucleation. These insights can contribute to the production of future therapeutic agents that can prevent or combat host colonization and persistence in biofilm-associated bacterial amyloids, but also offer promising prospects in nanobiotechnology, where there is an increasing interest in harnessing the physical properties and self-assembling nature of amyloids for biomaterials and nanotechnological purposes29–33.
Results
Kinetics of CsgA polymerization in bulk solution
We first focused on one of the key reported characteristics of in vitro CsgA fibrillation, and by extension amyloid formation in general, i.e. the presence of a lag phase in the kinetic read-out of bulk biophysical techniques. Indeed, following the fibrillation of monomeric CsgA by thioflavin T (ThT) fluorescence as a function of time showed the characteristic sigmoidal curve seen in nucleation-dependent polymerization reactions (Figure 1a, inset). An analogous curve was obtained using dynamic light scattering (DLS): plotting the temporal dependence of the total scattering intensity of polymerizing CsgA solutions yielded a sigmoidal curve that is functionally similar to those obtained for ThT fluorescence (Figure 1b, inset). From the collected correlation curves, we extracted a mean, apparent, hydrodynamic radius for these scattering particles of 2.8±1.0 nm at 5 minutes after desalting, which is consistent with the predicted hydrodynamic radius of 2.9 nm for monomeric, unfolded CsgA (Supplementary Results, Supplementary Figure 1). At 5 minutes, CsgA monomers were the predominant scattering species, which at later time points started to be replaced by particles ranging in size from hundreds of nm to multiple μm (shown at 2.5 hours in Supplementary Fig. 1). Plot as a function of time, we obtained a steady linear increase of the characteristic size (Figure 1b).
Figure 1. CsgA fibrillation in bulk solution.
(a) Tht fluorescence of a polymerizing CsgA solution (15 µM) cast into semi-log and linear (inset) coordinates; (b) total scattering intensity collected at 90° of 15 µM CsgA cast into semi-log and linear (inset) coordinates ; (c) negative-stain TEM image of a CsgA sample (15 µM) that was collected after 3.5 minutes of incubation; Inset: zoom-in of a single fiber; fibers measure 7.4±1.1 nm in width (sample mean±s.d.; n=100); (d) consecutive CD spectra of a 15 µM CsgA solution monitored every 45 minutes starting from 5 min (black) to 9 hours (light grey) after desalting, inset: CD signal at 220 nm as a function of time (the characteristic maximum at 195 nm for β-sheets is missing from our spectra due to light scattering effects at this wavelength).
The lag phase seen in the bulk growth curves has been suggested to represent the formation and conformational sampling of oligomeric complexes that lead to nucleation of the minimal oligomers that allow fiber extension21,34. Although not yet detectable by DLS or ThT fluorescence, a time series of transmission electron microscopy (TEM) images clearly demonstrated the presence of single fibers starting from the moments immediately after buffer-exchange, i.e. within 3.5 minutes of removing the denaturing agent (Figure 1c). These early fibers were 7.4±1.1 nm in width (n=100), compared to 7.8±1.2 nm (n=100) observed for native curli on bacterial cells (Supplementary Figure 2b), and increase in length over time to ultimately end in a tangled mesh of curli fibers at saturation (Supplementary Figure 2a). We found no evidence by TEM for the existence of any non-fibrous CsgA oligomers that were alluded to previously in literature26. Instead, we observed the presence of nascent fibers with curli-like morphology within our experimental sampling time (3.5 min) (Figure 1c). When using circular dichroism (CD) to follow the secondary structure evolution during CsgA fibrillation (Figure 1d), we saw a gradual shift from a minimum around 206 nm to a minimum around 220 nm, i.e. a transition from random coil to β-sheet. Keeping the wavelength fixed at 220 nm, we recorded a constant decrease of the molar ellipticity as a function of time, starting from the earliest measurable time point, and with a linear dependence over the entire observation period (Figure 1d, inset).
Curli growth kinetics at single fiber resolution
The high number density of fibers present in the earliest TEM images calls into question the sensitivity of the ThT assay towards the presence of short, single fibers. Such false negatives emphasize the potential risks associated with interpreting ThT data and underlie the need for independent methods to monitor CsgA fibrillation. Therefore we employed in situ atomic force microscopy (AFM) to provide a dynamic image of in vitro curli growth (Figure 2). When first looking at ex situ grown fibers, we observed a tangled mesh of single fibers that were devoid of any measurable substructure and which measured approximately 1.7±0.4 nm in height, although a smaller fraction of double height fibers was observed as well, measuring 3.3±0.9 nm (Figure 2a, Supplementary Fig. 2b). Although lateral interactions were observed, we found no evidence for a defined ultrastructure. Rather, curli conglomerates appeared composed of single fibers that form dispersed nodes of lateral and crossing interactions.
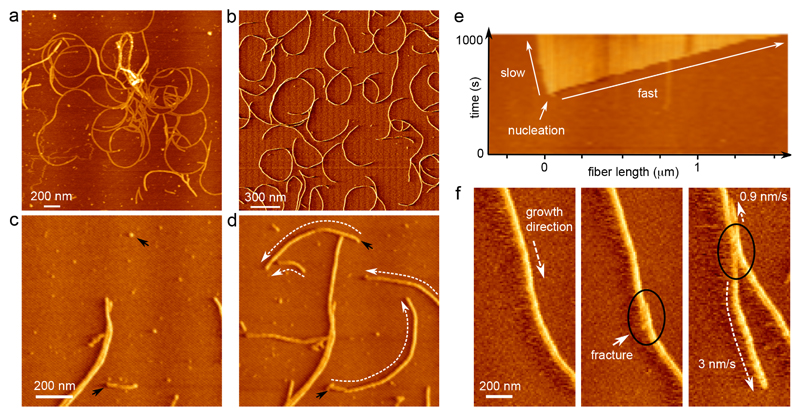
Figure 2. Curli show polar growth kinetics.
(a) Vertical deflection AFM image of the ultrastructure of CsgA curli grown ex situ and deposited onto mica after 2 h 30 min; (b) CsgA fibers nucleated and grown in situ; (c, d) time-lapse imaging reveals that fibers grow anti-clockwise, with an average radius of curvature of 548±149 nm (sample mean±s.d.; n=100) (black arrows indicate nucleation events, dashed white arrows indicate the direction of growth; images taken 200 s apart); (e) AFM kymograph showing the asymmetry in the rates of elongation of the two termini of a single CsgA fiber; (f) asymmetry in growth kinetics is reproduced when a fracture (black circle) is introduced into a single fiber by briefly increasing the tip-sample interaction force. Figures (b) – (e) correspond to the AFM error channel.
We then performed time-lapse AFM imaging on a system that starts out as a homogenous solution of CsgA monomers and then gradually transitioned into a network of mature fibers (Figure 2b, Supplementary Video 1). As also observed by TEM, single fibers were already present in the first images that were collected (dead-time 3 min). Nascent fibers rapidly elongated over the course of the observation period, whilst new fibers were constantly and independently being formed (Figure 2c,d). We observed no fiber branching events or fiber surface catalyzed secondary nucleation: single fibers did not give rise to multiple growth poles. We found that on the AFM substrate, curli elongation ceased when a fiber terminus made contact with another fiber – vertical bridging events were only rarely observed (Figure 2c,d). As fibers had a tendency to develop a curvature over time, isolated fibers often self-terminated by coiling in on themselves (Figure 2b).
Polar growth, fiber chirality and stop-and-go dynamics
Closer inspection of the collected time-lapse AFM movies revealed that the two termini of a single fiber were generally not equally active. Most fibers grew anisotropically, i.e. the rate of elongation of the leading fiber end was considerably larger than the rate of the trailing end. This growth polarity is best illustrated by constructing a kymograph pseudo-image, where in a single image the dynamic process is represented by extracting the pixel values along the trajectory of a fiber, and linearizing the extracted points into columns that are stacked for the consecutive frames of a single movie (Figure 2e, Supplementary Figure 3). With the kymograph axes now in units of time and space, it is easy to pinpoint the exact moment of nucleation, as well as to see the anisotropy in growth kinetics. We could locally fracture individual fibers by increasing the mechanical force applied by the tip while scanning (Figure 2f). The local defect resulted in two new growth poles that retained the directionality and elongation anisotropy of the parental fiber (Figure 2f). This demonstrated that growth polarity was not a result of extrinsic kinetic factors, but rather reflected the intrinsic structural polarity of the CsgA amyloid structure.
Native curli fibers that can grow unperturbed have the tendency to form curled structures, hence the name curli4,35. Solution grown fibers exhibited a radius of curvature of 269±52 nm (n=35) after deposition onto the AFM substrate (Figure 2a), which is in good correspondence to the curvature of in vivo grown fibers expressed on cells, i.e. 259±100 nm (n=105). The majority of in situ grown fibers elongated with a curvature of 548±149 nm (n=100) and with a positive chirality, i.e. curled fibers grew anti-clockwise on the substrate chosen in this work (mica) (Figure 2c,d). The tip growth rates were found independent of the radial angle along the curved fibers, indicating that our kinetic observations did not correspond to any preferred orientation(s) of fibers on the mica substrate.
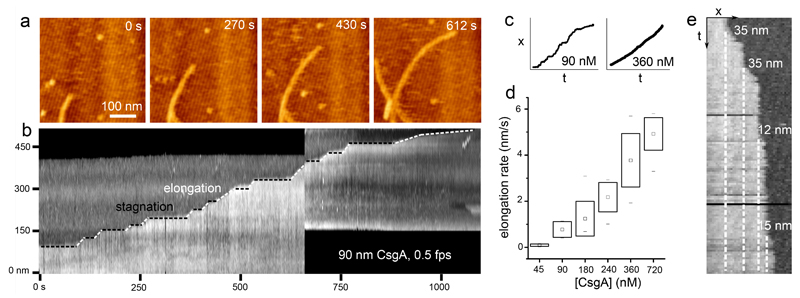
Using high-speed time-lapse AFM imaging, we could investigate the kinetics of individual fiber elongation with high temporal resolution (Figure 3). Scanning at 60 Hz, i.e. down to 2.2 seconds per frame, we detected significant variations in the instantaneous rate of elongation over the course of the observation period (Figure 3a, b, Supplementary Video 2). Fiber ends tended to accelerate and pause seemingly at random. These kinetic fluctuations are again best illustrated using a kymograph. At 90 nM CsgA, fibers did not exhibit steady-state kinetics, rather they displayed periods of steady growth, alternated with periods of stagnation (see Figure 3b, c). The minimal step-size observed between two stagnation episodes was 12±4 nm, which is expected to correspond to a few copies of CsgA (predicted dimensions36 of folded CsgA = 3.2 × 1.8 nm) (Figure 3e). The periods of steady burst growth suggested growth pauses are not due to diffusion-limited availability of CsgA monomers. We also found no correlation in terms of kinetics with any neighboring fibers or fiber orientation on the mica surface, so that gradients (thermal, concentration) or epitaxy could be ruled out as the cause for these variations in the burst rate. Instead, growth stagnations frequently went accompanied with an increased contrast at the fiber terminus suggesting a localized perturbation in fiber morphology (Supplementary Fig. 4b). When these temporary defects resolved into continued growth, this could occur with or without leaving a morphological “scar” at the site of the growth stagnation.
Figure 3. Curli display stop-and-go growth kinetics.
(a) High-speed time lapse imaging of a single growing fiber terminus (still images are frames of Supporting Video 3); (b) kymograph constructed from the fiber terminus followed in the upper panel reveals the stop-and-go dynamics of fiber growth; (c) representative traces of the fiber terminus location as a function of time at low and high CsgA concentration; (d) box-plot of the observed fiber elongation rate (robs, i.e. the global fiber elongation rate composed of periods of stagnation and burst elongation) averaged over 10 different fibers per concentration (box range = sample standard deviation; □ = sample mean; dashes show the minima and maxima of the dataset); (e) kymograph constructed from high speed AFM imaging at 45 nM CsgA shows the typical minimal detectable size of the fiber end displacements of 12±4 nm (mean elongation rate is 0.1±0.05 nm/s (see panel d); time-resolution: 10s). Figures (a), (b) and (e) correspond to the AFM error channel.
Growth fluctuations became dampened at higher concentrations of CsgA. The mean rate of fiber elongation ranged from 0.1 nm/s to 5 nm/s going from 45 nM to 720 nM CsgA concentration (Figure 3d). Remarkably, however, the increased average fiber elongation rates at higher CsgA concentration were primarily a result of shortened periods of stagnation, and reflect only a moderate increase in burst rates (Figure 3c, Supplementary Figure 4). We therefore identify two kinetic regimes of fiber elongation. At high CsgA concentration, fibers exhibited steady-state kinetics with an instantaneous rate of growth that was constant as a function of time. At low concentration, single fibers exhibited stop-and-go dynamics characterized by periods of burst growth that were interspersed with periods of stagnation. The transition from stop-and-go to steady-state kinetics was realized by two effects: (i) a gradual reduction of the time spent in the arrested state, and (ii) an increase of the elongation rate during periods of growth.
Vanishing nucleus size
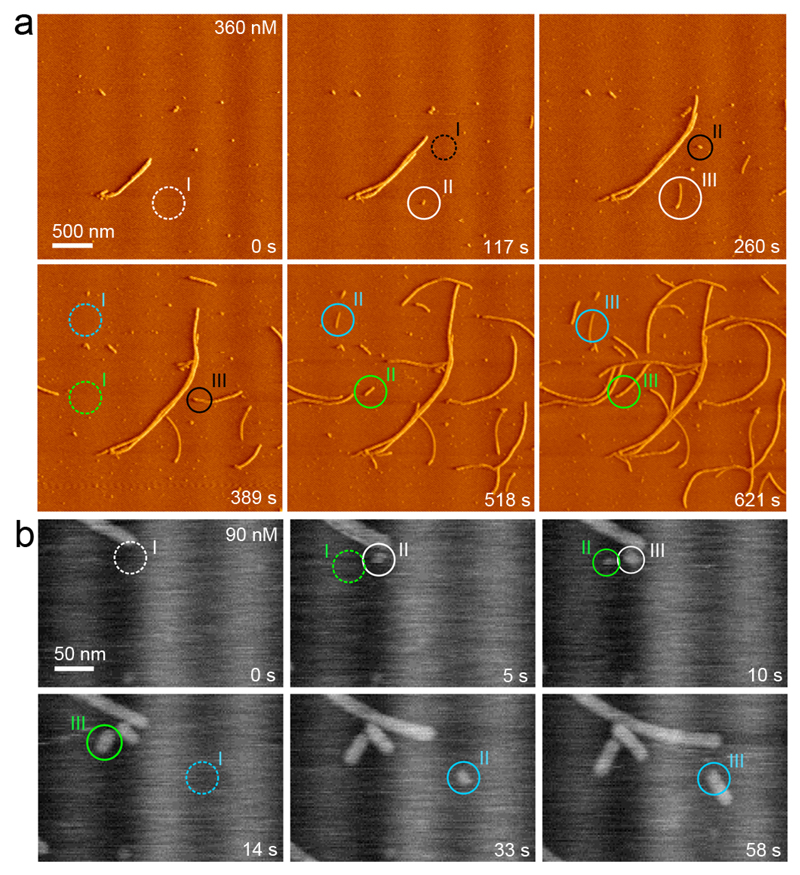
Next, we used intermediate (20 Hz) and high-speed (60 Hz) AFM imaging to map the nucleation pathway of single fibers at high temporal and spatial resolution. Still images of collected movies are shown in Figures 4a and b (See Supplementary Videos 1 and 3). New fiber fragments formed seemingly unprompted. There are three points worth emphasizing. First, the newly formed particles had a width and contrast that is indistinguishable from that of matured fibers also present in the field of view. Secondly, we observed non-fibrous, potentially oligomeric species on the mica surface. These particles, however, did not seem to be the precursors of new fiber fragments. Indeed, most nucleation events occurred at empty surface sites, and elongated into fibers within the resampling time of 2.1 s (Figure 4a, b). And thirdly, after careful inspection of numerous nucleation events we could not identify a minimal stable fragment size. The latter statement is based on the fact that there were no observations of newly formed fiber fragments that decreased in size and ultimately dissolved. Phrased in the language of nucleation theory, this means that the critical size of the system was either close to the dimensions of the monomers, or below the lateral resolution of our imaging setup (i.e. 10 nm laterally). Vanishing nucleus dimensions are a well-known phenomenon and typically occur in the regime of high supersaturation. We could estimate the supersaturation for our sample conditions by determining the CsgA equilibrium concentration Ce, i.e. the number density of monomers that remained in solution in contact with fully matured fibers at long timescales. When looking for excess monomeric CsgA in solution at presumed equilibrium (i.e. 2 weeks after desalting), Ce proved to be below the detection limit of our chosen concentration determination method (ELISA; see Supplementary Information). As a result, we could only estimate an upper limit of Ce, i.e. 50 nM. This was in correspondence with AFM, where we still measured low elongation rates (0.1 nm/s) at 45 nM (Figure 3d). To probe for any measurable dissociation, we transferred CsgA fibers from an equilibrated solution to 15 mM MES pH 6.0 and monitored the length of individual fibers as a function of time in AFM. We found no measurable decrease in the mean fiber length for a system that was assumed to be undersaturated. These data indicated that CsgA fibrillation occurred with a subnanomolar equilibrium concentration, and/or that CsgA dissociation from curli fibers was associated with a large activation barrier, i.e. incorporated CsgA protomers are kinetically trapped into the amyloid scaffold. The small nucleus size seen by AFM, the short nucleation induction times, and the lack of measurable protomer dissociation suggest that the minimal stable oligomer with curli-like amyloid properties, i.e. the nucleus, could be composed of only a very limited number of CsgA monomers, so that it may be isolated using conventional size-exclusion techniques. However, we only detected monomers and fibers in polymerizing CsgA solutions using silver staining SDS-PAGE or Western blot (Figure 5a). We hypothesize that the low number density of nuclei and their short lifetime (AFM demonstrates that the smallest fiber fragments grow out to 10s–100s of nanometers over the course of seconds–minutes) rendered them undetectable.
Figure 4. Curli originate from one-step, direct nucleation events.
(a) AFM monitoring of nucleation and growth of CsgA fibers in situ (see also Supporting Video 1; vertical deflection channel); (b) Following single fiber nucleation using high speed AFM (error signal): new fiber fragments form seemingly unprompted and have lateral dimensions that are identical to mature, single fibers (see also Supporting Video 2). In both panels, separate fibers are indicated in a different color, with pre-nucleation events encircled with a dashed line (I) and the consecutive events indicated by II and III.
Figure 5. CsgC binds curli and halts curli elongation.
(a) Normalized ThT fluorescence of an 8 µM CsgA solution supplemented with 1000:1, 500:1, 100:1, 1:1 CsgC respectively (from dark to light grey); (b) apparent hydrodynamic radii of CsgA as a function of time for a 14 µM CsgA solution (circles), supplemented with 100:1 CsgC at t=0 h (t0; triangles) and at tx=3 h (diamonds) determined using dynamic light scattering (as a guide, dashed lines give apparent rates of fiber growth with CsgC added at 0 or 3 hours ; (c) mean fiber length determined using TEM after 3.5 min incubation in an 8 µM CsgA solution alone (CsgA), or supplemented with 100:1 CsgC (+CsgC) (n=150); (d, e) elongation of single fibers as a function of time in a 90 nM CsgA solution alone (d) or supplemented with 10:1 CsgC (e); (f) Immuno-fluorescence microscopy demonstrating binding of CsgC to matured CsgA fibrils: brightfield view, CsgsA-His in the fibers (α-His) and bound CsgC-strep (α-Strep).
CsgC is an amyloid fiber capping agent
Once we characterized the formation of CsgA fibers in depth, we could start to gauge the effects of the natural inhibitor CsgC at the levels of both nucleation and elongation. When bulk amyloidogenesis was followed by ThT fluorescence, addition of CsgC was seen to slow down the buildup of fibrillar mass (Figure 5a), as seen previously25,26. CsgC’s inhibitory effect occurred at substoichiometric concentrations and appeared to approach saturation near a 100:1 CsgA:CsgC ratio. We did not identify a CsgC concentration threshold where fiber formation was inhibited indefinitely. Even in excess of CsgC, fibers with native morphology were formed, albeit after a prolonged incubation time of days (Supplementary Figure 5). It thus emerged that CsgC is not an absolute inhibitor of amyloidogenesis, but that it influences the kinetics with which a CsgA solution ends up in curli fibers, its thermodynamic sink. When the CsgA fibrillation time course was followed by DLS, the mean particle elongation rate (r) was drastically reduced in the presence of CsgC (rC) (Figure 5b). Strikingly, the reduction in elongation rate was equivalent whether CsgC was added at the start or at later stages of the polymerization reactions, i.e. This demonstrated that the observed reduction in elongation rate in presence of CsgC largely resulted from inhibiting fiber extension rather than from reduced or slowed fiber nucleation. Also when observed by TEM, the mean length of fibers nucleated and grown with CsgC present was markedly shorter compared to those formed in absence of CsgC (Figure 5c, Supplementary Figure 5). When measuring single fiber elongation rates in the presence of sub-stoichiometric levels of CsgC using AFM, we detected a significant decrease in the mean rate of elongation when CsgC was present at a molar ratio of 10:1 CsgA to CsgC (Figure 5d, e). Inspecting the fiber end-point trajectories as a function of time showed that individual fibers displayed long periods of stagnation, but occasionally did recover and continued to elongate further with burst velocities similar to the control sample (Figure 5d, e). The AFM and DLS results thus showed that CsgC acted as a decelerator of fiber elongation. The question remained whether CsgC achieved this retarding effect by lowering the effective concentration of growth-competent CsgA in solution, or by blocking fiber termini. Immuno-fluorescence imaging of matured CsgA fiber networks using fluorescently labeled CsgC demonstrated that CsgC could indeed bind to CsgA fibers (Figure 5f). Using dot blots, we confirmed that CsgC bound to CsgA fibers, but found no detectable binding to monomeric CsgA (Supplementary Figure 6a); as a proxy for CsgA monomers, we used buffer-exchanged CsgAslowgo, a mutant with reduced nucleation rates (Supplementary Figure 7)24. We therefore concluded that CsgC bound folded, but not unfolded CsgA. Unlike CsgA-only samples, Western blot analysis of a 5:1 CsgA:CsgC solution revealed the presence of a diminishing ladder of low molecular mass bands that corresponded to SDS-stable oligomeric species of CsgA with the approximate size of a dimer, trimer, and tetramer (Supplementary Figure 6b,c). Our combined observations suggested that the dampened curli elongation rate in presence of CsgC increased the number density of minimal curli fiber species (i.e. curli nuclei and nascent fibers), whereas under control conditions elongation more quickly removed minimal fiber fragments than could be replenished by nucleation, rendering these species experimentally intractable (see above).
Discussion
The resilience of bacterial biofilms is derived in part from the protective encapsulating features of the extracellular matrix in which the individual cells are embedded35. In this work we have focused on one of the principal components of many Gram-negative biofilms, i.e. functional amyloid fibers that are composed of the major curli subunit CsgA. Although curli biogenesis has been studied in depth using an array of biophysical methods, the molecular processes that take place at the various stages of the precipitation pathway remain unclear. To that end, we employed nanoscopic in situ imaging to follow CsgA polymerization at the single fiber level, and compare molecular events during functional amyloid formation with those previously observed during pathological amyloidogenesis. When observed in bulk solution, CsgA fibrillation shows an apparent lag phase that has been suggested to correspond to the formation of nuclei21. However, using TEM imaging, we observed nascent curli-like fibers already within minutes of removing denaturing conditions. Also, semi-log plots of ThT fluorescence and DLS time courses showed linear dependencies until the signal saturates at later times. These single exponential dependencies, combined with the CD data are compatible with the following picture: when CsgA is buffer-exchanged from denaturing to native conditions, it rapidly starts to associate into amyloid(-like) structures that grow as a function of time. The observed rapid nucleation is in line with theoretical considerations and mathematical fittings of experimental aggregation curves in Aβ amyloidogenesis, which show that primary nucleation occurs within milliseconds and likely retains a constant rate throughout the lag phase, where free monomer concentrations can be approximated as constant37,38.
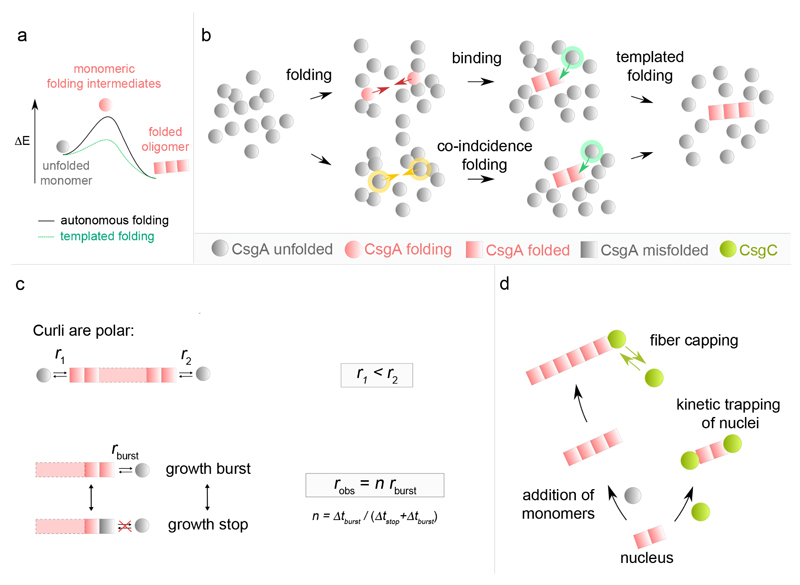
Looking at single fiber level, our AFM data showed that in vitro curli fiber nucleation is fast and direct, i.e. the system does not transition through an intermediate, non-amyloid, oligomeric state before ending up in the amyloid phase. Thus, the formation of productive CsgA oligomers appears inherently coupled to the adoption of the amyloid fold (see Figure 6a,b), where the minimal curli fragment that nucleates and which can induce templated CsgA folding may be as small as a folded CsgA dimer. Notably, the short SDS-resistant CsgA oligomers (dimer, trimer, tetramer, …) observed in conditions with excess elongation inhibitor C may represent such minimal species in the curli amyloid conformation, although we cannot exclude the possibility that the observed oligomers are off-pathway. On theoretical grounds, we foresee one of two possible pathways to a minimal curli fragment: (1) a “folding-binding pathway” in which transiently folded CsgA monomers can associate upon collision into a stable amyloid fragment that is rapidly extended by templated folding of new incoming subunits; or (2) a “co-incidence folding pathway” where contemporate folding of CsgA monomers leads to the formation of the minimal amyloid species that templates fiber extension (Figure 6b).
Figure 6. Integrated model for curli amyloidogenesis.
(a) Theoretical energy profile of the CsgA phase transition during curli assembly. During fibrillation or nucleation, CsgA monomers transition from an unfolded conformation into a lower energy, folded fiber-enclosed amyloid conformation. Monomer folding occurs via folding intermediates that are short-lived high-energy species under autonomous folding conditions. In presence of curli oligomers, templated monomer folding is cooperative and occurs via a lower energy transition state. Accordingly, fiber nucleation, i.e. the formation of a minimal stable curli oligomer is slow compared to fiber elongation. (b) Curli nucleation model. The formation of a minimal curli fragment is postulated to follow one of two pathways: (i) a “Folding-Binding pathway” in which transiently folded CsgA monomers collide (red arrows) and associate into a stable amyloid fragment that is rapidly extended by templated folding of new incoming subunits; or (ii) a “Co-incidence folding pathway” where contemporary folding of CsgA monomers (yellow arrows) leads to the formation of the minimal amyloid species that templates fiber extension. (c) Curli elongation is polar with fibrils displaying slow and fast growth rates (r1 and r2) at the two poles. Curli fibrils exhibit stop-and-go dynamics characterized by alternated periods of stagnation (Δtstop) and rapid elongation bursts (Δtburst). At high CsgA concentration, Δtstop becomes vanishingly small and robs approaches maximum or burst growth rate (rburst). (d) The curli inhibitor CsgC predominantly acts at the level of elongation, presumably by reversibly capping fibril growth poles, but can also kinetically trap nuclei by capping the growth surfaces.
If we consider the biological context in which curli are formed, one-step direct nucleation can be considered a logical route. In the event that unfolded CsgA monomers are secreted in the extracellular milieu where convective currents may exist, long nucleation induction times seem wasteful. Although more examples are needed, the absence of a lengthy induction time could be a defining trait of functional amyloids. The reduction of the kinetic and energetic barriers for amyloid formation increase the efficiency gain of the aggregation process which fits the rationale from an evolutionary perspective. This stands in sharp contrast to the amyloid transformation of natively folded proteins where the un- and refolding steps into cross β structures is the molecular origin of the induction time. For these cases, amyloid structures are an unwanted aberration and the large activation barriers associated with their formation are no evolutionary accident. The fast and direct nucleation of CsgA demonstrates that the apparent lag phase which is seen in bulk techniques should not be associated with a single molecular process. This point has been emphasized for pathological amyloids38, but is reiterated here for the case of functional amyloids due to its importance.
Our AFM imaging showed that curli display strong growth polarity, as well as stop-and-go dynamics at non-saturating CsgA concentration (Figure 6c). Polar growth and stop-and-go elongation seems to be emerging as a conserved feature of amyloid kinetics given the high number and diversity of systems for which it has been observed (see Supplementary Table 1): e.g. amylin39, Aβ25–3540, Aβ1–4041 and Aβ1–4212 aggregates, glucagon42 and α-synuclein43,44. We find but a single report of steady-state kinetics of individual amyloid fibers (β2-m and medC in ref45), but the low temporal resolution (˜5 min) of that study may have concealed underlying fluctuations. CsgA fibrillation is believed to correspond to the folding and lateral stacking of five ˜20-residue long pseudo-repeat regions (R1 to R5) as β-hairpins21–24,36. The relative β-aggregation propensity and kinetics have been shown to differ for the five repeats, with a decreasing order R5>R1>R3>>R2≈R4 23. How and if these localized differences in aggregation propensity relate to growth polarity is unknown. Stop-and-go dynamics in amyloid fibrillation has been reconciled with a quasi iso-energetic two-state model where fibril ends are either in a growing or blocked state, with the transition between the two related to a conformational change of the terminus epitope40,42. In curli, we observe that growth stagnation is frequently associated with a local increased image contrast, indicative of a structural polymorphism at the arrested growth tip. When resolving back into growth, the fiber elongates with a burst growth rate that is similar to that prior to the growth stagnation. This growth resolution can occur with complete healing of the temporary and localized structural defect (“scarless” stop resolution), or can leave a lasting structural polymorphism at the site of growth arrest (“scarred” stop resolution). These observations are in agreement with a model where the last incorporated subunit or subunits are in an altered conformation that serves as a poor template for the folding and inclusion of new subunits from the bulk solution. In CsgA, the structural defect possibly reflects a misfolding or lack of folding in part of the monomer pseudorepeats. Scarless resolution of the growth stops may occur by the dissociation of the aberrant subunit(s) or its (their) refolding to the native curli amyloid structure, thus restoring the templating surface and bringing the fiber back into the burst growth regime. Scarred stop resolution, on the other hand, may reflect instances where the non-native conformation is not resolved, but still allows a delayed folding and inclusion of a new subunit, which when adopting the native curli amyloid conformation, will restore the optimal growth template and burst growth regime.
Remarkably, the occurrence and length of growth pauses in curli was monomer concentration dependent, unlike what has been observed for Aβ25–35 epitaxy on mica40. This concentration-dependence suggests a mode of cooperation between CsgA monomers during incorporation that could be related to the fiber ultrastructure. Hints towards the nature of this cooperativity may be found in the minimal observed step size of growth (i.e. 12±4 nm), which is well beyond the dimensions expected for a single CsgA monomer (we measured a hydrodynamic radius of 3±1 nm for the unstructured species in solution using DLS). A possible explanation involves two hypotheses that require further study: (i) the unit cell of CsgA fibers does not correspond to a single CsgA monomer (e.g. Aβ1–42 46,47), and (ii) growth proceeds by the formation of unit cells, rather than by the incorporation of individual monomers.
The potency of CsgA to rapidly form amyloid structures at low concentration underlines the need for a biological failsafe mechanism in the periplasmic environment. Previous reports25,26 combined with our observations suggest that CsgC could indeed be a safeguard against amyloid formation in vivo. Based on in situ imaging and CsgA binding data, we propose that CsgC predominantly acts at the level of fiber growth, most likely by reversibly binding to fiber termini (Figure 6d). In doing so, it effectively acts as a fiber capping agent that blocks the further addition of new CsgA molecules to the fiber tip. Note that this model readily explains how CsgC can function at sub-stoichiometric concentrations without invoking any enzymatic or (un)foldase activity. For smaller CsgA:CsgC stoichiometries, the probability that a newly formed fiber fragment is capped by CsgC increases, leading to a net reduction of the mean particle size at a given instant. In the limit one expects to find that the earliest, minimal post-nucleation fibers become kinetically stabilized (Figure 6d), which was indeed confirmed here. The consequence of such a mechanism of inhibition is that the controlling parameter is the ratio of the number of active growth poles versus the number of CsgC molecules, rather than the absolute CsgA:CsgC ratio.
Online Methods
Protein purification
CsgA, CsgAslowgo (i.e. CsgA - Q49A/N54A/Q139A/N144A)24 and CsgC were cloned into pET22b via the NdeI site without their signal sequence but with a C-terminal tag, respectively a 6xHis-tag for CsgA and CsgAslowgo and a Strep-tag for CsgC. Expression was induced in BL21(DE3) ΔslyD cells for CsgA and CsgAslowgo and in BL21(DE3) cells for CsgC by addition of 1 mM IPTG after an OD600nm of 0.6 was reached. Cells were harvested by centrifugation at 5,000 g for 10 minutes after one (CsgA) or four hours (CsgC) of induction. For CsgA and CsgAslowgo purification, pellets were lysed overnight using lysis buffer (50 mM Kpi pH 7.2, 8 M GdnHCl) and the cell lysate was centrifuged at 40,000 g for 20 minutes at 20°C. After sonication and filtration the supernatant was loaded on a HisTrapTM FF column (GE Heathcare Life Sciences) equilibrated in 5 column volumes (CV) of lysis buffer. After washing in 10 CV buffer A (50 mM Kpi pH 7.2, 8 M Gnd HCl, 12.5 mM imidazole) the protein was eluted using buffer B (50 mM Kpi pH 7.2, 8 M Gnd HCl, 125 mM imidazole). Protein fractions were pooled (see Supplementary Figure 8a, b for purity analysis) and stored at –80°C. In order to prevent any unwanted amyloid formation in our CsgA stock solutions, all purification steps were performed under denaturing conditions (i.e. 8 M GdnHCl). This approach allows us to store CsgA in a monodisperse, monomeric form and gives control over the exact starting point of polymerization simply by buffer switching to native conditions, i.e. 15 mM MES pH 6.0. To remove guanidinium, ZebaTM Spin Desalting columns (7K MWCO) (Thermo Scientific) were used. We benchmark our CsgA system using a number of bulk biophysical methods that are well-established in the field, i.e. Thioflavin T (ThT) fluorescence, circular dichroism (CD) and dynamic light scattering (DLS) (Figure 1). To purify CsgC, harvested cells were resuspended and incubated for 20 minutes in lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1 mg/mL lysozyme, 50 μg/mL DNase, 1 mM MgCl2, 0.1 mg/mL AEBSF, 1 μg/mL leupeptin). Cell lysis was performed using a EmulsiFlex cell cracker (Glen Creston Ltd.) and the cell lysate was centrifuged at 40,000 g for 25 minutes at 4°C. The supernatant was loaded on a Strep-tactin column (IBA GmbH) and washed in 20 CV of wash buffer (50 mM Tris-HCl pH 8.0, 150 mM sodium chloride, 1 mM EDTA). The CsgC protein was eluted using elution buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 2.5 mM desthiobiotin). Fractions containing CsgC protein were concentrated using a spin concentrator with a 3 kDa cut-off to a final volume of 1 mL and loaded onto a Superdex 75 16/60 size exclusion column (GE Healthcare Life Sciences) equilibrated in 10 mM Tris-HCl pH 8.0, 150 mM NaCl. Fractions containing pure protein were pooled and stored at –20°C (see Supplementary Figure 8c, d for purity analysis).
Thioflavin T assays
ThT assays were performed using freshly desalted CsgA in 15 mM MES pH 6.0 in the presence or absence of CsgC (molar ratios between 1:1000 and 1:1). Protein samples were pipetted into a black flat-bottom 96-well microplate (Greiner Bio-One) in the presence of 50 µM Thioflavin T dye. Fluorescence measurements were performed using the Infinite 200 plate reader (Tecan) at 25°C with excitation at 430 nm and emission at 495 nm. Fluorescence readings were taken every 10 minutes and the plate was shaken for 5 seconds prior to each reading. Where indicated, the ThT fluorescence values were normalized over the maximum.
Circular dichroism spectroscopy
Far-UV CD spectra were recorder on a J-715 spectropolarimeter (Jasco). Continuous scans in the wavelength range 190 – 260 nm were taken using a 1 mm cuvette, a scan rate of 50 nm/min, a band width of 1.0 nm and a resolution of 0.5 nm. CsgA time course experiments were performed at room temperature in 15 mM MES pH 6.0 and a concentration of 12 µM. The temperature of the cuvette was kept at 20 °C using a thermoelectric Peltier device connected with a water bath.
Dynamic Light Scattering and hydrodynamic radii
Intensity correlation functions of freshly desalted CsgA solutions were collected at 20 °C in 10 mm cylindrical cuvettes at an angle of 90° employing an ALV-CGS-3 static and dynamic light scattering device (ALV GmbH) using a 22 mW He-Ne laser with a wavelength of 632.8 nm. Data were collected in a pseudo cross-correlation setup to minimize the contribution of dead time effects and PMT after-pulsing to the recorded signal. The digital correlator outputs the intensity autocorrelation function g2(τ)−1 with τ the delay time48. The function g2(τ) is connected to the electric field correlation function g1(τ) through the Siegert relation
where B is the baseline of the correlation function at infinite delay and β the function value at zero delay. For a monodisperse solution, g1(τ) is a single exponential decay g1(τ) = exp(−Γτ) with the decay rate Γ = Dq2 defined by the diffusion coefficient D of the particles and the magnitude of the scattering vector q = 4πn/λ sin (θ/2) at the scattering angle θ. The predicted hydrodynamic radius (Rh) for CsgA was calculated according to the sequence corrected Rh relation for intrinsically unfolded proteins: Rh = (APpro + B) (C|Q| + D) Shis*R0Nv, where Ppro is the fraction Pro residues, |Q| is the protein net charge and A = 1.24, B = 0.904, C = 0.00759, D = 0.963, Shis* = 0.901, R0 = 2.49, and v = 0.509 49. The minimal radius of folded CsgA is calculated using Rmin = 0.066 M1/3.
Negative stain transmission electron microscopy (TEM)
Desalted CsgA was incubated at room temperature in the presence or absence of CsgC. Samples for negative stain EM were prepared at different time points following guanidinium removal, by applying 2 µL of desalted protein solution on carbon-coated copper grids (Agar Scientific). After an incubation of 1 minute a wash step with deionized water was performed before staining with 1% (w/v) uranyl acetate. The earliest time point for TEM imaging was standardized to 3.5 min after the start of the desalting. The samples were imaged using a JEM-1400 electron microscope (JEOL Ltd.) equipped with a LaB6 cathode and operated at 120 kV. Images were recorded with a 4096 × 4096 pixel CMOS TemCam-F416 camera (TVIPS GmbH). Image J was used to measure fiber widths, either on in vitro grown fibers or on native curli on E. coli UGB1236 (n= 100) 50.
Dot blot
Two µL samples were spotted on a nitrocellulose membrane and air dried. Membrane blocking for non-specific binding was carried out with a 10% (w/v) skimmed milk solution in PBS for 10 min. CsgC was added (0.4 mM in 0.1% (w/v) milk in PBS) for 1h shaking at 37°C to allow binding. Bound CsgC was detected using a mouse anti-strep monoclonal antibody (AbD Serotec; MCA2489; 1:1000 dilution) as primary and an anti-mouse IgG alkaline phosphatase conjugated (Sigma-Aldrich; A3562; 1:5000 dilution) as secondary antibody.
(Immuno)fluorescence microscopy
Mature CsgA fibers were deposited onto poly-L-lysine treated microscope slides51 and slides were subsequently blocked for nonspecific binding by incubation with 5% (w/v) bovine serum albumin (BSA) for 10 min. The slides were then incubated at room temperature for 5 min with 3 μL 25 µM CsgC labeled with Alexa Fluor®488. Next, 10 μL of a mixture of an anti-6xHis antibody (AbD Serotec; MCA1396; 1:50 dilution) and an Alexa Fluor® 594-labeled goat anti-mouse antibody (Invitrogen; A11005; 1:20 dilution) in PBS was added for the staining of CsgA fibers. After 10 min incubation, slides were washed by flushing 3 times with 5 mL deionized water before examining them using a TE2000-U Nikon microscope.
ELISA CsgA equilibrium concentration
Mature CsgA fibers were centrifuged for 90 min at 20,000 g to pellet fibers. The supernatant was loaded on a 10 kDa or 30 kDa cutoff spin concentrator (Amicon®Ultra) and the flow through was probed for the presence of CsgA in ELISA. TEM samples were prepared to visually inspect for fibers. As a concentration standard, freshly desalted CsgA was used. 50 μL of two fold dilution series in MES were coated on 96-well Maxisorp microtiter plates (Nunc) for 30 min at 37°C. Wells were blocked for 45 min at 37°C with 10% skimmed milk powder in PBS prior to incubation with the primary anti-His mAb antibody (AbD Serotec; MCA1396; 1:500 dilution) for 1h at 37°C. Wells were subsequently washed and bound antibodies were detected by incubation with an anti-mouse IgG alkaline phosphatase conjugated secondary antibody (Sigma-Aldrich; A3562; 1:500 dilution) at 37°C for 1 h. Binding was revealed using p-dinitrophenylphosphatase (p-DNPP) as substrate. Absorbance values were measured at 405 nm.
Atomic Force Microscopy
High speed AFM imaging was performed in tapping mode using a Nanowizard III AFM (JPK Instruments AG) equipped with a high speed AFM head (version JPK-00178-H-12-0021). As a substrate for imaging we use 10mm muscovite disks (AFM mica disks V1 Agar Scientific) glued with two-component epoxy glue onto a glass support. Prior to sample loading, the mica was cleaved using sticky tape. Silicon nitride tips (DNP-S10) were used with a nominal tip radius of 10 nm and a spring constant of 0.06 N/m. Sample approach was performed in air to minimize the delay between CsgA injection and the onset of imaging. In order to minimize the force applied to the sample while scanning and to counter any drifts in the system, the set point voltage was continuously adjusted to the lowest level for which tip-sample contact was maintained. Supplementary movies were made by flattening the AFM images, exporting to png-format and loading into ImageJ as a stack. Drift correction was performed using the Template Matching and Slice Alignment ImageJ plugin tool52. Kymographs were then constructed in ImageJ by reslicing the image stack along a segmented line that follows the curvature of a single fibril (Supplementary Figure 3). Extracting the pixel values along the trajectory of a fiber, linearizing the extracted points into a column, and stacking the columns sideways for all the frames of a single movie provides an image where we move along space in the x direction and along time in the y direction.
Supplementary Material
Acknowledgments
MS and NVG acknowledge financial support by the FWO respectively under project G0H5316N and 1516215N. HR acknowledges financial support by VIB project grant PRJ9 and ERC grant 649082 BAS-SBBT. IVB is recipient of a PhD fellowship by the IWT. Work at the Université catholique de Louvain was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No [693630]), the National Fund for Scientific Research (FNRS), and the FNRS-WELBIO (Grant n°WELBIO-CR-2015A-05).
Footnotes
Author Contributions
IVB, WJ and NVG produced proteins samples, performed bulk fibrillation and CsgC activity experiments, and TEM. IVB, NVG and MS performed the biochemical assays. MS performed and analyzed AFM measurements, with help from CF, CV and YD. MS and HR supervised the study, analyzed data and wrote the paper, with contributions from all authors.
Competing Financial Interests
NVG and HR are inventors on PCT/EP2014/079319, describing the use of amyloid peptides for protein display.
Data Availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files) or are available from the corresponding author on reasonable request.
References
- 1.Chiti F, Dobson CM. Protein Misfolding, Functional Amyloid, and Human Disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Knowles TPJ, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 3.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid--from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295(1095–9203):851. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dueholm MS, et al. Functional amyloid in Pseudomonas. Mol Microbiol. 2010;77:1009–1020. doi: 10.1111/j.1365-2958.2010.07269.x. [DOI] [PubMed] [Google Scholar]
- 6.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 2012;8:e1002744. doi: 10.1371/journal.ppat.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oli MW, et al. Functional amyloid formation by Streptococcus mutans. Microbiology. 2012;158:2903–2916. doi: 10.1099/mic.0.060855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alteri CJ, et al. Mycobacterium tuberculosis produces pili during human infection. Proc Natl Acad Sci. 2007;104:5145–5150. doi: 10.1073/pnas.0602304104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toyama BH, Weissman JS. Amyloid structure: conformational diversity and consequences. Annu Rev Biochem. 2011;80:557–585. doi: 10.1146/annurev-biochem-090908-120656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichner T, Radford SE. A diversity of assembly mechanisms of a generic amyloid fold. Mol Cell. 2011;43:8–18. doi: 10.1016/j.molcel.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe-Nakayama T, et al. High-speed atomic force microscopy reveals structural dynamics of amyloid β1–42 aggregates. Proc Natl Acad Sci. 2016;113:5835–5840. doi: 10.1073/pnas.1524807113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 14.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 15.Tipping KW, van Oosten-Hawle P, Hewitt EW, Radford SE. Amyloid Fibres: Inert End-Stage Aggregates or Key Players in Disease? Trends Biochem Sci. 2015;40:719–727. doi: 10.1016/j.tibs.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652. doi: 10.1038/338652a0. 0028–0836. [DOI] [PubMed] [Google Scholar]
- 17.Collinson SK, Clouthier SC, Doran JL, Banser PA, Kay WW. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. JBacteriol. 1996;178(0021–9193):662. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dueholm MS, Albertsen M, Otzen D, Nielsen PH. Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PloS One. 2012;7:e51274. doi: 10.1371/journal.pone.0051274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Gerven N, Klein RD, Hultgren SJ, Remaut H. Bacterial Amyloid Formation: Structural Insights into Curli Biogensis. Trends Microbiol. 2015;23:693–706. doi: 10.1016/j.tim.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawthorne W, Rouse S, Sewell L, Matthews SJ. Structural insights into functional amyloid inhibition in Gram negative bacteria. Biochem Soc Trans. 2016;44:1643–1649. doi: 10.1042/BST20160245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu Q, et al. Solution NMR structure of CsgE: Structural insights into a chaperone and regulator protein important for functional amyloid formation. Proc Natl Acad Sci. 2016;113:7130–7135. doi: 10.1073/pnas.1607222113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Hammer ND, Chapman MR. The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem. 2008;283(0021–9258):21530. doi: 10.1074/jbc.M800466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Chapman MR. Sequence determinants of bacterial amyloid formation. J Mol Biol. 2008;380(1089–8638):570. doi: 10.1016/j.jmb.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans ML, et al. The Bacterial Curli System Possesses a Potent and Selective Inhibitor of Amyloid Formation. Mol Cell. 2015 doi: 10.1016/j.molcel.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor JD, et al. Electrostatically-guided inhibition of Curli amyloid nucleation by the CsgC-like family of chaperones. Sci Rep. 2016;6:24656. doi: 10.1038/srep24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ando T, Uchihashi T, Scheurin S. Filming biomolecular processes by high-speed atomic force microscopy. Chem Rev. 2014;114:3120–3188. doi: 10.1021/cr4003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao J, Dufrêne YF. Optical and force nanoscopy in microbiology. Nat Microbiol. 2016;1:16186. doi: 10.1038/nmicrobiol.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherny I, Gazit E. Amyloids: Not Only Pathological Agents but Also Ordered Nanomaterials. Angew Chem Int Ed. 2008;47:4062–4069. doi: 10.1002/anie.200703133. [DOI] [PubMed] [Google Scholar]
- 30.Knowles TPJ, Buehler MJ. Nanomechanics of functional and pathological amyloid materials. Nat Nanotechnol. 2011;6:469–479. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- 31.Van Gerven N, et al. Secretion and functional display of fusion proteins through the curli biogenesis pathway. Mol Microbiol. 2014 doi: 10.1111/mmi.12515. [DOI] [PubMed] [Google Scholar]
- 32.Chen AY, et al. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat Mater. 2014 doi: 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen PQ, Botyanszki Z, Tay PKR, Joshi NS. Programmable biofilm-based materials from engineered curli nanofibres. Nat Commun. 2014;5 doi: 10.1038/ncomms5945. [DOI] [PubMed] [Google Scholar]
- 34.Shammas SL, et al. A mechanistic model of tau amyloid aggregation based on direct observation of oligomers. Nat Commun. 2015;6:7025. doi: 10.1038/ncomms8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hung C, et al. Escherichia coli biofilms have an organized and complex extracellular matrix structure. mBio. 2013;4:e00645–00613. doi: 10.1128/mBio.00645-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian P, et al. Structure of a functional amyloid protein subunit computed using sequence variation. J Am Chem Soc. 2015;137:22–25. doi: 10.1021/ja5093634. [DOI] [PubMed] [Google Scholar]
- 37.Cohen SIA, et al. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc Natl Acad Sci. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arosio P, Knowles TPJ, Linse S. On the lag phase in amyloid fibril formation. Phys Chem Chem Phys. 2015;17:7606–7618. doi: 10.1039/c4cp05563b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldsbury C, Kistler J, Aebi U, Arvinte T, Cooper GJS. Watching amyloid fibrils grow by time-lapse atomic force microscopy1. J Mol Biol. 1999;285:33–39. doi: 10.1006/jmbi.1998.2299. [DOI] [PubMed] [Google Scholar]
- 40.Kellermayer MSZ, Karsai Á, Benke M, Soós K, Penke B. Stepwise dynamics of epitaxially growing single amyloid fibrils. Proc Natl Acad Sci. 2008;105:141–144. doi: 10.1073/pnas.0704305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ban T, et al. Direct Observation of Aβ Amyloid Fibril Growth and Inhibition. J Mol Biol. 2004;344:757–767. doi: 10.1016/j.jmb.2004.09.078. [DOI] [PubMed] [Google Scholar]
- 42.Ferkinghoff-Borg J, et al. Stop-and-go kinetics in amyloid fibrillation. Phys Rev E. 2010;82:010901. doi: 10.1103/PhysRevE.82.010901. [DOI] [PubMed] [Google Scholar]
- 43.Wördehoff MM, Bannach O, Shaykhalishahi H, Kulawik A, Schiefer S, Willbold D, Hoyer W, Birkmann EJ. Single fibril growth kinetics of α-synuclein. Mol Biol. 2015;427:1428. doi: 10.1016/j.jmb.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 44.Hoyer W, Cherny D, Subramaniam V, Jovin TM. Rapid Self-assembly of α-Synuclein Observed by In Situ Atomic Force Microscopy. J Mol Biol. 2004;340:127–139. doi: 10.1016/j.jmb.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 45.Ban T, Hamada D, Hasegawa K, Naiki H, Goto Y. Direct Observation of Amyloid Fibril Growth Monitored by Thioflavin T Fluorescence. J Biol Chem. 2003;278:16462–16465. doi: 10.1074/jbc.C300049200. [DOI] [PubMed] [Google Scholar]
- 46.Colvin MT, et al. Atomic Resolution Structure of Monomorphic Aβ42 Amyloid Fibrils. J Am Chem Soc. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wälti MA, et al. Atomic-resolution structure of a disease-relevant Aβ(1–42) amyloid fibril. Proc Natl Acad Sci. 2016;113:E4976–E4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berne BJ, Pecora R. Dynamic Light Scattering: With Applications to Chemistry, Biology, and Physics. Courier Dover Publications; 1976. [Google Scholar]
- 49.Marsh JA, Forman-Kay JD. Sequence determinants of compaction in intrinsically disordered proteins. Biophys J. 2010;98(10):2383. doi: 10.1016/j.bpj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D, Haagensen JAJ, Molin S, Prensier G, Arbeille B, Ghigo J-M. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol. 2004;51(3):659. doi: 10.1046/j.1365-2958.2003.03865.x. [DOI] [PubMed] [Google Scholar]
- 51.Pallesen L, Poulsen LK, Christiansen G, Klemm P. Chimeric FimH adhesin of type 1 fimbriae: a bacterial surface display system for heterologous sequences. Microbiology. 1995;141:2839. doi: 10.1099/13500872-141-11-2839. [DOI] [PubMed] [Google Scholar]
- 52.Tseng Q, Duchemin-Pelletier E, Deshiere A, Balland M, Guillou H, Filhol O, Théry M. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci. 2012;109(5):1506. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.