Summary
The immediate early gene Arc (also Arg3.1) produces rapid changes in synaptic properties that are linked to de novo translation. Here we develop a novel translation reporter that exploits the rapid maturation and “flash” kinetics of Gaussia luciferase (Gluc) to visualize Arc translation. Following glutamate stimulation, discrete Arc-Gluc bioluminescent flashes representing sites of de novo translation are detected within 15 s at distributed sites in dendrites, but not spines. Flashes are episodic, lasting ∼20 s, and may be unitary or repeated at ~minute intervals at the same sites. Analysis of flash amplitudes suggests they represent the quantal product of one or more polyribosomes, while inter-flash intervals appear random, suggesting they arise from a stochastic process. Surprisingly, glutamate-induced translation is dependent on Arc open reading frame. Combined observations support a model in which stalled ribosomes are reactivated to rapidly generate Arc protein.
Introduction
Activity-regulated cytoskeletal associated protein (Arc; Lyford et al., 1995; Arg3.1; Link et al., 1995) is an immediate early gene (IEG) that is required for learning and memory (Plath et al., 2006; Shepherd and Bear, 2011) and is involved in multiple forms of synaptic plasticity (Chowdhury et al., 2006; Jakkamsetti et al., 2013; Park et al., 2008; Plath et al., 2006; Shepherd et al., 2006; Tzingounis and Nicoll, 2006; Waung et al., 2008). In response to patterned neuronal activity, Arc mRNA is rapidly transcribed and traffics into dendrites, where it accumulates at regions of synaptic activity (Farris et al., 2014; Steward et al., 1998). Several lines of evidence indicate that Arc mRNA is translated following transport into dendrites. Arc mRNA associates with RNA granules and translation regulatory proteins, including fragile X mental retardation protein (FMRP) (Zalfa et al., 2003); activation of group 1 metabotropic glutamate receptor (mGluR) results in upregulation of Arc protein in dendrites within 5 min, which is blocked by translation inhibitors (Park et al., 2008; Waung et al., 2008); and de novo translation of Arc-Venus fusion mRNA is reported in dendrites (Tatavarty et al., 2012). Arc binds the TARP family of AMPA receptor chaperones (Zhang et al., 2015) and is required for the removal of synaptic AMPA receptors in the initial phase of mGluR-dependent long-term depression (mGluR-LTD) (Jakkamsetti et al., 2013; Park et al., 2008; Waung et al., 2008). Since mGluR-LTD is a synapse-specific form of plasticity (Waung and Huber, 2009) and dependent on de novo translation (Huber et al., 2000), effects of Arc protein must be rapid and highly localized. Moreover, studies of fragile X syndrome (FXS) indicate that Arc translation must be dynamically regulated by synaptic activity since loss of fragile X protein results in constitutive Arc expression that underlies synaptic deficits (Jakkamsetti et al., 2013; Park et al., 2008; Waung et al., 2008). Accordingly, Arc provides a physiologically relevant gene to study mechanisms of de novo protein translation in dendrites important for rapid synaptic plasticity.
Several strategies have been developed to visualize de novo protein synthesis (Aakalu et al., 2001; Butko et al., 2012; Dieterich et al., 2010; Gao et al., 2008; Ifrim et al., 2015; Ju et al., 2004; Kim et al., 2013; Leung et al., 2006; Lin et al., 2008; Lin and Tsien, 2010; Martin et al., 2005; Raab-Graham et al., 2006; Rodriguez et al., 2006; Schmidt et al., 2009; Smith et al., 2005; Tatavarty et al., 2012; Wang et al., 2009; Yu et al., 2006). A recent methodological advance (SunTag) engineers protein epitopes that bind and concentrate constitutively expressed GFP affinity ligands to visualize nascent polypeptides and overcomes limitations of time resolution consequent to requirement for GFP fluorescence maturation (Tanenbaum et al., 2014; Wang et al., 2016; Yan et al., 2016). As epitopes emerge from the ribosome, GFP binds to create a cumulative integrated signal of protein synthesis that can provide quantitative measures of translation initiation, elongation, and termination in cells. In combination with RNA epitopes and a second GFP, it is possible to simultaneously monitor mRNA trafficking and translation (Wang et al., 2016; Yan et al., 2016).
Here we develop an alternative approach that exploits Gaussia luciferase (Gluc) to create an instantaneous, binary signal of protein synthesis. Gluc is the smallest reported luciferase (19.9 kDa) and generates >1,000-fold stronger signal compared to Renilla luciferase or Firefly luciferase (Tannous et al., 2005). Gluc does not require cofactors such as ATP or Mg2+ and generates light immediately upon access to the substrate, coelenteraizine (CTZ) (Tannous et al., 2005). Importantly, Gluc light emission rapidly decays (flash kinetics) even in the continued presence of the substrate. We created a fusion construct of Arc and Gluc (Arc-Gluc) that is expressed in neurons and mimics the dendritic distribution of native Arc. We confirmed enzyme properties and established that following addition of CTZ, ∼90% of Arc-Gluc bioluminescence decays with an exponential time constant of <9 s. Studies indicate that individual molecules of Arc-Gluc generate a brief, consistent-intensity flash of light upon translation and are thereafter invisible. Thus, the de novo protein is resolved as immediate, brief flashes that can be unambiguously distinguished from the pre-existing protein. Studies using this reporter support a model in which glutamate-induced de novo translation of Arc-Gluc is mediated by reversal of stalled polyribosomes. Simultaneous quantitative analyses of multiple translation sites across entire dendritic arbors indicate that the protein is generated in discrete quantal bursts while the amount and timing of protein generation is indicative of stochastic processes.
Results
Generating an Arc Fusion Construct with Gluc
Gluc encodes a signal peptide and is secreted (Tannous et al., 2005). We reasoned that we might mask the signal sequence and prevent its secretion by inserting Gluc within the open reading frame (ORF) of Arc. We generated a fusion construct that includes the 5′ and 3′ UTR of murine Arc, Arc ORF, and Gluc ORF. Gluc was embedded 50 aa after the Arc translation start site (Figure 1A). After verifying the sequence, we confirmed the expression of Arc-Gluc protein in HEK293T cells by western blot (Figure 1B). To assess expression and possible toxicity in neurons, we co-transfected Arc-Gluc with EGFP into DIV (days in vitro) 13 cortical neurons and performed immunocytochemistry 24 hr later. Gluc and Arc antibody staining co-localized in permeabilized EGFP+ neurons (Figure 1C), indicating that the Arc-Gluc fusion protein was expressed and accumulated in dendrites and spines, similar to native and wild-type (WT) Arc trans-gene (Shepherd et al., 2006). No morphological changes were evident by EGFP filling of transfected neurons. Arc-Gluc was not detected on the surface of neurons (Figure 1D), indicating that Arc-Gluc fusion protein is retained within the cytoplasm. To assess possible secretion of Arc-Gluc, we measured luciferase activity in the medium normalized to lysate (secretion index) and compared with Gluc and Renilla luciferase in DIV 13 cortical neurons (Figure 1E). Gluc was robustly secreted into the medium, whereas Arc-Gluc and Renilla luciferase remained inside the cell.
Figure 1. Characterization of Arc-Gluc Fusion Protein.
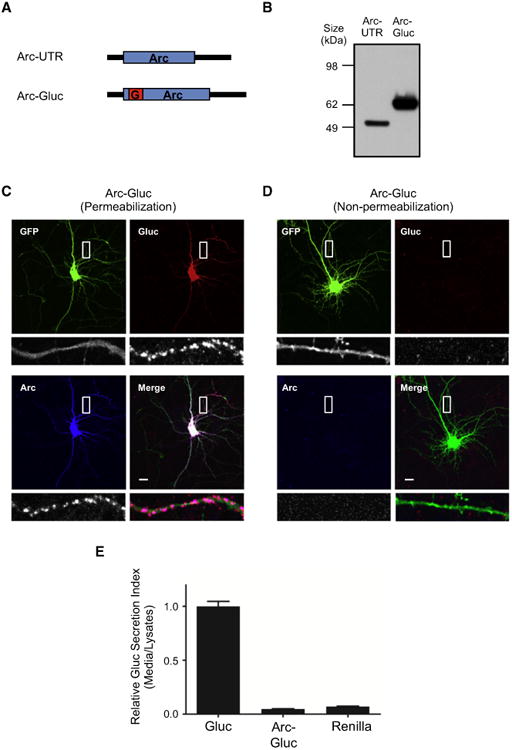
(A) Diagram of Arc-UTR and Arc-Gluc. Black bars,Arc UTR; blue box, Arc ORF; red box, Gaussia (Gluc)luciferase ORF.
(B) Arc-UTR and Arc-Gluc were transfected toHEK293T cells and lysates were blotted with Arcantibody.
(C and D) Immunocytochemistry of Arc-Gluc withEGFP. Bottom row shows the magnified picturesfrom the white square in the upper row. Scale bar,10 μm.
(C) Total staining after permeabilization.
(D) Surface staining without permeabilization.
(E) Relative secretion index of Gluc. Luciferaseactivity was measured from media and cell lysatesby spectrometry. Substrate, 40 μM coelenterazine(CTZ). Gluc, Gaussia luciferase; Renilla, Renillaluciferase (mean ± SEM, N = 4, 6, and 4, respectively).
Characterization of Arc-Gluc Expression and Bioluminescence
We examined luciferase activity of Gluc and Arc-Gluc expressed in HEK293T cells by spectrophotometry. Gluc generated bioluminescence upon addition of substrate to either cell lysate or culture medium. No bioluminescence was detected from HEK293T cells expressing Arc-UTR alone (Figure S1 A, available online). The relative bioluminescence of Gluc, Arc-Gluc, and Renilla luciferase was assayed in lysates of transfected cortical neurons. Luciferase activity of Arc-Gluc in cell lysates was ∼one-third of native Gluc, but 31 times greater than Renilla luciferase (Figure S1B). Gluc was previously reported to exhibit flash kinetics (Tannous et al., 2005). We confirmed that Gluc collected from transfected HEK293T cell medium creates a flash of bioluminescence when added to substrate (CTZ), followed by a smooth decay curve (Figure S1C). In order to avoid substrate depletion, we tested different concentrations of CTZ to find a saturating concentration (35.4 μM; data not shown) and used it for all experiments with neurons. The kinetic of luciferase activity was not changed by further addition of substrate (Figure S1D). Cumulative bioluminescence was linearly dependent on the amount of enzyme protein. We confirmed that Arc-Gluc bioluminescence was not altered by Ca2+ ion concentrations that span the physiological range (Figures S1E and S1F). We also determined that H2O2 or superoxide does not alter bioluminescence of Arc-Gluc (Figures S1G and S1H).
The flash kinetics of Gluc are potentially useful to distinguish newly synthesized protein from pre-existing protein. To better understand the parameters of flash bioluminescence, Arc-Gluc was expressed in HEK293T cells and luminescence was measured following addition of CTZ. Bioluminescence peaked within 1 s and the decay curve was fit with two exponential time constants of 8.75 s (∼90% total) and 490 s (Figure S1I). Our measured decay rate is faster than previously reported (Tannous et al., 2005). We simulated de novo translation of Arc-Gluc in a neuron containing pre-existing Arc-Gluc protein that had previously reacted with substrate and was no longer bioluminescent by adding fresh enzyme to the substrate containing reacted enzyme. Measured bioluminescence was not different than when fresh enzyme was added to the substrate without reacted enzyme (Figures S1J and S1K). This indicates that bioluminescence signals are not altered by reacted, pre-existing Gluc enzyme or its products. Moreover, dilution of a reacted enzyme mixture did not change bioluminescence per enzyme molecule, indicating that inhibition is not mediated in a concentration-dependent way by a diffusible product (Figures S1L and S1M). These data indicate that individual Gluc molecules act independently and support an interpretation that the amplitudes of bioluminescent signals can represent sums of independent Gluc molecules.
Monitoring the Local Translation of Arc-Gluc in Neurons
Arc-Gluc was co-transfected with EGFP into DIV 13-15 rat cortical neurons. After 30-36 hr, transfected neurons were identified by EGFP fluorescence (Figure 2A). We then switched the filterto an open channel to collect all emitted light from the chamber using a cooled electron multiplier CCD camera (see Supplemental Experimental Procedures for details of microscopy). We monitored the background signal for 1 min before applying the substrate and confirmed EGFP itself did not emit any detectable signal without excitation. Upon addition of the substrate, bioluminescent signal appearedinboth cell bodies and dendrites (Movie S1). The initial signal from pre-existing Arc-Gluc expression in dendrites declined below the detection limit within 3 min, while the time of decay of Arc-Gluc to baseline in soma was greater than 3 min. We continued to monitor the luciferase signal in dendrites for an additional 10 min, during which it remained below detection. We focused on Arc-Gluc in dendrites and empirically determined that 10 μM glutamate was sufficient to induce Arc-Gluc bioluminescence (see Experimental Procedures). We noted that glutamate-induced bioluminescence required healthy neurons, as assessed by growth and dendritic complexity (see Supplemental Experimental Procedures). EGFP fluorescence was recorded to image fine dendritic processes (soma is overexposed) as a template to localize Arc-Gluc bioluminescence (Figure 2A). We routinely monitored luciferase signal for 14 min after glutamate addition since this encompassed the major response. Glutamate in these conditions is reported to be nontoxic (Choi et al., 1987; Dugan et al., 1995). Bioluminescent punctate flashes lasting ∼20 s appeared in dendrites as early as 15 s after glutamate addition (Movie S1). To assess the location of puncta, we overlaid EGFP fluorescence images (Figure 2A). Puncta localized throughout dendritic processes were identified based on morphology. We manually selected all the puncta after addition of CTZ and monitored the bioluminescence intensity of each punctum. A plot of bioluminescence intensity versus time highlights the synchronous and intense signal upon initial addition of CTZ, as well as the time-dependent reduction to baseline that precedes the non-synchronous response to glutamate (Figure 2B). To assess the dependence of bioluminescence on de novo translation of Arc-Gluc, we treated cultures with the translation inhibitor, emetine, beginning 5 min before the recording and continuing to the end. Emetine did not prevent the initial bioluminescence response to CTZ (Figure 2C), consistent with this signal deriving from previously translated Arc-Gluc (Movie S2). By contrast, emetine completely blocked the gluta-mate-induced bioluminescent puncta (Figure 2C; Movie S2). These data support the conclusion that glutamate-induced biolu-minescent puncta are derived from the local de novo translation of Arc-Gluc.
Figure 2. De Novo Arc Translation Is Detected by Gluc.
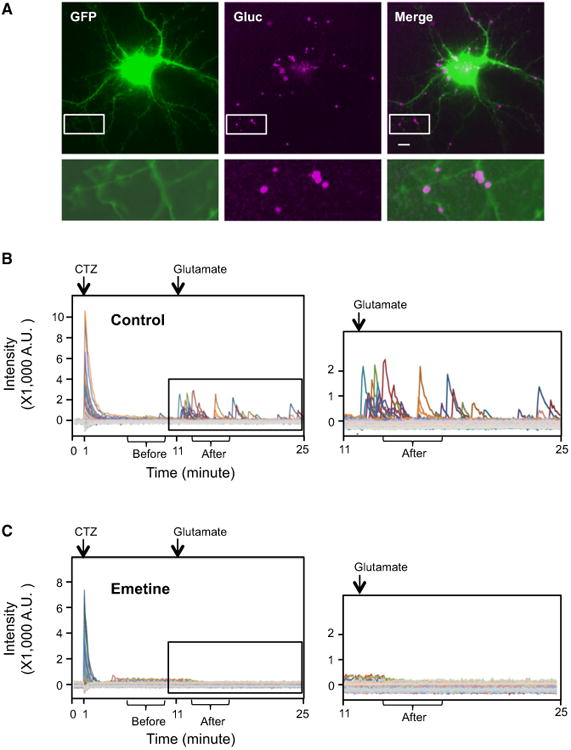
Substrate (CTZ) was added at 1 min and glutamate was added at 11 min.
(A) Gluc signals after glutamate addition were stacked and overlaid to EGFP signal. Bottom row shows the magnified pictures from the white square in the upper row. The intensity of EGFP signal in the magnified pictures is increased to show the dendritic morphology. Scale bar, 10 μm. (B and C) Discrete spikes were manually selected and individual signal intensities were plotted in different colors. The right is the magnified graph from the box in the left graph. (B) Control. (C) Emetine.
We adapted an automated image segmentation and analysis algorithm previously used to monitor calcium dynamics (Mukamel et al., 2009) to analyze puncta of luciferase bioluminescent signal in dendrites. Parameters for the automated algorithm were established that aligned well with manual inspection (comparing Figures 2A and 2B with Figures S2A and S2C). These parameters were fixed before the following experiments, and the cell body was manually masked to focus on events in dendritic processes. The image analysis algorithm was instructed to detect signals greater than six times the SD of background determined after the decay of bioluminescence signal from pre-existing Arc-Gluc. We did not include the initial 3 min signal from the pre-existing protein since it increased the calculated background used to determine threshold and is unrelated to de novo protein synthesis (Figures 2B and 2C). Once the algorithm acquired a punctum, we could track its history of intensity changes during the recording, including the first 3 min (Figure S2C), and measure the number of puncta and intensity of bioluminescence. Sites of glutamate-induced bioluminescence overlapped with a small subset of foci generated upon first addition of CTZ (co-localization rate=0.160±3.538, mean±SEM,5cells), suggesting that a major portion of pre-existing Arc-Gluc had trafficked away from sites of initial translation (Figure S2B). A comparison of the number of bioluminescent puncta acquired during an interval of 4 min before versus 4 min after glutamate stimulation indicated that glutamate increased the number of puncta (Figures 3A, S2C, and S2D). We confirmed using automated image analysis that emetine and anisomycin blocked glutamate-induced Arc-Gluc bioluminescence (Figures 3B and 3C).
Figure 3. The Effect of Pharmacological Agents and Arc cis Elements on Glutamate-Induced De Novo Translation of Arc.
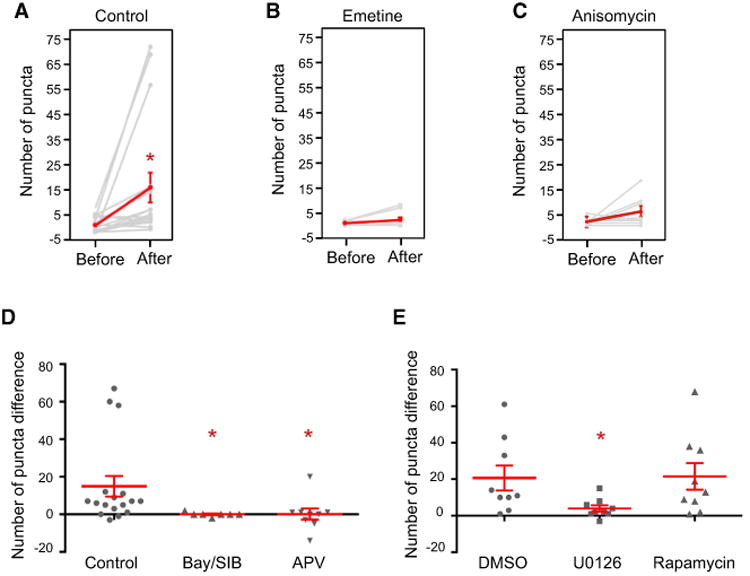
The number of dendritic Gluc puncta before (58–97 frames) and after (128–167 frames) glutamate. Indicated drugs or vehicles were added to cultures 5–30 min before addition of CTZ (16–41 min before addition of glutamate) and continuing to the end. (A–C) Each gray line represents an individual cell and red line represents the average of all cells (mean ± SEM).
(A) Control. p = 0.015 (N = 17).
(B) Emetine. p = 0.176 (N = 9).
(C) Anisomycin. p = 0.131 (N = 8).
(D and E) Population responses to drugs administered in (D) water vehicle (p = 0.006; N = 17, 7, and 9, respectively, Kruskal-Wallis test) and (E) DMSO vehicle (p = 0.034; N = 9, 9, and 9, respectively, Kruskal-Wallis test).
Signaling Pathways for Arc-Gluc Dendritic Translation
Group 1 mGluR and NMDA receptors (NMDARs) are implicated in de novo translation in neurons (Huber et al., 2000; Park et al., 2008; Scheetz et al., 2000).Toassess receptor dependence of glutamate-induced Arc-Gluc translation, we treated cells with group I mGluR antagonists (Bay36-7620, 25 μM; SIB1893, 50 μM) beginning 10 min before recording. Group I mGluR antagonists blocked glutamate-induced bioluminescent puncta in neurons expressing Arc-Gluc (Figure 3D). Group 1 mGluR antagonists did not block the response upon initial addi-tion of CTZ, providing a control that antagonists do not alter biolu-minescence from pre-existing Arc-Gluc protein. We also tested antagonists of NMDA (APV, 100 μM; Figures 3D and S2E) and AMPA (NBQX, 100 μM; data not shown) receptors and found they also blocked glutamate-induced puncta, but not biolumi-nescence from pre-existing Arc-Gluc protein. Co-dependence on glutamate receptors is consistent with a cooperative model in which NMDAR activation increases intracellular Ca2+ and thereby enhances group 1 mGluR responses (Kim et al., 2008).
Extracellular signal-regulated kinase (Erk) and mammalian target of rapamycin complex 1 (mTORC1) have been implicated in mGluR-dependent protein translation and synaptic plasticity (Bloomer et al., 2008; Panja et al., 2009). The relevant target of Erk for de novo protein synthesis is not known, while the mTORC1 pathway creates an active 4E complex important for translation initiation (Thoreen et al., 2012). To test the signaling pathway for Arc-Gluc translation, we treated cultures with U0126 (20 μM; Erk pathway inhibitor) or rapamycin (0.5 μM; mTORC1 pathway inhibitor) beginning 30 min before recording. U0126 blocked glutamate-induced Arc-Gluc translation, but rapamycin did not (Figure 3E). We confirmed that DMSO used to deliver drugs did not alter glutamate-induced Arc-Gluc biolu-minescence (Figure 3E). These findings indicate a selective role for Erk signaling in glutamate-induced translation of Arc-Gluc. Rapamycin is reported to block mGluR-LTD, and we infer this may involve proteins other than Arc.
Arc ORF Is Necessary and Sufficient for Glutamate-Induced Dendritic Translation of Arc-Gluc
We examined cis elements of Arc-Gluc mRNA that confer gluta-mate-induced protein expression by creating a set of constructs that deleted or swapped 5′ and 3′ UTRs and regions of the Arc ORF (Figure 4A). Because we sought to identify determinants of translation in dendrites independent of targeting of the mRNAs, we first assessed if mRNAs were present in dendrites 30–36 hr after transfection by performing in fluorescence in situ hybridization (FISH) with a probe for Gluc. All mRNAs appeared in somas and dendrites and showed asimilar uniform distribution (Figures 4B–4F and S3A). Notably, transgene mRNAs did not require UTR sequences to be present in dendrites. While the role of Arc UTR in dendritic trafficking has not been determined in native conditions, UTRs are implicated in the trafficking of other mRNAs (Gao et al., 2008; Kobayashi et al., 2005; Pinkstaff et al., 2001), and we infer that the uniform dendritic distribution of these transgenes may be consequent to overexpression. Nevertheless, their presence in dendrites afforded the opportunity to test if they are rapidly translated in response to glutamate.
Figure 4. Arc cis Elements on Glutamate-Induced De Novo Translation of Arc.
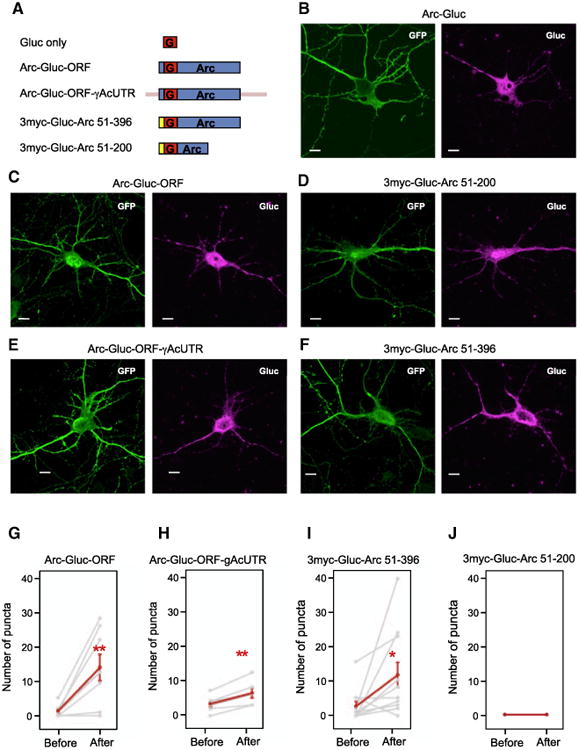
(A) Diagram of Gluc fusion constructs. Red box, Gluc ORF; blue box, Arc ORF; yellow box, repeated myc sequences; pink bar, γ-actin UTR.
(B–F) mRNA localization of Arc-Gluc fusion constructs. FISH (fluorescence in situ hybridization) detecting Gluc mRNA was performedafter Arc-Gluc fusion proteins and GFP were transiently overex-pressed in DIV13–15 cortical neurons. (B) Arc-Gluc. (C) Arc-Gluc-ORF. (D) 3myc-Gluc-Arc 51-200. (E) Arc-Gluc-ORF-γAcUTR.(F) 3myc-Gluc-Arc 51-396. Scale bar, 10 μm.
(G–J) The number of dendritic Gluc puncta before (58–97 frames) and after (128–167 frames) gluta-mate. Each gray line represents an individual cell and the red line represents the average of all cells (mean ± SEM).
(G) Arc-Gluc-ORF. p = 0.007 (N = 9).
(H) Arc-Gluc-ORF with γ-actin UTR. 5′ and 3′ UTRs of γ-actin were fused to Arc ORF with Gluc. p = 0.004 (N = 6).
(I) 3myc-Gluc-Arc 51-396. First 50 aa of Arc were switched to repeated myc sequences. p = 0.031 (N = 11).
(J) 3myc-Gluc-Arc 51-200. The fusion construct without Arc amino acids 201–396 did not show translational events (N = 8).
The construct in which the 5′ and 3′ UTRs of γ-actin were swapped for 5′ and 3′ UTRs of Arc (Arc-Gluc-ORF-γAcUTR; Figure 4A) showed glutamate-induced bioluminescent puncta in dendrites (Figure 4H). Moreover, glutamate-induced dendritic translation of Arc-Gluc-ORF was not different from that of Arc-Gluc (Figure 4G; Movie S3). This indicates that Arc ORF alone is sufficient to confer glutamate-induced translation. Translation of Gluc (without Arc ORF; Figure 4A) was not induced by gluta-mate (Figure S3B), although it is effectively translated in neurons since its luciferase activity in cell lysates was three times higher than Arc-Gluc (Figure S1B). A construct in which the first 50 amino acids of the Arc ORF preceding the Gluc ORF were replaced with repeated myc sequences (3myc-Gluc-Arc 51-396; Figure 4A) also showed glutamate-induced translation (Figure 4I; Movie S4), while a construct that deleted the C-terminal half of Arc ORF (3myc-Gluc-Arc 51-200; Figure 4A) did not (Figure 4J; Movie S5). Findings indicate that the sequence within the C-ter-minal half of Arc ORF confers glutamate-dependent translation in dendrites.
The absence of a requirement for 5′ and 3′ UTRs for gluta-mate-induced Arc translation and insensitivity to rapamycin (Figure 3E) suggested that translation initiation is not rate limiting. In further support, treatment of primary neuronal cultures with the translation initiation inhibitor pateamine A (Low et al., 2005) reduced S35 incorporation into protein, but did block glutamate receptor-induced increase of Arc protein (Figures S3C–S3E).
Distribution of Arc-Gluc Translation Sites within Dendrites
Sites of de novo Arc translation have been reported using GFP-based reporters (Tatavarty et al., 2012), but limitations of temporal resolution that may not distinguish sites of translation from trafficking prompted us to re-examine the issue. We used GFP-PSD95 to label spines and visualize dendrites and performed a wide-field analysis of the spatial frequency throughout entire neurons. We first measured the distance between bioluminescent Arc-Gluc puncta in dendrite segments after glutamate treatment. Five neurons with the most frequent events were used for analysis, resulting in an average of 8.9 ± 0.9 μm between puncta (mean ± SEM, n = 78, N = 29 dendrites, 5 cells; Figure 5B). Based on previously reported spine density in vivo (Moser et al., 1994; Seib and Wellman, 2003) and in dissociated cortical culture (Hayashi and Shirao, 1999), spines outnumber sites of de novo translation by a factor of 7–17. A similar number (14.9) was obtained using GFP-PSD95 to assess spine numbers in our cultures.
Figure 5. Localization of Glutamate-Induced De Novo Translation of Arc.
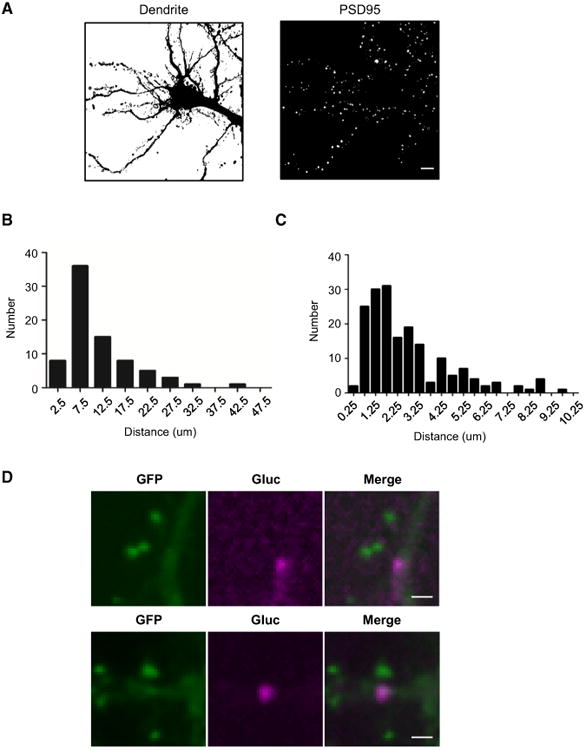
(A) An example of dendrite (left) and spine (GFP-PSD95+, right) segmentation.
(B) The distribution of distances between Arc-Gluctranslation sites in dendrites. The average distance between de novo translation sites is 8.9 ± 0.9 μm (mean ± SEM, n = 78, N = 29 dendrites, 5 cells).
(C) The distribution of distance between spines (GFP-PSD95+) and Arc-Gluc (n = 188, N = 6 cells).
(D) Two examples of localization of GFP-PSD95 and glutamate-induced Arc-Gluc. Scale bar, 2 μm.
We next asked if glutamate-induced Arc-Gluc foci could be detected in spines. GFP-PSD95 fluorescence images distinguished spines from dendrites, and the average spine diameter was 0.38 μm (see Supplemental Experimental Procedures), which is similar to the reported mean distance between the spine head and spine neck (Arellano et al., 2007). CCD camera images had a similar spatial resolution (0.254 μm/pixel) and were confirmed by manual inspection to be in register with fluorescence images. Therefore, it was possible to calculate the distance from the center of individual Arc-Gluc foci to the most proximal spine head marked by GFP-PSD95. In total, 80% of Arc-Gluc punctae were within 5 μm of a spine. However, only 3.4% ± 1.8% (mean ± SEM, n = 188, N = 6 cells) were closer than 0.4 μm (Figure 5C). Analysis of whether the center of Arc-Gluc foci overlapped with any pixel of spines determinedby GFP-PSD95 signal yielded a similar result, 3.4% ± 1.4% (mean ± SEM, n = 188, N = 6 cells). Examination of instances of spine-associated Arc-Gluc indicated that the majority of these located to the base of spines (Figure 5D) orin a dendritic shaft where the spine neck and GFP-PSD95 were superimposed along the Z axis of imaging (Figures S4A–S4C). There was no difference in the distribution of distances to spines comparing Arc-Gluc foci with a single event versus multiple events (“hotspots”; see below; Figure S4D). As an alternative method, we determined the statistical significance of co-localization of Arc-Gluc within spines by calculating the spine-to-dendrite area for each cell (Figure 5A) and found that the observed proportion of Arc-Gluc puncta that overlapped with spines was significantly lower than expected from a random distribution of Arc-Gluc (p = 0.005, N = 6 cells, paired t test, null distribution 7.9% ± 0.8%). These observations indicate that Arc-Gluc de novo translation is not detected at a significant level in spines.
FMRP plays an important role as a negative regulator of ribosomal translocation (Darnell et al., 2011; Muddashetty et al., 2007; Zalfa et al., 2007). Group 1 mGluR activation induces a rapid increase in FMRP binding to mRNAs (Narayanan et al., 2007), and fmr1–/– mice show deregulated Arc expression that contributes to disease phenotypes (Huber et al., 2000; Park et al., 2008). To assess the spatial distribution of FMRP relative to Arc-Gluc translation sites, we co-transfected Arc-Gluc and GFP-FMRP and overlaid the puncta of glutamate-induced Arc-Gluc with GFP-FMRP imaged before recording (Figure 6B). Again, we used FMRP segmentation to calculate co-localization of Arc-Gluc and FMRP (Figure 6A, right) and determined a co-localization rate of 16% ± 2% (mean ± SEM, n = 230, N = 6 cells). This is significantly higher than null distribution calculated using manually defined dendrites (Figure 6A, left) in each cell (Figure 6C), indicating that a portion of glutamate-induced Arc-Gluc foci co-localize with GFP-FMRP. However, it is notable that many of the glutamate-induced Arc-Gluc foci do not co-localize with GFP-FMRP. This might occur if GFP-FMRP effectively represses translation or if GFP-FMRP association with Arc-Gluc foci is dynamic since GFP-FMRP foci were recorded ∼15-20 min prior to addition of glutamate to induce Arc-Gluc puncta.
Figure 6. Co-localization of GFP-FMRP with Sites of Glutamate-Induced De Novo Translation of Arc.
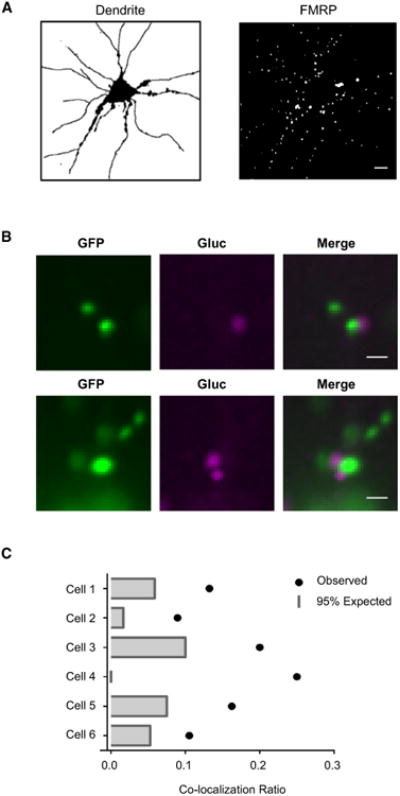
(A) An example of dendrite (left) and FMRP (right) segmentation.
(B) Two examples of localization of GFP-FMRP and glutamate-induced Arc-Gluc. Scale bar, 2 μm.
(C) Co-localization rate of Arc-Gluc and FMRP. Six independent experimentswere compared to null (bar) Arc-Gluc puncta.
Arc-Gluc Expression Reveals Quantal and Stochastic Properties of Glutamate-Induced Translation in Dendrites
Arc-Gluc bioluminescent signal is episodic, with variations in amplitude and timing between foci within the same neuron. Biochemical studies of Arc-Gluc bioluminescence (Figure S1) support the premise that differences in luminescence intensity are due primarily to differences in the amount of Arc-Gluc created. If ribo-some processivity attains a maximum rate of ∼20 bases/second (Wohlgemuth et al., 2011) and codon recognition sites of adjacent ribosomes are separated by ∼72 nucleotides (Brandt et al., 2009), then each Arc-Gluc mRNA polyribosome complex could generate 1 molecule of Arc-Gluc protein during a 6 s frame and 3-4 molecules during a flash lasting 3 frames (18 s). This contrasts with a plot of the amplitude of bioluminescent events (amplitude histogram) within an individual neuron, which reveals amplitudes spanning a range of ∼10-fold (Figures 7A and S5A-S5D). The broad range of intensities is difficult to reconcile with product generated by a single mRNA polyribosome and suggests that flashes may represent the products of multiple polyribosomes. Moreover, cumulative histograms from several neurons exhibited periodic peaks of intensity, which is consistent with the interpretation that flashes represent the translational generation of discrete numbers of Arc-Gluc molecules. Biochemical studies exclude cooperative effects between Arc-Gluc molecules that might give rise to variations in intensity (Figures S1J-S1M) and exclude the possibility that signals generated by glutamate receptors might directly induce or inhibit bioluminescence from pre-existing Arc-Gluc molecules since Gluc activity is not sensitive to Ca2+ in the physiological range (Figures S1E and S1F) and is not altered by H2O2 or superoxide (Figures S1G and S1H). Moreover, recordings from sites with repeated flashes (“hotspots,” below) often show large variations in amplitude that exclude the possibility that amplitude differences are due to local opacity of cultured neurons. Accordingly, we suggest that the microenvironment within individual sites serves to coordinate product generation from multiple polyribosome Arc-Gluc mRNAs.
Figure 7. Dynamics of Dendritic De Novo Arc-Gluc Translation Events.
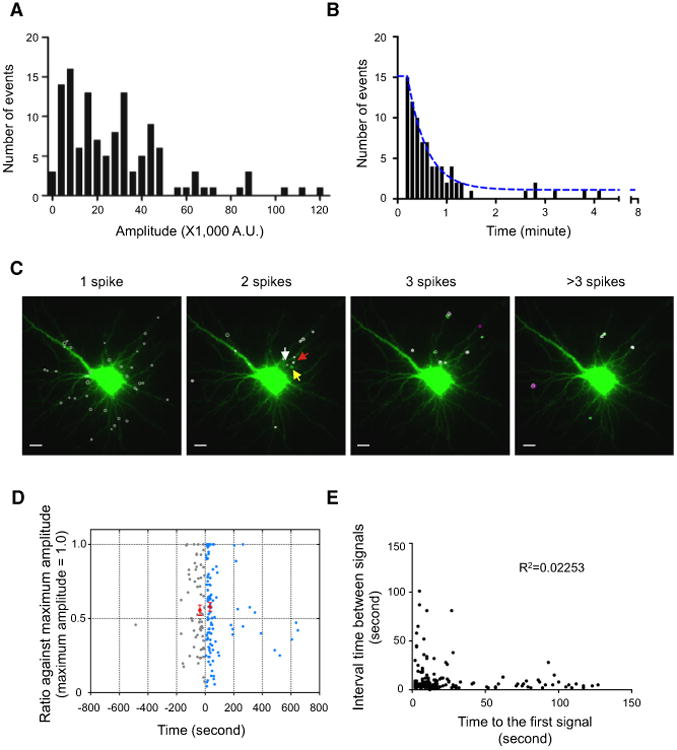
(A) The distribution of peak amplitude of spikes fromone example cell.
(B) Histogram of time interval between the spikesat hotspots. Blue line, nonlinear fit (K = 0.043, t½ =16.29 s, n = 82, N = 10 cells).
(C) An example showing the distribution of puncta with a single flash (one spike) and repeated flashes (translation hotspots; more than two spikes). Different colored circles represent the number of flashes at the same center of the signal (white, single; green, two; magenta, three flashes). Three arrows are marked for Movie S6. Scale bar, 10 μm.
(D) Relationship between consecutive peaks to that of maximum peak. Normalized intensity of consecutive peaks at translation hotspots with two flashes is plotted as a function of the time interval from maximum peak. The time frame is aligned such that the maximal peak time is set to zero. In 60% of cases, first peak is the maximal peak and the following peak is color coded in blue. When the first peak is followed by the maximum peak, it is plotted in gray on the left of the y axis. The two red circles represent the average of the smaller peak that occurred within 100 s of maximal peak (time interval after maximum peak = 33.2 ± 1.9 s, normalized amplitude = 0.58 ± 0.03, mean ± SEM, n=99, N= 12 cells; time interval before maximum peak = 36.8 ±3.4 s, normalized amplitude = 0.56 ± 0.04, mean ± SEM, n = 53, N = 12 cells). See also Figures S5F and S5G.
(E) Inter-flash interval relative to the time of the first flash after addition of glutamate (R2 = 0.2253, n = 158, N = 17 cells).
Two additional features of the timing of Arc-Gluc biolumines-cence are notable. The majority of the glutamate-induced Arc-Gluc bioluminescent signal occurred within 5 min of adding glutamate (Figures 2B, S2C, and S2D). However, the Arc-Gluc bioluminescent signal is not synchronous within a neuron, but rather spans a range from 1 min to 14 min (end of recording) (Figures S2C and S2D; Movie S1). We also noted that the Arc-Gluc bioluminescence signal was often repeated at the same site (Figures 7B and 7C; Movie S6). We defined a dendritic translation “hotspot” as an area where repeated peaks separated by more than two frames (>12 s) overlapped. About 25% of total signals were observed from hotspots (Table S1). Sites of repeated Arc-Gluc generation were dispersed throughout branches of dendrites (Figure 7C) and were not different in spatial distribution or bioluminescence intensity (Figure S5E) from sites with single Arc-Gluc generation. We reasoned that hotspots offer an opportunity to assess the dynamics and amplitude of “on” states of Arc-Gluc translation. We examined event pairs and plotted the time interval between a first and second event (Figure 7B). This analysis reveals that most second events occur within 1 min of the first and that their cumulative histogram is modeled by a single exponential function with t1/2 ∼30 s. We asked if the inter-flash interval or the amplitude of the second flash is related to the amplitude or timing of the first event. We plotted the time of the smaller amplitude event relative to the larger event set to time 0 (Figure 7D). Amplitudes were calculated from either the prior trough or estimated basal (Figure S5F), with similar results (Figure S5G). In 60%, the first peak was greater than following ones (Figure 7D). For most events, the amplitude did not correlate with the time interval between repeated signals (Figures 7D and S5G). This was evident whether the measured peak occurred before or after the maximum signal. Signals less than 20% of the maximum amplitude were clustered <50 s following the maximum signals, which could occur if stalled ribo-some mRNAs were depleted from the initial event. A second analysis examined the inter-flash interval relative to the time of the first flash after addition of glutamate. Again, there was no correlation (Figure 7E). These observations indicate that the amplitude and timing of individual flash events are not predicted by the prior state and appear essentially random. These properties are consistent with the notion that complex cell biological processes obey predictions of stochastic rather than deterministic systems when viewed at the level of individual molecules (Bressloff, 2014; Paulsson, 2004; Yu et al., 2006).
Discussion
We exploit the rapid kinetic properties of Gluc together with the high spatial resolution of EM-CCD imaging to reveal fundamental properties of de novo Arc synthesis in neurons. De novo translation sites are abundant within dendrites and are distributed at ∼10 μm intervals. Spines outnumber translation sites by ∼10-fold. De novo translation sites are occasionally observed at the base of spines labeled with GFP-PSD95, as anticipated from electron microscopy (EM) studies (Steward and Levy, 1982), but do not co-localize in spines at a frequency above a statistical threshold for random distribution. A model in which Arc translation is controlled at the level of dendritic segments rather than individual spines is consistent with Arc's observed synapse-specific action (Béïque et al., 2011) and a model that proposes Arc targets from sites of translation in dendrites to synapses based on binding to βCaMKII in its kinase-inactive state (inverse tagging) (Okuno et al., 2012). This model is also consistent with evidence that dendritic segments, rather than individual synapses, function as units of synaptic integration (Branco and Häusser, 2011; Palmer, 2014; Spruston, 2008). The co-dependence of Arc-Gluc de novo translation on group 1 mGluRs and NMDARs suggests that compartmentalized signaling from multiple synaptic inputs within the dendritic segment may coor-dinately control Arc translation.
Previous studies highlight the complexity of mechanisms that regulate Arc translation in neurons. Glutamate receptor-dependent upregulation of Arc protein that underlies mGluR-LTD in the hippocampus is dependent on eukaryotic elongation factor 2 kinase (eEF2K) (Park et al., 2008). Activated eEF2K phosphor-ylates its only known substrate elongation factor 2 (eEF2). eEF2 functions as part of the 80S complex to enhance ribosome trans-location (Spahn et al., 2004), and its phosphorylation inhibits translation of most mRNAs. However, Arc protein is upregulated in response to dynamic phosphorylation of eEF2. Arc is also up-regulated upon addition of low-dose cycloheximide, which acts to prevent ribosome elongation like phosphorylated eEF2 (Park et al., 2008). These observations suggest that rapid, glutamate receptor-induced Arc translation occurs in association with mechanisms that transiently inhibit ribosome translocation generally but that advantage certain mRNAs (including Arc). Arc translation is also regulated by FMRP, an RNA binding protein that can stall ribosomes of specific mRNAs (Darnell et al., 2011). Group 1 mGluRs activate S6K1, which phosphorylates FMRP and reduces its RNA binding and inhibition of translation (Levenga et al., 2010; Narayanan et al., 2007, 2008; Niere et al., 2012). Genetic deletion of fmr1 results in a loss of dynamic Arc expression that is replaced by steady-state expression and enhanced Arc-dependent, de novo protein synthesis-independent mGluR-LTD (Park et al., 2008).
Observations here support a model in which glutamate-induced translation of Arc-Gluc involves activation of stalled polyribosome Arc-Gluc mRNA complexes. This model rationalizes a contribution of Arc within the first 5 min after induction of mGluR-LTD (Park et al., 2008), as well as the priming effect of Arc expression on subsequent plasticity (Jakkamsetti et al., 2013). Flash bioluminescent puncta corresponding to de novo translated Arc-Gluc beginning within 15 s of glutamate stimulation is consistent with biochemical measures of ribosome processivity only if Arc-Gluc translation were initiated in a non-rate-limiting step prior to the stimulus such that upon glutamate stimulation, translation of ∼100 aa would be sufficient to generate Gluc. This model is supported by biochemical observations that mGluR induces translation from peptide-ribosome complexes and is insensitive to an inhibitor of translation initiation (Graber et al., 2013). Glutamate receptor-dependent upre-gulation of native Arc in cultured cortical neurons is insensitive to the initiation inhibitor pateamine A (Low et al., 2005) (Figures S3C-S3E). Our observation that glutamate-induced translation of Arc-Gluc mRNA does not require the 5′ and 3′ UTRs and is insensitive to mTORC1 inhibition provides further support for the notion that translation initiation is not rate limiting. A recent study provides precedent for regulation of ribosome stalling and re-initiation by coding sequence and a trans-acting factor (Gutierrez et al., 2013). We note that since Gluc is N-terminal to the presumed regulatory region of the Arc ORF, our model requires that Gluc not be active in the peptide-ribosome complex. Nascent peptides associate with chaperones and isomerases (Kramer et al., 2009) that may coordinate Gluc enzyme activity with release of mature protein from the ribosome complex. Alternatively, the presumed cis element in the Arc ORF might stall ribosomes at positions 5′ to Gluc. Interestingly, the region of the Arc ORF that is required for glutamate-induced translation extending from amino acid 200 to the C terminus evolved from a retrotransposon gene of the Ty3/Gypsy family and encodes a protein that is structurally related to retroviral capsid protein (Zhang et al., 2015). Retroviruses evade host translational control mechanisms, and it is possible that translational control mechanisms of Arc are a remnant of its retrotransposon origin. The dependence of Arc upregulation on FMRP and eEF2K (Park et al., 2008), together with their known role as regulators of ribosome elongation, suggests these signaling pathways may contribute to stalling or activation of stalled ribosomes that control Arc translation.
Wide-field imaging affords a comparison of many translation events in the same neuron. The cumulative histogram of time to generation of Arc-Gluc after glutamate stimulation establishes a half-life for the overall process, yet the timing of individual events appears random. Amplitudes are also widely variable even for repeated events at the same site. The observed episodic production of protein in neurons is similar to that reported in E. coli (Yu et al., 2006), but many features are notably distinct, including the stimulus dependence and the range of amplitudes. An explanation for the broad range of amplitudes at individual sites is that multiple polyribosome Arc-Gluc mRNA complexes can synchronize product generation. This is consistent with the notion that ribosomes stall at a specific sequence (Gutierrez et al., 2013), that events prior to activation of stalled ribosomes are not rate limiting, and that stochastic conditions within the microenvironment of individual sites of translation determine the timing and efficacy of ribosome re-activation. These stochastic properties may have implications for the timing and amount of Arc protein within dendritic segments and ultimately at individual synapses. Other aspects of synaptic transmission and plasticity display stochastic properties that have been rationalized in models of learning (Seung, 2003; Shouval and Kalantzis, 2005). For Arc, the potential for repetitive induction (Guzowski et al., 1999, 2006; Nakayama et al., 2015; Ramírez-Amaya et al., 2005), together with its molecular function to differentially act at inactive synapses (Kawata et al., 2014; Okuno et al., 2012), could resolve stochastic challenges to de novo protein synthesis-dependent memory consolidation.
Gluc is uniquely useful for its low background, high temporal resolution for new protein and compatibility with wide-field analysis. Our experiments overexpressed Arc-Gluc mRNA to study translation independently of mRNA trafficking and to achieve robust signals. Arc 3′ UTR confers non-sense-mediated decay (Giorgi et al., 2007) and will certainly contribute to natural regulation of Arc expression. Addition of Arc 3′ UTR to a translation reporter construct increases the distance of translation sites from the soma (Wang et al., 2016). Another limitation of the present approach is that it is not possible to determine the biolumines-cent signal of single Gluc molecules. By combing Gluc with SunTag methods, it should be possible to assess Gluc signal from defined numbers of mRNA molecules and thereby estimate absolute numbers of new protein molecules. Gluc, in combination with the protein epitope-GFP signal, could reveal trafficking of newly synthesized proteins and test targeting mechanisms of de novo Arc or other proteins.
Experimental Procedures
Cell Culture and Transfection
All animal procedures were approved by Johns Hopkins University Animal Care and Use Committee. Sprague-Dawley rat (Charles River, RRID: RGD_734476) cortical cultures from embryonic day 18 (E18) were prepared as reported previously (Rumbaugh et al., 2003). HEK293T cells were maintained in DMEM with 10% fetal bovine serum. Transfection was performed with Lipofectamine 2000 (Invitrogen) for neurons or FuGENE6 (Roche) for HEK293T cells according to manufacturer's instruction. For more information, see Supplemental Experimental Procedures.
Immunocytochemistry
Immunocytochemistry was performed as described (Shepherd et al., 2006) with some alterations. For surface labeling, antibodies were incubated without permeabilization. For more information, see Supplemental Experimental Procedures.
Time-Lapse Microscopy
Time-lapse images were captured with 5 s exposure and 1 s interval time with a cooled EM-CCD camera controlled by Slidebook software (Intelligent Imaging Innovations Inc.). Open channel was used to collect all emitted lights from the chamber for luciferase imaging. Acquisition of images was paused for CTZ and glutamate treatments and reinitiated 15 s after the treatment. For more information, see Supplemental Experimental Procedures.
Fish
The probes were generated by PCR using full ORF of pCMV-Gluc (NanoLight technology) and subcloned into pCR 2.1-TOPO vector (Invitrogen), and sequence was verified. Riboprobes were synthesized using in vitro transcription and digoxinin-labled UTP. Primary neuronsonthe coverslips were fixed by 4% paraformaldehyde followed by permeabilization and acetylation. After 2 hr of prehybridization, cells were incubated with probes for 18 hr. Cells were then washed and mounted for imaging.
Statistical Analysis
Statistical analysis was performedusing GraphPad Prism version 6, GraphPad Software. All data were analyzed by two-tailed, paired Student's t test unless otherwise stated. We used Kruskal-Wallis test for the multiple comparison of data (Figures 3A and 3B) since we cannot assume Gaussian distributions of translation events and each population did not show the same variation. For more information, see Supplemental Experimental Procedures.
Supplemental Information includes Supplemental Experimental Procedures, five figures, one table, and six movies and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2016.06.017.
Supplementary Material
Highlights.
De novo translation is imaged using Gaussia (Gluc) luciferase-Arc fusion
Glutamate-induced Gluc-Arc translation in dendrites is quantal and stochastic
Arc ORF is essential for rapid glutamate-induced de novo translation
Model of rapid de novo translation mediated by reversal of stalled polyribosomes
Acknowledgments
We thank Dr. Nicholas Guydosh and Rachel Green for helpful discussions. This work was supported by NIMH RO1 MH053608 (P.F.W.) and MH084020 (R.L.H.).
Footnotes
Author Contributions: Conceptualization, Y.N., S.P., and P.F.W.; Methodology, Y.N., S.P., and P.F.W.; Image Processing & Analysis, C.L.; Investigation, Y.N., S.P., D.-K.K., and J.M.P.; Writing – Original Draft, Y.N. and P.F.W.; Writing – Review & Editing, Y.N., S.P., C.L., S.S., R.L.H., and P.F.W.; Visualization, Y.N.; Funding Acquisition, R.L.H. and P.F.W.; Resources, S.S., R.L.H., and P.F.W.; Supervision, R.L.H. and P.F.W.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Arellano JI, Benavides-Piccione R, Defelipe J, Yuste R. Ultrastructure of dendritic spines: correlation between synaptic and spine morphologies. Front Neurosci. 2007;1:131–143. doi: 10.3389/neuro.01.1.1.010.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béïque JC, Na Y, Kuhl D, Worley PF, Huganir RL. Arc-dependent synapse-specific homeostatic plasticity. Proc Natl Acad Sci USA. 2011;108:816–821. doi: 10.1073/pnas.1017914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomer WA, VanDongen HM, VanDongen AM. Arc/Arg3.1 translation is controlled by convergent N-methyl-D-aspartate and Gs-coupled receptor signaling pathways. J Biol Chem. 2008;283:582–592. doi: 10.1074/jbc.M702451200. [DOI] [PubMed] [Google Scholar]
- Branco T, Häusser M. Synaptic integration gradients in single cortical pyramidal cell dendrites. Neuron. 2011;69:885–892. doi: 10.1016/j.neuron.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, Baumeister W. The native 3D organization of bacterial polysomes. Cell. 2009;136:261–271. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- Bressloff PC. Stochastic Processes in Cell Biology. Springer; 2014. [Google Scholar]
- Butko MT, Yang J, Geng Y, Kim HJ, Jeon NL, Shu X, Mackey MR, Ellisman MH, Tsien RY, Lin MZ. Fluorescent and photo-oxidizing TimeSTAMP tags track protein fates in light and electron microscopy. Nat Neurosci. 2012;15:1742–1751. doi: 10.1038/nn.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW, Maulucci-Gedde M, Kriegstein AR. Glutamate neurotoxicity in cortical cell culture. J Neurosci. 1987;7:357–368. doi: 10.1523/JNEUROSCI.07-02-00357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan LL, Bruno VM, Amagasu SM, Giffard RG. Glia modulate the response of murine cortical neurons to excitotoxicity: glia exacerbate AMPA neurotoxicity. J Neurosci. 1995;15:4545–4555. doi: 10.1523/JNEUROSCI.15-06-04545.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris S, Lewandowski G, Cox CD, Steward O. Selective localization of arc mRNA in dendrites involves activity- and translation-dependent mRNA degradation. J Neurosci. 2014;34:4481–4493. doi: 10.1523/JNEUROSCI.4944-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Tatavarty V, Korza G, Levin MK, Carson JH. Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol Biol Cell. 2008;19:2311–2327. doi: 10.1091/mbc.E07-09-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Graber TE, Hébert-Seropian S, Khoutorsky A, David A, Yewdell JW, Lacaille JC, Sossin WS. Reactivation of stalled polyribosomes in synaptic plasticity. Proc Natl Acad Sci USA. 2013;110:16205–16210. doi: 10.1073/pnas.1307747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippo-campal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Miyashita T, Chawla MK, Sanderson J, Maes LI, Houston FP, Lipa P, McNaughton BL, Worley PF, Barnes CA. Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc Natl Acad Sci USA. 2006;103:1077–1082. doi: 10.1073/pnas.0505519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Shirao T. Change in the shape of dendritic spines caused by overexpression of drebrin in cultured cortical neurons. J Neurosci. 1999;19:3918–3925. doi: 10.1523/JNEUROSCI.19-10-03918.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- Ifrim MF, Williams KR, Bassell GJ. Single-molecule imaging of PSD-95 mRNA translation in dendrites and its dysregulation in a mouse model of fragile X syndrome. J Neurosci. 2015;35:7116–7130. doi: 10.1523/JNEUROSCI.2802-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakkamsetti V, Tsai NP, Gross C, Molinaro G, Collins KA, Nicoletti F, Wang KH, Osten P, Bassell GJ, Gibson JR, Huber KM. Experience-induced Arc/Arg3.1 primes CA1 pyramidal neurons for metabo-tropic glutamate receptor-dependent long-term synaptic depression. Neuron. 2013;80:72–79. doi: 10.1016/j.neuron.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, Garner CC, Tsien RY, Ellisman MH, Malenka RC. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci. 2004;7:244–253. doi: 10.1038/nn1189. [DOI] [PubMed] [Google Scholar]
- Kawata S, Miyazaki T, Yamazaki M, Mikuni T, Yamasaki M, Hashimoto K, Watanabe M, Sakimura K, Kano M. Global scaling down of excitatory postsynaptic responses in cerebellar Purkinje cells impairs developmental synapse elimination. Cell Rep. 2014;8:1119–1129. doi: 10.1016/j.celrep.2014.07.014. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Jin Y, Kim J, Shin JH, Worley PF, Linden DJ. Transient upregulation of postsynaptic IP3-gated Ca release underlies short-term potentiation of metabotropic glutamate receptor 1 signaling in cerebellar Purkinje cells. J Neurosci. 2008;28:4350–4355. doi: 10.1523/JNEUROSCI.0284-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Sul JY, Helmfors H, Langel U, Kim J, Eberwine J. Dendritic glutamate receptor mRNAs show contingent local hotspot-depen-dent translational dynamics. Cell Rep. 2013;5:114–125. doi: 10.1016/j.celrep.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Yamamoto S, Maruo T, Murakami F. Identification of a cis-acting element required for dendritic targeting of activity-regulated cytoskeleton-associated protein mRNA. Eur J Neurosci. 2005;22:2977–2984. doi: 10.1111/j.1460-9568.2005.04508.x. [DOI] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenga J, de Vrij FM, Oostra BA, Willemsen R. Potential therapeutic interventions for fragile X syndrome. Trends Mol Med. 2010;16:516–527. doi: 10.1016/j.molmed.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MZ, Tsien RY. TimeSTAMP tagging of newly synthesized proteins. Curr Protoc Protein Sci. 2010;Chapter 26 doi: 10.1002/0471140864.ps2605s59. http://dx.doi.org/10.1002/0471140864.ps2605s59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MZ, Glenn JS, Tsien RY. A drug-controllable tag for visualizing newly synthesized proteins in cells and whole animals. Proc Natl Acad Sci USA. 2008;105:7744–7749. doi: 10.1073/pnas.0803060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci USA. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell. 2005;20:709–722. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Martin BR, Giepmans BN, Adams SR, Tsien RY. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- Moser MB, Trommald M, Andersen P. An increase in dendritic spine density on hippocampal CA1 pyramidal cells following spatial learning in adult rats suggests the formation of new synapses. Proc Natl Acad Sci USA. 1994;91:12673–12675. doi: 10.1073/pnas.91.26.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Kelić S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, Schnitzer MJ. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron. 2009;63:747–760. doi: 10.1016/j.neuron.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama D, Iwata H, Teshirogi C, Ikegaya Y, Matsuki N, Nomura H. Long-delayed expression of the immediate early gene Arc/Arg3.1 refines neuronal circuits to perpetuate fear memory. J Neurosci. 2015;35:819–830. doi: 10.1523/JNEUROSCI.2525-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J Biol Chem. 2008;283:18478–18482. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niere F, Wilkerson JR, Huber KM. Evidence for a fragile X mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered Arc translation and long-term depression. J Neurosci. 2012;32:5924–5936. doi: 10.1523/JNEUROSCI.4650-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno H, Akashi K, Ishii Y, Yagishita-Kyo N, Suzuki K, Nonaka M, Kawashima T, Fujii H, Takemoto-Kimura S, Abe M, et al. Inverse synaptic tagging of inactive synapses via dynamic interaction of Arc/Arg3.1 with CaMKIIβ. Cell. 2012;149:886–898. doi: 10.1016/j.cell.2012.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LM. Dendritic integration in pyramidal neurons during network activity and disease. Brain Res Bull. 2014;103:2–10. doi: 10.1016/j.brainresbull.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Panja D, Dagyte G, Bidinosti M, Wibrand K, Kristiansen AM, Sonenberg N, Bramham CR. Novel translational control in Arc-dependent long term potentiation consolidation in vivo. J Biol Chem. 2009;284:31498–31511. doi: 10.1074/jbc.M109.056077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, et al. Elongation factor 2and fragileX mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- Pinkstaff JK, Chappell SA, Mauro VP, Edelman GM, Krushel LA. Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc Natl Acad Sci USA. 2001;98:2770–2775. doi: 10.1073/pnas.051623398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Raab-Graham KF, Haddick PC, Jan YN, Jan LY. Activity-and mTOR-dependentsuppression ofKv1.1 channel mRNA translationinden-drites. Science. 2006;314:144–148. doi: 10.1126/science.1131693. [DOI] [PubMed] [Google Scholar]
- Ramírez-Amaya V, Vazdarjanova A, Mikhael D, Rosi S, Worley PF, Barnes CA. Spatialexploration-induced Arc mRNA and protein expression: evidence for selective, network-specific reactivation. J Neurosci. 2005;25:1761–1768. doi: 10.1523/JNEUROSCI.4342-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. Visualization of mRNA translation in living cells. J Cell Biol. 2006;175:67–76. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh G, Sia GM, Garner CC, Huganir RL. Synapse-associated protein-97 isoform-specific regulation of surface AMPA receptors and synaptic function in cultured neurons. J Neurosci. 2003;23:4567–4576. doi: 10.1523/JNEUROSCI.23-11-04567.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat Neurosci. 2000;3:211–216. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- Seib LM, Wellman CL. Daily injections alter spine density in rat medial prefrontal cortex. Neurosci Lett. 2003;337:29–32. doi: 10.1016/s0304-3940(02)01287-9. [DOI] [PubMed] [Google Scholar]
- Seung HS. Learning in spiking neural networks by reinforcement of stochastic synaptic transmission. Neuron. 2003;40:1063–1073. doi: 10.1016/s0896-6273(03)00761-x. [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Bear MF. New views of Arc, a master regulator of synaptic plasticity. Nat Neurosci. 2011;14:279–284. doi: 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic syn-aptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shouval HZ, Kalantzis G. Stochastic properties of synaptic transmission affect the shape of spike time-dependent plasticity curves. J Neurophysiol. 2005;93:1069–1073. doi: 10.1152/jn.00504.2004. [DOI] [PubMed] [Google Scholar]
- Smith WB, Starck SR, Roberts RW, Schuman EM. Dopaminergic stimulation of local protein synthesis enhances surface expression of GluR1 and synaptic transmission in hippocampal neurons. Neuron. 2005;45:765–779. doi: 10.1016/j.neuron.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jørgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruston N. Pyramidal neurons: dendritic structure and synaptic integration. Nat Rev Neurosci. 2008;9:206–221. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Tatavarty V, Ifrim MF, Levin M, Korza G, Barbarese E, Yu J, Carson JH. Single-molecule imaging of translational output from individual RNA granules in neurons. Mol Biol Cell. 2012;23:918–929. doi: 10.1091/mbc.E11-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzingounis AV, Nicoll RA. Arc/Arg3.1: linking gene expression to synaptic plasticity and memory. Neuron. 2006;52:403–407. doi: 10.1016/j.neuron.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, Martin KC. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324:1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Han B, Zhou R, Zhuang X. Real-time imaging of translation on single mRNA transcripts in live cells. Cell. 2016;165:990–1001. doi: 10.1016/j.cell.2016.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waung MW, Huber KM. Protein translation in synaptic plasticity: mGluR-LTD, fragile X. Curr Opin Neurobiol. 2009;19:319–326. doi: 10.1016/j.conb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth I, Pohl C, Mittelstaet J, Konevega AL, Rodnina MV. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos Trans R Soc Lond B Biol Sci. 2011;366:2979–2986. doi: 10.1098/rstb.2011.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Hoek TA, Vale RD, Tanenbaum ME. Dynamics of translation of single mRNA molecules in vivo. Cell. 2016;165:976–989. doi: 10.1016/j.cell.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SG, Bagni C. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wu J, Ward MD, Yang S, Chuang YA, Xiao M, Li R, Leahy DJ, Worley PF. Structural basis of arc binding to synaptic proteins: implications for cognitive disease. Neuron. 2015;86:490–500. doi: 10.1016/j.neuron.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


