Abstract
Objectives
To analyse and describe the clinical and genetic spectrum of Charcot-Marie-Tooth disease (CMT) caused by mutations in the neurofilament light polypeptide gene (NEFL).
Methods
Combined analysis of new patients with NEFL-related CMT, identified from those attending clinics at the participating institutions, and all previously reported cases from the literature.
Results
Five new unrelated patients with CMT carrying heterozygous NEFL mutations (N98S, P8R and L311P) were identified. Combined data from these cases and 62 kindreds from the literature revealed four common mutations (P8R, P22S, N98S and E396K) and three mutational hotspots accounting for 55% and 75% of kindreds, respectively. De novo mutations were identified in eight patients. Loss of large-myelinated fibres was a uniform feature in 21 sural nerve biopsies, and ‘onion bulb’ formations and/or thin myelin sheaths were observed in 67%. The neurophysiological phenotype was broad but most patients carrying the mutations E90K and N98S had all reported upper limb motor conduction velocities <38 m/s. Age of symptoms onset was ≤ 3 years in 25 cases. Pyramidal tract signs were described in 13 patients and seven patients were initially diagnosed with or tested for inherited ataxia. Patients with E90K and N98S frequently presented before age 3 years and developed hearing loss or other neurological features including ataxia or cerebellar atrophy on brain MRI.
Conclusions
NEFL-related CMT is clinically and genetically heterogeneous. Based on this study, however, we propose mutational hotspots and relevant clinical-genetic associations that may be helpful in the evaluation of NEFL sequence variants and the differential diagnosis with other forms of CMT.
Keywords: Charcot-Marie-Tooth disease, NEFL, neurofilament light polypeptide, whole-exome sequencing
INTRODUCTION
Charcot-Marie-Tooth disease (CMT) comprises a heterogeneous group of inherited neuropathies clinically characterised by progressive, distal-predominant weakness, amyotrophy and sensory loss. CMT is classically divided into demyelinating (CMT1) and axonal (CMT2) subtypes based on upper limb motor nerve conduction velocities (MNCV) below or above 38 m/s, respectively, although intermediate forms of CMT with MNCV of 25–45 m/s are also recognised.1,2 Whilst CMT1 is frequently caused by mutations affecting peripheral myelin proteins and CMT2 by mutations in genes related to neuronal and axonal functions, there is significant genetic overlap between CMT subtypes.1,3
Neurofilaments are neuron-specific intermediate filaments essential for the radial growth of axons during development and the maintenance of the axonal diameter.4 Like other intermediate filaments, neurofilaments are formed by polymerization of subunits, the neurofilament light (NFL), medium and heavy polypeptides, all of which contain three major domains: globular N-terminal head, α-helical central rod and C-terminal tail. The rod domain is required for co-assembly between subunits while the head and tail domains are involved in regulation of assembly, axonal transport and radial growth.4–7
Mutations in the NEFL gene encoding NFL were first identified as a cause of autosomal dominant CMT in 2000,8 and subsequent reports have confirmed the association of NEFL mutations with different phenotypes including CMT1 (designated CMT1F), CMT2 (designated CMT2E), dominant intermediate CMT and autosomal recessive CMT.9–18 Despite the growing number of reports about NEFL-related CMT published in the past 15 years, it is a rare form of inherited neuropathy, accounting for less than 1% of all CMT cases in large cohorts,19,20 and obvious genotype-phenotype correlations have not been established so far.21 Here we report five new patients with NEFL-related CMT and review all previously published cases to describe the clinical and genetic spectrum of the disease.
METHODS
Patients and clinical investigations
Patients with diagnosis of NEFL-related CMT were identified from those attending neuropathy clinics at the National Hospital for Neurology and Neurosurgery, Queen Square, London, UK, and the Department of Neurology, Carver College of Medicine, University of Iowa, Iowa, USA. All patients had undergone clinical and investigational assessments during the routine diagnostic process and were under follow-up at the time of the study. Skeletal muscle strength was manually tested and scored according to the Medical Research Council (MRC) grading system ranging from 0 (no movement) to 5 (normal strength). The CMT Examination Score, a composite subscale of the CMT Neuropathy Score (version 2) based on symptoms and signs, with scores ranging from 0 (normal) to 28 (worst score), was obtained in four of the patients.22 Laboratory tests, neurophysiological studies, MRI scans and nerve and muscle biopsy specimens were performed and analysed according to standard procedures.
Mutation identification
In four patients, NEFL mutations were identified by bidirectional Sanger sequencing during the diagnostic process or as part of a cohort-based study to determine the frequency of CMT subtypes. In one patient, a pathogenic NEFL mutation was identified through whole-exome sequencing followed by bidirectional Sanger sequencing validation. Sanger sequencing of parents’ samples were performed for three cases. Further methodological details are provided in appendix e-1 and table e-1.
Additional published cases
A systematic literature search to identify all previously published cases of NEFL-related CMT until July 1, 2016 was performed in PubMed using the terms: “NEFL”, “neurofilament light”, “neuropathy” and “Charcot-Marie-Tooth”. These cases and the patients from our cohort were combined in order to determine the clinical and genetic spectrum of NEFL-related CMT. All cases with at least a simple description of the phenotype (CMT subtype and familial vs. individual case) were included in the analysis of mutation frequencies. Only cases with a diagnosis of CMT and a more detailed clinical and neurophysiological description were included in the phenotypic analysis.
Internal whole-exome data
Whole-exome sequencing data from 202 patients with suspected hereditary CNS disorders investigated at the UCL Institute of Neurology, London, UK, was screened for non-synonymous NEFL sequence variants with a minor allele frequency ≤1% in the 1000 Genomes (http://www.1000genomes.org/) and the Exome Variant Server (http://evs.gs.washington.edu/EVS/) datasets.
Bioinformatic analyses
NEFL sequence variants are described in accordance with the recommendations of the Human Genome Variation Society (http://www.hgvs.org/mutnomen/) using GenBank NM_006158.4 as the reference sequence. Minor allele frequencies of all NEFL variants were obtained from the Exome Aggregation Consortium (ExAC) browser version 0.3 (http://exac.broadinstitute.org) using genomic coordinates from the human reference genome assembly 19 (GRCh37). Evolutionary conservation of nucleotides was assessed using phyloP (46 vertebrate basewise conservation) and GERP scores, which were accessed through the UCSC Genome Browser (https://genome.ucsc.edu/cgi-bin/hgGateway) using genomic coordinates from GRCh37. Grantham scores were used to assess the physicochemical nature of the amino acid substitutions.23 In silico analyses of sequence variants were performed using SIFT (http://sift.jcvi.org/), PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/) and CADD (http://cadd.gs.washington.edu/).
Ethics approval
All human studies were approved by the ethics committee of the National Hospital for Neurology and Neurosurgery and UCL Institute of Neurology, London, UK, and the Institutional Review Board of the Carver College of Medicine, University of Iowa, Iowa, USA. All patients and their relatives provided written informed consent prior to genetic testing.
RESULTS
Case series
Five unrelated patients with NEFL-related CMT were identified at the participating institutions. Clinical and investigational data is summarised in table 1 and a more detailed clinical description is provided in appendix e-1.
Table 1.
Clinical, neurophysiological, pathological and genetic features in our series (n=5)
| Patient | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Sex/age (years) | F/40 | M/34 | F/15 | M/53 | M/76 |
| NEFL mutation | N98S | N98S | N98S | P8R | L311P |
| Inheritance* | De novo | De novo | De novo | n/a | n/a |
| Ethnicity | British | British/Irish | Austrian/Russian | British | British |
| Age of onset | <1 year | <1 year | <2 years | Second decade | Sixth decade |
| Initial symptoms | Delayed motor milestones, finger contractures | Delayed motor milestones, hypotonia | Delayed motor milestones, optic nerve hypoplasia | Unsteadygait, tremor in hands restless legs | Unsteady gait, tremor in hands |
| Muscle atrophy UL | Distal | Distal | Distal | Hands | Hands |
| Muscle atrophy LL | Distal and proximal | Distal and proximal | Distal | Distal | Distal |
| Muscle weakness UL | Hands 0–2, forearms 1–4, proximal 4–5 | Hands 3, forearms 4–5 | Hands 4–5 | Hands 3–4 | Hands 4–5 |
| Muscle weakness LL | Distal 0, proximal 2–5 | Distal 1, proximal 4–5 | Distal 3–5 | Distal 0–1 | Distal 3–4 |
| Deep tendon reflexes | Absent | Absent | Absent | Absent | Absent |
| Pinprick sensation | Reduced to mid-forearms/above knees | Reduced to mid-forearms/above knees | Normal | Reduced to wrists/knees | Reduced to above ankles |
| Vibration sense | Reduced to left shoulder/costal margins | Reduced to shoulders/sternum | Reduced to elbows/ankles | Reduced to elbows/costal margins | Reduced to the knees |
| Position sense | Reduced to the ankles | Reduced to the ankle and knee | Normal | Normal | Reduced to the ankles |
| Limb ataxia | UL/LL, marked, worse with eyes closed | UL/LL, mild, only with eyes closed | No | No | No |
| Pyramidal signs | No | No | No | No | No |
| Romberg’s test | Positive | Positive | Positive | Positive | Positive |
| Gait pattern | Steppage, ataxic | Steppage, proximal LL weakness | Steppage, ataxic | Steppage | Stamping, ataxic |
| Pes cavus | Yes | Yes | Yes | Yes | No |
| SNHL | Since age 5, hearing aids | Since age 8, hearing aids | Since age 8, hearing aids | Subclinical, high frequency | n/a |
| Other features | Dysarthria, mild head tremor, broken-up smooth pursuits, tongue hemiatrophy, mild scoliosis, scapular winging | Dysarthria, mild head tremor, broken-up smooth pursuits, scapular winging | Mild tremor in hands, muscle cramps, urinary incontinence | Mild head tremor, mild tremor in UL, mild broken-up smooth pursuits, RLS, PLMS | Mild head tremor tremor in hands, thickened ulnar nerves |
| NCS | Demyelinating neuropathy | Demyelinating neuropathy | Demyelinating neuropathy | Neuropathy with axonal and demyelinating features | Neuropathy with axonal and demyelinating features |
| CMTES | 24 | 22 | 9 | 18 | n/a |
| CSF protein (g/L) | n/a | 0.55 | n/a | 0.39 | 0.76 |
| BAEPs | Absent | Absent right, abnormal left | Absent | Normal | n/a |
| Brain MRI | Mild cerebellar and medullary volume loss | Normal | n/a | Normal | Normal |
| Spinal cord MRI | Mild cervical cord volume loss | Mild cervical cord volume loss | n/a | Normal | Normal |
| Nerve pathology | n/a | Loss or large myelinated fibres, irregularly shaped fibres, thin myelin sheaths, onion bulbs | n/a | Loss of large myelinated fibres, irregularly shaped fibres, regeneration clusters | n/a |
| Muscle pathology | Neurogenic changes | Neurogenic changes | n/a | n/a | n/a |
Based on cosegregation analysis.
BAEPs, brainstem auditory evoked potentials; CMTES, Charcot-Marie-Tooth examination score; CK, plasma creatine kinase; distal, muscle groups below elbow/knee; F, female; LL, lower limbs; M, male; NCS, nerve conduction studies; PLMS, periodic limb movements disorder; proximal, muscle groups above elbow/knee; RLS, restless legs syndrome; SNHL, sensorineural hearing loss, UL = upper limbs
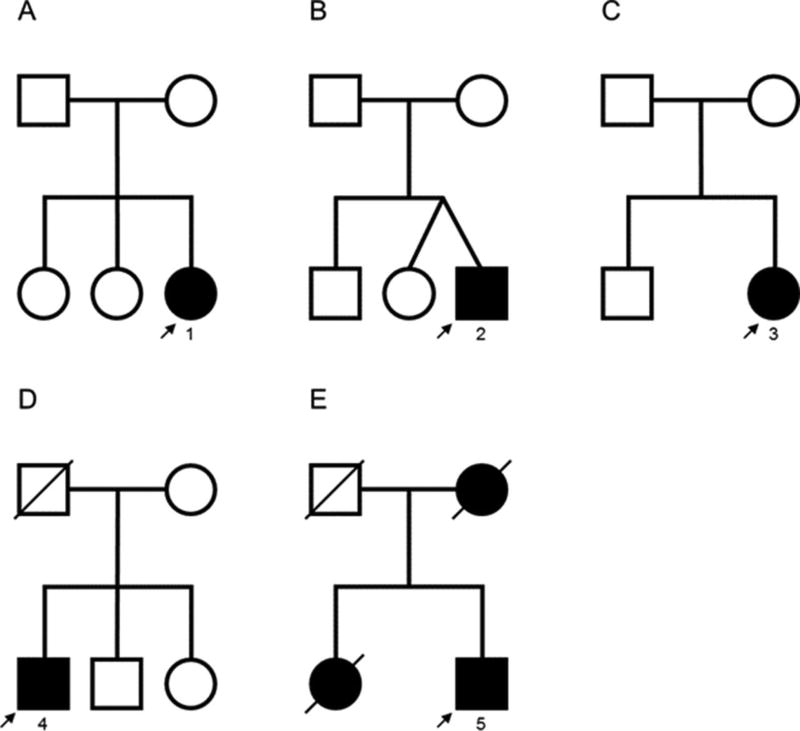
Patients 1, 2 and 3 (figure 1) carried the heterozygous pathogenic mutation N98S. All had a similar phenotype consisting of motor developmental delay, hypotonia and walking difficulties before age 3–4 years, bilateral hearing loss requiring hearing aids between ages 5–8 years, progressive distal greater than proximal motor impairment within the first two decades, and marked proprioceptive sensory deficit with positive Romberg’s test and gait ataxia. Patients 1 and 2, who had the longest follow-up, also developed proximal lower limb weakness and scapular winging, appendicular ataxia, and cerebellar features supported by neuro-otological examination or brain MRI (figure 2). Additional features included tongue hemiatrophy in patient 1, mild cervical spinal cord volume loss in patients 1 and 2, and optic nerve hypoplasia in patient 3. Neurophysiological studies were consistent with a length-dependent motor and sensory neuropathy with reduced or absent compound muscle action potential (CMAP) amplitudes and upper limb MNCV ranging from 23 to 35 m/s (corresponding CMAPs 1.5 mV 0.8 mV; table e-2). In patients 1 and 2, ulnar nerve CMAP waveforms were dispersed or complex on proximal stimulation. Co-segregation analyses indicated that the heterozygous mutation N98S occurred de novo in the three probands. Biological paternity was confirmed by short-tandem repeat analysis for patients 1 and 2 (results not shown; patient 3 was not tested).
Figure 1. Family trees of patients from our series.
The number corresponds to the patient number in the text and table 1.
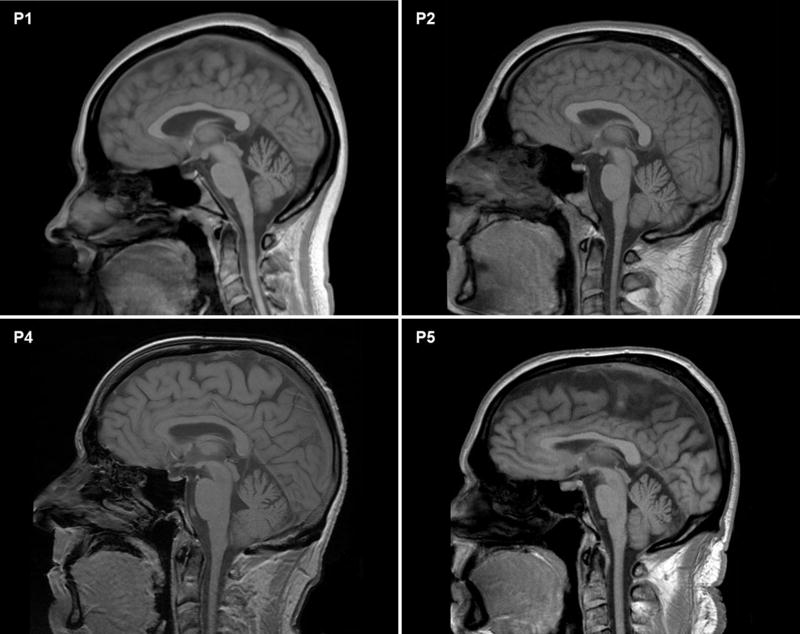
Figure 2. Brain MRI.
Cranial sagittal T1-weighted MR images showing mild cerebellar and medullary volume loss in patient 1 (P1) but not in patients 2 (P2), 4 (P4) and 5 (P5). Mild volume loss of cervical spinal cord can also be appreciated in patients P1 and P2.
Patient 4 carried the heterozygous pathogenic mutation P8R. He developed progressive distal motor and sensory impairment from the second decade along with restless legs syndrome and periodic limb movements disorder. Neurophysiological studies in his 40s revealed a length-dependent motor and sensory neuropathy with reduced or absent CMAP amplitudes and upper limb MNCV ranging from 31 to 44 m/s (corresponding CMAPs 1.7 mV; table e-2).
Patient 5 carried the novel heterozygous mutation L311P. He presented in the sixth decade with hand tremor followed with gait ataxia and muscle weakness and atrophy in the lower limbs. Neurophysiological studies confirmed a length-dependent motor and sensory neuropathy with reduced CMAP amplitudes in the lower limbs, upper limb MNCV ranging from 43 to 55 m/s (corresponding CMAPs 6.6 and 5.4 mV), and dispersion of CMAP waveforms in two nerve segments (table e-2).
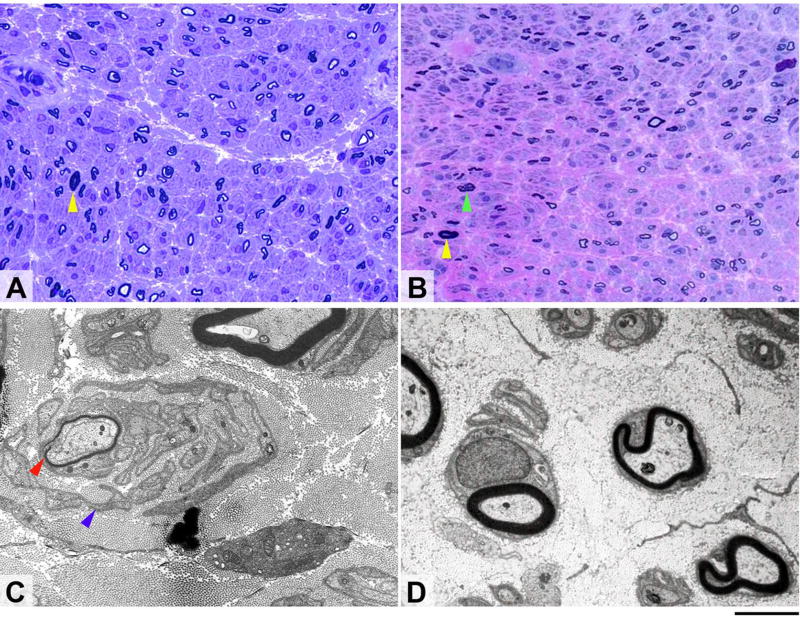
Sural nerve biopsies from patients 2 and 4 at ages 24 and 46, respectively, showed severe reduction in myelinated fibre density with complete absence of large myelinated fibres (figure 3A and B). Immunostaining for neurofilament proteins showed reduced density of axons, mainly of large axons. There was no evidence of active axonal degeneration. Definite regeneration clusters were observed in patient 4. Both biopsies contained irregularly-shaped fibres. Disproportionately thin myelin sheaths and onion bulbs, confirmed by electron microscopy imaging, were seen in patient 2 (figure 3C) but not in patient 4 (figure 3D).
Figure 3. Morphological appearances of sural nerve biopsies from patients 2 (A and C) and 4 (B and D).
Semi-thin resin preparations stained with toluidine blue (A) and methylene blue azure–basic fuchsin (B) show transverse sections of nerve fascicles in both of which there is severe widespread loss of large myelinated fibres (yellow arrows indicate occasional remaining large fibres). In both patients the small myelinated fibres are better preserved and several regeneration clusters are evident in case 2 (B, green arrow). Ultrastructural examination of the biopsy from the first patient (C) shows a thinly myelinated fibre (red arrow) surrounded by several layers of Schwann cell profiles (blue arrow) closely resembling an onion bulb, while electron microscopy in the other patient (D) confirms marked reduction of large myelinated fibres accompanied by endoneural fibrosis but shows no onion bulb formations. Scale bar: 30µm in A and B; 3µm in C and D.
Literature review
A systematic review of the literature revealed published data on 62 kindreds with NEFL-related CMT.8–18,24–48 These cases and the five unrelated patients from our cohort were combined for analysis.
Mutational spectrum
A total of 30 different NEFL mutations have been reported to date (tables 2 and e-3).8–18,24–48 For descriptive reasons, the mutations c.22_23delCCinsAG and c.23C>G, causing the same amino acid substitution (P8R), were classified as one single mutation. For two mutations (E186* and Q334P) there was no clinical data available. R421* has been reported in a family with an overlapping myopathic-neurogenic phenotype40 and also in association with CMT41 but no clinical details were available for the latter.
Table 2.
Mutation frequency, mode of transmission, age of onset and neurophysiological features (n=67 kindreds)
| NEFL amino acid change |
Exon | Domain | Subdomain | Kindreds, n (%) |
Familial cases,† n |
Individual cases, n |
Confirmed inheritance patterns‡ |
De novo mutation, n (cases) |
AOO, range (decade) |
AOO ≤3 years, n (cases) |
Median MNCV, range (m/s) |
Ulnar MNCV, range (m/s) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P8L | 1 | Head | – | 2 (3) | – | 2 | SP | 1 | 1 | 1 | 13–33 | 19 |
| P8Q | 1 | Head | – | 1 (1.5) | 1 | – | – | – | 1 | – | 21 | 33 |
| P8R | 1 | Head | – | 7 (10.4) | 27 | 3 | AD, SP | 1 | 1–3 | – | 23–41 | 30–49 |
| T21Afs*83 | 1 | Head | – | 1 (1.5) | – | 1 | – | – | – | – | – | – |
| P22R | 1 | Head | – | 1 (1.5) | 5 | – | AD | – | 1–2 | 1 | 22–29 | 24–25 |
| P22S | 1 | Head | – | 7 (10.4) | 71 | 1 | AD | – | 1–5 | – | 21–54 | 22–59 |
| P22T | 1 | Head | – | 2 (3) | 5 | – | AD | – | 2–3 | – | 21–36 | 33–39 |
| E90K | 1 | Head | – | 2 (3) | – | 2 | SP | 2 | 1 | 2 | 17–27 | 28 |
| L94P | 1 | Rod | Coil 1A | 1 (1.5) | 10 | – | AD | – | 1–2 | – | 35–38 | 29–40 |
| N98S | 1 | Rod | Coil 1A | 10 (14.9) | 4 | 8 | AD, SP | 4 | 1–2 | 11 | 18–35§ | 23–33§ |
| N98T | 1 | Rod | Coil 1A | 1 (1.5) | – | 1 | – | – | 1 | 1 | 44 | – |
| E140* | 1 | Rod | Coil 1B | 1 (1.5) | 2 | – | AR | – | 1 | – | 14 | – |
| A149V | 1 | Rod | Coil 1B | 1 (1.5) | – | 1 | – | – | 4 | – | – | – |
| E186* | 1 | Rod | Coil 1B | – | – | – | – | – | – | – | – | – |
| E210* | 1 | Rod | Coil 1B | 1 (1.5) | 4 | – | AR | – | 1 | 4 | 12–25 | 14–23 |
| Y265C | 1 | Rod | Coil 2A | 1 (1.5) | 7 | – | AD | – | – | – | – | – |
| L268R | 1 | Rod | Coil 2A | 1 (1.5) | 8 | – | AD | – | – | – | – | – |
| L268P | 1 | Rod | Coil 2A | 2 (3) | 17 | – | AD | – | 1–5 | – | 38–55 | 35–56 |
| L311P | 1 | Rod | Coil 2B | 1 (1.5) | 3 | – | – | – | 6 | – | 45–55 | 43–48 |
| C322_N326del | 1 | Rod | Coil 2B | 1 (1.5) | 11 | – | AD | – | 1 | – | 39–46 | 37–43 |
| Q332P | 1 | Rod | Coil 2B | 3 (4.5) | 21 | 1 | AD | – | 2–3 | – | 38–52 | – |
| L333P | 1 | Rod | Coil 2B | 1 (1.5) | – | 1 | – | – | – | – | – | – |
| Q334P | 1 | Rod | Coil 2B | – | – | – | – | – | – | – | – | – |
| L336P | 1 | Rod | Coil 2B | 1 (1.5) | – | 1 | – | – | – | – | – | – |
| I384F | 2 | Rod | Coil 2B | 1 (1.5) | 22 | – | AD | – | 1–4 | – | 34–46 | 32–46 |
| Y389C | 2 | Rod | Coil 2B | 1 (1.5) | 9 | – | AD | – | 6 | – | 48–49 | 50–52 |
| E396K | 3 | Rod | Coil 2B | 13 (19.4) | 63 | 3 | AD | – | 1–6 | 5 | 29–63 | 25–58 |
| R421* | 3 | Tail | Subdomain A | – | – | – | – | – | – | – | – | – |
| F439I | 3 | Tail | Subdomain A | 1 (1.5) | – | 1 | – | – | 3 | – | 45 | – |
| P440L | 3 | Tail | Subdomain A | 2 (3) | 3 | 1 | AD | – | 3 | – | – | – |
| Total/range | – | – | – | 67 (100) | 293 | 27 | – | 8 | 1–6 | 25 | 12–63 | 14–59 |
Families or isolated cases with positive family history.
Based on cosegregation analysis.
Proximal nerve segments excluded.
AD, autosomal dominant; AOO, age of disease onset; AR, autosomal recessive; MNCV, motor nerve conduction velocities; SP, sporadic.
For two mutations (E186* and Q334P), there was no clinical data available.40 43 R421* has been reported in a family with an overlapping myopathic–neurogenic phenotype39 and also in association with CMT40 but no clinical details were available for the latter.
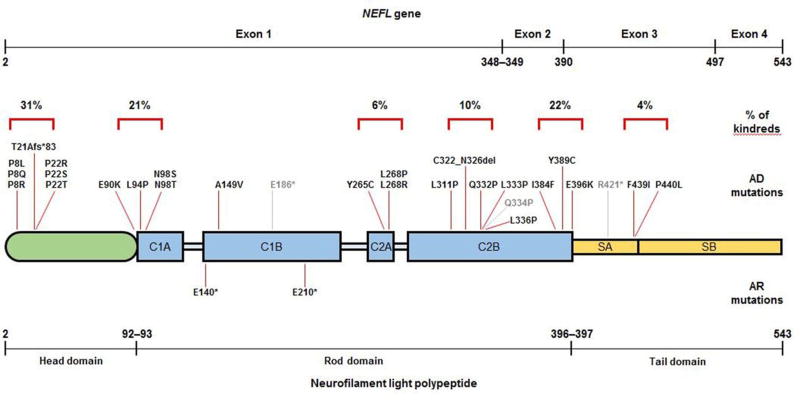
Of the remainder 27 mutations, 17 were private to a single kindred and 10 were observed in two or more kindreds. Twenty-four mutations were clustered in six regions of the NFL polypeptide (figure 4): the initial segment of the head domain (amino acid positions 8–22), the junction between the head and the rod domains (90–98), coil 2A (265–268), the mid portion of coil 2B (311–336), the end portion of coil 2B (384–396) and the tail subdomain A (421–440). Mutations in the head domain and the two ends of the rod domain accounted for 75% of kindreds and four common mutations within these regions (P8R, P22S, N98S and E396K) were observed in 55% of kindreds. Only three mutations (E140*, A149V and E210*) were located in coil 1B. No mutations were observed in linker regions of the rod domain or the tail subdomain B.
Of the remainder 27 mutations, 17 were private to a single kindred and 10 were observed in two or more kindreds. Twenty-four mutations were clustered in six regions of the NFL polypeptide (figure 4): the initial segment of the head domain (amino acid positions 8–22), the junction between the head and the rod domains (90–98), coil 2A (265–268), the mid portion of coil 2B (311–336), the end portion of coil 2B (384–396), and the tail subdomain A (421–440). Mutations in the head domain and the two ends of the rod domain accounted for 75% of kindreds and four common mutations within these regions (P8R, P22S, N98S, and E396K) were observed in 55% of kindreds. Only three mutations (E140*, A149v, and E210*) were located in coil 1B. No mutations were observed in linker regions of the rod domain or the tail subdomain B.
Figure 4. Localization of reported NEFL mutations in the neurofilament light polypeptide.
The size of the NEFL gene (cDNA; top line) and neurofilament light polypeptide (figure and bottom line) correspond to the number of bases and amino acids involved; numbers in the top line indicate base and numbers in the bottom line indicate amino acid position. Light blue = linker regions of the rod domain. Light grey = mutations for which there was no clinical information available. AD = autosomal dominant; AR = autosomal recessive; C1A, C1B, C2A and C2B = coil 1A, 1B, 2A and 2C subdomains; SA and SB = subdomains A and B.
A detailed analysis of all mutations, including conservation and pathogenicity prediction scores, is provided in table e-3. Pathogenicity for 23 of the 30 mutations was supported by co-segregation studies, existence of other mutations in the same amino acid position, detection in more than one kindred or functional studies (in vitro or animal models). This information was not available for the remaining seven mutations (E186*, L311P, L333P, Q334P, L336P, F439I and R421*); of these, only F439I is described in the EXaC dataset, being present in 13 alleles (minor allele frequency of 0.01%).
Inheritance pattern
Of the 67 kindreds with NEFL mutations, 27 correspond to individual cases and 40 correspond to families or familial cases. Of the latter, 29 had an autosomal dominant inheritance based on co-segregation analysis. This was not reported or was not available in nine additional families with probable autosomal dominant inheritance; only two of them, however, had private mutations (L311P reported in this study and P8Q). The inheritance pattern was autosomal recessive in two consanguineous families in which all affected individuals had homozygous nonsense mutations located in coil 1B (E140* and E210*).13,14
Of the 27 individual cases analysed, one was adopted and 26 were reported as isolated or sporadic. Eight of these patients had de novo mutations based on co-segregation analysis. Of note, 11 out of 15 patients with substitutions of amino acids E90 and N98 (E90K and N98S/T) were isolated/sporadic and six of them had confirmed de novo mutations.10,14
Neurophysiologic pattern
A neurophysiologic classification of the peripheral neuropathy was reported for 58 individual cases or families with the following distribution: CMT1 or demyelinating neuropathy in 18 kindreds (31%), CMT2 or axonal neuropathy in 23 kindreds (40%), and intermediate CMT or neuropathy with axonal and demyelinating features in 17 kindreds (29%).8,9,11–18,24–34,36,37,39,42,43,45,46,48
Upper limb nerve conduction data was available for 115 patients from 53 kindreds: 44 patients (38%) had all reported upper limb MNCV ≥38 m/s, 24 patients (21%) had upper limb MNCV ranging from <38 and ≥38 m/s, and 47 patients (41%) had all reported upper limb MNCV <38 m/s.10,11,13–18,24,25,27–33,35,36,38,39,46,47 Upper limb CMAP values were available in 21 patients from the latter group, of whom 18 had amplitudes ≥75% of lower limit of normal.
Analysis of the same upper limb MNCV data using 25 m/s and 45 m/s as cut-off values revealed that 12 patients (10%) had all reported values >45 m/s, 74 patients (64%) had all reported values between 25–45 m/s, and 9 patients (8%) had all reported values <25 m/s; the remaining 20 patients had upper limb MNCV in overlapping ranges.
Median and/or ulnar MNCV <20 m/s were detected in eight patients, five of them with recessive mutations (E140* and E210*) and three with dominant mutations (P8L, E90K and N98S).10,13,14,18 Of the four common NEFL mutations, only N98S was consistently associated with upper limb MNCV <38 m/s although MNCV in proximal nerve segments varied between 30.1 and 47.8 m/s in one kindred (classified as dominant intermediate-CMT).10,14,18,25,35,47
Sural nerve histopathology
Twenty-one sural nerve biopsies of patients with NEFL-related CMT, including two from our cohort, have been reported to date (table 3)10,11,13,14,16,26,28,29,32,34,39. Loss of large myelinated features was a uniform feature. Onion bulbs and thin or irregularly-folded myelin sheaths were observed in 14 biopsies (67%) and giant axons, with or without thin myelin sheaths, were found in six (29%).
Table 3.
Clinical and pathology features in patients with NEFL-related neuropathy (n=173)*
| All n=173 (%) |
P8R n=23 (%) |
P22S n=28 (%) |
N98S n=12 (%) |
E396K n=43 (%) |
|
|---|---|---|---|---|---|
| Clinical features | |||||
| Ataxia† | 22 (13) | 4 (17) | 3 (11) | 5 (42) | 4 (9) |
| Sensory ataxia‡ | 8 (5) | 1 (4) | – | 3 (25) | 1 (2) |
| Diagnosed with SCA/FRDA§ | 7 (4) | 1 (4) | – | 3 (25) | 1 (2) |
| Motor developmental delay | 18 (10) | – | – | 11 (92) | 1 (2) |
| Hearing loss/deafness | 17 (10) | – | – | 8 (67) | 4 (9) |
| Pyramidal signs/brisk reflexes | 13 (8) | – | – | – | 4 (9) |
| Head/limb tremor¶ | 11 (6) | 3 (13) | – | 3 (25) | – |
| Hypotonia in infancy | 8 (5) | – | – | 4 (33) | – |
| Claw hands/finger contractures | 9 (5) | – | 6 (21) | 2 (17) | 1 (2) |
| Dysarthria/abnormal speech | 6 (3) | 1 (4) | – | 3 (25) | 2 (5) |
| Early upper limb involvement | 5 (3) | – | – | 1 (8) | 1 (2) |
| Waddling gait | 5 (3) | – | – | 1 (8) | 3 (7) |
| Facial weakness | 3 (2) | – | – | 1 (8) | – |
| Scoliosis/hyperlordosis | 3 (2) | 2 (9) | – | 1 (8) | – |
| Broken-up smooth pursuits | 4 (2) | 1 (4) | – | 3 (25) | – |
| Nystagmus | 4 (2) | 1 (4) | – | 3 (25) | – |
| Intellectual impairment | 3 (2) | – | – | 2 (17) | – |
| Delayed/poor growth | 2 (1) | – | – | 2 (17) | – |
| Scapular winging | 2 (1) | – | – | 2 (17) | – |
| Laboratory | |||||
| Raised CK (≤1414 IU/L) | 14 (9) | – | – | 1 (10) | 10 (23) |
| Raised CSF protein (≤0.76 g/L) | 2 (1) | – | – | 1 (10) | – |
| Neurophysiology | |||||
| BAEP abnormal/absent | 20 (12) | – | – | 3 (30) | 11 (26) |
| VEP delayed | 3 (2) | – | – | – | – |
| MEP abnormal | 7 (4) | – | – | – | 7 (16) |
| MR imaging (n=17) | |||||
| Cerebellar volume loss | 4 (24) | – | – | 3 (75) | 1 (33) |
| Spinal cord volume loss | 3 (18) | – | – | 3 (75) | – |
| Thinning of corpus callosum | 2 (12) | – | – | – | – |
| Dilation of fourth ventricle | 1 (6) | – | – | – | 1 (33) |
| Sural nerve biopsy (n=21) | |||||
| Loss of large-myelinated fibres | 21 (100) | 1 (100) | 2 (100) | 2 (100) | 6 (100) |
| Regenerating clusters | 17 (81) | 1 (100) | 1 (50) | 1 (50) | 5 (83) |
| Onion bulbs | 12 (57) | – | 1 (50) | 1 (50) | 3 (50) |
| Thin myelin sheaths | 7 (33) | – | – | 1 (50) | 3 (50) |
| Irregularly shaped nerve fibres | 7 (33) | 1 (100) | – | 1 (50) | 1 (17) |
| Giant axons ± thin myelin sheaths | 6 (29) | – | 2 (100) | – | – |
| Irregularly folded myelin sheaths | 5 (24) | – | – | – | 3 (50) |
| Axonal degeneration | 4 (19) | – | – | – | 2 (33) |
| Muscle biopsy (n=4) | |||||
| Neurogenic features | 4 (100) | – | – | 2 (100) | 2 (100) |
| Myopathic features | 2 (50) | – | – | – | 2 (100) |
Features observed in two or more patients.
Any ataxia, including gait ataxia/imbalance, limb ataxia or ataxia not otherwise specified.
Described as sensory ataxia or positive Romberg’s test.
Diagnosed with or tested for spinocerebellar ataxia or Friedreich’s ataxia.
Any tremor, including head tremor, limb tremor or tremor not otherwise specified.
BAEP, brainstem auditory evoked potentials; CK, plasma creatine kinase; FRDA, Friedreich’s ataxia; MEP, motor evoked potentials; SCA, spinocerebellar ataxia; VEP, visual evoked potentials.
Values in brackets indicate percentages.
Phenotype-genotype associations
To date, a total of 173 individuals with NEFL-related CMT have been reported as clinically examined.8–18,24,25,28–39,45–47 The majority developed a classical CMT phenotype but some of them had atypical features including hearing loss, tremor, cerebellar features, and pyramidal tract signs (tables 3 and 4).
Table 4.
Selection of additional clinical features associated with ≥2 NEFL mutations
| Delayed walking/motor milestones | P8L, E90K, N98S, E210*, E396K |
| Disease onset≤3 years | P8L, P22R, E90K, N98S/T, E210*, E396K |
| Hearing loss | E90K, N98S/T, L268P, C322_N326del, E396K |
| Waddling gait | N98S, E140*, E396K |
| Facial weakness | N98S, Y265C, Q332P |
| Raised CK levels | N98S, Q332P, E396K |
| Tremor | P8L, P8R, N98S, L311P, C322_N326del |
| Ataxia | P8R, P22S, N98S, L311P, C322_N326del, Y389C, E396K |
| Diagnosed as SCA/FRDA | P8R, N98S, L311P, C322_N326del, E396K |
| Nystagmus/abnormal pursuits | P8R, N98S |
| Cerebellar atrophy | N98S, E396K |
| Dysarthria | P8R, N98S, E396K |
| Pyramidal tract signs | Y265C, Y389C, E396K, F439I, P440L |
| Scoliosis/hyperlordosis | P8R, N98S |
Abnormal pursuits, broken-up pursuit eye movements; ataxia, limb or gait ataxia, sensory ataxia, episodic ataxia or ataxia not otherwise specified; CK, creatine kinase; FRDA, Friedreich’s ataxia; SCA, spinocerebellar ataxia; tremor, head or limb tremor, or tremor not otherwise specified.
Early onset with delayed motor milestones
Twenty-five patients from 18 kindred had onset of symptoms at or before age 3 years.10,13,14,18,32,35,36,38,39,47 Of them, 18 patients were reported as having delayed walking or motor milestones with or without additional features such as hearing loss (11 patients), hypotonia (8 patients), intellectual impairment (3 patients), growth retardation (1 patient), or optic nerve hypoplasia (1 patient). Four dominant and one recessive mutation were associated with both early-onset and motor delay (P8L, E90K, N98S, E210* and E396K). With one exception, all patients with substitutions of amino acids E90 and N98 (E90K and N98S/T) had onset at age ≤3 years.
Pyramidal tract signs
A variable combination of brisk reflexes, extensor plantar responses and lower limb spasticity (sometimes simply described as upper motor neuron involvement), with or without evidence of prolonged central motor conduction times, has been observed in 13 patients from 7 kindreds.14,16,17,31,38,45 Except for the mutation Y265C, all mutations associated with pyramidal tract signs were located in end portion of the rod domain (Y389C and E396K) and the tail subdomain A (F439I and P440L).
Cerebellar features
Limb or gait ataxia (sensory, cerebellar, episodic or described as gait ataxia or wide-based gait) was reported in 22 patients but distinction between sensory or cerebellar ataxia based on the available data was not always possible.11,14–18,28–30 Seven patients carrying the mutations P8R, N98S, L311P, C322_N326del and E396K were initially diagnosed with or tested genetically for spinocerebellar ataxia or Friedreich’s ataxia, four of them with dysarthria and/or nystagmus. Cerebellar atrophy on brain MRI was observed in 4 patients with the mutations N98S and E396K (table 3).14,18,47
Raised creatine kinase (CK) levels and proximal weakness
Raised CK levels up to 1414 IU/L have been detected in 14 patients carrying the mutations N98S, Q332P and E396K.17,37,39 Of these patients, six had an element of proximal weakness in the upper or lower limbs or a waddling gait. This gait pattern is also described in patients with early-onset disease due to N98S and E396K.14 Myopathic features on EMG or muscle biopsy, however, have only been reported in two individuals with the E396K mutation and in one family with a myopathic-neurogenic phenotype carrying the mutation R421*.39,40
Screening for additional mutations
Based on the observation that patients with NEFL mutations may develop CNS features, we retrospectively reviewed whole-exome sequencing data from patients with pure or complicated hereditary spastic paraplegia (27 cases), early-onset ataxia (100 cases), and complex CNS syndromes (75 cases with a variable combination of ataxia, spasticity, epilepsy, extrapyramidal features and cognitive involvement), searching for rare or novel NEFL variants. Two heterozygous missense NEFL variants were identified: Q537R (c.1610A>G, rs377121179) and L336V (c.1006C>G, rs551896980). Both variants are present in population databases (minor allele frequency of 0.03% and 0.02%, respectively, in the EXaC dataset) and predicted to be tolerated or benign by SIFT and PolyPhen2. No other rare variants in NEFL were found.
DISCUSSION
In this study, we have reported five new cases of NEFL-related CMT and performed an analysis of the entire genetic and clinical spectrum of the disease based on data from the literature.
We observed that NEFL mutations are clustered in terms of frequency and location in six regions of the NFL protein, predominantly in the initial segment of the head domain, the junction between the head and rod domains and the end portion of the rod domain. The pathogenic relevance of these three regions is also supported by in vitro and animal studies. Substitution of amino acids P8 and P22 of NFL disrupts its ability to self-assemble into a filamentous network,49–52 affects the axonal transport of neurofilaments and mitochondria51,53,54 and impairs motor neuron viability in vitro.55 Transient transfection studies also indicate that T21Afs*83, E90K and N98S result in NFL assembly defects.12,50 In addition, mice models of the mutations N98S and E396K, located at the two ends of the rod domain, display an abnormal hindlimb posture together with muscle atrophy and reduced MNCV in the hNFLE396K mice and tremor in the NeflN98S/+ mice.56,57
Mutations affecting three other regions of NFL (coil 2A, mid portion of coil 2B and tail subdomain A) were less common. Six mutations involved the mid region of coil 2B, including Q332P, which has been detected in two families and causes similar defects in neurofilament assembly and transport as P8R in vitro.49,51,53 For four mutations located in this region (L333P, Q334P, L336P and L311P reported in this study), however, there was no co-segregation or functional data available. Three mutations were located in the tail subdomain A but co-segregation data was not reported for the family with CMT and R421* or for F439I, which has a minor allele frequency of 0.01%. Therefore, further research is needed to determine if these regions of NFL are mutational hotspots.
NEFL-related CMT presented with a broad clinical and neurophysiological phenotype. The mode of transmission was usually autosomal dominant although 40% of kindreds in our study were classified as sporadic or isolated cases and de novo mutations were confirmed in eight of them. Disease onset varied between the first and sixth decades of life, with 13% of patients having an early-onset presentation (≤3 years) often associated with delayed walking or motor milestones. Neurophysiologically, NEFL-related CMT was reported as axonal, demyelinating or mixed/intermediate in 40%, 31% and 29% of kindreds, respectively; descriptions were not uniform across the literature, but our analysis of published data also suggests that neuropathies with MNCV in the demyelinating or intermediate ranges are overall more frequent than those with upper limb MNCV >38 m/s. Although the reduced velocities may well be mainly explained by preferential loss of large-myelinated fibres confirmed in 21 sural nerve biopsies, additional features suggestive of demyelination and remyelination (onion bulbs and/or thin myelin sheaths) were observed in 67% of them.
Establishing definite genotype-phenotype correlations in patients with NEFL-related neuropathies is difficult given the significant number of private mutations and low prevalence of the disease. Pyramidal tract signs might be preferentially associated with mutations located towards the end of the NFL protein (end of rod domain and tail subdomain A) but this should be confirmed in future studies. Ataxia was described in association with the four common mutations (P8R, P22S, N98S and E396K) and raised CK levels was more frequently described in association with the mutation E396K.
We identified, however, an important clinical association with the mutations E90K and N98S (n = 14). Most patients with these mutations were isolated or sporadic (71%), six with confirmed de novo mutations (three from our cohort); presented before age 3 (93%) with delayed walking or motor milestones (86%); had all reported upper limb MNCV in the demyelinating range (86%); and developed additional features such as hearing loss (64%), ataxia with or without nystagmus or dysarthria (36%), hypotonia (29%) and less frequently intellectual impairment, tongue hemiatrophy, signs of vestibular dysfunction and optic nerve hypoplasia; cerebellar atrophy on brain MRI was confirmed in three cases. Patients carrying E90K and N98S, therefore, presented with a rather uniform syndrome for which we propose the acronym “DECADE” (demyelinating neuropathy based on distal upper limb MNCV with cerebellar signs and/or ataxia, deafness and delayed motor milestones), which also highlights the early onset of the disease. Although only six out of 14 patients conformed to the full syndrome, its recognition may aid in the differential diagnosis with other autosomal recessive cerebellar ataxias.18 Of note, NFL E90K or N98S mutants share a particular phenotype in vitro, different from that observed with substitutions of amino acid P8,50 and NeflN98S/+ mice display an early-onset phenotype with tremor and pathological changes in the CNS including neurofilament aggregates in the spinal cord and cerebellum.56
Beyond the above genotype-phenotype associations, identification of uncommon clinical features such as hearing loss, pyramidal tract signs or ataxia may be useful in the differential diagnosis of NEFL-related CMT with other forms of CMT. In our cohort of 202 patients with CNS syndromes we found no other potential NEFL mutations, which supports that these are unlikely to present with a predominantly CNS phenotype.
We acknowledge this study’s limitations related to the amount, completeness and heterogeneity of the data in the literature. Missing data may indeed reflect true negative data (i.e., absence of a clinical feature), false negative data due to lack of assessment or unreported data. Also, cases with atypical clinical characteristics may be overrepresented in the literature because of their particular interest. We believe, however, that the results of the present study may be helpful in the evaluation of novel NEFL sequence variants and the differential diagnosis with other CMT subtypes. Also, our study highlights potential aspects of interest to be evaluated in future studies.
Supplementary Material
ABBREVIATIONS
- CMAP
compound muscle action potential
- CMT
Charcot-Marie-Tooth disease
- CMT1
Charcot-Marie-Tooth disease type 1
- CMT2
Charcot-Marie-Tooth disease type 2
- CK
creatine kinase
- ExAC
Exome Aggregation Consortium
- MNCV
motor nerve conduction velocity
- NFL
neurofilament light polypeptide
References
- 1.Reilly MM, Rossor AM. Charcot-Marie-Tooth disease. In: Hilton-Jones D, Turner MR, editors. Neuromuscul Disord. 1. Oxford: Oxford University Press; 2014. pp. 61–74. [Google Scholar]
- 2.Nicholson G, Myers S. Intermediate forms of Charcot-Marie-Tooth neuropathy: a review. Neuromolecular Med. 2006;8:123–30. doi: 10.1385/nmm:8:1-2:123. [DOI] [PubMed] [Google Scholar]
- 3.Jerath NU, Shy ME. Hereditary motor and sensory neuropathies: Understanding molecular pathogenesis could lead to future treatment strategies. Biochim Biophys Acta. 2015;1852:667–78. doi: 10.1016/j.bbadis.2014.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Yuan A, Rao MV, Veeranna, Nixon RA. Neurofilaments at a glance. J Cell Sci. 2012;125:3257–63. doi: 10.1242/jcs.104729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yates DM, Manser C, De Vos KJ, Shaw CE, McLoughlin DM, Miller CC. Neurofilament subunit (NFL) head domain phosphorylation regulates axonal transport of neurofilaments. Eur J Cell Biol. 2009;88:193–202. doi: 10.1016/j.ejcb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Heins S, Wong PC, Muller S, Goldie K, Cleveland DW, Aebi U. The rod domain of NF-L determines neurofilament architecture, whereas the end domains specify filament assembly and network formation. The Journal of cell biology. 1993;123:1517–33. doi: 10.1083/jcb.123.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao MV, Campbell J, Yuan A, et al. The neurofilament middle molecular mass subunit carboxyl-terminal tail domains is essential for the radial growth and cytoskeletal architecture of axons but not for regulating neurofilament transport rate. The Journal of cell biology. 2003;163:1021–31. doi: 10.1083/jcb.200308076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mersiyanova IV, Perepelov AV, Polyakov AV, et al. A new variant of Charcot-Marie-Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. Am J Hum Genet. 2000;67:37–46. doi: 10.1086/302962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Jonghe P, Mersivanova I, Nelis E, et al. Further evidence that neurofilament light chain gene mutations can cause Charcot-Marie-Tooth disease type 2E. Ann Neurol. 2001;49:245–9. doi: 10.1002/1531-8249(20010201)49:2<245::aid-ana45>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 10.Jordanova A, De Jonghe P, Boerkoel CF, et al. Mutations in the neurofilament light chain gene (NEFL) cause early onset severe Charcot-Marie-Tooth disease. Brain. 2003;126:590–7. doi: 10.1093/brain/awg059. [DOI] [PubMed] [Google Scholar]
- 11.Fabrizi GM, Cavallaro T, Angiari C, et al. Giant axon and neurofilament accumulation in Charcot-Marie-Tooth disease type 2E. Neurology. 2004;62:1429–31. doi: 10.1212/01.wnl.0000120664.07186.3c. [DOI] [PubMed] [Google Scholar]
- 12.Leung CL, Nagan N, Graham TH, Liem RK. A novel duplication/insertion mutation of NEFL in a patient with Charcot-Marie-Tooth disease. Am J Med Genet A. 2006;140:1021–5. doi: 10.1002/ajmg.a.31242. [DOI] [PubMed] [Google Scholar]
- 13.Yum SW, Zhang J, Mo K, Li J, Scherer SS. A novel recessive Nefl mutation causes a severe, early-onset axonal neuropathy. Ann Neurol. 2009;66:759–70. doi: 10.1002/ana.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abe A, Numakura C, Saito K, et al. Neurofilament light chain polypeptide gene mutations in Charcot-Marie-Tooth disease: nonsense mutation probably causes a recessive phenotype. J Hum Genet. 2009;54:94–7. doi: 10.1038/jhg.2008.13. [DOI] [PubMed] [Google Scholar]
- 15.Choi BO, Koo SK, Park MH, et al. Exome sequencing is an efficient tool for genetic screening of Charcot-Marie-Tooth disease. Hum Mutat. 2012;33:1610–5. doi: 10.1002/humu.22143. [DOI] [PubMed] [Google Scholar]
- 16.Hashiguchi A, Higuchi Y, Nomura M, et al. Neurofilament light mutation causes hereditary motor and sensory neuropathy with pyramidal signs. J Peripher Nerv Syst. 2014;19:311–6. doi: 10.1111/jns.12102. [DOI] [PubMed] [Google Scholar]
- 17.Berciano J, Garcia A, Peeters K, et al. NEFL E396K mutation is associated with a novel dominant intermediate Charcot-Marie-Tooth disease phenotype. J Neurol. 2015;262:1289–300. doi: 10.1007/s00415-015-7709-4. [DOI] [PubMed] [Google Scholar]
- 18.Berciano J, Peeters K, Garcia A, et al. NEFL N98S mutation: another cause of dominant intermediate Charcot-Marie-Tooth disease with heterogeneous early-onset phenotype. J Neurol. 2016;263:361–9. doi: 10.1007/s00415-015-7985-z. [DOI] [PubMed] [Google Scholar]
- 19.Fridman V, Bundy B, Reilly MM, et al. CMT subtypes and disease burden in patients enrolled in the Inherited Neuropathies Consortium natural history study: a cross-sectional analysis. Journal of neurology, neurosurgery, and psychiatry. 2015;86:873–8. doi: 10.1136/jnnp-2014-308826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudnik-Schoneborn S, Tolle D, Senderek J, et al. Diagnostic algorithms in Charcot-Marie-Tooth neuropathies: experiences from a German genetic laboratory on the basis of 1206 index patients. Clin Genet. 2016;89:34–43. doi: 10.1111/cge.12594. [DOI] [PubMed] [Google Scholar]
- 21.De Jonghe P, Jordanova AK. Charcot-Marie-Tooth Neuropathy Type 2E/1F. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews® [Internet] Seattle (WA): University of Washington, Seattle; 2004. Apr 1, [Updated 2011 Oct 27] [Google Scholar]
- 22.Murphy SM, Herrmann DN, McDermott MP, et al. Reliability of the CMT neuropathy score (second version) in Charcot-Marie-Tooth disease. Journal of the peripheral nervous system : JPNS. 2011;16:191–8. doi: 10.1111/j.1529-8027.2011.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–4. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 24.Georgiou DM, Zidar J, Korosec M, Middleton LT, Kyriakides T, Christodoulou K. A novel NF-L mutation Pro22Ser is associated with CMT2 in a large Slovenian family. Neurogenetics. 2002;4:93–6. doi: 10.1007/s10048-002-0138-4. [DOI] [PubMed] [Google Scholar]
- 25.Yoshihara T, Yamamoto M, Hattori N, et al. Identification of novel sequence variants in the neurofilament-light gene in a Japanese population: analysis of Charcot-Marie-Tooth disease patients and normal individuals. J Peripher Nerv Syst. 2002;7:221–4. doi: 10.1046/j.1529-8027.2002.02028.x. [DOI] [PubMed] [Google Scholar]
- 26.Lus G, Nelis E, Jordanova A, et al. Charcot-Marie-Tooth disease with giant axons: a clinicopathological and genetic entity. Neurology. 2003;61:988–90. doi: 10.1212/wnl.61.7.988. [DOI] [PubMed] [Google Scholar]
- 27.Choi BO, Lee MS, Shin SH, et al. Mutational analysis of PMP22, MPZ, GJB1, EGR2 and NEFL in Korean Charcot-Marie-Tooth neuropathy patients. Hum Mutat. 2004;24:185–6. doi: 10.1002/humu.9261. [DOI] [PubMed] [Google Scholar]
- 28.Züchner S, Vorgerd M, Sindern E, Schroder JM. The novel neurofilament light (NEFL) mutation Glu397Lys is associated with a clinically and morphologically heterogeneous type of Charcot-Marie-Tooth neuropathy. Neuromuscul Disord. 2004;14:147–57. doi: 10.1016/j.nmd.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Fabrizi GM, Cavallaro T, Angiari C, et al. Charcot-Marie-Tooth disease type 2E, a disorder of the cytoskeleton. Brain. 2007;130:394–403. doi: 10.1093/brain/awl284. [DOI] [PubMed] [Google Scholar]
- 30.Miltenberger-Miltenyi G, Janecke AR, Wanschitz JV, et al. Clinical and electrophysiological features in Charcot-Marie-Tooth disease with mutations in the NEFL gene. Arch Neurol. 2007;64:966–70. doi: 10.1001/archneur.64.7.966. [DOI] [PubMed] [Google Scholar]
- 31.Butinar D, Starr A, Zidar J, Koutsou P, Christodoulou K. Auditory nerve is affected in one of two different point mutations of the neurofilament light gene. Clin Neurophysiol. 2008;119:367–75. doi: 10.1016/j.clinph.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin JS, Chung KW, Cho SY, et al. NEFL Pro22Arg mutation in Charcot-Marie-Tooth disease type 1. J Hum Genet. 2008;53:936–40. doi: 10.1007/s10038-008-0333-8. [DOI] [PubMed] [Google Scholar]
- 33.Bhagavati S, Maccabee PJ, Xu W. The neurofilament light chain gene (NEFL) mutation Pro22Ser can be associated with mixed axonal and demyelinating neuropathy. J Clin Neurosci. 2009;16:830–1. doi: 10.1016/j.jocn.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 34.Benedetti S, Previtali SC, Coviello S, et al. Analyzing histopathological features of rare charcot-marie-tooth neuropathies to unravel their pathogenesis. Arch Neurol. 2010;67:1498–505. doi: 10.1001/archneurol.2010.303. [DOI] [PubMed] [Google Scholar]
- 35.Baets J, Deconinck T, De Vriendt E, et al. Genetic spectrum of hereditary neuropathies with onset in the first year of life. Brain. 2011;134:2664–76. doi: 10.1093/brain/awr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin KP, Soong BW, Yang CC, et al. The mutational spectrum in a cohort of Charcot-Marie-Tooth disease type 2 among the Han Chinese in Taiwan. PLoS ONE. 2011;6:e29393. doi: 10.1371/journal.pone.0029393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlotter-Weigel B, Garcia-Angarita N, Rautenstrauss B. 20 Kongress des Wissenschaftlichen Beirates der Deutschen Gesellschaft für Muskelkranke eV. Ulm, Germany: 2011. Mutation in the neurofilament light chain gene (NEFL) associated with facial diplegia and marked elevation of creatine kinase. [Google Scholar]
- 38.Sivera R, Sevilla T, Vilchez JJ, et al. Charcot-Marie-Tooth disease: genetic and clinical spectrum in a Spanish clinical series. Neurology. 2013;81:1617–25. doi: 10.1212/WNL.0b013e3182a9f56a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elbracht M, Senderek J, Schara U, et al. Clinical and morphological variability of the E396K mutation in the neurofilament light chain gene in patients with Charcot-Marie- Tooth disease type 2E. Clin Neuropathol. 2014;33:335–43. doi: 10.5414/NP300742. [DOI] [PubMed] [Google Scholar]
- 40.Agrawal PB, Joshi M, Marinakis NS, et al. Expanding the phenotype associated with the NEFL mutation: neuromuscular disease in a family with overlapping myopathic and neurogenic findings. JAMA Neurol. 2014;71:1413–20. doi: 10.1001/jamaneurol.2014.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DiVincenzo C, Elzinga CD, Medeiros AC, et al. The allelic spectrum of Charcot-Marie-Tooth disease in over 17,000 individuals with neuropathy. Mol Genet Genomic Med. 2014;2:522–9. doi: 10.1002/mgg3.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manganelli F, Tozza S, Pisciotta C, et al. Charcot-Marie-Tooth disease: frequency of genetic subtypes in a Southern Italy population. J Peripher Nerv Syst. 2014;19:292–8. doi: 10.1111/jns.12092. [DOI] [PubMed] [Google Scholar]
- 43.Noto Y, Shiga K, Tsuji Y, et al. Nerve ultrasound depicts peripheral nerve enlargement in patients with genetically distinct Charcot-Marie-Tooth disease. J Neurol Neurosurg Psychiatry. 2015;86:378–84. doi: 10.1136/jnnp-2014-308211. [DOI] [PubMed] [Google Scholar]
- 44.Bird TD. Charcot-Marie-Tooth Neuropathy Type 1. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews® [Internet] Seattle (WA): University of Washington, Seattle; 1998. Aug 31, [Updated 2015 Mar 26] [Google Scholar]
- 45.Drew AP, Zhu D, Kidambi A, et al. Improved inherited peripheral neuropathy genetic diagnosis by whole-exome sequencing. Mol Genet Genomic Med. 2015;3:143–54. doi: 10.1002/mgg3.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pisciotta C, Bai Y, Brennan KM, et al. Reduced neurofilament expression in cutaneous nerve fibers of patients with CMT2E. Neurology. 2015;85:228–34. doi: 10.1212/WNL.0000000000001773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Gu LQ, Burnette WB, Li J. N98S mutation in NEFL gene is dominantly inherited with a phenotype of polyneuropathy and cerebellar atrophy. J Neurol Sci. 2016;365:46–7. doi: 10.1016/j.jns.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Werheid F, Azzedine H, Zwerenz E, et al. Underestimated associated features in CMT neuropathies: clinical indicators for the causative gene? Brain and behavior. 2016;6:e00451. doi: 10.1002/brb3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez-Olle R, Leung CL, Liem RK. Effects of Charcot-Marie-Tooth-linked mutations of the neurofilament light subunit on intermediate filament formation. J Cell Sci. 2002;115:4937–46. doi: 10.1242/jcs.00148. [DOI] [PubMed] [Google Scholar]
- 50.Perez-Olle R, Jones ST, Liem RK. Phenotypic analysis of neurofilament light gene mutations linked to Charcot-Marie-Tooth disease in cell culture models. Hum Mol Genet. 2004;13:2207–20. doi: 10.1093/hmg/ddh236. [DOI] [PubMed] [Google Scholar]
- 51.Perez-Olle R, Lopez-Toledano MA, Goryunov D, et al. Mutations in the neurofilament light gene linked to Charcot-Marie-Tooth disease cause defects in transport. J Neurochem. 2005;93:861–74. doi: 10.1111/j.1471-4159.2005.03095.x. [DOI] [PubMed] [Google Scholar]
- 52.Sasaki T, Gotow T, Shiozaki M, et al. Aggregate formation and phosphorylation of neurofilament-L Pro22 Charcot-Marie-Tooth disease mutants. Hum Mol Genet. 2006;15:943–52. doi: 10.1093/hmg/ddl011. [DOI] [PubMed] [Google Scholar]
- 53.Brownlees J, Ackerley S, Grierson AJ, et al. Charcot-Marie-Tooth disease neurofilament mutations disrupt neurofilament assembly and axonal transport. Hum Mol Genet. 2002;11:2837–44. doi: 10.1093/hmg/11.23.2837. [DOI] [PubMed] [Google Scholar]
- 54.Gentil BJ, Minotti S, Beange M, Baloh RH, Julien JP, Durham HD. Normal role of the low-molecular-weight neurofilament protein in mitochondrial dynamics and disruption in Charcot-Marie-Tooth disease. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:1194–203. doi: 10.1096/fj.11-196345. [DOI] [PubMed] [Google Scholar]
- 55.Zhai J, Lin H, Julien JP, Schlaepfer WW. Disruption of neurofilament network with aggregation of light neurofilament protein: a common pathway leading to motor neuron degeneration due to Charcot-Marie-Tooth disease-linked mutations in NFL and HSPB1. Hum Mol Genet. 2007;16:3103–16. doi: 10.1093/hmg/ddm272. [DOI] [PubMed] [Google Scholar]
- 56.Adebola AA, Di Castri T, He CZ, et al. Neurofilament light polypeptide gene N98S mutation in mice leads to neurofilament network abnormalities and a Charcot-Marie-Tooth Type 2E phenotype. Hum Mol Genet. 2015;24:2163–74. doi: 10.1093/hmg/ddu736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen H, Barry DM, Dale JM, Garcia VB, Calcutt NA, Garcia ML. Muscle pathology without severe nerve pathology in a new mouse model of Charcot-Marie-Tooth disease type 2E. Hum Mol Genet. 2011;20:2535–48. doi: 10.1093/hmg/ddr152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.