Abstract
Blood vessels in the mammalian skeletal system control bone formation and support haematopoiesis by generating local niche environments. While a specialized capillary subtype, termed type H, has been recently shown to couple angiogenesis and osteogenesis in adolescent, adult and ageing mice, little is known about the formation of specific endothelial cell populations during early developmental endochondral bone formation. Here, we report that embryonic and early postnatal long bone contains a specialized endothelial cell subtype, termed type E, which strongly supports osteoblast lineage cells and later gives rise to other endothelial cell subpopulations. The differentiation and functional properties of bone endothelial cells require cell-matrix signalling interactions. Loss of endothelial integrin β1 leads to endothelial cell differentiation defects and impaired postnatal bone growth, which is, in part, phenocopied by endothelial cell-specific laminin α5 mutants. Our work outlines fundamental principles of vessel formation and endothelial cell differentiation in the developing skeletal system.
Introduction
The skeletal system grows rapidly in embryonic and postnatal life, which requires tightly coordinated cell proliferation, differentiation and mineralization processes1,2 together with a substantial expansion of the local vasculature. Chondrocytes and bone-forming osteoblasts release vascular endothelial growth factor (VEGF) and stimulate angiogenesis through the activation of VEGF receptors in endothelial cells (ECs)3–6. Likewise, bone repair involves angiogenesis and osteoblast precursors enter fracture lesions along with invading blood vessels7. In addition to their essential transport function, vascular ECs release paracrine acting signalling factors that control growth and regeneration in various organs8–12. In the skeletal system, osteogenesis has been associated with a specific capillary EC subtype, termed type H, which shows high expression of the markers CD31/PECAM1 and Endomucin (CD31hi Emcnhi) and is found in the metaphysis and endosteum of postnatal long bone11,13. In addition to mediating angiogenic growth, type H ECs provide molecular signals acting on osteoprogenitor cells and thereby couple angiogenesis and osteogenesis. By contrast, type L (CD31lo Emcnlo) ECs, characterized by relatively low CD31 and Emcn expression, form the bone marrow sinusoidal vessel network and are not associated with osteoprogenitors expressing the transcription factor Osterix (Osx)11,13. Interestingly, impairment of the function of bone-degrading osteoclasts by cathepsin K (CTSK) inhibitors, a treatment that might help to prevent bone loss in osteoporosis and other disease conditions, led to an increase of CD31hi Emcnhi capillaries in mice, arguing that type H vessels might have therapeutic relevance14.
Extracellular matrix (ECM) molecules promote mineralization and regulate the behaviour of osteoblasts and of bone-degrading osteoclasts15–17. Cell-matrix interactions are frequently mediated by integrin receptors, composed of α and β subunits, that can bind a wide range of ECM proteins but also soluble factors and cell surface proteins18,19. Integrin β1, a subunit that can partner with 12 different α chains, is an important regulator of EC function. EC-specific inactivation of Itgb1, the gene encoding β1 integrin, is incompatible with embryonic angiogenesis and survival beyond midgestation20–22. Inducible genetic strategies have shown that integrin β1 controls arteriolar lumen formation23 as well as postnatal vessel growth and endothelial barrier function24.
While these previous studies have focused on well-established models of embryonic and postnatal angiogenesis, insight into endothelial cell-matrix interactions in the developing skeletal system is lacking. In addition, it is not known how different EC subtypes are specified during bone morphogenesis. By combining high-resolution confocal imaging, RNA sequencing and mouse genetics, we provide insight into these important questions and outline fundamental principles of vessel growth and EC specialization.
Results
Developmental vascularization of long bone
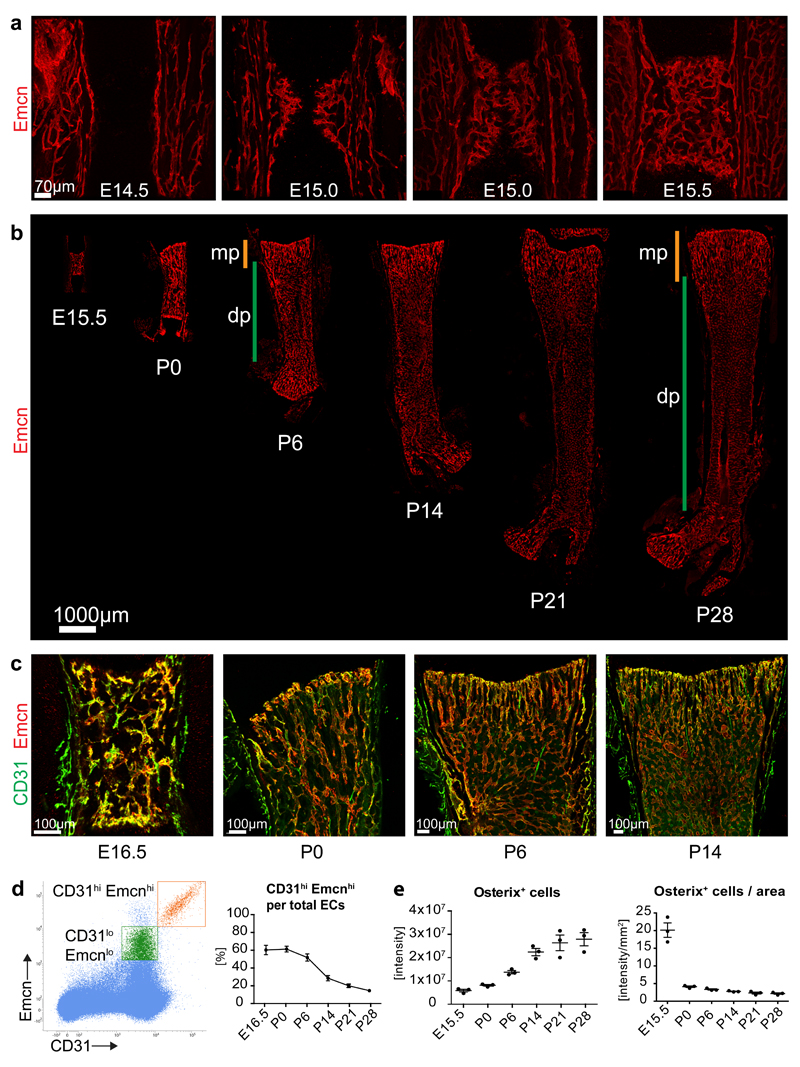
As reported previously3,5,7,25, vascularization of murine femur is initiated at embryonic day (E) 14.5. In a process resembling sprouting angiogenesis (Fig. 1a; Supplementary Fig. 1a), blood vessels entered the initially avascular cartilage containing hypertrophic chondrocytes and by E15.5 the primary ossification centre contained a highly branched vessel plexus (Fig. 1a, Supplementary Fig. 1a). Later, long bones expanded both longitudinally and radially, which was accompanied by expansion of the local vasculature (Figure 1b). From postnatal day (P) 6 onwards, morphologically distinct capillary populations became visible in the metaphysis and diaphysis (Figure 1b; Supplementary Fig. 1b). Metaphyseal capillaries displayed a column-like structure13, while the diaphysis contained a dense sinusoidal network surrounded by hematopoietic cells (Figure 1c; Supplementary Fig. 1b). Metaphyseal and diaphyseal capillary beds were interconnected by a transition zone, which contained sprouts and thin endothelial connections indicative of vascular remodelling (Supplementary Fig. 1b). As previously shown for 4-week-old long bone13, metaphyseal but not diaphyseal capillary ECs showed high expression of CD31 and Emcn at P0, P6 and P14 (Fig. 1c).
Figure 1. Developmental angiogenesis and EC subtypes in long bone.
a, Maximum intensity projections of confocal images showing Emcn-stained (red) ECs in E14.5, E15.0 and E15.5 femur. Note invading EC sprouts at E15.0 and establishment of a plexus at E15.5.
b, Confocal tile scans of Emcn-immunostained (red) E15.5, P0, P6, P14, P21 and P28 femur. Orange bars mark metaphysis (mp), green bars indicate diaphysis (dp).
c, Confocal images of CD31 (green) and Emcn (red) immunostained wild-type femur at the indicated stages. Note strong CD31 signal in ECs near postnatal growth plate and throughout the E16.5 vessel plexus.
d, Representative flow cytometry dot plot showing CD31hi Emcnhi (orange) and CD31lo Emcnlo ECs (green) in total bone marrow cells at P21. Gate settings exclude cells with very low Emcn expression (namely CD31+ hematopoietic cells and arterial ECs) from the analysis. Panel on the right shows the ratio of CD31hi Emcnhi among total (sum of CD31hi Emcnhi and CD31lo Emcnlo) ECs at the indicated developmental stages. Data represents mean± s.e.m. (n=3 individual experiments).
e, Quantitation of Osterix immunostaining signal intensity (artificial units) within the primary ossification centre, as measured in confocal tile scan images at given developmental stages (left). Graph on the right shows Osterix signal normalised to area of the primary ossification centre. Data represents mean± s.e.m. (n=3 individual mice per stage). Statistics source data are shown in Supplementary Table 6.
Previous work has indicated that type H EC abundance declines during adult life13. Flow cytometry analysis of long bone showed that the Emcnhi CD31hi EC fraction was highest around birth and gradually decreased during the first 4 weeks postnatally (Fig. 1d). During the same period, Osx+ cells expanded rapidly indicating bone growth (Fig. 1e; Supplementary Fig. 1c).
Characterization of a third bone EC subpopulation
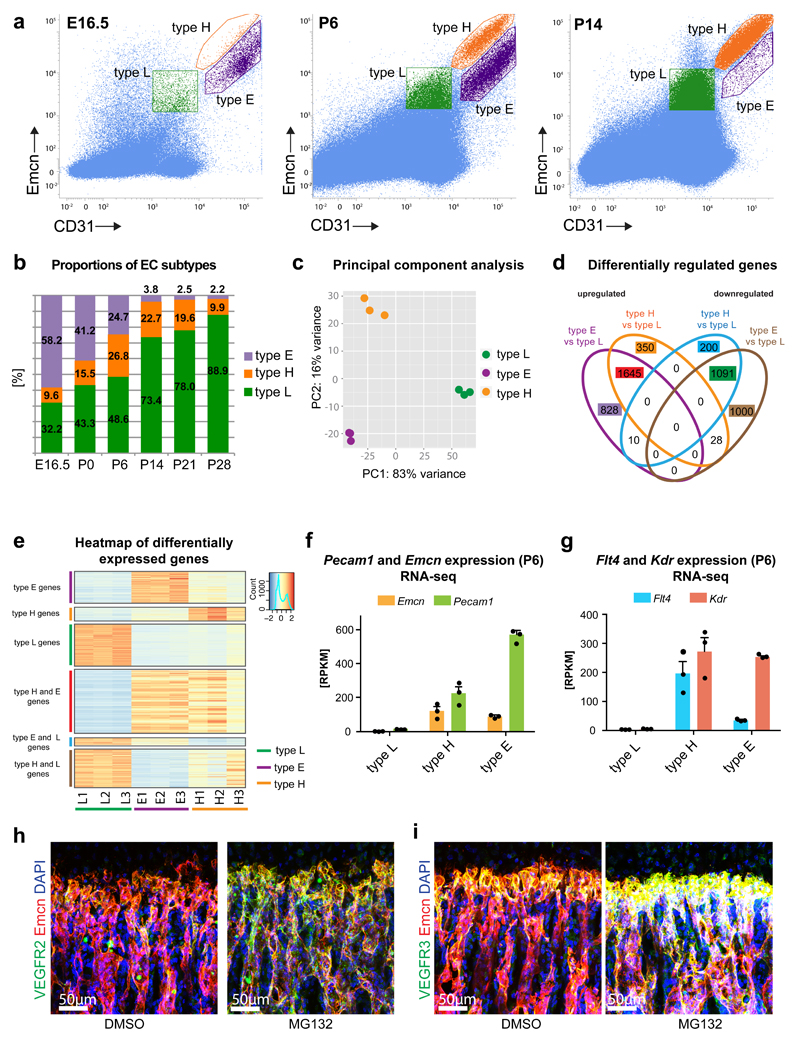
Flow cytometry analyses using the same gate settings as established for P21 (Fig. 1d) revealed the existence of a third, previously not described subpopulation of bone ECs (Fig. 2a). Emcnhi CD31hi clusters from E16.5, P6 and P14 femur were found in two distinct locations in two-dimensional plots and argued for a separate EC subset in addition to type H endothelium (Fig. 2a). We termed this population type E because of its high abundance in embryonic long bone at E16.5, while type H cells were comparably rare at this stage (Fig. 2b). Type E EC abundance decreased postnatally to 3.8% at P14 and 2.2% at P28, respectively. The fraction of type H ECs initially increased during early postnatal life, peaked at P6 and was already in decline at P14. In accordance with the high abundance of sinusoidal (type L) endothelium in adult bone13, Emcnlo CD31lo ECs increased continuously throughout all postnatal stages analysed and represented the major population after P6 (Fig. 2b).
Figure 2. Identification of a distinct EC subpopulation in early bone development.
a, Representative flow cytometry dot plots showing EC subsets with distinct expression of CD31 and Emcn in E16.5, P6 and P14 bone marrow. Gates indicating type L (green), type H (orange) and type E (purple) ECs are marked.
b, Bar chart showing proportions of EC subpopulations at different developmental stages.
c, Principal component analysis of RNA-sequencing data using the top 500 most variable genes across the samples. The first principal component (PC1) explained 83% of all variance; the second principal component (PC2) showed 16% of the variance between samples.
d, Venn diagram displaying the overlap of the significantly upregulated and downregulated genes between different EC populations in wild-type long bone at P6 identified by RNA-seq. FDR-adjusted p-value < 0.01 and absolute log2 fold change > 1.
e, Heat map illustration of gene expression values for differentially expressed genes in bone-derived type E, type H and type L ECs at P6. Each heat map displays data for the different sets of upregulated (red) and downregulated genes (blue) in Fig 2d. The data represented correspond to the normalized read counts scaled by row. Columns display data for each of the three replicates per cell subtype.
f, Expression of endothelial markers Pecam1 and Emcn in RNA sequencing samples. RPKM, reads per kilobase per million mapped reads. Data represents mean± s.e.m. (n=3 independent experiments).
g, RNA-seq-based relative expression levels of Kdr/Vegfr2 and Flt4/Vegfr3 transcripts in endothelial subpopulations at P6. Data represents mean± s.e.m. (n=3 independent experiments).
Statistics source data are shown in Supplementary Table 6.
h, i, Immunostaining for VEGFR2 or VEGFR3 (green) and Emcn (red) in sections of P21 wild-type femur after treatment with vehicle control (DMSO) or proteasome inhibitor (MG132) for 3 hours. MG132 strongly increased VEGF receptor levels in type H vessel columns. Nuclei, DAPI (blue).
To gain insight into their molecular properties, type H, E and L ECs were sorted by flow cytometry from P6 bones in triplicate. Principal component analysis of RNA-sequencing samples showed low variation within each sample group, while sample clustering indicated distinct profiles of individual EC populations (Fig. 2c). Expression profiles of type E and H ECs were more similar to each other than to type L. Differential gene expression analysis for every possible pairwise combination of samples, with a FDR-adjusted p-value < 0.01 and an absolute log2 fold change > 1 identified 3324 differentially expressed genes for type H vs. L, 4602 for type E vs. L, and 1619 for type E vs. H (Fig. 2d; Supplementary Fig. 2a). A high proportion of genes (1645) was up-regulated in both E and H subtypes relative to type L cells, while 828 and 350 genes were specifically higher in type E and H, respectively (Fig. 2e). RNA-sequencing results were confirmed by RT-qPCR for a panel of 14 genes. R2-values of 0.98 for type H vs. L, 0.93 for type E vs. L, and 0.90 for type E vs. H indicate high correlation of results (Supplementary Fig. 2b). RNA-seq results for Emcn and Pecam1 confirmed enrichment of these markers in type H and E ECs relative to type L (Fig. 2f). Consistent with the flow cytometry data, type E cells showed higher Pecam1 and lower Emcn transcription than type H ECs (Fig. 2a, f).
Analysis of selected markers showed high expression of Vegfr2/Kdr and Vegfr3/Flt4 in type H and E ECs relative to type L endothelium (Fig. 2g). Both genes encode VEGF receptors and regulate angiogenesis in numerous settings26–30. Surprisingly, anti-VEGFR2 immunostaining labelled column-shaped type H vessels less than the adjacent type L network (Supplementary Fig. 2c). VEGFR3 protein was also low on type H columns, whereas the most distal type H vessels next to the growth plate and the sinusoidal network were strongly stained (Supplementary Fig. 2d). As VEGF receptors undergo rapid turnover in the growing retinal vasculature31, mice were injected with the proteasome inhibitor MG132, which limits the availability of ubiquitin and interferes with protein degradation32. MG132 led to elevated VEGFR2 and VEGFR3 staining of type H vessel columns, whereas changes in diaphyseal vessels were comparably modest (Fig. 2h, i; Supplementary Fig. 2e, f). This validated the corresponding RNA-seq and RT-qPCR results and indicated high turnover and low steady state levels of VEGF receptors in type H vessels.
Functional properties of type E ECs
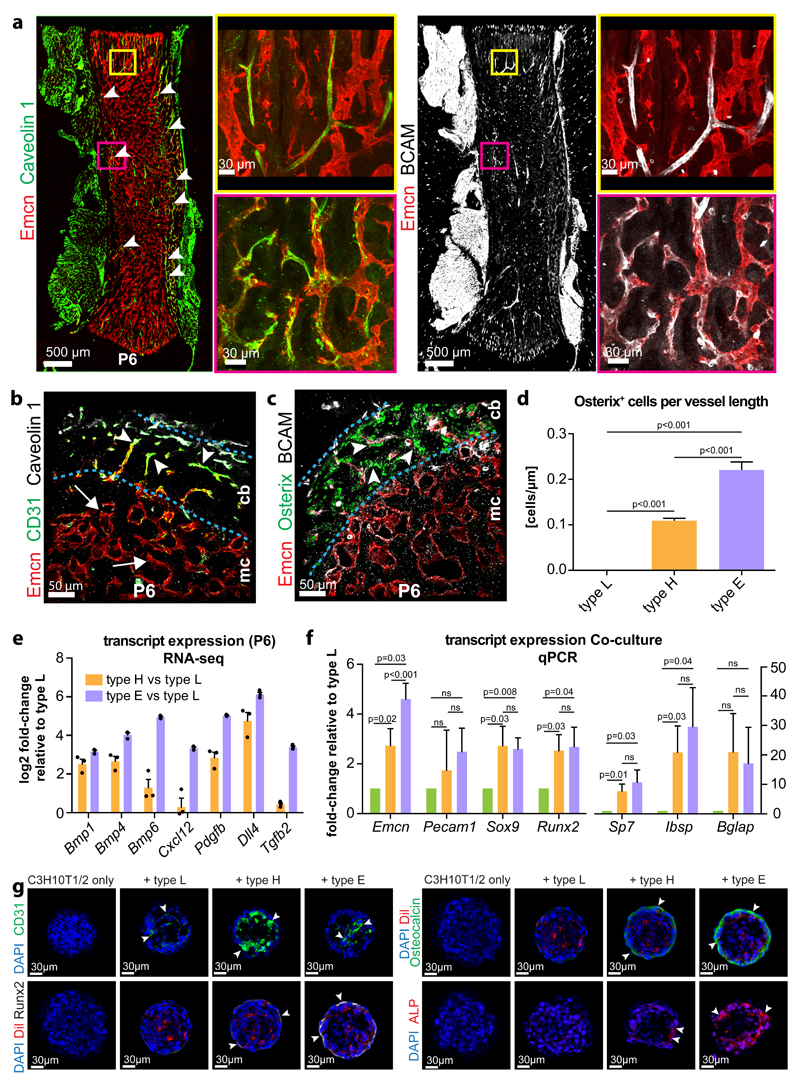
Further analysis and validation of RNA-seq results enabled the identification of markers that were expressed at high levels in type E endothelium relative to other EC subtypes, including Esm1, Kitl, Unc5b, Bcam, Cav1, and Apln (Supplementary Fig. 3a). In P6 femur, the type E-enriched markers Caveolin 1 and BCAM/CD239/Lutheran blood group glycoprotein, a specific receptor for laminin 511 in blood cells33, showed expression in Emcn-negative arterioles, which therefore did not correspond to type E ECs, as well as in Emcn+ capillaries in compact bone and endosteum (Fig. 3a-c). In E16.5 femur, most capillaries were CD31+ (Fig. 1c) and a fraction also showed expression of Caveolin 1 and BCAM (Supplementary Fig. 3b). Cross sections through P6 femur showed Caveolin 1+ or BCAM+ vessels in the endosteum and the adjacent compact bone (Fig 3b, c; Supplementary Fig. 3c). Interestingly, type E vessels were even more strongly associated with Osx+ osteoprogenitors than type H capillaries (Fig. 3c, d), which we attribute to high expression of bone morphogenetic proteins (BMPs) and of other factors controlling angiogenesis and osteogenesis (Fig. 3e). When co-cultured with murine mesenchymal C3H10T1/2 cells34, freshly isolated type H or type E ECs induced remodelling of cell spheroids and expression of osteoblast lineage markers (Fig. 3f, g; Supplementary Fig. 3d, e). In contrast, spheroids consisting of C3H10T1/2 cells and type L ECs lacked osteoblast lineage differentiation (Fig. 3f, g; Supplementary Fig. 3d, e).
Figure 3. Characterization of EC subpopulations in developing long bone.
a, Immunostaining for Emcn (red) and the type E markers Caveolin 1 (green) and BCAM (white) in P6 femur. Panels on the right show higher magnification of corresponding insets.
b, c, Transverse sections through P6 femur showing CD31 (green) and Caveolin 1-positive (white) vessels in compact bone (cb) (arrowheads) (b). By contrast, the bone marrow cavity (mc) contains Emcn-positive vessels with low CD31 signal (arrow). BCAM-positive vessels (white) in compact bone were associated with Osterix-expressing osteoprogenitors (green) (c).
d, Quantitation of Osterix positive cells in proximity to type L vessels in the marrow cavity, metaphyseal type H vessels and to type E vessels in compact bone normalized to vessel length. Data represents mean± s.e.m. (n=6 mice), (p<0.001 between all compared groups, two-tailed unpaired t-test).
e, RNA-seq-based expression levels of the indicated transcripts relative to expression in type L in freshly isolated P6 EC subpopulations. Data based on RPKM values obtained from RNA-seq. Data represents mean± s.e.m. (n=3 independent experiments). Statistics source data are shown in Supplementary Table 6.
f, Transcript expression in EC-C3H10T1/2 spheroid co-cultures (at day 7). Transcripts encoding Sox9, Runx2, Osteopontin (Sp7), integrin binding sialoprotein (Ibsp), and Osteocalcin (Bglap) were upregulated in cultures containing primary type H or type E ECs. Data represents mean± s.e.m. (n=7 independent experiments), two-tailed unpaired t-test.
g, Immunostaining of EC-C3H10T1/2 spheroids after 7 days in culture. Signals for Runx2 (white), Osteocalcin (green) and Alkaline phosphatase (red) were upregulated by CD31hi (green) type H and type E ECs (arrowheads) but not in the presence of type L ECs. Nuclei, DAPI (blue). ECs were pre-labeled by DiI (red) at the onset of the experiments.
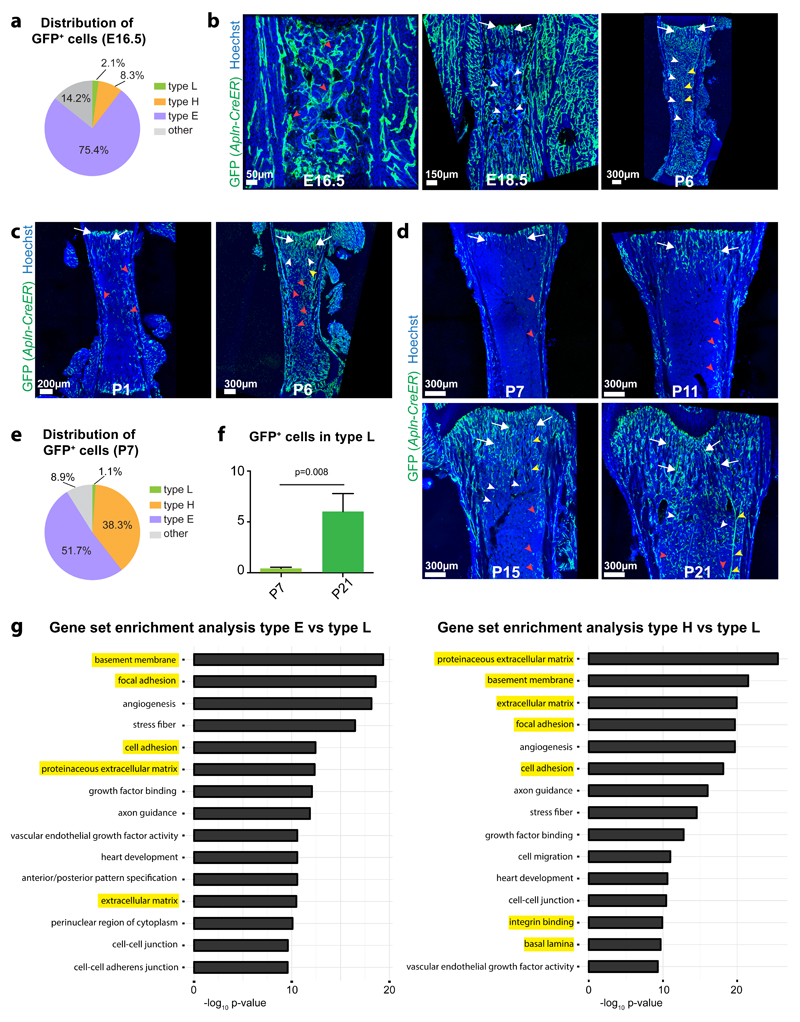
As Apelin (encoded by the Apln gene) was one of the markers enriched in type E ECs, we performed in vivo fate mapping with Apln-CreER knock-in mice35. Following a single administration of 4-hydroxy tamoxifen (4-OHT) at E15.5, Apln-CreER-induced expression of green fluorescent protein (GFP) under control of the R26-mT/mG Cre reporter36 labelled ECs throughout the E16.5 and E18.5 femur, which predominantly represented type E endothelium (Fig. 4a, b; Supplementary Fig. 4a). At P6, GFP+ ECs were highly abundant and had contributed to metaphyseal type H capillaries, type L sinusoids in the diaphysis and arteries (Fig. 4b). Following 4-OHT administration to Apln-CreER R26-mT/mG newborn animals (P0), GFP+ ECs were observed at P1 in type H capillaries near the growth plate and in endosteum and compact bone, i.e. in sites enriched in type E endothelium (Fig. 4c). At P6, GFP+ ECs were highly abundant in arteries and all capillary subtypes (Fig. 4c; Supplementary Fig. 4b). Induction of Apln-CreER R26-mT/mG animals with 4-OHT at P6 showed that labelled EC were initially concentrated in the distal metaphysis, endosteum and compact bone at P7, whereas a substantial expansion of the GFP+ endothelium into all capillary subtypes and arteries was seen at P11, P15 and P21 (Fig. 4d; Supplementary Fig. 4c). Flow cytometry of P6-injected Apln-CreER R26-mT/mG bone ECs at P7 showed enrichment of type E ECs in the GFP+ population, but in contrast to E16.5, the fraction of type H ECs was substantially increased (Fig. 4a, e). Analysis of P6-injected Apln-CreER R26-mT/mG mice at P7 and P21 indicated a significant increase in GFP+ type L ECs (Fig. 4f).
Figure 4. Hierarchy und molecular properties of bone capillary ECs.
a, e, Pie charts of the flow cytometry analysis of GFP+ cells in E16.5 (a) or P7 (e) long bone of Apln-CreER R26-mT/mG mice at 24 hours after 4-OHT administration. Note that the majority of labelled cells are type E ECs and only few GFP+ cells are type L ECs (n=5 mice for E16.5 and n=7 mice for P7).
b, c, d, Maximum intensity projections of tile scan confocal images showing GFP signal (green) in Apln-CreER R26-mT/mG femur at the indicated developmental stages after 4-OHT administration at E15.5 (b), P0 (c) or P6 (d). Nuclei, Hoechst (blue). GFP signal marks type E (red arrowheads) and a subset of type H ECs (arrows) one day after CreERT2 activation, whereas GFP+ ECs were seen in all capillary subpopulations (including type L, white arrowheads) and arteries (yellow arrowheads) at the respective later stages.
f, Flow cytometry analysis of GFP+ type L ECs in Apln-CreER R26-mT/mG mice at P7 and P21 after 4-OHT induction at P6. Note significant increase of labelled type L ECs. Data represents mean± s.e.m. (n=7 mice for P7 and n=8 mice for P21), (p=0.008, two-tailed unpaired t-test).
g, Gene set enrichment analysis for significantly differentially regulated genes in type E or type H ECs relative to type L ECs.
Together, these data show that type E vessels represent a spatially and temporally confined subpopulation of capillaries in developing bone. Characteristic features of type E ECs are their strong association with Osx+ cells and capacity to promote osteogenesis. Moreover, type E ECs can give rise to type H endothelium and both subtypes can differentiate into type L and arterial ECs during postnatal development.
Enrichment of cell-matrix adhesion molecules in type H and E ECs
To unravel the biological relevance of the RNA-seq dataset, we performed Generally Applicable Gene-set Enrichment for Pathway Analysis (GAGE)37. Altogether 78 gene sets displayed significant levels of upregulation in type E vs. L, while 76 gene sets were upregulated in type H vs. L ECs (q-value < 0.0001, Supplementary table 1, 2). Representation of the top 15 upregulated gene sets in type E vs. L and type H vs. L ECs showed a striking enrichment of extracellular matrix (ECM), basement membrane and cell adhesion components (Fig. 4g). In particular, RNA-seq and RT-qPCR data indicated high expression of Lama4 and, to a lower extent, Lama5 in type E and H ECs (Supplementary Fig. 4d). Both encode alpha subunits of heterotrimeric laminins that are central constituents of the endothelial basement membrane and control cell behaviour38. By contrast, Fn1 transcripts (encoding Fibronectin) were comparable in all three EC subtypes (Supplementary Fig. 4d). Transcripts for integrins were also enriched in type E and H ECs relative to type L (Supplementary Fig. 4e-g). Accordingly, strong vascular staining was seen for Laminin α4, Laminin α5, Fibronectin, and the integrin β1 subunit in P6 or P21 femoral metaphysis (Supplementary Fig. 4h-j). Anti-integrin β1 staining also decorated the type L endothelium of the diaphysis, whereas the interstitial matrix proteins Collagen type I and Osteopontin labelled trabecular bone in the metaphysis (Supplementary Fig. 4i, j).
The phenotype of bone ECs is controlled by endothelial integrin β1
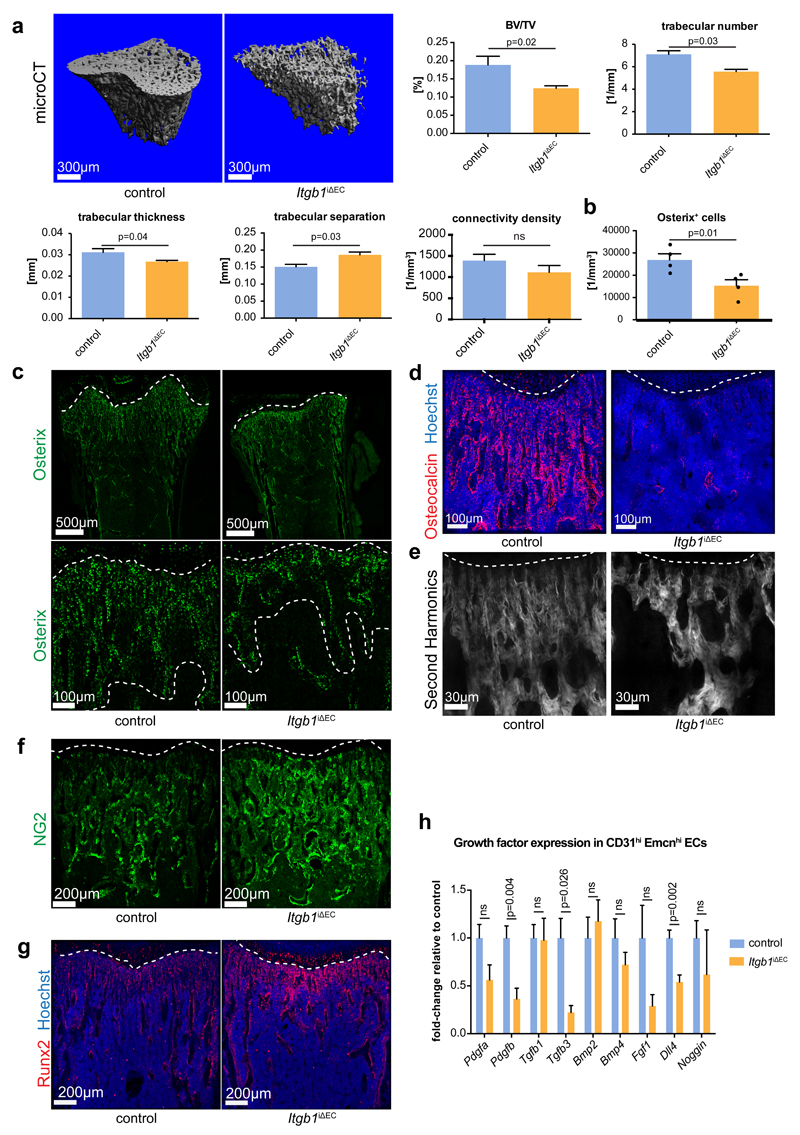
To analyse integrin function in the endothelium of developing bone, Cdh5(PAC)-CreERT2 transgenic animals39 were interbred with mice carrying loxP-flanked alleles of Itgb140. Cdh5(PAC)-CreERT2 activity is strictly EC-specific at P21 (Supplementary Fig. 5a, b) and all other stages investigated previously13. As tamoxifen administration during embryonic development led to rapid lethality of the resulting Itgb1iΔEC mutants24, only postnatal mutants generated by tamoxifen injection from P10-P12 were analysed. Efficient Itgb1 inactivation was confirmed by RT-qPCR of freshly isolated bone ECs at P21 (Supplementary Fig. 5c). 3 week-old Itgb1iΔEC mutants were smaller and had significantly lower body weight than control littermates (Supplementary Fig. 5d). Freshly isolated mutant femurs were 9% shorter than controls, displayed a dark red colour and contained a disorganized vasculature (Fig. 5a; Supplementary Fig. 5e, f). Type H vessels columns were much shorter, whereas the rest of the metaphyseal capillaries were highly branched and lacked a straight columnar organization (Fig. 5b, c). VEGFR3 immunostaining, with the distinct low steady state levels of the protein in type H EC columns (Supplementary Fig. 2d), highlighted the loss of Itgb1iΔEC columnar vessels (Fig. 5d, e). Reflecting the reduced expansion of Itgb1iΔEC long bone, the number of ECs was slightly but significantly increased despite reduced EC proliferation (Fig. 5f, g). Arteries, which connect to type H capillaries in metaphysis and endosteum13, were present both in Itgb1iΔEC and control femurs (Supplementary Fig. 5g).
Figure 5. Altered bone vasculature in EC-specific Itgb1 mutant mice.
a, Confocal images of 3 week-old Itgb1iΔEC and littermate control femurs stained for Emcn (red). Nuclei, Hoechst (blue).
b, High magnification images showing Emcn-immunostained vessel columns (marked by dashed lines) in the Itgb1iΔEC and control femoral metaphysis.
c, Number of branch points in Emcn-stained metaphyseal vessels. Data was normalized relative to 100% in control littermates and represents mean± s.e.m. (n=5 individual femurs per group), (p=0.02, two-tailed unpaired t-test).
d, Quantitation length of vessel area with low VEGFR3 immunostaining relative to total vessel area in Itgb1iΔEC and control femoral metapyhsis. Data represents mean± s.e.m. (n=3 individual femurs per group), (p=0.04, two-tailed unpaired t-test). Statistics source data are shown in Supplementary Table 6.
e, Confocal images of VEGFR3 (white) immunostaining in Itgb1iΔEC and control femoral sections. Dashed lines indicate upper and lower border of metaphysis.
f, g, Charts showing the flow cytometry analysis of total ECs (f) and proliferating (EdU+) cells per total ECs (g) number in P21 Itgb1iΔEC and Cre- littermate controls. Data in (f) was normalized relative to 100% in control littermates and represents mean± s.e.m. (n=16 mice), (p=0.03, two-tailed unpaired t-test). Data in (g) represents mean± s.e.m. (n=7 independent samples), (p=0.01, two-tailed unpaired t-test).
h, Flow cytometry analysis of CD31hi Emcnhi cells expressed as percent of total bone ECs. Data represents mean± s.e.m. (n=16 mice per group), (p=0.0004, two-tailed unpaired t-test).
i, Tile scan confocal overview pictures (top row) and magnified details of CD31-immunostained metaphyseal and diaphyseal vessels in 3 week-old Itgb1iΔEC and Cre-control femur. Note strong CD31 staining of Itgb1iΔEC diaphyseal vessels relative to control (arrowheads).
j, Flow cytometry analysis for EdU+ cells among total CD31hi Emcnhi ECs. Data represents mean± s.e.m. (n=7 mice per group), (p=0.003, two-tailed unpaired t-test).
k, Representative flow cytometry analyses of Itgb1iΔEC mice and littermate control bone marrow cells stained for Endomucin and CD31. Gates indicating type L (green), type H (orange) and type E (purple) ECs. Charts show quantification of type E ECs per total ECs in P21 Itgb1iΔEC relative to control bone. Data represents mean± s.e.m. (n=11 mice per group), (two-tailed unpaired t-test).
Flow cytometry analysis of Itgb1iΔEC femoral ECs showed an unexpected increase in CD31hi Emcnhi cells (Fig. 5h) and immunohistochemistry showed strong CD31 signal in diaphyseal sinusoidal vessels (Fig. 5i). Moreover, proliferation of mutant Emcnhi CD31hi ECs was strongly reduced, whereas no significant change was seen in the type L subset (Fig 5j; Supplementary Fig. 5h, i). Flow cytometry analysis of Itgb1iΔEC bone ECs indicated that the increase in Emcnhi CD31hi cells was also in part attributable to expansion of the type E subset (Fig. 5k). Together, this indicated that the loss of endothelial integrin β1 leads to substantial phenotypic changes in bone ECs. Significantly more ECs showed elevated expression of the markers CD31 and Emcn but lacked the normal morphological and functional properties of type E or H endothelium. These changes were accompanied by alterations in regional metabolic properties. The non-hypoxic zone in the distal metaphysis13 was shortened, whereas phosphorylated mitogen activated protein kinase (phospho-Erk1/2), a marker of high metabolic activity, was decreased (Supplementary Fig. 5j, k).
Regulation of osteogenesis by endothelial integrin β1
Bone angiogenesis and CD31hi Emcnhi ECs are tightly coupled to osteogenesis7,13. Micro–computed tomography (µCT) analysis of the P21 Itgb1iΔEC tibial metaphysis uncovered profound structural defects in bone formation (Fig. 6a). Mutants displayed significantly reduced bone volume, trabeculae number and trabecular thickness, whereas trabecular separation was increased. Connectivity density of trabecular bone was slightly but not significantly decreased (Fig. 6a). Accordingly, the abundance of Osx+ cells was strongly reduced in EC-specific Itgb1 mutants (Fig. 6b, c). Osteocalcin, a marker for mature osteocytes, was also profoundly decreased in Itgb1iΔEC mice (Fig. 6d). Analysis of Collagen type I and Osteopontin, which are potential binding partners of integrin β1, revealed reduction of those proteins. Mutants displayed lower staining intensities than control samples and the length of trabecular segments was reduced, which was confirmed by visualization of matrix-rich structures by second harmonic generation imaging using multi-photon microscopy (Fig. 6e; Supplementary Fig. 6a-c). Furthermore, immunostaining for the proteoglycan NG2/Cspg4 and the transcription factor Runx2, markers for primitive mesenchymal cells and preosteoblasts, respectively, were enhanced in Itgb1iΔEC femurs relative to littermate controls suggesting defective maturation of osteoblast progenitors (Fig. 6f, g). Thus, postnatal loss of endothelial integrin β1 leads to profound bone formation defects, which are presumably caused by loss of the pro-osteogenic properties of CD31hi Emcnhi ECs and reduced expression of potential paracrine regulators11,13 such as Pdgfb, Tgfb3 and Dll4 (Fig. 6h). Arguing that bone defects not caused by increased osteoclast activity, Calcitonin receptor and TRAP (Tartrate-resistant acid phosphatase) staining were reduced in Itgb1iΔEC bone sections (Supplementary Fig. 6d, e). Serum levels of parathyroid hormone and calcitonin, hormones controlling bone remodelling, were not significantly changed (Supplementary Fig. 6f).
Figure 6. Bone defects in EC-specific Itgb1 mutant mice.
a, Representative 3D reconstruction from µCT measurements of tibial metaphysis of 3 week-old Itgb1iΔEC and littermate control mice. Diagrams represent bone parameters measured in µCT analyses: bone volume/total volume (BV/TV) in percentage, trabeculae number in 1 per millimeter, trabecular thickness in millimeters, trabecular separation in millimeters, and connectivity density in 1 per cubic millimeter. Data represent mean± s.e.m. (n=6 mice), (p-values determined by two-tailed unpaired t-test).
b, Quantitation of metaphyseal Osx+ cells in Itgb1iΔEC mutant and Cre-negative littermate bone sections. Data represents mean± s.e.m. (n=4 individual femurs), (p=0.01, two-tailed unpaired t-test). Statistics source data are shown in Supplementary Table 6.
c, Tile scan and high magnification confocal images of Itgb1iΔEC and littermate control femurs stained for Osterix (green). Dashed lines indicate borders of metaphysis to growth plate and marrow cavity, respectively.
d, Itgb1iΔEC and littermate control femurs stained for Osteocalcin (red) and counterstained with Hoechst (blue).
e, High magnification of 2-photon second harmonic generation signals (white) of sections of P21 Itgb1iΔEC and control femurs. Dashed line indicates adjacent growth plate (top).
f, g, Maximum intensity projection of femoral sections from Itgb1iΔEC mutants and littermate controls stained for NG2 (f, green) or Runx2 (g, red). Nuclei in (g), Hoechst (blue).
h, RT-qPCR analysis of growth factor transcripts in freshly sorted Itgb1iΔEC CD31hi Emcnhi (orange) and control ECs (blue). Data represents mean± s.e.m. (n=9 mice per group), (p-values, two-tailed unpaired t-test). ns, not significant.
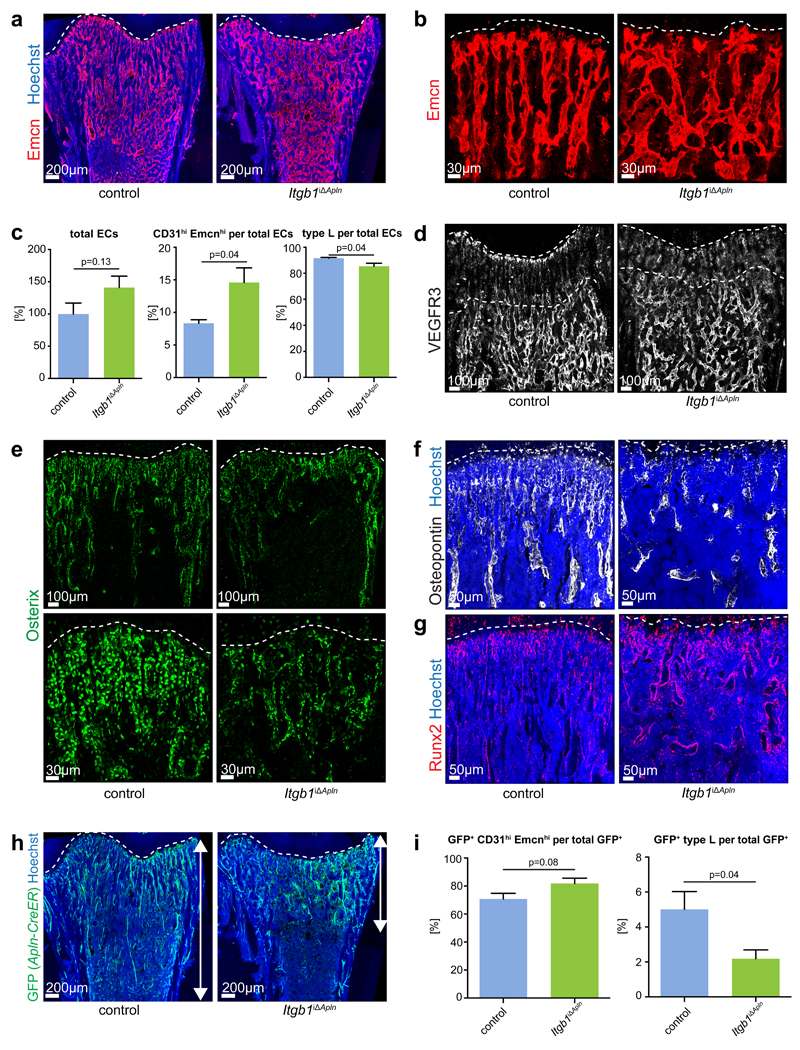
To address whether preferential gene inactivation in Emcnhi CD31hi ECs would reproduce defects seen in pan-endothelial mutants, Apln-CreER and Itgb1 conditional mice were interbred. Administration of tamoxifen from P10-P12 predominantly led to GFP expression in metaphyseal and endosteal type H ECs in Apln-CreER R26-mT/mG animals at P13, whereas no GFP signals were seen in the absence of tamoxifen (Supplementary Fig. 7a-c). As shown above, the initially labelled EC population expanded substantially and, by P21, GFP+ cells were seen in arteries, veins and throughout the different bone capillary networks (Supplementary Fig. 7d). Apln-CreER-controlled Itgb1iΔApln mutants generated with the same scheme of tamoxifen administration showed disorganization of the metaphyseal vasculature, disruption of type H capillary columns and the appearance of ectopic interconnections, shortening of the vessel region with low VEGFR3 immunostaining, and increases in total and CD31hi Emcnhi ECs with only minor changes in the type L subpopulation (Fig. 7a-d). In addition to these vascular alterations, Itgb1iΔApln mutants also phenocopied the reduction in Osx+ cells and osteopontin deposition, the increase in Runx2 preosteoblasts, and the reductions in body weight, femur length, mineralized bone and TRAP staining seen in pan-endothelial Itgb1iΔEC animals (Fig. 7e-g; Supplementary Fig. 7e-i). GFP+ cells, generated by the activation of the R26-mT/mG reporter in the Itgb1iΔApln background, were confined to the metaphysis and showed defective expansion into the diaphysis (Fig. 7h). Moreover, the abundance of type L ECs in the GFP-labelled population was significantly reduced in Itgb1iΔApln mutants (Fig. 7i) arguing that integrin β1 is required for the expansion of CD31hi Emcnhi ECs and their conversion into type L ECs.
Figure 7. Defects in Apln-CreER-generated Itgb1 mutants.
a, Confocal images of 3 week-old Itgb1iΔApln and littermate control femurs stained for Emcn (red). Nuclei, Hoechst (blue). Dashed line marks growth plate.
b, High magnification images showing Emcn-immunostained Itgb1iΔApln and control femoral metaphyseal vessels. Dashed line marks growth plate.
c, Charts of flow cytometry analysis of total ECs relative to 100% in control littermates, CD31hi Emcnhi cells per total ECs and type L cells per total ECs in P21 Itgb1iΔApln and Cre- littermate controls. Data represent mean± s.e.m. (n=6 mice per group), (p=0.13 for total ECs, p=0.04 for CD31hi Emcnhi cells and p=0.04 for type L ECs, two-tailed unpaired t-test).
d, Confocal images of VEGFR3 (white) immunostaining in Itgb1iΔApln and control femoral sections. Dashed lines indicate upper and lower border of metaphysis.
e, Overview and high magnification confocal images of Itgb1iΔApln and littermate control femurs stained for Osterix (green). Dashed lines indicate growth plate.
f, g, Confocal images of P21 Itgb1iΔApln and control femoral sections stained for Osteopontin (f, white) and Runx2 (g, red). Nuclei, Hoechst (blue). Dashed line marks growth plate.
h, Maximum intensity projections of tile scan confocal images showing Apln-CreER R26-mT/mG-generated GFP signal (green) in Itgb1iΔApln and littermate control femurs.
i, Bar charts showing ratio of CD31hi Emcnhi ECs and type L ECs in total GFP+ (Apln-CreER R26-mT/mG) cells in Itgb1Apln+ littermate control bone (n=6 mice per group), (p=0.08 for CD31hi Emcnhi ECs and p=0.04 for type L ECs, two-tailed unpaired t-test).
Role of matrix molecules in the specification of bone ECs
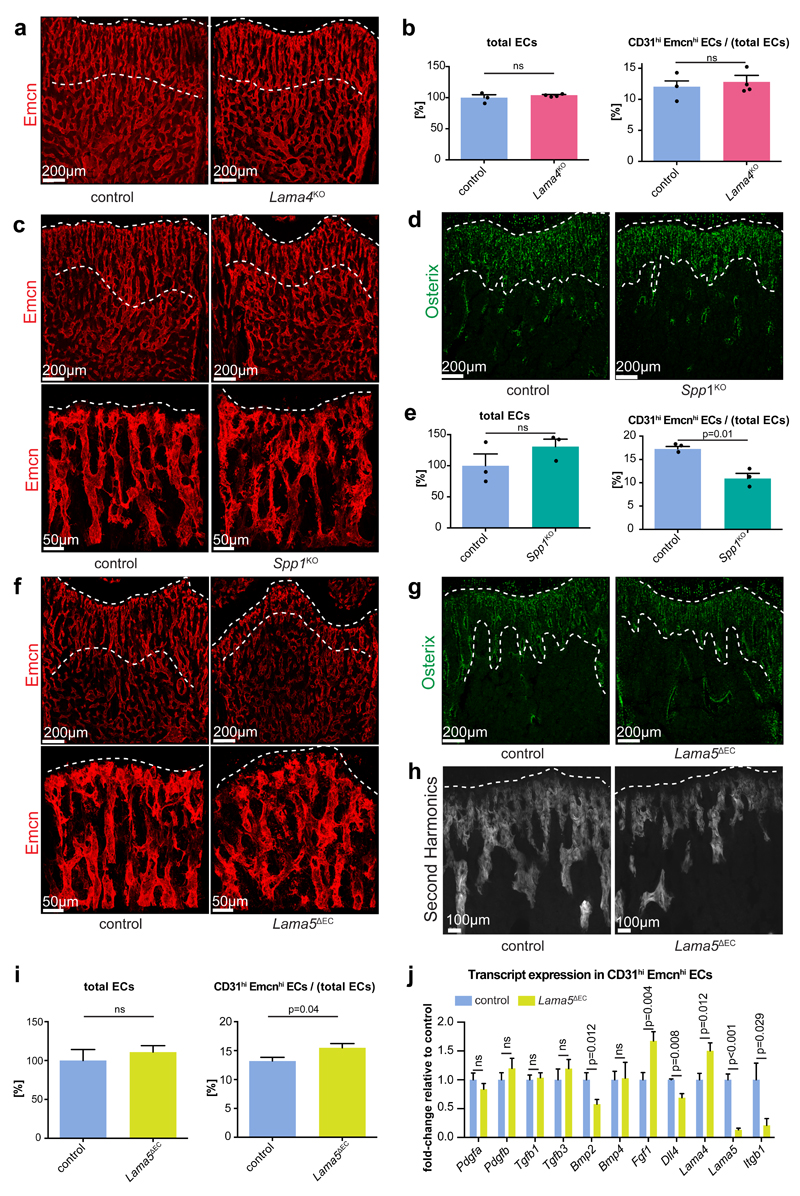
Given the numerous important roles of matrix proteins in bone formation15–17, we wanted to identify ECM components interacting with endothelial integrin β1. Global deletion of Lama4 did not result in any overt phenotype in P21 femur. Vascular organization was normal, the length of metaphyseal columnar vessels and Osx immunostaining were comparable to controls, and total and CD31hi Emcnhi bone ECs were not altered (Fig. 8a, b; Supplementary Fig. 8a-c).
Figure 8. Phenotypes of extracellular matrix mutants.
a, c, f, Overview and high magnification confocal images of Emcn-immunostained (red) femoral sections of 3 week-old Lama4KO (a), Spp1KO (c) and Lama5ΔEC (f) mice and respective controls, as indicated. Dashed lines indicate upper/lower border of column-like vessels in proximity of the growth plate (top).
b, e, i, Flow cytometry quantitation of total and fraction of CD31hi Emcnhi ECs in 3 week-old mutant and control bone. Data for total ECs was normalized to 100% (control) and represents mean± s.e.m. (b, n=4, p=0.49 for Lama4KO; e, n=3, p=0.25 for Spp1KO; i, n=7, p=0.72 for Lama5ΔEC mice). CD31hi Emcnhi data represent mean± s.e.m. (n=4, p=0.60 for Lama4KO; n=3, p=0.01 for Spp1KO; n=7, p=0.04 for Lama5ΔEC mice; two-tailed unpaired t-test; n represents number of mice per group). Statistics source data are shown in Supplementary Table 6.
d, g, Osterix (green) stained sections of Spp1KO (d) or Lama5ΔEC (g) femur and littermate controls. Dashed lines indicate upper and lower borders of trabecular region.
h, High magnification of 2-photon second harmonic generation signals (white) of thick sections (100µm) of P21 Lama5ΔEC and control femurs. Dashed line indicates adjacent growth plate.
j, RT-qPCR analysis of growth factor transcripts in freshly sorted Lama5ΔEC CD31hi Emcnhi (green) and control ECs (blue). Data represents mean± s.e.m. (n=10 mice per group), (p-values, two-tailed unpaired t-test). ns, not significant.
Osteopontin-deficient (Spp1KO) mice had weak defects resembling certain aspects of the Itgb1iΔEC phenotype. Metaphyseal vessel columns were shortened and branching between columns was increased, and Osx+ cells and the VEGFR3-low area within the metaphyseal vessel columns were slightly reduced (Figure 8c, d; Supplementary Fig. 8d). However, the total number of ECs was not significantly changed and the CD31hi Emcnhi subpopulation was decreased in Spp1KO long bone (Fig. 8e).
As global Lama5 knockout mice are embryonic lethal41, bone from constitutive EC-specific Lama5 mutants42 (Lama5ΔEC) were analysed, which showed defects resembling major aspects of Itgb1iΔEC mutants even though the overall phenotype was comparably milder. Vessel columns near the Lama5ΔEC growth plate were shortened, branching within the metaphyseal vasculature was increased, and the VEGFR3-low vessel area, Osx+ cells and femur length were reduced (Fig. 8f-h; Supplementary Fig. 8e-h). Mineralized bone, however, was not reduced significantly (Supplementary Fig. 8i, j). While the total number of Lama5ΔEC bone ECs was not substantially altered, the Emcnhi CD31hi fraction was significantly increased (Fig. 8i). The analysis of selected transcripts in freshly isolated ECs by RT-qPCR indicated strongly reduced expression of Lama5 and Itgb1 in the Lama5ΔEC bone endothelium (Fig. 8j). Expression of Lama4 and Fgf1 was increased, while levels of Bmp2 and Dll4 were slightly but significantly decreased.
These results indicate that disrupted cell-matrix interactions contribute, at least in part, to the defects in Itgb1 mutant bone. While it is likely that that several ECM molecules are relevant in this context, our data argue for roles of Osteopontin and the Laminin α5 subunit-containing Laminin 511 or 521 isoforms, which are produced by ECs43 and deposited in the subendothelial basement membrane.
Discussion
ECs are highly heterogeneous in terms of gene expression, morphology and functional specialization44,45. While differences between arterial and venous or lymphatic and blood vessel ECs are widely recognized, much less is known about the existence and specification of distinct capillary subsets. In kidney, glomerular capillaries are part of the renal ultrafiltration system, whereas peritubular capillaries surrounding nephron tubules participate in ion and mineral reabsorption46, 47. In adult brain, neurogenesis occurs in the adult subventricular zone and the hippocampal subgranule zone, which contain neural stem cells and vessels that are morphologically and functionally distinct from other brain capillaries48,49. Specific properties of distinct capillary subtypes are perhaps best documented for the skeletal system. Type H capillary ECs support osteogenesis, whereas type L ECs form the sinusoidal vessel network of the bone marrow cavity13. However, the molecular signatures of the different capillary EC subtypes and the processes controlling their specification have remained unknown. Our current study not only provides unbiased insight into the molecular properties of bone ECs but also establishes the existence of a third capillary EC subpopulation, termed type E, with high capacity to support perivascular Osx+ osteoprogenitors in embryonic and early postnatal bone. While type E cells predominate in early long bone, genetic lineage tracing supports the existence of type E-to-H and type H-to-L conversion processes, which accompany the rapid developmental expansion of bone and none marrow. Based on the sum of the findings presented here, we propose a developmental and functional hierarchy of bone ECs where type E cells occur upstream of type H and type L endothelium. Similar principles are likely to apply to other organ systems where, however, the identity, localization and functional roles of specific capillary EC subsets remain to be uncovered.
ECM molecules and cell-matrix receptors were particularly enriched in the type E and type H gene expression profiles. Integrin family cell-matrix receptors are well known for their important roles in a wide range of cell differentiation processes50,51. Our data establish that the β1 subunit is essential for the normal specification and function of bone EC subpopulations. In addition to matrix, integrins can bind and signal in response to growth factors and other secreted molecules52–55. However, Itgb1iΔEC defects were partially phenocopied in Lama5ΔEC and, to a lesser extent, in Spp1 global knockout mice, which argues for a role of cell-matrix signalling in bone EC specification. A caveat of the genetic approaches in our study is that they affect ECs throughout the body and might therefore potentially influence processes in the skeletal system indirectly. Nevertheless, the data presented here argue that integrin β1 and Laminin α5 are required in bone ECs in a cell-autonomous and direct fashion. Thus, our findings raise the possibility that changes in the matrix environment of bone, which are the result of congenital defects and other pathological conditions in human patients56–58, might potentially lead to disease-promoting alterations in the vasculature. Our identification and characterization of distinct, functionally specialized capillary EC subsets might facilitate the characterization of disease-associated changes in the future.
Methods
Mouse models
C57BL/6J mice were used for the analysis of wild-type bones. For embryonic stages, females were checked for presence of a vaginal plug in the morning.
Lineage tracing experiments were performed by mating Apln-CreER mice59 and R26-mG/mT reporter animals36. Cre activity and GFP expression were induced by intraperitoneal injection of 4-hydroxy tamoxifen (4-OHT) (H7904, Sigma). Induction was performed at E15.5 (750µg 4-OHT/female), P0 (15µg 4-OHT/pup) and P6 (25µg 4-OHT/pup). For postnatal analysis after embryonic induction, cesarean sections were performed at E19.5 and pups were raised by foster mothers. As the Apln-CreER knock-in allele disrupts the function of the X-linked Apln gene59, only females were analyzed.
For inducible and EC-specific genetic experiments, Cdh5(PAC)-CreERT2 transgenics39 were interbred with mice carrying loxP-flanked Itgb1 (Itgb1lox/lox) alleles40. Cre activity and gene inactivation were induced by intraperitoneal injection of 200µg tamoxifen (T5648, Sigma) from postnatal day 10 to 12. At the age of 3 weeks, femurs and tibiae were collected from Itgb1lox/lox Cdh5(PAC)-CreERT2T/+ mice and Cre- negative Itgb1lox/lox littermate controls.
The specificity of Cdh5(PAC)-CreERT2 transgenic animals for ECs in long bone was determined by crossing Cdh5(PAC)-CreERT2 mice with R26-mT/mG36 reporter mice. Cre activation was induced from P10 to P12 and mice were analyzed at P21. Furthermore, the Apln-CreER was introduced into the R26-mT/mG reporter background for tracking of Apln+ ECs with the same induction and analysis scheme.
For gene inactivation in Apln+ ECs, Apln-CreER was bred into the Itgb1lox/lox background. Gene inactivation was induced by tamoxifen administration from P10 to P12 (200µg/pup/day). Long bones were collected at the age of 3 weeks from Itgb1lox/lox Apln-CreERT/+ (Itgb1iΔApln) mice and Cre-negative Itgb1lox/lox littermate controls. Furthermore, Itgb1iΔApln mice were interbred with R26-mT/mG reporters. The resulting Itgb1lox/lox R26-mT/mGlox/+ Apln-CreERT/+ and Itgb1+/+ R26-mT/mGlox/+ Apln-CreERT/+ mice were analyzed at 3 weeks of age after tamoxifen administration from P10 to P12.
The role of endothelial Lama5 was investigated by crossing mice carrying loxP-flanked Lama5 alleles42 with Tek-Cre transgenics60. In addition, global Lama4-/- 61 and Spp1-/- knockout mice62 were used. Genotypes of mice were determined by PCR.
All animal experiments were performed in compliance with the relevant laws and institutional guidelines, were approved by local animal ethics committees and were conducted at the University of Münster and the MPI for Molecular Biomedicine with permissions granted by the Landesamt für Natur, Umwelt und Verbraucherschutz (LANUV) of North Rhine-Westphalia. Animals were combined in groups for experiments irrespective of their sex. The investigators were not blinded to allocation during experiments and outcome assessment. Experiments were not randomized and no statistical method was used to predetermine sample size, which was, instead, chosen according to previous experiments.
Proteasome inhibition, EdU labelling and hypoxia analysis
Three week-old wildtype mice were injected intraperitoneally with 50µg/g MG132 (Millipore, #474790) in DMSO (Sigma, D8418) or with DMSO only as a vehicle control. Three hours after injection femurs were dissected and processed for immunostaining as described above.
Proliferating cells in vivo were labelled by intraperitoneal injection of 500µg of EdU (Invitrogen) 2 hours before the analysis. Detection of proliferating cells in fixed bone sections and in flow cytometry analysis was achieved with staining according to the manufacturer’s instructions (Invitrogen).
Hypoxic cells were detected with the hypoxia probe pimonidazole (Pimo, Hypoxyprobe Inc.) according to the manufacturer’s instructions. Mice were intraperitoneally injected with 60 mg/ kg Pimo 2 hours before analysis.
Bone immunohistochemistry
Murine femurs and tibiae were dissected, cleaned from adjacent muscles, immediately transferred into cold 2% paraformaldehyde solution and fixed overnight at 4°C under rotation. Next, bones were transferred into 0.5M EDTA solution and kept overnight at 4°C under rotation for decalcification. On the following day, EDTA was exchanged for 20% sucrose and 1% polyvinylpyrrolidone (PVP) solution. After 24 hours at 4°C under rotation, bones were embedded and frozen in 8% gelatin (porcine) with 15% sucrose and 1.5% PVP. Finally, samples were cut into 100µm sections using low-profile blades on a Leica CM3050 cryostat.
For immunofluorescence staining, sections were air-dried at room temperature, rehydrated in PBS and permeabilised with 0.3% Triton X-100 for 15 minutes at room temperature. After washing with PBS, blocking solution (5% donkey serum + 0.1% Triton X-100) was applied for 20 minutes at room temperature. Following another PBS wash, sections were treated with primary antibodies over night at 4°C. The next day, sections were washed four times with PBS and incubated with secondary antibodies for 1.5 hours at room temperature. After five wash steps with PBS, sections were mounted using FluoroMount-G (Southern Biotech). Finally, cover slips were sealed with nail polish.
The following primary antibodies were used: rat anti-Endomucin (sc-65495, Santa Cruz, diluted 1:100), goat anti-CD31 conjugated to Alexa Fluor (AF) 488 (FAB3628G, R&D Systems, 1:50), rabbit anti-Osterix (c-22536-R, Santa Cruz, 1:500), goat anti-VEGFR2 (AF644, R&D Systems, 1:50), goat anti-VEGFR3 (AF743, R&D Systems, 1:50), rabbit anti-Caveolin 1 (#3238, Cell Signaling, 1:50), goat anti-BCAM (AF8299, R&D Systems, 1:50), chicken anti-GFP (ab13970, Abcam, 1:500), rabbit anti-GFP conjugated to AF488 (A21311, Invitrogen, 1:250), goat anti-Integrin beta 1 (AF2405, R&D Systems, 1:50), rabbit anti-Laminin α4 (serum 377, 1:500), rat anti-Laminin α5 (clone 4G6, 1:10), rabbit anti-Fibronectin (F3648, Sigma, 1:100), rabbit anti-Collagen Type1 (AB765P, Millipore, 1:100), goat anti-Osteopontin (AF808, R&D Systems, 1:100), α-SMA-Cy3 (C6198, Sigma, 1:50), mouse anti-hypoxyprobe conjugated to fluorescein isothiocyanate (FITC) (#FITC-Mab, Hydroxyprobe, 1:50), rabbit anti-phosphoERK1/2 (D13.14.4E, Cell Signaling, 1:50), rabbit anti- Osteocalcin (LS-C17044, LifeSpan BioSciences, 1:300), rabbit anti-NG2 (AB5320, Millipore, 1:100), rat anti- Runx2 (MAB2006, R&D Systems, 1:50), rabbit anti-Calcitonin receptor (ab11042, Abcam , 1:50), rabbit anti-TRAP (sc-28204, Santa Cruz, 1:50).
The following secondary antibodies were used: donkey anti-rat coupled to Cy3 (712-165-150, Jackson ImmunoResearch, 1:250), donkey anti-rat coupled to AF594 (A21209, Invitrogen, 1:250), chicken anti-rat coupled to AF647 (A21472, Invitrogen, 1:350), donkey anti-goat coupled to AF647 (A21447, Invitrogen, 1:350), donkey anti-goat coupled to AF488 (A11055, Invitrogen, 1:350), donkey anti-rabbit coupled to AF647 (A31573, Invitrogen, 1:350), donkey anti-rabbit coupled to AF488 (A21206, Invitrogen, 1:350), donkey anti-rabbit coupled to AF546 (A10040, Invitrogen, 1:350), donkey anti-chicken coupled to AF488 (703-545-155, Jackson ImmunoResearch, 1:350). Nuclei were counterstained with Hoechst33342 (H3570, Invitrogen, 1:1000).
EdU labelling was performed according to the manufacturer’s instructions (Invitrogen).
Dissection, processing, sectioning, staining, imaging and analysis for phenotypes of mutant mice and controls were always performed together under identical conditions.
Image acquisition and quantitative analysis
Stained bone sections were analysed at high resolution with a Leica TCS SP5 confocal microscope. Z-stacks of images were processed and 3D reconstructed with Imaris software (version 7.00, Bitplane). Photoshop CS5 and Illustrator CS5 (Adobe) were used for image processing. Quantitations were performed with Volocity (Version 6.0.1, Perkin Elmer) on high resolution images.
For Fig. 1e, the left graph shows the sum of the Osterix immunosignal within the primary ossification centre, as measured with Volocity software. The graph on the right shows the sum of the Osterix immunosignal normalized to the area of the primary ossification centre.
All images shown are representative for the respective staining in several experiments. Within one experiment laser excitation and confocal scanner detection were the same.
Flow Cytometry
Tibiae and femurs were dissected from mice of interest and pooled. All adjacent tissue was removed thoroughly. After removal of the epiphysis, bones were crushed in ice-cold PBS using mortar and pestle. To obtain a single-cell suspension, samples were digested with collagenase for 20 minutes at 37°C and filtered. Subsequently, samples were treated with blocking solution (1% FCS) for 30 minutes, washed with PBS and immunostained with rat anti-Endomucin antibody (sc-65495, Santa Cruz, 1:50) for 45 minutes on ice. After washing, cells were stained with goat anti-CD31 coupled to AF488 or allophycocyanin (APC) antibody (FAB3628G or FAB3628A, R&D Systems, 1:10) and donkey anti-rat antibody coupled to phycoerythrin (PE) (712-116-153, Jackson Immunoresearch, 1:100) or donkey anti-rat antibody coupled to DyLight405 (712-475-150, Jackson Immunoresearch, 1:200) for 45 minutes on ice. After thorough washing, cells were treated with DAPI to exclude dead cells. Finally, cells were acquired on BD FACSVerse flow cytometer and analysed using BD FACSuite Software (Version 1.0.5.3841, BD Bioscience). Cell sorting was performed with a FACS Aria II.
Labelling of proliferating cells with EdU (Invitrogen) was done according to the manufacturer’s instructions.
RNA sequencing and data analysis
Femurs and tibiae of twenty wild-type P6 mice were pooled and sorted by flow cytometry for type H, type E and type L ECs. RNA from 120,000 cells from each group was isolated using the RNeasy Plus Micro Kit (QIAGEN) according to the manufacturer’s instructions. RNA quality was checked using a 2100 BioAnalyzer (Agilent). 100ng of RNA were used for preparation of sequencing libraries with the TruSeq Stranded Total RNA Library Prep Kit (Illumina) according to the manufacturer’s instructions. Libraries were validated using a BioAnalyzer and quantified by qPCR. Libraries were diluted to a final concentration of 15pM for sequencing. The MiSeq Reagent Kit v3 (Illumina) was used for sequencing with a MiSeq (Illumina). The experiment was performed in triplicates.
Quality assessment
The quality assessment of raw sequence data of endothelial cell subtypes H, E and L was performed using FastQC (Version: FastQC 0.11.3, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). No samples were discarded from the analysis.
Alignment to reference sequence
Paired-end sequence reads were mapped to the mm10 mouse genome assembly (GRCm38) using TopHat-2 with the following settings (Version: tophat-2.0.13; [tophat2 –g 2])63. The mouse genome was downloaded from the iGenome portal (https://support.illumina.com/sequencing/sequencing_software/igenome.html). HTSeq was used to count the aligned reads on a per gene basis using the following settings (Version: HTSeq-0.6.1; [htseq-count --mode=intersection-nonempty--stranded=reverse]) 64. RNA sequencing data were uploaded to ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) with the accession number E-MTAB-4066.
Differential gene expression analysis
The count data were normalized using the Variance Stabilizing Transformation (VST) function from the DESeq2 package [reference 11 should be cited here]. Principal Component Analysis (PCA) was performed on transformed read counts using the top 500 most variable genes to assess the overall similarity between the samples65, 66. Differential gene expression analysis between the H vs L, E vs L and E vs H groups was performed using DESeq2. Differentially expressed genes were selected using a FRD-adjusted p-value cut-off < 0.01 and an absolute log2 fold change > 1. Gene symbols were annotated using biomaRt (BioConductor version 3.1). Lists of differentially regulated genes for the different contrasts are provided in Supplementary Tables 3–5.
Functional analysis
Gene Set Enrichment Analysis was performed using Generally Applicable Gene-set Enrichment (GAGE; BioConductor version 3.1)37. For functional annotation, we used gene-sets from org.Mm.eg.db, a genome-wide annotation package for mouse (Bioconductor, version 3.1). The analysis was performed based on one-on-one comparisons between H vs L and E vs L subtypes where L was always used as a control dataset. For this analysis, read counts were normalized as described in the GAGE vignette (Bioconductor, version 3.1). Significant GO terms were selected using an FDR-adjusted p-value < 0.0001. Lists of enriched GO terms in up-regulated gene-sets are provided in Supplementary Tables 1–2.
Quantitative RT-PCR
RNA was isolated from flow cytometrically sorted bone marrow endothelial cells using the RNeasy Plus Micro Kit (QIAGEN) according to the manufacturer’s instructions. iScript cDNA Synthesis Kit (BIO- RAD) was used to transcribe 100ng RNA per reaction into cDNA. The quantitative PCR (qPCR) was performed on an ABI PRISM 7900HT Fast Real Time PCR System (Applied Biosystems). The following FAM-conjugated TaqMan gene expression probes were used in combination with TaqMan Gene Expression Master Mix (Life Technologies): Flt4 (Mm00433337_m1), Pecam1 (Mm01242584_m1), Efnb2 (Mm01215897_m1), Notch1 (Mm00435245_m1), Notch3 (Mm01345646_m1), Bcam (Mm01321454_g1), Itgb1 (Mm01253233_m1), Esm1 (MM00469953_M1), Kitl (Mm00442972_m1), Unc5b (Mm00504054_m1), Cav1 (Mm00483057_m1), Itga5 (Mm00439797_m1), Itgav (Mm00434506_m1), Lama4 (Mm01193660_m1), Lama5 (Mm01222051_g1), Pdgfa (Mm01205760_m1), Pdgfb (Mm01298578_m1), Tgfb1 (Mm01178820_m1), Tgfb3 (Mm00436960_m1), Bmp2 (Mm01340178_m1), Bmp4 (Mm00432087_m1), Fgf1 (Mm00438906_m1), Dll4 (Mm00444619_m1), Noggin (Mm01297833_s1), Sox9 (Mm00448840_m1), Runx2 (Mm00501584_m1), Sp7 (Mm04209856_m1), Ibsp(Mm00492555_m1), Bglap (Mm03413826_mH).
VIC-conjugated ActB(4352341E) TaqMan probe was used to normalize gene expression. Per group at least six mice from three independent litters were analysed to obtain the relative expression differences.
ELISAs
Serum levels of parathyroid hormone and calcitonin were measured by quantitative enzyme-linked immunosorbent assay (ELISA) kits (CEA866Mu and CEA472Mu, Cloud-Clone Corp.) in blood samples from mutant and control mice.
Second harmonic generation (SHG) imaging using multi-photon microscopy
A TriM Scope II multi photon system from LaVision BioTec was used for SHG imaging to visualise anisotropic structures, such as collagen fibres. The microscope setup is a single beam instrument with an upright Olympus BX51 WI microscope stand equipped with high sensitive non-descanned detectors close to the objective lens. The TriM Scope II is fitted with a Coherent Scientific Chameleon Ultra II Ti:Sapphire laser (tuning range 680-1080 nm) and a Coherent Chameleon Compact OPO (automated wavelength extension from 1000 nm to 1600 nm). A 20x IR objective lens (Olympus XLUMPlanFl 20x/1.0W) with a working distance of 2.0 mm was used. SHG signals were detected using TiSa light at 850 nm, a 420/40 band pass filter and a blue-sensitive photomultiplier (Hamamatsu H67080-01). 3D-images were acquired and processed with LaVision BioTec ImSpector Software under Microsoft Windows. Overview images were reconstructed from a sequence of tiled 3D images with Image J.
Micro Computed Tomography
Dissected tibiae were cleaned from adjacent tissue, fixed overnight in 4% PFA at 4°C and analysed using µCT. Measurements were carried out by Scanco Medical AG, Switzerland. The chosen voxel size was 6μm in all three spatial dimensions. For each sample, 248 slices were evaluated, covering a total of 1.488 mm, X-ray voltage was 70 kVp, Intensity 114 μA, Integration Time 300 ms, Frame Averaging 4.
Cell culture experiments
Mouse C3H10T1/2 cells (#CCL-226, ATCC) were cultured in Dulbecco’s modified Eagle’s medium (Sigma), supplemented with 10% FBS (PAA Laboratories), 2mM L-glutamine, 100 U/ml of penicillin, and 100 mg/ml streptomycin (Invitrogen). Cultured cells were maintained at 37°C in a humidified 5% CO2 atmosphere.
For co-culture spheroids, freshly sorted EC subpopulations were combined with C3H10T1/2 cells by the hanging-drop method. In brief, equal numbers of suspended C3H10T1/2 cells and sorted ECs or an equal number of only C3H10T1/2 cells (5,000 cells per spheroid) were suspended in spheroid-formation media consisting of DMEM containing 1 % FBS, 0.05 % (w/v) polyvinyl alcohol (PVA; Sigma-Aldrich), N2 supplement and 0.25 % (w/v) methylcellulose (Sigma-Aldrich; M-0512, 4000 c.p.). Drops of 17µl cell suspension were placed onto each position of a 96 well plate lid. The lids were flipped on and placed on non-adherent round-bottom 96-well plates, which were kept at 37°C in a humidified 5% CO2 incubator overnight to generate spheroids. Forming spheroids were seeded into each well with a plate spin, and then cultured for 7 days with spheroid culture media of DMEM containing 1 % FBS, 0.05 % (w/v) polyvinyl alcohol (PVA; Sigma-Aldrich), N2 supplement.
For immunostaining, spheroids were embedded into 2% low melting agarose gel
For easy detection of ECs in spheroids, each endothelial population was labelled with the lipophilic fluorescent dye CM-DiI (1μM; Invitrogen) for 10 min at 37°C, and then for an additional 20 minutes at 4°C. Cells were then washed with complete culture medium twice. DiI-labelled ECs were combined with C3H10T1/2 cells as described above.
ALP staining was performed after 7 days of culture in spheroid culture media. ALP activity was detected with Leukocyte Alkaline Phosphatase Kit (Sigma 86R) following manufacturer's instructions.
Cell lines used in this study were tested for mycoplasma contamination, were not authenticated and are not listed in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample.
Statistics and reproducibility
Statistical analysis was performed using Graphpad Prism software or the R statistical environment (http://r-project.org). All data are presented as mean± s.e.m. unless indicated otherwise. Unpaired two tailed student t-tests were used to determine statistical significance. P< 0.05 was considered significant unless stated otherwise. All experiments were performed independently at least three times (unless noted otherwise in the respective figure legend) and respective data were used for statistical analyses. Sample sizes for each experiment are described in the respective figure legends, and source data from independent replicates are provided in Supplementary Table 6. No animals were excluded from analysis.
Supplementary Material
Acknowledgements
We thank H.-W. Jeong for his support in RNA-seq experiments, K. Kato for guidance on cryosectioning, and M. Stehling for expert advice in flow cytometry experiments. The Max Planck Society, the University of Muenster, the Cells in Motion (CiM) graduate school, the DFG cluster of excellence ‘Cells in Motion’ (L.S., J.M.V. and R.H.A), and the European Research Council (AdG 339409 AngioBone) have supported this study.
Footnotes
Author Contributions
U.H.L., M.E.P., J.M.V. and R.H.A. designed the study. R.E.-G., A.S. and J.M.V. performed bioinformatics analyses, M.G.B. the 2-photon microscopy, K.K.S. the MG132 inhibition experiments, J.M.K. the spheroid assays, and A.P.K. the ELISA experiments. A.P.K. also developed the FACS protocol for the isolation of bone ECs. J.DiR. and L.S. provided Lama4 and 5 mutant tissues, B.Z. the Apln-CreER mice. All other experiments were performed by U.H.L.; U.H.L., J.M.V. and R.H.A. wrote the manuscript.
Author Information
The authors declare that they have no competing financial interests.
Data availability
RNA-sequencing data are available through the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-4066.
Source data for Fig. 1e, 2f, 2g, 3e, 5d, 6b, 8b, 8f and Supplementary Fig. 2b, 3a, 4d, 4e, 4f, 4g, 7h, 8j have been provided as Supplementary Table 6. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 2.Long F, Ornitz DM. Development of the endochondral skeleton. Cold Spring Harb Perspect Biol. 2013;5:a008334. doi: 10.1101/cshperspect.a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eshkar-Oren I, et al. The forming limb skeleton serves as a signaling center for limb vasculature patterning via regulation of Vegf. Development. 2009;136:1263–1272. doi: 10.1242/dev.034199. [DOI] [PubMed] [Google Scholar]
- 4.Conen KL, Nishimori S, Provot S, Kronenberg HM. The transcriptional cofactor Lbh regulates angiogenesis and endochondral bone formation during fetal bone development. Dev Biol. 2009;333:348–358. doi: 10.1016/j.ydbio.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maes C, et al. Impaired angiogenesis and endochondral bone formation in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188. Mech Dev. 2002;111:61–73. doi: 10.1016/s0925-4773(01)00601-3. [DOI] [PubMed] [Google Scholar]
- 6.Maes C, et al. Increased skeletal VEGF enhances beta-catenin activity and results in excessively ossified bones. EMBO J. 2010;29:424–441. doi: 10.1038/emboj.2009.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maes C, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding BS, et al. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature. 2014;505:97–102. doi: 10.1038/nature12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding BS, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding BS, et al. Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell. 2011;147:539–553. doi: 10.1016/j.cell.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramasamy SK, Kusumbe AP, Wang L, Adams RH. Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature. 2014;507:376–380. doi: 10.1038/nature13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J, et al. Endothelial cell-derived angiopoietin-2 controls liver regeneration as a spatiotemporal rheostat. Science. 2014;343:416–419. doi: 10.1126/science.1244880. [DOI] [PubMed] [Google Scholar]
- 13.Kusumbe AP, Ramasamy SK, Adams RH. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature. 2014;507:323–328. doi: 10.1038/nature13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie H, et al. PDGF-BB secreted by preosteoclasts induces angiogenesis during coupling with osteogenesis. Nat Med. 2014;20:1270–1278. doi: 10.1038/nm.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alford AI, Kozloff KM, Hankenson KD. Extracellular matrix networks in bone remodeling. Int J Biochem Cell Biol. 2015;65:20–31. doi: 10.1016/j.biocel.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Marie PJ, Hay E, Saidak Z. Integrin and cadherin signaling in bone: role and potential therapeutic targets. Trends Endocrinol Metab. 2014;25:567–575. doi: 10.1016/j.tem.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Olsen BR, Reginato AM, Wang W. Bone development. Annu Rev Cell Dev Biol. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- 18.Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–280. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei L, et al. Endothelial expression of beta1 integrin is required for embryonic vascular patterning and postnatal vascular remodeling. Mol Cell Biol. 2008;28:794–802. doi: 10.1128/MCB.00443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlson TR, Hu H, Braren R, Kim YH, Wang RA. Cell-autonomous requirement for beta1 integrin in endothelial cell adhesion, migration and survival during angiogenesis in mice. Development. 2008;135:2193–2202. doi: 10.1242/dev.016378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanjore H, Zeisberg EM, Gerami-Naini B, Kalluri R. Beta1 integrin expression on endothelial cells is required for angiogenesis but not for vasculogenesis. Dev Dyn. 2008;237:75–82. doi: 10.1002/dvdy.21385. [DOI] [PubMed] [Google Scholar]
- 23.Zovein AC, et al. Beta1 integrin establishes endothelial cell polarity and arteriolar lumen formation via a Par3-dependent mechanism. Dev Cell. 2010;18:39–51. doi: 10.1016/j.devcel.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto H, et al. Integrin beta1 controls VE-cadherin localization and blood vessel stability. Nat Commun. 2015;6:6429. doi: 10.1038/ncomms7429. [DOI] [PubMed] [Google Scholar]
- 25.Zelzer E, et al. Skeletal defects in VEGF(120/120) mice reveal multiple roles for VEGF in skeletogenesis. Development. 2002;129:1893–1904. doi: 10.1242/dev.129.8.1893. [DOI] [PubMed] [Google Scholar]
- 26.Tammela T, et al. VEGFR-3 controls tip to stalk conversion at vessel fusion sites by reinforcing Notch signalling. Nat Cell Biol. 2011;13:1202–1213. doi: 10.1038/ncb2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benedito R, et al. Notch-dependent VEGFR3 upregulation allows angiogenesis without VEGF-VEGFR2 signalling. Nature. 2012;484:110–114. doi: 10.1038/nature10908. [DOI] [PubMed] [Google Scholar]
- 28.Lohela M, Bry M, Tammela T, Alitalo K. VEGFs and receptors involved in angiogenesis versus lymphangiogenesis. Curr Opin Cell Biol. 2009;21:154–165. doi: 10.1016/j.ceb.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol. 2006;7:359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- 30.Zarkada G, Heinolainen K, Makinen T, Kubota Y, Alitalo K. VEGFR3 does not sustain retinal angiogenesis without VEGFR2. Proc Natl Acad Sci U S A. 2015;112:761–766. doi: 10.1073/pnas.1423278112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama M, et al. Spatial regulation of VEGF receptor endocytosis in angiogenesis. Nat Cell Biol. 2013;15:249–260. doi: 10.1038/ncb2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Latonen L, Taya Y, Laiho M. UV-radiation induces dose-dependent regulation of p53 response and modulates p53-HDM2 interaction in human fibroblasts. Oncogene. 2001;20:6784–6793. doi: 10.1038/sj.onc.1204883. [DOI] [PubMed] [Google Scholar]
- 33.Kikkawa Y, Miner JH. Review: Lutheran/B-CAM: a laminin receptor on red blood cells and in various tissues. Connect Tissue Res. 2005;46:193–199. doi: 10.1080/03008200500344074. [DOI] [PubMed] [Google Scholar]
- 34.Reznikoff CA, Brankow DW, Heidelberger C. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res. 1973;33:3231–3238. [PubMed] [Google Scholar]
- 35.Liu Q, et al. Genetic targeting of sprouting angiogenesis using Apln-CreER. Nat Commun. 2015;6:6020. doi: 10.1038/ncomms7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 37.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yousif LF, Di Russo J, Sorokin L. Laminin isoforms in endothelial and perivascular basement membranes. Cell Adh Migr. 2013;7:101–110. doi: 10.4161/cam.22680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 2010;465:483–486. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- 40.Raghavan S, Bauer C, Mundschau G, Li Q, Fuchs E. Conditional ablation of beta1 integrin in skin. Severe defects in epidermal proliferation, basement membrane formation, and hair follicle invagination. J Cell Biol. 2000;150:1149–1160. doi: 10.1083/jcb.150.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miner JH, Cunningham J, Sanes JR. Roles for laminin in embryogenesis: exencephaly, syndactyly, and placentopathy in mice lacking the laminin alpha5 chain. J Cell Biol. 1998;143:1713–1723. doi: 10.1083/jcb.143.6.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song J, et al. Extracellular matrix of secondary lymphoid organs impacts on B-cell fate and survival. Proc Natl Acad Sci U S A. 2013;110:E2915–2924. doi: 10.1073/pnas.1218131110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorokin LM, et al. Developmental regulation of the laminin alpha5 chain suggests a role in epithelial and endothelial cell maturation. Dev Biol. 1997;189:285–300. doi: 10.1006/dbio.1997.8668. [DOI] [PubMed] [Google Scholar]
- 44.Scoazec JY, Racine L, Couvelard A, Flejou JF, Feldmann G. Endothelial cell heterogeneity in the normal human liver acinus: in situ immunohistochemical demonstration. Liver. 1994;14:113–123. doi: 10.1111/j.1600-0676.1994.tb00059.x. [DOI] [PubMed] [Google Scholar]
- 45.Rajotte D, et al. Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. J Clin Invest. 1998;102:430–437. doi: 10.1172/JCI3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott RP, Quaggin SE. Review series: The cell biology of renal filtration. J Cell Biol. 2015;209:199–210. doi: 10.1083/jcb.201410017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molema G, Aird WC. Vascular heterogeneity in the kidney. Semin Nephrol. 2012;32:145–155. doi: 10.1016/j.semnephrol.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Palmer TD, Willhoite AR, Gage FH. Vascular niche for adult hippocampal neurogenesis. J Comp Neurol. 2000;425:479–494. doi: 10.1002/1096-9861(20001002)425:4<479::aid-cne2>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 49.Culver JC, Vadakkan TJ, Dickinson ME. A specialized microvascular domain in the mouse neural stem cell niche. PLoS One. 2013;8:e53546. doi: 10.1371/journal.pone.0053546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 51.Danen EH, Sonnenberg A. Integrins in regulation of tissue development and function. J Pathol. 2003;201:632–641. doi: 10.1002/path.1472. [DOI] [PubMed] [Google Scholar]
- 52.Felcht M, et al. Angiopoietin-2 differentially regulates angiogenesis through TIE2 and integrin signaling. J Clin Invest. 2012;122:1991–2005. doi: 10.1172/JCI58832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hakanpaa L, et al. Endothelial destabilization by angiopoietin-2 via integrin beta1 activation. Nat Commun. 2015;6:5962. doi: 10.1038/ncomms6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emre Y, Imhof BA. Matricellular protein CCN1/CYR61: a new player in inflammation and leukocyte trafficking. Semin Immunopathol. 2014;36:253–259. doi: 10.1007/s00281-014-0420-1. [DOI] [PubMed] [Google Scholar]
- 55.Ivaska J, Heino J. Cooperation between integrins and growth factor receptors in signaling and endocytosis. Annu Rev Cell Dev Biol. 2011;27:291–320. doi: 10.1146/annurev-cellbio-092910-154017. [DOI] [PubMed] [Google Scholar]
- 56.Forlino A, Cabral WA, Barnes AM, Marini JC. New perspectives on osteogenesis imperfecta. Nat Rev Endocrinol. 2011;7:540–557. doi: 10.1038/nrendo.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tosi LL, Warman ML. Mechanistic and therapeutic insights gained from studying rare skeletal diseases. Bone. 2015;76:67–75. doi: 10.1016/j.bone.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 58.Tranquilli Leali P, et al. Bone fragility: current reviews and clinical features. Clin Cases Miner Bone Metab. 2009;6:109–113. [PMC free article] [PubMed] [Google Scholar]
- 59.Tian X, et al. Subepicardial endothelial cells invade the embryonic ventricle wall to form coronary arteries. Cell Res. 2013;23:1075–1090. doi: 10.1038/cr.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kisanuki YY, et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 61.Thyboll J, et al. Deletion of the laminin alpha4 chain leads to impaired microvessel maturation. Mol Cell Biol. 2002;22:1194–1202. doi: 10.1128/MCB.22.4.1194-1202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liaw L, et al. Altered wound healing in mice lacking a functional osteopontin gene (spp1) J Clin Invest. 1998;101:1468–1478. doi: 10.1172/JCI1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wold S, Esbensen K, Geladi P. Principal Component Analysis. Chemometrics and Intelligent Laboratory Systems. 1987;2:37–52. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.