Abstract
CRISPR/Cas9 has been successfully adapted for gene editing in malaria parasites including Plasmodium falciparum and Plasmodium yoelii. However, the reported methods were limited to editing one gene at a time. In practice, it is often desired to modify multiple genetic loci in a parasite genome. Here we described a CRISPR/Cas9 mediated genome editing method that allows successive modification of more than one gene in the genome of P. yoelii using an improved single-vector system (pYCm) we developed previously. Drug resistant genes encoding human dihydrofolate reductase (hDHFR) and a yeast bifunctional protein (yFCU) with cytosine deaminase (CD) and uridyl phosphoribosyl transferase (UPRT) activities in the plasmid allowed sequential positive (pyrimethamine, Pyr) and negative (5-fluorocytosine, 5FC) selections and generation of transgenic parasites free of the episomal plasmid after genetic modification. Using this system, we were able to efficiently tag a gene of interest (Pyp28) and subsequently disrupted two genes (Pyctrp and Pycdpk3) that are critical for ookinete motility individually. Disruption of the genes either eliminated (Pyctrp) or greatly reduced (Pycdpk3) ookinete forward motility in matrigel in vitro and completely blocked oocyst development in mosquito midgut. The method will greatly facilitate studies of parasite gene function, development, and disease pathogenesis.
Keywords: rodent malaria, genetic modifications, Pyctrp, Pycdpk3, yFCU
1. Introduction
Malaria remains one of the most serious infectious diseases worldwide, although great progresses have been made in studying parasite molecular biology, development, and mechanisms of disease pathology through development and application of new technologies and methods such as high-throughput genome sequencing, genome-wide linkage and association analyses, and genome editing [1–3]. However, genetic modification of a malaria parasite genome remains challenging because of introducing a plasmid vector through multiple layers of membrane and other technical issues [3]. Indeed, successful transfections of malaria parasites were not reported until mid-1990’s [4–6]. The efficiencies of these early transfection methods are relatively low, although various modifications to improve transfection efficiency have been reported [7, 8]. More recently, methods using zinc-finger nucleases (ZFNs) and CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/ CRISPR-associated protein 9) were successfully introduced to edit genomes of malaria parasites [9–13]. These methods appear to improve the efficiency of gene editing in both human P. falciparum and rodent P. yoelii parasites [11–15], although the efficiencies of traditional gene editing methods for rodent malaria parasites are generally higher than those of human malaria parasite P. falciparum [11–13].
For a typical CRISPR/Cas9-mediated editing experiment in malaria parasites, the Cas9/sgRNA complex first introduces double strand-break (DSB) at a specific locus in the genome. The DSB is then repaired by homologous recombination (HR) using exogenous DNA donor template because the error-prone nonhomologous end joining (NHEJ) pathway is absent in Plasmodium [16]. For editing the genome of P. falciparum , the Cas9/sgRNA expression cassette and donor DNA template were delivered into parasites in two separate vectors with each plasmid carrying a different drug resistance gene (selection marker) [11, 13–15]. After electroporation, parasites transfected with the vectors were enriched by selection of two drugs simultaneously [11, 13, 14]. For the rodent malaria parasite P. yoelii, a single vector system was used because limited independent selection markers were available for the parasites. In the one-vector plasmid (pYC) design, all components, including the genes encoding the Cas9 protein, the human dihydrofolate reductase (hDHFR) for positive selection with pyrimethamine (Pyr), the sgRNA, and the donor template DNA were included in the plasmid vector [12]. After electroporation, pYC-transfected parasites were enriched by Pyr selection. DSB at specific locus introduced by Cas9/sgRNA complex required only transient expression of Cas9 and sgRNA from pYC plasmid. Gene modification at the targeted locus could be generated by HR between genomic DNA and exogenous pre-designed donor DNA. The pYC has been successfully applied for gene deletion, gene tagging, and nucleotide replacement in the genome of P. yoelii [12].
In the resulting engineered parasites of P. yoelii, the hdhfr marker was maintained episomally instead of being integrated into the parasite genome. After first round gene editing, complete removal of the episomal plasmid containing hdhfr marker is a prerequisite for the sequential modification in the same parasite. Although the number of the episomal plasmid gradually decreases in the absence of selection drug, it generally takes several weeks or even months to completely remove the episome in the parasite population because of uneven partition of plasmids into the daughter cells during parasite asexual proliferation [17]. In our hands, the episomal DNA could still be detected in transgenic parasites >50 days after selection drug was removed. To overcome this drawback, here we added a selection marker yFCU into the pYC plasmid and performed negative selection to kill any parasite that carried the pYC plasmid (expressing the yfcu gene). We applied this updated plasmid pYCm system to tag a gene and then to delete two genes separately. We showed that inhibition of yFCU by 5-formylcytosine (5FC) could efficiently remove the episomal vector in pYCm transfected parasites completely after tagging the endogenous Pyp28 gene with mCherry. We then demonstrated that this episome-free modified parasite could be subsequently modified again by deleting two genes (Pyctrp and Pycdpk3, respectively). These improvements of the CRISPR/Cas9 plasmid provide a new alternative for multiple genetic modifications in the rodent malaria parasite P. yoelii.
2. Materials And Methods
2.1 Plasmid construction
To replace the positive selection marker hdhfr with the combined positive-negative selection marker hdhfr-yfcu in the original pYC plasmid [12], we amplified partial coding fragment of hdhfr-yfcu from plasmid GOMO that contains an hdhfr-yfcu fusion gene [18] using PCR primers p22/p23 (seen in Fig. S1A). Replacement of hdhfr in pYC plasmid with hdhfr-yfcu was performed using LIC method as described previously [19], generating a new plasmid pYCm. The procedure for generating construct for targeting gene tagging and gene deletion was as described previously [12]. To generate the pYCm vector for tagging gene PyP28 (Gene ID PY17X_0515900 in PlasmoDB) with mCherry, we amplified the C-terminal 621bp coding region of p28 as the left arm and 639bp from the 3’UTR region following translation stop codon as the right arm using primers listed in Table S1. DNA fragment encoding mCherry was inserted between the left and right arms in frame (Fig. S1B). One sgRNA was designed to target a site close to C-terminal of coding region of the PyP28 (Fig. S1B). To generate pYCm vector to interrupt the Pyctrp (Gene ID PY17X_0415800) gene, we amplified 495bp 5’ flanking genomic regions as left homologous arm and a 493bp coding region as right arms using PCR using primers listed in Table S1. The left and right arms were connected by linker sequence in the pYCm plasmid (Fig. S1C). One sgRNA was designed to target the site in the coding region to be deleted (Fig. S1C). The deletion of N-terminal partial coding region caused frame-shift mutation of the coding region and thus gene inactivation. A construct for deletion of the Pycdpk3 (Gene ID PY17X_0410700) was similarly constructed (Fig. S1D).
2.2. Malaria parasite and parasite transfection
All transfections were performed on the P. yoelii 17XNL strain. The Parasite was propagated in ICR mice (female, 5–6 weeks old) purchased from the Animal Care Center, Xiamen University. All mouse experiments were performed in accordance with approved protocols (XMULAC20140004) by the Committee of Care and Use of Laboratory Animals at the School of Life Sciences, Xiamen University. The procedures for parasite transfection, Pyr selection and cloning were as described previously [12]. Briefly, parasites were electroporated with purified circular plasmid DNA. Transfected parasites were immediately intravenously injected into a new mouse and placed under Pyr pressure (provided in drinking water at concentration 7mg/L) from day 2 post transfection. A small amount of blood sample was taken through tail clip daily and Giemsa-stained for infected red blood cells (iRBCs). Pyr resistant parasites usually appear 5–6 days during drug selection.
2.3. Negative selection with 5FC
The procedures for negative selection with 5FC were as described in [20] with minor modifications. Briefly, 5FC (Sigma) was prepared in water at a final concentration of 2.0 mg/ml and was provided to the animals in a dark drinking bottle. A naïve mouse receiving parasites with residual plasmid from previous Pyr selection was subjected to 5FC pressure for 8 days, with a change of drug at day 4.
2.4. DNA preparation and confirmation of genetic modifications
Blood samples from infected mice were collected from the orbital sinus, and RBCs were lysed using 1% saponin in PBS. Parasite genomic DNAs were isolated using DNeasy Blood kits (Qiagen) after washing off hemoglobin and were used in PCR amplifications. For gene deletion and gene tagging, targeted modification was confirmed by PCRs using two pairs of primer to detect 5’ and 3’ integrations (seen in Fig. S1). To estimate the amount of pYC and pYCm plasmid in the parasite populations, we used two independent primer pairs from the plasmid backbone to amplify the DNAs All the PCR primers used are listed in Table S1.
2.5. Ookinete culture in vitro
Ookinetes were prepared according to the procedure described previously [21]. Briefly, infected blood was injected intraperitoneously into mice that were made anemic by phenyl-hydrazine treatment three days ago. Approximately three days after infection, 200µl of gametocyte-containing infected blood was obtained from the orbital sinus and mixed immediately with 1ml ookinete culture medium. The mixture was incubated at 22°C for 20–24 hours to allow gametogenesis, fertilization and ookinete differentiation. Ookinetes formation was monitored by Giemsa staining of smears of the cultured cells. Cultured ookinetes were pelleted for 2 min at 5000 rpm, washed twice with PBS, and fixed with 4% paraformaldehyde/0.0075% glutaraldehyde on poly-L-lysine coated glass slides for 30 min. After 3 washes with PBS, cells were stained with 2 µg/ml (in PBS) Hoechst33342 (Sigma) to visualize nuclei. All images were captured and processed using identical settings in the Zeiss LSM 780 laser scanning confocal microscope with a 63×/1.49 NA oil objective. Results were obtained from three independent experiments.
2.6. Mosquito infection and observation of parasites in mosquitoes
For mosquito infection, 50 female Anopheles stephensi mosquitoes were allowed to feed on anaesthetized infected mice that carried comparable numbers of gametocytes as determined by Giemsa staining for 20 min. Mosquito midguts were dissected days 1 and 4 post infection. Midguts were washed twice with PBS and then fixed with 4% paraformaldehyde/0.0075% glutaraldehyde on slides for 30 min. After 3 washes in PBS, cells were stained with Hoechst33342. Ookinetes expressing mCherry, Hoechst33342-labeled parasite nuclei, and midgut epithelium cells are observed using Zeiss LSM 780 laser confocal microscope. Twenty mosquitos were dissected day 7 post-infection, and oocysts in the midguts were counted. Salivary glands were isolated from 20–25 dissected mosquitos day 14 days post infection, and sporozoites were counted under a microscope.
2.7. Ookinete motility assay
Ookinete motility was evaluated as previously described [21, 22]. All the operations were carried out in a temperature controlled room (22°C). Twenty microliters of the ookinete cultures were added to an equal volume of Matrigel (Corning) on ice, mixed thoroughly, applied to a slide, and sealed with nail varnish after addition of a coverslip. The slide was then allowed to set at 22°C for 30 minutes. After identifying a field containing ookinetes, time-lapse videos (1 frame every 20 sec, for 20 min) were taken to monitor ookinete movement using a 40× objective lens on a Nikon ECLIPSE E100 microscope fitted with an ISH500 digital camera controlled by the ISCapture v3.6.9.3_N software (Tucsen). Time-lapse movies were analyzed with Fiji and the Manual Tracking plugin. Speed of motility was calculated by dividing the distance of individual ookinete covered with the total tracking time. Results were obtained from three independent experiments.
3. Results
3.1. Construction of pYCm plasmid and double drug selections
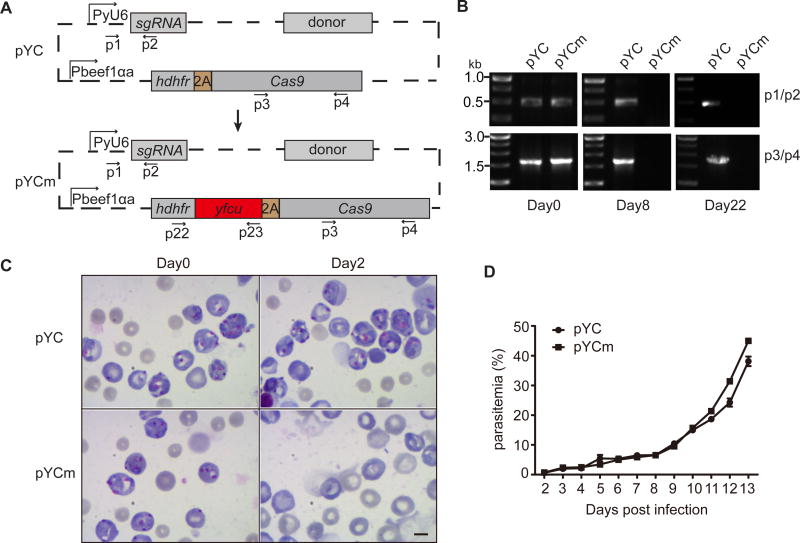
To facilitate removal of episcopal plasmid from parasites after first round of gene editing, we constructed a plasmid (pYCm) with positive and negative drug selection markers hdhfr-yfcu (Fig. 1A). We tested the feasibility of removing the pYCm episomal plasmid by sequential drug selections after electroporation of P. yoelii 17XNL with pYCm and pYC (as control) plasmids separately. Pyr resistant parasites emerged in both YC and pYCm transfected groups in 5–6 days after injection of parasites into mice, indicating proper introduction and replication of plasmids inside the parasite cells. Pyr was then replaced with 5FC to kill parasites with episomal plasmid pYCm expressing the yfcu gene. After 5FC treatment for 8 days, the episomal plasmid could not be detected in the pYCm transfected parasites (Fig. 1B), which suggests complete removal of the pYCm plasmid. In contrast, the pYC plasmid containing no yfcu gene could be easily detected 22 days after 5FC treatment (Fig. 1B).
Fig. 1.
Negative selection with 5FC removes pYCm plasmid episome in Plasmodium yoelii. A. Schematic of construction of pYCm plasmid vector expressing selection markers/genes encoding human dihydrofolate reductase (hdhfr) and a yeast gene encoding a bifunctional protein (yFCU) with cytosine deaminase (CD) and uridyl phosphoribosyl transferase (UPRT) activities driven by Pbeef1αa promoter. B. pYC and pYCm episome detection using PCR after sequential Pyr and 5FC selection. Primers (p) used are shown in (A) and listed in Table S1. C. Parasites completely lost pYCm episome via 5FC selection and become sensitive to Pyr selection again. Scale bar=5 µm. D. Asexual growth of pYC and pYCm transfected parasites after sequential Pyr and 5FC selection. Parasitemia was counted daily for 13 days after infection. The data points were means from three mice in each group.
We next treated the 5FC-selected parasite with Pry to functionally confirm the loss of episomal pYCm in the parasite population. Indeed, the double-selected pYCm parasites were sensitive to Pyr pressure again and were completely killed at day 2 (Fig. 1C), indicating no pYCm intracellularly. The pYC parasites grew normally under Pyr pressure, indicating the existing of episomes conferring drug resistance (Fig. 1C). We also compared the growth rates of both pYCm and pYC parasites after sequential Pyr and 5FC selection. We found comparable growth rates between the two parasite populations (Fig. 1D), suggesting that additional 5FC selection did not affect parasite growth in the mice.
3.2. Tagging endogenous P28 gene with mCherry
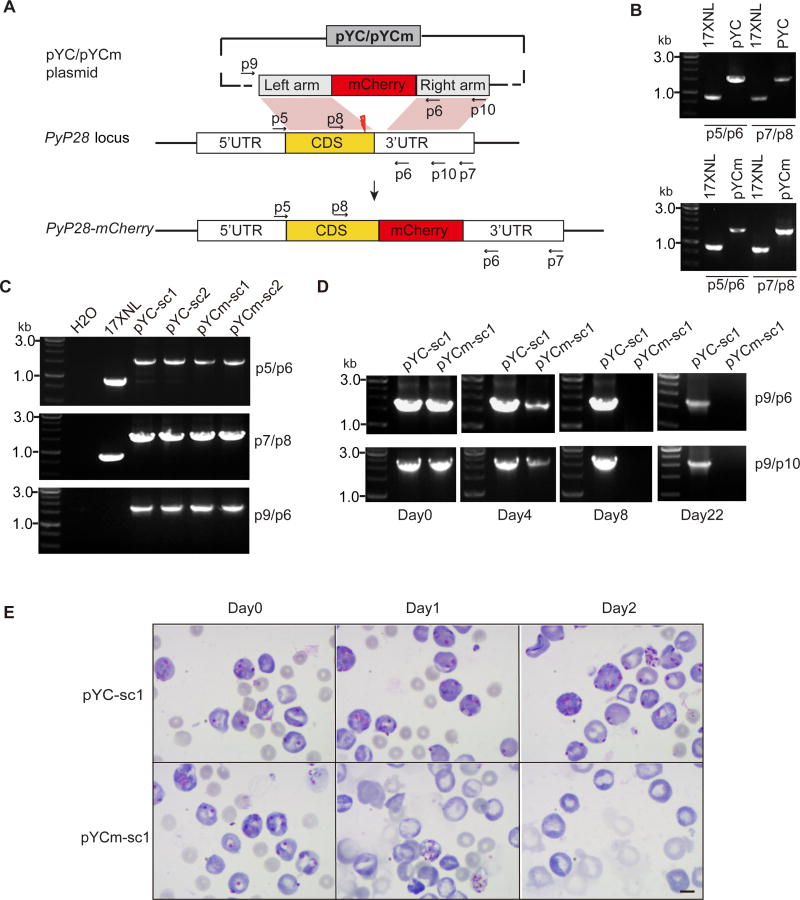
To apply the pYCm plasmid for genome editing, we first tagged the P. yoelii gene (PyP28) encoding a 28 kDa ookinete surface protein with mCherry gene encoding a red fluorescence protein. Plasmodium P28 protein is expressed abundantly in activated female gamete, zygote, ookinete, and young oocyst [23, 24]. Antibody-based immuno-detection of P28 protein expression was widely used in studies of parasite gametocyte activation and ookinete formation [25]. To tag the endogenous PyP28 gene with red fluorescence protein mCherry gene, we made two constructs based on the pYC and pYCm plasmids, respectively. The constructs contained a 621bp C-terminal region of the PyP28 gene followed by mCherry gene and a 639bp 3’-untranslated region (3-UTR) of PyP28 gene (Fig. 2A). After electroporation of P. yoelii 17XNL with the plasmids separately and injection of the transfected parasites into mice, we treated the mice with Pyr in drinking water for 6 days. In both pYC and pYCm transfected parasites, independent genomic recombinations with both left and right homologous arms were detected using PCR analysis (Fig. 2B). We then obtained two parasite clones from each transfection (pYC-sc1, pYC-sc2, pYCm-sc1 and pYCm-sc2) after limiting dilution cloning. All four clones had the expected replacement of endogenous PyP28 locus with donor template (Fig. 2C), indicating successful C-terminal tagging of the PyP28 gene with mCherry.
Fig. 2.
Tagging of endogenous PyP28 gene with mCherry gene in Plasmodium yoelii. A. pYCm plasmid construct for tagging PyP28 with mCherry. The plasmid contains donor template for homologous recombination repair after double strand break (DSB, thunderbolt) targeting PyP28-terminal was induced by Cas9/sgRNA complex. p indicates the positions of PCR primers used. B. PCR products detecting genomic integration of mCherry into genomes of the parasites transfected with pYC and pYCm, respectively. C. PCR detection of mCherry integration in cloned parasites. Primer p9/p6 pairs were used for monitoring the existence of episome plasmid. D. Detection of episome plasmid in parasite clones pYC-sc1 and pYCm-sc1 during 5FC selection. No plasmid was detected in the pYCm-sc1 clone day 8 post selection. E. Blood smears of mice infected with parasites transfected with pYC-sc1 or pYCm-sc1, respectively, and selected with 5FC. Clone pYCm-sc1 completely lost the episomal plasmid after 5FC selection and became Pyr sensitive (no parasite detected day 2). Scale bar=5 µm.
To remove the intracellular episomes containing hdhfr marker, 5FC selection was applied to parasite clones pYC-sc1 and pYCm-sc1. Amplification of DNAs showed reduction of plasmid DNA in the pYCm-sc1 parasites on day 4 post selection and absence of the plasmid on day 8 and day 22, but not the pYC-sc1 parasites (Fig. 2D). To confirm the complete loss of the plasmid, we treated 5FC-selected pYCm-sc1parasites with Pyr again and showed that the parasites were sensitive to Pyr. Indeed, no live 5FC-selected pYCm-sc1 parasite was found day 2 post Pyr treatment (Fig. 2E). In the pYC-sc1 group, parasites containing the pYC plasmid proliferated normally under the Pyr pressure (Fig. 2E). These results demonstrate that pYCm can be used to remove parasites with episomal plasmid carrying drug selection marker for further genetic modifications.
3.3. P28::mCherry parasite develop normally throughout the Plasmodium life cycle
We next evaluated the development of the parasite having mCherry tagged P28 in mice and in mosquito. Both asexual and sexual stages of the parasite appeared to develop normally and were comparable to those of wild type 17XNL parasite (Fig. S2A and S2B). Furthermore, the numbers of midgut oocysts at day 7 and salivary sporozoites at day 14 were similar between the 17XNL wild type and P28::mCherry parasites (Fig. S2C and S2D). We also obtained P28::mCherry blood stage parasites after mosquito biting (data not shown). All these data indicate that the engineered P. yoelii 17XNL P28::mCherry parasite develops normally throughout the life cycle.
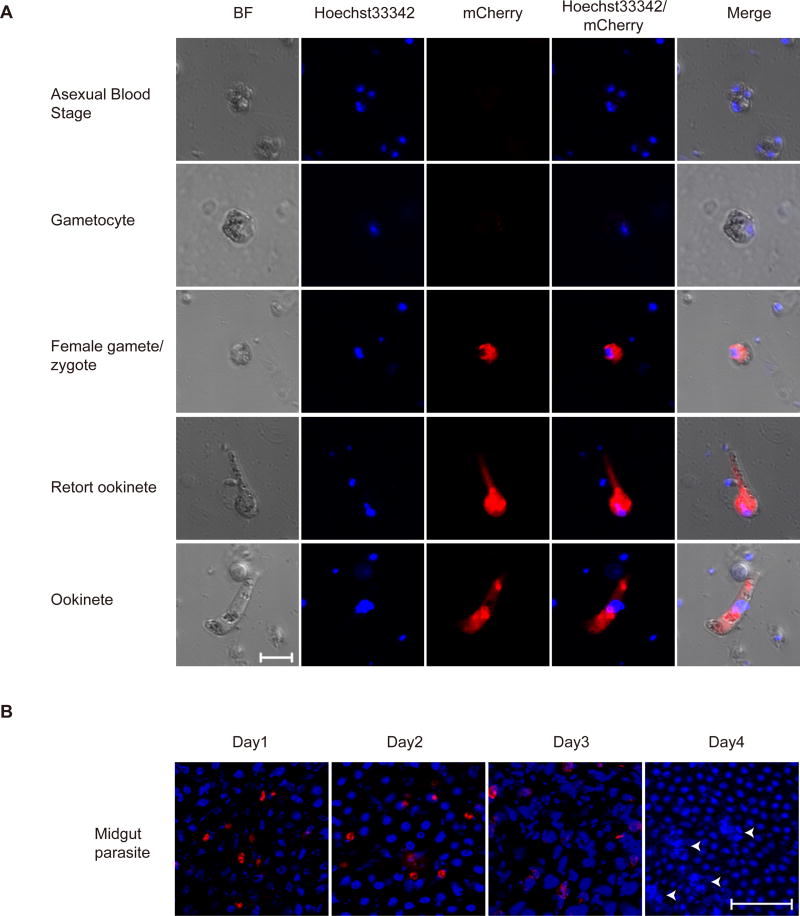
We next investigated the stage specific expression of mCherry reporter and detected no mCherry signal in asexual stages and gametocytes (Fig. 3A). After gametocyte activation, strong mCherry expression was observed in the cytoplasm in the female gametes, fertilized zygotes, and in the retort and mature ookinetes (Fig. 3A). Furthermore, we analyzed the mCherry expression in ookinetes in midguts of infected mosquito. Crescent-shaped mCherry-positive ookinetes were observed on the monolayer midgut epithelium, locating either in the lumen or basal side of midgut at day 1 post blood feeding (Fig. 3B). However, mCherry expression was completely lost in the oocysts at day 4 post infection. These results showed that P28::mCherry protein was expressed normally in this engineered parasite and is consistent with those observed using anti-P28 antibody staining [23–25].
Fig. 3.
Expression of mCherry tagged p28 during different stages of Plasmodium yoelii life cycle. A. Fluorescence microscopy observing of fixed parasites at the indicated stages of life cycle. Nuclei were stained with Hoechst33342 (blue). Results are representative of three independent experiments. Bar=5 µm. B. mCherry-expressing ookinetes and oocysts from infected mosquito midgut. Nuclei were stained with Hoechst33342 (blue). The white arrows mark oocysts in sporogony proliferation with DNA replication. Results are representative of three independent experiments. Bar=5 µm.
3.4. Sequential gene deletion in P28::mCherry parasite
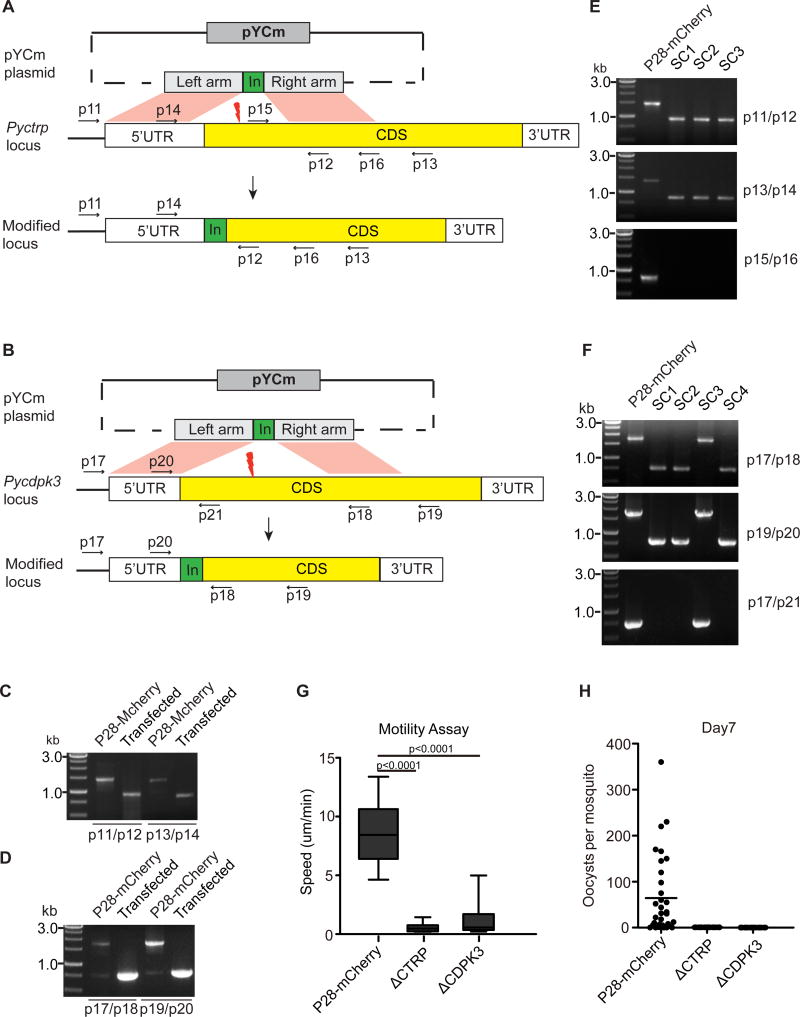
To test whether the P28::mCherry clone could be used for additional genome editing, we attempted to delete two genes (Pyctrp and Pycdpk3) in the genome of parasite separately. Pbctrp and Pbcdpk3 genes in Plasmodium berghei were previously disrupted, leading to complete loss or severe defect in ookinete motility, respectively, and absence of oocysts in the midgut [26–29]. We constructed two plasmids based on pYCm to disrupt the endogenous Pyctrp and Pycdpk3 gene in the P28::mCherry parasite, respectively (Fig. 4A and 4B). For both constructs, Pry resistant parasites emerged 6 to 7 days post transfection. Targeted integrations of left and right homologous arms were detected from both transfected parasites after PCR analysis (Fig. 4C and 4D). We finally obtained three cloned parasites with disrupted Pyctrp and three clones with disrupted Pycdpk3 gene after limiting dilution cloning (Fig. 4E and 4F).
Fig. 4.
Genetic deletion of Pyctrp or Pycdpk3 in cause defects in ookinete motility. A. pYCm construct for deleting the Pyctrp gene. B. pYCm construct for deleting Pycdpk3 gene. The plasmid contains donor template for homologous recombination repair after double strand break (DSB, thunderbolt) was induced by Cas9/sgRNA complex. p indicates the positions of PCR primers used. C. PCR analysis of 5’ and 3’ homologous integration in P28::mCherry parasite transfected with Pyctrp deleting plasmid. D. PCR analysis of 5’ and 3’ homologous integration in P28::mCherry parasite transfected with Pycdpk3 deleting plasmid. E. PCR analysis of clonal parasites with targeted Pyctrp deletion. Sc1-sc3 are three individual parasite clones. F. PCR analysis of clonal parasites with Pycdpk3 deletion. Sc1-sc4 are four individual parasite clones. G. Ookinetes gliding speeds of parental P28::mCherry, ΔPyctrp and ΔPycdpk3 parasites detected using Matrigel Motility assay. Speed of at least 20 individual cultured ookinetes was measured over a 20 min period. Living ookinete gliding was recorded as time-lapse videos (Videos S1–S3). H. Midgut oocyst counts 7 days after mosquito feeding with parasite-infected mice. Results are representative of two independent experiments.
To confirm the functional role of Pyctrp and Pycdpk3 in ookinete motility, ookinete gliding motility of the Pyctrp and Pycdpk3 mutants (ΔPyctrp and ΔPycdpk3) were investigated using Matrigel motility assay as previously reported [21]. Cultured ookinetes from 17XNL P28::mCherry parasite had a gliding speed of 5–13 um/min (Fig. 4G, and Video S1), which is similar to those of P. berghei ookinetes [21]. However, the ookinetes with disrupted Pyctrp displayed a complete defect in forward gliding motility (Fig. 4A, and Video S2), although ookinete bending was still observed (Video S2). Similarly, ookinetes without Pycdpk3 had a dramatically reduced forward gliding motility (Fig. 4A and Video S3). Furthermore, no midgut oocyst was observed in Anopheles stephensi mosquitos infected with ΔPyctrp and ΔPycdpk3 mutants 7 days after feeding (Fig. 4H). These results confirm functional disruption of the genes and support an essential role of CTRP and CDPK3 in ookinete motility in both rodent malaria parasites P. berghei and P. yoelii. The experiments also demonstrate successful modification of parasite genes after removal of a drug selection marker from prior gene editing.
4. Discussion
Functional investigations of malaria parasites often require multiple rounds of gene editing or editing more than one gene. For examples, it may be necessary to delete or to tag more than one copy of a multi-gene family. To confirm a specific gene function, it is often necessary to delete a gene first and then re-introduce the original or a modified gene back into the genome to partially or completely reinstall a gene function. However, successive modifications in the genome of the same parasite line are frequently hampered by limited availability of independent drug selectable markers, particularly, for rodent malaria parasites. Six selectable markers are now available for genome modification in P. falciparum [3]. For rodent malaria parasites, there are only three related selectable markers/genes available, including Toxoplasma DHFR-thymidylate synthase (tgDHFR-TS), P. berghei DHFR-thymidylate synthase (pbDHFR-TS), and hDHFR, all of which confers resistance to Pyr. The hDHFR also confers resistance to WR99210. Thus, for sequential editing in rodent malaria parasites, either tgDHFR-TS or pbDHFR-TS must be used first, and transgenic parasites are selected with Pyr. The parasites are then selected with WR99210 after introduction of the hDHFR marker. Development of methods to edit a parasite genome multiple times by re-using a selectable drug marker will greatly facilitate the investigation of gene functions. Our study developed and tested a CRISPR/Cas9-based re-useable selection system for two rounds of gene editing for rodent malaria parasite P. yoelii , which can also be modified and applied for other malaria parasites, including P. falciparum.
In our previous study, we developed a CRISPR/Cas9-based vector pYC to successfully modify P. yoelii genome, including gene deletion, gene tagging and nucleotide replacement [12]. In this system, homologous repair of DSB introduced by CRISPR/Cas9 complex requires transient expression of Cas9/sgRNA and hDHFR from the pYC vector without integration of the drug resistance gene (hdhfr) into the parasite genome. In theory, the lack of integration of drug resistance gene into parasite genome will allow removal of the episomal plasmid after removal of drug pressure. In practice, we consistently detected the episomal plasmid in transgenic parasites weeks or even months after removal of Pyr pressure, which is consistent with the results from a previous report in P. berghei parasite [17]. The results suggest that episomal plasmids may remain within a parasite population for a long time in the absence of drug pressure. To overcome the problem, we introduced into the pYC plasmid a second negative selection marker (yfcu/5FC) that has been widely used for recycling of selection marker and for successive modification of a parasite genome [20, 30]. We showed that negative selection by 5FC could completely remove the episomal plasmid from transfected parasites within a few days. Because the CRISPR/Cas9 system does not lead to integration of a drug marker into the genome, it is more efficient to obtain a parasite that is totally free of drug marker than the traditional methods that often result in vector integration into the genome.
We applied our plasmid vector to successfully tag the Pyp28 with mCherry gene, removed the plasmid with hdhfr gene after 5FC selection, and then used the same plasmid backbone to disrupt Pyctrp or Pycdpk3 gene. We also evaluated the expressions of the genes and the functional effects of the genes. Our results also showed that both the Pyctrp or Pycdpk3 genes are critical for ookinete motility and oocyst development in mosquito midgut, supporting the observations in P. berghei. These results advance our understanding of P. yoelii development in mosquito and may facilitate identification of reagents to block parasite transmission.
Supplementary Material
Acknowledgments
We are grateful to Dr. Olivier Silvie for providing plasmid pGOMO. This work was supported by grants from National Natural Science Foundation of China (grant numbers 81522027(Y.J.), 31401143(Y.J.) and 31501912(H.C.)), China Thousand Youth Talents Plan (Y.J.), the ‘863’ National High Technology Research and Development Program of China (2014AA020530, Y.J.), the ‘973’National Basic Research Program of China (2014CB744501 Y.J.), Fundamental Research Funds for the Central Universities of China-Xiamen University (20720160069 (Y.J.), 20720150165 (Y.J.) and 2013121033 (Y.J.)), Special Research Fund for the Doctoral Program of Higher Education of China (20130121120023 (Y.J.) and 20130121120024 (H.C.)), Scientific Research Foundation for Returned Scholars, Ministry of Education of China (H.C), the “111” Project of the Ministration of Education of China (B06016), and by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). The authors thank Cindy Clark, NIH Library Writing Center, for manuscript editing assistance.
Footnotes
References
- 1.Volkman SK, Neafsey DE, Schaffner SF, Park DJ, Wirth DF. Harnessing genomics and genome biology to understand malaria biology. Nature reviews Genetics. 2012;13(5):315–328. doi: 10.1038/nrg3187. [DOI] [PubMed] [Google Scholar]
- 2.Su X, Hayton K, Wellems TE. Genetic linkage and association analyses for trait mapping in Plasmodium falciparum. Nature reviews Genetics. 2007;8(7):497–506. doi: 10.1038/nrg2126. [DOI] [PubMed] [Google Scholar]
- 3.de Koning-Ward TF, Gilson PR, Crabb BS. Advances in molecular genetic systems in malaria. Nature reviews Microbiology. 2015;13(6):373–387. doi: 10.1038/nrmicro3450. [DOI] [PubMed] [Google Scholar]
- 4.van Dijk MR, Waters AP, Janse CJ. Stable transfection of malaria parasite blood stages. Science. 1995;268(5215):1358–1362. doi: 10.1126/science.7761856. [DOI] [PubMed] [Google Scholar]
- 5.van Dijk MR, Janse CJ, Waters AP. Expression of a Plasmodium gene introduced into subtelomeric regions of Plasmodium berghei chromosomes. Science. 1996;271(5249):662–665. doi: 10.1126/science.271.5249.662. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y, Kirkman LA, Wellems TE. Transformation of Plasmodium falciparum malaria parasites by homologous integration of plasmids that confer resistance to pyrimethamine. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(3):1130–1134. doi: 10.1073/pnas.93.3.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deitsch K, Driskill C, Wellems T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic acids research. 2001;29(3):850–853. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janse CJ, Franke-Fayard B, Mair GR, Ramesar J, Thiel C, Engelmann S, Matuschewski K, van Gemert GJ, Sauerwein RW, Waters AP. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Molecular and biochemical parasitology. 2006;145(1):60–70. doi: 10.1016/j.molbiopara.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Straimer J, Lee MC, Lee AH, Zeitler B, Williams AE, Pearl JR, Zhang L, Rebar EJ, Gregory PD, Llinas M, et al. Site-specific genome editing in Plasmodium falciparum using engineered zinc-finger nucleases. Nature methods. 2012;9(10):993–998. doi: 10.1038/nmeth.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNamara CW, Lee MC, Lim CS, Lim SH, Roland J, Nagle A, Simon O, Yeung BK, Chatterjee AK, McCormack SL, et al. Targeting Plasmodium PI(4)K to eliminate malaria. Nature. 2013;504(7479):248–253. doi: 10.1038/nature12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nature biotechnology. 2014;32(8):819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 12.Zhang C, Xiao B, Jiang Y, Zhao Y, Li Z, Gao H, Ling Y, Wei J, Li S, Lu M, et al. Efficient editing of malaria parasite genome using the CRISPR/Cas9 system. mBio. 2014;5(4):e01414–01414. doi: 10.1128/mBio.01414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner JC, Platt RJ, Goldfless SJ, Zhang F, Niles JC. Efficient CRISPR-Cas9-mediated genome editing in Plasmodium falciparum. Nature methods. 2014;11(9):915–918. doi: 10.1038/nmeth.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng CL, Siciliano G, Lee MC, de Almeida MJ, Corey VC, Bopp SE, Bertuccini L, Wittlin S, Kasdin RG, Le Bihan A, et al. CRISPR-Cas9-modified pfmdr1 protects Plasmodium falciparum asexual blood stages and gametocytes against a class of piperazine-containing compounds but potentiates artemisinin-based combination therapy partner drugs. Molecular microbiology. 2016;101(3):381–393. doi: 10.1111/mmi.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Tong Y, Pan J, Yang Y, Liu Q, Tan X, Zhao S, Qin L, Chen X. A redesigned CRISPR/Cas9 system for marker-free genome editing in Plasmodium falciparum. Parasites & vectors. 2016;9:198. doi: 10.1186/s13071-016-1487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirkman LA, Lawrence EA, Deitsch KW. Malaria parasites utilize both homologous recombination and alternative end joining pathways to maintain genome integrity. Nucleic acids research. 2014;42(1):370–379. doi: 10.1093/nar/gkt881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Dijk MR, Vinkenoog R, Ramesar J, Vervenne RA, Waters AP, Janse CJ. Replication, expression and segregation of plasmid-borne DNA in genetically transformed malaria parasites. Molecular and biochemical parasitology. 1997;86(2):155–162. doi: 10.1016/s0166-6851(97)02843-0. [DOI] [PubMed] [Google Scholar]
- 18.Manzoni G, Briquet S, Risco-Castillo V, Gaultier C, Topcu S, Ivanescu ML, Franetich JF, Hoareau-Coudert B, Mazier D, Silvie O. A rapid and robust selection procedure for generating drug-selectable marker-free recombinant malaria parasites. Scientific reports. 2014;4:4760. doi: 10.1038/srep04760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Evans RM. Ligation independent cloning irrespective of restriction site compatibility. Nucleic acids research. 1997;25(20):4165–4166. doi: 10.1093/nar/25.20.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orr RY, Philip N, Waters AP. Improved negative selection protocol for Plasmodium berghei in the rodent malarial model. Malaria journal. 2012;11:103. doi: 10.1186/1475-2875-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moon RW, Taylor CJ, Bex C, Schepers R, Goulding D, Janse CJ, Waters AP, Baker DA, Billker O. A cyclic GMP signalling module that regulates gliding motility in a malaria parasite. PLoS pathogens. 2009;5(9):e1000599. doi: 10.1371/journal.ppat.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brochet M, Collins MO, Smith TK, Thompson E, Sebastian S, Volkmann K, Schwach F, Chappell L, Gomes AR, Berriman M, et al. Phosphoinositide metabolism links cGMP-dependent protein kinase G to essential Ca(2)(+) signals at key decision points in the life cycle of malaria parasites. PLoS biology. 2014;12(3):e1001806. doi: 10.1371/journal.pbio.1001806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.del Carmen Rodriguez M, Gerold P, Dessens J, Kurtenbach K, Schwartz RT, Sinden RE, Margos G. Characterisation and expression of pbs25, a sexual and sporogonic stage specific protein of Plasmodium berghei. Molecular and biochemical parasitology. 2000;110(1):147–159. doi: 10.1016/s0166-6851(00)00265-6. [DOI] [PubMed] [Google Scholar]
- 24.Tomas AM, Margos G, Dimopoulos G, van Lin LH, de Koning-Ward TF, Sinha R, Lupetti P, Beetsma AL, Rodriguez MC, Karras M, et al. P25 and P28 proteins of the malaria ookinete surface have multiple and partially redundant functions. The EMBO journal. 2001;20(15):3975–3983. doi: 10.1093/emboj/20.15.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guttery DS, Poulin B, Ramaprasad A, Wall RJ, Ferguson DJ, Brady D, Patzewitz EM, Whipple S, Straschil U, Wright MH, et al. Genome-wide functional analysis of Plasmodium protein phosphatases reveals key regulators of parasite development and differentiation. Cell host & microbe. 2014;16(1):128–140. doi: 10.1016/j.chom.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuda M, Sakaida H, Chinzei Y. Targeted disruption of the plasmodium berghei CTRP gene reveals its essential role in malaria infection of the vector mosquito. The Journal of experimental medicine. 1999;190(11):1711–1716. doi: 10.1084/jem.190.11.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dessens JT, Beetsma AL, Dimopoulos G, Wengelnik K, Crisanti A, Kafatos FC, Sinden RE. CTRP is essential for mosquito infection by malaria ookinetes. The EMBO journal. 1999;18(22):6221–6227. doi: 10.1093/emboj/18.22.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishino T, Orito Y, Chinzei Y, Yuda M. A calcium-dependent protein kinase regulates Plasmodium ookinete access to the midgut epithelial cell. Molecular microbiology. 2006;59(4):1175–1184. doi: 10.1111/j.1365-2958.2005.05014.x. [DOI] [PubMed] [Google Scholar]
- 29.Siden-Kiamos I, Ecker A, Nyback S, Louis C, Sinden RE, Billker O. Plasmodium berghei calcium-dependent protein kinase 3 is required for ookinete gliding motility and mosquito midgut invasion. Molecular microbiology. 2006;60(6):1355–1363. doi: 10.1111/j.1365-2958.2006.05189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maier AG, Braks JA, Waters AP, Cowman AF. Negative selection using yeast cytosine deaminase/uracil phosphoribosyl transferase in Plasmodium falciparum for targeted gene deletion by double crossover recombination. Molecular and biochemical parasitology. 2006;150(1):118–121. doi: 10.1016/j.molbiopara.2006.06.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.