Abstract
Quantification of malondialdehyde (MDA) as a marker of lipid peroxidation is relevant for many research fields. We describe a new sensitive and selective method to measure free and total plasmatic MDA using derivatization with 2,4-dinitrophenylhydrazine (DNPH) and ultra-HPLC-high-resolution MS. Free and total MDA were extracted from minute sample amounts (10 μl) using acidic precipitation and alkaline hydrolysis followed by acidic precipitation, respectively. Derivatization was completed within 10 min at room temperature, and the excess DNPH discarded by liquid-liquid extraction. Quantification was achieved by internal standardization using dideuterated MDA as internal standard. The method’s lowest limit of quantification was 100 nM and linearity spanned greater than three orders of magnitude. Intra- and inter-day precisions for total MDA were 2.9% and 3.0%, respectively, and those for free MDA were 12.8% and 24.9%, respectively. Accuracy was 101% and 107% at low and high concentrations, respectively. In human plasma, free MDA levels were 120 nM (SD 36.26) and total MDA levels were 6.7 μM (SD 0.46). In addition, we show the applicability of this method to measure MDA plasma levels from a variety of animal species, making it invaluable to scientists in various fields.
Keywords: lipid peroxidation; fatty acid/oxidation; oxidized lipids; quantitation; mass spectrometry; derivatization; 2,4-dinitrophenylhydrazine; ultra-high-performance liquid chromatography-high-resolution mass spectrometry
Oxidative stress is a cellular state in which the production of reactive oxygen species (ROS) is in misbalance with the available antioxidants (1). Although ROS are constitutive by-products of cell metabolism, their production can be amplified in the presence of environmental perturbations (2). The presence of ROS can result in the oxidation of important biomolecules, such as DNA, proteins, and lipids, potentially leading to cell damage. One of the most studied oxidative injuries is that incurred by lipids due to their importance in the maintenance of cellular structure. Peroxidation of PUFAs results in lipid hydroperoxides, which, after breakdown, result in a variety of aldehydes, malondialdehyde (MDA) being the primary and best studied of these molecules (3).
The study and quantification of MDA as a marker of lipid peroxidation has been relevant for many research fields. In medical research, several diseases have been related to higher levels of MDA compared with healthy controls (4–6). In food sciences, quantification of MDA concentration has been frequently taken as a measure of quality of stored foodstuff (7). MDA levels have also been used as a toxicity marker of herbicides used in agriculture, such as paraquat (8). Recently, research in ecology, animal behavior, and evolution has also used MDA as a key marker of oxidative damage to investigate the physiological mechanisms underlying important evolutionary concepts, such as sexual selection and life-history trade-offs (9–11).
In biological matrices, MDA exists in both free and protein-bound forms, the latter representing the vast majority of total MDA (12–14). Several techniques have been developed to assess either total MDA or its free form, most often based on derivatization chemistry coupled with various separation and/or detection methods, including GC or LC, UV or fluorescence spectrophotometry, and MS (15–19). The most widely used derivatizing agent is thiobarbituric acid (TBA). This method is relatively easy to perform, but its specificity is low (20), as TBA reacts with other compounds naturally available in biological samples (20–23). Also, because derivatization with TBA requires high temperatures (95–100°C) under strongly acidic conditions, this method likely leads to an overestimation of MDA concentrations (23). Finally, consistent and significant peaks in blank samples result in an increased LOD, precluding the analysis of samples with low levels of MDA (24). Currently, an important need is the development of a method that allows specific and accurate quantification of MDA, while reducing sample volumes used. Specifically, a method that does not produce peaks in blank samples is of most importance for researchers working with limited sample volumes and/or naturally low concentrations of MDA.
To overcome the limitations of the TBA assay, more specific and reliable methods have been developed using other derivatizing agents, such as phenylhydrazine (PH), 2,4,6- trichlorophenylhydrazine, 3-nitrophenylhydrazine, pentafluorophenylhydrazine, and 2,4-dinitrophenylhydrazine (DNPH) (13, 18, 25). A clear advantage of hydrazine-based over TBA-based derivatization is the milder conditions needed for the reaction, which can be performed at room temperature. Specifically, derivatization with DNPH is completed after 10–30 min (12, 26), a much shorter time in comparison to TBA (1 h), PH (3 h), and trichlorophenylhydrazine (1 h) methods, and yields a stable MDA-DNPH derivative (12, 13). Chemicals used for the MDA-DNPH derivatization have minimal costs and accurate and specific quantification can be done using robust LC-UV (7, 12, 26) or LC-MS (27). However, one main disadvantage of most published protocols for quantification of MDA using DNPH is the high volume of sample required, from 50 μl (26) up to 1 ml (12), precluding analysis in studies where sample volumes are restrictively small.
Here we evaluated the use of ultra-HPLC-high-resolution MS (UHPLC-HRMS) for the quantification of both free and total plasmatic MDA, using DNPH as the derivatizing agent. While HRMS was once not considered suitable for quantitative analysis, new generation HRMS instruments have been shown to provide similar quantitative performances to triple quadrupoles (28). We show that our validated method allows the use of minute sample volumes and can be used for quantification of MDA in plasma from different vertebrate species, including human, rodent (mole-rat, Fukomys damarensis), bat (Carollia perspicillata), bird (Passer domesticus), and fish (Scolopsis bilineatus), making it of great interest to scientists in various fields, from biomedicine to ecology and evolution.
MATERIALS AND METHODS
Reagents and chemicals
Malondialdehyde tetrabutylammonium salt (MDA-salt), DNPH, sodium hydroxide (NaOH), and methyl tert-butyl ether (MTBE) were purchased from Sigma-Aldrich. [D2]1,1,3,3-tetra-ethoxypropane (D2-TEP) was obtained from Dr. Ehrenstorfer GmbH. Trichloroacetic acid (TCA) cyclohexane, and toluene were obtained from Merck KGaA. Hydrochloric acid (HCl; 37%), methanol (MeOH), and ethyl acetate (EtOAc) were purchased from Fisher Scientific. Hexane was purchased from Acros Organics. Acetonitrile (ACN), water, and formic acid used for LC-MS analyses were purchased at Biosolve. For semi-preparative isolation, HPLC grade ACN and analytical grade formic acid from Sigma-Aldrich and milli-Q water were employed.
Blood collection
Peripheral human blood was collected from healthy volunteers among the authors of this study (female and male, Caucasian) into a Sarstedt Monovette® with heparin as an anticoagulant and centrifuged straight away for 10 min at 4°C and 2,000 g.
Mole-rat (Fukomys damarensis) blood was collected into heparinized tubes from individuals under anesthesia from a prewarmed hind foot by using a sterile needle to puncture a vein. Blood was centrifuged straight away at room temperature for 5 min at 2,000 g.
Blood samples from bats (Carollia perspicillata) and house sparrows (Passer domesticus) were drawn from the antebrachial wing vein and from the alar vein, respectively, into heparinized CB300 Sarstedt Microvette® by puncturing with a sterile needle. Samples were transported to the laboratory on ice, where they were centrifuged for 5 min at 4°C and 2,000 g.
Fish (Scolopsis bilineatus) blood was collected from sedated individuals from the caudal vein using a sterile needle and a preheparinized syringe. Blood was centrifuged for 5 min at 2,000 g.
For all species, plasma was separated from the cellular component following centrifugation and stored at −80°C until analysis. For method validation, human plasma was split into 70 μl aliquots before freezing.
Human subjects provided written informed consent before sampling. Animal samples were collected as part of other ongoing studies. All animal procedures complied with Public Health Service policy on humane care and use of laboratory animals. Blood sampling procedures were approved by the University of Pretoria Animal Ethics Committee (Fukomys damarensis, EC093-14), the veterinary office of the Canton Fribourg (Carollia perspicillata, FR_2013_46 and FR_2014_44), the veterinary office of the Canton Bern (Passer domesticus, BE41/12 and WTH/g-525/14), and the Australian National University animal experimentation ethics committee (Scolopsis bilineatus, A2015/66).
Synthesis of dideuterated MDA
Dideuterated MDA (D2-MDA) was synthesized from D2-TEP by acidic hydrolysis. D2-TEP (30 μM) was incubated in 0.1 M HCl for 40 min at 40°C. The resulting D2-MDA (30 μM in 0.1 M HCl) was used as internal standard (IS) for MDA quantification (29).
Extraction of free and total MDA
For extraction of the free fraction of MDA, 10 μl of plasma were mixed with 40 μl of water, 10 μl of IS, and 150 μl of TCA 20% in a microcentrifuge tube to precipitate proteins. Samples were vortexed, sonicated for 30 s, and centrifuged (5 min, 25,000 g). The supernatant (190 μl) was collected and MDA was derivatized by incubating for 10 min with 19 μl of DNPH (5 mM in TCA 20%) at room temperature. The reaction was then stopped by adding 22 μl of NaOH 10 M and the resulting MDA-DNPH was extracted twice with 250 μl of a mixture of cyclohexane:toluene (1:1, v/v). Organic phases (two times 240 μl) were combined and evaporated in a centrifugal evaporator (Labconco) at 25°C for approximately 60 min. The residue was reconstituted in 100 μl of MeOH 50% and the resulting solution was sonicated, centrifuged, and transferred into an HPLC vial fitted with a 250 μl conical insert. The reconstituted solution represented a 10-fold dilution of the initial sample concentration.
For extraction of total MDA (free + bound MDA fractions), 10 μl of plasma were mixed with 40 μl of NaOH 1.25 M in a microcentrifuge tube and heated for 30 min at 60°C. Samples were cooled down in the refrigerator for 10 min and 10 μl of IS and 150 μl of TCA 20% were added. The rest of the procedure was performed as described for extraction of free MDA.
UHPLC-PDA-HRMS conditions
The analysis of MDA-DNPH was performed on an Acquity UPLCTM system coupled to both an eλ photodiode array (PDA) detector and a Synapt G2 QTOF mass spectrometer (Waters). The separation was carried out on an Acquity BEH C18 column (50 × 2.1 mm internal diameter, 1.7 μm particle size) at a flow rate of 0.4 ml min−1 in gradient mode. Mobile phases consisted of water + 0.05% formic acid (phase A) and ACN + 0.05% formic acid (phase B). The gradient program started at 2% B and increased linearly to 60.8% B in 3.0 min, then increased to 100% B in 0.4 min, was held at 100% B for 2.0 min before switching back to initial conditions and re-equilibrating for 1.5 min. The column temperature and that of the autosampler were both set to 25°C. A volume of 2.5 μl was injected in the so-called partial loop with needle overfill mode into the column, after which the autosampler needle was washed with 700 μl of “strong” wash (ACN:MeOH:isopropanol, 1:1:1, v/v) followed by 600 μl of “weak” wash (MeOH 20%). The UV detection range was set from 190 to 400 nm with a resolution of 1.2 nm and a frequency of 20 Hz. Absorbance maxima for DNPH and MDA-DNPH were 358 and 305 nm, respectively. The mass spectrometer was operated in electrospray positive ionization in full scan mode over a mass range of 85–600 Da (scan time 0.4 s). The enhanced duty cycle (EDC) mode was activated and centered on m/z 235. EDC increases the quadrupole transmission at and around the selected m/z value. TOF resolution at full width half maximum was about 20,000 at m/z 500. Source parameters were as follows: capillary voltage 2.8 kV, cone voltage 25 V, source temperature 120°C, desolvation gas flow and temperature 800 l h−1 and 450°C, respectively, cone gas flow 20 l h−1. Exact mass measurements (<2 ppm) were ensured by infusing a 500 ng ml−1 solution of leucine-enkephalin at 15 μl min−1 through the LocksprayTM probe. In addition, external calibration using a 0.5 mM sodium formate solution was performed every week. For MS/MS analysis, argon at a flow rate of 2.2 ml min−1 and a voltage ramp from 10 to 35 eV were used as collision gas and energy, respectively. The UHPLC flow was diverted from the mass spectrometer from 0.0 to 2.6 min and from 3.1 to 6.0 min. The entire system was controlled by MasslynxTM v.4.1. Peaks were automatically integrated using QuanlynxTM with a 0.1 min chromatographic window centered on the retention time of MDA-DNPH (2.87 min) and a 0.012 Da mass window centered on the (M+H)+ ion of MDA-DNPH (m/z 235.0462).
Isolation of MDA-DNPH
Twenty milligrams of MDA-salt and 40 mg of DNPH were dissolved into 100 ml of TCA 20%. The mixture was stirred for 30 min and partitioned twice with 100 ml hexane. Hexane phases were combined, evaporated, and reconstituted in 1.2 ml of MeOH 60%. The solution was filtered through a 0.22 μm hydrophilic polytetrafluoroethylene (PTFE) syringe filter and split in two equal parts. A volume of 500 μl was injected twice on a semi-preparative XTerra MS C18 column (150 × 19 mm internal diameter, 5 μm particle size). The semi-preparative HPLC system was composed of a 1525 EF pump (Waters), a manual Rheodyne valve fitted with a 500 μl loop, and a dual wavelength 2487 UV detector (Waters) equipped a semi-preparative UV cell (path length 3 mm). Gradient conditions using the same mobile phases as for analytical separation (see above) were as follows: 2% B for 5 min, 2–60% B from 5 to 63 min, 60–100% B from 63 to 65 min, holding at 100% B for 7 min. The flow rate was 5 ml min−1. Two wavelengths at 305 and 358 nm were monitored. Fractions were collected every minute in 9 ml glass tubes using a FC203B fraction collector (Gilson). Fractions containing most of the MDA-DNPH, which eluted between 57.5 and 59.5 min were combined and evaporated by centrifugal evaporation followed by lyophilization of the remaining water. Under these conditions, 2 mg of pure MDA-DNPH was obtained. The purity (>95%) was controlled by UV, MS, and NMR.
Method validation
MDA-DNPH was quantified by internal standardization using standard curves prepared from a stock solution of MDA-salt at 3.19 mM (1 mg ml−1) in milli-Q water. The stock solution was further diluted in milli-Q water to obtain calibration solutions at 64.27, 16.06, 8.03, 3.21, 0.80, and 0.32 μM. Standard solutions were subjected to the same conditions as the plasma samples (see extraction process above). A linear regression weighted by 1/x was applied.
An assay validation was carried out using human plasma regarding selectivity, linearity, lowest LODs (LLODs), lowest limits of quantification (LLOQs), intra-day and inter-day precisions, accuracy, recovery, matrix effects, process efficiency, and stability. Selectivity was determined by analyzing blank samples and plasma samples spiked with a known concentration of MDA-DNPH (1.28 μM) upon reconstitution. Linearity was assessed by using 10 standard solutions of MDA-DNPH from 1 to 5,000 ng ml−1 in MeOH 50%. LLODs and LLOQs were defined as the concentrations that produced signals 3 and 10 times higher than the noise level, respectively. Precision, accuracy, recovery, matrix effects, and process efficiency were all determined at low and high concentrations using the “free MDA” and the “total MDA” sample preparations, respectively. Spike solutions were obtained from dilutions in milli-Q water of the MDA stock solution to yield concentrations of 240 nM (similar to physiological levels of plasma free MDA) and 12.8 μM (similar to physiological levels of plasma total MDA). For evaluation of intra-day and inter-day precisions, a plasma sample was spiked, extracted, and analyzed over three consecutive days (five replicates per day). Precision was expressed in percentage of relative SD (RSD). Accuracy was determined by comparing nonspiked with spiked plasma samples and expressed as percentage of deviation between nominal and measured concentrations (n = 5). Extraction recovery was determined by comparing samples spiked with equivalent amounts of MDA-salt and MDA-DNPH before and after extraction, respectively, and the corresponding ratio calculated and expressed in percent (n = 5). Matrix effects were evaluated by comparing plasma samples spiked after extraction with MDA-DNPH with a stock solution of identical concentration (n = 5). Process efficiency was calculated as the product of recovery and matrix effects. Stability of samples stored over time was assessed by comparing freshly collected samples with aliquots of the same samples stored for 1 month at −80°C before analysis was performed.
RESULTS
Sample preparation
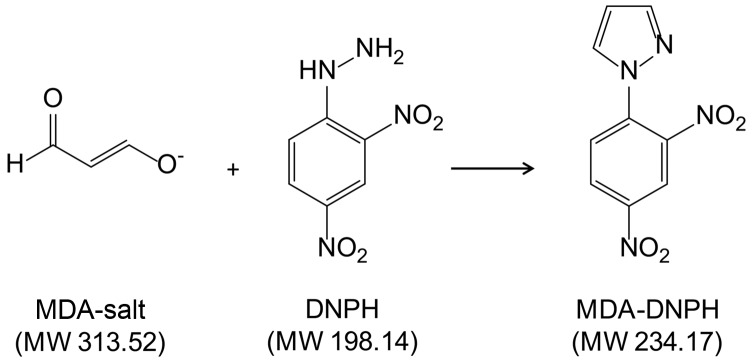
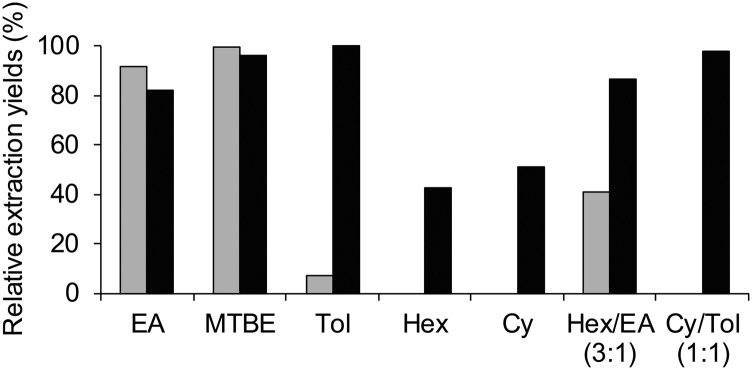
As an alternative to the frequently used but controversial TBA assay, derivatization of MDA with DNPH at room temperature has been shown to yield stable MDA-DNPH derivatives, which can be monitored by both UV and MS detections (12, 30). However, to our knowledge, it has never been investigated whether the MDA-salt is able to react quantitatively and exhaustively with DNPH (Fig. 1). We therefore first synthesized an MDA-DNPH standard according to conditions adapted from previous reports (12) and further isolated it by semi-preparative HPLC to yield a pure standard. All subsequent experiments were then performed relative to the MDA-DNPH standard to determine the different yields of derivatization and extraction. At this stage of the process, UV detection at 358 and 305 nm for DNPH and MDA-DNPH, respectively, was employed. We found that an incubation time of 10 min at room temperature was sufficient for a complete reaction between the MDA-salt and DNPH (added in excess). To eliminate the excess DNPH before analysis, liquid-liquid extraction (LLE) of the aqueous reaction mixture with different organic solvents was evaluated (Fig. 2). While relatively polar solvents, such as EtOAc or MTBE, extracted MDA-DNPH efficiently, they were not able to selectively remove DNPH. In contrast, nonpolar solvents, such as hexane and cyclohexane, totally prevented the extraction of DNPH, but yielded significantly lower amounts of MDA-DNPH. Finally, a 1:1 mixture of toluene:cyclohexane was found to provide excellent recovery of MDA-DNPH and complete removal of the DNPH in excess. We also tested solid phase extraction as a potential alternative to LLE. We found that 100% wettable polymeric-based sorbents, such as Oasis HLB, were able to irreversibly retain low amounts of DNPH, while still releasing MDA-DNPH under strong acidic conditions (TCA 5%). However, they exhibited a loading capacity too low to afford efficient removal of DNPH at the concentration employed for derivatization (data not shown).
Fig. 1.
Derivatization of MDA with DNPH yielding a stable MDA-DNPH derivative. The reaction was performed using the MDA-salt. MW, molecular weight.
Fig. 2.
Relative extraction yields for DNPH (gray bars) and MDA-DNPH (black bars) using various organic solvents and solvent mixtures. Extraction of the aqueous reaction mixture was performed twice with 250 μl of the mentioned solvents. Data are the mean of two to four replicates. EA, ethyl acetate; MTBE, methyl tert-butyl ether; Tol, toluene; Hex, hexane; Cy, cyclohexane.
Free and total MDA were extracted using acidic precipitation and alkaline hydrolysis followed by acidic precipitation, respectively. For the free fraction, TCA 20% was found to efficiently precipitate proteins while keeping MDA intact for further derivatization. For the total MDA, hydrolysis performed in 1 M NaOH at 60°C for 30 min gave the best results, in accordance with Pilz, Meineke, and Gleiter (12). To determine whether MDA was bound to components other than proteins, plasma samples were split into three equal parts and subjected to: i) “normal” protocol involving alkaline hydrolysis of bound MDA from intact plasma; ii) lipid extraction of plasma using a mixture of H2O:chloroform:MeOH (1:2:1, v/v) followed by alkaline hydrolysis; or iii) protein precipitation with ACN followed by resolubilization of the pellet in NaOH 1 M and alkaline hydrolysis. Results showed that proteins and lipids accounted for more than 90% and less than 10% of bound MDA, respectively, which confirms previous findings (12–14, 31).
Optimization of UHPLC-HRMS conditions
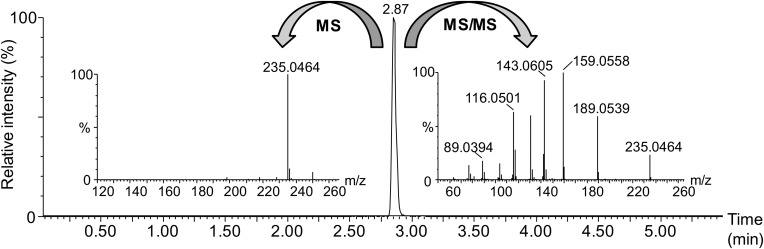
To our knowledge, no study has reportedly used UHPLC coupled to HRMS for measuring MDA in plasma samples. In the present study, we developed a very rapid gradient on an ethylene-bridged hybrid C18 column, which enabled the selective detection of MDA-DNPH as a sharp and single peak at a retention time of 2.87 min (Fig. 3), well after the solvent peak (retention time 0.25 min) and the DNPH peak (retention time 2.35 min). Total run time, including re-equilibration, did not exceed 6 min. Detection was achieved by quadrupole TOFMS in positive electrospray mode and yielded an intense (M+H)+ ion at m/z 235.0464 corresponding to the molecular formula C9H7N4O4 (error 0.2 mDa; calculated mass 235.0462 Da) (Fig. 3). Additional HRMS/MS data confirmed the identity of MDA-DNPH based on fragments at m/z 189.0539 (C9H7N3O2), 159.0558 (C9H7N2O), 143.0605 (C9H7N2), and 116.0501 (C8H6N) (Fig. 3). To obtain maximal sensitivity for MDA-DNPH, several parameters of the mass spectrometer, including capillary voltage (2.8 kV), cone voltage (25 V), desolvation gas flow (800 l h−1), and temperature (450°C), were optimized. The signal was further improved by using the EDC mode, which led to a 2-fold increase in signal-to-noise ratio, as compared with the “normal” mode. To construct extracted ion chromatograms, a mass extraction window of 0.012 Da centered on m/z 235.0462 was selected. Using optimized conditions, a 2.5 μl injection of a 6.5 nM MDA-DNPH solution gave a signal-to-noise ratio equal to 10. Based on values from the literature for MDA levels, we anticipated that this level of sensitivity should be sufficient for quantification of both free and total MDA in plasma.
Fig. 3.
Base peak intensity chromatogram and high resolution mass spectra of MDA-DNPH. Data were obtained from an MDA-DNPH standard (500 ng ml−1) on a quadrupole TOF mass spectrometer at a resolution of approximately 20,000 (full width half maximum at m/z 500). MS spectrum acquired at low collision energy (4 eV) yielded the (M+H)+ ion. MS/MS spectrum acquired at high collision energy (10–35 eV ramp) yielded typical fragments. For fragment annotation, refer to the text.
Internal standardization
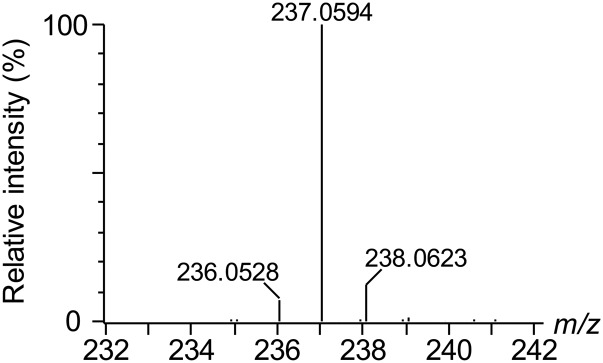
To overcome unwanted variation caused by reactions with nucleophilic species during derivatization, sample preparation loss, and matrix effects, D2-MDA was prepared from D2-TEP using a simple acidic hydrolysis step (29) and used as IS. We found that equivalent molar amounts of MDA-salt and freshly produced D2-MDA provided similar signals after derivatization using UHPLC-HRMS. The peak for D2-MDA-DNPH eluted at 2.86 min and the mass spectrum was dominated by an intense (M+H)+ ion at m/z 237.0594 matching with the molecular formula C9H5D2N4O4 (error 0.6 mDa; calculated mass 237.0588; Fig. 4). Because of the low mass difference (2 Da) between the unlabeled and labeled molecules, it was possible that the M+2 isotope of MDA-DNPH could interfere with the IS peak at high MDA concentrations. While this should not have altered the method’s precision and accuracy, as both samples and calibration standards would be equally affected, it might, however, degrade the linearity of the calibration curve. To ensure that the relative contribution of the M+2 isotope of MDA-DNPH to the D2-MDA-DNPH peak would remain negligible (<2% for the highest calibration standard at 64.27 μM), a high IS concentration of 30 μM was selected. At this concentration, no peak was detected in the mass spectrum for m/z 235.0462 (i.e., the protonated ion of MDA-DNPH), demonstrating the high purity of D2-TEP and, thereby, D2-MDA-DNPH (Fig. 4). D2-MDA was therefore considered a suitable IS for MDA quantification.
Fig. 4.
High resolution mass spectrum obtained for D2-MDA-DNPH at a final concentration of 3 μM. No detectable peak at m/z 235.0462 was observed.
Method validation
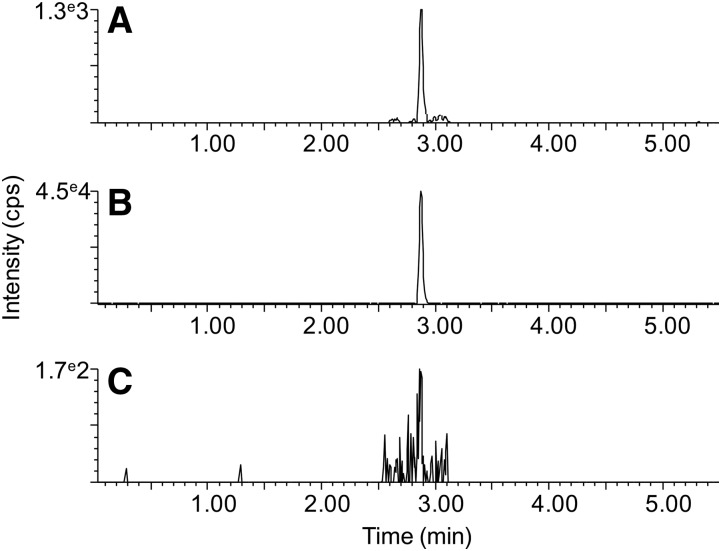
Validation was performed on 10 μl aliquots of human plasma for both free and total MDA using acidic precipitation and alkaline hydrolysis followed by acidic precipitation, respectively. The method was found to be highly selective for MDA-DNPH, as there were no interfering peaks for both free and total MDA in plasma samples (Fig. 5A, B). A blank sample prepared under identical conditions gave no detectable signal (Fig. 5C). Linearity was achieved from 1 ng ml−1 (4.3 nM) to 5,000 ng ml−1 (21.4 μM) (supplemental Table S1, supplemental Fig. S3). All points of the linear regressions obtained from a calibration curve built from the MDA-salt and subjected to the entire sample preparation process fell within ±15% of the nominal value (supplemental Table S2, supplemental Fig. S4). The instrumental LLOD and LLOQ obtained from the injection of standard solutions were 2.1 and 6.5 nM, respectively. The method LLOD and LLOQ, which take into account the dilution factor and recovery of the sample preparation, as well as the matrix effects in the MS source, were 32 and 100 nM, respectively. The results for precision, accuracy, recovery, matrix effects, and process efficiency are presented in Table 1. In brief, precision was excellent at high MDA concentrations (12.8 μM, within the range of expected total physiological levels) and good at low concentrations (240 nM, within the range of expected free physiological levels). At both levels of concentration, accuracy fell within acceptable ranges (80–120%). Extraction recovery was approximately 70%, due to losses during analyte extraction. Matrix effects were negligible and did not impact the MS response. The overall process efficiency was about 65–70%. Plasma samples stored for 1 month at −80°C did not show any significant deviation from freshly collected samples (t-test: n = 5; P = 0.97).
Fig. 5.
Typical chromatograms obtained for plasma and blank samples. Extracted ion chromatograms centered on m/z 235.0462 using a mass extraction window of 0.012 Da are displayed. A: Representative human plasma sample, free MDA fraction. B: Representative human plasma sample, total MDA fraction. C: Blank sample.
TABLE 1.
Assay validation parameters of MDA quantification in human plasma
| Sample Preparation | Concentration (μM) | Precision | Accuracy (%) | Recovery (%) | Matrix Effects (%) | Process Efficiency (%) | |
| RSD1 (%) | RSD2 (%) | ||||||
| Free MDA | 0.24 | 12.8 | 24.9 | 101 | 67 | 104 | 69 |
| Total MDA | 12.8 | 2.9 | 3.0 | 107 | 70 | 91 | 64 |
RSD1, intra-day precision (n = 5); RSD2, inter-day precision over three consecutive days (n = 15).
Taken together, these results demonstrate that the developed method is sensitive, precise, and accurate enough to measure both free and total MDA from minute volumes of plasma samples.
Analysis of plasma samples from different species
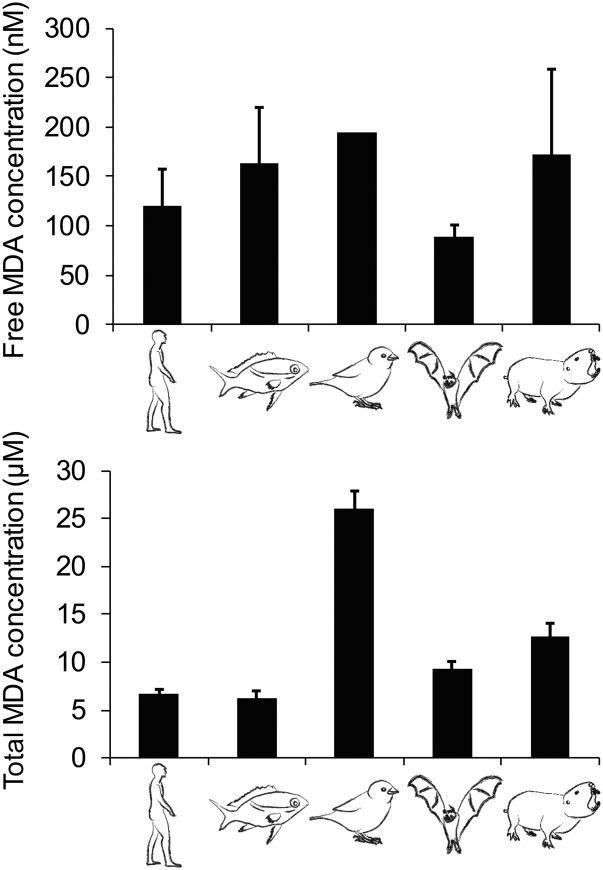
To illustrate the applicability of the developed method, free and total MDA levels were measured in plasma samples of humans as well as other vertebrates from various families and classes. In general, both free and total MDA levels were relatively similar among species with the exception of total MDA in house sparrow, which was much higher than in other species (Fig. 6). Free MDA concentrations ranged from 89 to 195 nM and total MDA concentrations from 6.2 to 26.0 μM. Average values for free and total MDA in human plasma were 120 nM (SD 36.26) and 6.7 μM (SD 0.46), respectively. MDA bound to proteins through the formation of Schiff base adducts (calculated as total minus free MDA), represented about 98% of the total MDA in humans and 97–99% in other species.
Fig. 6.
Levels of free (top) and total (bottom) MDA in plasma of humans and other vertebrates. The studied animal species were (from left to right) Homo sapiens (human), Scolopsis bilineatus (fish), Passer domesticus (bird), Carollia perspicillata (bat), and Fukomys damarensis (rodent). Data are from three replicates + SD, except for free MDA in house sparrow for which only one replicate was available.
DISCUSSION
This study reports a novel method based on UHPLC-HRMS for the quantification of MDA after derivatization with DNPH. Both free and total MDA were measured from minute amounts of plasma, thanks to the high sensitivity of HRMS. To measure free levels, at least 10 μl were needed, while for total MDA, volumes as low as 0.5–1 μl would be sufficient to reliably quantify MDA. In comparison, Pilz, Meineke, and Gleiter (12) attempted to measure free MDA as DNPH derivative by HPLC-UV starting from 1 ml of plasma, but levels were below detection limits. An MS-based approach thus represents a real advantage over UV- or fluorescence-based methods, in particular when sample volumes are restricted (e.g., from newborns, small animals, etc.). Another advantage of HRMS is its high selectivity due to the ability to set very narrow mass extraction windows (0.01–0.02 Da for quadrupole TOFMS) (32): plasma samples provided particularly clean chromatograms with a single peak corresponding to MDA-DNPH and low background noise (Fig. 5). The total chromatographic time was only 6 min, which increased throughput without compromising selectivity and sensitivity.
A key aspect of MDA analysis, in general, is the possible presence of artifactually produced MDA (24, 33), which can be revealed by preparing blank (water) samples. For instance, in our laboratory, the TBA assay usually generates amounts of MDA up to 500 nM in blank matrices subjected to the extraction process, precluding the analysis of free MDA and complicating the determination of total MDA for small plasma volumes. Using the described method based on derivatization with DNPH, we were able to keep MDA levels in blank samples below detection limits (Fig. 5). Nevertheless, it should be noted that very small amounts of MDA may occasionally still be present in blank samples and a series of measures should be taken to avoid interference, particularly during the analysis of free MDA in plasma samples. We determined that contamination of blanks occurs only during LLE with the organic solvent mixture; therefore, high quality solvents should be used and solutions regularly re-prepared. Moreover, the unwanted reaction of residual MDA with DNPH preferentially occurs at low pH, thus increasing the pH to 9–10 before solvent partitioning reduces the risk of contamination. It should be noted that the use of phase partitioning is not absolutely essential, as plasma samples prepared without LLE did not generate more background noise or matrix effects during detection. Yet, excess DNPH over an extended period can be harmful to both HPLC and MS systems, as it may crystallize in the injector and the source, respectively (L. Lachat, unpublished observations). We found that a 1:1 (v/v) mixture of cyclohexane and toluene efficiently removed over 99% of unbound DNPH, while providing very good recovery of MDA-DNPH. Moreover, it enabled the concomitant removal of TCA and other matrix compounds that could potentially affect the robustness of the method over time. We thus highly recommend the use of phase partitioning.
Previous methods for MDA analysis have either relied on external standardization (i.e., no IS) (12, 34) or internal standardization using methyl-MDA (26, 35) or isotopically labeled MDA (13, 25, 36). Isotopically labeled ISs represent the gold standard in LC-MS quantification of complex matrices because they show identical or near identical behavior during sample extraction, chromatography, and MS ionization, but display different masses from their unlabeled isotopomers, which enables their separation in the MS analyzer (37). In the present study, we used D2-MDA as IS and showed its suitability for the quantification of MDA by UHPLC-HRMS. A high IS concentration (30 μM) was used to avoid the relative contribution of the M+2 isotope of MDA-DNPH to the IS peak becoming substantial and possibly affecting the linearity and accuracy of the calibration curve. It should be noted that the M+2 isotope of MDA-DNPH is mostly caused by the presence of 13C2 and that it could theoretically be separated from D2-MDA-DNPH using high resolution MS because both ions have different exact masses (m/z 237.0588 and 237.0529, respectively). However, complete resolution of both peaks would require a much higher resolution (>80,000) than that afforded by our mass spectrometer (approximately 17,000 at m/z 235). In this respect, orbitrap-based systems may represent an attractive alternative to quadrupole TOF-based systems for MDA quantification in the future.
To our knowledge, this is the first study to assess MDA levels across different animal species and classes using the same method. The average levels found for free (120 nM) and total (6.7 μM) MDA in human plasma are within the range of previously found values (12–14, 26, 38), although different methods have been shown to yield quite different results (3, 12, 19, 25). These discrepancies were mainly observed for bound MDA and are likely due to different hydrolytic conditions during sample preparation (18). MDA levels in other species differed from those in humans, although they remained in a similar range. Yet, total MDA levels in the house sparrow constituted an exception, as they were about four times higher than in humans. These particularly high concentrations are possibly species, but not class, related, as Pérez-Rodríguez et al. (39) found large variations in MDA levels between adults of three different bird species.
CONCLUSIONS
Here, we present a highly sensitive and selective method to measure free and total MDA with good precision and accuracy, allowing the use of sample volumes that can be as small as 0.5–1 μl. We show that the method is applicable not only to human samples but also to a variety of other vertebrate species, potentiating its use for comparative studies. Finally, although our method has been validated for plasma samples, we believe that small adaptations can be done to allow measurement of free and total MDA in cell homogenate samples, such as red blood cells. Currently, the physiological meaning of the two forms of MDA remains elusive. Future research should therefore clarify this difference to allow accurate interpretation of free and total/bound MDA measurements.
Supplementary Material
Acknowledgments
The authors are grateful to the two anonymous reviewers, who helped improve the manuscript. The authors thank Edward Farmer for providing D2-TEP; Dominique Roche, Sandra Binning, and Zegni Triki for providing plasma samples from Scolopsis bilineatus; Magali Meniri for providing plasma samples from Carollia perspicillata; and Professor T. H. Clutton-Brock for access to Fukomys damarensis maintained by the Kalahari Research Trust, which has been supported by the European Research Council (Research Grant 294494 to T. H. Clutton-Brock since 1/7/2012), the University of Zurich, and the Mammal Research Institute at the University of Pretoria. The authors are grateful to Raquel Mendonça for providing the drawings of our study species.
Footnotes
Abbreviations:
- ACN
- acetonitrile
- D2-MDA
- dideuterated malondialdehyde
- DNPH
- 2,4-dinitrophenylhydrazine
- D2-TEP
- [D2]-1,1,3,3-tetra-ethoxypropane
- EDC
- enhanced duty cycle
- EtOAc
- ethyl acetate
- HCl
- hydrochloric acid
- IS
- internal standard
- LLE
- liquid-liquid extraction
- LLOD
- lowest LOD
- LLOQ
- lowest limit of quantification
- MDA
- malondialdehyde
- MDA-salt
- malondialdehyde tetrabutylammonium salt
- MeOH
- methanol
- MTBE
- methyl tert-butyl ether
- NaOH
- sodium hydroxide
- PDA
- photodiode array
- PH
- phenylhydrazine
- ROS
- reactive oxygen species
- RSD
- relative SD
- TBA
- thiobarbituric acid
- UHPLC-HRMS
- ultra-HPLC-high-resolution MS
This work was supported by Swiss National Science Foundation Grant PP00P3_139011 to F.H.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Sies H. 1985. Introductory remarks. In Sies H., editor. Academic Press, London: 1–8. [Google Scholar]
- 2.Halliwell B. 1994. Free radicals, antioxidants, and human disease: curiosity, cause, or consequence? Lancet. 344: 721–724. [DOI] [PubMed] [Google Scholar]
- 3.Del Rio D., Stewart A. J., and Pellegrini N.. 2005. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metab. Cardiovasc. Dis. 15: 316–328. [DOI] [PubMed] [Google Scholar]
- 4.Gönenç A., Ozkan Y., Torun M., and Simşek B.. 2001. Plasma malondialdehyde (MDA) levels in breast and lung cancer patients. J. Clin. Pharm. Ther. 26: 141–144. [DOI] [PubMed] [Google Scholar]
- 5.Madazli R., Benian A., Gümüştaş K., Uzun H., Ocak V., and Aksu F.. 1999. Lipid peroxidation and antoxidants in preeclampsia. Eur. J. Obstet. Gynecol. Reprod. Biol. 85: 205–208. [DOI] [PubMed] [Google Scholar]
- 6.Noberasco G., Odetti P., Boeri D., Maiello M., and Adezati L.. 1991. Malondialdehyde (MDA) level in diabetic subjects. Relationship with blood glucose and glycosylated hemoglobin. Biomed. Pharmacother. 45: 193–196. [DOI] [PubMed] [Google Scholar]
- 7.Marcincák S., Sokol J., Bystrický P., Popelka P., Turek P., Bhide M., and Máaté D.. 2004. Determination of lipid oxidation level in broiler meat by liquid chromatography. J. AOAC Int. 87: 1148–1152. [PubMed] [Google Scholar]
- 8.Bus J. S., Aust S. D., and Gibson J. E.. 1976. Paraquat toxicity: proposed mechanism of action involving lipid peroxidation. Environ. Health Perspect. 16: 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monaghan P., Metcalfe N. B., and Torres R.. 2009. Oxidative stress as a mediator of life history trade-offs: mechanisms, measurements and interpretation. Ecol. Lett. 12: 75–92. [DOI] [PubMed] [Google Scholar]
- 10.Metcalfe N. B., and Alonso-Alvarez C.. 2010. Oxidative stress as a life-history constraint : the role of reactive oxygen species in shaping phenotypes from conception to death. Funct. Ecol. 24: 984–996. [Google Scholar]
- 11.Costantini D. 2008. Oxidative stress in ecology and evolution: lessons from avian studies. Ecol. Lett. 11: 1238–1251. [DOI] [PubMed] [Google Scholar]
- 12.Pilz J., Meineke I., and Gleiter C.. 2000. Measurement of free and bound malondialdehyde in plasma by high-performance liquid chromatography as the 2,4-dinitrophenylhydrazine derivative. J. Chromatogr. B Biomed. Sci. Appl. 742: 315–325. [DOI] [PubMed] [Google Scholar]
- 13.Cighetti G., Debiasi S., Paroni R., and Allevi P.. 1999. Free and total malondialdehyde assessment in biological matrices by gas chromatography-mass spectrometry: what is needed for an accurate detection. Anal. Biochem. 266: 222–229. [DOI] [PubMed] [Google Scholar]
- 14.Steghens J. P., van Kappel A. L., Denis I., and Collombel C.. 2001. Diaminonaphtalene, a new highly specific reagent for HPLC-UV measurement of total and free malondialdehyde in human plasma or serum. Free Radic. Biol. Med. 31: 242–249. [DOI] [PubMed] [Google Scholar]
- 15.Esterbauer H., Schaur R. J., and Zollner H.. 1991. Chemistry and biochemistry of 4-hydroxynonenal, malondialdehyde and related aldehydes. Free Radic. Biol. Med. 11: 81–128. [DOI] [PubMed] [Google Scholar]
- 16.Giera M., Lingeman H., and Niessen W. M. A.. 2012. Recent advancements in the LC- and GC-based analysis of malondialdehyde (MDA): a brief overview. Chromatographia. 75: 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mateos R., and Bravo L.. 2007. Chromatographic and electrophoretic methods for the analysis of biomarkers of oxidative damage to macromolecules (DNA, lipids, and proteins). J. Sep. Sci. 30: 175–191. [DOI] [PubMed] [Google Scholar]
- 18.Sobsey C. A., Han J., Lin K., Swardfager W., Levitt A., and Borchers C. H.. 2016. Development and evaluation of a liquid chromatography–mass spectrometry method for rapid, accurate quantitation of malondialdehyde in human plasma. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1029–1030: 205–212. [DOI] [PubMed] [Google Scholar]
- 19.Kadiiska M. B., Gladen B. C., Baird D. D., Germolec D., Graham L. B., Parker C. E., Nyska A., Wachsman J. T., Ames B. N., Basu S., et al. 2005. Biomarkers of Oxidative Stress Study II: are oxidation products of lipids, proteins, and DNA markers of CCl4 poisoning? Free Radic. Biol. Med. 38: 698–710. [DOI] [PubMed] [Google Scholar]
- 20.Knight J. A., Robert K., and Mcclellan L.. 1988. Specificity of the thiobarbituric acid reaction: its use in studies of lipid peroxidation. Clin. Chem. 34: 2433–2438. [PubMed] [Google Scholar]
- 21.Gutteridge J. M., and Tickner T. R.. 1978. The characterisation of thiobarbituric acid reactivity in human plasma and urine. Anal. Biochem. 91: 250–257. [DOI] [PubMed] [Google Scholar]
- 22.Baumgartner W. A., Baker N., Hill V. A., and Wright E. T.. 1975. Novel interference in thiobarbituric acid assay for lipid peroxidation. Lipids. 10: 309–311. [DOI] [PubMed] [Google Scholar]
- 23.Halliwell B., and Chirico S.. 1993. Lipid peroxidation: its mechanisms, measurement, and significance. Am. J. Clin. Nutr. 57 (Suppl.): 715S–724S. [DOI] [PubMed] [Google Scholar]
- 24.Agarwal R., and Chase S. D.. 2002. Rapid, fluorimetric-liquid chromatographic determination of malondialdehyde in biological samples. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 775: 121–126. [DOI] [PubMed] [Google Scholar]
- 25.Yeo H. C., Helbock H. J., Chyu D. W., and Ames B. N.. 1994. Assay of malondialdehyde in biological fluids by gas chromatography-mass spectrometry. Anal. Biochem. 220: 391–396. [DOI] [PubMed] [Google Scholar]
- 26.Sim A. S., Salonikas C., Naidoo D., and Wilcken D. E. L.. 2003. Improved method for plasma malondialdehyde measurement by high-performance liquid chromatography using methyl malondialdehyde as an internal standard. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 785: 337–344. [DOI] [PubMed] [Google Scholar]
- 27.Douny C., Tihon A., Bayonnet P., Brose F., Degand G., Rozet E., Milet J., Ribonnet L., Lambin L., Larondelle Y., et al. 2015. Validation of the analytical procedure for the determination of malondialdehyde and three other aldehydes in vegetable oil using liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) and application to linseed oil. Food Anal. Methods. 8: 1425–1435. [Google Scholar]
- 28.Grund B., Marvin L., and Rochat B.. 2016. Quantitative performance of a quadrupole-orbitrap-MS in targeted LC-MS determinations of small molecules. J. Pharm. Biomed. Anal. 124: 48–56. [DOI] [PubMed] [Google Scholar]
- 29.Weber H., Chételat A., Reymond P., and Farmer E. E.. 2004. Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J. 37: 877–888. [DOI] [PubMed] [Google Scholar]
- 30.Manini P., Andreoli R., Sforza S., Dall’Asta C., Galaverna G., Mutti A., and Niessen W. M. A.. 2010. Evaluation of alternate isotope-coded derivatization assay (AIDA) in the LC-MS/MS analysis of aldehydes in exhaled breath condensate. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 878: 2616–2622. [DOI] [PubMed] [Google Scholar]
- 31.Hong Y. L., Yeh S. L., Chang C. Y., and Hu M. L.. 2000. Total plasma malondialdehyde levels in 16 Taiwanese college students determined by various thiobarbituric acid tests and an improved high-performance liquid chromatography-based method. Clin. Biochem. 33: 619–625. [DOI] [PubMed] [Google Scholar]
- 32.Glauser G., Grund B., Gassner A-L., Menin L., Henry H., Bromirski M., Schutz F., McMullen J., and Rochat B.. 2016. Validation of the mass-extraction-window for quantitative methods using liquid chromatography high resolution mass spectrometry. Anal. Chem. 88: 3264–3271. [DOI] [PubMed] [Google Scholar]
- 33.Lykkesfeldt J. 2001. Determination of malondialdehyde as dithiobarbituric acid adduct in biological samples by HPLC with fluorescence detection: comparison with ultraviolet-visible spectrophotometry. Clin. Chem. 47: 1725–1727. [PubMed] [Google Scholar]
- 34.Mateos R., Lecumberri E., Ramos S., Goya L., and Bravo L.. 2005. Determination of malondialdehyde (MDA) by high-performance liquid chromatography in serum and liver as a biomarker for oxidative stress. Application to a rat model for hypercholesterolemia and evaluation of the effect of diets rich in phenolic antioxidants from fruits. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 827: 76–82. [DOI] [PubMed] [Google Scholar]
- 35.Claeson K., Thorsén G., and Karlberg B.. 2001. Methyl malondialdehyde as an internal standard for the determination of malondialdehyde. J. Chromatogr. B Biomed. Sci. Appl. 751: 315–323. [DOI] [PubMed] [Google Scholar]
- 36.Schmid-Siegert E., Stepushenko O., Glauser G., and Farmer E. E.. 2016. Membranes as structural antioxidants recycling of malondialdehyde to its source in oxidation-sensitive chloroplast fatty acids. J. Biol. Chem. 291: 13005–13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thevenet D., Pastor V., Baccelli I., Balmer A., Vallat A., Neier R., Glauser G., and Mauch-Mani B.. 2017. The priming molecule β-aminobutyric acid is naturally present in plants and is induced by stress. New Phytol. 213: 552–559. [DOI] [PubMed] [Google Scholar]
- 38.Carbonneau M. A., Peuchant E., Sess D., Canioni P., and Clerc M.. 1991. Free and bound malondialdehyde measured as thiobarbituric acid adduct by HPLC in serum and plasma. Clin. Chem. 37: 1423–1429. [PubMed] [Google Scholar]
- 39.Pérez-Rodríguez L., Romero-Haro A. A., Sternalski A., Muriel J., Mougeot F., Gil D., and Alonso-Alvarez C.. 2015. Measuring oxidative stress: the confounding effect of lipid concentration in measures of lipid peroxidation. Physiol. Biochem. Zool. 88: 345–351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.