Abstract
Reactive oxygen species (ROS) arise through normal cellular aerobic respiration, and, in combination with external sources such as ionizing radiation, cigarette tar and smoke, and particulate matter generated by combustion, can have a profound negative effect on cellular macromolecules such as DNA that may lead to a number of human pathological disorders including accelerated aging and cancer. A major end product of ROS damage to DNA is the formation of apurinic/apyrimidinic (AP) sites, which without removal are known to halt mRNA and DNA synthesis, or act as non-coding lesions resulting in the increased generation of DNA mutations. In human cells, the major enzyme in correcting the deleterious effects of AP sites in DNA is through the participation of AP endonuclease (APE), which initiates the removal of baseless sites in DNA through the catalytic scission of the phosphodiester bond 5′ and adjacent to an AP site. Interestingly, APE also possesses an activity (Ref-1) that controls the redox status of a number of transcription factors including Fos and Jun. The means by which APE/Ref-1 is directed to carry out such disparate roles are unknown. The presence of a number of phosphorylation sites scattered throughout both functional domains of APE/Ref-1 however offered one possible mechanism that we reasoned could play a role in dictating how this protein responds to different stimuli. Here we show that the in vitro redox activity of APE/Ref-1 is stimulated by PKC phosphorylation. Furthermore, when human cells were exposed to the PKC activator phorbol 12-myristate 13-acetate, an increase in redox activity was observed that corresponded to an increase in the phosphorylation status of APE/Ref-1. Importantly, human cells exposed to the oxidizing agent hypochlorite, followed by methyl methanesulfanate, responded with an increase in redox activity by APE/Ref-1 that also involved an increase in PKC activity and a corresponding increase in the phosphorylation of APE/Ref-1. These results suggest that the ability of APE/Ref-1 to perform its in vivo redox function is correlated to its susceptibility to PKC phosphorylation that notably occurs in response to DNA damaging agents.
INTRODUCTION
Apurinic/apyrimidinic (AP) sites can arise in DNA under a variety of circumstances. For example, certain alkylating agents as well as free radicals are known to destabilize purines and increase the likelihood of their loss from DNA; in some cases these lesions can further lead to the formation of 3′ phosphate and phosphoglycolate residues that block DNA synthesis. AP sites can also be formed as an intermediate in base excision repair initiated by a DNA glycosylase. Notably, AP sites can also arise spontaneously, where it has been estimated that 10 000 purines and 200 pyrimidines are lost each 24 h cell cycle in human cells (1). If left unchecked, the presence of AP sites is known to block DNA synthesis, or lead to mutations (2).
In view of the deleterious consequences that baseless sites can have on genomic integrity, it is not surprising to find the ubiquitous presence of enzymes that act in their removal. The major activity, at least quantitatively, in human cells is an endonuclease (AP endonuclease or APE) that hydrolytically cleaves DNA 5′ and adjacent to an AP site to initiate its removal (3,4). A second activity is needed for the actual excision of the AP site, which is most likely accomplished through the action of a deoxyribophosphodiesterase. Importantly, DNA polymerase β possesses this activity (5). Since DNA polymerase β and APE have been shown to act together in human cells (6), it can be concluded that these two activities are critical for the repair of baseless sites in DNA.
The gene that encodes human APE has been cloned (7,8) and shown to have sequence similarities to the Exonuclease III family of APEs (7,8).
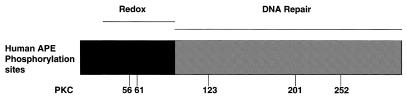
Several years after its cloning, human APE was discovered to also contain vigorous redox activity (Ref-1) through its ability to promote the binding of Jun/Jun homodimers or Fos/Jun heterodimers to a synthetic oligonucleotide containing an AP-1 binding site (9). The observed activation of AP-1 binding is through the use of a domain within APE that exists at the N-terminus of APE (Fig. 1) and is specifically involved in converting AP-1 transcription factors to a reduced, active state (10). This is accomplished by the use of a cysteine residue at position 65, the loss of which abolishes redox activity. More recent evidence indicates that AP-1 transcriptional activity is regulated by a direct association between APE/Ref-1 and the redox-active compound thioredoxin (11).
Figure 1.
Potential PKC phosphorylation sites possessed by APE/Ref-1.
A number of other transcription factors have since been identified that are also subject to redox regulation by APE/Ref-1. These include the p50 subunit of NF-κB (Nuclear Factor kappa B), CREB, Myb, HIF-Iα (12) and Pax-8 (13), a transcription factor important for the correct development of the thyroid gland.
Perhaps the most surprising find in regard to APE/Ref-1 is the recent discovery that it also interacts with the tumor suppressor protein p53, in which APE/Ref-1 acts in converting it from an inert form to an active form (14). The events in this case, however, appear to be both redox dependent and independent. It is important to note that p53, like APE/Ref-1, is an important factor in preserving genomic integrity under stressful conditions. For example, in response to DNA-damaging agents, p53 plays an integral role in either arresting cell-cycle progression or inducing apoptosis (15).
Attempts to alter the endogenous levels of APE/Ref-1 in response to stress, unlike p53, have for the most part met with little success. One exception is a study in which human cells exposed to different DNA damaging agents showed an increase in APE activity that corresponded with an increase in the amount of APE/Ref-1 (16). Specifically, when HeLa cells were first exposed to sublethal doses of the oxidizing agent hypochlorite for 12 h, followed by a second treatment with methyl methanesulfonate (1 h), an increase in both the amount of APE/Ref-1 and a corresponding rise in APE activity was detected in what was described as an ‘adaptive response’.
We have previously shown that APE/Ref-1 contains several putative phosphorylation sites including CK II and PKC (Fig. 1), and that the in vitro phosphorylation of APE/Ref-1 by CK II inhibited APE activity (17). However, when we turned to a number of pharmacological agents known to alter the in vivo expression of CK II, none were shown to reproducibly alter the in vivo expressed APE activity possessed by APE/Ref-1. Since we had not yet examined the redox activity of APE/Ref-1, we tested what effect the phosphorylation by CK II, CK I or PKC had on this activity. It was apparent from these early experiments that, while two of the three kinases had little if any effect on redox activity, PKC phosphorylation of APE/Ref-1 was found to enhance the in vitro binding of Fos and Jun to AP-1 oligonucleotides. Furthermore, we also show here that the conditions described above to elicit an adaptive response lead to a rise in PKC activity, which in turn leads to an increase in the phosphorylation of APE/Ref-1. The end result is an increase in redox activity without a change in the amount of the APE/Ref-1 protein.
MATERIALS AND METHODS
Cells and cell culture
Human myeloid leukemia cell line K562 was used in the studies described here since its expression of APE/Ref-1 is low when compared to other cell lines in our possession. Cells were cultured in RPMI-1640 (Sigma) supplemented with 10% fetal bovine serum (Sigma) and antibiotics (penicillin, 250 U/ml and streptomycin, 250 µg/ml; Gibco BRL) at 95% humidity/5% CO2 in air at 37°C.
Construction of an HA–APE/Ref-1 expression vector
Cells were stably transfected with a mammalian expression vector containing an influenza hemaglutinin (HA)-tagged APE/Ref-1. The HA tag is a nine amino acid (27 bp) addition that follows the ATG start codon: ATG TAC CCA TAC GAT GTG CCA GAT TAC GCT. Polymerase chain reaction (PCR) was used to introduce these base pairs onto the 5′ end of the APE/Ref-1 cDNA. A 5′ primer (GCGAATTCATGGGTAT GCTACACGGTCTAATGGCTCCGAAGCGTGGGAAAAAG with an EcoRI site) was designed that includes these 27 bp in addition to ~15 bp of the APE/Ref-1 and a restriction site, which was used to subclone the PCR fragment into a pcDNA vector. The 3′ primer (CGCCGCTCGAGTCACAGTGCTAGGTATAG with a XhoI site) was designed to anneal to the 3′ end of the APE/Ref-1 cDNA including the stop codon and a restriction site for sub-cloning. APE/Ref-1 was amplified by standard PCR protocols and resulted in APE/Ref-1 containing the HA tag at the amino end.
PKC assay
The PKC activity of cell lysates (5 µg) was tested using the PepTag® PKC assay kit (Promega) following the manufacturer’s instructions. The PepTag® C1 fluorescent peptide (R-L-S-R-T-L-S-V-A-A-K, 500 ng) was phosphorylated by PKC in lysates in reaction buffer (20 mM HEPES pH 7.4, 1.3 mM CaCl2, 1 mM DTT, 10 mM MgCl2 and 1 mM ATP) with 250 µM phosphotidyl serine. Phosphorylation changed the net charge of the peptide substrate from +1 to –1. The phosphorylated and unphosphorylated versions of the substrate were then separated on a 1% agarose gel at pH 8.0 in 50 mM Tris–HCl. The gel was photographed under UV light and the phosphorylated substrate was quantitated by a densitometer.
In vitro phosphorylation of GST–APE/Ref-1 with PKC
The conditions for overexpression and purification of GST–APE/Ref-1 were by previously published procedures (16). Purified GST–APE/Ref-1 (1.5 µg) was phosphorylated in vitro using 5 ng PKC (Promega) in buffer containing 20 mM HEPES pH 7.4, 1.3 mM CaCl2, 0.5 mM DTT, 5 mM MgCl2 and 500 µM ATP with or without 125 µM phosphotidyl serine. The reaction mixture was incubated at 30°C for 30 min. The resulting phosphorylated GST–APE/Ref-1 was stored on ice until use or dephosphorylated by the addition of 10 U of Lambda phosphatase (λ-PPase; New England Biolabs) in buffer containing 50 mM Tris–HCl (pH 7.5), 0.1 mM Na2EDTA, 5 mM DTT, 0.01% Brij 35 and 2 mM MnCl2. The reaction mixture was incubated at 30°C for 30 min, then stored on ice until its incorporation into in vitro assays described below. One unit of λ-PPase is defined as the amount of enzyme that hydrolyzes 1 nM of p-nitrophenyl phosphate (50 mM) per min at 30°C under the conditions described above. The phosphorylated and dephosphorylated GST–APE/Ref-1 proteins were dialyzed against ATP and/or λ-PPase in 20 mM HEPES, 20 mM KCl, 0.05% Triton X-100 and 1 mM DTT.
Drug treatment or 32P labeling of cells
Cells were seeded at 2 × 106 cells/ml 1 h prior to drug treatment or 32P labeling. For labeling cells with 32P inorganic phosphate, cells were washed twice with DMEM and seeded in DMEM 1 h prior to labeling.
HOCl and MMS treatment of wild-type and HA–APE/Ref-1 transfected K562 cells
Cells were treated with 130 nM HOCl (Sigma) for 12 h and then with 200 µM MMS (Fluka) for 1 h. Cells were harvested and lysates prepared as described above.
PMA treatment of HA–APE/Ref-1 transfected K562 cells
Cells were treated with 1.6 µM phorbol 12-myristate 13-acetate (PMA) (Sigma) for 2 h. Cells were harvested by centrifugation and washed twice with ice-cold TBS.
Preparation of cell lysates
Cells were resuspended in 200 µl of mild lysis buffer (1% NP-40, 150 mM NaCl, 10 mM Tris, pH 7.2, 2 mM EDTA, 50 mM NaF, 0.2 mM NaVanadate, 100 U/ml Aprotinin, 10 µg/ml leupeptin and a 100-fold dilution of Phosphatase Inhibitor cocktail; Sigma) and incubated on ice for 20 min. Particulates were removed by centrifugation at 15 000 r.p.m. for 30 min at 4°C. Supernatants were collected and protein amounts determined by the Bradford method.
Electrophoretic mobility shift assay (EMSA) of APE/Ref-1 redox activity
Gel shift Fos–Jun binding reactions (EMSA; 65 µl) contained: GST–APE/Ref-1 (1.5 µg; in vitro phosphorylated or unphosphorylated) and/or cell lysate (9 µg); oligonucleotide (1 ng; Santa Cruz Biotechnology) containing the cognate AP-1 binding site that was 5′ end labeled with [γ-32P]ATP (Amersham), 10 mM Tris, pH 7.5, 50 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 1 mM MgCl2, 4% glycerol and poly dI–dC (500 ng; Phamacia). After a 50 min incubation at 25°C, the reactions were electrophoresed on an 8% polyacrylamide gel, dried and exposed to X-ray film.
Immunoprecipitation
Cell lysates (400 µg) were diluted to a final volume of 100 µl with TETN250 buffer (25 mM Tris–HCl, pH 7.5, 5 mM EDTA, 250 mM NaCl and 1% Triton X-100) and precleared by Protein A Sepharose (Zymed) overnight at 4°C with agitation. A monoclonal antibody against APE/Ref-1 [Novus Biologicals, Littletan, CO; 1:200 in 100 µl TETN250 with 5% w/v BSA (Boehringer Mannheim)] was then added to the precleared lysate and incubated for 1 h at 4°C with agitation to form an immunocomplex. Protein A was added to the mixture and incubated for an additional 2 h at 4°C with agitation to form a Protein A-immunocomplex. The Protein A-immunocomplex was then collected by centrifugation at 10 000 g for 20 s. The pellet was washed once with TETN250 and once with TE buffer (10 mM Tris–HCl, pH 7.5, and 5 mM EDTA). SDS gel-loading buffer (50 µl; 50 mM Tris, pH 6.8, 2% SDS, 0.1% bromophenol blue and 10% glycerol) was added to the pellet and boiled for 3 min. The supernatant was transferred to a fresh tube after centrifugation at 10 000 g. The supernatant (15 µl) was electrophoresed on a 12% SDS–polyacrylamide gel. After electrophoresis, the gel was transferred to a nitrocellulose membrane (Schleicher and Schuell) for western blot analysis or dried and exposed to X-ray film.
Western blotting
Cell lysates (30 µg) or immunoprecipited APE/Ref-1 (15 µl) in SDS gel-loading buffer were electrophoresed on a 12% SDS–polyacrylamide gel. After electrophoresis, the gel was transferred to a nitrocellulose membrane. The membrane was blocked in blocking solution (0.5% non-fat dry milk in 1× PBS) for 1 h at room temperature with agitation. Monoclonal antibody against APE/Ref-1 (1:2000) was added to the blocking solution and incubated for 1 h at room temperature with agitation. The membrane was then washed twice (15 min each) with PBS. The washed membrane was incubated with HRP conjugated secondary antibody (1:3000; Zymed) in blocking solution. The membrane was washed twice (30 min each) with PBS and developed with ECL-plus western blot detection kit (Amersham).
RESULTS
Phosphorylation of APE/Ref-1 by PKC increases in vitro redox activity
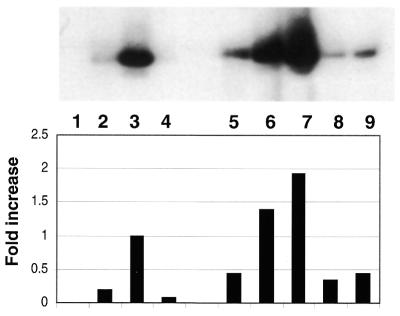
The redox activity possessed by APE/Ref-1 was measured in an in vitro assay that combined extracts of K562 cells (as a source of Fos and Jun), a 5′ end-labeled oligonucleotide containing the cognate AP-1 binding site and various forms of purified GST–APE/Ref-1. The results are presented in Figure 2, in which our original experiments showed that the addition of PKC-phosphorylated GST–APE/Ref-1 to reactions measuring redox activity (lane 5) exhibited less activity than that observed by the addition of unphosphorylated GST–APE/Ref-1 (lane 3). However, if the unincorporated ATP used in the PKC kinase reaction was removed by dialysis prior to redox reactions, phosphorylated GST–APE/Ref-1 in fact produced a modest increase in redox activity (lane 6) when compared to unphosphorylated GST–APE/Ref-1. If GST–APE/Ref-1 was phosphorylated in the presence of an activator/stabilizer of PKC, phosphatidyl serine (lane 7), even greater amounts of redox activity were observed. The addition of phosphatase to PKC-phosporylated GST–APE/Ref-1 (lanes 8 and 9), on the other hand, reduced the amount of redox activity seen in lanes 6 and 7. Since the phosphatases used to dephosphorylate purified GST–APE/Ref-1 could have also affected the phosphorylation status of Fos and Jun present in extracts of K562 cells, we added phosphatase inhibitors after de-phosphorylating GST–APE/Ref-1 and prior to its addition to extracts of K562 cells. No changes from that reflected in reactions resolved in lanes 8 and 9 were observed, confirming that the reduction in redox activity after phosphatase treatment was due to a loss of phosphate groups associated with APE/Ref-1, and not those associated with Fos and Jun.
Figure 2.
In vitro phosphorylation of GST–APE/Ref-1 by PKC increases redox activity as shown by EMSA. An extract of wild-type K562 cells (10 µg) was used in combination with 1.5 µg of purified GST–APE/Ref-1 or PKC-phosphorylated GST–APE/Ref-1. ATP (Promega) was 500 µM, and where indicated 125 µM of phosphotidyl serine was used. Lane 1, oligo alone; lane 2, K562 cell lysate, which was also present in the reactions shown in lanes 3–9; lane 3, purified GST–APE/Ref-1; lane 4, purified GST–APE/Ref-1 and ATP; lane 5, phosphorylated GST–APE/Ref-1 and ATP; lane 6, dialyzed, phosphorylated GST–APE/Ref-1; lane 7, dialyzed GST–APE/Ref-1 phosporylated in the presence of phosphotidyl serine; lane 8, reaction in lane 6, plus phosphatase; lane 9, reaction in lane 7, plus phosphatase
The inhibition of GST–APE/Ref-1 mediated redox activity by ATP was unexpected. Beyond its ability to adversely effect not only the redox activity of the phosphorylated form of GST–APE/Ref-1 (Fig. 2, lane 5), the unphosphorylated form of GST–APE/Ref-1 was also found to be sensitive to ATP inhibition (Fig. 2, lane 4).
Attempts using CK II and CK I to alter the in vitro redox activity of APE/Ref-1 were unsuccessful.
Human cells exposed to a pharmacological agent known to induce PKC results in increased phosphorylation of APE/Ref-1 and enhanced redox activity
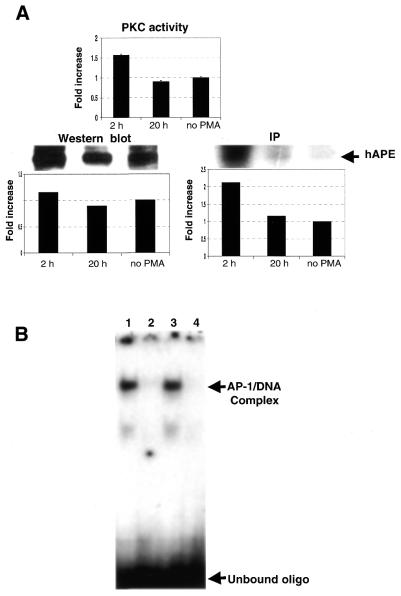
We next tested whether the in vivo phosphorylation status of APE/Ref-1 could be altered similar to that observed in vitro. In order to accomplish this, we exposed stably transfected K562 cells harboring APE/Ref-1 to PMA, an agent that is known to induce PKC in the short term (18). Human K562 cells were exposed to PMA for the time indicated (Fig. 3A), either in the presence or absence of 32P, lysed, and the extracts examined for changes in APE/Ref-1 and PKC. The results show that PMA exposure resulted in a modest increase in PKC activity (Fig. 3A) that was accompanied by a 2-fold increase in the amount of APE/Ref-1 phosphorylation. Western analysis using a monoclonal antibody against APE/Ref-1 showed, however, that the amount of APE/Ref-1 protein did not change either in the presence or absence of PMA (Fig. 3A).
Figure 3.
PMA treatment of APE/Ref-1 stably-transfected K562 cells results in increased amounts of APE/Ref-1 phosphorylation and enhanced redox activity. (A) A monoclonal antibody prepared against APE/Ref-1 was used to immunoprecipitate (IP) the overexpressed protein (and the endogenous expressed APE/Ref-1 as well), and to detect the amount of APE/Ref-1 by western blot as described in Materials and Methods. PKC activity was measured as described in Materials and Methods. (B) The methods for PMA exposure and detection of redox activity by EMSA are provided in the Materials and Methods section. Lane 1, cells exposed to PMA for 2 h; lane 2, immunodepletion of APE/Ref-1 from lysates of cells exposed to PMA for 2 h; lane 3, unexposed cells; and lane 4, immunodepletion of APE/Ref-1 in unexposed cells. The fold increase was quantitated by a densitometer.
We next questioned whether the in vivo redox activity of APE/Ref-1 could be altered using the conditions employed above. Human K562 cells were exposed to PMA for 2 h, lysed, combined with a 5′ end-labeled AP-1 oligonucleotide, and the products separated by EMSA. As can be seen in Figure 3B, short-term exposure to PMA (lane 1) led to an increase in redox activity of roughly 1.4-fold when compared to cells not exposed to PMA (lane 3).
The specificity of the complex formed in Figure 3B was determined in two ways. First, when cell lysates were immunoprecipitated with the APE/Ref-1 monoclonal antibody prior to conducting EMSAs, the AP-1 complex failed to form for both PMA treated cells (lane 2) as well as untreated cells (lane 4). The loss of DNA binding activity was also noted when a 100-fold excess of unlabeled AP-1 oligonucleotide was added to reactions prior to performing the EMSA (not shown). Taken together, the formation of the DNA binding complex observed in Figure 3B was dependent upon APE/Ref-1 and the AP-1 DNA binding site.
It should be noted that the preceding experiments with PMA showed no change in APE activity (not shown).
An ‘adaptive response’ to DNA damaging agents causes stimulation of PKC, increased phosphorylation of APE/Ref-1, and enhanced redox activity
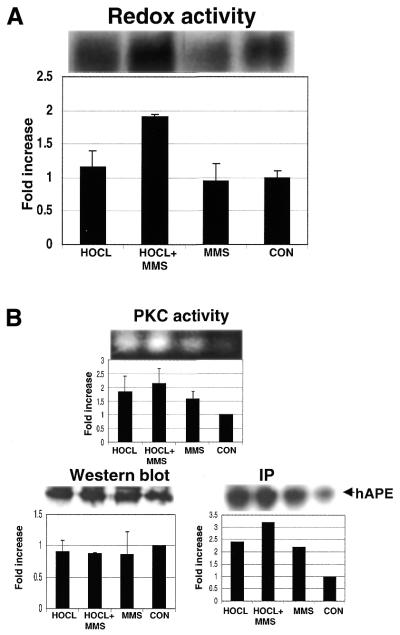
We next turned to a procedure that has been reported to increase the amount of endogenous APE activity (16) to see if the same experimental procedure could influence redox activity as well. Briefly, cells were challenged with a non-toxic dose of a DNA damaging agent, and then subsequently exposed to a secondary DNA damaging agent that, together, was described as eliciting a so-called ‘adaptive response’ in human cells. The agents used to elicit this response (16) were an original challenge with hypochlorite, followed by exposure to MMS. We attempted to execute the same type of experiment as previously described, and in addition we also included an analysis of the phosphorylation status of APE/Ref-1 and any redox changes that might result from exposure to the aforementioned DNA damaging agents. Specifically, stably transfected K562 cells were first exposed for 12 h to hypochlorite (130 nM), and then to MMS (200 µM) for an additional 1 h. Under these conditions, we found that the amount of APE/Ref-1 did not change (Fig. 4B), nor did the amount of APE activity (not shown). On the other hand, an increase in redox activity was evident (Fig. 4A). Furthermore, the increase in redox activity was correlated with an increase in the phosphorylation status of APE/Ref-1 (Fig. 4B). Both were highest in cells exposed to both hypochlorite and MMS. Exposure to hypochlorite and MMS also stimulated a large increase in PKC activity (Fig. 4B) that presumably was responsible for the increase in APE/Ref-1 phosphorylation.
Figure 4.
Exposure to hypochlorite and MMS leads to an increase in PKC activity, phosphorylation (as dewtermined by IP of APE/Ref-1), and redox activity possessed by APE/Ref-1. (A) The conditions for exposure of HA–APE/Ref-1 transfected K562 cells to hypochlorite and MMS are described in Materials and Methods. Redox activity was measured by EMSA. (B) The techniques used here are described in Materials and Methods.
The preceding experiments were also performed in untransfected wild-type K562 cells, which revealed an identical pattern of response as that seen in transfected K562 cells (not shown). The levels of redox activity in wild-type cells were however roughly 30% less than that observed in HA–APE/Ref-1 transfected cells, which should be expected if the source of increased redox activity brought on by DNA damaging agents was indeed due to changes in the phosphorylation status of APE/Ref-1 and not some other protein, or proteins.
DISCUSSION
We previously reported on the presence of a number of potential phosphorylation sites that are scattered throughout the APE/Ref-1 protein (17). In view of the diverse functions that are possessed by APE/Ref-1, we reasoned that these phosphorylation sites may play a role in regulating the protein:protein or protein:DNA interactions involved in redox and DNA repair. Although we were originally encouraged by our in vitro results that CK II phosphorylation might control APE activity (17), we have been unable to show any modification of APE/Ref-1 under a variety of in vivo conditions when the expression of CK II was altered. On the other hand, it appears that PKC phosphorylation does influence the redox activity possessed by APE/Ref-1, and that this conclusion is corroborated by both in vitro and in vivo studies.
The in vivo phosphorylation of APE/Ref-1 has been suggested, but not directly shown, in another study (19). The involvement of a single CK II site at position 121–125 and two CK I sites at positions 123–127 and 158–162 were the only potential phosphorylation sites identified by this study, all of which reside in the DNA repair domain of APE/Ref-1.
The results presented here directly show that the phosphorylation of APE/Ref-1 results in enhanced redox activity but, unlike those of Fritz and Kaina (19), point to the involvement of PKC. This conclusion was established under a variety of conditions that first began by showing that the in vitro binding of AP-1 to its cognate binding site was substantially enhanced in the presence of APE/Ref-1 that had been phosphorylated by PKC (Fig. 2). This result was especially evident when ATP was removed prior to performing EMSAs. The inhibition of the redox activity of APE/Ref-1 by ATP was unexpected since we would not have anticipated a nucleotide to interfere with protein:protein interactions involved in the redox control of Fos and Jun. The inhibition of the APE activity possessed by APE/Ref-1 by small molecules containing a DNA base has been observed previously (3), and seems reasonable since it may interfere with the binding of the repair enzyme to nucleotides present in DNA.
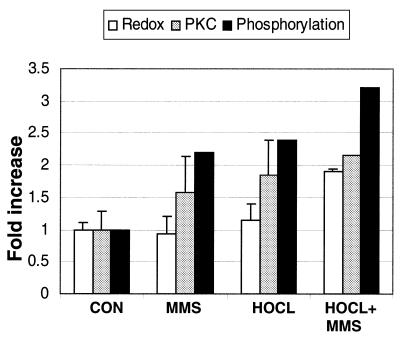
Our in vitro results showing that PKC stimulates the redox activity of APE/Ref-1 were corroborated by two in vivo assays. First, with the use of the pharmacological agent PMA, which is known to induce PKC activity shortly after cellular exposure, increases in redox activity over that for cells not exposed to PMA was observed (Fig. 3). Changes in redox activity were also tested in cells exposed to non-toxic doses of DNA damaging agents that previous studies had concluded elicited an adaptive response in human cells (16). The outcome of those earlier experiments concluded that exposure to hypochlorite (an oxidizer), and MMS (an agent that increases the presence of AP sites in DNA), produced an increase in the amount of APE/Ref-1 in exposed cells. An increase in APE activity was also observed. We have examined a variety of cell lines using the same techniques established by Ramana et al. (16) in an attempt to reproduce not only their observed changes in APE/Ref-1, but to also determine if those changes somehow correlated with alterations in the phosphorylation status of APE/Ref-1. The conditions chosen by Ramana et al. (16) in our hands did not yield results similar to theirs, but did point to an intriguing change in the redox activity possessed by APE/Ref-1. Specifically, long time exposure to the oxidant hypochlorite (12 h), followed by a 1 h incubation with MMS, resulted in cellular changes much like that observed for cells exposed to PMA. For example, exposure to hypochlorite and MMS brought about an increase in PKC activity (Fig. 4B). An increase in the phosphorylation of APE/Ref-1 was also observed. Lastly, elevated redox activity was generated by exposure to hypochlorite and MMS, yet the amount of APE/Ref-1 did not change from that observed in unexposed cells. These results are summarized in Figure 5, which shows that increases in PKC activity are correlated with a rise in APE/Ref-1 phosphorylation and enhanced redox activity that stimulates AP-1 DNA binding activity.
Figure 5.
Bar diagram summarizing the results presented in Figure 4 showing that increases in PKC activity are correlated with increases in both the phosphorylation and redox activity of APE/Ref-1.
Oxidative stress is known to elicit a translocation of cytoplasmic thioredoxin to the nucleus, where its direct association with APE/Ref-1 has been observed (11). This complex is thought to control redox-sensitive transcription factors such as AP-1. Importantly, a recent study shows that ionizing radiation elicits an increase in nuclear thioredoxin that is accompanied by an increase in AP-1 binding activity dependent on the presence of APE/Ref-1 (20).
It is known that oxidants stimulate PKC activity by reacting with its regulatory domain (21). This most likely played a role in producing the increases in PKC activity we observed in cells exposed to the oxidizing agent hypochlorite, which in turn leads to an increase in the phosphorylation of APE/Ref-1. This remains somewhat speculative, however, until we have specifically determined the APE/Ref-1 site(s) that is subject to in vivo phosphorylation. Furthermore, it remains possible that the phosphorylation of APE/Ref-1 has a more important function other than its effect on redox activity, such as it being linked to APE/Ref-1 translocation to the nucleus.
The results presented here nevertheless suggest the involvement of APE/Ref-1 in a signaling pathway that is mediated by a specific kinase that ultimately leads to the more effective redox binding of AP-1 transcription factors to their specific DNA binding sites. Notably, changes in the redox function of APE/Ref-1 appear to result from challenges brought on by agents known to damage DNA. This includes the ones used in this study, as well as the recent observation in cells exposed to ionizing radiation (20). It therefore appears that both functional domains of APE/Ref-1, although appearing to be catalytically diverse, may share a common characteristic in protecting and preserving the integrity of human DNA.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health to W.A.D. and M.R.K.
References
- 1.Lindahl T. and Andersson,A. (1972) Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry, 11, 3618–3623. [DOI] [PubMed] [Google Scholar]
- 2.Loeb L.A. and Preston,B.D. (1986) Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet., 20, 201–230. [DOI] [PubMed] [Google Scholar]
- 3.Kane C.M. and Linn,S. (1981) Purification and characterization of an apurinic/apyrimidinic endonuclease from HeLa cells. J. Biol. Chem., 256, 3405–3414. [PubMed] [Google Scholar]
- 4.Warner H.R., Demple,B.F., Deutsch,W.A., Kane,C.M. and Linn,S. (1980) Apurinic/apyrimidinic endonucleases in repair of pyrimidine dimers and other lesions in DNA. Proc. Natl Acad. Sci. USA, 77, 4602–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsumoto Y. and Kim,K. (1995) Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science, 269, 699–702. [DOI] [PubMed] [Google Scholar]
- 6.Bennett R.A., Wilson,D.M.,III, Wong,D. and Demple,B. (1997) Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc. Natl Acad. Sci. USA, 94, 7166–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demple B., Herman,T. and Chen,D.S. (1991) Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl Acad. Sci. USA, 88, 11450–11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robson C.N. and Hickson,I.D. (1991) Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E.coli xth (exonuclease III) mutants. Nucleic Acids Res., 19, 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xanthoudakis S., Miao,G., Wang,F., Pan,Y.C. and Curran,T. (1992) Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J., 11, 3323–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xanthoudakis S., Miao,G.G. and Curran,T. (1994) The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc. Natl Acad. Sci. USA, 91, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirota K., Matsui,M., Iwata,S., Nishiyama,A., Mori,K. and Yodoi,J. (1997) AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc. Natl Acad. Sci. USA, 94, 3633–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang L.E., Arany,Z., Livingston,D.M. and Bunn,H.F. (1996) Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem., 271, 32253–32259. [DOI] [PubMed] [Google Scholar]
- 13.Tell G., Pellizzari,L., Cimarosti,D., Pucillo,C. and Damante,G. (1998) Ref-1 controls pax-8 DNA-binding activity. Biochem. Biophys. Res. Commun., 252, 178–183. [DOI] [PubMed] [Google Scholar]
- 14.Jayaraman L., Murthy,K.G.K., Zhu,C., Curran,T., Xanthoudakis,S. and Prives,C. (1997) Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev., 11, 558–570. [DOI] [PubMed] [Google Scholar]
- 15.Levine A.J. (1997) p53, the cellular gatekeeper for growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- 16.Ramana C.V., Boldogh,I., Izumi,T. and Mitra,S. (1998) Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc. Natl Acad. Sci. USA, 95, 5061–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yacoub A., Kelley,M.R. and Deutsch,W.A. (1997) The DNA repair activity of human redox/repair protein APE/Ref-1 is inactivated by phosphorylation. Cancer Res., 57, 5457–5459. [PubMed] [Google Scholar]
- 18.Lu Z., Liu,D., Hornia,A., Devonish,W., Pagano,M. and Foster,D.A. (1998) Activation of protein kinase C triggers its ubiquitination and degradation. Mol. Cell. Biol., 18, 839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritz G. and Kaina,B. (1999) Phosphorylation of the DNA repair protein APE/REF-1 by CKII affects redox regulation of AP-1. Oncogene, 18, 1033–1040. [DOI] [PubMed] [Google Scholar]
- 20.Wei S.J., Botero,A., Hirota,K., Bradbury,C.M., Markovina,S., Laszlo,A., Spitz,D.R., Goswami,P.C., Yodoi,J. and Gius,D. (2000) Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res., 60, 6688–6695. [PubMed] [Google Scholar]
- 21.Gopalakrishna R. and Jaken,S. (2000) Protein kinase C signaling and oxidative stress. Free Radic. Biol. Med., 28, 1349–1361. [DOI] [PubMed] [Google Scholar]