Abstract
APOC3 is produced mainly by the liver and intestine and approximately half of plasma APOC3 associates with HDL. Though it was believed that APOC3 associates with HDL by simple binding to preexisting particles, recent data support that biogenesis of APOC3-containing HDL (APOC3-HDL) requires Abca1. Moreover, APOC3-HDL contributes to plasma triglyceride homeostasis by preventing APOC3 association with triglyceride-rich lipoproteins. Interestingly, APOC3-HDL also shows positive correlation with the morbidly obese phenotype. However, the roles of APOC3 in HDL functionality and adipose tissue metabolic activity remain unknown. Therefore, here we investigated the direct effects of APOC3 expression on HDL structure and function, as well as white adipose tissue (WAT) and brown adipose tissue (BAT) metabolic activity. C57BL/6 mice were infected with an adenovirus expressing human APOC3 or a recombinant attenuated control adenovirus expressing green fluorescent protein and blood and tissue samples were collected at 5 days postinfection. HDL was then analyzed for its apolipoprotein and lipid composition and particle functionality. Additionally, purified mitochondria from BAT and WAT were analyzed for uncoupling protein 1, cytochrome c (Cytc), and Cytc oxidase subunit 4 protein levels as an indirect measure of their metabolic activity. Serum metabolomic analysis was performed by NMR. Combined, our data show that APOC3 modulates HDL structure and function, while it selectively promotes BAT metabolic activation.
Keywords: cholesterol efflux, high density lipoprotein, lipidomics, metabolomics, brown adipose tissue, white adipose tissue
APOC3 is a 79 amino acid glycoprotein secreted by the liver and intestine (1) in a lipid-poor form (2). APOC3 plays an important role in plasma triglyceride metabolism because numerous epidemiological and animal studies have established a direct correlation of plasma APOC3 levels to plasma triglyceride levels, and an inverse relationship to the rate of postprandial triglyceride clearance (3–10). Moreover, many in vitro studies suggest a role of APOC3 in the catabolism of triglyceride-rich lipoproteins (11–19). Based on this important function of APOC3, silencing antisense oligonucleotides targeting Apoc3 expression are currently in clinical trials for the treatment of hypertriglyceridemia and its related pathologies (20).
Approximately half of plasma APOC3 is associated with HDL (21, 22) and the rest is distributed between VLDL and chylomicrons (21). Trace quantities of APOC3 may also be found in IDL and LDL particles (23, 24). Though initial studies suggested that association of APOC3 with HDL is a matter of simple binding of the lipid-poor protein to preexisting HDL particles and the result of a free APOC3 exchange between lipoproteins (21), more recent studies show that formation of APOC3-containing HDL (APOC3-HDL) requires the action of the lipid transporter, Abca1, in a fashion similar to Apoa1-containing HDL (25). Moreover, formation of APOC3-HDL is crucial for maintaining physiological plasma triglyceride levels, by sequestering away APOC3 and preventing its association with triglyceride-rich lipoproteins (25). Specifically, in the absence of Abca1, such as in the case of abca1-deficient (abca1−/−) mice infected with recombinant attenuated adenovirus expressing the human APOC3 protein and the green fluorescent protein (GFP) (AdGFP-APOC3), no APOC3-HDL could be formed (25). In that case, all plasma APOC3 accumulated only on VLDL, resulting in inhibition of lipolysis of VLDL triglycerides and hypertriglyceridemia, even at low plasma concentrations of APOC3 (25). In contrast, when Abca1 was expressed, APOC3 was preferentially associated with HDL, allowing only minimal presence of APOC3 on VLDL and subsequent normal lipolysis of VLDL triglycerides (25).
For years, HDL has been an intriguing lipoprotein that attracted the attention of biomedical community, mainly due to its important role in atheroprotection (26). The inverse correlation between HDL cholesterol (HDL-C) levels and the risk for developing coronary heart disease (27–32) suggested that high HDL-C levels in plasma are protective against the development of atherosclerosis. As a result, the vast majority of studies in the literature focused on the understanding of HDL-C in human pathology, a simplified approach to HDL that dates back to the early days when little was known about HDL structure and function. However, more recent data from experimental mice and clinical trials indicated that HDL particle functionality, as determined by its apolipoprotein and lipid content, is far more important in atheroprotection than HDL-C levels alone (30, 32). Of note, recent data indicate that the HDL apoproteome dictates its lipidome and both jointly regulate HDL particle functionality (33), suggesting that the understanding of the HDL proteome and the factors affecting it is crucial for successful improvement of HDL functionality.
In a clinical trial of morbidly obese subjects, we observed significant levels of APOC3-HDL particles in plasma immediately prior to bariatric surgery (34), while 6 months after the operation all HDL was mainly APOA1-HDL with only trace amounts of APOC3-HDL being present (34). This change in apolipoprotein content of HDL was associated with alterations in the antioxidant properties of HDL and plasma enzymatic activities of LCAT and cholesteryl ester transfer protein (CETP) (34). These clinical observations supported the interesting hypothesis that changes in APOC3 content of HDL associate with alterations in HDL particle functionality and overall body energy metabolism.
To test this hypothesis, here we investigated the effects of increased APOC3 expression on HDL structure and function, using adenovirus-mediated gene transfer of human APOC3 to C57BL/6 mice. Because our clinical data identified a correlation of APOC3-HDL with the morbidly obese phenotype (34), we also studied the effects of elevated plasma APOC3 levels on white adipose tissue (WAT) and brown adipose tissue (BAT) mitochondrial activity. Our combined data from biochemical, lipidomic, and metabolomic analyses indicate that APOC3 may alter apolipoprotein and lipid composition of HDL, triggering distinct changes in its properties. Moreover, it stimulates oxidative phosphorylation toward ATP production selectively in BAT.
MATERIALS AND METHODS
Animals
Forty C57BL/6 mice aged 16–18 weeks were allowed unrestricted access to food (standard diet) and water under a 12 h light/dark cycle (7:00 AM to 6:59 PM, light). Sample size was determined based on the desired power of statistical analysis, using an online statistical tool (http://www.stat.ubc.ca/~rollin/stats/ssize/n2.html). All animal experiments were conducted according to the European Union guidelines of the Protocol for the Protection and Welfare of Animals. The work was authorized by the Laboratory Animal Centre committee of the University of Patras Medical School and the Veterinary Authority of the Prefecture of Western Greece.
Construction of AdGFP-APOC3
The construction of AdGFP-APOC3 has been described previously (25). Briefly, construction was performed using the AdEasy-1 system (35), where the complete adenovirus genome, including the human genomic DNA sequences of APOC3 under the control of the CMV promoter, was constructed by homologous recombination in bacteria BJ-5183 cells (35), as described previously (25, 36–39). The resulting recombinant vector also contained the GFP gene under the independent control of a second CMV promoter, which enabled detection of the infected cells and tissues by green fluorescence. Correct recombinant clones were propagated in RecA DH5a cells and the recombinant vector was used to infect HEK293 cells (40), as described previously (39). As control, an empty recombinant attenuated adenovirus expressing only GFP (AdGFP) (39) was used.
Expansion and purification of AdGFP-APOC3
Following plaque identification and isolation, adenoviruses were expanded in HEK293 cells (40) and then purified by double CsCl ultracentrifugation, followed by dialysis and titration of the recombinant adenoviruses, as described previously (39).
Ectopic expression of APOC3 by adenovirus-mediated gene transfer
Mice were injected intravenously through the tail vein with doses of either 5 × 108 plaque-forming units (pfu) (AdGFP-APOC3_L) or 2 × 109 pfu of AdGFP or AdGFP-APOC3 (AdGFP-APOC3_H) virus, as indicated (25). Following 4 h of fasting, blood samples were collected from the tail vein 5 days postinjection.
Isolation and fractionation of plasma and serum by density gradient ultracentrifugation
Following 4 h of fasting, plasma and serum samples were isolated. Pooled plasma (0.4 ml) from each mouse group was fractionated by KBr density gradient ultracentrifugation over a 4 ml KBr (Sigma-Aldrich, St. Louis, MO) gradient (1.23 g/ml over 1.21 g/ml over 1.063 g/ml over 1.019 g/ml over saline), as described previously (25). For all analyses, equal volumes of all isolated HDL fractions were pooled together, except for the determination of the anti-inflammatory capacity of HDL, where single fractions of endotoxin-free serum lipoproteins were fractionated by a different density gradient (1.063 g/ml over 1.019 g/ml over 1.0063 g/ml) and separation of 4 ml of pooled serum samples was performed under aseptic conditions, as described previously (41).
Isolation of mitochondria
Mitochondria from BAT and WAT were isolated, as described previously (42). The protein concentration of each mitochondrial sample was determined using the DCTM protein assay kit (catalog number 500-0116; Bio-Rad) (41).
Determination of plasma and lipoprotein lipid levels
Total cholesterol levels were assessed spectrophotometrically in plasma samples and lipoprotein fractions by using the DiaSys Cholesterol FS kit (reference number 11300; Diagnostic Systems, GmbH, Holzheim, Germany) according to the manufacturer’s instructions and as described previously (25). Free cholesterol levels were determined spectrophotometrically using the free cholesterol E kit, CHOD-DAOS method (catalog number 435-35801, Wako Chemicals). Esterified cholesterol was determined by subtracting free cholesterol from total cholesterol and multiplying the difference by 1.67 to correct for the fatty acid moiety, as proposed by Chapman et al. (43). Triglyceride and phospholipid levels were also assessed spectrophotometrically in plasma samples and lipoprotein fractions by using the DiaSys Triglycerides FS kit (reference number 15710, Diagnostic Systems, GmbH) and the Lab Assay phospholipid determination kit (catalog number 296-63801, Wako Chemicals), respectively, according to manufacturers’ instructions (41, 44). Total protein in lipoprotein fractions was measured by the Lowry assay using the DCTM protein assay kit (catalog number 500-0116, Bio-Rad) (41).
Western blot analyses
For the semiquantitative measurement of human APOC3 and murine Apoc3, Apoa1, Apoa2, Apoe, Apoc1, and Apoc2 in lipoprotein fractions, Western blot analysis was performed as described previously (41, 44, 45). Goat anti-APOC3 (catalog number K74140G), anti-Apoa2 (catalog number K34001G), anti-Apoc1 (catalog number K74110G), and anti-Apoc2 (catalog number K59600R) antibodies from Meridian Life Science were used, respectively, as primary, and a rabbit anti-goat antibody (catalog number sc-2768, Santa-Cruz) as secondary. For the detection of Apoa1, Apoe, and Apoc3, rabbit anti-Apoa1 (catalog number AP23390PU-N, Acris), anti-Apoe (catalog number K23100R, Meridian Life Science), and anti-Apoc3 (catalog number ab55984, Abcam, and catalog number PAB5869, Abnova) were used, respectively, as primary and a goat anti-rabbit antibody (catalog number 7074, Cell Signaling, Danvers, MA) as secondary. Western blotting for cytochrome c (Cytc), uncoupling protein 1 (Ucp1), and Cytc oxidase subunit 4 (Cox4) was performed using rabbit anti-mouse antibodies (catalog number 4272, Cell Signaling; catalog number GTX10983, Acris, Herford, Germany; and catalog number 4844, Cell Signaling, respectively). SDS-PAGE of pure mitochondrial extracts was performed using 6 μg protein per sample for BAT and 15 μg protein per sample for WAT. Semiquantitative determination of the relative protein amounts was performed by Image J free software.
Real-time PCR analysis of gene expression
Total RNA was extracted from fresh frozen liver tissue using TRIzol reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions. Reverse transcription was performed using the PrimeScriptTM RT reagent kit from Takara Bio Inc. (catalog number RR037A; Otsu, Shiga, Japan). Real-time PCR was performed in a MicroAmp® 96-well reaction plate from Applied Biosystems (catalog number 4346906; Foster City, CA) using the KAPA SYBR® FAST Universal qPCR kit from Kapa Biosystems (catalog number KK4601; Woburn, MA) in an Applied Biosystems® StepOnePlus™ cycler. The primers used are shown in supplemental Table S1. Primers were synthesized by Eurofins Genomics (Ebersberg, Germany). Data were normalized for rps18 expression.
Lipidomic analysis of APOC3-HDL isolated from C57BL/6 mice infected with AdGFP-APOC3
APOC3-HDL samples were analyzed using the HILIC LC/MS/MS method. Briefly, lipids were separated on a Kinetex HILIC 150 × 2.1 mm, 2.6 μm column (Phenomenex, Torrance, CA) using a Prominence HPLC (Shimadzu). Elution was lipid polar head group dependent. Lipids were detected using scheduled multiple reaction monitoring on polar head group-specific fragments using a QTrap 4000 (AB Sciex) and were quantified using 23 internal standards and 37 calibration curves. An in-house developed R script was used to correct for isotopic contribution to multiple reaction monitoring signals. HDL from AdGFP-infected mice was used as control.
Electron microscopy analysis of HDL particles
Electron microscopy analysis of purified HDL particles was performed at the Laboratory of Electron Microscopy and Microanalysis of the University of Patras. Briefly, APOC3-HDL particles were visualized by transmission electron microscopy (TEM) at 200,000× magnification, then photographed and analyzed as described previously (34, 46). HDL from AdGFP-infected mice was used as control. By visual inspection, the distributions of HDL particle diameter and area for the samples were evidently skewed. Image analysis of TEM pictures was followed by unpaired t-test statistical analysis for comparison of particle diameters between APOC3-HDL and control-HDL samples. Image analysis of TEM pictures was performed with Image J software.
Nondenaturing two-dimensional electrophoresis analysis of APOC3-HDL isolated from C57BL/6 mice infected with AdGFP-APOC3
Nondenaturing two-dimensional electrophoresis analysis of APOC3-HDL was performed as described previously (34, 46). HDL from AdGFP-infected mice was used as control. The assignment of mobility to the various HDL subpopulations was based on the work of Asztalos et al. (47).
Lcat activity assay
Lcat enzymatic activity in total plasma samples from AdGFP- and AdGFP-APOC3-infected mice was measured using an Lcat activity assay fluorometric kit (catalog number 428900; Calbiochem®, San Diego, CA) according to manufacturer’s instructions and as described previously (34). Enzymatic activity was assessed by measuring the ratio of 390 nm (hydrolyzed fluorescent substrate) over 470 nm (intact fluorescent substrate) emission intensity following excitation at 320 nm 4 h after assay initiation.
Lipoprotein-associated phospholipase A2 activity assay
Lipoprotein-associated phospholipase A2 (Lp-pla2) activity was measured by a modification of the TCA precipitation procedure in plasma using [3H]platelet activating factor ([3H]PAF) (1-O-hexadecyl-2-[3H-acetyl]sn-glycero-3-phosphocholine) (10 Ci/mmol; DuPont-New England Nuclear, Boston, MA) as a substrate at a final concentration of 100 μM, as previously described (48). Briefly, 50 μl of diluted plasma [1:25 v/v with HEPES (pH 7.4) buffer, which consists of 4.2 mM HEPES, 137 mM NaCl, 2.6 mM KCl, and 2 mM EDTA] as the source of the enzyme or blanks (for determination of background) were mixed with assay buffer (HEPES) to a final volume of 90 μl. The reaction was allowed to proceed for 10 min at 37°C by addition of 10 μl of working substrate solution (2 mM [3H]PAF in 2.5 mg/ml BSA, prepared fresh daily). The reaction was terminated by addition of 20 μl ice-cold aqueous BSA solution (100 mg/ml) followed by vortex-mixing and incubation for 10 min at 4°C. Then, 80 μl of ice-cold TCA solution (20% TCA) were added and the mixture was incubated for an additional 30 min at 4°C. The samples were centrifuged at 6,000 g for 5 min at 4°C and 100 μl of supernatant were transferred to 2 ml of scintillation fluid. The amount of [3H] tracer was then quantitated in a liquid scintillation counter (Packard Tri-Card 2100). Lp-pla2 activity was expressed as nanomoles PAF degraded per milliliter of plasma per minute, as described previously (48).
HDL antioxidant capacity
For the assessment of the anti-oxidant capacity of APOC3-HDL, a modification of the dihydrorhodamine 123 method of Kelesidis et al. (49) was used, as described previously (33, 34). The oxidation rate of dihydrorhodamine in the presence of APOC3-HDL was assessed. HDL from AdGFP-infected mice (control-HDL) was used as control. The oxidation rate was calculated for each sample as the mean slope for the linear regression of fluorescence intensity between 10 and 60 min of quadruplicates and expressed as fluorescence units per minute: the lower the slope, the lower the substrate oxidation rate.
Total cholesterol efflux assay
The cholesterol efflux capacity of APOC3-HDL was measured using the RAW 264.7 macrophage cell line, as described previously (33). Total [14C]cholesterol input was calculated as the sum of [14C]cholesterol in the efflux medium plus [14C]cholesterol present in the cell lysate. The rate of HDL-stimulated [14C]cholesterol efflux was expressed as the ratio of total [14C]cholesterol effluxed onto APOC3-HDL and corrected for nonspecific [14C] tracer efflux measured in the absence of HDL. HDL from AdGFP-infected mice was used as control.
Effects of APOC3-HDL on LPS-induced inflammation in RAW 264.7 cells
The effects of APOC3-HDL on inflammation were assessed in the RAW 264.7 macrophage cell line, as described previously (41). Briefly, cells were seeded in 96-well plates (7 × 104 cells/well) and cultured in DMEM containing 10% FBS and 1% penicillin/streptomycin. After a 16 h incubation at 37°C, cultures were washed with serum-free DMEM medium and treated for 4 h with lipopolysaccharide (LPS) (100 ng/ml) that was preincubated for 30 min at 37°C with serum HDL. The amount of HDL used in the assay was defined by the total final concentration of HDL proteins added to the incubation medium. As control, cells were incubated with DMEM containing serum HDL in the absence of exogenous LPS. Following incubation, medium was collected and TNFα was determined by ELISA, as described previously (41).
Metabolomic analysis of serum isolated from C57BL/6 mice infected with AdGFP-APOC3
Two hundred microliters of each serum sample were mixed with 60 μl of D2O and 340 μl of phosphate buffer [1.5 M Na2HPO4 in water containing 4% NaN3 and 1 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (pH 7.4)]. Then, 550 μl of the resulting supernatant were transferred in a 5 mm NMR tube (Bruker BioSpin srl) for analysis. NMR serum metabolomic profiling was performed using a Bruker Avance III HD 700 MHz NMR spectrometer equipped with a TCI cryogenically cooled probe. Three 1H-NMR spectra were acquired for each sample applying standardized protocols (NOESY, CPMG, and J-resolved pulse sequences). Partial least squares discriminant analysis (PLS-DA) was performed as a supervised method for data reduction and pattern recognition. The quality of the PLS-DA model was determined by the goodness-of-fit parameter (R2) and the goodness-of-prediction parameter (Q2) following a 10-fold cross validation procedure. Univariate analysis was conducted by calculating the relative concentration through signal’s area integration of all successfully assigned metabolites across the 21 acquired spectra. P values <0.05 were determined using the nonparametric Wilcoxon rank-sum test. NMR data were acquired and processed using TopSpin 3.6 (Bruker BioSpin srl). Serum metabolite identification was performed using Chenomx NMR software (Profiler 8.1; Chenomx Inc., Canada), the public database (HMDB), and literature data (50, 51). Glucose levels were further assessed spectrophotometrically in serum samples using the DiaSys Glucose GOD FS kit (reference number 10021; Diagnostic Systems, GmbH).
Statistical analysis
All data sets were tested using the Kolmogorov-Smirnov and Shapiro-Wilk tests and were treated with parametric (P > 0.1) or nonparametric tests (P < 0.1) according to their deviation from normality. Data are reported as mean ± SEM. All statistical tests were performed using the GraphPad Prism 6 software.
RESULTS
Expression of human APOC3 in C57BL/6 mice by adenovirus-mediated gene transfer
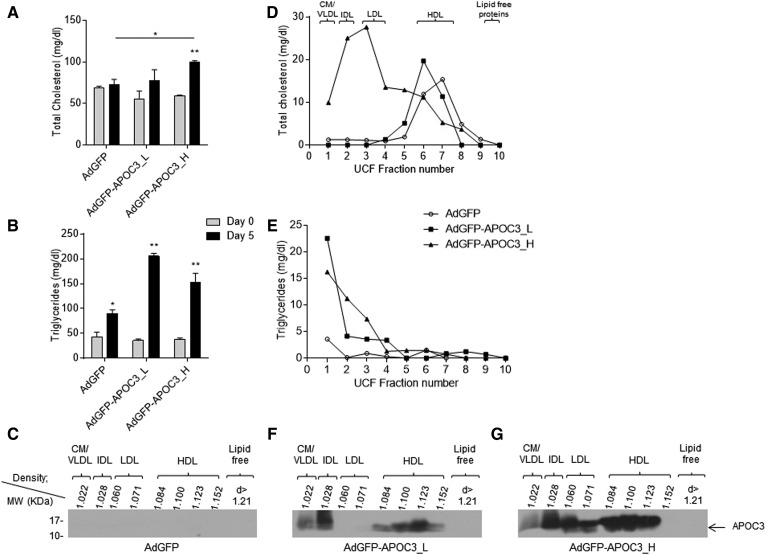
To produce HDL particles rich in APOC3 (APOC3-HDL), C57BL/6 mice were infected with either 5 × 108 pfu or 2 × 109 pfu of the recombinant adenovirus, AdGFP-APOC3, as described previously (25). To assess potential nonspecific effects of viral infection in subsequent analyses, an additional mouse group was also infected with 2 × 109 pfu of the control AdGFP virus (39). Analysis of plasma lipid levels in mice infected with 2 × 109 pfu of AdGFP-APOC3 showed that, 5 days postinfection, expression of APOC3 resulted in a significant increase of plasma total cholesterol and triglyceride levels, compared with the same mice prior to infection (day 0) or AdGFP-infected mice 5 days postinfection (Fig. 1A, B). This increase was associated with a marked increase in chylomicrons/VLDL, IDL, and LDL cholesterol and triglycerides and a shift of HDL-C distribution toward larger and less dense HDL particles (Fig. 1D, E). Interestingly, these mice also displayed elevated phospholipid and free cholesterol levels in their HDL. Mice infected with 5 × 108 pfu of AdGFP-APOC3 showed that, at this lower level of expression, APOC3 does not have any major effect on plasma cholesterol and phospholipid content, though it triggers an increase in plasma triglyceride levels due to accumulation of triglyceride-rich VLDL (Fig. 1A, B, D, E). The relative lipid content of APOC3-HDL expressed as milligrams of total HDL protein or weight percent content of total lipids in HDL is shown in Table 1. Western blot analysis of plasma lipoprotein fractions confirmed APOC3 expression in the mice infected with the two different doses of AdGFP-APOC3 (Fig. 1F, G). In mice infected with 5 × 108 pfu of adenovirus, APOC3 was found only on HDL and IDL/VLDL (Fig. 1F), while in mice infected with 2 × 109 pfu, APOC3 was distributed on all lipoprotein fractions (Fig. 1G), yielding a lipoprotein distribution reminiscent of many morbidly obese patients with BMI >50 prior to bariatric surgery (34). As expected, mice infected with AdGFP showed no detectable levels of human APOC3 in their plasma (Fig. 1C).
Fig. 1.
Total plasma cholesterol (A) and triglyceride (B) levels at the indicated days and density gradient ultracentrifugation cholesterol (D) and triglyceride (E) profiles of C57BL/6 mice 5 days postinfection with 2 × 109 pfu of the control adenovirus, AdGFP, or 5 × 108 pfu (AdGFP-APOC3_L) and 2 × 109 pfu (AdGFP-APOC3_H) of the recombinant adenovirus expressing human APOC3 (AdGFP-APOC3). Western blot analyses (C, F, G) of density gradient ultracentrifugation fractions from plasma of mice infected with 2 × 109 pfu AdGFP (C) or 5 × 108 pfu (F) and 2 × 109 pfu (G) AdGFP-APOC3 5 days postinfection. Data were analyzed using two-way ANOVA and are presented as mean ± SEM. CM, chylomicron. *P < 0.05, **P < 0.005.
TABLE 1.
Lipid composition of HDL
| AdGFP | AdGFP-APOC3_L | AdGFP-APOC3_H | P | |||
| AdGFP versus AdGFP-APOC3_L | AdGFP versus AdGFP-APOC3_H | AdGFP-APOC3_L versus AdGFP-APOC3_H | ||||
| Free cholesterol | 0.10 ± 0.013 (8.09%) | 0.23 ± 0.03 (13.68%) | 1.33 ± 0.029 (19.7%) | 0.0565 | 0.0007 | 0.0015 |
| Esterified cholesterol | 0.30 ± 0.015 (24.1%) | 0.52 ± 0.09 (31.01%) | 0.31 ± 0.02 (4.56%) | 0.1238 | 0.6094 | 0.1404 |
| Triglycerides | 0.02 ± 0.013 (1.61%) | 0.016 ± 0.003 (0.95%) | 0.23 ± 0.004 (3.38%) | 0.8271 | 0.0046 | 0.0007 |
| Phospholipids | 0.81 ± 0.001 (66.2%) | 0.92 ± 0.012 (54.35%) | 4.88 ± 0.415 (72.35%) | 0.0127 | 0.0102 | 0.0108 |
| Total lipids | 1.22 ± 0.01 | 1.7 ± 0.065 | 6.75 ± 0.46 | 0.0186 | 0.0069 | 0.0083 |
Data are expressed as milligrams of lipid per milligram of total HDL protein. Numbers in parenthesis indicate the weight percent content of total lipids in the HDL isolated from AdGFP- or AdGFP-APOC3-infected mice. AdGFP-APOC3_L and AdGFP-APOC3_H refer to mice infected with 5 × 108 or 2 × 109 pfu of AdGFP-APOC3, respectively.
APOC3 changes apolipoprotein composition of HDL
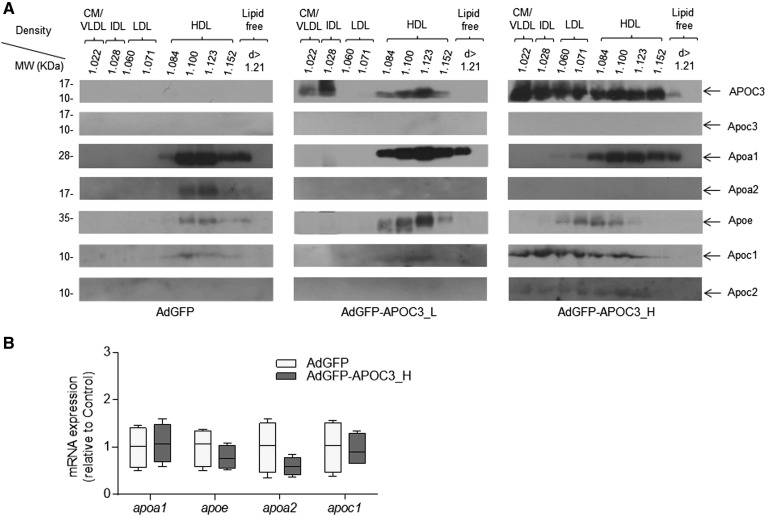
Fractionation of plasma samples by density gradient ultracentrifugation followed by Western blot analysis of lipoprotein fractions for human APOC3, Apoa1, Apoa2, Apoe, Apoc1, Apoc2, and murine Apoc3, showed that both low (5 × 108 pfu) and high (2 × 109 pfu) APOC3 expression result in qualitative and quantitative changes in apolipoprotein composition of HDL and other lipoprotein classes (Fig. 2A). As shown before (Fig. 1G) in mice infected with 2 × 109 pfu AdGFP-APOC3, APOC3 was distributed among all lipoprotein subclasses. Of note, expression of APOC3 resulted in substantial recruitment of Apoc1 on HDL, IDL, LDL, and VLDL. Trace amounts of Apoc2 were also observed in almost all lipoprotein fractions. Similarly, high APOC3 levels increased overall plasma Apoe content and led to formation of LDL and larger less dense Apoe-containing HDL particles. On the other hand, high APOC3 expression caused a modest reduction in Apoa1 of HDL, while Apoa2 was no longer detectable (Fig. 2A). Nondetectable levels of Apoa2 were also observed in mice infected with 5 × 108 pfu AdGFP-APOC3, though these mice contained only trace amounts of Apoc1 in their HDL, similar to control AdGFP-infected animals (Fig. 2A). The changes in apolipoprotein composition of HDL following expression of APOC3 were not due to changes in apolipoprotein mRNA expression in the liver (Fig. 2B), indicating that they were most likely due to APOC3-induced modifications in plasma lipoprotein assembly and metabolism.
Fig. 2.
Distribution of human APOC3, and murine Apoa1, Apoa2, Apoe, Apoc1, Apoc2, and Apoc3 in various lipoprotein fractions isolated by density gradient ultracentrifugation (A). CM, chylomicron. The densities of the various lipoprotein fractions are indicated. B: The relative hepatic mRNA expression of apoa1, apoe, apoa2, and apoc1 genes. Data were analyzed using Student’s t-test and are presented as median, minimum to maximum. AdGFP-APOC3_L and AdGFP-APOC3_H refer to mice infected with 5 × 108 pfu or 2 × 109 pfu of AdGFP-APOC3, respectively.
Negative staining TEM and nondenaturing two-dimensional electrophoresis analyses of HDL
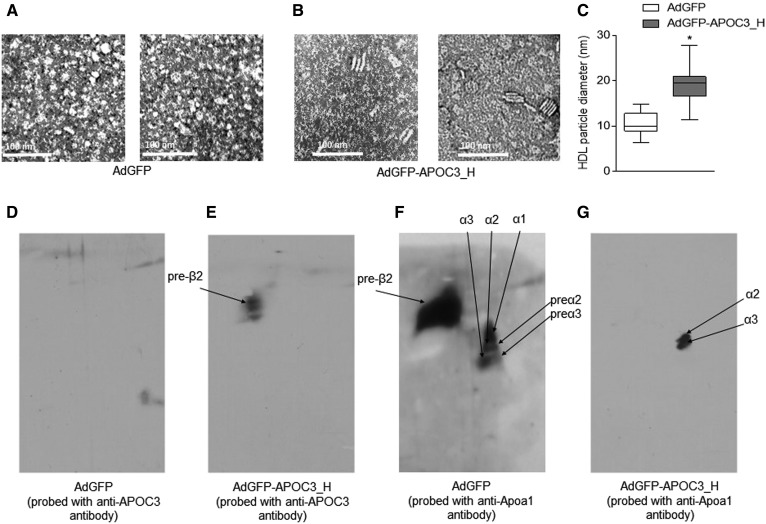
Because infection of mice with 2 × 109 pfu APOC3 led to an APOC3 distribution across all lipoprotein fractions and elevated plasma cholesterol and triglyceride levels, as observed in many morbidly obese patients with BMI >50 prior to bariatric surgery (34), in all subsequent experiments, we used this virus titer for our analyses. To study the effects of human APOC3 on HDL geometry and subpopulation distribution, we next isolated HDL from the plasma of infected mice and performed a qualitative negative staining TEM analysis coupled with nondenaturing two-dimensional electrophoresis analysis of pools of HDL fractions, as described in the Materials and Methods. As expected, negative TEM staining confirmed the presence of mainly spherical HDL particles in the HDL density fractions of plasma from AdGFP-infected mice (Fig. 3A). However, similar analysis revealed the presence of both discoidal and spherical HDL particles in the plasma of mice expressing APOC3 (Fig. 3B), in agreement with previous observations (25). APOC3-HDL particle diameter, expressed as median (interquartile range 25–75%) was calculated to be 19.4 nm (16.6–20.9 nm) and was significantly larger (P < 0.0001, unpaired t-test) than the diameter of control-HDL, calculated to be 10 nm (8.9–12.8 nm) (Fig. 3C).
Fig. 3.
Representative TEM analyses (A, B) and nondenaturing two-dimensional electrophoreses (D–G) of HDL from AdGFP-infected (A, D, F) and AdGFP-APOC3-infected mice (B, E, G). C: A semi-quantitative analysis of particle diameter of the HDL shown on TEM images (A, B). D, E: APOC3-containing HDL particles. F, G: Apoa1-containing HDL particles. Data were analyzed using Student’s t-test and are presented as median, minimum to maximum. *P < 0.05. AdGFP-APOC3_H refers to mice infected with 2 × 109 pfu of AdGFP-APOC3.
Nondenaturing two-dimensional electrophoresis analysis confirmed that all APOC3-HDL in mice infected with AdGFP-APOC3 were discoidal preβ2 particles (Fig. 3E). In agreement with the lack of human APOC3 expression in mice infected with AdGFP (Fig. 1C), no APOC3-HDL subpopulations could be detected in the plasma of these mice (Fig. 3D). Moreover, in the presence of APOC3, only spherical α2 and α3 Apoa1-containing HDL particles could be detected (Fig. 3G); while in the absence of APOC3 (mice infected with AdGFP), Apoa1-containing HDL particles were distributed in preβ2, preα2, preα3, α1, α2, and α3 (Fig. 3F). These findings indicate that in mice expressing APOC3, discoidal HDL contains APOC3, while spherical HDL contains Apoa1.
APOC3 expression triggers significant changes in HDL lipidome
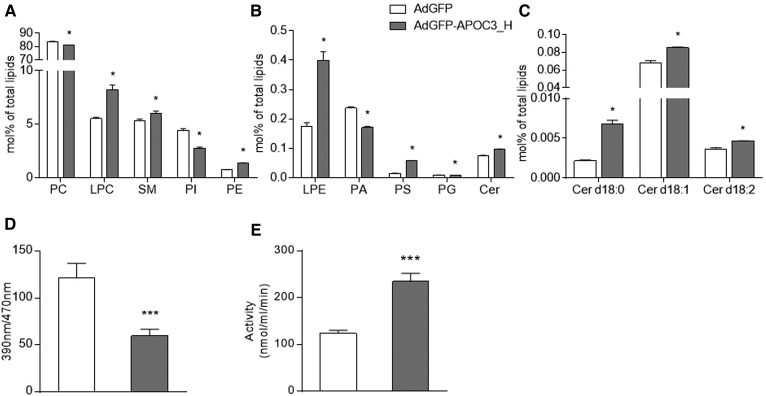
Comparative analysis of the lipidome of APOC3-HDL and control-HDL isolated from mice infected with AdGFP-APOC3 and AdGFP, respectively, showed significant differences in all tested lipid subclasses (Fig. 4A–C), suggesting that the presence of APOC3 results in the recruitment of different lipids on HDL. When expressed in molar percent of total lipids, phosphatidylcholine (PC), phosphatidylinositol (PI), phosphatidic acid (PA), and phosphatidylglycerol (PG) subclasses were reduced in APOC3-HDL. To the contrary, lysophosphatidylcholine (LPC), SM, phosphatidylethanolamine (PE), lysophosphatidylethanolamine (LPE), phosphatidylserine (PS), and ceramide (Cer) d18:0, Cer d18:1, and Cer d18:2 subclasses were found to be significantly increased in APOC3-HDL. The percent content of these lipid subclasses in the tested samples and the percent difference between them are shown in Table 2. The absolute lipid content of APOC3-HDL and control-HDL in the respective lipid classes and lipid species is summarized in supplemental Tables S2 and S3.
Fig. 4.
Lipidomic data showing major (A) and minor (B) lipid classes and Cer species (C) expressed as mole percent of total lipids in HDL. Data were analyzed using the U Mann-Whitney test and are presented as mean ± SEM.. *P < 0.05. D, E: Plasma enzymatic activities of Lcat and Lp-pla2, respectively. Data were analyzed using the U Mann-Whitney test (D) and Student’s t-test (E) and are presented as mean ± SEM. ***P < 0.001. AdGFP-APOC3_H refers to mice infected with 2 × 109 pfu of AdGFP-APOC3.
TABLE 2.
Comparative lipidome analysis of HDL isolated from AdGFP- and AdGFP-APOC3-infected mice
| Lipid Class | Major Fatty Acid Moieties | AdGFPa | AdGFP-APOC3_Ha | Percent Difference between AdGFP and AdGFP-APOC3_H |
| PC | 34:1/34:2/36:2/36:3/36:4/38:6 | 83.47 ± 0.44 | 80.90 ± 0.33 | −3 |
| LPC | 16:0/18:0 | 5.52 ± 0.13 | 8.24 ± 0.42 | 33 |
| SM | 34:1/42:2 | 5.33 ± 0.14 | 6.02 ± 0.21 | 11.4 |
| PI | 36:2/38:4 | 4.40 ± 0.17 | 2.75 ± 0.11 | −60.4 |
| PE | 34:2/36:2/38:6 | 0.74 ± 0.01 | 1.36 ± 0.03 | 45.1 |
| LPE | 18:0 | 0.18 ± 0.01 | 0.40 ± 0.03 | 56 |
| PA | 34:2/36:2/38:4 | 0.23 ± 0.004 | 0.17 ± 0.003 | −38.4 |
| PS | 36:1/40:6 | 0.015 ± 0.0002 | 0.06 ± 0.001 | 74 |
| PG | 34:1/36:2 | 0.009 ± 0.0008 | 0.007 ± 0.0001 | −26.2 |
| Cer d18:0 | 16:0/24:1 | 0.002 ± 0.0001 | 0.007 ± 0.0005 | 68 |
| Cer d18:1 | 16:0/22:0/24:0/24:1 | 0.07437 ± 0.003 | 0.09 ± 0.0007 | 20.2 |
| Cer d18:2 | 24:1 | 0.004 ± 0.0002 | 0.005 ± 0.00002 | 21.7 |
The percent difference between the two groups is also presented. AdGFP-APOC3_H refers to mice infected with 2 × 109 pfu of AdGFP-APOC3.
HDL content (mol% of HDL lipids).
APOC3-induced changes in HDL lipidome correlate with changes in plasma enzymatic activities
To determine whether the observed changes in the APOC3-HDL lipidome were associated with alterations in Lcat and Lp-pla2 activities, we next performed activity assays, as described in the Materials and Methods. As shown in Fig. 4, expression of APOC3 results in much lower Lcat activity (Fig. 4D), though Lp-pla2 activity (Fig. 4E) was increased, in agreement with previous in vitro reports (52, 53).
Antioxidant activity and total cholesterol efflux capacity of APOC3-HDL
Given the distinct structural differences between APOC3-HDL and control-HDL, we next sought to investigate how these differences might influence HDL functionality and, in particular, HDL antioxidant activity and total cholesterol efflux from RAW 264.7 macrophage cells in vitro.
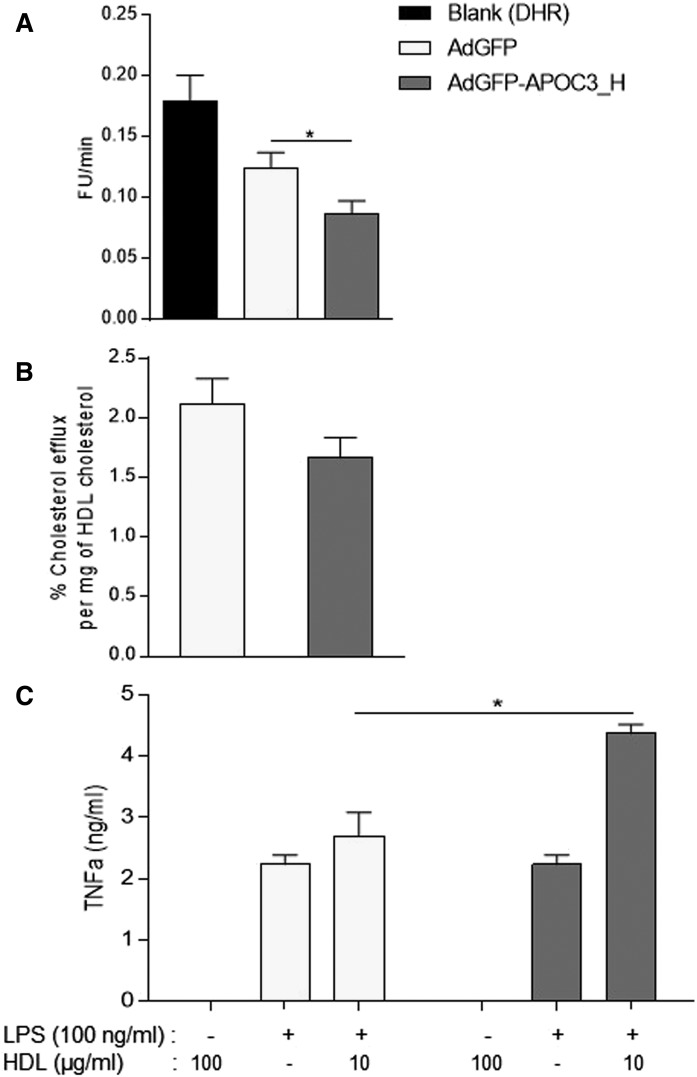
To determine the HDL antioxidant activity, HDL samples isolated from the two study groups were analyzed using a modification of the dihydrorhodamine assay (49), as described previously (34). As shown in Fig. 5A, APOC3-HDL demonstrated a significantly higher antioxidant function (i.e., greater inhibition of substrate oxidation) compared with control-HDL, when equal amounts of HDL-C from each group were used in the assay. Control-HDL also inhibited substrate oxidation, though to a much lesser extent.
Fig. 5.
Antioxidant potential (A) and cholesterol efflux capacity (B) of HDL isolated from AdGFP- and AdGFP-APOC3-infected mice. Data were analyzed using Student’s t-test and are presented as mean ± SEM. C: The effects of APOC3-HDL on LPS-induced (100 ng/ml) TNFα production in cultured RAW 264.7 macrophages. Data were analyzed using two-way ANOVA and are presented as mean ± SEM. *P < 0.05. AdGFP-APOC3_H refers to mice infected with 2 × 109 pfu of AdGFP-APOC3.
To assess the ability of APOC3-HDL and control-HDL to accept cholesterol from cells, we performed a total cholesterol efflux assay in the RAW 264.7 macrophage cell line that expresses Abca1, Abcg1, and scavenger receptor class B type I (Srb1), as described previously (41). As shown in Fig. 5B, when equal amounts of HDL-C from each group were used in the assay, a reduced capacity of APOC3-HDL to accept [14C]cholesterol from [14C]cholesterol-charged RAW 264.7 cells was observed.
Effects of APOC3-HDL on LPS-induced TNFα release from RAW 264.7 macrophage cells
To compare the effects of APOC3-HDL on inflammation, we employed the pertinent model of LPS-induced inflammation in RAW 264.7 macrophages, as described previously (41). In the absence of LPS, addition of 100 μg/ml HDL from each tested sample (i.e., APOC3-HDL and control-HDL) did not result in any significant production of TNFα, confirming that samples were properly prepared free of LPS. Stimulation of cells with LPS in the absence of HDL resulted in a significant production of TNFα in culture medium. When LPS stimulation was performed in the presence of APOC3-HDL, TNFα production was significantly induced compared with control-HDL, which showed no major pro- or anti-inflammatory effects (Fig. 5C).
APOC3 increases whole-body energy demand in mice
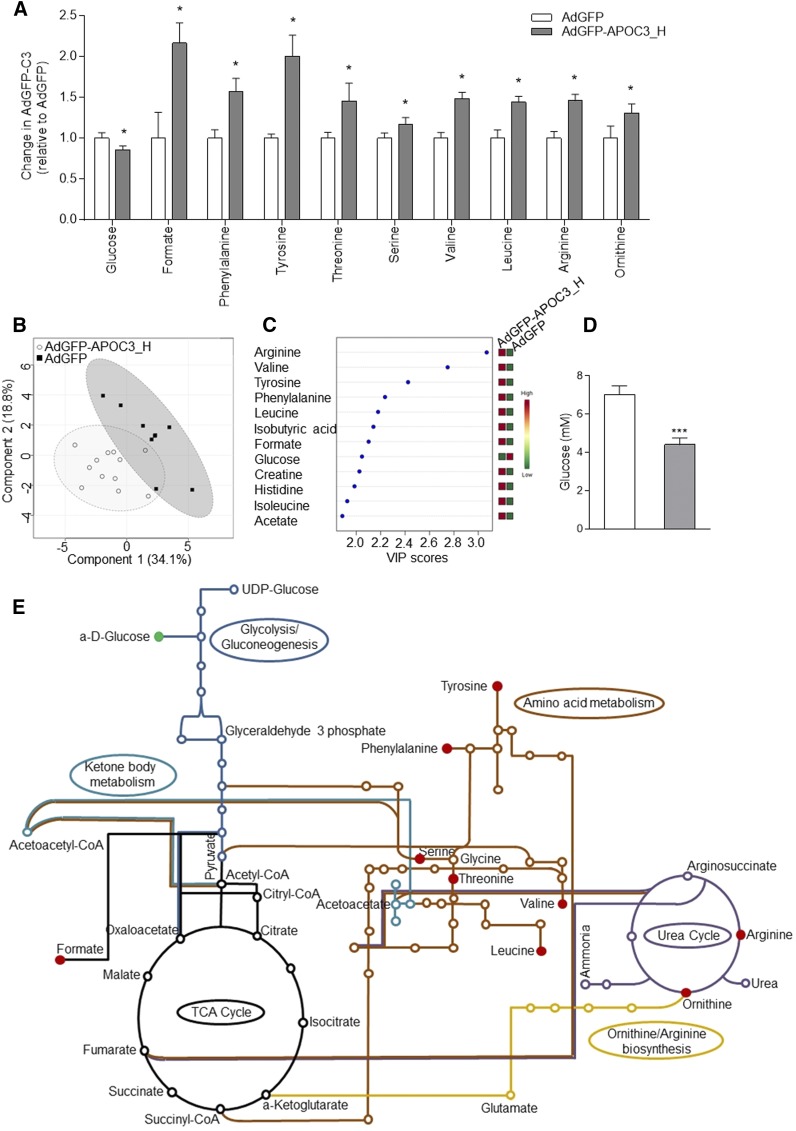
The clinical observation that APOC3-HDL shows a positive correlation with the morbidly obese phenotype (34) raised the possibility that APOC3-HDL may affect energy metabolism. To test this hypothesis, in the next set of experiments, we investigated the effects of APOC3 expression on serum metabolites linked to energy metabolism.
Briefly, typical CPMG spectra of serum samples from AdGFP-APOC3- or AdGFP-infected mice were used in order to identify signals of the low molecular weight metabolites. A total of 21 samples were screened: 9 derived from AdGFP-infected mice and 12 from AdGFP-APOC3 infected mice. Analysis and assignment of CPMG contributed greatly to the identification of key metabolites, which are valuable to enhance the accuracy of the classification presented below (Fig. 6A). The effects of APOC3 expression on serum metabolic profiles were examined by PCA and PLS-DA. The PLS-DA scores plot (Fig. 6B) showed a similar, but more pronounced, discrimination (accuracy, 85.7%; R2, 84.1%; Q2, 46.5%). To identify and rank signature metabolites corresponding to the variance in the metabolic profiles between AdGFP and AdGFP-APOC3 serum samples, we evaluated the variables of importance in projection (VIP scores >2.0) from the PLS-DA model (Fig. 6C).
Fig. 6.
Metabolomic analysis. A: Serum metabolites showing a statistically significant difference between AdGFP- and AdGFP-APOC3-infected mice. Data were analyzed using Wilcoxon rank-sum test and are presented as mean ± SEM. *P < 0.05. B: A multivariate analysis of CPMG serum spectra (PLS-DA score plot results) of AdGFP-infected (squares) and AdGFP-APOC3-infected (circles) mice. C: The top VIP scores followed by expression heat map from the PLS-DA model. D: Serum glucose levels as determined by a colorimetric glucose oxidase assay. E: Graphic representation of metabolic pathways involving the significant metabolites shown in (A). The map was created using the KEGG database (http://www.genome.jp/dbget-bin/www_bget?pathway:mmu01100). Red and green indicate increased and decreased metabolite levels, respectively. AdGFP-APOC3_H refers to mice infected with 2 × 109 pfu of AdGFP-APOC3.
A total of 38 metabolites were identified and quantified (in arbitrary units) from the 1H NMR CPMG spectra. The chemical shift and the statistically significant concentration changes of metabolites between the two groups are shown in Table 3. This analysis showed reduced levels of serum glucose that were further confirmed by a colorimetric glucose oxidase assay (Fig. 6A, D). Moreover, metabolomic analysis revealed elevated levels of citric acid cycle and urea cycle metabolites, suggesting enhanced metabolism of serum glucose toward energy production. Specifically, serum ornithine, formate, arginine, valine, phenylalanine, tyrosine, serine, threonine, and leucine were found elevated in the serum of AdGFP-APOC3-infected mice (Fig. 6A, C, E).
TABLE 3.
Serum metabolites showing a statistically significant difference between mice infected with 2 × 109 pfu of either AdGFP- or AdGFP-APOC3-infected mice
| Metabolites | Chemical Shift (δ) | Type | P | Changea |
| Glucose | 3.40 | d | 0.00871 | ↓ |
| Formate | 8.46 | s | 0.00090 | ↑ |
| Phenylalanine | 7.44 | m | 0.00337 | ↑ |
| Tyrosine | 7.20 | d | 0.00262 | ↑ |
| Threonine | 4.29…4.23 | m | 0.00306 | ↑ |
| Serine | 4.00…3.94 | m | 0.00754 | ↑ |
| Valine | 1.05…1.03 | d | 0.00431 | ↑ |
| Leucine | 0.97…0.95 | d | 0.00431 | ↑ |
| Arginine | 1.76…1.68 | m | 0.02063 | ↑ |
| Ornithine | 3.08…3.05 | t | 0.00058 | ↑ |
Signal multiplicity: s, singlet; d, doublet; t, triplet; m, multiplet.
Changes in AdGFP-APOC3-infected mice compared with AdGFP-infected mice
APOC3 expression selectively promotes mitochondrial energy metabolism in BAT
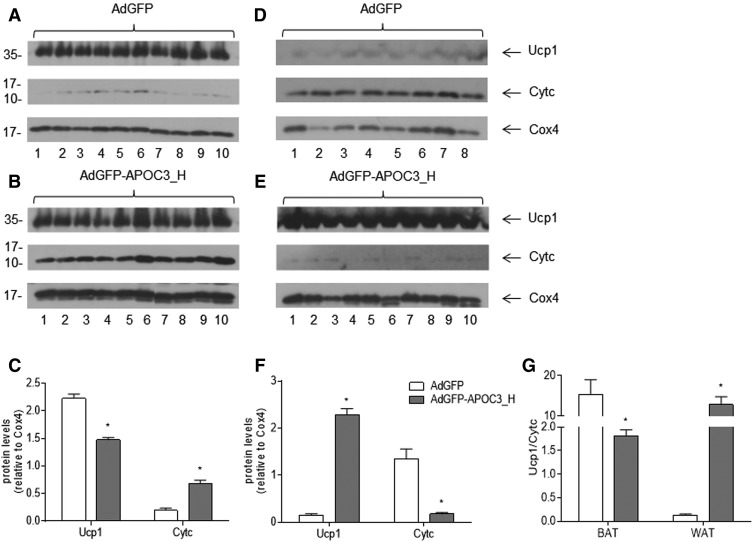
In an effort to validate the metabolomic data hinting that APOC3 expression may promote increased energy expenditure and energy demand, in the next set of experiments, mitochondria were isolated from BAT and WAT of mice infected with AdGFP-APOC3 or AdGFP, as described in the Materials and Methods. Western blot analysis of purified BAT mitochondrial fractions showed that expression of APOC3 stimulated a significant induction of mitochondrial Cytc levels, suggesting elevated oxidative phosphorylation (Fig. 7A–C). Moreover, it appears that oxidative phosphorylation is coupled with respiration toward ATP production due to an apparent relative decrease of Ucp1 when corrected for mitochondrial Cox4 (Fig. 7A–C, G), corroborating our metabolomic data.
Fig. 7.
Representative Western blot analyses and semiquantitative determination of Cytc and Ucp1 relative to Cox4 in mitochondrial extracts from BAT (A–C) and WAT (D–F) isolated from AdGFP- or AdGFP-APOC3-infected mice. G: The relative Ucp1/Cytc ratio in BAT and WAT, respectively. Data were produced from the same blots probed with the indicated antibodies and analyzed using Student’s t-test. Bars are expressed as mean ± SEM. *P < 0.05. AdGFP-APOC3_H refers to mice infected with 2 × 109 pfu of AdGFP-APOC3.
Similar analysis of WAT mitochondrial fractions showed that APOC3 expression resulted in substantial reduction of mitochondrial Cytc levels in WAT despite a very significant stimulation of Ucp1 expression. These findings indicate significantly reduced oxidative phosphorylation in WAT of AdGFP-APOC3-infected mice at 5 days post infection (Fig. 7D, E, G).
DISCUSSION
More than 30 years since the human Apoc3 nucleotide sequence was identified (54), our knowledge of APOC3 functions in relation to human health and disease still remains limited. The sole most widely accepted function of APOC3 is in atherosclerosis and, more precisely, in the regulation of plasma triglyceride levels through inhibition of plasma Lpl activity and catabolism of triglyceride-rich lipoproteins (11–19). Based on this function, Apoc3 antisense oligonucelotides are currently in clinical trials for the treatment of hypertriglyceridemia (20). However, little, if any, information exists on other functional roles of APOC3 in human pathology. In a work published previously, we showed that APOC3 associates with HDL in a process that requires lipid transporter Abca1 (25). Moreover, in a more recent clinical study in bariatric subjects, we found that markedly increased levels of plasma APOC3 correlate with the obese phenotype, though no mechanistic information exists to explain this correlation (34). To better understand the effects of APOC3 in morbidly obese patients, we infected mice with an adenovirus expressing human APOC3 and studied the effects of APOC3 on HDL structure and function, as well as WAT and BAT mitochondrial metabolic activity. We found that infection with 5 × 108 pfu of AdGFP-APOC3 resulted in an APOC3 lipoprotein distribution and lipid profile that was closer to the healthy human conditions (except for hypertriglyceridemia). In contrast, 2 × 109 pfu of AdGFP-APOC3 resulted in an APOC3 lipoprotein distribution and lipid profile that is closer to the one seen in morbidly obese humans (34); therefore, all subsequent analyses were performed with this dose of adenovirus.
Our results revealed that expression of APOC3 in C57BL/6 mice leads to the formation of APOC3-HDL particles that differ in their subpopulation distribution and apolipoprotein content from control-HDL (Figs. 2, 3). Our TEM analysis indicated that accumulation of APOC3 on HDL results in a mix of discoidal and spherical HDL particles of larger diameter (Fig. 3), a finding consistent with the increased phospholipid and free cholesterol content of APOC3-HDL (Table 1), as well as the reduced plasma Lcat activity (Fig. 4). Apoa1 was reduced, whereas Apoa2 was absent from APOC3-HDL. In contrast, Apoe, Apoc1, and Apoc2 appeared elevated (Fig. 2). A very modest induction in Apoc2 was also evident. Interestingly, the two-dimensional nondenaturing electrophoresis analysis confirmed that APOC3-HDL particles are mainly discoidal preβ2 (Fig. 3E) and distinct from Apoa1-HDL, which are mainly spherical α2 and α3 particles (Fig. 3G). This observation is in agreement with proteomic studies in centrifugally isolated HDL, suggesting that specific proteins may be found on distinct particle subspecies, which are differentially distributed across the HDL density spectrum (55–59).
In addition to quantitative changes in HDL apolipoprotein composition, our analysis also revealed qualitative changes because, in the presence of APOC3, Apoe is present only in less dense HDL particles (d 1.084 and 1.100), while more dense HDL particles primarily contain Apoa1. Of note, the lipid-free fraction (d > 1.21 g/ml) contains a significant amount of Apoa1 with only trace amounts of APOC3 present. Based on our previous findings that biogenesis of APOC3-HDL requires Abca1 (25), it is conceivable to hypothesize that APOC3 effectively competes with Apoa1 for Abca1. Such competition, however, is not evident for the other apolipoproteins, such as Apoe and Apoc1 (Fig. 2).
At this point, it is not clear what drives the effects of APOC3-expression on HDL apolipoprotein content and subspecies distribution. Our analysis did not reveal any measurable changes in the hepatic mRNA expression levels of these apolipoproteins, suggesting that the observed changes in the apolipoprotein content of HDL in the presence of APOC3 are posttranscriptional (Fig. 2). Likely possibilities include specific protein:protein interactions on the particle surface and/or the attraction of certain proteins to particular particle biophysical characteristics, such as lipid packing density or surface curvature. Alternatively, given that Apoe, Apoa1, Apoa2, Apoc1, Apoc2, and APOC3 are all capable of interacting with Abca1 to promote biogenesis of HDL-like particles (25, 60), it is possible that increased availability of APOC3 toward HDL formation in the plasma of mice infected with AdGFP-APOC3 may differentially influence the selectivity of Abca1 for these apolipoproteins.
Notably, the APOC3-induced modifications in HDL particle structure are not limited only to the apolipoprotein content, but extend to its lipidome. The data presented in Table 1 support that APOC3-HDL has a higher content of lipids per milligram of protein and significantly more free cholesterol, phospholipids, and triglycerides, which explains the differences in particle size and geometry between APOC3-HDL and control-HDL (Fig. 3). The reduced Lcat activity in the plasma of APOC3-expressing mice is consistent with the higher free cholesterol content of the HDL of these mice. Moreover, our comparative analysis of APOC3-HDL and control-HDL lipidome (Fig. 4 and Table 2) further confirms that expression of APOC3 results in the recruitment of different types and amounts of lipids on HDL. Interestingly, APOC3-HDL contains elevated levels of PS, though levels of PG, PI, and PA are reduced. These negatively charged lipids significantly impact the net surface charge of HDL, thereby modulating charge-dependent interactions with plasma enzymes, such as lipases, lipid transport proteins, extracellular matrix, and other protein components (61). The decreased levels of PC and increased levels of LPC, the product of PC hydrolysis by Lcat and Lp-pla2, in APOC3-HDL suggested that these HDL particles may be more suitable substrates for one or both of these enzymes, as proposed previously (52, 53). Surprisingly, despite an increase in LPC, our functional analysis showed reduced Lcat enzymatic activity in the plasma of mice infected with AdGFP-APOC3 (Fig. 4) that was further confirmed by our TEM analysis, providing an in vivo confirmation that APOC3 is indeed an inhibitor of Lcat, as previously suggested by in vitro studies (53). In contrast, Lp-pla2 activity was found to be increased, confirming in vitro data showing that APOC3 is, instead, an activator of this plasma enzyme. Moreover, this result suggests that the increased LPC content of APOC3-HDL is, instead, the result of Lp-pla2 activity on HDL. Because elevated Lp-pla2 activity is associated with increased risk for coronary heart disease (62), this in vivo finding provides additional evidence for the proatherogenic effects of APOC3.
The presence of elevated levels of SM, Cer d18:0, and Cer d18:1 in APOC3-HDL is consistent with the increased levels of triglyceride-rich lipoproteins in the plasma of mice infected with AdGFP-APOC3 because SM, Cer d18:0, and Cer d18:1 originate primarily from triglyceride-rich lipoproteins and only to a minor extent from nascent HDL (61, 63). In addition, Cer d18:0 and Cer d18:1 play important signaling roles in cellular survival, growth, and differentiation (61), suggesting that APOC3-HDL may be a more efficient signaling mediator than control-HDL in promoting these processes.
In an analogy with our previous observations with APOE- and APOA1-containing HDL (33), we also found here that changes in particle structure brought about by APOC3 correlate with significant alterations in particle functionality. Though some properties appeared improved, others deteriorated. Indeed, under the conditions of our experiment, APOC3-HDL was a poorer acceptor of [14C]cholesterol in RAW 264.7 cells, indicating reduced total cholesterol efflux, a key property for the effective unloading of free cholesterol from peripheral tissues and its shuttling back to the liver for catabolism. Moreover, it potentiates the effect of LPS on TNFα release in RAW 264.7 macrophage cells, suggesting a proinflammatory role in circulation. In contrast, APOC3-HDL has a much higher antioxidant capacity than control-HDL, suggesting a positive role in reducing oxidative stress (Fig. 5).
Moreover, our metabolomic data coupled with data from mitochondrial analyses indicate that expression of APOC3 in mice infected with AdGFP-APOC3 stimulates aerobic catabolism of glucose via the citric acid cycle for increased ATP production in BAT. Metabolomic data further show that, upon APOC3 expression, there is an increase in the levels of branched chain amino acids, which may be used for anaplerosis in the TCA cycle by yielding additional substrate (Fig. 6). This observation suggests that, in addition to glucose, APOC3 expression may stimulate protein degradation for energy production, as previously reported (64).
Mitochondrial Cytc is a biomarker of mitochondrial metabolic function (65). In adipose tissue, this function relates to ATP production when oxidative phosphorylation is coupled to respiration or thermogenesis marked by increased mitochondrial Ucp1 expression when oxidative phosphorylation is uncoupled from respiration (66). Our molecular analysis revealed a selective increase of Cytc in BAT, while Ucp1 levels were decreased (Fig. 7). These changes indicate that APOC3 stimulates oxidative phosphorylation, coupled with respiration for ATP production in BAT, further corroborating our metabolomic data.
The effects of APOC3 expression on BAT mitochondrial metabolic activity identified here may also explain the increased sensitivity of apoc3-deficient (apoc3−/−) mice toward diet-induced obesity that has been previously reported in the literature (67). It was shown that after 20 weeks of high-fat feeding, apoc3−/− mice had reduced plasma triglyceride levels and gained more body weight due to increased body lipid mass. Adipose tissue uptake of triglyceride-derived free fatty acids following lipolysis by Lpl was also significantly increased in these mice, while whole-body insulin sensitivity was decreased by 43% and endogenous glucose production was decreased by 25% (67). Apparently, APOC3 appears to be a critical regulator of processes associated with peripheral management of dietary lipids and their conversion into energy, maintaining a balance between tissue lipid uptake and catabolism. Our findings also raise the interesting possibility that the increased APOC3-HDL levels observed in morbidly obese subjects (34) may not represent a cause of obesity, but rather a protective mechanism triggered in response to nutrient surplus, aiming at promoting excess substrate oxidation by shifting BAT mitochondrial metabolism toward energy production.
Similar analysis in WAT revealed significantly reduced mitochondrial Cytc despite a measurable elevation of Ucp1 in APOC3-expressing mice, suggesting that APOC3 reduces WAT oxidative phosphorylation (Fig. 7) and raising the possibility of dysfunctional mitochondria in this tissue. Moreover, these data show that the increased metabolic activity identified by our metabolomic data may be attributed to BAT metabolic activation in mice expressing APOC3.
Our findings indicate that, in contrast to the previous view identifying it as detrimental to human health, APOC3 also possesses some very important properties, which should not be ignored in the designing of new pharmaceuticals aiming at APOC3. Apparently, APOC3 is another example of the ancient Greek saying “μέτρον άριστον” (everything in moderation). Reducing APOC3 levels to physiological may be beneficial for the treatment of some forms of hypertriglyceridemia; however, complete silencing of its expression may adversely affect other physiological processes crucial for human health.
Supplementary Material
Footnotes
Abbreviations:
- AdGFP
- recombinant attenuated control adenovirus expressing green fluorescent protein
- AdGFP-APOC3
- recombinant attenuated adenovirus expressing the human APOC3 protein and the green fluorescent protein under the independent control of two separate CMV promoters
- APOC3-HDL
- APOC3-containing HDL
- BAT
- brown adipose tissue
- Cer
- ceramide
- Cox4
- cytochrome c oxidase subunit 4
- Cytc
- cytochrome c
- GFP
- green fluorescence protein
- HDL-C
- HDL cholesterol
- LPC
- lysophosphatidylcholine
- LPE
- lysophosphatidylethanolamine
- Lp-pla2
- lipoprotein-associated phospholipase A2
- LPS
- lipopolysaccharide
- PA
- phosphatidic acid
- PAF
- platelet activating factor
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- pfu
- plaque-forming unit
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PLS-DA
- partial least squares discriminant analysis
- PS
- phosphatidylserine
- TEM
- transmission electron microscopy
- Ucp1
- uncoupling protein 1
- WAT
- white adipose tissue
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Shulman R. S., Herbert P. N., Fredrickson D. S., Wehrly K., and Brewer H. B. Jr. 1974. Isolation and alignment of the tryptic peptides of alanine apolipoprotein, an apolipoprotein from human plasma very low density lipoproteins. J. Biol. Chem. 249: 4969–4974. [PubMed] [Google Scholar]
- 2.McKeone B. J., Massey J. B., Knapp R. D., and Pownall H. J.. 1988. Apolipoproteins C-I, C-II, and C-III: kinetics of association with model membranes and intermembrane transfer. Biochemistry. 27: 4500–4505. [DOI] [PubMed] [Google Scholar]
- 3.Windler E. E., Kovanen P. T., Chao Y. S., Brown M. S., Havel R. J., and Goldstein J. L.. 1980. The estradiol-stimulated lipoprotein receptor of rat liver. A binding site that membrane mediates the uptake of rat lipoproteins containing apoproteins B and E. J. Biol. Chem. 255: 10464–10471. [PubMed] [Google Scholar]
- 4.Sehayek E., and Eisenberg S.. 1991. Mechanisms of inhibition by apolipoprotein C of apolipoprotein E-dependent cellular metabolism of human triglyceride-rich lipoproteins through the low density lipoprotein receptor pathway. J. Biol. Chem. 266: 18259–18267. [PubMed] [Google Scholar]
- 5.Aalto-Setälä K., Fisher E. A., Chen X., Chajek-Shaul T., Hayek T., Zechner R., Walsh A., Ramakrishnan R., Ginsberg H. N., and Breslow J. L.. 1992. Mechanism of hypertriglyceridemia in human apolipoprotein (apo) CIII transgenic mice. Diminished very low density lipoprotein fractional catabolic rate associated with increased apo CIII and reduced apo E on the particles. J. Clin. Invest. 90: 1889–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aalto-Setälä K., Weinstock P. H., Bisgaier C. L., Wu L., Smith J. D., and Breslow J. L.. 1996. Further characterization of the metabolic properties of triglyceride-rich lipoproteins from human and mouse apoC-III transgenic mice. J. Lipid Res. 37: 1802–1811. [PubMed] [Google Scholar]
- 7.de Silva H. V., Lauer S. J., Wang J., Simonet W. S., Weisgraber K. H., Mahley R. W., and Taylor J. M.. 1994. Overexpression of human apolipoprotein C-III in transgenic mice results in an accumulation of apolipoprotein B48 remnants that is corrected by excess apolipoprotein E. J. Biol. Chem. 269: 2324–2335. [PubMed] [Google Scholar]
- 8.Maeda N., Li H., Lee D., Oliver P., Quarfordt S. H., and Osada J.. 1994. Targeted disruption of the apolipoprotein C-III gene in mice results in hypotriglyceridemia and protection from postprandial hypertriglyceridemia. J. Biol. Chem. 269: 23610–23616. [PubMed] [Google Scholar]
- 9.Ebara T., Ramakrishnan R., Steiner G., and Shachter N. S.. 1997. Chylomicronemia due to apolipoprotein CIII overexpression in apolipoprotein E-null mice. Apolipoprotein CIII-induced hypertriglyceridemia is not mediated by effects on apolipoprotein E. J. Clin. Invest. 99: 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jong M. C., Dahlmans V. E., Hofker M. H., and Havekes L. M.. 1997. Nascent very-low-density lipoprotein triacylglycerol hydrolysis by lipoprotein lipase is inhibited by apolipoprotein E in a dose-dependent manner. Biochem. J. 328: 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oswald B., and Quarfordt S.. 1987. Effect of apoE on triglyceride emulsion interaction with hepatocyte and hepatoma G2 cells. J. Lipid Res. 28: 798–809. [PubMed] [Google Scholar]
- 12.Agnani G., Bard J. M., Candelier L., Delattre S., Fruchart J. C., and Clavey V.. 1991. Interaction of LpB, LpB:E, LpB:C-III, and LpB:C–III:E lipoproteins with the low density lipoprotein receptor of HeLa cells. Arterioscler. Thromb. 11: 1021–1029. [DOI] [PubMed] [Google Scholar]
- 13.Clavey V., Lestavel-Delattre S., Copin C., Bard J. M., and Fruchart J. C.. 1995. Modulation of lipoprotein B binding to the LDL receptor by exogenous lipids and apolipoproteins CI, CII, CIII, and E. Arterioscler. Thromb. Vasc. Biol. 15: 963–971. [DOI] [PubMed] [Google Scholar]
- 14.Mann C. J., Troussard A. A., Yen F. T., Hannouche N., Najib J., Fruchart J. C., Lotteau V., Andre P., and Bihain B. E.. 1997. Inhibitory effects of specific apolipoprotein C-III isoforms on the binding of triglyceride-rich lipoproteins to the lipolysis-stimulated receptor. J. Biol. Chem. 272: 31348–31354. [DOI] [PubMed] [Google Scholar]
- 15.Windler E., Chao Y., and Havel R. J.. 1980. Regulation of the hepatic uptake of triglyceride-rich lipoproteins in the rat. Opposing effects of homologous apolipoprotein E and individual C apoproteins. J. Biol. Chem. 255: 8303–8307. [PubMed] [Google Scholar]
- 16.Windler E., and Havel R. J.. 1985. Inhibitory effects of C apolipoproteins from rats and humans on the uptake of triglyceride-rich lipoproteins and their remnants by the perfused rat liver. J. Lipid Res. 26: 556–565. [PubMed] [Google Scholar]
- 17.Shelburne F., Hanks J., Meyers W., and Quarfordt S.. 1980. Effect of apoproteins on hepatic uptake of triglyceride emulsions in the rat. J. Clin. Invest. 65: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quarfordt S. H., Michalopoulos G., and Schirmer B.. 1982. The effect of human C apolipoproteins on the in vitro hepatic metabolism of triglyceride emulsions in the rat. J. Biol. Chem. 257: 14642–14647. [PubMed] [Google Scholar]
- 19.Kortz W. J., Schirmer B. D., Mansbach C. M., Shelburne F., Toglia M. R., and Quarfordt S. H.. 1984. Hepatic uptake of chylomicrons and triglyceride emulsions in rats fed diets of differing fat content. J. Lipid Res. 25: 799–804. [PubMed] [Google Scholar]
- 20.Gaudet D., Alexander V. J., Baker B. F., Brisson D., Tremblay K., Singleton W., Geary R. S., Hughes S. G., Viney N. J., Graham M. J., et al. . 2015. Antisense inhibition of apolipoprotein C-III in patients with hypertriglyceridemia. N. Engl. J. Med. 373: 438–447. [DOI] [PubMed] [Google Scholar]
- 21.Herbert P. N., Assmann G., Grotton J. A. M., and Fredrickson D. S.. 1999. Disorders of lipoprotein and lipid metabolism. In Stanbury J. B., Wyngaarden J. B., Fredrickson D. S., et al., editors. McGraw-Hill, New York: 589–651. [Google Scholar]
- 22.Onat A., Hergenc G., Sansoy V., Fobker M., Ceyhan K., Toprak S., and Assmann G.. 2003. Apolipoprotein C-III, a strong discriminant of coronary risk in men and a determinant of the metabolic syndrome in both genders. Atherosclerosis. 168: 81–89. [DOI] [PubMed] [Google Scholar]
- 23.Herbert P. N., Assmann G., Gotto A. M. Jr., and Fredrickson D. S.. 1982. Familial lipoprotein deficiency: abetalipoproteinemia, hypobetalipoproteinemia, and Tangier disease. In Stanbury J. B., Wyngaarden J. B., Fredrickson D. S., et al., editors. McGraw-Hill, New York: 589–651. [Google Scholar]
- 24.Zannis V. I., and Breslow J. L.. 1980. Two-dimensional maps of human apolipoproteins in normal and diseased states. In Radola B. J., editor. Walter de Gruyter, Berlin: 437–473. [Google Scholar]
- 25.Kypreos K. E. 2008. ABCA1 promotes the de novo biogenesis of apolipoprotein CIII-containing HDL particles in vivo and modulates the severity of apolipoprotein CIII-induced hypertriglyceridemia. Biochemistry. 47: 10491–10502. [DOI] [PubMed] [Google Scholar]
- 26.Constantinou C., Karavia E. A., Xepapadaki E., Petropoulou P. I., Papakosta E., Karavyraki M., Zvintzou E., Theodoropoulos V., Filou S., Hatziri A. , et al. 2016. Advances in high density lipoprotein physiology: surprises, overturns and promises. Am. J. Physiol. Endocrinol. Metab. 310: E1–E14. [DOI] [PubMed] [Google Scholar]
- 27.Gofman J. W., Glazier F., Tamplin A., Strisower B., and De L. O.. 1954. Lipoproteins, coronary heart disease, and atherosclerosis. Physiol. Rev. 34: 589–607. [DOI] [PubMed] [Google Scholar]
- 28.Delalla O. F., Elliott H. A., and Gofman J. W.. 1954. Ultracentrifugal studies of high density serum lipoproteins in clinically healthy adults. Am. J. Physiol. 179: 333–337. [DOI] [PubMed] [Google Scholar]
- 29.Miller G. J., and Miller N. E.. 1975. Plasma-high-density-lipoprotein concentration and development of ischaemic heart-disease. Lancet. 1: 16–19. [DOI] [PubMed] [Google Scholar]
- 30.Tsompanidi E. M., Brinkmeier M. S., Fotiadou E. H., Giakoumi S. M., and Kypreos K. E.. 2010. HDL biogenesis and functions: role of HDL quality and quantity in atherosclerosis. Atherosclerosis. 208: 3–9. [DOI] [PubMed] [Google Scholar]
- 31.Kypreos K. E., Gkizas S., Rallidis L. S., and Karagiannides I.. 2013. HDL particle functionality as a primary pharmacological target for HDL-based therapies. Biochem. Pharmacol. 85: 1575–1578. [DOI] [PubMed] [Google Scholar]
- 32.Karavia E. A., Zvintzou E., Petropoulou P. I., Xepapadaki E., Constantinou C., and Kypreos K. E.. 2014. HDL quality and functionality: what can proteins and genes predict? Expert Rev. Cardiovasc. Ther. 12: 521–532. [DOI] [PubMed] [Google Scholar]
- 33.Filou S., Lhomme M., Karavia E. A., Kalogeropoulou C., Theodoropoulos V., Zvintzou E., Sakellaropoulos G. C., Petropoulou P. I., Constantinou C., Kontush A., et al. . 2016. Distinct roles of apolipoproteins A1 and E in the modulation of high-density lipoprotein composition and function. Biochemistry. 55: 3752–3762. [DOI] [PubMed] [Google Scholar]
- 34.Zvintzou E., Skroubis G., Chroni A., Petropoulou P. I., Gkolfinopoulou C., Sakellaropoulos G., Gantz D., Mihou I., Kalfarentzos F., and Kypreos K. E.. 2014. Effects of bariatric surgery on HDL structure and functionality: results from a prospective trial. J. Clin. Lipidol. 8: 408–417. [DOI] [PubMed] [Google Scholar]
- 35.He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., and Vogelstein B.. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA. 95: 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chroni A., Kan H. Y., Kypreos K. E., Gorshkova I. N., Shkodrani A., and Zannis V. I.. 2004. Substitutions of glutamate 110 and 111 in the middle helix 4 of human apolipoprotein A-I (apoA-I) by alanine affect the structure and in vitro functions of apoA-I and induce severe hypertriglyceridemia in apoA-I-deficient mice. Biochemistry. 43: 10442–10457. [DOI] [PubMed] [Google Scholar]
- 37.Kypreos K. E., Teusink B., Van Dijk K. W., Havekes L. M., and Zannis V. I.. 2001. Analysis of the structure and function relationship of the human apolipoprotein E in vivo, using adenovirus-mediated gene transfer. FASEB J. 15: 1598–1600. [DOI] [PubMed] [Google Scholar]
- 38.Kypreos K. E., Morani P., Van Dijk K. W., Havekes L. M., and Zannis V. I.. 2001. The amino-terminal 1–185 domain of apoE promotes the clearance of lipoprotein remnants in vivo. The carboxy-terminal domain is required for induction of hyperlipidemia in normal and apoE-deficient mice. Biochemistry. 40: 6027–6035. [DOI] [PubMed] [Google Scholar]
- 39.Kypreos K. E., Van Dijk K. W., van Der Z. A., Havekes L. M., and Zannis V. I.. 2001. Domains of apolipoprotein E contributing to triglyceride and cholesterol homeostasis in vivo. Carboxyl-terminal region 203–299 promotes hepatic very low density lipoprotein-triglyceride secretion. J. Biol. Chem. 276: 19778–19786. [DOI] [PubMed] [Google Scholar]
- 40.Fallaux F. J., Kranenburg O., Cramer S. J., Houweling A., Van Ormondt H., Hoeben R. C., and van der Eb A. J.. 1996. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 7: 215–222. [DOI] [PubMed] [Google Scholar]
- 41.Petropoulou P. I., Berbee J. F., Theodoropoulos V., Hatziri A., Stamou P., Karavia E. A., Spyridonidis A., Karagiannides I., and Kypreos K. E.. 2015. Lack of LCAT reduces the LPS-neutralizing capacity of HDL and enhances LPS-induced inflammation in mice. Biochim. Biophys. Acta. 1852: 2106–2115. [DOI] [PubMed] [Google Scholar]
- 42.Constantinou C., Mpatsoulis D., Natsos A., Petropoulou P. I., Zvintzou E., Traish A. M., Voshol P. J., Karagiannides I., and Kypreos K. E.. 2014. The low density lipoprotein receptor modulates the effects of hypogonadism on diet-induced obesity and related metabolic perturbations. J. Lipid Res. 55: 1434–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapman M. J., Goldstein S., Lagrange D., and Laplaud P. M.. 1981. A density gradient ultracentrifugal procedure for the isolation of the major lipoprotein classes from human serum. J. Lipid Res. 22: 339–358. [PubMed] [Google Scholar]
- 44.Karavia E. A., Papachristou N. I., Sakellaropoulos G. C., Xepapadaki E., Papamichail E., Petropoulou P. I., Papakosta E. P., Constantinou C., Habeos I., Papachristou D. J., et al. . 2015. Scavenger receptor class B type I regulates plasma apolipoprotein E levels and dietary lipid deposition to the liver. Biochemistry. 54: 5605–5616. [DOI] [PubMed] [Google Scholar]
- 45.Karavia E. A., Hatziri A., Kalogeropoulou C., Papachristou N. I., Xepapadaki E., Constantinou C., Natsos A., Petropoulou P. I., Sasson S., Papachristou D. J., et al. . 2015. Deficiency in apolipoprotein A-I ablates the pharmacological effects of metformin on plasma glucose homeostasis and hepatic lipid deposition. Eur. J. Pharmacol. 766: 76–85. [DOI] [PubMed] [Google Scholar]
- 46.Kavo A. E., Rallidis L. S., Sakellaropoulos G. C., Lehr S., Hartwig S., Eckel J., Bozatzi P. I., Anastasiou-Nana M., Tsikrika P., and Kypreos K. E.. 2012. Qualitative characteristics of HDL in young patients of an acute myocardial infarction. Atherosclerosis. 220: 257–264. [DOI] [PubMed] [Google Scholar]
- 47.Asztalos B. F., Roheim P. S., Milani R. L., Lefevre M., McNamara J. R., Horvath K. V., and Schaefer E. J.. 2000. Distribution of ApoA-I-containing HDL subpopulations in patients with coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 20: 2670–2676. [DOI] [PubMed] [Google Scholar]
- 48.Tselepis A. D., Dentan C., Karabina S. A., Chapman M. J., and Ninio E.. 1995. PAF-degrading acetylhydrolase is preferentially associated with dense LDL and VHDL-1 in human plasma. Catalytic characteristics and relation to the monocyte-derived enzyme. Arterioscler. Thromb. Vasc. Biol. 15: 1764–1773. [DOI] [PubMed] [Google Scholar]
- 49.Kelesidis T., Currier J. S., Huynh D., Meriwether D., Charles-Schoeman C., Reddy S. T., Fogelman A. M., Navab M., and Yang O. O.. 2011. A biochemical fluorometric method for assessing the oxidative properties of HDL. J. Lipid Res. 52: 2341–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang H., Wang Y., Nicholson J. K., and Lindon J. C.. 2004. Use of relaxation-edited one-dimensional and two dimensional nuclear magnetic resonance spectroscopy to improve detection of small metabolites in blood plasma. Anal. Biochem. 325: 260–272. [DOI] [PubMed] [Google Scholar]
- 51.Psychogios N., Hau D. D., Peng J., Guo A. C., Mandal R., Bouatra S., Sinelnikov I., Krishnamurthy R., Eisner R., Gautam B., et al. . 2011. The human serum metabolome. PLoS One. 6: e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han X., Wang T., Zhang J., Liu X., Li Z., Wang G., Song Q., Pang D., Ouyang H., and Tang X.. 2015. Apolipoprotein CIII regulates lipoprotein-associated phospholipase A2 expression via the MAPK and NFkappaB pathways. Biol. Open. 4: 661–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishida H. I., Nakanishi T., Yen E. A., Arai H., Yen F. T., and Nishida T.. 1986. Nature of the enhancement of lecithin-cholesterol acyltransferase reaction by various apolipoproteins. J. Biol. Chem. 261: 12028–12035. [PubMed] [Google Scholar]
- 54.Levy-Wilson B., Appleby V., Protter A., Auperin D., and Seilhamer J. J.. 1984. Isolation and DNA sequence of full-length cDNA for human preapolipoprotein CIII. DNA. 3: 359–364. [DOI] [PubMed] [Google Scholar]
- 55.Karlsson H., Leanderson P., Tagesson C., and Lindahl M.. 2005. Lipoproteomics I: mapping of proteins in low-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 5: 551–565. [DOI] [PubMed] [Google Scholar]
- 56.Karlsson H., Leanderson P., Tagesson C., and Lindahl M.. 2005. Lipoproteomics II: mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 5: 1431–1445. [DOI] [PubMed] [Google Scholar]
- 57.Heller M., Stalder D., Schlappritzi E., Hayn G., Matter U., and Haeberli A.. 2005. Mass spectrometry-based analytical tools for the molecular protein characterization of human plasma lipoproteins. Proteomics. 5: 2619–2630. [DOI] [PubMed] [Google Scholar]
- 58.Rezaee F., Casetta B., Levels J. H., Speijer D., and Meijers J. C.. 2006. Proteomic analysis of high-density lipoprotein. Proteomics. 6: 721–730. [DOI] [PubMed] [Google Scholar]
- 59.Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., et al. . 2007. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117: 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Remaley A. T., Stonik J. A., Demosky S. J., Neufeld E. B., Bocharov A. V., Vishnyakova T. G., Eggerman T. L., Patterson A. P., Duverger N. J., Santamarina-Fojo S., et al. . 2001. Apolipoprotein specificity for lipid efflux by the human ABCAI transporter. Biochem. Biophys. Res. Commun. 280: 818–823. [DOI] [PubMed] [Google Scholar]
- 61.Kontush A., Lhomme M., and Chapman M. J.. 2013. Unraveling the complexities of the HDL lipidome. J. Lipid Res. 54: 2950–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenson R. S., and Stafforini D. M.. 2012. Modulation of oxidative stress, inflammation, and atherosclerosis by lipoprotein-associated phospholipase A2. J. Lipid Res. 53: 1767–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nilsson A., and Duan R. D.. 2006. Absorption and lipoprotein transport of sphingomyelin. J. Lipid Res. 47: 154–171. [DOI] [PubMed] [Google Scholar]
- 64.Yang F., Du J., Zhang H., Ruan G., Xiang J., Wang L., Sun H., Guan A., Shen G., Liu Y., et al. . 2017. Serum metabolomics of Burkitt lymphoma mouse models. PLoS One. 12: e0170896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hüttemann M., Lee I., Grossman L. I., Doan J. W., and Sanderson T. H.. 2012. Phosphorylation of mammalian cytochrome c and cytochrome c oxidase in the regulation of cell destiny: respiration, apoptosis, and human disease. Adv. Exp. Med. Biol. 748: 237–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nicholls D. G., and Locke R. M.. 1984. Thermogenic mechanisms in brown fat. Physiol. Rev. 64: 1–64. [DOI] [PubMed] [Google Scholar]
- 67.Duivenvoorden I., Teusink B., Rensen P. C., Romijn J. A., Havekes L. M., and Voshol P. J.. 2005. Apolipoprotein C3 deficiency results in diet-induced obesity and aggravated insulin resistance in mice. Diabetes. 54: 664–671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.