Abstract
Compartmentalization of metabolism into specific regions of the cell, tissue, and organ is critical to life for all organisms. Mass spectrometric imaging techniques have been valuable in identifying and quantifying concentrations of metabolites in specific locations of cells and tissues, but a true understanding of metabolism requires measurement of metabolite flux on a spatially resolved basis. Here, we utilize desorption ESI-MS (DESI-MS) to measure lipid turnover in the brains of mice. We show that anatomically distinct regions of the brain have distinct lipid turnover rates. These turnover measurements, in conjunction with relative concentration, will enable calculation of regiospecific synthesis rates for individual lipid species in vivo. Monitoring spatially dependent changes in metabolism has the potential to significantly facilitate research in many areas, such as brain development, cancer, and neurodegeneration.
Keywords: arachidonic acid, brain lipids, diet and dietary lipids, diagnostic tools, kinetics, molecular imaging
Lipid biosynthesis occurs in every cell and is a critical process for life. Lipids act as structural components, essential nutrients, and signals for processes as diverse as cell proliferation or apoptosis (1). Many lipids within the body are synthesized locally (2, 3). Certain classes of lipids, though, cannot be synthesized by mammals (4, 5) and some cells, like neurons, do not produce enough lipids to serve their needs. Therefore, dietary lipid availability and transport of lipids within the body are critical components of metabolism. Lipid transport is closely linked to multiple diseases, including heart disease and Alzheimer’s disease (6).
By measuring lipid abundance across a tissue surface, one can create images composed of lipid type and concentration (7–10). Imaging lipids within the structure of the brain and other organs has been used extensively to understand response to injury (11) and to detect cancers (7, 8, 12). Here, we have used the desorption ESI (DESI) imaging technique. In DESI, a fine spray of charged droplets in a high-velocity gas jet is directed toward a tissue sample (Fig. 1). Dissolved analytes from the tissue become ionized, and are pulled into the mass spectrometer (DESI-MS) (13, 14). The mass spectrometer records the mass-to-charge ratio and intensity as the spot where the spray hits the tissue is rastered over the tissue surface. By correlation of the time-dependent mass spectrometric data with the motion of the tissue on a computerized stage, an image can be constructed that displays the types and relative concentrations of analytes at specific positions in the tissue sample.
Fig. 1.
DESI is an ambient ionization mass spectrometric imaging technique. Electrospray droplets are formed when high voltage is applied to the solvent. Desorbed analyte is vacuumed into the mass spectrometer inlet, separated by m/z, and detected. After the sample is slowly rastered beneath the DESI source and mass spectrometer inlet, an image of the sample surface can be recreated from the mass spectral data.
Changes in lipid types or concentrations allow boundaries between tissues, as well as regions within the tissue, to be identified (7–10, 12). The spectra from modern high-resolution mass spectrometers are exceptionally information rich; but even so, positive identification of all ions in a spectrum is rarely possible and it is often difficult to unambiguously assign peaks to individual lipids (15, 16). As a result, many studies have been published using changes in unidentified features to differentiate between areas of an image (8, 9). Even with this limitation, DESI-MS imaging has recently been shown to improve the efficacy of cancer surgery by highlighting the boundaries between tumor and healthy tissue (7–10, 12).
An advantage of identifying and focusing on discrete lipid species, when they can be positively identified, is that we can assess the activity of biological pathways that produce those lipids. This is particularly useful in developing a rational intervention to prevent developmental defects and treat cancers or degenerative diseases. Trying to evaluate enzyme activity using only in vivo concentration is problematic because the body modifies synthesis and degradation rates to resist changes in concentration. Therefore, measurement of in vivo rates is the most sensitive method for detecting changes in metabolism (17) and may allow prognostic diagnosis of developing problems.
Here, we show that by using DESI in conjunction with metabolic labeling, we can measure turnover of individual lipid species in the mouse brain (Fig. 2). Further, we show that the turnover rates vary between physiologically recognizable structures within the brain. Although a large number of compounds can be detected in the mass spectra, in this proof-of-principle study, we limited the analysis to four identified lipids. In principle, this analysis can be conducted on large numbers of lipids simultaneously and will be particularly useful in detecting fast-growing tissues like cancer within the context of healthy tissues.
Fig. 2.
The workflow is shown here in chronological order. First, mice are fed 8% deuterated water over a course of different time periods to increase their deuterium concentration in the lipids within their bodies. Then the mice are euthanized, their organs are sliced to 50 μm thick, and the slices are thaw mounted on glass slides. The DESI source is rastered over each tissue slice over a period of approximately 2 h. The mass spectral data files are converted initially from .d files to a MATLAB data cube. Individual m/z peaks are selected to see the spatial and regioselective nature of individual lipids. Isotope ratio images reveal differing lipid turnover rates within the organ structure for individual lipids, which are measured using incorporation curves.
MATERIALS AND METHODS
Metabolic labeling
All experiments were performed under the approval of the Institutional Animal Care and Use Committees of Brigham Young University in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animals. Female mice (C57/Bl6:Spt2) between the ages of 13 and 16 months were housed in groups of five. To initiate the experiment, all mice received an intraperitoneal (IP) injection of saline deuterium oxide (0.9% w/v NaCl, 99.8% D2O) and 15 μl saline per gram body weight. After the IP bolus, mice were provided free access to food and deuterium-enriched drinking water (8% molar enrichment) for the remainder of the experiment (Fig. 2). This method caused an initial jump in deuterium enrichment up to two molar percent excess (MPE) to initiate the experiment, followed by a rise to five MPE over the next 10 days. Urine was collected up to 10 days during the labeling period to build a profile of deuterium enrichment in each animal (supplemental Fig. S1). At the designated time points (0, 1, 5, 10, 20, and 40 days post-IP injection), mice were anesthetized with carbon dioxide and then euthanized via cardiac puncture to collect blood. Brain tissue was immediately collected. Cells were removed from the blood plasma via centrifugation (10 min at 800 g at 4°C). Brain tissue and blood plasma were flash-frozen on dry ice and stored at −80°C until needed.
Measurement of deuterium enrichment
Blood plasma and urine isotopic enrichments were measured in duplicate for all time points. In preparation for this measurement, aliquots of each sample were diluted in duplicate (1:300 in 18 MΩ water) and placed into the caps of inverted sealed screw-capped vials for overnight distillation at 80°C. Similar to previous studies, the MPE deuterium in the distilled water was measured against a D2O standard curve using a cavity ring-down water isotope analyzer (Los Gatos Research, Los Gatos, CA) according to the published method (18).
Cryosectioning
Brains were cut coronally immediately after dissection and flash-frozen on dry ice. To obtain slices for DESI, frozen forebrains were adhered using VWR clear frozen section compound to the chuck of a cryostat, Thermo Scientific Microm HM550, and flash-frozen in liquid nitrogen. Fifty micron sections were sliced at −15°C. Sections were thaw-mounted onto VWR Superfrost Plus glass slides, flash-frozen on dry ice, and stored at −80°C. Prior to DESI imaging, the glass slides were dried at slight negative pressure for approximately 20 min at room temperature, as previously described (19).
DESI-MS data acquisition
Sample images were gathered using a MicrOTOF II (Bruker Daltonics, Billerica, MA) and a laboratory-constructed DESI source. DESI parameters are presented in Table 1. Together, those parameters created a solvent spot that desorbed lipids and fatty acids from the brain tissue, releasing them into the atmosphere, where they were sampled by an extension to the capillary inlet of the mass spectrometer.
TABLE 1.
Instrumental parameters for DESI image acquisition
| DESI source | |
| Emitter voltage | −5 kV |
| Solvent | 100% methanol |
| Solvent flow rate | 2.5–3.0 μl min−1 |
| N2 gas pressure | 160 psi |
| Emitter angle | 55 degrees |
| Emitter tip to surface distance | 1 mm |
| Emitter tip to inlet distance | 4 mm |
| Stage parameters | |
| x axis scan rate | 75 μm s−1 |
| y axis row spacing | 150 µm |
| Mass spectrometer parameters | |
| Ion mode | Negative |
| Mass range | m/z 200–900 |
| Acquisition rate | 1 spectrum s−1 |
| Averaging | 2 spectra |
| Inlet voltage | 500 V |
DESI image quality considerations
Acquisition of high-quality DESI images requires careful attention to a large number of experimental parameters and inevitably requires a compromise between sensitivity and spatial resolution (20, 21). The experimental parameters presented in Table 1 represent such a compromise. The nominal pixel size was 75 × 150 μm. Additional important considerations included the complete elimination of gases from the solvent, the shape of the mass spectrometer inlet extension, and the age of the sample tissue (21). Because no preventive measures were taken to keep the brain cells intact at the surface of the tissue slices during cryosectioning, the DESI-MS data represent an average of all membrane and cytosolic lipid populations.
Analysis workflow (Image Inspector)
Acquisition files from the Bruker MicrOTOF were converted from .d files to .imzML using ProteoWizard MSconvert (22, 23). The imzML was then converted to a data cube using a MatLab script adapted from the read_imzML script that was included as part of the omniSpect package published by the Fernandez group at Georgia Institute of Technology (24). It is worth noting that, despite attempts to ensure that imzML is a universal format, the imaging files derived from different proprietary manufacturers’ formats have idiosyncrasies that require attention when reading them into a custom software package. For example, the number of spectra in chronograms acquired by the Bruker instrument varies by one because of a lack of synchronization between an external start signal and the internal clocking of the data acquisition. We provide the imzML-to-cube script as a template for other users, but it has only been tested with Bruker data.
After creation, the brain data cube was then assembled into an image using a MatLab script we developed, called Image Inspector (Fig. 2). Image Inspector allows flexible manipulation of the data cube to examine and export mass spectra from individual pixels or regions of interest, to view and export concentration maps for selected masses, and to view and export maps of neutromer ratios. Portions of Image Inspector were also adapted from the omniSpect library. Areas defined by a region of interest tool were summed for export and calculation of isotope labeling. Turnover kinetics of individual brain regions in each image were calculated from the time-dependent ratio of labeled to unlabeled lipids. The code and readme for installation and usage of the Image Inspector tool are provided as supplemental information.
Identification of individual lipids
In order to verify the lipid identities, purified lipids were ordered from Sigma-Aldrich and used as standards: arachidonic acid (AA) (A3611-10MG); cis-4,7,10,1,16,19-DHA (53171-10MG); 1,2-diacyl-sn-glycero-3-phospho-L-serine (P7769-5MG); and L-α-phosphatidylinositol (PI) ammonium salt solution (P2517-5MG). While the AA, DHA, and PI standards were exact matches for the lipid identifications we tested, it is worth noting that the phosphatidylserine (PS) standard turned out to be a different PS species, with a lower molecular weight than the one we observed in our DESI brain scans. For DESI scanning, a few drops of standard in solution (∼10 mg/ml in chloroform) were placed onto VWR glass slides via pipette and fragmentation was accomplished as described below for the brain slices.
MS/MS fragmentation spectra (supplemental Fig. S3) were collected in negative ion mode for ions at m/z 303.25, 327.25, 834.5, and 885.6, using an Agilent QTOF mass spectrometer equipped with a DESI source. A metabolically unlabeled 50 μm-thick brain slice was scanned in targeted MS/MS mode three times at each of three different collision-induced dissociation fragmentation energies (10, 20, and 40 EV). Nitrogen was the collision gas. Each scan lasted approximately 3 min; during the scans, the brain slice was manually moved under the DESI spray (100% methanol at 3 ml/min) in order to maximize total ion counts. The capillary voltage was set at 1,000 V, and MS1 and MS2 data were collected at a maximum rate of four spectra per second (other settings: 250 MS per spectrum, narrow isolation width m/z ∼1.3). Fragmentation data at each collision energy were averaged to collect the MS/MS spectra (supplemental information); experimentally observed fragments for tissue and standards were initially compared against the literature (14) and then further analyzed using online tools from LipidMaps.org (25) and Metlin.Scripps.edu (26) to identify each of the lipid species.
Calculation of the number of deuterium sites
As we (27, 28) and others (29, 30) have previously described for peptides, independent turnover rates can be measured for multiple analytes simultaneously using MS and D2O labeling. The three main variables to consider when calculating the turnover rate (k) are the number of sites in each lipid that can incorporate deuterium from water (n), the D2O enrichment of the body water at the time the lipid was labeled (p), and the time-dependent labeled fraction (f) of the lipid being measured (28).
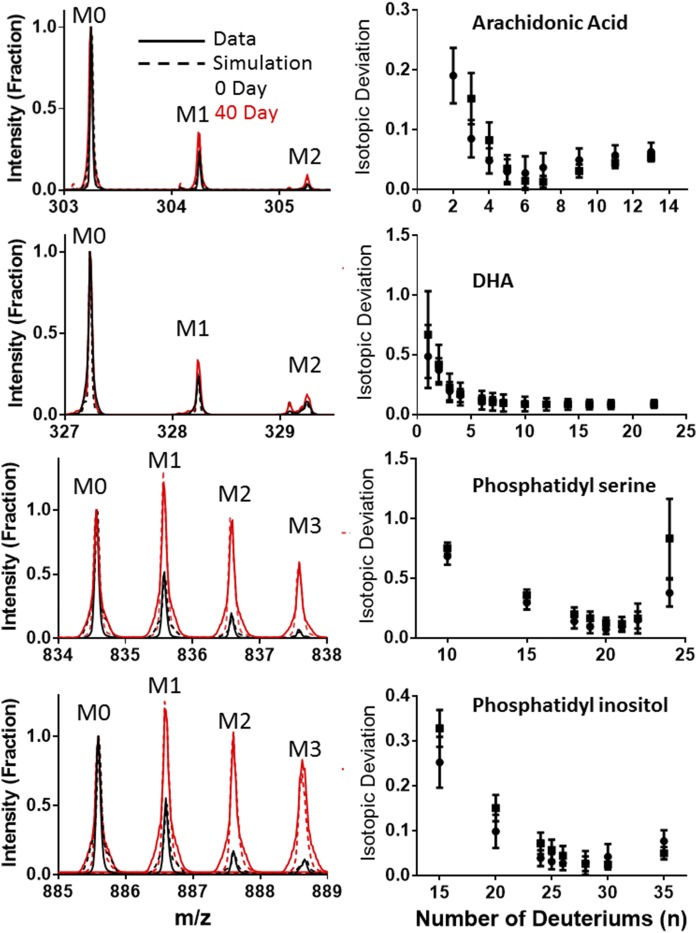
The number of covalent deuterium sites (n) is unique for each lipid (Table 2). This value was calculated using the isotope pattern for the metabolically labeled form of each identified lipid and the known D2O enrichment (Fig. 3). Highly labeled spectra for the individual lipids were collected by imaging brain tissue from mice collected after 40 days. We subtracted the unlabeled isotope pattern from the labeled spectra to isolate the specific changes due to the newly synthesized lipid. We found that using an experimentally determined unlabeled background improved the calculations. We then calculated the deviation, assuming a range of n values for the lipid. Three of the four lipids had a single best n, which minimized the deviation between theory and experiment for each of the isotope peaks within the isotope pattern (Fig. 3), as previously described (27, 28, 31). DHA had a very small overall change in the isotope pattern that was affected by interference from nearby molecules. We were not able to find a unique best n value for DHA.
TABLE 2.
The common name, observed ionic mass, and elemental composition of the four major singly charged ions observed in our DESI-MS spectra
| Lipid | m/z | Formula | n | k in CA | k in CO | Percent |
| AA* | 303.25 | C20H32O2 | 6 | 0.046 ± 0.005 | 0.055 ± 0.007 | 40 |
| DHA | 327.25 | C22H32O2 | ∼8 | 0.057 ± 0.019 | 0.045 ± 0.010 | 32 |
| PS* | 834.6 | C46H77NO10P | 21 | 0.041 ± 0.003 | 0.053 ± 0.001 | 100 |
| PI | 885.6 | C47H82O13P | 27 | 0.072 ± 0.005 | 0.081 ± 0.005 | 73 |
The asterisk (*) next to the name indicates a statistically significant difference in the turnover rate (k) of lipids between the caudoputamen (CA) and cortex (CO). The number of deuteriums (n) was the same regardless of the area of the brain. There were large differences in the amount of biosynthesized versus dietary lipid (percent) for different lipid species.
Fig. 3.
Comparison of experimental and optimized simulations of labeled and unlabeled spectra (left). The number of deuterium sites was derived by minimizing the deviation between the simulated and experimental spectra when the number of covalant deuterium sites was allowed to vary (right).
Calculating in vivo lipid turnover
Every time a new lipid is synthesized, it will incorporate protons and deuterons from the body water pool. The relative percentage of deuterium incorporated is dependent on the enrichment of the water at that time. The changing deuterium enrichment of the body water in these mice required that the effective deuterium enrichment be calculated based on the half-life of the analyte. Because human studies usually have an increasing nonlinear body water enrichment (28, 30), this study provided a good proof-of-principle test for the calculations that would be required for analyzing metabolically D2O-labeled human tissue samples. The enrichment of the body water for this calculation was defined as the average of the urine and plasma values for all mice at a time point (supplemental Fig. S1). This body water curve provided the time-dependent precursor enrichment for the calculation of the f values. The time-dependent change in normalized intensity ΔIx for each mass in the neutromer pattern was calculated according to equation 1, for both experimental and theoretical spectra. As the initial point for the Bayesian calculation of k, we calculated the theoretical ΔIx for the labeled molecule using the body water measured that day. The daily f values were then calculated using the single pool rise to plateau kinetic shown in equation 2. The half-life from this initial k value was then used to calculate the effective body water at each time point as the integral over that time period, which was used to calculate new theoretical ΔIx and a new f, as described previously for protein (28). These f values were used to fit a new k, which was used to calculate the new effective deuterium enrichment for the next round. This recursive optimization was conducted until the lipid half-life changed by less than 0.5 days.
| Eq. 1 |
| Eq. 2 |
We compared k values in the cortex and caudoputamen of the brain by averaging the spectra in those regions using Image Inspector, and then calculating k using the scipy package in python for the recursive nonlinear regression fit.
RESULTS
Establishment of lipid identities and n values
In the DESI spectra, four singly charged ions were consistently observed with significant signal intensities (Fig. 3). We compared the parent m/z and MS/MS fragmentation data (supplemental information) observed here against a literature source (14) and tentatively identified them as AA (C20:4 Δ5,8,11,14, m/z 303.25, C20H32O2), DHA (C22:6 Δ4,7,10,13,16,19, m/z 327.3, C22H32O2), PS (C40:6, m/z 834.6, C46H77NO10P), and PI (C38:4, m/z 885.6, C47H82O13P). Subsequent MS/MS fragmentation of purified lipid standards verified the identification of AA, DHA, PS, and PI (supplemental Fig. S3).
The calculated n value for each of the lipids (Fig. 3) is consistent with their individual biosynthetic pathways. AA and DHA are both conditionally essential fatty acids. They are synthesized from the essential fatty acids, linoleic acid (C18:2 D9,12) and linolenic acid (C18:3 D9,12, 15), respectively, but can also be obtained through the diet. Our results suggested that the addition of the C2 unit to make AA from linoleic acid included the covalent addition of six protons from water (supplemental Fig. S2). During the deuterium enrichment, these six positions incorporated deuterium, leading to the n = 6 value for this lipid. (Fig. 3). DHA was less clear because the spectra were noisier and the experimental ΔI was very small. The change at M2 for DHA was confounded in many spectra by overlapping peaks from unidentified species. Although there were multiple n values mathematically (Fig. 3), the similarity to the AA biological pathway suggested that an n = 8 value would be most appropriate (supplemental Fig. S2). It is highly likely that the PS and PI spectra represent two families of related structures where the unsaturated bonds are at multiple different positions in the fatty acyl tails. Our DESI fragmentation method was not capable of distinguishing between specific isomers that differed only in the placement of double bonds. However, in spite of this potential structural variation, we were able to calculate an n value of 21 for PS and 27 for PI (Fig. 3). The glycerol backbone and the head groups of these lipids are synthesized from glucose, which would allow each of the carbon hydrogen bonds to incorporate a deuterium (2). For PS, this means that up to eight deuterium positions are within the glycerol and serine. The 13 other deuterium sites are presumably distributed between the two fatty acyl tails. The inositol head group in PI and two fewer desaturations in the fatty acyl tails could easily account for the six more deuterium positions observed in this molecule (supplemental Fig. S2).
Spatially distinct changes in isotope ratio are specific to individual lipids and independent of concentration
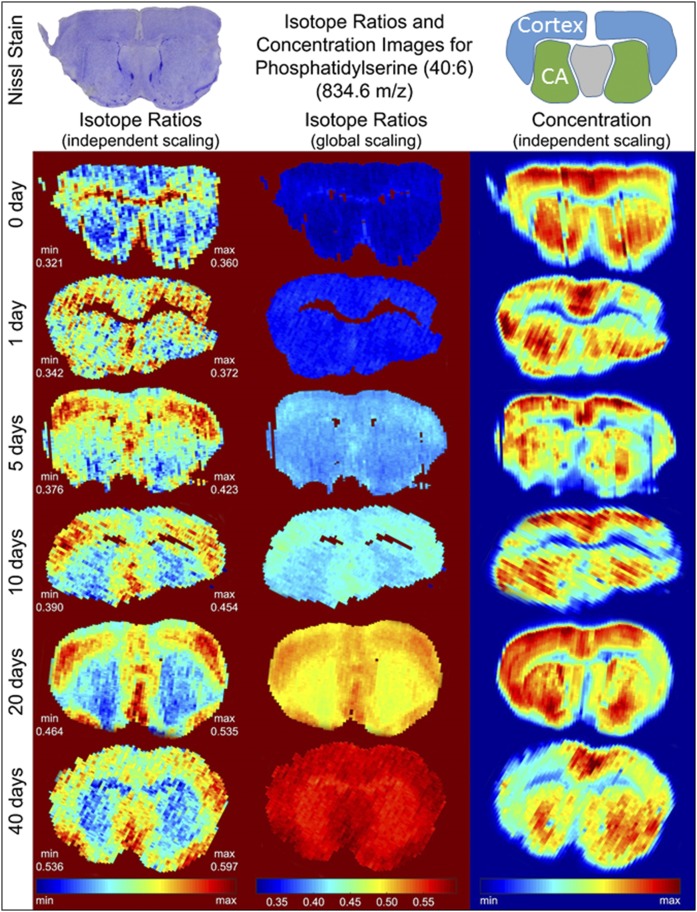
In DESI images, signal intensity at a given m/z value is a measure of the relative concentration of a particular compound (the neutromer distribution is a measure of the turnover of that compound). In addition to depending on the concentration of a compound in the tissue, DESI signals can be affected by a range of factors, including the tissue type, microscopic surface roughness, and changes in ambient conditions during image acquisition. Even with this inherent variability, the spatial distribution of lipid signal intensity was distinct for each targeted lipid and followed boundaries of the classical brain structures in a manner that was consistent with changes in concentration. Measurement of PS intensity in tissues collected throughout the metabolic labeling time course (Fig. 4, right column) showed that its concentration was consistently relatively high in the cortex and the caudoputamen and lower in the corpus callosum.
Fig. 4.
Concentration/isotope ratio images for PS. A representative stained tissue slice is shown (top left), with the areas of the cortex and caudoputamen (CA) highlighted in the cartoon (top right).
Comparison of the change in intensity with the change in isotope ratio for the four test compounds across an image showed that they are not necessarily linked for the different lipid species (Fig. 4, supplemental Fig. S4). Using PS as an example, if the change in isotope pattern was scaled linearly over the 40 days of the experiment (Fig. 4, global scaling), we saw similar changes across the entire brain. This similarity suggested that newly synthesized lipid is present across the entire organ. Changing the scaling of the image to emphasize the range within a single image (independent scaling) showed that there was a small, but measurably faster, rate in a subsection of the cerebral cortex and the septal nucleus. Importantly, the symmetry of the image was in agreement with the structure of the brain, reducing the possibility that this was a transient fluctuation in the image. The turnover measurement of each lipid was much more resistant to variations in ionization efficiency, because the heavy and light versions of the lipid are chemically identical and change similarly, preserving the ratio between them. This means that the turnover measurement is, in principle, more robust across an image than simple concentration measurements.
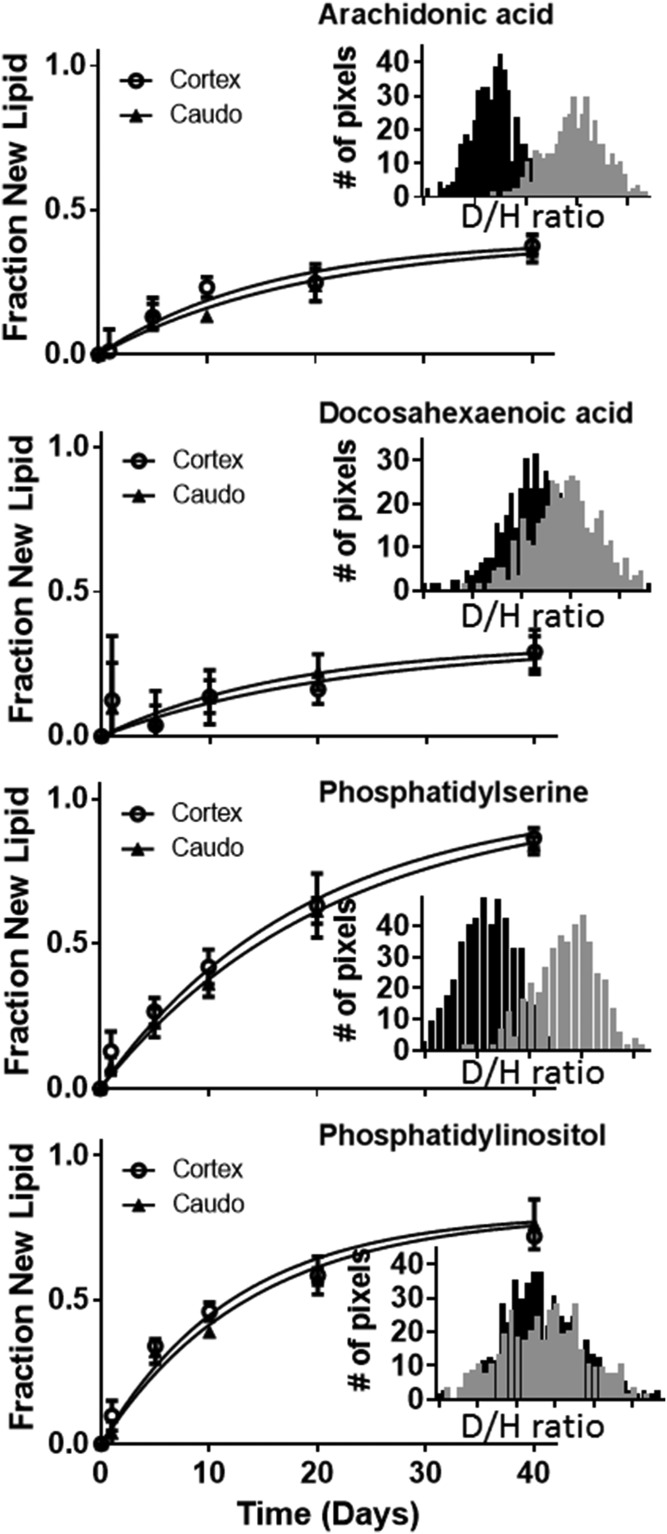
The other lipids all showed similar, yet unique, distributions in both concentration and turnover rate. AA, for example, was found at relatively high concentrations everywhere in the brain except the caudoputamen area (supplemental Fig. S4A). The rate of turnover was also slower in the caudoputamen. This is consistent with the idea that AA is synthesized and degraded locally. DHA was found at relatively high concentrations in the cortex, but there was a very small and somewhat noisy change in the isotope ratio, suggesting that turnover was minimal anywhere in the brain (supplemental Fig. S4B). This is not surprising, given that DHA is conditionally essential in the diet, but these results suggest that 65% of the DHA in the brain is sourced directly from the diet. PI has a similarly wide distribution of relatively high concentrations in the brain, except in the approximate area of the corpus callosum (supplemental Fig. S4C). Interestingly, the turnover of this lipid was fastest in the corpus callosum where the concentration was very low. This suggests that the lipid was either synthesized and immediately degraded in the area of the corpus callosum or synthesized and distributed to the rest of the brain from the corpus callosum.
Turnover calculations highlight differences in metabolism between lipids
Using the lipid-specific n value and the measured body water, we calculated the time-dependent percentage of the molecules that had been labeled. This allowed us to directly compare metabolic rates between the different lipids. We found that using the average body water curve and combining all the time-dependent measurements from the individual mice to constrain the turnover calculation dramatically improved the confidence in the fit. This was consistent with the fact that these mice were all genetically identical and of a similar age. Therefore, they could be considered biological replicates for these metabolic measurements.
As suggested by the images (Fig. 4, supplemental Fig. S4), we found that there was a small, but measureable, difference in turnover rate between different regions of the brain for the lipids AA and PS (Fig. 5, cortex vs. caudoputamen). PI turned over more rapidly than the others and did not have a significant difference between the cortex and caudoputamen (Table 2). Interestingly, the turnover kinetics also highlighted differences in the source of the lipids. For example, the incorporation curve for PS had a rate of approximately 5% per day and increased over the 40 day experiment such that ∼100% of the lipid was new by the final measurements. PS is evidentially formed from fully biosynthesized precursors, while the majority of AA and DHA are sourced from the diet. PS, AA, and DHA curves had similar rates of incorporation (∼5% per day), but the amount of labeled AA and DHA saturated at much lower amounts than PS (Table 2). The saturation percentage reflects the relative contributions of biosynthesis and dietary sources for each lipid species. De novo synthesis and dietary sources of lipids are known to both be critical for the development and health of the brain (4). The uniform kinetic plateau across the image for each lipid also suggests there is no spatial differentiation in the source, i.e., diet versus de novo synthesis.
Fig. 5.
Deuterium enrichment of all four lipids included was compared in different areas of the brain. The distribution of enrichment in individual pixels in the cortex (gray inset) versus the caudoputamen (black inset) varied according to lipid and area of the brain (day 20 is shown). Turnover rates were calculated using kinetic curves composed of three separate images from two mice at each time point.
DISCUSSION
We have shown that DESI can be used to image spatial regulation of metabolism in the mouse brain. Although any imaging technique could potentially be used to monitor metabolism using these methods (9). DESI is convenient as an imaging system because the sample preparation is fast and inexpensive, with simple atmospheric pressure ionization. A disadvantage of DESI is the variation in ionization due to chemical and/or physical variability in the tissue samples. Because the sniffer (Fig. 1) is held so close to the tissue surface, we have seen that if the 50 μm tissue slice has ragged edges or variable thickness, the sniffer position relative to the sample surface changes during a scan, causing biases in the image.
Imaging lipids is convenient because they ionize well in DESI and because lipid metabolism lies at the core of multiple important diseases, such as neurodegeneration (3, 4, 6, 32) and cancer (8, 17, 33). Improved identification of lipids is an important direction for future development in biochemistry because of their roles in metabolism and signaling. Identification of lipids is difficult; grouping mass spectral features into (sometimes broad) lipid classes is the current state of the art (15, 16). For this proof-of-principle study, we monitored the turnover of four previously identified lipids, but the techniques can be applied to any identifiable molecule.
There have been many excellent studies investigating metabolism of lipids in the brain (34). To our knowledge, we are the first to image metabolism of specific lipids. By identifying the individual lipids, we unlock the ability to investigate the metabolic pathways of each lipid. The in vivo turnover reports the rate-limiting step for metabolism. Interestingly, use of radiolabeled AA and DHA has shown that phospholipid incorporation and turnover occurs over a range of about 2–8% per day (35), in agreement with our observations. The use of radioactivity in the previous study precluded the assessment of the percent of the total pool that is synthesized. Using stable isotopes, the experiment measures turnover rate, as well as the relative contribution of dietary sources to the metabolism of the tissue. We observed that the PI turnover is faster than the PS, but that there is a portion of the PI pool within the brain that does not display turnover. This could be due to a dietary supply of PI or to brain cells that are preserving the PI in a specific structure. We favor the dietary supply hypothesis because this 80% limit was uniform across the brain and both AA and DHA, which are known to be sourced from the diet to supplement biosynthesis (4), show similar trends (supplemental Fig. S4).
Both biological and instrumental factors cause variations between images and in the symmetry of individual images. As mentioned above, changing ionization efficiency in the DESI source introduces variations that are particularly evident in the concentration images. The section of the brain that is imaged varies somewhat as well because it can be difficult to match the exact location of the tissue slice within the brain for each time point. The isotope analysis feature of Image Inspector is valuable for analysis of both biological and instrumental noise. In unlabeled samples it provides the analyst with an accuracy metric that can be used to evaluate image quality. Overlapping isotopic signatures can be identified by looking at the intensity patterns across the brain. For example, there was a significant change in the observed distribution of the m/z 888 neutromer of PI across the brain relative to the monoisotopic m/z 885 (supplemental Fig. S5). This was indicative of an overlapping lipid species, i.e., the low purity of the mass spectrum. Therefore, the kinetics calculations didn’t include the m/z 888 or heavier m/z neutromers. We also found that by monitoring bias in the neutromer pattern, we have rational criteria for setting signal thresholds for each analyte in each image. This is a continuous quality analysis that can be used to rapidly identify noise in the instrument or bias from closely overlapping signals, which can vary with time or tissue composition.
The next step in developing this kinetic imaging method will be to focus on improved quantitation in combination with the turnover of the individual lipids. This would allow investigators to distinguish between changes in synthesis, transport, or degradation of lipids. Expanding the imaging to a wide variety of other analytes, including other metabolites and proteins, would dramatically expand the utility of this method. This allows us to consider future experiments looking into the effect of diet on brain health and development or investigating the effect of cancer therapies that target lipid metabolism.
Supplementary Material
Acknowledgments
The authors would like to thank members of the Facundo Fernandez group at Georgia Institute of Technology, particularly Rachel Bennett Stryffeller and Martin Payne, for their assistance in designing and setting up the DESI imaging system. The authors would also like to thank Dr. Benjamin Bikman of the Physiology and Developmental Biology Department at Brigham Young University for donating the mice used in this study.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- DESI
- desorption ESI
- IP
- intraperitoneal
- MPE
- molar percent excess
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
This work was supported by Brigham Young University and in part by funding from the Fritz B. Burns Foundation.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Canals D., Jenkins R. W., Roddy P., Hernandez-Corbacho M. J., Obeid L. M., and Hannun Y. A.. 2010. Differential effects of ceramide and sphingosine 1-phosphate on ERM phosphorylation: probing sphingolipid signaling at the outer plasma membrane. J. Biol. Chem. 285: 32476–32485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allister C. A., Liu L. F., Lamendola C. A., Craig C. M., Cushman S. W., Hellerstein M. K., and McLaughlin T. L.. 2015. In vivo 2H2O administration reveals impaired triglyceride storage in adipose tissue of insulin-resistant humans. J. Lipid Res. 56: 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith S., Witkowski A., and Joshi A. K.. 2003. Structural and functional organization of the animal fatty acid synthase. Prog. Lipid Res. 42: 289–317. [DOI] [PubMed] [Google Scholar]
- 4.Guemez-Gamboa A., Nguyen L. N., Yang H., Zaki M. S., Kara M., Ben-Omran T., Akizu N., Rosti R. O., Rosti B., Scott E., et al. 2015. Inactivating mutations in MFSD2A, required for omega-3 fatty acid transport in brain, cause a lethal microcephaly syndrome. Nat. Genet. 47: 809–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raz R., and Gabis L.. 2009. Essential fatty acids and attention-deficit-hyperactivity disorder: a systematic review. Dev. Med. Child Neurol. 51: 580–592. [DOI] [PubMed] [Google Scholar]
- 6.Christensen K. D., Roberts J. S., Whitehouse P. J., Royal C. D., Obisesan T. O., Cupples L. A., Vernarelli J. A., Bhatt D. L., Linnenbringer E., Butson M. B., et al. ; REVEAL Study Group. 2016. Disclosing pleiotropic effects during genetic risk assessment for Alzheimer disease: a randomized trial. Ann. Intern. Med. 164: 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chughtai K., Jiang L., Greenwood T. R., Glunde K., and Heeren R. M.. 2013. Mass spectrometry images acylcarnitines, phosphatidylcholines, and sphingomyelin in MDA-MB-231 breast tumor models. J. Lipid Res. 54: 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberlin L. S., Margulis K., Planell-Mendez I., Zare R. N., Tibshirani R., Longacre T. A., Jalali M., Norton J. A., and Poultsides G. A.. 2016. Pancreatic cancer surgical resection margins: molecular assessment by mass spectrometry imaging. PLoS Med. 13: e1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louie K. B., Bowen B. P., McAlhany S., Huang Y., Price J. C., Mao J. H., Hellerstein M., and Northen T. R.. 2013. Mass spectrometry imaging for in situ kinetic histochemistry. Sci. Rep. 3: 1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy R. C., Hankin J. A., and Barkley R. M.. 2009. Imaging of lipid species by MALDI mass spectrometry. J. Lipid Res. 50 (Suppl.): S317–S322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hankin J. A., Farias S. E., Barkley R. M., Heidenreich K., Frey L. C., Hamazaki K., Kim H. Y., and Murphy R. C.. 2011. MALDI mass spectrometric imaging of lipids in rat brain injury models. J. Am. Soc. Mass Spectrom. 22: 1014–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oezdemir R. F., Gaisa N. T., Lindemann-Docter K., Gostek S., Weiskirchen R., Ahrens M., Schwamborn K., Stephan C., Pfister D., Heidenreich A., et al. 2012. Proteomic tissue profiling for the improvement of grading of noninvasive papillary urothelial neoplasia. Clin. Biochem. 45: 7–11. [DOI] [PubMed] [Google Scholar]
- 13.Takáts Z., Wiseman J. M., and Cooks R. G.. 2005. Ambient mass spectrometry using desorption electrospray ionization (DESI): instrumentation, mechanisms and applications in forensics, chemistry, and biology. J. Mass Spectrom. 40: 1261–1275. [DOI] [PubMed] [Google Scholar]
- 14.Wiseman J. M., Ifa D. R., Song Q., and Cooks R. G.. 2006. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew. Chem. Int. Ed. Engl. 45: 7188–7192. [DOI] [PubMed] [Google Scholar]
- 15.Gathungu R. M., Stavrovskaya I. G., Larrea P., Sniatynski M. J., and Kristal B. S.. 2016. Simple LC-MS method for differentiation of isobaric phosphatidylserines and phosphatidylcholines with deuterated mobile phase additives. Anal. Chem. 88: 9103–9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan E., and Reid G. E.. 2016. Chemical derivatization and ultrahigh resolution and accurate mass spectrometry strategies for “shotgun” lipidome analysis. Acc. Chem. Res. 49: 1596–1604. [DOI] [PubMed] [Google Scholar]
- 17.Metallo C. M., and Vander Heiden M. G.. 2013. Understanding metabolic regulation and its influence on cell physiology. Mol. Cell. 49: 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lis G., Wassenaar L. I., and Hendry M. J.. 2008. High-precision laser spectroscopy D/H and 18O/16O measurements of microliter natural water samples. Anal. Chem. 80: 287–293. [DOI] [PubMed] [Google Scholar]
- 19.Bennet R. V., Gamage C. M., and Fernandez F. M.. 2013. Imaging of biological tissues by desorption electrospray ionization mass spectrometry. J. Vis. Exp. 77: e50575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodzon-Kulakowska A., Drabik A., Ner J., Kotlinska J. H., and Suder P.. 2014. Desorption electrospray ionisation (DESI) for beginners–how to adjust settings for tissue imaging. Rapid Commun. Mass Spectrom. 28: 1–9. [DOI] [PubMed] [Google Scholar]
- 21.Dill A. L., Eberlin L. S., Costa A. B., Ifa D. R., and Cooks R. G.. 2011. Data quality in tissue analysis using desorption electrospray ionization. Anal. Bioanal. Chem. 401: 1949–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., et al. 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30: 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessner D., Chambers M., Burke R., Agus D., and Mallick P.. 2008. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 24: 2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parry R. M., Galhena A. S., Gamage C. M., Bennett R. V., Wang M. D., and Fernandez F. M.. 2013. omniSpect: an open MATLAB-based tool for visualization and analysis of matrix-assisted laser desorption/ionization and desorption electrospray ionization mass spectrometry images. J. Am. Soc. Mass Spectrom. 24: 646–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fahy E., Sud M., Cotter D., and Subramaniam S.. 2007. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 35: W606–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith C. A., O’Maille G., Want E. J., Qin C., Trauger S. A., Brandon T. R., Custodio D. E., Abagyan R., and Siuzdak G.. 2005. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 27: 747–751. [DOI] [PubMed] [Google Scholar]
- 27.Mathis A. D., Naylor B. C., Carson R. H., Evans E., Harwell J., Knecht J., Hexem E., Peelor F. F. III, Miller B. F., Hamilton K. L., et al. 2017. Mechanisms of in vivo ribosome maintenance change in response to nutrient signals. Mol. Cell Proteomics. 16: 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price J. C., Holmes W. E., Li K. W., Floreani N. A., Neese R. A., Turner S. M., and Hellerstein M. K.. 2012. Measurement of human plasma proteome dynamics with (2)H(2)O and liquid chromatography tandem mass spectrometry. Anal. Biochem. 420: 73–83. [DOI] [PubMed] [Google Scholar]
- 29.Kasumov T., Ilchenko S., Li L., Rachdaoui N., Sadygov R. G., Willard B., McCullough A. J., and Previs S.. 2011. Measuring protein synthesis using metabolic (2)H labeling, high-resolution mass spectrometry, and an algorithm. Anal. Biochem. 412: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam M. P., Wang D., Lau E., Liem D. A., Kim A. K., Ng D. C., Liang X., Bleakley B. J., Liu C., Tabaraki J. D., et al. 2014. Protein kinetic signatures of the remodeling heart following isoproterenol stimulation. J. Clin. Invest. 124: 1734–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naylor B. C., Porter M. T., Wilson E., Herring A., Lofthouse S., Hannemann A., Piccolo S. R., Rockwood A. L., and Price J. C.. 2017. DeuteRater: a tool for quantifying peptide isotope precision and kinetic proteomics. Bioinformatics. 33: 1514–1520. [DOI] [PubMed] [Google Scholar]
- 32.Lee C. Y., Tse W., Smith J. D., and Landreth G. E.. 2012. Apolipoprotein E promotes beta-amyloid trafficking and degradation by modulating microglial cholesterol levels. J. Biol. Chem. 287: 2032–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menendez J. A., and Lupu R.. 2007. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 7: 763–777. [DOI] [PubMed] [Google Scholar]
- 34.Bruce K. D., Zsombok A., and Eckel R. H.. 2017. Lipid processing in the brain: a key regulator of systemic metabolism. Front. Endocrinol. (Lausanne). 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rapoport S. I., Chang M. C., and Spector A. A.. 2001. Delivery and turnover of plasma-derived essential PUFAs in mammalian brain. J. Lipid Res. 42: 678–685. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.