Abstract
Osteopontin (OPN) is involved in different liver pathologies in which metabolic dysregulation is a hallmark. Here, we investigated whether OPN could alter liver, and more specifically hepatocyte, lipid metabolism and the mechanism involved. In mice, lack of OPN enhanced cholesterol 7α-hydroxylase (CYP7A1) levels and promoted loss of phosphatidylcholine (PC) content in liver; in vivo treatment with recombinant (r)OPN caused opposite effects. rOPN directly decreased CYP7A1 levels through activation of focal adhesion kinase-AKT signaling in hepatocytes. PC content was also decreased in OPN-deficient (OPN-KO) hepatocytes in which de novo FA and PC synthesis was lower, whereas cholesterol (CHOL) synthesis was higher, than in WT hepatocytes. In vivo inhibition of cholesterogenesis normalized liver PC content in OPN-KO mice, demonstrating that OPN regulates the cross-talk between liver CHOL and PC metabolism. Matched liver and serum samples showed a positive correlation between serum OPN levels and liver PC and CHOL concentration in nonobese patients with nonalcoholic fatty liver. In conclusion, OPN regulates CYP7A1 levels and the metabolic fate of liver acetyl-CoA as a result of CHOL and PC metabolism interplay. The results suggest that CYP7A1 is a main axis and that serum OPN could disrupt liver PC and CHOL metabolism, contributing to nonalcoholic fatty liver disease progression in nonobese patients.
Keywords: glycerolipids, nonalcoholic fatty liver disease, de novo lipogenesis
Osteopontin (OPN) is a multifunctional cytokine that is expressed in various tissues (1–3). In the liver, OPN expression is upregulated in response to inflammation and liver injury. In obesity, liver OPN expression correlates with the triacylglyceride (TG) content (1, 4, 5). OPN is increased in several models of liver fibrosis (6–8) and in hepatocellular carcinoma (9) in which the overexpression of OPN has been identified as a novel early marker (10). Its deficiency protects against obesity-induced hepatic steatosis and attenuates glucose production in mice (4) and neutralization of circulating OPN abrogates liver fibrogenesis in mice (8). Even though it plays a major role in liver diseases, OPN is also involved in the pathophysiology of other malignancies (11), such as breast (12) and lung cancer (13).
Altered lipid metabolism is a hallmark of liver disease. In nonalcoholic fatty liver disease (NAFLD) development, an imbalance between liver lipid input (lipid uptake and de novo synthesis) and lipid output (lipid catabolism and secretion) plays a key role (14). Altered phosphatidylcholine (PC) metabolism has also been linked with NAFLD development and progression. In certain conditions PC can constitute a source of TG (15). PC metabolism is also altered in a variety of tumors and cancer cells because of the role of PC in provision of membranes and lipid second messengers (16–18).
Liver PC and cholesterol (CHOL) metabolism are highly interconnected, both being bile lipids synthesized in liver. Experimental and clinical evidence has linked altered hepatic CHOL homeostasis and free CHOL accumulation in liver to nonalcoholic steatohepatitis (NASH) (19). Part of the liver CHOL is converted into bile acids (BAs), and some BA species are increased in the liver of patients with NASH, which could also play a role in NAFLD progression (20, 21).
Thus, here, we wanted to know whether OPN could directly dysregulate liver (and more specifically hepatocyte) lipid metabolism and elucidate the mechanism involved. The current work shows how OPN regulates the metabolic fate of liver acetyl-CoA as a result of CHOL and PC metabolism interplay; even more, it shows that OPN directly activates the focal adhesion kinase (FAK)-AKT signaling pathway in hepatocytes and decreases the cholesterol 7α-hydroxylase (CYP7A1) level. The results suggest that modulation of CYP7A1 protein levels by OPN represents a main axis in the dysregulated metabolic profile and that, in nonobese NAFL patients, circulating OPN could disrupt liver PC and CHOL metabolism that is involved in NAFLD progression.
MATERIALS AND METHODS
Human samples
This study comprised 32 patients on whom a liver biopsy was performed during laparoscopic cholecystectomy. Nineteen patients showed histological features of nonalcoholic fatty liver (NAFL) according to Brunt’s criteria (22). Among them, six were obese. Thirteen patients had histologically normal liver (NL) (Table 1). The study was performed in agreement with the Declaration of Helsinki and with local and national laws. The human ethics committee of the Santa Cristina University Hospital and the University of the Basque Country approved the study procedures and written informed consent was obtained from all patients before inclusion in the study.
TABLE 1.
Demographic metabolic biochemical and histological features of nonobese patients with NL, nonobese patients with NAFL, and obese patients with NAFL
| NL (n = 13) | NAFL (Nonobese) (n = 13) | NAFL (Obese) (n = 6) | |
| BMI (kg/m2) | 24.4 (17.9–29.4) | 26.0 (21.4–29.6) | 34.4 (31.2–35.4)a |
| Age (years) | 48.46 (28–78) | 52.9 (21–78) | 55.7 (34–77) |
| Male gender | 4 (30.8%) | 6 (46.2%) | 2 (33.3%) |
| TG (mg/dl) | 76.6 (54–100) | 128.1 (50–317)b | 122.2 (88–156)a |
| Glucose (mg/dl) | 93.9 (74–128) | 104.0 (78–177) | 101.0 (89–116) |
| CHOL (mg/dl) | 184.3 (144–246) | 201.2 (93–247) | 197.8 (183–224) |
| CHOL-HDL (mg/dl) | 49.7 (37–64) | 53.2 (33–69) | 47.7 (36–76) |
| ALT (IU/l) | 16.6 (6–32) | 34.6 (16–93)c | 20.5 (11–33) |
| AST (IU/l) | 17.2 (10–31) | 25.7 (13–43)c | 17.8 (13–23) |
| Alkaline phosphatase (IU/l) | 63.3 (38–138) | 87.5 (50–124)b | 80.3 (59–112) |
| Insulin (mU/ml) | 6.2 (3–14.7) | 6.9 (4.4–10) | 11.5 (8.4–15.8)c |
| HOMA-IR | 0.85 (0.4–2) | 0.92 (0.4–1.4) | 1.7 (1.2–2.1)c |
| Steatosis (%) | |||
| Grade 0 | 13 (100%) | ||
| Grade 1 | 11 (84.6%) | 4 (66.7%) | |
| Grade 2 | 2 (15.4%) | 2 (33.3%) | |
| Grade 3 |
Data are shown as mean (range) or percentage (%). Nonobese patients (BMI <30 kg/m2), obese patients (BMI ≥30 kg/m2). HOMA-IR, homeostatic model assessment-insulin resistance.
P ≤ 0.001, NL versus NAFL (non-obese) or NL versus NAFL (obese).
P ≤ 0.05, NL versus NAFL (non-obese) or NL versus NAFL (obese).
P ≤ 0.01, NL versus NAFL (non-obese) or NL versus NAFL (obese).
Animals
Ten- to twelve-week-old female OPN-KO mice and their WT littermates were provided by Jackson Laboratory. They were maintained on a rodent chow diet (Teklad global 18% protein rodent diet 2018S; Harlan Laboratories Inc.) and were housed in a temperature-controlled room with a 12 h light/dark cycle. Animal procedures were approved by the Ethics Committee for Animal Welfare of the University of the Basque Country UPV/EHU and were conducted in conformity with the European Union directives for animal experimentation.
Recombinant OPN and atorvastatin treatments
Recombinant (r)OPN (15 μg/mouse; R&D Systems, United Kingdom) or vehicle was injected via the tail vein on two alternate days. Twenty-four hours after the last injection, the mice were euthanized.
Intragastric atorvastatin (100 mg/kg) was provided to another group of animals during 2 weeks on alternate days. Twenty-four hours after the last dose, the mice were euthanized.
Quantification of lipids
After homogenization of liver tissue, lipids were extracted as described before (23). PC, phosphatidylethanolamine (PE), and CHOL were quantified as described previously (24). TGs were quantified using a commercially available kit (A. Menarini Diagnostics, Italy).
In vitro experiments
The liver cells were isolated by perfusion with collagenase and hepatocytes were purified by density centrifugation and selective adherence, as described (25). For metabolic studies, primary pure hepatocytes from OPN-deficient (OPN-KO) mice were treated with rOPN (32 nM) and hepatocytes were incubated with [3H]acetate (20 μM, 20 μCi/ml), as described (15). At the indicated times, cells and medium were separately recovered and lipids were extracted (26) and separated (24). PC, PE, TG, and CHOL content was quantified and the label incorporated into glycerophospholipids (GPLs) and CHOL was determined as described previously (15). For the glucose uptake measurement, hepatocytes were incubated with D-[3H]glucose (2.45 μM, 112 μCi/ml) as described (27). After 1 min of incubation, cells were collected and the label incorporated into the cells was determined by scintillation counter.
For signaling experiments, primary pure hepatocytes from WT mice were incubated with or without rOPN (150 nM). For inhibition of the FAK-AKT pathway, hepatocytes were preincubated for 1 h with the FAK inhibitor, Y15 (30 μM); for inhibition of the PI3K-AKT pathway, PI3K is positioned downstream of FAK, hepatocytes were preincubated with LY294002 (50 μM) for 1 h and then hepatocytes were incubated with or without rOPN (150 nM) in the presence or absence of the inhibitors for another 3 h.
BA quantification
BAs from liver samples were extracted using silica-based bonded phase cartridges (Sep-Pack Plus C18; Waters-Millipore, Madrid, Spain) and analyzed in an HPLC-tandem mass spectrometer (6410 Triple Quad LC/MS; Agilent Technologies, Santa Clara, CA), as previously reported (28), following a modification of the method described (29). Chromatographic separation was carried out with gradient elution using a Zorbax Eclipse XDB-C18 column (150 × 4.6 mm, 5 μm) at 35°C and a flow rate of 0.5 ml/min. Mobile phase (pH 4.6) was initially 80:20 methanol/water, and it was changed to 97:3 methanol/water over 9 min and then returned to 80:20 in 1 min. Both solvents contained 5 mM ammonium acetate and 0.01% formic acid. ESI in negative mode was used with the following conditions: gas temperature 350°C, gas flow 8 l/min, nebulizer 10 psi, capillary voltage 2,500 V. MS/MS acquisition was achieved in multiple reaction monitoring mode using the specific m/z transitions: [M-H]− ion to 80.2 for taurine-conjugated BAs and [M-H]− ion to 74 for glycine-conjugated BAs. Unfortunately, free BAs did not generate characteristic ion fragments, as reported before (29), and transition from unfragmented precursor molecular ions 407.1 to 407.1, 391.3 to 391.3, and 375.3 to 375.3 were selected for trihydroxylated, dihydroxylated, and monohydroxylated free BAs, respectively.
Analysis of 7α-hydroxy-4-cholesten-3-one levels in hepatocytes
The BA precursor, 7α-hydroxy-4-cholesten-3-one (C4), was determined in aqueous mouse hepatocyte lysates after acetonitrile precipitation/extraction (30) by a modification of an HPLC-MS/MS method (31).
Biochemical analysis
Serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) activities were measured using commercially available kits (Randox Laboratories, United Kingdom and Spinreact, Spain). Serum TG and CHOL were also measured using commercially available kits (A. Menarini Diagnostics, Spain).
Liver metabolomic analysis
Liver metabolomic analysis was performed by OWL (One Way Liver S. L.), as previously described by Barbier-Torres et al. (32). Briefly, four UPLC®/TOF-MS based platforms analyzing methanol, methanol/water, and chloroform/methanol liver extracts were combined. The methanol/water extract platform comprised the study of polar metabolites, such as vitamins, nucleosides, nucleotides, carboxylic acids, CoA derivatives, carbohydrate precursors/derivatives, and redox-electron-carriers. For this platform, a mixture of methanol/water (60:40, v/v) containing nonendogenous internal standards was added to liver tissue (50:1, v/w) and homogenized using a Precellys 24 grinder. After 1 h of incubation at −20°C, samples were centrifuged at 16,000 g for 15 min. The supernatant was collected and chloroform was added. The polar phase was then transferred to a clean tube for solvent evaporation. Dried extracts were dissolved in water and, after centrifugation, supernatants were transferred to vials for UPLC®-MS analysis. Data obtained with the UPLC®-MS were processed with the TargetLynx application manager for MassLynx (Waters Corp.), as detailed (15, 33). Intra- and inter-batch normalization followed the procedure described by van Vilsteren et al. (34). All the calculations were performed with R v2.13.0 (R Development Core Team, 2010).
Inmunoassays
For the immunoblots, samples were subjected to SDS-PAGE and proteins were transferred to Immobilon-P membranes. Western blotting was performed using different primary antibodies (supplemental Table S1).
RNA extraction, cDNA synthesis, and quantitative PCR
Total RNA was extracted using Trizol reagent (Invitrogen, Spain) and cDNAs were obtained by retrotranscription (SuperScript III RT; Invitrogen), following the manufacturers’ instructions.
cDNA was used for specific target amplification using the Qiagen Multiplex PCR Master; after 14 cycles of amplification (95°C 15 min, 14 cycles 95°C for 15 s, and 60°C for 4 min), amplified cDNA was treated with Exo I following Fluidigm protocol instructions, diluted 1:5 with low EDTA TE buffer, and loaded onto a 48.48 or 96.96 Dynamic Array integrated fluidic circuit. SsoFastTM EvaGreen® Supermix with Low ROX (Bio-Rad Laboratories) was used for amplification. The cycling program consisted of 1 min at 95°C, 35 cycles of 95°C for 5 s and 60°C for 20 s, followed by a melting curve.
The expression of the selected genes was measured by RT-quantitative (q)PCR using the BioMarkTM HD system in combination with Dynamic Array integrated fluidic circuits (Fluidigm Corporation) following Fluidigm’s Fast Gene Expression Analysis using EvaGreen on the BioMark HD system version D1 protocol. The sequences of the oligonucleotides used are detailed in the supplemental information (supplemental Table S2). Finally, the stability of candidate reference genes was analyzed with NormFinder algorithms and all the data analyses were performed using GenEx software. All reactions were performed in duplicate and expression levels were normalized to the average level of Gapdh, Ppia, Alb, and Actb in each sample.
Total protein measurements
Protein concentration was measured using commercially available bicinchoninic acid reagent (Thermo Fisher Scientific Inc., Spain).
Statistical analysis
Data are represented as mean ± SEM. Differences between groups were tested using the Student’s t-test and two-way ANOVA. Significance was defined as P < 0.05. The baseline characteristics of the patients used in this study were compared using the unpaired t-test or Mann-Whitney U test and correlation between serum OPN levels and lipids was done using the Pearson correlation test. These analyses were performed using GraphPad Prism software.
RESULTS
Serum OPN correlates with liver PC and CHOL concentration in nonobese patients with NAFL
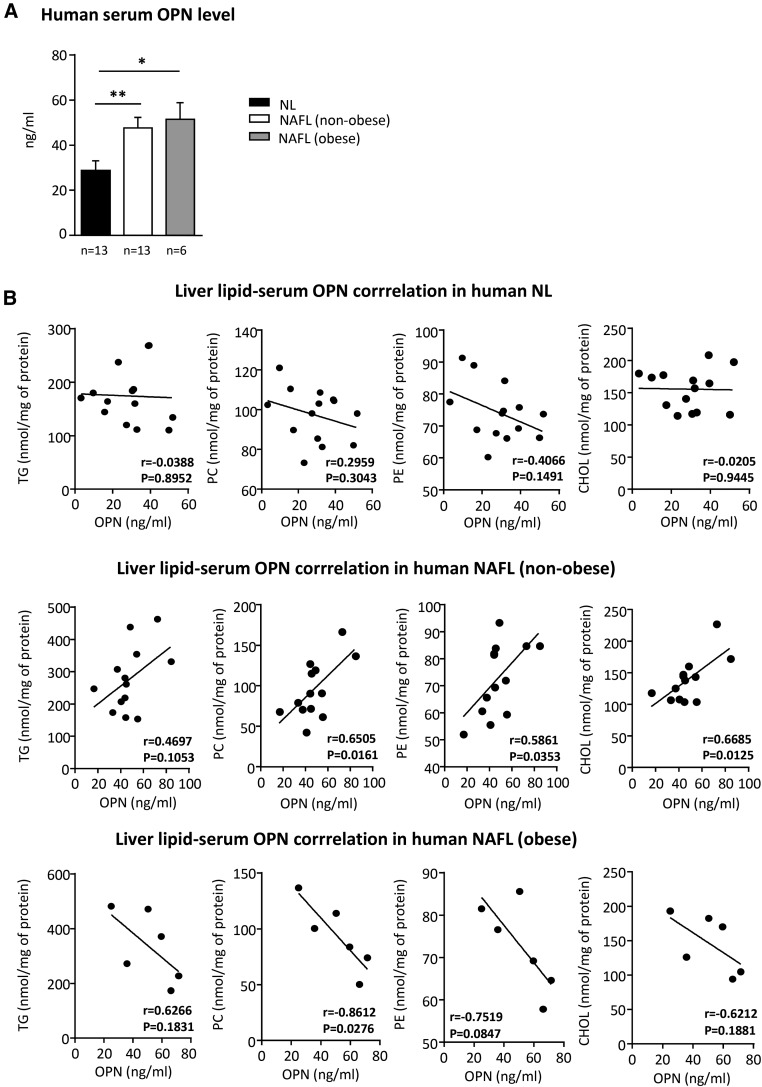
OPN is involved in different kinds of liver diseases in which metabolic deregulation is a hallmark. Thus, we wanted to know whether serum OPN could correlate with liver lipid concentration in nonobese subjects with NL and nonobese and obese patients with NAFL (Table 1). We found that in nonobese NAFL patients, in whom serum OPN FXR and LXR increased (Fig. 1A), serum OPN correlated positively with liver PC, liver PE, and liver CHOL concentration (Fig. 1B), while there was no correlation between serum OPN and liver TG concentration. No positive correlations were found in NL or obese NAFL patients, suggesting that extracellular OPN affects liver lipid metabolism differently depending on the metabolic status.
Fig. 1.
Serum OPN correlates with liver PC and CHOL content in nonobese NAFL patients. Liver samples from nonobese NL patients (n = 13), NAFL patients (n = 13), and obese NAFL patients (n = 6) were obtained by liver biopsy. A: Serum samples from NL and NAFL patients were obtained after a 12 h overnight fast and OPN levels were measured by ELISA. B: Lipids were extracted from homogenized livers and TG, PC, PE, and total CHOL were separated by TLC and quantified. Significant differences between NL and NAFL are denoted by *P < 0.05; **P < 0.01 (Student’s t-test). Correlation analysis between serum OPN and liver lipids was performed using the Pearson test.
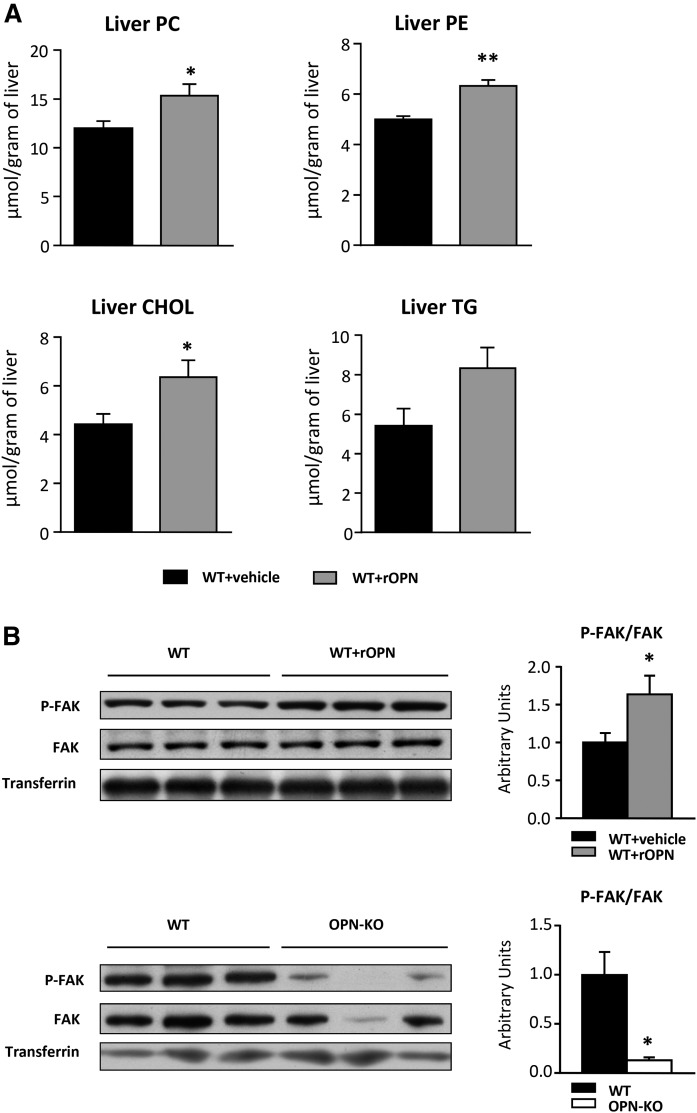
To analyze whether OPN directly induces changes in liver PC, PE, and CHOL concentration, rOPN was administered to WT mice. The results showed that liver PC, PE, and CHOL levels were increased in rOPN-treated WT mice, as compared with vehicle-treated mice (Fig. 2A). However, liver TG levels did not increase significantly (Fig. 2A), which is in concordance with the lack of correlation between serum OPN and liver TG levels in NAFL patients (Fig. 1B). To assess that rOPN was reaching functional levels in liver, activation of FAK, which activates in response to extracellular matrix-receptor interaction, was measured (Fig. 2B). Treatment with rOPN activated the FAK signaling pathway (Fig. 2B), which was inhibited in OPN-KO mice when compared with WT mice (Fig. 2B).
Fig. 2.
OPN regulates the PC and CHOL content in liver. rOPN-treated (15 μg/mouse) (WT+rOPN) or vehicle-treated WT mice (WT+vehicle), and OPN-KO mice and their control mice (WT) were used. A: Lipids were extracted from liver homogenate and PC, PE, CHOL, and TG were separated and quantified. B: Liver Tyr-397 phosphorylated FAK (P-FAK) and total FAK levels were assessed by immunoblotting using transferrin as loading control. Values are mean ± SEM of five animals per group. Significant differences between WT+vehicle and WT+rOPN are denoted by *P < 0.05 and **P < 0.01 (Student’s t-test).
Extracellular OPN directly modulates liver and hepatocyte CYP7A1 levels
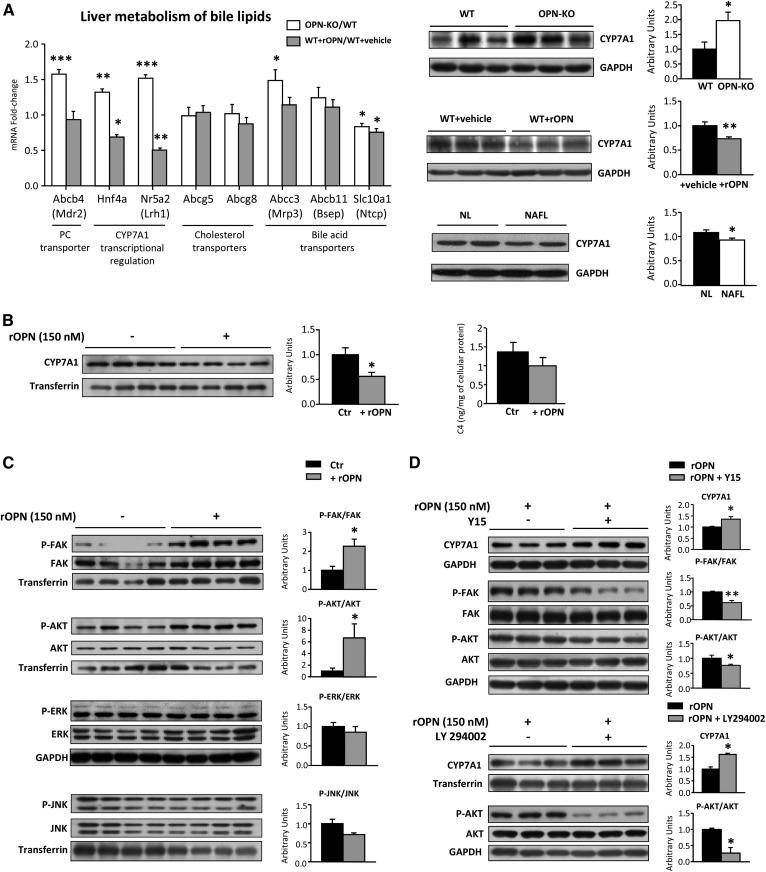
Extracellular OPN regulates concentration of liver PC and CHOL (Fig. 2A), both bile lipids. PC constitutes 90–95% of bile GPLs, so we wanted to explore whether OPN could modulate liver metabolism of bile lipids. For this, we used rOPN-treated mice, vehicle-treated mice, OPN-deficient mice that did not exhibit liver injury (Table 2), and their control mice. It has been firmly established that the multidrug resistance protein 2 (Mdr2) gene (Abcb4) is essential for biliary PC secretion in mice (35). The results showed that Abcb4 expression was upregulated in OPN-KO mice together with that of the nuclear receptors, Hnf4a and Nr5a2 (Fig. 3A), which cooperate in regulating CYP7A1 expression (36). In coherence, the administration of OPN in vivo decreased the expression of these nuclear receptors (Fig. 3A). CYP7A1 is also regulated by the activity of liver X receptor (LXR) and farnesoid X receptor (FXR). The expression of Abcg5 and Abcg8 (Fig. 3A), targets of LXR, and the expression of bile salt export pump (Bsep) (Abcb11) (Fig. 3A) and SHP (data not shown), targets of FXR, remained unaltered, suggesting no changes in FXR and LXR activity.
TABLE 2.
Liver and serum parameters of OPN-KO mice and their WT littermates and WT injected with rOPN and the corresponding control mice injected with the vehicle
| WT | OPN-KO | WT+Vehicle | WT+rOPN | |
| ALT (IU/l) | 17.3 ± 0.87 | 14.12 ± 0.61a | 12.15 ± 0.4 | 13.71 ± 1.1b |
| AST (IU/l) | 33.6 ± 7.4 | 28.6 ± 2.6 | 30.7 ± 0.33 | 35.3 ± 0.84 |
| Milligrams of protein per gram liver | 186.2 ± 8.3 | 231.5 ± 9.5a | 223.3 ± 24.9 | 175.4 ± 8.2a |
| Liver weight/body weight (%) | 3.91 ± 0.06 | 3.98 ± 0.09 | 4.06 ± 0.11 | 4.45 ± 0.15a |
| Serum TG (mmol/l) | 0.89 ± 0.14 | 0.88 ± 0.3 | 0.52 ± 0.14 | 0.62 ± 0.25 |
| Serum CHOL (mmol/l) | 2.40 ± 0.15 | 2.54 ± 0.18 | 1.59 ± 0.16 | 1.51 ± 0.12 |
Values are mean ± SEM from 5 to 26 animals per group.
P < 0.01 significant difference between OPN-KO and WT mice or between WT+rOPN and WT+vehicle.
P < 0.05 significant difference between OPN-KO and WT mice or between WT+rOPN and WT+vehicle.
Fig. 3.
OPN regulates BA metabolism. OPN-KO mice (n = 8), their control mice (WT) (n = 8), rOPN-treated mice (15 μg/mouse) (WT+rOPN) (n = 5), or vehicle-treated WT (WT+vehicle) (n = 5) mice were used. A: Hepatic mRNA of genes involved in liver metabolism of bile lipids were measured and represented as fold-change of OPN-KO/WT and WT+rOPN/WT+vehicle. Liver CYP7A1 content was assessed by immunoblotting in mice and in nonobese patients with NL and NAFL. GAPDH was used as loading control. B: Primary hepatocytes from WT mice were incubated with rOPN (150 nM) during 3 h. CYP7A1 levels in hepatocytes were assessed and transferrin was used as loading control. Analyses of the C4 levels in hepatocytes were assessed. C: Primary hepatocytes from WT mice were incubated with rOPN (150 nM) during 3 h and the FAK-AKT signaling pathway was analyzed. Transferrin was used as a loading control. D: Primary hepatocytes from WT mice were incubated with rOPN (150 nM) during 3 h in the presence or absence of the FAK inhibitors, Y15 (30 μM) or LY294002 (50 μM). Values are mean ± SEM. Significant differences between WT and OPN-KO, WT+vehicle and WT+rOPN, control (Ctr) and rOPN-treated hepatocytes, rOPN and rOPN+LY294002, and NL and NAFL patients are denoted by *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t-test).
The levels of CYP7A1 protein, in charge of the conversion of CHOL into BA, were higher in OPN-KO mice than in WT mice, but were decreased when WT mice were treated with rOPN (Fig. 3A). Liver levels of CYP7A1 were slightly decreased in nonobese patients with NAFL (Fig. 3A), in whom serum OPN correlated with liver CHOL, suggesting that the OPN-induced decrease in CYP7A1 could be, at least in part, in charge of the CHOL accumulation. In OPN-deficient mice, the increased BA synthesis was not linked with increased liver BA content, as compared with WT mice (Table 3). In fact, the content of some glycoconjugated and free BA in liver was decreased (Table 3), suggesting increased secretion of BA and/or decreased uptake. Concerning BA transporters, deficiency of OPN led to increased gene expression of Abcc3 and decreased expression of Slc10a1, while expression of Abcb11 was unaltered (Fig. 3A). rOPN did not affect the expression of these transporters (Fig. 3A).
TABLE 3.
Liver BAs in OPN-KO and WT mice
| Species | WT | OPN-KO | |
| BA | TOTAL | 396.57 ± 58.96 | 304.99 ± 31.64 |
| Tauroconjugated BA | TOTAL | 338.58 ± 55.43 | 266.35 ± 29.40 |
| TαMCA | 12.80 ± 3.02 | 12.36 ± 1.41 | |
| TβMCA | 94.98 ± 19.6 | 76.96 ± 10.77 | |
| TUDCA | 6.86 ± 1.29 | 6.59 ± 0.82 | |
| THCA | 0.06 ± 0.04 | 0.07 ± 0.02 | |
| THDCA | 2.44 ± 0.51 | 2.61 ± 0.36 | |
| TCA | 201.2 ± 30.45 | 146.35 ± 15.29 | |
| TCDCA | 8.13 ± 1.43 | 6.90 ± 1.02 | |
| TDCA | 11.83 ± 1.26 | 14.23 ± 1.60 | |
| TLCA | 0.28 ± 0.04 | 0.29 ± 0.05 | |
| Glycoconjugated BA | TOTAL | 0.54 ± 0.09 | 0.26 ± 0.05a |
| GUDCA | 0.01 ± 0.002 | 0.003 ± 0.001a | |
| GCA | 0.52 ± 0.09 | 0.24 ± 0.04a | |
| GCDCA | 0.003 ± 0.002 | 0.002 ± 0.001 | |
| GDCA | 0.01 ± 0.01 | 0.007 ± 0.003 | |
| Free BA | TOTAL | 57.4 ± 8.70 | 38.4 ± 6.21 |
| UDCA | 9.92 ± 1.51 | 6.57 ± 0.97 | |
| αMCA | 13.32 ± 2.24 | 8.10 ± 1.16a | |
| βMCA | 19.80 ± 2.80 | 10.97 ± 1.77a | |
| CA | 12.91 ± 2.95 | 11.54 ± 2.86 | |
| CDCA | 0.57 ± 0.1 | 0.47 ± 0.08 | |
| DCA | 0.18 ± 0.04 | 0.17 ± 0.02 | |
| HDCA | 0.70 ± 0.05 | 0.52 ± 0.07a | |
| LCA | 0.04 ± 0.01 | 0.04 ± 0.01 | |
| Secondary BA | TOTAL | 13.04 ± 1.3 | 15.24 ± 1.6 |
Values are mean ± SEM from eight animals per group and are expressed in nanomoles per gram of liver. CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; GCA, glycocholic acid; GCDCA, glycochenodeoxycholic acid; GDCA, glycodeoxycholic acid; GUDCA, glycoursodeoxycholic acid; HDCA, hyodeoxycholic acid; LCA, lithocholic acid; TCA, taurocholic acid; TCDCA, taurochenodeoxycholic acid; TDCA, taurodeoxycholic acid; THCA, taurohyocholic acid; THDCA, taurohyodeoxycholic acid; TLCA, taurolithocholic acid; TUDCA, tauroursodeoxycholic acid; TαMCA, tauro-α-muricholic acid; TβMCA, tauro-β-muricholic acid; UDCA, ursodeoxycholic acid; αMCA, α-muricholic acid; βMCA, β-muricholic acid.
P < 0.05, significant difference between OPN-KO and WT mice.
To elucidate whether extracellular OPN could directly modulate CYP7A1 levels in hepatocytes and to identify the mechanism involved, hepatocytes were treated with rOPN for 3 h. rOPN decreased CYP7A1 by nearly a 50% (Fig. 3B), and there was a trend toward decreased levels of C4, whose levels, even in serum, are considered an indirect biomarker of CYP7A1 activity (37) (Fig. 3B). To elucidate the mechanism involved in CYP7A1 downregulation and taking into account that several reports have involved AKT (38) and/or ERK and JNK (39, 40) in CYP7A1 transcription, activation of these pathways was investigated. The results showed that the FAK-AKT signaling pathway was induced after treatment with rOPN, whereas rOPN treatment did not affect phosphorylation of ERK or JNK (Fig. 3C). Inhibition of FAK-AKT signaling increased CYP7A1 in rOPN-treated hepatocytes; in addition, inhibition of the PI3K pathway, which is downstream of FAK, also increased CYP7A1 levels demonstrating a mechanism by which rOPN decreases CYP7A1 levels (Fig. 3D).
Deficiency in OPN results in increased cholesterogenesis and decreased de novo lipogenesis in hepatocytes
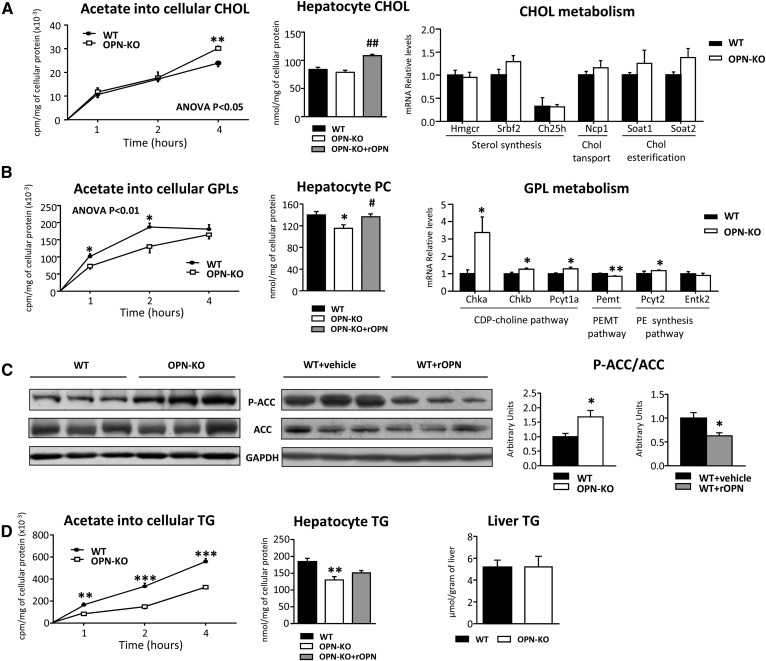
It has been previously suggested that newly synthesized CHOL is the preferred substrate for CYP7A1 (41), so we wondered whether changes in CYP7A1 levels in hepatocytes could be linked to altered de novo cholesterogenesis. For this, we used hepatocytes from OPN-KO mice, in which there was a chronic increase in CYP7A1 levels. The results showed that de novo CHOL synthesis was higher in the hepatocytes of OPN-KO mice than in the hepatocytes of WT mice, while the CHOL content in hepatocytes (Fig. 4A) or in liver (data not shown) maintained unaltered. However, treatment with rOPN, which induced CYP7A1 downregulation in liver (Fig. 3A) and hepatocytes (Fig. 3B), increased the CHOL content in the hepatocytes of OPN-KO mice (Fig. 4A). The analysis of the liver mRNA levels of the different genes involved in CHOL metabolism could not explain the increase in the cholesterogenesis observed in the OPN-KO mouse hepatocytes when compared with those of WT mice, reinforcing the involvement of CYP7A1 in the altered process.
Fig. 4.
OPN modulates de novo lipogenesis. OPN-KO mice (n = 8) and their control mice (WT) (n = 8), and rOPN-treated (15 μg/mouse) (WT+rOPN) (n = 5) or vehicle-treated WT (WT+vehicle) (n = 5) mice were used. A: Hepatocytes were isolated from the two strains and OPN-KO hepatocytes were treated with rOPN (32 nM; OPN-KO+rOPN). Lipids from primary hepatocytes were extracted and CHOL was separated and quantified. Hepatocytes isolated from both of the strains were incubated in medium supplemented with [3H]acetate for the indicated times. Lipids were extracted and the radioactivity incorporated into CHOL was assessed by a scintillation counter. Hepatic mRNA of genes involved in CHOL GPL metabolism was measured by RT-qPCR. B: PC content and de novo synthesis of GPLs in hepatocytes were assessed as described for CHOL. Hepatic mRNA of genes involved in GPL metabolism was measured by RT-qPCR. C: Liver Ser-65 phosphorylated ACC/total ACC (P-ACC/ACC) was assessed by immunoblotting using GAPDH as loading control. D: De novo TG synthesis in hepatocytes was assessed as described for CHOL. Lipids were extracted from hepatocytes and liver homogenates, lipids were separated and quantified. Values are mean ± SEM. Significant differences between OPN-KO and WT mice and between WT+vehicle and WT+rOPN mice are denoted by *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t-test) and differences between OPN-KO and OPN-KO+rOPN are denoted by #P < 0.05 and ##P < 0.01 (Student’s t-test). Statistical differences between OPN-KO and WT mice are also denoted by two-way ANOVA test.
Taking into account that the other bile lipid, PC, was increased in rOPN-treated mice (Fig. 2A) and that one of the metabolic pathways that controls liver PC concentration is de novo lipogenesis, de novo GPL synthesis and PC content were measured in OPN-KO hepatocytes. The incorporation of [3H]acetate into GPLs was lower in OPN-deficient hepatocytes than in WT hepatocytes (Fig. 4B). Also, PC content was decreased in OPN-deficient hepatocytes (Fig. 4B) and liver (Fig. 5D) as compared with WT mice. Treatment with rOPN restored PC levels in OPN-KO hepatocytes (Fig. 4B), which shows that extracellular OPN controls PC metabolism in liver and hepatocytes. Surprisingly, mRNA levels of genes involved in the CDP-choline pathway, the PE N-methyltransferase pathway, and the PE synthesis pathway were higher in the liver of OPN-KO mice than in WT mice (Fig. 4B). We observed that the decreased de novo GPL synthesis could be attributed, at least in part, to decreased acetyl-CoA carboxylase (ACC) activity, as demonstrated by the increased phosphorylation at Ser-79 of liver ACC (Fig. 4C), the rate-limiting enzyme in de novo synthesis of FAs. Conversely, rOPN-treated mice, in which PC content was increased, exhibited a decrease in liver ACC phosphorylation as compared with the vehicle-treated mice (Fig. 4C). The decreased incorporation of [3H]acetate into TG in OPN-KO hepatocytes (Fig. 4D) reinforced the idea of the lower lipogenesis in OPN-KO hepatocytes than in the WT hepatocytes. Even though this induces a decrease in hepatocyte TG content, liver TG content in OPN-KO liver was maintained unaltered and the rOPN treatment did not restore the hepatocyte TG content (Fig. 4D), showing that extracellular OPN does not modulate TG content in liver.
Fig. 5.
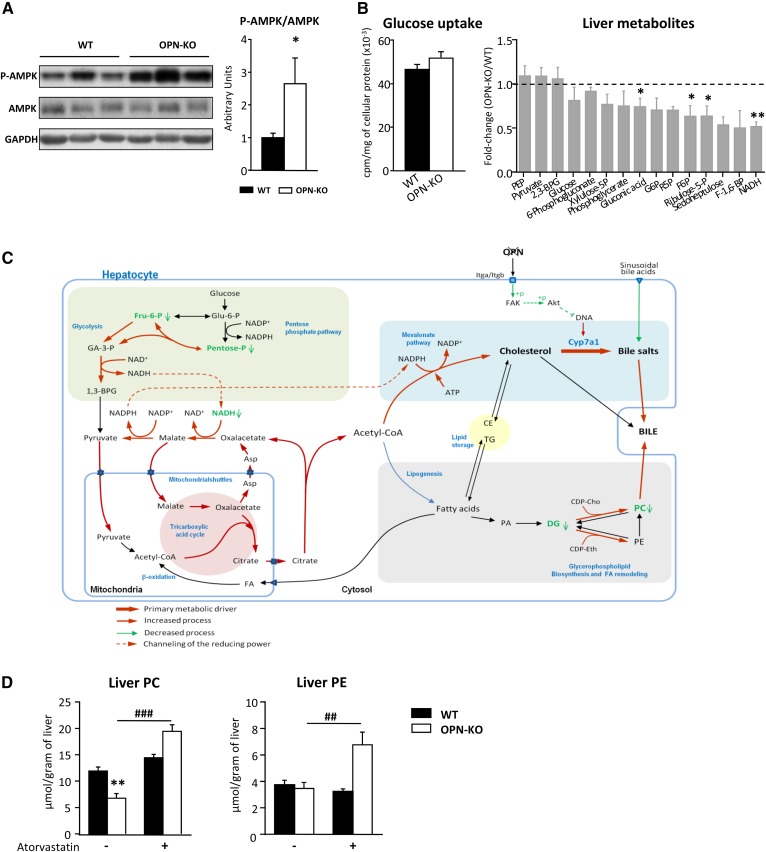
Atorvastatin treatment recovers the altered liver PC content in OPN-KO mice. WT (n = 8), OPN-KO (n = 8), and atorvastatin-treated (100 mg/kg) WT (n = 5) and OPN-KO (n = 5) mice were used. A: Thr-172 phosphorylated AMPK (P-AMPK) and total AMPK of liver extracts were assessed by immunoblotting using GAPDH as loading control. B: Primary hepatocytes were incubated with D-[3H]glucose for 1 min to measure the glucose uptake by scintillation counting. Liver metabolomic analysis was performed. C: Model of metabolic dysregulation in OPN-KO mouse liver. D: Lipids were extracted from liver homogenate and PC and PE were separated and quantified. Values are mean ± SEM. Significant differences between WT and OPN-KO mice are denoted by *P < 0.05, **P < 0.01 (Student’s t-test); differences due to the atorvastatin treatment are denoted by ##P < 0.01, ###P < 0.001 (Student’s t-test). Asp, aspartate; CDP-Cho, CDP-choline; CDP-Eth, CDP-ethanolamine; CE, cholesteryl ester; DG, diacylglyceride; Fru-6-P, fructose-6-phosphate; GA-3-P, glyceraldehyde-3-phosphate; Glu-6-P, glucose-6-phosphate; PA, phosphatidic acid.
Inhibition of cholesterogenesis recovers liver PC concentration in OPN-KO mice
CHOL de novo synthesis exhibits high demands of energy. Phosphorylation of the AMP-activated protein kinase (AMPK) catalytic subunit at Thr-172, required for its activation and phosphorylation of ACC (42), was higher in the OPN-KO mice than in the WT mice (Fig. 5A), suggesting a lower liver energy status. AMPK phosphorylation maintained unaltered in rOPN-treated mice (data not shown).
Taking into account the high demand of energy when de novo cholesterogenesis is increased, although glucose uptake maintained unchanged in hepatocytes of OPN-KO mice as compared with those from WT mice (Fig. 5B), the relative amount of NADH and some pentose phosphate pathway and glycolysis intermediates were lower in OPN-KO mouse liver than in WT mouse liver (Fig. 5B; Table 4), suggesting the increased flux of those metabolic pathways to provide the required acetyl-CoA.
TABLE 4.
Metabolomic analysis of livers from OPN-KO and WT mice
| Individual Notation | Common Name | P | Fold Change |
| D-gluconic acid | 3.85E-02 | 0.74 | |
| Fructose 6-phosphate | F6P | 4.30E-02 | 0.63 |
| D-ribose 5-phosphate | R5P | 2.84E-02 | 0.69 |
| Nicotinamide adenine dinucleotide reduced | NADH | 1.19E-03 | 0.39 |
| Theobromine | Methyl purines | 2.90E-03 | 9.58 |
Metabolomic analysis of livers from OPN-KO mice was performed and compared with that of their corresponding WT animals. “Individual notation” refers to the confirmed identification of the metabolites. Fold-changes and unpaired Student’s t-test Pvalues (or Welch’s t-test where unequal variances were found) were calculated for each comparison considering five animals per group.
Taken all together, we propose a model to explain how the OPN deficiency will lead to a whole metabolic dysregulation in hepatocytes (Fig. 5C). Lack of OPN will induce, through inhibition of FAK-AKT signaling, the upregulation of CYP7A1 protein levels and, thus, the conversion of CHOL into BA; OPN deficiency will promote the synthesis and secretion of BA. In such a condition, to respond to the high energy demands required for the CHOL synthesis, the pentose phosphate pathway, the tricarboxylic cycle, and the malate-aspartate shuttle will be more active than in the WT mice. The acetyl-CoA will preferentially be channeled toward the CHOL synthesis rather than to the de novo synthesis of FAs. Also, ACC will be inhibited because of the low energy status induced with the increased cholesterogenesis. The increased bile secretion will also promote PC secretion, which will contribute to the lower liver concentration in OPN-KO mice than in WT mice (Fig. 5C).
Finally, to demonstrate that OPN regulates the cross-talk between PC and CHOL metabolism, de novo CHOL synthesis was inhibited in vivo by providing intragastric atorvastatin to OPN-KO mice and PC and PE content was measured in liver. As expected, treatment with atorvastatin increased liver PC and PE concentration only in OPN-KO mice. It did not affect WT mice (Fig. 5D).
DISCUSSION
OPN, a multifunctional cytokine, expresses during inflammation and cancer in different tissues (43, 44). It plays a role in liver fibrogenesis (7) and in obesity-related hepatosteatosis (1). In mice, OPN neutralization diminishes liver fibrosis (8) and the inflammation induced in adipose tissue by a high fat diet (45). OPN-deficient mice are resistant to obesity-induced hepatosteatosis (4). Until now, there has been no information about the specific role of OPN in regulating liver lipid metabolism, when both increased OPN and disrupted lipid metabolism are components of liver disease progression. The bulk of studies have focused on the role of OPN in regulating immune cell function and activation of hepatic stellate cells. In this work, we provide evidence of a direct role of OPN in regulating lipid metabolism and conversion of CHOL into BA in hepatocytes and liver.
Most of the studies performed in animal models have been carried out using male mice. Fortunately, nowadays there are more and more works performed in males and females. Here, we selected female mice, with higher complexity in metabolic regulation, to add information about modulation of liver lipid metabolism in females.
Taking into account that in NAFLD development there could be a dysfunction in TG, PC, and PE metabolism (15), we investigated whether OPN was needed to maintain liver glycerolipid metabolism. We observed that extracellular OPN regulates PC metabolism; OPN deficiency in liver and hepatocytes was linked to decreased PC concentration and the addition of extracellular OPN in OPN-KO mouse hepatocytes restored the normal PC concentration. Also, rOPN administration to WT mice increased liver PC concentration.
It has been previously reported that the PCs synthesized through the CDP-choline pathway are enriched in medium-chain monounsaturated FAs (46), which are more frequently found in human bladder (47). OPN deficiency led to increased mRNA levels of enzymes involved in the CDP-choline pathway, but PC content was decreased in liver, so here we propose that the decreased liver PC concentration could be linked with altered BA metabolism, which is also dysregulated in morbidly obese patients with NASH (48). The fact that OPN deficiency led to increased liver mRNA levels of Abcb4, supported the hypothesis. Also, the results here demonstrated that extracellular OPN regulates the conversion of CHOL into BA because OPN deficiency led to increased liver CYP7A1 protein levels, while the in vivo treatment of WT mice with rOPN decreased it, a feature also found in NAFLD patients in which CHOL metabolism is dysregulated (49). It has been previously demonstrated in HCV-infected hepatocytes that the secreted OPN interacts with integrins and/or CD44 at the cell surface of hepatocytes, among other cell types, thus, inducing a signaling cascade through phosphorylation/activation of FAK and AKT (50). Here, we show that OPN, through regulation of the FAK-AKT signaling pathway, modulated CYP7A1 levels in hepatocytes. It has been reported that FoxO1, which is downstream of AKT, inhibited human reporter activity by blocking HNF4α transactivation activity (51). However, whether this mechanism could also regulate CYP7A1 transcription in mouse hepatocytes is something we do not know. The specific mechanism involved is still not fully understood and remains to be studied. Different combinations of integrins that bind to OPN are expressed by hepatocytes (52, 53). Among others, one of them is α9B1 (52). So, we believe that the OPN-integrin interaction will lead to the activation of FAK-AKT signaling in hepatocytes.
It has been previously suggested that newly synthesized CHOL is the preferred substrate for CYP7A1 (41); here, we show that in OPN-KO hepatocytes, cholesterogenesis was higher than in the WT hepatocytes and that in OPN-KO mouse liver, CYP7A1 levels were higher than in the WT levels, which supports this idea. De novo CHOL synthesis requires high energy demands; in OPN-KO mouse liver, relative amounts of NADH and some pentose phosphate pathway and glycolysis intermediates were lower than in WT mice, suggesting increased flux of those metabolic pathways to provide the required acetyl-CoA. As a counteraction, phosphorylation of the main metabolic sensor, AMPK, will be higher in OPN-KO mouse liver than in the WT mouse liver, which, together with the channeling of acetyl-CoA toward the CHOL synthesis, will contribute to the decreased FA and PC de novo synthesis. The fact that phosphorylation of AMPK maintained unaltered in rOPN-treated mouse liver, while ACC phosphorylation was decreased, supports the idea that other mechanisms are also involved in the regulation of ACC activity. Inhibition of cholesterogenesis in vivo with atorvastatin in OPN-KO mice restored liver PC concentration, which supports this idea of OPN-driven PC and CHOL metabolism interplay.
Here, we also show that serum OPN correlated positively with liver PC, PE, and CHOL concentration in matched samples of nonobese NAFL patients in which CYP7A1 protein levels were decreased, suggesting that accumulation of CHOL, mainly driven as a consequence of altered conversion into BA and secretion of bile, will also be related with accumulation of PC. It has been reported that CHOL is increased in NASH (54, 55). CHOL storage has also been associated with NAFLD progression and activation of fibrogenesis (19). Here, we demonstrated that OPN directly decreased CYP7A1 protein levels in hepatocytes, which suggests that this could lead to increased CHOL storage and NAFLD progression. There was no positive correlation between serum OPN and liver PC, PE, and CHOL concentration in NL patients or obese NAFL patients, suggesting that extracellular OPN will modulate liver lipid metabolism differently depending on the metabolic status.
In conclusion, extracellular OPN regulates the cross-talk among PC and CHOL metabolism and will drive the acetyl-CoA metabolic fate in liver. The results here suggest that modulation of CYP7A1 will be a main axis in this metabolic dysregulation and that, in the early stage of NAFL in nonobese patients, increased serum OPN could be involved in liver PC and CHOL metabolic disruption, which contributes to NAFLD progression.
Supplementary Material
Acknowledgments
The authors thank Jose Antonio López Gómez and Unidad de Formación e Investigación UFI11/20, University of the Basque Country UPV/EHU for technical support.
Footnotes
Abbreviations:
- ACC
- acetyl-CoA carboxylase
- ALT
- alanine aminotransferase
- AMPK
- AMP-activated protein kinase
- AST
- aspartate aminotransferase
- BA
- bile acid
- C4
- 7α-hydroxy-4-cholesten-3-one
- CHOL
- cholesterol
- CYP7A1
- cholesterol 7α-hydroxylase
- FAK
- focal adhesion kinase
- FXR
- farnesoid X receptor
- GPL
- glycerophospholipid
- LXR
- liver X receptor alpha
- NAFL
- nonalcoholic fatty liver
- NAFLD
- nonalcoholic fatty liver disease
- NASH
- nonalcoholic steatohepatitis
- NL
- normal liver
- OPN
- osteopontin
- OPN-KO
- osteopontin-deficient
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- rOPN
- recombinant osteopontin
- TG
- triacylglyceride
This work was supported by Basque Government Grant IT-336-10, Ministerio de Economía y Competitividad Grants SAF2013-40620-R and SAF2015-64352-R, Consejería de Educación, Junta de Castilla y León Grants SA015U13 and BIO/SA65/13, Fundación Samuel Solórzano Barruso Grants FS/7-2013 and FS/10-2014, and Instituto de Salud Carlos III Grant PI13/01299. Additional support was provided by the University of the Basque Country UPV/EHU in the form of a predoctoral fellowship to M.N-G. and a postdoctoral fellowship to J.L.G-R. The authors declare no competing financial, personal, or professional interests.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Bertola A., Deveaux V., Bonnafous S., Rousseau D., Anty R., Wakkach A., Dahman M., Tordjman J., Clement K., McQuaid S. E., et al. 2009. Elevated expression of osteopontin may be related to adipose tissue macrophage accumulation and liver steatosis in morbid obesity. Diabetes. 58: 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang K. X., and Denhardt D. T.. 2008. Osteopontin: role in immune regulation and stress responses. Cytokine Growth Factor Rev. 19: 333–345. [DOI] [PubMed] [Google Scholar]
- 3.Junaid A., Moon M. C., Harding G. E., and Zahradka P.. 2007. Osteopontin localizes to the nucleus of 293 cells and associates with polo-like kinase-1. Am. J. Physiol. Cell Physiol. 292: C919–C926. [DOI] [PubMed] [Google Scholar]
- 4.Kiefer F. W., Neschen S., Pfau B., Legerer B., Neuhofer A., Kahle M., Hrabe de Angelis M., Schlederer M., Mair M., Kenner L., et al. 2011. Osteopontin deficiency protects against obesity-induced hepatic steatosis and attenuates glucose production in mice. Diabetologia. 54: 2132–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahai A., Malladi P., Melin-Aldana H., Green R. M., and Whitington P. F.. 2004. Upregulation of osteopontin expression is involved in the development of nonalcoholic steatohepatitis in a dietary murine model. Am. J. Physiol. Gastrointest. Liver Physiol. 287: G264–G273. [DOI] [PubMed] [Google Scholar]
- 6.Syn W. K., Choi S. S., Liaskou E., Karaca G. F., Agboola K. M., Oo Y. H., Mi Z., Pereira T. A., Zdanowicz M., Malladi P., et al. 2011. Osteopontin is induced by hedgehog pathway activation and promotes fibrosis progression in nonalcoholic steatohepatitis. Hepatology. 53: 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Syn W. K., Agboola K. M., Swiderska M., Michelotti G. A., Liaskou E., Pang H., Xie G., Philips G., Chan I. S., Karaca G. F., et al. 2012. NKT-associated hedgehog and osteopontin drive fibrogenesis in non-alcoholic fatty liver disease. Gut. 61: 1323–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coombes J. D., Swiderska-Syn M., Dolle L., Reid D., Eksteen B., Claridge L., Briones-Orta M. A., Shetty S., Oo Y. H., Riva A., et al. 2015. Osteopontin neutralisation abrogates the liver progenitor cell response and fibrogenesis in mice. Gut. 64: 1120–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gotoh M., Sakamoto M., Kanetaka K., Chuuma M., and Hirohashi S.. 2002. Overexpression of osteopontin in hepatocellular carcinoma. Pathol. Int. 52: 19–24. [DOI] [PubMed] [Google Scholar]
- 10.Shang S., Plymoth A., Ge S., Feng Z., Rosen H. R., Sangrajrang S., Hainaut P., Marrero J. A., and Beretta L.. 2012. Identification of osteopontin as a novel marker for early hepatocellular carcinoma. Hepatology. 55: 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shevde L. A., and Samant R. S.. 2014. Role of osteopontin in the pathophysiology of cancer. Matrix Biol. 37: 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook A. C., Tuck A. B., McCarthy S., Turner J. G., Irby R. B., Bloom G. C., Yeatman T. J., and Chambers A. F.. 2005. Osteopontin induces multiple changes in gene expression that reflect the six “hallmarks of cancer” in a model of breast cancer progression. Mol. Carcinog. 43: 225–236. [DOI] [PubMed] [Google Scholar]
- 13.Wu J., Pungaliya P., Kraynov E., and Bates B.. 2012. Identification and quantification of osteopontin splice variants in the plasma of lung cancer patients using immunoaffinity capture and targeted mass spectrometry. Biomarkers. 17: 125–133. [DOI] [PubMed] [Google Scholar]
- 14.Fabbrini E., Sullivan S., and Klein S.. 2010. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology. 51: 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez-Uña M., Varela-Rey M., Cano A., Fernandez-Ares L., Beraza N., Aurrekoetxea I., Martinez-Arranz I., Garcia-Rodriguez J. L., Buque X., Mestre D., et al. 2013. Excess S-adenosylmethionine reroutes phosphatidylethanolamine towards phosphatidylcholine and triglyceride synthesis. Hepatology. 58: 1296–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daly P. F., Lyon R. C., Faustino P. J., and Cohen J. S.. 1987. Phospholipid metabolism in cancer cells monitored by 31P NMR spectroscopy. J. Biol. Chem. 262: 14875–14878. [PubMed] [Google Scholar]
- 17.Glunde K., Ackerstaff E., Mori N., Jacobs M. A., and Bhujwalla Z. M.. 2006. Choline phospholipid metabolism in cancer: consequences for molecular pharmaceutical interventions. Mol. Pharm. 3: 496–506. [DOI] [PubMed] [Google Scholar]
- 18.Ridgway N. D. 2013. The role of phosphatidylcholine and choline metabolites to cell proliferation and survival. Crit. Rev. Biochem. Mol. Biol. 48: 20–38. [DOI] [PubMed] [Google Scholar]
- 19.Caballero F., Fernandez A., De Lacy A. M., Fernandez-Checa J. C., Caballeria J., and Garcia-Ruiz C.. 2009. Enhanced free cholesterol, SREBP-2 and StAR expression in human NASH. J. Hepatol. 50: 789–796. [DOI] [PubMed] [Google Scholar]
- 20.Dasarathy S., Yang Y., McCullough A. J., Marczewski S., Bennett C., and Kalhan S. C.. 2011. Elevated hepatic fatty acid oxidation, high plasma fibroblast growth factor 21, and fasting bile acids in nonalcoholic steatohepatitis. Eur. J. Gastroenterol. Hepatol. 23: 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai Y. S., Chen W. C., Kuo T. C., Ho C. T., Kuo C. H., Tseng Y. J., Lu K. H., Lin S. H., Panyod S., and Sheen L. Y.. 2015. Mass-spectrometry-based serum metabolomics of a C57BL/6J mouse model of high-fat-diet-induced non-alcoholic fatty liver disease development. J. Agric. Food Chem. 63: 7873–7884. [DOI] [PubMed] [Google Scholar]
- 22.Brunt E. M., Kleiner D. E., Wilson L. A., Belt P., and Neuschwander-Tetri B. A.; NASH Clinical Research Network (CRN). 2011. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology. 53: 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 24.Ruiz J. I., and Ochoa B.. 1997. Quantification in the subnanomolar range of phospholipids and neutral lipids by monodimensional thin-layer chromatography and image analysis. J. Lipid Res. 38: 1482–1489. [PubMed] [Google Scholar]
- 25.Smedsrød B., and Pertoft H.. 1985. Preparation of pure hepatocytes and reticuloendothelial cells in high yield from a single rat liver by means of Percoll centrifugation and selective adherence. J. Leukoc. Biol. 38: 213–230. [DOI] [PubMed] [Google Scholar]
- 26.Folch J., Lees M., and Sloane Stanley G. H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 27.Baker R., Dowdall M. J., and Whittaker V. P.. 1976. The incorporation of radioactive fatty acids and of radioactive derivatives of glucose into the phospholipids of subsynaptosomal fractions of cerebral cortex. Biochem. J. 154: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nytofte N. S., Serrano M. A., Monte M. J., Gonzalez-Sanchez E., Tumer Z., Ladefoged K., Briz O., and Marin J. J.. 2011. A homozygous nonsense mutation (c.214C→A) in the biliverdin reductase alpha gene (BLVRA) results in accumulation of biliverdin during episodes of cholestasis. J. Med. Genet. 48: 219–225. [DOI] [PubMed] [Google Scholar]
- 29.Ye L., Liu S., Wang M., Shao Y., and Ding M.. 2007. High-performance liquid chromatography-tandem mass spectrometry for the analysis of bile acid profiles in serum of women with intrahepatic cholestasis of pregnancy. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 860: 10–17. [DOI] [PubMed] [Google Scholar]
- 30.Leníček M., Vecka M., Zizalova K., and Vitek L.. 2016. Comparison of simple extraction procedures in liquid chromatography-mass spectrometry based determination of serum 7alpha-hydroxy-4-cholesten-3-one, a surrogate marker of bile acid synthesis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1033–1034: 317–320. [DOI] [PubMed] [Google Scholar]
- 31.Steiner C., von Eckardstein A., and Rentsch K. M.. 2010. Quantification of the 15 major human bile acids and their precursor 7alpha-hydroxy-4-cholesten-3-one in serum by liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 878: 2870–2880. [DOI] [PubMed] [Google Scholar]
- 32.Barbier-Torres L., Delgado T. C., Garcia-Rodriguez J. L., Zubiete-Franco I., Fernandez-Ramos D., Buque X., Cano A., Gutierrez-de Juan V., Fernandez-Dominguez I., Lopitz-Otsoa F., et al. 2015. Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget. 6: 2509–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barr J., Caballeria J., Martinez-Arranz I., Dominguez-Diez A., Alonso C., Muntane J., Perez-Cormenzana M., Garcia-Monzon C., Mayo R., Martin-Duce A., et al. 2012. Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J. Proteome Res. 11: 2521–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Vilsteren F. G., Baskin-Bey E. S., Nagorney D. M., Sanderson S. O., Kremers W. K., Rosen C. B., Gores G. J., and Hobday T. J.. 2006. Liver transplantation for gastroenteropancreatic neuroendocrine cancers: defining selection criteria to improve survival. Liver Transpl. 12: 448–456. [DOI] [PubMed] [Google Scholar]
- 35.Smit J. J., Schinkel A. H., Oude Elferink R. P., Groen A. K., Wagenaar E., van Deemter L., Mol C. A., Ottenhoff R., van der Lugt N. M., and van Roon M. A.. 1993. Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell. 75: 451–462. [DOI] [PubMed] [Google Scholar]
- 36.Kir S., Zhang Y., Gerard R. D., Kliewer S. A., and Mangelsdorf D. J.. 2012. Nuclear receptors HNF4alpha and LRH-1 cooperate in regulating Cyp7a1 in vivo. J. Biol. Chem. 287: 41334–41341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gälman C., Arvidsson I., Angelin B., and Rudling M.. 2003. Monitoring hepatic cholesterol 7alpha-hydroxylase activity by assay of the stable bile acid intermediate 7alpha-hydroxy-4-cholesten-3-one in peripheral blood. J. Lipid Res. 44: 859–866. [DOI] [PubMed] [Google Scholar]
- 38.Li T., Ma H., and Chiang J. Y.. 2008. TGFbeta1, TNFalpha, and insulin signaling crosstalk in regulation of the rat cholesterol 7alpha-hydroxylase gene expression. J. Lipid Res. 49: 1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T., Jahan A., and Chiang J. Y.. 2006. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 43: 1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henkel A. S., Anderson K. A., Dewey A. M., Kavesh M. H., and Green R. M.. 2011. A chronic high-cholesterol diet paradoxically suppresses hepatic CYP7A1 expression in FVB/NJ mice. J. Lipid Res. 52: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiang J. Y. 2013. Bile acid metabolism and signaling. Compr. Physiol. 3: 1191–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viollet B., Guigas B., Leclerc J., Hebrard S., Lantier L., Mounier R., Andreelli F., and Foretz M.. 2009. AMP-activated protein kinase in the regulation of hepatic energy metabolism: from physiology to therapeutic perspectives. Acta Physiol. (Oxf.). 196: 81–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramaiah S. K., and Rittling S.. 2008. Pathophysiological role of osteopontin in hepatic inflammation, toxicity, and cancer. Toxicol. Sci. 103: 4–13. [DOI] [PubMed] [Google Scholar]
- 44.Zeyda M., Gollinger K., Todoric J., Kiefer F. W., Keck M., Aszmann O., Prager G., Zlabinger G. J., Petzelbauer P., and Stulnig T. M.. 2011. Osteopontin is an activator of human adipose tissue macrophages and directly affects adipocyte function. Endocrinology. 152: 2219–2227. [DOI] [PubMed] [Google Scholar]
- 45.Kiefer F. W., Zeyda M., Gollinger K., Pfau B., Neuhofer A., Weichhart T., Saemann M. D., Geyeregger R., Schlederer M., Kenner L., et al. 2010. Neutralization of osteopontin inhibits obesity-induced inflammation and insulin resistance. Diabetes. 59: 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeLong C. J., Shen Y. J., Thomas M. J., and Cui Z.. 1999. Molecular distinction of phosphatidylcholine synthesis between the CDP-choline pathway and phosphatidylethanolamine methylation pathway. J. Biol. Chem. 274: 29683–29688. [DOI] [PubMed] [Google Scholar]
- 47.Hay D. W., Cahalane M. J., Timofeyeva N., and Carey M. C.. 1993. Molecular species of lecithins in human gallbladder bile. J. Lipid Res. 34: 759–768. [PubMed] [Google Scholar]
- 48.Bechmann L. P., Kocabayoglu P., Sowa J. P., Sydor S., Best J., Schlattjan M., Beilfuss A., Schmitt J., Hannivoort R. A., Kilicarslan A., et al. 2013. Free fatty acids repress small heterodimer partner (SHP) activation and adiponectin counteracts bile acid-induced liver injury in superobese patients with nonalcoholic steatohepatitis. Hepatology. 57: 1394–1406. [DOI] [PubMed] [Google Scholar]
- 49.Min H. K., Kapoor A., Fuchs M., Mirshahi F., Zhou H., Maher J., Kellum J., Warnick R., Contos M. J., and Sanyal A. J.. 2012. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 15: 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iqbal J., McRae S., Mai T., Banaudha K., Sarkar-Dutta M., and Waris G.. 2014. Role of hepatitis C virus induced osteopontin in epithelial to mesenchymal transition, migration and invasion of hepatocytes. PLoS One. 9: e87464. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Li T., Kong X., Owsley E., Ellis E., Strom S., and Chiang J. Y.. 2006. Insulin regulation of cholesterol 7alpha-hydroxylase expression in human hepatocytes: roles of forkhead box O1 and sterol regulatory element-binding protein 1c. J. Biol. Chem. 281: 28745–28754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Couvelard A., Bringuier A. F., Dauge M. C., Nejjari M., Darai E., Benifla J. L., Feldmann G., Henin D., and Scoazec J. Y.. 1998. Expression of integrins during liver organogenesis in humans. Hepatology. 27: 839–847. [DOI] [PubMed] [Google Scholar]
- 53.Yokosaki Y., Tanaka K., Higashikawa F., Yamashita K., and Eboshida A.. 2005. Distinct structural requirements for binding of the integrins alphavbeta6, alphavbeta3, alphavbeta5, alpha5beta1 and alpha9beta1 to osteopontin. Matrix Biol. 24: 418–427. [DOI] [PubMed] [Google Scholar]
- 54.Puri P., Baillie R. A., Wiest M. M., Mirshahi F., Choudhury J., Cheung O., Sargeant C., Contos M. J., and Sanyal A. J.. 2007. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 46: 1081–1090. [DOI] [PubMed] [Google Scholar]
- 55.Puri P., Wiest M. M., Cheung O., Mirshahi F., Sargeant C., Min H. K., Contos M. J., Sterling R. K., Fuchs M., Zhou H., et al. 2009. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 50: 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.