Abstract
Lipoprotein (a) [Lp(a)] is characterized by apolipoprotein (a) [apo(a)] covalently bound to apolipoprotein B 100. It was described in human plasma by Berg et al. in 1963 and the gene encoding apo(a) (LPA) was cloned in 1987 by Lawn and colleagues. Epidemiologic and genetic studies demonstrate that increases in Lp(a) plasma levels increase the risk of atherosclerotic cardiovascular disease. Novel Lp(a) lowering treatments highlight the need to understand the regulation of plasma levels of this atherogenic lipoprotein. Despite years of research, significant uncertainty remains about the assembly, secretion, and clearance of Lp(a). Specifically, there is ongoing controversy about where apo(a) and apoB-100 bind to form Lp(a); which apoB-100 lipoproteins bind to apo(a) to create Lp(a); whether binding of apo(a) is reversible, allowing apo(a) to bind to more than one apoB-100 lipoprotein during its lifespan in the circulation; and how Lp(a) or apo(a) leave the circulation. In this review, we highlight past and recent data from stable isotope studies of Lp(a) metabolism, highlighting the critical metabolic uncertainties that exist. We present kinetic models to describe results of published studies using stable isotopes and suggest what future studies are required to improve our understanding of Lp(a) metabolism.
Keywords: apolipoproteins, atherosclerosis, clinical trials, low density lipoprotein/metabolism, lipoproteins/kinetics, human studies, lipoprotein (a), lipoprotein/kinetics, stable isotopes
Lipoprotein (a) [Lp(a)] is characterized by apo(a) covalently bound to apoB-100. It was described in human plasma by Berg in 1963 (1, 2). The human gene encoding apo(a) (LPA), located on chromosome 6q26-27, was cloned in 1987 by Lawn and colleagues (3). Lp(a) plasma concentrations are genetically determined by the LPA gene (4), with 30–70% (5) of the variability of plasma levels due to differences in the number of repeats in the DNA sequence encoding kringle IV type 2 (KIV-2). This results in a spectrum of circulating apo(a) protein isoforms from 1 to >40 KIV-2 repeats (3, 6–9) and a corresponding range of molecular masses that has been estimated to range from 300 to 800 kDa. There is an inverse relationship between the size of the apo(a) isoforms and Lp(a) plasma concentration. Additionally, sequence variants in LPA play a significant role in determining plasma Lp(a) levels (4). Large epidemiologic studies show a strong link between increases in Lp(a) plasma levels and risk of atherosclerotic cardiovascular disease (9–12), and Lp(a) is present in human atheroma (13, 14). More recently, genetic studies, including Mendelian randomization approaches, have provided convincing evidence that LPA is associated causally with coronary heart disease and the development of aortic stenosis (15–19).
Despite all that is known about the LPA gene and the structure of circulating Lp(a) (4), there is much uncertainty regarding the site(s) of the assembly and production of Lp(a), the stability of the bond between apo(a) and apoB in plasma, and the site(s) of Lp(a) clearance and degradation. Specifically, there is ongoing controversy about where apo(a) and apoB-100 bind to form Lp(a); which class of apoB-100 lipoproteins bind to apo(a) to create Lp(a); whether the binding of apo(a) is reversible, allowing apo(a) to bind to more than one apoB-100 lipoprotein during its lifespan in the circulation; and whether Lp(a) or apo(a) leave the circulation via the LDL receptor pathway (20–22) or through other potential pathways (22, 23).
The difficulties inherent in understanding Lp(a) metabolism have been previously reviewed by various investigators including Dieplinger (24) in 1999 and more recently by Hoover-Plow and Huang (25) and Lamon-Fava, Diffenderfer, and Marcovina (23). Importantly, comparison of available in vivo metabolic studies is confounded by the different approaches to isolating Lp(a), different labeling techniques, and different dietary and sampling protocols that have been used. Due to the difficulty in studying Lp(a) metabolism, each original research report described in the present review also includes an in-depth examination of the relevant literature available to the investigators at the time of their study. This review will focus on the past, current, and future use of stable isotopes in the study of Lp(a) metabolism, highlighting critical metabolic uncertainties that exist.
WHERE IS Lp(a) ASSEMBLED?
It is well established that the apo(a) and apoB components of Lp(a) are assembled in the liver. Most studies in cultured hepatocytes indicate that binding of apo(a) and apoB to form Lp(a) occurs either at the surface of the hepatocyte or in the media (which represents the extracellular space in vivo), rather than in the intracellular secretory pathway (26, 27), with the number of KIV-2 repeats determining the proportion of newly synthesized apo(a) that is secreted rather than degraded intracellularly, possibly by the proteasome (27). On the other hand, there are studies in cultured cells supporting intracellular assembly of Lp(a) (28, 29).There is also evidence supporting a role for translational efficiency of apo(a) mRNA in the regulation of apo(a) production (30).
From the authors’ perspective, studies in cultured cells support a model where the assembly of Lp(a) occurs on the surface of the liver or in an extra-hepatic space separated from plasma.
In contrast to the cellular data, three early in vivo stable isotope studies in humans, by Gaubatz et al. (31), Su et al. (32), and Frischmann et al. (33), provided support for the intracellular assembly of Lp(a). All of these studies used deuterated amino acids, but isolated Lp(a), apo(a), and apoB in Lp(a) by different methods. Importantly, they all observed similar rates of enrichment of plasma Lp(a)-apo(a) and Lp(a)-apoB and determined the fractional clearance rates (FCRs) for both moieties of Lp(a) to be about 0.25 pool/day, lower than that of LDL-apoB (which ranges from 0.3 to 0.5 pool/day). Although these data did not rule out assembly of Lp(a) from its components at sites on the surface of hepatocytes protected from circulating LDL, they argued strongly against the fusion of circulating free apo(a) with circulating apoB particles. However, Demant et al. (34) observed differences in the rise in enrichment of Lp(a)-apo(a) and Lp(a)-apoB and, using a more complex compartmental model to analyze their data, reported that about one-half of Lp(a) was assembled in the liver and one-half in plasma, where free apo(a) combined with circulating IDL and LDL. In vivo studies in genetically manipulated mice support both intracellular and extracellular assembly of Lp(a). Thus, crossing apo(a) transgenic mice with apoB transgenic mice resulted in the appearance of mature Lp(a) in the circulation concomitant with the disappearance of free apo(a) that was present in apo(a) transgenic mice, supporting the intracellular assembly of Lp(a) (35). In contrast, infusion of human LDL-apoB into mice transgenic for human apo(a) generated Lp(a) from circulating free apo(a), albeit under very nonphysiological conditions (36).
From the authors’ perspective, at the present time, in vivo studies of Lp(a) kinetics in humans support assembly of Lp(a) within the liver, at the surface of the liver, or in an extra-hepatic space separated from plasma.
WHICH apoB LIPOPROTEIN DOES apo(a) BIND TO WHEN IT FORMS Lp(a)?
If Lp(a) is assembled within the liver, at the surface of the liver, or just outside the liver in a “protected” space from which circulating LDL-apoB is excluded, which apoB-100 lipoprotein is involved? Because VLDL is the major apoB-100 lipoprotein secreted from the liver, several groups have examined the binding of apo(a) to VLDL or the presence of Lp(a) among lipoproteins isolated in the d < 1.006 range. The consensus of these studies is that, in vitro, apo(a) can bind to a range of lipoproteins (37) and that, in vivo, Lp(a) can be isolated from the d < 1.006 range in plasma of hypertriglyceridemic subjects and of individuals with normal fasting triglycerides (TGs) after they have ingested a high fat meal (37–40). However, Krempler et al. (41) found no evidence of a precursor-product relationship between VLDL or LDL and Lp(a) after injection of radiolabeled VLDL into normal subjects. In addition, when VLDL was infused into apo(a) transgenic mice, the appearance of circulating Lp(a) was significantly delayed compared with the infusion of LDL (35). If VLDL is not the lipoprotein to which apo(a) initially binds, then the nascent Lp(a) must arise from the binding of apo(a) to smaller apoB particles (IDL and/or LDL) and, if this does not occur in the circulation, the liver must be secreting one or both of these Lp(a) particle sizes. In fact, direct production of LDL-apoB has been observed in numerous human lipoprotein kinetic studies utilizing both exogenous and endogenous labeling approaches (42, 43, 43a). Those studies did not, however, examine Lp(a)-apo(a) and Lp(a)-apoB metabolism at the same time. We have unpublished data indicating that Lp(a) isolated at the density of VLDL and IDL (d < 1.019) has the same FCR as Lp(a) in the density range of LDL/HDL. Because the pool size of Lp(a) in the d < 1.019 fraction is very small, it cannot have the same FCR of the much larger pool of Lp(a) in the LDL/HDL density range and also be the precursor of the latter.
From the authors’ perspective, based on available data, the majority of Lp(a) must enter the circulation as a particle with a density greater than 1.019.
IS apo(a) IRREVERSIBLY BOUND TO apoB ON Lp(a)?
The earliest reported kinetic studies by Krempler and colleagues (44, 45) used radio-iodinated Lp(a) and found that the FCR for Lp(a) in normal subjects was 0.3 pool/day. They demonstrated equivalent decay curves for serum, purified Lp(a), and the fractionated protein components of Lp(a), indicative of equal clearance rates of Lp(a)-apo(a) and Lp(a)-apoB. However, Knight et al. (46), using radiolabeled Lp(a) isolated by a different method than used by Krempler, observed dissociation and rapid clearance of the Lp(a)-apo(a) originally bound to Lp(a)-apoB, with the generation of an LDL-like particle in plasma of normal subjects and individuals with familial hypercholesterolemia (FH). Knight’s result must be viewed with caution, as there is no evidence for a “significant” pool of free apo(a) in the circulation and, if dissolution of the apo(a)-apoB complex in Lp(a) was followed by rapid clearance of free apo(a) [leaving behind an LDL, as suggested by Knight et al. (46)], the FCR of that “Lp(a),” which would essentially be an LDL labeled in apoB, would be similar to the FCR of radiolabeled LDL in normal subjects that Knight et al. (46) studied simultaneously.
As noted above, Demant et al. (34), using stable isotopes, found that the FCRs for Lp(a)-apo(a) and Lp(a)-apoB were similar, indicating that, independent of the site of formation of Lp(a), Lp(a)-apo(a) and Lp(a)-apoB remained linked until they left the circulation together and were degraded. The results of this study have been replicated recently (33). Two additional studies from one group, using stable isotopes, found differences in the rates of enrichment of Lp(a)-apo(a) and Lp(a)-apoB. In the first, Jenner et al. (47) used a lectin-based isolation method and found FCRs of 0.22 and 0.46 pool/day for Lp(a)-apo(a) and Lp(a)-apoB, respectively. In the second study, Diffenderfer et al. (48) used immunoprecipitation of Lp(a) from plasma and reported FCRs of 0.10 and 0.26 pool/day for Lp(a)-apo(a) and Lp(a)-apoB-100. The results of these two studies are incompatible with an irreversible combination event for apo(a) and apoB-100 in Lp(a). Indeed, the authors offered a model in which apo(a) recycles on and off VLDL [forming a TG-rich Lp(a) at least once before leaving plasma] (48). Of note, the subjects studied by Jenner et al. (47) had baseline lipid levels similar to subjects in other studies in which FCRs for Lp(a)-apo(a) and Lp(a)-apoB-100 were the same (32, 33, 35), whereas the four subjects studied by Diffenderfer et al. (48) were significantly hypertriglyceridemic at baseline.
From the authors’ perspective, at the present time, conflicting kinetic data in the literature do not allow us to draw a firm conclusion regarding the stability of the apo(a)-apoB complex during the entire lifespan of Lp(a). The absence of a significant pool of free apo(a) in plasma, however, argues against recycling of apo(a) within the circulation. Future studies that examine larger cohorts and can determine the individual kinetics of each apo(a) isoform and the apoB associated with each of those isoforms will be necessary to answer this important question. Crucial to the solution of this problem is the isolation of Lp(a) that is completely free of other apoB-containing lipoproteins.
DOES PRODUCTION OR FRACTIONAL CLEARANCE DETERMINE THE LEVELS OF Lp(a) IN BLOOD?
From the earliest studies using radiolabeled tracers, plasma levels of Lp(a) have correlated very strongly with the production rates (PRs) of Lp(a), with little or no relationship between levels and the FCR of Lp(a). The opposite was true for correlations focused on LDL metabolism, where the LDL apoB-100 FCR and not the PR were closely related to LDL levels in blood. The importance of production was confirmed when Rader and colleagues published results of two studies using radio-iodinated Lp(a), which examined the important role of apo(a) isoform size on the rate of appearance of Lp(a) in plasma (49, 50). Importantly, while one study demonstrated that the inverse relationship between apo(a) isoform size and plasma Lp(a) concentration was due to differences in apo(a) production rather than clearance, the second revealed that rates of apo(a) production determined differences in plasma levels of Lp(a) in people with the same apo(a) isoform size. In recent studies using stable isotopes, significant correlations between both the FCR and PR of Lp(a) and plasma concentrations were reported by Jenner et al. (47), whereas Diffenderfer et al. (48) only observed a correlation between the PR and plasma Lp(a) levels.
From the authors’ perspective, the results published to date indicate that, in individuals not receiving any lipid-altering medications, both fractional clearance and production play roles in the regulation of Lp(a) levels in blood. Overall, however, the PR is more closely related to plasma concentrations of Lp(a) than is the FCR.
The studies described above highlight three key questions about the metabolism of Lp(a) that remain unanswered. First, does covalent binding of apo(a) to apoB occur outside or within the circulation? Second, is the covalent bond, once formed, irreversible during the lifetime of apo(a)? Third, how is Lp(a) cleared from the circulation?
Lp(a) KINETIC MODELING
Physiological considerations
We have approached the first two questions by developing a series of mechanistic models. These models serve as guidance to understand how stable isotope labeling kinetic studies can be used to address questions of Lp(a) assembly and its production and clearance in the circulation. The models assume that all proteins synthesized by the liver have a common precursor pool of their amino acids and, therefore, the plateau of enrichment reached by a rapidly turning over protein, such as VLDL-apoB, during infusion of a stable isotope of an amino acid can be used as the precursor enrichment for all other liver-derived proteins (51–53). What is not always recognized is the need to account for additional commonalities when different molecules are being studied together on a unique lipoprotein particle. In studies of apoB and TG kinetics in VLDL (54), our multi-compartmental model required that, although the TG:apoB ratio could vary among different VLDL subfractions (ranging from a large value in the largest subfraction to smaller values down the delipidation cascade), the rate constants (or FCRs) for apoB and TG have to be the same for any given VLDL pool. This approach was taken even earlier by others who modeled apoB and TG together (55) and by us when modeling off- and on-statin LDL-apoB kinetics data using three radiotracers (56).
What distinguishes a combined model for Lp(a)-apoB and Lp(a)-apo(a) from other combined models, such as for VLDL-apoB and VLDL-TG, is that there is a 1:1 molar ratio of apoB to apo(a) on every Lp(a) particle (unlike apoB:TG ratios, which vary among VLDL particles). This fixed molar ratio means that the number of molecules of apoB in an Lp(a) pool must equal that of apo(a). Furthermore, if there is no recycling of apo(a) or of apoB within Lp(a), the FCR in any Lp(a) pool must be the same for apoB and for apo(a) because the exit of one Lp(a) particle means the exit of one and only one molecule each of apoB and apo(a). It follows then that, because, in each Lp(a) pool, the mass and FCR of apoB and apo(a) are equal, the flux in and out of that pool must also be the same for apoB and apo(a). If one accepts these premises, analyzing Lp(a) tracer kinetics data requires the use of a combined model for apoB and apo(a) with identical molar masses, fluxes, and FCRs for apoB and apo(a) in each Lp(a) pool. A logical question then is how enrichment curves for Lp(a)-apoB and Lp(a)-apo(a) can be different, as seen in some published studies (47, 48, 57), if their kinetic parameters are the same in every Lp(a) pool. The answer lies in possible differences in the synthetic pathways, i.e., the precursor pools of apo(a) and apoB for assembly of Lp(a) can have different isotopic enrichment.
Below, we explore the possibilities suggested by earlier work for Lp(a) assembly: 1) apo(a) and apoB bind inside the liver; 2) apo(a) binds extracellularly to apoB lipoproteins in the bloodstream; and 3) there is recycling of Lp(a)-apo(a) or Lp(a)-apoB. The resulting four models are explained below in the context of a primed constant infusion study of a stable isotope of an amino acid. We provide the simplest models for each possibility. We understand that more complexity than is offered by these four models is possible, i.e., there could be combinations of models. Additionally, although we include the possibility that apo(a) can exit the liver alone and bind to apoB in the circulation, we have not allowed apo(a) and apoB to separate in the circulation. As a result of the latter assumption, when our models include recycling of either apo(a) or apoB, this occurs within the liver, after uptake of an intact Lp(a). We based this decision on the lack of data supporting a significant pool of free apo(a) in the circulation. We are aware of convincing data that a small fraction of Lp(a) has been isolated as free apo(a) from serum (58) and fragments of apo(a) can be found in urine (59), but we have not included these in our models.
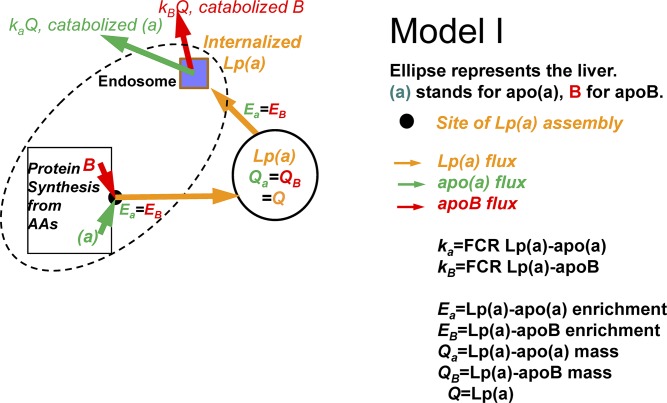
Model I
In this scenario, Lp(a)-apoB and Lp(a)-apo(a) enrichment curves in plasma are identical and, thus, they have the same FCR, as observed by Gaubatz et al. (31), Su et al. (32), and Frischmann et al. (33). In this model, Lp(a) particles are assembled within or at the surface of the liver, as newly synthesized apo(a) binds to newly synthesized apoB. The precursor is the hepatic amino acid pool for both Lp(a)-apoB and Lp(a)-apo(a), which, combined with identical masses and FCRs, means that the Lp(a)-apoB and Lp(a)-apo(a) enrichment curves are identical as are their FCRs (Fig. 1).
Fig. 1.
Model I: apo(a) and apoB combine to form Lp(a) either within the liver, at the surface of the liver, or just outside the liver in a compartment protected from circulating plasma. Lp(a)-apo(a) and Lp(a)-apoB have the same enrichments, FCRs, and fluxes.
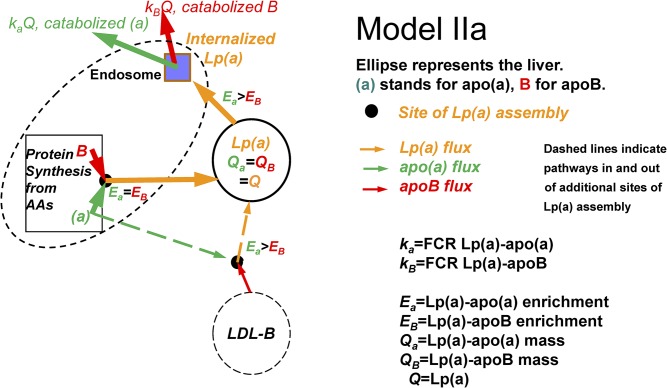
Model II
Lp(a)-apoB has a lower enrichment curve in plasma than Lp(a)-apo(a) and has a smaller slope, as in the study by Demant et al. (34). There are two possible inferences in this case: In model IIa, we can infer that Lp(a) particles are made from newly synthesized apo(a) binding partly to newly synthesized apoB (in which case the precursor is the hepatic amino acid pool for both) and partly to an apoB-lipoprotein that is already circulating. The latter Lp(a) particles, formed in plasma in the model, have apoB at a much lower enrichment than apo(a). The higher the relative contribution of circulating apoB-lipoproteins (e.g., LDL-apoB) to the assembly of the Lp(a), the lower the Lp(a)-apoB enrichment curve will be compared with the enrichment curve of Lp(a)-apo(a). As demonstrated by Demant et al. (29), data from the apo(a) and apoB slopes of the enrichments provide the relative contributions of these proteins to circulating Lp(a) that is assembled in the liver and one that derives from apo(a) binding to a circulating apoB-lipoprotein. Despite the existence of two sources of Lp(a) with different enrichments in this model, there is only a single catabolic pathway into the liver with full degradation of both Lp(a)-apo(a) and Lp(a)-apoB, meaning that their FCRs and fluxes are equal (Fig. 2).
Fig. 2.
Model IIa: apo(a) and apoB combine to form Lp(a) at the same site(s) as in model I, but some apo(a) is also secreted from the liver and combines with an apoB-lipoprotein in the circulation (dashed lines). Lp(a) apoB will have a lower enrichment than Lp(a) apo(a), with the same FCR and flux for Lp(a) apo(a) and Lp(a) apoB.
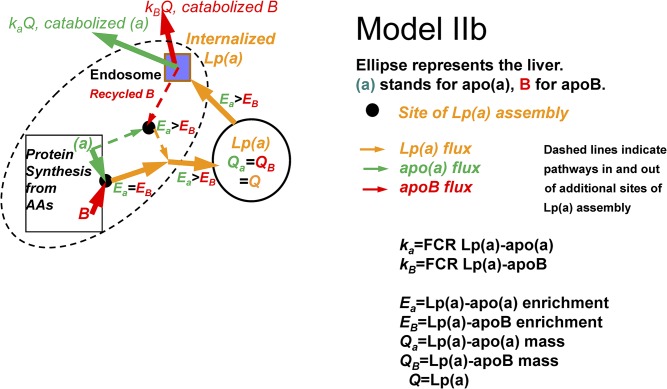
In model IIb, we infer an alternative basis for an Lp(a)-apoB enrichment curve that is lower than the Lp(a)-apo(a) curve. Following liver uptake of circulating Lp(a) particles, the Lp(a)-apo(a) is degraded while at least some of the Lp(a)-apoB can recycle and bind to newly synthesized apo(a). Because the Lp(a) entering the liver from the circulation is a mixture of recently labeled Lp(a) and “older” unlabeled Lp(a) that was circulating prior to the start of the stable isotope infusion, any Lp(a) generated by the binding of newly synthesized labeled apo(a) to a mixture of newly synthesized apoB and apoB derived from circulating Lp(a) that was taken up by the liver will have an average enrichment of apoB lower than that of apo(a). This inference means that there is a higher flux and a higher FCR for Lp(a)-apo(a) than for Lp(a)-apoB. It is not possible to choose between model IIa and model IIb ( ) from the tracer studies alone. However, cell culture studies indicate secretion of free apo(a) into the circulation is unlikely, making the recycling model (model IIb) more likely as the explanation of lower enrichment in the Lp(a)-apoB curve compared with the Lp(a)-apo(a) curve.
Fig. 3.
Model IIb: apo(a) and apoB combine to form Lp(a) at the same site(s)as in model I, but some Lp(a)-apoB is recycled to the Lp(a) assembly site after a circulating Lp(a) is taken up by the liver (dashed lines). Lp(a)-apo(a) that is taken up by the liver is all degraded. Lp(a) apoB will have a lower enrichment than Lp(a) apo(a), with Lp(a) apo(a) having higher FCR and flux than Lp(a) apoB.
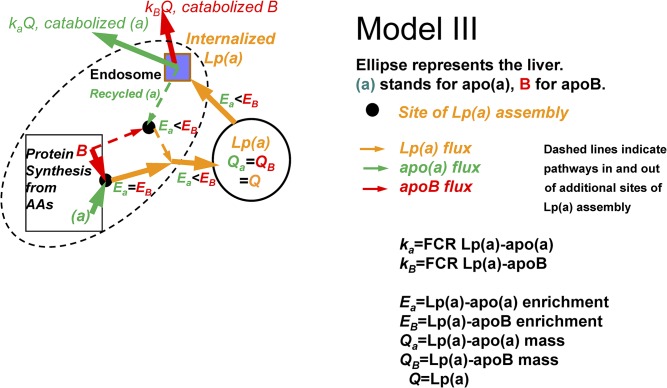
Model III
Lp(a)-apoB has a higher enrichment curve in plasma than Lp(a)-apo(a) and has a larger slope, as observed by Jenner et al. (47) and Diffenderfer et al. (48). There are two possible inferences theoretically, as in models IIa and IIb. However, the analog to model IIa, which would have newly secreted apoB-lipoproteins binding to apo(a) in the circulation, requires a large free apo(a) pool in the circulation (if it was small, it would rapidly achieve the precursor liver amino acid enrichment). Because there is no evidence for such a pool, only one inference, analogous to model IIb is possible. We can infer that, following liver uptake of circulating Lp(a), the apoB is degraded while at least some of the apo(a) can recycle and bind to newly synthesized apoB. This is the opposite of model IIb; but now, because the Lp(a) entering the liver from the circulation is at a much lower enrichment than the newly synthesized apo(a), any Lp(a) generated by the binding of newly synthesized labeled apoB to the mixture of labeled and unlabeled apo(a) will have an average enrichment of apo(a) lower than that of apoB. This inference means that there is a higher flux and a higher FCR for Lp(a)-apoB than Lp(a)-apo(a) ( ). This appears to be the favored interpretation in Diffenderfer et al. (48). Importantly, a very recent paper by Sharma et al. (22) demonstrated degradation of Lp(a)-apoB, but recycling of Lp(a)-apo(a), in cultured liver cells.
Fig. 4.
Model III: apo(a) and apoB combine to form Lp(a) at the same site(s) as in model I, but some Lp(a)-apo(a) is recycled to the Lp(a) assembly site after a circulating Lp(a) is taken up by the liver (dashed lines). Lp(a)-apoB that is taken up by the liver is all degraded. Lp(a)-apoB has a higher enrichment than Lp(a)-apo(a) and also a higher FCR and flux.
In summary, in all cases, the steady state masses of Lp(a)-apoB and Lp(a)-apo(a) are equal [as they must be because there is one apo(a) and one apoB on each Lp(a)]. In models I and IIa, the fluxes of Lp(a)-apoB and Lp(a)-apo(a) are equal, and the FCRs of Lp(a)-apoB and Lp(a)-apo(a) are equal. In model IIb, the flux and FCR of Lp(a)-apo(a) are larger than those of Lp(a)-apoB. In model III, the flux and FCR of Lp(a)-apoB are larger than those of Lp(a)-apo(a).
From the authors’ perspective, the models provided above attempt to clarify the types of data that have led previous investigators studying Lp(a)-apo(a) and Lp(a)-apoB to draw differing conclusions regarding the origin of Lp(a), the stability of the apo(a)-apoB bond in the Lp(a) particle, and the question of whether there is recycling of either Lp(a)-apo(a) or Lp(a)-apoB. It is important to note that stable isotope studies of apo(a) kinetics that use only the enrichment of apo(a), can provide valid data for the FCR of Lp(a) in plasma only if model I or model IIa is correct. If there is recycling of either apoB or apo(a), as in models IIb and III, then there is no single FCR for Lp(a), and individual enrichment data for Lp(a)-apo(a) and Lp(a)-apoB must be generated and used to calculate the individual FCRs of these components of Lp(a). Identification of the “correct” model will offer critical insights into targets for Lp(a) lowering.
HOW IS Lp(a) CLEARED FROM THE CIRCULATION?
After more than 30 years of work by many investigators using cells, animal models, and humans, we continue to have great uncertainty about the pathway(s) involved in determining the clearance of Lp(a). This topic was presented in a previous review in this series (60), and is an essential component of this review as well. Lp(a) levels are increased in people with FH, and Kraft et al. (61) reported that for Lp(a) with the same allele sizes, the concentration was dependent on the gene dose of LDL receptors. This study and others point to an important role for the LDL receptor in the clearance of Lp(a) from the circulation. However, drugs that clearly increase LDL receptor number, including statins, bile acid-binding resins, and ezetimibe, do not affect plasma Lp(a) concentrations.
One would expect that kinetic studies, which actually provide FCRs, would have answered this question. The earliest studies, performed by Krempler and colleagues (44, 45), used radio-iodinated Lp(a) and found that the FCRs for Lp(a) in normal subjects were, as noted earlier in the review, about 0.3 pool/day: 30% lower than the FCR they observed for LDL apoB-100 (44, 45). In one patient with homozygous FH, the FCR of Lp(a) was not different from those in normal subjects (45). Despite the differences in Lp(a) and LDL apoB-100 metabolism in their first study (44) and the similarities between Lp(a) and LDL FCRs measured in a homozygous FH subject in their second study, the authors concluded that their initial kinetic studies did provide support for the LDL receptor pathway as the major clearance mechanism for Lp(a). Their conclusion hinged strongly on their in vitro studies of the binding and uptake of 125LDL and 125Lp(a) by fibroblasts from normal and homozygous FH individuals, which showed similar effects of the presence or absence of LDL receptors between the two lipoproteins. In contrast, Maartmann-Moe and Berg (62) reported results indicating very different mechanisms for the binding and uptake of LDL and Lp(a) in fibroblasts.
A decade later, Knight et al. (46) also using radio-iodinated Lp(a), studied normal and FH patients. In the normal group, Lp(a) FCRs were similar to those reported by Krempler and colleagues (44, 45) and almost 50% slower than the FCRs for LDL apoB-100 in the normal subjects. In contrast, there were no differences between the FCRs for Lp(a) and LDL apoB in the FH group; they were both the same as the Lp(a) FCRs in the normal subjects (44, 46). Knight et al. (46) also provided evidence suggesting that a remnant of Lp(a) with physical characteristics of LDL was generated in plasma, offering a solution to the problem of Lp(a) clearance, i.e., there may be more than one pathway involved, with the LDL receptor taking an Lp(a) remnant that lacks apo(a) out of the circulation while intact Lp(a) is cleared by other pathways. In this scenario, both the level of LDL receptor activity and the degree of apoB dissociation from Lp(a) would be determinants of the apparent Lp(a) FCR. As we noted above, however, there is no evidence for a significant pool of free apo(a) in the circulation and if dissolution of the apo(a)-apoB complex in Lp(a) was followed by very rapid clearance of free apo(a) leaving behind an LDL-apoB, as suggested by Knight et al. (46), the stable isotope studies in normal subjects would have FCRs for Lp(a) that were similar to those for LDL-apoB.
The results in nonhuman studies have been conflicting as well. In 1990, Hofmann et al. (63) reported that Lp(a) clearance was accelerated in LDL receptor transgenic mice. This group also demonstrated high affinity binding of Lp(a) to the LDL receptor. In 1993, Liu et al. (64), using LDL receptor-deficient Watanabe rabbits, observed a much lower FCR for Lp(a) in Watanabe rabbits compared with normal Japanese White rabbits, although there was an even greater difference between the FCRs for LDL in the two groups of rabbits. Soon after these two reports, Rader et al. (65) used radiolabeled tracers to examine the effects of FH on the FCRs of LDL apoB-100 and Lp(a). They observed that, whereas the removal of LDL apoB was reduced in heterozygous FH patients and even more in homozygous FH patients compared with normal controls, the FCRs of Lp(a) were the same across all three groups (65). In a follow-up study, Cain et al. (66) demonstrated similar FCRs for radiolabeled Lp(a) in wild-type and LDL receptor-deficient mice. In the same study, these investigators found that unlabeled apo(a) was a strong competitor for Lp(a) clearance, suggesting that the apo(a) component, rather than apoB-100, was involved in clearance of Lp(a).
In the era of stable isotope studies, the differences in FCRs for Lp(a) and LDL apoB-100 have persisted. Importantly, despite different methods of isolation of Lp(a) for determination of enrichment and differences in sampling and feeding protocols during the kinetic studies, the FCRs for Lp(a) have been consistently reduced compared with those for LDL apoB-100, with a range of FCRs for Lp(a) from about 0.10 to 0.35. Of note, a nontracer non-steady state study in FH patients undergoing LDL-apheresis reported by Parhofer et al. (67) yielded an FCR for Lp(a) of 0.16 pool/day, similar to results from several tracer kinetic studies.
Our understanding of the role of the LDL receptor in Lp(a) catabolism can be informed by studying the effects of drugs that affect LDL receptor number. We recently reported a 19% reduction in Lp(a) levels in normal subjects receiving the PCSK9 inhibitor, alirocumab, which was associated with a trend toward an increase in the FCR of Lp(a) of 25% without any change in PR (53). The Lp(a) FCR change was small compared with the 80% increase in the LDL apoB FCR. A possible explanation for this finding might be that if Lp(a) was being cleared only by the LDL receptor, Lp(a) must have a much lower affinity for the LDL receptor. On further consideration, however, it is clear that the percent increase of ligand binding to a receptor should reflect the increase in available receptors, independent of the affinity of the ligand for that receptor. Thus, although Lp(a) may, in general, have a lower affinity for the LDL receptor than LDL, an increase in the number of LDL receptors should have equal relative effects on the FCRs of Lp(a) and LDL. A more likely explanation for our result is that Lp(a) is cleared by more than one receptor pathway. In this instance, if only the LDL receptors are increased, the increase in the Lp(a) FCR would reflect the proportion of Lp(a) cleared by LDL receptors versus other receptors, whereas the increase in FCR for LDL would be directly dependent on the increase in LDL receptors. The studies by Sharma et al. (22) indicated that a plasminogen receptor is involved in Lp(a) uptake by human hepatoma cells. Romagnuolo and colleagues reported that adding supra-physiological levels of PCSK9 to HepG2 cells reduced surface LDL receptors and the internalization of Lp(a) (68). A possible role of PCSK9 in the clearance of Lp(a) has been suggested by the work of Tavori et al. (69) that demonstrated binding of PCSK9 to both Lp(a) and LDL; these findings have also been recently reviewed (70).
To add further uncertainty to this issue, we have found that mipomersen (apoB antisense) treatment in normal subjects lowered Lp(a) by 21% due to an increase in the FCR of apo(a) by 27% (unpublished observations). At the same time, mipomersen reduced levels of LDL by increasing the LDL apoB FCR by 30%, suggesting that the LDL receptor pathway was involved to similar degrees in the changes in both Lp(a) and LDL. In contrast, we have also observed that reductions of 35% in Lp(a) during treatment of normal individuals with the CETP inhibitor, anacetrapib, were associated with a reduced PR without changes in the FCR (unpublished observations). Anacetrapib increased the FCR of LDL apoB by 27% in the same study, suggesting that there was no link between increased LDL receptors and reductions in Lp(a) levels. These results, unfortunately, do not add clarity to the question of the role of the LDL receptor in Lp(a) clearance from the circulation. The conflicting data for the role of the LDL receptor has led investigators to examine the binding of Lp(a) to other characterized lipoprotein receptors. A detailed review of these studies was recently published by Hoover-Plow and Huang (25).
Early studies by Kostner and colleagues pointed to the kidney as playing a role in Lp(a) clearance (71, 72). These investigations identified various fragments of apo(a) not bound to apoB that accounted for about 1–3% of the total apo(a) in plasma. Importantly, the plasma concentration of Lp(a) increases in patients with renal disease, while the urinary excretion of apo(a) fragments decreases, suggesting that the kidney is required for the degradation of some apo(a). In a small kinetic study by Frischmann et al. (57), the FCRs of Lp(a)-apo(a) and Lp(a)-apoB in hemodialysis patients were significantly reduced compared with healthy subjects; plasma Lp(a) concentrations were nonsignificantly increased.
From the authors’ perspective, at present, we do not believe that the existing data support any single receptor pathway for the clearance of Lp(a) from the circulation. The uniformly observed lower FCRs for Lp(a) versus LDL do not rule out a role for the LDL receptor, but if the latter is involved, then either Lp(a) has a lower affinity for the LDL receptor than LDL or the LDL receptor is only one of several receptors involved in Lp(a) clearance. The affinity of Lp(a) for other receptors, along with the proportion of Lp(a) cleared via each of those receptors, will determine the background FCR of Lp(a) in an individual, as well as the change in FCR when the number of LDL receptors increases. The unraveling of this complex system will clearly require further studies. Unfortunately, without a physiologic animal model of Lp(a) metabolism, this will be very difficult.
CONCLUSIONS
Lp(a) is linked to increased risk for cardiovascular disease and the development of aortic stenosis. This risk and the availability of novel treatments that indirectly and directly (73–75) lower plasma levels of Lp(a) highlight the need to better understand the pathways of assembly, production, and clearance of this lipoprotein. Even after 35 years of excellent work by numerous investigators, additional studies of plasma Lp(a), Lp(a)-apo(a), and Lp(a)-apoB metabolism, together with cellular and murine models of Lp(a) assembly and uptake, will be needed to fully characterize the regulation of Lp(a) levels in the circulation. Future studies of the kinetics of Lp(a) and, more specifically, Lp(a)-apo(a) and Lp(a)-apoB must start with purified Lp(a) that is free of contamination by other apoB-lipoproteins. Studies of the kinetics of Lp(a) isolated in the densities of VLDL, IDL, LDL, and HDL will provide information crucial to addressing several of the questions regarding the origin and clearance of Lp(a), but this will require efficient recovery of Lp(a) from isolated fractions, something that has eluded investigators to the present time. Finally, detailed characterization of the Lp(a) lipidome and proteome (76) would allow examination of the role of Lp(a) composition in the regulation of its metabolism.
Footnotes
Abbreviations:
- FCR
- fractional clearance rate
- FH
- familial hypercholesterolemia
- KIV-2
- kringle IV type 2
- Lp(a)
- lipoprotein (a)
- PR
- production rate
- TG
- triglyceride
This work was supported by National Heart, Lung, and Blood Institute Grant R35 HL135833 and Foundation for the National Institutes of Health Grants 1UL1 TR001873 and KL2 TR001874. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Berg K. 1963. A new serum type system in man-the LP system. Acta Pathol. Microbiol. Scand. 59: 369–382. [DOI] [PubMed] [Google Scholar]
- 2.Witztum J. L., and Ginsberg H. N.. 2016. Lipoprotein (a): coming of age at last. J. Lipid Res. 57: 336–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLean J. W., Tomlinson J. E., Kuang W. J., Eaton D. L., Chen E. Y., Fless G. M., Scanu A. M., and Lawn R. M.. 1987. cDNA sequence of human apolipoprotein(a) is homologous to plasminogen. Nature. 330: 132–137. [DOI] [PubMed] [Google Scholar]
- 4.Boerwinkle E., Leffert C. C., Lin J., Lackner C., Chiesa G., and Hobbs H. H.. 1992. Apolipoprotein(a) gene accounts for greater than 90% of the variation in plasma lipoprotein(a) concentrations. J. Clin. Invest. 90: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Utermann G. 1999. Genetic architecture and evolution of the lipoprotein(a) trait. Curr. Opin. Lipidol. 10: 133–141. [DOI] [PubMed] [Google Scholar]
- 6.Utermann G., Menzel H. J., Kraft H. G., Duba H. C., Kemmler H. G., and Seitz C.. 1987. Lp(a) glycoprotein phenotypes. Inheritance and relation to Lp(a)-lipoprotein concentrations in plasma. J. Clin. Invest. 80: 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandholzer C., Hallman D. M., Saha N., Sigurdsson G., Lackner C., Császár A., Boerwinkle E., and Utermann G.. 1991. Effects of the apolipoprotein(a) size polymorphism on the lipoprotein(a) concentration in 7 ethnic groups. Hum. Genet. 86: 607–614. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt K., Noureen A., Kronenberg F., and Utermann G.. 2016. Structure, function, and genetics of lipoprotein (a). J. Lipid Res. 57: 1339–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enkhmaa B., Anuurad E., Zhang W., Tran T., and Berglund L.. 2011. Lipoprotein(a): genotype-phenotype relationship and impact on atherogenic risk. Metab. Syndr. Relat. Disord. 9: 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danesh J., Collins R., and Peto R.. 2000. Lipoprotein(a) and coronary heart disease. Meta-analysis of prospective studies. Circulation. 102: 1082–1085. [DOI] [PubMed] [Google Scholar]
- 11.Wilde C. 2003. LP (a) risk factor # IV, inherited. In Hidden Causesof Heart Attack and Stroke: Inflammation, Cardiology’s New Frontier. Abigon Press, Studio City, CA: 182–183. [Google Scholar]
- 12.Emerging Risk Factors Collaboration, Erqou S., Kaptoge S., Perry P. L., Di Angelantonio E., Thompson A., White I. R., Marcovina S. M., Collins R., et al. 2009. Lipoprotein(a) concentration and the risk of coronary heart disease, stroke, and nonvascular mortality. JAMA. 302: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pepin J. M., O’Neil J. A., and Hoff H. F.. 1991. Quantification of apo[a] and apoB in human atherosclerotic lesions. J. Lipid Res. 32: 317–327. [PubMed] [Google Scholar]
- 14.Hoff H. F., O’Neil J., and Yashiro A.. 1993. Partial characterization of lipoproteins containing apo[a] in human atherosclerotic lesions. J. Lipid Res. 34: 789–798. [PubMed] [Google Scholar]
- 15.Clarke R., Peden J. F., Hopewell J. C., Kyriakou T., Goel A., Heath S. C., Parish S., Barlera S., Franzosi M. G., Rust S., et al. 2009. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med. 361: 2518–2528. [DOI] [PubMed] [Google Scholar]
- 16.Afshar M., Kamstrup P. R., Williams K., Sniderman A. D., Nordestgaard B. G., and Thanassoulis G.. 2016. Estimating the population impact of Lp(a) lowering on the incidence of myocardial infarction and aortic stenosis-brief report. Arterioscler. Thromb. Vasc. Biol. 36: 2421–2423. [DOI] [PubMed] [Google Scholar]
- 17.Nordestgaard B. G., and Langsted A.. 2016. Lipoprotein (a) as a cause of cardiovascular disease: insights from epidemiology, genetics, and biology. J. Lipid Res. 57: 1953–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saleheen D., Haycock P. C., Zhao W., Rasheed A., Taleb A., Imran A., Abbas S., Majeed F., Akhtar S., Qamar N., et al. 2017. Apolipoprotein(a) isoform size, lipoprotein(a) concentration, and coronary artery disease: a Mendelian randomisation analysis. Lancet Diabetes Endocrinol. 5: 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thanassoulis G., Campbell C. Y., Owens D. S., Smith J. G., Smith A. V., Peloso G. M., Kerr K. F., Pechlivanis S., Budoff M. J., Harris T. B., et al. ; CHARGE Extracoronary Calcium Working Group. 2013. Genetic associations with valvular calcification and aortic stenosis. N. Engl. J. Med. 368: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dieplinger H., and Utermann G.. 1999. The seventh myth of lipoprotein(a): where and how is it assembled? Curr. Opin. Lipidol. 10: 275–283. [DOI] [PubMed] [Google Scholar]
- 21.Koschinsky M. L., and Marcovina S. M.. 2004. Structure-function relationships in apolipoprotein(a): insights into lipoprotein(a) assembly and pathogenicity. Curr. Opin. Lipidol. 15: 167–174. [DOI] [PubMed] [Google Scholar]
- 22.Sharma M., Redpath G. M., Williams M. J. A., and McCormick S. P. A.. 2017. Recycling of apolipoprotein(a) after PlgRKT-mediated endocytosis of lipoprotein(a). Circ. Res. 120: 1091–1102. [DOI] [PubMed] [Google Scholar]
- 23.Lamon-Fava S., Diffenderfer M. R., and Marcovina S. M.. 2014. Lipoprotein(a) metabolism. Curr. Opin. Lipidol. 25: 189–193. [DOI] [PubMed] [Google Scholar]
- 24.Dieplinger H. 1999. Lipoprotein(a): the really bad cholesterol?. Biochem. Soc. Trans. 27: 439–447. [DOI] [PubMed] [Google Scholar]
- 25.Hoover-Plow J., and Huang M.. 2013. Lipoprotein(a) metabolism: potential sites for therapeutic targets. Metabolism. 62: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White A. L., Rainwater D. L., Hixson J. E., Estlack L. E., and Lanford R. E.. 1994. Intracellular processing of apo(a) in primary baboon hepatocytes. Chem. Phys. Lipids. 67–68: 123–133. [DOI] [PubMed] [Google Scholar]
- 27.Lobentanz E. M., Krasznai K., Gruber A., Brunner C., Muller H. J., Sattler J., Kraft H. G., Utermann G., and Dieplinger H.. 1998. Intracellular metabolism of human apolipoprotein(a) in stably transfected Hep G2 cells. Biochemistry. 37: 5417–5425. [DOI] [PubMed] [Google Scholar]
- 28.Bonen D. K., Hausman A. M., Hadjiagapiou C., Skarosi S. F., and Davidson N. O.. 1997. Expression of a recombinant apolipoprotein(a) in HepG2 cells. Evidence for intracellular assembly of lipoprotein(a). J. Biol. Chem. 272: 5659–5667. [DOI] [PubMed] [Google Scholar]
- 29.Edelstein C., Davidson N. O., and Scanu A. M.. 1994. Oleate stimulates the formation of triglyceride-rich particles containing apoB100-apo(a) in long-term primary cultures of human hepatocytes. Chem. Phys. Lipids. 67–68: 135–143. [DOI] [PubMed] [Google Scholar]
- 30.Zysow B. R., Lindahl G. E., Wade D. P., Knight B. L., and Lawn R. M.. 1995. C/T polymorphism in the 5′ untranslated region of the apolipoprotein(a) gene introduces an upstream ATG and reduces in vitro translation. Arterioscler. Thromb. Vasc. Biol. 15: 58–64. [DOI] [PubMed] [Google Scholar]
- 31.Gaubatz J. W., Nava M. N., Guyton J. R., Hoffman A. S., Opekun A. R., Hachey D. L., and Morrisett J. D.. 1993. Metabolism of apo(a) and apoB-100 in human lipoprotein(a). In Drugs Affecting Lipid Metabolism. Catapano A. L., Gotto A. M. Jr., Smith L. C., et al., editors. Springer, Dordrecht, The Netherlands: 161–167. [Google Scholar]
- 32.Su W., Campos H., Judge H., Walsh B. W., and Sacks F. M.. 1998. Metabolism of Apo(a) and ApoB100 of lipoprotein(a) in women: effect of postmenopausal estrogen replacement. J. Clin. Endocrinol. Metab. 83: 3267–3276. [DOI] [PubMed] [Google Scholar]
- 33.Frischmann M. E., Ikewaki K., Trenkwalder E., Lamina C., Dieplinger B., Soufi M., Schweer H., Schaefer J. R., Konig P., Kronenberg F., et al. 2012. In vivo stable-isotope kinetic study suggests intracellular assembly of lipoprotein(a). Atherosclerosis. 225: 322–327. [DOI] [PubMed] [Google Scholar]
- 34.Demant T., Seeberg K., Bedynek A., and Seidel D.. 2001. The metabolism of lipoprotein(a) and other apolipoprotein B-containing lipoproteins: a kinetic study in humans. Atherosclerosis. 157: 325–339. [DOI] [PubMed] [Google Scholar]
- 35.Chiesa G., Hobbs H. H., Koschinsky M. L., Lawn R. M., Maika S. D., and Hammer R. E.. 1992. Reconstitution of lipoprotein(a) by infusion of human low density lipoprotein into transgenic mice expressing human apolipoprotein(a). J. Biol. Chem. 267: 24369–24374. [PubMed] [Google Scholar]
- 36.Linton M. F., Farese R. V. Jr., Chiesa G., Grass D. S., Chin P., Hammer R. E., Hobbs H. H., and Young S. G.. 1993. Transgenic mice expressing high plasma concentrations of human apolipoprotein B100 and lipoprotein (a). J. Clin. Invest. 92: 3029–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaffinger D., Schuelke J., Kim C., and Scanu A. M.. 1991. Relationship between apo[a] isoforms and Lp[a] density in subjects with different apo[a] phenotype: a study before and after a fatty meal. J. Lipid Res. 32: 679–683. [PubMed] [Google Scholar]
- 38.Ooi E. M., Watts G. F., Chan D. C., Pang J., Tenneti V. S., Hamilton S. J., McCormick S. P., Marcovina S. M., and Barrett P. H.. 2015. Effects of extended-release niacin on the postprandial metabolism of Lp(a) and ApoB-100-containing lipoproteins in statin-treated men with type 2 diabetes mellitus. Arterioscler. Thromb. Vasc. Biol. 35: 2686–2693. [DOI] [PubMed] [Google Scholar]
- 39.McConathy W. J., Trieu V. N., Koren E., Wang C. S., and Corder C. C.. 1994. Triglyceride-rich lipoprotein interactions with Lp(a). Chem. Phys. Lipids. 67–68: 105–113. [DOI] [PubMed] [Google Scholar]
- 40.Bersot T. P., Innerarity T. L., Pitas R. E., Rall S. C. Jr., Weisgraber K. H., and Mahley R. W.. 1986. Fat feeding in humans induces lipoproteins of density less than 1.006 that are enriched in apolipoprotein (a) and that cause lipid accumulation in macrophages. J. Clin. Invest. 77: 622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krempler F., Kostner G., Bolzano K., and Sandhofer F.. 1979. Lipoprotein (a) is not a metabolic product of other lipoproteins containing apolipoprotein B. Biochim. Biophys. Acta. 575: 63–70. [DOI] [PubMed] [Google Scholar]
- 42.Parhofer K. G., and Barrett H. R.. 2006. What we have learned about VLDL and LDL metabolism from human kinetics studies. J. Lipid Res. 47: 1620–1630. [DOI] [PubMed] [Google Scholar]
- 43.Ginsberg H. N., Le N. A., Short M. P., Ramakrishnan R., and Desnick R. J.. 1987. Suppression of apolipoprotein B production during treatment of cholesteryl ester storage disease with lovastatin. Implications for regulation of apolipoprotein B synthesis. J. Clin. Invest. 80: 1692–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Reyes-Soffer G., Moon B., Hernandez-Ono A., Dionizovik-Dimanovski M. , Jimenez J., Obunike J., Thomas T., Ngai C., Fontanez N., Donovan D. S., et al. 2016. Complex effects of inhibiting hepatic apolipoprotein B100 synthesis in humans. Sci. Transl. Med. 8: 323ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krempler F., Kostner G. M., Bolzano K., and Sandhofer F.. 1980. Turnover of lipoprotein (a) in man. J. Clin. Invest. 65: 1483–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krempler F., Kostner G. M., Roscher A., Haslauer F., Bolzano K., and Sandhofer F.. Studies on the role of specific cell surface receptors in the removal of in man. 1983. J. Clin. Invest. 71: 1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knight B. L., Perombelon Y. F., Soutar A. K., Wade D. P., and Seed M.. 1991. Catabolism of lipoprotein(a) in familial hypercholesterolaemic subjects. Atherosclerosis. 87: 227–237. [DOI] [PubMed] [Google Scholar]
- 47.Jenner J. L., Seman L. J., Millar J. S., Lamon-Fava S., Welty F. K., Dolnikowski G. G., Marcovina S. M., Lichtenstein A. H., Barrett P. H., deLuca C., et al. 2005. The metabolism of apolipoproteins (a) and B-100 within plasma lipoprotein (a) in human beings. Metabolism. 54: 361–369. [DOI] [PubMed] [Google Scholar]
- 48.Diffenderfer M. R., Lamon-Fava S., Marcovina S. M., Barrett P. H., Lel J., Dolnikowski G. G., Berglund L., and Schaefer E. J.. 2016. Distinct metabolism of apolipoproteins (a) and B-100 within plasma lipoprotein(a). Metabolism. 65: 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rader D. J., Cain W., Zech L. A., Usher D., and Brewer H. B. Jr. 1993. Variation in lipoprotein (a) concentrations among individuals with the same apolipoprotein (a) isoform is determined by the rate of lipoprotein (a) production. J. Clin. Invest . 91: 443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rader D. J., Cain W., Ikewaki K., Talley G., Zech L. A., Usher D., and Brewer H. B. Jr. 1994. The inverse association of plasma lipoprotein(a) concentrations with apolipoprotein(a) isoform size is not due to differences in Lp(a) catabolism but to differences in production rate. J. Clin. Invest. 93: 2758–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Millar J. S., Reyes-Soffer G., Jumes P., Dunbar R. L., deGoma E. M., Baer A. L., Karmally W., Donovan D. S., Rafeek H., Pollan L., et al. 2015. Anacetrapib lowers LDL by increasing ApoB clearance in mildly hypercholesterolemic subjects. J. Clin. Invest. 125: 2510–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reyes-Soffer G., Millar J. S., Ngai C., Jumes P., Coromilas E., Asztalos B., Johnson-Levonas A. O., Wagner J. A., Donovan D. S., Karmally W., et al. 2016. Cholesteryl ester transfer protein inhibition with anacetrapib decreases fractional clearance rates of high-density lipoprotein apolipoprotein A-I and plasma cholesteryl ester transfer protein. Arterioscler. Thromb. Vasc. Biol. 36: 994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reyes-Soffer G., Pavlyha M., Ngai C., Thomas T., Holleran S., Ramakrishnan R., Karmally W., Nandakumar R., Fontanez N., Obunike J. C., et al. 2017. Effects of PCSK9 inhibition with alirocumab on lipoprotein metabolism in healthy humans. Circulation. 135: 352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagashima K., Lopez C., Donovan D., Ngai C., Fontanez N., Bensadoun A., Fruchart-Najib J., Holleran S., Cohn J. S., Ramakrishnan R., et al. 2005. Effects of the PPARgamma agonist pioglitazone on lipoprotein metabolism in patients with type 2 diabetes mellitus. J. Clin. Invest. 115: 1323–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adiels M., Packard C., Caslake M. J., Stewart P., Soro A., Westerbacka J., Wennberg B., Olofsson S. O., Taskinen M. R., and Boren J.. 2005. A new combined multicompartmental model for apolipoprotein B-100 and triglyceride metabolism in VLDL subfractions. J. Lipid Res. 46: 58–67. [DOI] [PubMed] [Google Scholar]
- 56.Berglund L., Witztum J. L., Galeano N. F., Khouw A. S., Ginsberg H. N., and Ramakrishnan R.. 1998. Three-fold effect of lovastatin treatment on low density lipoprotein metabolism in subjects with hyperlipidemia: increase in receptor activity, decrease in apoB production, and decrease in particle affinity for the receptor. Results from a novel triple-tracer approach. J. Lipid Res. 39: 913–924. [PMC free article] [PubMed] [Google Scholar]
- 57.Frischmann M. E., Kronenberg F., Trenkwalder E., Schaefer J. R., Schweer H., Dieplinger B., Koenig P., Ikewaki K., and Dieplinger H.. 2007. In vivo turnover study demonstrates diminished clearance of lipoprotein(a) in hemodialysis patients. Kidney Int. 71: 1036–1043. [DOI] [PubMed] [Google Scholar]
- 58.Gries A., Nimpf J., Nimpf M., Wurm H., and Kostner G. M.. 1987. Free and Apo B-associated Lpa-specific protein in human serum. Clin. Chim. Acta. 164: 93–100. [DOI] [PubMed] [Google Scholar]
- 59.Kostner K. M., Maurer G., Huber K., Stefenelli T., Dieplinger H., Steyrer E., and Kostner G. M.. 1996. Urinary excretion of apo(a) fragments. Role in apo(a) catabolism. Arterioscler. Thromb. Vasc. Biol. 16: 905–911. [DOI] [PubMed] [Google Scholar]
- 60.Kostner K. M., and Kostner G. M.. 2017. Lipoprotein (a): a historical appraisal. J. Lipid Res. 58: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kraft H. G., Lingenhel A., Raal F. J., Hohenegger M., and Utermann G.. 2000. Lipoprotein(a) in homozygous familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 20: 522–528. [DOI] [PubMed] [Google Scholar]
- 62.Maartmann-Moe K., and Berg K.. 1981. Lp(a) lipoprotein enters cultured fibroblasts independently of the plasma membrane low density lipoprotein receptor. Clin. Genet. 20: 352–362. [DOI] [PubMed] [Google Scholar]
- 63.Hofmann S. L., Eaton D. L., Brown M. S., McConathy W. J., Goldstein J. L., and Hammer R. E.. 1990. Overexpression of human low density lipoprotein receptors leads to accelerated catabolism of Lp(a) lipoprotein in transgenic mice. J. Clin. Invest. 85: 1542–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu R., Saku K., Kostner G. M., Hirata K., Zhang B., Shiomi M., and Arakawa K.. 1993. In vivo kinetics of lipoprotein(a) in homozygous Watanabe heritable hyperlipidaemic rabbits. Eur. J. Clin. Invest. 23: 561–565. [DOI] [PubMed] [Google Scholar]
- 65.Rader D. J., Mann W. A., Cain W., Kraft H. G., Usher D., Zech L. A., Hoeg J. M., Davignon J., Lupien P., Grossman M., et al. 1995. The low density lipoprotein receptor is not required for normal catabolism of Lp(a) in humans. J. Clin. Invest. 95: 1403–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cain W. J., Millar J. S., Himebauch A. S., Tietge U. J., Maugeais C., Usher D., and Rader D. J.. 2005. Lipoprotein [a] is cleared from the plasma primarily by the liver in a process mediated by apolipoprotein [a]. J. Lipid Res. 46: 2681–2691. [DOI] [PubMed] [Google Scholar]
- 67.Parhofer K. G., Demant T., Ritter M. M., Geiss H. C., Donner M., and Schwandt P.. 1999. Lipoprotein (a) metabolism estimated by nonsteady-state kinetics. Lipids. 34: 325–335. [DOI] [PubMed] [Google Scholar]
- 68.Scipione C. A., Sayegh S. E., Romagnuolo R., Tsimikas S., Marcovina S. M., Boffa M. B., and Koschinsky M. L.. 2015. Mechanistic insights into Lp(a)-induced IL-8 expression: a role for oxidized phospholipid modification of apo(a). J. Lipid Res. 56: 2273–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tavori H., Christian D., Minnier J., Plubell D., Shapiro M. D., Yeang C., Giunzioni I., Croyal M., Duell P. B., Lambert G., et al. 2016. PCSK9 association with lipoprotein(a). Circ. Res. 119: 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lambert G., Thedrez A., Croyal M., Ramin-Mangata S., Couret D., Diotel N., Nobécourt-Dupuy E., Krempf M., LeBail J. C., Poirier B., et al. 2017. The complexity of lipoprotein (a) lowering by PCSK9 monoclonal antibodies. Clin. Sci. 131: 261–268. [DOI] [PubMed] [Google Scholar]
- 71.Karádi I., Romics L., Pálos G., Domán J., Kaszás I., Hesz A., and Kostner G. M.. 1989. Lp(a) lipoprotein concentration in serum of patients with heavy proteinuria of different origin. Clin. Chem. 35: 2121–2123. [PubMed] [Google Scholar]
- 72.Cauza E., Kletzmaier J., Bodlaj G., Dunky A., Herrmann W., and Kostner K.. 2003. Relationship of non-LDL-bound apo(a), urinary apo(a) fragments and plasma Lp(a) in patients with impaired renal function. Nephrol. Dial. Transplant. 18: 1568–1572. [DOI] [PubMed] [Google Scholar]
- 73.Lippi G., and Targher G.. 2012. Optimal therapy for reduction of lipoprotein(a). J. Clin. Pharm. Ther. 37: 1–3. [DOI] [PubMed] [Google Scholar]
- 74.Gencer B., Kronenberg F., Stroes E. S., and Mach F.. 2017. Lipoprotein(a): the revenant. Eur. Heart J. 38: 1553–1560. [DOI] [PubMed] [Google Scholar]
- 75.Tsimikas S. 2017. A test in context: lipoprotein(a): diagnosis, prognosis, controversies, and emerging therapies. J. Am. Coll. Cardiol. 69: 692–711. [DOI] [PubMed] [Google Scholar]
- 76.von Zychlinski A., Williams M., McCormick S., and Kleffmann T.. 2014. Absolute quantification of apolipoproteins and associated proteins on human plasma lipoproteins. J. Proteomics. 106: 181–190. [DOI] [PubMed] [Google Scholar]