Abstract
Background
Direct-acting antivirals (DAAs) have greatly improved the treatment of HCV infection. To improve response and prevent resistance, combination regimens have been the focus of clinical development. Regimens are often first assessed in vitro, with most combination studies to date using subgenomic replicon systems, which do not replicate the complete HCV life cycle and preclude study of entry and assembly inhibitors. Infectious full-length HCV systems have been developed and are being used to test drug efficacy.
Methods
Using cell-based HCV Con1b replicon and an infectious full-length HCV (HCVcc-Luc) infection system, we systematically tested the synergy, additivity or antagonism of combinations of protease, NS5A and nucleotide NS5B inhibitor classes as well as the combination of these DAAs with host-targeting agent cyclosporin A or non-antibody entry inhibitor (S)-chlorcyclizine. Two computational software packages, MacSynergyII and CalcuSyn, were used for data analysis.
Results
Combinations between different classes showed good consistency across the two viral assay systems and two software platforms. Combinations between NS5A and nucleotide NS5B inhibitors were synergistic, while combinations of protease inhibitors with the other two classes were additive to slightly antagonistic. As expected, combinations of antivirals of the same class were additive. Combination studies between these DAA classes and cyclosporin A or (S)-chlorcyclizine demonstrated additive to synergistic effects and highly synergistic effects, respectively. Combinations of these drugs did not show any added or unexpected cytotoxicity.
Conclusions
Our results show that in vitro combination studies of anti-HCV DAAs in the HCVcc system may provide useful guidance for drug combination designs in clinical studies. We also demonstrate that these DAAs in combination with host-targeting agents or entry inhibitors may improve HCV treatment response.
Introduction
HCV is a positive-stranded RNA virus infecting over 200 million people worldwide. Chronic HCV infection is a leading cause of hepatocellular carcinoma and a leading indication for liver transplantation in developed countries. Effective vaccination for HCV is not yet available. For many years, combination antiviral therapies utilizing pegylated interferon-α (PEG-IFN) and ribavirin (RBV) were the first line of care [1–3]. Recently, the US Food and Drug Administration (FDA) has approved direct-acting antivirals (DAAs), including the protease, NS5A and polymerase inhibitors tel-aprevir, boceprevir, simeprevir, paritaprevir, ledipasvir, daclatasvir, ombitasvir, sofosbuvir and dasabuvir for use in combination or with PEG-IFN and RBV. Many regimens demonstrated improved viral clearance in patients infected with HCV genotype-1 [4–6]. In light of HCV’s high rate of developing mutations that confer drug resistance, combination therapies of DAAs have been crucial for the development of effective regimens for chronic HCV infection [7].
To facilitate selective development of such regimens, it is therefore important to investigate the potential synergistic or antagonistic effects of such combinations first with a robust in vitro HCV model system. Most in vitro combination studies published to date use the subgenomic replicon system to evaluate combinations of certain novel agents with well-developed antivirals that have completed advanced clinical trials or obtained FDA approval [8–10]. Recently, more combination studies have utilized an infectious HCV system [11–14]. However, replicon systems cannot reproduce the entire infectious HCV life cycle. In addition, the replicon-containing cells are highly selected and may not reflect the native cellular environment for HCV replication in vivo. A cell-based infection assay using infectious HCV with full-length HCV genomes is more biologically relevant to in vivo infection. It also expands the testing of agents to target entry and assembly phases of the HCV life cycle. To date, no systematic combination tests between the major HCV antiviral classes of HCV protease, NS5A and NS5B polymerase inhibitors in a true infection system have been conducted. A side-by-side comparison of combinations between replicon and infection systems in drug combination tests is also lacking in the current literature.
Here we present a study of systematic combination of HCV antivirals utilizing a previously described full-length HCV infection system. The system relies on a Renilla luciferase reporter inserted into a full-length HCV genome (HCVcc-Luc) for detection of viral replication [15,16]. We tested this method in comparison with the HCV replicon assay. Various DAAs were tested in the models, such as the protease inhibitor, NS5A inhibitor and nucleotide NS5B inhibitor classes. Antivirals with the same mechanism of action were also evaluated in combination as controls. We subsequently applied the system to test a series of combinations of these DAAs with cyclosporin A, a host-targeting agent (HTA) and chlorcyclizine, a recently described entry inhibitor [17,18]. For data analysis, we utilized two different software packages which base their calculation on alternative definitions of drug interaction and non-interaction: MacSynergyII (Bliss independence model) and CalcuSyn (Loewe additivity model). Both concepts of drug interaction have been widely applied and well established in the evaluation of drug combinations [11,19,20].
Methods
Antiviral agents, viruses and cell lines
Antiviral agents included two active site protease inhibitors telaprevir (Selleckchem, Houston, TX, USA) and boceprevir (ChemScene, Monmouth Junction, NJ, USA), NS5A inhibitor daclatasvir (Selleckchem), two nucleotide NS5B polymerase inhibitors, sofosbuvir (Advanced Chemblocks, Burlingame, CA, USA) and 2′-C-methylcytidine (US Biological, Salem, MA, USA), host-targeting cyclophillin inhibitor cyclosporin A (Sigma-Aldrich, St Louis, MO, USA) and entry-inhibitor (S)-chlorcyclizine (NCATS, Bethesda, MD, USA).
The HCVcc-Luc infectious virus consisted of a full-length J6/JFH-1 HCV with insertion of a luciferase reporter gene at the 3′ end of the p7 gene [16].
Huh7.5.1 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fischer Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Serum Source International, Charlotte, NC, USA), 100 IU/ml penicillin, and 100 μg/ml streptomycin in 5% CO2, at 37°C. A stably expressing Con1b replicon cell line with luciferase reporter under the direction of 5′-NTR for quantification described previously [21,22] was grown in the same condition as Huh7.5.1 cells with the addition of 500 μg/ml G-418.
Efficacy assays
Cells were seeded in 96-well plates (104 cells/well) and cultured overnight in the above-mentioned culture medium in absence of G-418. In the infectious efficacy assay, Huh7.5.1 cells underwent simultaneous infection at a multiplicity of infection of 0.05 and treatment with a matrix of concentrations of two compounds. No viral inoculum was used in the Con1b replicon efficacy assay.
Eight or nine concentrations of each agent were used, for a total matrix of 72 concentration combinations. Concentration ranges were chosen to include at least one point of approximately 0% inhibition and at least one point of approximately 100% inhibition for each drug alone. Concentrations were in ½ log10 increments (for example, 1 μM, 0.316 μM, 0.1 μM and so on), unless the individual dose–response curves were too steep (>70% change in inhibition between adjacent ½ log10 increments). In such cases, the assay was repeated with ¼ log10 increments to more accurately define dose–response relationships. After 48 h, a Renilla luciferase assay kit (Promega, Madison, WI, USA) was used to measure HCV inhibition, normalized to DMSO-control treatment. All experiments were conducted in triplicate.
Cytotoxicity assay
Cytotoxicity was assessed in parallel assays under the same conditions as in the HCVcc-Luc or Con1b replicon efficacy assay using a luminescence-based ATP sensor (ATPlitc 1-Step Kit; PerkinElmer, Waltham, MA, USA).
Data analysis
Experimental data were analysed according to two drug combination models, Bliss independence and Loewe additivity.
Bliss independence and MacSynergyII
Data were analysed using the software MacSynergyII [23]. From experimental individual drug effects, the program calculates a theoretically expected ‘additive’ combined-drug effect if the drugs act independently, for each combination of agents at specific concentrations. From (1−A)(1−B)=(1−C), the theoretical additive combined-drug effect, C, is determined when A and B are the experimental individual drug effects. The program then compares this theoretically expected additive effect to the experimental combined-drug effect. If the experimental antiviral activity significantly deviates (95% confidence level) from the theoretical additive effect at any concentration combination, MacSyncrgyII quantitates that synergy or antagonism. This can be described by a surface plot of peaks and valleys of synergy and antagonism values, with the log volumes (LV) of these peaks and valleys informing a quantitative measure of synergy or antagonism over the sum of the concentrations tested. LV ≥2, −2< LV <2 and LV ≤−2 indicate synergy, additive effect and antagonism, respectively.
Loewe additivity and CalcuSyn
Data were also analysed using the software CalcuSyn (BioSoft, Ferguson, MO, USA), which draws on the methodology of Chou and Talaly [24]. The program takes the drug concentrations required to produce a given effect in an experimental combination and compares them to the drug concentrations that would be needed individually to achieve that same effect. Inputting concentrations and effect values from individual antiviral agent treatments, we identified the concentration at 50% efficacy (EC50) for each agent. We then selected the concentration that was closest to the approximated EC50 of each drug (for example, for EC50s of 0.29 μM and 0.9 μM, we select 0.316 μM and 1 μM, respectively) and took the ratio of those concentrations. We subsequently inputted the concentrations and effect values for all experimental combinations that had the same ratio of drug concentrations. Using this, CalcuSyn interpolates the drug concentrations needed in combination at that ratio to produce effects of 50%, 75% and 90% inhibition. It compares these combined drug concentrations with the concentrations from the two drugs’ individual dose–effect curves needed to achieve 50%, 75% and 90% inhibition. The extent of synergy or antagonism is reported in a combination index (CI) value, calculated by CI = ac/ai + bc/bi, where ac and bc are experimental drug concentrations needed to achieve a stated effect, and ai and bi are the individual drug concentrations needed to achieve that effect according to individual drug dose curves. CI >1.1, 1.1≥ CI ≥0.9 and CI <0.9 indicate synergy, additive effect and antagonism, respectively.
The two software packages CalcuSyn and MacSynergyII diagnose synergy and antagonism based on two different definitions of non-interaction on which the alternative concepts of interaction are based. It is possible that the results generated from the two analysis methods sometimes may differ depending on the mechanisms of drug action. One should take this into consideration in interpreting the results.
Results
Comparison of HCVcc-Luc infection assay and replicon assay in evaluating combinations of DAAs
We systematically tested the effects on HCV inhibition of combinations of antivirals of different DAA classes, including protease inhibitors telaprevir and boceprevir, NS5A inhibitor daclatasvir, and nucleotide NS5B polymerase inhibitor sofosbuvir in both HCVcc-Luc infection and Con1b replicon systems. Results in CalcuSyn will be discussed first, followed by results in MacSyncrgyII, in order to facilitate the comparison of the two viral assay platforms. Table 1 shows EC50 and concentration at 50% cytotoxicity (CC50) values of the antivirals from individual dose–response curves in both systems.
Table 1.
In vitro activity of HCV antivirals
| Antiviral name | EC50, μM | CC50, μM | ||
|---|---|---|---|---|
|
|
|
|||
| HCVcc-Luc (2a) | Con1b replicon | HCVcc-Luc (2a) | Con1b replicon | |
| Daclatasvir | 0.00004 ±0.00002 | 0.00004 ±0.00002 | 45 ±1 | 22 ±1 |
| Telaprevir | 0.20 ±0.04 | 0.55 ±0.05 | >100 | 97.8 ±0.9 |
| Boceprevir | 0.22 ±0.01 | 0.62 ±0.01 | >100 | >100 |
| Sofosbuvir | 0.060 ±0.002 | 0.4 ±0.1 | >100 | >100 |
| 2′-C-methylcytidine | 1.4±0.4 | 1.61 ±0.07 | >100 | >100 |
| Cyclosporin A | 0.14 ±0.02 | 0.62 ±0.01 | >100 | >100 |
| (S)-CCZ | 0.024 ±0.009 | N/A | 33 ±2 | N/A |
Data shown represents means of results of ≥3 independent tests ±SEM. CCZ, chlorcyclizine; CC50, concentration at 50% cytotoxicity; EC50, concentration at 50% efficacy; N/A, not applicable.
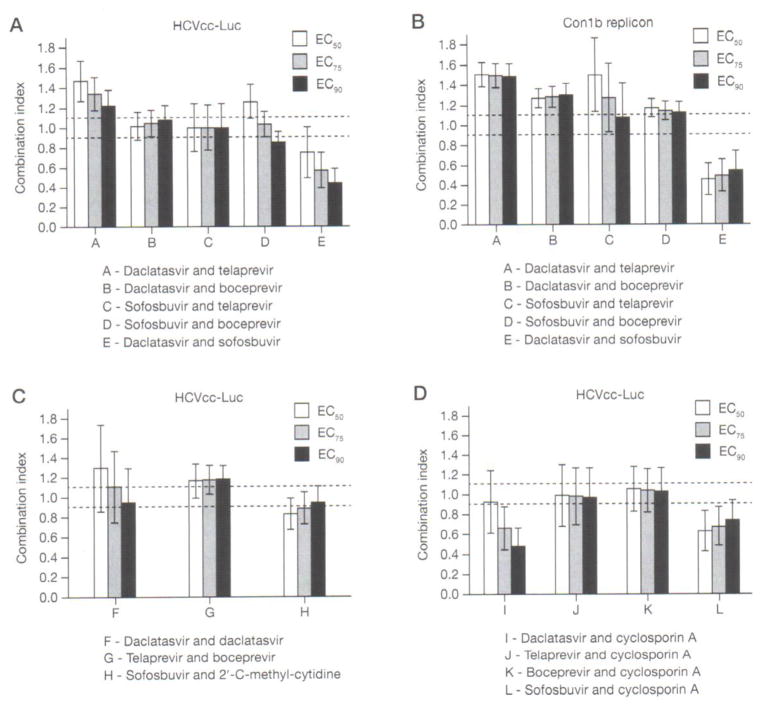
Combinations of daclatasvir with telaprevir in HCVcc-Luc and Con1b replicon yielded CIs in CalcuSyn representing slight-to-moderate antagonism (1.23–1.47) and moderate antagonism (1.48–1.50), respectively, at EC50, EC75, and EC90 (Figure 1A and Table 2). The combinations resulted in the same level of synergy or antagonism (additive, slight, moderate, major) or differing only by 1 level. The combination of daclatasvir and boceprevir demonstrated additivity (1.02–1.08) in HCVcc-Luc and slight antagonism (1.26–1.29) in Con1b replicon, when analysed in CalcuSyn (Figure 1B and Table 2). For combinations of sofosbuvir and telaprevir or boceprevir in the HCVcc-Luc and Con1b replicon systems, CalcuSyn interaction assessments generated results varying widely from 0 to 2 levels. CI values for sofosbuvir with telaprevir were additive (1.00) and additive to moderately antagonistic (1.07–1.49) for the HCVcc-Luc and replicon systems, respectively (Figure 1A, 1B and Table 2). Sofosbuvir and boceprevir were slightly synergistic to slightly antagonistic (0.85–1.26) and slightly antagonistic (1.12–1.17) in HCVcc-Luc and Con1b systems, respectively (Figure 1A, 1B and Table 2). The combination of daclatasvir and sofosbuvir was moderate to highly synergistic in the HCVcc-Luc synergy assay with CI values 0.44–0.75 and highly synergistic in replicons with CI 0.45–0.54 (Figure 1A, 1B and Table 2).
Figure 1. Combination index of combinations between different classes of DAAs or DAAs with cyclosporin A.
Huh7.5.1 cells infected with HCVcc-Luc or Con1b replicon cell lines were treated with agents in serial dilutions to form a matrix of concentrations. Both were analyzed by Renilla luciferase activity. Combinations of different classes of direct acting antivirals (DAAs) were tested in Huh7.5.1 cells infected with (A) full-length HCVcc-Luc virus and (B) subgenomic Con1b replicons. Combinations between (C) DAAs of the same class or between (D) DAAs and host-targeting agent cyclosporin A were also tested in HCVcc-Luc-infected cells. Projected combination index (CI) SE of concentration at 50%, 75% and 90% efficacy (EC50, EC75 and EC90) from the means of experiments in triplicates are shown. Area between dotted lines represents additivity (0.9 ≤CI ≤ 1.1). Exact CI values are shown in Tables 2 and 3.
Table 2.
Comparison of combinations of DAAs of different classes in HCVcc-Luc and Con1b replicon assays and combination study of DAAs of same class in HCVcc-Luc assay
| Virus type | ID | Antiviral name | MacSyncrgy II
|
CalcuSyn
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Synergism LV |
Antagonism LV |
Conclusiona | |||||||
| EC50 CI | EC75 CI | EC90 CI | Conclusionb | ||||||
| HCVcc-Lucc | A | Daclatasvir and telaprevir | 0.13 | −1.95 | () Additive | 1.47 ±0.20 | 1.34 ±0.16 | 1.23 ±0.15 | (−) Minor antagonism |
| HCVcc-Lucc | B | Daclatasvir and boceprevir | 3.61 | 0 | (+) Minor synergy | 1.01 ±0.14 | 1.05 ±0.13 | 1.08 ±0.14 | () Additive |
| HCVcc-Lucc | C | Sofosbuvir and telaprevir | 4.1 | −1.27 | (+) Minor synergy | 1.00 ±0.24 | 1.00 ±0.23 | 1.01 ±0.23 | () Additive |
| HCVcc-Lucc | D | Sofosbuvir and boceprevir | 0.42 | 0 | () Additive | 1.26 ±0.17 | 1.03 ±0.12 | 0.85 ±0.10 | () Additive |
| HCVcc-Lucc | E | Daclatasvir and sofosbuvir | 2.11 | 0 | (+) Minor synergy | 0.75 ±0.26 | 0.57 ±0.18 | 0.44 ±0.14 | (+++) Major synergy |
| Con1b repliconc | A | Daclatasvir and telaprevir | 0.4 | −0.94 | () Additive | 1.50 ±0.12 | 1.49 ±0.12 | 1.48 ±0.13 | (−) Minor antagonism |
| Con1b repliconc | B | Daclatasvir and boceprevir | 1.39 | −1 | () Additive | 1.26 ±0.09 | 1.28 ±0.10 | 1.29 ±0.12 | (−) Minor antagonism |
| Con1b repliconc | C | Sofosbuvir and telaprevir | 0.02 | −4.83 | (−) Minor antagonism | 1.49 ±0.36 | 1.26 ±0.34 | 1.07 ±0.34 | (−) Minor antagonism |
| Con1b repliconc | D | Sofosbuvir and boceprevir | 0 | −1.36 | () Additive | 1.17 ±0.09 | 1.14 ±0.09 | 1.12 ±0.11 | (−) Minor antagonism |
| Con1b repliconc | E | Daclatasvir and sofosbuvir | 19.08 | −0.03 | (+++) Major synergy | 0.45 ±0.16 | 0.49 ±0.16 | 0.54 ±0.19 | (++) Moderate synergy |
| HCVcc-Lucd | F | Daclatasvir and daclatasvir | 3.06 | −1.77 | (+) Minor synergy | 1.30 ±0.44 | 1.11 ±0.36 | 0.95 ±0.35 | () Additive |
| HCVcc-Lucd | G | Telaprevir and boceprevir | 1.77 | 0 | () Additive | 1.17 ±0.17 | 1.18 ±0.15 | 1.18 ±0.14 | (−) Minor antagonism |
| HCVcc-Lucd | H | Sofosbuvir and 2′-C-methylcytidine | 0.16 | −0.47 | () Additive | 0.83 ±0.16 | 0.89±.0.15 | 0.95 ±0.16 | (+) Minor synergy |
Extent of synergy or antagonism in MacSynergyII is defined according to absolute value of log volume (LV) as follows: () additive, LV <2; (+/−) minor, 2< LV <5; (++/−−) moderate, 5≤ LV <9; (+++/−−−) major, LV ≥9.
Extent of synergy and antagonism in CalcuSyn is defined as follows: (+++) major synergy, combination index (CI) ≤0.7; (++) moderate synergy, 0.7≤ CI <0.8; (+) minor synergy, 0.8≤ CI <0.9; () additive, 0.9≤ CI <1.1; (−) minor antagonism, 1.1≤ CI <1.3; (−−) moderate antagonism, 1.3≤ CI <3.0; (−−−) major antagonism, CI ≥3.0.
LV (95% confidence level) and CI (±SE)of combinations of different classes of direct-acting antivirals (DAAs) and with HCVcc-Luc and Con1b rcplicons.
LV (95% confidence level) and CIs (±SE) of combinations of DAAs of the same class in HCVcc-Luc. Additivity is defined as a combinatorial effect equal to the sum of each drug’s effect alone. Raw luciferase values of all combinations in triplicate were inputted into MacSynergyII and % inhibition values of combinations at a fixed ratio of concentration of two drugs inputted into CalcuSyn, except those exhibiting >25% toxicity.
In MacSynergyII, combinations between daclatasvir and telaprevir or boceprevir yielded additive-level log volumes for both the HCVcc-Luc and replicon systems. Sofosbuvir with telaprevir or boceprevir straddled additivity, resulting in additivity, minor synergism or minor antagonism across the two HCV systems in MacSynergyII. Daclatasvir and sofosbuvir combinations were slightly synergistic in HCVcc-Luc and highly synergistic in replicon when analysed with MacSynergyII. These results show general similarity with small differences in determination of the level of synergism or antagonism between analyses of data using either CalcuSyn or MacSynergyII (Table 2).
To test whether combinations of these drugs have any cytotoxicity at the concentrations used, parallel ATPlite assays were performed. No toxicity was observed except at the highest concentration of a single drug in a single test, which was excluded from analysis in CalcuSyn and MacSynergyII.
Combination studies of DAAs of the same class in HCVcc-Luc assay
Additivity represents a situation in which the combination of two drugs results in effects no greater or less than the addition of their individual effects. Although the definition of additivity and the expected result varies depending on which concept of drug non-interaction is used, additivity would be expected in combinations of drugs of the same mechanism of action or in combinations of a drug with itself [25]. To validate this hypothesis in the HCVcc-Luc system, we tested such combinations. As expected, combinations between daclatasvir with itself, protease inhibitors telaprevir and boceprevir, and nucleotide polymerase inhibitors sofosbuvir and 2′-C-methylcytidine yielded additive or nearly additive (minor synergy or minor antagonism) CIs of 0.95–1.30, 1.17–1.18 and 0.83–0.95, respectively (Figure 1C and Table 2). MacSynergyII yielded log volumes indicating additivity for the latter two combinations (Table 2). As MacSynergyII calculates additivity assuming the two drugs bind to dissimilar sites, it becomes nonsensical to test a drug combination with itself [23,26,27], For completeness, the result of minor synergism for the combination of daclatasvir with itself is still reported in Table 2. No toxicity was observed in the parallel ATPlite cytotoxicity assay at the concentrations tested.
Combination studies of DAAs and an HTA in HCVcc-Luc assay
Combinations involving host-targeting agents (HTAs) provide another possibility aside from DAAs for IFN-free regimens, especially as host-targets provide a higher barrier to resistance and pan-genotypic activity [3]. Cyclosporin A, an immunosuppressive agent and cyclophillin inhibitor, has been demonstrated to have antiviral activity against HCV replication [28–30]. Therefore, cyclosporin A was tested as an HTA in this study. In HCVcc-Luc, in vitro combinations of cyclosporin A with daclatasvir or sofosbuvir were slightly to moderately synergistic, with CI values 0.49–0.93 and 0.63–0.74, respectively, and MacSynergyII synergy log volumes were 2.27 and 7.22, respectively (Figure 1D and Table 3). Combinations between cyclosporin A and telaprevir or boceprevir were additive in CalcuSyn, with CIS of 0.97–0.99 and 1.03–1.06, respectively. This near additivity resulted again in MacSynergyII with minor synergism for the telaprevir combination with log volume 2.17 and additivity for the boceprevir combination with log volume 1.83 (Table 3). No toxicity was observed at the concentrations tested.
Table 3.
Combination studies of DAAs, cyclosporin A and (S)-CCZ in HCVcc-Luc assay
| Virus type | ID | Antiviral name | MacSynergyII
|
CalcuSyn
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Synergism LV |
Antagonism LV |
Conclusiona | |||||||
| EC50 CI | EC75 CI | EC90 CI | Conclusionb | ||||||
| HCVcc-Lucc | I | Daclatasvir and cyclosporin A | 2.27 | −0.33 | (+) Minor synergy | 0.93 ±0.31 | 0.67 ±0.22 | 0.49 ±0.18 | (++) Moderate synergy |
| HCVcc-Lucc | J | Telaprevir and cyclosporin A | 2.17 | −0.47 | (+) Minor synergy | 0.99 ±0.31 | 0.98 ±0.28 | 0.97 ±0.29 | () Additive |
| HCVcc-Lucc | K | Boceprevir and cyclosporin A | 1.83 | −0.09 | () Additive | 1.06 ±0.23 | 1.04 ±0.22 | 1.03 ±0.24 | () Additive |
| HCVcc-Lucc | L | Sofosbuvir and cyclosporin A | 7.22 | −0.11 | (++) Moderate synergy | 0.63 ±0.20 | 0.68 ±0.19 | 0.74 ±0.20 | (++) Moderate synergy |
| HCVcc-Lucd | N/A | Daclatasvir and (S)-CCZ | 11.76 | 0 | (+++) Major synergy | 0.31 ±0.18 | 0.43 ±0.21 | 0.63 ±0.26 | (+++) Major synergy |
| HCVcc-Lucd | N/A | Telaprevir and (S)-CCZ | 11.90 | −0.59 | (+++) Major synergy | 0.67 ±0.19 | 0.64 ±0.16 | 0.01 ±0.16 | (+++) Major synergy |
| HCVcc-Lucd | N/A | Boceprevir and (S)-CCZ | 17.00 | −1.21 | (+++) Major synergy | 0.33 ±0.09 | 0.32 ±0.09 | 0.36 ±0.12 | (+++) Major synergy |
| HCVcc-Lucd | N/A | Sofosbuvir and (S)-CCZ | 30.56 | 0 | (+++) Major synergy | 0.16 ±0.03 | 0.20 ±0.04 | 0.27 ±0.05 | (+++) Major synergy |
| HCVcc-Lucd | N/A | 2′-C-methylcytidine and (S)-CCZ | 5.66 | −0.05 | (++) Moderate synergy | 0.36 ±0.09 | 0.34 ±0.07 | 0.35 ±0.07 | (+++) Major synergy |
| HCVcc-Lucd | N/A | Cyclosporin A and (S)-CCZ | 40.80 | −2.16 | (+++) Major synergy | 0.39 ±0.14 | 0.24 ±0.07 | 0.26 ±0.06 | (+++) Major synergy |
Extent of synergy or antagonism in MacSynergyII is defined according to absolute value of log volume (LV) as follows: () additive, LV <2; (+/−) minor, 2≤ LV <5; (++/−−) moderate, 5≤ LV <9; (+++/−−−) major, LV ≥9.
Extent of synergy and antagonism in CalcuSyn is defined as follows: (+++) major synergy, combination index (CI) ≤0.7; (++) moderate synergy, 0.7≤ CI <0.8; (+) minor synergy, 0.8≤ CI <0.9; () additive, 0.9≤ CI <1.1; (−) minor antagonism, 1.1≤ CI <1.3;(−−) moderate antagonism, 1.3≤ CI <3.0; (−−−) major antagonism, CI ≥3.0.
LV (95% confidence level) and CI (±SE) of combinations of direct-acting antivirals (DAAs) with host-targeting agent (HTA) cyclosporin A in HCVcc-Luc.
LV (95% confidence level) and CIs (±SE)of combinations of previously described entry inhibitor (S)-chlorcyclizinc (CCZ) with DAAs and HTA cyclosporin A, in HCVcc-Luc. Raw luciferase values of combinations in triplicate were inputted into MacSynergyII and % inhibition values of combinations at a fixed ratio of concentration of two drugs inputted into CalcuSyn, except those exhibiting >25% toxicity. N/A, not applicable.
Combination studies of DAAs and an entry inhibitor in HCVcc-Luc assay
A major advantage of HCVcc-Luc infection over the replicon system is its ability to assess the combinatorial effect of HCV inhibitors targeting all stages of the HCV replication cycle. We and others previously reported the entry-related antiviral activity of FDA-approved antihistamine chlorcyclizine (CCZ) [17,18]. In the current study, we performed an extensive analysis of the combination of DAAs and CCZ in accordance with the commonly used fixed ratio and fitting method of CalcuSyn to calculate the projected CI values at 50%, 75% and 90% effective concentrations. Combinations between (S)-CCZ and daclatasvir, telaprevir, boceprevir, sofosbuvir, 2′-C-methylcytidine or cyclosporin A were highly synergistic with CI values ranging from 0.16 to 0.67 (Figure 2 and Table 3). In Table 3, we also report the moderate to major synergy in MacSynergyll. No toxicity was observed at the concentrations tested.
Figure 2. Combination index of combinations of entry inhibitor (S)-chlorcyclizine with DAAs or with cyclosporin A.
Huh7.5.1 cells infected with HCVcc-Luc were simultaneously treated with (S)-chlorcyclizine in combination with inhibitors as indicated in the figure at effective concentrations of 50% (EC50). 75% (EC75) and 90% (EC90), respectively. Projected combination index (CI) ± SE at EC50, EC75 and EC90 from the means of experiments in triplicates are shown. Area between dotted lines represents additivity (0.9 ≤ CI ≤ 1.1). Exact CI values are shown in Table 3.
Discussion
Because of the importance of combination drug regimens against HCV to prevent resistance and the high cost to clinically evaluate all combinations, there is a need for robust in vitro systems for analysis of antiviral combinations. The commonly used method of subgenomic replicons does not represent the complete HCV life cycle and precludes investigation of entry and assembly inhibitors [21,22]. A full-length infectious system would capture all stages of the HCV life cycle, and therefore could be used to test HCV inhibitor combinations involving every aspect of HCV life cycle. A systematic combination of three classes of DAAs in a full-length HCV cell culture system compared to a replicon system has not been conducted to date. A comprehensive study of these drug combinations in vitro using the HCVcc infection system may provide useful information for clinical drug combination designs and future anti-HCV drug development.
In the current study, we analysed the results of drug combination studies using the HCVcc-Luc system in five aspects. First was a systematic study of various combinations of three different classes of DAAs (protease inhibitors, NS5A inhibitors and nucleotide NS5B inhibitors). Second was comparison of combinations of these three different classes of DAAs in our infectious system to a subgenomic replicon system. Third was to test if combinations of compounds of the same class or a compound with itself were additive. Fourth was to analyse synergy, additivity or antagonism using two different software packages, CalcuSyn and MacSynergyII that are based on two different concepts of drug non-interaction. Last, we verified no added or unexpected cytotoxicity in the combinations used in parallel ATPlite assays.
In our study of telaprevir, boceprevir, daclatasvir and sofosbuvir, only the combination of NS5A inhibitor (daclatasvir) and nucleotide NS5B inhibitor (sofosbuvir) was synergistic. Combinations between protease inhibitors and NS5A inhibitors or nucleotide NS5B inhibitors exhibited additivity in general and even some antagonistic effects despite the differences in the drug target. Consistency over two protease inhibitors verified that the effect is indeed real for this class of DAA. Previous combination studies of protease inhibitors with NS5A inhibitors or nucleotide NS5B inhibitors using the Chou-Talaly method [24] in subgenomic replicons have shown mixed results from moderately antagonistic to moderately synergistic CI values [10,31–33].
The differences between studies may be methodological. We had observed especially steep changes in the dose-response curves for telaprevir and boceprevir in experiments, such as a >70% difference between the two concentrations of 1 μM and 0.316 μM in telaprevir. Due to these steep changes in inhibition, we suspected that more intermediate concentrations are needed to generate adequate data for the independent dose-response curves. from which Chou and Talaly’s method in CalcuSyn draws upon for calculation of CI values. To prevent any errors that might result from poor curve fitting we repeated the combinations in ¼ log10 concentration increments to yield greater resolution of independent drug curves. Indeed, in the case of the theoretically additive combination of two protease inhibitors, telaprevir and boceprevir, ½ log10 dilutions resulted in synergistic CI values from 0.53–0.82 and synergy log volume 7.14, whereas ¼ log10 dilutions yielded near-additivity as expected, with CIs of 1.17–1.18 and synergy log volume 1.77. This highlights the variability of results in in vitro combination tests based on drug dose-responsiveness and selected dilution factors. Future studies should make note to ensure that one should select dilution factors capable of yielding good resolution of the drug’s dose–response curve. Based on our results, we therefore believe that protease inhibitors act mostly in additive effect with the other two DAA classes.
The reason for a lack of synergy between protease inhibitors and NS5A or NS5B inhibitors is not apparent. It is known that inhibition of NS3/4a protease activity prevents cleavage of the HCV polyprotein, resulting in a reduction of NS5A and NS5B protein levels. Thus the action of protease inhibitors may not be totally independent of NS5A and NS5B inhibitors. For example, the reduction in NS5A and NS5B protein levels from inhibition of NS3/4a and polyprotein processing may result in reduced effectiveness of NS5A and nucleotide NS5B inhibitors due to a reduction in the concentration of their targets. Recently it has also been shown that telaprevir may have additional inhibitory effects on HCV RNA synthesis independent of its effect on reducing HCV non-structural protein levels [34]. These possibilities would therefore suggest against a synergistic effect and in favour of additive or antagonistic effects.
In general, our results showed systematic near-agreement in synergistic, additive or antagonistic effects between the replicon and infectious virus system with combinations of protease inhibitors, NS5A inhibitors and NS5B inhibitors. Combinations yielded deviation of no more than one level of synergism or antagonism (additive, minor, moderate, major) between the infectious and replicon systems in CalcuSyn, and only a few deviated by two levels in MacSynergyII. It is likely that one level of difference in synergy or antagonism is insignificant for these software tools, as theoretically additive combinations of DAAs of the same class were sometimes one level off the additive effect (minor synergism and minor antagonism). Making note of this variation, results in in vitro combination studies should be regarded with caution, unless results indicate strong synergy, such as in the case of combinations of replication inhibitors with entry inhibitor (S)-CCZ in this study.
For variations greater than one level of synergy, the differences could be attributed to the fact that the HCVcc-Luc system represents more steps of the HCV life cycle than the replicon. However, it is also important to consider that the HCVcc-Luc system used in this study is based on a genotype-2a clone, whereas the replicon system used is based on a genotype-lb clone. It is well known that drugs may have different efficacy depending on genotype both in vitro and in vivo [4,31]. The differences in results between the two systems could be in part due to differences in genotype. Nonetheless, the functional relevance of testing drugs of different or same targets in combination should still remain valid regardless of the genotype of the virus. Therefore, it is still of interest from the perspective of dissecting drug mechanism of action to test combination in both a full-length infectious virus and a replicon virus.
We applied our system to test a previously described non-antibody entry inhibitor, chlorcyclizine [17,18] and demonstrated a highly synergistic effect of combinations with active site protease inhibitors, NS5A inhibitors, nucleotide NS5B inhibitors and cyclosporin A. Recently, Xiao et al. [35] showed synergistic effects in combinations between antibodies against HCV entry factors and different classes of DAAs with methods and software packages similar to those in our study, though their study focused on entry inhibitors in the HCVcc-Luc system. Collectively our studies support the potential promise of developing combination therapies utilizing HCV entry and replication inhibitors.
Our study compares the accuracy of an infectious HCV virus system for analysis of the combinatorial effect of HCV antivirals to a replicon HCV cell culture system. In studying the three DAA classes, we found only the combination of NS5A and NS5B inhibitors to be synergistic in HCVcc-Luc and Con lb replicons, while the combinations of protease inhibitors with NS5A or nucleotide NS5B inhibitors were additive to antagonistic. It is interesting that our in vitro findings are correlated with recent in vivo data in clinical studies of drug combinations. Combination of NS5B polymerase inhibitor sofosbuvir and an NS5A inhibitor appears to be highly effective, whereas combinations of other classes tend to require 3–4 drugs (protease inhibitor, NS5A inhibitor, non-nucleoside analogue NS5B inhibitor and ribavirin) to achieve high clinical efficacy [5]. The combination of a replication inhibitor with an entry inhibitor, like (S)-CCZ, demonstrates strong synergy in vitro, supporting future development of such combination regimens. Finally our study supports that this HCVcc-Luc system designed for in vitro combination studies of new HCV antivirals, including drugs targeting other stages of the HCV life cycle, is promising in guiding the efficacy studies of drug combinations in vivo.
Acknowledgments
Funding was provided by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Disclosure statement
TJL, SH and ZH, are named on patent application related to this work: US Provisional Patent Application 62/011,462, ‘Heterocyclic compounds and methods of use thereof’. BL. and HJY declare no competing interests.
References
- 1.Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 2.Liang TJ. Current progress in development of hepatitis C virus vaccines. Nat Med. 2013;19:869–878. doi: 10.1038/nm.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheel TKH, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang TJ, Ghany MG. Current and future therapies for hepatitis C virus infection. N Engl J Med. 2013;368:1907–1917. doi: 10.1056/NEJMra1213651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster DP, Klenerman P, Dusheiko GM. Hepatitis C. Lancet. 2015;385:1124–1135. doi: 10.1016/S0140-6736(14)62401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferenci P, Bernstein D, Lalezari J, et al. ABT-450/r–ombitasvir and dasabuvir with or without ribavirin for HCV. N Engl J Med. 2014;370:1983–1992. doi: 10.1056/NEJMoa1402338. [DOI] [PubMed] [Google Scholar]
- 7.Vermehren J, Sarrazin C. The role of resistance in HCV treatment. Best Pract Res Clin Gastroenterol. 2012;26:487–503. doi: 10.1016/j.bpg.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Lin K, Perni RB, Kwong AD, Lin C. VX-950, a novel hepatitis C virus (HCV) NS3-4A protease inhibitor, exhibits potent antiviral activities in HCV replicon cells. Antimicrob Agents Chemother. 2006;50:1813–1822. doi: 10.1128/AAC.50.5.1813-1822.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toma S, Yamashiro T, Arakaki S, et al. Inhibition of intracellular hepatitis C virus replication by nelfinavir and synergistic effect with interferon-α. J Viral Hepat. 2009;16:506–512. doi: 10.1111/j.1365-2893.2009.01102.x. [DOI] [PubMed] [Google Scholar]
- 10.Bilello JP, Lallos LB, McCarville JF, et al. In vitro activity and resistance profile of samatasvir, a novel NS5A replication inhibitor of hepatitis C virus. Antimicrob Agents Chemother. 2014;58:4431–4442. doi: 10.1128/AAC.02777-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Einav S, Sobol HD, Gehrig E, Glenn JS. The hepatitis C virus (HCV) NS4B RNA binding inhibitor clemizole is highly synergistic with HCV protease inhibitors. J Infect Dis. 2010;202:65–74. doi: 10.1086/653080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu H, Wong-Staal F, Lee H, et al. Evaluation of ITX 5061, a scavenger receptor B1 antagonist: resistance selection and activity in combination with other hepatitis C virus antivirals. J Infect Dis. 2012;205:656–662. doi: 10.1093/infdis/jir802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahid MN, Turek M, Xiao F, et al. The postbinding activity of scavenger receptor class B type I mediates initiation of hepatitis C virus infection and viral dissemination. Hepatology. 2013;57:492–504. doi: 10.1002/hep.26097. [DOI] [PubMed] [Google Scholar]
- 14.Lupberger J, Duong FH, Fofana I, et al. Epidermal growth factor receptor signaling impairs the antiviral activity of interferon-alpha. Hepatology. 2013;58:1225–1235. doi: 10.1002/hep.26404. [DOI] [PubMed] [Google Scholar]
- 15.Hu Z, Lan KH, He S, et al. Novel cell-based hepatitis C virus infection assay for quantitative high-throughput screening of anti-hepatitis C virus compounds. Antimicrob Agents Chemother. 2014;58:995–1004. doi: 10.1128/AAC.02094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80:1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He S, Lin B, Chu V, et al. Repurposing of the antihistamine chlorcyclizine and related compounds for treatment of hepatitis C virus infection. Sci Transl Med. 2015;7:282ra249. doi: 10.1126/scitranslmed.3010286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chamoun-Emanuelli AM, Pécheur E-I, Chen Z. Benzhydrylpiperazine compounds inhibit cholesterol-dependent cellular entry of hepatitis C virus. Antiviral Res. 2014;109:141–148. doi: 10.1016/j.antiviral.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 19.Tallarida RJ. Quantitative methods for assessing drug synergism. Genes Cancer. 2011;2:1003–1008. doi: 10.1177/1947601912440575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panigrahi R, Hazari S, Chandra S, et al. Interferon and ribavirin combination treatment synergistically inhibit HCV internal ribosome entry site mediated translation at the level of polyribosome formation. PLoS ONE. 2013;8:e72791. doi: 10.1371/journal.pone.0072791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Matsumura T, Hu Z, Kato T, et al. Amphipathic DNA polymers inhibit hepatitis C virus infection by blocking viral entry. Gastroenterology. 2009;137:673–681. doi: 10.1053/j.gastro.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nanda SK, Herion D, Liang TJ. Src homology 3 domain of hepatitis C virus NS5A protein interacts with Bin1 and is important for apoptosis and infectivity. Gastroenterology. 2006;130:794–809. doi: 10.1053/j.gastro.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 23.Prichard MX, Shipman C., Jr A three-dimensional model to analyze drug-drug interactions. Antiviral Res. 1990;14:181–205. doi: 10.1016/0166-3542(90)90001-n. [DOI] [PubMed] [Google Scholar]
- 24.Chou TC, Talaly P. A simple generalized equation for the analysis of multiple inhibitions of Michaelis-Menten kinetic systems. J Biol Chem. 1977;252:6438–6442. [PubMed] [Google Scholar]
- 25.Tallarida RJ. Drug synergism: its detection and applications. J Pharmacol Exp Tber. 2001;298:865–872. [PubMed] [Google Scholar]
- 26.Geary N. Understanding synergy. Am] Physiol Endocrinol Metab. 2013;304:E237–E253. doi: 10.1152/ajpendo.00308.2012. [DOI] [PubMed] [Google Scholar]
- 27.Berenbaum MC. What is synergy? Pharmacol Rev. 1989;41:93–141. [PubMed] [Google Scholar]
- 28.Zeisel MB, Lupberger J, Fofana I, Baumert TF. Host-targeting agents for prevention and treatment of chronic hepatitis C – perspectives and challenges. J Hepatol. 2013;58:375–384. doi: 10.1016/j.jhep.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 29.Arai M, Tsukiyama-Kohara K, Takagi A, Tobita Y, Inoue K, Kohara M. Resistance to cyclosporin A derives from mutations in hepatitis C virus nonstructural proteins. Biochem Biophys Res Commun. 2014;448:56–62. doi: 10.1016/j.bbrc.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 30.Morohashi K, Sahara H, Watashi K, et al. Cyclosporin A associated helicase-like protein facilitates the association of hepatitis C virus RNA polymerase with its cellular cyclophilin B. PLoS ONE. 2011;6:el8285. doi: 10.1371/journal.pone.0018285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao M, Nettles RE, Belema M, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465:96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelosi LA, Voss S, Liu M, Gao M, Lemm JA. Effect on hepatitis C virus replication of combinations of direct-acting antivirals, including NS5A inhibitor daclatasvir. Antimicrob Agents Chemother. 2012;56:5230–5239. doi: 10.1128/AAC.01209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottwein JM, Jensen SB, Li YP, et al. Combination treatment with hepatitis C virus protease and NS5A inhibitors is effective against recombinant genotype 1a, 2a, and 3a viruses. Antimicrob Agents Chemother. 2013;57:1291–1303. doi: 10.1128/AAC.02164-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGivern DR, Masaki T, Lovell W, Hamlett C, Saalau-Bethell S, Graham B. Protease inhibitors block multiple functions of the NS3/4A protease-helicase during the hepatitis C virus life cycle. J Virol. 2015;89:5362–5370. doi: 10.1128/JVI.03188-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao F, Fofana I, Thumann C, et al. Synergy of entry inhibitors with direct-acting antivirals uncovers novel combinations for prevention and treatment of hepatitis C. Gut. 2015;64:483–494. doi: 10.1136/gutjnl-2013-306155. [DOI] [PMC free article] [PubMed] [Google Scholar]