Abstract
Mammalian cells recognize virus-derived nucleic acids using a defined set of intracellular sensors including the DNA sensors cyclic GMP–AMP (cGAMP) synthase (cGAS) and interferon gamma (IFNγ)-inducible protein 16 (IFI16) as well as viral RNA receptors of the retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) family. Following innate immune recognition, these sensors launch an immune response that is characterized by the transcriptional upregulation of many antiviral molecules, including proinflammatory cytokines, chemokines, and IFN-stimulated genes. Recent studies have demonstrated that the signal transduction initiated by these sensors is sophisticatedly regulated by post-translational modifications (PTMs) resulting in a robust yet ‘tunable’ cytokine response to maintain immune homeostasis. Here we summarize recent advances in our understanding of how PTMs and regulatory enzymes control the signaling activity of RLRs, cGAS, and IFI16 as well as their proximal adaptor proteins.
Trends
Positive feedforward regulatory mechanisms serve as an important means of signal amplification to ensure an effective innate immune response. However, negative regulatory circuits are essential for the prevention of premature or overactive proinflammatory responses, which could have harmful consequences for the host organism.
Phosphorylation and different types of polyubiquitin chains, particularly K63-linked ubiquitination, are important for fine-tuning signaling initiated by intracellular viral RNA and DNA receptors.
Acetylation, glutamylation, and deamidation of innate immune sensors or components in their signaling pathways also dynamically modulate antiviral cytokine induction.
Insight into the molecular mechanisms and regulatory enzymes that modulate innate sensing pathways may lead to therapeutics to boost antiviral immunity or dampen proinflammatory/autoimmune responses.
Innate Sensing Pathways and Control of Immune Homeostasis
Innate immune mechanisms provide the immediate defense of a mammalian host organism against viral infection. Key steps in this immediate immune response are the detection of the virus and the subsequent initiation of transcriptional events that result in antiviral gene expression. Pattern recognition receptors (PRRs) (also frequently called ‘innate immune sensors’) recognize pathogen-associated molecular patterns (PAMPs), such as viral nucleic acids, proteins, and carbohydrates, with molecular features that are distinct from host-derived molecules.
The individual families of PRRs can be categorized based on their subcellular location into membrane-bound sensors such as C-type lectin receptors (CLRs) and Toll-like receptors (TLRs) and the intracellularly localized NOD-like receptors (NLRs), AIM2-like receptors (ALRs), and RLRs (reviewed in 1, 2, 3). Furthermore, a group of structurally unrelated intracellular DNA sensors has recently been identified, most prominently cGAS of the oligoadenylate synthase (OAS) family and IFI16 of the PYRIN superfamily (reviewed in [4]). Besides their different subcellular localization, PRRs also recognize distinct PAMPs. Within the RLR family, RIG-I recognizes viral RNA that harbors short double-stranded RNA (dsRNA) stretches and a 5′-triphosphate or 5′-diphosphate moiety, while melanoma differentiation-associated protein 5 (MDA5) detects longer dsRNA or viral RNA aggregates 1, 5. In vitro studies and studies in mice demonstrated that recognition of viral RNA PAMPs by RIG-I and MDA5 confers resistance to a wide spectrum of RNA viruses, including flaviviruses, influenza viruses, rhabdoviruses, paramyxoviruses, and picornaviruses. In addition, RLRs are involved in detecting DNA virus infections, a mechanism that, in the case of RIG-I, relies on the recognition of RNA polymerase III-generated viral transcripts, at least for some DNA viruses (e.g., adenoviruses, Epstein–Barr virus).
RIG-I and MDA5 are DExD/H-box-containing RNA helicases with similar structures: both possess two N-terminal caspase activation and recruitment domains (CARDs) for initiating downstream signaling and a central helicase domain and carboxy-terminal domain (CTD) [in the case of RIG-I also called the ‘repressor domain’ (RD)], which are both required for RNA binding. Although they recognize different ligands, RIG-I and MDA5 interact with a common adaptor protein, mitochondrial antiviral signaling protein (MAVS) (also known as IPS-1, CARDIF, or VISA) which serves as a scaffold for protein complex assembly at the mitochondria and mitochondria-associated membranes (MAMs) to trigger an antiviral program [6]. It has been demonstrated that the interaction of RIG-I with the chaperone protein 14-3-3ɛ mediates RIG-I translocation from the cytosol to mitochondria and MAMs to initiate signaling [7]. MAVS then recruits the IKK-related serine/threonine kinases TANK-binding kinase 1 (TBK1) and IKKɛ, which phosphorylate and activate IRF3 or IRF7. Furthermore, the tripartite IKKα–IKKβ–IKKγ complex is activated by MAVS, ultimately resulting in nuclear factor kappa B (NF-κB) activation. NF-κB and IRFs then translocate into the nucleus and cooperatively induce the gene expression of type I IFNs (IFNα subtypes and IFNβ), proinflammatory cytokines (e.g., tumor necrosis factor superfamily proteins and interleukins) and chemokines, for the initiation of the antiviral response.
More recently, cGAS and IFI16 have been identified as intracellular DNA sensors that mediate a type I IFN response via the recruitment and activation of their shared adaptor protein stimulator of IFN genes (STING) (also known as MITA or TMEM173) 8, 9, 10. Following recognition of viral dsDNA –derived from either a DNA virus (e.g., herpesviruses) or retroviral infection – cytoplasmic cGAS catalyzes the formation of the second messenger cGAMP, which binds to and activates STING. The binding of cGAMP to STING results in the translocation of the adaptor protein from the endoplasmic reticulum to the Golgi and perinuclear sites. Activated STING dimerizes and binds to TBK1, resulting in IRF3 activation and IFNα/β gene expression. The sensor IFI16 is reportedly localized in both the cytoplasm and the nucleus and detects viral dsDNA via its hematopoietic expression, IFN-inducible nature, and nuclear localization (HIN) domain followed by signaling to STING to initiate an IFN-mediated antiviral response. In addition to its ability to elicit type I IFN induction, IFI16 also leads to IL-1β secretion via activation of caspase 1-dependent inflammasomes (reviewed in [4]).
PTMs are key regulatory events in which proteins are enzymatically modified to reversibly modulate the activity, subcellular localization, conformation, and/or protein–protein interactions of the target protein. Thus, PTMs are crucial for the dynamic regulation of signal transduction pathways. Each step of the intracellular pathogen sensing pathways is regulated by a complex network of PTMs resulting in a controlled yet effective antiviral cytokine response. In particular, ubiquitination (described in Box 1 ) and phosphorylation events are critical for fine-tuning the RLR-, cGAS-, and IFI16-mediated antiviral response.
Box 1. Protein Ubiquitination Types and Their Functions.
In its simplest form, ubiquitin can be attached to the target protein as a single moiety, resulting in monoubiquitination, or multiubiquitination occurs if several monoubiquitin units are attached. Furthermore, ubiquitin itself can be ubiquitinated, resulting in the formation of ubiquitin chains attached to the target protein, and these oligo- or polyubiquitin moieties can be further subdivided into homotypic (single linkage type) or heterotypic (mixed linkage type) chains. Three enzymes, ubiquitin activating (E1), ubiquitin conjugating (E2), and ubiquitin ligase (E3), act sequentially to covalently attach ubiquitin to a substrate. In particular, substrate specificity is in large part directed by E3 ligases, and mammals encode hundreds of such ligases allowing for specific targeting of ubiquitin modifications. Protein ubiquitination has widespread implications in the regulation of cellular processes including protein processing and trafficking and signal transduction (reviewed in 72, 73).
For polyubiquitination, it is the specific linkage type that determines the fate of the target protein. Polyubiquitin chains are formed when the C-terminal glycine of one ubiquitin molecule forms a covalent bond with one of the seven internal lysine residues of another ubiquitin molecule. This gives rise to seven different polyubiquitination linkage types: K6-, K11-, K27-, K29-, K33-, K48-, and K63-linked ubiquitination. In addition, M1 (or linear) polyubiquitin chains can be formed when the C terminus of one ubiquitin molecule binds to the N-terminal methionine of the other ubiquitin molecule. The first-identified and most well-characterized linkage type is K48-linked polyubiquitination, which primarily functions in proteasomal degradation of proteins. Other linkage types, such as K11- and K29-linked ubiquitin chains, can reportedly also function as marks to discard unwanted proteins via proteasomal degradation. K63-linked polyubiquitination modulates numerous cellular processes including autophagy, innate immunity, DNA repair, and endocytosis. Much less is understood about the function of other, noncanonical polyubiquitination types; however, recent studies suggested that they play a role in regulating diverse cell signaling pathways including NF-κB activation, autophagy, and immunity (reviewed in [72]). For example, K6-linked polyubiquitin has been implicated in mitochondrial homeostasis and mitophagy. In addition to promoting proteolysis, K11-linked polyubiquitin chains regulate cell division and in mixed-linkage chains together with K63-linked ubiquitin trigger MHC I (major histocompatibility complex I) internalization. The role of K27-linked polyubiquitination remains largely enigmatic, while K33-linked chains have been implicated in post-Golgi membrane protein trafficking. Finally, M1-linked ubiquitination events are critical for the positive regulation of NF-κB signal activation and inflammation 74, 75.
Furthermore, a structurally related yet functionally diverse subset of ubiquitin-like proteins (UBLs) has emerged. These UBLs include SUMO, ISG15, FAT10, and NEDD8, which regulate the activity, interactome, or transport of the target protein rather than triggering its proteasomal degradation.
In the case of protein phosphorylation, kinases phosphorylate the target protein typically at a serine, threonine, or tyrosine residue to regulate its function by inducing conformational changes or modulating the specific recruitment of other proteins to the phosphomotif.
Other, less well-studied PTMs that modulate innate immunity are acetylation, glutamylation, and deamidation. Acetylation, a process catalyzed by acetyltransferases, results in the introduction of an acetyl group, typically on lysine residues. This modification occurs commonly in the proteome and plays an important role in numerous biological processes including cell cycle regulation, chromatin remodeling, and immunity. Glutamylation, which was first implicated in tubulin modification, is the addition of a glutamate to the γ-carboxyl group of a glutamic acid in the target protein. Similar to ubiquitination the sequential addition of glutamates to the target protein can lead to a polyglutamate chain. Protein deamidation, the process in which an amide functional group is removed from the target protein (typically at an asparagine or glutamine residue), has been associated with protein degradation. However, recent studies also unveiled a role for deamidation in innate immune signaling. Protein ubiquitination, phosphorylation, acetylation, and glutamylation are reversed by deubiquitinases (DUBs), phosphatases, deacetylases, and deglutamylases, respectively, allowing for tight regulation of pathogen sensing and immune signaling.
In this Review we provide an update on the role of PTMs in the regulation of intracellular sensors of viral infection and their proximal adaptor proteins. PTMs are also critical in the regulation of downstream effectors of the innate immune signaling cascade such as IRF3 and 7, TRAF3, and TBK1, which have previously been reviewed elsewhere. Here we describe in detail the role of phosphorylation, ubiquitination, and other PTMs in both positively and negatively regulating the signaling activity or protein abundance of RLRs, cGAS, and IFI16 as well as their adaptors MAVS and STING. We also discuss how regulatory networks are crucial for launching an effective antiviral response while preventing immunopathology caused by overactivation of innate immunity.
PTMs that Control RLR–MAVS-mediated Signaling
Modulation of RLR Activity by Ubiquitination and Phosphorylation
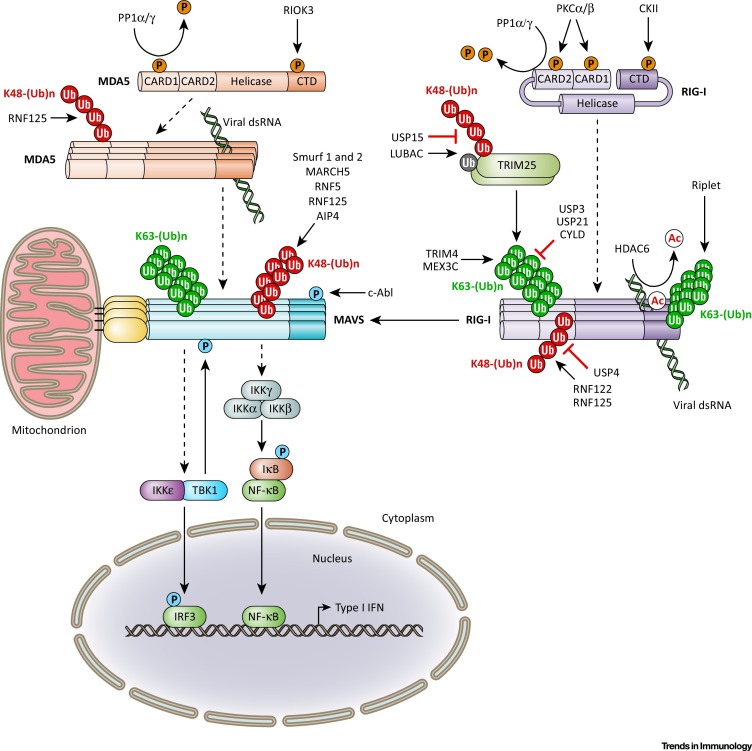
Recent studies demonstrated that RLR–MAVS-dependent signal transduction is precisely controlled by several PTMs. Among them, reversible Ser/Thr phosphorylation and K63- and K48-linked polyubiquitination are critical ‘on/off switch’ signals to control proinflammatory cytokine induction triggered by RLRs (Figure 1 ). A key regulator of the RIG-I-mediated IFN response is the ubiquitin E3 ligase TRIM25, which modifies RIG-I with K63-linked polyubiquitination [11]. TRIM25 [also termed estrogen-responsive finger protein (Efp)] is a member of the TRIM family of RING E3 ligases, which are important regulators of PRR signaling pathways (Box 2 ). RIG-I ubiquitination by TRIM25 induces RIG-I oligomerization and its interaction with MAVS to induce antiviral gene expression. Following viral RNA binding and exposure of the tandem CARDs, TRIM25 is recruited to the first CARD of RIG-I and then attaches covalent K63-linked polyubiquitin chains to K172 in the second CARD 11, 12. Crystallization of the two CARD domains of RIG-I, referred to as RIG-I 2CARD, bound to K63-diubiquitin showed that the CARDs are stabilized into a tetrameric ‘lock-washer’ conformation in which three chains of K63-diubiquitin are bound along the outer rim of the helical trajectory to assemble a stable 2CARD tetramer [13]. In vitro cell-free experiments further showed that unanchored K63-linked polyubiquitin generated by TRIM25 can also activate RIG-I by stabilizing the signaling-active RIG-I tetramer 13, 14, but further investigation is warranted to determine the role and contribution of unanchored K63-polyubiquitin to RIG-I activation in an infected cell. Recent studies also provided important insight into the activation mechanism of TRIM25: TRIM25 dimerization mediated by its RING and coiled-coil domains (CCD) assembles the catalytically active form of TRIM25 essential for RIG-I activation 15, 16.
Figure 1.
Post-translational Modifications (PTMs) that Modulate the Retinoic Acid-Inducible Gene I (RIG-I)-like receptor (RLR)–Mitochondrial Antiviral Signaling Protein (MAVS) Signaling Pathway. Schematic of the signaling pathway triggered by the intracellular viral double-stranded RNA (dsRNA) sensors RIG-I and melanoma differentiation-associated protein 5 (MDA5) and their shared adaptor MAVS localized at the mitochondrion. Activation of this pathway leads to the gene expression of type I interferons (IFNs) and other proinflammatory cytokines via TANK-binding kinase 1 (TBK1) and IKK family kinases and downstream transcription factors, primarily IRF3 and nuclear factor kappa B (NF-κB). This figure further illustrates the PTMs and responsible enzymes that modulate the signaling activity or protein abundance of RIG-I, MDA5, and MAVS. The details of the post-translational control mechanisms and specific residues modified in RIG-I, MDA5, and MAVS are described in the text. Unbroken arrows indicate direct effects or signaling events. Broken arrows indicate events that are indirect or that have not yet been completely established. Abbreviations: Ac, acetylation; CARD, caspase activation and recruitment domain; CTD, carboxy-terminal domain; P, phosphate; Ub, ubiquitin; K63-(Ub)n, K63-linked ubiquitination; K48-(Ub)n, K48-linked ubiquitination. Red/orange signifies inhibitory PTMs; blue/green indicates positive regulatory PTMs.
Box 2. Regulation of Innate Immunity by TRIM E3 Ligases.
TRIM (tripartite motif) family proteins are characterized by the RBCC motif, which comprises an N-terminal RING (really interesting new gene) domain that confers E3 ligase enzymatic activity, one or two B-box domains of largely unknown function, and a central CCD that mediates homo- or hetero-oligomerization. In addition, most TRIM proteins have a unique C-terminal domain (e.g., a SPRY, PHD-Bromo, or ARF domain) that mediates specific protein–protein interactions or has enzymatic activity 76, 77. Recent studies demonstrated that many of the ∼80 human TRIM proteins are important regulators of PRR-mediated signal transduction and cytokine responses [78], with TRIM25 being the first TRIM protein identified that directly targets an innate immune sensor; namely, the intracellular viral RNA sensor RIG-I [11]. TRIM25 and several other TRIM proteins promote PRR signaling through catalysis of non-degradative polyubiquitin linkage types (e.g., K63- or K33-linked ubiquitination), which facilitates the multimerization, stabilization, and/or protein–protein interactions of the target protein, ultimately enhancing its signal-transducing activity (reviewed in [79]). Some TRIM proteins also negatively regulate PRR-mediated cytokine induction; in these cases by mediating classical degradative K48-linked ubiquitination of substrate proteins and targeting them for proteasomal degradation. Besides their regulatory roles in innate signaling pathways, TRIM proteins act as antiviral restriction factors by directly targeting viral components 76, 77. For example, TRIM5α targets the capsid of many retroviruses, including HIV-1, and TRIM19 [also known as promyelocytic leukemia (PML)] inhibits the replication of many RNA viruses and DNA viruses as part of PML nuclear bodies/ND10 compartments. Recent studies indicated that some TRIM proteins play dual roles by acting both as ‘effectors’ that neutralize viral infection and as ‘sensors’ that induce innate signal transduction. For example, TRIM21 recognizes antibody-opsonized virus particles leading to proteasomal degradation of the virus [80]. In addition, following recognition of the viral pathogen, TRIM21 initiates a signaling cascade that leads to NF-κB activation and gene expression of proinflammatory cytokines [81]. Finally, some TRIM proteins regulate fundamental cellular processes such as gene transcription, the cell cycle, or autophagy that are associated with diverse pathological conditions (e.g., neurodegenerative disorders and cancer), but can also influence innate signaling and antiviral defense (reviewed in 82, 83).
Besides TRIM25, several other E3 ligases have been identified that modify RIG-I with K63-polyubiquitin chains, including Riplet/RNF135, TRIM4, and MEX3C. Furthermore, several host-encoded DUBs remove K63-linked polyubiquitin from RIG-I to balance proinflammatory responses (described in Table 1 ). The importance of covalent K63-linked ubiquitination of RIG-I for its activation is further supported by the finding that several viruses have evolved ways to inhibit the K63-polyubiquitin-dependent activation of RIG-I. These strategies can be grouped into two major categories: (i) viral targeting of the E3 ligases TRIM25 and Riplet; and (ii) virus-encoded DUBs that actively cleave the RIG-I K63-linked ubiquitin chains (described in more detail in Box 3 ).
Table 1.
Overview of the Regulation of Intracellular Innate Sensors and Their Adaptors by Ubiquitination and Deubiquitinationa
| Modifying enzyme | Type of ubiquitination | Target molecule | Regulatory effect | Refs | |
|---|---|---|---|---|---|
| E3 ubiquitin ligases | TRIM25 | K63-linked ubiquitination | RIG-I | Promotes RIG-I oligomerization, RIG-I–MAVS interaction and thereby antiviral gene expression | 11, 12, 13, 14 |
| Riplet/RNF135 | K63-linked ubiquitination | RIG-I | Promotes RIG-I signaling, likely through opening up the RIG-I conformation and facilitating TRIM25-mediated ubiquitination of the CARDs | 58, 59, 60 | |
| TRIM4 | K63-linked ubiquitination | RIG-I | Promotes RIG-I signaling and IFN gene expression | [61] | |
| MEX3C | K63-linked ubiquitination | RIG-I | Promotes RIG-I signaling and IFN gene expression | [62] | |
| RNF122 | K48-linked ubiquitination | RIG-I | Induces proteasomal degradation of RIG-I, dampening the innate immune response | [27] | |
| RNF125 | K48-linked ubiquitination | RIG-I, MDA5, MAVS | Induces proteasomal degradation of RIG-I, MDA5, and MAVS, suppressing IFN induction | 25, 26 | |
| LUBAC | Monoubiquitination and M1- and K48-linked ubiquitination | TRIM25 | Induces TRIM25 degradation, dampening TRIM25–RIG-I signaling as a negative feedback mechanism | [63] | |
| ? | K63-linked ubiquitination | MAVS | Facilitates recruitment of IKKɛ and thereby NF-κB activation and IFNβ induction | [37] | |
| Smurf1 and 2 | K48-linked ubiquitination | MAVS | Induces proteasomal degradation of MAVS | 64, 65 | |
| RNF5 | K48-linked ubiquitination | MAVS | Induces proteasomal degradation of MAVS | [66] | |
| AIP4/ITCH | K48-linked ubiquitination | MAVS | Induces proteasomal degradation of MAVS | [67] | |
| MARCH5 | K48-linked ubiquitination | MAVS | Induces proteasomal degradation of MAVS | [38] | |
| TRIM32 | K63-linked ubiquitination | STING | Promotes TBK1 recruitment and thereby STING-mediated IFN induction | [48] | |
| TRIM56 | K63-linked ubiquitination | STING | Promotes TBK1 recruitment and thereby STING-mediated IFN induction | [47] | |
| AMFR | K27-linked ubiquitination | STING | Promotes TBK1 recruitment and thereby STING-mediated IFN induction | [49] | |
| RNF5 | K48-linked ubiquitination | STING | Promotes STING degradation | [50] | |
| RNF26 | K11-linked ubiquitination | STING | Prevents STING degradation, thereby promoting STING signaling | [51] | |
| DUBs | USP3 | Removal of K63-linked ubiquitination | RIG-I | Downregulates RIG-I-mediated IFN induction as a negative feedback mechanism | [68] |
| CYLD | Removal of K63-linked ubiquitination | RIG-I, TBK1, IKKɛ | Prevents premature innate immune activation in uninfected cells | 69, 70 | |
| USP21 | Removal of K63-linked ubiquitination | RIG-I | Downregulates the RIG-I-mediated IFN response during viral infection | [71] | |
| USP4 | Removal of K48-linked ubiquitination | RIG-I | Stabilizes RIG-I and thereby prolongs IFN production | [28] | |
| USP15 | Removal of K48-linked ubiquitination | TRIM25 | Stabilizes TRIM25 and thereby induces a sustained cytokine response | [29] | |
| EIF3S5 | Removal of K48-linked ubiquitination | STING | Stabilizes STING | [52] |
a This table summarizes the E3 ubiquitin ligases and DUBs that regulate RLR–MAVS- or STING-mediated immune responses. For each modifying enzyme, the specific linkage type of ubiquitination catalyzed or removed is noted along with the target protein and the resulting effect on the antiviral innate immune response.
Box 3. Viral Manipulation of PTMs to Dysregulate Innate Signaling.
The essential role of PTMs in regulating PRR-mediated immunity is underscored by findings that many viral pathogens manipulate the PTMs of PRRs or their downstream signaling molecules for efficient virus replication (reviewed in detail in [84]). Several viruses target the ubiquitin E3 ligases TRIM25 and Riplet to block the K63-polyubiquitin-dependent activation of RIG-I. Influenza virus utilizes its non-structural protein 1 (NS1), which directly interacts with TRIM25 and Riplet thereby blocking the K63-ubiquitination of RIG-I 85, 86. The protease NS3/4A of HCV cleaves Riplet, thereby inhibiting RIG-I activation [59]. Dengue virus (DenV) blocks the TRIM25-mediated RIG-I ubiquitination in a viral protein-independent manner: the subgenomic RNA of an epidemic strain of DenV, termed ‘sfRNA’, binds to TRIM25 and interferes with its stabilization by USP15 [87]. Other viruses, such as Kaposi's sarcoma-associated herpesvirus (KSHV), severe acute respiratory syndrome coronavirus (SARS-CoV), foot-and-mouth disease virus, nairoviruses, and arteriviruses, encode DUBs to actively remove K63-polyubiquitin chains from RIG-I (reviewed in [84]). Measles and Nipah viruses, which are members of the Paramyxoviridae family, prevent the PP1α/γ-dependent dephosphorylation of RLRs through two different mechanisms. The non-structural V protein from these viruses interacts with and sequesters PP1α/γ thereby inhibiting MDA5 S88 dephosphorylation and hence the activation of this receptor [88]. In dendritic cells, binding of measles virus to DC-SIGN prevents the assembly of an active PP1 complex thereby blocking both RIG-I and MDA5 dephosphorylation [89]. Another escape strategy of viruses is to induce the K48-linked ubiquitin-dependent proteasomal degradation of RLRs or downstream molecules. For example, Sendai virus and VSV upregulate the lectin family protein Siglec-G thereby promoting the interaction of the E3 ligase c-Cbl with RIG-I, inducing its degradation [90], while SARS-CoV usurps AIP4 to degrade MAVS [91].
Analogous to the blocking of RIG-I K63-linked ubiquitination, several viral pathogens manipulate the K63-linked polyubiquitination of STING to avoid recognition through intracellular DNA sensing pathways. For example, the polymerase of hepatitis B virus (HBV) and the papain-like proteases of several coronaviruses bind to STING and prevent its K63-linked ubiquitination to evade antiviral immunity (reviewed in [84]). IFI16 signaling is antagonized by the pUL97 kinase of human cytomegalovirus (HCMV) [92]. pUL97 interacts with and phosphorylates IFI16 thereby inducing translocation of IFI16 from the nucleus, the site of sensing of HCMV DNA, to multivesicular bodies in the cytoplasm.
In contrast to RIG-I, studies supporting K63-linked ubiquitination of the MDA5 CARDs are conflicting 14, 17. Given that MDA5 cooperatively assembles into a filament along the length of dsRNA to initiate innate signaling (reviewed in [18]), it is likely that MDA5 activation, in contrast to that of RIG-I, does not require K63-polyubiquitin modification for multimerization and signal amplification.
While K63-linked ubiquitination leads to RIG-I activation following virus infection, Ser/Thr phosphorylation of the CARD and CTD keeps RIG-I and also MDA5 in the signaling-repressed state in uninfected cells 19, 20, 21. RIG-I CARD phosphorylation at S8 and T170 induced by PKCα/β, and at S788 in the CTD mediated by casein kinase II (CKII), impedes RIG-I activation in uninfected cells 22, 23. Similarly, MDA5 phosphorylation at S88 (in the first CARD) and S828 (in the CTD), the latter mediated by RIO kinase 3 (RIOK3), keeps MDA5 inactive in uninfected cells 21, 24. Following viral infection, RIG-I and MDA5 form a complex with the phosphatase PP1α or PP1γ, which induces dephosphorylation of the RIG-I and MDA5 CARDs [21]. Biochemical studies showed that the dephosphorylated CARDs of RIG-I and MDA5 have a high binding affinity for MAVS and efficiently trigger IFN induction 21, 22. Dephosphorylation is likely to induce a conformational change within the tandem CARDs allowing binding of effector proteins. In support of this, mutants of RIG-I in which S8 and T170 are mutated to non-phosphorylatable alanine residues strongly bind to TRIM25 and exhibit robust CARD ubiquitination; by contrast, phosphomimetic RIG-I CARD mutants had a low binding affinity for TRIM25 and were not efficiently modified with K63-linked polyubiquitin 19, 20, 22.
The protein stability of RLRs and their upstream regulatory proteins is also modulated by classical K48-linked ubiquitination, which leads to their degradation by the proteasome (Figure 1 and Table 1). Several E3 ligases including RNF122 and RNF125 have been implicated in the degradation of these innate immune molecules, attenuating RLR signal transduction 25, 26, 27. Conversely, several DUBs stabilize RIG-I, MDA5, and MAVS, or critical regulatory proteins upstream of RLRs, by removing K48-linked ubiquitin chains (described in Table 1). For example, USP4 stabilizes RIG-I through removal of K48-linked polyubiquitin, thereby prolonging IFN production [28]. Moreover, a recent study identified the USP15–TRIM25 dyad as a pivotal regulatory complex in proinflammatory and antiviral responses [29]. Mechanistically, USP15 removes K48-linked ubiquitination from the TRIM25 SPRY domain thereby stabilizing TRIM25 and inducing a sustained cytokine response.
Modulation of RLR Activity by Ubiquitin-like Proteins, Acetylation, and Deamidation
RIG-I is also modified with UBL proteins that regulate its innate signaling function. Modification of RIG-I with ISG15 reportedly inhibits RIG-I signaling, while SUMOylation promotes innate signal activation by RIG-I and also MDA5 30, 31, 32. Furthermore, HLA-F adjacent transcription 10 (FAT10), a UBL protein that is uniquely found in mammals, dampens the RIG-I-mediated proinflammatory response through noncovalent binding to the activated RIG-I 2CARD, sequestering RIG-I from mitochondria [33]. More detailed insights into the physiological role of UBL modifications of RLRs in controlling antiviral and proinflammatory responses are required, however.
Recently, two independent studies demonstrated that RIG-I undergoes reversible acetylation, which regulates RIG-I-mediated immunity to RNA virus infection 34, 35. Acetylation of RIG-I at its CTD (with K909 as the primary acetylation site) inhibits the ability of RIG-I to sense viral RNA and form signaling-active homo-oligomers. Inversely, deacetylation of RIG-I by histone deacetylase 6 (HDAC6) promotes RIG-I signaling and antiviral restriction, unveiling HDAC6 as an important upstream regulator of RIG-I. It is possible that acetylation of K909 affects specific hydrogen bonds formed between RIG-I and the 5′-triphosphate group of RNA ligands thereby preventing RNA recognition by RIG-I. Conversely, binding of a ligand may facilitate the open RIG-I conformation, allowing HDAC6 binding and, ultimately, initiation of innate signaling. These studies also examined the role of HDAC6 in the antiviral response to RNA virus infection. Cells depleted of endogenous HDAC6 exhibited enhanced permissiveness for hepatitis C virus (HCV) or influenza virus and reduced innate immune responses. HDAC6-deficient mice exhibited more rapid onset of disease during acute infection with West Nile virus and greater replication of vesicular stomatitis virus (VSV) compared with wild-type mice.
A midotransferase-mediated deamidation, an understudied protein modification in the immune response, has been shown to regulate RIG-I signaling during herpesvirus infection [36]. In this study the authors identified RIG-I as an interaction partner of the herpesviral glutamine amidotransferase (vGAT) by mass spectrometry. vGAT, which lacks intrinsic amidotransferase activity, recruits the cellular phosphoribosylformylglycinamidine synthetase (PFAS) and together the two proteins induce the deamidation of RIG-I at Q10, N245, and N445. Deamidation of RIG-I by the vGAT–PFAS complex leads to the activation of RIG-I, which is likely to occur through a conformational change caused by the conversion of these neutral glutamine and asparagine residues to negatively charged glutamate or aspartate. Intriguingly, deamidation-mediated RIG-I activation is hijacked by herpesviruses to evade innate immunity. Viral pathogens also manipulate other PTMs in RIG-I and also MDA5 to dampen cell-intrinsic immunity (Box 3).
Modulation of MAVS-mediated Signaling by Ubiquitination and Phosphorylation
K63-linked polyubiquitination is also a critical modification in the control of MAVS signaling activity (Figure 1 and Table 1). Following RNA virus infection, K500 in MAVS is conjugated with K63-linked polyubiquitin, which leads to the recruitment of IKKɛ to mitochondria, ultimately inducing NF-κB activation and IFNβ transcriptional activation [37].
MAVS protein stability is intricately regulated by K48-linked ubiquitination and several ubiquitin E3 ligases have been identified that induce the ubiquitin-dependent degradation of MAVS, including RNF5, AIP4/ITCH, and SMAD ubiquitin regulatory factor (Smurf) 1 and 2 (see also Table 1). Furthermore, the mitochondria-resident E3 ligase MARCH5 was identified as a MAVS regulator [38]. Following viral infection, MARCH5 binds to MAVS aggregates and modifies K7 and K500 in MAVS with K48-linked polyubiquitin, promoting the proteasomal degradation of the adaptor. MARCH5-deficient immune cells exhibited elevated IFN responses and lower RNA virus replication compared with wild-type cells [38].
At least one protein has been identified that counteracts the K48-linked ubiquitination of MAVS, stabilizing it for sustained IFN production [39]. TRIM44, which is an atypical TRIM protein because it has no RING E3 ligase domain but has a ZF-UBP domain, inhibits the PCBP2/AIP4-induced ubiquitination of MAVS. However, the precise mechanism by which TRIM44 blocks MAVS ubiquitination and the role of its ZF-UBP domain, which may have putative DUB activity, remains to be determined.
MAVS is further regulated by reversible phosphorylation/dephosphorylation (Figure 1). In contrast to RIG-I and MDA5, for which serine/threonine phosphorylation inhibits their signal-transducing activities, MAVS phosphorylation at two conserved serine and threonine clusters by TBK1 or IKKs promotes innate signaling [40]. The negative charge induced by phosphorylation allows MAVS to bind to a positively charged surface in IRF3, facilitating IRF3 recruitment and its activation by TBK1. Intriguingly, the phosphorylation-dependent activation mechanism is also conserved in the innate adaptor proteins STING and TRIF, promoting cGAS- and TLR-mediated signaling [40]. Furthermore, the tyrosine kinase c-Abl was identified as a positive regulator of MAVS-dependent signaling, as silencing or pharmacological inhibition of c-Abl abrogated antiviral IFNβ production triggered by MAVS [41]. Although the precise residue in MAVS that is phosphorylated by c-Abl remains to be determined, T9 may be a potential phosphorylation candidate site as it has recently been identified as a critical residue that promotes MAVS activation and propagation of antiviral signaling [42].
Post-translational Control of cGAS-, IFI16-, and STING-mediated Signaling
Modulation of cGAS and IFI16 Activity by Phosphorylation, Glutamylation, and Acetylation
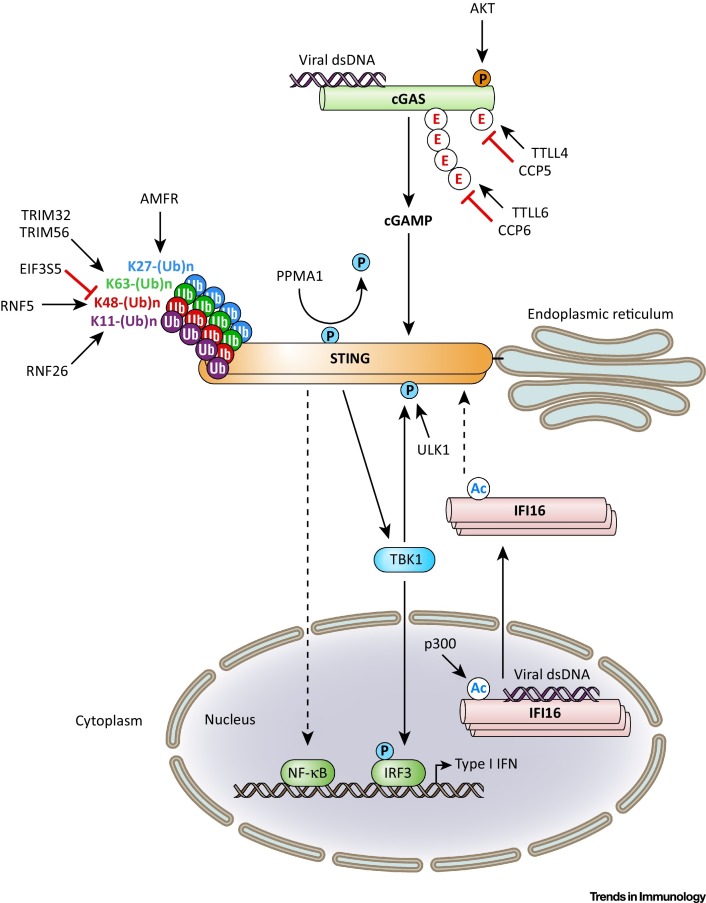
Regulatory PTMs also play a crucial role in DNA sensing pathways, controlling the sensing and/or signaling activity of intracellular receptors and their adaptor protein STING (Figure 2 ). Compared with RLR regulation by PTMs, much less is known about the PTMs that directly modulate the activity of cGAS or IFI16; however, recent studies have identified an important role of phosphorylation, acetylation, and glutamylation in controlling these sensors. Similarly to RLR regulation by kinase-dependent serine/threonine phosphorylation, which renders RIG-I and MDA5 inactive, phosphorylation of cGAS at S291 (mouse) or S305 (human) by the kinase Akt [also known as protein kinase B (PKB)] negatively impacts the antiviral immune response by suppressing cGAS enzymatic activity [43]. This phosphorylation-dependent dampening mechanism is usurped by viruses to inhibit IFNβ-dependent antiviral responses. Specifically, infection by herpes simplex virus 1 (HSV-1) activates Akt leading to cGAS phosphorylation and inhibition. A phosphorylation-resistant cGAS mutant protein effectively promoted IFNβ production and restricted HSV-1 replication [43]. Furthermore, poly- and monoglutamylation of cGAS by tubulin tyrosine ligase-like enzyme (TTLL) 6 and 4, respectively, negatively regulates the cGAS-triggered antiviral response through distinct mechanisms: polyglutamylation of cGAS at E272 inhibits its DNA-binding capacity, whereas monoglutamylation at E302 prevents efficient synthase activity [44]. Conversely, the carboxypeptidases CCP5 and CCP6 perform nonredundant functions to remove the monoglutamate or polyglutamate moieties synthesized by TTLL4 and TTLL6, respectively.
Figure 2.
Post-translational Modifications that Control Innate Immunity Mediated by Cyclic GMP–AMP (cGAMP) Synthase (cGAS), Interferon Gamma (IFNγ)-Inducible Protein 16 (IFI16), and Stimulator of IFN Genes (STING). This figure illustrates the post-translational modifications and regulatory enzymes that play a critical role in balancing the innate immune response mediated by the intracellular DNA sensors cGAS and IFI16 as well as their adaptor protein STING. Following recognition of viral double-stranded DNA (dsDNA), cGAS produces the second messenger cGAMP, which then binds to and induces STING activation characterized by dimerization and its translocation from the endoplasmic reticulum (ER) to the Golgi and perinuclear sites (not shown). Similarly, in response to viral DNA binding IFI16 oligomerizes and translocates from the nucleus to the cytoplasm to activate STING. The two signals then result in the activation of TANK-binding kinase 1 (TBK1) and IRF3 as well as nuclear factor kappa B (NF-κB), evoking the gene expression of type I IFNs. Many of the described steps in cGAS, IFI16, and STING activation are delicately controlled by regulatory enzymes that mediate the reversible phosphorylation, ubiquitination, glutamylation, or acetylation of these proteins. Unbroken arrows indicate direct effects or signaling events. Broken arrows indicate events that are indirect or that have not yet been completely established. Abbreviations: Ac, acetylation; E, glutamylation; P, phosphate; Ub, ubiquitin; K11-(Ub)n, K11-linked ubiquitination; K27-(Ub)n, K27-linked ubiquitination; K48-(Ub)n, K48-linked ubiquitination; K63-(Ub)n, K63-linked ubiquitination. Red/orange signifies inhibitory PTMs; blue/green/purple indicates positive regulatory PTMs.
PTMs can also modulate the subcellular localization of intracellular viral DNA sensors. Acetylation of the nuclear localization signal of IFI16 promotes the translocation of IFI16 from the nucleus to the cytoplasm 45, 46. Acetylation of IFI16 is mediated by the acetyltransferase p300 following recognition of herpesvirus genomes in the nucleus. Acetylated IFI16 induces STING activation and IFNβ production. Furthermore, acetylation of IFI16 is critical for its association with inflammasomes and ensuing IL-1β production [46].
Modulation of STING Activity by Phosphorylation and Different Linkage Types of Polyubiquitin
A recent series of studies demonstrated that STING is also extensively modified by PTMs, including degradative and non-degradative polyubiquitin types as well as phosphorylation (Figure 2); however, how these different modifications dynamically regulate the activity of STING is not yet completely understood.
The E3 ubiquitin ligases TRIM32 and TRIM56 target STING for K63-linked ubiquitination at K150 (additionally, TRIM32 targets K20, K224, and K236), facilitating the recruitment of TBK1 to STING as a means of positive regulation 47, 48. Overexpression of both of these E3 ligases enhanced STING-dependent IFNβ production while knockout of either abrogated cytokine responses. Thus, the relative contribution of TRIM32 and TRIM56 to STING activation, and whether the two enzymes act in concert or in a temporally distinct fashion, remains to be elucidated. Another atypical polyubiquitination type that positively regulates STING activity is K27-linked polyubiquitination catalyzed by the autocrine motility factor receptor (AMFR) [49]. AMFR in complex with insulin-induced gene 1 (INSIG1) modifies STING such that TBK1 is recruited. Conversely, STING signaling is dampened by RNF5, which modifies STING at K150 with K48-linked polyubiquitin, promoting STING degradation at the mitochondria during the course of viral infection [50]. RNF26 catalyzes K11-linked polyubiquitination of STING at the same residue, competing with RNF5 to ubiquitinate STING [51]. Thus, ubiquitination of K150 by RNF26 prevented the attachment of K48-polyubiquitin chains by RNF5 and thereby STING degradation.
As ubiquitination is reversible in nature, DUBs also modulate the signaling activity of STING. Inactive rhomboid protein 2 (iRhom2) stabilizes STING by recruiting eukaryotic translation initiation factor 3, subunit 5 (EIF3S5), which functions as a DUB to remove K48-linked polyubiquitin from STING [52]. iRhom2 reportedly stimulates STING signaling through a second mechanism that is not dependent on modulating PTMs of STING; it promotes trafficking of STING from the endoplasmic reticulum to perinuclear membrane regions by facilitating the interaction of STING with the translocon-associated protein TRAPβ.
Serine phosphorylation further fine-tunes the signaling activity of STING, and at least two kinases – TBK1 and UNC51-like kinase-1 (ULK1) – reportedly mediates STING phosphorylation. Phosphorylation of STING at residue S366 by TBK1 leads to IRF3 binding and activation 40, 53; however, phosphorylation of activated STING at the same residue by ULK1 reportedly suppresses IRF3 activation [54]. Further research is warranted to resolve these apparently conflicting results and to investigate whether specific regulatory molecules associated with each of the two kinases, or additional modifications on STING that occur simultaneously with S366 phosphorylation, determine the functional outcome of STING S366 phosphorylation.
At least one additional residue in STING, residue S358, has been reported to undergo phosphorylation, although the kinase responsible for this phosphorylation event is currently unknown [55]. Protein phosphatase, Mg2+/Mn2+ dependent 1A (PPM1A) dephosphorylates STING at S358 to prevent STING aggregation and antiviral signaling, which supports the concept that phosphorylation plays a positive role in STING activation [55]. Recently, it was shown that the ribosomal protein S6 kinase 1 (S6K1) promotes the signal transduction of the cGAS–STING–TBK1–IRF3 axis [56]. S6K1 interacts with phosphorylated STING and TBK1 in a cGAS–cGAMP-dependent manner to form a ternary signaling complex that is necessary for IRF3 activation. Although the kinase activity of S6K1 was not required for this mechanism, the kinase domain was critical for the STING- and TBK1-dependent phosphorylation of IRF3 and activation of antiviral immunity.
Intriguingly, in addition to these host mechanisms to modulate intracellular DNA surveillance pathways, viral pathogens manipulate the PTMs of cGAS, IFI16, or STING to escape immune recognition (Box 3).
Concluding Remarks
Regulation of the innate immune system is coordinated by a myriad of host enzymes that modify key innate signaling molecules to fine-tune antiviral and proinflammatory cytokine responses. In many cases these regulatory enzymes (e.g., E3 ligases, kinases, phosphatases, acetyltransferases) not only regulate innate immunity but fulfill many other functions in the cell. Therefore, it will be important to address what determines the specificity of these enzymes towards their innate target proteins. Furthermore, several innate immune sensors, such as RLRs, are already modified by PTMs in uninfected cells, suggesting that these modifications maintain immune homeostasis before virus infection. Following viral infection, ‘stimulatory signals’ must exist that activate the enzymes responsible for removing these inhibitory ‘marks’. It will be important to identify the specific stimuli that activate upstream immunomodulatory enzymes during viral infection. Moreover, while K63-linked ubiquitination has emerged as a pivotal modification regulating innate signaling, little is known about other, noncanonical polyubiquitin linkage types that may also play important roles in regulatory immune circuits (see Outstanding Questions). As most molecular mechanisms of immunoregulation have been studied in human cells, it remains to be addressed whether the same or similar PTMs and enzymes regulate the respective innate sensing pathways in different species. Related to this point, it is still unknown whether regulatory mechanisms are common or differ between different cell types. Finally, for a given innate signaling protein many PTMs have been reported; however, how they dynamically work together to modulate target protein activity needs further investigation.
The multifaceted control of immune surveillance pathways is crucial to prevent uncontrolled cytokine production, which may result in premature or constant immune activation and potentially harmful consequences for the host organism. While SNPs in key molecules of pathogen recognition pathways have been associated with several autoimmune diseases, including systemic lupus erythematosus and interferonopathies such as Aicardi–Goutières syndrome [57], it is largely unknown whether proinflammatory disorders can arise from mutations that directly affect PTMs or the interaction or expression of regulatory enzymes. It is tempting to speculate that our knowledge of the regulatory enzymes and PTMs that are critically involved in the regulation of innate sensing pathways may be translated into means of boosting cytokine responses as a potential therapeutic strategy against viral infection.
Outstanding Questions.
What role do less well-characterized non-degradative polyubiquitin linkage types (e.g., K6-, K27-, K29-, and K33-linked ubiquitination) play in the regulation of pathogen sensing pathways? Do glycosylation, acylation, and palmitoylation also regulate innate signaling?
How do PTMs ‘crosstalk’ to each other to dynamically activate or turn off the signal transduction pathways initiated by innate immune sensors?
For a given innate immune protein, do specific PTMs act in a temporal manner? Do the signal strength and the type of stimulus (e.g., a specific virus) determine which PTM event occurs and when?
What is the physiological role of the individual upstream regulatory enzymes in different species and cell types?
How is the activity of regulatory enzymes (e.g., kinases, ubiquitin E3 ligases, DUBs) modulated? What are the stimuli that activate upstream immunomodulatory enzymes during viral infection?
Are SNPs in key regulatory enzymes associated with autoimmune diseases or interferonopathies or do they lead to the differential susceptibility of humans to viral infection?
How can our knowledge of immunomodulatory circuits in intracellular innate sensing pathways be translated into the development of therapeutics for infectious diseases or disorders caused by dysregulation of proinflammatory responses?
Acknowledgments
The authors apologize to all colleagues for not being able to discuss and cite all papers related to this topic due to space constraints. Work in the Gack laboratory is funded by National Institutes of Health (NIH) grants R01 AI087846, R01 AI127774, and R21 AI118509 as well as an award from the PML Consortium.
References
- 1.Goubau D. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J., Chen Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Dempsey A., Bowie A.G. Innate immune recognition of DNA: a recent history. Virology. 2015;479-480:146–152. doi: 10.1016/j.virol.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlee M. Master sensors of pathogenic RNA – RIG-I like receptors. Immunobiology. 2013;218:1322–1335. doi: 10.1016/j.imbio.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vazquez C., Horner S.M. MAVS coordination of antiviral innate immunity. J. Virol. 2015;89:6974–6977. doi: 10.1128/JVI.01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H.M. The mitochondrial targeting chaperone 14-3-3ɛ regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe. 2012;11:528–537. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L. Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unterholzner L. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gack M.U. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 12.Gack M.U. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peisley A. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez J.G. Mechanism of TRIM25 catalytic activation in the antiviral RIG-I pathway. Cell Rep. 2016;16:1315–1325. doi: 10.1016/j.celrep.2016.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez J.G. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2494–2499. doi: 10.1073/pnas.1318962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu B. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 18.del Toro Duany Y. MDA5-filament, dynamics and disease. Curr. Opin. Virol. 2015;12:20–25. doi: 10.1016/j.coviro.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gack M.U. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J. Virol. 2010;84:3220–3229. doi: 10.1128/JVI.02241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nistal-Villan E. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J. Biol. Chem. 2010;285:20252–20261. doi: 10.1074/jbc.M109.089912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wies E. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maharaj N.P. Conventional protein kinase C-alpha (PKC-α) and PKC-β negatively regulate RIG-I antiviral signal transduction. J. Virol. 2012;86:1358–1371. doi: 10.1128/JVI.06543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Z. Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J. Virol. 2011;85:1036–1047. doi: 10.1128/JVI.01734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takashima K. RIOK3-mediated phosphorylation of MDA5 interferes with its assembly and attenuates the innate immune response. Cell Rep. 2015;11:192–200. doi: 10.1016/j.celrep.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 25.Arimoto K-I. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao Q. A non-canonical role of the p97 complex in RIG-I antiviral signaling. EMBO J. 2015;34:2903–2920. doi: 10.15252/embj.201591888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W. RNF122 suppresses antiviral type I interferon production by targeting RIG-I CARDs to mediate RIG-I degradation. Proc. Natl. Acad. Sci. U.S.A. 2016;113:9581–9586. doi: 10.1073/pnas.1604277113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L. USP4 positively regulates RIG-I-mediated antiviral response through deubiquitination and stabilization of RIG-I. J. Virol. 2013;87:4507–4515. doi: 10.1128/JVI.00031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pauli E-K. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci. Signal. 2014;7:ra3. doi: 10.1126/scisignal.2004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim M.J. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J. Virol. 2008;82:1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi Z. SUMOylation of RIG-I positively regulates the type I interferon signaling. Protein Cell. 2010;1:275–283. doi: 10.1007/s13238-010-0030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu J. MDA5 is SUMOylated by PIAS2β in the upregulation of type I interferon signaling. Mol. Immunol. 2011;48:415–422. doi: 10.1016/j.molimm.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen N.T. Ubiquitin-like modifier FAT10 attenuates RIG-I mediated antiviral signaling by segregating activated RIG-I from its signaling platform. Sci. Rep. 2016;6:23377. doi: 10.1038/srep23377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi S.J. HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG-I. EMBO J. 2016;35:429–442. doi: 10.15252/embj.201592586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H.M. Regulation of retinoic acid inducible gene-I (RIG-I) activation by the histone deacetylase 6. EBioMedicine. 2016;9:195–206. doi: 10.1016/j.ebiom.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He S. Viral pseudo-enzymes activate RIG-I via deamidation to evade cytokine production. Mol. Cell. 2015;58:134–146. doi: 10.1016/j.molcel.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paz S. Ubiquitin-regulated recruitment of IκB kinase epsilon to the MAVS interferon signaling adapter. Mol. Cell. Biol. 2009;29:3401–3412. doi: 10.1128/MCB.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoo Y.S. The mitochondrial ubiquitin ligase MARCH5 resolves MAVS aggregates during antiviral signalling. Nat. Commun. 2015;6:7910. doi: 10.1038/ncomms8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang B. Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J. Immunol. 2013;190:3613–3619. doi: 10.4049/jimmunol.1202507. [DOI] [PubMed] [Google Scholar]
- 40.Liu S. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 41.Song T. c-Abl tyrosine kinase interacts with MAVS and regulates innate immune response. FEBS Lett. 2010;584:33–38. doi: 10.1016/j.febslet.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 42.Wen C. Identification of tyrosine-9 of MAVS as critical target for inducible phosphorylation that determines activation. PLoS One. 2012;7:e41687. doi: 10.1371/journal.pone.0041687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo G.J. Akt kinase-mediated checkpoint of cGAS DNA sensing pathway. Cell Rep. 2015;13:440–449. doi: 10.1016/j.celrep.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia P. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat. Immunol. 2016;17:369–378. doi: 10.1038/ni.3356. [DOI] [PubMed] [Google Scholar]
- 45.Li T. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl. Acad. Sci. U.S.A. 2012;109:10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ansari M.A. Herpesvirus genome recognition induced acetylation of nuclear IFI16 is essential for its cytoplasmic translocation, inflammasome and IFN-β responses. PLoS Pathog. 2015;11:e1005019. doi: 10.1371/journal.ppat.1005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuchida T. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity. 2014;41:919–933. doi: 10.1016/j.immuni.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 50.Zhong B. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 51.Qin Y. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog. 2014;10:e1004358. doi: 10.1371/journal.ppat.1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo W.W. iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat. Immunol. 2016;17:1057–1066. doi: 10.1038/ni.3510. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka Y., Chen Z.J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konno H. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z. PPM1A regulates antiviral signaling by antagonizing TBK1-mediated STING phosphorylation and aggregation. PLoS Pathog. 2015;11:e1004783. doi: 10.1371/journal.ppat.1004783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang F. S6K–STING interaction regulates cytosolic DNA-mediated activation of the transcription factor IRF3. Nat. Immunol. 2016;17:514–522. doi: 10.1038/ni.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barrat F.J. Importance of nucleic acid recognition in inflammation and autoimmunity. Annu. Rev. Med. 2016;67:323–336. doi: 10.1146/annurev-med-052814-023338. [DOI] [PubMed] [Google Scholar]
- 58.Oshiumi H. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J. Biol. Chem. 2009;284:807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 59.Oshiumi H. A distinct role of Riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 2013;9:e1003533. doi: 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oshiumi H. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe. 2010;8:496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 61.Yan J. TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J. Mol. Cell Biol. 2014;6:154–163. doi: 10.1093/jmcb/mju005. [DOI] [PubMed] [Google Scholar]
- 62.Kuniyoshi K. Pivotal role of RNA-binding E3 ubiquitin ligase MEX3C in RIG-I-mediated antiviral innate immunity. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5646–5651. doi: 10.1073/pnas.1401674111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inn K-S. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol. Cell. 2011;41:354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y. Ndfip1 negatively regulates RIG-I-dependent immune signaling by enhancing E3 ligase Smurf1-mediated MAVS degradation. J. Immunol. 2012;189:5304–5313. doi: 10.4049/jimmunol.1201445. [DOI] [PubMed] [Google Scholar]
- 65.Pan Y. Smurf2 negatively modulates RIG-I-dependent antiviral response by targeting VISA/MAVS for ubiquitination and degradation. J. Immunol. 2014;192:4758–4764. doi: 10.4049/jimmunol.1302632. [DOI] [PubMed] [Google Scholar]
- 66.Zhong B. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J. Immunol. 2010;184:6249–6255. doi: 10.4049/jimmunol.0903748. [DOI] [PubMed] [Google Scholar]
- 67.You F. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat. Immunol. 2009;10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 68.Cui J. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Cell Res. 2014;24:400–416. doi: 10.1038/cr.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Friedman C.S. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin W. Syndecan-4 negatively regulates antiviral signalling by mediating RIG-I deubiquitination via CYLD. Nat. Commun. 2016;7:11848. doi: 10.1038/ncomms11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fan Y. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J. Exp. Med. 2014;211:313–328. doi: 10.1084/jem.20122844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swatek K.N., Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yau R., Rape M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 74.Iwai K. Linear ubiquitin chains: NF-κB signalling, cell death and beyond. Nat. Rev. Mol. Cell Biol. 2014;15:503–508. doi: 10.1038/nrm3836. [DOI] [PubMed] [Google Scholar]
- 75.Walczak H. Generation and physiological roles of linear ubiquitin chains. BMC Biol. 2012;10:23. doi: 10.1186/1741-7007-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ozato K. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rajsbaum R. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J. Mol. Biol. 2014;426:1265–1284. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Versteeg G.A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Davis M.E., Gack M.U. Ubiquitination in the antiviral immune response. Virology 479-480. 2015:52–65. doi: 10.1016/j.virol.2015.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mallery D.L. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21) Proc. Natl. Acad. Sci. U.S.A. 2010;107:19985–19990. doi: 10.1073/pnas.1014074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McEwan W.A. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat. Immunol. 2013;14:327–336. doi: 10.1038/ni.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hatakeyama S. TRIM proteins and cancer. Nature Rev. Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 83.Kimura T. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J. Cell Biol. 2015;210:973–989. doi: 10.1083/jcb.201503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chan Y.K., Gack M.U. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016;14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gack M.U. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rajsbaum R. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012;8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Manokaran G. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davis M.E. Antagonism of the phosphatase PP1 by the measles virus V protein is required for innate immune escape of MDA5. Cell Host Microbe. 2014;16:19–30. doi: 10.1016/j.chom.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mesman A.W. Measles virus suppresses RIG-I-like receptor activation in dendritic cells via DC-SIGN-mediated inhibition of PP1 phosphatases. Cell Host Microbe. 2014;16:31–42. doi: 10.1016/j.chom.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen W. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell. 2013;152:467–478. doi: 10.1016/j.cell.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 91.Shi C.S. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dell’Oste V. Innate nuclear sensor IFI16 translocates into the cytoplasm during the early stage of in vitro human cytomegalovirus infection and is entrapped in the egressing virions during the late stage. J. Virol. 2014;88:6970–6982. doi: 10.1128/JVI.00384-14. [DOI] [PMC free article] [PubMed] [Google Scholar]