Abstract
The heterogeneous dynamics of clonal lineages within a cell population, in aggregate, shape both normal and pathological biological processes. Studies of clonality typically relate the fitness of clones to their relative abundance, thus requiring long-term experiments and limiting conclusions about the heterogeneity of clonal fitness in response to perturbation. We present, for the first time, a method that enables a dynamic, global picture of clonal fitness within a mammalian cell population. This novel assay allows facile comparison of the structure of clonal fitness in a cell population across many perturbations. By utilizing high-throughput imaging, our methodology provides ample statistical power to define clonal fitness dynamically and to visualize the structure of perturbation-induced clonal fitness within a cell population. We envision that this technique will be a powerful tool to investigate heterogeneity in biological processes involving cell proliferation, including development and drug response.
Keywords: Cell proliferation, Imaging, Systems biology, Clonal lineages, Cancer, Development
INTRODUCTION
Proliferation of a cell population describes, at its most basic level, the process of cell numbers changing over time. How this population size is governed is fundamentally important in both normal (Hipfner and Cohen, 2004) and pathologic processes (Sporn and Harris, 1981) such as tissue homeostasis, cell differentiation, degenerative diseases and cancer progression. Identifying changes in proliferation can be a powerful means to measure genetic or pharmacologic perturbations. Indeed, high-throughput screening of cell proliferation is often used to capture the effects of cellular perturbations, including drug potency (Barretina et al., 2012). Thus, accurate metrics to quantify proliferation are essential to understanding perturbation-induced cell responses.
Historically, each data point in these in vitro assays is the average of the proliferative response of thousands of cells in a perturbagen-treated well, at single or few time points, in comparison to untreated control. However, it is becoming increasingly clear that heterogeneity within a cell population can have profound influence on outcomes (Huang, 2009). Heterogeneity itself, built in a cell population, can be a biological strategy for adaptation to stressful environments. Well known cases include clonal subpopulations of tumor cells evading therapy (Brock et al., 2009), or bet-hedging bacteria (Balaban et al., 2004).
Proliferation is also a dynamic process that, by its very nature, would be best quantified as a rate (e.g., population doublings per unit of time). In contrast, it has become common practice to quantify effects of perturbations on cell population size as fold change with respect to control at one or few time points, insufficient to produce a reliable rate.
It is well established that polyclonal cell populations, display variable fitness at the clonal level in certain microenvironments (Dexter et al., 1978; Nowell, 1976) and provides a mechanism for how cell populations adapt to stress. For example, human colorectal cancers grown as tumor xenografts in mice have been shown to be comprised of clones with unique fitness properties that affect response to treatment (Kreso et al., 2013). Clonal differences observed in vivo at the genetic and phenotypic levels have also been recapitulated in vitro (Anaka et al., 2013) indicating that cultured cells can be used to query relevant clonal differences. However, differences in clonal fitness have generally been considered in qualitative terms, relative to the size of the population. Thus, studies of clonal fitness variation would benefit from an approach that quantitatively assesses clonal differences over time, that is the dynamics and heterogeneity of polyclonal responses.
Since few if any current assays provide information on these two key aspects of cell proliferation, heterogeneity and dynamics, we developed the clonal Fractional Proliferation (cFP), a novel high-throughput imaging method to directly quantify the clonal fitness within a cell population as a rate of proliferation in response to perturbation. This approach builds on our previous studies that determine the relative contribution of different cell fates to overall population dynamics (Tyson et al., 2012). We demonstrate that, by sparse plating and high-throughput measurements of colony size, cFP effectively captures clonal dynamics in a single novel metric of fitness, the perturbation-induced proliferation (PIP) rate. Because PIP rate is defined by a rate of change, it is a scalable, dynamic metric of clonal fitness. Measured in bulk, the PIP rate distributions reveal perturbation-induced changes to the structure of clonal fitness across various conditions.
MATERIALS AND METHODS
Cell culture and reagents
PC9 cells were obtained as a gift from William Pao (Vanderbilt University School of Medicine). A375 cells were obtained from ATCC. Cells were fluorescently labeled as previously described with Histone-H2B-mRFP (Tyson et al., 2012) and Geminin-mAg (Sakaue-Sawano et al., 2008). PC9 cells were cultured in RPMI 1640 (obtained from ATCC) media supplemented with 10% fetal bovine serum. A375 cells were cultured in DMEM media (Gibco) supplemented with 10% FBS. All cells were cultured in CO2-, temperature-controlled and humidified incubators. Cells were confirmed negative for mycoplasma before use. Cycloheximide was obtained from Sigma. Other chemical reagents (trametinib, SB203580, and PLX4720, and anisomycin (Abcam; all others, Selleckchem) were solubilized in DMSO at a stock concentration of 10 mM. All chemicals were stored at −20°C.
Immunocytochemistry
Immunofluorescence detection of EGFR utilized an EGFR antibody obtained from Cell Signaling Technologies and Alexa Fluor 647-conjugated secondary antibody from Life Technologies. Cells were grown in a 96-well imaging plate (BD Biosciences) and fixed using 4% paraformaldehyde-PBS for ten minutes at the indicated time, washed in PBS, and stored in PBS at 4°C. Cells were permeabilized with a blocking buffer containing 0.3% Triton-X and 5% normal goat serum. Primary antibody (1:100, Calbiochem) incubation went overnight at 4C. Cells were washed three times with PBS, and then secondary antibody (1:1,000) was added for 1 hour in blocking buffer. Cells were counterstained with Hoechst 33342 (Invitrogen, 1:10,000 in PBS, 15 min) and imaged. Single-cell quantitation of EGFR intensity was performed using CellAnimation (Georgescu et al., 2012).
clonal Fractional Proliferation (cFP) assay
Assay conditions are described in the legend to Supplementary Figure 3. Briefly, subconfluent cells are split and seeded at optimized single-cell density into 96-well cell culture imaging plates (BD Biosciences). For PC9, the ideal conditions are 40 cells seeded per well in a final volume of 100 µL growth media. Plates are kept in tissue culture incubators for six days to allow single cells to expand into single-cell derived colonies. After this period, all wells are imaged and the experimental perturbation is added immediately thereafter. The plates are returned to the tissue culture incubators and all wells of the plate are imaged daily until the end of the experiment.
Image registration and processing
To prepare raw images for analysis, images were sequentially organized into spatially registered image montages and temporally assembled into stacks (Fig. 1c and Supplementary Note 1) using the freely-available ImageJ (Rasband, 2008) software (http://imagej.nih.gov/ij/). Subsequent image processing scripts were applied on a per-colony basis (Supplementary Note 2). Raw and processed images were each stored for reference.
Figure 1.
Schematic of clonal Fractional Proliferation experimental workflow. (a) Cells are seeded at single-cell density into microtiter imaging plates. Single cells are allowed to proliferate for 6 days in full growth media to expand into colonies. Once colonies have reached an optimal size, cells are imaged and then the experimental perturbation is immediately added. Subsequently, each well is imaged daily until the end of the experiment. (b) Spatially- and temporally registered images facilitate tracking of individual colonies. Time-ordered stacks of image montages allow sequential measurements of colony cell numbers during drug treatment. (c) Fluorescent images of cell nuclei for single-cell derived PC9 colonies. Sequential images show that single-cells form colonies when cultured in full growth media (Untreated). Tracking colonies after addition of cycloheximide (Chx, 500 ng/ml) or DMSO (control), individual colonies shows drug-induced changes in cell number over time.
Generation of discrete sublines
PC9 cells were isolated as single cells and expanded in drug-free media until frozen stocks could be obtained. Sublines were cultured for less than ten passages to ensure consistency across experiments.
Statistical analysis
All statistical analysis was performed using the R statistical software (R-project.org). Linear model fits were performed using the lm function applied to the data points indicated in the text. Adjusted R2 values were calculated from the lm function. Pearson correlation coefficients were calculated using cor.test. And skew normal distribution fits for the immunofluorescence data were generated using the selm function contained within the SN package (Azzalini, 2005) (http://azzalini.stat.unipd.it/SN, version 1.0).
RESULTS
To quantify clonal fitness globally within a cell population, we devised the clonal Fractional Proliferation (cFP) assay, which tracks, in parallel, clonal proliferation in response to perturbation (Fig. 1a). We chose to represent clonal fitness by the proliferation of single-cell derived colonies, because this yields a dynamic metric based on direct measurements of clonal behavior. To facilitate accurate, high-throughput tracking of clones over time, we implemented the CellaVista High-End imager (Synentec) and a lentivirally-introduced nuclear marker (H2B-mRFP) (Tyson et al., 2012).
In setting up the cFP assay, we aimed to balance several prerequisites. We reasoned that if the cell population response to a perturbation is composed of clones with variable fitness, then it is imperative to quantify as many clones as feasible per experiment in order to obtain a representative sample of the range and frequency (or diversity) of clonal fitness. Additionally, individual colonies must contain a sufficiently high number of cells prior to treatment in order to minimize error introduced by small cell number counts, especially if the perturbation induces a decline in cell number within a clone. However, to ensure that colonies are in fact clonally derived, the cell population must be sparsely plated and the assay terminated prior to colony confluence. On balance, we found that using a 96-well plate format (Fig. 1a), and plating ~40 cells/well are optimal initial conditions to obtain single-cell derived colonies (Fig. 1b). After plating, the colonies are allowed to grow for six days in complete growth media, and subsequently subjected to a continuous experimental perturbation, during which the entire well is imaged daily for ten days. Plating efficiency is sufficiently high so that information on ~200 colonies per experimental condition can be obtained from 3-8 replicate wells.
Tracking of colonies throughout the duration of the perturbation requires that images must be both spatially and temporally registered (Fig. 1c). To this end, we use the freely available ImageJ (Rasband, 2008) software (http://imagej.nih.gov/ij/, version 1.48i; Supplementary Note 1) to generated time-series image montages of individual wells. First, the subset of all images belonging to a single well at a single time point is considered. Then, images are spatially ordered based on acquisition time and converted to a stitched image montage of the entire well. This step is repeated for all time points and all the montages are ordered by acquisition time, resulting in an image stack of wells over the course of the experimental treatment (Fig. 1c).
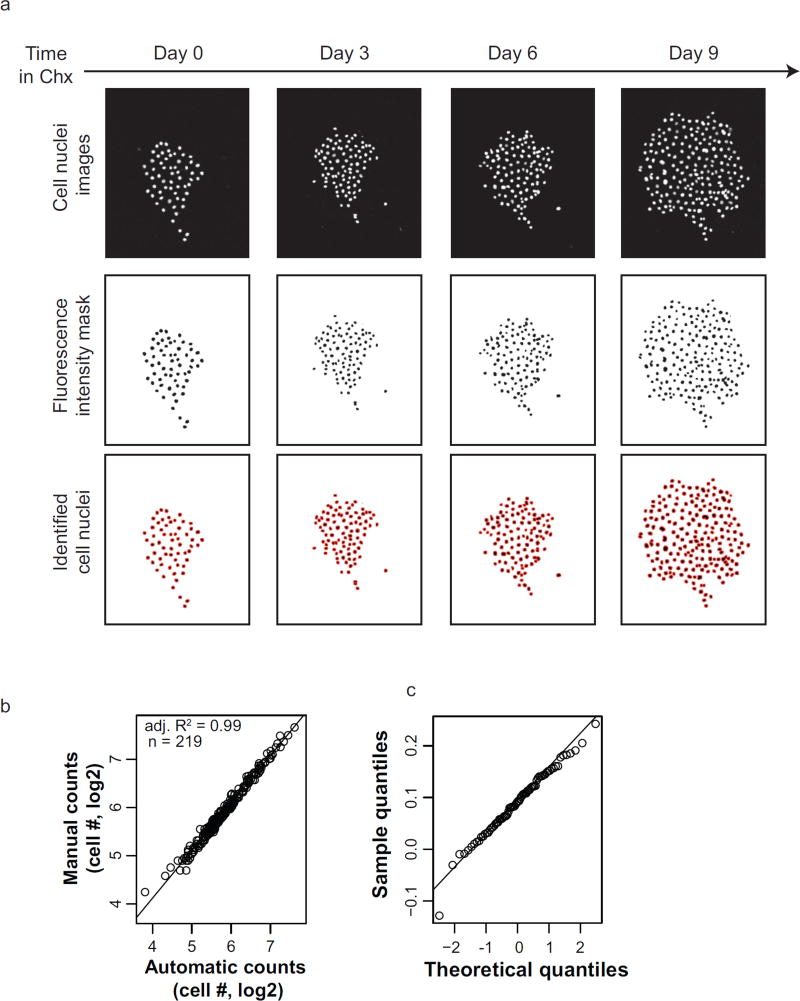
To quantify the proliferation dynamics of individual clones, we developed an image processing workflow using ImageJ to count the total number of cell nuclei at each time point from registered image stacks (Supplementary Note 2). Fluorescence intensity masks were generated for each colony by applying a user-defined threshold to the pixel intensity histogram computed from all images unique to that colony. These masks show that the fluorescence from cell nuclei is sufficiently high above background to detect cells. However, colonies remain under-segmented, compared to manual counting. To correct for this, the watershed segmentation algorithm was implemented to distinguish nearby nuclei. Then the “analyze particles” command scans the image stack for elements with optimized morphological parameters and returns the number of identified cell nuclei and the corresponding images (Fig. 2a).
Figure 2.
Validation of cFP image processing. (a) Top row: Images of a representative colony throughout cycloheximide (Chx, 500 ng/ml) treatment at the indicated days. Middle row: Binary mask generated in ImageJ using the same intensity threshold at all time points. Bottom row: Identified cells after image processing. (b) Automated counting of cell nuclei appropriately quantifies colony cell number. Manual cell counts from colony images at various time points (n = 219) are used as a reference to validate the automatically measured colony cell numbers. The superimposed line represents the linear model fit for the data, with the corresponding adjusted R-squared value (adj. R2) (c) Q-Q plot of the residuals of the linear model fit in used in (b). There is insufficient bias to conclude that automated image analysis is inappropriate for colonies of certain size.
This technique was used to serially identify cell nuclei throughout a representative experiment. To quantify how well the automated image segmentation represented the actual cell number at each time point, we manually counted the total cell nuclei per colony at 219 total time points. The manual counts are highly correlated (adjusted R2 = 0.99) with the automated cell counts (Fig. 2b) showing that automated cell counting is an efficient and faithful representation of changes in colony cell number. Additionally, the residual errors of the linear model fit do not show evidence of bias (Fig. 2c) and the standard deviation of the residuals is over fifty times smaller than the smallest cell number of any colony.
Having validated the ability to quantify colony size by cFP, we set out to measure the clonal fitness variability within a population in response to the drug cycloheximide, an inhibitor of protein translation well known to severely stunt cell proliferation(Liu et al., 2010). We tracked two representative colonies in PC9, an epithelial lung cancer cell line, for six days untreated, followed by three days in the presence or absence of 500 ng/ml cycloheximide. In the absence of cycloheximide (day -6–0), single cells steadily grow to form colonies (Fig. 1c). The addition of cycloheximide stunts the increase in cell number relative to control and the colony sizes rapidly diverge.
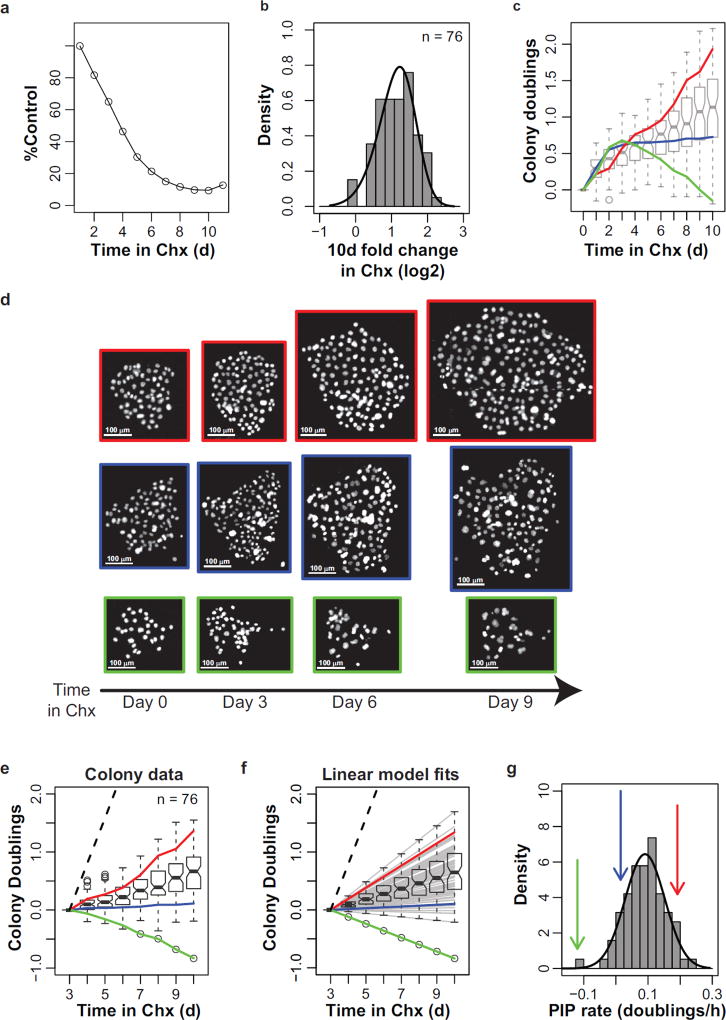
A conventional way to visualize cell proliferation is by plotting growth inhibition, or the percentage of treated cells relative to control cells. We thus plotted the time-course growth inhibition of PC9 cells in response to cycloheximide using images from a cFP experiment (Fig. 3a). Adding cycloheximide appears to progressively reduce the cell number until the cells are ~85% growth inhibited after 8 days. However, the distribution of relative colony size after ten days in cycloheximide reveals that most colonies continue to increase in size during the course of the experiment (Fig. 3b). Thus, with the cFP assay the actual dynamics of cycloheximide inhibition are revealed, i.e., a reduction in proliferation rate. Interestingly the standard deviation of colony cell number progressively increases, which indicates divergent clone-to-clone behavior over time (Supplementary Fig. 1a).
Figure 3.
Perturbation-induced proliferation (PIP) rates capture clonal fitness. (a) Cycloheximide (Chx, 500 ng/ml) reduces PC9 proliferation relative to control. Each data point represents the average number of cycloheximide-treated cells relative to control (n = 4 wells from a cFP experiment). (b) Histogram of the per-colony relative log2 fold change in cell number after 10 days of cycloheximide treatment. Curve represents the best fit using a skew-normal distribution. (c) Cycloheximide-induced colony dynamics. Colony doublings represents the log2-transformed cell number per colony normalized to the initial size of that colony. Boxplots show the interquartile range of cycloheximide-treated colony doublings at each time point. Colored lines display the dynamics of three representative colonies in d. (d) Images of H2B-mRFP labeled cell nuclei for the representative colonies in c. Scale bars = 100 µm (e) Colony dynamics, same as in c, except colonies are normalized to the colony size after 3 days of treatment. Lines and boxplots are the same as in c. (f) Linear model fits of colony dynamics simplify the colony doublings data. The linear model fit of each colony response from 3 days on is shown as a gray line. The estimated linear model fits of the representative colonies from c are shown as colored lines, as previously described. Boxplots represent the interquartile range of the expected values for colony doublings at each time point.
To investigate this time-dependent increase of clonal variability more in depth, we examined the distribution of individual colony responses. Consistent with the overall fold-increase in colony size throughout the duration of the experiment (Fig. 3b), the median colony size at each time point progressively increases over time in response to cycloheximide (Fig. 3c). Additionally, the interquartile range of colony sizes also increases over time (Fig. 3c, boxplots) supporting the possibility that divergence of colony sizes may be due to an underlying biological correlate, such as clonal fitness. Indeed, individual colonies display unique dynamics in response to cycloheximide (representative colony dynamics are quantified by colored lines in Figure 3c and visualized in Figure 3d).
This apparent increase in clone-to-clone variability over time was surprising. However, we noticed that, although colony growth dynamics appear initially complex (i.e., nonlinear on a log2 scale), after three days of treatment they achieve a steady linear rate. We reasoned that this steady rate of proliferation for each colony could simplify interpretation of the dynamic colony response data, by eliminating the confounding factor of observations taken at multiple time points. That is, the variability in the size of the colonies would increase because of divergent rates of proliferation, not because of underlying “diversifying factors”. We therefore normalized the colony sizes to the population doublings at three days on a per-colony basis (Fig. 3e). Each of the representative colonies appears to be linear from 3–10 days. Indeed, the R-squared value of ninety-five percent of pooled cycloheximide and control colonies was above 0.8 (Supplementary Fig. 1b). In summary, the rate of proliferation of a colony provides an absolute metric for the fitness of that clone in the presence of a perturbation, independent of comparison to untreated control.
To test how well the assumption of linear colony proliferation matches the data, we estimated the linear model fit for each colony using all population doubling data from day 3 onward. Plotting the model fits shows that the estimated behavior of colonies (Fig. 3f) approximates the normalized colony proliferation (Fig. 3e). Both the representative colonies (colored lines) and the range of colony responses (gray lines and boxplots) are similar between the normalized colony data (Fig. 3e) and the plot of the linear model fits (Fig. 3f). Thus the steady proliferation rate of a colony appears appropriate to summarize clonal behavior. Therefore we coin the proliferation rate obtained from 3d onward as the Perturbation-Induced Proliferation (PIP) rate.
Because the PIP rate estimates the overall ability of a clonal lineage to survive, or perish, and expand, or regress, in size over time, it can be considered as a metric of clonal fitness. Indeed, the change in colony size after 10d is highly correlated with PIP rate (Supplementary Fig. 1c), further validating the use of PIP rates to capture clonal fitness. An advantage of using a single parameter to describe clone behavior is that it enables an easy display of the clonal variability within a cell population. In response to cycloheximide, clonal PIP rates within PC9 are well approximated by a skew-normal distribution of PIP rates (Kolmogorov-Smirov test p=0.99; high p-value indicates insufficient statistical power to reject the fit), described by three parameters, µ, σ, and α (Supplementary Table 1). Because cFP was designed to measure clonal variability within a cell population, we interpret µ as the average clonal fitness of the population, whereas σ and α represent the variability of clonal fitness. Taken together, we interpret the distribution of PIP rates as the structure of clonal fitness of a cell population in the presence of a given perturbation.
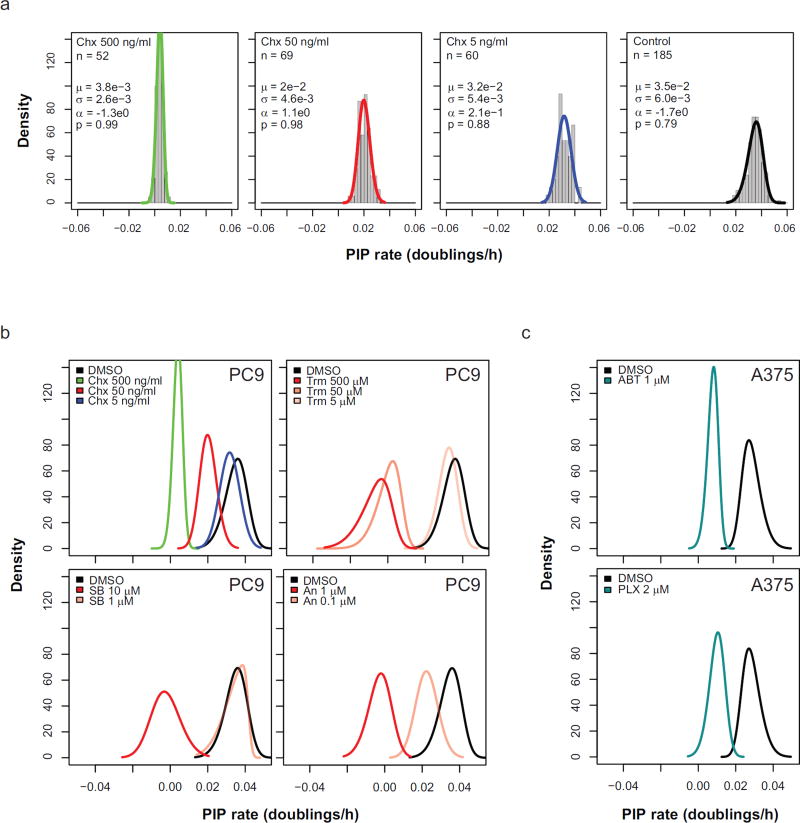
We next aimed to use cFP to describe the concentration-dependent effects of cycloheximide treatment. We treated PC9 cells at various concentrations of cycloheximide for a 10d time course (Fig. 4a). The distribution of PIP rates at each concentration could be fit appropriately with a normal distribution (Kolmogorov–Smirnov test p>0.05; high p-value indicates insufficient statistical power to exclude the distribution fit). As expected, the mean PIP rate of PC9 treated with 500 ng/ml cycloheximide (µ = 3.8e-3 doublings/h) is reduced relative to the control (µ = 3.5e-2 doublings/h). Additionally, with increasing concentrations of cycloheximide, the mean PIP rate is progressively reduced. An unexpected finding is that the variability of PIP rates (σ) in PC9 is altered by cycloheximide treatment. The variability of PIP rates decreases with increasing concentrations of cycloheximide relative to control as indicated by the narrowing of the density distribution (Fig. 4a). Thus, changes in the distribution fit parameters both accurately reflect the experimental data and provide intuition on how perturbations influence both the mean and variability of intra-population clonal fitness.
Figure 4.
Measuring the clonal structure in response to perturbations. (a) Concentration dependent effects of cycloheximide treatment. Histograms of PIP rates are shown for PC9 colonies at decreasing concentrations (left to right). Curves represent the estimated skew-normal probability distribution with the indicated values. Kolmogorov-Smirnov test p-values are shown. High p-value indicates there is insufficient evidence to reject the fit (b). Upper-left, The skew-normal fits (same color) from a are superimposed to illustrate the concentration-dependent effects of cycloheximide without the data. Concentration dependent effects are similarly shown for trametinib (Trm), SB203580 (SB), and anisomycin (An). (c) Skew normal fits are displayed for A375 cells treated with either ABT-737 (ABT) or PLX4720 (PLX).
To visualize the changes in shape of clonal PIP rates directly, we overlaid the distribution fits for each concentration (Fig. 4b; colors matched to the distributions shown in Fig. 4a). By comparing the fits alone, the effects on the PIP rate distribution shape can be easily seen relative to other conditions. The progressive decrease in both the clonal PIP rate mean and variability induced by increasing cycloheximide concentration (Fig. 4a) are evident by the PIP rate distribution shifting to the left and becoming narrower (Fig. 4b upper left). Similar trends were observed in B-Raf mutant A375 melanoma cells when treated with either PLX4720, a specific B-Raf kinase inhibitor (Tsai et al., 2008) or ABT-737 (Rooswinkel et al., 2012), a BH3 mimetic that sequesters pro-survival Bcl2 family member proteins (Fig. 4c). Thus comparing the shapes of PIP rate distributions quickly summarizes how perturbations shape the clonal fitness profile within a cell population.
We next sought to examine the generality of our findings in the context of other perturbations. We chose to use trametinib (Gilmartin et al., 2011), a small molecule kinase inhibitor that specifically inhibits the activity of MEK, a protein kinase involved in growth factor signal transduction downstream of Ras (Shaul and Seger, 2007). Similar to cycloheximide, increasing trametinib concentrations induce a step-wise reduction in mean PIP rate (Fig. 4b, upper right). However, the decreasing mean is paired with an increasing variability of clonal PIP rates, evidenced by a left-shift and broadening of the PIP rate distribution. Another perturbagen, anisomycin, an antibiotic known to inhibit protein synthesis and activate stress-activated protein kinases (Ferreiro et al., 2010), progressively shifts the distribution to the left, but does not change the clonal PIP rate variability (Fig. 4b, lower right). In the case of SB203580, a specific inhibitor (Ferreiro et al., 2010) of the stress activated protein kinase p38, high doses shift the distribution to the left and broaden it, but this effect appears to happen as an all-or-none response (Fig. 4b, lower left). Thus cFP reveals the concentration-dependent effects of perturbations to the clonal structure of a cell population.
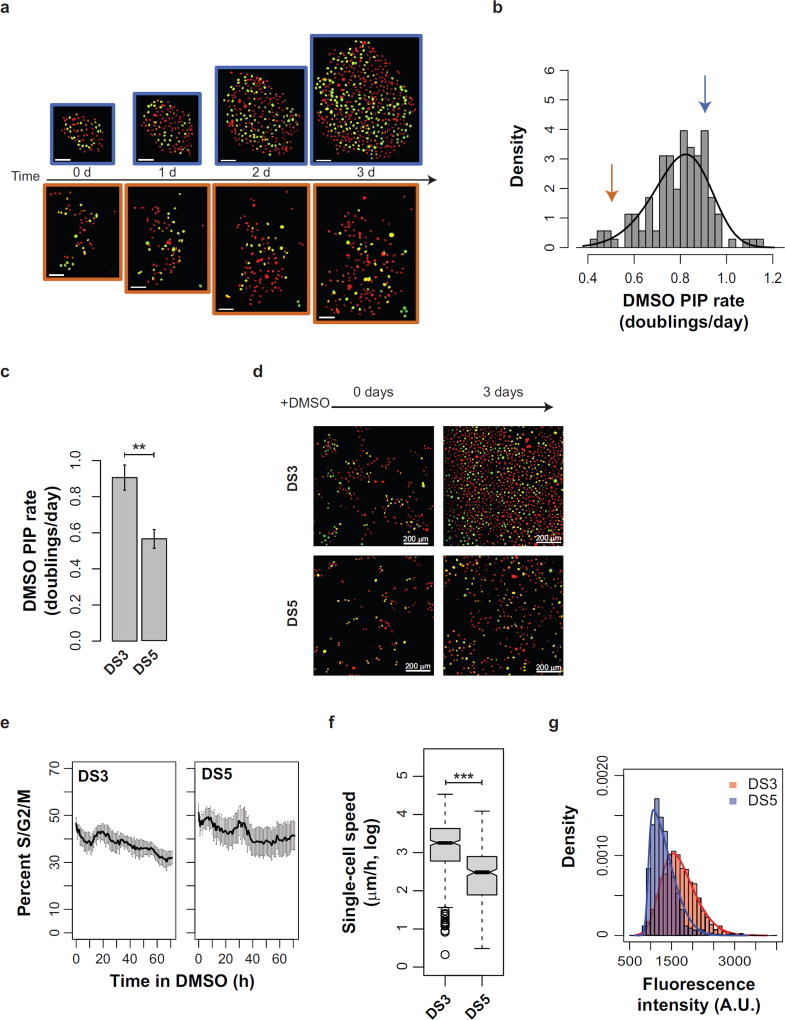
Heretofore we have defined clonal fitness by the proliferation rate of a clone. However, several aspects of fitness are not evident by colony growth alone. For example, different cellular phenotypes – such as motility, morphology, cell cycle progression, etc. – may contribute to fitness in a given microenvironment (Anderson et al., 2006). Indeed, colonies can display a variety of cell morphologies that are similar within a colony, yet vary broadly between colonies (Supplementary Fig. 2). In order to study cell cycle progression in the context of clonal fitness, we expressed in PC9 cells geminin-mAg (Sakaue-Sawano et al., 2008), a live-cell fluorescent marker of cell cycle position that is stable stably expressed after a cell has passed the G1/S transition and maintained until the cell undergoes mitosis (detectable in S/G2/M phase). To characterize inter-clonal differences, we chose to study DMSO-treated colonies because they display high PIP rate clonal variability (Fig. 4b). We then examined both cell cycle position (geminin-mAg; pseudocolored green) and the total cell nuclei (H2B-mRFP; pseudocolored red) in representative colonies. Geminin-mAg is readily detectable throughout the experiment in colonies exhibiting linear proliferation over a 3 d DMSO treatment (R2 = 0.99 for both). These two colonies (Fig. 5a) displayed PIP rates above (Fig. 5b blue arrow corresponding to the blue-outlined colony in Fig. 5a) and below (Fig. 5b orange arrow) the mean clonal PIP rate (Fig. 5b). The faster colony had a PIP rate of 3.8e-2 doublings/h, while the slower colony had a PIP rate of 2.1e-2 doublings/h.
Figure 5.
Investigating PIP rates using discrete sublines. (a) PC9 colonies with unique proliferation rates cultured in DMSO imaged at the indicated time points. All cells are fluorescently labeled with both Histone H2B-RFP (red pseudocolor) and geminin-mAg (green pseudocolor). Green cell nuclei indicate cells that have passed the G1/S transition; red cell nuclei mark cells that have not. Scale bars = 100 microns. (b) Distribution PC9 of DMSO-treated PIP rates. Arrows indicate the PIP rate of the color-matched colonies from a. (c) Quantitation of proliferation rates for two representative PC9 discrete sublines, DS3 and DS5 (values are the average of triplicate wells from n = 3 experiments). Error bars = SE. (d) Representative images from the experiments in c. (e) Percent of cells in S/G2/M phase (geminin-mAg) during DMSO treatment (data are the average of triplicate wells in n=3 experiments). Error bars = SE. (f) Quantitation of single-cell speed for DS3 and DS5 treated with DMSO. (g) Quantitation of single-cell EGFR expression from immunofluorescence images using CellAnimation. Curves represent the best fit using a skew normal distribution. * p < 0.05; ** p < 0.01; *** p < 0.001.
While cFP is ideal for measuring clonal behavior en masse, it is limited in characterizing individual clones because of experimental difficulty in matching clones across experiments. To examine clonal differences in more detail, we generated a panel of single cell-derived PC9 discrete sublines (PC9 DS). We selected two sublines, PC9-DS3 and PC9-DS5, for further analysis with DMSO PIP rates (Fig. 5c; PC9-DS3 PIP = 3.8e-2 doublings/h; and PC9-DS5 PIP = 2.3e-2 doublings/h) that closely matched those of the representative colonies in 5a–b). Interestingly, the PIP rates of DS3 and DS5 were highly statistically significant (p = 1.4e-3) across three experiments. The DS sublines provide an advantage in that they enable parallel experimentation using higher cell numbers (Fig. 5d) and are more amenable to high time frequency imaging. Using the same image processing methodology used to quantify all cell nuclei, we then measured the number of geminin-mAg positive cells in DS3 and DS5 hourly relative to the total cell number (Fig. 5e). Both clones maintain a high level of geminin-mAg throughout the experiment, suggesting that asymmetry in cell cycle progression does not mediate the differences in PIP rate between clones.
Another notable difference between the clones in Figure 5a is that the orange clone appears to have a higher spacing between individual cells (Supplementary Fig. 2). This may reflect differences in single-cell motility. We used CellAnimation (Georgescu et al., 2012), a freely-available cell tracking software, to examine single-cell speeds using a dataset of DS3 and DS5 cells imaged at twelve-minute time intervals. The average single-cell speed for DS3 is significantly faster than that of DS5 (Fig. 5f; p < 1e-16).
To begin to examine molecular differences between DS3 and DS5, we performed immunostaining of EGFR on each, since EGFR is constitutively active in PC9 cells (Chin et al., 2008). Using CellAnimation (Georgescu et al., 2012) we quantified the single-cell EGFR intensity. For each clone, the individual fluorescence levels of single cells matched a skew-normal distribution (Fig. 5g). Comparing the distributions, DS3 had a higher median single-cell EGFR expression level than DS5 and a greater standard deviation. Taken together, these data demonstrate that pairing the DS sublines with the colonies can reveal the relationship between clonal fitness, population stability, and single-cell phenotypes.
DISCUSSION
A fundamental question in biology is to understand how and why heterogeneous responses arise even within an isogenic cell population. Several key features are important when considering an assay to measure heterogeneity. First, if it is capable of differentiating between subpopulations, does it provide intuition on how that heterogeneity is structured? How does the total heterogeneity change over time and to what extent are cell behaviors remembered across cell generations? How do perturbations shift the fitness landscape of a cell population? While each of these features is important, to our knowledge no other assays collect this information together. To address this gap, we devised clonal Fractional Proliferation (cFP), a high-throughput cell imaging methodology to measure the structure of clonal fitness of a cell population in response to perturbation (Fig. 1 and Supplementary Fig 3). By directly tracking clonal cell populations, cFP quantifies the fitness of clones as the perturbation-induced proliferation (PIP) rate (Fig. 3). With this simplification, cFP can measure PIP rates in high-throughput to measure perturbation induced changes in clonal structure (Fig. 4)
Live-cell microscopy and flow cytometry are commonly used experimental systems to study heterogeneity in a population. Flow cytometry reveals the distribution of a biological trait due to high sample throughput. Repeat measurements can infer how a heterogeneous population evolves in time by the ergodic hypothesis (Brock et al., 2009), and state-of-the-art techniques can even extract growth rate (Zuleta et al., 2014), but fundamentally the same cell cannot be tracked over time. Live-cell microscopy, by contrast, resolves heterogeneous cell behavior dynamically (Tyson et al., 2012), but the difficulty of tracking single cells often limits the sample throughput and the duration of the experiment. By quantifying heterogeneity as clonal proliferation rates, cFP overcomes both of these limitations. Accurate image processing (Fig. 2) enables high-throughput measurements of heterogeneity. Daily imaging of individual colonies allows for long term experiments (Fig. 1b), while requiring orders of magnitude fewer measurements than needed for tracking an equivalent number of single cells over time(Tyson et al., 2012).
The landscape of clonal fitness is key to understand how a cell population will evolve in a new environment (Gordo and Campos, 2013). Often differential clonal fitness is thought of in terms of rare somatic mutations involved in cancer progression (Nowell, 1976) or mutations in microorganisms allowing survival in stressful environments (Gordo et al., 2012). However increasing evidence from studies utilizing genetic barcoding indicates that both the structure and the dynamics of clonal fitness are greater than previously appreciated both in vitro (Porter et al., 2014) and in vivo (Kreso et al., 2013). While these approaches give an unprecedented view of clonal behavior over time, they are limited in that clonal fitness is reported as the fraction of the total populations. That is, because, the measurements are relative, it is difficult to know the actual cellular phenotypes within clones. For example, a decrease in the relative proportion of clones could be due to either cell death, or quiescence, or reduced proliferation, or a combination of these (Tyson et al., 2012). By contrast, cFP quantifies clonal fitness directly as the PIP rate of individual clones. Thus the clonal structure can be represented as probability distributions of a functional trait, namely proliferation, and the dynamic behavior of clones can be determined without long-term selection (Fig. 4). Moreover, because clonal fitness is presented as rates, cFP outputs data should be amenable to mathematical models of population dynamics (Michor et al., 2005). We envision cFP as a powerful functional complement to genetic analyses of clonal dynamics.
Because each treatment condition is approximated by a probability distribution (Fig. 4), we view the distribution itself as a reflection of the clonal structure. That is, though cFP does not physically measure every clone in a population, it gives an estimate of what the phenotypic distribution would be if every clone were sampled. The meaning of this distribution may give insight into the adaptation of a cell population to stress and should generate novel biological hypotheses. We expect this to be especially informative to linking molecular signaling gradients to the shape of the distribution (Fig. 4b).
By necessity, the information obtained for each clone is limited in cFP (Supplementary Fig. 3). The sublines (Fig. 5c) represent a complementary system to cFP capable of studying clonal behavior in more detail. We demonstrate that clones with a stable fitness (Fig. 5c) can be further interrogated for single-cell information including cell cycle progression, motility, and protein expression levels (Fig. 5d–g). We envision that this type of information will broaden our definition of clonal fitness to include additional cell phenotypes. Additionally, through mixing experiments, the sublines should help to address limitations of cFP, such as assumptions that fitness is clone autonomous.
In summary, we describe clonal Fractional Proliferation, a high-throughput imaging method to capture the structure and dynamics of clonal fitness in response to perturbation. By sparse plating and quantitation of single-cell-derived colonies over time, cFP directly tracks the proliferation of many clones within a cell population. Because clones achieve a steady proliferation rate in response to perturbation, their proliferation can be summarized by a novel dynamic metric, the perturbation-induced proliferation (PIP) rate. Measuring clonal PIP rates repeatedly reveals an underlying structure to clonal fitness. Thus cFP enables quantitation of the full landscape of clonal fitness heterogeneity within a cell population without long-term selection. This gives an unprecedented view of perturbation-induced changes to the clonal structure of a cell population across conditions. We envision that this approach will have broad applicability in studying processes driven by the proliferation of heterogeneous cell populations, such as tissue homeostasis, embryonic development, cell reprogramming and cancer progression.
Supplementary Material
Acknowledgments
Contract grant sponsor: National Institutes of Health;
Contract grant number: 5U54CA113007-07
We would like to thank William Pao, (Vanderbilt), for the PC9 cell line and A. Miyawaki, (RIKEN Brain Science Institute) for the mAG-geminin plasmid. This work has been funded by the National Institutes of Health (5U54CA113007-07, VQ; F31-CA165840, PLF), Uniting Against Lung Cancer (13020513, DRT), and VICTR STARBRITE.
References
- Anaka M, Hudson C, Lo P-H, Do H, Caballero OL, Davis ID, Dobrovic A, Cebon J, Behren A. Intratumoral genetic heterogeneity in metastatic melanoma is accompanied by variation in malignant behaviors. BMC Med Genomics. 2013;6:40. doi: 10.1186/1755-8794-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AR, Weaver AM, Cummings PT, Quaranta V. Tumor Morphology and Phenotypic Evolution Driven by Selective Pressure from the Microenvironment. Cell. 2006;127:905–915. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- Azzalini A. The Skew-normal Distribution and Related Multivariate Families*. Scand J Stat. 2005;32:159–188. [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock A, Chang H, Huang S. Non-genetic heterogeneity--a mutation-independent driving force for the somatic evolution of tumours. Nature Reviews Genetics. 2009;10:336–342. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- Chin TM, Quinlan MP, Singh A, Sequist LV, Lynch TJ, Haber DA, Sharma SV, Settleman J. Reduced Erlotinib sensitivity of epidermal growth factor receptor-mutant non-small cell lung cancer following cisplatin exposure: a cell culture model of second-line erlotinib treatment. Clin. Cancer Res. 2008;14:6867–6876. doi: 10.1158/1078-0432.CCR-08-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter DL, Kowalski HM, Blazar BA, Fligiel Z, Vogel R, Heppner GH. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 1978;38:3174–3181. [PubMed] [Google Scholar]
- Ferreiro I, Joaquin M, Islam A, Gomez-Lopez G, Barragan M, Lombardía L, Domínguez O, Pisano DG, Lopez-Bigas N, Nebreda AR, et al. Whole genome analysis of p38 SAPK-mediated gene expression upon stress. BMC Genomics. 2010;11:144. doi: 10.1186/1471-2164-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu W, Wikswo JP, Quaranta V. CellAnimation: an open source MATLAB framework for microscopy assays. Bioinformatics. 2012;28:138–139. doi: 10.1093/bioinformatics/btr633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin AG, Bleam MR, Groy A, Moss KG. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clinical Cancer. 2011 doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- Gordo I, Perfeito L, Sousa A. Fitness effects of mutations in bacteria. Journal of molecular microbiology. 2012 doi: 10.1159/000332747. [DOI] [PubMed] [Google Scholar]
- Gordo I, Campos PRA. Evolution of clonal populations approaching a fitness peak. Biol. Lett. 2013;9:20120239. doi: 10.1098/rsbl.2012.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipfner DR, Cohen SM. Connecting proliferation and apoptosis in development and disease. Nat. Rev. Mol. Cell Biol. 2004;5:805–815. doi: 10.1038/nrm1491. [DOI] [PubMed] [Google Scholar]
- Huang S. Non-genetic heterogeneity of cells in development: more than just noise. Development. 2009;136:3853–3862. doi: 10.1242/dev.035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A, Kreso A, O'Brien CA, van Galen P, Gan OI, Notta F, Brown AM, Ng K, Ma J, Wienholds E, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang J-M, Zhang SS, Liu X-Y, Liu DX. Induction of cell cycle arrest at G1 and S phases and cAMP-dependent differentiation in C6 glioma by low concentration of cycloheximide. BMC Cancer. 2010;10:684. doi: 10.1186/1471-2407-10-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michor F, Hughes TP, Iwasa Y, Branford S, Shah NP, Sawyers CL, Nowak MA. Dynamics of chronic myeloid leukaemia. Nature. 2005;435:1267–1270. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Porter SN, Baker LC, Mittelman D, Porteus MH. Lentiviral and targeted cellular barcoding reveals ongoing clonal dynamics of cell lines in vitro and in vivo. Genome Biol. 2014;15:R75. doi: 10.1186/gb-2014-15-5-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. 2008 ( http://rsbweb.nih.gov/ij/)
- Rooswinkel RW, van de Kooij B, Verheij M, Borst J. Bcl-2 is a better ABT-737 target than Bcl-xL or Bcl-w and only Noxa overcomes resistance mediated by Mcl-1, Bfl-1, or Bcl-B. Cell Death Dis. 2012;3:e366. doi: 10.1038/cddis.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A. Visualizing Spatiotemporal Dynamics of Multicellular Cell-Cycle Progression. Cell. 2008 doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Shaul YD, Seger R. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochimica Et Biophysica Acta (BBA)-Molecular Cell. 2007 doi: 10.1016/j.bbamcr.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Sporn MB, Harris ED. Proliferative Diseases. The American Journal of Medicine. 1981;70:1231–1236. doi: 10.1016/0002-9343(81)90832-9. [DOI] [PubMed] [Google Scholar]
- Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, Bremer R, Gillette S, Kong J, Haass NK, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proceedings of the National Academy of Sciences. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson DR, Garbett SP, Frick PL, Quaranta V. Fractional proliferation: a method to deconvolve cell population dynamics from single-cell data. Nat. Methods. 2012;9:923–928. doi: 10.1038/nmeth.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuleta IA, Aranda-Díaz A, Li H, El-Samad H. Dynamic characterization of growth and gene expression using high-throughput automated flow cytometry. Nat. Methods. 2014;11:443–448. doi: 10.1038/nmeth.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.