Abstract
Aim:
This study aims to isolate, characterize, and in silico evaluate of anticancer polyphenols from different parts of Syzygium alternifolium.
Materials and Methods:
The polyphenols were isolated by standard protocol and characterized using Fourier-transform infrared (FT-IR), High performance liquid chromatography - Photodiode array detector coupled with Electrospray ionization - mass spectrometry (MS/MS). The compounds were elucidated based on retention time and molecular ions (m/z) either by [M+H]+/[M-H]− with the comparison of standard phenols as well as ReSpect software tool. Furthermore, absorption, distribution, metabolism, and excretion (ADME)/toxicity properties of selected phenolic scaffolds were screened using OSIRIS and SwissADME programs, which incorporate toxicity risk assessments, pharmacokinetics, and rule of five principles. Molecular docking studies were carried out for selected toxicity filtered compounds against breast cancer estrogen receptor a (ERa) structure (protein data bank-ID: 1A52) through AutoDock scoring functions by PyRx virtual screening program.
Results:
The obtained results showed two intensive peaks in each polyphenol fraction analyzed with FT-IR, confirms O-H/C-O stretch of the phenolic functional group. A total of 40 compounds were obtained, which categorized as 9 different classes. Among them, flavonol group represents more number of polyphenols. In silico studies suggest seven compounds have the possibility to use as future nontoxic inhibitors. Molecular docking studies with ERa revealed the lead molecules unequivocally interact with Leu346, Glu353, Leu391, Arg394, Gly521, Leu525 residues, and Phe404 formed atomic π-stacking with dihydrochromen-4-one ring of ligands as like estrodial, which stabilizes the receptor structure and complicated to generate a single mutation for drug resistance.
Conclusion:
Overall, these results significantly proposed that isolated phenolics could be served as potential ER mitigators for breast cancer therapy.
Keywords: Estrogen receptor a, Fourier-transform infrared, high performance liquid chromatography- photodiode array detectors - electrospray ionization - mass spectrometry/mass spectrometry, molecular docking, polyphenols, Syzygium alternifolium
INTRODUCTION
The term polyphenol was derived from the Greek word “polus” means many and “phenol” means a chemical structure formed by one or more hydroxyl groups fortified with an aromatic hydrocarbon group [1]. Polyphenols, a group of plant secondary metabolites have increasing considerations in the fields of biological systems to cure different diseases, act as natural antioxidants, have nourishing and play a big part to human beings’ health. These are widespread in apples, berries, coffee, cocoa, onions, tea, and wine [2]. Based on number of phenol units, the plant phenolics were mainly divided into simple phenols and polyphenols. Polyphenols were again divided into two groups, i.e., non-flavonoids and flavonoids. Simple phenols, phenyl alcohols, stilbenes, chalcones, and lignans were categorized under the nonflavonoid compounds. Flavones, flavonols, flavanones, flavanols, dihydroflavonols, anthocyanins, proanthocyanidins, and isoflavones were categorized under the flavonoid group.
They arranged from simple single aromatic ring to complex polymers [3]. Tropical medicinal plants, green leafy vegetables, thick colored fruits, and wines were wealthiest wellspring of polyphenols. The previous studies experienced to the isolation and characterization of polyphenols with the assistance of various chromatographic and mass spectroscopic techniques like high performance liquid chromatography - electrospray ionization - mass spectrometry (HPLC-ESI-MS/MS) in different medicinal plants such as Rhus verniciflua [4], Citrus limetta [5], Juglans regia [6], and Annona cherimola [7]. HPLC is a significant tool for separation of a mixture of polyphenolics in a desired manner and ESI coupled with MS/MS can produce the ions from macromolecules and can fragment them for identification. In this study, polyphenolic compounds were separated by HPLC - PDA detector. Because the equivocal of compounds was effectively separated and is superior in the virtue of flavan-3-ols, flavonoids, nonflavonoids, and their derivatives. HPLC-PDA coupled with ESI-MS/MS was chosen as a sought apparatus for correct identification of polyphenols in this study.
In the recent past, various researchers derived polyphenols from various medicinal plants to prove different biological activities such as anti-inflammatory [8], antibacterial [9], anticancer [10], antihyperglycemic [11], antimutagenic [12], antioxidant [13], hepatoprotective [14], and wound healing activities [15]. Thereafter, the exponential biologically active polyphenols were isolated from processed foods/drinks such as vanillic acid, gallic acid, caffeic acid, ferulic acid, and hydroxyphenylacetic acid from Origanum vulgare, Camellia sinensis, Prunus virginiana, Thymus vulgaris, and Olea europaea, respectively, showed synergistic biological activities [16]. The plant Syzygium alternifolium belongs to the family Myrtaceae and is locally known as mogi/adavi neredu. This plant inhabited to high altitude hilly top areas of Tirumala hills, part of the Eastern Ghats, Andhra Pradesh, India, and is recently categorized under the endangered state by IUCN-red data book [17]. The ethnobotanical studies state that stem bark powder was utilized for the treatment of external wounds [18] and oral intake regulate blood sugar level [19]. Fruit powder was used for the treatment of diabetes [20] and diarrhea [21]. The previous enormous evidence revealed that leaf part of the plant has anticancer [22], antimicrobial [23], antioxidant [24], hypoglycemic and antihyperglycemic activities [25]. The earlier qualitative and quantitative studies of secondary metabolites from S. alternifolium purported that rich in phenols [26].
However, isolation, characterization, and toxicity evaluation of polyphenols from S. alternifolium are still disputable. Hence, the present work was conducted to isolate and to characterize the polyphenols through Fourier-transform infrared (FT-IR), HPLC-PDA-ESI-MS/MS from stem bark, leaf and fruit parts of S. alternifolium. The pharmacokinetics assets and toxicological etiologies of isolated polyphenols were characterized using in silico tools like virtual screening and molecular docking approaches established against estrogen receptor a (ERa) (protein data bank [PDB]-ID: 1A52) ligand binding domain to potentiate the plausible recognized lead scaffolds as for future anticancer therapeutics.
MATERIALS AND METHODS
Chemicals
The high purity Milli q-MilliPak water (Merck water solutions, France) was used for the preparation of chemicals and ultra-pure Milli q-LCPak water for HPLC analysis. Polyvinylpolypyrrolidone was procured from Himedia Laboratories, India. HPLC grade dichloromethane, acetone, methanol, formic acid, and NaOH were purchased from Molychem Laboratories, India. 0.1 mM concentration of stock solution was prepared using 18 standard polyphenols (data not shown) were used as reference compounds for identification of polyphenols. The obtained pseudomolecular ions (m/z values) were cross checked with available previous literature as well as liquid chromatography (LC)/MS database developed by ReSpect-Riken MSn spectral database [27].
Collection and Extraction of Plant Materials
Matured plant parts such as stem bark, leaves, and fruits were collected from the Nagatheertham area of Tirumala Hills and authenticated with the help of herbarium (voucher no. 121) deposited in Department of Botany, Sri Venkateswara University, Tirupati. The collected plant materials were washed and shade dried up to 15-20 days at room temperature (37°C). Then, grounded with the help of a blender and sieved it for further studies. Extraction of polyphenols from various parts of S. alternifolium was followed by the method of Magalhães et al. [28].
HPLC-PDA-ESI-MS/MS Instrumentation
The chromatographic separation of polyphenols was analyzed using HPLC (Shimadzu lab solutions, Kyoto, Japan) equipped with a LC-20 AD pump, detection with SPD-20A PDA and ultraviolet-visible detectors. The LC solution data acquisition software was retrieved from Shimadzu, Kyoto, Japan, and was installed in the Hewlett Packard system for recording of chromatography and its integrated data. For mass analysis, bench top Triple Quadrupole mass spectrometer (Quattro Micro manufactured by Waters Company, Manchester, UK) was equipped with an ESI source, operated by Masslynx version 4.1 software program.
Chromatographic Conditions
Agilent XDB C18 (150 × 4.6 mm, 5 µm) column was used for separation of polyphenols. The mobile phase consists of 0.1% formic acid in 70% methanol for 25-30 min recording time of the column at 210 nm with a speed of 1.0 ml/min at 25°C. Sample injection was performed with the help of Rheodyne 7725 injection valve via 20 µl loop and pH of the mobile phase was adjusted to 3.0 using a Dolphin pH meter. The MS acquisition was performed using ESI in positive and negative modes. For negative mode [M-H]− spectral range was recorded from 100 to 900 m/z range, while in the positive mode [M+H]+ spectral range was recorded from 50 to 750 m/z. The parameters were set as 0.5 s interval period, 10,000 amu/s scan speed of flow rate, heat block and DL temperature was adjusted to 200°C, DL voltage 4.5 kV, qarray voltage 1.0 V, RF voltage 90 V, detection gain at 1.0 kV were maintained, and N2 gas was used as a nebulizer gas at the speed of 1.5 L/min.
Computational Analysis
The pharmacokinetics were exceptionally crucial and advancement strategies for identification of therapeutic potential molecular candidates, especially in ethnopharmacology. The analysis of pharmacokinetics through in vitro and in vivo approaches was captivating a lot of time and more expensive process [29]. Thus, we applied in silico programs for molecular screening and the properties computation of confined polyphenols from S. alternifolium.
Absorption, distribution, metabolism, excretion (ADME)/Tox properties
Initially, toxicity risk assessments such as mutagenic, tumorigenic, irritant, reproductive effects along with fragment-based drug-likeness and the overall drug score of polyphenols were predicted using the OSIRIS property explorer program [30]. The bioactive properties of lead molecules were envisaged using molinspiration server (www.molinspiration.com). Furthermore, the physicochemical properties, n-octanol/water partition coefficient, pharmacokinetics, drug-likeness, and synthetic accessibility of molecules were anticipated within the adaptable range through GB/SA approach by using robust SwissADME server [31]. From the above auspicious consequences, the filtered potential nontoxic compounds were utilized for further screening and docking approaches.
Ligands preparation
The recognized small molecules were retrieved with three dimensional (3D) structure data file format from PubChem database [32]. The protonation state of ligands was charged to neutral position (pH=7) for outline and sustains the hydrogen bond formation. The leads comprises stereochemical clashes were optimized through conjugate gradient energy minimization by using visual molecular dynamics v1.9.1 tool [33] by applying CHARMM27 force fields with an exclusive topology and parameters acquired from SwissParam server [34]. Furthermore, relaxed ligands were converted into AutoDock ligand (PDBQT) format and arranged as a spreadsheet by PyRx virtual screening module [35].
Receptor preparation
Recent enormous investigations reported that polyphenolic compounds were profoundly inhibited the breast cancer cell proliferation and malignant tumor growth through estrogen mediated effects. In the present examination, we screened the isolated compounds against human ERa by using molecular docking approaches. Here, ERa (PDB ID: 1A52) [36] crystal structure was downloaded from PDB (http://www.rcsb.org/pdb). Furthermore, protein structure was neutralized by addition of polar hydrogens to the side chains and main chain. Subsequent to ensuring substance precision, protein 3D structure was subjected to energy minimization to control the crystallography conflicts by applying GROMOS96 force fields. They optimize the bonds, angles, torsions, nonbonded, and electrostatic potentials by using Swiss-PDB Viewer v4.1 software package [37]. The final protein energy was reduced to 0.01 kcal/mol−1 energy consistency. Furthermore, the streamlined protein was used as possibly permitted structure for virtual screening and docking simulations.
Virtual Screening and Molecular Docking Studies
Moreover, to portray the potential dynamic site in prepared protein, we utilized AutoLigand module implicit in AutoDock tools. By applying AutoDock force fields, habilitated pocket was generated for binding of ligands [38]. Pocket grid was generated using AutoGrid module and grid dimensions set as center_x = 101.1269, center_y = 23.015, center_z = 97.0783 (xyz axis Å3) and grid points set as 25×25×25 Å3 with 0.375 Å grid spacing. Initially, we screened segregated polyphenolic compounds onto the ligand binding domain (LBD) of ERa though AutoDock Vina program in PyRx software [35]. In this contest, we used default Lamarckian genetic algorithm parameters and empirical free energy function as scoring algorithms and docked each ligand with 300 maximum exhaustiveness runs against protein grid. The top-ranked ligands were again re-docked by using a flexible docking approach with three replication frameworks as past docking strategies. Finally, the resulted lead phenolics have greater probability for profoundly utilized as templates for ERa as anticancer therapeutics.
RESULTS
FT-IR Analysis
The first and foremost method to know the isolated fractions with functional group analysis was by FT-IR instrument. From the FT-IR consequences, a total number of 6-8 peaks were obtained from each fraction [Figure 1]. Among those, the peaks at 3328.51 cm−1 and 1238.39 cm−1 of stem bark fraction I; 3361.83 cm−1, 1235.55 cm−1, and 1093.48 cm−1 of stem bark fraction II; 3334.42 cm−1 and 1236.38 cm−1 of leaf fraction I; 3354.87 cm−1 and 1093.91 cm−1 of leaf fraction II; 3337.25 cm−1 and 1236.70 cm−1 of fruit fraction I; 3357.99 cm−1, 1235.63 cm−1, and 1093.44 cm−1 of fruit fraction II corresponds to O-H/C-O stretch of phenols. These results paved a clear way for further HPLC-ESI-MS/MS analysis.
Figure 1.
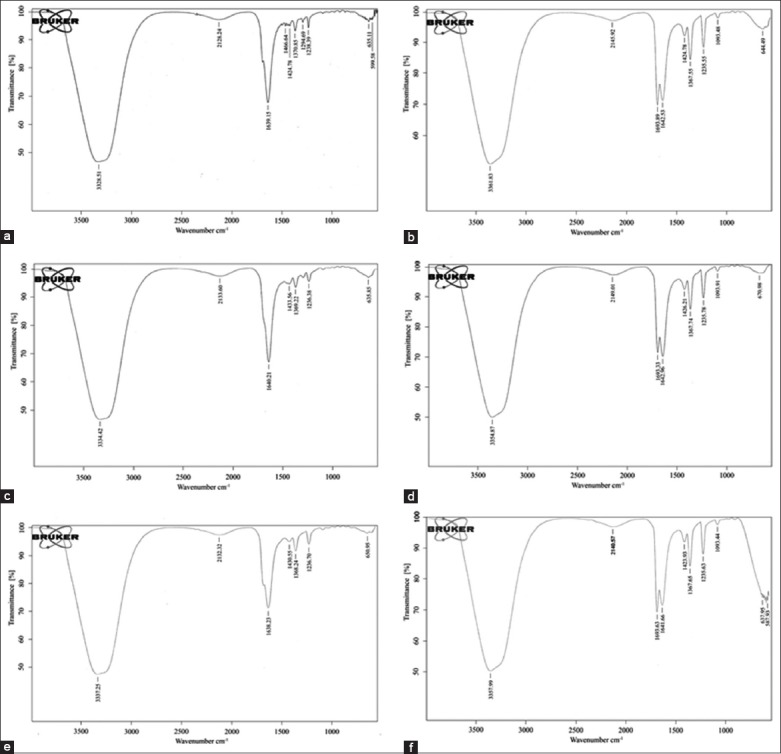
Fourier-transform infrared spectroscopic analysis of isolated polyphenolic fractions (a) stem bark fraction I, (b) stem bark fraction II, (c) leaf fraction I, (d) leaf fraction II, (e) fruit fraction I, (f) fruit fraction II
Identification of Polyphenols
Identification of polyphenols from stem bark
The identification of polyphenols was done based on their retention time and mass spectra (MS, m/z) determined using HPLC coupled with triple quadrupole mass spectrometer in positive and negative ion modes. These data were tabulated as retention time, peak area (%), m/z values, molecular weight, molecular formula, and name of the compound. Four peaks were obtained from both positive and negative ion modes of stem bark fraction-I. Here in the case of positive [M+H]+ mode showed retention time at 1.40 (m/z 111), 2.45 (m/z 121), 6.03 (m/z 165), and 7.30 (m/z 431) were identified as kaempferol, flavanone, epicatechin, and homoorientin, respectively [Table 1 and Figure 2 ai]. In negative [M-H]− mode, the retention time at 1.40 (m/z 113), 2.45 (m/z 227), 6.03 (m/z 317), and 7.30 (m/z 329) were identified as palatinose monohydrate, kaempferol-3-glucoside, myricitrin, and syringetin-3-O-galactoside, respectively [Table 1 and Figure 2 aii].
Table 1.
HPLC-ESI-MS/MS characterized polyphenols isolated from different parts of S. alternifolium
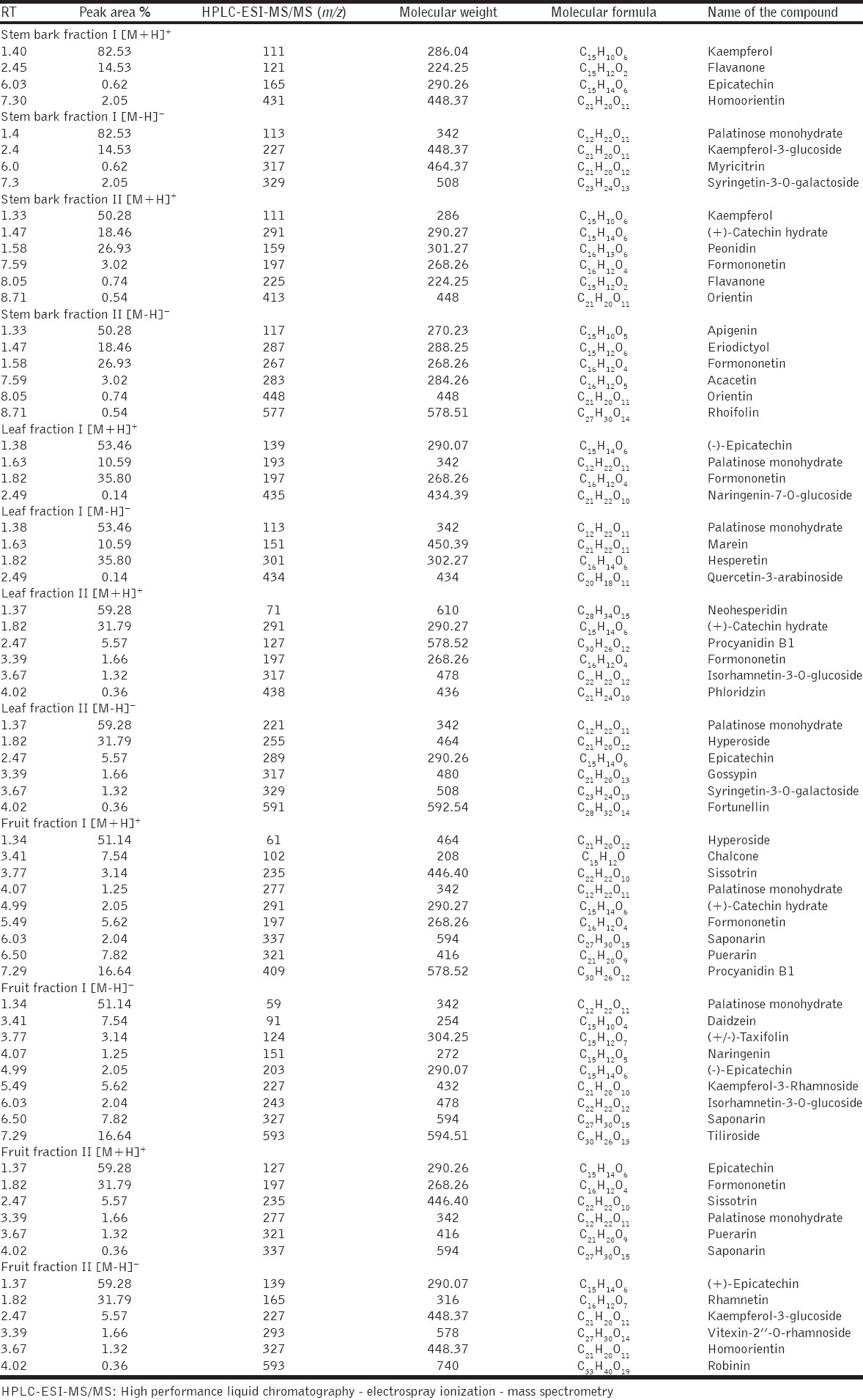
Figure 2.
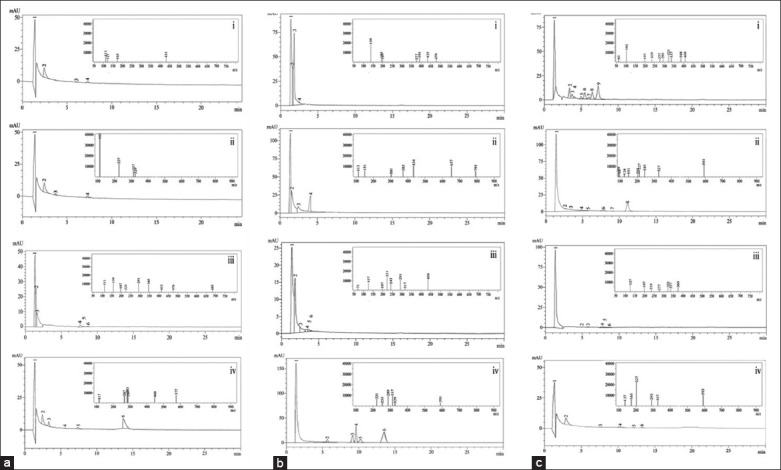
(a-c) High performance liquid chromatography chromatograms of a stem bark b leaf c fruit polyphenols. Inset image belongs to respective mass spectra (i) fraction I positive mode [M+H]+, (ii) fraction I negative mode [M-H]−, (iii) fraction II positive mode [M+H]+, (iv) fraction II negative mode [M-H]−
Whereas in the case of fraction-II positive [M+H]+ mode of stem bark gives 6 peaks with the retention time at 1.33 (m/z 111), 1.47 (m/z 291), 1.58 (m/z 159), 7.59 (m/z 197), 8.05 (m/z 225), and 8.71 (m/z 413) were identified as kaempferol, (+)-catechin hydrate, peonidin, formononetin, flavanone, and orientin [Table 1 and Figure 2 aiii]. In negative [M-H]− mode, the retention time at 1.33 (m/z 117), 1.47 (m/z 287), 1.58 (m/z 267), 7.59 (m/z 283), 8.05 (m/z 448), and 8.71 (m/z 577) were identified as apigenin, eriodictyol, formononetin, acacetin, orientin, and rhoifolin, respectively [Table 1 and Figure 2 aiv].
Identification of polyphenols from leaf
The leaf part of the fraction-I gives 4 peaks in both positive and negative modes. Here, the positive [M+H]+ mode demonstrated retention time at 1.38 (m/z 139), 1.63 (m/z 193), 1.82 (m/z 197), and 2.49 (m/z 435) were identified as (-)-epicatechin, palatinose monohydrate, formononetin, and naringenin-7-O-glucoside, respectively [Table 1 and Figure 2 bi]. In negative [M-H]− mode, the retention time at 1.38 (m/z 113), 1.63 (m/z 151), 1.82 (m/z 301), and 2.49 (m/z 434) were identified as palatinose monohydrate, marein, hesperetin, and quercetin-3-arabinoside [Table 1 and Figure 2 bii]. Whereas in the case of leaf part of the fraction-II gives 6 peaks in both positive [M+H]+ and negative [M-H]− modes. In positive [M+H]+ mode the retention time at 1.37 (m/z 71), 1.82 (m/z 291), 2.47 (m/z 127), 3.39 (m/z 197), 3.67 (m/z 317), and 4.02 (m/z 438) were identified as neohesperidin, (+)-catechin hydrate, procyanidin B1, formononetin, isorhamnetin-3-O-glucoside, and phloridzin (Table 1 and Figure 2 biii). In negative [M-H]− mode, the retention time at 1.37 (m/z 221), 1.82 (m/z 255), 2.47 (m/z 289), 3.39 (m/z 317), 3.67 (m/z 329), and 4.02 (m/z 591) were identified as palatinose monohydrate, hyperoside, epicatechin, gossypin, syringetin-3-O-galactoside, and fortunellin [Table 1 and Figure 2 biv].
Identification of polyphenols from fruit
The fruit part of the fraction-I gives 9 peaks in both positive and negative modes. Here, in the positive [M+H]+ mode, the retention time at 1.34 (m/z 61), 3.41 (m/z 102), 3.77 (m/z 235), 4.07 (m/z 277), 4.99 (m/z 291), 5.49 (m/z 197), 6.03 (m/z 337), 6.50 (m/z 321), and 7.29 (m/z 409) were identified as hyperoside, chalcone, sissotrin, palatinose monohydrate, (+)-catechin hydrate, formononetin, saponarin, puerarin, and procyanidin B1, respectively [Table 1 and Figure 2 ci]. Whereas in negative [M-H]− mode, the retention time at 1.34 (m/z 59), 3.41 (m/z 91), 3.77 (m/z 124), 4.07 (m/z 151), 4.99 (m/z 203), 5.49 (m/z 227), 6.03 (m/z 243), 6.50 (m/z 327), and 7.29 (m/z 593) were identified as palatinose monohydrate, daidzein, (±)-taxifolin, naringenin, (-)-epicatechin, kaempferol-3-rhamnoside, isorhamnetin-3-O-glucoside, saponarin, and tiliroside [Table 1 and Figure 2 cii]. In fruit part of the fraction-II gives 6 peaks in both positive and negative modes. Here in the case of positive [M+H]+ mode, the retention time at 1.33 (m/z 127), 4.90 (m/z 197), 5.73 (m/z 235), 7.66 (m/z 277), 8.01 (m/z 321), and 8.71 (m/z 337) were identified as epicatechin, formononetin, sissotrin, palatinose monohydrate, puerarin, and saponarin, respectively [Table 1 and Figure 2 ciii]. Whereas in the case of negative [M-H]− mode, the retention time at 1.33 (m/z 139), 4.90 (m/z 165), 5.73 (m/z 227), 7.66 (m/z 293), 8.01 (m/z 327), and 8.71 (m/z 593) were identified as (-)-epicatechin, rhamnetin, kaempferol-3-glucoside, vitexin-2’’-O-rhamnoside, homoorientin, and robinin [Table 1 and Figure 2 civ].
Pharmacokinetics and Structure-based Virtual Screening
ADME/Tox evaluations
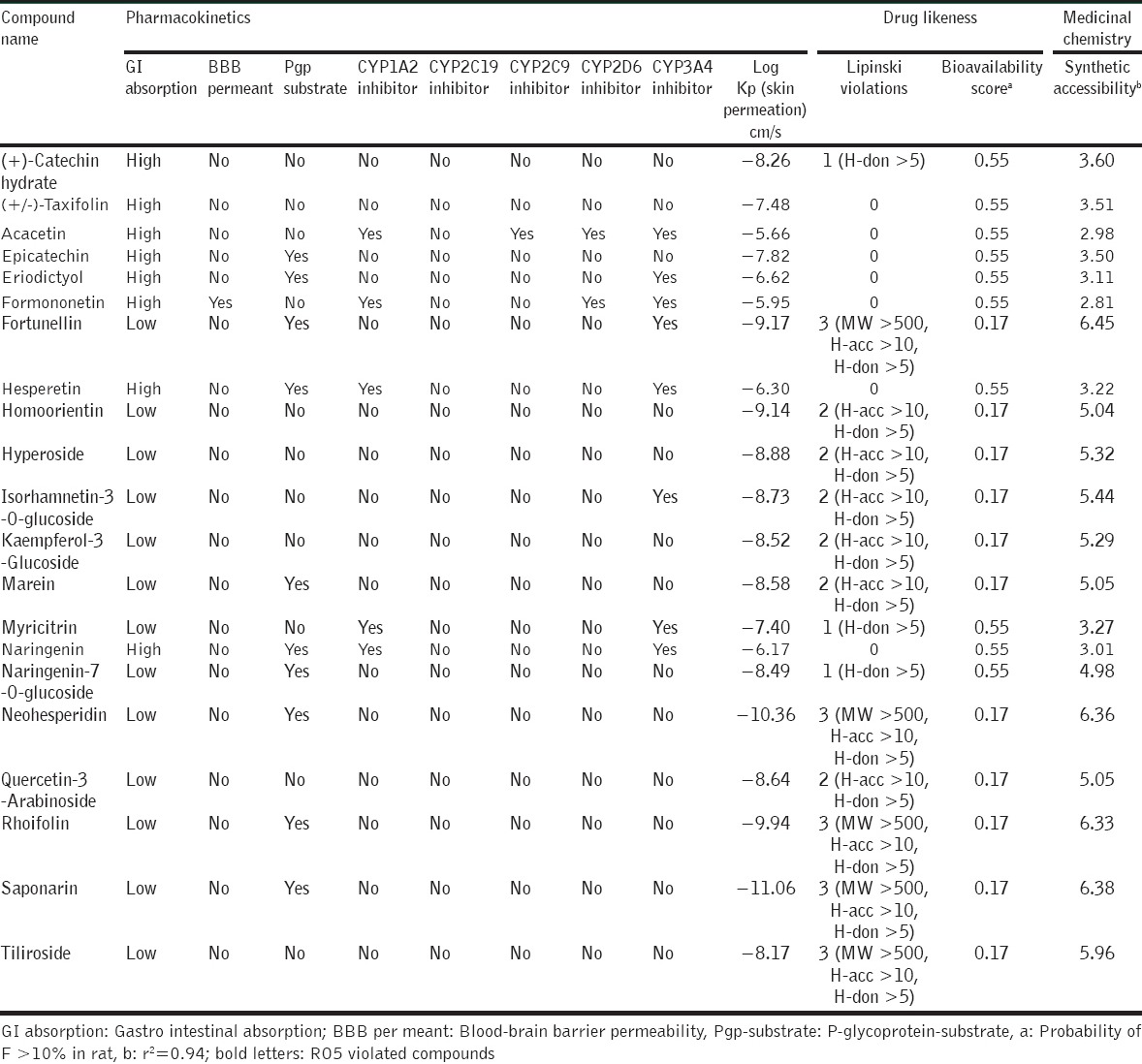
Among the 40 isolated polyphenols from S. alternifolium the 7 compounds such as (+)-epicatechin, epicatechin, flavanone, kaempferol-3-rhamnoside, palatinose monohydrate, syringetin-3-O-galactoside, and vitexin-2’’-O-rhamnoside have repeated scaffolds and unknown chirality. Due to this, these compounds were discarded for further analysis. Finally, 33 lead phenolics were listed based on principle fragments. Furthemore, they subjected to initial toxicity assessments through in silico pharmacokinetic screening strategies. The chemical scaffolds were retrieved from PubChem database and illustrated in the OSIRIS Property Explorer program, which computes drug-relevant properties of compounds and provides results as color coded features. From the 33 phenolics, apigenin, chalcone, kaempferol, orientin, rhamnetin, and robinin have serious mutagenic properties. The compound sissotrin showed strong tumorigenic effect. The compounds daidzein, gossypin, phloridzin, procyanidin B1, and puerarin consist of reproductive effects [Table 2]. Therefore, these 12 compounds are not profitable for therapeutic usage. The other bioactivity scores against druggable targets such as G protein–coupled receptors ligands, kinase inhibitors, ion channel modulators, nuclear receptor ligands, and enzyme inhibitors were predicted from molinspiration server, revealed that all the compounds act as prominent drug candidates for above targets receptors [Table 2]. The ADME evaluations of drugs were considerably more vital for potential therapeutic ligands prophecy. Here, broadly used Lipinski rule of five (RO5) [39] strategies was implemented for ADME property prediction by SwissADME tool. The physicochemical features of lead scaffolds uncovered that there were only 07 compounds obey the RO5 values. Other 14 compounds such as (+)-catechin hydrate, fortunellin, homoorientin, hyperoside, isorhamnetin-3-O-glucoside, kaempferol-3-glucoside, marein, myricitrin, naringenin-7-O-glucoside, neohesperidin, quercetin-3-arabinoside, rhoifolin, saponarin, and tiliroside are violating the RO5 principles [Tables 3a and b]. Further, the pharmacokinetics resources like drug-likeness and synthetic accessibility of respected lead candidates were likewise analyzed [Table 3b].
Table 2.
Toxicity and bioavailability properties of isolated S. alternifolium phenolic scaffolds
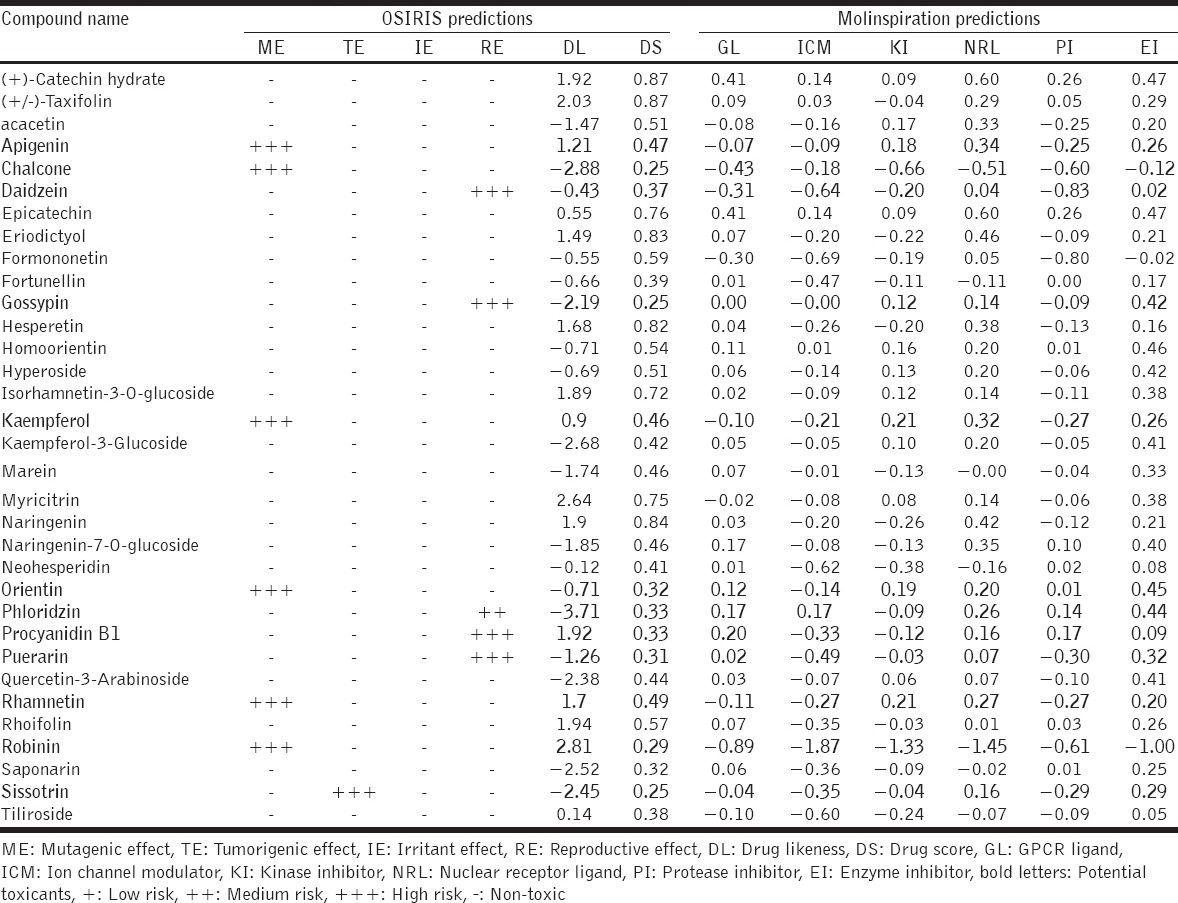
Table 3a.
Physicochemical properties of toxicity filtered lead compounds
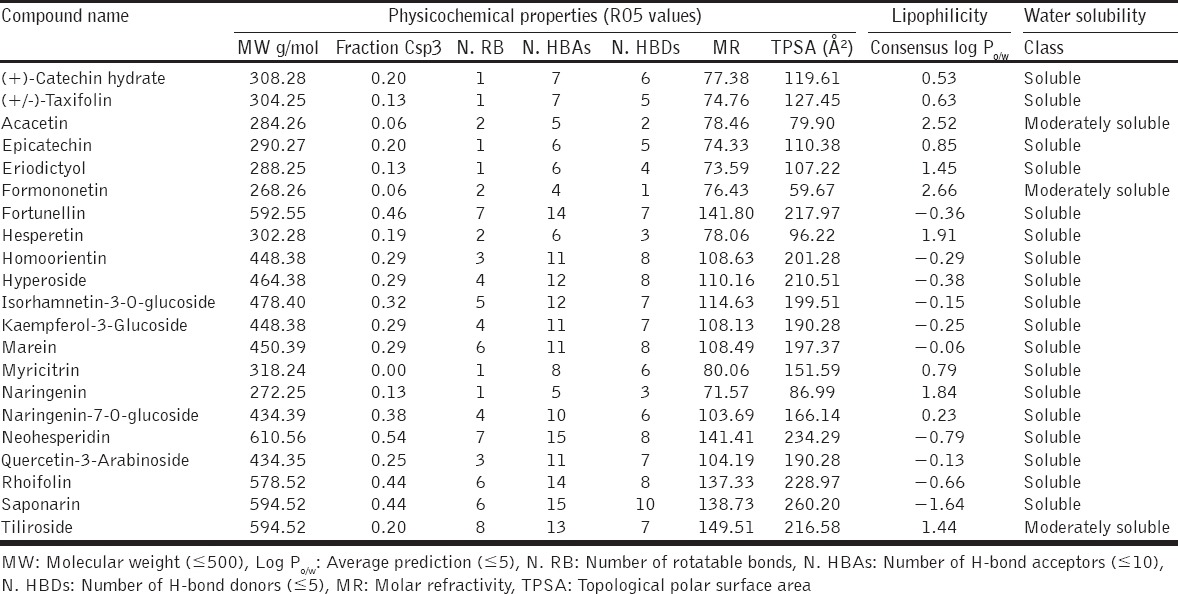
Table 3b.
ADME, RO5 violations, bioavailability and synthetic accessibility properties of selected lead compounds
Virtual screening and binding mode analysis
Recent in vitro studies reveal that S. alternifolium plant species have great antioxidant and anticancer activities. The previous reports positively revealed that the ERa has identified as a most significant therapeutic target specifically in breast cancer therapy. Hence, the investigations of phenolic scaffolds that can bind to ERa an interesting area for recognition of potential druggable lead compounds. In this study, we used virtual screening and molecular docking methods for 7 screened polyphenols against human ERa (PDB ID: 1A52) LBD with Autodock scoring functions by PyRx virtual screening program. Before going to docking, protein and ligands structure protonation states were adjusted to flexible point (pH=7) via structure optimization algorithms. The docking simulations found that all the ligands were prominently interacting with the core cavity of ERa active site like estradiol with functional residues [Figure 3]. As per docking log files, the compounds naringenin (439246), eriodictyol (440735), (+/-)-taxifolin (439533), (-)-epicatechin (72276), formononetin (5280378), acacetin (5280442), and hesperetin (72281) showed −8.9, −8.9, −8.7, −8.6, −7.4, −7.2, and −7.2 kcal/mol−1 ΔGbinding energies, respectively [Table 4].
Figure 3.
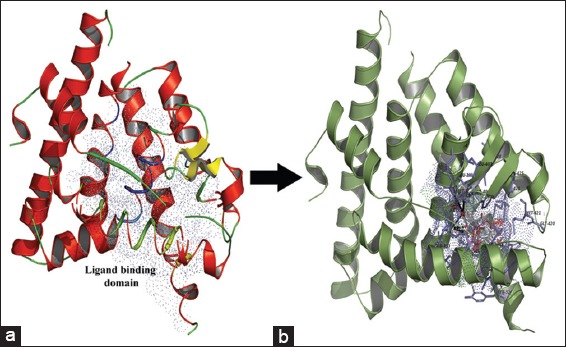
The secondary structure view of ligand binding domain of Estrogen receptor with potential isolated phenolics (a) secondary structure (red = helix, green = loops, yellow = sheets) with domain cavity (blue color dotted surface), (b) the structural alignment of lead scaffolds potentially fix in AutoLigand predicted domain cavity (blue color lines = functional residues, blue dotted surface = functional cavity, sticks = phenolic scaffolds)
Table 4.
AutoDock binding energy scoring values and interacting residues of selected polyphenols with ERαLBD
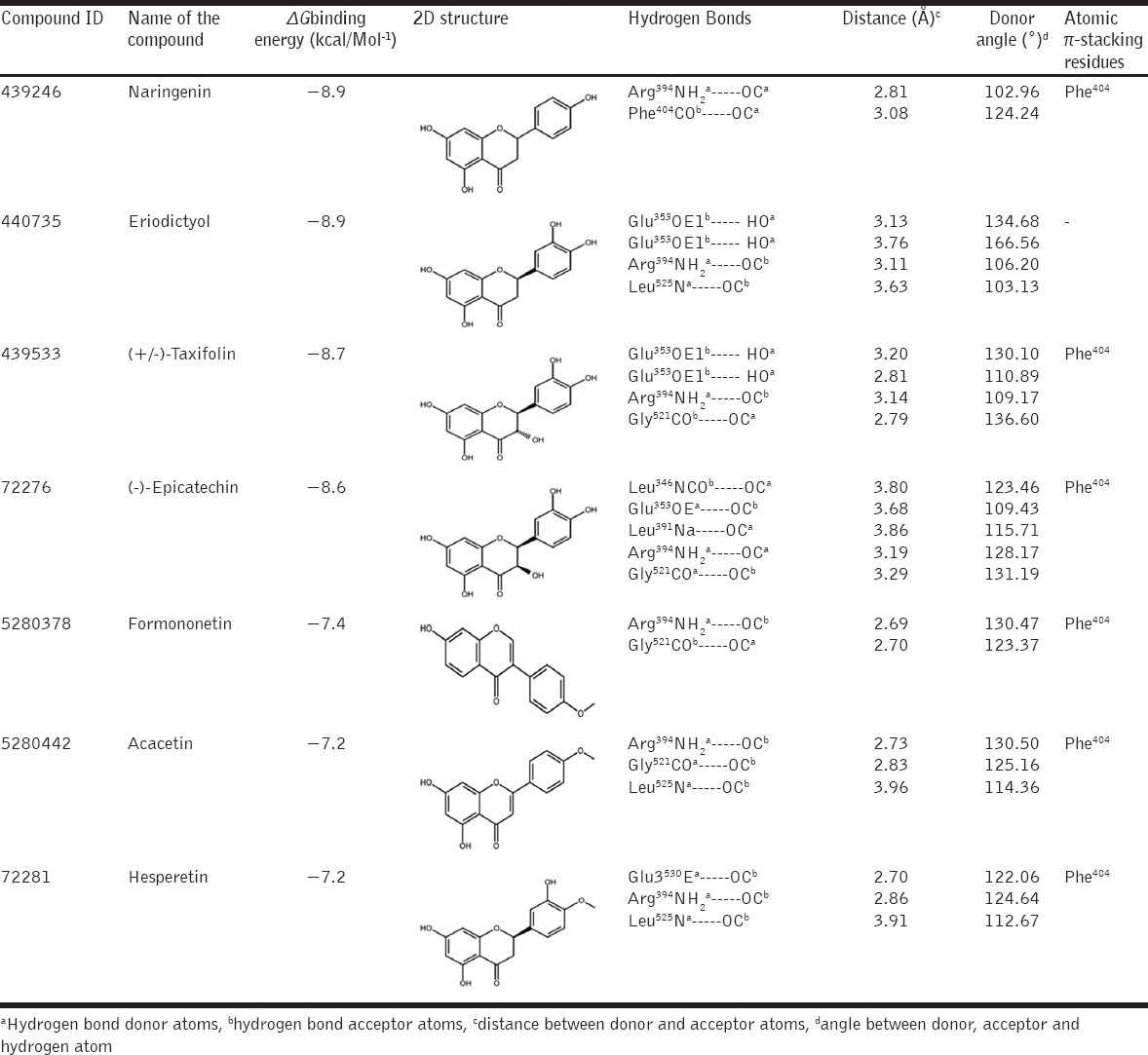
The compound naringenin framed two H-bonds with Arg394 and Phe404 residues with 2.81, 3.08 Å bond distances. In H-bond, Arg394 and dihydrochromen-4-one both acts as H-bond donors and mutually shares the electrons. In another bond, Phe404 bound as H-bond acceptor with donor dihydrochromen-4-one ring stacks and one additional atomic π-stacking was observed [Figure 4a]. The compound eriodictyol formed four H-bonds with Glu353, Arg394, and Leu525 residues with 3.13, 3.76, 3.11, 3.63 Å bond distances. Here, Glu353 acts as H-bond acceptor and it pulls electrons from CO group atoms of dihydrochromen-4-one ring stacks, Arg394 and Leu525 both are acts as H-bond donors to CO group atoms of dihydrochromen-4-one and dihydroxyphenyl ring stack, respectively, and Phe404 bound as hydrophobic residue [Figure 4b]. The compound (+/-)-taxifolin bound like eriodictyol and formed four H-bonds with Glu353, Arg394, and Gly521 residues with 3.13, 3.76, 3.11 and 3.63 Å bond distances. Here, Glu353 acts as H-bond acceptor and it pulls electrons from CO group atoms of dihydrochromen-4-one ring stacks, Arg394 and Gly521 both are acts as H-bond donors and acceptors to CO group atoms of dihydrochromen-4-one and dihydroxyphenyl ring stacks, respectively, Phe404 framed one atomic π-stacking with ring stacks of dihydrochromen-4-one ring stacks [Figure 4c]. The compound (-)-epicatechin bound like eriodictyol, (+/-)-taxifolin and frame five H-bonds with Leu346, Glu353, Leu391, Arg394 and Gly521 residues with 3.80, 3.68, 3.86, 3.19 and 3.29 Å bond distances. Leu391 and Arg394 bound as donors, their shares electrons to CO group atoms of dihydrochromen-4-one chain, respectively. Leu346, Glu353 and Gly521 acts as H-bond acceptor, and they pull electrons from CO group atoms of dihydrochromen-4-one and dihydroxyphenyl ring stacks, the Phe404 framed one atomic π-stacking with ring stacks of dihydrochromen-4-one ring stacks [Figure 4d].
Figure 4.
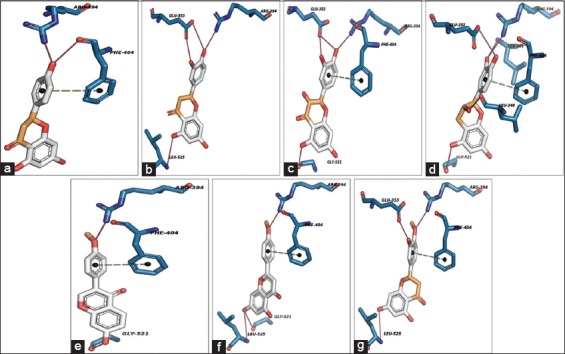
Molecular interaction analysis of best potential phenolics, (a) naringenin, (b) eriodictyol, (c) (+/-)-taxifolin, (d) (-)-epicatechin, (e) formononetin, (f) acacetin and (g) hesperetin (red = H-bond interactions, dotted lines = atomic π-contacts)
The compound formononetin formed two H-bonds with Arg394 and Gly521 residues of ERLBD with 2.69 and 2.70 Å bond distances. Arg394 and Gly521 both are acts as H-bond donors and acceptors to CO group atoms of dihydrochromen-4-one and dihydroxyphenyl ring stacks, respectively, Phe404 framed one atomic π-stacking with ring stacks of dihydrochromen-4-one ring stacks [Figure 4e]. The compound acacetin built three H-bonds with Arg394, Gly521, and Leu525 residues with 2.73, 2.83, and 3.96 Å of bond distances. Arg394 and Leu525 both are acts as H-bond donors to CO group atoms of dihydrochromen-4-one and dihydroxyphenyl ring stack. Gly521 bound as the acceptor and it pull electrons from CO group atoms of dihydroxyphenyl ring stacks and Phe404 framed one atomic π-stacking with ring stacks of dihydrochromen-4-one ring stacks [Figure 4f]. The compound hesperetin formed three H-bonds with Glu353, Arg394 and Leu525 residues of ERLBD with 2.70, 2.86 and 3.91 Å of bond distances. Arg394 and Leu525 both are acts as H-bond donors to CO group atoms of dihydrochromen-4-one and dihydroxyphenyl ring stacks. Glu353 acts as H-bond acceptor and it pulls electrons from CO group atoms of dihydrochromen-4-one and Phe404 framed one atomic π-stacking with ring stacks of dihydrochromen-4-one ring stacks [Figure 4g]. Structural superimposition of all lead scaffolds to LBD of ERa showed that functional Glu353, Arg394, Phe404 and other hydrophobic residues are participating to unique H-bond acceptor, H-bond donors and van der Waals contacts (π-stacking) formation, which are crucial for estrogenic 3-hydroxyl group interaction, water-mediated H-bond formation and receptor fixed in specific positions for hormone specificity [Figure 5].
Figure 5.
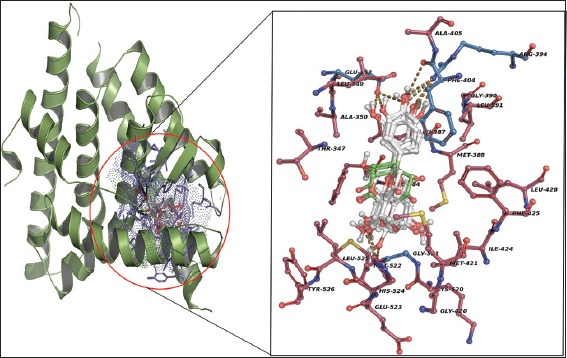
Structural superimposition of ligands with in AutoLigand catalytic cleft of estrogen receptor a, red color sticks = hydrophobic contacts, yellow dotted lines = H-bond interactions, blue color sticks = functional cleft residues, center aligned sticks = ligands
DISCUSSION
A total number of 40 compounds were identified from stem bark, leaf, and fruit parts of the plant. Among them, the compound kaempferol from stem bark fraction I and II of positive [M+H]+ mode shows a noteworthy peak area of percentage. Kaempferol was the principle compound found in Brassica crops. Higher intake of Kaempferol reduces coronary heart disease, has strong antioxidant activity and suppress the growth of human gut cancer cell lines [40]. The compounds palatinose monohydrate from fraction I and apigenin from fraction II of stem bark extract of negative [M-H]− mode indicated highest peak area of percentage. The compound apigenin was also identified from Acanthophora spicifera as principle compound, possesses potent analgesic, anti-inflammatory, and antiproliferative activities [41]. The compounds (-)-epicatechin and neohesperidin show the highest peak area in leaf part of fraction I and II of positive [M+H]+ mode. The compound (-)-epicatechin was found as the main compound in grapes and cocoa [42]. Whereas, neohesperidin as main compound in citron fruits [43] has anticancer activity. Whereas in the case of leaf fraction I and II of negative [M-H]− mode, the palatinose monohydrate showed astounding peak percentage. The compounds hyperoside and epicatechin from fruit fraction I and II of positive [M+H]+ mode show the highest peak area of percentage. The same type of result was obtained from Eucalyptus globulus showed hyperoside as the main compound and act as synergistic antimicrobial and antioxidant activity [44]. The compound epicatechin was isolated from tea leaves as the principle compound has antioxidant activity [45]. The compounds palatinose monohydrate and (-)-epicatechin from fraction I and II of negative mode of fruit part, respectively, showed an elevate peak area of percentage.
Compounds such as kaempferol, flavanone, peonidin, and myricitrin of the positive [M+H]+ mode and apigenin, acacetin, eriodictyol, orientin, and rhoifolin of negative [M-H]− mode were solely obtained from stem bark of the plant. The compounds neohesperidin, phloridzin and naringenin-7-O-glucoside of positive [M+H]+ mode, marein, hesperetin, gossypin, quercetin-3-arabinoside, fortunellin of negative [M-H]− mode were obtained from leaf part of the plant only. Whereas the compounds, chalcone, sissotrin, puerarin and saponarin of positive [M+H]+ mode, daidzein, (+/-)-taxifolin, naringenin, rhamnetin, kaempferol-3-rhamnoside, robinin, tiliroside, vitexin-2’’-O-rhamnoside of negative [M-H]− mode were obtained only from fruit part of the plant. The compounds, Syringetin-3-O-galactoside was obtained from stem bark and leaf parts, homoorientin and kaempferol-3-glucoside were obtained from stem bark and fruit parts, (-)-epicatechin, hyperoside, isorhamnetin-3-O-glucoside and procyanidin B1 were obtained from leaf and fruit parts of the plant. The compounds such as (+)-catechin hydrate, epicatechin, formononetin, and palatinose monohydrate were obtained from all the parts of the plant.
Based on structural complexity, the obtained polyphenols were arranged from simple to polymeric polyphenols, viz., flavones, flavonols, flavanones, flavanols, dihydroflavonols, anthocyanins, proanthocyanidins, isoflavones (flavonoid), and chalcones (nonflavonoid). The isolated compounds from S. alternifolium, apigenin, acacetin, homoorientin, orientin, rhoifolin, fortunellin, vitexin-2’’-o-rhamnoside, and saponarin are belonging to flavone class of flavonoid compounds. These are yellow colored flavonoid compounds having 2- phenylchromen-4-one backbone, soluble in water and ethanol. They found predominantly in cereals and herbs. They have hydroxy, carbonyl, and conjugated double bond functional groups to make possible ways for subsequent reactions [46]. The compounds, kaempferol, rhamnetin, kaempferol-3-glucoside, hyperoside, quercetin-3-arabinoside, kaempferol-3-rhamnoside, myricitrin, syringetin-3-o-galactoside, isorhamnetin-3-o-glucoside, gossypin, robinin, and tiliroside are belonging to flavonol class of flavonoid compounds. These are colorless flavonoid compounds structurally represent C6-C3-C6 system, possessing two benzene rings joined by a linear three carbon chain. They differ from other flavonoids by having hydroxyl group at 3 positions of C ring attributes antioxidant, anti-inflammatory, and anticancer properties [47].
Flavanones are contrasted with flavonols, by having a chiral C2 center, C2-C3 saturated bond, phenol B ring, and most of them are nonplanar in nature, which have several to multi hydroxylated groups either by glycosylated or methylated [48]. From the isolated compounds, flavanone, naringenin, eriodictyol, hesperetin, naringenin-7-O-glucoside, neohesperidin and palatinose monohydrate comes under flavanone class of flavonoid compounds. Flavanols are one of the major subclasses of flavonoids, which are structurally similar with anthocynadins. Chemically they differ from other flavonoids by lacking of oxygen group at 4 positions and presence of double bond between 2 and 3 positions of C ring is the contrasting characters. The presence of hydroxyl group at 3 position creates two centers of asymmetry for polymerization of flavanols to give brown pigments, which are rich sources in green tea and cocoa [49]. The compounds epicatechin, (-)-epicatechin and (+)-catechin hydrate comes under the flavanol class of flavonoid compounds. The compound (+/-)-taxifolin comes under dihydroflavonol class of flavonoid compound, which is the subclass of flavonols, structurally similar with flavonols [50]. The compound peonidin comes under anthocyanin group of flavonoid compounds derived from phenylalanine, which are synthesized in cytosol, stored in vacuoles and imparts different colors to flowers, fruits and vegetables. Structurally they contain falvylium cation linked either by hydroxyl or methoxyl groups and have one or more sugars [51].
Proanthocyanidins represent the second most abundant class of natural polyphenolics, which are widely distributed in various parts of bark, berries, flowers, fruits, and seeds; they give protection from microorganisms. They have a complex chemical structure being oligomers or polymers. Based on the bonding between monomers of proanthocyanidins, it may forms, either of B-type or A-type of structures [52]. The compound procyanidin B1 comes under proanthocyanidin group of tannin compound. From the isolated compounds, daidzein, formononetin, puerarin, and sissotrin comes under the isoflavone class of polyphenols, which are colorless polyphenols, predominant in legumes, especially in soybean plants has a significant impact on human health.
They are structurally similar to other flavonoids. However, differ in linking of the B ring to second position of the C ring, it may link via the third position of C ring. This structure is similar to an estrogen that is why they are also known as phytoestrogens [53]. Chalcones are nonflavonoid class of polyphenolic compounds have 1,3-diaryl-2-propen-1-one as backbone structure, found in fruits and vegetables. Chalcones display dimer, oligomer, diels-alder adducts and other conjugates, which differ from other flavonoids by displaying open chain with three carbon molecules, binds to A and B ring instead of C ring [54]. From the isolated compounds chalcone, phloridzin and marein are belonging to chalcone class of flavonoid compounds.
The pre-ADME approaches of pharmacological candidates screening are vital potentiated towards moderating different diseases. Here, the in silico ADME/Tox analysis is the profitable approach and safe to examine the promising drugs recognition within low time and cost [29]. The previous outcomes evidenced that plant polyphenols were strongly acted as antagonist or agonists for different therapeutic targets to ameliorate the various diseases [55]. However, in vivo and in vitro studies revealed the plant phenols consists some toxicological properties [56]. The plant S. alternifolium has enormous medicinal and bioavailability features [57]. Here, the adaptable polyphenols are isolated and characterized from S. alternifolium through in vitro and in silico approaches are considered as our theme line object. After distinguished polyphenols from S. alternifolium, pharmacokinetics and lead-likeness properties of compounds are implemented to optimize the ADME toxicological features by using computational programmings. Overall two filtration schemes reveal that 7 potential polyphenols such as naringenin, eriodictyol, (+/-)-taxifolin, (-)-epicatechin, formononetin, acacetin, and hesperetin are obeying the RO5 principles and have a reliable ADME features without toxicity. Here, we put our efforts to screen nontoxic next generation druggables for reducing the breast carcinoma. Some RO5 fluctuations of phenolics are acceptable; however, the resulted compounds are potentiate for further in vitro analysis; only versatile compounds are considered in our study.
Molecular docking simulations of virtually screened compounds against the predicted druggable pocket of the ERa structure showed that binding affinities occupied in between −8.9 and −7.2 kcal/mol−1 energies. The three replication methods indicated that naringenin, eriodictyol, (+/-)-taxifolin, (-)-epicatechin, formononetin, acacetin, and hesperetin were bound with −8.9, −8.9, −8.7, −8.6, −7.4, −7.2, and −7.2 kcal/mol−1 ΔG binding energies within the core cavity of receptor, respectively. From the docking consequences, the specificity of screened phenolic compounds resulted that they possibly repositioning the estradiol and has strong binding affinities with ERa like naringenin. Molecular interaction profiles reveal that compounds are from probable hydrogen bonds, atomic π-contacts, salt bridges like reference drug estradiol. Henceforth, they may serve as inhibitors to ERa for mitigating the breast invasive carcinoma. From the previous studies, naringenin inhibits or antagonize the respective biochemical or biosynthesis action of estrogen substrates by estrogen-mediated mechanism. It potentially reduces the breast carcinoma by preventing and suppresses the transforming growth factor beta 1 secretion [58] and protein kinase-C activation inhibition [59]. Moreover, naringenin, eriodictyol, (+/-)-taxifolin, (-)-epicatechin prompts the apoptosis in cancer cells through associated inhibition of fatty acid biosynthesis [60]. Isoflavone and formononetin are arresting the cell cycle through interferes with several cell signaling pathways to reduce breast cancer cell viability [61]. Acacetin prompts the apoptosis in breast cancer cells through interactive with various cell signaling pathways [62]. The flavonone, hesperetin, and naringenin have highest inhibitory potency in aromatase-expressing MCF-7 tumor cell viability [63]. Overall, our docking outcomes strongly agree with the previous enormous evidences. Therefore, these hopeful leads are reliable for further clinical and preclinical approaches. Thus, these compounds could use as anticancer agents against potential therapeutic targets toward various malignant disorders.
CONCLUSION
In this study, an investigation has been made to isolate polyphenols from the medicinal plant S. alternifolium and to characterize those with FT-IR and extended with HPLC-PDA-ESI-MS/MS. Our selected protocol and characterization tools were best suitable for isolation of polyphenols from S. alternifolium. Overall, a total number of 40 compounds were obtained, and most of them are belongs to flavonol class of polyphenolics. The result of this study screened out a selective number of compounds has anticancer activity. There is a good phenomenon about the polyphenols, acts as natural anticancerous agents. However, the isolation and standardization of polyphenols are meager in the present scenario. This study extends the use of natural polyphenols available in S. alternifolium in cancer-related diseases, especially in the case of breast cancer. The present computational methodologies revealed that the 7 phenolics consist of best drug-like properties, virtual screening, and molecular docking studies against ER indicate ligands potentially interact with functional cleft residues by forming prominent H-bonds and atomic π-contacts with flexible residues. The obtained results were agreed with previous evidence. Thus, overall in silico strategies indicate that the lead scaffolds could be served as future anticancer agents to ameliorate the breast carcinoma.
ACKNOWLEDGMENTS
The authors, Pulicharla Yuganthar and Konidala Kranthi Kumar, wish to acknowledge the University Grants Commission-Basic Science and Research and Rajiv Gandhi National Fellowship, New Delhi, respectively, for providing fellowships.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Khoddami A, Wilkes MA, Roberts TH. Techniques for analysis of plant phenolic compounds. Molecules. 2013;18:2328–75. doi: 10.3390/molecules18022328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kyle JA, Duthie GG. In: Flavonoids in foods in flavonoids. Chemistry, Biochemistry and Applications. Andersen O, Markhamv K, editors. Boca Raton: CRC Press; 2006. p. 219. [Google Scholar]
- 3.Crozier A, Jaganath IB, Clifford MN. Dietary phenolics: Chemistry, bioavailability and effects on health. Nat Prod Rep. 2009;26:1001–43. doi: 10.1039/b802662a. [DOI] [PubMed] [Google Scholar]
- 4.Kim KH, Moon E, Choi SU, Kim SY, Lee KR. Polyphenols from the bark of Rhus verniciflua and their biological evaluation on antitumor and anti-inflammatory activities. Phytochemistry. 2013;92:113–21. doi: 10.1016/j.phytochem.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Rodríguez-Rivera MP, Lugo-Cervantes E, Winterhalter P, Jerz G. Metabolite profiling of polyphenols in peels of Citrus limetta Risso by combination of preparative high-speed countercurrent chromatography and LC-ESI-MS/MS. Food Chem. 2014;158:139–52. doi: 10.1016/j.foodchem.2014.02.077. [DOI] [PubMed] [Google Scholar]
- 6.Grace MH, Warlick CW, Neff SA, Lila MA. Efficient preparative isolation and identification of walnut bioactive components using high-speed counter-current chromatography and LC-ESI-IT-TOF-MS. Food Chem. 2014;158:229–38. doi: 10.1016/j.foodchem.2014.02.117. [DOI] [PubMed] [Google Scholar]
- 7.García-Salas P, Gómez-Caravaca AM, Morales-Soto A, Segura-Carretero A, Fernández-Gutiérrez A. Identification and quantification of phenolic and other polar compounds in the edible part of Annona cherimola and its by-products by HPLC-DAD-ESI-QTOF-MS. Food Res Int. 2015;78:246–57. doi: 10.1016/j.foodres.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Oliveira RB, Chagas-Paula DA, Secatto A, Gasparoto TH, Faccioli LH, Campanelli AP, et al. Topical anti-inflammatory activity of yacon leaf extracts. Rev Bras Farmacogn. 2013;23:497–505. [Google Scholar]
- 9.Junqueira-Gonçalves MP, Yáñez L, Morales C, Navarro M, A Contreras R, Zúñiga GE. Isolation and characterization of phenolic compounds and anthocyanins from Murta (Ugni molinae Turcz.) Fruits. Assessment of antioxidant and antibacterial activity. Molecules. 2015;20:5698–713. doi: 10.3390/molecules20045698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen V, Staub RE, Baggett S, Chimmani R, Tagliaferri M, Cohen I, et al. Identification and analysis of the active phytochemicals from the anti-cancer botanical extract Bezielle. PLoS One. 2012;7:e30107. doi: 10.1371/journal.pone.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo KH, Ra JE, Lee SJ, Lee JH, Kim SR, Lee JH, et al. Anti-hyperglycemic activity of polyphenols isolated from barnyard millet (Echinochloa utilis L.) And their role inhibiting a-glucosidase. J Korean Soc Appl Biol Chem. 2015;58:571–9. [Google Scholar]
- 12.Pellati F, Bruni R, Righi D, Grandini A, Tognolini M, Pio Prencipe F, et al. Metabolite profiling of polyphenols in a Terminalia chebula Retzius ayurvedic decoction and evaluation of its chemopreventive activity. J Ethnopharmacol. 2013;147:277–85. doi: 10.1016/j.jep.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 13.Cai H, Xie Z, Liu G, Sun X, Peng G, Lin B, et al. Isolation, identification and activities of natural antioxidants from Callicarpa kwangtungensis Chun. PLoS One. 2014;9:e93000. doi: 10.1371/journal.pone.0093000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eid HH, Labib RM, Hamid NS, Hamed MA, Ross SA. Hepatoprotective and antioxidant polyphenols from a standardized methanolic extract of the leaves of Liquidambar styraciflua L. Bull Fac Pharm. 2015;53:117–27. [Google Scholar]
- 15.Kisseih E, Lechtenberg M, Petereit F, Sendker J, Zacharski D, Brandt S, et al. Phytochemical characterization and in vitro wound healing activity of leaf extracts from Combretum mucronatum Schum. & Thonn: Oligomeric procyanidins as strong inductors of cellular differentiation. J Ethnopharmacol. 2015;174:628–36. doi: 10.1016/j.jep.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Motilva MJ, Serra A, Macià A. Analysis of food polyphenols by ultra high-performance liquid chromatography coupled to mass spectrometry: An overview. J Chromatogr A. 2013;1292:66–82. doi: 10.1016/j.chroma.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Saha D, Ved D, Ravikumar K, Haridasan K. Syzygium alternifolium. The IUCN Red list of Threatened Species. Switzerland: IUCN; 2015. [Google Scholar]
- 18.Karuppusamy S, Muthuraja G, Rajasekaran KM. Lesser known ethno medicinal plants of Alagar Hills, Madurai district of Tamil Nadu, India. Ethnobot Leaflets. 2009;13:1426–33. [Google Scholar]
- 19.Sudhakar A, Ramesh C, Nagaraju N, Vedavathy S, Murthy KS. Pharmacognostical studies on stem & fruit of Syzygium alternifolium (Wight) Walp-an endemic to South Eastern Ghats, India. Asian J Biochem Pharm Res. 2012;1:127–38. [Google Scholar]
- 20.Savithramma N, Yugandhar P, Haribabu R, Sivaprasad K. Validation of indigenous knowledge of Yanadi tribe and local villagers of Veyilingala Kona - A sacred grove of Andhra Pradesh, India. J Pharm Sci Res. 2014;6:382–8. [Google Scholar]
- 21.Savithramma N, Yugandhar P, Lingarao M. Ethnobotanical studies on Japali hanuman Theertham - A sacred grove of Tirumala Hills, Andhra Pradesh, India. J Pharm Sci Res. 2014;6:83–8. [Google Scholar]
- 22.Komuraiah B, Srinivas C, Kumar AN, Srinivas VN, Venu CH, Kumar JK, et al. Isolation of phytochemicals from anticancer active extracts of Syzygium alternifolium Walp. Leaf. Pharmacogn J. 2014;6:83–5. [Google Scholar]
- 23.Raju VV, Ramesh M, Narsau ML, Kumar MM. Antimicrobial activity of the plant Syzygium alternifolium. Asian J Chem. 2007;19:4923–4. [Google Scholar]
- 24.Sreelathadevi RK, Sreenivasulu P, Basha SK. Antioxidant activity and total polyphenols content of certain high valued medicinal plants of Tirumala Hills, Andhra Pradesh. Indian J Plant Sci. 2013;2:93–8. [Google Scholar]
- 25.Ramohan A, Prasad KV, Sharma JA. Hypoglycemic and anti hyperglycemic activity of Syzygium alternifolium (Wt.) Walp. Leaf extracts in normal and diabetic rats. Int J Drug Dev Res. 2010;2:27–32. [Google Scholar]
- 26.Yugandhar P, Savithramma N. Spectroscopic and chromatographic exploration of different phytochemical and mineral contents from Syzygium alternifolim (Wt.) Walp. An endemic, endangered medicinal tree taxon. J Appl Pharm Sci. 2017;7:73–85. [Google Scholar]
- 27.Sawada Y, Nakabayashi R, Yamada Y, Suzuki M, Sato M, Sakata A, et al. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: A plant-specific MS/MS-based data resource and database. Phytochemistry. 2012;82:38–45. doi: 10.1016/j.phytochem.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Magalhães PJ, Vieira JS, Gonçalves LM, Pacheco JG, Guido LF, Barros AA. Isolation of phenolic compounds from hop extracts using polyvinylpolypyrrolidone: Characterization by high-performance liquid chromatography-diode array detection-electrospray tandem mass spectrometry. J Chromatogr A. 2010;1217:3258–68. doi: 10.1016/j.chroma.2009.10.068. [DOI] [PubMed] [Google Scholar]
- 29.Singh SS. Preclinical pharmacokinetics: An approach towards safer and efficacious drugs. Curr Drug Metab. 2006;7:165–82. doi: 10.2174/138920006775541552. [DOI] [PubMed] [Google Scholar]
- 30.Sander T, Freyss J, von Korff M, Reich JR, Rufener C. OSIRIS, an entirely in-house developed drug discovery informatics system. J Chem Inf Model. 2009;49:232–46. doi: 10.1021/ci800305f. [DOI] [PubMed] [Google Scholar]
- 31.Daina A, Michielin O, Zoete V. iLOGP: A simple, robust, and efficient description of n-octanol/water partition coefficient for drug design using the GB/SA approach. J Chem Inf Model. 2014;54:3284–301. doi: 10.1021/ci500467k. [DOI] [PubMed] [Google Scholar]
- 32.Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44:D1202–13. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. 27-8. [DOI] [PubMed] [Google Scholar]
- 34.Zoete V, Cuendet MA, Grosdidier A, Michielin O. SwissParam: A fast force field generation tool for small organic molecules. J Comput Chem. 2011;32:2359–68. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 35.Pradeepkiran JA, Kumar KK, Kumar YN, Bhaskar M. Modeling, molecular dynamics, and docking assessment of transcription factor rho: A potential drug target in Brucella melitensis 16M. Drug Des Devel Ther. 2015;9:1897–912. doi: 10.2147/DDDT.S77020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanenbaum DM, Wang Y, Williams SP, Sigler PB. Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains. Proc Natl Acad Sci U S A. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 38.Harris R, Olson AJ, Goodsell DS. Automated prediction of ligand-binding sites in proteins. Proteins. 2008;70:1506–17. doi: 10.1002/prot.21645. [DOI] [PubMed] [Google Scholar]
- 39.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 40.Cartea ME, Francisco M, Soengas P, Velasco P. Phenolic compounds in Brassica vegetables. Molecules. 2010;16:251–80. doi: 10.3390/molecules16010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El Shoubaky GA, Abdel-Daim MM, Mansour MH, Salem EA. Isolation and Identification of a flavone apigenin from marine red alga Acanthophora spicifera with antinociceptive and anti-inflammatory activities. J Exp Neurosci. 2016;10:21–9. doi: 10.4137/JEN.S25096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mojzer EB, Hrncic MK, Skerget M, Knez Z, Bren U. Polyphenols: Extraction methods, antioxidative action, bioavailability and anticarcinogenic effects. Molecules. 2016;21:1–38. doi: 10.3390/molecules21070901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venturini N, Barboni T, Curk F, Costa J, Paolini J. Volatile and flavonoid composition of the peel of Citrus medica L. Var. Corsican fruit for quality assessment of its liqueur. Food Technol Biotechnol. 2014;52:403–10. doi: 10.17113/ftb.52.04.14.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dezsi S, Badarau AS, Bischin C, Vodnar DC, Silaghi-Dumitrescu R, Gheldiu AM, et al. Antimicrobial and antioxidant activities and phenolic profile of Eucalyptus globulus Labill. And Corymbia ficifolia (F. Muell.) K.D. Hill & L.A.S. Johnson leaves. Molecules. 2015;20:4720–34. doi: 10.3390/molecules20034720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dreosti IE. Antioxidant polyphenols in tea, cocoa, and wine. Nutrition. 2000;16:692–4. doi: 10.1016/s0899-9007(00)00304-x. [DOI] [PubMed] [Google Scholar]
- 46.Singh M, Kaur M, Silakari O. Flavones: An important scaffold for medicinal chemistry. Eur J Med Chem. 2014;84:206–39. doi: 10.1016/j.ejmech.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 47.Kim JD, Liu L, Guo W, Meydani M. Chemical structure of flavonols in relation to modulation of angiogenesis and immune-endothelial cell adhesion. J Nutr Biochem. 2006;17:165–76. doi: 10.1016/j.jnutbio.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Constantin RP, Nascimento GS, Constantin RP, Salgueiro CL, Bracht A, Ishii-Iwamoto EL, et al. Citrus flavanones affect hepatic fatty acid oxidation in rats by acting as prooxidant agents. Biomed Res Int. 2013;2013:1–12. doi: 10.1155/2013/342973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hollman PC, Arts IC. Flavonols, flavones and flavanols-nature, occurrence and dietary burden. J Sci Food Agric. 2000;80:1081–93. [Google Scholar]
- 50.Tang LK, Chu H, Yip WK, Yeung EC, Lo C. An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New Phytol. 2009;181:576–87. doi: 10.1111/j.1469-8137.2008.02692.x. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–49. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- 52.Lin GM, Lin HY, Hsu CY, Chang ST. Structural characterization and bioactivity of proanthocyanidins from indigenous cinnamon (Cinnamomum osmophloeum) J Sci Food Agric. 2016;96:4749–59. doi: 10.1002/jsfa.7802. [DOI] [PubMed] [Google Scholar]
- 53.Zhang YC, Schwartz SJ. In: Analysis of isoflavones in soy foods. Current Protocol in Food Analytical Chemistry. Wrolstad RE, Acree TE, An H, Decker EA, Penner MH, Reid DS, et al., editors. New York: John Wiley & Son; 2003. [Google Scholar]
- 54.Aksoz BE, Ertan R. Chemical and structural properties of chalcones I. FABAD J Pharm Sci. 2011;36:223–42. [Google Scholar]
- 55.Lounnas V, Ritschel T, Kelder J, McGuire R, Bywater RP, Foloppe N. Current progress in structure-based rational drug design marks a new mindset in drug discovery. Comput Struct Biotechnol J. 2013;5:e201302011. doi: 10.5936/csbj.201302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pandey KB, Rizvi SI. Plant polyphenols as dietary antioxidants in human health and disease. Oxid Med Cell Longev. 2009;2:270–8. doi: 10.4161/oxim.2.5.9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yugandhar P, Haribabu R, Savithramma N. Synthesis, characterization and antimicrobial properties of green-synthesised silver nanoparticles from stem bark extract of Syzygium alternifolium (Wt.) Walp 3. Biotech. 2015;5:1031–9. doi: 10.1007/s13205-015-0307-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gustafsson Sheppard N, Heldring N, Dahlman-Wright K. Estrogen receptor-a, RBCK1, and protein kinase C ß1 cooperate to regulate estrogen receptor-a gene expression. J Mol Endocrinol. 2012;49:277–87. doi: 10.1530/JME-12-0073. [DOI] [PubMed] [Google Scholar]
- 59.Zhang F, Dong W, Zeng W, Zhang L, Zhang C, Qiu Y, et al. Naringenin prevents TGF-b1 secretion from breast cancer and suppresses pulmonary metastasis by inhibiting PKC activation. Breast Cancer Res. 2016;18:1–16. doi: 10.1186/s13058-016-0698-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brusselmans K, Vrolix R, Verhoeven G, Swinnen JV. Induction of cancer cell apoptosis by flavonoids is associated with their ability to inhibit fatty acid synthase activity. J Biol Chem. 2005;280:5636–45. doi: 10.1074/jbc.M408177200. [DOI] [PubMed] [Google Scholar]
- 61.Li S, Dang Y, Zhou X, Huang B, Huang X, Zhang Z, et al. Formononetin promotes angiogenesis through the estrogen receptor alpha-enhanced ROCK pathway. Sci Rep. 2015;5:1–17. doi: 10.1038/srep16815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shim HY, Park JH, Paik HD, Nah SY, Kim DS, Han YS. Acacetin-induced apoptosis of human breast cancer MCF-7 cells involves caspase cascade, mitochondria-mediated death signaling and SAPK/JNK1/2-c-Jun activation. Mol Cells. 2007;24:95–104. [PubMed] [Google Scholar]
- 63.Ye L, Chan FL, Chen S, Leung LK. The citrus flavonone hesperetin inhibits growth of aromatase-expressing MCF-7 tumor in ovariectomized athymic mice. J Nutr Biochem. 2012;23:1230–7. doi: 10.1016/j.jnutbio.2011.07.003. [DOI] [PubMed] [Google Scholar]


