Abstract
Although telomerase activity is known to be regulated mainly at the level of transcription of the human telomerase catalytic subunit (hTERT) gene, the molecular mechanism underlying tumor-specific expression of telomerase remains unclear. Emerging evidence suggests that reversible acetylation of nucleosomal histones and the resultant changes in the chromatin structure are important processes in gene transcription. In particular, histone deacetylase (HDAC) inhibitors activate the transcription of certain genes by altering the acetylation status of nucleosomal histones. The present study examines the effects of HDAC inhibitor on hTERT gene transcription. Treatment with tricostatin A (TSA) induced significant activation of hTERT mRNA expression and telomerase activity in normal cells, but not in cancer cells. Transient expression assays revealed that TSA activates the hTERT promoter. Furthermore, the proximal 181 bp core promoter of hTERT, which contains two c-Myc and five Sp1 sites, was determined to be the responsible element. Overexpression of Sp1 enhanced responsiveness to TSA, and mutation of Sp1 sites, but not c-Myc sites, of the core promoter of hTERT abrogated this activation. Introduction of the dominant-negative form of the Sp family inhibited TSA activation. These results indicate that HDAC inhibitor activates the hTERT promoter in normal cells, in which Sp1 plays a key role. This finding suggests one way whereby histone deacetylation may be involved in silencing the hTERT gene in normal cells.
INTRODUCTION
Telomeres are essential elements that protect chromosome ends from degradation and ligation, thereby contributing to chromosomal stability (1). Telomeres undergo progressive shortening with cell division due to the inability of DNA polymerase to completely replicate the ends of chromosome DNA (2). The critical shortening of telomeres with cell division induces replicative senescence. Further division of cells beyond senescence results in a serious loss of telomeres, with the result being chromosomal instability. End-to-end fusions and dicentric or multicentric chromosomes are formed, and a cellular crisis occurs. Telomerase is a specialized ribonucleoprotein polymerase that directs the de novo synthesis of telomeric repeats at chromosome ends (1). Telomerase is not active in most somatic tissues, but is widely activated in cancer cells (3). Telomerase activation is thought to be required for cells to continuously divide beyond replicative senescence, and may therefore be a critical step in cellular immortality and carcinogenesis.
Three major subunits comprising the human telomerase complex have been identified. The most important component responsible for the enzymatic activity of telomerase is human telomerase reverse transcriptase (hTERT) (4,5). Many studies have found that hTERT is preferentially expressed in malignant tumors, but not expressed in normal tissues, and that hTERT expression is closely associated with telomerase activity in each sample (4–6). Recently, it was shown that introduction of the TERT gene into telomerase-negative cells led to telomerase expression, telomere elongation and to an extension of cellular lifespans (7). These findings suggest that hTERT is a rate-limiting determinant of telomerase enzymatic activity.
Expression of hTERT is known to be regulated mainly at the transcriptional level. Cloning of hTERT promoter sequences enabled us to analyze transcriptional regulation of the hTERT gene (8). Several transcription factors regulating hTERT transcription have been identified. Among them, c-Myc and Sp1 are the major activators of hTERT transcription, of which binding sites are located within the proximal core promotor (9,10). Expression of c-Myc and Sp1 is known to be up-regulated during the process of carcinogenesis, likely causing telomerase activation during carcinogenesis (10). However, these factors are also expressed in normal cells lacking telomerase activity, and mechanisms of transcriptional silencing must therefore be present in these cells.
Remodeling of chromatin and nucleosome organization is a key factor in the physiological control of transcription. Post-transcriptional modifications of histones have been implicated in the physiological control of chromatin structure (11). Acetylation of the lysine residue of nucleosomal histones is assumed to lead to local chromatin decondensation, resulting in increasing accessibility of particular DNA regions for RNA polymerase complexes. Histone acetylation is a dynamic process catalyzed by histone acetyltransferase (HAT) and histone deacetylase (HDAC). There are multiple acetyltransferase and deacetylase enzymes acting as activators and repressors of promoters within the cells. Recently, it was demonstrated that several transcription factors, such as Mad, can repress transcription by recruiting HDACs to specific promoters (12–14). In addition, HDAC1 has been shown to mediate transcriptional repression via the Sp1 binding sites (15). Given that the core promoter of hTERT contains E-boxes that bind to Mad as well as multiple Sp1 sites, the possibility is suggested that histone acetylation is involved in transcripitonal regulation of hTERT. Here, we show that HDAC inhibitor can induce hTERT transcription in normal cells, and that Sp1 plays a crucial role in this regulation. These findings may explain one mechanism of promoter silencing of the hTERT gene in normal cells.
MATERIALS AND METHODS
Cell cultures
Normal human renal cortical epithelial cells, normal human foreskin fibroblasts and normal human foreskin keratinocytes were purchased from Clonetics (San Diego, CA) and were grown in special media in accordance with the manufacturer’s protocol. C33A and ME180 cervical cancer cells were obtained from the American Type Culture Collection and were incubated in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum. All cell lines were incubated at 37°C with or without Tricostatin A (TSA) (Sigma) in the presence of 5% CO2.
RNA PCR analysis
The expression of hTERT mRNA was analyzed by semi-quantitative RT–PCR amplification as previously described (6). Total RNA was isolated from the cells using Isogen (Nippon Gene, Japan) according to the manufacturer’s protocol. cDNA was synthesized from 1 µg of RNA using the RNA PCR kit version 2 (TaKaRa, Ohtsu, Japan) with random primers. PCR products were resolved by electrophoresis in 7% polyacrylamide gels and stained with SYBR green I (FMC BioProducts, Rockland, ME). The efficiency of cDNA synthesis from each sample was estimated by PCR with glyceraldehyde-3-phosphate dehydrogenase-specific primers as described (6).
TRAP assay
Telomeric repeat amplification protocol (TRAP) assay was performed as described previously with some modifications (6). The PCR products were electrophoresed on a 12% polyacrylamide gel and visualized with SYBR Green I Nucleic Acid Gel Stain (FMC BioProducts, Rockland, ME). In our assay conditions, we confirmed that the band intensity of telomerase ladders, measured by NIH image software, was linearly correlated with the content of cell extracts applied within a range of between 0.1 and 2 µg. Therefore, we did not use internal standards to normalize relative telomerase activity.
Plasmid construction
pGL3-3328 and pGL3-181 are hTERT promoter-luciferase reporter plasmids in which 3328 and 181 bp sequences upstream of the transcription start site (numbered as +1) as well as a 77 bp 5′-untranslated region are cloned upstream of the luciferase gene (LUC) in pGL3-Basic (Promega) at MluI and BglII sites (8). Sp1-MT1-5 and Myc-MT1+2 are mutant reporter plasmids, in which five Sp1 binding sites and two c-Myc binding sites in the core promoter are abrogated by substitution mutations in pGL3-181, respectively (9). pG5-Luc is a reporter plasmid that contains five Gal4 binding sites upstream of E1B minimal promoter and the TATA box (16). pM-Sp1 is an expression vector for Sp1-Gal4 binding domain fusion protein (16). pCMV-DNSp3 is an expression vector for the dominant-negative form of Sp3, which is known to abrogate the function of the Sp family (16).
Transient expression assays
Cells were incubated in 24-well dishes and were transfected with 1 µg of various reporter plasmids alone or together with the same amounts of effector plasmids using LipofectAMINE (Gibco, Gaithersburg, MD) according to the manufacturer’s protocol, and incubated in the absence or presence of tricostatin A (TSA) at various concentrations for 24 h. The cells were then harvested and luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI), in which Renilla luciferase plasmids were co-transfected as controls to standardize transfection efficiency. All experiments were performed at least three times for each plasmid and the results represent average relative luciferase activity.
RESULTS
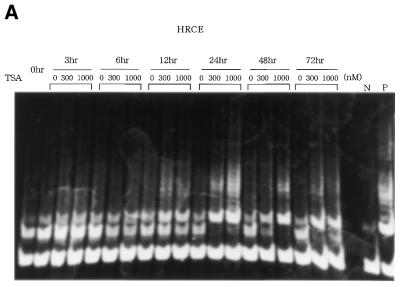
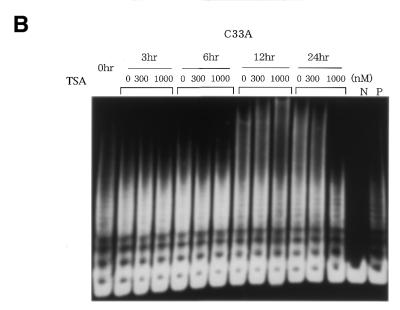
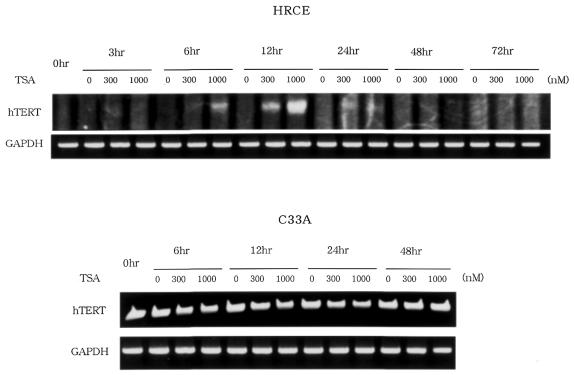
We first examined the effects of HDAC inhibitor, TSA, on telomerase activity in normal cells. Normal renal cortical epithelial cells (HRCE) are telomerase-negative, but treatment of these cells with TSA at 300 or 1000 nM resulted in significant induction of telomerase activity, which peaked at 24 h (Fig. 1A). Similar results were obtained using normal foreskin fibroblasts (data not shown). In contrast, the same treatment did not affect the levels of telomerase activity in cancer cell lines C33A (Fig. 1B) or SiHa cells (data not shown). RT–PCR assays were then performed to examine changes in hTERT mRNA expression. TSA treatment led to a significant induction of hTERT mRNA expression, which peaked at 12 h after, in normal cells, but not in cancer cells (Fig. 2). This result was consistent with the results of TRAP assays.
Figure 1.
TRAP assays to detect change in telomerase activity in cells treated with TSA. (A) TSA induced telomerase activity in normal HRCE cells. (B) No change in telomerase activity was observed in C33A cervical cancer cells. N, negative control using lysis buffer only. P, positive control using extracts from cancer cell lines with telomerase activity.
Figure 2.
RT–PCR assays to detect change in hTERT mRNA expression in cells treated with TSA. TSA significantly induced hTERT mRNA expression in normal HRCE cells. No change in expression was observed in C33A cancer cells.
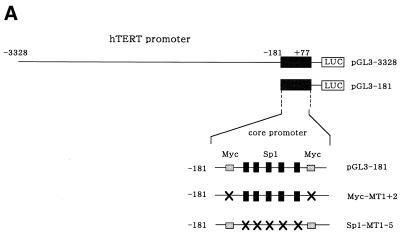
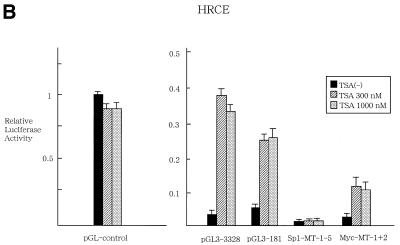
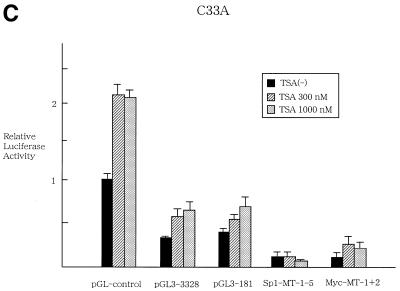
Transient expression assays were subsequently performed to examine whether or not TSA regulates transcriptional activity of hTERT promoter. HRCE cells were transfected with luciferase reporter plasmid containing 3.3 kb hTERT promoter (pGL3-3328) (Fig. 3A) and treated with TSA at 300 or 1000 nM. At 48 h after treatment, cells were harvested and luciferase assays were performed. TSA treatment led to >10-fold activation of transcriptional activity of hTERT promoter (Fig. 3B). A 5′-deletion of the 3.3 kb promoter (pGL3-2000, pGL3-1375 and pGL3-378) did not affect responsiveness to TSA (data not shown), and even the 181-bp proximal core promoter (pGL3-181) exhibited 5-fold activation of transcriptional activity in response to TSA treatment, suggesting that then core promoter is responsible for TSA regulation. Interestingly, mutations introduced into Sp1 sites within core promoter (Sp1-MT1-5) abrogated TSA activation of the hTERT promoter. Mutation introduced into c-Myc binding sites (Myc-MT1+2) did not alter responsiveness to TSA. To examine the promoter specificity of TSA regulation, reporter plasmids containing SV40 LT enhancer/promoter (pGL3-control) were tested for TSA regulation. Transcriptional activity of pGL3-control was not affected by treatment with TSA (Fig. 3A), suggesting that this regulation is promoter specific. In contrast to normal cells, TSA activation of hTERT promoter was marginal in cancer cells (Fig. 3C). Only 2-fold activation of luciferase activity was observed in pGL3-3328 and pGL3-181 in response to treatment of C33A cells with TSA.
Figure 3.
Transient expression assays to examine the transcriptional regulation of hTERT promoter by TSA. (A) Luciferase reporter plasmids used in the assays. c-Myc and Sp1 sites within a core promoter of hTERT are shown. Mutated sites are described as crossed-out. LUC, luciferase gene. HRCE (B) and C33A (C) cells were transfected with luciferase reporter plasmids and incubated with or without TSA. pGL3-control reporter plasmid was used to confirm the promoter specificity of TSA. At 24 h after treatment, cells were harvested and luciferase assays were performed. Relative luciferase activity of each reporter plasmid is shown, which is normalized to that of the pGL3-control untreated with TSA (given as 1.0). Bars, SD.
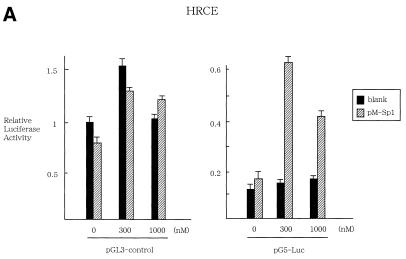
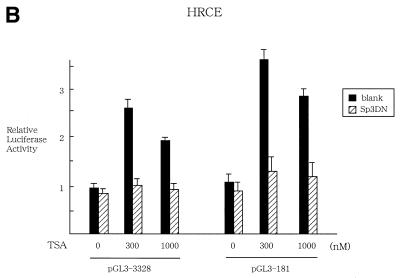
To further clarify the role of Sp1 in TSA regulation, transient expression assays were performed, in which HRCE cells were transfected with pG5-Luc alone or together with pM-Sp1 in the presence or absence of TSA at 300 or 1000 nM (Fig. 4A). The former (pG5-Luc) is a luciferase reporter plasmid that contains Gal4 binding sites upstream of E1B minimal promoter and the TATA box, and the latter (pM-Sp1) is an expression vector for Sp1-Gal4 binding domain fusion proteins. In the absence of Sp1/Gal4 expression, TSA did not activate transcription of pG5-Luc. However, when Sp1/Gal4 was overexpressed, treatment with TSA resulted in ∼3- or 4-fold activation of transcriptional activity of pG5-Luc, suggesting that Sp1 is required for TSA-induced activation of promoter. This is due to the specific function of Sp1, given that no significant transactivation was observed when pGL3-control plasmids lacking Gal4 binding sites were used as reporter plasmids. To further define the role of Sp1, transient expression assays were performed in which HRCE cells were transfected in the absence or presence of TSA with pGL3-181 and pCMV-DNSp3, which express the dominant-negative form of Sp3, which is known to abrogate the function of the Sp family (Fig. 4B). Expression of the dominant-negative form of Sp3 significantly abrogated the responsiveness to TSA. These findings suggest that the Sp family plays a crucial role in TSA regulation of hTERT promoter in normal cells.
Figure 4.
Transient expression assays to examine the role of Sp1 in TSA regulation of promoter. (A) HRCE cells were transfected with pG5-Luc containing Gal4 sites or pGL3-control together with pM-Sp1 effector plasmids, which expresses Sp1/Gal4 fusion proteins, and were incubated with or without TSA. Relative luciferase activity of each reporter plasmid is shown, which is normalized to that of pGL3-control untreated with TSA (given as 1.0). (B) HRCE cells were transfected with hTERT-promoter reporter plasmids together with pCMV-DNSp3, which expresses the dominant-negative form of Sp3, and were incubated with or without TSA. At 24 h after treatment, cells were harvested and luciferase assays were performed. Relative luciferase activity is shown, which is normalized to that of pGL3-3328 untreated with TSA (given as 1.0). Bars, SD.
DISCUSSION
The present findings clearly demonstrated that one of the HDAC inhibitors, TSA, activates telomerase through transcriptional activation of hTERT promoter in normal renal cells and skin fibroblasts. We also examined keratinocytes from various origins, and confirmed similar activation of the hTERT promoter by TSA treatment, although the extent of activation varied (data not shown). In contrast, no significant activation was observed in cervical cancer C33A and other cell lines, such as ME180 or Saos2 cells (data not shown). These findings suggested that TSA action is specific to normal cells, and futher suggested a mechanism whereby HDAC(s) is involved in telomerase silencing in normal cells. In the course of this study, one published report appeared demonstrating regulation of hTERT promoter by TSA, in which the role of Sp1 remained to be elucidated (17).
We determined that the TSA-responsive element is localized within the 181 bp-proximal core promoter, which contains two E-boxes and five GC boxes. Recent studies have demonstrated that the Mad/Max complex, which binds E-boxes, interacts with Sin3A and can recruit HDACs. This suggests the involvement of these sites in TSA-induced activation of the hTERT promoter (12–14). However, mutation at these sites did not affect responsiveness to TSA in luciferase reporter assays. Moreover, this activation was abrogated by mutation at Sp1 sites. Furthermore, overexpression of Sp1 enhanced responsiveness to TSA, and introduction of the dominant-negative form of the Sp family reduced TSA activation of the hTERT promoter. These findings suggest that Sp1, but not Mad, is one component of TSA-dependent activation of the hTERT promoter. Various different HDAC agents have been identified in mammalian cells. These enzymes are found in high-molecular complexes associated with adapter proteins like SIN3 and nuclear corepressor N-CoR (13,14,18). Recently, it was demonstrated that several mammalian transcription factors, including Mad, YY1 and hormone-dependent nuclear receptors, can repress transcription by recruiting HDACs to specific promoters (12–14,19,20). In particular, HDAC1 is known to repress transcription by direct binding to Sp1 (15). It is therefore possible that Sp1 functions as a cofactor that recruits HDAC onto the hTERT promoter, and thereby represses transcriptional activity in normal cells.
We have previously shown that Sp1 is the major transcriptional activator of hTERT. Up-regulation of the Sp1 family is thought to be critical for transactivation of hTERT during carcinogenesis (9). However, the present findings are inconsistent with this hypothesis. While Sp1 contributes to transactivation of hTERT, it might be involved in silencing this promoter in normal cells, possibly by recruiting HDACs. While Sp1 was shown to interact with HDAC1, it is also known to interact with some other transactivating factors, and Sp1-dependent gene regulation is altered by competition between HDACs and these factors (15). Furthermore, compelling evidence suggests that Sp1 interacts with a p300 coactivator possessing intrinsic HAT activity (21). It is therefore possible that Sp1 interacts with various factors that have HAT or HDAC activity, and that this switching explains the different actions of Sp1 on the hTERT promoter between normal and cancerous cells.
Recent experimental advances in the development of anticancer drugs have suggested that HDAC inhibitors have applications as targets for a novel anticancer drug. Some HDAC inhibitors have been shown to have remarkable anti-tumor activity in vitro, and are now under clinical investigation (22). One mechanism of these anti-tumor activities is undoubtedly due to the induction of tumor suppressor genes, such as p21, possibly through an interaction between Sp1/3 and factors possessing HDAC activity (16). However, our findings suggest the need for caution when anticipating the action of these drugs, such as telomerase activation in normal cells.
In summary, we demonstrated that an HDAC inhibitor, TSA, activates telomerase through the transcriptional activation of hTERT promoter in normal cells. In this regulation, Sp1 plays a causative role. These findings provide an important insight into the molecular mechanism of telomerase regulation in normal and cancerous cells, and further suggest a way whereby HDAC inhibitor may induce expression of anticipated genes, other than tumor suppressor genes.
Acknowledgments
ACKNOWLEDGEMENTS
This study was supported in part by a Grant-in-Aid for the Second Term Comprehensive 10-year Strategy for Cancer Control from the ministry of Health and Welfare, Japan.
References
- 1.Harley C.B., Futcher,A.B. and Greider,C.W. (1990) Telomeres shorten during ageing of human fibroblasts. Nature, 345, 458–460. [DOI] [PubMed] [Google Scholar]
- 2.Watson J.D. (1972) Origin of concatemeric T7 DNA. Nature, 239, 197–201. [DOI] [PubMed] [Google Scholar]
- 3.Kim N.W., Piatyszek,M.A., Prowse,K.R., Harley,C.B., West,M.D., Ho,P.L., Coviello,G.M., Wright,W.E., Weinrich,S.L. and Shay,J.W. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- 4.Meyerson M., Counter,C.M., Eaton,E.N., Ellisen,L.W., Steiner,P., Caddle,S.D., Ziaugra,L., Beijersbergen,R.L., Davidoff,M.J., Liu,Q. et al. (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell, 90, 785–795. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura T.M., Morin,G.B., Chapman,K.B., Weinrich,S.L., Andrews,W.H., Lingner,J., Harley,C.B. and Cech,T.R. (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science, 277, 955–959. [DOI] [PubMed] [Google Scholar]
- 6.Takakura M., Kyo,S., Kanaya,T., Tanaka,M. and Inoue,M. (1998) Expression of human telomerase subunits and correlation with telomerase activity in cervical cancer. Cancer Res., 58, 1558–1561. [PubMed] [Google Scholar]
- 7.Bodnar A.G., Ouellette,M., Frolkis,M., Holt,S.E., Chiu,C.P., Morin,G.B., Harley,C.B., Shay,J.W., Lichtsteiner,S. and Wright,W.E. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- 8.Takakura M., Kyo,S., Kanaya,T., Hirano,H., Takeda,J., Yutsudo,M. and Inoue,M. (1999) Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res., 59, 551–557. [PubMed] [Google Scholar]
- 9.Kyo S., Takakura,M., Kanaya,T., Taira,T., Kanaya,T., Itoh,H., Yutsudo,M., Ariga,H. and Inoue,M. (2000) Sp1 cooperates with c-Myc to activate transcription of human telomerase reverse transcriptase (hTERT) gene. Nucleic Acids Res., 28, 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu K.J., Grandori,C., Amacker,M., Simon-Vermot,N., Polack,A., Lingner,J. and Dalla-Favera,R. (1999) Direct activation of TERT transcription by c-MYC. Nature Genet., 21, 220–224. [DOI] [PubMed] [Google Scholar]
- 11.Stein G.S., Montecino,M., van Wijnen,A.J., Stein,J.L. and Lian,J.B. (2000) Nuclear structure–gene expression interrelationships: implications for aberrant gene expression in cancer. Cancer Res., 60, 2067–2076. [PubMed] [Google Scholar]
- 12.Hassig C.A., Fleischer,T.C., Billin,A.N., Schreiber,S.L. and Ayer,D.E. (1997) Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell, 89, 341–347. [DOI] [PubMed] [Google Scholar]
- 13.Heinzel T., Lavinsky,R.M., Mullen,T.M., Sonderstrom,M., Laherty,C.D., Torchia,J., Yang,W.M., Brard,G., Ngo,D., Davie,J.R. et al. (1997) A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature, 387, 43–48. [DOI] [PubMed] [Google Scholar]
- 14.Laherty C.D., Yang,W.M., Sun,J.M., Davie,J.R., Seto,E. and Eisenman,R.N. (1997) Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell, 89, 349–356. [DOI] [PubMed] [Google Scholar]
- 15.Doetzlhofer A., Rotheneder,H., Lagger,G., Koranda,M., Kurtev,V., Brosch,G., Wintersberger,E. and Seiser,C. (1999) Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol., 19, 5504–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sowa Y., Orita,T., Minamikawa-Hiranabe,S., Mizuno,T., Nomura,H. and Sakai,T. (1999) Sp3, but not Sp1, mediates the transcriptional activation of the p21/WAF1/Cip1 gene promoter by histone deacetylase inhibitor. Cancer Res., 59, 4266–4270. [PubMed] [Google Scholar]
- 17.Cong Y.S. and Bacchetti,S. (2000) Histone deacetylation is involved in the transcriptional repression of hTERT in normal human cells. J. Biol. Chem., 275, 35665–35668. [DOI] [PubMed] [Google Scholar]
- 18.Alland L., Muhle,R., Hou,H., Protes,J., Chin,L., Schreider Agus,N. and DePinho,R.A. (1997) Role for N-CoR and histone deacetylase in Sin3-mediated transcripitonal repression. Nature, 387, 49–55. [DOI] [PubMed] [Google Scholar]
- 19.Spencer T.E., Jenster,G., Burcin,M.M., Allis,C.D., Zhou,J., Mizzen,C.A., McKenna,N.J., Onate,S.A., Tsai,S.Y., Tsai,M.J. and O’Malley,B.W. (1997) Steroid receptor coactivator-1 is a histone acetyltransferase. Nature, 389, 194–198. [DOI] [PubMed] [Google Scholar]
- 20.Nagy L., Kao,H.Y., Chakravarti,D., Lin,R.J., Hassig,C.A., Ayer,D.E., Schreiber,S.L. and Evans,R.M. (1997) Nuclear receptor repression mediated by a complex containing SMRT, mSin3A and histone deacetylase. Cell, 89, 373–380. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki T., Kimura,A., Nagai,R. and Horikoshi,M. (2000) Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells, 5, 29–41. [DOI] [PubMed] [Google Scholar]
- 22.Marks P.A., Richon,V.M. and Rifkind,R.A. (2000) Histone deacetylase inhibitors: Inducers of differentiation or apoptosis of transformed cells. J. Natl Cancer Inst., 92, 1210–1216. [DOI] [PubMed] [Google Scholar]