Abstract
Ulva flexuosa, one kind of green tide algae, has outbroken in the Yellow Sea of China during the past ten years. In the present study, we sequenced the chloroplast genome of U. flexuosa followed by annotation and comparative analysis. It indicated that the chloroplast genomes had high conservation among Ulva spp., and high rearrangement outside them. Though U. flexuosa was closer to U. linza than U. fasciata in phylogenetic tree, the average Ka/Ks between U. flexuosa and U. linza assessed by 67 protein-coding genes was higher than those between U. flexuosa and other species in Ulva spp., due to the variation of psbZ, psbM and ycf20. Our results laid the foundation for the future studies on the evolution of chloroplast genomes of Ulva, as well as the molecular identification of U. flexuosa varieties.
Introduction
The macroalgae representing the cosmopolitan green algae Ulva flexuosa Wulfen species have been found in Europe since the mid-1800s [1], which are very common and regularly encountered in fresh waters, slightly saline waters, and estuaries [2], containing the characteristics of transitional species. As one of the original species in green tide outbreak in Yellow Sea of China since 2008, U. flexuosa has attracted global attention for its fast growth if given appropriate nutrient supply, with maximum growth rate of 73% d-1 at room temperature under light intensity of 70 μmol·m-2·s -1 [3]. On the other hand, culturists in Brazilian are interested in the sulfated polysaccharide ulvan which triggers plant defenses against disease and can be obtained from U. flexuosa within a production cycle of 15 days [4].
U. flexuosa belongs to Ulvophyceae, with thalli composed of uninucleate cells, and is difficult to be set apart from species among Ulva spp., since the latter is often sheet like in external morphology [5]. Furthermore, the vegetative cells arrangement of the mature thalli show tendency towards distortions determined by the calcium carbonate crystals which increase the difficulty for identification [6, 7]. Then, the rRNA ITS region, 5S and partial rbcL gene sequences are applied to demonstrate sequence variation within species or even subspecies [2, 8]. The chloroplast genome sequencing may provide more molecular markers.
In this study, we present the first complete chloroplast genome of U. flexuosa. The genome was sequenced, assembled, and annotated as circular-mapping DNA molecules. Additionally, we analyzed the repeat sequence using MISA, Tandem Repeats Finder, and REPuter. Furthermore, U. flexuosa chloroplast genome was compared to others using phylogenomic investigation, Mauve and blastn program provided by EMBOSS to assess the monophyly of the Ulvophyceae and relationships of ulvophytes with other core chlorophyta clades.
Materials and methods
Sampling and species identification
Samples of green tide algae were collected from the southern Yellow Sea near Rudong sea area, Jiangsu (N32°31′04.10″ E121°25′11.99″), where lavers breed every year and no endangered or protected species is involved. So no specific permissions were required for these locations and activities. After repeated washing with seawater, the samples were dried in the shade to the moisture content of 30–40%. Then, they were transported to the laboratory in an insulated specimen box at 4°C.
Healthy individual algae were selected to be washed several times with sterile seawater autoclaved at 121°C for 15 minutes. After removing surface attachments, they were cultured in VSE medium [9] for 2d at 24°C under a 12:12 h LD photoperiod and photon flux 130–160 μmolm-2 s-1.
After being put into the white porcelain tray with a small amount of seawater, algae were gently expanded with a brush to exhibit the main external form. Slices, made through freehand transverse section, were placed under an Olympus optical microscope for observation.
Algae matching the description of U. flexuosa were used to extract DNA using the DNAsecure Plant Kit (Tiangen Biotech, Beijing) [10] and subjected to PCR amplification using primers specific for ITS [11] and 5S rDNA [12]. The amplified sequences were sent to Shanghai Biology Engineering Technology Service Co., Ltd. for shotgun sequencing.
DNA sequencing and assembly
After comparison, the chloroplast genome of Ulva sp. UNA00071828 [13] was used as a model for primer sequence design (S1 Table). After preliminary experiment, the primers were re-integrated. Then the whole genome was divided into 15 segments (S2 Table) for PCR amplification under the following conditions: predenaturation at 94°C for 1 min, 35 cycles of amplification at 94°C for 30 s, 55°C for 30 s and 68°C for 8 min, followed by a final extension at 68°C for 15 min. The PCR reaction mixture contained 0.5μl of DNA polymerase (5U/μl), 5μl of Buffer (Mg2+ Plus) (10×), 8μl of dNTP Mixture (2.5mM each), 0.2–1.0μM (final concentration) of forward primer, 0.2–1.0μM (final concentration) of reverse primer, and purified chloroplast DNA (<1 μ g). RNase-free water was added to increase the final reaction volume to 50 μl. Wherein Takara LA Taq polymerase was used for fragments longer than 4,000 bp, Takara Ex Taq was used for the others. The PCR products of long segments were broken with ultrasound, connected to vector PMD19-T, and transformed into E. coli DH5a. Positive clones were selected for shotgun sequencing by ABI3730 sequencer. The correctness of the assembly was validated further by mapping all raw sequence reads to the assembly using phred V1.09 program [14].
Species verification of U. flexuosa
In order to verify species U. flexuosa by comparison of rbcL and tufA genes, Chloroplast rbcL and tufA genes in Ulva spp. were extracted from genebank (S3 Table) followed by blastn with corresponding U. flexuosa genes using DNAMAN. Poorly aligned positions were removed using the Gblocks server [15, 16] with the least stringent settings: allowing smaller final blocks, gap positions within the final blocks, less strict flanking positions and many contiguous non-conserved positions. Gblocks removed 87 of 1430 positions for rbcL and 458 of 1231 positions for tufA, 1343pb and 773bp alignments were remained respectively for phylogenetic analysis. A UPGMA tree was made by MEGA 6.06 [17] for each alignment with number of differences genetic distance model and 1000 bootstrap replicates [18]. Blidingia minima (AF387109) and Monostroma grevillei (GU183089) were used as outgroups for rbcL, while B. minima (HQ610329) and Monostroma sp. (HQ610262) were used for tufA. Additionally, the pairwise distances (bp) were calculated to aid in the identification of this Ulva species.
Genome annotation and codon usage analysis
The CpGAVAS web service was used to annotate the U. flexuosa chloroplast genome [19]. Cutoffs for the E-values of BLASTN and BLASTX were 1e-10. The number of top hits included in the reference gene sets for annotation after the pre-filtering step was 10. Meanwhile, tRNA genes were identified using tRNAscan-SE [20] and ARAGORN [21]. Manual corrections on the positions of the start and stop codons, and for the intron/exon boundaries were aided by blastn with other chloroplast genomes in Ulvophyceae. The inverted repeat (IR) sequences were predicted by Gepard-1.40 analysis [22]. Moreover, the circular chloroplast genome map of U. flexuosa was drawn using Organellar Genome DRAW [23]. Furthermore, codon usage and GC content were analyzed using the Cusp and Compseq programs provided by EMBOSS [24]. Final genome assembly and genome annotation results were deposited in GenBank (accession number: KX579943).
Repeat sequence analysis
SSRs were detected using MISA Perl Script available at “http://pgrc.ipk-gatersleben.de/misa/”, with the following thresholds: eight repeat units for mononucleotide SSRs, four repeat units for di- and trinucleotide repeat SSRs, and three repeat units for tetra-, penta-, and hexa-nucleotide repeat SSRs. Tandem repeats were analyzed using Tandem Repeats Finder [25] with parameter settings of two for matches and seven for mismatches and indels. The minimum alignment score and maximum period size were set at 50 and 500, respectively. All the identified repeats were manually verified and nested or redundant results were removed. REPuter [26] was employed to identify the IRs in U. flexuosa by forward, reverse, complementary and palindromic alignment. The minimal repeat size was set at 30 bp, and the cutoff for similarities among the repeat units was set at 100%.
Phylogenomic analysis
A total of 67 complete chloroplast DNA sequences belonging to algae were obtained from Genbank. For the phylogenetic analysis, 24 nucleotide sequences shared among all these 67 species and U. flexuosa were aligned by Clustal-Omega with default parameters. The 24 genes are atpA, atpB, atpE, atpF, petB, petD, petG, psaB, psbB, psbD, psbE, psbF, psbH, psbJ, psbK, psbL, psbN, rbcL, rpl2, rpl20, rpl36, rpoA, rps8 and ycf3. The appropriate model of evolution for each gene determined by jModelTest 2.1 was GTR+I+G, which was reassessed by PAUP 4.0a152 (P value = 1 − (99/100) = 0.010000). Then, the evolutionary history was inferred using the Maximum Likelihood method implemented in RaxML. The detailed parameters were “raxmlHPC-PTHREADS -f a -x 12345 -# 1000 -m GTRGAMMA -s dna.phy -n nex -p 12345 -q part.txt -T 6”. The tree with the final ML Optimization Likelihood (-548221.173374) was shown. The significance level for the phylogenetic tree was assessed by bootstrap testing with 1000 replications. Finally, MrBayes were used to calculate Bayesian posterior probabilities for another line of statistical support, in which the main parameters are: ngen = 2000000, samplefreq = 100, nchains = 4, nst = 6, rates = gamma. The results were published to TreeBASE <treebase.org> (submission number: 21293).
Rearrangements in genomes
Genome rearrangement was aligned with progressive Mauve by Mauve 2.4.0 [27]. 10 species were applied including U. flexuosa (KX579943), U. linza (NC_030312), U. fasciata (NC_029040), Ulva sp. UNA00071828 (KP720616), Pseudendoclonium akinetum (NC_008114), Gloeotilopsis sterilis (NC_025538), Tydemania expeditionis (NC_026796), Bryopsis plumosa (NC_026795), Bryopsis hypnoides (NC_013359) and Ostreobium sp. (KU979013) [28, 29].
Comparative genome analysis
Genome synteny was analyzed between the chloroplast genomes of U. flexuosa (KX579943) and those of B. hypnoides (NC_013359), B. plumosa (NC_026795), Gloeotilopsis planctonica (KX306824), Gloeotilopsis sarcinoidea (KX306821), G. sterilis (NC_025538), Ostreobium sp. (KU979013), P. akinetum (NC_008114), T. expeditionis (NC_026796), Ulva fasciata (NC_029040), Ulva linza (NC_030312), and Ulva sp. UNA00071828 (KP720616) using blastn program provided by EMBOSS. The detailed parameters were “blastn -query genome1.fasta -db genome2.fasta -out result.o -outfmt 7”. The homologous regions were visualized using a web-based genome synteny viewer GSV [30].
Results
Species verification of U. flexuosa
Four samples of algae were identified as most likely belonging to U. flexuosa, based on morphology and ITS sequence. The bright green thallus is tubular at the base but flattened above. It grows into long, narrow, ruffled blades two cell layers thick, 0.7–1 cm wide and up to 6–18 cm tall. Cells are 11–15 μm in diameter. Additionally, each cell contains 1–3 pyrenoids, or even 4–5 occasionally (Fig 1). The four samples had highest sequence similarity with U. flexuosa (AB097646.1, HM031176.1), with 99.7% ITS sequence identity.
Fig 1. Morphological characteristics of U. flexuosa.
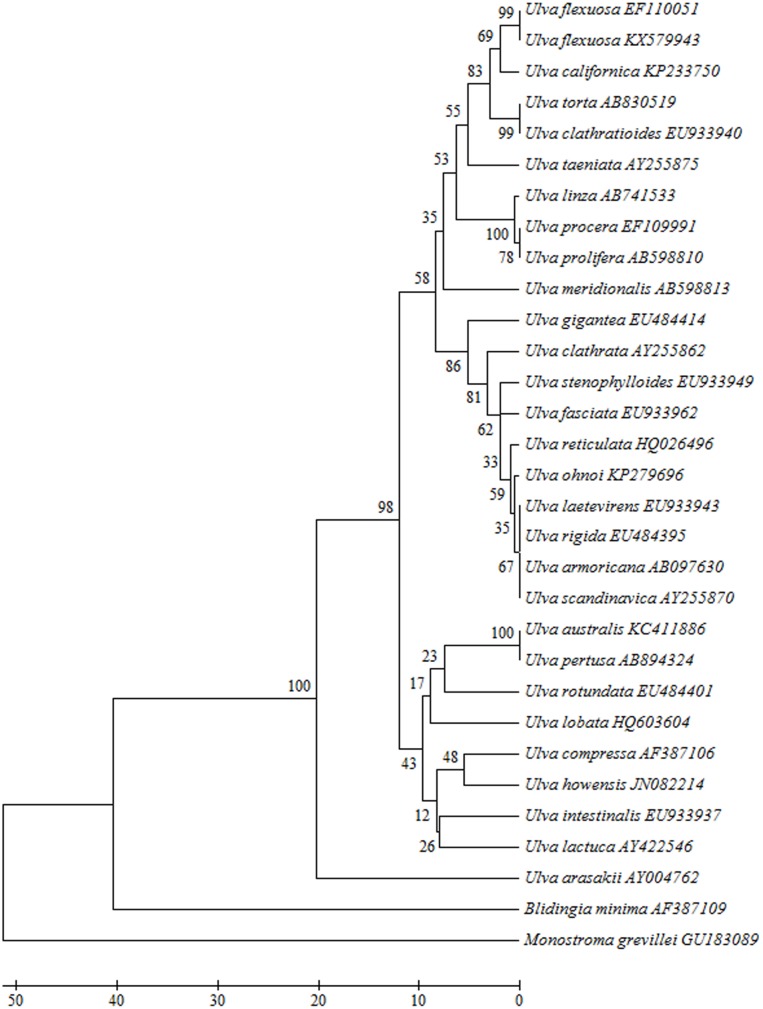
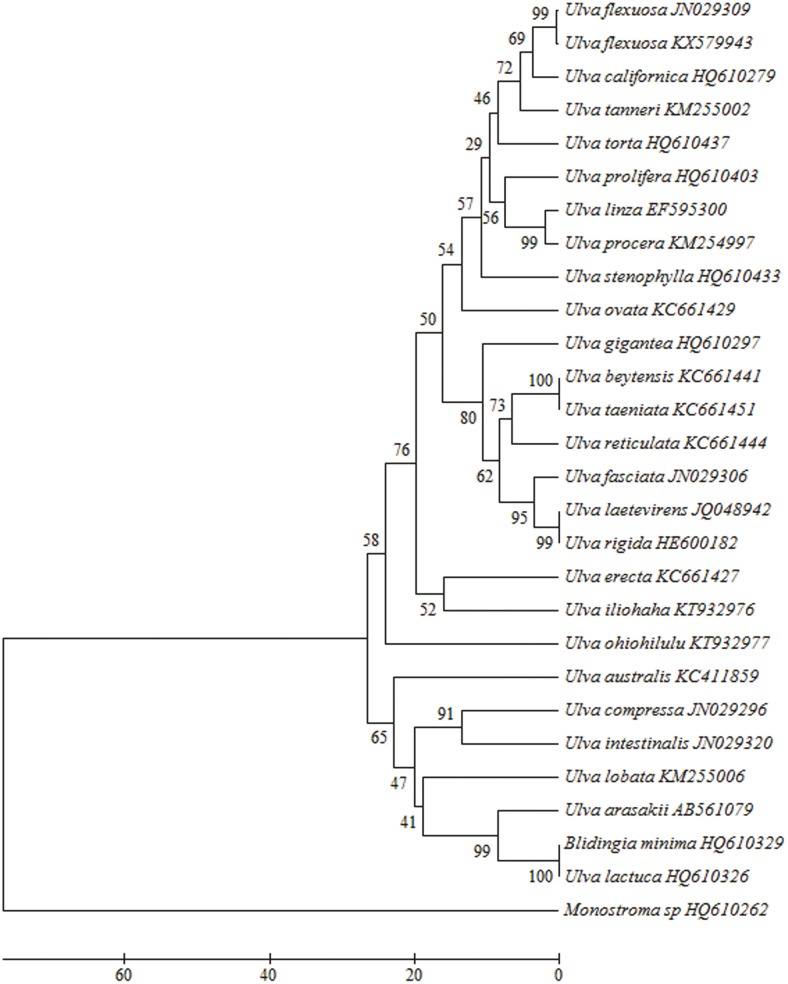
The chloroplast rbcL (EF110051) and tufA (JN029309) sequences from genus Ulva species in NCBI had highest consistency with U. flexuosa samples in this report according to UPGMA phylogenetic tree, both with credibility of 99% (Figs 2 and 3). The pairwise distances were zero and one (S1 and S2 Fig). Combined with its morphological characteristics, it could be confirmed as U. flexuosa.
Fig 2. Evolutionary relationships of taxa from Ulva based on the rbcL gene.
Fig 3. Evolutionary relationships of taxa from Ulva based on the tufA gene.
General features of the U. flexuosa chloroplast genome
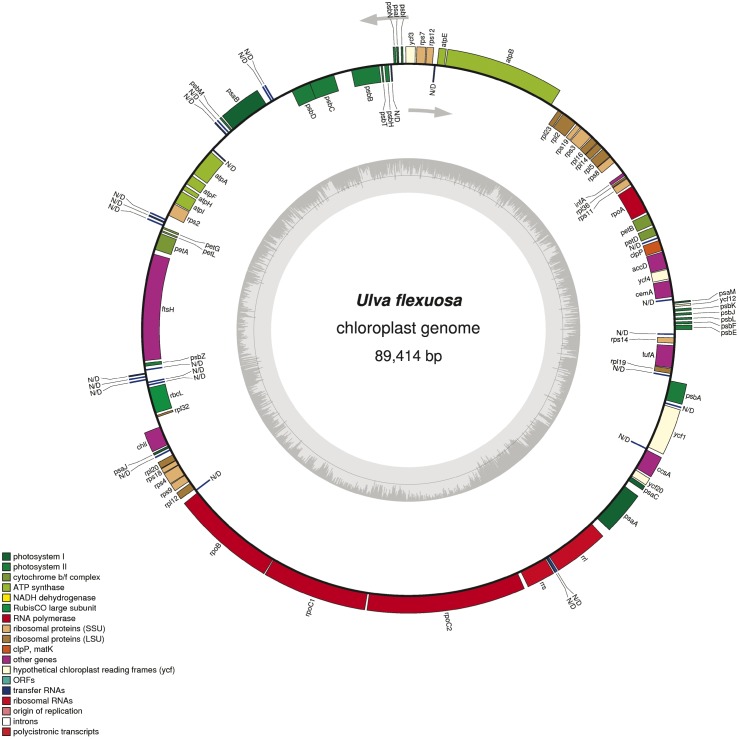
The complete chloroplast genome sequence is 89414 bp long with no IR region. Moreover, a total of 99 genes were identified, including 71 protein coding genes, 26 tRNA genes, and 2 genes for large and small ribosomal RNA (rrl and rrs). In addition to the conserved genes, two conserved open reading frames were determined (Table 1). The general structure and locations of the 99 genes in the chloroplast genome are depicted in Fig 4. No intron has been predicted. Overall, 72.0% of the U. flexuosa chloroplast genome sequence is composed of genes that code proteins. The overall GC content of the U. flexuosa chloroplast genome is 25.0%, whereas the protein-coding regions account for 26.5%. Within the protein-coding regions, the GC contents for the first, second and third positions of the codons occupy 35.62%, 31.34% and 12.43%, respectively.
Table 1. Genes predicted in the chloroplast genome of U. flexuosa.
| Category of genes | Group of genes | Name of genes |
|---|---|---|
| Self-replication | tRNA genes | 26 trn genes |
| Small subunit of ribosome | rps11, rps12, rps14, rps18, rps19, rps2, rps3, rps4, rps7, rps8, rps9 | |
| Large subunit of ribosome | rpl12, rpl14, rpl16, rpl19, rpl2, rpl20, rpl23, rpl32, rpl36, rpl5 | |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| rRNA genes | rrl(23S), rrs(16S) | |
| Genes for photosynthesis | Subunits of ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI |
| Subunits of cytochrome b/f complex | petA, petB, petD, petG, petL | |
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ, psaM, ycf3, ycf4 | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ, ycf12 | |
| Subunit of rubisco | rbcL | |
| Subunit of protochlorophyllide reductase | chlI | |
| Other genes | Subunits of Acetyl-CoA-carboxylase | accD |
| C-type cytochrome synthesis gene | ccsA | |
| Envelope membrane protein | cemA | |
| Protease | clpP | |
| Elongation factor | tufA | |
| Zinc metalloprotease | ftsH | |
| Translation initiation factor | infA | |
| Genes of unknown function | Conserved open reading frames | ycf1, ycf20 |
Fig 4. Schematic representation of the U. flexuosa chloroplast genome using OGDRAW.
The predicted genes are shown and colors represent functional classifications, which are shown at the left bottom. The genes drawn outside the circle are transcribed counterclockwise. The inner circle shows the GC content. IR regions are not found according to Gepard-1.40 analysis.
The codon usage and codon-anticodon recognition pattern of the U. flexuosa chloroplast genome are summarized in S4 Table. The 29 tRNA genes include codons corresponding to all 20 amino acids that are necessary for biosynthesis. UUA (encoding Leu) is the most frequently used codon, followed by AAA (Lys), AAU (Asn), AUU (Ile), UUU (Phe), GAA (Glu), UAU (Tyr), GGU (Gly) and GAU (Asp). These nine codons account for 50% of all codons, and the third base pair for all of these is either A or U. In addition, optimal codons for all amino acids end either in A or U, with the exception of Met and Trp, both of which have a single codon (AUG and UGG). All the results are consistent with the high AT content of U. flexuosa chloroplast DNA (75.0%).
Repeat and SSR analysis
SSRs are valuable molecular markers of high-degree variations within the same species and have been used in population genetics and polymorphism investigation [31]. We analyzed the occurrence, type, and distribution of SSRs in the U. flexuosa chloroplast genome and the distribution of SSRs in 11 other chloroplast genomes belonging to Ulvophyceae. In total, 236 SSRs were identified in the U. flexuosa chloroplast genome (S5 Table and Table 2). Among these SSRs, the majority consisted of mono- and di- nucleotide repeats, which were found 167 and 40 times, respectively. Tri- (19), tetra- (9), hexa- nucleotide repeat sequences (1) were found with lower frequency. This observed pattern is similar to those observed in 3 chloroplast genomes of other species belonging to Ulvaceae (Table 3). All the mononucleotide repeat sequences consisted of A/T repeats. Similarly, most dinucleotide repeat sequences consisted of AT/AT repeats (92.5%) (S5 Table). Our findings are in agreement with the previous findings that the chloroplast SSRs are generally composed of short polyA or polyT repeats and rarely contain tandem G or C repeats [32]. In this study, we also analyzed the locations of 29 tri-, tetra- and hexa- nucleotides in the chloroplast genome, and the results are shown in Table 2. Among these nucleotides, 15 are localized in the intergenic regions, 13 are in the coding regions, and the last one is stretching across intergenic regions and coding regions.
Table 2. Distribution of tri-, tetra-, penta- and sex- nucleotide SSR loci in the chloroplast genome of U. flexuosa.
| SSR type | SSR sequence | Start | End | Location |
|---|---|---|---|---|
| tri | (TAA)4 | 704 | 715 | IGSa(psbL-psbJ) |
| tri | (ATA)4 | 28544 | 28555 | CDSb(psbD) |
| tri | (ATA)6 | 32962 | 32979 | IGS(trnH-trnS) |
| tri | (ATA)4 | 36822 | 36833 | IGS(atpH-atpI) |
| tri | (ATT)4 | 44068 | 44079 | CDS(ftsH) |
| tri | (TAA)4 | 47408 | 47419 | CDS(trnE),IGS(trnE-trnM) |
| tri | (TAA)4 | 47423 | 47434 | IGS(trnE-trnM) |
| tri | (TAA)5 | 47438 | 47452 | IGS(trnE-trnM) |
| tri | (TAA)4 | 47465 | 47476 | IGS(trnE-trnM) |
| tri | (ATT)6 | 51077 | 51094 | IGS(chlI-psaJ) |
| tri | (AAT)4 | 60077 | 60088 | CDS(rpoC1) |
| tri | (TAA)4 | 60436 | 60447 | CDS(rpoC1) |
| tri | (TTA)4 | 61640 | 61651 | CDS(rpoC1) |
| tri | (ATA)4 | 67042 | 67053 | CDS(rpoC2) |
| tri | (ATA)4 | 69097 | 69108 | CDS(rpoC2) |
| tri | (ATA)4 | 71404 | 71415 | CDS(rpoC2) |
| tri | (TAA)4 | 71651 | 71662 | CDS(rpoC2) |
| tri | (CAA)4 | 78378 | 78389 | CDS(psaA) |
| tri | (TAA)4 | 89011 | 89022 | IGS(rps14-trnG) |
| tetra | (AATT)3 | 9084 | 9095 | IGS(infA-rps8) |
| tetra | (CTTT)3 | 25471 | 25482 | IGS(psbB-psbC) |
| tetra | (AATT)3 | 29352 | 29363 | IGS(psbD-trnM) |
| tetra | (TAAT)3 | 41783 | 41794 | CDS(ftsH) |
| tetra | (ATAA)3 | 46377 | 46388 | IGS(ftsH-psbZ) |
| tetra | (AAAT)3 | 72259 | 72270 | CDS(rpoC2) |
| tetra | (GTAG)3 | 76586 | 76597 | CDS(rrl) |
| tetra | (TTTA)3 | 80831 | 80842 | IGS(psaA-psaC) |
| tetra | (ATTA)3 | 86695 | 86706 | IGS(psaA-trnT) |
| sex | (TAATTT)3 | 10101 | 10118 | IGS(rps8-rpl5) |
aintergenic spacer region.
bcoding sequences.
Table 3. Distributions of SSRs in the 11 chloroplast genomes belonging to Ulvophyceae.
| Species Name | Accession | Inverted Repeat | Number of SSRs | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | Total | |||
| Bryopsis hypnoides | NC_013359 | No | 257 | 32 | 1 | 7 | 2 | 1 | 300 |
| Bryopsis plumosa | NC_026795 | No | 124 | 18 | 1 | 6 | 1 | 1 | 151 |
| Gloeotilopsis planctonica | KX306824 | No | 650 | 14 | 6 | 11 | 1 | 3 | 685 |
| Gloeotilopsis sarcinoidea | KX306821 | No | 806 | 25 | 7 | 14 | 0 | 17 | 869 |
| Gloeotilopsis sterilis | NC_025538 | Yes | 302 | 18 | 5 | 14 | 1 | 2 | 342 |
| Ostreobium sp | KU979013 | No | 67 | 20 | 0 | 5 | 0 | 0 | 92 |
| Pseudendoclonium akinetum | NC_008114 | Yes | 445 | 40 | 8 | 10 | 2 | 1 | 506 |
| Tydemania expeditionis | NC_026796 | No | 101 | 26 | 4 | 3 | 0 | 3 | 137 |
| Ulva fasciata | NC_029040 | No | 162 | 34 | 16 | 14 | 3 | 1 | 230 |
| Ulva linza | NC_030312 | No | 174 | 38 | 19 | 7 | 4 | 5 | 247 |
| Ulva sp | KP720616 | No | 166 | 38 | 20 | 18 | 10- | 5 | 247 |
Using Tandem Repeats Finder, five repeats were detected in the U. flexuosa chloroplast genome. All of these repeats were between 10 and 15 bp in length. The intergenic spacers between rpoC2 and rrs, psbB and psbC possessed two copies of the longest tandem repeats, respectively (15 bp). Most of the repeated sequences were located in the intergenic spacer besides one repeat which was located among coding sequences of rpoC2 (Table 4).
Table 4. Repeat sequences identified in the chloroplast genome of U. flexuosa.
| Repeat Number | Repeat size (bp) | Type | Location | Repeat Unit sequence |
|---|---|---|---|---|
| 1 | 10 | T | IGSa (trnI-psbH) | (ATTATTTTAT)3 |
| 2 | 13 | T | CDSb(rpoC2) | (AATTTAAAGAAAA)2 |
| 3 | 15 | T | IGS(rpoC2-rrs) | (GTTTTAATATAAATT)2 |
| 4 | 15 | T | IGS(psbB-psbC) | (AAATTAAAAAATAT)2 |
| 5 | 12 | T | IGS(trnV-rpoB) | (ATTATTAATTAA)2 |
| 6 | 31 | F | IGS(trnE-trnM) | TAATAATATTAATAATAATAATATTAATAAT |
| 7 | 30 | F | IGS(rpl32-chlI) | TTTATTAATATATACTAAAATTATTAATTA |
aintergenic spacers.
bcoding sequences.
Two forward repeats were identified using REPuter with a size cutoff of 30 bp, which were between trnE and trnM (31 bp), rpl32 and chlI (30 bp) (Table 4).
Phylogenomic analyses
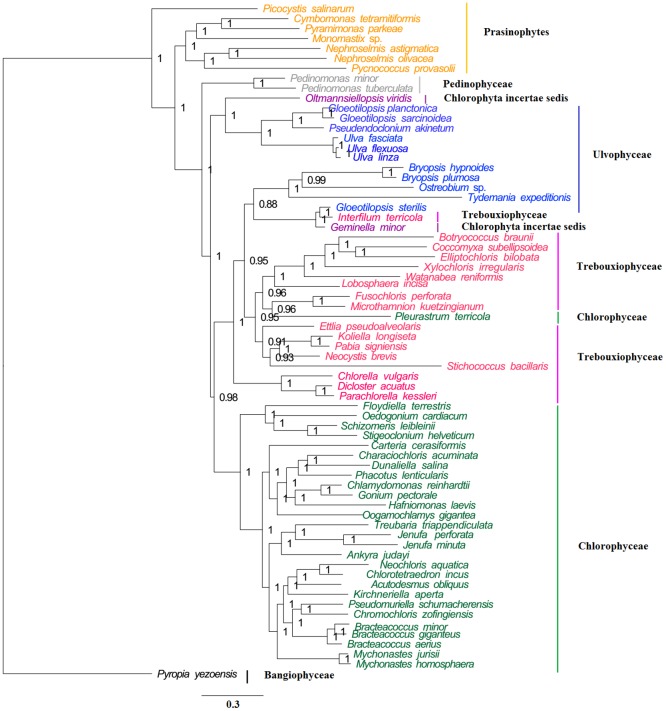
To determine the phylogenetic position of U. flexuosa in Chlorophyta, 67 complete chloroplast genome sequences were obtained from the Genebank database, which belonged to Chlorophyceae (28), Trebouxiophyceae (17), Ulvophyceae (10), Prasinophytes (7), Pedinophyceae (2), Chlorophyta incertae sedis (2) and Bangiophyceae (1) respectively. The number shown in the parenthesis represents the number of species in the corresponding clade. To conduct phylogenetic analysis, we extracted 24 protein sequences, which were present among all the 68 chloroplast genomes. After alignment there were a total of 21534 positions in the final dataset. The phylogenetic tree showed that U. flexuosa and U. linza gathered into a cluster (Fig 5, S3 Fig).
Fig 5. Bayesian 50% majority rule consensus tree based on 24 nucleotide sequences of 68 strains representing the Chlorophyta.
The 24 genes were atpA, atpB, atpE, atpF, petB, petD, petG, psaB, psbB, psbD, psbE, psbF, psbH, psbJ, psbK, psbL, psbN, rbcL, rpl2, rpl20, rpl36, rpoA, rps8 and ycf3. The main parameters were: ngen = 2000000, samplefreq = 100, nchains = 4, nst = 6, rates = gamma. The Bayesian posterior probabilities were given at the nodes. The tree was rooted to Pyropia yezoensis.
Rearrangements in the Chlorophyte cpDNA
Genome rearrangement analysis for the ten cpDNAs Mauve alignment from Ulvophyceae showed there was no significant difference in genomes rearrangement within Ulva spp. (Fig 6). However, the organellar genomes out of Ulva spp. across Ulvophyceae were highly rearranged comparing to U. flexuosa.
Fig 6. Mauve alignments of U. flexuosa cpDNA (top of alignment) with those of 9 chlorophytes.
Genome synteny analysis
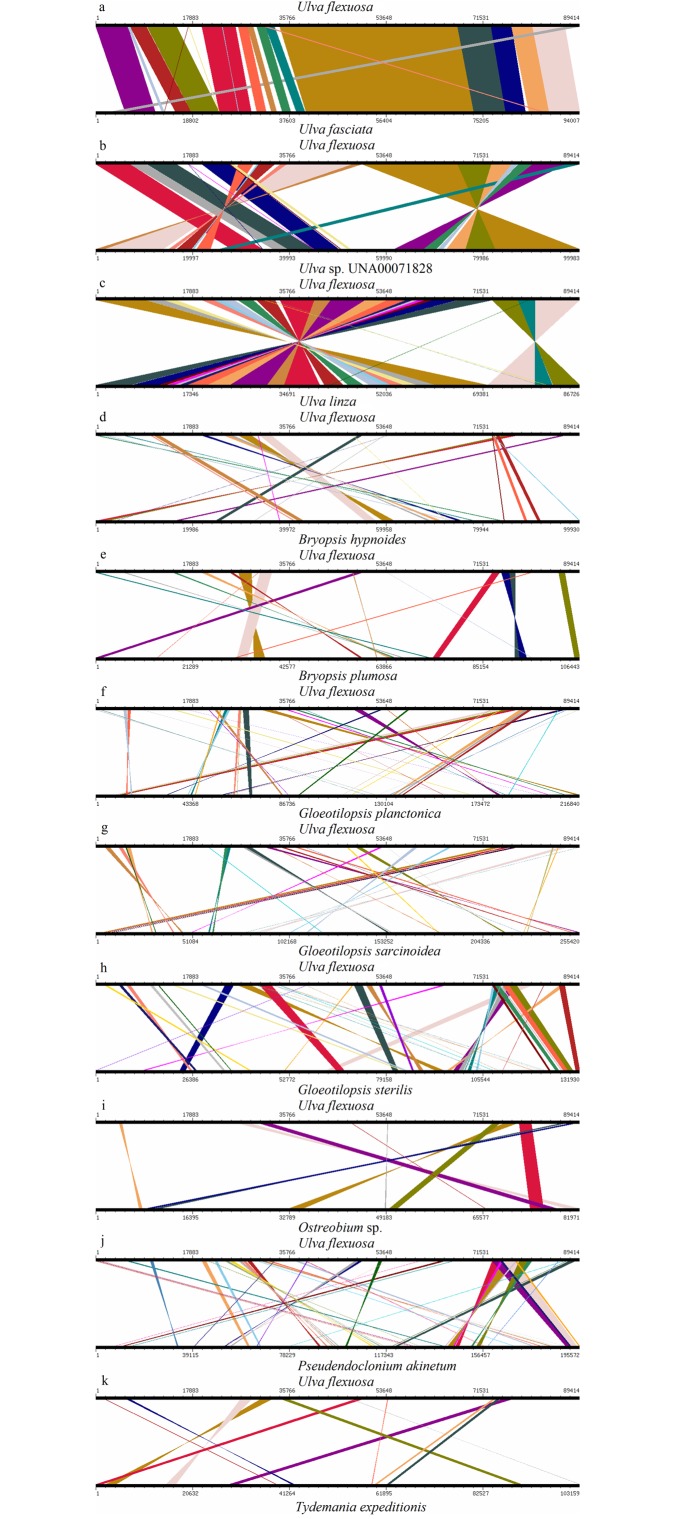
Genome synteny was identified between the chloroplast genomes of U. flexuosa and 11 other species. The 11 species were classified into two groups based on degree of genome sequence similarity relative to the U. flexuosa chloroplast genome. The first group included U. fasciata, Ulva sp. UNA00071828 and U. linza. The chloroplast genome of this group was high conserved compared with that of U. flexuosa (Fig 7a–7c), with similarity above 91%. The second group included B. hypnoides, B. plumosa, G. planctonica, G. sarcinoidea, G. sterilis, Ostreobium sp., P. akinetum and T. expeditionis with similarity below 30% relative to U. flexuosa (Fig 7d–7k). These results suggested that genomes had high conservation among Ulva spp.
Fig 7. Comparative genomic analyses of 12 chloroplast genomes.
The chloroplast genome of U. flexuosa was aligned with those of 12 species. Each horizontal black line represents a genome. The species names are shown next to the corresponding line. The conserved regions are bridged by lines.
Discussion
It indicated that the main genes of chloroplast genomes had high conservation among Ulva spp. which was monophyletic clade in Fig 5, with bayesian posterior probabilities (PP) of 1.0 and RAxML bootstrap support values (ML) of 100. These genomes might function similarly in photosynthesis, which were in line with their simultaneous originating, floating and outbreaking as green tide in the Yellow Sea of China. Meanwhile, the SSRs among U. flexuosa was also similar to those observed in three reported chloroplast genomes of other species belonging to Ulvaceae (Table 3), which provided potential molecular markers for Ulva spp. identification. Furthermore, there were no significant difference in genomes rearrangement (Fig 6) and genome synteny within cpDNAs from Ulva spp. (Fig 7). In a word, the Ulva spp. are conserve and usually monophyletic in phylogenetic tree [5].
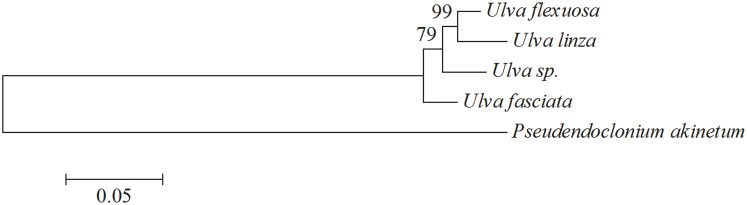
Correlational studies [5, 33] within Ulva spp. are rarely seen, since few related genomes are reported to illustrate the evolutionary relationship [5, 13, 34, 35]. U. flexuosa is a cosmopolitan green algae encountered in fresh waters to saline waters [2], which may take on the characteristics of transition state and be supposed as a coordinate points for evolution [5]. In order to compare U. flexuosa with reported Ulva spp. in terms of chloroplast genome, we then made another phylogenetic tree for evolutionary analysis, including four kinds of species from Ulva spp. (U. flexuosa, U. fasciata, U. linza and Ulva sp. UNA00071828) and one exogenous species (Monomastix sp.) from prasinophytes. Since Ulvophyceae, Trebouxiophyceae, and Chlorophyceae shared a common ancestor which may have been prasinophyte-like [5]. 67 protein sequences (S6 Table) shared among all these five species were aligned using the ClustalW2 program. It was indicated that U. flexuosa was clustered with U. linza (Fig 8) in the phylogenetic tree, standing for the maximum homology between them. This was consistent with the phylogenetic tree in Fig 5 that U. flexuosa was clustered with U. linza (PP of 1.0 and ML of 100).
Fig 8. Molecular phylogenetic analysis of Ulva genus.
The tree was constructed with the sequences of 67 coding genes present in all five species (U. flexuosa, U. fasciata, U. linza, Ulva sp. and Monomastix sp.) using the Maximum Likelihood method. Monomastix sp. was used as outgroup. Bootstrap supports were calculated from 1000 replicates.
The close relationships between U. flexuosa and U. linza are supported by morphological and life history similarities to a certain degree. For example, the bright green thallus of two algae are tubular at the base but flattened above, in which cells are 11–15 μm in diameter. Besides, they are both alternate generations. However, we can still distinguish them easily according to the difference in morphology, since U. linza is linear or lanceolate banding, with wavy folds in the margin, while it is not the case in U. flexuosa (Fig 1). Thus nuclear genome may be the main factor which shapes the algae.
Molecular markers, including SSR, tandem repeats and forward repeats, are important supplement of morphology for species identification. These repeats were mainly distributed among intergenic spacers (S5 Table, Tables 2 and 4), which were variate more easily than coding sequences. The vast majority of repetitive sequence was composed of A/T, which was consistent with high A/T content (75%) in the genome. Therefore, whether high A/T content is related to the high variation is worthy studying.
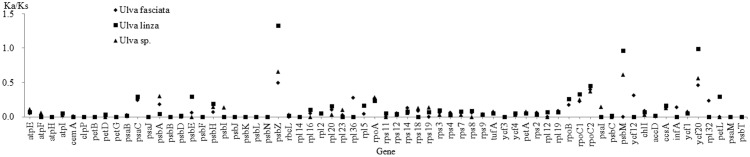
In order to find differential gene in U. flexuosa apart from other species among Ulva spp., a selection force and substitution rate assessment by 67 protein-coding genes (S6 Table) extracted from four species (U. flexuosa, U. fasciata, U. linza and Ulva sp. UNA00071828) using the KaKs_Calculator Toolbox 2.0 (https://sourceforge.net/projects/kakscalculator2) were performed using gamma-series methods of γ-NG and standard genetic code [36]. Taken U. flexuosa as control, most of Ka/Ks values were below one, which meant purify selection and conservative, wherein those of some genes (atp1, atp6, cob, cox2, cox3, nad6, nad7, rpl5, rps3 and rps10) in U. linza were zero. However, Ka/Ks values of psbZ from U. linza were 1.3, meaning positive selection, and those of psbM and ycf20 were near 1.0, meaning neutral evolution (Fig 9).
Fig 9. Comparison of selection forces (Ka/Ks) of the 67 common protein-coding genes in three-species matrix.
As gene of high variability, psbZ, psbM and ycf20 are potential molecular markers to identify U. flexuosa from Ulva spp. Besides, psbZ and psbM both belong to the subunits of photosystem II, involved in photosynthesis of photon absorption and energy transfer (Table 1). In previous research, we found that light and temperature were important factors which affected growth rate of green tide algae. For example, as high temperature species, the daily growth rate of U. flexuosa (including seedlings and adults) were higher than U. prolifera, U. compressa and U. linza under 25–35°C, or below 70 μmol·m-2·s-1 of light intensity [3]. Whether they are regulated by psbZ and psbM may worth studying in the next step.
Conclusions
In the present study, we have: (1) sequenced the chloroplast genome of U. flexuosa; (2) annotated the chloroplast genome including ORFs and genetic code; (3) identified SSR, tandem repeats and forward repeats of the genome; (4) carried out a phylogenetic analysis of the 68 chloroplast genomes based on 24 conserved genes; (5) compared the chloroplast genome rearrangement of 10 species; (6) analyzed the synteny of U. flexuosa and 12 other species in Chlorophyte; and finally (7) comprehensively compared the chloroplast genome homology of four Ulva species. We found that organellar genomes in other species across Ulvophyceae were high rearranged comparing with U. flexuosa, while those among Ulva spp. were high conserved. Though U. flexuosa was closer to U. linza than U. fasciata and Ulva sp. UNA00071828 in phylogenetic tree, the average Ka/Ks between U. flexuosa and U. linza assessed by 67 protein-coding genes was higher than those between U. flexuosa and other species in Ulva genus, due to the variation of psbZ, psbM and ycf20. Future studies may include range of light, temperature and salinity on photosynthesis of U. flexuosa based on chloroplast genomes and transcriptome, as well as molecular identification of Ulva spp.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(TIF)
(TIF)
The 24 genes were atpA, atpB, atpE, atpF, petB, petD, petG, psaB, psbB, psbD, psbE, psbF, psbH, psbJ, psbK, psbL, psbN, rbcL, rpl2, rpl20, rpl36, rpoA, rps8 and ycf3. The appropriate model of evolution for each gene determined by jModelTest 2.1 was GTR+I+G, which was reassessed by PAUP 4.0a152 (P value = 1 − (99/100) = 0.010000). Then, the evolutionary history was inferred using the Maximum Likelihood method implemented in RaxML. The detailed parameters were “raxmlHPC-PTHREADS -f a -x 12345 -# 1000 -m GTRGAMMA -s dna.phy -n nex -p 12345 -q part.txt -T 6”. The tree with the final ML Optimization Likelihood (-548221.173374) was shown. The significance level for the phylogenetic tree was assessed by bootstrap testing with 1000 replications. The RAxML bootstrap support values were given at the nodes. The tree was rooted to Pyropia yezoensis.
(TIF)
Acknowledgments
We thank Shanghai MAP Biotech Co., Ltd. for technical assistance and the reviewers for providing helpful feedback.
Data Availability
All relevant data are within the paper and its supporting information files.
Funding Statement
This work was supported by National Natural Science Foundation of China (41576163), National Key R&D Program of China (2016YFC1402105), Key Laboratory of Integrated Marine Monitoring and Applied Technologies for Harmful Algal Blooms, S.O.A., (MATHAB2017010), and General Financial Grant from the China Postdoctoral Science Foundation (2015M572743). The funders did not play a role in the study design, data collection and analysis, decision to publish, or manuscript preparation.
References
- 1.Rybak AS, Czerwoniec A. Differentiation of inland Ulva flexuosa Wulfen (Ulvaceae, Chlorophyta) from Western Poland. Oceanol Hydrobiol St. 2015; 44: 393–409. [Google Scholar]
- 2.Rybak A, Czerwoniec A, Gabka M, Messyasz B. Ulva flexuosa (Ulvaceae, Chlorophyta) inhabiting inland aquatic ecosystems: molecular, morphological and ecological discrimination of subspecies. Eur J Phycol. 2014; 49: 471–485. [Google Scholar]
- 3.Cui JJ, Zhang JH, Huo YZ, Zhou LJ, Wu Q, Chen LP, et al. Adaptability of free-floating green tide algae in the Yellow Sea to variable temperature and light intensity. Mar Pollut Bull. 2015; 101: 660–666. doi: 10.1016/j.marpolbul.2015.10.033 [DOI] [PubMed] [Google Scholar]
- 4.Castelar B, Reis RP, Calheiros ACD. Ulva lactuca and Ulva flexuosa (Chlorophyta, Ulvophyceae) cultivation in Brazilian tropical waters: recruitment, growth, and ulvan yield. J Appl Phycol. 2014; 26: 1989–1999. [Google Scholar]
- 5.Cocquyt E, Verbruggen H, Leliaert F, De Clerck O. Evolution and cytological diversification of the green seaweeds (Ulvophyceae). Mol Biol Evol. 2010; 27: 2052–2061. doi: 10.1093/molbev/msq091 [DOI] [PubMed] [Google Scholar]
- 6.Messyasz B, Czerwik-Marcinkowska J, Uher B, Rybak A, Szendzina L, Pikosz M. Ulva flexuosa subsp pilifera (Chlorophyta, Ulvophyceae) from the Wielkopolska region (West Poland): a new observation on the ultrastructure of vegetative cells. Oceanol Hydrobiol St. 2013; 42: 209–215. [Google Scholar]
- 7.Messyasz B, Czerwik-Marcinkowska J, Massalski A, Uher B, Rybak A, Szendzina L. Morphological and ultrastructural studies on Ulva flexuosa subsp pilifera (Chlorophyta) from Poland. Acta Soc Bot Pol. 2013; 82: 157–163. [Google Scholar]
- 8.Zhang J, Liu C, Yang L, Gao S, Ji X, Huo Y, et al. The source of the Ulva blooms in the East China Sea by the combination of morphological, molecular and numerical analysis. Estuar Coast Shelf S. 2015; 164: 418–424. [Google Scholar]
- 9.Ott FD. Synthetic media and techniques for the xenic cultivation of marine algae and flagellate. Virginia J Science. 1965; 16: 205–218. [Google Scholar]
- 10.Chen H, Zhou D, Luo G, Zhang S, Chen J. Macroalgae for biofuels production: progress and perspectives. Renew Sust Energ Rev. 2015; 47: 427–437. [Google Scholar]
- 11.Coat G, Dion P, Noailles MC, Reviers BD, Fontaine JM, Berger-Perrot Y, et al. Ulva armoricana (Ulvales, Chlorophyta) from the coasts of Brittany (France). II. Nuclear rDNA ITS sequence analysis. Eur J Phycol. 1998; 33: 81–86. [Google Scholar]
- 12.Shimada S, Yokoyama N, Arai S, Hiraoka M. Phylogeography of the genus Ulva (Ulvophyceae, Chlorophyta), with special reference to the Japanese freshwater and brackish taxa. J Appl Phycol. 2009; 20: 529–539. [Google Scholar]
- 13.Melton JT, Leliaert F, Tronholm A, Lopez-Bautista JM. The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp UNA00071828 (Ulvophyceae, Chlorophyta). Plos One. 2015; 10: e0121020 doi: 10.1371/journal.pone.0121020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee WH, Vega VB. Heterogeneity detector: finding heterogeneous positions in Phred/Phrap assemblies. Bioinformatics. 2004; 20: 2863–2864. [DOI] [PubMed] [Google Scholar]
- 15.Talavera G, Castresana J Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biol. 2007; 56: 564–577. [DOI] [PubMed] [Google Scholar]
- 16.Castresana J Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000; 17: 540–552. [DOI] [PubMed] [Google Scholar]
- 17.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenstein J Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985; 39: 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- 19.Chang L, Shi L, Zhu Y, Chen H, Zhang J, Lin X, et al. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 2012; 13: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005; 33: 686–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004; 32: 11–16. doi: 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krumsiek J, Arnold R, Rattei T Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007; 23: 1026–1028. doi: 10.1093/bioinformatics/btm039 [DOI] [PubMed] [Google Scholar]
- 23.Lohse M, Drechsel O, Bock R OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007; 52: 267–274. [DOI] [PubMed] [Google Scholar]
- 24.Rice P, Longden I, Bleasby A EMBOSS: the european molecular biology open software suite. Trends in Genetics Tig. 2000; 16: 276–277. [DOI] [PubMed] [Google Scholar]
- 25.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999; 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001; 29: 4633–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. Plos One. 2010; 5:e11147 doi: 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitfield J Mathematics of evolution and phylogeny. edited by olivier gascuel. Brief Bioinform. 2008; 9: 547–549. [Google Scholar]
- 29.Braga MDV, Willing E, Stoye J Double cut and join with insertions and deletions. J Comput Biol. 2011; 18: 1167–1184. [DOI] [PubMed] [Google Scholar]
- 30.Revanna KV, Chiu CC, Bierschank E, Dong Q. GSV: a web-based genome synteny viewer for customized data. BMC Bioinformatics. 2011; 12: 316–316. doi: 10.1186/1471-2105-12-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue J, Wang S, Zhou SL Polymorphic chloroplast microsatellite loci in nelumbo (Nelumbonaceae). Am J Bot. 2012; 99: 240–244. [DOI] [PubMed] [Google Scholar]
- 32.Kuang DY, Wu H, Wang YL, Gao LM, Zhang SZ, Lu L Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome. 2011; 54: 663–673. [DOI] [PubMed] [Google Scholar]
- 33.Sun L, Fang L, Zhang Z, Chang X, Penny D, Zhong B. Chloroplast phylogenomic inference of green algae relationships. Sci Rep. 2015; 6: 20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leliaert F, Lopez-Bautista JM The chloroplast genomes of Bryopsis plumosa and Tydemania expeditiones (Bryopsidales, Chlorophyta): compact genomes and genes of bacterial origin. BMC Genomics. 2015; 16: 204 doi: 10.1186/s12864-015-1418-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, et al. The chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007; 318: 245–250. doi: 10.1126/science.1143609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nei M, Gojobori T Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986; 3: 418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(TIF)
(TIF)
The 24 genes were atpA, atpB, atpE, atpF, petB, petD, petG, psaB, psbB, psbD, psbE, psbF, psbH, psbJ, psbK, psbL, psbN, rbcL, rpl2, rpl20, rpl36, rpoA, rps8 and ycf3. The appropriate model of evolution for each gene determined by jModelTest 2.1 was GTR+I+G, which was reassessed by PAUP 4.0a152 (P value = 1 − (99/100) = 0.010000). Then, the evolutionary history was inferred using the Maximum Likelihood method implemented in RaxML. The detailed parameters were “raxmlHPC-PTHREADS -f a -x 12345 -# 1000 -m GTRGAMMA -s dna.phy -n nex -p 12345 -q part.txt -T 6”. The tree with the final ML Optimization Likelihood (-548221.173374) was shown. The significance level for the phylogenetic tree was assessed by bootstrap testing with 1000 replications. The RAxML bootstrap support values were given at the nodes. The tree was rooted to Pyropia yezoensis.
(TIF)
Data Availability Statement
All relevant data are within the paper and its supporting information files.