Abstract
The mRNA of the intronless, single-copy CCAAT/enhancer-binding protein-β (C/EBPβ) gene encodes several isoforms that have truncated transcription activation domains. This occurs by the alternative translational initiation (ATI) at multiple AUG start sites. The C/EBPβ mRNA has four in-frame AUGs and an internal out-of-frame AUG associated with a small open reading frame (sORF). Initiation of translation at the in-frame AUGs forms 40-kDa (AUG-1), 35-kDa (AUG-2), 20-kDa (AUG-3) and 8.5-kDa (AUG-4) isoforms. We show that in COS-1 cells the 20-kDa isoform is not a product of proteolysis of the higher molecular weight isoforms. The sORF contains an AUG and termination signal that may produce the oligopeptide MPPAAARRL. Our studies suggest that ATI involves three mRNA structural features: (i) the cap structure, (ii) the context of the Kozak sequences that flank the AUG and (iii) the integrity of the sORF. We propose that formation of C/EBPβ isoforms is accomplished by a leaky ribosomal scanning mechanism that facilitates ATI of multiple internal AUGs.
INTRODUCTION
The intronless genes of two of the CCAAT/enhancer-binding trans-activating proteins, α and β (C/EBPα) and (C/EBPβ), each encode a single mRNA species (1). These mRNAs are translated to produce several isoforms that have truncated transcription activation domains and specific transcriptional regulatory functions (2–4). It has been proposed that these isoforms are produced by a leaky ribosomal scanning mechanism that facilitates the alternative translational initiation (ATI) at multiple AUG initiation codons within the open reading frame (ORF) of the single mRNA.
Two of the C/EBP mRNA structural features may play important roles in the ATI mechanism: (i) the context of the nucleotide sequences that flank the AUG, i.e. the Kozak sequences, and (ii) the presence of a small open reading frame (sORF) within the mRNA. The C/EBPα and C/EBPβ mRNAs contain multiple AUG translational initiation sites whose Kozak sequences vary from least optimal to most optimal, and sORFs within their mRNAs that function in the regulation of ATI from multiple internal AUGs (2,3,5–12). One mechanism proposes that the C/EBPα sORF functions as a cis-acting inhibitor of translation that is overcome during such biological functions as terminal differentiation of hepatocytes (13,14), adipocytes (15) and intestinal epithelial cells (16). Similarly, it has been proposed that the C/EBPβ sORF also acts as a repressor of the synthesis of the 35-kDa isoform (17). Alternatively, we and others have proposed that the C/EBPβ sORF regulates the induction of the 20-kDa isoform (5,7,8,10) in response to cell-specific or physiological stimuli, such as viral infection (18), inflammatory agents (19) or terminal differentiation (20).
In previous studies, we and others identified multiple molecular weight forms of C/EBPβ proteins (isoforms) in the mouse liver (3,5,10) and tissue culture cells including hepatocytes (7), CHO cells (8), adipocytes (21), HeLa cells (17) and virus infected cells (22). Characterization of these isoforms revealed that they have truncated transcription activation domains. The fact that only one C/EBPβ mRNA is transcribed from the intronless gene suggests that these isoforms are produced by ATI of the first (AUG-1), second (AUG-2) and third (AUG-3) in-frame AUGs within the full-length mRNA (5,8,10). The locations of these downstream AUGs would account for the molecular masses of the isoforms detected by western or southwestern analyses, which further supports the mechanism of ATI.
The formation of the C/EBPβ isoforms has also been attributed to proteolytic cleavage of the high molecular weight isoform (12,17). It has been proposed that C/EBPα regulates low molecular weight C/EBPβ isoform production through activation of calpain, a proteolytic enzyme that cleaves C/EBPβ (12,17). Recently, it has been shown that mutation of AUG-3, the initiation site of the 20-kDa isoform, in a C/EBPβ expression vector transfected into COS-1 cells, abolishes the synthesis of this isoform (8,10). This expression vector continues to synthesize an abundance of both the 40-kDa (AUG-1) and 35-kDa (AUG-2) isoforms. These data suggest that the high molecular weight isoforms, which are abundant in these transfected cells, are not proteolytically cleaved to form the 20-kDa isoform (8,10). However, the data do not completely rule out proteolytic cleavage, as conversion of AUG-3 to UUG replaces the methionine with a leucine, which may affect the ability of the proteolytic enzyme to cleave the protein.
We now present data to support our hypothesis that multiple C/EBPβ isoforms are produced by ATI at multiple AUG start sites, and demonstrate a role for the C/EBPβ sORF in the regulation of ATI of C/EBPβ mRNA. We report here the results of our analyses of specific mutations of (in-frame) AUG start sites, and mutation of the sORF-AUG on the production of C/EBPβ isoforms. Furthermore, we hypothesize that this occurs by a ribosomal scanning mechanism in which ribosomal initiation occurs at specific AUG start sites.
MATERIALS AND METHODS
Construction of expression vectors
Oligonucleotides used to construct expression vectors are listed in Table 1.
Table 1. Expression vector mutations are listed under Constructs and the oligonucleotides used are listed under Oligonucleotides column.
| Constructs |
Oligonucleotides |
Sequences |
| mtsORF | AN1 | 5′-CCGCGTTCATGCACCGCCTGCTGGCCTGG-3′ (ATG-1 underlined) |
| AN2 | 5′-cagcaggcggtgcatgaacgcggggcc-3′ | |
| AN11 | 5′-GACGCAGCTTGCCTCCC-3′ (mutated ATG of sORF underlined) | |
| AN12 | 5′-CAAGCTGCGTCCCAGGC-3′ | |
| AN5 | 5′-GCCGCCGCCCGCCGC-3′ | |
| AN6 | 5′-GGCGGCGGCGGGAGG-3′ | |
| AN7 | 5′-CTTTAGACC-3′ (stop codon of sORF underlined) | |
| AN8 | 5′-CATGGGTCTAAAGGCGGCG-3′ | |
| mtAUG-1 | Beta-MT35/30-P1 | 5′-CCGCGGGCCCCGCGTTCTTGCACCGC-3′ (ATG-1 changed to TTG) |
| Beta-P30-P | 5′-GCTCGTAGTAGAAGTTGG-3′ | |
| mtAUG-2 | Beta-WT-P | 5′-CGGAGCCCGCGGGCCCCGCG-3′ |
| Beta-MT35/30-P2 | 5′-AAGTTGGCCACTTCCAAGGGTCTAAAG-3′ (ATG-2 changed to TTG) | |
| AUG-1(K) | Beta-ATG-1K | 5′-CAGCGGAGCCCGCGGGCCCCGCGGCCATGGAC CGCCTGC-3′ (Kozak ATG-1 underlined) |
| Beta-P30-P | 5′-GCTCGTAGTAGAAGTTGG-3′ | |
| Spacer112 | Beta-BN-P1 | 5′-GCTTGGATCCTCTAGAGTCCGAGC-3′ (BamHI site underlined) |
| Beta-BN-P2 | 5′-TGATACTAGTCTAAAGGCGGCGCGCGGC-3′ (SpeI site underlined) | |
| Beta-Spacer112-P1 | 5′-GGCGGCTAGCCGAGAGACTTCTCGCGGAGCTG-3′ (NheI site underlined) | |
| Beta-Spacer112-P2 | 5′-TGAACCATGGGTTCGGTAGGAAAAGGAGCGAGT TTTG-3′ (NcoI site underlined) | |
| sORF(K2) | Beta-WT-P | 5′-CGGAGCCCGCGGGCCCCGCG-3′ |
| ORF(K)-P1 | 5′-CATGGTGGCTCCCAGGCC-3′ (Kozak ATG of sORF underlined) | |
| ORF(K)-P2 | 5′-GGAGCCACCATGGCTCCCGCCG-3′ (Kozak ATG of sORF underlined) | |
| Beta-P30-P | 5′-GCTCGTAGTAGAAGTTGG-3′ | |
| sORF(G8) | Beta-WT-P | 5′-CGGAGCCCGCGGGCCCCGCG-3′ |
| ORF(K)-P1 | 5′-CATGGTGGCTCCCAGGCC-3′ (Kozak ATG of sORF underlined) | |
| ORF(K)-G-F | 5′-GGAGCCACCATGGGCGGCGGTGGCGGCGGTGG CGGCGACTACAAAG-3′ (Kozak ATG of sORF underlined) | |
| Beta-P30-P | 5′-GCTCGTAGTAGAAGTTGG-3′ | |
| ORF(K)-G | 5′-AGTTGGCCACTTCCATGGGTCTAGCCGCCACCG CCGCCAC-3′ | |
| NewAUG | Beta-WT-P | 5′-CGGAGCCCGCGGGCCCCGCG-3′ |
| NewAUG1 | 5′-TCTCCGACGCCATGGCCGACGAC-3′ | |
| NewAUG2 | 5′-GTCGTCGGCCATGGCGTCGGAGA-3′ | |
| Csp-P | 5′-GCTTGCAGTCCGCGGG-3′ | |
| δ-wt | Delta-F1 | 5′-CGCCGACTGCCGGGACTACAAAGACGATGACG ATAAATAACG-3′ (Flag tag underlined) |
| Delta-F2 | 5′-CGCGTTATTTATCGTCATCGTCTTTGTAGTCCCGGCAGTCGG-3′ | |
| (β-sORFwt)-C/EBPδ or (β-sORFmt)-C/EBPδ | Delta-1 | 5′-ACGCCGCCATGGGCGCCGCGCTTTTCAGCCTG-3′ (NcoI site underlined) |
| Delta-2 | 5′-CGGCGGCCATGGTGTCAATGTAGGCGCTGAAG-3′ |
Construction of expression vectors pCMV-C/EBPβ-wt and pCMV-C/EBPβ-mtAUG-3 has been described previously (10). All of the nucleotide primers used to make additional expression vectors are listed in Table 1. All constructs were confirmed by restriction enzyme digestion and sequence analysis.
The expression vector pCMV-C/EBPβ-mtsORF has a mutation in the initiation codon of the sORF. It was constructed by annealing and ligating the following oligonucleotide pairs: AN1–AN2, AN11–AN12, AN5–AN6 and AN7–AN8. The mutant fragment was then ligated into pCMV-C/EBPβ-wt.
Several expression vectors were constructed by replacing wild-type fragments with fragments containing specific mutations generated by PCR. These include the specific AUG initiation codon mutations in pCMV-C/EBPβ-wt and pCMV-C/EBPβ-mtsORF, pCMV-C/EBPβ-AUG-1K, and pCMV-C/EBPβ-Spacer 112. The mutations and primers used in these constructions are summarized in Table 1.
Some expression vectors were constructed by a strategy of two-step PCR that involves two pairs of PCR primers that overlap each other in the region to be modified. Fragments with specific mutations generated by this technique were also cloned into the appropriate expression vector to replace the wild-type sequence. Expression vectors pCMV-C/EBPβ-sORF(K2) and pCMV-C/EBPβ-NewAUG were constructed in this manner. The mutations and primers used in these constructions are summarized in Table 1.
Construction of pCMV-C/EBPδ-wt was achieved by inserting an EcoRI–BamHI fragment containing the C/EBPδ coding region and 3′-UTR of pMSV-C/EBPδ (1) into the EcoRI–EcoRV sites of pCMB-B (10) and by inserting two annealed oligonucleotides containing Flag tag sequence, Delta-F1 and Delta-F2, into the NarI site. PCMV-C/EBPδ-AUG-3 containing the sequence from the third AUG to the 3′-UTR of C/EBPδ was made by cloning the NcoI–XbaI fragment including these sequences into pFLAG-CMV-2 (Eastman Kodak Co., Rochester, NY). We have also made expression vectors with a wild-type or mutated C/EBPβ sORF placed in front of the C/EBPδ gene. To accomplish this, we introduced a mutation at the first AUG of C/EBPδ producing an NcoI restriction site using primers Delta-1 and Delta-2. A C/EBPβ fragment containing the 5′-UTR, sORF (wild-type or mutated), and a partial coding region was inserted into this site immediately upstream of the first AUG of C/EBPδ, producing pCMV-(β-sORFwt)-C/EBPδ and pCMV-(β-sORFmt)-C/EBPδ.
For in vitro transcription and translation, two constructs, pT7-C/EBPβ-wt and pT7-C/EBPβ-mtAUG-3, were made by inserting BamHI fragments containing C/EBPβ coding regions from the appropriate pCMV expression vector into pGEM4z (Promega, Madison, WI).
Cell culture, transfection, and preparations of nuclear protein
Transfection of expression vectors into COS-1 cells was done as previously described with some modifications (5). Briefly, 1 day after plating COS-1 cells at a density of 1 × 106 cells per dish (100 mm), 10 µg of each construct DNA was transfected by the DNA–calcium phosphate coprecipitation method. After 48 h, the cells were harvested and divided into two parts, one for preparation of nuclear proteins and another for preparation of total RNA. Nuclear proteins were prepared as described by Schreiber et al. (23).
Antisera
Antisera specific for the Flag tag sequences were purchased from Eastman Kodak.
Western blot analyses
Western immunoblot procedure was performed as described by An et al. (5). To more accurately compare the translation from multiple transfected expression vectors, protein loading was adjusted to reflect variations in vector mRNA pool levels. Total RNA was isolated from transfected cells and vector mRNA pool size determined by northern analysis (5). Protein loading for western analysis was then adjusted to reflect translation from an equivalent mRNA pool. Autoradiograms of western analyses were quantified by densitometry using MultiImage light cabinet and analysis software (Alpha Innotech, San Leandro, CA).
In vitro transcription and translation
In vitro transcription reactions (50 µl) had 5 µg of linearized pT7-C/EBPβ-wt or pT7-C/EBPβ-mtAUG-3 as template, 10 mM DTT, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM UTP, 0.05 mM GTP or 0.5 mM GTP ± the cap analog, m7G (5′)ppp(5′) G, and 40 U of T7 RNA polymerase, and was incubated for 60 min at 37°C (Promega). After digestion with RNase-free DNase, transcripts were phenol–chloroform extracted, and precipitated by addition of 1/20 vol of 3.5 M sodium acetate and 2.5 vol of 100% ethanol. RNA pellets were washed in 70% ethanol, resuspended in 10 µl of DEPC-treated H2O, electrophoresed on 1.5% of denaturing agarose gel and quantitated by DU-64 Spectrophotometer. In vitro translation reactions were conducted for 60 min at 30°C in 25 µl reactions containing 17.5 µl of rabbit reticulocyte lysate (Promega), 0.5 µg of RNA and 0.02 mCi of [35S]methionine (ICN). In vitro translation aliquots were resolved in 12% SDS–PAGE and the gels were dried and autoradiographed at –80°C.
Affinity identification and isolation of capped C/EBPβ mRNA
The GST/eIF-4E fusion protein was expressed and purified as previously described (24). Total RNA was isolated from COS-1 cells as previously described (25). Total poly(A)-RNA was purified using an mRNA separator kit (ClonTech) utilizing an oligo(dT)-cellulose spin column.
Affinity selection was performed via a protocol referred to as the CAPture method (24). To select for capped mRNAs, the GST/eIF-4E fusion protein was first coupled to glutathione agarose (Sigma) according to the manufacturer’s recommendations. mRNAs were selected by using a 200 µl volume of packed resin. RNA preparations were resuspended in a final volume of 400 µl reaction buffer prior to being passed over the cap column. Reaction binding buffer consisted of 0.5× BB (1× BB = 10 mM KHPO4, pH 8.0; 165 mM KC1; 2 mM EDTA; 5% glycerol), 7 mM DTT, 100 U RNasin (Promega), 1.3% polyvinyl alcohol (Sigma), and was supplemented with 82.5 mM KC1. RNA preparations [capped and uncapped C/EBPβ mRNA, and poly(A)-RNA] were passed over the affinity column, collected, and passed over the column again four successive times. The fifth pass was collected and labeled as flow through. The column was then washed with 4 vol of 400 µl 1× BB (these were collected and labeled wash #1–#4), followed by one wash with 400 µl of 50 µM GTP in 1× BB (and labeled GTP Wash). Capped mRNA was then eluted twice with 2× 400 µl volumes of 500 µM m7GTP in 1× BB. Remaining bound RNA was stripped with 1× 400 µl volume of HCB (1.0 M KC1, 20 mM HEPES, 0.2 mM EDTA, 0.5 mM PMSF). The collections were ethanol-precipitated, resuspended in buffer and analyzed by northern blot with C/EBPβ probe (5).
Determination of polysome distribution of the C/EBPβ mRNA
Polysomes were isolated from livers of control and lipopolysaccharide (LPS)-treated mice as described previously (5,25). LPS-treated mice were sacrificed 6 h after an IP injection of 50 µg LPS. The isolated polysomes were separated on a 0.5–1.5 M sucrose gradient (Beckman SW40 rotor, 35 000 r.p.m., 90 min, 4°C). Fractions were collected, phenol–chloroform–isoamyl alcohol (25:24:1)-extracted, and ethanol-precipitated. The RNA from each fraction was resuspended in buffer and further analyzed on formaldehyde–agarose gels, transferred to nitrocellulose membranes and hybridized (5). After hybridization with the C/EBPβ-specific probe, the membranes were stripped and reprobed with a mouse albumin cDNA clone. The relative amount of a specific mRNA in each fraction is normalized to a value of 1.0 in the fraction with the greatest hybridization signal.
RESULTS
Does the mRNA cap structure affect translation of C/EBPβ mRNA?
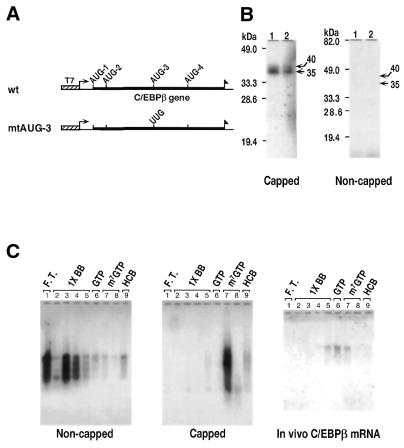
Our hypothesis states that the synthesis of different C/EBPβ isoforms occurs by ATI at specific AUG sites within a single mRNA. It is well established that ATI occurs on certain viral mRNAs (27–31) and some eukaryotic mRNAs (32–34) by internal ribosomal entry, and that this occurs on mRNAs that are not capped. However, there are also reports of ATI of capped eukaryotic mRNAs, presumably by the ribosomal scanning mechanism (34). In these experiments we ask whether the murine C/EBPβ mRNA requires a capped structure for its translation. We used the T7 in vitro transcription system to prepare wild-type C/EBPβ mRNA (C/EBPβ-wt) and C/EBPβ mRNA in which AUG-3, the initiation site for p20C/EBPβ, was mutated to UUG (Fig. 1A). The mRNAs, which contain Flag sequences at their C-termini, were isolated by gel electrophoresis and translated in a rabbit reticulocyte lysate translation system to compare the level of translation of the capped versus non-capped mRNAs. The 35S-labeled translation products were immunoprecipitated using anti-Flag antibody, resolved by polyacrylamide gel electrophoresis and visualized by autoradiography (Fig. 1B). The data clearly show significantly greater translation of the capped C/EBPβ mRNA than of the non-capped mRNA. Both the 40- and 35-kDa isoforms are produced, while the 20-kDa isoform is not produced (Fig. 1B). Failure to produce the 20-kDa isoform suggests that the factors required for its formation may not be present in this in vitro translation system. We conclude that the efficient in vitro translation of C/EBPβ requires a capped mRNA. These data do not, however, demonstrate whether the mRNA coded for by our C/EBPβ expression vector is capped. To demonstrate this we constructed an eIF-4E affinity column to resolve capped from uncapped C/EBPβ mRNA from transfected COS-1 cells. The data in Figure 1C show that the elution of T7-in vitro transcribed uncapped and capped C/EBPβ mRNAs occurs with 1× BB and m7GTP, respectively. The data also show that the mRNA coded for by the C/EBPβ expression vector in COS-1 cells is bound by the eIF-4E column and is eluted by GTP and m7GTP. Our data suggest that the in vivo synthesized C/EBPβ is capped and that translation of this C/EBPβ mRNA involves entry of the 40S ribosomal subunit at the mRNA cap site.
Figure 1.
Analysis of C/EBPβ mRNA. (A) Schematic presentation of the C/EBPβ wild-type (pT7-βmtAUG3) constructs used to produce mRNAs by in vitro transcription (B). (B) C/EBPβ wild-type and mutant mRNAs were prepared using the constructs shown in (A). In vitro translation of capped and uncapped wild-type (lanes 1) and mutant (lanes 2) mRNAs are shown. The reaction (25 µl) was started by addition of 1 µg mRNA in the presence of [35S]methionine, and the translation products were resolved by 12% SDS–PAGE. The gels were dried and autoradiographed. (C) C/EBPβ mRNA was analyzed by affinity selection of capped mRNA via the 5′-cap structure. The affinity matrix was generated by binding GST/elF-4E fusion protein to gluthathione agarose. Specific elution of bound mRNA was achieved using saturating amounts of the cap analog m7GTP. The results are shown for non-capped and capped in vitro transcribed mRNA and poly-(A) RNA from COS-1 cells transfected with pCMV-C/EBPβ. Column fractions were collected, concentrated, and subjected to agarose gel electrophoresis and northern blot analysis using a C/EBPβ-specific hybridization probe. Lane, 1, flow through; lanes 2–5, 1× BB washers; lane 6, 1× BB containing 50 µM GTP; lanes 7 and 8, 1× BB containing 50 µM m7GTP; lane 9, HCB wash.
Polysomal distribution of C/EBPβ mRNA in control and LPS treated mouse livers
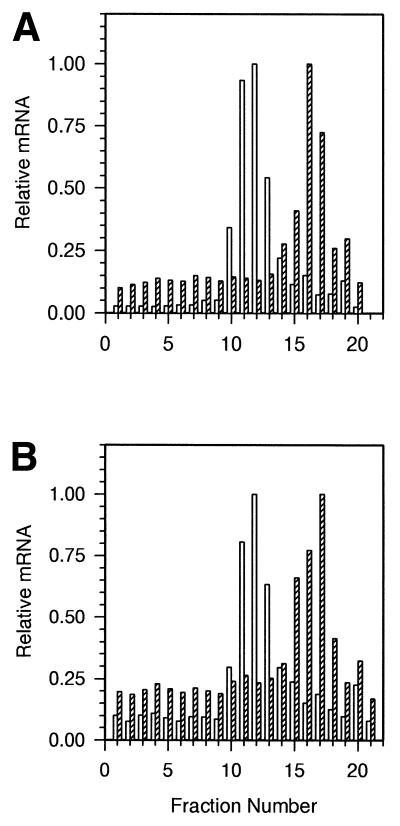
Our previous studies have shown that the p20C/EBPβ isoform is induced in mouse liver in response to bacterial LPS (5,10) and that p20C/EBPβ makes up a significant portion of the total C/EBPβ protein pool. In these experiments, we localized the C/EBPβ mRNA in size-fractionated polysomes from control and LPS-treated livers, in order to determine whether there is a shift in the sedimentation of polysome-associated C/EBPβ mRNA in cells predominantly synthesizing the low molecular weight isoform. We postulated that the size of some polysomes associated with C/EBPβ mRNA would decrease significantly with polysomes from LPS-treated livers (p20C/EBPβ) if internal ribosomal entry occurred, and that we should be able to detect a biphasic C/EBPβ polysomal distribution: those that synthesize the high molecular weight isoforms versus those synthesizing the 20-kDa isoform. Alternatively, if 40S ribosomal entry occurs at the 5′-cap, as expected, such a shift in polysomal distribution might not occur. The distribution of polysomal C/EBPβ mRNA relative to the distribution of polysomal albumin mRNA in sucrose gradients showed that C/EBPβ mRNA-associated polysomes from control and LPS-treated mouse livers sediment to the same location (Fig. 2A and B) so there is no detectable change in the size of the polysomes synthesizing the high molecular weight (40- and 35-kDa) or low molecular weight (20-kDa) isoforms. Our results suggest that internal ribosomal entry does not occur in the ATI of the C/EBPβ mRNA in response to LPS.
Figure 2.
A northern blot analysis of the polysomal distribution of C/EBPβ and albumin mRNAs from (A) control mouse livers which make the 40- and 35-kDa isoforms and (B) LPS-treated mouse livers in which the 20-kDa isoform is induced. Open bars show C/EBPβ and hatched bars show albumin mRNA distribution. RNA abundance in each fraction is normalized to a value of 1.0 in the most abundant fraction.
Initiation at internal AUG sites and regulation by the sORF
Our model proposes that the observed molecular weight isoforms of C/EBPβ are produced by ATI at multiple internal AUG sites and further that initiation at the out-of-frame sORF AUG will be involved in the regulation of AUG site usage (5,8,10). To further support our hypothesis we analyzed the translation products of a series of plasmids, with either single or multiple mutations at the AUG sites (Fig. 3). In addition, we examined translation products from plasmids with a mutated sORF AUG site alone and in combination with the mutated internal AUGs. Each of the expression vectors contained a Flag tag at the C-terminal end, allowing peptides originating from the expression vectors to be detected on immunoblot. Protein loading for western analysis was normalized to the expression vector mRNA pool to more accurately reflect translation from an equivalent mRNA pool. Any observed effect would, therefore, represent a difference in translation rather than a change in transfection efficiency or mRNA pool size.
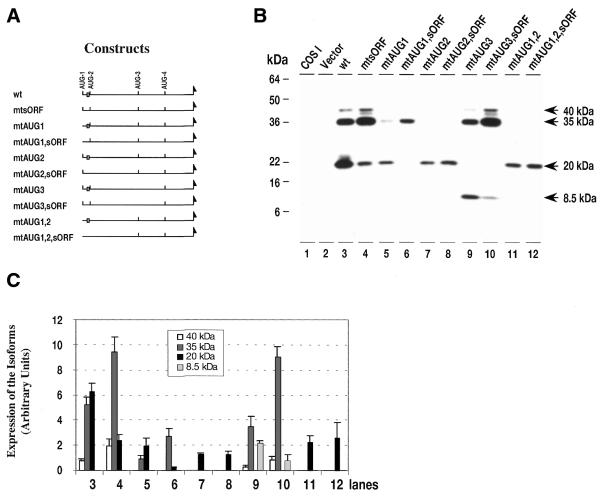
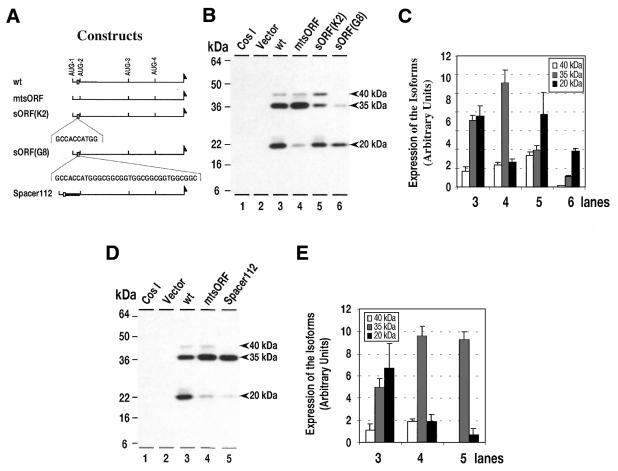
Figure 3.
Western analysis of the translation products of wild-type C/EBPβ expression vector (pCMV-C/EBPβ-wt), expression vectors with mutations at multiple AUG start sites, and mutation of the sORF-AUG. (A) Maps of the expression vectors used in (B). The open box denotes the sORF; (B) western blot analysis of proteins from COS-1 cells: lane 1, proteins from control, non-transfected COS-1 cells; lane 2, proteins from cells transfected with control vector (pCMV); lanes 3–12, proteins from cells transfected with the vector indicated. (C) Comparison of the pool levels of C/EBPβ isoforms produced by each expression vector. Protein loading was adjusted to reflect variations in vector mRNA pool levels, and translation from an equivalent mRNA pool level. Data for three independent transfections are shown as mean ± SD. The numbering of the grouped bars corresponds to the lanes in (B).
Using the anti-Flag antibodies, we detected 40-, 35- and 20-kDa isoforms synthesized by the C/EBPβ wild-type vector (Fig. 3B, lane 3). The relative pool level of each isoform is consistent with the predicted strength of the context of its Kozak sequence (see Fig. 7). Mutation of any of the AUG sites, but AUG-1 and AUG-2 in particular, resulted in an overall reduction in C/EBPβ pool size (Fig. 3B and C, lanes 5 and 7). Mutation of AUG-1 completely abolishes the 40-kDa isoform pool, while both the 35- and 20-kDa pools are reduced (Fig. 3B and C, lane 5). Mutation of AUG-2 abolishes the 35- and 40-kDa pools while the 20-kDa pool remains unchanged (Fig. 3B and C, lane 7). Finally, mutation of AUG-3 completely abolishes the 20-kDa pool, and results in a decrease of the 35- and 40-kDa pools relative to the wild-type pools. Interestingly, a new 8.5-kDa isoform is observed (Fig. 3B and C, lane 9). We attribute this isoform to initiation at an in-frame AUG start site in the DNA binding domain of C/EBPβ mRNA, which would give rise to a protein with a predicted molecular weight of 8.5 kDa. This isoform is never seen in vivo, nor is it seen in COS-1 cells transfected with wild-type expression vector. It appears, therefore, that the ribosome that bypass mutated AUG-3 proceed to initiate at AUG-4. A mutation of AUG-1 and AUG-2 results in the formation of the 20-kDa isoform only (Fig. 3B and C, lane 11). These data are consistent with similar recent studies showing initiation at these multiple downstream AUG sites (8).
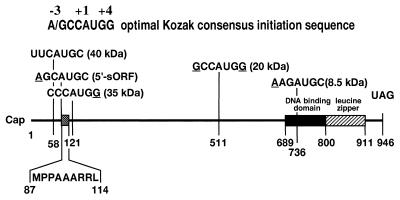
Figure 7.
A map of the C/EBPβ mRNA. The map shows the positions of the AUG initiation sites within the mRNA and the Kozak sequences relative to A (+1) in each AUG. The molecular masses of each isoform initiated at specific AUGs are designated in parentheses at each in-frame AUG site. MPPAAARRL is the amino acid sequence of the sORF.
An additional component of our hypothesis is that AUG initiation site selection will be regulated, at least in part, by initiation of translation at the sORF downstream of AUG-1. To test this hypothesis, we analyzed the isoforms produced in COS-1 cells transfected with wild-type and AUG-mutated C/EBPβ expression vectors in which the sORF-AUG was mutated to UUG. When the sORF initiation site alone is mutated (Fig. 3B and C, lane 4) the pools of 40- and 35-kDa isoforms are increased while the 20-kDa pool is significantly reduced. Unlike mutation of the in-frame AUG sites, mutation of the sORF did not reduce the overall C/EBPβ protein pool, whereas the relative abundance of each isoform, as noted above, changed dramatically (Fig. 3B and C, lane 4). Therefore, in analyzing the interaction of sORF and internal AUG mutations, we compared the double mutations against the corresponding internal, in-frame, AUG mutation. With the AUG-1 plus sORF mutation (Fig. 3B and C, lane 6), the 35-kDa isoform increased and the 20-kDa decreased (Fig 3B and C, compare lanes 5 and 6), similar to but significantly less intense than the sORF mutation alone (Fig. 3B and C, lanes 3 and 4). Mutation of AUG-2 in combination with the sORF mutation produces no change, and only the 20-kDa isoform is observed (Fig. 3B and C, lanes 7 and 8). Mutation of the sORF in combination with a mutation in AUG-3 increases the 40- and 35-kDa pools and results in the appearance of the 8.5-kDa isoform (Fig. 3B and C, lanes 9 and 10). Finally, in combination with the AUG-1 and AUG-2 double mutant, mutation of the sORF has no effect on the 20-kDa pool size (Fig. 3B and C, lanes 11 and 12). These results are consistent with the hypothesis that initiation at the sORF has a significant influence on initiation at the downstream AUG-2 and AUG-3 sites.
Role of the sORF-AUG in initiation at downstream AUGs
The potential role of the sORF in regulating translation initiation at AUG-2 and AUG-3 was further examined using C/EBPβ expression vectors in which the sORF-AUG was converted to an optimal Kozak sequence [Fig. 4A; sORF(K2)] and one in which an optimal Kozak sORF-AUG was combined with an altered sORF sequence [Fig. 4A; sORF(G8)].
Figure 4.
Western analysis of nuclear extracts from COS-1 cells transfected with wild-type C/EBPβ expression vector (pCMV-C/EBPβ-wt); expression vectors with mutations of the sORF-AUG and in which a 112-nt spacer was inserted between the termination site of the sORF and AUG-2. (A) Maps of the expression vectors used in these experiments. Vector mtsORF has a mutation changing the sORF-AUG to -UUG, sORF(K2) has a mutation changing the sORF to a perfect Kozak sequence, sORF(G8) is altered to produce a sORF peptide of one methionine and eight glycines rather than the normal MPPAAARRL, and spacer 112 has a 112-nt spacer inserted between the termination site of the sORF and AUG-2. (B) Western blot analysis of proteins from COS-1 cells: lane 1, proteins from non-transfected cells; lane 2, proteins from cells transfected with pMCV control vector; lanes 3–6, proteins from cells transfected with the expression vector indicated. (C) Summary of protein pool levels of C/EBPβ isoforms from several independent experiments as shown in (B), mean ± SD. The numbering of the grouped bars corresponds to the lanes in (B). (D) Western blot analysis of proteins from COS-1 cells, lanes labeled as in (B). (E) Summary of protein pool levels of C/EBPβ isoforms from several independent experiments as shown in (D), mean ± SD. The numbering of the grouped bars corresponds to the lanes in (D).
As noted earlier, mutation of the sORF to a non-functional initiation codon increased the 35-kDa (AUG-2) and decreased the 20-kDa (AUG-3) pools (Fig. 3B and C, lane 4). If, however, the sORF-AUG is converted to an optimal Kozak, the 35-kDa (AUG-2) pool is significantly decreased while the 20-kDa (AUG-3) pool is unchanged (Fig. 4B and C, lane 5). An expression vector that would produce a sORF peptide of one methionine and eight glycines rather than the normal MPPAAARRL was also examined. Alteration of the sORF peptide produces a dramatic overall decrease in the pool levels of all C/EBPβ isoforms, although the 20-kDa isoform is least affected (Fig. 4B and C, lane 6).
An additional expression vector was produced in which a 112-nt spacer was inserted between the termination site for the sORF and AUG-2. Insertion of a 112-nt spacer between the sORF termination codon and AUG-2 results in an increase of the 35-kDa pool and decrease in the 20-kDa pool (Fig. 4D and E, lane 5). This progressive increase in the 35-kDa and decrease in 20-kDa pools is similar to the result seen when the sORF initiation codon is mutated (Fig. 4B and C, lane 4 versus Fig. 4D and E, lane 5).
Interaction of the sORF with downstream initiation sites
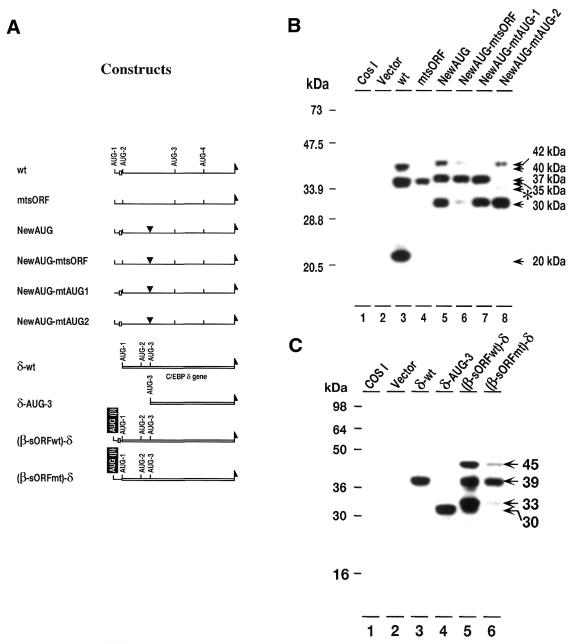
The specificity of the interaction between the sORF and downstream translation initiation sites was examined in two ways. First, a new initiation site (new AUG) was introduced between AUG-2 and AUG-3 and the effects of a sORF mutation on expression from this initiation site was examined (Fig. 5A and B). Second, a C/EBPβ–C/EBPδ chimera was constructed which contains the sORF from C/EBPβ and a silent downstream AUG from C/EBPδ (Fig. 5A and C).
Figure 5.
Functional analysis of the C/EBPβ sORF in alternate translational initiation. (A) Maps of the expression vectors used. The NewAUG vectors have a new in-frame AUG initiation codon inserted either alone, or in combination with mutations in the sORF start site (NewAUG-mtsORF), AUG-1 (NewAUG-mtAUG1) or AUG-2 (NewAUG-mtAUG2). Also shown are the maps for a series of chimeric constructs in which the C/EBPδ (in front of C/EBPδ AUG-1). Both the wild-type (β-sORFwt-δ) and mutant (β-sORFmt-δ) sORF were used. (B) Western blot analysis of proteins from COS-1 cells: lane 1, proteins from non-transfected cells; lane 2, proteins from cells transfected with pMCV control vector; lanes 3–8, proteins from cells transfected with the expression vector indicated. (C) Western blot analysis of proteins from COS-1 cells: lane 1, proteins from non-transfected cells; lane 2, proteins from cells transfected with pMCV control vector; lanes 3–6, proteins from cells transfected with the expression vector indicated.
Insertion of a new downstream AUG site initiates translation of a new 30-kDa isoform (Fig. 5B, lane 5). The expression vectors which include the new AUG produce slightly higher apparent molecular weight peptides at 42 and 37 kDa instead of 40- and 35- kDa (Fig. 5B, lane 5). Mutations of AUG-1 and AUG-2, however, abolish these transcripts demonstrating that they are initiated at AUG-1 and AUG-2, respectively (Fig. 5B, lanes 7 and 8). In the presence of a functional sORF, the new AUG expression vector directs synthesis of isoforms from AUG-1 (42-kDa) and AUG-2 (37-kDa), and the new AUG (30-kDa). When the sORF is mutated the pool of 42- and 30-kDa isoforms are greatly reduced while the 37-kDa isoform pools remains unchanged (Fig. 5B, lane 6). This is similar to the effect of a sORF mutation on the 40- and 20-kDa isoforms (Fig. 5B, lane 4) initiated at AUG-1 and AUG-3, respectively.
Sequence analysis of the C/EBPδ mRNA has shown that there are three potential AUG start sites that would produce isoforms of 39-, 33- and 30-kDa (Fig. 5C). However, C/EBPδ does not have a sORF like that in C/EBPβ and does not produce low molecular weight isoforms (Fig. 5C, lane 3). Therefore, we constructed a series of C/EBPβ–C/EBPδ chimeras in which the AUG-1 and sORF-AUG of C/EBPβ were linked to the 5′-end of the C/EBPδ gene (Fig. 5A). The wild-type C/EBPδ expression vector produces only the 39-kDa isoform from AUG-1 (Fig. 5C, lane 3). When the AUG-1 and AUG-2 sequences are deleted a 30-kDa C/EBPδ isoform is produced (Fig. 5C, lane 4). Insertion of the C/EBPβ AUG-1-sORF fragment produces 45-, 39- and 33-kDa isoforms from the C/EBPβ-AUG-1, C/EBPδ-AUG-1 and C/EBPδ-AUG-2, respectively (Fig. 5C, lane 5). These data indicate that initiation at C/EBPδ-AUG-2 can occur when the C/EBPβ sORF is inserted upstream of this AUG site and that the sORF plays a role in initiation at the site. Mutation of the sORF-AUG in wild-type C/EBPβ results in the severe decrease of initiation at C/EBPβ-AUG-1 and C/EBPδ-AUG-2 (Fig. 5C, lane 6).
Is the 20-kDa C/EBPβ isoform a product of proteolysis?
Although our data strongly indicate that the 40- and 35-kDa C/EBPβ isoforms are the result of ATI, there have been reports that the 20-kDa isoform is a product of proteolytic cleavage of these high molecular weight proteins (12,17). Furthermore, it has been reported that m-calpain, which specifically cleaves several transcription factors while leaving the binding and dimerization domains intact, may be the proteolytic enzyme that produces the 20-kDa isoform (12,17,35). To determine whether COS-1 cells contain factors which can catalyze the proteolysis of C/EBPβ to a 20-kDa product we allowed nuclear extracts of COS-1 cells containing high molecular weight C/EBPβ isoforms to incubate at 37°C for up to 48 h (Fig. 6, lanes 4–9). Although the extracts were kept at 37°C, degradation of the high molecular weight isoforms did not give rise to a 20-kDa isoform. Furthermore, we used the C/EBPβ expression vector in which the AUG-1 site was converted to an optimal Kozak sequence in order to eliminate any change in amino acid sequence at that site that might alter the specificity of a proteolytic enzyme. This vector favors initiation at AUG-1 (i.e. synthesis of the 40-kDa isoform) over initiation at AUG-2 (i.e. synthesis of the 35-kDa isoform; Fig. 6, lanes 4–9). These data suggest that although C/EBPβ degradation occurs over time, there is no indication of a specific 20-kDa product of degradation in COS-1 cells.
Figure 6.
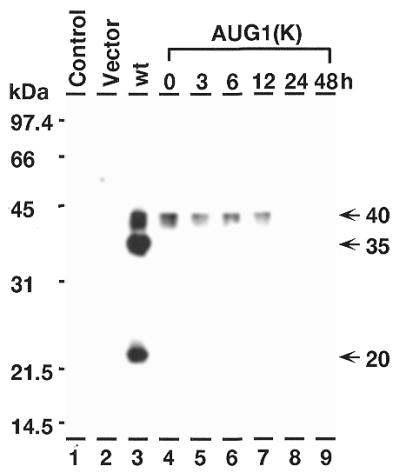
Stability of high molecular weight isoforms of C/EBPβ. Western blot analysis of proteins from COS-1 cells. Lane 1, proteins from non-transfected cells; lane 2, proteins from cells transfected with pMCV control vector; lane 3, pCMV-C/EBPβ-wt; lanes 4–9, pCMV-C/EBPβ-AUG-1K (AUG-1 has been converted to an optimal Kozak sequence). The extracts in lanes 5–9 have been incubated in at 37°C for the number of hours indicated before electrophoresis.
DISCUSSION
We have previously shown the presence of multiple C/EBPβ molecular weight isoforms in the livers of young mice, and dramatic changes in pool levels and relative isoform abundance in response to LPS treatment (5,10). The isoforms described in those studies are indistinguishable from those reported here. The data presented here show that COS-1 cells, which do not synthesize C/EBPβ mRNA, also exhibit differential translation from multiple AUG sites of a C/EBPβ mRNA transcribed from an expression vector. These products may be similar to those produced in vivo (5,10).
The mouse C/EBPβ gene is transcribed into a single mRNA species that, upon translation, gives rise to a number of different molecular weight isoforms (Fig. 7). Specific downstream AUG translation start sites within the C/EBPβ mRNA would be predicted to give rise to proteins with the observed molecular weights (Fig. 7). We propose that these isoforms are produced by ATI in which the 40S ribosomal subunit enters at the 5′-cap and, by a leaky ribosomal scanning mechanism, initiates translation at various downstream AUG sites. The C/EBPβ mRNA also contains an out-of-frame sORF immediately upstream of AUG-2, and we propose that this sORF is important in regulating the choice of potential downstream AUG sites (AUG-2 and AUG-3). Finally, we propose that this choice of AUG site is tightly regulated in vivo, and in the case of C/EBPβ is responsive to stress factors such as LPS (5,10). The studies presented here support our hypothesis that the formation of C/EBPβ isoforms occurs by ATI at multiple AUG sites within the C/EBPβ mRNA, and that this differential initiation occurs via the leaky ribosomal scanning mechanism and that the sORF plays a role in the regulation of choice of initiation site (5,8,10,36,37).
Using expression vectors mutated at each of the AUG start sites strongly supports our hypothesis that these AUGs are functional initiation sites for translation of C/EBPβ isoforms. Mutation of each start site abolished synthesis of the corresponding isoform. Mutation of AUG-2 abolished the 35-kDa isoform allowing continued expression of the 20-kDa isoform, however the 40-kDa isoform was also abolished in this mutant. This indicates that an additional factor, perhaps mRNA structure, may be affecting initiation at AUG-1 in this mutant vector. When AUG-1 and AUG-2 are both mutated, only the 20-kDa isoform is produced, confirming that AUG-3 is a functional initiation site, and inconsistent with the 20-kDa isoform originating via cleavage of the 40- or 35-kDa isoforms. When AUG-3 was mutated, a small amount of an 8.5-kDa isoform was produced. This isoform, which lacks DNA-binding activity, is probably translated from AUG-4, in the C/EBPβ DNA-binding domain. It has not, however, been detected in vivo, nor is it seen in COS-1 cells transfected with wild-type expression vector. The data suggest the 8.5-kDa isoform is only made when AUG-3 is mutated, and that some ribosomes bypass the mutated AUG-3 and proceed to initiate at AUG-4. It is interesting that the 8.5-kDa isoform lacks DNA-binding activity (10) and upon dimerization with intact C/EBPs, could potentially serve as a repressor of C/EBP regulated genes. We conclude that each of the C/EBPβ isoforms is produced by ATI at a specific AUG start site (AUG-1, -2, -3 and -4) and that this may occur by the ribosomal scanning mechanism (36,37).
In COS-1 cells, the wild-type expression vector initiates translation preferentially at the AUG-2 start site, producing predominantly the 35-kDa isoform. We observed a similar pattern in mouse livers in vivo (5,10). According to our hypothesis, then, most of the 40S subunits ignore AUG-1 and proceed to initiate at AUG-2 (Fig. 7). The frequency of initiation at a particular AUG site is dependent upon the context of the Kozak sequence (36,37). The sequences at AUG-1 and AUG-2 are not optimal Kozak sequences, and so may facilitate leaky ribosomal scanning at these sites as some ribosomes will ignore such sub-optimal sites and undergo ATI at AUG-3 which has an optimal Kozak sequence. Consistent with this idea, mutation to a perfect Kozak sequence at AUG-1 favored initiation at this site, reduced initiation at AUG-2 and eliminated initiation at AUG-3 (Fig. 6). These results suggest that a major factor in the mechanism of ATI of C/EBPβ mRNA, particularly initiation site selection, involves the context of the Kozak sequences.
Our results indicate that the 20-kDa isoform is not a product of proteolytic cleavage of the high molecular weight isoforms. An expression vector with mutated AUG-1 and AUG-2 produced only the 20-kDa isoform, indicating that AUG-3 is functional. When AUG-3 was mutated, no 20-kDa isoform is detected, consistent with ATI rather than proteolytic cleavage. However, in the AUG-3 mutation the amino acid sequence is altered and the specificity for a processing protease such as calpain might be abolished. However, when AUG-1 is mutated to an optimal Kozak sequence, no 20-kDa isoform was detected, even in the presence of an abundance of 40- and 35-kDa isoforms. Since in this case the amino acid sequence at AUG-3 is not changed, this reinforces our conclusion that this low molecular weight isoform is not a product of proteolytic cleavage. Finally, when the 40-kDa isoform produced by the optimal Kozak expression vector is incubated at room temperature the protein undergoes degradation but no 20-kDa peptide is produced. From these results we conclude that the 20-kDa isoform is not the product of proteolytic cleavage.
We have also proposed that the C/EBPβ sORF, located upstream of the 35-kDa (AUG-2) start site, plays a role in regulating translation at AUG-3, the 20-kDa start site. Mutation of the sORF-AUG to -UUG resulted in a decrease in the 20-kDa isoform and a corresponding increase in 40- and 35-kDa isoforms, consistent with decreased initiation at AUG-3 and increased initiation at AUG-1 and AUG-2. These results lead us to conclude that the sORF is an important factor in the regulation of ATI at the multiple C/EBPβ initiation sites. Thus, the integrity of the sORF or the expression of the oligopeptide is an important factor in the regulation of ATI at AUG-1, AUG-2 and AUG-3.
Regulation of initiation site selection through the sORF could be accomplished through regulation of initiation at the sORF-AUG or might involve the sORF peptide itself. Mutation of the sORF-AUG to an optimal Kozak sequence thereby enhancing initiation at the sORF greatly reduces initiation at AUG-2, which is immediately downstream. This indicates that initiation at the sORF might affect ribosomal reinitiation at the AUG-2 located immediately downstream of the sORF terminator. Additionally, when the distance between the sORF terminator and AUG-2 is increased by the insertion of 112 nt, the 35-kDa pool is increased and the 20-kDa pool is decreased, consistent with an increased initiation at AUG-2. This also is in agreement with our proposed interference with initiation at AUG-2 when the sORF is expressed. However, when the nucleotide sequence and the resulting amino acid sequence of the sORF peptide is changed, the overall expression is greatly reduced, indicating a potential role for the sORF peptide in C/EBPβ expression. However, the effects of changed nucleotide sequence on mRNA structure or regulatory factor binding cannot be ruled out.
C/EBPδ, like C/EBPβ, has three in-frame downstream AUG sites, which are potential initiation start sites. However, unlike C/EBPβ, the downstream C/EBPδ AUGs are not active; only a single 39-kDa isoform is produced. Interestingly, C/EBPδ does not have a sORF, as is found in C/EBPβ. To test whether this is an important factor in failure of C/EBPδ to make smaller isoforms we placed the C/EBPβ sORF in front of the AUG-1 of C/EBPδ. The C/EBPβ–sORF–C/EBPδ chimera produces 45-, 39- and 33-kDa isoforms, consistent with initiation at C/EBPβ-AUG-1, C/EBPδ-AUG-1 and C/EBPδ-AUG-2, respectively. Mutation of the C/EBPβ sORF-AUG in the chimera greatly reduced initiation at C/EBPβ-AUG-1 (45-kDa) and C/EBPδ-AUG-2 (33-kDa), which also occurs with the same mutation in C/EBPβmtsORF. Similar experiments were performed with C/EBPβ expression vectors into which a new AUG site had been inserted between AUG-2 and AUG-3. In this expression vector, initiation at the new AUG was regulated by the sORF in a manner identical to AUG-3. Thus, the upstream C/EBPβ sORF can regulate initiation at an otherwise inactive downstream AUG initiation site in C/EBPδ mRNA or any in-frame AUG initiation site introduced into the mRNA.
ATI can occur via two different mechanisms, leaky ribosomal scanning, as we propose for C/EBPβ mRNA, or internal ribosomal entry at multiple AUG sites, as occurs in various viral systems and some eukaryotic mRNAs (28,38,39). Our data are consistent with a leaky ribosomal scanning model which would require a 5′-capped mRNA for 40S ribosomal entry, followed by scanning the mRNA until a specific AUG was selected for initiation. For example, the in vitro translation of C/EBPβ mRNA in a rabbit reticulocyte lysate system indicates that maximal translation of C/EBPβ mRNA requires a cap structure; affinity chromatography with a cap-binding eIF-4E column indicates that the transcribed mRNAs of C/EBPβ transfected cells are capped. Furthermore, we are unable to detect a shift in the size of polysomes associated with C/EBPβ mRNA under conditions where the 20-kDa isoform is the primary product compared with 35-kDa synthesizing polysomes. Such a shift would be expected if ATI was occurring via an internal ribosomal entry mechanism. In other studies in our laboratory we have been able to detect such shifts in polysome size distribution (25). Finally, our results indicate that the 40S subunits ignore the first and/or second AUG and proceed to initiate 60S entry at the downstream AUGs. Thus, the cap structure, the context of the Kozak sequences and the sORF may play a key role in this selection process.
The location of the sORFs is a key structural feature that functions in the regulation of choice among the translational start sites within the C/EBPα and C/EBPβ mRNAs (7,8,17). One mechanism by which these sORFs may exert their function is to serve as a start site for the synthesis of oligopeptides which conditions ribosomes for leaky scanning by allowing some of the ribosomes to ignore the first downstream AUG codon and proceed to the next initiation site (5,7,8,10). An alternative mechanism has been proposed in which the sORF acts in cis to inhibit C/EBPα and C/EBPβ translation (17). This is based on the observation that mutation of the sORF completely abolishes translational repression of C/EBPα. In contrast, our studies show that in C/EBPβ mRNA, the wild-type sORF correlates with a high level of initiation at the downstream AUG-3 (20-kDa), and that mutation of the sORF-AUG results in abolishing initiation at this site. Thus, although the integrity of the sORF, as indicated by a requirement for a functional AUG, is important in the regulation of translation initiation, the basis for the repressor effect in C/EBPα and activator effect in C/EBPβ mRNAs is unclear, especially because mutation of the sORF also affects initiation upstream.
Some cases have been described where sORFs stimulate translation. For example, mutation of sAUG-2 of the retinoic acid receptor β2 results in reduced reporter expression (40,41). To explain this, it is proposed that when this sORF is translated, the protein-synthesizing 80S ribosome is better equipped than a scanning 40S subunit to penetrate stable secondary structures within a 5′-UTR. After terminating at the sORF stop codon, ribosomes could re-initiate at the next downstream AUG. Similarly, mutation of the C/EBPβ sORF-AUG reduces AUG-3 (20-kDa) isoform expression, suggesting that the integrity of the sORF-AUG is essential for initiation at AUG-3 in the COS-1 cell. Furthermore, the role of the C/EBPβ sORF-AUG in the initiation of a downstream AUG is strongly indicated by its ability to induce initiation of C/EBPδ AUG-2 in the C/EBPβ–C/EBPδ chimera.
Our studies with mouse liver have shown a significant level of differential initiation of translation at the three AUG sites in C/EBPβ mRNA (5,10). Initiation at AUG-1 and AUG-3 are low whereas initiation at AUG-2 is highly favored in the young adult mouse livers, in vivo. Furthermore, AUG-3 is strongly induced in aged liver as well as by stress factors such as LPS (5,10), heat shock (42) and heavy metals (43). Our present studies show a high level of translational initiation in COS-1 cells at the AUG-3 site in the absence of any stress factor. This significant difference suggests that the ATI of C/EBPβ mRNA may be regulated by tissue-specific factors or physiological state (transformation or proliferation). This is suggested by studies which showed that in HeLa cells, another transformed cell line, the C/EBPβ sORF represses initiation at the downstream AUG-2 (35-kDa) site, and that mutation of the sORF-AUG results in derepression of initiation at AUG-2, while no initiation of AUG-3 was detected under either condition (17). Our studies have shown that in COS-1 cells initiation at both AUG-2 and AUG-3 is high, and that mutation of sORF-AUG causes an increase of initiation of AUG-2 and a reduction of initiation at AUG-3. Thus, the level of factors that regulate ATI and, more specifically, initiation or repression at AUG-3, may be tissue- or cell type-specific.
The importance of the interaction of trans-acting regulatory proteins with cis-acting RNA binding sites is suggested by the observation that the 5′-UTR of c-myc mRNA may inhibit translation from downstream initiation sites (34,44–47). More specifically, it is the 240 nt restrictive element within exon 1 of murine c-myc that inhibits translation of both c-myc and heterologous mRNAs in rabbit reticulocyte and wheat germ extracts (46). These data suggest that specific regulatory factors are required to allow translation of full-length c-myc to occur in vivo.
Although the synthesis of C/EBPβ isoforms has been attributed to ATI at multiple AUG start sites, there have been reports that the formation of low molecular weight isoforms is due to specific proteolytic cleavage of the high molecular weight isoforms (12,17). It was reported that a specific cell lysis protocol was essential for preventing low molecular weight isoforms, and that calpain protease cleaves recombinant C/EBPβ to form a protein indistinguishable from p20C/EBPβ. Thus, truncated forms of C/EBPβ may arise in certain cases from proteolysis rather than ATI. It has been proposed that both proteolytic cleavage and ATI give rise to the 20-kDa isoform, and that C/EBPα plays a key role in the induction of proteases that catalyze this proteolysis (12). In a C/EBPα (–/–) null mouse mutant, the predominance of p20C/EBPβ is attributed to ATI, while in C/EBPα (+/+) formation of p20C/EBPβ is attributed to proteolysis of the higher molecular weight isoform. Our data indicate that in mouse livers in vivo, or in COS-1 cells transfected with a C/EBPβ expression vector, p20C/EBPβ is produced by an ATI mechanism.
The intronless C/EBPβ gene codes for a single mRNA that serves as a template for synthesis of multiple isoforms having truncated trans-activation domains and specific transcriptional regulatory functions. Our studies indicate that the synthesis of C/EBPβ isoforms by ATI provides a genetic diversity for these single copy genes at the level of translational regulation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr David Konkel for review of the manuscript. Publication No. 106 supported by U.S.P.H.S. grant PO2 AG10514 awarded by the National Institute on Aging.
References
- 1.Cao Z., Umeck,R.M. and McKnight,S.L. (1991) Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev., 5, 1538–1552. [DOI] [PubMed] [Google Scholar]
- 2.Lin F.-T., MacDougald,O.A., Diehl,A.M. and Lane,M.D. (1993) A 30 kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: Transcriptional activator lacking antimitotic activity. Proc. Natl Acad. Sci. USA, 90, 9606–9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ossipow V., Descombes,P. and Schibler,U. (1993) CCAAT/enhancer binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc. Natl Acad. Sci. USA, 90, 8219–8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams S.C., Baer,M., Dillner,A.J. and Johnson,P.F. (1995) CRP2 (C/EBPβ) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J., 14, 3170–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An M.R., Hsieh,C.-C., Reisner,P.D., Rabek,J.P., Scott,S.G., Kuninger,D.T. and Papaconstantinou,J. (1996) Evidence for posttranscriptional regulation of C/EBPα and C/EBPβ isoforms expression during the lipopolysaccharide-mediated acute-phase response. Mol. Cell. Biol., 16, 2295–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess-Beusse B.L., Timochenko,N. and Darlington,G.J. (1999) CCAAT/enhancer binding protein alpha (C/EBPalpha) is an important mediator of mouse C/EBPβ protein isoform production. Hepatology, 29, 597–601. [DOI] [PubMed] [Google Scholar]
- 7.Calkhoven C.F., Bouwman,P.R.J., Snippe,L. and Geert,A.B. (1994) Translation start sight multiplicity of the CCAAT/enhancer binding protein α mRNA is dictated by a small 5′ open reading frame. Nucleic Acids Res., 22, 5540–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calkhoven C.F., Muller,C. and Leutz,A. (2000) Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev., 14, 1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 9.Descombes P. and Schibler,U. (1991) A liver-specific transcription activator protein, LAP and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Genes Dev., 67, 569–579. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh C.-C., Xiong,W., Xie,Q., Rabek,J.P., Scott,S.G., An,M.R., Reisner,P.D., Kuninger,D.T. and Papaconstantinou,J. (1998) Effects of age on the posttranslational regulation of CCAAT/enhancer binding protein α and CCAAT/enhancer binding protein β isoform synthesis in control and LPS-treated livers. Mol. Biol. Cell, 9, 1479–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephans J.M. and Pekala,P.H. (1992) Transcriptional repression of the C/EBPα and GLUT 4 genes in 3T3-L1 adipocytes by tumor necrosis factor-α. J. Biol. Chem., 267, 13580–13584. [PubMed] [Google Scholar]
- 12.Welm A.L., Timochenko,N.A. and Darlington,G.J. (1999) C/EBPα regulates generation of C/EBPβ isoforms through activation of specific proteolytic cleavage. Mol. Cell. Biol., 19, 1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landschulz W.H., Johnson,P.F., Adashi,E.Y., Graves,B.J. and McKnight,S.L. (1988) Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev., 2, 786–800. [DOI] [PubMed] [Google Scholar]
- 14.Williams S.C., Cantwell,C.A. and Johnson,P.F. (1991) A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev., 5, 1553–1567. [DOI] [PubMed] [Google Scholar]
- 15.Christy R.J., Yang,V.W., Ntambi,J.M., Geiman,D.E., Landschulz,W.H., Friedman,A.D., Nakabeppu,Y., Kelly,T.J. and Lane,M.D. (1989) Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev., 3, 1323–1335. [DOI] [PubMed] [Google Scholar]
- 16.Chitra C. and Gordon,J.L. (1993) Cell lineage-specific and differentiation dependent patterns of CCAAT/enhancer binding protein α expression in the gut epithelium of normal and transgenic mice. Proc. Natl Acad. Sci. USA, 90, 8871–8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lincoln A.J., Monczak,Y., Williams,S.C. and Johnson,P.F. (1998) Inhibition of CCAAT/enhancer-binding protein α and β translation by upstream open reading frame. J. Biol. Chem., 273, 9552–9560. [DOI] [PubMed] [Google Scholar]
- 18.Jamaluddin M., Garofalo,R., Ogra,P.L. and Brasier,A.R. (1996) Inducible translational regulation of the NF-IL6 transcription factor by respiratory syncytial virus infection in pulmonary epithelial cells. J. Virol., 70, 1554–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akira S. and Kishimoto,T. (1992) IL-6 and NF-IL6 in acute-phase response and viral infection, Immunol. Rev., 127, 25–50. [DOI] [PubMed] [Google Scholar]
- 20.Nalbant D., Williams,S.C., Stocco,D.M. and Khan,S.A. (1998) Luteinizing hormone-dependent gene regulation in Leydig cells may be mediated by CCAAT/enhancer-binding protein-beta. Endocrinology, 139, 272–279. [DOI] [PubMed] [Google Scholar]
- 21.Clarkson R.-W.E., Chen,C.M., Harrison,S., Wells,C., Muscat,A.E.O. and Waters,M.J. (1995) Early responses of trans-acting factors to growth hormone in preadipocytes; differential regulation of CCAAT enhancer-binding protein-P (C/EBPβ) and C/EBPδ. Mol. Endocrinol., 9, 108–120. [DOI] [PubMed] [Google Scholar]
- 22.Sears R.C. and Sealy,L. (1994) Multiple forms of C/EBPβ bind to the EFII enhancer sequence in the Rous sarcoma virus long terminal repeat. Mol. Cell. Biol., 14, 4855–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schreiber E., Matthias,P., Muller,M.M. and Schaffner,W. (1989) Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res., 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edery I., Chu,L.L., Sonenberg,N. and Pelletier,J. (1995) An efficient strategy to isolate full-length cDNAs based on an mRNA cap retention procedure (CAPture). Mol. Cell. Biol., 15, 3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabek J.P., Scott,S., Hsieh,C.-C., Reisner,P.D. and Papaconstantinou,J. (1998) Regulation of LPS-mediated induction of C/EBP delta gene expression in livers of young and aged mice. Biochim. Biophys. Acta, 1398, 137–147. [DOI] [PubMed] [Google Scholar]
- 26.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 27.Glass M.J. and Summers,D.H. (1993) Identification of a trans-acting activity from liver that stimulates hepatitis A virus translation in vitro. Virology, 193, 1047–1050. [DOI] [PubMed] [Google Scholar]
- 28.Hellen C.U., Witherell,G.W., Schmid,M., Shin,S.H., Pestova,T.V., Gil,A. and Wimmer,E. (1993) A cytoplasmic 57 kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl Acad. Sci. USA, 90, 7642–7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson R.J. (1996) A comparative view of initiation site selection mechanisms in Translational Control. In Hershey,J.W.B., Mathews,M.B. and Sonnenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. pp 71–112.
- 30.Martinez-Salas E., Saiz,J.-C., Davilla,M., Belsham,G.J. and Domingo,E. (1993) Distinct repertoire of antigenic variants of foot-and-mouth disease virus in the presence or absence of immune selection. J. Virol., 67, 3748–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelletier J. and Sonnenberg,N. (1988) Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature, 334, 320–325. [DOI] [PubMed] [Google Scholar]
- 32.Vagner S., Gensac,M.-C., Maret,A., Bayard,F., Amalric,F., Prats,H. and Prats,A.-C. (1995) Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal ribosomal entry of ribosomes. Mol. Cell. Biol., 15, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van der Velden A.W. and Thomas,A.A.M. (1999) The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int. J. Biochem. Cell Biol., 31, 87–106. [DOI] [PubMed] [Google Scholar]
- 34.Willis A.E. (1999) Translational control of growth factor and proto-oncogene expression. Int. J. Biochem. Cell Biol., 31, 73–86. [DOI] [PubMed] [Google Scholar]
- 35.Watt F. and Malloy,L. (1993) Specific cleavage of transcription factors by the thiol protease, m-calpain. Nucleic Acids Res., 21, 5092–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. (1989) The scanning model for translation: an update. J. Cell Biol., 108, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozak M. (1995) Adherence to the first AUG rule when a second AUG codon follows closely upon the first. Proc. Natl Acad. Sci. USA, 92, 2662–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johannes G., Carter,M.S., Eisen,M.B., Brown,P.A. and Sarnow,P. (1999) Identification of eukaryotic mRNAs that are translated at reduced cap binding complex e-IF4F concentrations using a cDNA microarray. Proc. Natl Acad. Sci. USA, 96, 13118–13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang S.K. and Wimmer,E. (1990) Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev., 4, 1560–1572. [DOI] [PubMed] [Google Scholar]
- 40.Reynolds K., Zimmer,A.M. and Zimmer,A. (1996) Regulation of RARβ2 mRNA expression: Evidence for an inhibitory peptide encoded in the 5′-untranslated region. J. Cell Biol., 134, 827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmer A., Zimmer,A.M. and Reynolds,K. (1994) Tissue-specific expression of the retinoic acid receptor-β2: Regulation by short open reading frames in the 5′-non-coding region. J. Cell Biol., 127, 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yiangou M., Paraskeva,E., Hsieh,C.-C., Markou,E., Victoratos,P., Scouras,Z. and Papaconstantinou,J. (1998) Induction of a subgroup of acute phase protein genes in mouse liver by hyperthermia. Biochim. Biophys. Acta, 1396, 191–206. [DOI] [PubMed] [Google Scholar]
- 43.Yiangou M., Scott,S.G., Rabek,J.P., An,M.R., Xiong,W. and Papaconstantinou,J. (2001) Effects of mercuric chloride on the regulation of expression of the acute phase response components α1-acid glycoprotein and C/EBP transcription factors. Biochim. Biophys. Acta, 1518, 47–56. [DOI] [PubMed] [Google Scholar]
- 44.Butnick N.Z., Miyamoto,C., Chizzonite,R., Cullen,B.R., Ju,G. and Skalka,A.M. (1985) Regulation of the human c-myc gene: 5 non-coding sequences do not affect translation. Mol. Cell. Biol., 5, 3009–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nanbru C., Lafon,I., Audigier,S., Gensae,M., Vagner,S., Huez,G. and Prats,A.-C. (1997) Alternative translation of the proto-oncogene c-myc by an internal ribosomal entry site. J. Biol. Chem., 272, 32061–32066. [DOI] [PubMed] [Google Scholar]
- 46.Parkin N., Darveau,A., Nicholson,R. and Sonenberg,N. (1988) Cis-acting translational effects of the 5′ noncoding region of c-myc mRNA. Mol. Cell. Biol., 8, 2875–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoneley M., Paulin,F.E., Le Quesne,J.P., Chappell,S.A. and Willis,A.E. (1998) C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene, 16, 423–428. [DOI] [PubMed] [Google Scholar]