Abstract
Rationale
Post-mitotic cells such as cardiomyocytes appear to be particularly susceptible to proteotoxic stimuli, and large, proteinaceous deposits are characteristic of the Desmin Related Cardiomyopathies and crystallin cardiomyopathic diseases. Increased activity of protein clearance pathways in the cardiomyocyte such as proteasomal degradation and autophagy have proven to be beneficial in maintaining cellular and cardiac function in the face of multiple proteotoxic insults, holding open the possibility of targeting these processes for the development of effective therapeutics.
Objective
Here we undertake an unbiased, total genome screen for RNA transcripts and their protein products that impact on aggregate accumulations in the cardiomyocytes.
Methods and Results
Primary mouse cardiomyocytes that accumulate aggregates as a result of a mutant αB crystallin causative for human Desmin Related Cardiomyopathy were used for a total genome-wide screen to identify gene products that impacted on aggregate formation. We infected cardiomyocytes using a short hairpin RNA lentivirus library in which the mouse genome was represented. The screen identified multiple candidates in a number of cell signaling pathways that were able to mediate significant decreases in aggregate levels.
Conclusions
Subsequent validation of one of these candidates, Janus kinase 1 (Jak1), a tyrosine kinase of the non-receptor type, confirmed the usefulness of this approach in identifying previously unsuspected players in proteotoxic processes.
Keywords: cardiomyopathy, heart failure, Janus kinase, autophagy, proteasome
Subject Terms: Basic Science Research, Animal Models of Human Disease, Cardiomyopathy
INTRODUCTION
Proteotoxicity is defined as an impairment of cellular function as a result of protein misfolding or aggregation.1,2 Although it probably occurs in most or all cells, its potential pathogenic implications are particularly relevant to post-mitotic cell populations such as those found in brain and heart. For example, many of the neurodegenerative diseases are characterized by accumulation of misfolded and/or aggregated proteinaceous accumulations.3–5 When these accumulations of misfolded protein occur largely in post-mitotic cells, unless the cell can autonomously clear them through various protein quality control (PQC) pathways such as the proteasome or autophagy, the aggregates can eventually impair normal cellular functions and lead to pathology. As such, protein aggregates are a hallmark of many diseases, including neurodegeneration,6,7 diabetes,8,9 cystic fibrosis10,11 and, as we and others have shown, heart failure.12,13 While these phenotypes have been well studied and the pathogenic mechanisms at least partially defined, effective therapeutic approaches have proven more elusive.
Some of the first evidence for the importance of cardiac proteotoxicity was observed in desmin-related myopathy, a progressive myopathic disease culminating in skeletal muscle wasting, cardiomyopathy and premature death.14 Although multiple gene mutations can lead to this disease, the mutations in the intermediate filament protein desmin or its molecular chaperone, αB-crystallin (CryAB) have been most intensively studied. Mutations in these proteins render the filamentous desmin architecture unstable, and electron dense, granulofilamentous protein aggregates accumulate within the cytoplasm of cardiomyocytes.15 Upon expressing mutated CryAB (CryABR120G) exclusively in cardiomyocytes of transgenic mice, we were able to assess the cardiac specific effects.16,17 Multiple investigators showed that deficits in proteasomal processing18,19 as well as autophagy were associated with the progressive accumulation of misfolded proteins within the cardiomyocytes. In the CryABR120G hearts, these deficits led to mitochondrial dysfunction and declining cardiac function, resulting in death by heart failure at or before 7 months.17 Restoring normal levels of a PQC pathway such as autophagy delayed both the onset of overt disease and increased the probability of survival over time. These improvements were accompanied by decreased protein aggregate levels and improved cardiac function.20 In fact, by upregulating autophagy, we were able to clear or significantly reduce aggresomal aggregates,21 decrease toxic pre-amyloid oligomer load, restore cardiac function and prolong life.20,22
Pathological accumulation of misfolded proteins may occur by common or related pathways in diverse cell types and across different diseases. For example, the accumulation of large perinuclear aggregates, or aggresomes, is found in both the neurodegenerative disorders23 and in certain types of heart failure where the underlying pathologies are amplified by proteotoxic stimuli.24 This not only implies convergent pathogenic pathways across disease presentations, but also suggests that therapeutic avenues capable of decreasing aggregate load, either by preventing their accumulation or increasing cell autonomous clearance, might work across the spectrum of proteotoxic disease phenotypes.
Modulating the different pathways that cells use for PQC is becoming a focus for developing new therapies in multiple diseases, including cancer and heart failure.25,26 However, our basic understanding of the system-wide communication networks that control these processes and talk to one another are woefully incomplete. Driven by these conceptual difficulties, we have devised and carried out an unbiased total genomic screen to find novel genes that can prevent or clear protein aggregates in a model of cardiac proteotoxicity. Using CryABR120G-expressing mouse cardiomyocytes as the substrate, we used an inhibitory RNA library approach by systematically infecting the cells with pools of short hairpin (sh)RNA lentivirus that provided multiple coverage of approximately 16000 mouse genes.27,28 The subsequent validation of a selected primary candidate identified novel interacting proteins that were able to significantly impact on aggregate levels in the cardiomyocytes.
METHODS
Neonatal Rat or Mouse Cardiomyocyte Isolation
Hearts were harvested from 1–1.5 day old pups and incubated in 0.05% trypsin overnight at 4°C. Upon removal of trypsin, hearts were rinsed with ice-cold αMEM and hearts were dissociated with 260 units/heart collagenase in αMEM for 40 minutes at 37°C. The solution was triturated to disperse cell clumps and passed through a 40 μm strainer. The homogenate was then centrifuged for 5 minutes at 1500 rpm and the pellet resuspended in αMEM containing 10% FBS and DNAse. A preplating step was done to selectively isolate cardiomyocytes from the cardiac fibroblasts. Ten ml of cell suspension were added to uncoated 10 cm2 plates and incubated for 40 minutes at 37°C in a 5% CO2 incubator. The medium was removed and transferred to a conical fermenter. 1×104 cells were added to each well of a gelatin-coated clear-bottomed black 96 well plate.
Neonatal Rat Cardiomyocyte (NRVM) siRNA transfection
NRVMs were transfected one day after plating with either siJak1 (50 nmol/L, Thermo Fisher Scientific, s136789 and s136791) or scrambled siRNA (50 nmol/L, Thermo Fisher Scientific, 4390846) in OptiMEM (Thermo Fisher Scientific) for 5 hours. After that, cells were infected with different adenovirus encoding CryABR120G and virus constructs as indicated in the figure legends. After 48–72 hours, cells were processed for either immunohistochemical analyses or lysed in SDS lysis buffer (3% SDS, 30 mmol/L Tris base, pH 8.8, 5 mmol/L EDTA, 30 mmol/L NaF, 10% glycerol, 1 mmol/L DTT), sonicated and used for immunoblotting. For immunoblotting, 50 μg protein was loaded onto 4–15% Bio-Rad Mini-PROTEAN Precast Gels, transferred to PVDF membranes with Bio-Rad Tris/Glycine buffer and stained with antibodies (α-crystallin B – Enzo Life Sciences, clone 1B6.1-3G4: p62 – ProGen, GP62-C: LC3 – Novus Biologicals, NB100-2331: α-actinin – Sigma-Aldrich a-7811). For the longer culture times and cytotoxicity determinations (Figure 5E-G), NRVMs were transfected 20 hours after plating with 50 nM scrambled siRNA or siJak1 and transduced with adenovirus encoding wildtype CryAB or mutant CryABR120G 5 hours after transfection. Medium was changed 24 hours after transfection to DMEM 2% FBS (including 5 μM AraC to inhibit fibroblast growth) and then left for 6 days (7 days in total) until measurement of lactate dehydrogenase release using the Roche Cytotoxicity Detection KitPlus (LDH; #04-744-926-001). Absorbance was measured at OD 492 nm. For determination of Jak1 signaling (Figure 4C and D), p-Stat3 antibody and Stat3 antibody (Cell Signaling #9145 and #12640, respectively) were used.
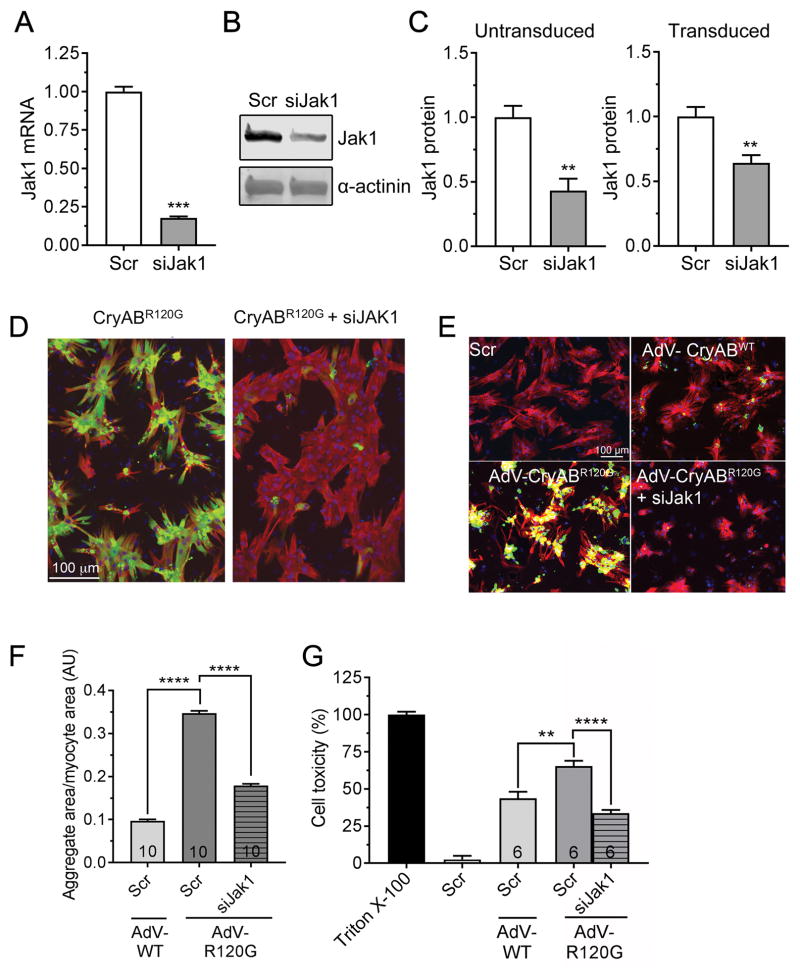
Figure 5. siJak1 knockdown in NRVM.
A, siRNA-mediated Jak1 knockdown in NRVMs was done as described in Methods and RNA quantitated via qPCR. B, Protein was isolated from the transfected cultures and assayed for Jak1 via Western blotting as described in Methods. C, The efficacy of Jak1 knockdown by siRNA was assayed with and without transfection of the cells with CryABR120G-containing adenovirus after 48–72 hours. D, Decreased Jak1 decreases aggregate load in NRVMs. Reduction of Jak1 levels resulted in a significant reduction in aggregate content after 48–72 hours. Data are depicted as mean ± SEM with **P<0.01 and ***P<0.001, unpaired Student’s t-test (n=3–4). E, NRVMs were transfected with scrambled siRNA (scr) or siJak1 and transduced with adenovirus (AdV) encoding wildtype CryAB or CryABR120G or left untransduced and aggregate accumulations visualized and F, quantitated after 7 days. G, Cytotoxicity measured 7 days after transduction. In E, nuclei are depicted in blue, GFP-CryAB in green and cardiac troponin I (cardiomyocyte marker) in red. Cell toxicity was measured using the lactate dehydrogenase assay as detailed in Methods. Triton X-100 was used as 100% and untransduced cells as 0%. Data are presented as mean ± SEM with **P<0.01 and ****P<0.0001, one-way ANOVA, Tukey’s post-hoc analysis. The number of images (F) or wells (G) is indicated in the histograms’ bars.
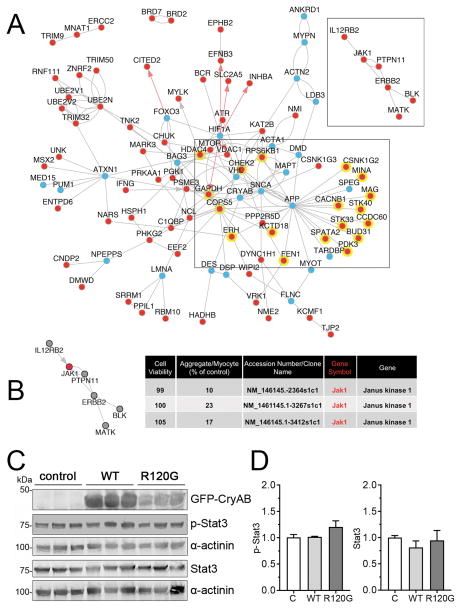
Figure 4. Systems biology prioritization of proteotoxicity regulators and Jak1 signaling.
We first used the OMIM and EntrezGene databases to identify proteotoxicity-annotated genes, and then extended the identified gene sets to related biological processes using ToppGene and AltAnalyze. AltAnalyze NetPerspective network combining proteotoxicity related genes and primary hits with shared direct protein-protein (BIOGRID, grey lines), pathway (WikiPathways, KEGG, grey arrows) or transcriptional regulatory (PAZAR database, red arrows) were visualized. A, A large proportion of primary hits (red dots) have reported interactions with known proteotoxicity genes (blue dots), including 18, which interact with amyloid precursor protein (APP): APP interacting genes are highlighted with a yellow background. A side cluster emerged centered about Jak1 (shown). B, Jak1 knockdown resulted in >75% reduction in aggregates using 3 individual shRNAs directed at the same gene transcript, with no effect to cell viability. A side cluster emerged centered about Jak1 (shown). C, NRVMs were transduced with adenovirus encoding either GFP-CryABWT or GFP-CryABR120G or left untransduced (control; C). Five days after transduction, cells were harvested for protein extraction. A, Representative Western blots stained for p-Stat3 and Stat3. α-actinin was used as a loading control. D, p-Stat3 and Stat3 quantification. Data are presented as mean ± SEM with n=5.
Lentiviral Infection
Short hairpin (sh)RNA constructs were transfected into HEK-293T cells for high-throughput lentivirus production. Lentiviruses (50 μl) were added to each well of 96 well plates 6 hours after plating. Lentiviruses were left in contact with cells overnight before rinsing cells with fresh 2% FBS in DMEM media. Cells were allowed to recover for 6 hours before adenovirus infection.
Adenoviral Infection
Adenovirus containing CryAB-Turbo Green Fluorescent Protein (tGFP) was added at a multiplicity of infection = 10 in serum-free DMEM and infected for 2 hours. After 2 hours, media was removed and replenished with DMEM containing 2% FBS and 1X antibiotic-antimycotic. Cells were fixed 4 days after CryAB infection.
Alamar Blue Assay
To assess lentiviral transduction efficiency, we treated cells that were or were not infected with lentivirus with puromycin; cells infected with lentivirus should be protected from the drug due to expression of a puromycin resistance gene. After emptying the well, 50 μl of 20% Alamar blue was added to each well and incubated for 1 hour at 37°C. Alamar blue fluorescence was measured on a Synergy II plate reader (λex – 530nm/25; λem = 590nm/35)
Screening Protocol
Ninety-six well black clear-bottomed plates were pre-coated with gelatin and allowed to air dry. Primary neonatal mouse cardiomyocytes were plated at 1×104 cells well in 100 μl of 10% FBS in αMEM and allowed to attach for 5 hours. After 6 hours, 70 μl of media was removed and replaced with 50 μl lentivirus stock and infected overnight. For control wells, 50 μl of media or scrambled (no target) lentivirus was added. After the incubation, 50 μl was removed from each well and replaced with 100 μl 4% FBS DMEM and the cells were allowed to recover for 5 hours. After 5 hours, 100 μl was removed and replaced with 100 μl of CryABR120G-tGFP adenovirus in serum-free, antibiotic-free DMEM at a multiplicity of infection = 10. Cells were incubated at 37°C for 2 hours, after which 100 μl of 4% FBS DMEM was added to halt the infection. At this point, 100 μl Torin 1 (1 μM) was added to the appropriate control well. After 48 hours, 150 μl was removed from each well and replaced with 150 μl puromycin (2 μg/ml) in 2% FBS DMEM except for non-puromycin treated controls. Forty-eight hours later, wells were emptied, rinsed with PBS and fixed with 100 μl 4% paraformaldehyde in PBS for 15 minutes. Wells were subsequently washed 2x with PBS and stored at 4°C until imaged.
Counterstaining and Imaging
After fixation, the plates were counterstained to identify cardiomyocytes and nuclei. Cells were permeabilized with 0.5% Triton X-100 in PBS (PBST) for 20 minutes followed by two, 10 minute washes with PBS. Cells were blocked with 3% BSA in PBST for 30 minutes at room temperature. Mouse troponin I antibody (Millipore MAB1691) was added at 1:2000 in blocking solution for 1 hour. Antibody was removed and the cells washed twice for 10 minutes with PBST, followed by incubation with 1:400 secondary antibody (donkey anti-mouse Alexa Fluor 586; Life Technologies A10037) for 1 hour at room temperature. Antibody was removed and the cells washed twice for 10 minutes each with PBST. Cells were incubated with DAPI (1:2000 in PBS) for 10 minutes at room temperature. After two washes with PBST, plates were sealed and stored until imaged. Imaging was performed on the BioTek Cytation 3 at room temperature using an automated plate stacker. Sixteen images were acquired using a 10X air objective above the center of the well, avoiding the edge of the wells. Physical separation between images was 800 μm (vertical) × 1100 μm (horizontal) in a 4 × 4 array. Images were acquired in three channels: DAPI (λem – 377nm, λex – 447nm; LED Power = 10, Integration time 100 ms, Gain 6.3), GFP (λem – 469nm, λex – 525nm; LED Power = 5, Integration time 28ms, Gain 0), RFP (λem – 531nm, λex 593nm; LED Power = 10, Integration time 601 ms, Gain 15.6).
Bioinformatics
In silico approaches were used to interrogate the candidate lists against database information of other known pathways. We applied a literature driven network analysis to identify genes from our screen associated with proteotoxicity-associated pathways. The OMIM and EntrezGene databases were used to identify annotated proteotoxicity genes. We then extended those gene sets to related biological processes using ToppGene [PMID:19465376] and AltAnalyze [PMID:20513647]. The network generated with AltAnalyze (NetPerspective interface) was based on ChIP-target interactions from the PAZAR database, protein-protein interactions from BIOGRID and pathway connections from the KEGG and WikiPathways for literature associated proteotoxicity genes and primary hits.
RESULTS
The High Throughput, Genome-Wide Screen
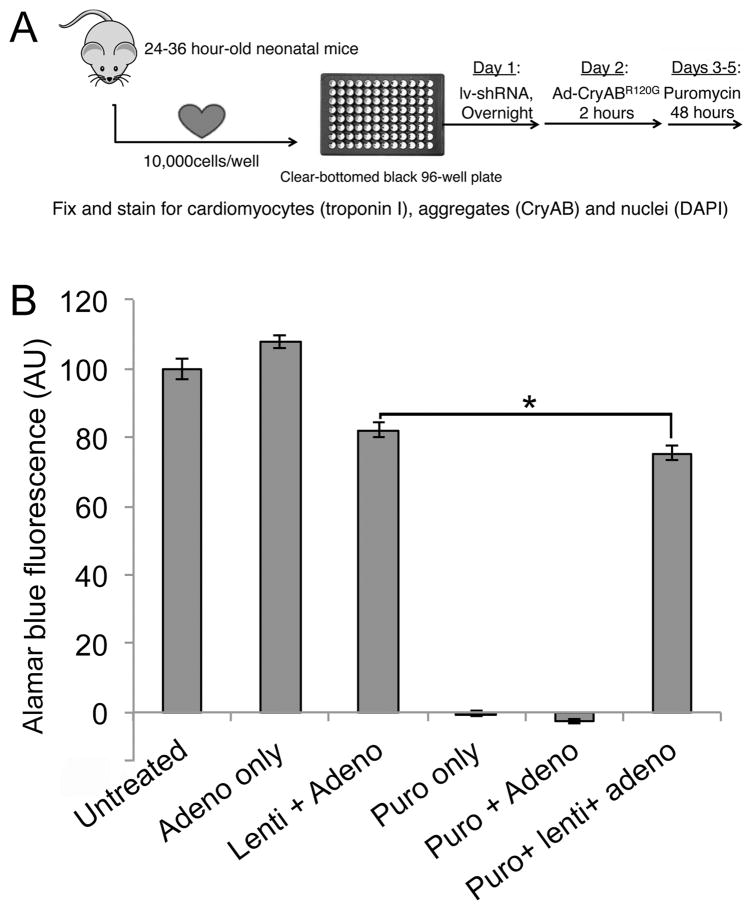
We utilized a genome-wide RNA knockdown approach to screen for gene products that impacted on aggregate formation in a cardiomyocyte model of proteotoxicity using adenovirus-driven expression of CryABR120G. The screen is schematically outlined in Figure 1A. When cultured under suitable conditions, primary neonatal mouse cardiomyocytes are non-dividing cells that beat spontaneously and synchronously, displaying well-defined sarcomeres. In order to detect aggregate formation that could be analyzed using high throughput imaging techniques, we cloned a turbo-GFP (tGFP) fusion tag onto either normal, wild type- (CryABWT-tGFP) or mutant-CryAB (CryABR120G-tGFP) and created replication deficient adenoviruses using the AdEasy® system. Genetic knockdown was achieved using the MISSION RNAi shRNA library, version 1.0, targeted against the mouse genome. The initial sets of shRNA lentivirus for mouse and human were set up through the RNAi Consortium, an initiative between Harvard, MIT, Washington and Columbia Universities.27 These libraries were then made available commercially. Lentivirus short hairpin (sh)RNA screening has now been successfully carried out in multiple cell lines and in primary cells as well. The library contains shRNAs targeting ~16,000 genes, in which each gene transcript is targeted by 3–5 distinct shRNAs in order to achieve a measure of internal verification during the primary screen. These constructs were then used to create lentiviruses expressing each shRNA. In addition to the sequence encoding the hairpin, the lentivirus construct contains a puromycin-resistance gene so that only those cells infected by the lentivirus will be viable after selection. To determine the infectivity of neonatal mouse cardiomyocytes, we used a non-targeted shRNA containing a scrambled DNA sequence (scr). Cells were infected for 96 hours before measuring cell viability using Alamar Blue (ThermoFisher) staining, with untreated cells used as a control (Figure 1B). Puromycin treatment of uninfected cells resulted in 100% cell death, which was not affected by adenoviral infection using the CryABR120G construct. Upon infection of shRNA, cell viability was largely preserved, indicating efficient cardiomyocyte infection by lentivirus. After puromycin treatment there was a small but significant (7%) decrease in cell viability in the presence of both adenovirus and lentivirus, suggesting that lentivirus infected 93% of the cell population (Figure 1B). Subsequently, we included puromycin treatment in our protocol to ensure the uninfected cells were eliminated from the analyses.
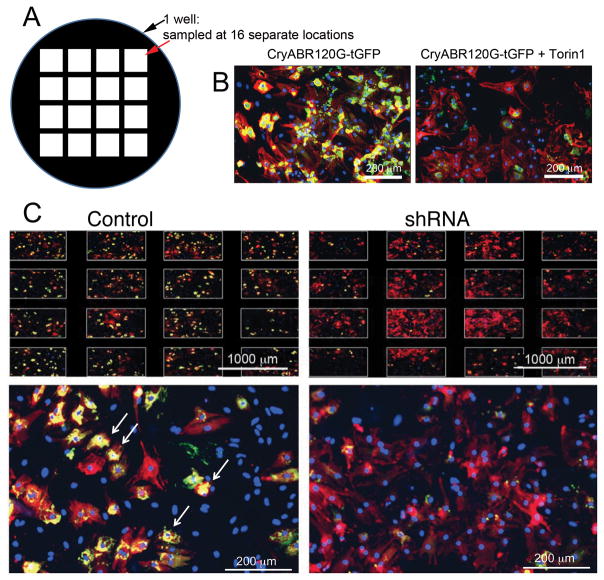
Figure 1. High throughput screen.
A, Screening protocol. Primary cardiomyocytes were harvested from 1–1.5 day old neonatal mice and plated (1×104 cells/well) in 96 well plates. Cells were infected with lentivirus overnight and, after 5 hours of recovery, infected with adenoviruses expressing the aggregate reporter (CryABR120G-tGFP) for 2 hours. After 48 hours, puromycin was added to remove uninfected cells and cells were fixed after an additional 48hrs and counterstained for imaging. B, Viability assay in the presence of viral infection and puromycin selection. The neonatal cardiomyocytes were infected with the indicated constructs. Each lentiviral shRNA construct contained a puromycin-resistance gene to gauge infectivity, as cells infected with lentivirus will be resistant to puromycin treatment. Treating uninfected or adenoviral infected cells resulted in 100% cell death, whereas infection with lentivirus largely preserved cell viability. The small (7%) decrease in Alamar Blue Fluorescence as a measure of cell viability (*P< 0.05), represents a small fraction of cells uninfected by lentivirus, which can be eliminated via puromycin selection.
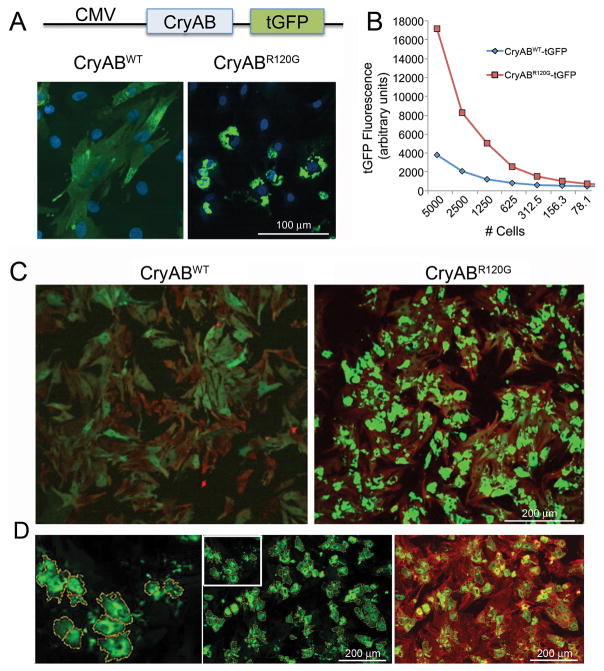
To determine if CryABR120G expression could provide a robust signal for aggregation using a high throughput imaging approach we infected cardiomyocytes with the adenovirus constructs containing either CryABWT-tGFP or CryABR120G-tGFP (Figure 2A). As expected, infection with CryABWT-tGFP resulted in diffuse cytoplasmic fluorescence due to distribution of a cytoplasmic chaperone. In contrast, expression of CryABR120G-tGFP resulted in the accumulation of perinuclear aggregates that yielded a 5-fold enhancement in fluorescence intensity relative to the CryABWT-tGFP -infected cells (Figure 2A and 2B). Staining with troponin I to identify the cardiomyocytes demonstrated that cell-type specific aggregate accumulation could be robustly measured by automated imaging of fluorescence intensity (Figure 2C and 2D), providing the basis for designing an analytical profiling algorithm amenable to high throughput techniques.
Figure 2. Aggregate induction and validation of the signal:noise ratio for CryABR120G-driven aggregate formation in primary cardiomyocytes.
A, High levels of either normal or mutant CryAB expression were driven off of the cytomegalovirus promoter after placement of the indicated construct into adenovirus. B, Quantitation of the relative signal strengths resulting from either CryABWT or CryABR120G was determined using immunofluorescence detected as a result of tGFP expression. C, Primary cardiomyocytes were plated at optimum densities for signal:noise generation, infected with either normal or mutant CryAB and the cells fixed and stained 3–4 days after infection. Cardiomyocytes were identified by immunofluorescent detection of troponin I (red) showing that >90% of the cardiomyocytes contained easily detectable (> 4-fold cytoplasmic CryABWTtGFP signal) CryAB-positive aggregates. D, Using BioTek Cytation 3 software, the algorithm defined aggregated protein on the basis of predefined filters and encircled them, creating yellow masks. To ensure measured aggregates were within cardiomyocytes as opposed to cardiac fibroblasts or endothelial contaminants, hierarchical detection was done using a cardiomyocyte-specific counterstain, TnI (red), such that only those cells where green aggregates were associated with red signal (green-over-red hierarchy) were scored and measured.
We then undertook an automated, high throughput screen using the primary cardiomyocytes that were infected with adenovirus carrying the aggregate reporter construct (CryABR120-tGFP). After optimizing the basic components needed for the Screen (Online Supplement, Table I; available at http://circres.ahajournals.org), the cells were plated robotically into 225, 96 well, black bottomed, microwell plates as detailed in Methods. All dilutions, washes, incubations and screenings were automated in order to reduce well-to-well and plate-to-plate variation. Each well was infected with shRNA lentivirus, the goal being at least 5-fold coverage for each transcript and each well in each plate was sampled in 16 different locations in order to rule out localized effects (Figure 3). The RNAi Consortium’s shRNA library was designed to minimize off-target effects, and the redundancy of multiple shRNAs able to target different regions of each gene’s transcript allows for internal validation.27 For the initial screen, we pooled these multiple shRNA lentiviruses, such that 3–5 different shRNAs targeting different regions of the target gene were expressed in the cardiomyocytes simultaneously. After lentivirus infection, the cells were allowed to grow, selected for infection via puromycin resistance (Figure 1 and Methods), fixed and then stained with troponin I antibody for cardiomyocyte identification, CryAB antibody for aggregate detection and DAPI to identify nuclei when needed. A hierarchal algorithm was used to quantitate puncta, with the first iteration detecting the aggregates and the second iteration keeping only those aggregates over a red (troponin I-containing) field (Figure 3). Imaging filters were set conservatively so that maximum signal to noise ratios were conserved between each plate. Each plate also contained a positive control that utilized the compound Torin1, a mechanistic target of rapamycin inhibitor (Figure 3B), which potently induces autophagy at both the early (autophagosome formation) and late (lysosomal degradation) steps.29 In test platings, Torin1 produced a reliable reduction in CryAB aggregates by between 50–70% and was used as an internal positive control for each plate to score a “hit” event. A BioTek Cytation 3 was used for data acquisition and high-content imaging. This device was also equipped with a plate loading tower, allowing image acquisition to occur unattended on a continuous basis.
Figure 3. Inhibitory RNA knockdown of specific genes leads to decreased aggregate load in cardiomyocytes.
A, Each well in each multi-welled plate was sampled in 16 separate locations and only those plates showing uniform cardiomyocyte density and aggregates were selected for further analyses. B, At least 1 well/plate was also treated with Torin1 to induce autophagy and confirm that a lack of aggregates could be scored on the particular plate. C, Typical data for those infections scored as primary “hits” for clones able to decrease aggregate load in cardiomyocytes subjected to the proteotoxic stimulus of CryABR120G expression. Arrows point to examples of regions that the algorithm scored as positive for aggregates. Shown is a plate of mixed cardiomyocytes and fibroblasts in which the “Control” panel was infected with Ad-CryABR120G but empty lentivirus. Shown in the “shRNA” panels are similar wells: an example of a well scored as a hit is shown. Note the staining across the different sampled areas of the well (top) and a striking decrease in aggregate content (yellow staining). Cardiomyocytes are stained red (TnI antibody) and nuclei blue (DAPI).
We set an initial threshold of 50% aggregate reduction (measured by area of aggregate/cardiomyocyte) for hit identification. The primary screen with the pooled library yielded 2,433 hits. For initial validation, we separated the pooled clones into 3–5 lentiviruses containing single shRNA clones and infected the cardiomyocytes with these single clones. In order to pass validation, at least one clone had to reduce aggregate content by > 50%. Analysis of the validation screen data yielded a total of 236 hits, constituting approximately 1.5% of the genome (Online Supplement, Table II).
Off-target effects can be sequence-dependent or independent. Sequence independent effects can relate to generalized cellular responses (eg, overall decrease in metabolic rate) while sequence dependent effects can occur as a result of mismatched silencing characteristics for the shRNAs: this was minimized by the design of the RNAi Consortium library but even so, for a genome-wide screen it has been estimated that “hits” scored as such on the basis of a single shRNA would consist of 3,362 off-target effects with only 20 true positives.30 However, this drops to only nine off-target hits if three unique shRNAs against the same target mRNA are all positive. Our initial, post-screen validations therefore focused on those clones that scored positive for >1–2 shRNAs against a single target transcript. This decreased the hits from 236 to 14 and these clones are identified in Table II (Online Supplement).
A literature-driven in silico network analysis was subsequently used to further prioritize identified screen targets. Using multiple pathway databases (Methods), we identified an expanded set of proteotoxicity-associated genes, based on the literature. This analysis included genes associated with protein degradation and protein misfolding. Combined with genes from our screen, we derived a network containing multiple known proteotoxic effectors that interacted with our primary hits (Figure 4A). A substantial number of the genes identified in the screen have reported physical interactions with known proteotoxicity genes, including 18 that physically interact with amyloid precursor protein, a known causative agent in Alzheimer’s disease. In addition, we used this method to identify gene clusters that were able to independently reduce proteotoxic inclusions in cardiomyocytes, but had no known links to protein degradation or proteotoxicity.
While our in silico, literature-and gene ontology-based analyses identified a number of intriguing pathways (Figure 4A), additional filtering was necessary to reduce the candidate number to a manageable size for experimentally based, low throughput validation. Two criteria constituted this next filter: 1) Minimal documentation of protein or pathway(s) actions in the heart, in order to enhance the potential novelty of the candidate and its impact in the cardiac system, 2) The protein or pathway would be present in different cell types and conserved throughout evolution, pointing to its general importance.
Janus kinase 1 (Jak1) satisfied all the criteria. Three separate shRNA lentivirus scored positive in the initial screen (Figure 4B) and reduced aggregated protein content by >75%, without affecting cell viability. Moreover, Jak1 has reported direct and indirect associations with 5 additional candidate genes (Figure 4A). This tyrosine kinase is part of the JAK/Signaling Transducer and Activator of Transcription (STAT) signaling pathway, whose members are widely distributed throughout different cell types and organ systems.31,32 Jak1 is a membrane protein whose kinase activity targets STAT proteins that are subsequently translocated to the nucleus to target specific gene transcription. Although most intensively studied for their important roles in cytokine signaling, modulating the immune response and oncogenesis, mutations in the proteins present in this pathway, including Jak1, cause a number of different diseases,33,34 and are responsible for numerous hepatocellular adenomas.35 Additionally, the pathway is evolutionarily conserved and functions in transducing extracellular stimuli to the transcriptional apparatus.36 However, while Jak1 is expressed in both heart and muscle (GeneCard database), Jak1’s role in normal and diseased cardiomyocytes is relatively unexplored. In order to determine if CryAB expression had any direct effects on Jak1 signaling, NRVMs were transduced with adenovirus encoding either GFP-CryABWT or GFP-CryABR120G or left untransduced. Five days after transduction, cells were harvested for protein extraction and a downstream target of Jak1 signaling, pstat3 quantitated (Figure 4). Wild type expression was approximately 1.7 times that of CryABR120G in order to see if simply overexpressing the protein at relatively high levels had an effect as well. Overexpression of neither the normal or mutant proteins led to a statistically significant increase in Jak signaling. We therefore chose to further validate Jak1’s effects on cardiomyocyte aggregate accumulation.
Multiple short inhibitory (si)RNAs were tested for their efficacy in decreasing Jak1 expression in the infected cardiomyocytes: we chose one that resulted in 75–80% knockdown at low micromolar concentrations (Figure 5A). siJak1 transfection efficiently reduced Jak1 protein as well (Figure 5B) although reduction at the protein levels was somewhat diminished when the cultures were co-transfected with CryABR120G adenovirus as well (Figure 5C). Despite the only partial knockdown, and as expected on the basis of the initial screening data, treatment of CryABR120G-infected cultures showed efficient abrogation of aggregate accumulation (Figure 5D).
To determine the effects of Jak1 knockdown for longer periods, a similar set of experiments was carried out but the cells were allowed to remain in culture for a total of 7 days to see if aggregates would begin to accumulate again. The data show that siJak1 treatment was effective during this time period as well, with no aggregates apparent in the cultures (Figure 5E and F). Importantly, cell toxicity was also significantly decreased at the end of the 7 days as well (Figure 5G).
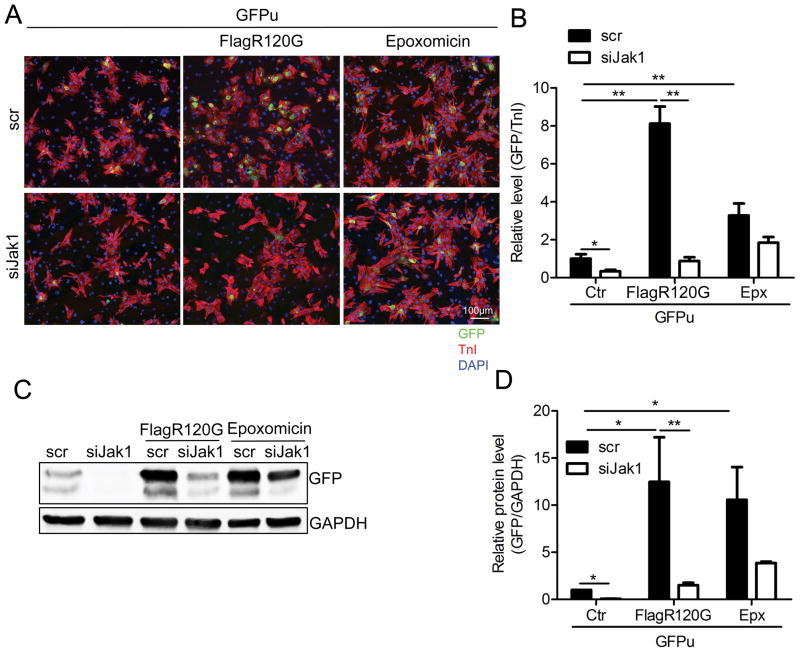
To begin to gain some insight into the processes responsible for the observed decreased aggregate load as a result of decreased Jak activity, we asked if either proteasomal or autophagic flux increased as a result of Jak1 inhibition. We first explored possible alterations in proteasomal activity by taking advantage of a well-defined fluorescent reporter construct capable of detecting proteasomal flux.37,38 This inverse reporter, GFPu, is ubiquitously degraded when the proteasome is active, but, upon inhibition of proteasomal flux, GFP levels are conserved and the fluorescent signal can be easily detected. We coinfected NRVMs with adenovirus containing Flag tagged CryABR120G and GFPu-containing adenovirus and subsequently transfected the cells with siRNA for Jak1 (Figure 6). Infection with CryABR120G significantly increased GFP accumulation, a result consistent with previous data showing that the proteotoxic environment decreased proteasomal flux (Figure 6A and B).39 siJak RNA transfection significantly reduced GFP levels, indicating that proteasomal flux was significantly increased in the cells, potentially impacting the accumulation of misfolded proteins and the subsequent aggregate formation and accumulation. siRNA transfection also decreased GFP in cultures that had not been transfected with CryABR120G indicating that decreasing Jak activity in a non-proteotoxic environment resulted in improved UPS performance as well. We also treated the cardiomyocytes for 15 hours with 250 nM epoxomicin, a proteasome inhibitor. As expected, the GFP level was increased after this treatment. Jak1 knockdown only slightly decreased GFP levels after this treatment but its effect did not reach statistical significance, confirming the effective inhibition of the proteasome with the drug (Figure 6).
Figure 6. Proteasomal activity is enhanced after Jak1 knockdown.
NRVMs were co-infected with adenoviruses containing Flag-CryABR120G and GFPu (a proteasome activity reporter). The cells were then transfected with siJak1 or a scrambled siRNA (scr) for 4 days and treated with the proteasome inhibitor epoxomicin for 15 hours. At 5 days post-infection, the cells were collected. A, Immunofluorescence with the cardiomyocyte marker TnI (red). Nuclei were counterstained with DAPI (blue). B, The relative GFP level in cardiomyocytes was quantitated using NIS-elements software. Control cells (ctr) were transfected with GFPu adenovirus only. *P<0.05, **P<0.01. C, Western blot analysis of GFP expression level. GAPDH was used as a loading control. D, The Western blot was quantitated. *P<0.05, **P<0.01.
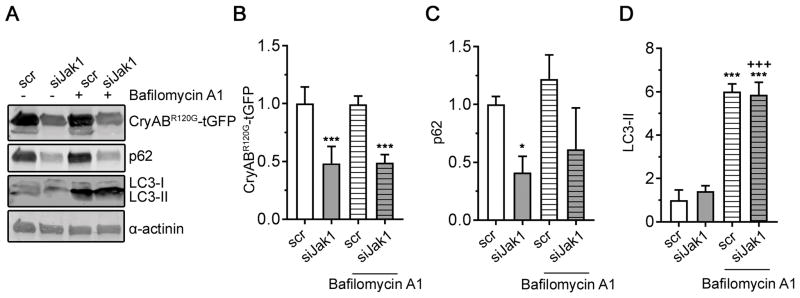
Similarly, we explored if the process of autophagy was affected by decreasing Jak1 activity, as we have shown that autophagic flux is decreased in CryABR120G hearts and this decrease is at least partially responsible for increased aggregate load.20,21 Increased autophagy can clear pre-existing aggregates,20 decreasing proteotoxic load and cell toxicity. To determine whether Jak1 knockdown induces autophagy and thus, reduces CryABR120G-tGFP protein aggregate levels, we transfected NRVMs with a siRNA targeting Jak1 and subsequently infected them with an adenovirus encoding CryABR120G-tGFP. Protein levels of p62 and LC3-II, which can indicate increased autophagy, were measured before and after treatment with the lysosomal inhibitor Bafilomycin A1, which is used to determine autophagic flux (Figure 7). Jak1 knockdown decreased CryABR120G-tGFP and p62 protein levels, confirming lower aggregate content. LC3-II protein levels did not differ between scr- and siJak1-transfected cells before and after Bafilomycin A1 treatment, suggesting that Jak1 knockdown did not affect autophagic flux. Taken together, the data in Figures 6 and 7 suggest that Jak1 knockdown reduces CryABR120G aggregation primarily by activation of proteasomal activity, presumably by preventing initial aggregate formation rather than by clearing pre-existing aggregates through autophagy.
Figure 7. Jak1 knockdown does not increase autophagic flux.
Cardiomyocytes were transfected with either scr or siJak1 and subsequently infected with CryABR120G-tGFP as detailed in Methods. Forty-eight hours after transfection, cells were treated with 30 nmol/L Bafilomycin A1 for 2 hours before being harvested in SDS lysis buffer (Methods). A, Representative Western blot of indicated proteins. Quantitation of CryABR120G-tGFP in B, p62 in C, and LC3-II in D. Data were normalized to α-actinin and are depicted as mean ± SD with *P<0.05 and ***P<0.001 against scr and +++P<0.001 against siJak1, one-way ANOVA with Tukey’s post-hoc analysis. n=3–4.
DISCUSSION
Cardiomyocytes, and therefore cardiac function and hemodynamics are uniquely sensitive to proteotoxic stimuli and subsequent protein aggregation. While preventing misfolded protein accumulation is key to every cell’s health and requires constant surveillance, post-mitotic cells are at particular risk for proteotoxic insult. In the presence of accumulating misfolded proteins, mitotic cells can remove them through asymmetric division, thus preferentially placing the aggregate load into one of the two daughter cells and effectively preserving one cell.40 Post-mitotic cells are unable to divide away the protein load, rendering the entire organ susceptible to proteotoxic insult. In this respect, cardiomyocytes are similar to neurons,41 as they are both largely post-mitotic cell populations in in an essential organ. Thus, proteotoxic insults can be particularly damaging; loss of neurons or cardiomyocytes via cell death can lead to neurodegeneration and heart failure, respectively. As proliferation of adult cardiomyocytes42,43 and neurons,41 are both rare, both cell types have developed sophisticated and inclusive PQC mechanisms. Although we have found that manipulation of autophagy can be particularly beneficial in our Desmin Related Cardiomyopathy and other proteotoxic models,44 other aspects of PQC are affected during CryABR120G disease as well.45 Thus we decided to cast our net as widely as possible and utilized a genome-wide scan using shRNA knockdown for any gene product that resulted in significant decreases in cardiomyocyte aggregate load. We used neonatal cardiomyocytes as they rapidly accumulate the proteotoxic aggregates (Figure 1), contain all PQC systems and robustly tolerate infection with adenovirus containing the CryABR120G construct. A potential limitation of the system is that, in the whole animal, proteotoxicity does not present until mid-adulthood and thus, formally, the processes under study are not exactly mimicking the situation in vivo, particularly as we used NRVMs.
With the discovery and rapid utilization of shRNA for generalized genome-wide screens in Drosophila, Caenorhabditis and Planaria, the stage was set for their utilization in mammals via infection. We think that the mouse is particularly well suited for the shRNA-based, high throughput screening approach. The mouse is genetically accessible and our ability to manipulate the genome renders it suitable for validating novel effectors of a particular gene or process using organ-specific or cell type specific substrates. If a well-designed screen can be implemented, new players in a particular process or new gene/drug therapeutics can be identified. For unambiguous verification, sophisticated phenotyping analyses are widely available for genetically modified mice as well so that organ function and interactions can also be subsequently defined.
During the development phase of the screen, we attempted to construct a number of stable cell lines expressing CryABR120G-tGFP, but were unable to obtain stable aggregate production in non-quiescent cells, presumably due to asymmetric cell division; that is, after their initial formation the aggregates were selectively divided into a daughter cell, resulting in cultures that did not exhibit the aggregate phenotype to an extent where a satisfactory signal to noise ratio could be obtained. We eventually chose to use primary neonatal mouse cardiomyocytes. This choice was made for several reasons: 1) robustness of signal, 2) ease of reporter induction and 3) ability to directly relate our findings to cardiac disease. The limitations of using primary cells include needing moderate numbers of mice and batch-to-batch variation. However, we were able to establish both external and internal controls on a plate by plate basis and all experimental wells were normalized to those controls, such that the normalized data from different plates could be compared.
With the virus infection protocols in place, we optimized the other relevant parameters to ensure the screen was robust and reproducible and the data could be compared between plates and wells. This required optimizing parameters such as plate format, cell number, timing of viral infection, false negative minimization, fixation time, incubation parameters and cardiomyocyte counterstaining (Online Supplement, Table I). However, while necessary to ensure high-quality assay data, a well-designed and operational high throughput screen is, in our view, only one of the essential determinants for success. Rather, what dictates the screen’s ultimate success is a well-designed plan for candidate nomination, validation and follow-up experimentation. The experiments that need to be carried out include: (a) a retest of the individual shRNAs in the initial pool that scores a “hit” in the first pass to eliminate false-positives and potential off-target effects, (b) assays designed to determine specificity, (c) bioinformatic-based in silico analyses to determine potentially interesting relationships and identify a limited number of genes for subsequent, low throughput assays and eventually d) creation of animal models suitable for testing the candidates’ efficacy in reversing aggregate formation along with the proteotoxic compromise in cardiac function.
The identification and subsequent validation of Jak1 as a protein able to modulate aggregate load was surprising and illustrates the power of an unbiased, genome wide screen. Jak1 is well known as an important player in interleukin, interferon, colony-stimulating factors, hormones and cytokine signaling and altered Jak activity has been linked to a number of different cancers such as acute myeloid leukemia46 and solid organ malignancies.47 The Jak/STAT pathway(s) remain the archetypal membrane-to-nuclear gene transcription paradigm, where Jak1 is associated at the membrane with a cytokine receptor and responds to receptor binding by activating its tyrosine kinase activity. Subsequently, it undergoes both autophosphorylation and phosphorylates the cytoplasmic tail of the receptor, making the associated site accessible for STAT docking. Upon docking, STAT is also phosphorylated, dimerizes and is subsequently transposed in that form to the nucleus where transcription of specific gene targets is initiated.
Currently more than twenty-five Jak inhibitors are being tested in different trials, highlighting the intense interest in these pathways as therapeutic targets and roxolitinib, a Jak1/2 inhibitor, has been approved by the FDA for treatment of myelofibrosis and polycythemia vera.48 But there are, to our knowledge, no data that bear directly on the pathway’s relationship to proteinaceous aggregate accumulation or clearance. Using the human chronic myeloid leukemia cell line K-562, an association between ruxolitinib treatment and enhanced autophagy as determined by LC3B-II puncta formation was recently documented.49 We had previously shown in the CryABR120G cardiomyopathic hearts that increased autophagy is sufficient to decrease aggregate load in both cell culture and the intact animal.20,21 Another study dealt with pathogenesis associated with α-synuclein accumulation. Accumulation of aggregated α-synuclein is a hallmark of Parkinson’s disease, an age-related neurodegenerative condition. Using a model of α-synuclein expression to induce neuroinflammation and neurodegeneration it was found that AZD1480 treatment, which reduces STAT1/3 activation, effectively reduced both the inflammatory process and cell degeneration.50 We have noted striking parallels in our model of CryABR120G-induced heart failure and the neurodegenerative diseases and the fact that STAT1/3 can effectively prevent dopaminergic neuron degeneration in vivo in a model of α-synuclein overexpression, by suppressing pathogenic inflammatory responses and other pathogenic signaling is intriguing, pointing the way towards undertaking a more comprehensive analysis of the potential role of these pathways in cardiac proteotoxicity as well. However, the effect of AZD1480 on Lewy body accumulations was not determined.
Although proteotoxicity is increasingly being recognized as an important pathogenic process in a variety of human diseases, we are only just beginning to understand its underpinnings and potential therapeutic targets. We think that a series of genome-wide, unbiased screens for candidates that impact on these processes would be of considerable value for generating new hypotheses and uncovering unsuspected players in both defined and undefined pathways. This approach is currently underway; we are currently performing genome-wide overexpression of the mouse genome to determine which genes can be activated to reduce accumulation of proteotoxic inclusions. While these approaches cannot provide a mechanistic understanding of data, they do provide a basis for hypothesis generation and subsequent mechanistic studies. In this initial report, we have established and validated a screen and carried out a preliminary analysis on one of the initial candidates. Our data point to the value of this approach as they uncovered JAK/STAT signaling as a potentially intersecting pathway in the heart. The data also offer a new group of potential candidates for others to study as well (Online Supplement, Table II).
In an attempt to begin to approach the mechanism by which a Jak pathway intersects with aggregate load, we asked whether the effects of Jak1 knockdown were mediated primarily at the early stages of aggregate accumulation, when misfolded proteins are targeted for proteasomal degradation, or at the later stages, when the large, granulofilamentous aggregates can only be cleared via autophagy or alternative clearance mechanisms as yet undefined, as they are too large to fit into the restricted bore of the proteasome. Due to the limited time in culture that is inherent in the primary cells, this screen could not distinguish between aggregate formation and clearance. Nor did it provide any mechanistic evidence of how Jak1 was intersecting with proteasomal or autophagic clearance of misfolded proteins or aggregates. However, our data do point to the upregulation of proteasomal flux via Jak1 inhibition and, considering the nature of the cells used for the screen, this is not surprising. Although aggregates can form relatively quickly,51,52 the screen was carried out in cells undergoing active aggregate formation rather than in a post-accumulation period (Figure 1A). Thus, although this particular screen could not distinguish between proteins that might function during the accumulation period rather than acting on aggregate clearance because of the relatively short times involved, we expected the screen would be biased towards the aggregate formation phases of the process. Thus, screening for factors that might enhance clearance will necessarily involve infection with the shRNA libraries after aggregate formation has occurred and primary cardiomyocytes are not suitable for such a screen. These alternative screens are in the active planning stages in the laboratory.
However, even with the current screen’s limitation, we isolated a number of interesting and unexpected candidates. Experiments dealing with loss of function of the pathway in the heart as well as dosing CryABR120G mice with Jak1 inhibitors under normal and proteotoxic conditions should provide some insights as to the precise mechanism by which Jak1 inhibition impacts on aggregate load in a proteotoxic environment.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What is known?
The process of cellular self digestion, autophagy, is critical for normal cell function and is often altered in cardiovascular disease but a complete understanding of how the cardiomyocyte clears large aggregates is lacking.
Unbiased, total genome-wide screens are useful for identifying unknown and unexpected candidates that impact on general cellular processes such as autophagy.
What New Information Does This Article Contribute?
We outline a coherent rationale and method for setting up and successfully executing an unbiased, genome-wide screen for identifying gene products that impact on aggregated protein clearance in the cardiomyocyte.
We validate the importance of one of the identified candidates, Jak1, in playing an unexpected role in autophagic clearance.
We present a database identifying additional candidates for the process, providing the community with a resource for future experimentation
As a result of disturbances in protein homeostasis emanating from a variety of stimuli including disease, protein misfolding occurs and can result in the formation of proteinaceous aggregates. This cellular response is observed in many failing hearts and the aggregates can be cleared by autophagy and clearing aggregated proteins can be beneficial. We developed a high throughput, genome-wide screen using short hairpin RNA knockdown via lentivirus infection, to identify novel participants in cardiomyocyte clearance processes. Cardiomyocytes were first transfected with adenovirus carrying a mutated αB crystallin, which causes Desmin Related Cardiomyopathy and is characterized by the accumulation of large, proteinaceous aggregates. The cells were subsequently transfected with lentivirus carrying short hairpin RNA that in aggregate, provided knockdown of the total genome complement. The screen identified gene products that prevented aggregation when chronically challenged with a proteotoxic stimulus. Candidates were selected on the basis of the screen and its effectiveness demonstrated by validation of one of the candidates, Jak1, whose role in the process had not been previously known. Our data demonstrate the effectiveness of the general approach for studying aggregate formation and clearance and provide a source of candidates that materially impact on the processes for future study.
Acknowledgments
SOURCES OF FUNDING
This work was supported by National Institutes of Health grants P01HL69779, P01HL059408, R01HL05924, R011062927 and a Trans-Atlantic Network of Excellence grant from Le Fondation Leducq (J.R.)
Nonstandard Abbreviations and Acronyms
- CryAB
αB crystallin
- CryABWT
Wild type CryAB
- CryABR120G
Mutated CryAB
- Jak1
Janus kinase 1
- NRVM
Neonatal rat ventricular cardiomyocytes
- tGFP
Turbo green fluorescent protein
- STAT
Signaling Transducer and Activator of Transcription
- PQC
Protein quality control
- shRNA
short hairpin RNA
- siRNA
short inhibitory RNA
- scr
scrambled siRNA
Footnotes
DISCLOSURES
No potential conflicts of interest were disclosed
References
- 1.Hightower LE. Heat shock, stress proteins, chaperones, and proteotoxicity. Cell. 1991;66:191–197. doi: 10.1016/0092-8674(91)90611-2. [DOI] [PubMed] [Google Scholar]
- 2.Douglas PM, Cyr DM. Interplay between protein homeostasis networks in protein aggregation and proteotoxicity. Biopolymers. 2010;93:229–236. doi: 10.1002/bip.21304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 4.Christie NT, Lee AL, Fay HG, Gray AA, Kikis EA. Novel polyglutamine model uncouples proteotoxicity from aging. PLoS One. 2014;9:e96835. doi: 10.1371/journal.pone.0096835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox D, Carver JA, Ecroyd H. Preventing alpha-synuclein aggregation: the role of the small heat-shock molecular chaperone proteins. Biochim Biophys Acta. 2014;1842:1830–1843. doi: 10.1016/j.bbadis.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Bemporad F, Chiti F. Protein misfolded oligomers: experimental approaches, mechanism of formation, and structure-toxicity relationships. Chem Biol. 2012;19:315–327. doi: 10.1016/j.chembiol.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trikha S, Jeremic AM. Clustering and Internalization of Toxic amylin oligomers in pancreatic cells require plasma membrane cholesterol. J Biol Chem. 2011;286:36086–36097. doi: 10.1074/jbc.M111.240762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zemva J, Schubert M. Central insulin and insulin-like growth factor-1 signaling: implications for diabetes associated dementia. Curr Diabetes Rev. 2011;7:356–366. doi: 10.2174/157339911797415594. [DOI] [PubMed] [Google Scholar]
- 10.Aleksandrov LA, Jensen TJ, Cui L, Kousouros JN, He L, Aleksandrov AA, et al. Thermal stability of purified and reconstituted CFTR in a locked open channel conformation. Protein Expr Purif. 2015;116:159–166. doi: 10.1016/j.pep.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du K, Karp PH, Ackerley C, Zabner J, Keshavjee S, Cutz E, et al. Aggregates of mutant CFTR fragments in airway epithelial cells of CF lungs: new pathologic observations. J Cyst Fibros. 2015;14:182–193. doi: 10.1016/j.jcf.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Klevitsky R, Huang W, Glasford J, Li F, Robbins J. AlphaB-crystallin modulates protein aggregation of abnormal desmin. Circ Res. 2003;93:998–1005. doi: 10.1161/01.RES.0000102401.77712.ED. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Osinska H, Dorn GW, 2nd, Nieman M, Lorenz JN, Gerdes AM, et al. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–2407. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 14.van Spaendonck-Zwarts KY, van Hessem L, Jongbloed JD, de Walle HE, Capetanaki Y, van der Kooi AJ, et al. Desmin-related myopathy. Clin Genet. 2011;80:354–366. doi: 10.1111/j.1399-0004.2010.01512.x. [DOI] [PubMed] [Google Scholar]
- 15.Vicart P, Caron A, Guicheney P, Li Z, Prevost M-C, Faure A, et al. A Missense mutation in the αB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Osinska H, Gerdes AM, Robbins J. Desmin filaments and cardiac disease: establishing causality. J Card Fail. 2002;8:S287–292. doi: 10.1054/jcaf.2002.129279. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Osinska H, Klevitsky R, Gerdes AM, Nieman M, Lorenz J, et al. Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Robbins J. Proteasomal and lysosomal protein degradation and heart disease. J Mol Cell Cardiol. 2014;71:16–24. doi: 10.1016/j.yjmcc.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhuiyan MS, Pattison JS, Osinska H, James J, Gulick J, McLendon PM, et al. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest. 2013;123:5284–5297. doi: 10.1172/JCI70877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pattison JS, Osinska H, Robbins J. Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes. Circ Res. 2011;109:151–160. doi: 10.1161/CIRCRESAHA.110.237339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maloyan A, Gulick J, Glabe CG, Kayed R, Robbins J. Exercise reverses preamyloid oligomer and prolongs survival in alphaB-crystallin-based desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2007;104:5995–6000. doi: 10.1073/pnas.0609202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka M, Kim YM, Lee G, Junn E, Iwatsubo T, Mouradian MM. Aggresomes formed by alpha-synuclein and synphilin-1 are cytoprotective. J Biol Chem. 2004;279:4625–4631. doi: 10.1074/jbc.M310994200. [DOI] [PubMed] [Google Scholar]
- 24.Sandri M, Robbins J. Proteotoxicity: An underappreciated pathology in cardiac disease. J Mol Cell Cardiol. 2014;71:3–10. doi: 10.1016/j.yjmcc.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meijering RA, Henning RH, Brundel BJ. Reviving the protein quality control system: therapeutic target for cardiac disease in the elderly. Trends Cardiovasc Med. 2015;25:243–247. doi: 10.1016/j.tcm.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Xia D, Ye Y. In search of a cure for proteostasis-addicted cancer: a AAA target revealed. Cancer Cell. 2015;28:550–552. doi: 10.1016/j.ccell.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral rnai library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 28.Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci U S A. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mtorc1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sigoillot FD, King RW. Vigilance and Validation: Keys to success in RNAi screening. ACS Chem Biol. 2011;6:47–60. doi: 10.1021/cb100358f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang YN, Baik EJ. Jak-Stat pathway and myogenic differentiation. Jakstat. 2013;2:e23282. doi: 10.4161/jkst.23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quintas-Cardama A, Verstovsek S. Molecular Pathways: Jak/Stat pathway: mutations, inhibitors, and resistance. Clin Cancer Res. 2013;19:1933–1940. doi: 10.1158/1078-0432.CCR-12-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brosius FC, 3rd, He JC. Jak inhibition and progressive kidney disease. Curr Opin Nephrol Hypertens. 2015;24:88–95. doi: 10.1097/MNH.0000000000000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menet CJ, Mammoliti O, Lopez-Ramos M. Progress toward Jak1-selective inhibitors. Future Med Chem. 2015;7:203–235. doi: 10.4155/fmc.14.149. [DOI] [PubMed] [Google Scholar]
- 35.Pilati C, Zucman-Rossi J. Mutations leading to constitutive active Gp130/Jak1/Stat3 pathway. Cytokine Growth Factor Rev. 2015;26:499–506. doi: 10.1016/j.cytogfr.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 36.O’Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The Jak-Stat pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–328. doi: 10.1146/annurev-med-051113-024537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bence NF, Bennett EJ, Kopito RR. Application and analysis of the GFPu family of ubiquitin-proteasome system reporters. Methods Enzymol. 2005;399:481–490. doi: 10.1016/S0076-6879(05)99033-2. [DOI] [PubMed] [Google Scholar]
- 38.Gupta MK, Gulick J, Liu R, Wang X, Molkentin JD, Robbins J. Sumo E2 enzyme Ubc9 is required for efficient protein quality control in cardiomyocytes. Circ Res. 2014;115:721–729. doi: 10.1161/CIRCRESAHA.115.304760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121:3689–3700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rujano MA, Bosveld F, Salomons FA, Dijk F, van Waarde MAWH, van der Want JJL, et al. Polarised Asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 2006;4:e417. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhardwaj RD, Curtis MA, Spalding KL, Buchholz BA, Fink D, Bjork-Eriksson T, et al. Neocortical neurogenesis in humans is restricted to development. Proc Natl Acad Sci U S A. 2006;103:12564–12568. doi: 10.1073/pnas.0605177103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Berlo JH, Kanisicak O, Maillet M, Vagnozzi RJ, Karch J, Lin SC, et al. C-Kit+ Cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pattison JS, Robbins J. Autophagy and proteotoxicity in cardiomyocytes. Autophagy. 2011;7:1259–1260. doi: 10.4161/auto.7.10.16882. [DOI] [PubMed] [Google Scholar]
- 45.McLendon PM, Robbins J. Proteotoxicity and cardiac dysfunction. Circ Res. 2015;116:1863–1882. doi: 10.1161/CIRCRESAHA.116.305372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang Z, Zhao Y, Mitaksov V, Fremont DH, Kasai Y, Molitoris A, et al. Identification of somatic Jak1 mutations in patients with acute myeloid leukemia. Blood. 2008;111:4809–4812. doi: 10.1182/blood-2007-05-090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loh ML, Tasian SK, Rabin KR, Brown P, Magoon D, Reid JM, et al. A Phase 1 Dosing study of ruxolitinib in children with relapsed or refractory solid tumors, leukemias, or myeloproliferative neoplasms: a children’s oncology group phase 1 consortium study (Advl1011) Pediatr Blood Cancer. 2015;62:1717–1724. doi: 10.1002/pbc.25575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bryan JC, Verstovsek S. Overcoming treatment challenges in myelofibrosis and polycythemia vera: The Role of Ruxolitinib. Cancer Chemother Pharmacol. 2016;77:1125–1142. doi: 10.1007/s00280-016-3012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bagca BG, Ozalp O, Kurt CC, Mutlu Z, Saydam G, Gunduz C, et al. Ruxolitinib induces autophagy in chronic myeloid leukemia cells. Tumour Biol. 2016;37:1573–1579. doi: 10.1007/s13277-015-3947-4. [DOI] [PubMed] [Google Scholar]
- 50.Qin H, Buckley JA, Li X, Liu Y, Fox TH, 3rd, Meares GP, et al. Inhibition of the Jak/Stat pathway protects against alpha-synuclein-induced neuroinflammation and dopaminergic neurodegeneration. J Neurosci. 2016;36:5144–5159. doi: 10.1523/JNEUROSCI.4658-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, et al. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101:10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanbe A, Osinska H, Villa C, Gulick J, Klevitsky R, Glabe CG, et al. Reversal of amyloid-induced heart disease in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2005;102:13592–13597. doi: 10.1073/pnas.0503324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.